Abstract
The causative bacterium of Lyme disease, Borrelia burgdorferi, expanded from an undetected human pathogen into the etiologic agent of the most common vector‐borne disease in the United States over the last several decades. Systematic field collections of the tick vector reveal increases in the geographic range and prevalence of B. burgdorferi–infected ticks that coincided with increases in human Lyme disease incidence across New York State.
We investigate the impact of environmental features on the population dynamics of B. burgdorferi. Analytical models developed using field collections of nearly 19,000 nymphal Ixodes scapularis and spatially and temporally explicit environmental features accurately explained the variation in the nymphal infection prevalence of B. burgdorferi across space and time.
Importantly, the model identified environmental features reflecting landscape ecology, vertebrate hosts, climatic metrics, climate anomalies and surveillance efforts that can be used to predict the biogeographical patterns of B. burgdorferi–infected ticks into future years and in previously unsampled areas.
Forecasting the distribution and prevalence of a pathogen at fine geographic scales offers a powerful strategy to mitigate a serious public health threat.
Synthesis and applications. A decade of environmental and tick data was collected to create a model that accurately predicts the infection prevalence of Borrelia burgdorferi over space and time. This predictive model can be extrapolated to create a high‐resolution risk map of the Lyme disease pathogen for future years that offers an inexpensive approach to improve both ecological management and public health strategies to mitigate disease risk.
Keywords: Borrelia burgdorferi, environmental change, infection prevalence, Lyme disease, population dynamics, predictive models, tick‐borne bacterium, vector‐borne pathogen
A decade of environmental and tick data was collected to create a model that accurately predicts the infection prevalence of Borrelia burgdorferi over space and time. This predictive model can be extrapolated to create a high‐resolution risk map of the Lyme disease pathogen for future years that offers an inexpensive approach to improve both ecological management and public health strategies to mitigate disease risk.

1. INTRODUCTION
Pathogens that naturally transmit among wildlife and are infectious to humans persist as a recurrent threat to public health (Jones et al., 2008). The recent emergence and re‐emergence of diseases ascribed to zoonotic pathogens is frequently connected to anthropogenic activity and global climate change, but the mechanistic processes are not well understood (Aguirre & Tabor, 2008; Aluwong & Bello, 2010; Chaves & Koenraadt, 2010; Lashley, 2004; Suzán et al., 2008). Environmental impacts on the life cycle of pathogens have been proposed as mechanistic factors precipitating zoonotic disease outbreaks (Fouque & Reeder, 2019; Petrosillo, 2019; Sleeman et al., 2009). Identifying the environmental features that promote changes in the geographic range or prevalence of pathogens can indicate ecological processes exacerbating disease risk. Importantly, these environmental features can then be used to predict the future range or increases in prevalence. This, in combination with the knowledge acquired about the ecology of the pathogen, can be crucial components of effective disease mitigation strategies (Diuk‐Wasser et al., 2010; Gage et al., 2008; Guerra et al., 2002; Kaplan et al., 2010; Khatchikian et al., 2011; Patz et al., 2008). We use a decade of spatio‐temporal data to quantify the relationship between the environment and the medically relevant Borrelia burgdorferi bacterium in a predictive biogeographic model.
The emergence of Lyme disease in the United States over the last three decades has occurred in areas with rapid and dramatic environmental change (CDC, 2019). These environmental changes are thought to impact the ecology and geographic range of the causative pathogen, B. burgdorferi, leading to the observed changes in Lyme disease. As an obligate parasite vectored by the ectothermic blacklegged tick Ixodes scapularis between wildlife hosts, the pathogen life cycle involves the integration of multiple ecological processes, including the impacts of climate, landscape and human activity on the bacterium and its animal hosts.
Suitable habitat conditions for both the pathogen and the tick vector have been described separately for multiple regions in North America and Europe (Johnson et al., 2016; Medlock et al., 2013; Slatculescu et al., 2020). Elevation, distance to water, precipitation, winter temperature and landscape are commonly identified as factors impacting pathogen abundance (Diuk‐Wasser et al., 2012; Eisen et al., 2016; James et al., 2013; Ostfeld et al., 2006; Slatculescu et al., 2020). For example, reforestation, land‐use change and plant communities are thought to alter animal communities in ways that increase densities of B. burgdorferi reservoir hosts leading to increases in pathogen prevalence in the northeastern United States (Allan et al., 2003; Brownstein et al., 2005; Kilpatrick et al., 2017; Levi et al., 2012; LoGiudice et al., 2003; Ostfeld et al., 2006; Wood & Lafferty, 2013). Similarly, climatic features such as air temperature and snowfall are thought to affect tick survival rates (Diuk‐Wasser et al., 2012; Eisen et al., 2016; Linske et al., 2019; Slatculescu et al., 2020). Despite advances in knowledge of features impacting B. burgdorferi populations, the Lyme disease burden continues to increase nationwide suggesting current management strategies may not adequately encompass pathogen ecology (Kilpatrick et al., 2017).
Analysing data collected during the dynamic phase of geographic expansion and prevalence increases, combined with environmental metadata, permit high‐resolution modelling that incorporates environmental heterogeneity and ecological processes (Bunnell et al., 2003; Cord & Rödder, 2011; He et al., 2015; Nicholson & Mather, 1996; Slatculescu et al., 2020). Ecological processes vary geographically due to environmental heterogeneity at both coarse and fine scales (Pickett & Cadenasso, 1995; Stein et al., 2014; Wiens et al., 1993). The accuracy and statistical power to identify and quantify the effect of environmental features on population dynamics increases with the temporal duration and spatial expanse of collected empirical data collected. The prevalence of B. burgdorferi (defined by nymphal infection prevalence, NIP) was measured annually using standardized field collections (2009–2019) from locations across New York State (NYS). This large geographic region has experienced recent and rapid changes in climate and landscape (NYS Department of Environmental Conservation, 2014; O'Connor et al., 2021; USGCRP, 2018; USGS, 2018b) and is representative of B. burgdorferi‐endemic region in the northeastern US (CDC, 2019; US EPA, 2015). The aim of this study was to use fine‐scale spatio‐temporal data collected during the dynamic phases of NIP increases and range expansions to build and validate a predictive biogeographic model.
Ascertaining how spatio‐temporal patterns of multi‐host pathogens correspond with environmental features in a natural ecosystem facilitates an understanding behind the mechanisms of potential drivers of population dynamics and realized geographic ranges. Identification of such features can guide experiments that identify causative relationships between the environment and pathogen populations. We acquired hundreds of environmental features that were previously suggested to be important to the B.burgdorferi transmission cycle. We developed spatio‐temporal biogeographic models to investigate the potential effect of environmental features on NIP. Understanding the ecological processes underlying pathogen population dynamics during the emergence of zoonotic disease can improve gaps of knowledge in current control strategies and inspire broadly applicable ecological management, while being used to predict future disease risk.
2. MATERIALS AND METHODS
2.1. Data
Ticks were collected April–December from 2008 to 2019 and tested for B. burgdorferi with multiplex PCR by experienced NYSDOH personnel (Prusinski et al., 2014). Nymphal I. scapularis ticks were collected at 283 publicly accessible forested sites across 56 (of 62) counties in NYS. Tick abundance and occurrence generally increased throughout NYS as previously described (Tran et al., 2020). NIP represents the prevalence of B. burgdorferi‐infected nymphal I. scapularis as Lyme disease is caused almost exclusively by B. burgdorferi in the eastern United States and is most commonly transmitted to humans through this tick life stage (CDC, 2020). Tick sampling was heterogeneous over time and space, with some sites visited multiple times each year while other sites visited only every 2–3 years. Additional analyses were performed on model residuals to ensure that the model accounted for any spatial and temporal correlations in the data (Tran et al., 2020; see Appendix S1). Sampling protocol at all sites was independent of sampling success and consisted of standardized dragging, flagging and walking surveys using 1 m2 of white flannel or canvas along irregular transects (Prusinski et al., 2014). Varying numbers of ticks were tested from each site due to the variable number of ticks collected per site. Tick collections were conducted on public lands, thus generally not requiring licences nor permits, and ethical approval was not necessary for invertebrate vector species. Further information about tick collection and pathogen testing can be found in Appendix S1.
