Abstract
Genetically based natural resistance to brucellosis in cattle provides for novel strategies to control zoonotic diseases. Bovine NRAMP1, the homologue of a murine gene (Bcg), has been identified as a major candidate for controlling the in vivo resistant phenotype. We developed an in vitro model for expression of resistance- and susceptibility-associated alleles of bovine NRAMP1 as stable transgenes under the regulatory control of the bovine NRAMP1 promoter in the murine RAW264.7 macrophage cell line (Bcgs) to analyze the regulation of the NRAMP1 gene and its role in macrophage function. We demonstrated that the 5′-flanking region of bovine NRAMP1, despite the lack of TATA and CAAT boxes, has a functional promoter capable of driving the expression of a transgene in murine macrophages. A polymorphism within a microsatellite in the 3′ untranslated region critically affects the expression of bovine NRAMP1 and the control of in vitro replication of Brucella abortus but not Salmonella enterica serovar Dublin. We did not observe any differences in the production of NO by resting or gamma interferon (IFN-γ)- and IFN-γ–lipopolysaccharide (LPS)-treated transfected cell lines, yet the resistant transfected cell lines produced significantly less NO than other cell lines, following stimulation with LPS at 24 and 48 h.
Losses attributable to infectious diseases continue to impede the livestock industries, despite widespread application of antimicrobials, vaccination, quarantine, and other traditional disease control measures. An alternative approach to the control of zoonotic infections in cattle is genetic disease resistance, which is the inherent capacity of a previously unexposed animal to resist a virulent challenge (1,34). Genetic studies with mice have demonstrated that a gene called natural resistance-associated macrophage protein gene 1 (Nramp1, formerly the Lsh/Ity/Bcg gene) controls innate resistance and susceptibility to Mycobacterium bovis BCG, Leishmania donovani, Salmonella enterica serovar Typhimurium, and several atypical mycobacteria (D. J. Bradley, Letter, Nature 250:353–354, 1974; 23, 27, 48). The Nramp1 gene mediates activity of macrophages against intracellular parasites during the early stages of infection (6).
In cattle, natural resistance to Brucella abortus is heritable, and its frequency (20% of cross-bred cattle) can be increased dramatically in one generation of selective breeding; the genetic analysis of classical breeding studies is consistent with two or more genes controlling the resistant phenotype (58). Extensive similarities between bovine macrophage function and resistance to brucellosis and murine macrophage function and resistance to Salmonella, Mycobacterium, and Leishmania have been documented (9, 33, 49, 57). Specifically, macrophages from cattle that are genetically resistant to a stringent in vivo challenge with virulent B. abortus S2308 are superior in their capacity to control the intracellular replication of not only B. abortus but also Salmonella enterica serovar Dublin and M. bovis BCG (52). The bovine homologue to the murine Nramp1 gene (designated bovine NRAMP1) was cloned, and the cDNA was sequenced (21). It is predicted to encode a protein that has 12 transmembrane segments with one hydrophilic N-terminal region containing an SH3-binding motif located at the cytoplasmic surface. The gene product contains one potential N-linked glycosylation site and four protein kinase C phosphorylation sites on serine. A 20-amino-acid transport motif is located between the predicted transmembrane domains 8 and 9 and is conserved in murine and human Nramp1. Comparison of human and murine Nramp1 and bovine NRAMP1 predicted protein sequences indicates a remarkable degree of homology (86.9% identical residues between murine and bovine; 88.6% identical residues between human and bovine). Bovine NRAMP1 is expressed primarily in macrophages of the reticuloendothelial system. The point mutation within the coding region of murine Nramp1 which distinguishes the resistant and susceptible alleles of the mouse Bcg gene (59) has never been documented in cattle or other mammalian species in which the Nramp1 homologue has been sequenced. However, in cattle, single-stranded conformational analysis (ssCA) disclosed a highly significant (P = 0.0089) association of a polymorphic (GT)n microsatellite (where n is 13, 14, 15, or 16) located in the 3′ untranslated region (3′UTR) of bovine NRAMP1 with cattle naturally resistant to brucellosis (1). Of the original 11 in vivo-resistant cattle tested by ssCA, 9 were homozygous for the (GT)13 allele while 2 had the (GT)13-(GT)14 genotype. By contrast, of the original 11 in vivo-susceptible cattle tested, only 2 were homozygous for the (GT)13 allele while the others had at least one (GT)14, (GT)15, or (GT)16 allele. These data support the hypothesis that bovine NRAMP1 is one of the major genes determining resistance and susceptibility to brucellosis.
Considering the multigenic determination of resistance and/or susceptibility to bovine brucellosis, we designed an experiment that would isolate the bovine NRAMP1 gene from the overall genetic background of cattle. The purpose of this study was to transfect the resistance-associated allele and one of the susceptibility-associated alleles of bovine NRAMP1 into the murine RAW264.7 (Bcgs) macrophage cell line, their expression being driven by the bovine NRAMP1 promoter. Stable transfectants were analyzed for their expression of the bovine transgene and their capacity to control the intracellular replication of Salmonella serovar Dublin and B. abortus, prototype intracellular pathogens, as well as their ability to produce nitric oxide (NO) following stimulation with lipopolysaccharide (LPS) and gamma interferon (IFN-γ).
MATERIALS AND METHODS
Parental cell line.
RAW264.7 cells, a mouse macrophage (Bcgs) cell line established from the ascites of a tumor induced in a male BALB/c mouse by the intraperitoneal injection of Abelson murine leukemia virus, were purchased from the American Type Culture Collection (Rockville, Md.). The cells were grown in RPMI 1640 (Life Technologies, Gaithersburg, Md.) supplemented with 10% fetal calf serum (HyClone Laboratories, Logan, Utah) in 25-cm2 culture flasks (Corning Costar Corporation, Cambridge, Mass.).
Transformation protocol.
Twenty-four hours prior to electroporation (56), the cells were split 1:6 in fresh culture medium. Five million cells were added to 10 μg of linearized plasmid DNA in a total volume of 50 μl of phosphate-buffered saline and transferred to a 0.4-cm gap electroporation cuvette (BTX, San Diego, Calif.). The cuvettes were left to equilibrate at room temperature for 5 min before electroporation at 750 V/cm, 1,050 μF, and 129 Ω in an ECM 600 electroporation system (BTX). Dilutions of the transfected cells were seeded in 90-mm petri dishes and allowed to recover for 48 h when selection was applied with G418 (Life Technologies) at a concentration of 400 μg/ml. Approximately 10-day-old colonies were isolated with cloning rings (Bellco Glass, Inc., Vineland, N.J.) and expanded.
Isolation of the bovine NRAMP1 promoter.
A bovine bacterial artificial chromosome (BAC) library prepared from a purebred Angus bull in the pBeloBac 11 vector (8) was kindly provided by Scott Davis, Department of Animal Science, Texas A&M University. It was screened using a PCR-based strategy with a primer pair designed from the third exon of bovine NRAMP1. Positive clones were digested with NotI and run on a 1% agarose gel to document the size of the insert. Then, a Southern blot prepared with DNA from positive clones digested with EcoRI, HindIII, and EcoRV was hybridized with a probe homologous to the third exon of bovine NRAMP1. The same blot was also hybridized with a 1.2-kb probe, which is composed of the second exon and intron and the third exon, to identify a digestion fragment containing the 5′-flanking region of bovine NRAMP1. A 2.6-kb HindIII fragment was detected and subcloned into pBluescript, and both strands of the DNA were sequenced by the dideoxy method (53).
Construction of pΔ3Bo-GFP.
