Abstract
Ras proteins are implicated in some of the most common life‐threatening cancers. Despite intense research during the past three decades, progress towards small‐molecule inhibitors of mutant Ras proteins still has been limited. Only recently has significant progress been made, in particular with ligands for binding sites located in the switch II and between the switch I and switch II region of K‐Ras4B. However, the structural diversity of inhibitors identified for those sites to date is narrow. Herein, we show that hydrazones and oxime ethers of specific bis(het)aryl ketones represent structurally variable chemotypes for new GDP/GTP‐exchange inhibitors with significant cellular activity.
Keywords: hydrazone, oxime ether, Ras/SOS inhibitor, GTPase; K-Ras4B.
Ras proteins are implicated in some of the most common life‐threatening cancers. Despite intense research over the past 30 years, progress towards small‐molecule inhibitors of mutant Ras proteins is limited. Herein we show that hydrazones and oxime ethers of specific bis(het)aryl ketones represent structurally variable chemotypes for new GDP/GTP‐exchange inhibitors with significant cellular activity.

Introduction
Cancer is one of the most common cause of death in industrialised countries, only second to cardiovascular diseases. Due to demographic changes in most of these industrialised countries, the number of new cases of cancer continues to rise significantly. In addition to prostate cancer and tumors of the mammary gland, colorectal cancer is one of the most common cancers. [1] Point mutations in the Ras gene are detectable in every fourth tumor. K‐Ras4B mutations are responsible for ∼85 % of all Ras‐driven human cancers, followed by the isoforms N‐Ras (∼12 %) and H‐Ras (∼3 %). [2] The incidence of a K‐Ras4B mutation in colorectal cancer is >50 %, in pancreatic tumors even >90 %.[ 3 , 4 ] In malignant melanoma, which is responsible for about 80 % of skin cancer deaths worldwide, mutations in the Ras‐signaling pathway decisively determine the course of the disease, the prognosis, and the success of the patient's therapy. [5] Despite considerable efforts to develop antitumor drugs directed against oncogenic Ras, only limited progress has been made in the past decades. A major reason for the failure of those direct targeting approaches is that Ras proteins interact with their effectors via large protein‐protein surface interactions (PPIs), which are notoriously difficult to address with small‐molecule inhibitors. [6]
Ras‐GTPases belong to the superfamily of GTP‐binding proteins that are involved in numerous cellular processes such as cell growth, cell regulation, and signal transduction. Ras proteins toggle between an inactive, GDP‐bound and an active, GTP‐bound state (Figure 1).[ 7 , 8 ] In the active state Ras‐proteins interact with various effectors and modulate the Raf‐MEK‐Erk and the PI3K‐Akt‐mTor pathway. [9] Ras proteins show weak activity as a GTPase, which intrinsically inactivates Ras by hydrolysis of GTP. The nucleotide exchange in Ras‐proteins is highly regulated by guanine exchange proteins (GEFs) and GTPase activation proteins (GAPs). The GEFs such as ‘son of sevenless’ (SOS) catalyse the release of GDP and thus act as activators, while GAPs increase the intrinsic Ras‐GTPase activity and thereby inactivate Ras proteins (Figure 1a). Switching between the GTP‐bound active and GDP‐bound inactive form is accompanied by significant conformational changes of the Ras protein (Figure 1b and 1c). Structural studies on GEF and GAP binding domains revealed two regions including residues 30–40 (switch‐I) and residues 60–76 (switch‐II) which are highly relevant for the GDP/GTP‐exchange process. [10] In the GTP‐bound state, Thr35 and Gly60 form hydrogen bonds with the γ‐phosphate and hold the switch‐I and switch‐II regions in their active states. Upon GTP‐hydrolysis, the γ‐phosphate is released and the switch regions return to the flexible conformation present in the GDP‐bound state. Most oncogenic mutations are found at positions Gly12, Gly13, and Gln61. Those point mutations impair GTPase activity and GAP‐mediated GTP‐hydrolysis, thus shifting the conformational equilibrium of RAS towards the GTP‐loaded active state. [9] This activates additional signaling pathways that deregulate crucial cellular functions such as proliferation, migration, differentiation, and cell death.
Figure 1.
Structure of Ras‐proteins and interactions with GEF and SOS proteins. a) Toggling of Ras between active and inactive state. b) SI/SII‐binding site is closed in ligand‐free state (PDB: 4EPR). c) SI/II‐binding site is open in ligand‐bound state (PDB: 4EPY).
Over the past three decades several strategies to inactivate oncogenic K‐Ras4B have been investigated with rather limited success.[ 11 , 12 , 13 , 14 , 15 , 16 , 17 ] Only recently, a first direct Ras inhibitor (AMG510, Sotorasib) which targets (GDP)K‐Ras4BG12C covalently has entered the market.[ 18 , 19 , 20 ] MRTX1133 (5), another structurally related compound, is currently in clinical development as a non‐covalent G12D Ras‐inhibitor of the SII‐site. [21]
One focus in search for new Ras inhibitors is on a Ras/SOS‐interaction site between the SI‐ and SII‐switches. In the ligand‐free state, this binding site forms a narrow pocket stabilised by a network of H‐bonds between the OH‐group of Y71 as well as D54, R41, and S39. Upon ligand binding, this network is broken and the binding site opens to form a channel, allowing for larger ligands to be accommodated (Figure 1b and 1c). Ligands of this particular binding site can interact either with inactive (GDP)K‐Ras4B – by blocking SOS‐binding and subsequent GDP release – or with GTP‐bound active K‐Ras4B which interrupts downstream signaling. [22] Both modes of action inhibit aberrant downstream signaling of Ras proteins. Targeting the SOS‐mediated GDP/GTP‐exchange represents a particular attractive strategy because this process remains intact in almost all K‐Ras4B mutants except for G13D which shows a considerably accelerated GDP/GTP‐exchange and thus faster reactivation. [23] However, the presence of this binding site both in wildtype (wt) and oncogenic K‐Ras4B constitutes a formidable challenge for the development of drugs interfering selectively only with mutant Ras. Additionally, the structural diversity of published ligands interfering with the SI/SII‐site is limited, with indole and azole building blocks prevailing (Figure 2).[ 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 ]
Figure 2.
Known ligands for the K‐RAS4B SI/II‐ and SII‐ interaction sites.
4,4’‐Dihydroxybenzophenone (DHBP, 9) nitrophenylhydrazones have been described to show significant activity against a variety of tumor cell lines. [33] The most active compound, DHBP 2,4‐dinitrophenyl‐hydrazone 7 a (Figure 3) has been used in a clinical study as a topical agent against cutaneous cancer lesions. Compound 7 a (A‐007) was well tolerated and no cytotoxic properties were observed during comparative preclinical toxicology studies. [34] Histochemical analysis of biopsies of human skin topically treated with A‐007 (7 a) led to the conclusion that the anticancer activity is caused by upregulation of CD45+ lymphocyte cell surface receptors. [35] However, no biochemical mode of action studies were published and thus it remains somewhat unclear as to whether CD45+ upregulation is in fact the basic mode of action for A‐007 (7 a). In a recent paper, we already demonstrated that DHBP (9), the scaffold of A‐007, binds to Rheb at a site related to the SI/SII‐site of Ras proteins. [36] Based on those previous results, we were interested in studying DHBP hydrazones as well as related scaffolds in more detail for their potential as a structurally different class of ligands for the SI/SII‐ binding site of mutant K‐Ras4B.
Figure 3.
4,4’‐Dihydroxyenzophenone (9) and derivatives tested for GDP/GTP‐exchange inhibition.
Results and Discussion
DHBP scaffold‐based Ras/SOS inhibitors
Not completely unexpected, DHBP (9) showed only weak activity in a Ras/SOS‐exchange assay (Table 1) similar to DCAI (1). [26] On the contrary, the known dinitrophenyl hydrazone 7 a (Figure 3) exhibited a significant inhibition of GDP/GTP‐exchange comparable to reference compound 2 which is by a factor of 5 to 8 better than DCAI. This finding suggests an additional mode of action for dinitrophenylhydrazone 7 a besides the postulated upregulation of CD45+ cell surface receptors. However, compound 7 a proves to be nonselective while halogen phenylhydrazone 7 c shows at least moderate selectivity for the oncogenic K‐Ras4BG12D mutant (Table 1).
Table 1.
GDP/GTP exchange and cellular activities,
|
(GDP)K‐Ras4B/SOS exchange (IC50 [μM]) |
|
Cellular activities (IC50 [μM]) |
||||||
|---|---|---|---|---|---|---|---|---|
|
|
G12D |
WT |
WT %CRTL (3 mM) |
SNU‐1 (G12D) |
NCI‐H441 (G12 V) |
HCT15 (G13D) |
H358 (G12 C) |
A549 (G12S) |
|
Benchmark compounds | ||||||||
|
1 |
1748±504 |
1559±490 |
84 |
33.0±1.6 |
37.0±0.1 |
n.d. |
n.d. |
34.4±1.3 |
|
2 |
308±8 |
508±244 |
n.d. |
9.5±1.9 |
27.7±5.8 |
14.3±2.7 |
20.2±3.2 |
16.2±2.5 |
|
3[ b] |
19.1±4.7 |
26.9±13.6 |
89 |
>30 |
>30 |
>30 |
>30 |
>30 |
|
4 |
1520±367 |
>3000 |
98 |
7.2±1.2 |
11.1±0.7 |
6.0±0.3 |
12.6±0.3 |
12.9±3.2 |
|
9 |
1214±116 |
1333±104 |
n.d. |
187±19 |
>300 |
n.d. |
n.d. |
>300 |
|
DHBP hydrazones | ||||||||
|---|---|---|---|---|---|---|---|---|
|
7 a |
236±46 |
306±49 |
26 |
16.7±1.2 |
22.8±4.2 |
18.9±0.6 |
20.9±0.8 |
16.1±1.5 |
|
7 b |
829[a] |
1826±295 |
86 |
26.6±9.9 |
30.9±5.8 |
25.1±10.6 |
37.6±0.9 |
34.4±3.1 |
|
7 c |
413±28 |
1036±427 |
83 |
23.7±14.4 |
24.7±8.3 |
17.6±13.4 |
33.7±0.6 |
28.7±4.7 |
|
7 d |
475[a] |
1295±417 |
84 |
31.2±6.8 |
29.3±4.1 |
27.4±4.5 |
40.2±1.0 |
30.1±9.0 |
|
7 e |
628±170 |
1067±312 |
90 |
16.7±1.9 |
13.4±2.7 |
12.5±1.4 |
15.1±1.2 |
11.9±0.7 |
|
7 f |
1907±98 |
2608±225 |
98 |
71.1±11.3 |
65.8±37.3 |
n.d. |
n.d. |
67.0±1.8 |
|
DHBP sulfonyl hydrazones | ||||||||
|---|---|---|---|---|---|---|---|---|
|
8 a |
1307±389 |
1031±371 |
91 |
92.1±12.0 |
157±54 |
113±13 |
109±3 |
105±2 |
|
8 b |
1288±307 |
1617±565 |
89 |
154±30 |
260±45 |
219±43 |
161±9 |
184±12 |
|
8 c |
583±146 |
1048±405 |
91 |
81.0±26.3 |
102±16 |
85.0±9.2 |
69.8±2.4 |
71.3±6.5 |
|
8 d |
850±120 |
2219±519 |
93 |
88.7±23.9 |
105±8 |
96.6±14.5 |
82.8±8.4 |
91.8±7.4 |
|
8 e |
255[a] |
298±196 |
n.d. |
46.8±1.7 |
58.4±6.9 |
51.1±1.2 |
49.2±0.9 |
49.8±2.1 |
|
8 f |
858±55 |
1653±731 |
92 |
110±24 |
179±43 |
112±10 |
104±3 |
111±10 |
|
8 g[ b] |
>3000 |
1500±572 |
86 |
>30 |
>30 |
>30 |
>30 |
>30 |
|
DHBP hydrazide | ||||||||
|---|---|---|---|---|---|---|---|---|
|
10 |
>3000 |
>3000 |
93 |
173±1 |
158±3 |
n.d. |
n.d. |
112±3 |
|
DHBP oxime ether | ||||||||
|---|---|---|---|---|---|---|---|---|
|
11 |
431±22 |
1332±343 |
98 |
41.7±4.2 |
62.2±4.8 |
43.9±1.8 |
58.6±0.5 |
54.3±3.6 |
Data are shown as mean±SD of independent repeats (2–5 times). [a] Single measurements due to limited substance availability. [b] Due to substance limitations or solubility problems testing was not possible at higher concentrations.
In general, halogens such as F (7 c), Cl (7 d) or the strongly electron‐withdrawing CF3‐group (7 e) increase overall activity (Table 1) compared to the unsubstituted phenyl hydrazone 7 b. Nevertheless, it is not possible to replace the phenyl ring by an electron‐deficient pyridine residue (7 f) or by a benzoic acid hydrazide (10). The SAR of sulfonyl hydrazones is similar to that of hydrazones. Electron‐donating groups reduce, whereas halogen substituents increase activity in the Ras/SOS‐exchange assay. Overall, the sulfonyl hydrazones appear to be less active compared to the DHBP hydrazones with the exception for the nitro‐derivative 8 e which exhibits the same activity in the Ras/SOS‐exchange assay as dinitrophenylhydrazone 7 a, however at the expense of any selectivity. Inhibition of Ras/SOS‐exchange is not limited to the hydrazone motif as indicated by oxime 11, which shows similar activity as hydrazone 7 c and an improved selectivity for the oncogenic G12D form.
Notably, a weak IC50 value in the Ras/SOS assay does not necessarily imply low cellular activity since additional binding of the test compound to active GTP‐bound Ras is not detected in the Ras/SOS‐exchange assay. This is demonstrated by benchmark compound 4, a specific (GTP)Ras‐binder, which exhibits high activity against all tumor cell lines tested (Table 1) but is almost inactive in the Ras/SOS‐assay. A similar effect is observed for our compounds presented here. MST (microscale thermophoresis) measurements indicate that the DHBP phenylhydrazones bind to both GDP and GTP K‐Ras4B. Thus, the significantly higher cellular activity compared to the Ras/SOS assay data may be attributed to concomitant (GTP)K‐Ras4B binding.
Reference compound 2, a (GDP)K‐Ras4B ligand, exhibited an excellent single‐digit micromolar activity against SNU‐1 cells (gastric adenocarcinoma). A somewhat weaker activity was observed for NCI‐H441 (papillary adenocarcinoma of the lung) cells. Fluorophenylhydrazone 7 c and chlorophenyl hydrazone 7 d exhibited IC50 values between 20–30 μM for all tested cell lines. Trifluoromethylphenyl hydrazone 7 e was identified as the strongest inhibitor in this series of compounds with IC50 values in the same range or even better than benchmark compounds 2 and 4.
It remains somewhat unclear as to whether the lower activity of the two p‐halogenophenyl hydrazones 7 c and 7 d compared to trifluoromethylphenyl hydrazone 7 e is caused by a reduced target affinity or by the limited cell culture medium stability of 2.2 % (7 c) and 0.4 % (7 d). Furthermore, hydrolysis of hydrazones 7 c and 7 d causes the release of a toxic arylhydrazine and 4,4’‐dihydroxybenzophenone (DHBP), a known pan assay interference compound (PAINS) as it exhibits a multitude of more or less specific biological effects, including endocrine disrupting activities.[ 37 , 38 , 39 ] Thus, we cannot exclude that the cellular activity observed for both compounds is partially based on off‐target effects. In contrast, trifluoromethyl hydrazone 7 e was found to be completely stable during a 24 h period.
