Abstract
Campylobacter, a major cause of food‐borne gastroenteritis worldwide, colonize the gastrointestinal tract of a wide range of animals, being birds the main reservoir. The mechanisms involved in the interaction of Campylobacter with the different hosts are poorly understood. The cytolethal distending toxin, encoded in the cdtABC operon, is considered a pivotal virulence factor during human infection. Differences in the prevalence of cdtABC genes in Campylobacter isolates from three distinct origins (wild birds, broiler chickens and humans) prompted us to further characterize their allelic variability. The sequence of cdtABC is highly conserved among broiler and human isolates. A high diversity of cdtABC alleles was found among wild bird isolates, including several alleles that do not produce any functional CDT. These results suggest that specific variants of the cdtABC operon might define the host range of specific Campylobacter jejuni isolates. Moreover, our data indicate that PCR methodology is inaccurate to characterize the prevalence of the cdt genes, since negative PCR detection can be the result of divergences in the sequence used for primer design rather than indicating the absence of a specific gene.
Keywords: allelic variability, Campylobacter, cytolethal distending toxin, PCR detection, WGS
Impacts.
The sequence of cdtABC operon, encoding the cytolethal distending toxin in Campylobacter jejuni, is highly conserved among broiler and human isolates. Instead, a high diversity is found among wild bird isolates, including several alleles that do not produce any functional CDT.
Sequence analysis reveals that the cdtABC operon seems to accumulate DNA modifications, particularly deletions, especially detected among wild bird isolates.
The data obtained indicate that PCR methodology is inaccurate to characterize the prevalence of the cdt genes, since negative PCR detection can be the result of divergences in the sequence used for primer design rather than indicating the absence of the specific nucleotide sequences of a specific gene.
1. INTRODUCTION
Campylobacter is a major enteropathogen worldwide, causing the most common zoonoses reported in humans in the European Union, the United States and Australia (European Food Safety Authority and European Centre for Disease Prevention and Control, 2021; Kaakoush et al., 2015). In most cases, campylobacteriosis is self‐limiting gastroenteritis. Severe cases, although rare, occur mostly among children younger than 5 years, elderly and immunocompromised patients (Kaakoush et al., 2015). Moreover, infection by Campylobacter can trigger the autoimmune polyneuropathic disorder Guillain–Barré syndrome (Koga et al., 2006). Campylobacter jejuni is the species reported to cause most of the campylobacteriosis cases in humans (European Food Safety Authority and European Centre for Disease Prevention and Control, 2021).
Campylobacter jejuni pathogenicity remains poorly understood. Among the array of bacterial products that contribute to its virulence, the cytolethal distending toxin (CDT) seems to play a pivotal role in the interaction of Campylobacter with the host. CDT blocks cell division, causing cell cycle arrest at the G2 stage prior to mitosis (Whitehouse et al., 1998). CDT is a heteromeric AB‐type genotoxin produced by several Gram‐negative bacterial pathogens, including Campylobacter, enteropathogenic Escherichia coli, Salmonella enterica serovar Typhi, Haemophilus duceryi and Shigella dysenteriae (Jinadasa et al., 2011). The toxin consists of three subunits CdtA, CdtB and CdtC. CdtB is the active subunit whereas CdtA and CdtC are required for binding and delivery of CdtB within the target cell by a process that is still poorly understood (Lara‐Tejero & Galán, 2001). CdtA/CdtC binds to the lipid rafts, cholesterol‐rich microdomains within the host cell membranes (Lai et al., 2016). CdtB has DNaseI‐like activity producing DNA double‐strand breaks in the host cell that causes arrest of the cell cycle, cellular distension and cell death (Smith & Bayles, 2006). Although CDT is considered a crucial virulence factor during human infection, it remains unclear its role during the colonization of natural hosts, such as birds.
Campylobacter CDT toxin is encoded in the polycistronic operon cdtABC. Most Campylobacter epidemiological studies reporting prevalence of genes coding for potential virulence factors, including CDT, are based in the use of PCR for gene detection. Very disparate data on cdtABC prevalence among C. jejuni isolates from human patients suffering of gastroenteritis has been reported using this methodology. Some studies indicate a high prevalence of the cdtA, cdtB and cdtC genes, whereas other reports much lower prevalence, ranging from 100% (50/50) to 15% (3/20; Bang et al., 2003; de Melo et al., 2021; Iglesias‐Torrens et al., 2018; Koolman et al., 2015; Pickett et al., 1996; Weis et al., 2014).
The allelic variability of the cdtABC genes in Campylobacter isolates from three distinct origins (wild birds, broiler chickens and humans) has been characterized. The present study aimed to evaluate the diversity of cdt genes among the isolates from different origins, and the possible role that this variability could play in Campylobacter host range specificity.
2. MATERIALS AND METHODS
2.1. Bacterial strains and culture conditions
The C. jejuni strains used in this study are part of a strain collection, previously described (Iglesias‐Torrens et al., 2018), composed of isolates obtained from faeces of three different sources: human suffering symptomatic gastroenteritis (50), broiler chicken (50) and wild birds (50). Wild bird isolates were obtained from different species: yellow‐legged gulls (Larus michahellis) from Barcelona (YLGB) and from Medes Islands (YLGM); Audouin's gulls (Larus audouinii) from Ebro Delta (AGD) and Alboran Islands (AGA); feral pigeons (Columba livia) from Barcelona (FP); common ravens (Corvus corax) from Barcelona (CR); white storks (Ciconia ciconia) from Lleida (WS); and norther shoveler (Spatula clypeata) from Ebro Delta (NS). Isolates were cultured onto Columbia blood agar (CBA) plates (Sharlau) and incubated at 42°C for 48 hr in microaerophilic conditions (CampyGen, Oxoid).
2.2. PCR amplification
Genomic DNA was extracted from cultures grown on CBA plates using the InstaGene Matrix Kit (Bio‐Rad Laboratories). PCR reactions (PCR Master Mix x2, ThermoScientific) were performed using 35 ng of DNA as a template and the specific primers for amplification of cdtA, cdtB and cdtC (Table 1) previously used in epidemiological studies (Martínez et al., 2006) and based in the genomic sequence of the reference C. jejuni strain 81–176 Amplified PCR products from cdtB gene of representative isolates showing different electrophoretic migration were purified using the E.Z.N.A Cycle Pure Kit (OMEGA Bio‐tek) and sequenced using the same primers as those used to generate the fragment.