Environmental data predicted to influence pathogen prevalence were compiled from multiple public databases including: climatic data from PRISM Climate Group (2018); biodiversity indices and hydrographical features data from the NYS Geographical Information Systems (GIS) Clearinghouse (New York State, 2014); vertebrate animal and fire history from the NYS Department of Environmental Conservation (2021); bird data from the North American Breeding Bird Survey (BBS) (USGS, 2018a); landscape data from the USGS National Land Cover Database (USGS, 2018b); elevation data from the USGS National Elevation Dataset (USGS, 2022); and road data and human population data from the US Census Bureau (2022).
Many of the included environmental features impact multiple biotic processes that can affect pathogen prevalence. That is, many features are likely to indirectly impact NIP. For example, anthropogenic land use practices impact the vertebrate community composition which affect the availability of tick bloodmeals and behaviour (Burtis et al., 2016; Ostfeld et al., 2006). Environmental features can be classified into the following general categories (see Table S1):
Landscape factors: It includes natural features such as elevation, recent fire history, measures of forest patches within a certain radius around sampling locations, biodiversity indices, ecological zone classification, landcover, man‐made features such as distance to roads and hydrological features, the nearest road classification, designation as a critical environmental area and proportion of urban, forest, shrub or other.
Vertebrate hosts: It includes annual counts for turkey, bear, deer and human population at the county level and annual counts for bird at the state level.
Surveillance conditions/efforts: It includes relative humidity and ambient air temperature that are specific to a location at the time of sampling, and hours spent collecting and distance surveyed.
Climate measures: It includes monthly temperature, precipitation, humidity proxies, cumulative degree days (degrees above 0°C during different seasons and tick life stages—larval (Jan–Aug) and nymphal (Jan–May)), degree days below 0°C in the winter (Dec–Feb). These climate features were also calculated for 1 and 2 years prior to nymphal collection for each location. Climatic anomalies, the difference between a climate measure and the 1981–2010 average (observation − mean [time series]), were also calculated.
2.2. Statistical methods
Models were built relating the number of infected and uninfected nymphal ticks at each collection site to the collection of environmental features using a binomial regression model with a logistic link function (see Appendix S1). Logistic regression modelling permits interpretable model parameters. Uncertainty in the estimates of NIP due to differences in sample sizes among sites is accounted for in the model by incorporating the binomial distribution parameters into the development of the statistical model. Interaction terms were not included in this model to limit the ratio of predictors to observations as there was not enough data to include higher‐order terms. The model was trained on data collected from 2009 to 2018 with data from 2019 reserved for out‐of‐sample model validation. Training data consisted of more than 16,700 ticks that were tested for B. burgdorferi collected from 238 different sites in 55 counties of NYS. More than 4,500 ticks were tested for B. burgdorferi at 158 different sites in 47 counties in 2019. A bi‐directional stepwise algorithm was applied for variable selection. The different combinations of features were assessed using k‐fold cross‐validation. Data subsets for cross‐validation were based on the training data with a single year being rotated as the hold‐out year for a total of k = 10 folds. Based on Akaike's information criterion (AIC) and root‐mean‐square‐error (RMSE), the final model was selected and refitted on all the training data and then externally validated using 2019 data. The stepwise search strategy is sensitive to multicollinearity such that highly correlated predictors are unlikely to be included in the final model (Chowdhury & Turin, 2020).
The model was trained on sites with more than 10 ticks tested. NIP estimate accuracy improves with increasing number of ticks tested, which better captures the underlying tick infection rates by accounting for stochasticity associated with tick sampling. Limiting the 2009–2019 dataset to sites where more than 10 ticks were tested resulted in 18,972 nymphal ticks over 158 sites in 49 counties included in model training and testing. Model accuracy was assessed by comparing model estimates of NIP versus the observed proportions of infected ticks at each site in 2019. NIP estimated by the model for each site was considered accurate if the estimate was within the 95% binomial confidence interval around the observed number of infected and uninfected ticks collected from that site. The confidence interval was calculated using the Clopper–Pearson exact binomial interval with the observed proportion of infected ticks at each site. The statistical significance of the environmental features in the model were determined using Wald tests. Predictive accuracy of the model was determined by out‐of‐sample predictions to 2019 data, which was not used in the model fitting described above. Using out‐of‐sample model evaluation protects against model overfitting (Lever et al., 2016). The purpose of this model is to predict NIP in the future. Although features in the model could be used to generate hypotheses about causative ecological drivers, their inclusion in the model should not be considered as evidence of causation.
2.3. Nymphal infection prevalence of Borrelia burgdorferi across New York State
The model was used to create a predictive B. burgdorferi NIP map for 2019 across NYS (Appendix S2.2), except over hydrological features and New York City (NYC), at a resolution of approximately 0.5 km (0.00513 degrees at the equator). Pathogen infection rates (B.b.) were categorized as follows: low (B.b. < 0.1 proportion of ticks infected), medium (0.1 < B.b. < 0.2), high (0.2 < B.b. ≤ 0.5) and very high (B.b. > 0.5).
3. RESULTS
Nearly 19,000 nymphal ticks were tested from more than 550 visits to 158 unique sites between 2009 and 2019. More than 10 nymphal ticks were collected and tested from all 550 site visits included in the analyses. B. burgdorferi was detected in at least one nymphal tick from 155 of the 158 sites. NIP increased between 2009 and 2019 in many sites although the rate of increase varied considerably among sites and among regions (Figure 1). The rate of increase in NIP was greatest in regions with few infected ticks in 2009 (south‐central and western NYS) and considerably less pronounced in regions with initially greater numbers of infected ticks (Hudson Valley and central NYS). However, NIP in some sites with relatively high NIP in 2009 increased at greater rates and there was considerable variation in the change in NIP among sites and year‐to‐year (see Appendix S2.1). Changes in annual median NIP across the state are only weakly correlated with changes in annual median tick densities (r = 0.02).
FIGURE 1.
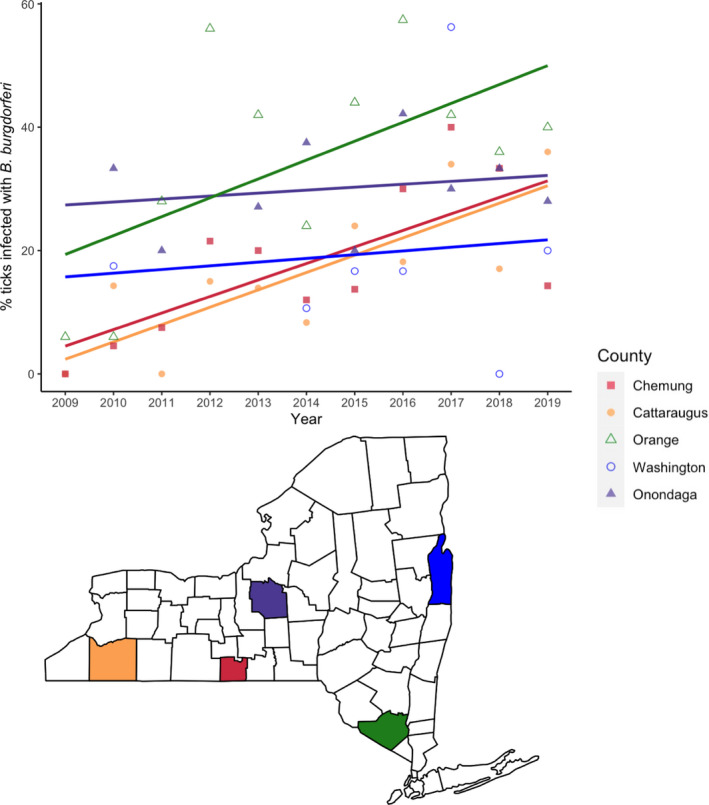
The prevalence of Borrelia burgdorferi‐infected nymphs increased non‐uniformly across NYS between 2009 and 2019. Five representative collection sites from different counties (each depicted by a unique colour and shape) illustrate the disparate temporal patterns of NIP among sites. NIP increased in these sites over the decade in which collections were made, but at different rates based on the slope of best‐fit line. Sites with initially low NIP increased at the fastest rates (south‐central and western NYS). In contrast, NIP generally increased at moderate rates in regions with established pathogen populations. These representative sites were sampled almost every year of the study period and thus capture pathogen population dynamics at each site most accurately. Points represent the observed annual median infection rate for each site.