The pronounced activity of oxime 11 already observed in the Ras/SOS‐exchange assay is confirmed by cellular assays with the best activities found for the mutant cell lines G12D and G13D. It is important to note that benzyloxime ether 11 was completely stable in cell culture medium and 92 % stable in human plasma solution over 24 h, which precludes the release of DHBP as a source of unspecific activity.
The most promising compounds were further examined on their ability to disrupt Ras‐PI3K‐Akt and Ras‐Raf‐MEK‐Erk downstream signaling in SNU‐1 cells (Table 2). Ras‐specific inhibitors are expected to invoke reduction of pAkt and/or pErk while not affecting tAkt or tErk protein levels. For comparison, all compounds were also tested on the Ras‐independent malignant melanoma cell‐line A375 carrying a BRAF V600E mutation. Reference compound 4 proved to be a selective inhibitor of Act phosphorylation in oncogenic SNU‐1 cells with around tenfold lower activity on the Erk pathway. However, the Erk pathway was also significantly inhibited in A375 cells. Hydrazone 7 e disappointingly only had a weak effect on Akt/Erk phosphorylation. Thus, the cellular activity of this compound must be attributed to off‐target effects. In contrast, p‐fluorophenyl hydrazone 7 c exhibited significant activity and a selectivity by a factor of five on the Akt pathway in SNU‐1 cells, while A375 cells remained unaffected. The strongest effect was found for DHBP oxime ether 11 which inhibited pAkt formation in SNU‐1 cells with an IC50 value of 47.7 μM. In A375 cells no reduction of pAkt was observed. Despite its selectivity for the Akt pathway in oncogenic SNU‐1 cells, oxime ether 11 also exhibited activity on the Erk pathway in Ras‐wildtype A375 cells similar to reference compound 4.
Table 2.
HTRF assay of downstream phosphorylation in SNU‐1 and A375‐cells (IC50 [μM]).
|
|
|
pErk |
tErk |
pAkt |
tAkt |
|---|---|---|---|---|---|
|
4 |
SNU‐1 |
124 |
>1000 |
14.5 |
527 |
|
|
A375 |
74.7 |
>1000 |
>1000 |
>1000 |
|
7c |
SNU‐1 |
455 |
>550 |
95.3 |
>1000 |
|
|
A375 |
415 |
491 |
>1000 |
>1000 |
|
7e |
SNU‐1 |
520 |
>1000 |
431 |
>1000 |
|
|
A375 |
>1000 |
>1000 |
>1000 |
>1000 |
|
8c |
SNU‐1 |
240 |
>875 |
96.6 |
586 |
|
|
A375 |
203 |
255 |
>1000 |
>1000 |
|
11 |
SNU‐1 |
218 |
>1000 |
47.7 |
475 |
|
|
A375 |
48.8 |
77.2 |
>1000 |
749 |
In order to develop a binding‐model, NMR‐studies of (GDP)K‐Ras4B complexes with hydrazone 7 c and oxime 11 were carried out. Residues experiencing NMR chemical shift perturbations (CSPs) of at least 2σ confidence level were used for Haddock‐based molecular docking calculations and subsequent cluster analyses. [40] For hydrazone 7 c, Haddock calculations with NMR CSPs found in the SI/II‐region revealed one highly relevant cluster (−24.7±1.7 kcal/mol, RMSD: 3.0±1.6) with 102 poses out of a total of 188 used for clustering. The main cluster exactly matches the SI/SII‐site known for ligands 1–3. Hydrazone 7 c does not intrude with one of the two phenol residues into the lipophilic pocket but rather utilises the 4‐fluorophenyl ring which is fixed by a π‐cation interaction to Lys5 (Figure 4a). One phenol ring is stabilised in its site by a π‐cation interaction to Arg41 and a hydrogen‐bond to Asp38 while the other phenol ring apparently points outside the pocket. A second, lower populated (55 poses) and less well‐defined cluster binds to the switch I region. Hydrazone 7 c is bound to this site through a π‐π interaction of the fluorophenyl ring to Tyr40 and another π‐π interaction of one phenol ring to Tyr32. Noteworthy, significant CSPs were also observed for the nucleotide binding pocket. This could indicate an allosteric binding effect of 7 c into the SI/SII‐pocket as previously described. [41]
Figure 4.
Haddock‐docking models of hydrazone 7c (a) and oxime 11 (b) to (GDP)K‐Ras4Bwt (PDB code: 6MBT). Dark blue surface areas: NMR CSPs with 2σ confidence level. Pale blue areas: NMR CSPs with shifts with 1σ confidence level.a rather low Glide docking‐score of −3.9 kcal/mol compared to −9.3 kcal/mol for the SIIP‐binding site. One phenol ring protrudes into the SI/SII‐pocket pocket and engages in a π‐cation interaction with Lys5, while the surface‐located second phenol fragment is stabilised by a hydrogen bond to Glu62. Apparently, the oxime benzyl ether fragment does not interact with the SI/SII‐site which could explain its low Glide docking‐score.
Most remarkably, Haddock calculations of oxime 11 demonstrated that the largest and also lowest‐scoring cluster (−22.7±2.5 kcal/mol, RMSD: 0.4±0.2), combining 66 out of 190 poses, binds into the same switch II pocket described for the nanomolar Ras‐inhibitor MRTX1133 (Figure 4b, 5). [21] A refinement of the Haddock SIIP‐cluster using the docking software Glide revealed an excellent docking score of −9.3 kcal/mol with all Glide poses adopting the same orientation in the binding‐pocket. The oxime ether 11 is stabilised within the SII‐site by two hydrogen bonds from the phenolic hydroxy‐groups to Ala59 and His95 as well as an additional π‐cation interaction with Arg68 (Figure 4b). The benzyl ether fragment protrudes into a deep lipophilic pocket surrounded by Val79, Tyr71, Met72, Phe78, and Ile100. In this preferred orientation one phenolic hydroxy group is in proximity to the β‐phosphate. Appropriate modifications at this position should allow for additional interactions directly with the phosphate binding site. The SII‐binding site is present primarily in the GDP‐bound K‐Ras form. Only when the amino acids Thr35 and Gly60 are not coordinated by the γ‐phosphate of (GTP)K‐Ras, the pocket‐constituting amino acids become flexible enough to accommodate a small molecule ligand. [9] Therefore, no clusters were observed for the SII‐site in Haddock calculations with the GTP‐bound 4DSO crystal structure. Those findings are corroborated by the results of an MST assay which showed a significant preference of oxime 11 for (GDP)K‐Ras4B (KD(GDP) 89 μM/KD(GTP) 602 μM).
The Haddock cluster (43 poses, RMSD: 1.4±0.2) with the second‐best docking score (−14.9 kcal/mol) actually addresses the canonical SI/SII‐site identified as the primary target for hydrazone 7 c and reference compounds 1–3. A refinement of the Haddock poses resulted in one consensus orientation of oxime 11 in the SI/SII‐pocket, however, with a rather low Glide docking‐score of ‐3.9 kcal/mol compared to ‐9.3 kcal/mol for the SIIP‐binding site. One phenol ring protrudes into the SI/SII‐pocket pocket and engages in a π‐cation interaction with Lys5, while the surface‐located second phenol fragment is stabilised by a hydrogen bond to Glu62. Apparently, the oxime benzyl ether fragment does not interact with the SI/SII‐site which could explain its low Glide docking‐score.
Ras/SOS inhibitors with tailor‐made scaffolds
In spite of the promising Ras‐activity, the DHBP hydrazones are potentially critical structure elements due to their hydrolytic instability. A solution to the stability issue is the replacement of the hydrazone linkage by a stable oxime ether as realised in compound 11. [42] The low affinity of DHBP to K‐Ras4B and potentially adverse biological effects as a known pan‐assay interference compound can be addressed by designing scaffolds that bind stronger and more specifically to the SI/SII‐binding site. Thus, extensive molecular dockings were performed. Overall, a virtual library of 360.000 compounds was generated consisting of two different aromatic and/or heteroaromatic building blocks which were linked by a set of sp2 and sp3 hybridised central fragments (Figure 5a). In silico screenings were performed with K‐Ras4BG12D (PDB code: 4DSO) using the docking software Glide in HTS modus. The top scoring 10.000 poses were docked again in standard precision‐mode and the highest scoring 1.000 poses were finally docked in extra precision‐mode. The top‐ten in silico hits were used for similarity searches in the structure data base GDB‐13 (970 million compounds) and GDB‐17 (166 billion compounds), however, without noticeable improvements of docking scores. Finally, three high‐scoring scaffolds (Figure 5b) were chosen for synthesis.
Figure 5.
Design of the in silico screening library. a) Top: bridging element with attachment points in green; bottom: aryl substituents with attachment points in green; marked in blue are the positions at which either a hydroxy or an amino group was attached. b) Top‐scoring in silico hits.
The new scaffold 12 was prepared convergently from hydroxy benzimidazole and hydroxy benzofuran building blocks which were coupled in one of the final steps prior to protecting group removal (Scheme 1). The MEM‐protected benzimidazoles 17 and 18 were obtained as a mixture of two stable tautomers in a two‐step sequence starting from 2‐amino‐3‐nitrophenol (15). Ring closure to hydroxy benzimidazole 16 was achieved by a reductive cyclisation following a procedure published by Shen et al. [43]
Scheme 1.
Synthesis of in‐silico‐docking hit 12: (a) H2, 10 % Pd/C, HC(OEt3), AcOH, MeOH, 51 %. (b) MEMCl, DIPEA, THF, 0 °C ‐ rt, 81 %. (c) NaH, PMBCl, DMSO, 0 °C ‐ rt, 22–80 %. (d) BrCH2COOEt, Cs2CO3, DMSO, 100 °C, 46–86 %. (e) MeNHOMe×HCl, NEt3, EDCI, DMAP, DCM, 25 %. (f) mixture of 23/24, n‐BuLi, THF, −78 °C, 85 %. (g) 10 % TFA in DCM, then 1 : 1 EtOH/4 M HCl in dioxane, rt, 19 %.
As starting material for the synthesis of the PMB‐protected hydroxybenzofuranoic acid 21 2,3‐dihydroxybenzaldehyde (19) was chosen (Scheme 1). Monoprotection succeeded in varying yields via formation of the bis‐phenolate using 2 eq. NaH and subsequent alkylation of the more nucleophilic meta‐phenolate with one eq. of 4‐methoxybenzyl chloride.[ 44 , 45 ] A Rap‐Stoermer cyclisation of the mono PMB‐protected aldehyde 20 with ethyl bromoacetate afforded benzofuranoic acid 21 in up to 86 % yield. [46] Subsequent coupling of N,O‐dimethyl hydroxylamine with EDCI generated the Weinreb amide 22 in sufficient yield for further coupling with lithium benzimidazolide to provide a tautomeric mixture of ketones 23 and 24 in a yield of 76 %. Due to the strongly acidic conditions required for the concomitant removal of the PMB‐ and MEM‐protecting groups in the final step, only low but sufficient yields of the target ketone 12 were obtained. [47] The regioisomer 13 was prepared analogously to Scheme 1 with the corresponding 5‐hydroxybenzimidazole as building block.
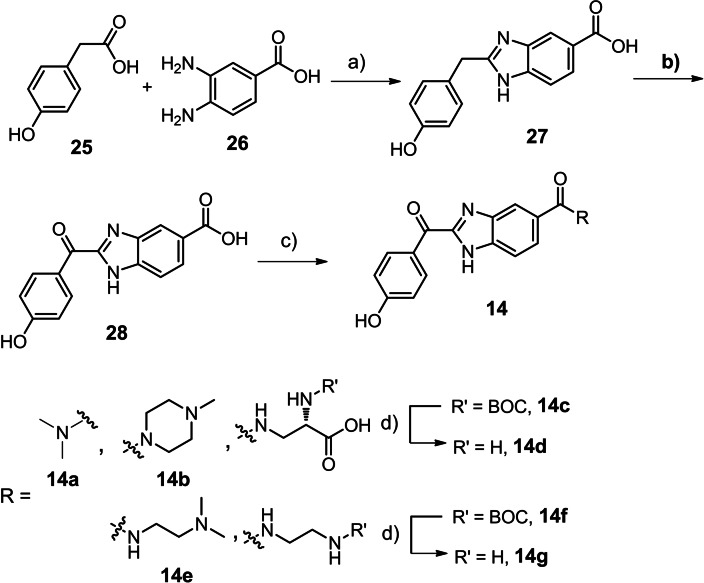
The straightforward synthesis of the benzimidazole phenol hybrid ketones 14 starts with the hydrochloric acid‐catalysed condensation of 2‐(4‐hydroxyphenyl)acetic acid and 3,4‐diaminobenzoic acid (Scheme 2). [48] Benzimidazole 27 was oxidised to ketone 28 with air in the presence of sulfur. [49] Amide couplings were accomplished either with EDC/Oxyma (14 f) or with HATU (14 a, 14 b, 14 e) followed by Boc‐group removal with hydrochloric acid in dioxane. [50] In general, in the series of benzimidazole phenol scaffolds most of the active structures (Table 3, Figure 5) were either unselective (14 a) or had an inverted selectivity in favor of wildtype K‐Ras4B (14 c, 14 g).
Scheme 2.
Synthesis of benzimidazole phenol ketones: (a) HCl, 120 °C, 6 h, 70 %. (b) sulfur, DMF, 90 °C, 7 d, 80 %. (c) EDC⋅HCl, oxyma, DIPEA, dioxane, 12 h for 14 f; HATU, DIPEA, DMF, 0 °C to rt., 12 h; 14a, 71 %; 14b, 65 %; 14c, 30 %; 14e, 52 %; 14 f, 67 %; 14 g, 99 %. (d) HCl, dioxane, rt., quant.
Table 3.
GDP/GTP‐exchange and cellular activities of new scaffolds and derivatives.
|
(GDP)K‐Ras4B/SOS exchange (IC50 [μM]) |
|
Cellular activities (IC50 [μM]) |
||||||
|---|---|---|---|---|---|---|---|---|
|
|
G12D |
WT |
WT %CRTL 3 mM |
SNU‐1 (G12D) |
NCI‐H441 (G12 V) |
HCT15 (G13D) |
H358 (G12 C) |
A549 (G12S) |
|
Benzimidazole benzofuran scaffold | ||||||||
|
12 |
340±72 |
1099±381 |
75 % |
34.5±2.1 |
47.4±3.0 |
37.7±0.4 |
37.7±1.3 |
45.3±1.5 |
|
13 |
409±32 |
334±5 |
n.d. |
52.7±0.3 |
54.5±0.2 |
n.d. |
n.d. |
50.3±2,0 |
|
29 |
769 [a] |
993±388 |
n.d. |
30.5±0.2 |
42.3±7.6 |
25.6±3.7 |
31.4±1.2 |
32.8±7.3 |
|
Benzimidazole phenol scaffold | ||||||||
|---|---|---|---|---|---|---|---|---|
|
14 a |
320±17 |
304 [a] |
n.d. |
|
|
|
|
|
|
14 b |
>3000 |
>3000 |
70 |
>300 |
>300 |
n.d. |
n.d. |
>300 |
|
14 c |
1069±130 |
493±31 |
77 |
>300 |
>300 |
n.d. |
n.d. |
>300 |
|
14 d |
1453±348 |
1758±215 |
70 |
>300 |
>300 |
n.d. |
n.d. |
>300 |
|
14 e |
>3000 |
>3000 |
77 |
265±14 |
>300 |
n.d. |
n.d. |
>300 |
|
14 f[ a] |
356[a] |
>3000 |
95 |
>30 |
>30 |
n.d. |
n.d. |
>30 |
|
14 g |
776±57 |
380±59 |
69 |
>300 |
>300 |
n.d. |
n.d. |
>300 |
|
30 |
873±294 |
1017±119 |
89 |
75.7±3.4 |
145±37 |
n.d. |
n.d. |
103±5 |
|
31 |
1272±99 |
1965[a] |
95 |
|
|
|
|
|
Data are shown as mean±SD of independent repeats (2–5 times). [a] Single measurements due to limited substance availability. [b] Due to substance limitations or solubility problems testing was not possible at higher concentrations.
