Abstract
Fully substituted peptide/[60]fullerene hexakis‐adducts offer an excellent opportunity for multivalent protein recognition. In contrast to monofunctionalized fullerene hybrids, peptide/[60]fullerene hexakis‐adducts display multiple copies of a peptide in close spatial proximity and in the three dimensions of space. High affinity peptide binders for almost any target can be currently identified by in vitro evolution techniques, often providing synthetically simpler alternatives to natural ligands. However, despite the potential of peptide/[60]fullerene hexakis‐adducts, these promising conjugates have not been reported to date. Here we present a synthetic strategy for the construction of 3D multivalent hybrids that are able to bind with high affinity the E‐selectin. The here synthesized fully substituted peptide/[60]fullerene hybrids and their multivalent recognition of natural receptors constitute a proof of principle for their future application as functional biocompatible materials.
Keywords: Fullerenes, Glycomimetic, Lectin, Multivalency, Peptides
Multivalent ligand presentation is a powerful strategy for the development of specific binders and inhibitors. Peptide/[60]fullerene hybrids have now been synthesized that exploit the complete substitution of the fullerene scaffold to afford globular structures presenting twelve copies of a peptide ligand for the recognition of E‐selectin.

Introduction
The artificially engineered multivalent presentation of biomolecular ligands offers an excellent opportunity to improve both the binding affinities and the distribution and stability of protein binders and inhibitors.[ 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 ] Among the different available platforms, the hexakis‐adduct of [60]fullerene has shown great potential as a bio‐compatible globular nanocarbon scaffold for three dimensional (3D) multivalent presentation of glycan ligands.[ 15 , 16 , 17 ] Protein‐glycan recognition events are behind critical biological processes including viral infection, cellular communication and metastatic development.[ 18 , 19 ] Instead of highly selective enthalpic interactions, the specificity in this flow of chemical information emerges from entropic recognition events that employ promiscuous multivalent binders. To achieve the required selectivity, Nature often capitalizes on chiral recognition by stereoselective multivalent glycan oligomers. [20] One example is the interaction between sialylated oligosaccharides with proteins of the family of the selectins, which has been shown to be implicated in different key recognition mechanisms such as immune cell extravasation or tumor cell metastasis.[ 18 , 21 ] However, the chemical synthesis of these saccharide ligands [22] can be challenging and could delay the development of synthetic surrogates. [23]
Monodisperse nanometric‐sized platforms are emerging as hybrid systems for protein selective recognition.[ 24 , 25 ] [60]Fullerene functionalization with Th octahedral geometry can be efficiently achieved through direct Bingel‐Hirsch cyclopropanation reaction by addition of suitably substituted malonates. [26] This procedure allows the preparation of a [60]fullerene hexakis‐adduct appended with 12 alkyne moieties, whose further regioselective modification can be efficiently carried out by copper‐catalyzed alkyne‐azide cycloaddition (CuAAC). [27]
Furthermore, the introduction of a copper‐free strain‐promoted alkyne‐azide cycloaddition (SPAAC)[ 28 , 29 , 30 ] strategy overcomes the copper‐related limitations for biological applications. [31] A recent modified version of SPAAC, under mild microwave irradiation (MW) conditions (50 °C), allows the very efficient fullerene functionalization in short times (30 min) to afford highly substituted biocompatible hybrid systems (e.g., glycofullerenes). [32] Stepwise synthesis of [5 : 1]‐hexa‐adducts of [60]fullerene and further development in orthogonal click additions have allowed the asymmetric substitution of A10B [60]fullerenes to combine multivalent ligands together with probes or therapeutics agents.[ 33 , 34 ] Over the last years, this multistep synthesis of functionalized fullerenes has shown great potential for blocking the carbohydrate recognition domain of the receptor for different emerging viruses such as Ebola,[ 35 , 36 ] Zika, [33] and Dengue. [33]
Recent advances in the screening and in vitro optimization of potential selective binders based on nucleic acids, peptides, and antibody or antibody mimetics [37] constitute a highly promising new scenario for the fullerene multivalent presentation of biomolecular scaffolds. For example, the use of the systematic evolution of ligands by exponential enrichment technology (SELEX) [38] for nucleic acids, or display platforms (phage display, yeast display, ribosome display and so on)[ 37 , 39 , 40 , 41 ] for peptides and proteins currently allow the directed evolution of high affinity binders for almost any conceivable target. In this regard, the possible coupling of the fullerene scaffold with peptide binders, developed by phage display optimization (Nobel prize 2018),[ 40 , 42 ] will offer a unique opportunity for the design of universal nanoplatforms for selective protein receptor recognition. However, despite the strong potential of this strategy,[ 43 , 44 ] the multistep, complete functionalization of the fullerene scaffold by defined peptide oligomers has not yet been experimentally achieved.
In this paper, we describe the first synthesis of a fully substituted peptide/[60]fullerene hexakis‐adduct by combining solid phase peptide synthesis and fullerene click chemistry. In contrast to previously reported fullerenes monofunctionalized or randomly derivatized with peptides or protein pendants,[ 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 ] this work constitutes the first example of a fully substituted fullerene/peptide hybrid hexakis‐adduct for multivalent protein recognition. The results reported here confirm the potential of the [60]fullerene for the multivalent presentation of robust and easily accessible peptide ligands as a viable alternative to natural ligands (Figure 1). The here described synthetic pathway and optimized reaction conditions, based on hexakis‐adducts of [60]fullerene appended with cyclooctynes for SPAAC, allowed the obtention of well‐defined monodisperse fullerene/peptide hybrids in quantitative yield by using mild conditions, and avoiding additional purification steps.
