Abstract
The direct binding of bacteria to platelets may be an important virulence mechanism in the pathogenesis of infective endocarditis. We have previously described Staphylococcus aureus strain PS12, a Tn551-derived mutant of strain ISP479, with reduced ability to bind human platelets in vitro. When tested in an animal model of endocarditis, the PS12 strain was less virulent than its parental strain, as measured by bacterial densities in endocardial vegetations and incidence of systemic embolization. We have now characterized the gene disrupted in PS12 and its function in platelet binding. DNA sequencing, Southern blotting, and PCR analysis indicate that PS12 contained two Tn551 insertions within the clumping factor A (ClfA) locus (clfA). The first copy was upstream from the clfA start codon and appeared to have no effect on ClfA production. The second insertion was within the region encoding the serine aspartate repeat of ClfA and resulted in the production of a truncated ClfA protein that was secreted from the cell. A purified, recombinant form of the ClfA A region, encompassing amino acids 40 through 559, significantly reduced the binding of ISP479C to human platelets by 44% (P = 0.0001). Immunoprecipitation of recombinant ClfA that had been incubated with solubilized platelet membranes coprecipitated a 118-kDa platelet membrane protein. This protein does not appear to be glycoprotein IIb. These results indicate that platelet binding by S. aureus is mediated in part by the direct binding of ClfA to a novel 118-kDa platelet membrane receptor.
The binding of bacteria to platelets is thought to be a major virulence determinant in the pathogenesis of infective endocarditis (28). Platelet binding may be an important mechanism for initiating endocardial infection, since platelets attached to damaged valve surface may serve as binding foci for organisms circulating in the blood (10, 30, 36). In addition, the adhesion of bacteria to platelets may be important for the subsequent development of mature vegetations (7). Bacteria on the valve surface may recruit circulating platelets to that site, resulting in the further deposition of platelets. These events, in conjunction with bacterial proliferation, eventually lead to the formation of macroscopic endocardial vegetations.
Previous studies have shown that Staphylococcus aureus can bind to platelets in vitro (1, 29). This binding is likely to be mediated in part by soluble bridging molecules that link the organism to the platelet surface. S. aureus binds a number of soluble host proteins that may serve as bridging molecules, including fibrinogen, fibronectin, thrombospondin, and von Willebrand factor (3, 15, 16, 18). Since these proteins can also bind activated platelets, it is likely that these molecules could contribute to staphylococcal binding to human platelets in vivo.
S. aureus has also been shown to bind human platelets directly. The mechanisms for direct binding have not been characterized extensively, but it is likely that such binding is multifactorial. Our previous studies have indicated that binding may be mediated in part by surface carbohydrates (including the staphylococcal capsule) and one or more surface proteins (36). Recent studies have also shown that staphylococcal protein A can bind platelet gC1qR, a receptor that is expressed on the surface of activated platelets (24). Thus, this latter interaction may be an additional mechanism for the direct attachment of staphylococci to platelets on the endocardial surface.
Although the binding of S. aureus to platelets may occur by multiple processes, the impact of the above interactions on the pathogenesis of endocarditis is largely unknown. Our previous studies have focused on the role of direct binding of S. aureus to unactivated platelets in the pathogenesis of bacterial endocarditis. These studies have described a mutant of S. aureus strain ISP479, derived by transposon mutagenesis, with both decreased direct binding to platelets in vitro and reduced virulence in an animal model of endocarditis (29). Rabbits infected with mutant strain PS12 had lower densities of S. aureus within vegetations and lower rates of systemic embolization than those of rabbits infected with the parental strain. These results demonstrated that the direct binding of S. aureus to platelets may be an important mechanism for the pathogenesis of endocarditis.
To better define the molecular basis of direct platelet binding by S. aureus, we have now characterized extensively the mutation within strain PS12. Our results indicate that staphylococcal clumping factor A (ClfA) (21), recognized initially for its role in fibrinogen binding, also mediates direct binding to human platelets. As discussed below, ClfA appears to bind a novel 118-kDa protein on the platelet membrane.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The bacterial strains, plasmids, and primers used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Bacterial strain, plasmid, or primer | Genotype or description | Reference or source |
|---|---|---|
| S. aureus strains | ||
| ISP479C | Derivative of 8325-4 cured of the Tn551 delivery plasmid pI258 | 26 |
| PS12 | Low-platelet binding mutant of ISP479 | 29 |
| Newman | Wild-type strain | 8 |
| DU5852 | Newman clfA::Tn917 | 21 |
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 λ−gyrA96 relA1 | 35 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | 27 |
| MC1061 | F−araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL(Strr)hsdR2(rK− mK−) mcrA mcrB1 | 33 |
| Plasmids | ||
| pBluescript SK(−) | E. coli cloning vector | Stratagene |
| pET28A | T7 expression vector | Novagen |
| pLTV1 | Shuttle vector containing Tn917 | |
| pRS401 | pBluescriptSK(−) containing a Tn551-PS12 XhoI fragment cloned into the XhoI site of the vector | |
| pRS402 | Variant of pRS401 from which a 3.5-kb PstI fragment had been deleted | |
| pRS404 | pET28A containing the clfA sequence corresponding to amino acids 40-559 as an in-frame N-terminal fusion with the His6 and T7 tags | |
| Primers | ||
| DOCF1 | 5′-CGCGGATCCAGTGAAAATAGTGTTACGCAATCT-3′; forward primer for cloning ClfA40-559a | 25 |
| DOCR1 | 5′-CGCAAGCTTCTCTGGAATTGGTTCAATTTC-3′; reverse primer for cloning ClfA40-559 | 25 |
| TNEND | 5′-TCGATACAAATTTCTCGTGCGCTCGGACCCC-3′; Tn551 left and right end primer | |
| DSF | 5′-TTTTGCTGCTTCAGTGTCAGA-3′; forward primer for ORF downstream of clfA | |
| DSR | 5′-GGCTGAAGATGAGGCTTTGA-3′; reverse primer for ORF downstream of clfA |
ClfA40-559, ClfA with amino acids 40 to 559.
Media and antibiotics.
