Abstract
Genes not only control traits of their carrier organism (known as direct genetic effects or DGEs) but also shape their carrier's physical environment and the phenotypes of their carrier's social partners (known as indirect genetic effects or IGEs). Theoretical research has shown that the effects that genes exert on social partners can have profound consequences, potentially altering heritability and the direction of trait evolution. Complementary empirical research has shown that in various contexts (particularly in animal agriculture) IGEs can explain a large proportion of variation in specific traits. However, little is known about the general prevalence of IGEs. We conducted a reciprocal cross‐fostering experiment with two genetic lineages of the clonal raider ant Ooceraea biroi to quantify the relative contribution of DGEs and IGEs to variation in brain gene expression (which underlies behavioural variation). We found that thousands of genes are differentially expressed by DGEs but not a single gene is differentially expressed by IGEs. This is surprising given the highly social context of ant colonies and given that individual behaviour varies according to the genotypic composition of the social environment in O. biroi. Overall, these findings indicate that we have a lot to learn about how the magnitude of IGEs varies across species and contexts.
Keywords: ants, behavior/social evolution, indirect genetic effects, transcriptomics
1. INTRODUCTION
When individuals interact, the phenotype of one individual depends not only on its own genotype, but also on the genotypes of its social partners. Such between‐individual genotype‐phenotype interactions are known as indirect genetic effects (IGEs) (Griffing, 1967). The presence of IGEs implies that the genotype‐phenotype relationship is not one‐to‐one, as assumed in standard evolutionary models, but many‐to‐many, and the evolutionary consequences of this are profound. Theoretical work has shown that consideration of IGEs can substantially increase estimates of heritability and can alter the rate and direction of evolution (Bijma et al., 2007; Bijma & Wade, 2008; Griffing, 1967; Moore et al., 1997). The importance of IGEs has also been demonstrated empirically in studies on livestock, where these effects have been harnessed to improve breeding programmes. IGEs account for 42% of feed intake and 27% of growth rate in domestic pigs (Bergsma et al., 2008), and an IGE‐based breeding programme in White Leghorns reduced annual mortality from 68% to 9% over four generations while increasing individual productivity (Muir, 1996). Furthermore, IGEs are of potential medical relevance. They account for 29% of variation in health‐related traits in mice, and influence human dietary traits and mental health (Baud et al., 2017; Xia et al., 2020).
Indirect genetic effects are likely to be particularly relevant to behavioural phenotypes (and their underlying gene expression profiles) because behaviour is inherently responsive to social context. Several studies using social model species have identified IGEs on behaviour‐related gene expression. Hundreds of genes were found to be differentially expressed in the brains of honeybees according to whether the social environment comprised bees of a “high hygienic” or a “low hygienic” genotype (Gempe et al., 2012). IGEs have also been found to strongly affect gene expression in the socially polymorphic system of the red imported fire ant, where some colonies have only one queen while others have multiple queens. Variation in queen number is controlled by a supergene, and workers either cull or tolerate supernumerary queens according to the frequencies of different supergene haplotypes in the colony (Ross & Keller, 2002). Whole‐body worker gene expression profiles were found to associate more strongly with the genotypic composition of the colony than with the genotype of the individual at the supergene (Wang et al., 2008).
While the important contribution of IGEs to behaviourally relevant gene expression has been shown in several specific contexts, little is known about the prevalence of these effects across different species or social contexts. Here, we quantify how much of variation in brain gene expression can be explained by IGEs from adult nestmates versus direct genetic effects (DGEs) in the clonal raider ant Ooceraea biroi. We use O. biroi as a model because of its unusual reproductive system. O. biroi has lost its queen caste, and instead, unmated workers reproduce via parthenogenesis. Consequently, the species comprises multiple distinct genetic lineages with lineage‐specific behavioural patterns (e.g., in degree of task specialization and level of extranidal activity (Kronauer et al., 2012; Oxley et al., 2014; Trible et al., 2020; Ulrich et al., 2021)). Using two of these lineages (“B” and “M” (Trible et al., 2020)) we performed a reciprocal cross‐fostering experiment, transplanting newly‐eclosed larvae so that the fostered individuals experienced their adoptive social environment during development and as adults. We performed RNA‐seq on individual brains of fostered individuals (variation in brain gene expression is most relevant to behavioural variation) and show that thousands of genes are differentially expressed according to the genotype of the focal individual. Surprisingly, however, not a single gene was differentially expressed according to the genotype of the individuals constituting the social environment. This result suggests that the magnitude of IGEs may vary considerably between species and social context, and that IGEs may be negligible even in highly social contexts.