Figure 1.

Different steps in the molecular scaffold construction: a) In vitro identification of a mimetic (IELLQAR) of a natural ligand (sLeX ); b) Development of a highly substituted peptide/[60]fullerene hybrid; c) Multivalent and selective recognition of E‐selectin (dark‐teal receptors) by the fullerene hybrids, with the potential application of blocking the attachment of sLeX ‐overexpressing circulating tumor cells (gray cells) to the activated endothelium (pink cells) with the subsequent inhibition of metastasis.
A selective peptide binder, previously identified by phage display to target the lectin binding domain of the E‐selectin, [54] was employed to achieve the experimental proof of principle of fullerene‐mediated peptide multivalent presentation and selective protein recognition. E‐selectin is a well known receptor with multivalent ligands that is involved in cancer cell trafficking and that can be potentially used as a target for diagnosis and treatment.[ 18 , 21 , 55 ] Therefore, it constitutes an optimal receptor for this study. The [60]fullerene hexakis‐adduct was equipped with twelve peptide subunits that were specifically recognized by the protein receptor (Figure 1). The surface plasmon resonance (SPR) binding characterization confirmed the importance of the peptide to fullerene linker orientation for the suitable presentation of chemical information on the globular carbon template. Asymmetric A10B [60]fullerenes fluorescently labelled at the focal point with a Rhodamine probe were also synthesized and employed to study the specific recognition of the E‐selectin surface receptors in the membrane of endothelial cells.
Results and Discussion
Design, synthesis and SPR studies of peptide ligands
Selectin adhesion receptors were chosen as excellent candidates for peptide/[60]fullerene multivalent specific recognition. Although the sialylated oligosaccharides sialyl Lewis X (sLeX ) and its isomer sialyl Lewis A (sLeA ) are the natural ligands of selectins, their weak binding constant (in the low mM range) [54] can be improved by their protein or lipid‐supported multivalent presentation.[ 14 , 54 , 56 ]
Nevertheless, the overexpression of the E‐selectin receptors at the surface of cytokine activated vascular endothelium, [57] a potential receptor site for circulating tumor cells, makes the synthesis of such inhibitors a primary target, which has been mainly approached by antibodies, thio‐aptamers, and non‐natural saccharides. [55] Phage display in vitro directed evolution has allowed the identification of a simple linear peptide sequence (IELLQAR) that interacts specifically with selectins, with an order of magnitude higher affinity than sLeX , and that can be employed to inhibit sLeX ‐dependent metastasis.[ 54 , 58 ]
Therefore, we hypothesized that the quantitative coupling of this peptide to the hexakis‐adduct of the fullerene core would afford a biocompatible globular scaffold for the 3D presentation of multiple peptide copies for protein recognition (Figure 2). It should be noted that this peptide was simply selected as an excellent example to demonstrate the potential functional capacities of fully substituted peptide fullerene adducts.
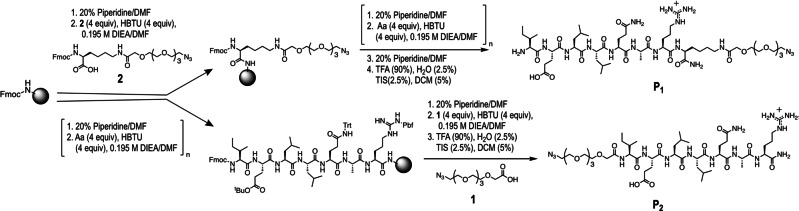
Figure 2.
Synthetic analysis of these peptide/[60]fullerene platforms. SPAAC reaction between peptides modified with an azide group: the IELLQARK (active peptide) and ISELRSE (negative control peptide) and the cyclooctyne groups of C60 fullerene hexakis‐adducts to give rise to the symmetric hybrids: the active F1 (C60‐(IELLQARK)12) and the control FC (C60‐(ISELRSE)12), as well as the asymmetric Rhodamine B fullerene adducts: the active F1 RB (RB‐C60‐(IELLQARK)10) and control FC RB (RB‐C60‐(ISELRSE)10).
To synthesize IELLQAR peptide/[60]fullerene adducts, the peptide must be conjugated to the multivalent platform. To determine the influence of the linker in the binding affinity of the IELLQAR peptide to the E‐selectin, we considered a head or tail peptide connection by anchoring to the fullerene either: i) the free amino group of the isoleucine residue located at the peptide N‐terminus, or ii) an amino group located on an additional new lysine residue introduced at the peptide C‐terminus (Figure 3). The linker connector was based on a short hydrophilic tetraethyleneglycol moiety equipped with two orthogonal functionalization sites: a carboxylic acid and a terminal azido group (1). This compound allowed both the conjugation to the peptide by amide bond formation and the subsequent attachment to the [60]fullerene cyclooctyne adduct via SPAAC click reaction. Two different peptide sequences were firstly synthesized by Fmoc solid‐phase, namely: IELLQARK‐(PEG)4‐N3 (P1 ) and N3‐(PEG)4‐IELLQAR (P2 ) (Figures 3, S6 and S7). For the synthesis of the N‐terminus functionalized P2 , the polyethylene glycol linker (1) was conjugated in the last step of the solid phase synthesis strategy (Figures 3 and S7). To prepare the C‐terminus functionalized peptide P1 , it was necessary to use a new lysine amino acid, previously modified with the corresponding oligoethylene glycol linker (2). This customized lysine (2) was anchored to the resin as first amino acid derivative during the solid phase synthesis of P1 (Figures 3 and S6). After HPLC purification, the final oligoethyleneglycol modified peptides (P1 and P2 ) showed an excellent purity and water solubility.