Brain heart infusion (BHI) and Luria-Bertani (LB) broth and agar were used as culture media for S. aureus and Escherichia coli, respectively. Erythromycin was used at a concentration of 15 μg/ml for selection of S. aureus and at 500 μg/ml for E. coli when these strains contained the erm gene. Kanamycin and ampicillin were used at a concentration of 50 μg/ml where appropriate.
DNA sequencing.
Chromosomal DNA was prepared from lysostaphin-treated PS12 strain as described previously (2), digested with XhoI, and ligated into pBluescript SK(−) that had been digested with the same enzyme. The Tn551-PS12 junction was sequenced using an ABI automated system (Applied Biosciences). Initial sequencing reactions were performed using the T7 and T3 primers. Subsequent reactions were done using primers based on derived staphylococcal sequences.
Southern blotting.
S. aureus chromosomal DNA was digested with selected restriction enzymes and transferred to Biodyne A membrane (Pall Corporation), using the Turboblotter Transfer System (Schleicher & Schuell Inc.). A 3.9-kb gel-purified fragment of Tn917, which has 98% homology to Tn551 (determined by sequence alignment), was labeled with digoxigenin (DIG/Genius System; Roche Diagnostics) for use as a probe. Hybridization was performed in a high-stringency buffer at 42°C, and then washing and imaging by chemiluminescence were done, according to the manufacturer's instructions.
Protein extraction.
Cell wall proteins were isolated from S. aureus by the method of Hartford et al. (12). In brief, cultures of bacteria grown overnight were washed in phosphate-buffered saline (PBS) and incubated for 30 min at 37°C in protoplasting buffer (50 mM Tris-HCl, 30% [wt/vol] raffinose, 145 mM NaCl [pH 7.5]) containing complete protease inhibitor (Roche Diagnostics), DNase I (80 μg/ml; Sigma), and lysostaphin (20 μg/ml; Sigma). The protoplasts were removed by centrifugation (12,000 × g for 10 min), and the protein concentration in the supernatant was determined using the BCA Protein Assay Reagent kit (Pierce). To isolate proteins from culture supernatants, 900 μl of spent media was precipitated with trichloroacetic acid, washed twice with 1 ml of acetone, and suspended in 20 μl of PBS.
SDS-PAGE and Western immunoblotting.
Proteins were prepared for electrophoresis in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and separated through 7.5% polyacrylamide gels (19). The proteins were then transferred electrophoretically to BioTrace NT membranes (Pall Corporation) by semidry transfer in Towbin buffer (31). The membranes were treated with a casein-based solution (blocking reagent; Roche Diagnostics) and then incubated overnight at 4°C with polyclonal rabbit anti-ClfA region A antiserum (1:1,000; kindly provided by T. J. Foster, Trinity College, Dublin, Ireland). Goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase was used to detect ClfA by chemiluminescence (Roche Diagnostics).
Northern (RNA) blot hybridization.
S. aureus RNA was prepared and blotted with the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. Cultures were lysed in 100 μl of a solution containing 10 mM Tris-HCl and 1 mM EDTA (pH 8.0) supplemented with lysozyme (3 mg/ml; Sigma), lysostaphin (20 μg/ml), and Anti-RNase (15 U; Ambion). Samples were separated via electrophoresis through a 1% agarose–0.66% formaldehyde gel in MOPS [3-(N-morpholino)propanesulfonic acid] running buffer. The RNA was transferred to positively charged nylon membranes (Roche Diagnostics) as described above. Hybridization was performed under high-stringency conditions, with PCR-generated, digoxigenin-labeled DNA probes, followed by washing and imaging by chemiluminescence, according to the manufacturer's instructions.
Assay for binding of staphylococci to soluble fibrinogen.
The ability of S. aureus to clump in the presence of soluble human fibrinogen was assessed by a turbidometric assay. Cultures of S. aureus were grown in BHI broth for 18 h at 37°C, washed as described above, and suspended in HEPES buffer (20 mM HEPES, 140 mM NaCl, 1 mM EDTA [pH 7.5]) to a final concentration of approximately 108 CFU/ml. Four hundred and fifty microliters of the diluted culture were added to siliconized glass tubes, placed in an aggregometer (Chronolog, Havertown, Pa.) and stirred continuously at 37°C. After a transmittance baseline was established, human fibrinogen (final concentration of 50 μg/ml; Sigma) was added to the tube, and the mixture was monitored for changes in light transmission. Clumping was detected as an increase in light transmission, with suspensions of bacteria alone and PBS serving as standards for minimal and maximal light transmission, respectively. All assays were performed at least three times.
Assay for binding of staphylococci to immobilized fibrinogen.
The binding of S. aureus to immobilized fibrinogen was quantified by an agar overlay assay. S. aureus was prepared as described above, washed twice in HEPES buffer, sonicated for 15 s to disperse clumps, and adjusted to a final concentration of approximately 103 CFU/ml. One milliliter of diluted culture was added to 25-mm-diameter culture wells that had been coated overnight with 1 ml of fibrinogen in PBS (50 μg/ml). The plates were incubated at 4°C for 1 h on a rotary shaker, followed by washing with agitation (three times for 5 min each time) to remove nonadherent bacteria. Two milliliters of BHI medium containing 0.5% agar were then added to the wells and allowed to solidify. After incubation of the plates overnight at 37°C, the number of colonies in each well was determined, and binding was expressed as a percentage of inoculum. All assays were performed at least six times.
Expression and purification of recombinant protein ClfA with amino acids 40 through 559 (rClfA).
A PCR-amplified fragment of the clfA gene (corresponding to the A region of the protein) was cloned into pET28A (Novagen). Recombinant proteins produced from this construct contain an N-terminal extension of six histidine residues. After transforming E. coli BL21(DE3) with the resultant plasmid, the protein was expressed and purified, using the His-Bind Purification kit (Novagen), according to the manufacturer's instructions. Subsequent to elution, the recombinant protein was dialyzed against PBS containing 10% glycerol and then stored at −70°C.
Assay for staphylococcal binding to platelet monolayers.