All restriction enzymes were purchased from Promega (Madison, Wis.) or Boehringer Mannheim (Indianapolis, Ind.). All plasmids were transfected into chemically competent E. coli (Stratagene, La Jolla, Calif.), and colonies were screened by blue-white phenotyping and/or restriction digestion; mini- and maxipreparations of plasmid DNA were performed using commercially available kits (Promega; Qiagen, Chatsworth, Calif.). Gel purifications of DNA were performed using the QIAEX II Gel Extraction Kit (Qiagen).
An expression vector carrying the green fluorescent protein (GFP) reporter gene under the control of the bovine NRAMP1 promoter (pΔ3Bo-GFP) was constructed in pcDNA3 (Invitrogen, San Diego, Calif.). A 734-bp cassette containing the humanized GFP locus was excised from pGreen Lantern (Life Technologies). The 2,138-bp fragment of the bovine NRAMP1 promoter region used to construct all expression vectors in this study contained 1,484 bp 5′ upstream of the transcription start site as well as the first intron. The construct encoding a fusion protein between the 5′ end of bovine NRAMP1 and GFP was cloned into pcDNA3 in which the endogenous viral promoter (pCMV) had been deleted.
Construction of pΔ3Bo-R.
An expression vector carrying the resistance-associated allele of the bovine NRAMP1 gene under the control of the bovine NRAMP1 promoter (pΔ3Bo-R) was constructed in pcDNA3. The cDNA encoding the resistance-associated allele of the bovine NRAMP1 gene was cloned downstream of the bovine NRAMP1 promoter fragment; the resulting construct was inserted into pcDNA3 in which the pCMV promoter had been deleted.
Construction of pΔ3Bo-S16 by site-directed mutagenesis.
The (GT)13 microsatellite in the 3′UTR of the resistance-associated allele of the bovine NRAMP1 gene was mutated into a (GT)16 repeat by site-directed mutagenesis (18). The entire cDNA encoding the resistance-associated allele of NRAMP1 was subcloned into pT7 Blue (Novagen, Madison, Wis.). A 5′-phosphorylated selection primer was designed from the pT7 Blue sequence so that it would convert the unique XmnI site into an EcoRV site (Select-P: 5′ CCCGAAGAACGATATC CAATGATGAGC 3′). On the same strand of the plasmid template, a 5′-phosphorylated mutagenic primer (Mut-P: 5′ CCCTCTCCGTCTTGCTGTGCATGCACACACACACACACA CACACACACACACACACCCTTGTCTGGCAGGCCAGTG 3′) was designed to insert three additional GT repeats in the (GT)13 microsatellite. One hundred ng of plasmid DNA was denatured at 100°C for 3 min in the presence of 100 ng of both the selection and mutagenic primers in a total volume of 20 μl containing 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 50 mM NaCl; the tube was then chilled on ice for 5 min and centrifuged briefly to collect condensation. The mutant DNA strand was synthesized with 2 to 4 U of T4 DNA polymerase (Clontech, Palo Alto, Calif.) and 4 to 6 U of T4 DNA ligase (Novagen) at 37°C for 2 h in a total volume of 30 μl containing a nucleotide mixture at optimum concentrations; the enzymes were inactivated at 75°C for 10 min, and the reaction mixture was cooled to room temperature. The NaCl concentration in the sample was then adjusted in order to get complete digestion with 20 U of XmnI at 37°C for 2 h. Following the digestion, an aliquot of the sample was introduced into chemically competent BMH71-18 mutS Escherichia coli, a DNA mismatch repair-deficient mutant (Clontech). The transformed bacteria were incubated for 1 h at 37°C in 1 ml of Luria broth and then grown overnight in a total volume of 5 ml of Luria broth containing 100 μg of ampicillin/ml. DNA from the mixed plasmid pool was extracted using the Wizard Plus Minipreps DNA purification system (Promega). One hundred nanograms of pool plasmid DNA was digested with 10 U of XmnI for 2 h at 37°C, and after the addition of another 10 U of XmnI, the sample was incubated for an additional 1 h at 37°C. For the final transformation, an aliquot of the reaction mixture was introduced into chemically competent E. coli. Several colonies were grown overnight for plasmid DNA extraction; the plasmid DNA was digested in two separate reactions with XmnI and EcoRV and analyzed on an agarose gel. Clones that had lost the unique XmnI site and acquired an additional EcoRV site were submitted for automated sequencing to verity the mutated sequence. The plasmid insert from several pooled, mutated clones was then substituted for the corresponding fragment in pΔ3Bo-R to create pΔ3Bo-S16.
RNase protection assays.
Plasmid templates used for the generation of antisense RNA probes were purified with the Qiagen Plasmid Maxi kit and linearized with the appropriate restriction enzyme. Following two consecutive phenol:chloroform:isoamyl alcohol extractions and an ethanol precipitation, the pellets were washed with 75% ethanol, air dried, and resuspended in RNase-free distilled water. High-specific-activity, radiolabeled antisense RNA probes were produced using the MAXIscript in vitro transcription kit (Ambion, Austin, Tex.) and gel purified. The bovine NRAMP1-R and -S RNase protection assay (RPA) probes were designed from the 3′UTR of the gene and had only 47% homology with the corresponding fragment in the murine Nramp1 gene. Total RNA was extracted from 107 cultured cells using the RNAqueous phenol-free total RNA isolation kit (Ambion), quantitated at A260, aliquoted, and stored at −80°C. RPAs were carried out using the RPA II RNase Protection Assay kit (Ambion), using 15 μg of total RNA and following the recommendations of the manufacturer. The protected fragments were displayed on 5% polyacrylamide–8 M urea gels that were dried and exposed to X-ray film at −80°C with intensifying screens.
In vitro killing assays.
In vitro macrophage killing assays were performed essentially as described (50), using virulent strains of B. abortus and Salmonella serovar Dublin. The day prior to the assay, all cells were washed in G418-free medium, harvested by scraping, counted, pelleted, and resuspended in fresh G418-free medium. Ten thousand cells were then added to each of three wells of two tissue culture 60-well Terasaki HLA plates (Nunc, Inc., Naperville, Ill.). One plate was used for harvesting bacteria at the starting time (T0) while the other was used at the end of the assay (T6 and T12, respectively, for Salmonella serovar Dublin and B. abortus). The plates were centrifuged at 750 × g for 5 min and incubated at 37°C for at least 18 h. Aliquots of each bacterial stock were stored at −80°C and used only once. The day of the assay, serial dilutions of the inoculum were plated to verify the viability of the bacteria and the concentration of the inoculum. The monolayers of macrophages were then challenged with bacteria at different multiplicities of infection (MOIs); the time allowed for phagocytosis was 30 min for Salmonella and 2 h for Brucella. Extracellular bacteria were then killed with gentamicin at 25 μg/ml for 1 h at 37°C. After three washes were carried out, the bacteria were harvested from one of the plates for bacterial counting at the start of the assay. In the other plate, the cells were washed, refed fresh medium, and incubated at 37°C. At the end of the assay, the bacteria were harvested, and serial dilutions were plated in duplicate. The cells were visually inspected at regular time points during the infection. As controls, bacteria were grown in triplicate wells without macrophages to assess their viability; also, each assay included a control to verify the activity of the antibiotic. Bacterial survival was calculated as the ratio of the total number of viable intracellular bacteria at the end of the assay to the total number of viable intracellular bacteria at the start of the assay. Each killing assay included the parental cells as a reference standard.
Measurement of NO production.
