Abstract
Aims
The reporting of tumour cellularity in cancer samples has become a mandatory task for pathologists. However, the estimation of tumour cellularity is often inaccurate. Therefore, we propose a collaborative workflow between pathologists and artificial intelligence (AI) models to evaluate tumour cellularity in lung cancer samples and propose a protocol to apply it to routine practice.
Methods and results
We developed a quantitative model of lung adenocarcinoma that was validated and tested on 50 cases, and a collaborative workflow where pathologists could access the AI results and adjust their original tumour cellularity scores (adjusted‐score) that we tested on 151 cases. The adjusted‐score was validated by comparing them with a ground truth established by manual annotation of haematoxylin and eosin slides with reference to immunostains with thyroid transcription factor‐1 and napsin A. For training, validation, testing the AI and testing the collaborative workflow, we used 40, 10, 50 and 151 whole slide images of lung adenocarcinoma, respectively. The sensitivity and specificity of tumour segmentation were 97 and 87%, respectively, and the accuracy of nuclei recognition was 99%. One pathologist's visually estimated scores were compared to the adjusted‐score, and the pathologist's scores were altered in 87% of cases. Comparison with the ground truth revealed that the adjusted‐score was more precise than the pathologists' scores (P < 0.05).
Conclusion
We proposed a collaborative workflow between AI and pathologists as a model to improve daily practice and enhance the prediction of tumour cellularity for genetic tests.
Keywords: artificial intelligence, digital pathology, deep learning, lung adenocarcinoma, tumour cellularity
Introduction
The rapid growth of artificial intelligence (AI) in recent years, especially deep learning (DL), has provided significant advancements in numerous fields, including medicine. The convolutional neural network (CNN) has emerged as the most suitable method for medical image analysis. 1 , 2 , 3 , 4 Trials using CNNs to assist physicians with diagnosis, treatment or even prognosis have produced extremely promising results. 5 , 6 , 7 , 8 , 9
In pathology, CNNs have been used to analyse various tissues and detect tumour regions to support histopathological diagnosis. 10 , 11 , 12 , 13 , 14 , 15 Studies demonstrate that CNNs can provide judgements equivalent to those of pathologists or even exceed them in certain tasks. 12 , 14 However, the implementation of AI in pathology is challenging because of the high rate of false positives and false negatives and the complex procedures used to optimise the balance between them. 16 Thus, a collaboration between pathologists and DL may be the most suitable approach to overcome these obstacles, as we still do not completely understand the nature of the ‘black box’ inside the DL training process. 17 The implementation of DL in the clinical workflow, including evidence of its safety for patient healthcare, is an important issue that needs to be addressed. 18 , 19
For decades, lung cancer has had the lowest survival rate among all major types of cancers in humans. 20 There have been revolutionary developments in cancer treatment, such as ‘personalised’ molecular therapies and the introduction of checkpoint inhibitors, with several studies presenting encouraging evidence of their efficacy. 21 , 22 As reported in individual studies and molecular testing guidelines, the accurate detection of tumour cell percentage in tissue specimens has been recognised as an important pre‐analytical variable for EGFR and KRAS mutation testing. 23 , 24 , 25 The minimum required percentage of tumour cells in a sample is dependent upon the analytical sensitivity of the platform conducting the tests, and varies considerably between the platforms. 25 , 26 Thus, the evaluation of tumour cellularity (also known as tumour purity or tumour fraction) by pathologists, i.e. the percentage of tumour cells in the sample, is considered critical. However, recent studies have reported that high variability and low reproducibility exist among individual pathologists. 27 , 28 , 29 , 30 The major goals of tumour cellularity assessment for molecular testing are to determine whether the specimen is adequate for molecular testing; to determine if a lack of identified mutations is due to an insufficient amount of tumour cells (cellularity below sensitivity of a platform); and to determine whether there is subclonal heterogeneity within a tumour when mutation allele frequency is low. Therefore, an effective and objective method for the precise estimation of tumour cellularity is urgently needed.
Studies have been conducted to detect, discriminate subtypes of and predict the mutations of lung cancers using the histological features of tumour cells. 9 , 18 , 31 , 32 , 33 , 34 , 35 , 36 However, to the best of our knowledge, no study has focused upon measuring tumour cellularity using DL – which is expected to aid pathologists in accurately determining the tumour purity for molecular testing. In this report, we develop a DL algorithm and evaluate a collaborative process of modification by pathologists as a clinically applicable protocol and investigate if it improves the quality of tumour cellularity counts.
Materials and methods
Study cohorts
The current study protocols were approved by the Institutional Review Board of Nagasaki University Hospital (#190218282 for the creation, validation and testing of the AI‐score model and #190311162 for testing the collaborative workflow). Both protocols were published on the Nagasaki University Hospital Clinical Research Center website for opt‐out. We designed the study in three phases: algorithm development, testing the AI results (AI‐score) and testing the collaborative workflow (Supporting information, Figure S1). In the algorithm development phase, a CNN model was constructed and validated to measure tumour cellularity. For testing the AI‐score the model was tested on 50 cases, and in testing the collaborative workflow, its efficacy was evaluated in 151 different cases. For the algorithm development and AI‐score testing phases, 100 haematoxylin and eosin (H&E)‐stained transbronchial biopsy (TBB) slides (one slide per case), diagnosed with lung adenocarcinoma, were collected from Nagasaki University Hospital, Nagasaki, Japan. Of these, 50 whole slide images (WSIs) were used as a data set for model development, and the other 50 slides were used to test the AI‐score. For the testing study of the collaborative workflow, 151 slides were used. These 151 slides included not only TBB, but also other modalities such as core needle biopsy (CNB), surgical resection, transbronchial needle aspiration (TBNA) and cell block. The 50 slides for testing the AI‐score were scanned using an Aperio Scanscope CS2 digital slide scanner (Leica, Wetzlar, Germany Biosystems, Buffalo Grove, IL, USA) with a 40× objective lens (0.25 μm/pixel). The 151 slides for testing the collaborative workflow were scanned using an Ultra Fast Scanner (Philips, Amsterdam, the Netherlands) with a 40× objective lens (0.25 μm/pixel). Afterwards, digital slides were imported into HALO version 2.2 (Indica Labs, Albuquerque, NM, USA), which included HALO AI (CNN VGG network) and the HALO Image Analysis program.
