Abstract
Two major aflatoxin-producing strains are Aspergillus flavus and Aspergillus niger. Probiotic bacteria have been identified as a potential means to fight aspergilli and reduce the availability of aflatoxin (AFs) as well as other food contaminants. In this study, the potential of ABRIIFBI-6 and ABRIIFBI-7 strains to inhibit the growth of aspergilli was investigated. Both strains survived in the simulated gastrointestinal conditions and inhibited the growth of Aspergillus significantly. Auto-aggregation ranged from 67.4 ± 1.9 for ABRIIFBI-6 to 75.8 ± 2.3% for ABRIIFBI-7, and hydrophobicity ranged from 57.3 ± 1.6 to 61.2 ± 1.4% for ABRIIFBI-6 and ranged from 51.2 ± 1.4 to 55.4 ± 1.8% for ABRIIFBI-7. The ranges of coaggregation with Staphylococcus aureus were 51.3 ± 1.7 and 52.4 ± 1.8% for ABRIIFBI-6 and ABRIIFBI-7, respectively, while coaggregation with Bacillus cereus was 57.9 ± 2.1 and 49.3 ± 1.9% for ABRIIFBI-6 and ABRIIFBI-7, respectively. Both strains indicated remarkable sensitivity to clinical antibiotics. According to the analysis of the identified potential probiotics, the findings of this study could significantly contribute to the understanding of the probiotic potential of LAB in dairy products in order to access their probiotic characterization for use as biocontrol of aflatoxin-producing species.
Subject terms: Biotechnology, Microbiology
Introduction
Moldy contamination of basic foods like cereal has been identified as one of the most important food safety issues. This is owing to the acute and chronic harm they cause to people and animals. Because of their biological and metabolic impacts on living systems, Aspergillus flavus and Aspergillus niger are very significant1. As secondary metabolites, A. flavus and A. niger, as well as other Aspergillus species, produce aflatoxins2. Molds that produce aflatoxin are found all over the world and can thrive on a variety of food and feed commodities during production, processing, storage, and transportation3. Aflatoxin production in food is controlled by a variety of factors, including temperature and humidity4. When these molds contaminate crops, particularly in hot and humid conditions, customers incur health issues. Aflatoxin is a potent carcinogen and mutagen with hepatotoxic and immunosuppressive properties, as well as the ability to disrupt various metabolic activities, resulting in liver and kidney damage5. B1, B2, G1, and G2 are the four most frequent aflatoxins discovered in mold-infested food crops. When nursing animals consume aflatoxin B1 (AFB1) via feed, it is physically changed in the body and released into the milk as aflatoxin M1 (AFM1)6. Furthermore, since aflatoxin is not digested by animals, its remains may be found in their meat7. Aflatoxin is one of the most dangerous food pollutants because even low doses are mutagenic and can cause hepatotoxicity and teratogenicity8. AFB1 is the most dangerous of the four primary forms of aflatoxins, and it has been classified as a human carcinogen by the International Agency for Research on Cancer (IARC) because it causes hepatic cancer9.
Government bodies all throughout the globe monitor the level of aflatoxin contamination in food8. The maximum allowed level for AFB1 in the European Union is 2 ppb; in the United States, it is 20 ppb10. Because of AFB1’s high thermal and mechanical stability, detoxification of contaminated food products is difficult. The concentration of aflatoxin in cereal grains is unaffected by processing11. There are various physical and chemical methods to prevent fungal and aflatoxin contamination, but these methods are non-biodegradable and must be applied to the product via heat, ionizing radiation, or pesticide and fungicide spraying. However, it causes both sensory changes and irreversible environmental damage12.
As a result, an effective, ecologically acceptable, and customizable approach for preventing aflatoxin contamination of food crops is required. Biological control, which includes the use of bacterial probiotic strains, has shown to be an effective method of achieving the same outcomes. Probiotics are among the nutritional supplements with the highest worldwide growth. Their commercial success in food is much greater. Yogurt is the most common source of probiotics, but many other foods, including as cheese, fruit juices, and cereals, are becoming more popular13,14.
Probiotic microorganisms, according to FAO/WHO, are “living microorganisms that, when consumed in appropriate proportions, provide health benefits to the host”15–17. This definition highlights the core of probiotics while addressing a broad variety of bacteria and uses. The definition distinguishes between live microbes used as processing aids or sources of useful compounds and those administered primarily for their health benefits. Although commensals in the gut are frequently the source of probiotic strains, they cannot be called probiotics until these strains are isolated and characterized and a credible case for their health effects is presented. As a result, the term “probiotic” has been restricted in some countries because it is deemed misleading to consumers in the absence of approved health claims17.