Only compound 14 f expressed good Ras/SOS‐exchange inhibition and a pronounced selectivity. Interestingly, cleavage of the Boc group to amine 14 g resulted in a decrease in activity and an unwanted reversal of selectivity. Expectedly, the highest‐scoring in silico hit 12, consisting of a benzimidazole and a benzofuran building block, showed best activity of all new scaffolds (Table 3, Figure 5) and a moderate selectivity for the G12D mutant. Surprisingly, regioisomer 13 which differs from scaffold 12 only by the position of one hydroxy group, was found somewhat less active and unselective. Obviously, even minimal structural changes are sufficient to influence decisively the Ras/SOS‐exchange activity but even more the selectivity for wildtype or oncogenic K‐Ras4B forms.
Unfortunately, it is not possible to simply transfer the SAR of the DHPB derivatives to the new scaffolds. Both 4‐fluorophenyl‐hydrazones 30 and 31 as well as the oxime 29 turned out somewhat weaker GDP/GTP‐exchange inhibitors of the G12D mutant than the parent ketones (Figure 6). Disappointingly, all compounds, containing the benzimidazole phenol scaffold were inactive in the tumor cell assays (Table 3). On the other side, the high‐scoring in silico scaffolds 12 and its regioisomer 13 expressed significantly improved activities in the cell assays compared to DHBP from which they were deduced.
Figure 6.
Hydrazone and oxime derivatives of new dihetaryl ketone related scaffolds.
MST measurements suggested approximately equally strong binding to GDP and GTP K‐Ras4B of ketone 12 (KD(GDP) 59 μM/KD(GTP) 34 μM) which provides a rationale for the higher cellular activities compared to the Ras/SOS‐exchange assay (Figure 6).
Oxime ether 29 expressed improved cellular activities in comparison to the unmodified scaffold 12 and DHBP oxime ether 11. The activity level of compound 29 against Ras mutant cell lines is in the range of 4‐fluorophenylhydrazone 7 c, but with complete hydrolytic stability over a 24 h period.
Disappointingly, we were not able to develop a binding model for oxime ether 29. Due to its limited solubility, NMR data of sufficient quality could not be acquired. However, the bisheteroaryl ketone 12 gave several significant chemical shift perturbations (CSPs) in a 2D 1H‐15N‐HSQC NMR spectrum with K‐Ras4B in the region of the SI/II‐site and additionally around the nucleotide binding site (Figure 7). Haddock calculations revealed 181 poses which were attributed to ten clusters. As expected, the poses of the energetically most favoured cluster (Haddock‐score: −18.3±2.7 kcal/mol, 34 poses) correspond with the SI/SII‐site. The highest populated but energetically less favoured cluster 2 (−15.7±0.7 kcal/mol, 66 poses) occupies a site adjacent to the nucleotide binding site (Figure 7d), involving several residues of the switch I region. The cluster poses obtained from the Haddock calculations were refined by Glide‐docking and subsequent MM‐GBSA calculations. [51] In addition to the wildtype, other relevant Ras structures (4EPY, 4DSO, 6ZL5) with different mutations and GDP/GTP loadings were included for those calculations. For all K‐Ras4B forms, we consistently found that the poses with the most favourable binding‐energies and docking‐scores showed the benzofuran moiety placed inside the pocket. This orientation is stabilised by a hydrogen bond with Ser39 and a π‐cation interaction with Lys5. The carbonyl group of ketone 12 is oriented towards the pocket wall formed by Gln70, Tyr71, and Thr74 which results in a further hydrogen bond between the benzimidazole hydroxy group and Glu62 (Figure 7c). In addition, NMR‐titration experiments show an 18‐fold stronger binding of the bis‐heteroaromatic scaffold 12 (KD 0.5±0.1 mM) to K‐Ras4B than the starting‐point DHBP (KD 9.1±3.9 mM).
Figure 7.
Compound 12 binding to K‐Ras4B. a) Haddock clusters 1 (dark green, SI/SII‐site) and 2 (light green, SI‐site) projected onto (GDP)K‐Ras4Bwt (PDB code: 6MBT). CSPs with 2σ are colored dark blue, CSPs with 1σ confidence are marked in pale blue. b) refined high‐scoring pose of compound 12 (green) in the SI/SII‐binding site. c) Ligand‐protein interactions with SI/SII‐site. d) Ligand‐protein interactions with SI.
Conclusion
In recent years, the SI/II‐ and SIIP‐binding sites have attracted particular attention as targets for small molecule inhibitors of mutant K‐Ras4B. Amongst others, one critical feature associated with those binding sites is the limited structural diversity of ligands published to date. In this paper, we demonstrated significant cellular activities of hydrazone 7 c as well as oxime ethers 11 and 29 against Ras tumour cell lines. The cellular activity of hydrazone 7 c and oxime 11 in particular may be attributed to their ability to preferentially disrupt Ras‐PI3K‐Akt downstream signaling. NMR studies provide evidence for DHBP hydrazone 7 c and bis‐hetaryl ketone 12 to bind to the SI/SII‐site of K‐Ras4B, whereas the DHBP oxime ether 11 preferably interacts with the SIIP‐site, which is also targeted by MRTX1133 (5), a clinical development product. Similar to MRTX1133, our three‐armed, Y‐like oximes 11 and 29 are able to address three different areas within their pockets. Moreover, the new scaffold 12 shows an 18‐fold higher affinity to the SI/SII‐site than DHBP from which it was derived.
However, with the identification of the nanomolar SIIP‐ligand MRTX1133 there is a growing question as to whether the SI/SII‐ binding site remains a prime target, since even the strongest published ligands 3 and 4 as well as our best compounds exhibit cellular activities only in the low micromolar range.
Nevertheless, based on our findings we are convinced that our initial hits have the potential to provide high‐affinity SIIP‐ligands with improved selectivity through systematic optimization.
Experimental Section
Chemical syntheses
Abbreviations: AcOH, acetic acid; Cyh, cyclohexane; DCM, dichloromethane; DCVC, dry column vacuum chromatography; DIPEA, N,N‐diisopropylethylamine; DMF, N,N‐dimethylformamide; DMSO, dimethyl sulfoxide; EDCI, 1‐ethyl‐3‐(3‐dimethyl‐aminopropyl)carbodiimide; MEMCl, 2‐methoxyethoxymethyl chloride; rt, room temperature; TFA, trifluoroacetic acid; THF, tetrahydrofuran.
General: IR spectra were recorded on a Bruker ALPHA FTIR spectrometer. 1H and 13C NMR spectra were recorded on Bruker Avance III 600 and Bruker Avance 400 spectrometers operating at 600 and 400 MHz (1H) respectively and 150 and 100 MHz (13C), respectively. Accurate mass determinations were achieved with a Bruker micrOTOF mass spectrometer. The reactions were monitored by TLC carried out on Macherey Nagel silica gel plates (60F‐254) or Merk silica gel 60 RP‐18 F254 plates using UV light and an aqueous solution of KMnO4, K2CO3, NaOH and heat as the visualizing agent. HPLC‐MS analyses: Bruker micrOTOF (ESI‐MS)+ Agilent 1100 Series; HPLC‐column: Perfect Sil Target ODS‐3 HD, 100×4.6 mm×5 μm; gradient 90 % water (5 mm NH4OAc) to 90 % MeCN in 24 min; flow 1.5 mL/min; 220 nm or 254 nm, positive or negative mode. Shimadzu LCMS‐2020+Shimadzu Prominence‐i LC‐2030 C 3D Plus; HPLC‐column: Shim‐pack GISS C18 1.9 μm 50×2.1 mm; gradient 95 % water (0.1 % HCOOH) to 95 % MeCN (0.1 % HCOOH) in 16 min; flow 400 μL/min; 200–800 nm; ESI: positive or negative. Reagents and solvents were purchased from commercial sources and used without further purification, unless otherwise stated. CH2Cl2 was dried with a MB‐SPS‐800 solvent purification system. MeOH was redistilled from magnesium turnings. Reactions were stirred magnetically under an argon atmosphere unless otherwise stated. All final compounds are >95 % pure by HPLC analysis, except compounds 13 (87 %) and 8 c (89 %), which were difficult to purify either due to a tautomeric equilibrium (13) or substance limitation (8 c).
General procedure for the synthesis of hydrazine derivatives
All hydrazones, acyl‐, and sulfonyl‐hydrazones, as well as oxime 11 were prepared according to standard literature procedures.[ 33 , 52 , 53 , 54 , 55 , 56 ]
Method A: The hydrazine derivative R1‐NHNH2 (3.45 mmol, 1.5 eq) was dissolved at 50 °C in a mixture of MeOH (6 mL) and AcOH (400 μL, 7.00 mmol, 3.00 eq). Subsequently, a solution of DHBP (500 mg, 2.33 mmol) in 6 mL MeOH was added. The reaction mixture was then stirred at 50 °C until the reaction was completed. The reaction mixture was concentrated to one quarter of the total volume and the residue was taken up in EtOAc and NaHCO3 solution (3 %). The phases were separated and the aqueous phase extracted with EtOAc (3×10 mL). After drying of the combined organic phases over Na2SO4, the solvent was removed and the crude product obtained was purified by column chromatography.
Method B: The corresponding hydrazine derivative R1‐NHNH2 (1.40 mmol, 1.5 eq) was dissolved at 50 °C in a mixture of MeOH (4 mL) and conc. sulfuric acid (270 μL, 5.00 mmol, 5.36 eq). Subsequently, a solution of DHBP (200 mg, 0.93 mmol) in 4 mL MeOH was added. The reaction mixture was then stirred at 50 °C until the reaction was completed. The reaction mixture was concentrated to one quarter of the total volume and the residue was taken up in 6.5 mL water. The precipitated solid was filtered off and washed with NaHCO3 solution (3 %). The raw product was purified by column chromatography.
4,4′‐((2‐(2,4‐Dinitrophenyl)hydrazinylidene)methylene)diphenol (7 a)
Compound 7 a was obtained from 2,4‐dinitrophenylhydrazine hydrochloride (329 mg, 1.40 mmol) and DHBP (200 mg, 934 μmol) according to method B. Yield 187 mg (51 %). Purity >99 %. 1H NMR (600 MHz, DMSO‐d6 ) δ 11.15 (1H, s), 10.06 (1H, s), 9.99 (1H, s), 8.80 (1H, d, J=2.64 Hz), 8.38 (1H, dd, J=9.63, 2.55 Hz), 8.15 (1H, d, J=9.60 Hz), 7.49 (2H, d, J=8.70 Hz), 7.23 (2H, d, J=8.52 Hz), 7.02 (2H, d, J=8.52 Hz), 6.82 (2H, d, J=8.76 Hz). 13C NMR (600 MHz, DMSO‐d6 ) δ 162.3, 159.7, 158.8, 155.9, 144.0, 136.8, 130.0, 129.9, 129.5, 127.6, 123.0, 121.7, 116.5, 116.4, 115.4. IR (cm−1): =3372 (w), 3277 (w), 1654 (w), 1609 (s), 1586 (w). LCMS (ESI): tR=10.1 min; m/z (% rel. intensity): 212.1 (18.3), 395.1 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C19H13N4O6 +: 393.0841 [M+H]+; found: 393.0841.
4,4′‐((2‐henylhydrazinylidene)methylene)‐diphenol (7 b)
Compound 7 b was obtained from phenylhydrazine (340 μL, 3.45 mmol) and DHBP (500 mg, 2.33 mmol) according to method A. Yield 363 mg (46 %). Purity >99 %. 1H NMR (600 MHz, DMSO‐d6) δ 9.74 (1H, bs, 9.54 (1H, bs), 8.69 (1H, s), 7.28 (2H, d, J=8.76 Hz), 7.18 (4H, m), 7.09 (2H, d, J=8.52 Hz) 6.94 (2H, d, J=8.64 Hz), 6.72 (2H, d, J=8.64 Hz). 13C NMR (600 MHz, DMSO‐d6) δ 157.7, 157.6, 144.9, 144.7, 130.2, 130.1, 128.5, 127.7, 123.4, 121.7, 116.1, 115.0, 114.1. IR (cm−1): =3321 (m), 1591 (m), 1493 (s), 1429. LCMS (ESI): tR=10.5 min; m/z (% rel. intensity): 339.1 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C19H16ClN2O2 +: 339.0895 [M+H]+; found: 339.0895.
4,4′‐((2‐(4‐Fluorophenyl)hydrazinylidene)‐methylene)diphenol (7 c)
Compound 7 c was obtained from 4‐fluorphenylhydrazine hydrochloride (569 mg, 3.50 mmol) and DHBP (500 mg, 2.33 mmol) according to method A. Yield 268 mg (36 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d6 ) δ 9.73 (1H, s), 9.51 (1H, s), 8.52 (1H, s), 7.28 (2H, d, J=8.76 Hz), 7.17 (2H, m), 7.09 (2H, d, J=8.56 Hz), 6.99 (2H, t, J=8.90 Hz), 6.94 (2H, d, J=8.56 Hz), 6.72 (2H, J=8.80 Hz). 13C NMR (400 MHz, DMSO‐d6 ) δ 157.7, 157.4, 156.9, 154.6, 144.1, 142.5, 130.2, 127.6, 123.5, 116.1, 115.3, 115.1, 115.0, 113.7, 113.6. 19F NMR (DMSO‐d6 ) δ −126.49 (1F, sept., J=4.56 Hz). IR (cm−1): =3348 (w), 3305 (w), 3037 (w), 2997 (w), 2789 (w), 2669 (w), 2596 (w), 1607 (m), 1592 (w). LCMS (ESI): tR=9.6 min; m/z (% rel. intensity): 229.1 (19.2), 323.1 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C19H16FN2O2 +: 323.1190 [M+H]+; found: 323.1190.
4,4′‐((2‐(4‐Chlorophenyl)hydrazinylidene)‐methylene)diphenol (7 d)
Compound 7 d was obtained from 4‐chlorophenylhydrazine hydrochloride (627 mg, 3.50 mmol) and DHBP (500 mg, 2.33 mmol) according to method A. Yield 363 mg (46 %). Purity >99 %. 1H NMR (600 MHz, DMSO‐d6 ) δ 9.74 (1H, bs), 9.54 (1H, bs), 8.69 (1H, s), 7.28 (2H, d, J=8.76 Hz), 7.18 (4H, m), 7.09 (2H, d, J=8.52 Hz) 6.94 (2H, d, J=8.64 Hz), 6.72 (2H, d, J=8.64 Hz). 13C NMR (600 MHz, DMSO‐d6 ) δ 157.7, 157.6, 144.9, 144.7, 130.2, 130.1, 128.5, 127.7, 123.4, 121.7, 116.1, 115.0, 114.1. IR (cm−1): =3321 (m), 1591 (m), 1493 (s). LCMS (ESI): tR=10.5 min; m/z (% rel. intensity): 339.1 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C19H16ClN2O2 +: 339.0895 [M+H]+; found: 339.0895.