2. MATERIALS AND METHODS
Laboratory stock colonies of lineages M and B were synchronized such that eggs hatched into larvae on the same day across colonies (reproduction is cyclic in O. biroi colonies, with all ants tending to lay eggs on the same day, and with eggs hatching into larvae on the same day (Ravary & Jaisson, 2002; Tsuji & Yamauchi, 1995)). On the day of larval hatching, larvae were collected and placed in groups of nine into Petri dishes that each contained nine ants of either lineage B or lineage M. Small groups occur naturally in this species (field‐collected colonies generally contain a dozen to a few hundred individuals (Ravary & Jaisson, 2002; Trible et al., 2020; Tsuji & Yamauchi, 1995), have high fitness, and exhibit complex social behaviour (Chandra et al., 2021; Ulrich et al., 2018)). The fostering of newly‐hatched larvae ensured that individuals experienced their adoptive social environment throughout larval and pupal development. We monitored the experimental subcolonies daily (at 09.00 AM) to check for adult eclosion. The first of the fostered individuals to eclose as an adult was sampled on the morning of eclosion. These samples (“day 1”) were used to investigate how much variation in brain gene expression was determined by direct and indirect genetic effects during development. When the second individual eclosed as an adult, the rest of the fostered brood were removed. This removal allowed for the focal adult to experience a genetically homogeneous social environment. The subcolonies were then maintained at 25°C, 60% RH, and under a 12 h:12 h light:dark cycle. The focal adult was sampled after eight days (“day 8”), which is long enough to allow interaction with social partners but not so long that some of the subcolonies would switch to the reproductive phase and introduce another source of variation. The eight‐day‐old individuals were used to investigate the direct and indirect genetic effects on brain gene expression from across development and adult life.
We initially established 20 experimental subcolonies per condition. Several subcolonies failed, and final sample sizes are reported in Table 1.
TABLE 1.
Sample sizes for the four conditions at two time‐points
| Focal of genotype B | Focal of genotype M | |
|---|---|---|
| Environment of genotype B | Day_1 = 7 | Day_1 = 14 |
| Day_8 = 10 | Day_8 = 12 | |
| Environment of genotype M | Day_1 = 9 | Day_1 = 20 |
| Day_8 = 10 | Day_8 = 14 |
2.1. RNA extraction, library preparation, and sequencing
Upon collection, ants were anaesthetized on ice and brains were dissected out in 1× PBS, and homogenized in 1 ml of TRIzol reagent with ceramic beads. Homogenized samples were incubated for 5 min at room temperature (RT). Chloroform (200 μl) was added, samples were vortexed, and then incubated for 5 min at RT. Samples were centrifuged (25 s at 13,523 g and 4°C) and the upper aqueous layer (~500 μl) was transferred to a new tube with Isopropanol (650 μl) and Glycogen blue (1 μl; RNAse‐free, Invitrogen, 15 mg/ml, no. AM9516). Samples were vortexed and incubated overnight at −20°C. Samples were then centrifuged (30 s at 18,407 g at 4°C), the supernatant was discarded, and EtOH (1 ml at 80%) was added. Samples were then vortexed and centrifuged (5 min at 18,407 g at 4°C). The supernatant was discarded and EtOH (1 ml at 70%) was added. Samples were then vortexed and centrifuged again (5 min at 18,407 g at 4°C). The supernatant was removed and the pellet was allowed to dry (15–20 s) at RT. The pellets were resuspended in nuclease‐free water. The extractions were performed in six batches. To prevent batch effects from confounding IGEs or DGEs, the four conditions for the day 1 samples were extracted in different batches, and the day 8 samples were approximately blocked across two batches.
The KAPA stranded mRNASeq library preparation kit (no. KK8421) was used for library preparation. Samples were sequenced (single‐end) across seven lanes of an Illumina HiSeq 2500, blocking samples of different conditions across the lanes.