The regression model with the greatest predictive accuracy correctly estimated NIP with 93.7% of the training data (Figure 2a). Model accuracy improved with more ticks tested per site, with inaccurate predictions generally at sites where 15 or fewer ticks were tested. Inaccurate predictions were not biased toward over or underpredictions (2.8% vs. 3.5%). Model residuals exhibited no departure from normality nor autocorrelation indicating that any spatial or temporal autocorrelation inherent in the dataset was accounted for by model covariates (see Appendix S1). Model predictions to sites visited in 2019, data not used in model training, were accurate but slightly lower than training data accuracy (Figure 2b). Nevertheless, the accuracy of model predictions to the out‐of‐sample dataset indicates that the environmental features included in this model capture the spatial and interannual heterogeneity necessary to accurately predict NIP into future years. Furthermore, NIP predictions were as accurate for sites visited for the first time in 2019 as for those sampled previously, demonstrating the spatial and temporal predictive power of the environmental features included in the model.
FIGURE 2.
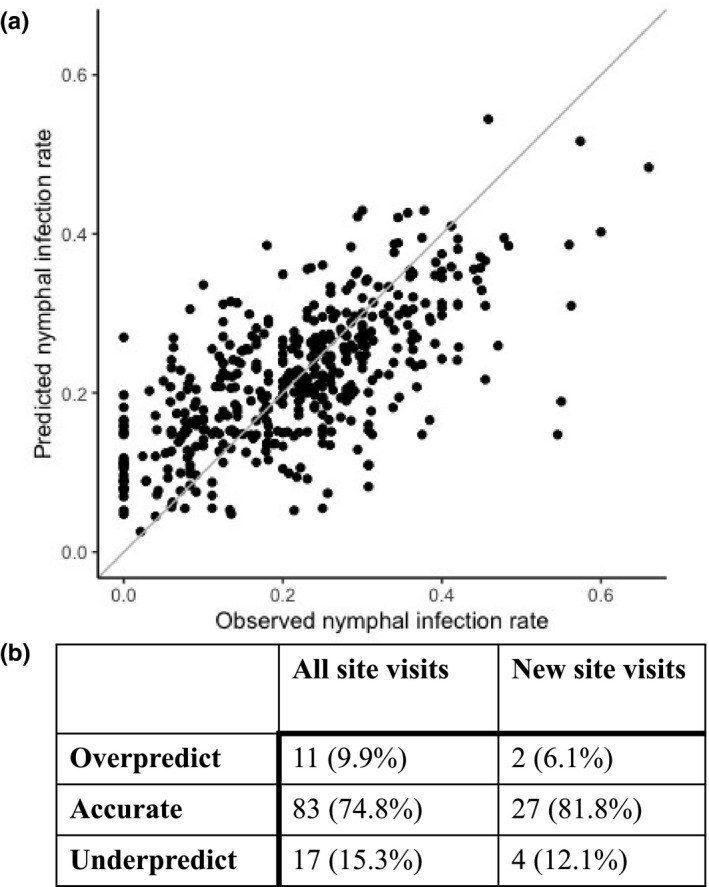
Nymphal infection prevalence is accurately predicted by the statistical model. (a) The model accurately predicts NIP across space and time for 457 visits to 129 sites between 2009 and 2018. The distance to the 1:1 line denotes the predictive accuracy, with closer distances being more accurate. The number of ticks tested per site ranged from 11 to 80 (median = 33). (b) The statistical model can predict NIP into a future year across geographic space. The model accurately predicts to an out‐of‐sample dataset (2019), to both new sites and into a future year across 111 visits to 97 sites. The predictive accuracy of the model to the 29 sites sampled for the first time in 2019 (n = 867 ticks) was comparable with sites represented in the training dataset despite fewer ticks tested at these new sites (medians of 19 and 50 ticks tested, respectively).
The regression covariates in the statistical model included temporal, geographic, seasonal, climatic and landscape features (Table 1). Most of the 47 covariates included in the model were statistically significant (X = 40, p < 0.05). Several competing models investigated during feature selection exhibited accuracy comparable with that of the final best‐fitting models. Each competing model differed from the best‐fitting model by substituting features in the best‐fitting models with highly correlated features. The coefficients estimated for these correlated features were nearly identical among these models, suggesting that the cross‐validated features selected are robust and issues of collinearity were eliminated during the feature selection procedure. Features measuring climate (X = 33), including both absolute measures and deviations from a 30‐year average (1981–2010), and landscape features (X = 11) comprised the majority of the model covariates. Only rarely were both features measuring the absolute values of local climate and deviations from the baseline average from the same seasonal and temporal period included in the model. However, in the three cases where both the absolute and deviation measures were included, the regression coefficients had different signs.
TABLE 1.
Environmental factors of Borrelia burgdorferi prevalence
| B. burgdorferi NIP model [X = 47] | Directionality | p‐value |
|---|---|---|
| (a) Surveillance efforts [1] | ||
| Nymph density per person‐hour of collection | (+) | ** |
| (b) Landscape ecology [11] | ||
| Latitude | (−) | *** |
| Elevation | (−) | *** |
| Ecological zone classification a | *** | |
| Watershed biodiversity score | (−) | *** |
| Indicator of critical zone a | (+) | *** |
| Number of fires (previous year) | (−) | *** |
| Other landcover (not forest, urban, shrub) | (+) | *** |
| Forest patch connectivity within 500 m radius | (+) | ** |
| Distance to nearest hydrographical feature | (+) | ** |
| Road type of nearest road a | (+) | * |
| Patch density within 50 m radius (edge length per unit area) | (−) | |
| (c) Vertebrate hosts [2] | ||
| Human population | (−) | *** |
| Bird population (migrant/non‐breeder status) | (−) | * |
| (d) Climatic measures (raw values) b [17] | ||
| Monthly maximum temperature | ||
| Jun (previous year) | (−) | *** |
| Oct (2 years prior) | (−) | *** |
| Monthly mean temperature | ||
| Jun (current year) | (+) | *** |
| Mar (previous year) | (−) | *** |
| Oct (previous year) | (+) | |
| Monthly total precipitation | ||
| Jan (current year) | (+) | |
| Mar (previous year) | (+) | |
| Oct (previous year) | (+) | ** |
| Mar (2 years prior) | (−) | *** |
| Monthly maximum vapour pressure deficit | ||
| Mar (previous year) | (+) | |
| Oct (2 years prior) | (+) | *** |
| Monthly minimum vapour pressure deficit | ||
| Mar (current year) | (+) | *** |
| Jun (current year) | (−) | *** |
| Mar (previous year) | (−) | *** |
| Degree days below 0°C (winter) | ||
| Winter (current year) | (−) | *** |
| Winter (previous year) | (+) | *** |
| Winter (2 years prior) | (+) | *** |
| (e) Climatic anomalies (deviations from historical mean from 1981 to 2010 baseline) [16] | ||
| Monthly maximum temperature anomalies | ||
| Oct (2 years prior) | (+) | *** |
| Monthly mean temperature anomalies | ||
| Jun (2 years prior) | (+) | *** |
| Monthly minimum temperature anomalies | ||
| Oct (current year) | (+) | ** |
| Jan (2 years prior) | (−) | * |
| Jun (2 years prior) | (−) | *** |
| Monthly total precipitation anomalies | ||
| Jan (current year) | (−) | *** |
| Mar (current year) | (−) | *** |
| Jun (2 years prior) | (+) | |
| Monthly max. vapour pressure deficit anom. | ||
| Jan (2 years prior) | (−) | *** |
| Mar (2 years prior) | (−) | *** |
| Jun (2 years prior) | (−) | *** |
| Monthly minimum vapour pressure deficit anom. | ||
| Jan (previous year) | (−) | ** |
| Mar (previous year) | (+) | *** |
| Jan (2 years prior) | (−) | |
| Monthly mean dew point temperature anomalies | ||
| Jan (current year) | (+) | ** |
| Oct (previous year) | (−) | ** |
Notes: t‐test *** < 0.001, ** < 0.01, * < 0.05; (−) = negative relationship; (+) = positive relationship.