4,4′‐((2‐(4‐(Trifluoromethyl)phenyl)‐hydrazinylidene)methylene)diphenol (7 e)
Compound 7 e was obtained from 4‐trifluoromethylphenylhydrazine (500 mg, 2.84 mmol) and DHBP (304 mg, 1.42 mmol) according to method A. Yield 518 mg (98 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d6 ) δ 9.76 (1H, s), 9.59 (1H, s), 9.06 (1H, s), 7.47 (2H, d, J=8.72 Hz), 7.30–7.33 (4H, m), 7.10 (2H, d, J=8.48 Hz), 6.94 (2H, d, J=8.52 Hz), 6.74 (2H, d, J=8.72 Hz). 13C NMR (400 MHz, DMSO‐d6 ) δ 157.9, 148.7, 146.5, 130.3, 129.9, 128.0,126.0 (q, J=3.31 Hz), 123.3, 116.0, 115.0, 112.3. 19F NMR (376 MHz, DMSO‐d6 ) δ −59.2 (3F, s). IR (cm−1): =3335 (w), 1609 (m), 1528 (w), 1508 (m), 1476 (w). LCMS (ESI): tR=10.1 min; m/z (% rel. intensity): 373.1 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C20H16F3N2O2 + [M+H]+ 373.1158; found: 373.1157.
4,4′‐((2‐(Pyridin‐2‐yl)hydrazinylidene)‐methylene)diphenol (7 f)
Compound 7 f was obtained from 2‐hydrazinepyridine (153 mg, 1.40 mmol) and DHBP (200 mg, 934 μmol) according to method A. Yield 134 mg (47 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d6 ) δ 9.89 (1H, s), 9.66 (1H, s), 8.08 (1H, s), 8.01 (1H, ddd, J=4.89, 1.85, 0.81 Hz), 7.67 (1H, ddd, J=8.42, 7.26, 1.80 Hz), 7.36 (2H, d, J=8.76 Hz), 7.33 (1H, d, J=8.40 Hz), 7.14 (2H, d, J=8.56 Hz), 6.99 (2H, d, J=8.60 Hz), 6.76 (3H, m). 13C NMR (400 MHz, DMSO‐d6 ) δ 158.1, 158.1, 156.3, 147.6, 146.9, 138.1, 129.9, 129.1, 128.0, 122.4, 116.3, 115.3, 115.1, 106.6. IR (cm−1): =3407 (w), 3305 (m), 3012 (w), 2786 (w), 2667 (w), 2258 (w), 1602 (m), 1574 (s), 1501 (s). LCMS (ESI): tR=7.8 min; m/z (% rel. intensity): 306.1 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C18H16N3O2 +: 306.1237 [M+H]+; found: 306.1237.
General procedure for the synthesis of aryl sulfonyl hydrazides
Hydrazine monohydrate (560 μL, 11.5 mmol) was added dropwise at 0 °C to a solution of the corresponding aryl sulfonyl chloride (4.60 mmol) in 20 mL THF. The mixture was stirred at 0 °C for 30 minutes and then 12 mL EtOAc were added. The organic phase was washed with sat. NaCl solution (3×20 mL), dried over Na2SO4 and then slowly added to cyclohexane (25 mL) with constant stirring. The precipitating solid was filtered off and residual solvent was removed, yielding the clean product.
4‐Chlorobenzenesulfonohydrazide
4‐Chlorobenzenesulfonohydrazide was obtained from 4‐chlorbenzolsulfonylchloride (970 mg, 4.60 mmol) according to the general procedure. Yield 900 mg (95 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d6 ) δ 8.50 (1H, s), 7.80 (2H, td, J=9.12, 2.26 Hz), 7.68 (2H, td, J=9.20, 2.26 Hz), 3.99 (2H, bs). 13C NMR (400 MHz, DMSO‐d6 ) δ 137.5, 137.1, 129.5, 129.1. IR (cm−1): =3387 (w), 3346 (w), 3282 (m), 3202 (w), 3090 (w), 2857 (w), 1613 (w), 1585 (w), 1573 (w), 1474 (m). HRMS (ESI): m/z calcd for C6H7ClN2O2SNa+: 228.9809 [M+Na]+; found: 228.9809.
4‐(Trifluoromethoxy)‐benzenesulfono‐hydrazide
4‐(Trifluoromethoxy)benzenesulfono‐hydrazide was obtained from 4‐trifluoro‐methoxybenzoylsulfonyl chloride (531 mg, 2.00 mmol) according to the general procedure. Yield 500 mg (98 %). Purity >99 %. 1H NMR (400 MHz, CDCl3) δ 7.98 (2H, d, J=8.92 Hz), 7.38 (2H, dd, J=8.94 Hz), 3.53 (2H, bs). 13C NMR (400 MHz, CDCl3) δ 153.0, 135.0, 130.6, 121.2. 19F NMR (400 MHz, CDCl3) δ −56.72 (s). IR (cm−1): =3315 (m), 3185 (w), 1619 (w), 1588 (w), 1491 (w). HRMS (ESI): m/z calcd for C7H8F3N2O3S+: 257.0202 [M+H]+; found: 257.0205.
4‐Nitrobenzenesulfonohydrazide
4‐Nitrobenzenesulfonohydrazide was obtained from 4‐nitrobenzoylsulfonyl chloride (1.00 g, 4.51 mmol) according to the general procedure. Yield 829 mg (85 %). Purity >99 % 1H NMR (400 MHz, DMSO‐d6 ) δ 8.74 (1H, s), 8.42 (2H, d, J=8.96 Hz), 8.05 (2H, d, J=8.96 Hz), 4.35 (2H, bs). 13C NMR (400 MHz, DMSO‐d6 ) δ 149.7, 144.2, 129.2, 124.2. IR (cm−1): =3394 (w), 3363 (w), 3219 (w), 3103 (w), 3077 (w), 2980 (w), 2143 (w), 1956 (w), 1607 (w), 1522 (s), 1478 (w). HRMS (ESI): m/z calcd for C6H8N3O4S+: 218.0230 [M+H]+; found: 218.0232.
4‐Cyanobenzenesulfonohydrazide
4‐Cyanobenzenesulfonohydrazide was obtained from 4‐cyanobenzoylsulfonyl chloride (485 mg, 2.41 mmol) according to the general procedure. Yield 437 mg (92 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d6 ) δ 8.68 (1H, s), 8.09 (2H, d, J=8.72 Hz), 7.96 (2H, d, J=8.72 Hz), 4.31 (2H, bs). 13C NMR (400 MHz, DMSO‐d6 ) δ 142.7, 133.1, 128.4, 117.8, 114.9. IR (cm−1): =3365 (w), 3215 (w), 3091 (w), 3036 (w), 2981 (w), 2239 (w), 1604 (w). HRMS (ESI): m/z calcd for C7H8N3O2S+: 198.0332 [M+H]+; found: 198.0333.
General procedure for the synthesis of sulfonyl hydrazone derivatives
p‐Toluene sulfonic acid monohydrate (3.8 mg, 20 μmol) was added to a solution of DHBP (428 mg, 2.00 mmol) and the corresponding aryl sulfonyl hydrazide (2.00 mmol) in 3.5 mL EtOH. The reaction mixture was heated at 85 °C in a pressure vial until completion. After cooling to room temperature, the solvent was evaporated.
N′‐(bis(4‐hydroxyphenyl)methylene)benzene‐sulfonohydrazide (8 a)
Compound 8 a was obtained from benzoylsulfonyl hydrazide (344 mg, 2.00 mmol) according to the general procedure and purification by column chromatography. Yield 509 mg (69 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d6 ) δ 10.09 (1H, s), 9.81 (1H, s), 9.75 (1H, s), 7.91 (2H, d, J=7.00 Hz), 7.63 (3H, m), 7.08 (2H, d, J=8.64 Hz), 7.03 (2H, d, J=8.48 Hz), 6.86 (2H, d, J=8.52 Hz), 6.69 (2H, d, J=8.68 Hz). 13C NMR (400 MHz, DMSO‐d6 )161.2, 158.9, 158.3, 155.5, 139.0, 132.7, 130.4, 129.1, 128.8, 127.7, 115.3, 115.0. IR (cm−1): =3384 (w), 3183 (w), 2981 (w), 1598 (m), 1552 (w), 1509 (m). LCMS (ESI): tR=7.7 min; m/z (% rel. intensity): 227.1 (2.5), 369.1 (100.0) [M+H]+, 590.1 (2.7). HRMS (ESI): m/z calcd for C19H17N2O4S+: 369.0904 [M+H]+; found: 369.0903.
N′‐(bis(4‐hydroxyphenyl)methylene)‐4‐methoxybenzenesulfonohydrazide (8 b)
Compound 8 b was obtained from 4‐methoxybenzoylsulfonyl hydrazide (417 mg, 2.00 mmol) according to the general procedure and purification by precipitation in Cyh/EtOAc. Yield 152 mg (19 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d 6) δ 9.89 (1H, s), 9.80 (1H, bs), 9.75 (1H, bs), 7.83 (2H, d, J=8.88 Hz), 7.13 (2H, d, J=8.96 Hz), 7.10 (2H, d, J=8.76 Hz), 7.02 (2H, d, J=8.52 Hz), 6.86 (2H, d, J=8.56 Hz), 6.69 (2H, d, J=8.72 Hz), 3.84 (3H, s). 13C NMR (400 MHz, DMSO‐d 6) δ 162.5, 158.9, 158.2, 155.2, 130.7, 130.4, 129.9, 129.1, 128.6, 123.2, 115.4, 115.0, 114.0, 55.6. IR (cm−1): =3137 (m), 2968 (w), 2940 (w), 1595 (s), 1578 (w), 1509 (w). LCMS (ESI): tR=3.8 min; m/z (% rel. intensity): 399.1 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C20H19N2O5S+: 399.1009 [M+H]+; found: 399.1011.
N′‐(bis(4‐hydroxyphenyl)methylene)‐4‐chlorobenzenesulfonohydrazide (8 c)
Compound 8 c was obtained from 4‐chlorobenzoylsulfonyl hydrazide (413 mg, 2.00 mmol) according to the general procedure and purification by column chromatography. Yield 81 mg (10 %). Purity 89 %. 1H NMR (400 MHz, DMSO‐d 6) δ 10.19 (1H, s), 9.84 (1H, s), 9.80 (1H, s), 7.91 (2H, d, J=8.56 Hz), 7.70 (2H, d, J=8.60 Hz), 7.10 (2H, d, J=8.60 Hz), 7.04 (2H, d, J=8.44 Hz), 6.87 (2H, d, J=8.52 Hz), 6.70 (2H, d, J=8.68 Hz). 13C NMR (400 MHz, DMSO‐d 6) δ 159.0, 158.3, 156.1, 137.8, 137.8, 130.5, 129.7, 129.2, 129.1, 128.5, 123.2, 115.4, 115.0. IR (cm−1): =3331 (m), 3194 (w), 1607 (w), 1598 (m), 1566 (w), 1508 (m), 1475 (w). LCMS (ESI): tR=8.7 min; m/z (% rel. intensity): 403.0 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C19H16ClN2O4S+: 403.0514 [M+H]+; found: 403.0513.
N′‐(bis(4‐hydroxyphenyl)methylene)‐4‐bromobenzenesulfonohydrazide (8 d)
Compound 8 d was obtained from 4‐bromobenzoylsulfonyl hydrazide (500 mg, 1.99 mmol) according to general procedure. Purification by column chromatography. Yield 92 mg (10 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d6) δ 10.18 (1H, s), 9.81 (1H, s), 9.77 (1H, s), 7.81–7.86 (4H, m), 7.09 (2H, d, J=8.72 Hz), 7.03 (2H, d, J=8.56 Hz), 6.86 (2H, d, J=8.65 Hz), 6.70 (2H, d, J=8.76 Hz). 13C NMR (400 MHz, DMSO‐d6 ) δ 159.0, 158.2, 156.0, 138.2, 131.9, 130.5, 129.7, 129.1, 128.4, 126.7, 123.1, 115.3, 115.0. IR (cm−1): =3486 (w), 3353 (w), 3222 (w), 2925 (w), 1703 (w), 1609 (m), 1592 (w), 1573 (w), 1511 (m), 1470 (w). LCMS (ESI): tR=8.7 min; m/z (% rel. intensity): 449.0 (100.0) [M+H]+, 895.0 (17.4) [2 M+H]+. HRMS (ESI): m/z calcd for C19H16N2O4S+: 447.0009 [M+H]+; found: 447.0019.
N′‐(bis(4‐hydroxyphenyl)methylene)‐4‐nitrobenzenesulfonohydrazide (8 e)
Compound 8 e was obtained from 4‐nitrobenzoylsulfonyl hydrazide (434 mg, 2.00 mmol) according to the general procedure and purification by recrystallisation in toluene/iPrOH. Yield 641 mg (78 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d6 ) δ 10.47 (1H, s), 9.84 (1H, s), 9.79 (1H, s), 8.45 (2H, d, J=8.88 Hz), 8.16 (2H, d, J=8.84 Hz), 7.11 (2H, d, J=8.72 Hz), 7.05 (2H, d, J=8.52 Hz), 6.87 (2H, d, J=8.56 Hz), 6.70 (2H, d, J=8.72 Hz). 13C NMR (400 MHz, DMSO‐d6 ) δ 159.1, 158.3, 156.8, 149.9, 144.4, 130.5, 129.3, 129.3, 128.3, 124.2, 123.1, 115.3, 115.0. IR (cm−1): =3438 (m), 3373 (w), 3231 (w), 3105 (w), 1608 (w), 1599 (m), 1584 (w), 1525 (s), 1509 (w). LCMS (ESI): tR=8.3 min; m/z (% rel. intensity): 414.1 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C19H16N3O6S+: 414.0754 [M+H]+; found: 414.0754.
N′‐(bis(4‐hydroxyphenyl)methylene)‐4‐(trifluoromethoxy)benzenesulfonohydrazide (8 f)
Compound 8 f was obtained from 4‐trifluoromethoxybenzoyl hydrazide (384 mg, 1.50 mmol) according to the general procedure and purification by recrystallisation in toluene/iPrOH. Yield 229 mg (34 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d 6) δ 10.23 (1H, s, 9.83 (1H, s), 9.78 (1H, s), 8.03 (2H, d, J=8.88 Hz), 7.63 (2H, d, J=8.12 Hz), 7.08 (2H, d, J=8.72 Hz), 7.04 (2H, d, J=8.56 Hz), 6.87 (2H, d, J=8.60 Hz), 6.69 (2H, d, J=8.76 Hz). 13C NMR (400 MHz, DMSO‐d 6) δ 159.0, 158.3, 156.2, 151.1, 137.9, 132.0, 130.5, 130.4, 129.2, 128.4, 123.1, 121.1, 115.3, 115.0. 19F NMR (376 MHz, DMSO‐d 6) δ −56.7. IR (cm−1): =3137 (m), 2797 (w), 2731 (w), 1608 (w), 1595 (m). LCMS (ESI): tR=9.3 min; m/z (% rel. intensity): 453.1 (100.0) [M+H]+, 717.2 (20.7). HRMS (ESI): m/z calcd for C20H16F3N2O5S+: 453.0727 [M+H]+; found: 453.0728.