Many grains have long been fermented with probiotic bacteria to increase their nutritional content while also acting as antimicrobial agents, making them safer to consume. The production of organic acids and bacteriocins by probiotic lactic acid bacteria (LAB) has been linked to antibacterial activity18. Some of these LABs, particularly those belonging to the genus Lactobacillus, have been shown to inhibit fungal growth. Antifungal compounds such as phenyl lactic acid, p-hydroxyl phenyl lactic acid, and other antifungal cyclic dipeptides are produced as a result19. This property is provided by the various antifungal chemicals produced primarily at the beginning of the process. In addition to its antifungal properties, LAB has been shown to bind and eliminate aflatoxin in vitro and in vivo18.
Probiotics have the potential to reduce aflatoxin absorption because some LAB cell wall components aid in the binding and removal of aflatoxin from solution. As a result, aflatoxins are less accessible and are removed from probiotic cell walls more quickly. Toxin elimination must occur within living cells, because mycotoxins can be eliminated by microbes and their metabolites20. The bacteria colonized within regular fermented yogurt were isolated and identified as capable probiotic microorganisms in this study. The purpose of this research is to investigate how effective two Lactobacillus probiotic bacteria are at suppressing aflatoxin production.
Materials and methods
LAB strains and culture condition
Fresh traditional fermented yogurt samples from different parts of Iran were collected. The samples were brought on ice to the laboratory of food biotechnology at the Agricultural Biotechnology Research Institute of Iran (ABRII), Tabriz, Iran. To separate the LAB from the sample, inoculate 50 mL of de Man Rogosa and Sharpe broth (MRS, Merck, Darmstadt, Germany) with 25 g/L NaCl and incubate at 37 °C for 48 h. After the incubation period, all cultures were separately drawn on MRS agar plates and anaerobically incubated at 37 °C for 48 h in an anaerobic vessel equipped with an anaerobic gas generation kit. Different forms of colonies were isolated from cultured MRS agar plates. After morphological and biochemical analysis (Gram stain, catalase activity, cell morphology, and spore formation), gram-positive and catalase-negative bacilli and cocci were identified as LAB isolates.
Acid and bile resistance
To test acid resistance, MRS broth with pepsin enzyme (3 mg/mL) was applied as a medium. With 1.0 N HCl, the pH of the broth was changed to 2.5, and broth (pH 7.0) was utilized as a control. In addition, the broth was inoculated for 3 h to determine the optical density (OD) at 600 nm. Resistance to low pH was calculated by the following formula:
Strains with survival rates of more than 80% were chosen for further study21. The growth potential of the isolate was tested in the presence of 0.3% w/v bile salts. The samples were inoculated for 4 h at 37 °C, and the OD620 of the samples was evaluated to ensure cell viability. The resistivity of oxgall was calculated using the equation below21:
Inhibition of aflatoxin-producing aspergilli by LAB isolates
The modified overlay method was used to assess the inhibition of aflatoxin-producing Aspergillus flavus and Aspergillus niger22 by the two selected LAB isolates. To accomplish this, selected LAB were streaked on MRS agar plates and incubated in an anaerobic container at 37 °C for 48 h. The soft potato dextrose agar (PDA, 75 wt%) was then prepared and overlaid on the MRS plates with fungus spores of defined inoculation size (9.5 × 104 spores/mL) as determined by counting on the Neubauer hemocytometer. The plates were then incubated aerobically at 30 °C for 7 days. Each LAB isolate was tested twice against each Aspergillus strain, with one control for each. They were looking for discrete zones of inhibition surrounding the bacterial streaks, which they called “positive impact zones”.
Antimicrobial activity
An agar well-diffusion experiment was used to determine the antibacterial properties of isolates. For this purpose, Mueller–Hinton agar was used as a medium and some different indicator bacteria, including Escherichia coli (PTCC 1276), Salmonella enterica (PTCC-1709), Listeria monocytogenes (PTCC 1163), Staphylococcus aureus (PTCC 1764), Bacillus cereus (PTCC 1539), Streptococcus iniae (PTCC 1887), Shigella flexneri (ATCC 9199), Klebsiella pneumoniae (ATCC 43816), and Yersinia enterocolitica (ATCC 23715) were used. To make wells in the medium, a sterile, pasteurized pipette was utilized.
Finally, 100 mL of isolate supernatants were poured into each well, and the plates were incubated at 37 °C for 24 h. After the incubation period, the measured inhibitory halo zone diameters were statistically examined23.
Antibiotic susceptibility
The disc diffusion method was used to test isolates susceptibility to many high-consumption and therapeutically significant antibiotics (see Table 1). The isolates were swabbed on solidified MRS medium, antibiotic discs were placed on the medium, and it was incubated overnight at 37 °C. Finally, the size of the suppression zone around the disc was measured with a digital caliper24.
Table 1.