Categorical variable.
For temperature, vapour pressure deficit and mean dew point temperature, monthly refers to the daily value averaged over all days in the month.
The accuracy of model predictions to data that are out‐of‐sample both temporally (data from 2019 were not used to train models) and spatially (sites sampled for the first time in 2019) suggest that environmental features can predict NIP at previously unvisited sites across NYS (Figure 3). NIP varies over geographic space and is heavily influenced by landscape topography. Three broad regions characterized by higher elevation are predicted to have lower B. burgdorferi NIP, likely due to shared environmental features. These predicted results are corroborated by the field collection data showing gradual changes in pathogen infection rate over the study period (Figure 1).
FIGURE 3.
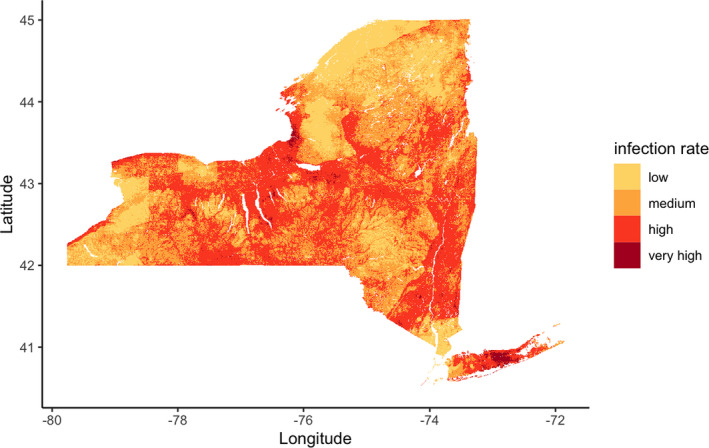
Estimated Borrelia burgdorferi‐nymphal infection prevalence in 2019: NIP can be predicted at least 1 year in the future and at very fine spatial scales. The spatio‐temporal variation of NIP can be visualized in unsampled locations with the validated environmental model. NIP predictions in this figure are at a resolution of approximately 500 m and rates are defined as low (NIP = 0–0.1), medium (0.1–0.2), high (0.2–0.5) and very high (>0.5).
4. DISCUSSION
The causative pathogen of human Lyme disease, B. burgdorferi, was not detected in North America for centuries (Hoen et al., 2009; Marshall III et al., 1994) before becoming the etiologic agent of the most common vector‐borne disease in the United States (CDC, 2019). Increases Lyme disease incidence observed in NYS over the last several decades coincided with a general increase in B. burgdorferi prevalence in nymphal ticks (Figure 1). Data from field collections revealed that the median annual NIP increased from 5.0% [0%–11.1%] in 2009 to 25.3% [20.0%–28.0%] in 2019. This change in NIP cannot be attributed to changes in tick population sizes as tick population size dynamics are poorly correlated with NIP (r = 0.02). A statistical model developed using spatio‐temporal variation in environmental features and field data collected during recent pathogen population growth and range expansion (2009–2018) predicts the variation in pathogen prevalence patterns across the state and among years. Importantly, this environmental model accurately predicted the prevalence of B. burgdorferi in future years (2019) and in previously unsampled areas (Figure 2b). The identification of environmental features and development of a powerful predictive tool is useful in mitigation strategies to address this public health threat.
The statistical model relating environmental features to NIP successfully predicted to sites that are outside the range of data used to train the model. That is, the model accurately predicted NIP in 2019, data not used to train the model. Furthermore, prediction accuracy was similar for sites that were visited prior to 2019 as well as sites that were not. The model also accurately predicted NIP in sites sampled in 2019 with environmental feature values outside of the training data range in some cases. For example, the model accurately predicted to sites at elevations that exceeded the range of the training data (5–560 m, median 170 m). However, some inaccurate predictions can be attributed to environmental feature values beyond the range observed in the training data (i.e. temperature and precipitation). This limitation highlights the importance of long‐term data collection to increase the environmental variability observed in the training dataset for model development. The overall accuracy of the out‐of‐sample predictions suggests that the environmental features can be used to account for B. burgdorferi population dynamics.
Interpretation of both the model predictions and environmental drivers should be approached with caution. For example, there is inherent temporal and spatial autocorrelation in NIP as the proportion of infected ticks is related to NIP in prior years and in nearby locations (Kitron & Kazmierczak, 1997; Schauber et al., 2005). Such autocorrelation can be challenging when applying the model to extrapolate outside of the training data. Although autocorrelation could have influenced prediction accuracy during model validation, it cannot account for the out‐of‐sample predictive accuracy to sites that were sampled for the first time in 2019. This out‐of‐sample accuracy suggests that the model successfully accounted for any temporal or spatial autocorrelation and did not suffer from overfitting to the data despite the large number of model predictors. Furthermore, cross‐validation during model development also girds against overfitting and demonstrates the predictive accuracy to out‐of‐sample years (Wenger & Olden, 2012). Lastly, the lack of autocorrelation in the model residuals increases confidence in the regression model (see Appendix S1). Although the predictors in statistical models cannot be interpreted as causal drivers, the identified features can be used to guide future hypothesis‐driven experiments to determine causation.
The model identified many environmental features that collectively predict the spatio‐temporal dynamics of B. burgdorferi prevalence. Pathogen prevalence is contingent upon the complex network of ecological interactions between the environment, vertebrate hosts and vector populations but only first‐order terms were considered in the model. Identifying the relationship between these environmental features and NIP is further complicated as many features likely indirectly impact B. burgdorferi. While environmental features impact tick population dynamics, the spatio‐temporal variation in NIP cannot be attributed to the impacts of the identified environmental features on tick populations as (a) the spatio‐temporal variation in NIP is only weakly correlated to the spatio‐temporal variation in nymphal tick densities (r = 0.02) and (b) nymphal density has been accounted for by including it as a model predictor (Khatchikian et al., 2012; Tran et al., 2020). The variance explained by the environmental features included in the model likely impact populations of vertebrate hosts that amplify or dilute NIP (Brisson et al., 2008; LoGiudice et al., 2003; Randolph & Dobson, 2012). The environmental features included in this model can serve to generate macroscale hypotheses about how anthropogenic impacts on the environment affects the emergence of diseases. Including the geospatial patterns and ecological processes that affect both the pathogen as well as the tick vector will improve models and assessment of Lyme disease risk.