Algorithm development phase
We enrolled 50 TBB slides as a data set for DL model development. Subsequently, the model was validated on the 10 cases of the validation data set. We used HALO Image Analysis (Indica Labs) to detect all nuclei in the tissue for measuring the tumour cellularity (i.e. tumour cell percentage).
Testing study of the DL model
An alternative set of 50 TBB WSIs was used to test the model by comparing pathologist and AI‐generated predictions. First, a representative fragment of each case was selected. Four pathologists, J.F., A.B., Y.K. and H.P., estimated the tumour cellularity by reviewing the virtual slides through conventional visual review. The tumour regions identified by the DL model constructed in the algorithm development phase were subsequently combined with the data obtained from the HALO Image Analysis software to calculate the percentage of nuclei detected within the highlighted tumour areas, thus evaluating the overall tumour cellularity (Figure 1). Based on the level of accuracy perceived by a group of pathologists, these DL model‐generated tumour cellularity estimates were sorted into three groups: ‘good’ (near perfect recognition of tumour cells and non‐tumour cells), ‘fair’ (with minor errors, needing modifications by pathologists) and ‘poor’ (the error exceeds accurate recognition). Representative images are shown in Supporting information, Figure S2. Thereafter, a ground truth was established using a combination of the pathologists’ annotations for tumour regions and the HALO Image Analysis detection of nuclei for tumour cellularity. A statistical comparison between the results of the DL model and pathologists was performed based on the deviation (absolute value of the difference) between these results and the ground truth.
Figure 1.
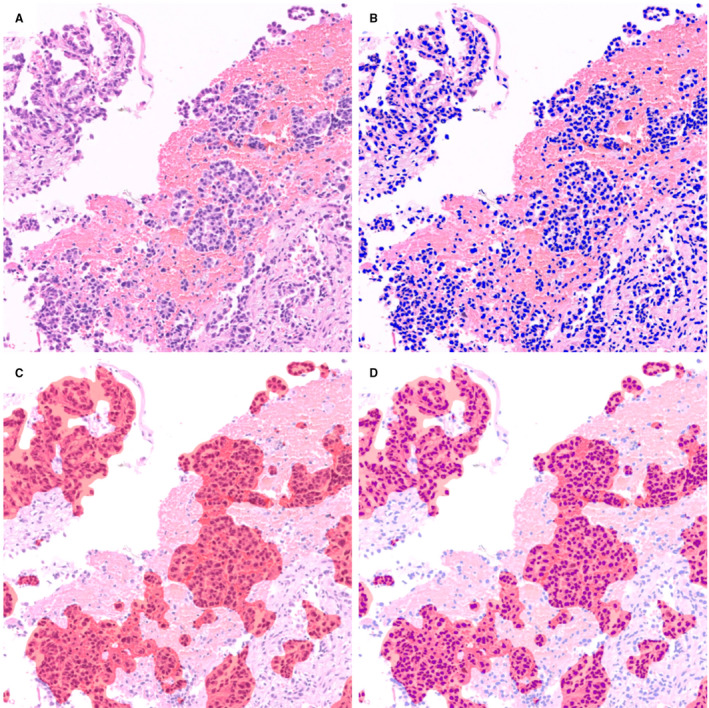
Calculation of tumour cellularity using two algorithms. Original haematoxylin and eosin (H&E) image (A) was analysed using two distinct algorithms. Nuclei on the H&E image were masked as blue markers (B) and tumour clusters were segmented as red masking (C). Combining (B) with (C), the total number of nuclei (number of blue markers) and the number of tumour nuclei (number of blue markers within red mask) could be obtained (D), thus we can calculate the tumour cellularity of samples.
To compare different scanners, 20 cases of TBB were scanned by both a Aperio Scanscope CS2 digital slide scanner and a Philips Ultra Fast scanner, and the quality of the segmentation between the two scanners was compared.
Testing the collaborative workflow between pathologists and the DL model
From April 2019 to September 2020, a total of 151 biopsies and surgical resections of pulmonary adenocarcinoma cases from three institutes (Nagasaki University Hospital, Kameda Medical Center and Awaji Medical Center) were enrolled into this study. All three institutions are fully digitised, i.e. glass slides are scanned before assigning the case to a pathologist. 18 The designed workflow is presented in Figure 2. Initially, the WSIs with suspected adenocarcinoma containing the highest number of tumour cells were selected as per the pathologists’ assessment. These selected images were downloaded by a member of the analysis team and converted to the pyramid TIFF format following anonymisation. For certain cases, the images were cropped. The tumour cells were annotated to enclose the region of interest (ROI) (Figure 2). The WSIs were evaluated using the trained algorithm and the results were shared at sign‐out sessions, where the pathologists were blinded to the results of the AI analysis and were asked to estimate the tumour cellularity of the specimens; their answers were averaged to produce the path‐score. Subsequently, the pathologists visually reviewed the results of the AI analysis represented by the automated tumour detection map and nuclear detection overlay (AI‐score). The pathologists determined the final tumour cellularity (adjusted‐score) by adjusting the AI‐score as they deemed fit. As described earlier, the quality of AI analysis was categorised into three levels; the representative images for each level are presented in Supporting information, Figure S2. For the cases categorised as ‘good’ by multiple pathologists during a sign‐out session, the AI‐scores were included in the pathology report. Regarding research ethics, we followed the guidelines for digital pathology usage published by the Japanese Society of Pathology and included the AI‐score in pathology reports only after validation by a laboratory developed test. 37
Figure 2.