Staphylococcal binding to human platelet monolayers was assayed as described previously (29). In brief, 18-h-old cultures of S. aureus were washed twice in HEPES buffer, sonicated for 15 s to disperse clumps, and adjusted to a final concentration of A600 of 0.15 (approximately 107 CFU/ml). Fifty microliters of each suspension was added to individual 6-mm-diameter microtiter wells that had been previously coated with fixed, inactive human platelet monolayers and blocked with the above casein-based solution to reduce nonspecific binding. The plates were incubated at 4°C for 1 h, and the wells were washed three times for 5 min with HEPES buffer to remove nonadherent bacteria. The wells were then treated with 50 μl of trypsin (1 mg/ml) for 30 min at room temperature to release the bound bacteria from the platelets. The number of bound bacteria was determined by plating serial dilutions of the recovered organisms onto blood agar plates, and binding was expressed as a percentage of inoculum.
To assess the ability of ClfA to block the binding of S. aureus to human platelets, monolayers were treated with increasing concentrations of purified rClfA (0 to 50 μg/ml; 60 min, 4°C). Aliquots of ISP479C were prepared as described above and added to the platelet monolayers. As a control, parallel studies were done using platelets that had not been exposed to rClfA. Platelet binding under both conditions was then assessed as described above. Six binding values were recorded for each strain per assay, with each strain tested on multiple occasions.
Isolation of platelet membranes.
Platelet membranes were prepared by glycerol lysis and gradient centrifugation (11). In brief, 1 U of human platelet concentrate (obtained from the Blood Centers of the Pacific, San Francisco, Calif.) was centrifuged (900 × g, 22°C, 10 min). The resultant platelet pellets were washed in buffer A (10 mM sucrose, 130 mM NaCl, 4 mM monobasic sodium phosphate, 7.5 mM dibasic sodium phosphate, 5 mM sodium citrate dihydrate [pH 6.7]). The platelets were then suspended in 10 ml of buffer B (85 mM Tris-Cl, 965 mM NaCl, 857 mM glucose, 10 mM EDTA, 100 mM EGTA [pH 7.4]), layered onto 30 ml of a 0 to 40% linear gradient made in buffer C (1:10 dilution of buffer B), and centrifuged first at 1,500 × g for 30 min and then at 5,900 × g for 10 min. The glycerol was removed, and the pellet was lysed in 4 to 5 volumes of buffer C containing complete protease inhibitor. The sample was centrifuged (5,900 × g, 10 min) to remove unlysed platelets, and the supernatant was applied to a sucrose step gradient (10 ml of 33% sucrose on 5 ml of 66% sucrose in buffer C). After centrifugation in an SW28 rotor (90 min, 63,000 × g, 4°C), the membranes were removed, dialyzed against PBS containing 10% glycerol, and stored at −70°C.
Coimmunoprecipitation.
Platelet membranes (25 μg of total protein) were labeled with sulfosuccinimidyl-6-(biotinamido)hexanoate (Pierce) according to the manufacturer's instructions and then solubilized in binding buffer (PBS, 10% glycerol, 1.5% Triton X-100). The solubilized membrane proteins were incubated with 10 μg of rClfA in binding buffer (4°C, 60 min), followed by the addition of 30 μl of protein A-Sepharose that had been preincubated with 5 μg of anti-His monoclonal antibody (Amersham Pharmacia). This antibody specifically recognizes the histidine fusion tag on recombinant proteins. As controls for nonspecific binding, solubilized platelet membranes were also incubated separately with washed protein A-Sepharose alone or protein A-Sepharose coupled with anti-His. After further incubation (4°C, 60 min), the protein complexes were washed (three times with 1 ml of PBS–100 mM NaCl–0.1% Triton X-100, once with 1 ml of PBS–0.1% SDS, and once with 1 ml of PBS) and boiled in SDS-PAGE sample buffer to release the bound platelet membrane proteins. Following electrophoresis and transfer to Biotrace NT membranes, the recovered proteins were analyzed by Western blotting using a monoclonal antibody specific for glycoprotein IIb (GPIIb) (SZ22; Immunotech) or by probing with streptavidin conjugated to horseradish peroxidase.
Statistical methods.
Differences in platelet and fibrinogen binding were compared using the Student t test. P values of less than 0.05 were considered significant.
RESULTS
Identification of the gene interrupted in S. aureus PS12.
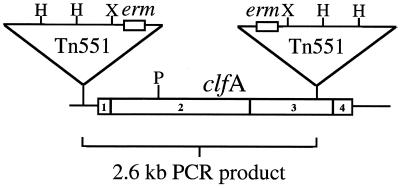
To determine the site of the Tn551 insertion within the PS12 chromosome, XhoI fragments of the PS12 genome were cloned into pBluescript SK(−). E. coli MC1061 was transformed with the ligation products by electroporation, followed by plating on LB agar supplemented with 500 μg of erythromycin per ml to select for clones containing plasmids with the erm gene of Tn551 and flanking PS12 DNA. Electrophoretic analysis of plasmid DNA from 10 randomly selected, erythromycin-resistant clones indicated that all 10 constructs contained inserts of 6.2 kb. One such plasmid (pRS401) was chosen for further characterization. DNA sequencing from the T7 and T3 primer sites on this plasmid indicated that two copies of the erm gene were present, with one located adjacent to each of the primer sites. Additional sequencing of pRS401 was performed using a primer (TNEND) that specifically hybridized with both ends of Tn551 (which are identical). Amplification of the plasmid insert in this manner produced two fluorescent signals at each nucleotide position, further suggesting that more than one copy of Tn551 was present. To circumvent this problem, we removed the 3.5-kb PstI fragment adjacent to the T3 primer site of pRS401, thereby producing the construct pRS402. Sequencing of pRS402 from the T3 site indicated that the chromosomal fragment cloned into the construct was a portion of the clumping factor gene clfA, which encodes a major fibrinogen binding protein of S. aureus (21). Sequencing of the entire insert in pRS401 was then completed using primers derived from the clfA sequence. The resultant sequence data confirmed that two copies of Tn551 had inserted in the PS12 chromosome. One copy is located upstream of the clfA start codon, and a second copy is inserted in a region encoding serine aspartate repeats (SD repeat) of the protein (Fig. 1).
FIG. 1.