TABLE 1.
Primers used in this work
| Primer | Sequence 5′–3′ | PCR product (bp) | |
|---|---|---|---|
| cdtA | Fw | CTATTACTCCTATTACCCCACC | 422 |
| Rv | AATTTGAACCGCTGTATTGCTC | ||
| cdtB | Fw | AGGAACTTTACCAAGAACAGCC | 531 |
| Rv | GGTGGAGTATAGGTTTGTTGTC | ||
| cdtC | Fw | ACTCCTACTGGAGATTTGAAAG | 339 |
| Rv | CACAGCTGAAGTTGTTGTTGGC | ||
2.3. Genome sequencing
Whole genome sequencing was performed for a selection of 47 C. jejuni isolates, including 12 isolates from broiler, 16 from human and 19 from wild birds. The selection was performed in order to sequence isolates from different origins, sequence types virulence and antibiotic resistance profiles (Iglesias‐Torrens et al., 2018). The DNA extraction was carried out with the Wizard DNA‐Purification kit (Promega). The DNA extracted was send to LifeSequencing (Parc Científic, Universitat de València, Spain) to proceed with the DNA library preparation using the Nextera XT DNA Preparation kit and the NexSeq 500 platform. The BioProject has been deposited at NCBI GenBank, accession number PRJNA850915. To find the cdtABC nucleotide sequences in each genome we looked for the primers used in previous amplifications by doing the following. We used the “DNAStringSet” function from the Biostrings package (Pagès et al., 2022) in the R (R Core Team, 2020) programming environment (v.3.0.2) to read the primers and genomes in R. The “vmatchPattern” function was used to find all occurrences of the primers with at most 1 mismatch. The start and end positions and the corresponding strand were recorded.
2.4. Phylogenetic analysis
Evolutionary analyses were conducted with MEGA X (MEGA v10.1.8) (Kumar et al., 2018). The evolutionary history was inferred using the Neighbour‐Joining method (Saitou & Nei, 1987). The evolutionary divergence between cdtABC sequences was inferred by estimating the distance matrix via the Maximum Composite Likelihood method (Tamura et al., 2004).
2.5. Ethics approval
No ethical approval was obtained because this study did not involve human subjects, animals or tissue samples and only involved non‐invasive procedures.
3. RESULTS
3.1. The prevalence of the cdtA , cdtB and cdtC genes is highly variable among Campylobacter jejuni isolates from wild birds
The prevalence of genes coding for cdtA, cdtB and cdtC in a collection of 150 isolates of C. jejuni from human suffering symptomatic grastroenteritis, broiler chiken and wild birds (see Materials and methods for details) was determined by PCR. The results indicated that the distribution of the cdt genes was very variable. The three cdt genes were found in all broiler chicken and human isolates. In contrast to what it was observed among human and broiler isolates, 22 out of 50 isolates from wild birds were negative for the detection of one or two of the cdt genes (Figure 1), and prevalence varies for each gene, being 58%, 94% and 90% for cdtA, cdtB and cdtC, respectively (Figure 1). The highest variability was detected for the cdtA gene. Hence, 14 isolates were cdtA negative, three isolates were cdtAB negative, four isolates were cdtAC negative and one was negative for cdtC.
FIGURE 1.
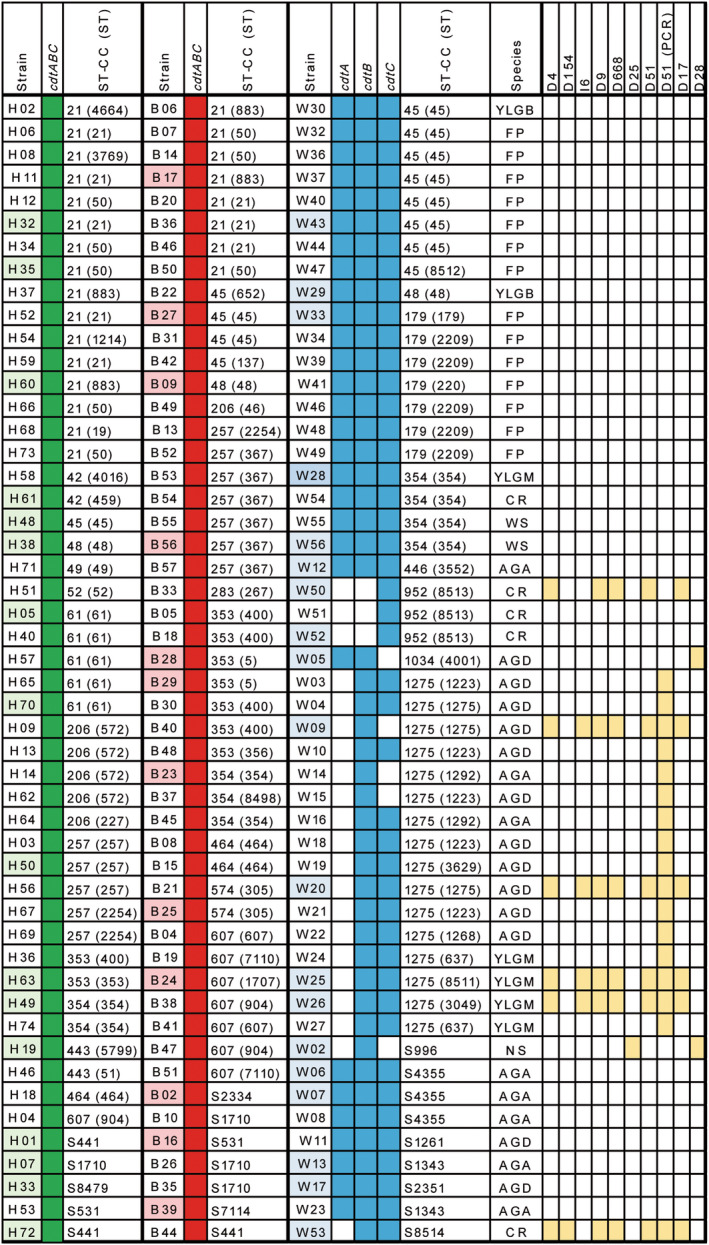
Collection of Campylobacter jejuni isolates from human patients (H), broiler chicken (B) and wild birds (W). The strains are organized attending to the host and ST clonal complexes (ST‐CC). Sequence types are also indicated (ST). The species of the wild bird strains are specified (YLGB: yellow‐legged gulls from Barcelona; YLGM: yellow‐legged gulls from Medes Islands; AGD: Audouin's gulls from Ebro Delta: AGA: Audouin's gulls from Alboran Islands; FP: feral pigeons; CR: common ravens; WS: white storks; NS: northern shoveler). cdtA, cdtB and cdtC positive strains by PCR amplification are indicated in colour background (green for human isolates, red for broiler chicken isolates and blue for wild bird isolates). The different nucleotide deletions and insertions determined by WGS or PCR analysis are indicated in yellow. The shadowed strain names (clear green, pink and clear blue) indicate the strains that the complete cdtABC operon sequence have been determined.