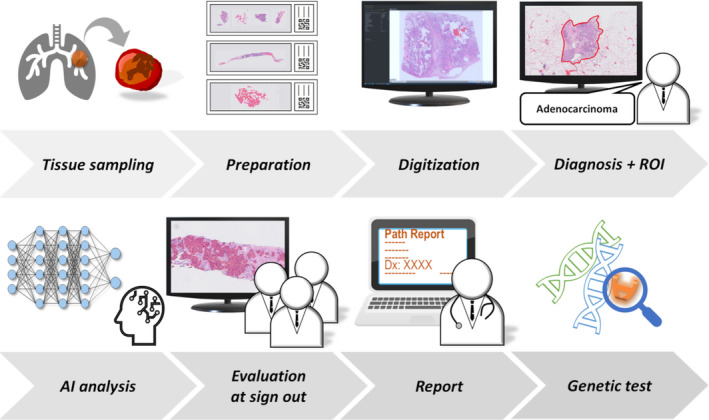
Synergistic workflow between pathologists and artificial intelligence (AI) model. The proposed approach significantly improved the pathologists’ workflow by enabling them to diagnose using digital images. The approach comprised routine operation of the AI model and evaluation of AI analysis results by pathologists. Starting with tissue sampling, specimen preparation and digitisation, the cases diagnosed as adenocarcinoma by pathologists were assigned regions of interest (ROIs) as necessary and subjected to analysis of tumour cellularity by AI. After the analysis, the cases were evaluated and modified by pathologists in a sign‐out session. This process will be followed by reporting the adjusted‐score and selecting the appropriate genetic test.
Adjusted‐score
The cases requiring further major or minor adjustment of the AI‐score were mathematically processed. For instance, in Figure 3, the recognition of tumour cells by AI was considered to be 30% less than the actual value. Therefore, based on the consensus of the pathologists, we added 30% to the AI‐score to obtain the adjusted‐score. In cases judged as ‘poor’ by attending pathologists, we did not refer to the AI‐score and used the path‐score without adjustment (Supporting information, Figure S2). Before applying any modification to the 151 collaborative workflow testing cases, 37 cases from the DL model testing sets were used as a practice set to train pathologists.
Figure 3.
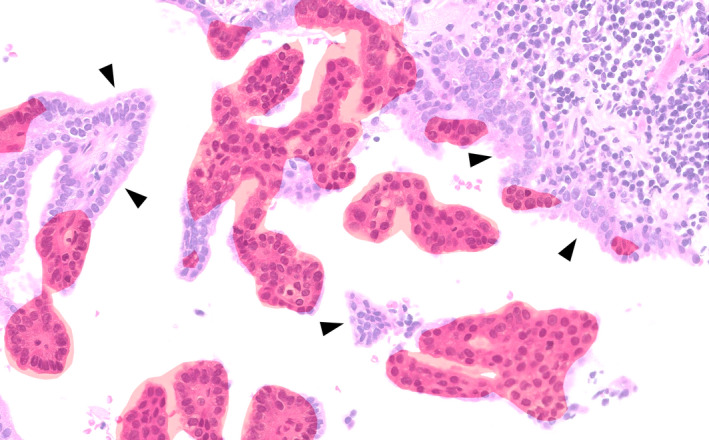
Artificial intelligence (AI)‐generated carcinoma segmentation and pathologist adjustments. Red masked regions were segmented and identified as carcinoma by AI. Certain areas were missed by AI (arrowhead). In such cases, pathologists had to include the missing percentage in AI's calculation of tumour cellularity (AI‐score) to create the adjusted‐score. In this region of interest (ROI), pathologists determined that approximately 30% was required to be added to the AI‐score. Thus, 30% of the tumour area was missed by the segmentation algorithm. In practice, this decision was applied at the whole slide image (WSI) level.
To validate the adjusted‐score, 20 adjusted cases were randomly selected and the tumour cells were manually annotated. The original H&E slides were destained with hydrochloric acid/ethanol solution and immunohistochemically restained with a cocktail of antibodies for thyroid transcription factor 1 (TTF‐1) and napsin A (ADC cocktail; Pathology Institute Corp., Toyama, Japan), 38 which are widely used markers for pulmonary adenocarcinoma. The immunostained slides were rescanned using a Philips Ultra Fast Scanner. Annotations for individual tumour cells were applied based on simultaneous observation of the immunostained slides and the original H&E images (Supporting information, Figure S3). All annotation data were verified by the expert pulmonary pathologist (J.F.). Ultimately, the cell count algorithm was applied to the annotated area, and the tumour cellularity values from those 20 cases were used as the ground truth. These numbers were compared to the original path‐score and the adjusted‐score.
Supplementary materials and methods, including details on algorithm development and validation, the architecture of the deep learning model and statistical analysis, are provided in the Supporting information files.
Results
Algorithm development and testing the AI‐score
To validate the proposed DL software in the algorithm development phase, a model with a cross‐entropy of 0.16 was adopted as the test model. The tumour cellularity of the 40 training WSIs ranged from 0.7 to 46.4%, with a median of 11.7%. The validation set contained 10 WSIs that were divided into 14 611 patches of 0.01 mm2 to match the size of the training patches. Among these 14 611 patches, 7771 were classified by the DL model as positive for the tumour regions and 7630 regions corresponded with the pathologists’ positive annotation, i.e. true positives. Conversely, 6024 of 6840 patches classified as negative by the DL model were true negatives. The overall sensitivity, specificity and accuracy were 97.1, 87.0 and 93.5%, respectively. To evaluate the nuclear recognition, the HALO Image Analysis results were compared with the pathologists’ exact manual count in 10 ROIs, which revealed an accuracy of 98.5%.
In evaluating tumour cellularity in 50 randomly selected cases, the mean deviation from the ground truth among the four participating pathologists was 15%, whereas the mean deviation of the results obtained by the proposed DL model from the ground truth was 6%. Among these 50 cases, 29, 12 and 9 were categorised as good, fair and poor, respectively. In 19 of the 29 cases categorised as ‘good’, the DL model outperformed all the participating pathologists (Figure 4, Supporting information, Table S1). The mean deviation of the DL model in these ‘good’ cases was 3%, whereas the mean deviation for the pathologist estimations was 16%. For the 12 ‘fair’ cases, the DL model deviated 4% on average from the ground truth, whereas the pathologists deviated by 14% on average. For the nine ‘poor’ cases, the mean deviations were 15 and 14% for the DL model and pathologists, respectively. The results show that the consensus judgement of pathologists generally deviated from ground truth by approximately 15%. Supplementary results on false positives and false negatives are provided in the Supporting information files.
Figure 4.