Figure 3.
Peptide IELLQARK‐(PEG)4‐N3 (P1 ) was prepared by adding a new lysine amino acid previously modified with the corresponding oligoethylene glycol linker (2) at the C‐terminus in the beginning of the synthesis. Peptide N3‐(PEG)4‐IELLQAR (P2 ) was prepared by incorporating the polyethylene glycol linker (1) at the N‐terminus at the end of the solid phase synthesis (see section 3 in the Supporting Information).
To determine which one of the two different anchoring sites of the IELLQAR peptide (N‐ or C‐terminal linkers) would afford the optimal presentation geometry, we sought to initially study the interaction of these monovalent peptides (P1 and P2 ) with the E‐selectin protein by surface plasmon resonance (SPR) direct binding experiments (see Supporting Information, section 6).
Binding data were collected at a flow rate of 30 μL min−1 at increasing concentrations of the peptides P1 , and P2 , and the sLeX pentasaccharide natural ligand (Figure S40). Prior to analysis, binding tests with E‐selectin at different flow rates were performed to discard mass transport effects that could influence the sensorgrams shape. The SPR sensorgrams obtained were fit to a Langmuir 1 : 1 binding mode, and the kinetic and binding constants (k on (k a), k off (k d)) were calculated by dose‐response experiments on the E‐selectin surface (Figure S40). Kinetic evaluation of the resulting curves was performed, and a dissociation constant (K D) value was obtained for the two peptides P1 , P2 , and sLeX (Table 1).
Table 1.
Kinetic constants determined by SPR.
|
|
k a [1/M s][a] |
k d [1/s][a] |
K D=k d/k a [M][a] |
|---|---|---|---|
|
sLeX |
5.34×106 |
9.33×102 |
1.75×10−4 |
|
P1 |
44.8±2.14 |
2.31×10−3±4.91×10−4 |
5.16×10−5 ±1.22×10−6 |
|
P2 |
26.4±2.45 |
7.79×10−2±3.81×10−3 |
2.95×10−3±1.97×10−4 |
|
F1 |
1.01×104±1.87×103 |
5.85×10−4±1.84×10−4 |
6.04×10−8±2.65×10−8 |
|
F1 RB |
1.37×104±6.28×103 |
3.34×10−3±1.89×10−3 |
2.54×10−7±1.06×10−7 |
[a] The kinetic constants obtained correspond to the average of the kinetic constants for each of the concentrations for sLeX , P1 , P2 , F1 and F1 RB .
The C‐terminus modified peptide P1 showed the best binding affinity to the E‐selectin receptor with a K D of 5.16×10−5 M, which was almost one order of magnitude lower than that of the natural ligand sLeX (K D=1.75×10−4 M). In contrast, the N‐terminus modified peptide P2 showed a significant drop in the affinity for the E‐selectin receptor (K D=2.95×10−3 M), which confirmed that the attachment orientation of peptide to the fullerene scaffold can significantly interfere with the peptide binding to the E‐selectin. Therefore, these results remark that for hexakis‐adducts of [60]fullerene with peptides, the linkers should be independently optimized for each particular peptide, and also confirmed the C‐terminus functionalized peptide P1 as the best candidate for further development of this peptide/[60]fullerene hybrid system.
Synthesis of Chemically Modified [60]Fullerenes
The synthetic route to obtain symmetric hexakis‐adduct F1 decorated with twelve peptides P1 is depicted in Figure 4. The previous phage display optimization indicated that similar peptides to that of the hit peptide sequence (IELLQAR) could retain certain binding activity. [54] Therefore, a negative control peptide, PC , was designed with a random hydrophilic sequence to prevent it from affecting the solubility of the macromolecules and from interacting with the E‐selectin. Thus, the N3‐(PEG)4‐ISELRSE sequence was also synthesized and coupled to the fullerene in the same conditions, to prepare a non‐binding control FC fullerene (Figures 4 and S14). As starting material we used the cyclooctyne [60]fullerene derivative 3 [32] in order to use the SPAAC click reaction. Thus, a DMSO solution of the cyclooctyne building block 3 and the corresponding peptide functionalized with an azide (1 equiv per cyclooctyne unit) was heated under MW irradiation at 50 °C for 30 min, as we previously reported for the synthesis of nanoballs. [33] The purification of the multivalent systems was performed by precipitation/centrifugation processes with a mixture of CHCl3/hexane as solvent. In order to obtain the corresponding fluorescent labelled peptide/[60]fullerene hybrids for cellular studies while maintaining the copper‐free strategy, an asymmetric A10B [60]fullerene derivative bearing 10 cyclooctynes and a Rhodamine B moiety at the focal point was prepared according to the procedure represented in Figure 5. Starting from asymmetric hexakis‐adduct 5, [33] we carried out the replacement of the chlorine atom by an azido group followed by SPAAC reaction with Rhodamine B derivative 8 under MW irradiation, which afforded the asymmetric compound 9 in excellent yield. Compound 8 was efficiently obtained by esterification of Rhodamine B and alcohol 7. NMR characterization experiments confirmed the attachment of one single fluorescent Rhodamine per fullerene scaffold (Figure S26). Esterification of derivative 9 with carboxylic acid 10, [32] yielded compound 11, which was employed as building block to react with peptides P1 and PC , leading to fluorescent labelled peptide/[60]fullerenes F1 RB and FC RB , respectively, in quantitative yield. Purification of the obtained derivatives F1 RB and FC RB was carried out following the previous methodology as described for the peptide/[60]fullerenes F1 and FC .