Thirteen out of the fourteen cdtA‐negative isolates belonged to the clonal complex ST‐1,275 which has been described to be highly predominant among wild birds (Hughes et al., 2009; Sheppard et al., 2009). These isolates were collected from Audouin's gulls and yellow‐legged gull from three distant location in the Spanish Mediterranean coast (Medes Islands, Ebro Delta, and Alboran Island). The remaining cdtA‐negative isolate belonged to the singleton ST‐8514 and it was collected from a common raven. Three out of four cdtAC‐negative isolates from Audouin's gulls from Ebro Delta and Alboran Island also belonged to the CC ST‐1275. The last cdtAC‐negative isolate belonged to the singleton ST‐996 and was collected from a northern shoveler in Ebro Delta. The three cdtAB‐negative belonged to the CC ST‐952 and were collected from common ravens. The cdtC‐negative isolate belonged to the CC ST‐1034 and was collected from an Audouin's gull in Ebro Delta. Although all cdt gene negative isolates were collected from wild birds, it should be point out that C. jejuni isolates positive for the three cdt genes were also collected from wild birds at the same geographical locations. Our data show that a high diversity in the prevalence of the cdt genes was observed among C. jejuni isolates from wild birds when compared with isolates from broilers and humans.
When considering the ST‐complex determined for each isolate, in most cases, the isolates belonging to a specific ST‐complex shared the same pattern of cdtABC prevalence (Figure 1). The only exception was within the ST‐1275 complex which was the most represented ST among wild birds (16 out of 50 isolates). All the ST‐1,275 complex isolates were cdtA negative whereas only three of them were cdtC negative by PCR.
3.2. Different cdtB alleles detected among the wild bird isolates
A puzzling observation was done during the PCR genotyping analysis of the different cdt genes. Most isolates PCR‐positive for cdtB depicted a band that migrated as expected for the canonical cdtB sequence of the reference strain 81–176 (531 bp). However, some isolates showed a band that migrated more than expected, indicating that the PCR amplified band carried a shorter sequence (Figure 2a). These results suggested the presence of different cdtB alleles among distinct isolates. Hence, from the 46 cdtB‐positive wild bird isolates, 17 apparently carried the shorter cdtB allele, representing 36.9% of the cdtB‐positive isolates. Sixteen isolates belonged to the CC ST‐1275 and the remaining isolate to the singleton ST‐8514 (Figure 2b). Remarkably, all the isolates that carried a shorter cdtB allele were PCR‐negative for cdtA. It should be noticed that 100% of the broiler and human isolates carry a cdtB allele that by PCR amplification resembled the canonical allele of 81–176. The amplicons from the reference strain 81–176, the isolates W02, W54, carrying an apparently canonical allele; and W20, W25, W53, showing a shorter cdtB allele, were sequenced (Figure 2c).The amplicons from W20, W25 and W53 showed the same feature when compared to 81–176 amplicon, a deletion of 51 nucleotides, resulting in an amplicon of 480 nucleotides (as compared with 531 bp amplicon from 81–176), that explained the shorter band detected by PCR. From now, we will refer to this cdtB allele as cdtB 480 . Interestingly, in the genome of the strain 81–176, the 51 bp deleted sequence from cdtB 480 allele was flanked by two 10 bp direct repeats (Figure 2c), suggesting that the cdtB 480 allele was most probably generated by a site‐specific recombination event.
FIGURE 2.

Identification of the cdtB 480 allele. (a) PCR amplification of cdtB gene from strains 81–176 (lane 1), W50 (lane 2), W54 (lane 3) and W54 (lane 4). The amplification of the housekeeping gltA gene was used as internal control. (b) Isolates that carry the cdtB 480 allele. (c) Sequences obtained from the PCR amplification of strains carrying the cdtB 81‐176 or the cdtB 480 allele. Shadow square indicates the 10 bp direct repeats flanking the 51 bp deletion.
3.3. Whole‐genome sequencing reveals a high diversity in the cdtABC locus among wild bird isolates
To further characterize the diversity within the cdtABC locus, whole‐genome sequencing (WGS) was performed for 47 C. jejuni isolates, including 12 isolates from broiler, 16 from human and 19 from wild birds. The sequence of the cdtABC locus was characterized (Figure S1, Appendix S1). Surprisingly, sequences of the three ORFs of the cdtABC operon were found in all 19 isolates from wild birds, including isolates that were negative by PCR for cdtA (5 isolates), cdtAB (2 isolates), cdtAC (2 isolates) and cdtC (1 isolate; Figure 1). Sequence alignment of the cdtABC sequences from all 47 sequenced isolates and the reference strain 81–176, revealed that 38 of the isolates carried a cdtABC operon of similar length to the cdtABC 81‐176 , whereas 9 isolates, all of them from wild birds, had suffer drastic DNA modifications, resulting in shorter cdtABC operons (Figure S1, Appendix S1).