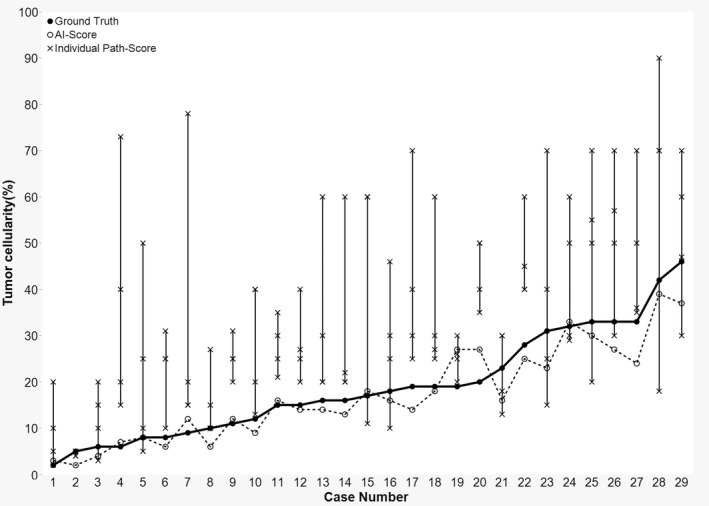
Interobserver variability and inconsistency of pathologist's tumour cellularity estimates in 29 cases assessed as ‘good’ in the 50 testing cases. A line plot displaying ground truth (solid line), artificial intelligence (AI)‐score (dotted line) and individual path‐score (vertical line). In 19 of 29 cases, AI‐score outperformed all participating pathologists’ assigned path‐score.
According to the evaluation of the segmentation quality between the Aperio Scanscope CS2 digital slide scanner and the Philips Ultra Fast scanner in 20 cases, the slides scanned by the Philips Ultra Fast scanner were categorised as ‘good’ in 85% of the cases (17 of 20), while the ones scanned by the Aperio Scanscope CS2 digital slide scanner were categorised as ‘good’ in 65% of the cases (13 of 20). There were no slides categorised as ‘poor’ in either group. The Pearson's correlation coefficient for the AI‐score was high between the two groups (r = 0.89, P < 0.001). Supporting information, Figure S4 shows the comparative images between these two scanners.
Assessing the level of classification in the collaborative workflow
A total of 151 samples were analysed in the testing phase of this study. Of these samples, 26 were acquired from Nagasaki University Hospital, 111 from Kameda Medical Center and 14 from Awaji Medical Center (Supporting information, Figure S1). By consensus of the participating pathologists, the AI segmentation of the samples was labelled by pathologists as follows: 80 good (53%), 38 fair (25%) and 33 poor (22%). AI‐scores of the 80 ‘good’ cases were included in the pathology reports. Data from the practice set, collected from 37 cases (from the data set used for testing the AI‐score), displayed a tendency for overestimation (i.e. ≥ 1% increments) by an individual pathologist (overestimation in 21 of 37). The data for the 151 collaborative workflow testing cases displayed a similar frequency of over‐ and underestimation: overestimation in 67 of 151 cases (44.3%) and underestimation in 65 of 151 cases (43%), as depicted in Supporting information, Figure S6. Based on the generalised Wilcoxon test, the pathologist tended to significantly overestimate the tumour cellularity to a greater extent in the first 37 samples used as a practice set than in the 151 samples for testing the collaborative workflow afterwards (P < 0.005).
The pathologist who led this study was involved in the sign‐out of all the 151 cases enrolled herein. In the first 20 cases evaluated, the pathologist tended to overestimate the tumour cellularity by more than 20%, resulting in a large deviation between the individual path‐score and adjusted‐score. However, after referring to the AI segmentation data from the 20 cases, the pathologist realised that they tended to overestimate (Figure 5). Based on logistic regression analysis, the pathologist's score did not tend to deviate from the adjusted‐score by more than ±20% after case 20 (P = 0.019). This pathologist's path‐score deviated from the adjusted‐score in 132 of 151 samples, which signified that the AI segmentation data caused the pathologist to reconsider their first estimate of the tumour cellularity in 87% of the samples.
Figure 5.
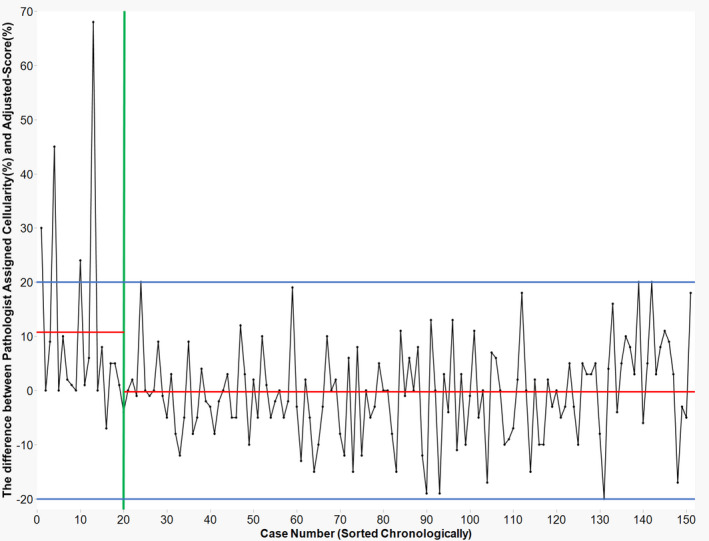
Improvement of cellularity estimation. Chronological variation of deviation between adjusted‐score and path‐score for an individual pathologist. In the early stage, overestimation exceeding 20% deviation from adjusted‐score appeared in certain cases, but the path‐score stabilised. Although the pathologists supervised the artificial intelligence (AI), they could similarly learn from it. These data not only implied that the human‐in‐the‐loop workflow effectively improved the pathologist's assessment but also highlighted the requirement of AI aid (despite increased trials of cellularity estimates, the pathologist's score varied).Red lines: the average difference between individually assigned cellularity and adjusted‐score. Separate averages are shown for case numbers 1–19 (average = 10.95), and from 20 on (average = −0.598). Blue lines: ±20% is shown as a cut‐off range in percentage difference. Green line: case number 20.