N′‐(bis(4‐hydroxyphenyl)methylene)‐4‐cyanobenzenesulfonohydrazide (8 g)
Compound 8 g was obtained from 4‐cyanobenzoylsulfonyl hydrazide (296 mg, 1.50 mmol) according to the general procedure and purification by column chromatography. Yield 392 mg (66 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d6 ) δ 10.40 (1H, s), 9.84 (1H, s), 9.80 (1H, s), 8.12 (2H, d, J=8.60 Hz, 8.06 (2H, d, J=8.60 Hz), 7.08 (2H, d, J=8.76 Hz), 7.04 (2H, d, J=8.56 Hz), 6.87 (2H, d, J=8.60 Hz), 6.70 (2H, d, J=8.76 Hz). 13C NMR (400 MHz, DMSO‐d6 ) δ 159.1, 158.3, 156.6, 143.0, 133.0, 130.5, 129.2, 128.5, 128.3, 123.1, 117.8, 115.3, 115.2, 115.0. IR (cm−1): =3329 (w), 3144 (w), 2246 (w), 1608 (w), 1594 (m), 1566 (w), 1512 (w), 1439 (w). LCMS (ESI): tR=7.8 min; m/z (% rel. intensity): 394.1 (100.0) [M+H]+, 590.1 (2.7). HRMS (ESI): m/z calcd for C20H16N3O4S+: 394.0856 [M+H]+; found: 394.0857.
N′‐(Bis(4‐hydroxyphenyl)methylene)‐benzohydrazide (10)
To a hot solution of benzhydrazide (389 mg, 2.80 mmol) in 4 mL EtOH was added a solution of DHBP (500 mg, 2.33 mmol) in 4 mL EtOH. The mixture was stirred overnight and after removal of the solvent the raw product was separated by column chromatography. Yield 572 mg (74 %). Purity >99 %. 1H NMR (600 MHz, DMSO‐d6 ) δ 9.91 (1H, s), 9.84 (2H, bs), 7.59 (2H, bs), 7.52 (1H, dd, J=7.29 Hz), 7.43–7.46 (2H, m), 7.38 (2H, bs), 7.19 (2H, d, J=8.22 Hz), 6.96 (2H, d, J=8.40 Hz), 6.79 (2H, d, J=7.56 Hz). 13C NMR (600 MHz, DMSO‐d6 ) δ 162.5, 159.2, 158.4, 156.5, 133.9, 131.4, 130.2, 129.4, 128.6, 127.0, 122.7, 116.0, 115.1. IR (cm−1): =3322 (m), 1626 (m), 1606 (m), 1501 (s). LCMS (ESI): tR=6.7 min; m/z (% rel. intensity): 333.1 (100.0) [M+H]+, 665.2 (31.2) [2 M+H]+. HRMS (ESI): m/z calcd for C20H17N2O3 +: 333.1234 [M+H]+; found: 333.1235.
Bis(4‐hydroxyphenyl)methanone‐O‐benzyl oxime (11)
To a solution of O‐benzylhydroxylamine (820 μL, 7.00 mmol) in 7 mL acetic acid sodium acetate (330 mg, 4.02 mmol) and DHBP (500 mg, 2.33 mmol) were sequentially added. After about one hour of reaction time at room temperature, the reaction mixture was added to water and neutralised with sat. NaHCO3 solution. The neutralised solution was extracted with EtOAc (10 mL×3) and the combined organic phases were washed with sat. NaCl solution and dried over Na2SO4. After evaporation of the solvent, the product was purified by column chromatography. Yield 553 mg (74 %). Purity >99 %. 1H NMR (400 MHz, DMSO‐d6 ) δ 9.71 (2H, s), 7.35 (4H, m), 7.29 (1H, m), 7.19 (2H, d, J=8.72 Hz), 7.14 (2H, d, J=8.60 Hz), 6.81 (2H, d, J=8.64 Hz), 6.74 (2H, d, J=8.72 Hz), 5.11 (2H, s). 13C NMR (400 MHz, DMSO‐d6 ) δ 158.6, 157.7, 156.3, 138.3, 130.3, 129.1, 128.2, 127.7, 127.5, 127.2, 123.6, 115.1, 114.7, 75.2. IR (cm−1): =3366 (w), 2980 (w), 2928 (w), 1608 (m), 1510 (s). LCMS (ESI): tR=9.3 min; m/z (% rel. intensity): 320.1 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C20H18NO3 +: 320,1281 [M+H]+; found: 320.1281.
2‐Hydroxy‐3‐(4‐methoxybenzyl)‐benzaldehyde (20)
NaH (60 %, 246 mg, 6.15 mmol) was suspended in dry DMSO (4 mL) and a solution of 2,3‐dihydroxybenzaldehyde (19, 340 mg, 2.46 mmol) in 2 mL DMSO was added dropwise under cooling with ice. After 1.5 h, 4‐methoxybenzyl chloride (334 μL, 2.46 mmol) was added dropwise and the resulting mixture was stirred at rt for 3 h. The reaction was stopped by slow addition of aqueous HCl (1 m) solution. The weakly acidic solution was extracted with CH2Cl2 and the combined organic layers were dried over Na2SO4. The crude product obtained after evaporation was purified by column chromatography to give benzaldehyde 20 (506 mg, 80 %). Purity >99 %. 1H NMR (600 MHz, CDCl3) δ 11.03 (1H, s), 9.92 (1H, s), 7.37 (2H, d, J=8.58 Hz), 7.19 (1H, dd, J=1.35, 7.83 Hz), 7.14 (1H, d, J=7.92 Hz), 6.90 (3H, m), 5.12 (2H, s), 3.81 (3H, s). 13C NMR (400 MHz, CDCl3) δ 196.6, 159.7, 152.6, 147.4, 129.3, 128.7, 125.4, 121.4, 119.6, 114.2, 71.5, 55.4. IR (cm−1): =2930 (w), 2842 (w), 1650 (m), 1612 (w), 1585 (w), 1512 (m), 1454 (m). LCMS (ESI): tR=8.5 min, m/z (% rel. intensity): 136.0 (40.9), 257.1 (100.0) [M−H]−. HRMS (ESI): m/z calcd for C15H13O4 −: 257.0819 [M‐H]−; found: 257.0815.
7‐(4‐Methoxybenzyl)benzofurane‐2‐carboxylic acid (21)
Cs2CO3 (11.7 g, 36.0 mmol) was added to a solution of 2‐hydroxy‐3‐(4‐methoxybenzyl)‐benzaldehyde (20, 3.00 g, 11.6 mmol) in 19 mL dry DMSO. Afterwards the mixture was heated to 100 °C. Then bromoacetic acid ethyl ester (1.35 mL, 12.2 mmol) was slowly added, and the solution changed color from greenish brown to beige. After stirring overnight at 100 °C, the reaction mixture was acidified with 1 m HCl solution and extracted with EtOAc. The combined organic phases were dried with Na2SO4 and, after removal of drying agent and solvent, impurities were removed by column chromatography. Product 21 was obtained in 86 % yield (2.97 g) as a brown solid. Purity 70 %. 1H NMR (400 MHz, DMSO‐d6 ) δ 7.63 (1H, s), 7.44 (2H, d, J=8.72 Hz), 7.32 (1H, dd, J=7.76, 1.08 Hz), 7.24 (1H, t, J=7.80 Hz), 7.17 (1H, dd, J=7.94, 1.06 Hz), 6.97 (2H, d, J=8.72 Hz), 5.21 (2H, s), 3.76 (3H, s). 13C NMR (400 MHz, DMSO‐d6 ) δ 160.0, 159.2, 146.2, 144.7, 144.4, 129.9, 128.5, 128.3, 124.5, 114.8, 113.9, 113.7, 110.5, 69.9, 55.1. IR (cm−1): =2932 (m), 1682 (s), 1585 (m), 1512 (s), 1490 (w). LCMS (ESI): tR=4.3 min, m/z (% rel. intensity): 297.1 (100.0) [M−H]−, 595.2 (18.1) [2 M−H]−. HRMS (ESI): m/z calcd for C17H13O5 −: 297.0768 [M−H]−; found: 297.0768.
N‐Methoxy‐7‐(4‐methoxybenzyl)‐N‐methyl‐benzofurane‐ 2‐carboxamide (22)
To a solution of 7‐(4‐methoxybenzyl)‐benzofurane‐2‐carboxylic acid (21, 2.00 g, 6.70 mmol) and N,O‐dimethylhydroxylamine hydrochloride (687 mg, 7.04 mmol) in 20 mL dry CH2Cl2 were added NEt3 (1.87 mL, 13.4 mmol), DMAP (819 mg, 6.70 mmol), and EDCI (1.35 g, 7.04 mmol) at 0 °C. The mixture was stirred overnight at rt, then diluted with CH2Cl2 and water was added. The organic phase was removed and the aqueous phase was extracted two more times with CH2Cl2. The combined organic phase was washed with water and brine and afterwards dried over Na2SO4. After filtration and removal of the solvent, the raw product was purified by column chromatography (576 mg, 25 %). Purity 89 %. 1H NMR (600 MHz, CDCl3) δ 7.47 (1H, s), 7.41 (2H, d, J=8.52 Hz), 7.25 (1H, d, J=8.46 Hz), 7.15 (1H, t, J=7.86 Hz), 6.94 (1H, d, J=7.86 Hz), 6.90 (2H, d, J=8.64 Hz), 5.26 (2H, s), 3.84 (3H, s), 3.81 (3H, s), 3.41 (3H, s). 13C NMR (600 MHz, CDCl3) δ 159.8, 159.5, 147.1, 145.0, 144.9, 129.2, 129.2, 128.9, 124.1, 114.9, 113.9, 113.4, 111.2, 71.0, 61.7, 55.3, 33.6. IR (cm−1): =2959 (w), 2935 (w), 2840 (w), 1640 (s), 1610 (w), 1590 (w), 1552 (m). LCMS (ESI): tR=9.6 min, m/z (% rel. intensity): 342.1 (65.8) [M+H]+, 700.3 (100.0) [2 M+NH4]+. HRMS (ESI): m/z calcd for C19H19NO5Na+: 364.1155 [M+Na]+; found: 364.1160.
1H‐Benzimidazol‐7‐ol (16)
2‐amino‐3‐nitrophenol (15, 2.00 g, 13.0 mmol) was dissolved in 40 mL MeOH and triethyl orthoformate (4.32 mL, 26.0 mmol), a drop of AcOH, and Pd/C (10 %, 200 mg, 0.19 mmol) were added. The mixture was stirred for 24 hours under a hydrogen atmosphere. Then the catalyst was filtered off over Celite and the solvent was removed. After column chromatographic separation (DCVC), benzimidazole 16 was obtained in 51 % yield (880 mg). Purity >99 %. 1H NMR (600 MHz, DMSO‐d6 ) δ 12.33 (1H, bs), 9.78 (1H, bs), 8.06 (1H, s), 7.01 (1H, d, J=7.84 Hz), 6.96 (1H, dd, J=7.72, 7.72 Hz), 6.57 (1H, d, J=8.32 Hz). 13C NMR (400 MHz, DMSO‐d 6) δ 140.4, 122.5, 106.4. IR (cm−1): =3220 (s), 3111 (w), 2357 (w), 1744 (w), 1635 (w), 1593 (s). LCMS (ESI): tR=2.5 min, m/z (% rel. intensity): 133.1 (100.0) [M−H]−. HRMS (ESI): m/z calcd for C7H5N2O−: 133.0407 [M−H]−; found: 133.0408.
7‐(2‐Methoxyethoxymethoxy)‐1‐(2‐methoxyethoxymethyl)benzimidazole and 4‐(2‐Methoxyethoxymethoxy)‐1‐ (2‐methoxyethoxymethyl)‐benzimidazole (17/18)
A solution of 1H‐benzimidazol‐7‐ol (16, 800 mg, 5.96 mmol) in 20 mL THF was added dropwise at 0 °C to a suspension of NaH (60 %, 596 mg, 14.9 mmol) in 5 mL THF. Subsequently, 10 mL of degassed DMF were added. After 20 min, MEMCl (1.43 mL, 12.5 mmol) was slowly added and after another 30 min, the ice bath was removed. Stirring was continued overnight at rt. Then the reaction was stopped by addition of water. The aqueous phase was saturated with NaCl, separated from the organic phase, and then extracted twice with EtOAc. The combined organic phases were dried with Na2SO4. The product mixture obtained after evaporation was separated by column chromatography to afford two tautomeric products in a total yield of 81 % (1.50 g, 17: 46 %, 18: 35 %). Purity >95 %. Isomer 17: 1H NMR (400 MHz, CDCl3) δ 7.95 (1H, s), 7.23 (1H, dd, J=7.80, 7.80 Hz), 7.19 (1H, dd, J =1.34, 8.14 Hz), 7.04 (1H, dd, J=7.44, 1.36 Hz), 5.60 (2H, s), 5.54 (2H, s), 3.91–3.93 (2H, m), 3.54–3.57 (4H, m), 3.47–3.50 (2H, m), 3.35 (3H, s), 3.35 (3H, s). 13C NMR (400 MHz, CDCl3) δ 149.1, 142.0, 135.3, 134.5, 124.5, 108.0, 104.2, 94.1, 75.0, 71.7, 71.6, 67.9, 67.8, 59.0, 58.9. IR (cm−1): =2923 (w), 2880 (w), 2819 (w), 1712 (w), 1620 (w), 1590 (w), 1495 (m), 1455 (w). LCMS (ESI): tR=4.8 min, m/z (% rel. intensity): 311.2 (100.0) [M+H]+, 621.3 (2.7) [2 M+H]+. HRMS (ESI): m/z calcd for C15H22N2O5Na+: 333.1421 [M+Na]+; found: 333.1420. Isomer 18 : 1H NMR (400 MHz, CDCl3) δ 8.00 (1H, s), 7.45 (1H, dd, J=0.62, 8.10 Hz), 7.18 (1H, dd, J=8.06, 8.06 Hz), 7.05 (1H, d, J=7.80 Hz), 5.84 (2H, s), 5.42 (2H, s), 3.86–3.88 (2H, m), 3.60–3.62 (2H, m), 3.55–3.58 (2H, m), 3.46–3.48 (2H, m), 3.37 (3H, s), 3.33 (3H, s). 13C NMR (400 MHz, CDCl3) δ 145.6, 144.5, 143.8, 123.5, 123.2, 114.0, 108.3, 93.9, 76.2, 71.7, 71.6, 68.1, 67.6, 59.0. IR (cm−1): =3092 (w), 2924 (w), 2881 (w), 2819 (w), 1711 (w), 1615 (w), 1590 (w), 1497 (m), 1455 (w). LCMS (ESI): tR=4.7 min, m/z (% rel. intensity): 311.1 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C15H22N2O5Na+: 333.1421 [M+H]+; found: 333.1425.