The optimal tree of the evolutionary analyses of the cdtABC operons (Figure 3a) showed that most of the operons from broiler and human isolates formed a very compact cluster (cluster I in Figure 3a). Only four wild bird isolates were found scattered within this cluster I, whereas most of the cdtABC operons from wild bird isolates were distant from the broiler/human cluster. The four wild bird isolates founded within the broiler/human cluster I were W12, a ST‐446 isolated from AGA; W29 a ST‐48 isolated from YLGB and W28 and W56, two ST‐354 isolated from YLGM and WS, respectively. The four isolates were determined as cdtABC positive by PCR detection.
FIGURE 3.
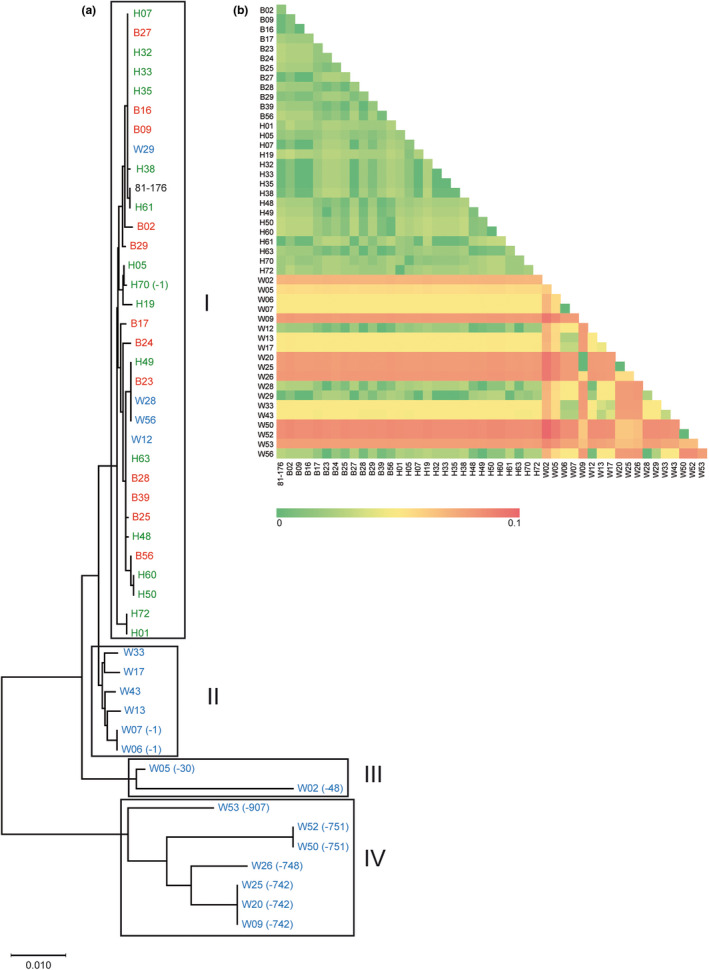
(a) Evolutionary relationships of 47 cdtABC sequences obtained from the WGS, inferred using the Neighbour‐Joining method. The optimal tree with the sum of branch length = 0.01161943 is shown. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 2,181 positions in the final dataset. (b) Evolutionary divergence between sequences. The number of base substitutions per site from between sequences are shown. Analyses were conducted using the Maximum Composite Likelihood model. Codon positions included were 1st + 2nd + 3rd + Noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 2,206 positions in the final dataset.
The rest of cdtABC operons from wild birds were grouped in three different clusters. Cluster II was characterized by carrying no deletions or 1 bp deletion, cluster III by having deletions shorter than 50 bp in total and cluster IV by carrying several deletions causing an overall deletion of more than 700 bp.
The evolutionary divergence between cdtABC sequences was also estimated and the resulting heat map showed the differences between most of the cdtABC operons from wild bird isolates respect those isolated from broiler and human (Figure 3b). As expected, similar clustering of the cdtABC operons as observed in the optimal tree (Figure 3a) was found.
The single nucleotide polymorphism (SNP) distribution respect to the cdtABC 81‐176 for the 38 isolates with a presumably intact cdtABC operon, corresponding to cluster I and II (Figure 3a), is shown in Figure 4a. Overall, it was manifest that cdtA accumulated higher number of SNPs as compared with cdtB and cdtC. The frequency of nucleotide substitution for C. jejuni isolates from broilers, humans and wild birds, was 0.48%, 0.53% and 0.53% for cdtA, 0.25%, 0.16% and 0.65% for cdtB, and 0.06%, 0.09% and 0.77% for cdtC, respectively (Figure 4b). Moreover, it was also patent that a group of wild bird isolates (W06, W07, W13, W17, W33, W43), corresponding to cluster II, carried cdtB and cdtC genes with a higher degree of variation than the isolates from cluster I. The results point out that the cdtABC operons from wild birds showed a higher variability (0.64%) as compared with the cdtABC operons from broiler and human isolates (0.28% in both groups; Figure 4b). The nucleotide substitution frequency was also calculated for the arbitrarily chosen ciaB, cadF and glnA genes, revealing similar divergence among the three groups of isolates.
FIGURE 4.
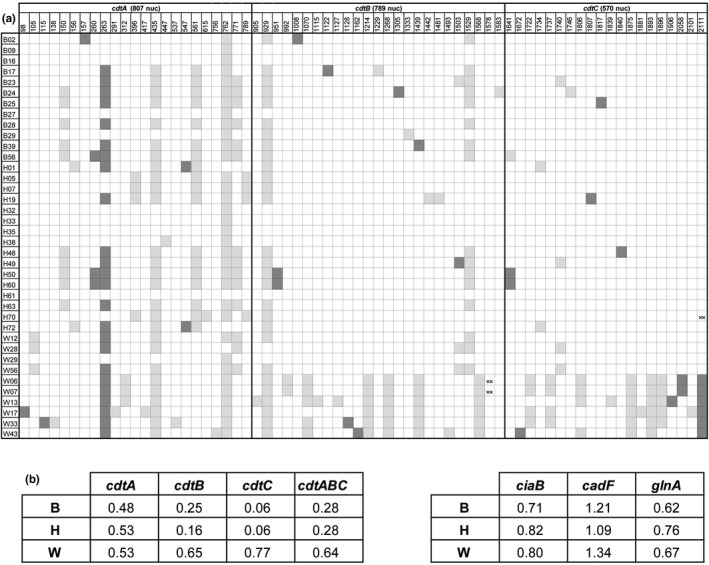
Single nucleotide polymorphism. (a) Distribution of SNPs in cdtA, cdtB and cdtC as compared to the cdtABC 81‐176 for the 38 isolates with a presumably intact cdtABC operon, corresponding to cluster I and II of the optimal tree (Figure 3a). Squares in light grey indicate silent mutations, dark grey squares indicate missense mutations and asterisks indicates nonsense mutations. (b) Frequency of nucleotide substitution for cdtA, cdtB, cdtC and cdtABC. The frequency determined for the indicated arbitrary chosen genes is also shown. In all cases sequences were compared to the sequence of the reference strain 81–176.