Comparison with ground truth
The individual tumour cluster‐level annotations in the 20 randomly selected ground truth cases numbered 4527. This ground truth was established by manually counting the tumour nuclei on H&E‐stained slides while referencing the same slide restained with TTF‐1/napsin A. The average number of annotations was 226.35 per sample (range = 33–526). The mean deviation between the ground truth and adjusted‐score was 3.08%. The adjusted‐score was within 5% of the ground truth in 80% of the test cases (16 of 20) and within 10% in all the cases. The mean deviation of the pathologist's path‐score to ground truth was 7.12%, confirming that the adjusted‐score was generally closer to the ground truth (P = 0.009, Wilcoxon's signed‐rank test) (Figure 6, Supporting information, Figure S7). The adjusted‐score was also superior to the scores obtained by the consensus of the pathologists (P = 0.032, Wilcoxon's signed‐rank test). Among the 20 test cases, the AI segmentation data was categorised as ‘good’ in four cases, ‘fair’ in 12 cases and ‘poor’ in four cases. The median deviation from the ground truth in each level was 3.2, 2.1 and 4.0%, respectively (Supporting information, Figure S8).
Figure 6.
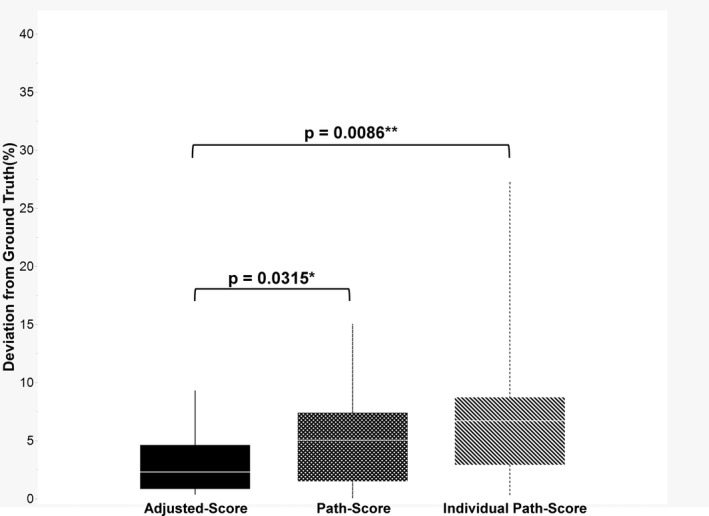
Deviation from the ground truth. The box‐plot displaying adjusted‐score was significantly closer to the ground truth than both the path‐score and individual path‐score for a pathologist. The proposed artificial intelligence (AI)‐based tumour cellularity adjustment attained less than 10% deviation in all the cases and less than 5% deviation in three‐quarters of the cases.
Assessment of the level of classification per sampling modality and among institutions
The proportion of samples categorised by the model as ‘good’ for each sampling method was 53% for TBB, 39% for CNB, 57% for surgical, 93% for TBNA and 33% for cell block (Table 1). The mean adjustment from AI‐score to adjusted‐score for each modality was −19.33% for cell block, +2.53% for CNB, −0.29% for surgical, +1.41% for TBB and +33% for TBNA (Table 2). False positives were highly prominent in the cell block samples (standard deviation = 31%), and the individual cells contained in the cell block samples could not be easily identified, especially mesothelial cells and macrophages floating in the pleural fluid in the thoracic cavity (Tables 1 and 2).
Table 1.
Classification level per sampling method
| Sampling | n | Good | Fair | Poor |
|---|---|---|---|---|
| Transbronchial biopsy | 85 | 52.9% (45) | 24.7% (21) | 22.4% (19) |
| Core needle biopsy | 38 | 39.5% (15) | 34.2% (13) | 26.3% (10) |
| Surgical resection | 7 | 57.1% (4) | 42.9% (3) | – |
| TBNA | 15 | 93.3% (14) | 6.7% (1) | – |
| Cell block | 6 | 33.3% (2) | – | 66.7% (4) |
| Total | 151 | 53% (80) | 25% (38) | 22% (33) |
TBNA, transbronchial needle aspiration.
Table 2.
Mean adjustment from AI‐score per sampling method
| Sampling | Mean adjustment | Standard deviation |
|---|---|---|
| Transbronchial biopsy | 1.41 | 7.38 |
| Core needle biopsy | 2.53 | 6.85 |
| Surgical resection | −0.29 | 9.96 |
| TBNA | 2.33 | 4.15 |
| Cell block | −19.33 | 30.90 |
TBNA, transbronchial needle aspiration.
The percentage of samples rated as ‘good’ from each institution ranged from 48 to 73% (Table 3). The percentage of samples rated ‘good’ was significantly higher in cases from Nagasaki University Hospital than those from Kameda Medical Center (P < 0.05). However, when pooling ‘good’ and ‘fair’ samples, no significant differences were observed among institutions: 88% of Nagasaki University Hospital samples (23 of 26), 76% of Kameda Medical Center samples (84 of 111) and 79% of Awaji Medical Center samples (11 of 14) were labelled either ‘good’ or ‘fair’.
Table 3.
Classification level across three institutions
| Institute* | Total cases | Good | Fair | Poor |
|---|---|---|---|---|
| Nagasaki University Hospital | 26 | 73.1% (19) | 15.4% (4) | 11.5% (3) |
| Kameda Medical Center | 111 | 47.7% (53) | 27.9% (31) | 24.3% (27) |
| Awaji Medical Center | 14 | 57.1% (8) | 21.4% (3) | 21.4% (3) |
| Total | 151 | 53% (80) | 25% (38) | 22% (33) |
All institutes included in this study used a Philips Ultra Fast scanner.
Discussion
We developed a DL image analysis model that yielded tumour cellularity values by integrating a tumour region segmentation model and a nuclear counting algorithm. In addition, we developed a trial workflow to compare the pathologists’ estimates before and after referring to the AI segmentation data.
In our study, pathologists were able to refer to the AI‐score and adjust their original consensus estimates (path‐score) to provide more accurate estimations. In particular, 87% of the 151 test samples involved a pathologist altering the earlier estimation after referring to AI data. Thus, this is a valid reason for incorporating AI into daily practice to provide a more accurate pathological diagnosis. We demonstrated that AI‐aided diagnosis improved the pathologists’ judgement. This primarily materialised in the form of improving large overestimations in the early stages. However, this adjustment increased the proportion of underestimations, which did not exhibit significant improvement regardless of the increased case experience (Figure 5). As human judgement is limited in this context, this may be a rationale for recommending AI‐aided diagnosis. It proved that even for suboptimal accuracy of the AI, the pathologists could visually assess the results of the segmentation model and adjust the calculation of the tumour percentage to obtain a more accurate prediction of the ground truth. Upon referring to the AI data, the human could extract more accurate data, indicating that the collaboration was significant. AI may have limited accuracy in recognition of the cancer cells, which is a common occurrence in pathological investigations. The pathologists make the final decisions, but this collaboration between AI and pathologists – also known as a type of human‐in‐the‐loop modelling 17 , 39 , 40 – is a promising direction for the future of pathological diagnosis and related tasks.