Schematic representation of the S. aureus clumping factor gene (clfA) including relevant subdomains. The relevant subdomains are indicated as follows: 1, Signal sequence; 2, fibrinogen binding A region; 3, SD repeat; 4, wall anchor (LPXTG) and membrane-spanning domain. The positions and orientations of the Tn551 insertions are indicated. Abbreviations: H, HindIII; P, PstI; X, XhoI.
Confirmation of transposon copy number.
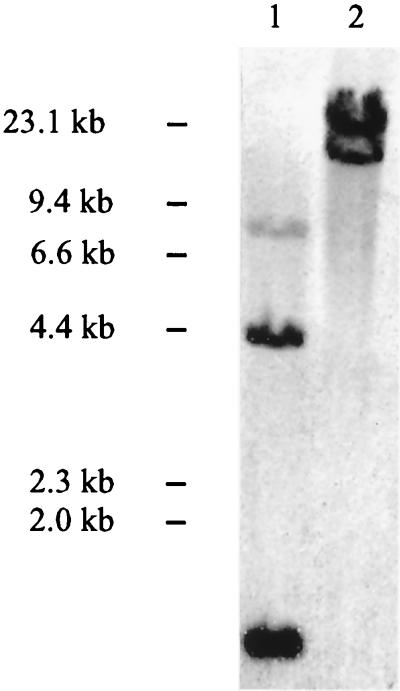
To further establish that two copies of the transposon had inserted in the S. aureus PS12 chromosome, Southern blotting was performed, using a probe specific for Tn551. This 3.9-kb probe encompasses 75% of the transposon sequence and overlaps both of the HindIII sites within Tn551. As we described previously (29), the probe appeared to hybridize with three bands in the HindIII digest of PS12 chromosomal DNA, suggesting that a single copy of the transposon was present. However, analysis of the DNA sequence flanking clfA indicated that HindIII digestion of this region could produce fragments of similar size. We were concerned, therefore, that Southern blotting of such digests could underestimate the number of Tn551 insertions within this region. For this reason, we examined a PS12 chromosomal DNA that had been treated with a different endonuclease. When examined by Southern blotting, the same probe hybridized with two bands in a PstI digest of the PS12 DNA (Fig. 2). Since Tn551 contains no PstI site, these latter results indicate that two copies of the transposon were inserted within the chromosome of PS12, thus confirming the DNA sequence data.
FIG. 2.
Southern blots of S. aureus PS12 chromosomal DNA digested with HindIII (lane 1) or PstI (lane 2). Chromosomal digests were hybridized with a digoxigenin-labeled, 3.9-kb fragment of Tn917 and imaged by chemiluminescence.
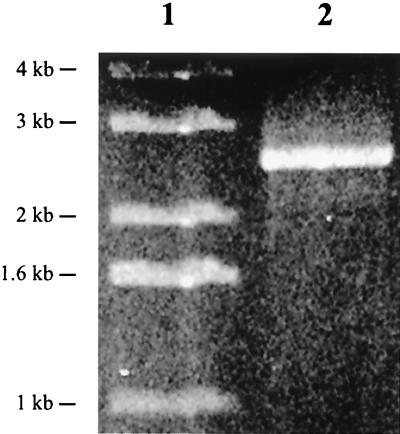
Sequencing also indicated that the two copies of Tn551 were inserted in opposite orientations within the S. aureus PS12 chromosome and separated by 2.6 kb. To confirm that two copies of Tn551 were present, a single primer (TNEND) corresponding to the end of the transposon was used to amplify PS12 chromosomal DNA by PCR. Since one primer was used in the reaction, a product could be obtained only if two copies of the transposon were present. As predicted, amplification produced a single fragment of 2.6 kb (Fig. 3), confirming that two copies of the transposon were inserted in opposite orientations within the PS12 genome.
FIG. 3.
PCR amplification of S. aureus PS12 chromosomal DNA. A single primer that recognizes the end of Tn551 was used to amplify chromosomal DNA from PS12. Lane 1, size standards; lane 2, single 2.6-kb product generated from the reaction.
Characterization of the effect of transposon insertion on protein expression.
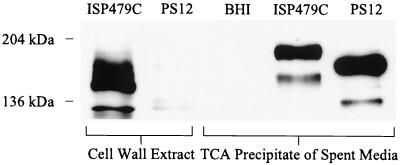
To assess the effects of the two transposon insertions on ClfA production, we examined the surface expression and extracellular secretion of ClfA by the parental and mutant strains. Western blot analysis of cell wall extracts probed with anti-ClfA region A antibodies indicated that clumping factor was present in the cell wall of S. aureus ISP479C. As noted previously with ClfA recovered from lysostaphin extracts (20), the protein migrated as two distinct bands with molecular masses of approximately 130 and 185 kDa. In contrast, little or no clumping factor was associated with the cell wall of PS12. Probing spent culture media of ISP479C in the same manner demonstrated that ClfA was present, though with an apparent increase in molecular mass. Western blotting of the culture supernatant from the mutant revealed that a truncated form of the protein was produced and released into the medium (Fig. 4). These results indicate that ClfA is still synthesized by PS12 but the protein is smaller and not retained in the wall of the organism. The production of truncated ClfA further indicates that the downstream copy of Tn551, which is inserted with the coding region, is responsible for the reduced platelet binding observed with strain PS12.
FIG. 4.
Localization of ClfA from S. aureus ISP479C and PS12. Western blot analysis of cell wall extracts and spent culture media precipitates from ISP479C and PS12. An equivalent volume of uninoculated media (BHI) was included as a control. The proteins were probed with antiserum raised against the A region of ClfA. Compared with the parental strain, ClfA was nearly undetectable in the cell wall extracts of PS12. Note the truncated form of the protein detected in the culture supernatants of PS12. TCA, trichloroacetic acid.
Disruption of clfA is not transcriptionally polar.