3.4. Several Campylobacter jejuni isolates from wild birds carry nonfunctional cdtABC operons
The analysis of the nucleotide sequence of the cdtABC operons from wild bird isolates showed that this operon is highly variable and prompted to accumulate DNA modifications. When considering deletion and insertion larger than 3 bp, several modifications were identified within the cdtABC operon. Remarkably, the modifications occurred within the cdtA and cdtB genes. No deletions and insertions larger than 3 bp were found within cdtC (Figure 5a, Figure S1, Appendix S1). Among cdtA genes, four deletions were detected, D4, D9, D25 and D154 of 4, 9, 25 and 154 bp, respectively. An insertion of 6 bp (I6) was also identified in four cdtA alleles. Within the cdtB gene three specific deletions were detected D17, D28 and the above described D51. Interestingly, six isolates carried a large deletion of 668 bp (D668) that affected both cdtA and cdtB genes.
FIGURE 5.
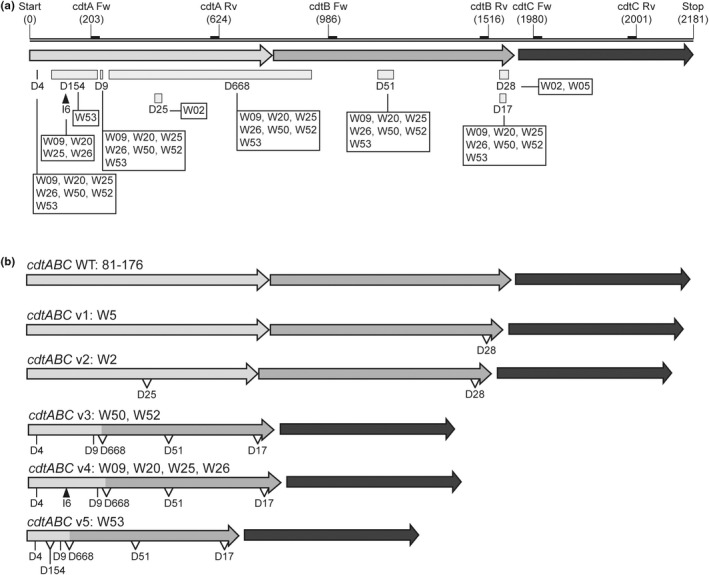
Rearrangements detected within the cdtABC operons. (a) Schematic representation of the cdtABC gene cluster. The arrows indicate the sequences of the three cdtA, cdtB and cdtC genes. The relative position of primers used for PCR amplification of each gene is indicated. The different deletions and insertions found are represented, indicating the number of nucleotides affected and the wild bird isolates that carries each of them. (b) Representation of the five variants of the cdtABC operon identified as compared with the cdtABC operon of strain 81–176. The deletions and insertions determined in each of the variants are showed, as well as the isolates where have been characterized.
Considering the mentioned modifications, five cdtABC operon variants were identified (Figure 5b). The cdtABC v1 has only a deletion (D28) within cdtB. The cdtABC v2 seems to derive from variant 1 since in addition to D28, an additional deletion (D25) was found in cdtA. The other three variants underwent drastic modifications, as they accumulate at least five different deletions within cdtA and cdtB, including the large D668. The variant 3 was detected in two isolates. The variant 4 which is identical to variant 3 but with an insertion of 6 bp, is the most prevalent (detected in four isolates). The variant 5, similar to variant 3 but carrying the deletion (D154) was detected in one isolate.
The production of the CDT subunits was predicted from the nucleotide sequence of the different cdt alleles (Table 2, Figures S2, S3 and S4, Appendix S1). All the isolates from broiler and human origin, with the only exception of H70, expressed apparently full CDT subunits with a theoretical size of 268, 265 and 189 amino acids for cdtA, cdtB and cdtC, respectively. The cdtC sequence of H70 isolate carried a deletion causing a nonsense mutation, and a predicted protein 18 AA shorter (Figure 4a and Table 2). High diversity was again found among the wild bird isolates (Table 2). The wild bird isolates grouped in clusters I and II seems to carry functional cdtABC since the predicted proteins are of same length as the one produced by the 81–176 strain. The only exception was the isolates W06 and W07 that carried a cdtB gene encoding a slightly shorter protein of 259 amino acids. The cdtABC from the two isolates in cluster III encoded a shorter CdtB toxin, since had a theoretical length of 145 and 179 residues for W02 and W05, respectively. Similarly, the CdtC subunit from both isolates was also shorter with 82 and 40 as compared with the 189 residues of CdtC18‐176. CdtA was apparently expressed in W05 whereas was non‐expressed in W02 by the presence of a codon stop in the third codon.
TABLE 2.