Figure 4.
Synthesis of peptide/[60]fullerenes F1 and FC via SPAAC reaction between hexakis‐adduct 3 and peptide azides P1 and PC , respectively. Reagents and conditions: 3 (1 equiv), P1 or PC (12 equiv), DMSO, MW 50 °C, 30 min (for F1 from P1 and for FC from PC , quant.).
Figure 5.
Synthesis of labelled peptide/[60]fullerenes F1 RB and FC RB . Reagents and conditions: i) NaN3, DMF, 60 °C, 3 days (quant.); ii) EDC⋅HCl, DMAP, CH2Cl2/DMF, r.t., overnight (quant.); iii) 6 (1 equiv), 8 (2 equiv), DMSO, MW 50 °C, 30 min (99 %); iv) 9 (1 equiv), 10 (15 equiv), DCC, DPTS, CH2Cl2/DMF, r.t., overnight (99 %); v) 11 (1 equiv), P1 or PC (10 equiv), DMSO, MW 50 °C, 30 min (for F1 RB from P1 , 99 %, and for FC RB from PC , quant.).
Characterization of the four final compounds was performed by FTIR, NMR, and XPS. Due to the lack of regioselectivity in the thermal cycloaddition to asymmetric cyclooctynes, a mixture of both regioisomers is obtained in all cases, giving rise to duplication of some signals in the NMR spectra. In particular, 13C NMR characterization of fullerene derivatives is useful for demonstrating the full functionalization of the cyclooctyne residues as well as the octahedral symmetry of the structure. The absence of the typical signals corresponding to the Csp of the cyclooctyne (at 100.1 ppm and 92.8 ppm) (e.g., compare Figure S30 with S34 and S37) together with the lack of the signal of the CH2 bound to the azido group at ≈50 ppm indicated that the cycloaddition and the subsequent purification steps have been adequately carried out, respectively. Also, four different signals are observed for the two quaternary carbons of the triazole ring fused to the cyclooctyne moieties corresponding to the two formed regioisomers at δ ≈134 and 144 ppm for the major isomer and δ ≈134 and 143 ppm for the minor isomer. Also, only two signals appear for the sp2 carbons of the C60 cage at δ ≈141 and 145, together with the signal for the two sp3 carbons (at ≈69 ppm) of the C60 core, thus providing evidence for the high Th symmetry of the compounds (Figures S16, S19, S34 and S37).
While MS spectra of all intermediate hexakis‐adducts showed the expected molecular ion peaks, those for final compounds F1 , FC , F1 RB and FC RB were difficult to obtain, and the molecular ion peak could not be observed due to the very high molecular mass of the compounds and high level of fragmentation. Therefore, XPS analysis was additionally employed to confirm the elemental composition of the final peptide/[60]fullerenes (see section 4 in the Supporting Information). The registered full scanned spectra give us information about the atoms present in their molecular structure. As expected, contributions steaming from the core levels of C 1s (284.6 eV), N 1s (398.6 eV), and O 1s (531.6 eV), were the only elements present in the analysis. Their relative abundances are collected in Table 2 and are in agreement with the theoretical calculated values for each compound. The S 2p doublet (S 2p1/2 and S 2p3/2) for the labelled peptide fullerenes F1 RB and FC RB feature is missing in the survey spectra due to the low resolution of XPS for this atom and its low atomic presence in the molecule. Moreover, the chemical state of C 1s and N 1s has been analysed in a high‐resolution spectrum. The C 1s was deconvoluted into three symmetrical (Gaussian‐Lorentzian curves) major components corresponding to C=C/C−C at 283.9 eV, C−O/C−N at 285.2 eV and N−C=O/C=O/O−C=O/C=N at 287.0 eV, presenting similar behaviour in all compounds (Figures S17b, S20b, S35b, and S38b). Analysing the N 1s peak decomposition, we observed three main components, as depicted in Figures S17c, S20c, S35c, and S38c. One major peak was observed at around 399.0 eV, assigned to the amide groups (N−C=O) characteristic of the peptide chain. The C−N peak was found at lower binding energies (398.3 eV) and is assigned, among others, to the two N atoms bound to C of the triazole ring. Finally, the third and minor component (≈7 %) with binding energy of near 400 eV belong to the third nitrogen of the triazole ring N−N−N. Note that, in all the analysed samples, no azido group peak (≈405.0 eV) is observable, confirming the lack of unbound organic molecule and the quantitative formation of the triazole ring.
Table 2.