2.2. Gene expression analysis
Reads were mapped using the O. biroi genome (GCF_003672135.1_Obir_version_5.4) and annotation (GCF_003672135.1_Obir_version_5.4_genomic.gtf) with STAR version 2.7.8a (Dobin et al., 2013). First, genome indices were generated using ‐‐runMode genomeGenerate with ‐‐sjdbOverhang 100 and ‐‐genomeSAindexNbases 12. Mapping was then run with ‐‐twopassMode Basic. Mapped reads were counted with FeatureCounts, with ‐Q 10 (Subread version 2.0.2 (Liao et al., 2013)). After mapping and counting, we obtained 16.9 ± 3.16 million reads per individual (mean ± SD). Before running the differential expression analysis, we filtered out genes with an average number of reads <10 across all samples (this removed 4353 out of 13,755 genes). We used DESeq2 (which is based on a negative binomial generalized linear model) to identify genes that were differentially expressed by time point (model: age). We then looked within time point to identify genes that were differentially expressed according to (i) direct genetic effects, while controlling for indirect genetic effects (model: genotype of social environment + genotype of focal), and (ii) indirect genetic effects, while controlling for direct genetic effects (model: genotype of focal + genotype of social environment). We additionally considered all samples together to investigate how many genes are differentially expressed by (i) direct genetic effects when controlling for indirect genetic effects and age, and (ii) indirect genetic effects when controlling for direct genetic effects and age. We reran these models adding terms for technical batch effects (sequencing lanes and extraction groups), and then again to test for an interaction between direct and indirect genetic effects (model: age + lane + extraction group + genotype of focal + genotype of social environment + genotype of focal:genotype of social environment).
In DESeq2, significant differential expression is assessed with a Wald test, and the Benjamini‐Hochberg procedure is used to obtain multiple‐testing adjusted p‐values. Genes are considered as significantly differentially expressed if the adjusted p‐value is < .1, which is default in DESeq2.
3. RESULTS AND DISCUSSION
There were substantial differences in the brain gene expression profiles of 1‐ and 8‐day‐old ants. The two time points clearly separated along the first principal component of a PCA, with 7,370 genes (78% of the genes that passed the low‐counts filter) being significantly differentially expressed (3645 genes were up regulated at day 8 and 3725 genes were upregulated at day 1; Figure 1). There was clearly more scatter at the earlier time point. To quantify this, we calculated the standard deviation in expression for each gene in the two age groups separately and found that there was ~37% more variation in the brain gene expression profiles of the 1‐day‐old than in the 8‐day‐old ants (mean difference in standard deviation of normalized expression per gene between ants of the two time points = 91 copies; Wilcoxon rank‐signed test p‐value < .001). This difference is probably because brains undergo rapid changes in gene expression during and immediately after eclosion (Guerrero‐Peña et al., 2021; Myers, 2003), and so the 24 h of variance in age within each time point introduced larger transcriptomic variation among the 1‐day‐old than among the 8‐day‐old ants.
FIGURE 1.
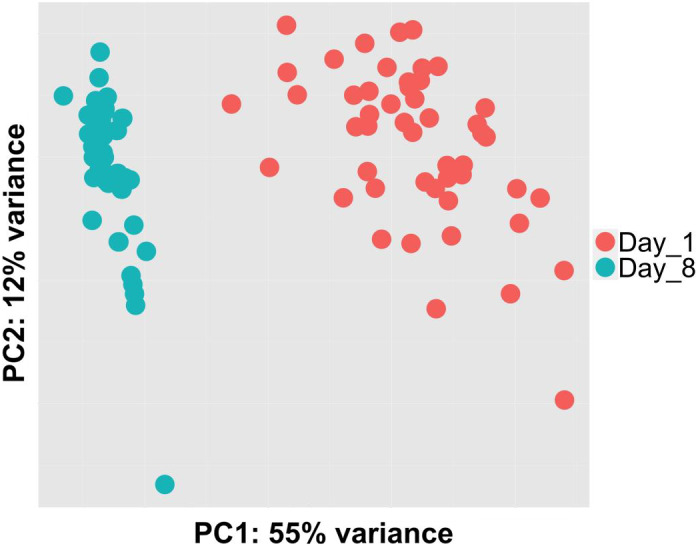
Age had a considerable effect on brain gene expression. There were clear differences in the brain gene expression profiles of adults on the first day of emergence, and 8 days post‐emergence. Colour indicates age.