Summary of the results of PCR amplification, sequence homology with the primers used, deletions and insertions determined, total bp and predicted AA from cdtA, cdtB and cdtC genes of wild bird isolates, grouped in clusters as defined in Figure 3a
| PCR | Primers | Deletions | Insertions | bp | AA | |||
|---|---|---|---|---|---|---|---|---|
| cdtA | WT | 81–174 | POS | 22/22 22/22 | 807 | 268 | ||
| I | W12 | POS | 22/22 22/22 | 807 | 268 | |||
| W28 | POS | 22/22 22/22 | 807 | 268 | ||||
| W29 | POS | 22/22 22/22 | 807 | 268 | ||||
| W56 | POS | 22/22 22/22 | 807 | 268 | ||||
| II | W06 | POS | 22/22 22/22 | 807 | 268 | |||
| W07 | POS | 22/22 22/22 | 807 | 268 | ||||
| W13 | POS | 22/22 22/22 | 807 | 268 | ||||
| W17 | POS | 22/22 22/22 | 807 | 268 | ||||
| W33 | POS | 22/22 22/22 | 807 | 268 | ||||
| W43 | POS | 22/22 22/22 | 807 | 268 | ||||
| III | W02 | NEG | 20/22 20/22 | D25, 1(−1) | 6 (+1) | 787 | 1 | |
| W05 | POS | 22/22 22/22 | 807 | 268 | ||||
| IV | W09 | NEG | 20/22 ND | D4, D9, D668 | 1 (+1), 1 (+6) | 567 | 11 | |
| W20 | NEG | 20/22 ND | D4, D9, D668 | 1 (+1), 1 (+6) | 567 | 11 | ||
| W25 | NEG | 20/22 ND | D4, D9, D668 | 1 (+1), 1 (+6) | 567 | 11 | ||
| W26 | NEG | 20/22 ND | D4, D9, D668 | 1 (+1), 1 (+6) | 256 | 11 | ||
| W50 | NEG | 20/22 ND | D4, D9, D668 | 1 (+1) | 250 | 11 | ||
| W52 | NEG | 20/22 ND | D4, D9, D668 | 1 (+1) | 250 | 11 | ||
| W53 | NEG | ND/ND | D4, D154, D9, D668, 1 (−1) | 94 | 11 | |||
| cdtB | WT | 81–174 | POS | 22/22 22/22 | 798 | 265 | ||
| I | W12 | POS | 22/22 22/22 | 798 | 265 | |||
| W28 | POS | 22/22 22/22 | 798 | 265 | ||||
| W29 | POS | 22/22 22/22 | 798 | 265 | ||||
| W56 | POS | 22/22 22/22 | 798 | 265 | ||||
| II | W06 | POS | 22/22 22/22 | 1 (−1) | 797 | 259 | ||
| W07 | POS | 22/22 22/00 | 1 (−1) | 797 | 259 | |||
| W13 | POS | 22/22 22/22 | 798 | 265 | ||||
| W17 | POS | 22/22 22/22 | 798 | 265 | ||||
| W33 | POS | 22/22 22/22 | 798 | 265 | ||||
| W43 | POS | 22/22 22/22 | 798 | 265 | ||||
| III | W02 | POS | 22/22 22/22 | D28 | 2 (+1) | 772 | 145 | |
| W05 | POS | 22/22 21/22 | D28 | 1 (+1) | 771 | 179 | ||
| IV | W09 | POS | 20/22 21/22 | D51, D17 | 1 (+1) | 607 | NO | |
| W20 | POS | 20/22 21/22 | D51, D17 | 1 (+1) | 606 | NO | ||
| W25 | POS | 20/22 21/22 | D51, D17 | 1 (+1) | 606 | NO | ||
| W26 | POS | 20/22 21/22 | D51, D17, 1 (−2), 2 (−1) | 1 (+1) | 601 | NO | ||
| W50 | NEG | 20/22 20/22 | D51, D17, 1 (−1) | 2 (+1) | 605 | NO | ||
| W52 | NEG | 20/22 20/22 | D51, D17, 1 (−1) | 2 (+1) | 605 | NO | ||
| W53 | POS | 21/22 20/22 | 2 (−1), D51, D17 | 602 | NO | |||
| cdtC | WT | 81–174 | POS | 22/22 22/22 | 570 | 189 | ||
| I | W12 | POS | 22/22 22/22 | 570 | 189 | |||
| W28 | POS | 22/22 22/22 | 570 | 189 | ||||
| W29 | POS | 22/22 22/22 | 570 | 189 | ||||
| W56 | POS | 22/22 22/22 | 570 | 189 | ||||
| II | W06 | POS | 22/22 22/22 | 570 | 189 | |||
| W07 | POS | 22/22 22/22 | 570 | 189 | ||||
| W13 | POS | 22/22 22/22 | 570 | 189 | ||||
| W17 | POS | 22/22 22/22 | 570 | 189 | ||||
| W33 | POS | 22/22 22/22 | 570 | 189 | ||||
| W43 | POS | 21/22 22/22 | 570 | 189 | ||||
| III | W02 | NEG | 19/22 21/22 | 1 (−3) 2 (−1) | 3 (+1) | 568 | 82 | |
| W05 | NEG | 19/22 21/22 | 1 (−3) 2 (−1) | 2 (+1) | 567 | 40 | ||
| IV | W09 | NEG | 22/22 19/22 | 2 (−2) | 567 | 44 | ||
| W20 | POS | 22/22 19/22 | 2 (−2) | 567 | 44 | |||
| W25 | POS | 22/22 19/22 | 2 (−2) | 567 | 44 | |||
| W26 | POS | 22/22 20/22 | 1 (−1), 2 (−2) | 1 (+1) | 566 | 11 | ||
| W50 | POS | 21/22 21/22 | 1 (−1), 2 (−2) | 565 | 11 | |||
| W52 | POS | 21/22 21/22 | 1 (−1), 2 (−2) | 565 | 11 | |||
| W53 | POS | 21/22 22/22 | 2 (−1) | 1 (+2) | 568 | 122 |
The sequence of the cdtABC operon from the the seven C. jejuni isolates from cluster IV indicated that these isolates did not produce any functional CDT toxin since apparently any of the three subunits could be produced. SNPs and the deletions and insertions in the sequences caused early stop codons within cdtA and cdtC, and the accumulated deletions did not allow generation of a CdtB protein.