Antibiotic susceptibility of selected strains against the high consumption antibiotics performed by disk diffusion assay.
| Strains | Antibiotic susceptibility (Clear zone (mm)) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | V | CFM | FEB | K | SXT | TE | CP | AM | E | CC | GM | C | CN | CRO | |
| ABRIIFBI-6 | 19 (S) | 11 (R) | 21 (S) | 23 (S) | 5 (R) | 18 (S) | 20 (S) | 15 (I) | 22 (S) | 29 (S) | 16 (I) | 12 (S) | 18 (S) | 21 (S) | 24 (S) |
| ABRIIFBI-7 | 22 (S) | 10 (R) | 24 (S) | 21 (S) | 3 (R) | 21 (S) | 28 (S) | 14 (I) | 19 (S) | 26 (S) | 17 (I) | 11 (S) | 20 (S) | 22 (S) | 21 (S) |
S streptomycin, V vancomycin, CFM cefixime, FEB cefepime, K kanamycin, SXT sulfamethoxazole, TE tetracycline, CP ciprofloxacin, AM ampicillin, E erythromycin, CC clindamycin, GM gentamycin, C chloramphenicol, CN cephalexin, CRO ceftriaxon.
Erythromycin results based on R ≤ 13 mm; I: 13–23 mm; S ≥ 23 mm.
Gentamycin results based on R ≤ 6 mm; I: 7–9 mm; S ≥ 10 mm.
Vancomycin results based on R ≤ 12 mm; I: 12–13 mm; S ≥ 13 mm.
I: intermediate (zone diameter, 12.5–17.4 mm); R: resistant (zone diameter, ≤ 12.4 mm); S: susceptible (zone diameter, ≥ 17.5).
Cell surface hydrophobicity
As previously mentioned, isolates’ capacity to adhere to xylene and toluene was tested24. The The test was run three times and was expressed by the following formula:
where A0 is the absorbance at 600 nm before xylene and toluene are added and A1 is the absorbance after xylene and toluene have been added for 4 h.
Autoaggregation assay
The ability of isolates to adhere to xylene and toluene was examined, as previously reported. The percentage of auto-aggregation was measured using this formula:
where A0 represents the absorbance at time t = 0, and at represents the absorbance at time t.
Coaggregation assay
Based on the method used, isolates were aggregated against S. aureus and B. cereus25. The coaggregation rate was calculated based on the following formula:
Biofilm formation
The ability of isolates to produce biofilms was assessed using Gomez et al., method 26, with some modifications. Sterile 6-well tissue culture plates were filled with 5 mL of MRS broth supplemented with 500 mL of overnight isolate (107–108 CFU/mL). For this purpose, the cultures were anaerobically incubated at 37 °C for 48 h. After this, the wells were gently washed three times with 5 mL of sterile distilled water. The adherent bacteria were then fixed using 3 mL of methanol for 15 min, after which the plates were emptied and dried at room temperature. Then 3 mL of a 2% (v/v) crystal violet solution was poured into the wells and left at room temperature for 5 min. Finally, 2 mL of 33% (v/v) glacial acetic acid was used to lyse the dye from the adherent cells, and the OD of each well was measured at 595 nm with a plate reader (Bio-Rad, Hercules, CA, USA)26.
Molecular identification
Genomic DNA extraction
Total genomic DNA was isolated from the isolates using a technique developed in our lab. A single colony of each isolate was inserted in a 0.2 PCR tube, and 20 L of lysis buffer was added to the tube for this purpose. The tubes were vortexed gently until the contents were homogenous, and then left at room temperature for 1 h. They were then incubated for 10 min at 85 °C in a thermal cycler PTC 200 (MJC Research, Waltham, USA). After this time, the tubes were filled with 150 L of deionized water and centrifuged at 8000×g for 5 min. Finally, the upper phase containing genomic DNA was taken out of the tubes and poured into new ones. The genomic DNA was kept in the refrigerator until it was needed27.
Amplification of 16S-rRNA gene by polymerase chain reaction (PCR)
Using LAB-specific universal primers, genomic DNA samples from the isolates were amplified in a PTC 200 thermal cycler23. The following temperature profile was used for amplification. First, DNA denaturation at 95 °C for 5 min is followed by denaturation at 94 °C for 60 s, annealing at 59 °C for 60 s, elongation at 72 °C for 60 s, and 32 cycles of the final elongation step at 72 °C for 5 min. PCR products were separated using electrophoresis on a 0.8% (w/v) agarose gel and stained with ethidium bromide.
16S-rRNA gene sequencing
The 16S rRNA gene (1544 bp) PCR products were produced using the aforementioned primer set. Macrogene Corporation, based in South Korea, sequenced the PCR products. The sequences were then examined using the National Center for Biotechnology Information’s BLAST program. (http://www.ncbi.nlm.nih.gov/BLAST).
Statistical analysis
To determine the significant differences between the parameters of each isolate (P ≤ 0.05), analysis of variance (ANOVA) and Duncan’s test were used. Moreover, Excel 2013 (Microsoft Corporation) and SPSS (IBM SPSS Statistics 20) were used for formal statistical analysis.
Results
Morphological and biochemical assays
A total of 31 rod- and cocci-shaped colonies were produced on culture media and identified as LAB isolates using gram-positive and catalase-negative tests. For acid and bile resistance testing, each colony was propagated independently.