Climatic features from multiple timepoints within and across years suggest multiple ecological processes affecting B. burgdorferi population dynamics. For example, total monthly precipitation from different times of the year and from years prior to sampling capture short‐term interannual and seasonal climatic effects (Table 1d). Climatic anomalies (site‐specific deviations from the long‐term average at that location) permit comparisons of present conditions relative to the past that give insight into historical effects caused by climate change. The climatic anomaly features included in the model suggest that both climate change, in addition to the absolute values of climatic features, affect B. burgdorferi population dynamics. For example, the extent of the increase in summer temperatures at collection sites compared with past temperatures (1981–2010) at those sites is associated with an increase in NIP (Table 1e). The use of abiotic features was particularly advantageous, despite the expectation that they indirectly affect NIP, because such features are readily measurable and permit predictive models. Of note, climate likely impacts biotic systems beyond 2 years preceding tick collections, an important question for future studies. A comprehensive understanding of the ecological consequences of climate change can guide future research using short‐term climatic forecasts to predict pathogen population dynamics.
B. burgdorferi prevalence is associated with landscape features that measure habitat disturbance (i.e. recent fires and roads). Prior studies demonstrate a positive association between human Lyme disease risk and habitat fragmentation, which alter vertebrate community composition and subsequently increases pathogen population sizes (Allan et al., 2003; Simon et al., 2014). In contrast, the model indicates that NIP is negatively associated with landscape features representing habitat disturbance like recent wildfire, patch connectivity, human population size and critical environment designations. However, some features like wildfire affect the continuity of fauna and can increase the abundance of small mammal and generalist species, which are thought to provide more optimal pathogen conditions (Roberts et al., 2015). This apparent contradiction may be explained by the positive association between habitat fragmentation and tick population sizes (Khatchikian et al., 2012; Talbot et al., 2019; Tran et al., 2020). Tick abundance has significantly increased throughout NYS which is a strong predictor of human Lyme disease risk regardless of B. burgdorferi prevalence (Tran et al., 2020).
The historical and current geospatial trends in B. burgdorferi populations mirrors many of the dynamic geographical patterns of Lyme disease cases in NYS (CDC, 2021). Field collections from 2009 to 2019 revealed that large regional increases in tick infection rates (Figure 1) parallel dramatic increases in Lyme disease cases and the range expansion into western NYS (CDC, 2021). For example, NIP and Lyme disease cases increased rapidly in areas where B. burgdorferi was only recently detected and remained high where B. burgdorferi populations have existed for decades (CDC, 2019). Although both NIP and the density of infected nymphs (DIN) have been used as indicators for Lyme disease risk, our predictive model specifically focused on NIP predictions (LoGiudice et al., 2003; Ogden & Tsao, 2009). Using NIP instead of DIN avoids the compound uncertainty inherently involved in estimating the population size of ticks, which requires accounting for day‐of‐collection factors (e.g. weather), and offers the advantage of using observed tick density as a model predictor (Tran et al., 2020). This qualitative association between the distribution of pathogen populations and human disease demonstrates the value of investigating the ecological processes driving pathogen population growth.
Discerning the environmental impacts on pathogen prevalence and disease risk can guide public health strategies. The influence of environmental factors on NIP suggests that land management strategies and preventative behaviour, in addition to methods targeting transmission in animal hosts, can be important mitigation strategies (70). For example, acorn production correlates with Lyme disease incidence but estimating acorn abundance over a large scale for landscape management is challenging even with the advances in remote sensing techniques (Ostfeld et al., 2006). Accurate NIP models that incorporate easily accessible data such as climate and landscape geography facilitate the production of annual risk maps at high spatial resolution. Public health warnings can be provided near high‐risk areas or specifically managing critical environmental areas that are within a certain distance from a body of water, roads or a fragmented forest habitat. Furthermore, fire management to reduce tick population sizes is a commonly proposed landscape management technique but poorly understood (Guo & Agusto, 2022). Given that the impact of fire is likely additive and indirect, multivariate regression models encompassing landscape type and distance to fire provide further insight into possible relationships between different environmental features. Importantly, it is possible that the results could be extrapolated to collect ticks in regions with similar climate and landscape outside NYS. This method affords an inexpensive and rapid approach to address a future forecasted with more frequent zoonotic disease outbreaks (Jones et al., 2008).
AUTHOR CONTRIBUTIONS
Tam Tran, Dustin Brisson, Melissa Prusinski conceived the study; Tam Tran, Dustin Brisson, Shane Jensen designed the study, carried out the statistical analyses, interpreted the results and drafted the manuscript; Melissa Prusinski coordinated tick data collection; Melissa Prusinski, Jennifer White, Richard Falco, Vanessa Vinci, Wayne Gall, Keith Tober, Jamie Haight, JoAnne Oliver, Lee Ann Sporn, Lisa Meehan, Elyse Banker, P. Backenson collected field data and carried out the pathogen testing; all authors revised the manuscript, gave final approval for publication and are accountable for the work performed therein.
FUNDING INFORMATION
This work was supported by the NYSDOH, the National Institutes of Health (AI097137, AI142572, AI137433, T32‐AI55400, F31AI133871), the Centers for Disease Control and Prevention (U01CK000509) and the Burroughs Wellcome Fund (1012376). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the CDC.
CONFLICT OF INTEREST
The authors have no conflicts of interest to disclose.
Supporting information
Appendix S1
Figure S3
Table S1
ACKNOWLEDGEMENTS
We thank the NYS Department of Environmental Conservation, NYS Department of Parks, Recreation and Historic Preservation and park managers for granting land use. We thank these individuals for assistance in collection, identification and/or molecular testing including students of Paul Smith's College, Jake Sporn and boat launch stewards of the Adirondack Watershed Institute, Lauren Rose, Alexis Russell, Collin O'Connor, Nicholas Piedmonte, Anna Perry, Joshua Rosansky, Konrad Fondrie, Kaitlin Driesse, Michael Suatoni, Margaret Mahoney, Michelle Wemette, Sandra Beebe, Kayla Mehigan, Emily Haner, Franz Soiro, Morgan Thorne, Kate Turcotte, Jacob Miller, Joseph Prusinski Jr., Jennifer DeLorenzo, Lauren DeLorenzo, James Sherwood, John Howard, Rachel Reichel, Marly Katz, Adam Rowe, Jean Stella, Donald Campbell, Daniella Giardina, Melissa Stone, Thomas Mistretta, R.C. Rizzitello, Emily Gicewicz, Christopher Murphy, David Rice, Melissa Fierke and associates with the SUNY ESF, Colgate University, Claire Hartl and others from SUNY Brockport, Niagara County DOH, Erie County DOH, Cornell Cooperative Extension of Onondaga County, Scott Campbell, Michael Santoriello, Christopher Romano and Suffolk County DOH and Ilia Rochlin and Moses Cucura and Suffolk County Vector Control. We thank Michael Benjamin for use of his photos.
Tran, T. , Prusinski, M. A. , White, J. L. , Falco, R. C. , Kokas, J. , Vinci, V. , Gall, W. K. , Tober, K. J. , Haight, J. , Oliver, J. , Sporn, L. A. , Meehan, L. , Banker, E. , Backenson, P. B. , Jensen, S. T. , & Brisson, D. (2022). Predicting spatio‐temporal population patterns of Borrelia burgdorferi, the Lyme disease pathogen. Journal of Applied Ecology, 59, 2779–2789. 10.1111/1365-2664.14274
Handling Editor Andrew Park
Contributor Information
Tam Tran, Email: mytamtran@gmail.com.