The cell lines tested for the production of NO included one each of the bovine NRAMP1-R and -S16 transgenic lines, a vector-transfected line, and the parental cells. The cells were seeded in 96-well microtiter plates in triplicate (105 cells/well) and left untreated or treated with murine recombinant IFN-γ (1 U/ml; R&D Systems, Minneapolis, Minn.), LPS from E. coli (5 ng/ml; Sigma, St. Louis, Mo.), or IFN-γ (1 U/ml) plus LPS (5 ng/ml). Each experiment also included control cells that were pretreated with NG-monomethyl-l-arginine (300 μM; Sigma) and then stimulated with IFN-γ and LPS. After 24 and 48 h, the amount of NO2− in the culture supernatants was measured using the Greiss reagent; for each cell line, the total concentration of NO2− was measured from the mean absorbance at 550 nm of triplicate samples. Three independent experiments were performed.
Nucleotide sequence accession number.
The complete sequence of bovine NRAMP1 is available in GenBank under accession number 321467.
RESULTS
We have produced stable transfectants of RAW264.7 cells, a murine macrophage cell line expressing the resistance- and susceptibility-associated alleles of the bovine NRAMP1 gene under the control of the bovine NRAMP1 promoter. We have isolated two plasmid clones containing the genomic sequence of bovine NRAMP1 from a BAC library. The clones both contained 90-kb inserts and displayed identical restriction patterns, indicating that these independent clones are in fact identical. PCR analysis using primer sets from both the 5′ and 3′ regions of the bovine NRAMP1 cDNA indicated that the inserts span the entire NRAMP1 genomic sequence. A 2.6-kb HindIII restriction fragment from one of the clones was subcloned and sequenced; this fragment contained 1,484 bp of sequence upstream of the transcription start site (Fig. 1). The analysis of this 1,484-bp fragment indicated that there were no consensus TATA or CAAT boxes, but there are two striking features of a functional promoter including the following: (i) enrichment in G+C dinucleotides with about 66% overall G+C content upstream of the ATG start codon (within 140 bp of nucleotide position −87 to +53) and (ii) the presence of a number of consensus transcription factor binding sites. In addition, a wide variety of potential transcription factor consensus sequences were found located in the first and second intron. Comparison of the sequence upstream of the transcription start site between human and murine Nramp1 and bovine NRAMP1 indicated a high degree of homology between positions −80 and −257 of the 5′ promoter region, with 58% and 68% nucleotide identity between bovine and murine and bovine and human genes, respectively. Of particular interest is the fact that several well-documented transcription factor consensus motifs are precisely conserved in this segment, including binding sites for interferon response element (IRE), NF–interleukin-6, PU.1, polyomavirus enhancer A-site 3, the W element, and NF-κB.
FIG. 1.
Sequence of the 5′-flanking region, exon 1, intron 1, exon 2, intron 2, and part of exon 3 of bovine NRAMP1. The transcription start site in exon 1 is shown by the first capital boldface G with +1. The putative binding sequence motifs for transcription factors are underlined with the name of the corresponding factor shown. The exon sequences are indicated by boldface uppercase letters. The complete sequence is available in GenBank under the temporary accession number bankit321467.
In a preliminary experiment, we tested the capacity of a 2.2-kb fragment of the bovine NRAMP1 promoter region containing 1,484 bp 5′ upstream of the transcription start site as well as the first intron to drive the expression of a transgene in murine macrophages. For this purpose, we constructed a fusion protein between the 5′ end of the bovine NRAMP1 gene and a humanized clone of GFP, both under the regulatory influence of the bovine NRAMP1 promoter construct (pΔ3Bo-GFP). Very weak GFP activity was observed in several stably transfected RAW264.7 clones under UV illumination. Four of them were harvested and expanded, and the presence or absence of expression of GFP mRNA was determined by RPA (Fig. 2). The message for GFP was easily detectable in all four clones with visual GFP activity.
FIG. 2.
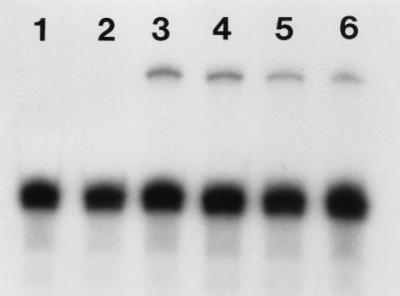
RPA analysis for expression of GFP under the control of the bovine NRAMP1 promoter in pΔ3Bo-GFP transfectants of RAW264.7 cells. The message for GFP is easily detectable in all four clones with visual GFP activity (lanes 3 through 6); no signal for GFP is obtained with the parental cells (lanes 1 and 2). The lower dominant band is β-actin. The respective sizes of the protected fragments are 308 (GFP) and 250 (β-actin; positive control) bases. Exposure is 3 h.
Next, RAW264.7 cells were transfected with eukaryotic expression vectors in which the expression of the resistance- and susceptibility-associated alleles of bovine NRAMP1 is driven by the same bovine NRAMP1 promoter region (pΔ3Bo-R and pΔ3Bo-S16, respectively). Sixteen stably transfected clones of each region were harvested and expanded. The expression of the transgene was determined by RPA in eight randomly selected clones. When we tested eight NRAMP1-R stable transfectants by RPA (Fig. 3), five of the eight clones tested displayed a readily detectable signal for bovine NRAMP1 after a short exposure; after an overnight exposure, all eight clones were positive, albeit expressing various levels of the transgene. Three clones (R2, R5, and R6) expressing the highest levels of the transgene were selected for further analysis.
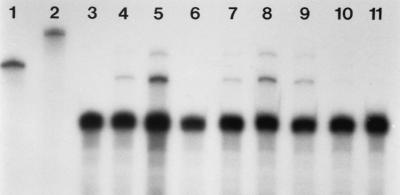
FIG. 3.
RPA analysis for expression of bovine NRAMP1-R in stable RAW264.7 transfectants. Lane 3, parental RAW264.7 cells; lanes 4 through 11, stable bovine NRAMP1-R transfected clones. A protected fragment of the expected size is observed for the clones in lanes 4, 5, 7, 8, and 9; no signal is obtained with the parental cells (lane 3). The presence of a larger protected fragment can be attributed to the presence of secondary structure in the antisense probe. The lower dominant band is β-actin. The respective sizes of the protected fragments are 289 (NRAMP1-R) and 250 (β-actin; positive control) bases. Full-length antisense probes for β-actin and NRAMP1-R are shown in lanes 1 and 2. Exposure is 3 h.
The construct encoding the susceptibility-associated allele (pΔ3Bo-S16) was derived from pΔ3Bo-R by site-directed mutagenesis. We chose to use the (GT)16 variant of the microsatellite because (i) in a previous analysis of the association between in vivo resistance and susceptibility of outbred and unrelated cattle to experimentally induced brucellosis and the documented polymorphism within the polymorphic microsatellite in the bovine NRAMP1 3′UTR, 4 of 11 susceptible cattle had the (GT)16 allele while none of the resistant cattle displayed this allele and (ii) the (GT)16 allele is the longest polymorphic variant of this microsatellite that has been documented in cattle to date. The resulting construct (pΔ3Bo-S16) was used to transfect RAW264.7 cells, and the expression of the transgene was determined by RPA in eight randomly selected clones; because of the low level of expression of the transgene, the RPA was repeated on those clones expressing the highest levels of NRAMP1-S16 (Fig. 4). The level of expression of the bovine NRAMP1-S16 gene was three- to sixfold lower than that observed in the NRAMP1-R clones, thus suggesting a critical role for the polymorphic microsatellite in determining the level of steady-state mRNA accumulation of NRAMP1. The three clones expressing the highest levels of bovine NRAMP1-S16 (S5, S6, and S7) were selected for further analysis.