Acid and bile tolerance
After 3 h of incubation at pH 2.5 and 4 h of incubation in 0.3% oxgall, the survival rates of the tested strains varied from 9.44 to 100% and 16.08 to 100%, respectively. According to the findings, most isolates survived between 29 and 63 percent after 3 h of incubation at pH 2.5 and 4 h of incubation in 0.3% oxgall, respectively. Only two isolates (ABRIIFBI-6 and ABRIIFBI-7) demonstrated more than 90% acid and bile resistance, with survival rates of 91 and 96% at pH 2.5 and 92 and 98% in 0.3% bile oxgall, respectively. As a result, these two isolates were chosen for further investigation.
Antifungal activity
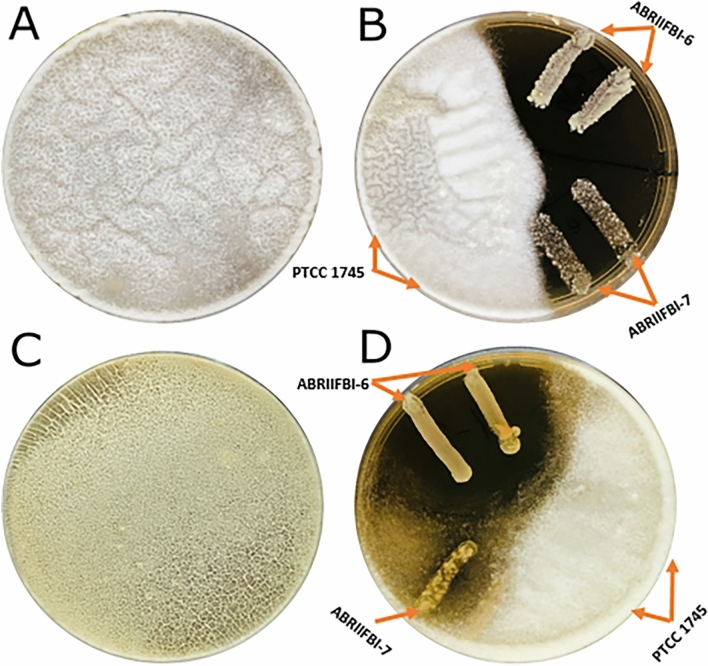
The antifungal activity results are shown in Fig. 1. According to the findings, both A. niger and A. flavus covered the entire plate in the absence of probiotics. The potential probiotics ABRIIFBI-6 and ABRIIFBI-7 were able to inhibit the growth of A. flavus, whereas PTCC 1745, used as a control, was unable to do so. The potential probiotic ABRIIFBI-6 was able to inhibit the growth of A. niger similarly to A. flavus, but ABRIIFBI-7 only slightly inhibits the growth of A. niger. Furthermore, strain PTCC-1745, used as a control, could not inhibit the growth of both aspergilli.
Figure 1.
Antifungal activity of potential probiotic strains against Aspergillus flavus and Aspergillus niger is shown in Figure. (A) A. flavus covered the entire plate in the absence of probiotics. (B) The potential probiotics ABRIIFBI-6 and ABRIIFBI-7 were able to inhibit the growth of A. flavus, whereas PTCC 1745, used as a control, was unable to do so. (C) A. niger covered the entire plate in the absence of probiotics. (D) The potential probiotic ABRIIFBI-6 was able to inhibit the growth of A. niger similarly to A. flavus, but ABRIIFBI-7 only slightly inhibit the growth of A. niger. Furthermore, strain PTCC 1745, used as a control, was unable to inhibit the growth of both aspergilli.
Antimicrobial activity
The antibacterial activity of the tested strains against a variety of gram-positive and gram-negative bacteria is listed in Table 2. Based on results, both strains showed a broad spectrum of antagonistic activity against gram-positive indicator bacteria, including Listeria monocytogenes, Staphylococcus aureus, Bacillus cereus, and Streptococcus iniae, whereas they had no effect on gram-negative bacteria, such as Klebsiella pneumoniae, Escherichia coli, Yersinia enterocolitica, Shigella flexneri, and Salmonella enterica. The maximum zones of inhibition of ABRIIFBI-6 and ABRIIFBI-7 strains were observed for Streptococcus iniae (26.3 ± 0.6 mm and 25.5 ± 0.8 mm, respectively).
Table 2.
Antimicrobial activity of isolates against the pathogenic bacteria.
| Strain | Antimicrobial activity (clear zone (mm)) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli | Salmonella enterica | Listeria monocytogenes | Staphylococcus aureus | Bacillus cereus | Streptococcus iniae | Shigella flexneri | Klebsiella pneumoniae | Yersinia enterocolitica | |
| ABRIIFBI-6 | 0 | 0 | 21.9 ± 0.6ab | 24.2 ± 0.1a | 23.7 ± 0.9a | 26.3 ± 0.6a | 0 | 0 | 0 |
| ABRIIFBI-7 | 0 | 0 | 22.2 ± 0.5a | 17.7 ± 0.7bc | 22.8 ± 0.8a | 25.5 ± 0.8a | 0 | 0 | 0 |
Values are mean ± standard deviation. Different lowercase superscript letters are significantly different (P ≤ 0.05). All experiments were repeated twice.