Relative abundances of C, N and O determined by XPS for peptide/[60]fullerenes F1 , FC , F1 RB and FC RB .
|
|
Atomic percentage [%][a] |
Molecular formula |
||
|---|---|---|---|---|
|
|
C |
N |
O |
|
|
F1 |
65.8 (66.0) |
13.7 (14.1) |
20.5 (19.9) |
C954H1536N204O288 |
|
FC |
64.3 (63.8) |
12.1 (12.7) |
23.5 (23.5) |
C846H1308N168O312 |
|
F1 RB |
67.2 (66.8) |
13.1 (13.5) |
19.8 (19.7) |
C869H1365N175O256S |
|
FC RB |
64.2 (64.9) |
11.8 (12.1) |
23.9 (23.0) |
C779H1175N145O276S |
[a] Theoretical values are given in brackets.
Analysis of F1 in solution by dynamic light scattering (DLS) showed two size populations (≈8 nm and 200 nm) depending on the aqueous buffer employed (Figure S39a). Scanning transmission electron microscopy (STEM) and atomic force microscopy (AFM) also revealed size distributions (8, 25 nm) consistent with the size of individual and small assemblies of the peptide fullerene hybrid macromolecules (Figure S39b and S39c).
Surface Plasmon Resonance Binding Assays
SPR biosensor direct binding assays were performed to compare the binding of the different peptide/[60]fullerene conjugates to E‐selectin. A high capacity sensorchip was functionalized with E‐selectin following the same procedure as previously described, and binding data were collected at a flow rate of 30 μL min−1 at increasing fullerene concentrations (see Supporting Information, section 6). As expected, the more complex binding profiles of the sensorgrams suggested a multivalent nature for the binding of the peptide/[60]fullerene hybrids to the chip supported protein (Figure 6 and Figure S41). As previously described in the literature,[ 59 , 60 ] a separate kinetic analysis of guest multivalent binding was carried out at diluted concentrations for the initial association and late dissociation binding events. The sensorgrams of the binding peptide/[60]fullerene conjugates (F1 and F1 RB ) showed a two‐phase dissociation process (t>240 s), which consisted in an initial fast dissociation phase followed by an extremely slow dissociation phase. A higher degree of heterogeneity in the binding events was also found at early dissociations times (t=240–360 s, Figure 6), but late dissociation times (t>600 s) showed a good fitting to a 1 : 1 Langmuir binding model. Kinetic evaluation of the early binding region (t<50 s) showed a linear dependence during the association phase, which could also be fitted to a 1 : 1 Langmuir binding model (Figure S41). The kinetic profile of the sensorgrams at short times showed a fast association at low concentrations (<12.5 μM) that was reduced at higher ligand concentrations, which was consistent with a multivalent binding of the peptide/[60]fullerene hybrid system.
Figure 6.

SPR sensorgrams of hybrid fullerenes a) F1 , and b) F1 RB at (12.5–0.78) μM binding to E‐selectin functionalized surfaces (representative graphics for low density (LD) functionalization, see Supporting Information). The association times below 50 s (yellow lines) and dissociation times longer than 600 s (red lines) used for constant determination (Table 1, Supporting Information) are shown.
The analysis of the SPR curves showed that the conjugate F1 , with twelve peptide units at the fullerene surface, binds E‐selectin with a dissociation constant in the nanomolar range (K D=6.04×10−8 M), indicating a multivalent effect resulting in an affinity enhancement (β) factor of 71 (see section 6 in the Supporting Information). [61] The affinity of this binding peptide/[60]fullerene conjugate F1 increases up to 3 orders of magnitude relative to its monomeric counterpart, which validates the functional capacity of the [60]fullerene hexakis‐adduct to display multiple copies of a peptide ligand and establish multivalent interactions with the corresponding suitable protein receptor (Figure 6). Despite multivalent interactions were also observed for the fluorescent asymmetric fullerene, a decrease in the binding constant (K D=2.54×10−7 M) was obtained for F1 RB , where two peptide copies have been substituted by a rhodamine fluorescence probe (Figures 5, 6 and S41). As expected, control experiments with the non‐specific hydrophilic peptide modified fullerenes FC and FC RB showed no apparent binding to the E‐selectin receptor (Figure S42).
Cellular Studies
After the synthesis and the SPR binding studies to the E‐selectin, in vitro cell studies were carried out to confirm the potential capacity of the new peptide/[60]fullerene conjugates F1 RB and FC RB to recognize surface receptors in living cells. The human umbilical vein endothelial cell line (HUVEC) was selected as a model for human endothelial cells expressing the E‐selectin. [62] This cell line can be activated by tumor necrosis factor alpha (TNFα) that induces the overexpression of membrane adhesion molecules such as the E‐selectin (Figure S43). As confirmed by SPR, the specific E‐selectin binding of the fluorescent adduct F1 RB was in a suitable concentration range for potential in vitro cell binding assays. Therefore, the potential selective cellular adhesion of the fluorescent targetable adduct F1 RB versus the negative control fullerene FC RB were both tested with activated and non‐activated HUVEC cells (Figure 7).
Figure 7.

Cellular assay. a) Confocal microscopy images of HUVEC cells (top row) or TNFα‐treated HUVEC cells (bottom row) incubated with 10 μM of F1 RB (left column) or FC RB (right column). Scale bar: 25 μm. b) Flow cytometry assay of HUVEC cells incubated with 10 μM of F1 RB or FC RB , pretreated (gray checkered bars) or not (smooth gray bars) with 10 ng mL−1 of TNFα. Data presented as mean of the median fluorescence intensity of three replicates±standard deviation.