FIG. 4.
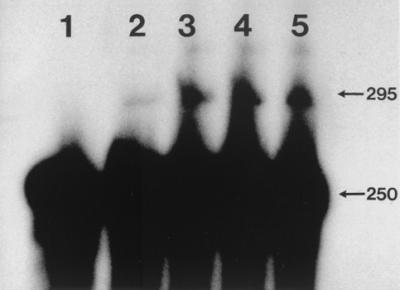
RPA analysis for expression of bovine NRAMP1-S16 in stable RAW264.7 transfectants (lanes 2 through 5). The protected fragments for actin (250 bases) and bovine NRAMP1-S16 (295 bases) are indicated. Lane 1, parental cell line. Exposure is 12 h.
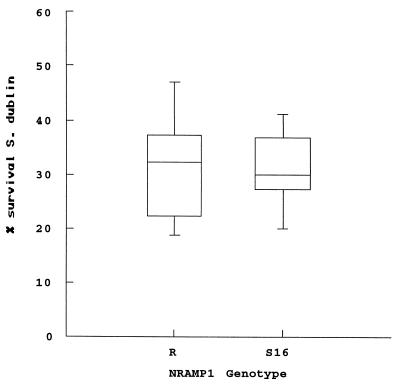
We then used highly standardized in vitro killing assays to assess the capacity of the parental cells and the transgenic lines to control the intracellular replication of a highly virulent strain of Salmonella serovar Dublin. The cells were infected with various MOIs (5, 10, and 20), and bacterial survival was determined after 6 h. The results indicate that (i) the parental cells are permissive for Salmonella serovar Dublin replication and (ii) the NRAMP1-R and NRAMP1-S16 clones, while superior to the parental cells in their capacity to control Salmonella serovar Dublin, restrict the intracellular growth of Salmonella serovar Dublin equally well (Fig. 5). At all MOIs used, we did not observe a significant difference in the uptake of Salmonella between the parental and transfected cell lines.
FIG. 5.
Survival of Salmonella serovar Dublin in three NRAMP1-R- or NRAMP1-S16-transfected clones of the murine RAW264.7 macrophage cell line. Bacterial survival is expressed as a percentage of survival in the parental RAW264.7 cells. The box plot shows the median and quartiles from two independent experiments; in each experiment, the cell lines were assayed in triplicate, and the survival rates were pooled for the three clones from each genotype. The cells were challenged at a MOI of 20. There is no significant difference in survival rates between the three R and the three S16 clones.
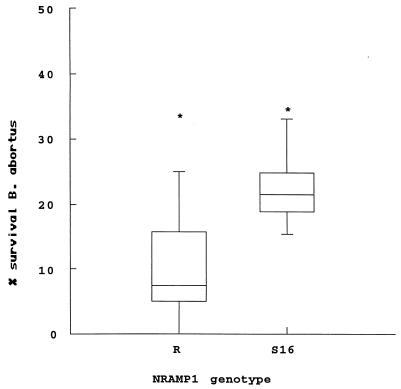
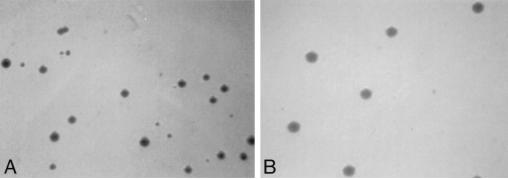
Next, we assessed the capacity of the parental cells and the transfected cell lines to restrict the growth of B. abortus S2308, a fully virulent strain. Since in preliminary experiments with a variety of MOIs and times allowed for phagocytosis the RAW264.7 cells proved to be poorly phagocytic for Brucella, we used a standard MOI of 100 and extended the time allowed for phagocytosis from 30 min to 2 h. The results of the B. abortus killing assays are summarized in Fig. 6. These results indicate that (i) the bovine NRAMP1-R and NRAMP1-S16 transgenic cells are superior in their ability to control the intracellular replication of B. abortus when compared to the parental cells and (ii) the difference in survival rates between the three R and three S16 clones, when analyzed by the Mann-Whitney test, is statistically significant (P < 0.05). However, in all the experiments we performed, we also observed marked differences (up to 20-fold) in the number of viable intracellular organisms at T0 in the parental and transfected cell lines. While these lower levels of intracellular infection may influence overall bacterial survival in these cell lines, it is important to note that in preliminary assays at similar levels of infection the parental cells consistently had higher rates of survival than those observed in the transfected lines. In addition, there were reproducible differences in the morphology of the bacterial colonies grown from lysates of the parental cells and the transfected cell lines at T0 and T12. While the size of the colonies recovered from the parental cells did not differ from those observed in the bacterial controls or the serial dilutions of the bacterial stock, a majority (60% or more) of the colonies grown from the transgenic cells were reproducibly smaller with a wide distribution of colony sizes ranging from 50 to <10% of expected colony size after 4 days of incubation (Fig. 7). When incubated for an additional 3 to 4 days, the smaller colonies reached normal size. Also, when the smaller colonies were subcultured, the phenotype persisted.
FIG. 6.
Survival of B. abortus in three NRAMP1-R or NRAMP1-S16 transfected clones of the murine RAW264.7 macrophage cell line. Bacterial survival is expressed as the percentage of survival in the parental RAW264.7 cells. The box plot shows the median, quartiles, and outside values (*) from three independent experiments; in each experiment, the cell lines were assayed in triplicate, and the survival rates were pooled for the three clones from each genotype. The difference in survival rates between the three R and the three S16 clones is significant at P < 0.05.
FIG. 7.
Colony morphology of B. abortus recovered from NRAMP1-R transgenic (A) and RAW264.7 (B) cells. B. abortus was grown on tryptic soy agar plates for 4 days. Note the normal appearance of B. abortus colonies (B) and the wide variation in colony size (A).
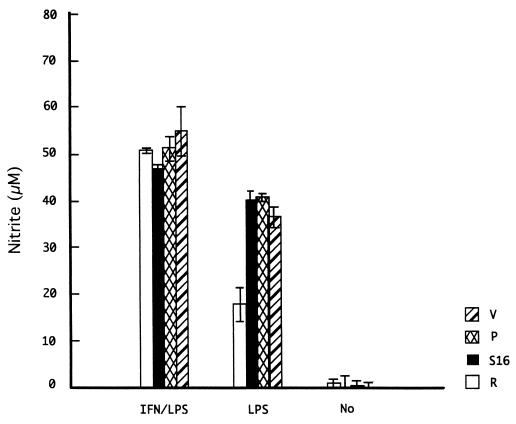
Finally, we assessed the production of NO in the transgenic lines by measuring levels of inorganic nitrite in culture supernatants. In these assays, we compared one each of the bovine NRAMP1-R and -S16 transgenic cell lines to vector-transfected cells and the parental cells. The cells were left untreated or stimulated with IFN-γ, LPS, or IFN-γ plus LPS for 24 and 48 h. Only with LPS stimulation at both 24 and 48 h did we observe a difference between the cell lines: the R cell line produced significantly lower levels of NO at both time points than the S16 line and both control cell lines (Fig. 8). We did not observe significant differences between the cell lines under resting conditions or with IFN-γ (data not shown) or IFN-γ–LPS stimulation.
FIG. 8.
Nitrite levels in supernatants from bovine NRAMP1-R (R), NRAMP1-S16 (S16), vector-transfected RAW264.7 cells (V), and parental RAW264.7 cells (P) left untreated (No) or stimulated for 48 h with LPS or IFN-γ–LPS. In each experiment, the cells were assayed in triplicate, and the results are represented as the mean ± standard deviation of three independent experiments.