At both time points, many genes were differentially expressed according to the genotype of the focal individual (i.e., DGE). In 1‐day‐old ants, 1966 genes (20.9%) were significantly differentially expressed between the brains of lineage M and lineage B ants (while controlling for social environment), and in 8‐day‐old ants 1268 genes (13.5%) were differentially expressed (Figure 2a,c). At both time points approximately half of the significantly differently expressed genes were upregulated in individuals of each genotype (Figures 2b,d). In striking contrast, not a single gene was differentially expressed according to the genotypic composition of the social environment (i.e., IGE) when controlling for the genotype of the focal individual at either time point (Figures 2a,c). Similarly, when considering data from both time points together, 1622 genes (17.3%) were differentially expressed by DGEs while controlling for age and IGEs, while 0 genes were differentially expressed by IGEs when controlling for age and DGEs. When additionally controlling for extraction batch and sequencing lane 770 genes (8.2%) were differentially expressed by DGEs and 0 genes were differentially expressed by IGEs (both of these technical batch effects were small: seven genes were differentially expressed by extraction group, and 42 by sequencing lane when controlling for age, genotype of focal individual, and genotype of the social environment). These findings suggest that indirect genetic effects do not explain variation in brain gene expression in O. biroi. We cannot rule out that with a larger sample size we would have detected genes differentially expressed by IGEs. However, given the number of genes differentially expressed by DGEs, we can estimate that DGEs have at minimum a 1000‐fold greater impact on brain transcriptomic variation than IGEs from adult ants under our experimental conditions.
FIGURE 2.
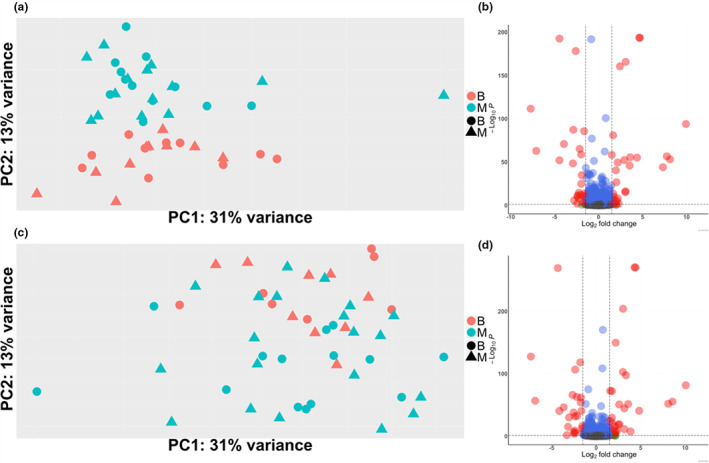
Brain gene expression depenjudded on direct and not indirect genetic effects. In both PCAs colour indicates individual genotype, and shape indicates the genotype of the individuals constituting the social environment. (a) Brains of 8‐day‐old ants, where there is clear clustering according to the genotype of the focal individual. (b) Volcano plot of genes differentially expressed according to the genotype of the focal individual among 8‐day‐old ants. (c) Brains of 1‐day‐old ants, where there is weak clustering according to the genotype of the focal individual. (d) Volcano plot of genes differentially expressed according to the genotype of the focal individual among the 1‐day‐old ants.
This finding is surprising for two reasons. First, there is evidence for IGEs on adult behaviour in O. biroi. The proportion of time that an individual of a given genotype spends away from the nest (a proxy for foraging) varies according to the genotypic composition of the social environment (Teseo et al., 2014; Ulrich et al., 2021; S. L. Jud, D. Knebel, Y. Ulrich, Unpublished data). We additionally observed IGEs on the duration of larval development time, showing that the behavioural differences between workers of different genotypes are relevant for larval development. Larvae raised by B workers took significantly longer to develop than larvae raised by M workers, which is consistent with B workers spending less time foraging (lm: larval development duration ~genotype of larvae + genotype of workers. Larval genotype p‐value = .295, adult genotype p‐value .00709; Figure S1) (S. L. Jud, D. Knebel, Y. Ulrich, Unpublished data). Several recent studies have reported a noncorrespondence between behavioural patterns and patterns of brain gene expression. In the red imported fire ant, adult behaviour varies according to the genotypes of individuals present in the social environment and yet cross‐fostering young adults showed no significant effect of the social environment on brain gene expression (Arsenault et al., 2022). In the paper wasp Polistes dominula, all workers exhibit substantial changes in brain gene expression if the queen is removed from the colony but only few individuals transition to exhibit queen‐like phenotypes (Taylor et al., 2021). One possible explanation for these apparent inconsistencies between behavioural patterns and brain gene expression profiles is that general behavioural heuristics (e.g., leave the nest to collect food when larval demand passes a given threshold, or develop into a queen if others appear relatively smaller or weaker) are associated with broad‐scale gene expression patterns in the brain, but that the actual performance of a specific behaviour is associated with comparatively subtle changes in brain gene expression which are not readily detectable with a bulk‐sequencing approach.