Large interobserver variability was present in the estimates between the pathologists (Figure 4, Supporting information, Table S1), similar to that reported in several prior studies, 27 , 28 , 29 , 30 which challenged consensus‐building. In this study, we adopted the average scores marked by the pathologists as the consensus. Although this average score was an improvement over individual pathologists’ scores, the collaborative method between the physician and the AI was ultimately the most accurate method. In clinical applications of AI, pathologists must be able to interpret the results and provide input. We overlaid the segmentation data on the WSI, reviewed the tumour cellularity percentages, verified the level of nuclear recognition and modified the AI‐scores. Pathologists could enhance their practice by utilising the AI model. Moreover, the differences in staining and specimen preparation techniques between laboratories are some of the major barriers to the adoption of AI. 41 , 42 This was confirmed, as the model yielded the best results on samples acquired from the institution at which training was conducted (Table 3), as well as a higher frequency of errors on samples from other institutions. However, input from a pathologist was able to compensate for these minor errors. We examined whether the accuracy of the adjusted‐score varied with the decreasing classification level, but no clear deterioration was observed (Supporting information, Figure S8). This indicated that the specimen preparation and staining procedures conducted at various institutions did not significantly impact the tumour nuclei count obtained using the proposed model, and the AI model can be used at any institution. This notion was further extended by our subgroup analysis using two different WSI scanners. Interestingly, our study showed that the levels of tumour cell recognition between the two modalities were identical and showed a high correlation.
There are certain limitations to this study. Although the annotations of the training data were highly accurate at the region level, the number of cases was small. Secondly, for testing the collaborative workflow, the WSI scanner used was changed, but the AI model was not adjusted to match the scanner. We did not train the model with additional data extracted by a Philips Ultra Fast Scanner to improve detection. Thirdly, cross‐validation for the 50 cases – 40 training and 10 validation – was not performed due to limitations in our ability to modify the code of the HALO‐AI model, as per our licensing agreement. Fourthly, this study was conducted using only digital data from lung adenocarcinoma tumours.
In conclusion, we developed an AI model and a human‐in‐the‐loop collaborative workflow to evaluate tumour cellularity in lung adenocarcinoma. This study demonstrated that the proposed model could more accurately determine tumour cellularity than pathologists’ consensus alone, and additionally implied that pathologists can learn from the implementation of AI. The collaboration between AI and pathologists can result in a synergistic, positive feedback loop in which each side improves the other.
Conflicts of interest
J.F. receives research funding from N Lab. Co. Ltd. The other authors declare that there are no conflicts of interest.
Ethics approval statement
The study protocol was approved by the Institutional Review Board of Nagasaki University Hospital (#190218282, #190311162).
Supporting information
Figure S1. Flowchart of study dataset
The study dataset was designed in three phases: the algorithm development phase (training and validation), testing the AI‐Score, and testing the collaborative workflow. This flowchart includes the number of each dataset, types of sampling, name of scanners, year, and the institution's name.WSI: whole slide image; TBB: transbronchial biopsy; CNB: core needle biopsy; Surgical: surgical resection; TBNA: transbronchial needle aspiration; Nagasaki Univ.: Nagasaki University Hospital; Kameda Med. Ctr.: Kameda Medical Center; Awaji Med. Ctr.: Awaji Medical Center; IHC: immunohistochemical.
Figure S2. Representative images with red classification mapping judged as good, fair, and poor by pathologists.
The segmentation level of each of the cases was judged as one of the three levels: good (A‐L), fair (M‐P), and poor (Q‐T). (10x)
Figure S3. Creation of ground truth based on simultaneous observations of H&E slides and immunostained slides.
In 20 cases, pathologist‐supervised annotations were collected to compile a ground truth dataset.
(A) Original H&E image.
(B) Re‐stained slide of original H&E specimen with TTF‐1 & napsin A cocktail.
(C) Segmentation output by AI model.
(D) Ground truth established by meticulous annotation with reference to immunostained slides. (40x).
Figure S4. Comparison of different scanners.
The level of segmentation when using different scanners was evaluated. (A), (C), and (E) were the images scanned by the Aperio Scanscope CS2 digital slide scanner, and (B), (D), and (F) were scanned by the Philips Ultra Fast scanner. The segmentation levels of both (A) and (B) were categorized as ‘good’; (C) was assigned as ‘good’, but (D) was assigned as ‘fair’ because of the several false negative areas. Meanwhile, (E) was assigned as ‘fair’, while (F) was assigned as ‘good’ since (E) had much more false‐positive areas than (F).
Figure S5. Examples of false positives and false negatives.
Bronchial epithelium (A), alveolar macrophages (B), lymphocyte infiltration/aggregation (C), tracheal cartilage (D), and anthracotic pigments (E) were detected as false positives in certain instances. Invasive mucinous adenocarcinoma (F) tended to possess weakly atypical nuclei and could result in false negatives. Upon weighing the number of these false positives and false negatives, the pathologists considered the extent of correction required in the AI‐Score. (40x)
Figure S6. Trends in the practice and testing phases.
These plots show the deviation between an individual pathologist's assigned scores (Path‐Score) and the final Adjusted‐score for 151 testing cases (A), and 37 cases as a practice set for pathologists to evaluate collaborative workflow (B) in the early phase when AI was implemented into the cell count workflow. The line on both plots represents when Path‐Score exactly equals the Adjusted‐score. Although the pathologist tended to overestimate the cellularity at first (points above the line), the frequency of overestimation and underestimation was almost even after gaining experience with the AI.
Figure S7. Deviation from ground truth in 20 cases.
Line plot showing the ground truth (solid line), Adjusted‐score (dotted line), and Path‐Score (vertical line). As observed, the interobserver variability in pathologists' estimations was resolved by adjusting AI‐Score.
Figure S8. Deviation from ground truth per classification level.