DISCUSSION
Bovine NRAMP1, the bovine homologue of a murine gene controlling natural resistance to antigenically and taxonomically unrelated intracellular pathogens, has been identified as one of the major candidates for controlling natural resistance and/or susceptibility to bovine brucellosis in cattle (21). The resistant phenotype in natural resistance to bovine brucellosis is encoded by two or more genes; in addition to bovine NRAMP1, BoLA, the bovine major histocompatibility complex, is a major candidate gene system believed to control resistance and/or susceptibility to bovine brucellosis. To study the function of bovine NRAMP1 and because of the multigenic determination of resistance and/or susceptibility to bovine brucellosis, we designed an experimental system in which the bovine NRAMP1 gene is isolated from the overall genetic background of cattle by transfecting it into the murine RAW264.7 (Bcgs) macrophage cell line, its expression driven by the bovine NRAMP1 promoter. We selected the RAW264.7 cell line because of its Bcgs background and because it can be easily transfected by electroporation and stable lines can be established (56). In this line of experiments, we cloned and characterized the 5′-flanking region of bovine NRAMP1. The 5′-flanking region as well as the first and second introns of bovine NRAMP1 contain motifs for transcription factors that may play a role in the macrophage-specific expression of NRAMP1 (NF-κB, NF–interleukin-6, and PU.1) and those related to IFN-γ-inducible gene expression (interferon repressor factor, gamma activation site, and IRE-1 and -2) (20, 22, 37, 42, 44, 60). The high degree of homology between the human and mouse Nramp1 and bovine NRAMP1 sequences in the −80 to −257 segment of the 5′-flanking region indicates that this segment may serve as a basal functional promoter (7, 26).
A bovine NRAMP1-GFP reporter construct transfected into the RAW264.7 cell line showed only weak GFP activity under UV illumination, but the mRNA for GFP could be easily detected by RPA. These results indicate that (i) the 5′-flanking region of bovine NRAMP1 is a functional promoter despite the lack of TATA and CAAT boxes and (ii) the expression of a transgene in the murine RAW264.7 macrophage cell line can be directed from a bovine promoter that has been shown to be active in bovine macrophages. In addition, this promoter construct may be suited for other applications, since it has been shown to be active exclusively in professional phagocytes (12); to this effect, it can be further streamlined by mapping essential elements required for tissue-specific expression. The low level of GFP activity under UV light is attributable to the fact that GFP is a relatively insensitive reporter due to lack of enzymatic amplification (61); about 1 μM cytosolic GFP (105 to 106 molecules per cell) is necessary to show up over an autofluorescent background.
Also, it appears that the bovine NRAMP1 promoter is relatively position independent. This further attribute makes the bovine NRAMP1 promoter attractive for transgenic applications in which the chromosome position effect may interfere with the expression or inducibility of a transgene (47).
The level of expression of the bovine NRAMP1-S16 gene in transfected RAW264.7 macrophages was three- to sixfold lower than that of the resistance-associated allele. Since the NRAMP1-R and -S16 constructs were identical except for the presence of three additional GT repeats in the 3′UTR of the S16 construct, it is reasonable to conclude that the polymorphic microsatellite within the 3′UTR of bovine NRAMP1 may influence either the rate of transcription or the stability of the bovine NRAMP1 mRNA. A role for microsatellites in the regulation of gene expression has previously been proposed (D. G. King, Letter, Science 263:595–596, 1994; 17, 32, 43), and the role of the 3′UTR in the posttranscriptional control of mRNA stability and translation has been well documented. Specifically, AU-rich elements have been mapped in the 3′UTR of transcripts encoding cytokines, oncoproteins, and growth and transcription factors (11) and are strongly associated with the regulation of mRNA stability (13,14). In addition, a paradigm for translational regulation by the AU-rich elements has been established for a number of mRNAs (39). More recently, Seiler-Tuyns et al. (54) have provided evidence that, in the case of the human tumor necrosis alpha (TNF-α) mRNA, the 3′UTR also participates in modulating gene expression at the transcriptional level. Because of the random selection process, it is highly unlikely that we selected a whole set of clones that expressed only low levels of the transgene. We cannot exclude the possibility that the promoter region that we used in both constructs may be lacking a regulatory element essential for high levels of expression of the S16 allele of bovine NRAMP1; this promoter region was isolated from a BAC clone in which the bovine NRAMP1 gene had the (GT)13, resistance-associated configuration. Further experiments investigating the rate of transcription of both transgenes as well as the stability of their mRNAs will be necessary to better characterize the role of this polymorphic microsatellite in determining NRAMP1 gene function. In addition, a thorough analysis of the entire genomic sequence of bovine NRAMP1 in cattle that are genetically resistant and susceptible to bovine brucellosis is crucial for a better understanding of the regulation of NRAMP1 expression. We were unable to measure bovine NRAMP1 protein levels because of the unavailability of an anti-bovine NRAMP1 antibody of sufficient sensitivity and specificity that would have been suitable for example for a Western blot analysis.
We selected three each of the R and S16 transgenic cell lines for in vitro killing assays, using highly virulent strains of Salmonella serovar Dublin and B. abortus. In order to minimize interassay variability, the number of clones tested in each assay was limited and the parental, nontransfected cells were included in every assay as an internal control. Thus, we express the survival in each clone tested for each assay as a percentage of the survival obtained with the parental cells in the same assay. When the transfected cell lines were tested for their capacity to control the intracellular replication of Salmonella serovar Dublin, they proved to be significantly superior to the parental cell line at all MOIs. These results indicate that (i) the protein encoded by the bovine NRAMP1 transgene is functional in RAW264.7 macrophages, (ii) the NRAMP1 gene plays a role in antimicrobial mechanisms of macrophages directed at controlling Salmonella serovar Dublin in vitro, and (iii) the bovine NRAMP1 gene can complement the mutated murine Bcgs allele.
From these killing assays, it also appears that the polymorphism within the 3′UTR of NRAMP1, while strongly influencing the level of steady-state accumulation of the NRAMP1 mRNA, does not affect the capacity of the S cell lines to control Salmonella serovar Dublin in vitro when compared to the R lines. While previous work has demonstrated that macrophages from cattle that are naturally resistant to bovine brucellosis are superior in their capacity to kill both B. abortus and Salmonella serovar Dublin in vitro when compared to macrophages from susceptible cattle, the existence of a state of genetic resistance to a virulent in vivo challenge with Salmonella serovar Dublin has not been documented in these cattle. Our data support the notion that NRAMP1 plays a role in macrophage killing mechanisms directed at Salmonella serovar Dublin but suggest that the difference in in vitro killing observed between cattle that are resistant and susceptible to brucellosis may be unrelated to NRAMP1. Considering the importance of Salmonella as both a bovine pathogen and a zoonotic agent, future work should be directed at providing a more-thorough understanding of the genetic and molecular mechanisms involved in host defense against Salmonella species.