(Strong ≥ 20 mm), (Moderate < 20 mm > 10 mm), and (Weak ≤ 10 mm).
Antibiotic susceptibility
The tested antibiotics were cefepime (30 g), streptomycin (10 g), sulfamethoxazole (2 g), cefixime (5 g), tetracycline (30 g), kanamycin (30 g), ampicillin (10 g), erythromycin (15 g), clindamycin (2 g), gentamycin (10 g), chloramphenicol (30 g), Both strains were sensitive to most of the antibiotics that were tested, but they were resistant to vancomycin, intermediate to clindamycin, and intermediate to ciprofloxacin, as shown in Table 1.
Cell surface hydrophobicity
The cell surface hydrophobicity percentages were 61.2 ± 1.4 and 57.3 ± 1.6% for ABRIIFBI-6 and 55.4 ± 1.8 and 51.2 ± 1.4% for ABRIIFBI-7 with toluene and xylene, respectively (Table 3).
Table 3.
Molecular name, accession numbers, hydrophobicity, autoaggregation, and coaggregation ability of evaluated strains.
| LAB strains | Accession numbers | Hydrophobicity (%) | Autoaggregation (%) | Coaggregation (%) | ||
|---|---|---|---|---|---|---|
| With toluene | With xylene | St. aureus | B. cereus | |||
| Limosilactobacillus fermentum ABRIIFBI-6 | OM666657 | 61.2 ± 1.4b | 57.3 ± 1.6a | 67.4 ± 1.9bc | 51.3 ± 1.7a | 57.9 ± 2.1a |
| Limosilactobacillus fermentum ABRIIFBI-7 | OM698829 | 55.4 ± 1.8a | 51.2 ± 1.4bc | 75.8 ± 2.3a | 52.4 ± 1.8a | 49.3 ± 1.9b |
Values are mean ± standard deviation. Different lowercase superscript letters are significantly different (P ≤ 0.05). All experiments were repeated twice.
Autoaggregation assay
The autoaggregation capacity of both selected strains is shown in Table 3. The values of autoaggregation ability of strains ABRIIFBI-6 and ABRIIFBI-7 were 67.4 ± 1.9 and 75.8 ± 2.3%, respectively, after 4 h of incubation.
Coaggregation assay
The findings of the coaggregation ability of both tested strains are shown in Table 3. The coaggregation percentages of strain ABRIIFBI-6 were 51.3 ± 1.7 and 57.9 ± 2.1% with S. aureus and B. cereus, respectively, whereas the coaggregation percentages of strain ABRIIFBI-7 were 52.4 ± 1.8 and 49.3 ± 1.9% with S. aureus and B. cereus.
Biofilm formation
Both strains were able to create biofilms, according to the results. Strain ABRIIFBI-6 showed the strongest biofilm formation value, with OD595 = 1.86, whereas strain ABRIIFBI-7 showed the weakest biofilm formation value, with OD595 = 0.19.
Molecular identification
The phenotypic characterization of the selected LAB strains was validated using 16S rRNA gene sequencing. Amplification of the 16S rRNA genes confirmed that both strains belonged to the genus Limosilactobacillus fermentum. They were submitted to the NCBI GeneBank with accession numbers OM666657 and OM698829 for ABRIIFBI-6 and ABRIIFBI-7, respectively.
Discussion
The current investigation clearly reveals that ABRIIFBI-6 and ABRIIFBI-7 strains have an antifungal impact on aflatoxigenic fungal isolates. Nonetheless, ABRIIFBI-6 was more effective than ABRIIFBI-7 in preventing the development of aflatoxin-producing aspergilli strains in vitro. The current study’s findings coincide with those of other researchers that tested Lactobacillus species comparable to those employed in this study but with other LAB strains in the in vitro growth control of Aspergillus spp. and other fungal strains28. Lactobacillus strains may limit fungal growth rate by producing secondary metabolites such as organic acids, bacteriocins, and hydrogen peroxide29.
It has been shown that selecting suitable potential probiotic strains that are resistant to bile salts and have a high tolerance for acidic conditions is critical in order to colonize the upper gut more effectively24,30. In our study, we isolated and identified two promising potential probiotic strains, ABRIIFBI-6 and ABRIIFBI-7, and showed that they were capable of growing at a low acidic pH and surviving in the presence of 0.3% bile oxgall bile in our experiment.