Two sets of HUVEC cells were cultured in parallel, in the presence or in the absence of TNFα, to induce E‐selectin overexpression in one of the groups. Both groups of HUVECs were then incubated with the fluorescently labelled binding F1 RB and the control FC RB peptide/[60]fullerene conjugates for 30 min (see Supporting Information, section 7). The cells were thoroughly washed with heparin and HKR buffer to remove any potential excess of fullerene adducts and were then observed by live‐cell confocal microscopy (Figure 7). The resulting micrographs confirmed an excellent selectivity of the interaction of the F1 RB conjugate towards the TNFα treated HUVEC cells (Figure 7). Residual fluorescence was observed in non‐treated HUVEC cells that did not express the E‐selectin receptor. The corresponding control experiments with the control FC RB fullerene conjugate showed residual fluorescence in both untreated and TNFα treated HUVEC cells, which confirmed the selectivity of the cellular binding process. Flow cytometry quantification validated the selectivity of the peptide/[60]fullerene adhesion (Figure 7).
Conclusion
The objective of this study was to synthesize and experimentally demonstrate the potential of a fully substituted peptide/[60]fullerene conjugate for the 3D multivalent presentation of the chemical information of peptide oligomers for protein recognition. This proof of principle is secured by a detailed collection of spectroscopic characterization of the conjugates and the corresponding E‐selectin binding experiments in model surfaces and living cells. In this work, solid phase peptide synthesis and a copper‐free SPAAC click chemistry approach were combined for the efficient preparation of highly substituted peptide/[60]fullerene conjugates. The globular structure of the resulting [60]fullerene hexakis‐adducts allowed the maximum spacing between the twelve IELLQAR copies in a minimal multivalent nanoplatform for protein recognition. Surface plasmon resonance unambiguously confirmed the multivalent specific E‐selectin binding, which was also validated in living cells models. These results support the potential of fullerene conjugates for the multivalent presentation of peptide ligands with specific information for protein recognition.
The here described peptide/[60]fullerene conjugates constitute an original new strategy for multivalent recognition that for the first time exploits the synthetic simplicity, the stability and the versatility of peptide binders supported on fully substituted fullerene templates. We hope that this report will pave the way for the synthesis of a new range of promising fully substituted peptide/fullerene hybrid materials with potential applications in protein targeting and biomedical sciences.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This work was partially supported by the Spanish Agencia Estatal de Investigación (AEI) [SAF2017‐89890‐R, PCI2019‐103400, PID2020‐117143RB‐I00, PID2020‐114653RB‐I00 and PID2020‐115120GB‐I00], Xunta de Galicia (ED431C 2017/25 and Centro singular de investigación de Galicia accreditation 2019–2022, ED431G 2019/03) and the European Commission (EC) (European Regional Development Fund‐ERDF). J.M. thanks the ERC‐STG (DYNAP, 677786), ERC‐POC (TraffikGene, 838002), Xunta de Galicia (Oportunius Program) and Human Frontier Science Programme Young Investigator Grant (RGY0066/2017) for funding. J.J.R. received a Beatriz Galindo Grant (BEAGAL18‐00051) by the Spanish Ministerio de Universidades. I.G. received predoctoral fellowships (ED481A‐2018/116 and FPU17/00941). J.C.‐G. thanks the Comunidad de Madrid Atracción de Talento program (2018‐T2/BMD‐10275). Figures created with BioRender.com.
Dedicated to Professor Luis Echegoyen on occasion of his 70th birthday
I. Gallego, J. Ramos-Soriano, A. Méndez-Ardoy, J. Cabrera-González, I. Lostalé-Seijo, B. M. Illescas, J. J. Reina, N. Martín, J. Montenegro, Angew. Chem. Int. Ed. 2022, 61, e202210043; Angew. Chem. 2022, 134, e202210043.
Contributor Information
Dr. Beatriz M. Illescas, Email: beti@ucm.es.
Dr. Jose J. Reina, Email: josejuan.reina@uma.es.
Prof. Dr. Nazario Martín, Email: nazmar@ucm.es.