In B. abortus killing assays, the microbicidal capacity of the R and S16 cell lines was significantly superior to that of the parental cells, thus suggesting a role for NRAMP1 in resistance to Brucella. Similar to the murine J774A.1 macrophages, adequate levels of infection of the RAW264.7 cells with B. abortus were achieved at a high MOI only (35). It has been reported that efficient phagocytosis of Brucella by mouse macrophages, contrary to bovine macrophages, requires opsonization by specific antibodies (10, 28). Reproducible differences in the number of viable intracellular organisms at T0 in the parental and transfected cell lines tend to implicate the bovine NRAMP1 gene in the process of phagocytosis of B. abortus. This finding is not incompatible with the previous demonstration that the differential capacity of macrophages from Bcgr and Bcgs mice to control the intracellular replication of S. enterica serovar Typhimurium is linked not to phagocytosis of the organism but rather to increased bacterial killing (41). In our experiments, levels of phagocytosis of Salmonella serovar Dublin were comparable in the parental and transgenic cell lines. Also, Radzioch et al. (51), when comparing two widely used bone marrow-derived murine macrophage cell lines, showed that B10S (Bcgs) macrophages phagocytosed Mycobacterium smegmatis better at 0.5, 1, and 2 h, but not at 3 or 6 h, than B10R (Bcgr) macrophages. It is very unlikely that the recovery of smaller numbers of viable intracellular bacteria at T0 from the transfected cell lines is due to the rapid killing of B. abortus.
The morphology and growth characteristics of the bacterial colonies grown from lysates of the transgenic cell lines are reminiscent of changes seen in colonies of B. abortus when exposed to conditions of acid shock in vitro (40) and may provide important insight into the putative function of the NRAMP1 protein in that they could implicate NRAMP1 either directly or indirectly in the phagosome acidification and/or phagosome-lysosome fusion processes inside macrophages. Nramp1 encodes an integral membrane protein that shares structural characteristics with ion channels and transporters (29). In addition, Nramp1 has been found not expressed at the plasma membrane but rather localized to the late endocytic compartments of resting macrophages in a Lamp1 (lysosome-associated membrane protein 1)-positive compartment. Upon phagocytosis, Nramp1 is recruited to the membrane of the phagosome and remains associated with this structure during its maturation to phagolysosome. The targeting of Nramp1 from endocytic vesicles to the phagosomal membrane supports the hypothesis that Nramp1 controls the replication of intracellular parasites by altering the intravacuolar environment of the microbe-containing phagosome, possibly by controlling divalent cation concentrations at that site (25). The Nramp1 gene has been implicated previously in the process of phagosome-lysosome fusion in murine macrophages when it was shown that the hydrolytic activity of macrophages, as measured by the capacity of lysosomes to fuse with and transfer active hydrolytic enzymes to phagosomes in which Mycobacterium avium resides, is an expression of the Bcg gene and that this phenomenon is a key antibacterial activity responsible for growth restriction of M. avium (15). The generation of an acidic interior is believed to play an essential role in the microbicidal activity of phagosomes (31), and recent work by Hackam et al. (30) suggests that Nramp1 may directly or indirectly influence the intraphagosomal pH to alter microbial proliferation. Also, Govoni et al. (24) have shown with a transfection model of RAW264.7 macrophages that transfection of wild-type Nramp1 into Bcgs RAW 264.7 cells abrogates the intracellular replication of S. enterica serovar Typhimurium and that the increased antibacterial activity is linked to increased phagosomal acidification.
The survival rate of B. abortus was significantly different between the three R and three S16 clones (P < 0.05). Thus, it appears that the polymorphic microsatellite affects the rate of transcription or stability of the bovine NRAMP1 mRNA, which in turn may result in reduced amounts of bovine NRAMP1-expressed protein and differential killing of B. abortus by cells transfected with the R and S16 alleles of bovine NRAMP1. These results are in accordance with the strong association observed between the in vivo phenotype of cattle naturally resistant and/or susceptible to bovine brucellosis and the ssCA polymorphism for bovine NRAMP1.
While the putative role of NRAMP1 in phagosome acidification provides an attractive explanation for our observations, we cannot exclude a possible role for reactive oxygen and nitrogen intermediates, considering their importance in macrophage antimicrobial strategies directed at Brucella and Salmonella species (2, 16) and the crucial role played by Nramp1 in the generation of these radicals (6). While Brucella is highly susceptible to reactive oxygen intermediates but less susceptible to reactive nitrogen intermediates, the ability of the respiratory burst and nitric oxide synthase to synergistically kill microbial pathogens such as Salmonella is well documented. Therefore, we wanted to assess the production of NO in our transgenic cell lines under various stimulation regimens. Surprisingly, both the R and S16 cell lines did not produce higher levels of NO than the two control (Bcgs) cell lines, following any of the treatments. For example, increased nitrite release after IFN-γ–LPS stimulation has been documented in RAW264.7 cells after in vitro transfection of the Bcgr allele (4). However, Radzioch et al. (52) showed with a Bcgr macrophage cell line in which the expression of Nramp1 had been downregulated by a ribozyme that the production of NO was markedly lower after IFN-γ stimulation but not if the treatment with IFN-γ occurred in the presence of LPS, suggesting that IFN-γ- but not LPS-induced activation is affected by the inhibition of the Nramp1 gene. After 24 and 48 h of LPS stimulation, the R cell line produced significantly lower levels of NO than the S16 cell line and the two controls. These results may seem unexpected in light of the superior anti-Brucella activity of the NRAMP1-R transgenic cell lines. Yet, a series of experiments with a mouse macrophage model of Brucella suis infection (28) supports the possibility that in mice, NO favors the elimination of Brucella, provided that IFN-γ and anti-Brucella antibodies are present, i.e., following expression of acquired immunity. Furthermore, Jiang et al. (36) showed that while the murine J774A.1 macrophage cell line has anti-Brucella activities which are enhanced by IFN-γ activation, it does not produce NO when activated with IFN-γ and infected with B. abortus. They also demonstrated that the IFN-γ-induced enhancement of anti-Brucella activities by murine peritoneal macrophages was inhibited by the addition of anti-TNF-α antibodies to the cultures, indicating that TNF-α is necessary for full expression of macrophage anti-Brucella activities. This is interesting considering that in RAW264.7 cells, endogenous NO has a negative feedback effect on TNF-α synthesis in vitro, mainly by reducing the half-life of TNF-α mRNA (19, 55). These findings underscore the complexity of the relationship between the host and intracellular parasites that cannot be reduced to the expression of a single gene or the production of a single antimicrobial factor. Macrophages utilize a wide array of molecules and mediators in an attempt to control the replication of the parasite, while the pathogen has evolved strategies to evade these antimicrobial mechanisms and avoid detection by the immune system.
Although the bovine NRAMP1 gene is a major candidate gene controlling natural resistance to brucellosis, it has been shown recently that under field conditions it does not determine resistance and susceptibility to infection with M. bovis (3). This finding is in accordance with recent studies with mice that have failed to establish a role for Nramp1 in protection against virulent Mycobacterium tuberculosis, despite the fact that a role for Nramp1 in controlling multiplication of M. bovis BCG vaccine within murine macrophages is well documented both in vivo and in vitro (45, 46). However, a new locus with a major effect on tuberculosis susceptibility, designated sst1 (for susceptibility to tuberculosis 1), has been mapped to a 9-centimorgan (cM) interval on mouse chromosome 1, 10 to 19 cM distal to Nramp1 (38). Similarly, in humans, genetic variants of Nramp1 account for only a small proportion of the overall genetic component of natural resistance to tuberculosis suggested by twin studies (5).
Further verification of our findings and attempts to repeat them in bovine macrophage cell lines are essential, since they could provide the basis for novel strategies aimed at controlling zoonotic diseases in cattle. Also, our in vitro model should greatly facilitate future work on the regulation and biology of bovine NRAMP1.
ACKNOWLEDGMENTS
This work was supported by the State of Texas Cattle and Deer Tuberculosis Management Plan, contract 1120168409; the USDA Agricultural Research Service National Animal Disease Center, cooperative agreement 53–3625-6–154; and the Texas Agricultural Experiment Station, project TEXO8409.