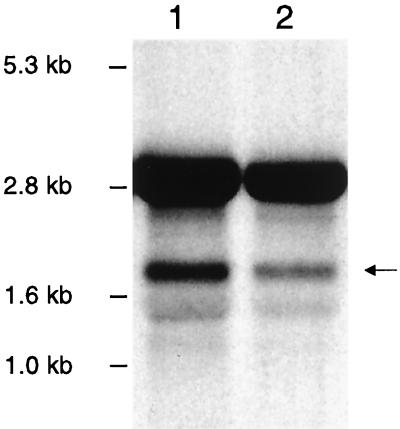
To confirm that the reduced platelet binding associated with disruption of clfA was not due to polar effects, we examined the transcription of the open reading frame (ORF) immediately downstream of clfA. This ORF was identified from the staphylococcal genome database at the University of Oklahoma, and based on that sequence, a 543-bp DNA probe specific for a portion of the coding sequence of the downstream ORF was generated by PCR. When RNA prepared from ISP479C was examined by Northern blotting, no hybridization with the probe was observed. The inability to detect transcripts by Northern blotting of RNA extracted from ISP479C was not wholly unexpected. Previous studies examining the expression of genes encoded by ISP479 had encountered similar difficulties, necessitating the use of another strain (34). For this reason, we then assessed the expression of the downstream ORF in S. aureus strain Newman. An isogenic variant of Newman (strain DU5852) in which clfA was disrupted by Tn917 was also assessed. When RNA from these strains was hybridized with the above 543-bp probe, transcripts of 1.6 kb were readily visible in both the Newman and DU5852 strains (Fig. 5). Thus, the transposon insertion within clfA does not alter downstream gene expression, indicating that the reduced platelet binding seen in PS12 is likely due to disruption of clfA and not a result of polar effects.
FIG. 5.
Northern blot analysis of total RNA isolated from S. aureus strains Newman (lane 1) and DU5852 (lane 2) grown to mid-exponential phase and hybridized with a digoxigenin-labeled probe generated from the ORF immediately downstream of clfA. Note the band at approximately 1.8 kb (arrow) for both samples, representing the transcript downstream of clfA. 23S and 16S rRNA can also be seen at approximately 2.8 and 1.5 kb, respectively.
Binding of S. aureus to fibrinogen.
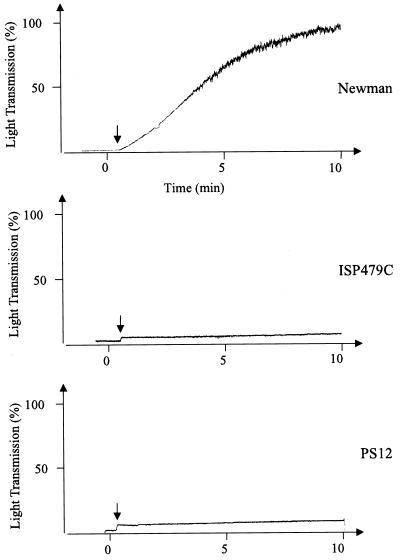
It was surprising to find that clfA was disrupted in PS12, since we had observed previously that the binding of soluble, radiolabeled fibrinogen by strains PS12 and ISP479C were comparable (29). To assess the fibrinogen binding properties of these strains in greater detail, we compared their ability to clump in the presence of soluble fibrinogen, as measured by a turbidimetric assay. Strain Newman, in which ClfA was first identified, was used as a positive control. Addition of fibrinogen to Newman in suspension produced rapid clumping, with maximal (100%) light transmission occurring within 10 min (Fig. 6). In contrast, addition of fibrinogen to strain ISP479C or PS12 produced no change in light transmission (Fig. 6), indicating that no clumping had occurred.
FIG. 6.
Clumping of S. aureus by soluble human fibrinogen. Arrows indicate time points when fibrinogen was added to suspensions of strain Newman, ISP479C, or PS12. The samples were monitored continuously in an aggregometer for turbidity. Clumping was detected as an increase in light transmission.
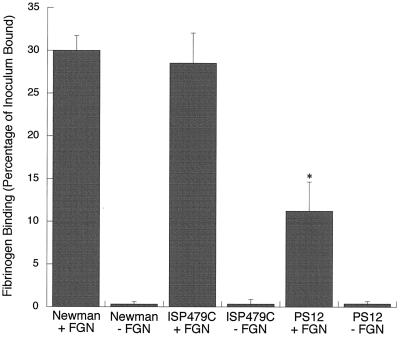
We then compared strains Newman, ISP479C, and PS12 for their binding to fibrinogen that had been immobilized in plastic wells. For Newman, 30.0% ± 1.73% of the inoculum was bound to immobilized fibrinogen after 1 h (Fig. 7). Of interest, ISP479C bound 28.52% ± 3.52% of the inoculum under the same conditions, a level comparable to that of Newman binding. However, only 11.22% ± 3.43% of the PS12 inoculum was bound, i.e., a 59% reduction in binding relative to ISP479C (P < 0.0001, n = 9). For all strains, the levels of binding to fibrinogen were significantly higher than binding to plastic wells treated with casein alone (inoculum-bound values were 0.31% ± 0.29% for Newman, 0.33% ± 0.53% for ISP479C, and 0.38% ± 0.28% for PS12). These results indicate that ClfA expressed by ISP479C appears to bind immobilized fibrinogen preferentially. Mutation of clfA, as has occurred in PS12, results in a loss of this binding.
FIG. 7.
Binding of S. aureus to immobilized human fibrinogen. The levels of binding of strains Newman, ISP479C, and PS12 to human fibrinogen immobilized in tissue culture wells (+FGN) were compared. Casein-blocked wells that had not been coated with fibrinogen (−FGN) served as controls for nonspecific binding. Results are expressed as the means ± standard deviations of nine experiments. The asterisk indicates statistical significance (P < 0.0001) compared with the binding of ISP479C.
Effects of rClfA treatment on the binding of S. aureus to human platelets.