[4‐(2‐Methoxyethoxymethoxy)‐1‐(2‐methoxyethoxymethyl)‐benzimidazol‐2‐yl]‐[7‐[(4‐methoxyphenyl)methoxy]benzofuran‐2‐yl]methanon and [7‐(2‐Methoxyethoxy‐methoxy)‐ 1‐(2‐methoxyethoxy‐methyl)benzimidazol‐ 2‐yl]‐[7‐[(4‐methoxy‐phenyl)methoxy]benzofuran‐ 2‐yl]methanon (23/24)
The mixture of benzimidazole isomers 17 (44 %) and 18 (56 %) (327 mg, 1.05 mmol) was dissolved in 6 mL dry THF and added slowly at −78 °C to freshly titrated n‐BuLi solution (2.1 m in n‐hexane, 510 μL, 1.06 mmol). The solution was stirred for 2 h at this temperature. Then, a solution of N‐methoxy‐7‐(4‐methoxybenzyl)‐N‐methyl‐benzofurane‐2‐carboxamide 22 (300 mg, 879 μmol) in 2.5 mL dry THF was added. After 4 h water was added slowly to the reaction mixture at rt. The aqueous phase was extracted several times with EtOAc and the combined organic layers were washed with water and brine. After drying over Na2SO4, the solvent was removed and the product mixture was separated by column chromatography. The two isomers were obtained in 85 % overall yield (23: 125 mg, 24 % 24: 269 mg, 52 %). Purity 90 % Isomer 23: 1H NMR (400 MHz, CDCl3) δ 8.49 (1H, s), 7.59 (1H, dd, J=8.22, 0.74 Hz), 7.44 (2H, d, J=8.68 Hz), 7.36 (1H, dd, J=7.88, 0.92 Hz), 7.29 (1H, dd, J=8.06, 8.06 Hz), 7.19 (1H, dd, J=7.90, 7.90 Hz), 7.19 (1H, dd, J=8.00, 0.72 Hz), 7.02 (1H, dd, J=7.88, 0.88 Hz), 6.92 (2H, d, J=8.68 Hz), 6.39 (2H, s), 5.47 (2H, s), 5.30 (2H, s), 3.91–3.93 (2H, m), 3.82 (3H, s), 3.66–3.68 (2H, m), 3.57–3.60 (2H, m), 3.40–3.43 (2H, m), 3.39 (3H, s), 3.25 (3H, s). 13C NMR (400 MHz, CDCl3) δ 174.0, 159.7, 151.7, 146.7, 146.0, 145.2, 144.0, 129.6, 129.5, 128.8, 126.5, 124.7, 124.6, 121.2, 116.3, 115.6, 114.1, 113.0, 111.0, 94.1, 75.7, 71.7, 71.7, 71.3, 68.3, 68.0, 59.2, 59.0, 55.4. IR (cm−1): =2925 (m), 2837 (w), 1647 (m), 1613 (w), 1590 (m), 1552 (m), 1514 (m), 1487 (m). LCMS (ESI): tR=12.8 min, m/z (% rel. intensity): 591.2 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C32H34N2O9Na+: 613.2157 [M+Na]+; found: 613.2158. Isomer 24: 1H NMR (600 MHz, CDCl3) δ 8.56 (1H, s), 7.44 (2H, d, J=8.58 Hz), 7.37 (4H, m), 7.20 (1H, dd, J=7.86, 7.86 Hz), 7.13 (1H, d, J=7.86 Hz), 7.02 (1H, d, J=7.86 Hz), 6.91 (2H, J=8.58 Hz), 6.15 (2H, s), 5.65 (2H, s), 5.29 (2H, s), 3.97–3.98 (2H, m), 3.82 (3H, s), 3.72–3.74 (2H, m), 3.59–3.60 (2H, m), 3.47–3.48 (2H, m), 3.38 (3H, s), 3.31 (3H, s). 13C NMR (400 MHz, CDCl3) δ 173.8, 159.7, 151.5, 150.6, 145.2, 144.7, 138.2, 133.5, 129.6, 129.5, 128.8, 127.7, 124.7, 121.4, 116.3, 114.1, 112.9, 109.2, 105.6, 94.7, 75.0, 71.8, 71.6, 71.2, 68.3, 59.2, 59.1, 55.4. IR (cm−1): =2925 (w), 2836 (w), 1644 (m), 1612 (w), 1590 (m), 1551 (m), 1512 (m), 1487 (m), 1471 (w). LCMS (ESI): tR=12.2 min, m/z (% rel. intensity): 591.2 (100.0) [M+H]+. HRMS (ESI): m/z calcd for C32H34N2O9Na+: 613.2157 [M+Na]+; found: 613.2168.
(7‐Hydroxy‐1H‐benzimidazol‐2‐yl)‐(7‐hydroxybenzofuran‐2‐yl)methanon (12)
Step A: A mixture of isomers 23 and 24 (150 mg, 254 μmol) was dissolved in 10 mL dry CH2Cl2 followed by dropwise addition of 1.1 mL TFA. After complete addition, the volatiles were removed in vacuo and the mixture was chromatographed. Step B: The chromatographed product from the first reaction step (85 mg) was dissolved in 1 mL EtOH and added to 1 mL 4 m HCl in dioxane. The mixture was first stirred overnight and then refluxed for 7 h. After completion of the reaction, sat. NaHCO3 solution was added and the mixture was extracted with EtOAc. Column chromatographic separation followed by preparative HPLC afforded product 12 (14 mg) in 19 % yield over two steps. Purity >99 %. 1H NMR (400 MHz, DMSO‐d 6) δ 8.95 (1H, s), 7.39 (1H, dd, J=7.84, 0.96 Hz), 7.20 (1H, dd, J=7.82 Hz), 7.19 (1H, dd, J=7.94 Hz), 7.06 (1H, d, J=7.96 Hz), 7.00 (1H, dd, J=7.76, 1.00 Hz), 6.70 (1H, dd, J=7.72, 0.76 Hz). 13C NMR (400 MHz, DMSO‐d 6) δ 171.7, 150.1, 149.9, 145.9, 144.9, 143.3, 137.6, 132.5, 128.9, 126.6, 125.0, 120.7, 114.3, 114.2, 107.3, 104.4. IR (cm−1): =3117 (w), 2961 (s), 2800 (w), 1644 (m), 1602 (w), 1565 (s), 1541 (w). LCMS (ESI): tR=7.4 min), m/z (% rel. intensity): 295.1 (100.0) [M+H]+, 611.1 (1.7) [2 M+H]+. HRMS (ESI): m/z calcd for C16H11N2O4 +: 295.0713 [M+H]+; found: 295.0712.
(5‐Hydroxy‐1H‐benzo[d]imidazol‐2‐yl)(7‐hydroxybenzofuran‐ 2‐yl)methanone (13)
Yield: 60 mg. Purity 87 %. 1H NMR (600 MHz, DMSO‐d 6) δ 8.13 (s, 1H, H‐), 7.63 (d, J=8.8 Hz, 1H, H‐), 7.22 (d, J=7.9 Hz, 1H, H−), 7.09 (t, J=7.8 Hz, 1H, H‐), 7.06–7.00 (m, 2H, H−), 6.94 (d, J=7.7 Hz, 1H, H−). 13C NMR (151 MHz, DMSO) δ 169.3 (s), 158.6 (s), 151.3 (s), 146.5 (s), 143.8 (s), 143.3 (s), 135.8 (s), 130.4 (s), 129.7 (s), 126.8 (s), 120.5 (s), 119.6 (s), 119.0 (s), 116.4 (s), 115.9 (s), 99.3 (s). IR (cm−1): =2980 (m), 1651 (m), 1632 (w), 1596 (w), 1565 (s). LCMS (ESI): tR=6.4 min.; m/z (% rel. intensity): 295.0 (100) [M+H]+. HRMS (ESI): m/z calcd for C16H11N2O4 +: 295.0713[M+H]+; found: 295.0715.
2‐(4‐Hydroxybenzyl)‐1H‐benzo[d]imidazole‐5‐carboxylic acid (27)
A mixture of 3,4‐diaminobenzoic acid (26, 1.00 g, 6.57 mmol), 4‐hydroxyphenylacetic acid (25, 2.50 g, 20 mmol) and hydrochloric acid (15 mL, 4 M) were stirred for about 10 minutes and then heated at 120 °C for 6 h. The precipitate was filtered off and dried. The crude product was recrystallized from hydrochloric acid (5 %) and filtrated yielding the product 27 (1.23 g, 70 %) as blue solid. Purity >99 %. 1H NMR (600 MHz, DMSO‐d6 ) δ 8.33–8.16 (m, 1H), 8.05 (dd, J=8.6, 1.5 Hz, 1H), 7.82 (d, J=8.6 Hz, 1H), 7.41–7.18 (m, 2H), 6.93–6.54 (m, 2H), 4.44 (s, 2H). 13C NMR (151 MHz, DMSO‐d6 ) δ 166.6, 157.11, 155.5, 134.0, 131.0, 130.4, 127.9, 126.3, 123.5, 115.8, 115.4, 113.9, 31.4. IR (cm−1): =3589 (m), 3272 (w), 2968 (w), 2912 (w), 2714 (s), 1678 (s), 1631 (w), 1610 (m), 1564 (m). LCMS (ESI): tR=3.2 min, m/z (%rel. intensity): 269.1 (100) [M+H]+. HRMS (ESI): m/z calcd for C15H13N2O3 +: 269.0919 [M+H]+; found: 269.0921.
2‐(4‐Hydroxybenzoyl)‐1H‐benzo[d]‐imidazole‐5‐carboxylic acid (28)
A mixture of compound 27 (1.44 g, 5.37 mmol) and sulfur (1.37 g, 5.37 mmol) in DMF (100 mL) was heated at 90 °C for 7 days. After cooling, the sulfur was filtered off and washed with DMF. The solvent was removed under reduced pressure. The crude product was dissolved in a sodium hydroxide solution and washed with Et2O, hexane and DCM. The product (1.21 g, 80 %) was precipitated with conc. hydrochloric acid, filtered off and dried to afford compound 28 as a yellow solid. Purity >99 %. 1H NMR (600 MHz, DMSO‐d6 ) δ 8.50 (d, J=8.7 Hz, 2H), 8.34 (d, J=0.8 Hz, 1H), 7.96 (dd, J=8.6, 1.5 Hz, 1H), 7.80 (d, J=8.7 Hz, 1H), 7.11–6.94 (m, 2H). 13C NMR (151 MHz, DMSO‐d6 ) δ 180.9, 167.5, 163.6, 149.8, 140.3, 137.7, 133.9, 126.9, 126.5, 125., 119.1, 116.5, 115.6. IR (cm−1): =3412 (w), 3227 (w), 1678 (m), 1635 (w), 1576 (s). LCMS (ESI): tR=5.3 min, m/z (% rel. intensity): 283.1 (100) [M+H]+. HRMS (ESI): m/z calcd for C15H11N2O4 +: 283.0713 [M+H+]; found: 283.0714.
(2‐(4‐Hydroxybenzoyl)‐1H‐benzo[d]imidazol‐5‐yl)(4‐methylpiperazin‐1‐yl)methanone (14 a)
A mixture of compound 28 (100 mg, 0.35 mmol), HATU (162 mg, 0.43 mmol) and DIPEA (183 mg, 1.42 mmol) in DMF (4 mL) was stirred for 20 min. at 0 °C. Then methylpiperazine (53 mg, 0.53 mmol) in DMF (1 mL) was added. The mixture was stirred at rt for 12 h. After that time, the solvent was removed under reduced pressure. The remaining residue was solved in EtOAc and water. The aqueous phase was extracted with EtOAc and DCM. The combined organic phases were dried over Na2SO4 and concentrated in vacuo. Column chromatography on silica gel (DCM to EtOH) afforded the desired product (84 mg, 52 %) as a yellow solid. Purity >99 %. 1H NMR (400 MHz, CD3COOD) δ 8.25–8.15 (m, 2H), 7.88 (d, J=0.7 Hz, 1H), 7.81 (d, J=8.5 Hz, 1H), 7.49 (dd, J=8.5, 1.5 Hz, 1H), 7.04–6.84 (m, 2H), 4.95–3.16 (m, 8H), 3.00 (s, 3H). 13C NMR (101 MHz, CD3COOD) δ 183.7, 172.5, 162.7, 149.7, 138.7, 138.4, 134.2, 129.8, 127.4, 124.3, 117.2, 117.0, 115.8, 53.30, 43.3. IR (cm−1): =3078 (s), 2938 (w), 2867 (w), 2805 (w), 1598 (s), 1482 (w), 1435 (m). LCMS (ESI): tR=3.7 min, m/z (%rel. intensity): 365.0 (100) [M+H]+. HRMS (ESI): m/z calcd for C20H21N4O3 +: 365.1608 [M+H]+; found: 365.1607.
(S)‐2‐((tert‐Butoxycarbonyl)amino)‐3‐(2‐(4‐hydroxybenzoyl)‐ 1H‐benzo[d]imidazole‐5‐carboxamido) propanoic acid (14 b)
DIPEA (581 mg, 4.50 mmol) was added to an ice cooled solution of starting material 28 (423 mg, 1.50 mmol) and HATU (627 mg, 1.65 mmol) in DMF (5 mL) after 30 min. (S)‐3‐amino‐2‐((tert‐butoxycarbonyl)amino)‐propanoic acid (306 mg, 1.50 mmol) in DMF (1 mL) was added after additional 30 min. at 0 °C. The mixture was stirred for 12 h and then quenched with water. The solvent was removed under reduced pressure. Column chromatography on silica gel (Cyh/DCM 95 : 5 to DCM to Ethanol) afforded the desired product (210 mg, 30 %) as a yellow solid. Purity >99 %. 1H NMR (400 MHz, CD3OD) δ 8.50–8.41 (m, 2H), 8.24 (s, 1H), 7.84 (d, J=8.6 Hz, 1H), 7.73 (d, J=8.6 Hz, 1H), 6.99–6.85 (m, 2H), 4.37 (dd, J=19.9, 15.3 Hz, 1H), 3.79 (ddd, J=21.3, 13.5, 6.4 Hz, 2H), 1.40 (s, 9H). 13C NMR (101 MHz, CD3OD) δ 183.5, 175.5, 170.7, 164.7, 158.0, 151.6, 141.3, 140.0, 134.9, 131.7, 128.5, 124.8, 118.3, 116.9, 116.4, 80.7, 55.9, 43.4, 28.7. IR (cm−1): =3191 (w), 3069 (w), 2976 (m), 1685 (w), 1635 (w), 1576 (m), 1504 (w), 1477 (w). LCMS (ESI): tR=6.0 min, m/z (%rel. intensity): 469.3 (100) [M+H]+. HRMS (ESI): m/z calcd for C23H25N4O7 +: 469.1718 [M+H]+; found: 469.1719.
(S)‐2‐Amino‐3‐(2‐(4‐hydroxybenzoyl)‐1H‐benzo[d]imidazole‐ 6‐carboxamido)‐propanoic acid (14 c)
HCl in dioxane (1 mL, 4 M) was added to solution of compound 14 b (40 mg, 85.4 μmol) in dioxane (2 mL). The solution was stirred at rt for 12 h. The solvent was removed under reduced pressure yielding the product (36 mg, 96 %) as a yellow solid. Purity: >99 %. 1H NMR (400 MHz, DMSO‐d6 ) δ 8.98 (t, J=5.6 Hz, 1H), 8.65 (d, J=4.5 Hz, 3H), 8.53 (d, J=8.8 Hz, 2H), 8.38 (d, J=0.7 Hz, 1H), 7.96 (dd, J=8.6, 1.5 Hz, 1H), 7.77 (d, J=8.6 Hz, 1H), 7.01 (d, J=8.8 Hz, 2H), 4.13 (dd, J=10.3, 5.0 Hz, 1H), 3.84 (t, J=5.7 Hz, 2H). 13C NMR (101 MHz, DMSO‐d6 ) δ 181.0, 169.2, 167.2, 163.4, 149.8, 139.4, 138.3, 133.8, 129.9, 126.6, 124.1, 117.5, 115.8, 115.5, 52.4, 39.5. IR (cm−1): =2980 (m), 1732 (w), 1693 (w), 1646 (w), 1594 (s), 1557 (w). LCMS (ESI): tR=3.9 min, m/z (% rel. inten.): 369.0 (100) [M+H]+. HRMS (ESI): m/z calcd for C18H16N4O5Na+: 391.1013 [M+Na]+; found: 391.1012.