Second, the focal individuals were fostered on the day that they eclosed from eggs, and so experienced the adoptive social environment across larval and pupal development. In O. biroi, as in other social insects, the brood is completely dependent on the social care provided by adults. Moreover, in social insects, differences in provisioning regimes can have profound consequences on gene expression (Vojvodic et al., 2015) and the developmental fate of females (queens and workers). In O. biroi, ants of different genotypes have been found to interact differently with brood (dedicating different proportions of their time to nursing), meaning that IGEs would be expected to influence development, and therefore brain gene expression upon eclosion (Teseo et al., 2014; Ulrich et al., 2021; Vojvodic et al., 2015).
In general, IGEs should increase in magnitude with group size (Baud et al., 2017; Bijma et al., 2007). However, this is unlikely to have a significant effect in O. biroi. Since these ants are not capable of individual recognition, what matters is the proportion of time spent in contact with individuals of given genotypes. In the nest, the workers are in constant contact – clumped together at maximal density – and so increases in group size should have only a marginal effect on the proportion of time individuals spend in contact with others of particular genotypes.
In conclusion, our study revealed an absence of IGEs in a context where they could be expected. This apparent absence suggests that IGEs may play a more minor role in determining patterns of brain gene expression and consequent behaviour and can be more system‐specific than previous evidence would suggest.
AUTHOR CONTRIBUTIONS
Conceptualisation: Tomas Kay, Yuko Ulrich, Max Reuter, and Laurent Keller; Investigation: Tomas Kay, Christine La Mendola, and Giacamo Alciatore; Original draft preparation: Tomas Kay, Laurent Keller; Writing & reviewing: All authors; Funding Acquisition: Laurent Keller; Resources: Laurent Keller, Yuko Ulrich.
CONFLICT OF INTEREST
The authors declare no competing interests.
Supporting information
Figure S1
ACKNOWLEDGEMENTS
We thank three anonymous reviewers for their useful comments on this study was funded by the European Research Council (ERC Advanced Grant, “resiliANT,” no. 741491) and the Swiss NSF. Open access funding provided by Universite de Lausanne.
Kay, T. , Alciatore, G. , La Mendola, C. , Reuter, M. , Ulrich, Y. , & Keller, L. (2022). A complete absence of indirect genetic effects on brain gene expression in a highly social context. Molecular Ecology, 31, 5602–5607. 10.1111/mec.16686
Handling Editor: Tatiana Giraud
DATA AVAILABILITY STATEMENT
Raw RNA‐sequencing data have been deposited in NCBI's Gene Expression Omnibus under accession number GSE210246. Metadata and code are available on GitHub at https://github.com/MO‐Katy/IGEvDGE_ClonalRaiderAnt.
REFERENCES
- Arsenault, S. V. , Riba‐Grognuz, O. , Shoemaker, D. , Hunt, B. G. & Keller, L. (2022). Direct and indirect effects of a social supergene. Authorea. 10.22541/au.165406692.26329813/v1 [DOI] [PubMed] [Google Scholar]
- Baud, A. , Mulligan, M. K. , Casale, F. P. , Ingels, J. F. , Bohl, C. J. , Callebert, J. , Launay, J. M. , Krohn, J. , Legarra, A. , Williams, R. W. , & Stegle, O. (2017). Genetic variation in the social environment contributes to health and disease. PLoS Genetics, 13, e1006498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsma, R. , Kanis, E. , Knol, E. F. , & Bijma, P. (2008). The contribution of social effects to heritable variation in finishing traits of domestic pigs (Sus scrofa). Genetics, 178, 1559–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijma, P. , Muir, W. M. , & Van Arendonk, J. A. (2007). Multilevel selection 1: Quantitative genetics of inheritance and response to selection. Genetics, 175, 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijma, P. , & Wade, M. J. (2008). The joint effects of kin, multilevel selection and indirect genetic effects on response to genetic selection. Journal of Evolutionary Biology, 21, 1175–1188. [DOI] [PubMed] [Google Scholar]
- Chandra, V. , Gal, A. , & Kronauer, D. J. (2021). Colony expansions underlie the evolution of army ant mass raiding. Proceedings of the National Academy of Sciences, USA, 118, e2026534118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A. , Davis, C. A. , Schlesinger, F. , Drenkow, J. , Zaleski, C. , Jha, S. , Batut, P. , Chaisson, M. , & Gingeras, T. R. (2013). STAR: ultrafast universal RNA‐seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gempe, T. , Stach, S. , Bienefeld, K. , & Beye, M. (2012). Mixing of honeybees with different genotypes affects individual worker behavior and transcription of genes in the neuronal substrate. PLoS One, 7, e31653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing, B. (1967). Selection in reference to biological groups I. Individual and group selection applied to populations of unordered groups. Australian Journal of Biological Sciences, 20, 127–140. [PubMed] [Google Scholar]