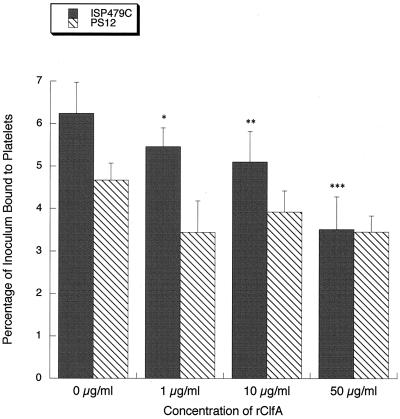
To further establish the role of ClfA in mediating platelet binding by S. aureus, we examined the effects of purified rClfA on the adherence of the bacteria to platelet monolayers. In control studies with strain PS12, pretreatment of platelet monolayers with 1 μg of rClfA per ml reduced platelet binding from 4.67% ± 0.40% to 3.44% ± 0.38% of the inoculum (Fig. 8). No further reduction in binding was seen at higher concentrations of the recombinant protein. In contrast, platelet binding by strain ISP479C was progressively reduced by increasing concentrations of rClfA. Moreover, the levels of inhibition observed were statistically significant at all concentrations tested. At 50 μg of rClfA per ml, platelet binding was reduced from 6.24% ± 0.73% to 3.51% ± 0.77% of the inoculum, representing a relative reduction of 44%. Of note, this concentration of rClfA reduced platelet binding by ISP479C to levels that were comparable to those observed with PS12, when tested under the same conditions. These results demonstrate that ClfA can specifically block platelet binding by ISP479C, indicating that it is a platelet adhesin.
FIG. 8.
Inhibition of staphylococcus-platelet binding by rClfA. Strains ISP479C and PS12 were added to immobilized platelet monolayers pretreated with 0 to 50 μg of rClfA per ml. Binding is expressed as a percentage of the inoculum (mean ± standard deviation). ISP479C binding to platelets was significantly reduced at all rClfA concentrations tested compared with untreated platelets (0 μg/ml) (∗, P = 0.05; ∗∗, P = 0.02; ∗∗∗, P < 0.0001) (n = 6). Note that at 50 μg/ml of rClfA, binding of ISP479C is reduced to that of PS12.
Detection of a ClfA receptor on the platelet membrane.
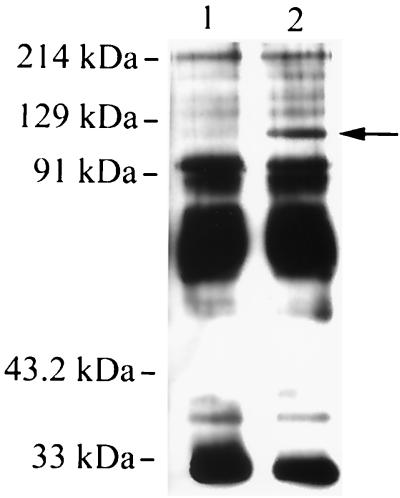
To assess the binding sites on platelets for ClfA, solubilized platelet membrane proteins were incubated with rClfA, followed by immunoprecipation with anti-His that had been coupled to protein A. As a control for nonspecific binding, parallel experiments were done in which rClfA was omitted. In these control studies, several platelet membrane proteins in the 30-to-100-kDa range were nonspecifically recovered (Fig. 9, lane 1). However, when platelet membrane proteins were first incubated with rClfA and then treated with anti-His bound to protein A, an additional membrane protein of approximately 118 kDa was precipitated (Fig. 9, lane 2).
FIG. 9.
Coimmunoprecipitation of rClfA and platelet membrane proteins. Biotinylated platelet membranes were solubilized and immunoprecipitated, using an anti-His monoclonal antibody, in the absence (lane 1) or presence (lane 2) of rClfA. The immunoprecipitates were separated by SDS-PAGE, transferred to membranes, and visualized by chemiluminescence using streptavidin conjugated with horseradish peroxidase. In addition to several proteins that were precipitated nonspecifically, note the additional 118-kDa protein coprecipitated by rClfA (arrow).
Of the major platelet membrane proteins characterized to date, GPIIb has a molecular mass (115 to 130 kDa) closest to 118 kDa (4). To assess if GPIIb was the 118-kDa protein recovered by coimmunoprecipitation with rClfA, we probed the immunoprecipitated material by Western blotting, using a monoclonal antibody specific for GPIIb. In control studies using solubilized platelet membrane proteins, GPIIb was readily detected with this monoclonal antibody. However, no GPIIb was detected in proteins precipitated with rClfA. These results indicate that the 118-kDa protein is not GPIIb.
DISCUSSION
ClfA has been characterized as a major fibrinogen binding protein of S. aureus (21). The protein itself has a modular structure that includes the N-terminal A region containing the fibrinogen binding site, and an EF hand domain mediating Ca2+ binding (20, 22, 25). Immediately adjacent to the A region is the SD repeat that is thought to extend the receptor portion of the protein away from the cell wall (12). The protein also contains an LPXTG domain, which allows ClfA to be covalently cross-linked to the bacterial peptidoglycan, at its C-terminal end (21). In strain Newman, ClfA is able to readily bind both soluble fibrinogen and fibrinogen immobilized onto surfaces (21).
In addition to binding fibrinogen, our studies indicate that ClfA can also contribute to platelet binding by S. aureus. Our sequencing data have shown that the low platelet binding phenotype of strain PS12 is linked to disruption of clfA by transposon insertion within the SD repeat region of the gene. This mutation results in the production of a truncated form of ClfA, which appears to have lost its cell wall-anchoring domain. As a consequence, little ClfA remains associated with the bacterial surface. Disruption of clfA by transposon insertion had no effect on the transcription of the ORF immediately downstream from the insertion site, indicating that the reduction in platelet binding associated with this mutation was not due to polar effects.
Further evidence that ClfA mediates platelet binding by S. aureus was seen in our studies in which rClfA was assessed for its ability to inhibit this interaction. In these experiments, addition of rClfA significantly reduced platelet binding by strain ISP479C at all concentrations tested. This inhibition was proportional to the amount of rClfA added to the reaction mixture, with 50 μg of the recombinant protein per ml reducing ISP479C binding by 44%. Of note, the level of platelet binding by ISP479C observed at this concentration of rClfa was comparable to that seen with PS12 tested under identical conditions. These results indicate that the quantitative difference in platelet binding between these two strains can be attributed to differences in ClfA surface expression. The small reduction of PS12 binding produced by rClfA may be explained by our finding that this strain is not totally devoid of cell wall-associated ClfA (Fig. 4). It is possible, therefore, that PS12 continues to have low-level, ClfA-mediated platelet binding, which was blocked by the recombinant protein.