N‐(2‐(Dimethylamino)ethyl)‐2‐(4‐hydroxybenzoyl)‐ 1H‐benzo[d]imidazole‐5‐carboxamide (14 d)
To a mixture of starting material 28 (60 mg, 0.21 mmol), HATU (97 mg, 0.26 mmol) and DIPEA (110 mmol, 0.85 mmol) in DMF (4 mL) was added 1,1‐dimethyl‐ethylendiamine (28 mg, 0.32 mmol). The mixture was stirred at rt for 12 h and then quenched with NH4Cl‐solution. The solvent was removed under reduced pressure. Purification of the residue by column chromatography on silica gel (DCM to EtOH) afforded the desired product (39 mg, 52 %) as a yellow solid. Purity >99 %. 1H NMR (600 MHz, CD3OD) δ 8.52–8.40 (m, 2H), 8.27 (s, 1H), 7.86 (dd, J=8.6, 1.6 Hz, 1H), 7.73 (d, J=8.6 Hz, 1H), 7.01–6.88 (m, 2H), 3.72 (t, J=6.2 Hz, 2H), 3.10 (t, J=6.2 Hz, 1H), 2.72 (s, 6H). 13C NMR (151 MHz, CD3OD) δ 183.4, 170.6, 164.8, 151.8, 141.1, 140.3, 134.9, 131.2, 128.5, 124.7, 118.5, 116.9, 116.4, 58.8, 44.4, 37.4. IR (cm−1): =3066 (m), 2966 (w), 1635 (w), 1544 (m). LCMS (ESI): tR=4.1 min, m/z (%rel. intensity): 353.0 (100) [M+H]+. HRMS (ESI): m/z calcd for C19H21N4O3 +: 353.1608 [M+H]+; found: 353.1609.
tert‐Butyl‐(2‐(2‐(4‐hydroxybenzoyl)‐1H‐benzo[d]imidazole‐5‐ carboxamido)‐ethyl)carbamate (14 e)
A solution of educt 28 (120 mg, 0,43 mmol) in dioxane (10 mL) was reacted for 10 min. with EDC⋅HCl (85 mg, 0.45 mmol) and Oxyma (69 mg, 0.45 mmol). After that time boc‐ethylendiamine (68 mg, 0.43 mmol) and DIPEA (66 mg, 0.51 mmol) were added and the mixture was stirred for 12 h at rt. Then water was added and the aqueous phase was extracted with EtOAc. The organic phase was washed with sat. NaHCO3, dried (Na2SO4) and concentrated in vacuo. Purification by column chromatography on silica gel (DCM to EtOH) afforded the desired product (120 mg, 67 %) as a yellow solid. Purity >99 %. 1H NMR (600 MHz, DMSO‐d6 ) δ 8.53–8.38 (m, 2H), 8.20 (s, 1H), 7.77 (d, J=8.6 Hz, 1H), 7.72 (d, J=8.6 Hz, 1H), 6.98–6.79 (m, 2H), 3.34 (t, J=6.1 Hz, 2H), 3.15 (t, J=6.0 Hz, 2H), 1.28 (s, 9H). 13C NMR (151 MHz, DMSO‐d6 ) δ 182.7, 168.7, 163.7, 157.5, 150.8, 134.8, 131.1, 127.8, 124.6, 116.3, 79.6, 40.8, 40.3, 28.9. IR (cm−1): =1691 (m), 1625 (w), 1603 (m), 1579 (w). LCMS (ESI): tR=6.4 min, m/z (%rel. intensity): 425.2 (100) [M+H]+. HRMS (ESI): m/z calcd for C22H24N4O5Na+: 447.1639 [M+Na]+; found: 447.1640.
N‐(2‐Aminoethyl)‐2‐(4‐hydroxybenzoyl)‐1H‐benzo[d]imidazole‐5‐carboxamide (14 f)
HCl in dioxane (4 mL, 4 M) was added to a solution of compound 14 e (80 mg, 0.19 mmol) in dioxane (4 mL). The solution was stirred at rt for 12 h. The solvent was removed under reduced pressure yielding the product (74 mg, 99 %) as a yellow solid. Purity >99 %. 1H NMR (400 MHz, CD3OD) δ 8.38–8.33 (m, 2H), 8.29 (s, 1H), 7.88 (dd, J=8.6, 1.5 Hz, 1H), 7.79 (d, J=8.6 Hz, 1H), 7.03–6.95 (m, 2H), 3.77 (t, J=5.8 Hz, 2H), 3.36–3.27 (m, 2H). 13C NMR (101 MHz, CD3OD) δ 183.9, 171.4, 164.2, 151.2, 140.7, 139.9, 134.9, 130.7, 128.3, 125.1, 118.6, 116.9, 116.5, 40.9, 38.8. IR (cm−1): =3310 (w), 2899 (w), 1651 (w), 1628 (w), 1606 (w), 1578 (s). LCMS (ESI): tR=3.9 min, m/z (%rel. inten.): 325.1 (100) [M+H]+. HRMS (ESI): m/z calcd for C17H17N4O3 +: 325.1295 [M+H]+; found: 325.1292.
Biomolecular NMR Spectroscopy
Multidimensional NMR spectra were acquired as previously published.[ 32 , 41 , 57 ] All spectra were recorded on Bruker DRX 600 and AVANCE III HD 700 spectrometers with Watergate solvent suppression at 298 K. The following Bruker pulse programmes for the recording the NMR data were used: 1D 1H (zggpw5), 2D 1H‐15N HSQC (hsqcfpf3gpphwg), and 2D 1H‐15N TROSY (trosyetf3gpsi). The interscan delay was set to 1 s. Typically, samples of 0.4 mM [U‐15N]‐enriched protein were measured applying one‐ or two‐dimensional NMR spectra. All samples included 0.4 mM protein in PBS buffer at pH 7.4, 1 mM DTT, 10 % D2O, and DSS for referencing. The acquisition parameters were as follows. 1D 1H: T=298 K, NS=128, TD (1H)=32768. 2D 1H‐15N HSQC: T=298 K, NS 16, TD (1H)=2048, TD (15N)=256. 2D 1H‐15N TROSY: T=298 K, NS=32, TD (1H)=2048, TD (15N)=128.
Computational methods
Haddock calculations
Docking calculations were performed on the Haddock web‐server version 2.4. The input consisted of the protein PDB and its corresponding ligand. The active protein residues were set according to the NMR‐derived chemical shift perturbations (CSPs) exceeding the 2σ confidence level (Figure 8). The passive residues were assigned by default as surface residues surrounding the active ones. The respective ligand as a whole was set as the active residue. Prior to docking, the PDB‐file 6MBT was modified to ensure atom type descriptions be compatible with Haddock. This included the magnesium ion and the removal of all hydrogens which were then re‐introduced by the Haddock webserver. The following residues were selected as active for each docking:
Figure 8.
NMR chemical shift perturbation (CSP) analysis for K‐Ras bound to GDP upon titration with compounds 7c, 11, and 12. Weighted CSPs plotted versus the amino acid sequence are shown on the left‐hand side, while representative regions of 2D 1H–15N HSQC spectra are displayed on the right‐hand side.
Docking results presented in Figure 4 (7 c)
6MBT [(GDP)K‐Ras4Bwt], active residues: Lys5, Leu6, Val29, Glu31, Ile55, Leu56, Met67, Thr74, Gly75, Leu79.
Docking results presented in Figure 4 (11)
6MBT [(GDP)K‐Ras4Bwt], active residues: Lys5, Val29, Glu31, Ile46, Gly48, Cys51, Ile55, Leu56, Ala59, Met67, Arg73, Thr74, Gly75, Asp92, Ile93, His95, Tyr96, Lys104, Val112, Asn116, His166.
Docking results presented in Figure 7 (12)
6MBT [(GDP)K‐Ras4Bwt], active residues: Lys5, Asn26, Val29, Tyr32, Asp47, Ile55, Leu56, Asp57, Ala66, Met67, Met72, Thr74, Gly75, Asp105, Asn116.
In silico docking
All calculations were done with the Schrödinger molecular modeling suite (Maestro version 11.5). For in silico dockings all ligands were prepared with the LigPrep tool. Protonation states and tautomers were calculated for pH±2 using Epik. The raw (GDP)K‐Ras4Bwt PDB file (code: 6MBT) was optimized for in silico dockings with the protein preparation wizard. Missing amino acid side chains and loops were added with the software tool Prime. Protonation and tautomerization states were optimized with Epik for pH±1. After removal of water molecules beyond 5 Å distance from heavy atoms, an energy minimization of the protein to RMSD≤0.3 Å with OPLS2005 or OPLS3 was performed. For in silico dockings Glide, a component of the Schrödinger package, was used with standard settings. Large libraries were screened in HTS modus, followed by SP and XP runs. For optimization of Haddock calculations XP (extra precision) mode was applied. MM‐GBSA calculations were performed with Prime using the output files of Glide runs. Solvation model: VSGB, force filed: OPLS3, protein flexibility: 0 Å.
Biological testing
Nucleotide exchange assay
Inactive, GDP‐bound K‐Ras is incubated with SOS1 and GTP. K‐Ras is transferred to its active GTP‐bound state which leads to a release of GDP. The K‐Ras bound GTP is hydrolysed to GDP even in the absence of the corresponding GAP protein. In the GDP Glo Bioluminescent GDP detection assay for glycosyltransferases (Promega, Madison, WI, USA) used, converts GDP to ATP which can be quantified using a luciferase/luciferin reaction. The resulting luminescence signal is then measured with a suitable microplate reader. This assay was developed for K‐Ras wild type protein as well as for the G12D mutant. Exact conditions are summarised in Tables 4 and 5. For every sample, 5 μL K‐Ras working solution in assay buffer (50 mM HEPES pH 7,5, 4 mM MgCl2, 2 mM EGTA, 0,01 % Brij35, 1 mM TCEP) were transferred into a suitable assay plate (e. g. Greiner #784075). The test compound was added with an Echo acoustic dispenser (Beckman Coulter, Brea, CA, USA) in a concentration range from 3000 μM to 3 μM (8‐point dilution). After addition of the test compound 5 μL of SOS1‐GTP mix in assay buffer were added. The reaction mix was incubated over night at room temperature followed by the addition of 10 μL GPD detection reagent. After a second incubation period of 1 h at room temperature the luminescence signal was measured with an Envision spectrophotometer (Perkin Elmer, Waltham, MA, USA). IC50 values were determined from the sigmoidal dose response curves with the software Quattro Workflow (Quattro GmbH, Munich, Germany). Control Assay for Nucleotide Exchange Assay
Table 4.
Reagents, stock concentrations and final assay concentrations used for K‐Ras.
|
Reagents |
Stock concentration |
Working concentration |
Final assay concentration |
Supplier |
|---|---|---|---|---|
|
K‐Ras |
206.7 μM |
100 nM |
50 nM |
Dortmund Protein Facility |
|
SOS1 |
47.8 μM |
9.2 nM |
4.6 nM |
Dortmund Protein Facility |
|
GTP |
5 mM |
2 μM |
1 μM |
Bellbrook Labs |
Table 5.
Reagents, stock concentrations and final assay concentrations for K‐Ras(G12D).
|
Reagents |
Stock concentration |
Working concentration |
Final assay concentration |
Supplier |
|---|---|---|---|---|
|
K‐Ras(G12D) |
174.5 μM |
100 nM |
50 nM |
Dortmund Protein Facility |
|
SOS1 |
47.8 μM |
60.4 nM |
30.2 nM |
Dortmund Protein Facility |
|
GTP |
5 mM |
2 μM |
1 μM |
Bellbrook Labs |
To filter out compounds which interfere with the Nucleotide Exchange Assay independent from KRas and SOS1 a control assay was developed. For this, GDP was titrated and detected with the GDP Glo Bioluminescent GDP detection assay for glycosyltransferases (Promega, Madison, WI, USA) to generate a luminescence signal comparable to the positive control of the Nucleotide Exchange Assay. Compounds were checked for assay interference by performing dose response curves in the same concentration range as for the Nucleotide Exchange Assay. For every sample, 10 μL GDP (125 nM final assay concentration) in assay buffer (50 mM HEPES pH 7,5, 4 mM MgCl2, 2 mM EGTA, 0,01 % Brij35, 1 mM TCEP) was transferred into a suitable assay plate (e. g. Greiner #784075). The test compound was added with an Echo acoustic dispenser (Beckman Coulter, Brea, CA, USA) in a concentration range from 3000 μM to 3 μM (8‐point dilution). The reaction mix was incubated over night at room temperature followed by the addition of 10 μL GPD detection reagent. After a second incubation period of 1 h at room temperature the luminescence signal was measured with an Envision spectrophotometer (Perkin Elmer, Waltham, MA, USA). IC50 values were determined from the sigmoidal dose response curves with the software Quattro Workflow (Quattro GmbH, Munich, Germany).
CellTiter‐Glo assay
The CellTiter‐Glo Luminescent Cell Viability Assay (Promega) is a homogeneous method of determining the number of viable cells in culture. It is based on quantification of ATP, indicating the presence of metabolically active cells. On day 1 25 μL of the cell suspension are seeded at a cell number that assure assay linearity and optimal signal intensity. After incubation for 24 h at 37 °C/5 % CO2‐compounds dissolved in DMSO are added at different concentrations by Echo Liquid Handling Technology. Cells are further incubated in humidified chambers for 72 h at 37 °C and 5 % CO2. Cells treated with the compound vehicle DMSO are used as positive controls and cells treated with 10 μM staurosporine serve as negative controls.
At day 5–72 h after compound addition ‐ the CellTiter Glo Reagent is prepared according to the instructions of the kit (Promega Inc.). Thereon, mixture and assay plates are equilibrated at room temperature for 20 min. Equal volumes of the reagent‐medium‐mixture is added to the volume of culture medium present in each well. The plates are mixed at ∼300 rpm for 2 minutes on an orbital shaker. The microplates are then incubated at room temperature for 10 minutes for stabilization of the luminescent signal. Following incubation, the luminescence is recorded on a Victor microplate reader (Perkin Elmer) using a 200 ms integration time. The data is then analyzed with Excel using the XLFIT Plugin (dose response Fit 205) for IC50‐determination.
As quality control the Z'‐factor is calculated from 16 positive and negative control values. Only assay results showing a Z'‐factor≥0.5 are used for further analysis.
HTRF assay
SNU‐1 cells harbouring the KRASG12D mutation (primary carcinoma of the stomach), were grown in RPMI medium with 10 % FBS (fetal bovine serum) and 1 % L‐Glut (L‐Glutamine). Cells were plated in white small volume cell culture 384 well microplates (Greiner Bio One GmbH) at a density of 50.000 cells/well/6 μL in RPMI medium with 1 % L‐Glut (serum‐free medium). The plated cells were placed in a high humidity incubator (37 °C, 5 % CO2 and 90 % humidity) to avoid evaporation of the small amount of medium for 3 hours before the compounds were added. With the Echo520 Liquid Handler the cells were treated with a 3‐fold 8‐point serial dilution of the compounds, with a top final concentration of 330 μM and a final DMSO concentration of 1 %. After compound transfer cells incubated for 1 hour in the high humidity incubator before the cells were treated with 3 μl of 9 nM EGF diluted in serum‐free medium (final EGF concentration in the well 3 nM) for 15 minutes. The lysis buffer from the Phospho‐ERK 1/2 (Thr202/Tyr204)‐ and the Total ERk 1/2 LANCE Ultra TR‐FRET Cellular Detection Kit (PerkinElmer Inc.) was prepared to a 4‐fold concentration in water. Then 3 μl of the prepared lysis buffer were added to each well and the plates were shaken at 300 rmp for 30 minutes at room temperature. During this incubation antibody detection mix was prepared according to the manufacturers protocol. In the final step 3 μl of antibody detection mix was added to each well. The plates were sealed with an aluminium sticky foil and incubated over night at room temperature for 20 hours. The detection was performed with the EnVision 2104 Multilabel Reader.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
The authors thank I. Polanz, S. Bettinger, and A. Siebert for measuring HRMS and routine NMR spectra as well as Stefanie Pütz for expert technical support. This work was supported by the European Regional Development Fund and the German province of North Rhine‐Westphalia (EFRE‐0800945, LS‐1‐2‐001a, EFRE‐0800947, LS‐1‐2‐001b, EFRE‐0800950, LS‐1‐2‐001c; RIST). Open Access funding enabled and organized by Projekt DEAL.