REFERENCES
- 1.Adams L G, Templeton J W. Genetic resistance to bacterial diseases of animals. Rev Sci Tech. 1998;17:200–219. doi: 10.20506/rst.17.1.1085. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin C L, Jiang X, Fernandes D M. Macrophage control of Brucella abortus: influence of cytokines and iron. Trends Microbiol. 1993;1:99–104. doi: 10.1016/0966-842x(93)90115-8. [DOI] [PubMed] [Google Scholar]
- 3.Barthel R, Piedrahita J A, McMurray D N, Payeur J, Baca D, Suarez Guemes F, Perumaalla V S, Ficht T A, Templeton J W, Adams L G. Pathologic findings and association of Mycobacterium bovis infection with the bovine NRAMP1 gene in cattle from herds with naturally occurring tuberculosis. Am J Vet Res. 2000;61:1140–1144. doi: 10.2460/ajvr.2000.61.1140. [DOI] [PubMed] [Google Scholar]
- 4.Barton C H, Whitehead S H, Blackwell J M. Nramp transfection transfers Ity/Lsh/Bcg-related pleiotropic effects on macrophage activation: influence on oxidative burst and nitric oxide pathways. Mol Med. 1995;1:267–279. [PMC free article] [PubMed] [Google Scholar]
- 5.Bellamy R, Beyers N, McAdam K P, Ruwende C, Gie R, Samaai P, Bester D, Meyer M, Corrah T, Collin M, Camidge D R, Wilkinson D, Hoal-Van Helden E, Whittle H C, Amos W, van Helden P, Hill A V. Genetic susceptibility to tuberculosis in Africans: a genome-wide scan. Proc Natl Acad Sci USA. 2000;97:8005–8009. doi: 10.1073/pnas.140201897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwell J M, Barton C H, White J K, Roach T I, Shaw M A, Whitehead S H, Mock B A, Searle S, Williams H, Baker A M. Genetic regulation of leishmanial and mycobacterial infections: the Lsh/Ity/Bcg gene story continues. Immunol Lett. 1994;43:99–107. doi: 10.1016/0165-2478(94)00161-8. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell J M, Barton C H, White J K, Searle S, Baker A M, Williams H, Shaw M A. Genomic organization and sequence of the human NRAMP gene: identification and mapping of a promoter region polymorphism. Mol Med. 1995;1:194–205. [PMC free article] [PubMed] [Google Scholar]
- 8.Cai L, Taylor J F, Wing R A, Gallagher D S, Woo S S, Davis S K. Construction and characterization of a bovine bacterial artificial chromosome library. Genomics. 1995;29:413–425. doi: 10.1006/geno.1995.9986. [DOI] [PubMed] [Google Scholar]
- 9.Campbell G A, Adams L G. The long-term culture of bovine monocyte-derived macrophages and their use in the study of intracellular proliferation of Brucella abortus. Vet Immunol Immunopathol. 1992;34:291–305. doi: 10.1016/0165-2427(92)90171-l. [DOI] [PubMed] [Google Scholar]
- 10.Campbell G A, Adams L G, Sowa B A. Mechanisms of binding of Brucella abortus to mononuclear phagocytes from cows naturally resistant or susceptible to brucellosis. Vet Immunol Immunopathol. 1994;41:295–306. doi: 10.1016/0165-2427(94)90103-1. [DOI] [PubMed] [Google Scholar]
- 11.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cellier M, Shustik C, Dalton W, Rich E, Hu J, Malo D, Schurr E, Gros P. Expression of the human NRAMP1 gene in professional primary phagocytes: studies in blood cells and in HL-60 promyelocytic leukemia. J Leukoc Biol. 1997;61:96–105. doi: 10.1002/jlb.61.1.96. [DOI] [PubMed] [Google Scholar]
- 13.Chen C Y, Del Gatto-Konczak F, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- 14.Chen C Y, Shyu A B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 15.de Chastellier C, Frehel C, Offredo C, Skamene E. Implication of phagosome-lysosome fusion in restriction of Mycobacterium avium growth in bone marrow macrophages from genetically resistant mice. Infect Immun. 1993;61:3775–3784. doi: 10.1128/iai.61.9.3775-3784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Groote A M, Ochsner U A, Shiloh M U, Nathan C, McCord J M, Dinauer M C, Libby S J, Vazquez-Torres A, Xu Y, Fang F C. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deka R, Shriver M D, Yu L M, Jin L, Aston C E, Chakraborty R, Ferrell R E. Conservation of human chromosome 13 polymorphic microsatellite (CA)n repeats in chimpanzees. Genomics. 1994;22:226–230. doi: 10.1006/geno.1994.1369. [DOI] [PubMed] [Google Scholar]
- 18.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 19.Eigler A, Moeller J, Endres S. Exogenous and endogenous nitric oxide attenuates tumor necrosis factor synthesis in the murine macrophage cell line RAW 264.7. J Immunol. 1995;154:4048–4054. [PubMed] [Google Scholar]
- 20.Faisst S, Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992;20:3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng J, Li Y, Hashad M, Schurr E, Gros P, Adams L G, Templeton J W. Bovine natural resistance associated macrophage protein 1 (Nramp1) gene. Genome Res. 1996;6:956–964. doi: 10.1101/gr.6.10.956. [DOI] [PubMed] [Google Scholar]
- 22.Fujita T, Ohno S, Yasumitsu H, Taniguchi T. Delimitation and properties of DNA sequences required for the regulated expression of human interferon-beta gene. Cell. 1985;41:489–496. doi: 10.1016/s0092-8674(85)80022-2. [DOI] [PubMed] [Google Scholar]
- 23.Goto Y, Nakamura R M, Takahashi H, Tokunaga T. Genetic control of resistance to Mycobacterium intracellulare infection in mice. Infect Immun. 1984;46:135–140. doi: 10.1128/iai.46.1.135-140.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Govoni G, Canonne-Hergaux F, Pfeifer C G, Marcus S L, Mills S D, Hackam D J, Grinstein S, Malo D, Finlay B B, Gros P. Functional expression of Nramp1 in vitro in the murine macrophage line RAW264.7. Infect Immun. 1999;67:2225–2232. doi: 10.1128/iai.67.5.2225-2232.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govoni G, Gros P. Macrophage NRAMP1 and its role in resistance to microbial infections. Inflamm Res. 1998;47:277–284. doi: 10.1007/s000110050330. [DOI] [PubMed] [Google Scholar]
- 26.Govoni G, Vidal S, Cellier M, Lepage P, Malo D, Gros P. Genomic structure, promoter sequence, and induction of expression of the mouse Nramp1 gene in macrophages. Genomics. 1995;27:9–19. doi: 10.1006/geno.1995.1002. [DOI] [PubMed] [Google Scholar]
- 27.Gros P, Skamene E, Forget A. Genetic control of natural resistance to Mycobacterium bovis (BCG) in mice. J Immunol. 1981;127:2417–2421. [PubMed] [Google Scholar]
- 28.Gross A, Spiesser S, Terraza A, Rouot B, Caron E, Dornand J. Expression and bactericidal activity of nitric oxide synthase in Brucella suis-infected murine macrophages. Infect Immun. 1998;66:1309–1316. doi: 10.1128/iai.66.4.1309-1316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruenheid S, Pinner E, Desjardins M, Gros P. Natural resistance to infection with intracellular pathogens: the Nramp1 protein is recruited to the membrane of the phagosome. J Exp Med. 1997;185:717–730. doi: 10.1084/jem.185.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hackam D J, Rotstein O D, Zhang W, Gruenheid S, Gros P, Grinstein S. Host resistance to intracellular infection: mutation of natural resistance-associated macrophage protein 1 (Nramp1) impairs phagosomal acidification. J Exp Med. 1998;188:351–364. doi: 10.1084/jem.188.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hackam D J, Rotstein O D, Zhang W J, Demaurex N, Woodside M, Tsai O, Grinstein S. Regulation of phagosomal acidification. Differential targeting of Na+/H+ exchangers, Na+/K+-ATPases, and vacuolar-type H+-ATPases. J Biol Chem. 1997;272:29810–29820. doi: 10.1074/jbc.272.47.29810. [DOI] [PubMed] [Google Scholar]