The adhesion qualities of lactobacilli strains have been linked to cell surface hydrophobicity, autoaggregation, and coaggregation and have been demonstrated to be necessary for the protection and colonization of the alimentary canal. A minimum hydrophobicity of 40% is a crucial criterion for potential probiotic strains31,32. In this line, our results showed a high hydrophobicity of the selected strains toward toluene and xylene. They also had substantial autoaggregation (ABRIIFBI-6: 67.4 ± 1.9 and ABRIIFBI-7: 75.8 ± 2.3%), indicating that these two dairy strains could probably be appropriate for both animal and human consumption.
Because of safety concerns, the acquired isolates were further examined for antibiotic resistance capabilities. Antibiotics are used as a major tool by the medical industry to combat diverse diseases. However, many pathogens are resistant to antibiotics and present significant danger to the treatment of nosocomial and community-acquired infections33. Thus, determining the antibiotic resistance of Lactobacillus strains is a critical criterion for selecting suitable potential probiotics, as commercially available probiotics with antibiotic-resistant genes may be passed on to pathogens in the intestine. In our study, we tested the security of ABRIIFBI-6 and ABRIIFBI-7 strains by determining their susceptibilities to fifteen antibiotics. Our results showed resistance of the two selected strains to vancomycin and kanamycin but not to the other antibiotics. This is consistent with previous research that found many Lactobacillus species to have high natural resistance to vancomycin and kanamycin33–35.
Bacterial biofilms are an important part of understanding how bacteria adapt to stress and colonize diverse environments. We discovered that the two tested dairy strains (ABRIIFBI-6 and ABRIIFBI-7) have the ability to form biofilms on the bottom of plates. The antimicrobial activity of LAB has been described by Tulumoglu et al.36.
Our results demonstrated the antifungal properties of ABRIIFBI-6 and ABRIIFBI-7 strains on aflatoxigenic fungal strains. The potential probiotics ABRIIFBI-6 and ABRIIFBI-7 were able to inhibit the growth of A. flavus, whereas PTCC 1745, used as a control, was unable to do so. The potential probiotic ABRIIFBI-6 was able to inhibit the growth of A. niger better than ABRIIFBI-7 which only slightly inhibit the growth of A. niger. Based on this finding, it could be suggested that ABRIIFBI-6 was the most effective strain in suppressing the growth of Aspergilli in vitro. Antifungal activity of strains inhibited both Aspergillus parasiticus expansion and thus the formation of aflatoxins36. Lactobacillus plantarum, Lactobacillus fermentum, Lactobacillus brevis, and Lactococcus spp. have in vitro antifungal activities on aflatoxigenic fungal isolates in proportions similar to those seen in this study. Our findings are consistent with those of Magnusson et al., who investigated Lactobacillus species similar to ours but with different LAB strains and controlled the growth of Aspergillus spp. and other fungal strains in vitro37,38.
We showed that the effect of the probiotic bacteria suspensions used in this study is extensive, each of them being active against a different Aspergillus spp. The formation of secondary metabolites by Lactobacillus strains on fungal species could possibly be the reason for their inhibition of expansion rates. Lactic and acetic acids are the most common results of LAB’s carbohydrate fermentation. These acids pass through the membranes of target species in their undissociated hydrophobic state, lowering cytoplasmic pH and inducing cell death38,39. Despite the lack of compelling evidence that protein molecules play a role in growth inhibition, several authors discovered that several carboxylic acid strains developed antifungal metabolites that were sensitive to proteolytic enzymes22.
In summary, our study demonstrated that strains ABRIIFBI-6 and ABRIIFBI-7 could be used as potential probiotics. We found that these selected strains could have potent antibacterial properties against gram-positive bacteria and two important species of Aspergillus. So, they could be used as biocontrols for testing in vivo to find out what health benefits they might have and as new probiotic strains in the food industry.
Acknowledgements
The financial support of the Agricultural Biotechnology Research Institute of Iran (ABRII) is gratefully acknowledged.
Author contributions
Y.M.: writing & doing tests. R.M.: data analysis & revising. M.A.H.: revising, designing experiment. Y.N.: project administration.
Funding
This work was supported by the Agricultural Biotechnology Research Institute of Iran [Grant Numbers 12-05-05-006-99014-990345].
Data availability
The datasets generated and/or analysed during the current study are available in the NCBI GeneBank repository, Accession Numbers OM666657 and OM698829.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tai B, Chang J, Liu Y, Xing F. Recent progress of the effect of environmental factors on Aspergillus flavus growth and aflatoxins production on foods. Food Qual. Saf. 2020;4:21–28. doi: 10.1093/fqsafe/fyz040. [DOI] [Google Scholar]
- 2.Ismail A, et al. Aflatoxin in foodstuffs: Occurrence and recent advances in decontamination. Food Res. Int. 2018;113:74–85. doi: 10.1016/j.foodres.2018.06.067. [DOI] [PubMed] [Google Scholar]
- 3.Organization, W. H. & Cancer, I. A. for R. on. Some naturally occurring substances: Food items and constituents, heterocyclic aromatic amines and mycotoxins. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol. 56 (1993).