Dr. Javier Montenegro, Email: javier.montenegro@usc.es.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Lauster D., Klenk S., Ludwig K., Nojoumi S., Behren S., Adam L., Stadtmüller M., Saenger S., Zimmler S., Hönzke K., et al., Nat. Nanotechnol. 2020, 15, 373–379. [DOI] [PubMed] [Google Scholar]
- 2. Gallego I., Rioboo A., Reina J. J., Díaz B., Canales Á., Cañada F. J., Guerra-Varela J., Sánchez L., Montenegro J., ChemBioChem 2019, 20, 1400–1409. [DOI] [PubMed] [Google Scholar]
- 3. Juanes M., Lostalé-Seijo I., Granja J. R., Montenegro J., Chem. Eur. J. 2018, 24, 10689–10698. [DOI] [PubMed] [Google Scholar]
- 4. Kwon S. J., Na D. H., Kwak J. H., Douaisi M., Zhang F., Park E. J., Park J. H., Youn H., Song C. S., Kane R. S., et al., Nat. Nanotechnol. 2017, 12, 48–54. [DOI] [PubMed] [Google Scholar]
- 5. Martinez-Veracoechea F. J., Frenkel D., Proc. Natl. Acad. Sci. USA 2011, 108, 10963–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W. P., Schneidman-Duhovny D., Zhang C., Shi Y., Science 2020, 370, 1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Budhadev D., Poole E., Nehlmeier I., Liu Y., Hooper J., Kalverda E., Akshath U. S., Hondow N., Turnbull W. B., Pöhlmann S., et al., J. Am. Chem. Soc. 2020, 142, 18022–18034. [DOI] [PubMed] [Google Scholar]
- 8. Blattes E., Vercellone A., Eutamene H., Turrin C.-O., Theodorou V., Majoral J.-P., Caminade A.-M., Prandi J., Nigou J., Puzo G., Proc. Natl. Acad. Sci. USA 2013, 110, 8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Richards S.-J., Gibson M. I., JACS Au 2021, 1, 2089–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zelikin A. N., Stellacci F., Adv. Healthcare Mater. 2021, 10, 2001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Insua I., Montenegro J., Chem 2020, 6, 1652–1682. [Google Scholar]
- 12. Kanfar N., Bartolami E., Zelli R., Marra A., Winum J.-Y., Ulrich S., Dumy P., Org. Biomol. Chem. 2015, 13, 9894–9906. [DOI] [PubMed] [Google Scholar]
- 13. Perets E. A., Indrasekara A. S. D. S., Kurmis A., Atlasevich N., Fabris L., Arslanoglu J., Analyst 2015, 140, 5971–5980. [DOI] [PubMed] [Google Scholar]
- 14. Fasting C., Schalley C. A., Weber M., Seitz O., Hecht S., Koksch B., Dernedde J., Graf C., Knapp E.-W., Haag R., Angew. Chem. Int. Ed. 2012, 51, 10472–10498; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 10622–10650. [Google Scholar]
- 15. Illescas B. M., Rojo J., Delgado R., Martín N., J. Am. Chem. Soc. 2017, 139, 6018–6025. [DOI] [PubMed] [Google Scholar]
- 16. Nierengarten J.-F., Chem. Commun. 2017, 53, 11855–11868. [DOI] [PubMed] [Google Scholar]
- 17. Castro E., Garcia A. H., Zavala G., Echegoyen L., J. Mater. Chem. B 2017, 5, 6523–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith B. A. H., Bertozzi C. R., Nat. Rev. Drug Discovery 2021, 20, 217–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mendes A. C., Baran E. T., Reis R. L., Azevedo H. S., Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013, 5, 582–612. [DOI] [PubMed] [Google Scholar]
- 20. Laine R. A., Glycobiology 1994, 4, 759–767. [DOI] [PubMed] [Google Scholar]
- 21. Harjes U., Nat. Rev. Cancer 2019, 19, 301–301. [DOI] [PubMed] [Google Scholar]
- 22. Reina J. J., Rioboo A., Montenegro J., Synthesis 2018, 50, 831–845. [Google Scholar]
- 23. Ley K., Trends Mol. Med. 2003, 9, 263–268. [DOI] [PubMed] [Google Scholar]
- 24. Ramos-Soriano J., Rojo J., Chem. Commun. 2021, 57, 5111–5126. [DOI] [PubMed] [Google Scholar]
- 25. Jiménez Blanco J. L., Benito J. M., Ortiz Mellet C., García Fernández J. M., J. Drug Delivery Sci. Technol. 2017, 42, 18–37. [Google Scholar]
- 26. Hirsch A., in Fullerenes Relat. Struct. (Ed.: Hirsch A.), Springer, Berlin, Heidelberg, 1999, pp. 1–65. [Google Scholar]
- 27. Nierengarten I., Nierengarten J.-F., Chem. Rec. 2015, 15, 31–51. [DOI] [PubMed] [Google Scholar]
- 28. Agard N. J., Prescher J. A., Bertozzi C. R., J. Am. Chem. Soc. 2004, 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- 29. Agard N. J., Baskin J. M., Prescher J. A., Lo A., Bertozzi C. R., ACS Chem. Biol. 2006, 1, 644–648. [DOI] [PubMed] [Google Scholar]
- 30. Baskin J. M., Prescher J. A., Laughlin S. T., Agard N. J., Chang P. V., Miller I. A., Lo A., Codelli J. A., Bertozzi C. R., Proc. Natl. Acad. Sci. USA 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ornelas C., Broichhagen J., Weck M., J. Am. Chem. Soc. 2010, 132, 3923–3931. [DOI] [PubMed] [Google Scholar]