- Guerrero‐Peña, L. , Suarez‐Bregua, P. , Méndez‐Martínez, L. , García‐Fernández, P. , Tur, R. , Rubiolo, J. A. , Tena, J. J. , & Rotllant, J. (2021). Brains in metamorphosis: Temporal transcriptome dynamics in hatchery‐reared flatfishes. Biology, 10, 1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronauer, D. J. , Pierce, N. E. , & Keller, L. (2012). Asexual reproduction in introduced and native populations of the ant Cerapachys biroi . Molecular Ecology, 21, 5221–5235. [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G. K. , & Shi, W. (2013). featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Moore, A. J. , Brodie, E. D., III , & Wolf, J. B. (1997). Interacting phenotypes and the evolutionary process: I. direct and indirect genetic effects of social interactions. Evolution, 51, 1352–1362. [DOI] [PubMed] [Google Scholar]
- Muir, W. M. (1996). Group selection for adaptation to multiple‐hen cages: selection program and direct responses. Poultry Science, 75, 447–458. [DOI] [PubMed] [Google Scholar]
- Myers, E. M. (2003). The circadian control of eclosion. Chronobiology International, 20, 775–794. [DOI] [PubMed] [Google Scholar]
- Oxley, P. R. , Ji, L. , Fetter‐Pruneda, I. , McKenzie, S. , Li, C. , Hu, H. , Zhang, G. , & Kronauer, D. J. (2014). The genome of the clonal raider ant Cerapachys biroi . Current Biology, 24, 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravary, F. , & Jaisson, P. (2002). The reproductive cycle of thelytokous colonies of Cerapachys biroi Forel (Formicidae, Cerapachyinae). Insectes Sociaux, 49, 114–119. [Google Scholar]
- Ross, K. , & Keller, L. (2002). Experimental conversion of colony social organization by manipulation of worker genotype composition in fire ants (Solenopsis invicta). Behavioral Ecology and Sociobiology, 51, 287–295. [Google Scholar]
- Taylor, B. A. , Cini, A. , Wyatt, C. D. , Reuter, M. , & Sumner, S. (2021). The molecular basis of socially mediated phenotypic plasticity in a eusocial paper wasp. Nature Communications, 12, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teseo, S. , Châline, N. , Jaisson, P. , & Kronauer, D. J. (2014). Epistasis between adults and larvae under‐ lies caste fate and fitness in a clonal ant. Nature Communications, 5, 1–8. [DOI] [PubMed] [Google Scholar]
- Trible, W. , McKenzie, S. K. , & Kronauer, D. J. (2020). Globally invasive populations of the clonal raider ant are derived from Bangladesh. Biology Letters, 16, 20200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, K. , & Yamauchi, K. (1995). Production of females by parthenogenesis in the ant, Cerapachys biroi . Insectes Sociaux, 42, 333–336. [Google Scholar]
- Ulrich, Y. , Kawakatsu, M. , Tokita, C. K. , Saragosti, J. , Chandra, V. , Tarnita, C. E. , & DJC, K. (2021). Response thresholds alone cannot explain empirical patterns of division of labor in social insects. PLoS Biology, 19, e3001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich, Y. , Saragosti, J. , Tokita, C. K. , Tarnita, C. E. , & Kronauer, D. J. (2018). Fitness benefits and emergent division of labour at the onset of group living. Nature, 560, 635–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojvodic, S. , Johnson, B. R. , Harpur, B. A. , Kent, C. F. , Zayed, A. , Anderson, K. E. , & Linksvayer, T. A. (2015). The transcriptomic and evolutionary signature of social interactions regulating honey bee caste development. Ecology and Evolution, 5, 4795–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Ross, K. G. , & Keller, L. (2008). Genome‐wide expression patterns and the genetic architecture of a fundamental social trait. PLoS Genetics, 4, e1000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, C. , Canela‐Xandri, O. , Rawlik, K. , & Tenesa, A. (2021). Evidence of horizontal indirect genetic effects in humans. Nature Human Behaviour, 5, 399–406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Data Availability Statement
Raw RNA‐sequencing data have been deposited in NCBI's Gene Expression Omnibus under accession number GSE210246. Metadata and code are available on GitHub at https://github.com/MO‐Katy/IGEvDGE_ClonalRaiderAnt.