- 4.Assaf JC, Khoury AE, Chokr A, Louka N, Atoui A. A novel method for elimination of aflatoxin M1 in milk using Lactobacillus rhamnosus GG biofilm. Int. J. Dairy Technol. 2019;72:248–256. doi: 10.1111/1471-0307.12578. [DOI] [Google Scholar]
- 5.Liu Y, Chang C-CH, Marsh GM, Wu F. Population attributable risk of aflatoxin-related liver cancer: Systematic review and meta-analysis. Eur. J. Cancer. 2012;48:2125–2136. doi: 10.1016/j.ejca.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Safari N, et al. The potential of plant-based bioactive compounds on inhibition of aflatoxin B1 biosynthesis and down-regulation of aflR, aflM and aflP genes. Antibiotics. 2020;9:728. doi: 10.3390/antibiotics9110728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benkerroum N. Aflatoxins: Producing-molds, structure, health issues and incidence in Southeast Asian and Sub-Saharan African countries. Int. J. Environ. Res. Public Health. 2020;17:1215. doi: 10.3390/ijerph17041215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Fels-Klerx HJ, Liu C, Battilani P. Modelling climate change impacts on mycotoxin contamination. World Mycotoxin J. 2016;9:717–726. doi: 10.3920/WMJ2016.2066. [DOI] [Google Scholar]
- 9.Agriopoulou S, Stamatelopoulou E, Varzakas T. Advances in occurrence, importance, and mycotoxin control strategies: Prevention and detoxification in foods. Foods. 2020;9:137. doi: 10.3390/foods9020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Udomkun P, et al. Innovative technologies to manage aflatoxins in foods and feeds and the profitability of application—A review. Food Control. 2017;76:127–138. doi: 10.1016/j.foodcont.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuda H, Miyamoto T. Guidelines for the evaluation of probiotics in food. Report of a joint FAO/WHO working group on drafting guidelines for the evaluation of probiotics in food Guidelines for the evaluation of probiotics in food. Report of a joint FAO/WHO working group on dr. Food Sci. Technol. Res. 2010;16:87–92. doi: 10.3136/fstr.16.87. [DOI] [Google Scholar]
- 12.Yerlikaya O. Starter cultures used in probiotic dairy product preparation and popular probiotic dairy drinks. Food Sci. Technol. 2014;34:221–229. doi: 10.1590/fst.2014.0050. [DOI] [Google Scholar]
- 13.Champagne CP, da Cruz AG, Daga M. Strategies to improve the functionality of probiotics in supplements and foods. Curr. Opin. Food Sci. 2018;22:160–166. doi: 10.1016/j.cofs.2018.04.008. [DOI] [Google Scholar]
- 14.Zendeboodi F, Khorshidian N, Mortazavian AM, da Cruz AG. Probiotic: Conceptualization from a new approach. Curr. Opin. Food Sci. 2020;32:103–123. doi: 10.1016/j.cofs.2020.03.009. [DOI] [Google Scholar]
- 15.Taghizadeh Moghaddam S, Javadi A, Matin AA. Reduction of bisphenol A by Lactobacillus acidophilus and Lactobacillus plantarum in yoghurt. Int. J. Dairy Technol. 2020;73:737–742. doi: 10.1111/1471-0307.12706. [DOI] [Google Scholar]
- 16.Liew W-P-P, Nurul-Adilah Z, Than LTL, Mohd-Redzwan S. The binding efficiency and interaction of Lactobacillus casei shirota toward aflatoxin B1. Front. Microbiol. 2018;9:1503. doi: 10.3389/fmicb.2018.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill C, et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 18.Peltonen K, El-Nezami H, Haskard C, Ahokas J, Salminen S. Aflatoxin B1 binding by dairy strains of lactic acid bacteria and bifidobacteria. J. Dairy Sci. 2001;84:2152–2156. doi: 10.3168/jds.S0022-0302(01)74660-7. [DOI] [PubMed] [Google Scholar]
- 19.Corassin CH, Bovo F, Rosim RE, Oliveira C. Efficiency of Saccharomyces cerevisiae and lactic acid bacteria strains to bind aflatoxin M1 in UHT skim milk. Food Control. 2013;31:80–83. doi: 10.1016/j.foodcont.2012.09.033. [DOI] [Google Scholar]
- 20.Ondiek W, et al. Removal of aflatoxin b1 and t-2 toxin by bacteria isolated from commercially available probiotic dairy foods. Food Sci. Technol. Int. 2022;28:15–25. doi: 10.1177/1082013220987916. [DOI] [PubMed] [Google Scholar]
- 21.Kouhi F, Mirzaei H, Nami Y, Khandaghi J, Javadi A. Potential probiotic and safety characterisation of Enterococcus bacteria isolated from indigenous fermented Motal cheese. Int. Dairy J. 2022;126:105247. doi: 10.1016/j.idairyj.2021.105247. [DOI] [Google Scholar]
- 22.Schnürer J, Magnusson J. Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci. Technol. 2005;16:70–78. doi: 10.1016/j.tifs.2004.02.014. [DOI] [Google Scholar]