- 32. Ramos-Soriano J., Reina J. J., Pérez-Sánchez A., Illescas B. M., Rojo J., Martín N., Chem. Commun. 2016, 52, 10544–10546. [DOI] [PubMed] [Google Scholar]
- 33. Ramos-Soriano J., Reina J. J., Illescas B. M., de la Cruz N., Rodríguez-Pérez L., Lasala F., Rojo J., Delgado R., Martín N., J. Am. Chem. Soc. 2019, 141, 15403–15412. [DOI] [PubMed] [Google Scholar]
- 34. Ramos-Soriano J., Reina J. J., Illescas B. M., Rojo J., Martín N., J. Org. Chem. 2018, 83, 1727–1736. [DOI] [PubMed] [Google Scholar]
- 35. Rodríguez-Pérez L., Ramos-Soriano J., Pérez-Sánchez A., Illescas B. M., Muñoz A., Luczkowiak J., Lasala F., Rojo J., Delgado R., Martín N., J. Am. Chem. Soc. 2018, 140, 9891–9898. [DOI] [PubMed] [Google Scholar]
- 36. Muñoz A., Sigwalt D., Illescas B. M., Luczkowiak J., Rodríguez-Pérez L., Nierengarten I., Holler M., Remy J.-S., Buffet K., Vincent S. P., et al., Nat. Chem. 2016, 8, 50–57. [DOI] [PubMed] [Google Scholar]
- 37. Simeon R., Chen Z., Protein Cell 2018, 9, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Darmostuk M., Rimpelova S., Gbelcova H., Ruml T., Biotechnol. Adv. 2015, 33, 1141–1161. [DOI] [PubMed] [Google Scholar]
- 39. Ullman C. G., Frigotto L., Cooley R. N., Briefings Funct. Genomics Proteomics 2011, 10, 125–134. [DOI] [PubMed] [Google Scholar]
- 40. Smith G. P., Petrenko V. A., Chem. Rev. 1997, 97, 391–410. [DOI] [PubMed] [Google Scholar]
- 41. Sergeeva A., Kolonin M. G., Molldrem J. J., Pasqualini R., Arap W., Adv. Drug Delivery Rev. 2006, 58, 1622–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mimmi S., Maisano D., Quinto I., Iaccino E., Trends Pharmacol. Sci. 2019, 40, 87–91. [DOI] [PubMed] [Google Scholar]
- 43. Ruiz-Santaquiteria M., Illescas B. M., Abdelnabi R., Boonen A., Mills A., Martí-Marí O., Noppen S., Neyts J., Schols D., Gago F., et al., Chem. Eur. J. 2021, 27, 10700–10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li H., Zhang B., Lu X., Tan X., Jia F., Xiao Y., Cheng Z., Li Y., Silva D. O., Schrekker H. S., et al., Proc. Natl. Acad. Sci. USA 2018, 115, 4340–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Svitova A., Braun K., Popov A. A., Dunsch L., ChemistryOpen 2012, 1, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fillmore H. L., Shultz M. D., Henderson S. C., Cooper P., Broaddus W. C., Chen Z. J., Shu C.-Y., Zhang J., Ge J., Dorn H. C., et al., Nanomedicine 2011, 6, 449–458. [DOI] [PubMed] [Google Scholar]
- 47. Shi J., Wang B., Wang L., Lu T., Fu Y., Zhang H., Zhang Z., J. Controlled Release 2016, 235, 245–258. [DOI] [PubMed] [Google Scholar]
- 48. Tanzi L., Terreni M., Zhang Y., Eur. J. Med. Chem. 2022, 230, 114104. [DOI] [PubMed] [Google Scholar]
- 49. Bosi S., Da Ros T., Spalluto G., Prato M., Eur. J. Med. Chem. 2003, 38, 913–923. [DOI] [PubMed] [Google Scholar]
- 50. Rivera-Nazario D. M., Pinzón J. R., Stevenson S., Echegoyen L. A., J. Phys. Org. Chem. 2013, 26, 194–205. [Google Scholar]
- 51. Barron A. R., J. Enzyme Inhib. Med. Chem. 2016, 31, 164–176. [DOI] [PubMed] [Google Scholar]
- 52. Jennepalli S., Pyne S. G., Keller P. A., RSC Adv. 2014, 4, 46383–46398. [Google Scholar]
- 53. Pantarotto D., Bianco A., Pellarini F., Tossi A., Giangaspero A., Zelezetsky I., Briand J.-P., Prato M., J. Am. Chem. Soc. 2002, 124, 12543–12549. [DOI] [PubMed] [Google Scholar]
- 54. Fukuda M. N., Ohyama C., Lowitz K., Matsuo O., Pasqualini R., Ruoslahti E., Fukuda M., Cancer Res. 2000, 60, 450–456. [PubMed] [Google Scholar]
- 55. Jubeli E., Moine L., Vergnaud-Gauduchon J., Barratt G., J. Controlled Release 2012, 158, 194–206. [DOI] [PubMed] [Google Scholar]
- 56. Gout S., Tremblay P.-L., Huot J., Clin. Exp. Metastasis 2008, 25, 335–344. [DOI] [PubMed] [Google Scholar]
- 57. Bevilacqua M., Stengelin S., Gimbrone M., Seed B., Science 1989, 243, 1160–1165. [DOI] [PubMed] [Google Scholar]
- 58. Zhang J., Nakayama J., Ohyama C., Suzuki M., Suzuki A., Fukuda M., Fukuda M. N., Cancer Res. 2002, 62, 4194–4198. [PubMed] [Google Scholar]
- 59. Munoz E. M., Correa J., Riguera R., Fernandez-Megia E., J. Am. Chem. Soc. 2013, 135, 5966–5969. [DOI] [PubMed] [Google Scholar]
- 60. Salvadó M., Reina J. J., Rojo J., Castillón S., Boutureira O., Chem. Eur. J. 2017, 23, 15790–15794. [DOI] [PubMed] [Google Scholar]
- 61. Krishnamurthy V. M., Estroff L. A., Whitesides G. M., Fragment-based Approaches in Drug Discovery, Wiley-VCH, Weinhiem, 2006, pp. 11–53. [Google Scholar]
- 62. Thornhill M. H., Li J., Haskard D. O., Scand. J. Immunol. 1993, 38, 279–286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.