Melissa A. Prusinski, Email: melissa.prusinski@health.ny.gov.
DATA AVAILABILITY STATEMENT
Data available via the Dryad Digital Repository https://doi.org/10.5061/dryad.tqjq2bvzb (Tran et al., 2022). Relevant R code and tick collection data that were previously published can be found at MendeleyData (doi: https://doi.org/10.17632/rtd52gnbyy.1) with further detail in Supplementary Materials in (Tran et al., 2020).
REFERENCES
- Aguirre, A. A. , & Tabor, G. M. (2008). Global factors driving emerging infectious diseases. Annals of the New York Academy of Sciences, 1149, 1–3. 10.1196/annals.1428.052 [DOI] [PubMed] [Google Scholar]
- Allan, B. F. , Keesing, F. , & Ostfeld, R. S. (2003). Effect of forest fragmentation on Lyme disease risk. Conservation Biology, 17(1), 267–272. 10.1046/j.1523-1739.2003.01260.x [DOI] [Google Scholar]
- Aluwong, T. , & Bello, M. (2010). Emerging diseases and implications for millennium development goals in Africa by 2015—An overview. Veterinaria Italiana, 46(2), 137–145. [PubMed] [Google Scholar]
- Brisson, D. , Dykhuizen, D. E. , & Ostfeld, R. S. (2008). Conspicuous impacts of inconspicuous hosts on the Lyme disease epidemic. Proceedings of the Royal Society B: Biological Sciences, 275(1631), 227–235. 10.1098/rspb.2007.1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein, J. S. , Skelly, D. K. , Holford, T. R. , & Fish, D. (2005). Forest fragmentation predicts local scale heterogeneity of Lyme disease risk. Oecologia, 146(3), 469–475. 10.1007/s00442-005-0251-9 [DOI] [PubMed] [Google Scholar]
- Bunnell, J. E. , Price, S. D. , Das, A. , Shields, T. M. , & Glass, G. E. (2003). Geographic information systems and spatial analysis of adult Ixodes scapularis (Acari: Ixodidae) in the middle Atlantic region of the USA. Journal of Medical Entomology, 40(4), 570–576. 10.1603/0022-2585-40.4.570 [DOI] [PubMed] [Google Scholar]
- Burtis, J. C. , Sullivan, P. , Levi, T. , Oggenfuss, K. , Fahey, T. J. , & Ostfeld, R. S. (2016). The impact of temperature and precipitation on blacklegged tick activity and Lyme disease incidence in endemic and emerging regions. Parasites & Vectors, 9(1), 606. 10.1186/s13071-016-1894-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . (2019). Lyme disease surveillance and available data | CDC. Centers for Disease Control and Prevention. Retrieved from https://www.cdc.gov/lyme/stats/survfaq.html [Google Scholar]
- CDC . (2020). Transmission of Lyme disease | CDC. Centers for Disease Control and Prevention. Retrieved from https://www.cdc.gov/lyme/transmission/index.html [Google Scholar]
- CDC . (2021). Lyme disease data and surveillance | CDC. Centers for Disease Control and Prevention. Retrieved from https://www.cdc.gov/lyme/datasurveillance/index.html [Google Scholar]
- Chaves, L. F. , & Koenraadt, C. J. M. (2010). Climate change and highland malaria: Fresh air for a hot debate. The Quarterly Review of Biology, 85(1), 27–55. 10.1086/650284 [DOI] [PubMed] [Google Scholar]
- Chowdhury, M. Z. I. , & Turin, T. C. (2020). Variable selection strategies and its importance in clinical prediction modelling. Family Medicine and Community Health, 8(1), e000262. 10.1136/fmch-2019-000262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cord, A. , & Rödder, D. (2011). Inclusion of habitat availability in species distribution models through multi‐temporal remote‐sensing data? Ecological Applications, 21(8), 3285–3298. 10.1890/11-0114.1 [DOI] [Google Scholar]
- Diuk‐Wasser, M. A. , Hoen, A. G. , Cislo, P. , Brinkerhoff, R. , Hamer, S. A. , Rowland, M. , Cortinas, R. , Vourc'h, G. , Melton, F. , Hickling, G. J. , Tsao, J. I. , Bunikis, J. , Barbour, A. G. , Kitron, U. , Piesman, J. , & Fish, D. (2012). Human risk of infection with Borrelia burgdorferi, the Lyme disease agent, in eastern United States. The American Journal of Tropical Medicine and Hygiene, 86(2), 320–327. 10.4269/ajtmh.2012.11-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diuk‐Wasser, M. A. , Vourc'h, G. , Cislo, P. , Hoen, A. G. , Melton, F. , Hamer, S. A. , Rowland, M. , Cortinas, R. , Hickling, G. J. , Tsao, J. I. , Barbour, A. G. , Kitron, U. , Piesman, J. , & Fish, D. (2010). Field and climate‐based model for predicting the density of host‐seeking nymphal Ixodes scapularis, an important vector of tick‐borne disease agents in the eastern United States. Global Ecology and Biogeography, 19(4), 504–514. 10.1111/j.1466-8238.2010.00526.x [DOI] [Google Scholar]
- Eisen, R. J. , Eisen, L. , Ogden, N. H. , & Beard, C. B. (2016). Linkages of weather and climate with Ixodes scapularis and Ixodes pacificus (Acari: Ixodidae), enzootic transmission of Borrelia burgdorferi, and Lyme disease in North America. Journal of Medical Entomology, 53(2), 250–261. 10.1093/jme/tjv199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouque, F. , & Reeder, J. C. (2019). Impact of past and on‐going changes on climate and weather on vector‐borne diseases transmission: A look at the evidence. Infectious Diseases of Poverty, 8(1), 51. 10.1186/s40249-019-0565-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage, K. L. , Burkot, T. R. , Eisen, R. J. , & Hayes, E. B. (2008). Climate and vectorborne diseases. American Journal of Preventive Medicine, 35(5), 436–450. 10.1016/j.amepre.2008.08.030 [DOI] [PubMed] [Google Scholar]
- Guerra, M. , Walker, E. , Jones, C. , Paskewitz, S. , Cortinas, M. R. , Stancil, A. , Beck, L. , Bobo, M. , & Kitron, U. (2002). Predicting the risk of Lyme disease: Habitat suitability for Ixodes scapularis in the north Central United States. Emerging Infectious Diseases, 8(3), 289–297. 10.3201/eid0803.010166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, E. , & Agusto, F. B. (2022). Baptism of fire: Modeling the effects of prescribed fire on Lyme disease. Canadian Journal of Infectious Diseases and Medical Microbiology, 2022, e5300887. 10.1155/2022/5300887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K. S. , Bradley, B. A. , Cord, A. F. , Rocchini, D. , Tuanmu, M.‐N. , Schmidtlein, S. , Turner, W. , Wegmann, M. , & Pettorelli, N. (2015). Will remote sensing shape the next generation of species distribution models? Remote Sensing in Ecology and Conservation, 1(1), 4–18. 10.1002/rse2.7 [DOI] [Google Scholar]
- Hoen, A. G. , Margos, G. , Bent, S. J. , Diuk‐Wasser, M. A. , Barbour, A. , Kurtenbach, K. , & Fish, D. (2009). Phylogeography of Borrelia burgdorferi in the eastern United States reflects multiple independent Lyme disease emergence events. Proceedings of the National Academy of Sciences of the United States of America, 106(35), 15013–15018. 10.1073/pnas.0903810106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, M. C. , Bowman, A. S. , Forbes, K. J. , Lewis, F. , Mcleod, J. E. , & Gilbert, L. (2013). Environmental determinants of Ixodes ricinus ticks and the incidence of Borrelia burgdorferi sensu lato, the agent of Lyme borreliosis, in Scotland. Parasitology, 140(2), 237–246. 10.1017/S003118201200145X [DOI] [PubMed] [Google Scholar]