4. DISCUSSION
In this research, the existing variability within the cdtABC operon has been studied in a collection of C. jejuni isolates from three different origins: broiler, human and wild birds. Our data shows that the cdtABC operon seems to be highly conserved among broiler and human isolates whereas a great diversity was found among wild bird isolates as shown by the prevalence of the three cdt genes by PCR, the fragment length polymorphism of the cdtB amplicon and the cdtABC sequence analysis from a selected group of isolates. The high conservation of cdtABC among broiler and human C. jejuni isolates is consistent with being the chicken meat the most common transmission route of C. jejuni to humans via food cross‐contamination. The cdtABC operon from most of the wild bird isolates showed modifications when compared with the cdtABC 81‐176 operon highly prevalent among broiler, suggesting that the cdtABC 81‐176 does not seem to promote efficient colonization among bird other than chicken. Evolutionary analyses of the cdtABC sequences grouped some wild bird isolates (W12, W28, W29 and W56) with the human and broiler isolates, which is not surprising since cross‐transmission among the three ecological niches studied has been demonstrated (Hald et al., 2016). W12, W28 and W56 isolates belong to clonal complexes ST‐354 and ST‐446 that have been previously associated with broiler chicken and humans (Cobo‐Díaz et al., 2021; Jolley et al., 2018). We do not find isolates of this clonal complexes among the wild bird isolates grouped in clusters II, III or IV. W29, also grouped with human and broiler, was isolated from a sea gull (L. audouinii) in the city of Barcelona. Most wild birds that live in urban environment have feeding habits that include feeding in urban refuse dumps.
Among the isolates carrying cdtABC operons that did not suffer drastic DNA modifications a higher number of SNPs were detected in cdtA as compared with cdtB and cdtC. Interestingly, cdtA was the most commonly missing gene when the prevalence of the three cdt genes was determined by PCR. In a previous report, it was shown that addition of CdtB and CdtC to human culture cells has a cytotoxic action as effective as when the three subunits were added, whereas addition of CdtA and CdtB did not induce any damage, suggesting that CdtA is not essential for the CDT action (Smith & Bayles, 2006). The frequency of nucleotide substitution for cdtA is similar among the isolates from clusters I and II independently of its origin, whereas cdtB and cdtC showed low variability among human and broiler and much higher among wild birds. These results suggest a correlation between certain cdtABC alleles and efficient colonization of specific hosts.
Analysis of the complete sequence of the cdtABC operons provide an explanation to the negative detection of cdt genes by PCR. All isolates that were negative for one or more cdt genes carries cdtABC alleles were deletions or SNPs affect the hybridization with the specific primers used. Our data indicate that PCR amplification fails when at least two SNPs are found within the primer‐hybridizing sequence. These results question the convenience of using PCR‐genotyping to determine gene prevalence of the cdt genes.
Several deletions were found among cdtABC operon from wild bird isolates defining five different allelic variants. The deletions D51 and D668 were previously reported in isolates from human patients (AbuOun et al., 2005; Kabir et al., 2011). In our isolate collection, we did not detect the indicated deletions among the isolates from broiler or human patients. In AbuOun et al. (2005), the only C. jejuni strains carrying the D51 and D668 were isolated from patients with underlying health problems that could induce an immunocompromised status. In fact, those patients develop Campylobacter bacteremia. The same deletions (D51 and D668) together with D9 and D17 were detected in C. jejuni isolates from crows (Sen et al., 2018). We detected those deletions not only in crow isolates but also in isolates from different wild birds, belonging to different clonal complex, and living in different ecological niches.
Our data indicates that isolates circulating among wild birds have specific characteristics, regarding the cdtABC operon, which differs from the isolates colonizing broiler and human. These findings suggest that cdtABC might be a cellular factor involved in host specificity attending to the high degree of conservation among broiler and the relevant differences detected among the wild bird isolates. Moreover, the fact that many wild bird isolates carry apparently nonfunctional CDT toxin suggest that C. jejuni from wild bird strains might be less virulent to humans.
AUTHOR CONTRIBUTIONS
P.G, E.M., Y. I‐T., F.N., C.S‐O.A, C.M. and C.B. contributed to conceptualization and formal analysis. P.G, Y. I‐T., E.M., C.M. and C.B. performed the investigation. C.S‐O.A performed the WGS analysis. The original draft was written by C.B. and C.M. All the authors reviewed the manuscript. C.B. and C.M. contributed to project administration and funding acquisition. All the authors have read and agreed to the published version of the manuscript.
FUNDING INFORMATION
This research was funded by the Spanish Ministry of Economy and Competitiveness (grant AGL2013‐45339R), the Spanish Ministry of Science, Innovation and Universities (MCIU), State Bureau of Investigation (AIE) and European Regional Development Fund (FEDER) (grant PGC2018‐096958‐B‐I00) and the Catalonian government (grant 2017SGR499). During this work, a trainee teaching and research staff supported PG grant (Universitat de Barcelona).
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Appendix S1
ACKNOWLEDGEMENTS
We deeply thank Francesc Mestres (University of Barcelona) for his helpful assistance with the evolutionary analysis and the fruitful discussion about the results described in this manuscript.
Guirado, P. , Iglesias‐Torrens, Y. , Miró, E. , Navarro, F. , Attolini, C.‐O. , Balsalobre, C. , & Madrid, C. (2022). Host‐associated variability of the cdtABC operon, coding for the cytolethal distending toxin, in Campylobacter jejuni . Zoonoses and Public Health, 69, 966–977. 10.1111/zph.12994
Contributor Information
Carlos Balsalobre, Email: cbalsalobre@ub.edu.
Cristina Madrid, Email: cmadrid@ub.edu.
DATA AVAILABILITY STATEMENT
Research data are not shared.