S. Jeuken, O. Shkura, M. Röger, V. Brickau, A. Choidas, C. Degenhart, D. Gülden, B. Klebl, U. Koch, R. Stoll, J. Scherkenbeck, ChemMedChem 2022, 17, e202200392.
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.
References
- 1.“COSMIC – Catalogue of Somatic Mutations in Cancer.” can be found under https://cancer.sanger.ac.uk/cosmic.
- 2. Indarte M., Puentes R., Maruggi M., Ihle N. T., Grandjean G., Scott M., Ahmed Z., Meuillet E. J., Zhang S., Lemos R., Du-Cuny L., Layng F. I. A. L., Correa R. G., Bankston L. A., Liddington R. C., Kirkpatrick L., Powis G., Cancer Res. 2019, 79, 3100–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simanshu D. K., Nissley D. V., McCormick F., Cell 2017, 170, 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shin S.-M., Choi D.-K., Jung K., Bae J., Kim J.-S., Park S.-W., Song K.-H., Kim Y.-S., Nat. Commun. 2017, 8, 15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schadendorf D., Fisher D., Garbe C., Gershenwald J. E., Grob J.-J., Halpern A., Herlyn M., Marchetti M. A., McArthur G., Ribas A., Roesch A., Hauschild A., Nat Rev Dis Primers 2015, 1, 15003. [DOI] [PubMed] [Google Scholar]
- 6. Cox A. D., Fesik S. W., Kimmelman A. C., Luo J., Der C. J., Nat. Rev. Drug Discovery 2014, 13, 828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buhrman G., Holzapfel G., Fetics S., Mattos C., PNAS 2010, 107, 4931–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gorfe A. A., Grant B. J., McCammon J. A., Structure 2008, 16, 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu S., Jang H., Muratcioglu S., Gursoy A., Keskin O., Nussinov R., Zhang J., Chem. Rev. 2016, 116, 6607–6665. [DOI] [PubMed] [Google Scholar]
- 10. Fang H., Zhang Y., Bos P. H., Chambers J. M., Dupont M. M., Stockwell B. R. K., Biochemistry 2019, 58, 2542–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moore A. R., Rosenberg S. C., McCormick F., Malek S., Nat. Rev. Drug Discovery 2020, 19, 533–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ostrem J. M. L., Shokat K. M., Nat. Rev. Drug Discovery 2016, 15, 771–785. [DOI] [PubMed] [Google Scholar]
- 13. Papke B., Der C. J., Science 2017, 355, 1158–1163. [DOI] [PubMed] [Google Scholar]
- 14. Teng K. W., Tsai S. T., Hattori T., Fedele C., Koide A., Yang C., Hou X., Zhang Y., Neel B. G., O'Brian J. P., Koide S., Nat. Commun. 2021, 12, 2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whyte D. B., Kirschmeier P., Hockenberry T. N., Nunez-Oliva I., James L., Catino J. J., Bishop W. R., Pai J. K., J. Biol. Chem. 1997, 272, 14459–14464. [DOI] [PubMed] [Google Scholar]
- 16. Franz M., Mörchen B., Degenhart C., Gülden D., Shkura O., Wolters D., Koch U., Klebl B., Stoll R., Helfrich I., Scherkenbeck J., ChemMedChem 2021, 16, 2504–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zimmermann G., Papke B., Ismail S., Vartak N., Chandra A., Hoffmann M., Hahn S. A., Triola G., Wittinghofer A., Bastiaens P. I. H., Waldmann H., Nature 2013, 497, 638–642. [DOI] [PubMed] [Google Scholar]
- 18. Lanman B. A., Allen J. R., Allen J. G., Amegadzie A. K., Ashton K. S., Booker S. K., Chen J. J., Chen N., Frohn M. J., Goodman G., Kopecky D. J., Liu L., Lopez P., Low J. D., Ma V., Minatti A. E., Nguyen T. T., Nishimura N., Pickrell A. J., Reed A. B., Shin Y., Siegmund A. C., Tamayo N. A., Tegley C. M., Walton M. C., Wang H.-L., Wurz R. P., Xue M., Yang K. C., Achanta P., Bartberger M. D., Canon J., Hollis L. S., McCarter J. D., Mohr C., Rex K., Saiki A. Y., Miguel T. S., Volak L. P., Wang K. H., Whittington D. A., Zech S. G., Lipford J. R., Cee C. J., J. Med. Chem. 2020, 63, 52–65. [DOI] [PubMed] [Google Scholar]
- 19. Bar-Sagi D., Knelson E. H., Sequist L. V., Nat. Can. 2020, 1, 25–27. [DOI] [PubMed] [Google Scholar]
- 20. Goebel L., Müller M. P., Goody R. S., Rauh D., RSC Med. Chem. 2020, 11, 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.X. Wang, S. Allen, J. F. Blake, V. Bowcut, D. M. Briere, A. Calinisan, J. R. Dahlke, J. B. Fell, J. P. Fischer, R. J. Gunn, J. Hallin, J. Laguer, J. D. Lawson, J. Medwid, B. Newhouse, P. Nguyen, J. M. O'Leary, P. Olson, S. Pajk, L. Rahbaek, M. Rodriguez, C. R. Smith, T. P. Tang, N. C. Thomas, D. Vanderpool, G. P. Vigers, J. G. Christensen, M. A. Marx, J. Med. Chem .2022, 65, 3123–3133. [Online early access]. DOI: 10.1021/acs.jmedchem.1c01688. Published Online: December 10, 2021.
- 22. Gasper R., Wittinghofer F., Biol. Chem. 2020, 401, 143–163. [DOI] [PubMed] [Google Scholar]
- 23. Hunter J. C., Manandhar A., Carrasco M. A., Gurbani D., Gondi S., Westover K. D., Mol. Cancer Res. 2015, 13, 1325–1335. [DOI] [PubMed] [Google Scholar]
- 24. Taveras A. G., Remiszewski S. W., Doll R. J., Cesarz D., Huang E. C., Kirschmeier P., Pramanik B. N., Snow M. E., Wang Y.-S., del Rosario J. D., Vibulbhan B., Bauer B. B., Brown J. E., Carr D., Catino J., Evans C. A., Girijavallabhan V., Heimark L., James L., Liberles S., Nash C., Perkins L., Senior M. M., Tsarbopoulos A., Ganguly A. K., Aust R., Brown E., Delisle D., Fuhrman S., Hendrickson T., Kissinger C., Love R., Sisson W., Villafranca E., Webber S. E., Bioorg. Med. Chem. 1997, 5, 125–133. [DOI] [PubMed] [Google Scholar]
- 25. Ganguly A. K., Wang Y. S., Pramanik B. N., Doll R. J., Snow M. E., Taveras A. G., Remiszewski S., Cesarz D., del Rosario J., Vibulbhan B., Brown J. E., Kirschmeier P., Huang E. C., Heimark L., Tsarbopoulos A., Girijavallabhan V. M., Aust R. M., Brown E. L., DeLisle D. M., Fuhrman S. A., Hendrickson T. F., Kissinger C. R., Love R. A., Sisson W. A., Villafranca E., Webber S. E., Biochemistry 1998, 37, 15631–15637. [DOI] [PubMed] [Google Scholar]
- 26. Maurer T., Garrenton L. S., Oh A., Pitts K., Anderson D. J., Skelton N. J., Fauber B. P., Pan B., Malek S., Stokoe D., Ludlam M. J. C., Bowman K. K., Wu J., Giannetti A. M., Starovasnik M. A., Mellman I., Jackson P. K., Rudolph J., Wang W., Fang G., PNAS 2012, 109, 5299–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun Q., Burke J. P., Phan J., Burns M. C., Olejniczak E. T., Waterson A. G., Lee T., Rossanese O. W., Fesik S. W., Angew. Chem. Int. Ed. 2012, 51, 6140–6143; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 6244–6247. [Google Scholar]
- 28. Kessler D., Gmachl M., Mantoulidis A., Martin L. J., Zoephel A., Mayer M., Gollner A., Covini D., Fischer S., Gerstberger T., Gmaschitz T., Goddwin C., Greb P., Häring D., Hela W., Hoffmann J., Karolyi-Oezguer J., Knesl P., Kornigg S., Koegl M., Kousek R., Lamarre L., Moser F., Munico-Martinez S., Peinsipp C., Phan J., Rinnenthal J., Sai J., Salamon C., Scherbantin Y., Schipany K., Schnitzer R., Schrenk A., Sharps B., Siszler G., Sun Q., Waterson A., Wolkerstorfer B., Zeeb M., Pearson M., Fesik S. W., McConnell D. B., PNAS 2019, 116, 15823–15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Quevedo C. E., Cruz-Migoni A., Bery N., Miller A., Tanaka T., Petch D., Bataille C. J. R., Lee L. Y. W., Fallon P. S., Tulmin H., Ehebauer M. T., Fernandez-Fuentes N., Russel A. J., Carr S. B., Phillips S. E. V., Rabbitts T. H., Nat. Commun. 2018, 9, 3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cruz-Migoni A., Canning P., Quevedo C. E., Bataille C. J. R., Bery N., Miller A., Russel A. J., Phillips S. E. V., Carr S. B., Rabbitts T. H., PNAS 2019, 116, 2545–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McCarthy M. J., Pagba C. V., Prakash P., Naji A. K., van der Hoeven D., Liang H., Gupta A. K., Zhou Y., Cho K.-J., Hancock J. F., Gorfe A. A., ACS Omega 2019, 4, 2921–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schöpel M., Jockers K. F. G., Düppe P. M., Autzen J., Potheraveedu V. N., Ince S., Yip K. T., Heumann R., Herrmann C., Scherkenbeck J., Stoll R., J. Med. Chem. 2013, 56, 9664–9672. [DOI] [PubMed] [Google Scholar]
- 33. Morgan L. R., Thangaraj K., LeBlanc B., Rodgers A., Wolford L. T., Hooper C. L., Fan D., Jursic B. S. J., J. Med. Chem. 2003, 46, 4552–4563. [DOI] [PubMed] [Google Scholar]
- 34. Morgan L. R., Rodgers A. H., Fan D., Soike K., Ratterree M., Sartin B. W., Harrison T. J., In Vivo 1997, 11, 29–38. [PubMed] [Google Scholar]
- 35. Morgan L. R., Jursic B. S., Hooper C. L., Neumann D. M., Thangaraj K., LeBlanc B., Bioorg. Med. Chem. Lett. 2002, 12, 3407–3411. [DOI] [PubMed] [Google Scholar]
- 36. Schöpel M., Jockers K. F. G., Düppe P. M., Autzen J., Potheraveedu V. N., Ince S., Yip K. T., Heumann R., Herrmann C., Scherkenbeck J., Stoll R., J. Med. Chem. 2013, 56, 9664–9672. [DOI] [PubMed] [Google Scholar]
- 37. Gao C., He H., Qiu W., Zheng Y., Chen Y., Hu S., Zhao X., Environ. Sci. Technol. 2021, 55, 1953–1963. [DOI] [PubMed] [Google Scholar]
- 38. Pouliot M., Jeanmart S., J. Med. Chem. 2016, 59, 497–503. [DOI] [PubMed] [Google Scholar]
- 39. Yang Z.-Y., Dong J., Yang Z.-J., Lu A.-P., Hou T.-J., Cao D.-S., J. Chem. Inf. Model. 2020, 60, 2031–2043. [DOI] [PubMed] [Google Scholar]
- 40. van Zundert G. C. P., Rodrigues J. P. G. L. M., Trellet M., Schmitz C., Kastritis P. L., Karaca E., Melquiond A. S. J., van Dijk M., de Vries S. J., Bonvin A. M. J. J., J. Mol. Biol. 2015, 428, 720–725. [DOI] [PubMed] [Google Scholar]
- 41. Schöpel M., Shkura O., Seidel J., Kock K., Zhong X., Löffek S., Helfrich I., Bachmann H. S., Scherkenbeck J., Herrmann C., Stoll R., Int. J. Mol. Sci. 2018, 19, 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kalia J., Raines R. T., Angew. Chem. Int. Ed. 2008, 47, 7523–7526; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2008, 120, 7633–7636. [Google Scholar]
- 43. Liu Z., Li H., Zhao Q., Shen J., Heterocycles 2008, 75, 1907–1911. [Google Scholar]
- 44. Han S., Zhang F.-F., Qian H.-Y., Chen L.-L., Pu J.-B., Xie X., Chen J.-Z., Eur. J. Med. Chem. 2015, 93, 16–32. [DOI] [PubMed] [Google Scholar]
- 45. Morandi B., Mariampillai B., Carreira E. M., Angew. Chem. Int. Ed. 2011, 50, 1101–1104; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2011, 123, 1133–1136. [Google Scholar]
- 46. Gensini M., Altamura M., Dimoulas T., Fedi V., Giannotti D., Giuliani S., Guidi A., Harmat N. J. S., Meini S., Nannicini R., Pasqui F., Tramontana M., Triolo A., Maggi C. A., ChemMedChem 2010, 5, 65–78. [DOI] [PubMed] [Google Scholar]
- 47. Yan L., Kahne D., Synlett 1995, 523–524. [Google Scholar]
- 48. Sun F., Yin Z., Wang Q.-Q., Sun D., Zeng M.-H., Kurmoo M., Angew. Chem. Int. Ed. 2013, 52, 4538–4543; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 4636–4641. [Google Scholar]
- 49. Zhan Z., Ma H., Cui X., Jiang P., Pu J., Zhang Y., Huang G., Org. Biomol. Chem. 2019, 17, 5148–5152. [DOI] [PubMed] [Google Scholar]
- 50. Subirós-Funosas R., El-Faham A., Albericio F., Org. Synth. 2013, 90, 306–315. [Google Scholar]
- 51. Genheden S., Ryde U., Expert Opin. Drug Discovery 2015, 10, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mokhtari J., Naimi-Jamal M. R., Hamzeali H., Dekamin M. G., Kaupp G., ChemSusChem 2009, 2, 248–254. [DOI] [PubMed] [Google Scholar]
- 53. Liu C., Ding L., Guo G., Liu W., Yang Fl., Org. Biomol. Chem. 2016, 14, 2824–2827. [DOI] [PubMed] [Google Scholar]
- 54. Xia Y., Hu F., Xia Y., Liu Z., Ye F., Zhang Y., Wang J., Synthesis 2017, 49, 1073–1086. [Google Scholar]
- 55. Wasylenko W. A., Kebede N., Showalter B. M., Matsunaga N., Miceli A. P., Liu Y., Ryzhkov L. R., Hadad C. M., Toscano J. P., J. Am. Chem. Soc. 2006, 128, 13142–13150. [DOI] [PubMed] [Google Scholar]
- 56. Li Y., Yang Z., Zhou M., He J., Wand X., Wu Y., Wang Z., J. Mol. Struct. 2017, 1130, 818–828. [Google Scholar]
- 57. Schöpel M., Herrmann C., Scherkenbeck J., Stoll R., FEBS Lett. 2016, 590, 369–375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.