Three‐line plots showing Ground truth (solid line), Adjusted‐score (dotted line), and individual Path‐Score (vertical line) categorized as per the samples judged as good (A), fair (B), and poor (C), respectively.
Table S1. Inter‐observer variability of Path‐Score in 29 cases judged as good in the 50 cases used for AI‐Score testing.
Acknowledgements
This study was supported by the JPNP20006 project commissioned by the New Energy and Industrial Technology Development Organisation (NEDO).
Data availability statement
Detailed data will be provided by the corresponding author upon the reasonable request." cd_value_code="text
References
- 1. Aggarwal R, Sounderajah V, Martin G et al. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta‐analysis. NPJ Digital Med. 2021; 4; 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Çallı E, Sogancioglu E, van Ginneken B, van Leeuwen KG, Murphy K. Deep learning for chest X‐ray analysis: A survey. Med. Image Anal. 2021; 72; 102125. [DOI] [PubMed] [Google Scholar]
- 3. Hinton G. Deep learning‐a technology with the potential to transform health care. JAMA 2018; 320; 1101–1102. [DOI] [PubMed] [Google Scholar]
- 4. Litjens G, Kooi T, Bejnordi BE et al. A survey on deep learning in medical image analysis. Med. Image Anal. 2017; 42; 60–88. [DOI] [PubMed] [Google Scholar]
- 5. Bychkov D, Linder N, Turkki R et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 2018; 8; 3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fan J, Wang J, Chen Z, Hu C, Zhang Z, Hu W. Automatic treatment planning based on three‐dimensional dose distribution predicted from deep learning technique. Med. Phys. 2019; 46; 370–381. [DOI] [PubMed] [Google Scholar]
- 7. Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 2019; 29; 102–127. [DOI] [PubMed] [Google Scholar]
- 8. Nielsen A, Hansen MB, Tietze A, Mouridsen K. Prediction of tissue outcome and assessment of treatment effect in acute ischemic stroke using deep learning. Stroke 2018; 49; 1394–1401. [DOI] [PubMed] [Google Scholar]
- 9. Yu KH, Zhang C, Berry GJ et al. Predicting non‐small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016; 7; 12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Med. Image Comput. Comput. Assist. Interv. 2013; 16; 411–418. [DOI] [PubMed] [Google Scholar]
- 11. Cruz‐Roa A, Basavanhally A, González F et al. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. Proc. SPIE Med. Imaging 2014; 9041; 904103. [Google Scholar]
- 12. Ehteshami Bejnordi B, Veta M, Johannes van Diest P et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017; 318; 2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ertosun MG, Rubin DL. Automated grading of gliomas using deep learning in digital pathology images: A modular approach with ensemble of convolutional neural networks. Proc. Am. Med. Informat. Associat. Annu. Symp. 2015; 2015; 1899. [PMC free article] [PubMed] [Google Scholar]
- 14. Litjens G, Sánchez CI, Timofeeva N et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016; 6; 26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pham HHN, Futakuchi M, Bychkov A, Furukawa T, Kuroda K, Fukuoka J. Detection of lung cancer lymph node metastases from whole‐slide histopathologic images using a two‐step deep learning approach. Am. J. Pathol. 2019; 189; 2428–2439. [DOI] [PubMed] [Google Scholar]
- 16. Bandi P, Geessink O, Manson Q et al. From detection of individual metastases to classification of lymph node status at the patient level: The CAMELYON17 challenge. IEEE Trans. Med. Imaging 2019; 38; 550–560. [DOI] [PubMed] [Google Scholar]
- 17. Uegami W, Bychkov A, Ozasa M et al. MIXTURE of human expertise and deep learning‐developing an explainable model for predicting pathological diagnosis and survival in patients with interstitial lung disease. Mod. Pathol. 2022; 35; 1083–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sakamoto T, Furukawa T, Lami K et al. A narrative review of digital pathology and artificial intelligence: focusing on lung cancer. Transl. Lung Cancer Res. 2020; 9; 2255–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van der Laak J, Litjens G, Ciompi F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021; 27; 775–784. [DOI] [PubMed] [Google Scholar]
- 20. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J. Clin. 2021; 71; 7. [DOI] [PubMed] [Google Scholar]
- 21. Herzberg B, Campo MJ, Gainor JF. Immune checkpoint inhibitors in non‐small cell lung cancer. Oncologist 2017; 22; 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yuan M, Huang LL, Chen JH, Wu J, Xu Q. The emerging treatment landscape of targeted therapy in non‐small‐cell lung cancer. Signal Transduct. Target. Ther. 2019; 4; 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen H, Luthra R, Goswami RS, Singh R, Roy‐Chowdhuri S. Analysis of pre‐analytic factors affecting the success of clinical next‐generation sequencing of solid organ malignancies. Cancer 2015; 7; 1699–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindeman NI, Cagle PT, Aisner DL et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Thorac. Oncol. 2018; 13; 323. [DOI] [PubMed] [Google Scholar]
- 25. Thunnissen E, Kerr KM, Herth FJ et al. The challenge of NSCLC diagnosis and predictive analysis on small samples. Practical approach of a working group. Lung Cancer 2012; 76; 1–18. [DOI] [PubMed] [Google Scholar]
- 26. Fan X, Furnari FB, Cavenee WK, Castresana JS. Non‐isotopic silver‐stained SSCP is more sensitive than automated direct sequencing for the detection of PTEN mutations in a mixture of DNA extracted from normal and tumor cells. Int. J. Oncol. 2001; 18; 1023–1026. [DOI] [PubMed] [Google Scholar]
- 27. Lhermitte B, Egele C, Weingertner N et al. Adequately defining tumor cell proportion in tissue samples for molecular testing improves interobserver reproducibility of its assessment. Virchows Arch. 2017; 470; 21–27. [DOI] [PubMed] [Google Scholar]
- 28. Mikubo M, Seto K, Kitamura A et al. Calculating the tumor nuclei content for comprehensive cancer panel testing. J. Thorac. Oncol. 2020; 15; 130–137. [DOI] [PubMed] [Google Scholar]
- 29. Smits AJ, Kummer JA, de Bruin PC et al. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod. Pathol. 2014; 27; 168–174. [DOI] [PubMed] [Google Scholar]
- 30. Viray H, Li K, Long TA et al. A prospective, multi‐institutional diagnostic trial to determine pathologist accuracy in estimation of percentage of malignant cells. Arch. Pathol. Lab. Med. 2013; 137; 1545–1549. [DOI] [PubMed] [Google Scholar]