REFERENCES
- AbuOun, M. , Manning, G. , Cawthraw, S. A. , Ridley, A. , Ahmed, I. H. , Wassenaar, T. M. , & Newell, D. G. (2005). Cytolethal distending toxin (CDT)‐negative Campylobacter jejuni strains and anti‐CDT neutralizing antibodies are induced during human infection but not during colonization in chickens. Infection and Immunity, 73, 3053–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang, D. D. , Nielsen, E. M. , Scheutz, F. , Pedersen, K. , Handberg, K. , & Madsen, M. (2003). PCR detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from Danish pigs and cattle and cytolethal distending toxin production of the isolates. Journal of Applied Microbiology, 94, 1003–1014. [DOI] [PubMed] [Google Scholar]
- Cobo‐Díaz, J. F. , González del Río, P. , & Álvarez‐Ordóñez, A. (2021). Whole resistome analysis in Campylobacter jejuni and C. coli genomes available in public repositories. Frontiers in Microbiology, 12, 1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo, R. T. , Dumont, C. F. , Braz, R. F. , Monteiro, G. P. , Takeuchi, M. G. , Lourenzatto, E. C. A. , & Dos Santos, J. P. (2021). Genotypical relationship between human and poultry strains of Campylobacter jejuni . Current Microbiology, 788(78), 2980–2988. [DOI] [PubMed] [Google Scholar]
- European Food Safety Authority and European Centre for Disease Prevention and Control . (2021). The European Union One Health 2019 Zoonoses Report. EFSA Journal, 19, e06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hald, B. , Skov, M. N. , Nielsen, E. M. , Rahbek, C. , Madsen, J. J. , Wainø, M. , Chrièl, M. , Nordentoft, S. , Baggesen, D. L. , & Madsen, M. (2016). Campylobacter jejuni and Campylobacter coli in wild birds on Danish livestock farms. Acta Veterinaria Scandinavica, 58, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, L. A. , Bennett, M. , Coffey, P. , Elliott, J. , Jones, T. R. , Jones, R. C. , Lahuerta‐Marin, A. , Leatherbarrow, A. H. , McNiffe, K. , Norman, D. , Williams, N. J. , & Chantrey, J. (2009). Molecular epidemiology and characterization of Campylobacter spp. isolated from wild bird populations in northern England. Applied and Environmental Microbiology, 75, 3007–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias‐Torrens, Y. , Miró, E. , Guirado, P. , Llovet, T. , Muñoz, C. , Cerdà‐Cuéllar, M. , Madrid, C. , Balsalobre, C. , & Navarro, F. (2018). Population structure, antimicrobial resistance, and virulence‐associated genes in campylobacter jejuni isolated from three ecological niches: Gastroenteritis patients, broilers, and wild birds. Frontiers in Microbiology, 9, 1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinadasa, R. N. , Bloom, S. E. , Weiss, R. S. , & Duhamel, G. E. (2011). Cytolethal distending toxin: A conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology, 157, 1521–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley, K. A. , Bray, J. E. , & Maiden, M. C. J. (2018). Open‐access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Research, 3, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush, N. O. , Castaño‐Rodríguez, N. , Mitchell, H. M. , & Man, S. M. (2015). Global epidemiology of Campylobacter infection. Clinical Microbiology Reviews, 28, 687–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir, S. M. L. , Kikuchi, K. , Asakura, M. , Shiramaru, S. , Tsuruoka, N. , Goto, A. , Hinenoya, A. , & Yamasaki, S. (2011). Evaluation of a cytolethal distending toxin (cdt) gene‐based species‐specific multiplex PCR assay for the identification of Campylobacter strains isolated from diarrheal patients in Japan. Japanese Journal of Infectious Diseases, 64, 19–27. [PubMed] [Google Scholar]
- Koga, M. , Gilbert, M. , Takahashi, M. , Li, J. , Koike, S. , Hirata, K. , & Yuki, N. (2006). Comprehensive analysis of bacterial risk factors for the development of Guillain‐Barré syndrome after Campylobacter jejuni enteritis. The Journal of Infectious Diseases, 193, 547–555. [DOI] [PubMed] [Google Scholar]
- Koolman, L. , Whyte, P. , Burgess, C. , & Bolton, D. (2015). Distribution of virulence‐associated genes in a selection of Campylobacter isolates. Foodborne Pathogens and Disease, 12, 424–432. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. , & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C. K. , Chen, Y. A. , Lin, C. J. , Lin, H. J. , Kao, M. C. , Huang, M. Z. , Lin, Y. H. , Chiang‐Ni, C. , Chen, C. J. , Lo, U. G. , Lin, L. C. , Lin, H. , Hsieh, J. T. , & Lai, C. H. (2016). Molecular mechanisms and potential clinical applications of Campylobacter jejuni cytolethal distending toxin. Frontiers in Cellular and Infection Microbiology, 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara‐Tejero, M. , & Galán, J. E. (2001). CdtA, CdtB, and CdtC form a tripartite complex that is required for cytolethal distending toxin activity. Infection and Immunity, 69, 4358–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, I. , Mateo, E. , Churruca, E. , Girbau, C. , Alonso, R. , & Fernández‐Astorga, A. (2006). Detection of cdtA, cdtB, and cdtC genes in Campylobacter jejuni by multiplex PCR. International Journal of Medical Microbiology, 296, 45–48. [DOI] [PubMed] [Google Scholar]
- Pagès, H. , Aboyoun, P. , Gentleman, R. , DebRoy, S. (2022). Biostrings: Efficient manipulation of biological strings. R package version 2.64.0 . https://bioconductor.org/packages/Biostrings
- Pickett, C. L. , Pesci, E. C. , Cottle, D. L. , Russell, G. , Erdem, A. N. , & Zeytin, H. (1996). Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene. Infection and Immunity, 64, 2070–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R‐project.org/ [Google Scholar]
- Saitou, N. , & Nei, M. (1987). The neighbor‐joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sen, K. , Lu, J. , Mukherjee, P. , Berglund, T. , Varughese, E. , & Mukhopadhyay, A. K. (2018). Campylobacter jejuni colonization in the crow gut involves many deletions within the cytolethal distending toxin gene cluster. Applied and Environmental Microbiology, 84, e01893‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, S. K. , Dallas, J. F. , Macrae, M. , Mccarthy, N. D. , Gormley, F. J. , Strachan, N. J. C. , Ogden, I. D. , Maiden, M. C. , & Forbes, K. J. (2009). Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6, 134, 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. L. , & Bayles, D. O. (2006). The contribution of cytolethal distending toxin to bacterial pathogenesis. Critical Reviews in Microbiology, 32, 227–248. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Nei, M. , & Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor‐joining method. Proceedings of the National Academy of Sciences of the United States of America, 101, 11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, A. M. , Miller, W. A. , Byrne, B. A. , Chouicha, N. , Boyce, W. M. , & Townsend, A. K. (2014). Prevalence and pathogenic potential of Campylobacter isolates from free‐living, human‐commensal American crows. Applied and Environmental Microbiology, 80, 1639–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse, C. A. , Balbo, P. B. , Pesci, E. C. , Cottle, D. L. , Mirabito, P. M. , & Pickett, C. L. (1998). Campylobacter jejuni cytolethal distending toxin causes a G2‐phase cell cycle block. Infection and Immunity, 66, 1934–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Appendix S1
Data Availability Statement
Research data are not shared.