These results, in conjunction with our immunoprecipitation data, indicate that ClfA can mediate the direct binding of S. aureus to human platelets. Thus, it appears that ClfA is a multifunctional protein, a property that has recently been described for a number of bacterial adhesins. For example, the S. aureus fibronectin binding protein FnbpA has been shown to also interact with fibrinogen (32). This binding occurs through a domain resembling the A region of ClfA that is distinct from the fibronectin binding region of the adhesin. In addition, protein A has been shown to bind IgG, the gC1qR protein on platelet membranes, and von Willebrand's factor (13, 24). The binding sites within protein A that mediate these interactions have yet to be defined. Other examples of multifunctional proteins can be found in the viridans group streptococci, including CshA, SspA, and SspB from Streptococcus gordonii, and FimA from Streptococcus parasanguis (5, 9, 23). Therefore, it appears that numerous bacterial adhesins possess multiple and at times overlapping functions. The impact of multifunctional surface components on virulence is not known. However, it is conceivable that such multipurpose adhesins may enhance microbial survival under different conditions in vivo.
It was surprising to find that we had isolated a mutant with a disruption of the clfA gene, since previous characterization of PS12 indicated that strains PS12 and ISP479C bound comparable amounts of soluble, radiolabeled fibrinogen (29). In our current study, when we measured binding of soluble fibrinogen by a turbidimetric assay, both the parental and mutant strains exhibited minimal clumping in response to fibrinogen. This suggests that ClfA, at least when expressed by ISP479C, has minimal binding to soluble fibrinogen. Since S. aureus is known to produce other fibrinogen binding proteins (32) it is possible that fibrinogen binding observed in our previous studies was mediated by these other surface components.
In contrast to our findings with soluble fibrinogen, strains ISP479C and PS12 did differ significantly in their binding to immobilized fibrinogen. Compared with the parental strain, PS12 had markedly reduced binding to immobilized fibrinogen. It appears, therefore that ClfA expressed by ISP479C binds immobilized fibrinogen preferentially. The selective recognition of soluble versus immobilized forms of a receptor has been described for other organisms. For example, the phase-dependent binding of S. gordonii antigen I/II to its ligands is thought to correctly target the organism for colonization (17). Although a mechanism has not been described for this selective binding by antigen I/II, sequence data indicates how this differential recognition may occur in ClfA. The fibrinogen binding A region of clumping factor has been identified and includes at minimum amino acids 221 to 550 (20). The clfA gene in ISP479C has a single base pair change (C→T) in this region, resulting in the replacement of an alanine for a valine at position 228. In fact, this single alteration is the only difference between the clfA genes in strains Newman and ISP479C. Although this change does not completely abrogate binding, it may affect the ability of the protein to interact with fibrinogen under different conditions.
As for the binding site on platelets for ClfA, our coimmunoprecipitation studies indicate that clumping factor binds to human platelets directly via a 118-kDa membrane protein. The identity of this protein is uncertain. Although fibrinogen can be found on the surface of activated platelets (14), none of its subunits has a molecular mass similar to that of the membrane protein identified in our studies (6). Of the characterized platelet membrane proteins, including GPIa, GPIbα, GPIIa, GPIIbα, and GPIIIa, the only receptor with a molecular mass close to 118 kDa is the GPIIbα subunit of the integrin αIIbβ3 (4). However, our Western blotting studies using a monoclonal antibody specific for GPIIb failed to detect the protein in the ClfA coimmunoprecipitates, indicating that GPIIb is not the ClfA receptor. It is possible that the ClfA receptor is a known platelet membrane protein that had atypical electrophoretic mobility due to the experimental conditions used in our studies. If so, then these studies reveal a previously uncharacterized function for that protein. It is also possible that this 118-kDa receptor is a previously unrecognized platelet membrane component. Studies to further characterize this ClfA binding protein are now in progress.
ACKNOWLEDGMENTS
This work was supported in part by grants AI41513 (P.M.S.), AI39108 (A.S.B.), and AI37142 (A.L.C.) from the National Institutes of Health and by the Merit Review Program of the Department of Veterans Affairs.
We thank L. I. Kupferwasser, Gordon Archer, and Barbara Bensing for helpful discussions.
REFERENCES
- 1.Bayer A S, Sullam P M, Ramos M, Li C, Cheung A L, Yeaman M R. Staphylococcus aureus induces platelet aggregation via a fibrinogen-dependent mechanism which is independent of principal platelet glycoprotein IIb/IIIa fibrinogen-binding domains. Infect Immun. 1995;63:3634–3641. doi: 10.1128/iai.63.9.3634-3641.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung A L, Krishnan M, Jaffe E A, Fischetti V A. Fibrinogen acts as a bridging molecule in the adherence of Staphylococcus aureus to cultured human endothelial cells. J Clin Invest. 1991;87:2236–2245. doi: 10.1172/JCI115259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford N, Authi K S, Hack N. Isolation and characterization of platelet membranes prepared by free flow electrophoresis. Methods Enzymol. 1992;215:5–20. doi: 10.1016/0076-6879(92)15048-h. [DOI] [PubMed] [Google Scholar]
- 5.Demuth D R, Duan Y, Brooks W, Holmes A R, McNab R, Jenkinson H F. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol Microbiol. 1996;20:403–413. doi: 10.1111/j.1365-2958.1996.tb02627.x. [DOI] [PubMed] [Google Scholar]
- 6.Doolittle R F. Fibrinogen and fibrin. Annu Rev Biochem. 1984;53:195–229. doi: 10.1146/annurev.bi.53.070184.001211. [DOI] [PubMed] [Google Scholar]