- 23.Nami Y, Haghshenas B, Khosroushahi AY. Molecular identification and probiotic potential characterization of lactic acid bacteria isolated from human vaginal microbiota. Adv. Pharm. Bull. 2018;8:683. doi: 10.15171/apb.2018.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haghshenas B, et al. Isolation and characterization of probiotics from dairies. Iran. J. Microbiol. 2017;9:234. [PMC free article] [PubMed] [Google Scholar]
- 25.Mosallaie F, Jooyandeh H, Hojjati M, Fazlara A. Biological reduction of aflatoxin B1 in yogurt by probiotic strains of Lactobacillus acidophilus and Lactobacillus rhamnosus. Food Sci. Biotechnol. 2020;29:793–803. doi: 10.1007/s10068-019-00722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gómez NC, Ramiro JMP, Quecan BXV, de Melo Franco BDG. Use of potential probiotic lactic acid bacteria (LAB) biofilms for the control of Listeria monocytogenes, Salmonella typhimurium, and Escherichia coli O157: H7 biofilms formation. Front. Microbiol. 2016;7:863. doi: 10.3389/fmicb.2016.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nami Y, Haghshenas B, Yari Khosroushahi A. Effect of psyllium and gum Arabic biopolymers on the survival rate and storage stability in yogurt of Enterococcus durans IW3 encapsulated in alginate. Food Sci. Nutr. 2017;5:554–563. doi: 10.1002/fsn3.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Fadeel A, Shalaby SO, Arafa MM, Khater AA. Biological control of some types of mycotoxins by probiotic bacteria and yeast strains. Egypt. J. Dairy Sci. 2022 doi: 10.21608/ejds.2022.171199.1008. [DOI] [Google Scholar]
- 29.Abdolmaleki K, et al. Emerging technologies in combination with probiotics for aflatoxins removal: An updated review. Int. J. Food Sci. Technol. 2022;57:5712–5721. doi: 10.1111/ijfs.15926. [DOI] [Google Scholar]
- 30.Nami Y, Lornezhad G, Kiani A, Abdullah N, Haghshenas B. Alginate-Persian gum-prebiotics microencapsulation impacts on the survival rate of Lactococcus lactis ABRIINW-N19 in orange juice. LWT. 2020;124:109190. doi: 10.1016/j.lwt.2020.109190. [DOI] [Google Scholar]
- 31.Del Re B, Sgorbati B, Miglioli M, Palenzona D. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 2000;31:438–442. doi: 10.1046/j.1365-2672.2000.00845.x. [DOI] [PubMed] [Google Scholar]
- 32.Nami Y, et al. Probiotic properties of Enterococcus isolated from artisanal dairy products. Front. Microbiol. 2019;10:300. doi: 10.3389/fmicb.2019.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moellering RC, Graybill JR, McGowan JE, Corey L. Antimicrobial resistance prevention initiative an update: Proceedings of an expert panel on resistance. Am. J. Med. 2007;120:S4–S25. doi: 10.1016/j.amjmed.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Chang LIU, Zhang Z-Y, Ke D, Jian-Ping Y, Xiao-Kui GUO. Antibiotic resistance of probiotic strains of lactic acid bacteria isolated from marketed foods and drugs. Biomed. Environ. Sci. 2009;22:401–412. doi: 10.1016/S0895-3988(10)60018-9. [DOI] [PubMed] [Google Scholar]
- 35.Mahjoory Y, Aliasgharzad N, Moghaddam G, Bybordi A. Long-term application of manure alters culturable soil microbial populations and leads to occurrence of antibiotic resistant bacteria. Soil Sediment Contam. Int. J. 2021;31:1–15. [Google Scholar]
- 36.Tulumoglu S, et al. Probiotic properties of lactobacilli species isolated from children’s feces. Anaerobe. 2013;24:36–42. doi: 10.1016/j.anaerobe.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Magnusson J, Schnürer J. Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad-spectrum proteinaceous antifungal compound. Appl. Environ. Microbiol. 2001;67:1–5. doi: 10.1128/AEM.67.1.1-5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palaniswamy SK, Govindaswamy V. In-vitro probiotic characteristics assessment of feruloyl esterase and glutamate decarboxylase producing Lactobacillus spp. isolated from traditional fermented millet porridge (kambu koozh) LWT-Food Sci. Technol. 2016;68:208–216. doi: 10.1016/j.lwt.2015.12.024. [DOI] [Google Scholar]
- 39.Besser M, et al. Impact of probiotics on pathogen survival in an innovative human plasma biofilm model (hp BIOM) J. Transl. Med. 2019;17:1–9. doi: 10.1186/s12967-019-1990-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the NCBI GeneBank repository, Accession Numbers OM666657 and OM698829.