- Johnson, T. L. , Bjork, J. K. H. , Neitzel, D. F. , Dorr, F. M. , Schiffman, E. K. , & Eisen, R. J. (2016). Habitat suitability model for the distribution of Ixodes scapularis (Acari: Ixodidae) in Minnesota. Journal of Medical Entomology, 53(3), 598–606. 10.1093/jme/tjw008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, K. E. , Patel, N. G. , Levy, M. A. , Storeygard, A. , Balk, D. , Gittleman, J. L. , & Daszak, P. (2008). Global trends in emerging infectious diseases. Nature, 451(7181), 990–993. 10.1038/nature06536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, L. , Kendell, D. , Robertson, D. , Livdahl, T. , & Khatchikian, C. (2010). Aedes aegypti and Aedes albopictus in Bermuda: Extinction, invasion, invasion and extinction. Biological Invasions, 12(9), 3277–3288. 10.1007/s10530-010-9721-z [DOI] [Google Scholar]
- Khatchikian, C. , Sangermano, F. , Kendell, D. , & Livdahl, T. (2011). Evaluation of species distribution model algorithms for fine‐scale container‐breeding mosquito risk prediction. Medical and Veterinary Entomology, 25(3), 268–275. 10.1111/j.1365-2915.2010.00935.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatchikian, C. E. , Prusinski, M. , Stone, M. , Backenson, P. B. , Wang, I.‐N. , Levy, M. Z. , & Brisson, D. (2012). Geographical and environmental factors driving the increase in the Lyme disease vector Ixodes scapularis . Ecosphere, 3(10), 1–18. 10.1890/ES12-00134.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick, A. M. , Dobson, A. D. M. , Levi, T. , Salkeld, D. J. , Swei, A. , Ginsberg, H. S. , Kjemtrup, A. , Padgett, K. A. , Jensen, P. M. , Fish, D. , Ogden, N. H. , & Diuk‐Wasser, M. A. (2017). Lyme disease ecology in a changing world: Consensus, uncertainty and critical gaps for improving control. Philosophical Transactions of the Royal Society B: Biological Sciences, 372(1722), 20160117. 10.1098/rstb.2016.0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitron, U. , & Kazmierczak, J. J. (1997). Spatial analysis of the distribution of Lyme disease in Wisconsin. American Journal of Epidemiology, 145(6), 558–566. 10.1093/oxfordjournals.aje.a009145 [DOI] [PubMed] [Google Scholar]
- Lashley, F. R. (2004). Emerging infectious diseases: Vulnerabilities, contributing factors and approaches. Expert Review of Anti‐Infective Therapy, 2(2), 299–316. 10.1586/14787210.2.2.299 [DOI] [PubMed] [Google Scholar]
- Lever, J. , Krzywinski, M. , & Altman, N. (2016). Model selection and overfitting. Nature Methods, 13(9), 703–704. 10.1038/nmeth.3968 [DOI] [Google Scholar]
- Levi, T. , Kilpatrick, A. M. , Mangel, M. , & Wilmers, C. C. (2012). Deer, predators, and the emergence of Lyme disease. Proceedings of the National Academy of Sciences of the United States of America, 109(27), 10942–10947. 10.1073/pnas.1204536109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linske, M. A. , Stafford, K. C. , Williams, S. C. , Lubelczyk, C. B. , Welch, M. , & Henderson, E. F. (2019). Impacts of deciduous leaf litter and snow presence on nymphal Ixodes scapularis (Acari: Ixodidae) overwintering survival in coastal New England, USA. Insects, 10(8), 227. 10.3390/insects10080227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoGiudice, K. , Ostfeld, R. S. , Schmidt, K. A. , & Keesing, F. (2003). The ecology of infectious disease: Effects of host diversity and community composition on Lyme disease risk. Proceedings of the National Academy of Sciences of the United States of America, 100(2), 567–571. 10.1073/pnas.0233733100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall III, W. F. , Telford III, S. R. , Rys, P. N. , Rutledge, B. J. , Mathiesen, D. , Malawista, S. E. , Spielman, A. , & Persing, D. H. (1994). Detection of Borrelia burgdorferi DNA in museum specimens of Peromyscus. The Journal of Infectious Diseases, 170(4), 1027–1032. 10.1093/infdis/170.4.1027 [DOI] [PubMed] [Google Scholar]
- Medlock, J. M. , Hansford, K. M. , Bormane, A. , Derdakova, M. , Estrada‐Peña, A. , George, J.‐C. , Golovljova, I. , Jaenson, T. G. T. , Jensen, J.‐K. , Jensen, P. M. , Kazimirova, M. , Oteo, J. A. , Papa, A. , Pfister, K. , Plantard, O. , Randolph, S. E. , Rizzoli, A. , Santos‐Silva, M. M. , Sprong, H. , … Van Bortel, W. (2013). Driving forces for changes in geographical distribution of Ixodes ricinus ticks in Europe. Parasites & Vectors, 6(1), 1. 10.1186/1756-3305-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- New York State . (2014). NYS GIS clearinghouse . Data. Retrieved from https://gis.ny.gov/
- Nicholson, M. C. , & Mather, T. N. (1996). Methods for evaluating Lyme disease risks using geographic information systems and geospatial analysis. Journal of Medical Entomology, 33(5), 711–720. 10.1093/jmedent/33.5.711 [DOI] [PubMed] [Google Scholar]
- NYS Department of Environmental Conservation . (2014). History of state forest program. Department of Environmental Conservation. Retrieved from https://www.dec.ny.gov/lands/4982.html [Google Scholar]
- NYS Department of Environmental Conservation . (2021). Deer and bear harvests . Retrieved from https://www.dec.ny.gov/outdoor/42232.html
- O'Connor, C. , Prusinski, M. A. , Jiang, S. , Russell, A. , White, J. , Falco, R. , Kokas, J. , Vinci, V. , Gall, W. , Tober, K. , Haight, J. , Oliver, J. , Meehan, L. , Sporn, L. A. , Brisson, D. , & Backenson, P. B. (2021). A comparative spatial and climate analysis of human granulocytic anaplasmosis and human babesiosis in New York state (2013–2018). Journal of Medical Entomology, 58(6), 2453–2466. 10.1093/jme/tjab107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden, N. H. , & Tsao, J. I. (2009). Biodiversity and Lyme disease: Dilution or amplification? Epidemics, 1(3), 196–206. 10.1016/j.epidem.2009.06.002 [DOI] [PubMed] [Google Scholar]
- Ostfeld, R. S. , Canham, C. D. , Oggenfuss, K. , Winchcombe, R. J. , & Keesing, F. (2006). Climate, deer, rodents, and acorns as determinants of variation in Lyme‐disease risk. PLoS Biology, 4(6), e145. 10.1371/journal.pbio.0040145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patz, J. A. , Olson, S. H. , Uejio, C. K. , & Gibbs, H. K. (2008). Disease emergence from global climate and land use change. Medical Clinics of North America, 92(6), 1473–1491. 10.1016/j.mcna.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Petrosillo, N. (2019). Emerging infections and future threats. Erciyes Medical Journal, 41(2), 130–135. 10.14744/etd.2019.57805 [DOI] [Google Scholar]