- 31. Coudray N, Ocampo PS, Sakellaropoulos T et al. Classification and mutation prediction from non‐small cell lung cancer histopathology images using deep learning. Nat. Med. 2018; 24; 1559–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khosravi P, Kazemi E, Imielinski M, Elemento O, Hajirasouliha I. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine 2018; 27; 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Q, Wang X, Liang F et al. A Bayesian hidden Potts mixture model for analyzing lung cancer pathology images. Biostatistics 2019; 20; 565–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luo X, Yin S, Yang L et al. Development and validation of a pathology image analysis‐based predictive model for lung adenocarcinoma prognosis—A multi‐cohort study. Sci. Rep. 2019; 9; 6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luo X, Zang X, Yang L et al. Comprehensive computational pathological image analysis predicts lung cancer prognosis. J. Thorac. Oncol. 2017; 12; 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang H, Chen L, Cheng Z et al. Deep learning‐based six‐type classifier for lung cancer and mimics from histopathological whole slide images: A retrospective study. BMC Med. 2021; 19; 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. The Japanese Society of Pathology, Digital Pathology Guideline , 2018; [Accessed July 31, 2022]: Available from: https://pathology.or.jp/jigyou/pdf/guideline‐20190326.pdf
- 38. Whithaus K, Fukuoka J, Prihoda TJ, Jagirdar J. Evaluation of napsin a, cytokeratin 5/6, p63, and thyroid transcription factor 1 in adenocarcinoma versus squamous cell carcinoma of the lung. Arch. Pathol. Lab. Med. 2012; 136; 155–162. [DOI] [PubMed] [Google Scholar]
- 39. Bodén ACS, Molin J, Garvin S, West RA, Lundström C, Treanor D. The human‐in‐the‐loop: an evaluation of pathologists' interaction with artificial intelligence in clinical practice. Histopathology 2021; 79; 210–218. [DOI] [PubMed] [Google Scholar]
- 40. Budd S, Robinson EC, Kainz B. A survey on active learning and human‐in‐the‐loop deep learning for medical image analysis. Med. Image Anal. 2021; 71; 102062. [DOI] [PubMed] [Google Scholar]
- 41. Janowczyk A, Zuo R, Gilmore H, Feldman M, Madabhushi A. HistoQC: an open‐source quality control tool for digital pathology slides. JCO Clin. Cancer Inform. 2019; 3; 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schömig‐Markiefka B, Pryalukhin A, Hulla W et al. Quality control stress test for deep learning‐based diagnostic model in digital pathology. Mod. Pathol. 2021; 34; 2098–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Flowchart of study dataset
The study dataset was designed in three phases: the algorithm development phase (training and validation), testing the AI‐Score, and testing the collaborative workflow. This flowchart includes the number of each dataset, types of sampling, name of scanners, year, and the institution's name.WSI: whole slide image; TBB: transbronchial biopsy; CNB: core needle biopsy; Surgical: surgical resection; TBNA: transbronchial needle aspiration; Nagasaki Univ.: Nagasaki University Hospital; Kameda Med. Ctr.: Kameda Medical Center; Awaji Med. Ctr.: Awaji Medical Center; IHC: immunohistochemical.
Figure S2. Representative images with red classification mapping judged as good, fair, and poor by pathologists.
The segmentation level of each of the cases was judged as one of the three levels: good (A‐L), fair (M‐P), and poor (Q‐T). (10x)
Figure S3. Creation of ground truth based on simultaneous observations of H&E slides and immunostained slides.
In 20 cases, pathologist‐supervised annotations were collected to compile a ground truth dataset.
(A) Original H&E image.
(B) Re‐stained slide of original H&E specimen with TTF‐1 & napsin A cocktail.
(C) Segmentation output by AI model.
(D) Ground truth established by meticulous annotation with reference to immunostained slides. (40x).
Figure S4. Comparison of different scanners.
The level of segmentation when using different scanners was evaluated. (A), (C), and (E) were the images scanned by the Aperio Scanscope CS2 digital slide scanner, and (B), (D), and (F) were scanned by the Philips Ultra Fast scanner. The segmentation levels of both (A) and (B) were categorized as ‘good’; (C) was assigned as ‘good’, but (D) was assigned as ‘fair’ because of the several false negative areas. Meanwhile, (E) was assigned as ‘fair’, while (F) was assigned as ‘good’ since (E) had much more false‐positive areas than (F).
Figure S5. Examples of false positives and false negatives.
Bronchial epithelium (A), alveolar macrophages (B), lymphocyte infiltration/aggregation (C), tracheal cartilage (D), and anthracotic pigments (E) were detected as false positives in certain instances. Invasive mucinous adenocarcinoma (F) tended to possess weakly atypical nuclei and could result in false negatives. Upon weighing the number of these false positives and false negatives, the pathologists considered the extent of correction required in the AI‐Score. (40x)
Figure S6. Trends in the practice and testing phases.
These plots show the deviation between an individual pathologist's assigned scores (Path‐Score) and the final Adjusted‐score for 151 testing cases (A), and 37 cases as a practice set for pathologists to evaluate collaborative workflow (B) in the early phase when AI was implemented into the cell count workflow. The line on both plots represents when Path‐Score exactly equals the Adjusted‐score. Although the pathologist tended to overestimate the cellularity at first (points above the line), the frequency of overestimation and underestimation was almost even after gaining experience with the AI.
Figure S7. Deviation from ground truth in 20 cases.
Line plot showing the ground truth (solid line), Adjusted‐score (dotted line), and Path‐Score (vertical line). As observed, the interobserver variability in pathologists' estimations was resolved by adjusting AI‐Score.
Figure S8. Deviation from ground truth per classification level.
Three‐line plots showing Ground truth (solid line), Adjusted‐score (dotted line), and individual Path‐Score (vertical line) categorized as per the samples judged as good (A), fair (B), and poor (C), respectively.
Table S1. Inter‐observer variability of Path‐Score in 29 cases judged as good in the 50 cases used for AI‐Score testing.
Data Availability Statement
Detailed data will be provided by the corresponding author upon the reasonable request." cd_value_code="text