- 32.Hamada H, Petrino M G, Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci USA. 1982;79:6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harmon B G, Adams L G, Templeton J W, Smith R. Macrophage function in mammary glands of Brucella abortus-infected cows and cows that resisted infection after inoculation of Brucella abortus. Am J Vet Res. 1989;50:459–465. [PubMed] [Google Scholar]
- 34.Hutt F B. Genetic resistance to disease in domestic animals. Ithaca, N.Y: Comstock Publishing Associates; 1958. Resistant big animals; pp. 67–99. [Google Scholar]
- 35.Jiang X, Baldwin C L. Effects of cytokines on intracellular growth of Brucella abortus. Infect Immun. 1993;61:124–134. doi: 10.1128/iai.61.1.124-134.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang X, Leonard B, Benson R, Baldwin C L. Macrophage control of Brucella abortus: role of reactive oxygen intermediates and nitric oxide. Cell Immunol. 1993;151:309–319. doi: 10.1006/cimm.1993.1241. [DOI] [PubMed] [Google Scholar]
- 37.Klemsz J M, McKercher S R, Celada A, Van Beveren C, Maki R A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 38.Kramnik I, Dietrich W F, Demant P, Bloom B R. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2000;97:8560–8565. doi: 10.1073/pnas.150227197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kruys V, Marinx O, Shaw G, Deschamps J, Huez G. Translational blockade imposed by cytokine-derived UA-rich sequences. Science. 1989;245:852–855. doi: 10.1126/science.2672333. [DOI] [PubMed] [Google Scholar]
- 40.Lin J, Ficht T A. Protein synthesis in Brucella abortus induced during macrophage infection. Infect Immun. 1995;63:1409–1414. doi: 10.1128/iai.63.4.1409-1414.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lissner C R, Swanson R N, O'Brien A D. Genetic control of the innate resistance of mice to Salmonella typhimurium: expression of the Ity gene in peritoneal and splenic macrophages isolated in vitro. J Immunol. 1983;131:3006–3013. [PubMed] [Google Scholar]
- 42.Lowenstein C J, Alley E W, Raval P, Snowman A M, Snyder S H, Russell S W, Murphy W J. Macrophage nitric oxide synthase gene: two upstream regions mediate induction by interferon gamma and lipopolysaccharide. Proc Natl Acad Sci USA. 1993;90:9730–9734. doi: 10.1073/pnas.90.20.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lue N F, Buchman A R, Kornberg R D. Activation of yeast RNA polymerase II transcription by a thymidine-rich upstream element in vitro. Proc Natl Acad Sci USA. 1989;86:486–490. doi: 10.1073/pnas.86.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsusaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Transcription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci USA. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medina E, North R J. Evidence inconsistent with a role for the Bcg gene (Nramp1) in resistance of mice to infection with virulent Mycobacterium tuberculosis. J Exp Med. 1996;183:1045–1051. doi: 10.1084/jem.183.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medina E, North R J. Genetically susceptible mice remain proportionally more susceptible to tuberculosis after vaccination. Immunology. 1999;96:16–21. doi: 10.1046/j.1365-2567.1999.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmiter R D, Brinster R L. Transgenic mice. Cell. 1985;41:343–345. doi: 10.1016/s0092-8674(85)80004-0. [DOI] [PubMed] [Google Scholar]
- 48.Plant J, Glynn A A. Natural resistance to Salmonella infection, delayed hypersensitivity and Ir genes in different strains of mice. Nature. 1974;248:345–347. doi: 10.1038/248345a0. [DOI] [PubMed] [Google Scholar]
- 49.Price R E, Templeton J W, Smith III R, Adams L G. Ability of mononuclear phagocytes from cattle naturally resistant or susceptible to brucellosis to control in vitro intracellular survival of Brucella abortus. Infect Immun. 1990;58:879–886. doi: 10.1128/iai.58.4.879-886.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qureshi T, Templeton J W, Adams L G. Intracellular survival of Brucella abortus, Mycobacterium bovis BCG, Salmonella serovar Dublin, and Salmonella typhimurium in macrophages from cattle genetically resistant to Brucella abortus. Vet Immunol Immunopathol. 1996;50:55–65. doi: 10.1016/0165-2427(95)05492-8. [DOI] [PubMed] [Google Scholar]
- 51.Radzioch D, Hudson T, Boule M, Barrera L, Urbance J W, Varesio L, Skamene E. Genetic resistance/susceptibility to mycobacteria: phenotypic expression in bone marrow derived macrophage lines. J Leukoc Biol. 1991;50:263–272. doi: 10.1002/jlb.50.3.263. [DOI] [PubMed] [Google Scholar]
- 52.Radzioch D, Kramnik I, Skamene E. Molecular mechanisms of natural resistance to mycobacterial infections. Circ Shock. 1994;44:115–120. [PubMed] [Google Scholar]
- 53.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seiler-Tuyns A, Dufour N, Spertini F. Human tumor necrosis factor-alpha gene 3′ untranslated region confers inducible toxin responsiveness to homologous promoter in monocytic THP-1 cells. J Biol Chem. 1999;274:21714–21718. doi: 10.1074/jbc.274.31.21714. [DOI] [PubMed] [Google Scholar]
- 55.Sinha B, Eigler A, Baumann K H, Greten T F, Moeller J, Endres S. Nitric oxide downregulates tumour necrosis factor in mRNA in RAW 264.7 cells. Res Immunol. 1998;149:139–150. doi: 10.1016/s0923-2494(98)80297-6. [DOI] [PubMed] [Google Scholar]
- 56.Stacey K J, Ross I L, Hume D A. Electroporation and DNA-dependent cell death in murine macrophages. Immunol Cell Biol. 1993;71:75–85. doi: 10.1038/icb.1993.8. [DOI] [PubMed] [Google Scholar]
- 57.Templeton J W, Adams L G. Natural resistance to bovine brucellosis. In: Adams L G, editor. Advances in brucellosis research. College Station, Tex: Texas A&M University Press; 1990. pp. 144–150. [Google Scholar]
- 58.Templeton J W, Estes D M, Price R E, Smith III R, Adams L G. : Allan Teal (ed.), Proceedings of the 4th World Congress Genetic Applications to Livestock Production, 23 to 27 July 1990. Edinburgh, Scotland: University of Edinburgh Press; 1990. Immunogenetics of natural resistance to bovine brucellosis pp. 396–399. [Google Scholar]
- 59.Vidal S M, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 60.Xie Q W, Whisnant R, Nathan C. Promoter of the mouse gene encoding calcium-independent nitric oxide synthase confers inducibility by interferon gamma and bacterial lipopolysaccharide. J Exp Med. 1993;177:1779–1784. doi: 10.1084/jem.177.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zlokarnik G, Negulescu P A, Knapp T E, Mere L, Burres N, Feng L, Whitney M, Roemer K, Tsien R Y. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science. 1998;279:84–89. doi: 10.1126/science.279.5347.84. [DOI] [PubMed] [Google Scholar]