- 7.Durack D T. Experimental bacterial endocarditis. IV. Structure and evolution of very early lesions. J Pathol. 1975;115:81–89. doi: 10.1002/path.1711150204. [DOI] [PubMed] [Google Scholar]
- 8.Duthie E S, Lorenz L L. Staphylococcal coagulase: mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 9.Fenno J C, Shaikh A, Spatafora G, Fives-Taylor P. The fimA locus of Streptococcus parasanguis encodes an ATP-binding membrane transport system. Mol Microbiol. 1995;15:849–863. doi: 10.1111/j.1365-2958.1995.tb02355.x. [DOI] [PubMed] [Google Scholar]
- 10.Gong K, Wen D Y, Ouyang T, Rao A T, Herzberg M C. Platelet receptors for the Streptococcus sanguis adhesin and aggregation-associated antigens are distinguished by anti-idiotypical monoclonal antibodies. Infect Immun. 1995;63:3628–3633. doi: 10.1128/iai.63.9.3628-3633.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmon J T, Greco N J, Jamieson G A. Isolation of human platelet plasma membranes by glycerol lysis. Methods Enzymol. 1992;215:32–36. doi: 10.1016/0076-6879(92)15050-m. [DOI] [PubMed] [Google Scholar]
- 12.Hartford O, Francois P, Vaudaux P, Foster T J. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Mol Microbiol. 1997;25:1065–1076. doi: 10.1046/j.1365-2958.1997.5291896.x. [DOI] [PubMed] [Google Scholar]
- 13.Hartleib J, Kohler N, Dickinson R B, Chhatwal G S, Sixma J J, Hartford O M, Foster T J, Peters G, Kehrel B E, Herrmann M. Protein A is the von Willebrand factor binding protein on Staphylococcus aureus. Blood. 2000;96:2149–2156. [PubMed] [Google Scholar]
- 14.Hawiger J, Timmons S. Binding of fibrinogen and von Willebrand factor to platelet glycoprotein IIb-IIIa complex. Methods Enzymol. 1992;215:228–243. doi: 10.1016/0076-6879(92)15067-m. [DOI] [PubMed] [Google Scholar]
- 15.Herrmann M, Hartleib J, Kehrel B, Montgomery R R, Sixma J J, Peters G. Interaction of von Willebrand factor with Staphylococcus aureus. J Infect Dis. 1997;176:984–991. doi: 10.1086/516502. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann M, Suchard S J, Boxer L A, Waldvogel F A, Lew P D. Thrombospondin binds to Staphylococcus aureus and promotes staphylococcal adherence to surfaces. Infect Immun. 1991;59:279–288. doi: 10.1128/iai.59.1.279-288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkinson H F. Genetics of Streptococcus sanguis. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: ASM Press; 2000. pp. 287–294. [Google Scholar]
- 18.Joh D, Wann E R, Kreikemeyer B, Speziale P, Hook M. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 1999;18:211–223. doi: 10.1016/s0945-053x(99)00025-6. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.McDevitt D, Francois P, Vaudaux P, Foster T J. Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol Microbiol. 1995;16:895–907. doi: 10.1111/j.1365-2958.1995.tb02316.x. [DOI] [PubMed] [Google Scholar]
- 21.McDevitt D, Francois P, Vaudaux P, Foster T J. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 22.McDevitt D, Nanavaty T, House-Pompeo K, Bell E, Turner N, McIntire L, Foster T, Hook M. Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur J Biochem. 1997;247:416–424. doi: 10.1111/j.1432-1033.1997.00416.x. [DOI] [PubMed] [Google Scholar]
- 23.McNab R, Holmes A R, Clarke J M, Tannock G W, Jenkinson H F. Cell surface polypeptide CshA mediates binding of Streptococcus gordonii to other oral bacteria and to immobilized fibronectin. Infect Immun. 1996;64:4204–4210. doi: 10.1128/iai.64.10.4204-4210.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen T, Ghebrehiwet B, Peerschke E I. Staphylococcus aureus protein A recognizes platelet gC1qR/p33: a novel mechanism for staphylococcal interactions with platelets. Infect Immun. 2000;68:2061–2068. doi: 10.1128/iai.68.4.2061-2068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Connell D P, Nanavaty T, McDevitt D, Gurusiddappa S, Hook M, Foster T J. The fibrinogen-binding MSCRAMM (clumping factor) of Staphylococcus aureus has a Ca2+-dependent inhibitory site. J Biol Chem. 1998;273:6821–6829. doi: 10.1074/jbc.273.12.6821. [DOI] [PubMed] [Google Scholar]
- 26.Smeltzer M S, Hart M E, Iandolo J J. Phenotypic characterization of xpr, a global regulator of extracellular virulence factors in Staphylococcus aureus. Infect Immun. 1993;61:919–925. doi: 10.1128/iai.61.3.919-925.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 28.Sullam P M. Host-pathogen interactions in the development of bacterial endocarditis. Curr Opin Infect Dis. 1994;4:304–309. [Google Scholar]
- 29.Sullam P M, Bayer A S, Foss W M, Cheung A L. Diminished platelet binding in vitro by Staphylococcus aureus is associated with reduced virulence in a rabbit model of infective endocarditis. Infect Immun. 1996;64:4915–4921. doi: 10.1128/iai.64.12.4915-4921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullam P M, Payan D G, Dazin P F, Valone F H. Binding of viridans group streptococci to human platelets: a quantitative analysis. Infect Immun. 1990;58:3802–3806. doi: 10.1128/iai.58.11.3802-3806.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wann E R, Gurusiddappa S, Hook M. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J Biol Chem. 2000;275:13863–13871. doi: 10.1074/jbc.275.18.13863. [DOI] [PubMed] [Google Scholar]
- 33.Wertman K F, Wyman A R, Botstein D. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene. 1986;49:253–262. doi: 10.1016/0378-1119(86)90286-6. [DOI] [PubMed] [Google Scholar]
- 34.Wolz C, Pohlmann-Dietze P, Steinhuber A, Chien Y T, Manna A, van Wamel W, Cheung A. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol Microbiol. 2000;36:230–243. doi: 10.1046/j.1365-2958.2000.01853.x. [DOI] [PubMed] [Google Scholar]
- 35.Woodcock D M, Crowther P J, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith S S, Michael M Z, Graham M W. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeaman M R, Sullam P M, Dazin P F, Norman D C, Bayer A S. Characterization of Staphylococcus aureus-platelet binding by quantitative flow cytometric analysis. J Infect Dis. 1992;166:65–73. doi: 10.1093/infdis/166.1.65. [DOI] [PubMed] [Google Scholar]