- Pickett, S. T. A. , & Cadenasso, M. L. (1995). Landscape ecology: Spatial heterogeneity in ecological systems. Science, 269(5222), 331–334. 10.1126/science.269.5222.331 [DOI] [PubMed] [Google Scholar]
- Prism Climate Group . (2018). Prism climate data. Oregon State University. Retrieved from http://www.prism.oregonstate.edu/ [Google Scholar]
- Prusinski, M. A. , Kokas, J. E. , Hukey, K. T. , Kogut, S. J. , Lee, J. , & Backenson, P. B. (2014). Prevalence of Borrelia burgdorferi (Spirochaetales: Spirochaetaceae), Anaplasma phagocytophilum (Rickettsiales: Anaplasmataceae), and Babesia microti (Piroplasmida: Babesiidae) in Ixodes scapularis (Acari: Ixodidae) collected from recreational lands in the Hudson Valley Region, New York State. Journal of Medical Entomology, 51, 226–236. 10.1603/me13101 [DOI] [PubMed] [Google Scholar]
- Randolph, S. E. , & Dobson, A. D. M. (2012). Pangloss revisited: A critique of the dilution effect and the biodiversity‐buffers‐disease paradigm. Parasitology, 139(7), 847–863. 10.1017/S0031182012000200 [DOI] [PubMed] [Google Scholar]
- Roberts, S. L. , Kelt, D. A. , van Wagtendonk, J. W. , Miles, A. K. , & Meyer, M. D. (2015). Effects of fire on small mammal communities in frequent‐fire forests in California. Journal of Mammalogy, 96(1), 107–119. 10.1093/jmammal/gyu011 [DOI] [Google Scholar]
- Schauber, E. M. , Ostfeld, R. S. , & Jr, A. S. E. (2005). What is the best predictor of annual Lyme disease incidence: Weather, mice, or acorns? Ecological Applications, 15(2), 575–586. 10.1890/03-5370 [DOI] [Google Scholar]
- Simon, J. A. , Marrotte, R. R. , Desrosiers, N. , Fiset, J. , Gaitan, J. , Gonzalez, A. , Koffi, J. K. , Lapointe, F.‐J. , Leighton, P. A. , Lindsay, L. R. , Logan, T. , Milord, F. , Ogden, N. H. , Rogic, A. , Roy‐Dufresne, E. , Suter, D. , Tessier, N. , & Millien, V. (2014). Climate change and habitat fragmentation drive the occurrence of Borrelia burgdorferi, the agent of Lyme disease, at the northeastern limit of its distribution. Evolutionary Applications, 7(7), 750–764. 10.1111/eva.12165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatculescu, A. M. , Clow, K. M. , McKay, R. , Talbot, B. , Logan, J. J. , Thickstun, C. R. , Jardine, C. M. , Ogden, N. H. , Knudby, A. J. , & Kulkarni, M. A. (2020). Species distribution models for the eastern blacklegged tick, Ixodes scapularis, and the Lyme disease pathogen, Borrelia burgdorferi, in Ontario, Canada. PLoS ONE, 15(9), e0238126. 10.1371/journal.pone.0238126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman, J. M. , Howell, J. E. , Matthew Knox, W. , & Stenger, P. J. (2009). Incidence of hemorrhagic disease in white‐tailed deer is associated with winter and summer climatic conditions. EcoHealth, 6(1), 11–15. 10.1007/s10393-009-0220-6 [DOI] [PubMed] [Google Scholar]
- Stein, A. , Gerstner, K. , & Kreft, H. (2014). Environmental heterogeneity as a universal driver of species richness across taxa, biomes and spatial scales. Ecology Letters, 17(7), 866–880. 10.1111/ele.12277 [DOI] [PubMed] [Google Scholar]
- Suzán, G. , Marcé, E. , Giermakowski, J. T. , Armién, B. , Pascale, J. , Mills, J. , Ceballos, G. , Gómez, A. , Aguirre, A. A. , Salazar‐Bravo, J. , Armién, A. , Parmenter, R. , & Yates, T. (2008). The effect of habitat fragmentation and species diversity loss on hantavirus prevalence in Panama. Annals of the New York Academy of Sciences, 1149(1), 80–83. 10.1196/annals.1428.063 [DOI] [PubMed] [Google Scholar]
- Talbot, B. , Slatculescu, A. , Thickstun, C. R. , Koffi, J. K. , Leighton, P. A. , McKay, R. , & Kulkarni, M. A. (2019). Landscape determinants of density of blacklegged ticks, vectors of Lyme disease, at the northern edge of their distribution in Canada. Scientific Reports, 9(1), 16652. 10.1038/s41598-019-50858-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, T. , Prusinski, M. A. , White, J. L. , Falco, R. C. , Kokas, J. , Vinci, V. , Gall, W. K. , Tober, K. J. , Haight, J. , Oliver, J. , Sporn, L. A. , Meehan, L. , Banker, E. , Backenson, P. B. , Jensen, S. T. , & Brisson, D. (2022). Data from: Predicting spatio‐temporal population patterns of Borrelia burgdorferi, the Lyme disease pathogen. Dryad Digital Repository, 10.5061/dryad.tqjq2bvzb [DOI] [PMC free article] [PubMed]
- Tran, T. , Prusinski, M. A. , White, J. L. , Falco, R. C. , Vinci, V. , Gall, W. K. , Tober, K. , Oliver, J. , Sporn, L. A. , Meehan, L. , Banker, E. , Backenson, P. B. , Jensen, S. T. , & Brisson, D. (2020). Spatio‐temporal variation in environmental features predicts the distribution and abundance of Ixodes scapularis . International Journal for Parasitology, 51, 311–320. 10.1016/j.ijpara.2020.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Census Bureau . (2022). Data . Census.Gov. Retrieved from https://www.census.gov/data
- US EPA . (2015). Level III and IV ecoregions of the continental United States [data and tools]. EPA United States Environmental Protection Agency. Retrieved from https://www.epa.gov/eco‐research/level‐iii‐and‐iv‐ecoregions‐continental‐united‐states [Google Scholar]
- USGCRP . (2018). Fourth National Climate Assessment. U.S. Global Change Research Program. Retrieved from https://nca2018.globalchange.govhttps://nca2018.globalchange.gov/chapter/18 [Google Scholar]
- USGS . (2018a). North American Breeding Bird Survey. BBS—USGS Patuxent Wildlife Research Center. Retrieved from https://www.pwrc.usgs.gov/bbs/ [Google Scholar]
- USGS . (2018b). National Land Cover Database . Retrieved from https://www.usgs.gov/centers/eros/science/national‐land‐cover‐database?qt‐science_center_objects=0#qt‐science_center_objects
- USGS . (2022). The National Map. National Geospatial Program. Retrieved from https://www.usgs.gov/core‐science‐systems/national‐geospatial‐program/national‐map [Google Scholar]
- Wenger, S. J. , & Olden, J. D. (2012). Assessing transferability of ecological models: An underappreciated aspect of statistical validation. Methods in Ecology and Evolution, 3(2), 260–267. 10.1111/j.2041-210X.2011.00170.x [DOI] [Google Scholar]
- Wiens, J. A. , Stenseth, N. C. , Van Horne, B. , & Ims, R. A. (1993). Ecological mechanisms and landscape ecology. Oikos, 66(3), 369–380. 10.2307/3544931 [DOI] [Google Scholar]
- Wood, C. L. , & Lafferty, K. D. (2013). Biodiversity and disease: A synthesis of ecological perspectives on Lyme disease transmission. Trends in Ecology & Evolution, 28(4), 239–247. 10.1016/j.tree.2012.10.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Figure S3
Table S1
Data Availability Statement
Data available via the Dryad Digital Repository https://doi.org/10.5061/dryad.tqjq2bvzb (Tran et al., 2022). Relevant R code and tick collection data that were previously published can be found at MendeleyData (doi: https://doi.org/10.17632/rtd52gnbyy.1) with further detail in Supplementary Materials in (Tran et al., 2020).


