Abstract
Background
Liver cancer stem cells (LCSCs) are a subpopulation of tumor cells that can drive cancer initiation and relapses. Because of their significance, researchers are looking for biomarkers that characterize or regulate LCSCs so that they can be used as targets for the diagnosis and treatment of chronic liver diseases and hepatocellular carcinoma (HCC).
Methodology
Six groups of patients having hepatitis C virus (HCV), HCV + cirrhosis, HCV + HCC, hepatitis B virus (HBV), HBV + cirrhosis, or HBV + HCC, in addition to a control group, were subjected to the measurement of LCSCs levels and analysis of miR-1290 and miR-1825 expression.
Results
The percentages of the CD133/EpCAM-expressing LCSCs were increased in viral hepatitis and cirrhosis groups, compared to the control group. HCC patients had the highest percentages of LCSCs. CD133/EpCAM-expressing cells showed significant correlations with stemness-associated miRNAs; miR-1290 and miR-1825. Also, the miR-1290 and miR-1825 were significantly up-regulated in viral hepatitis-associated cirrhosis and HCC groups. Moreover, in HCV + HCC, miR-1290 and miR-1825 expression was significantly positively correlated with tumor size and number. However, only miR-1825 could distinguish between HCV- and HBV-associated HCC groups. MiR-1290 exhibited the highest sensitivity and specificity for detecting HCC, followed by miR-1825 and CD133/EpCAM-expressing LCSCs.
Conclusions
These findings indicate the relevance of CD133/EpCAM-expressing cells in the pathogenesis of liver cirrhosis and HCC developed as a consequence of either chronic HCV or HBV infection. Accordingly, CD133/EpCAM-expressing cells, miR-1290, and miR-1825, could serve as promising diagnostic and prognostic biomarkers as well as therapeutic targets in patients suffering from liver cirrhosis or HCC.
Keywords: Cancer stem cell, Hepatitis B virus, Hepatitis C virus, Hepatocellular carcinoma, Liver cirrhosis, miRNA
1. Introduction
Chronic viral illness is one of the most frequent causes of hepatic fibrosis, distortion of the hepatic architecture, and ultimate progression to liver cirrhosis which has several co-morbid consequences, including hepatic decompensation and hepatocellular carcinoma (HCC) [1,2]. HCC is thought to be the 3rd most frequent cause of cancer-related mortality, with an annual rate of ∼750.000 death/year and a 5-year overall survival of < 15% globally [3,4]. It is anticipated that this rate will continue to escalate over time due to a 20-year delay in HCC formation from the time of acquiring hepatitis C virus (HCV) infection, being the most eminent risk factor. It has been demonstrated that the rates of incidence and mortality are relatively comparable around the world, revealing the short-term lethality of this tumor, especially when diagnosed at late stages impeding effective treatment strategies [5].
Although advances in non-invasive imaging technologies have contributed to improving the characterization of hepatic lesions in HCC patients, the diagnostic accuracy of these techniques is primarily dependent on the size of HCC nodules, and they are insensitive to small nodules. Also, CT scans expose the patients to the risks of radiation. Even though MRI is a highly effective method for detecting HCC, its utilization in cancer diagnosis is constrained by the high cost of the necessary equipment. As a result, it is critical to discover novel biomarkers for the early detection of HCC [6].
It has been demonstrated that liver cancer is originated from liver stem cells, present in the adult liver tissue. Also, it has been found that these cells are responsible for the chemotherapeutic resistance and recurrence of cancer. In HCC, liver cancer stem cells (LCSCs) that express molecular markers [e.g. CD133, CD90, CD44, CD13, and epithelial cell adhesion molecule (EpCAM)], exhibit resistance to radiotherapy and chemotherapy in vitro and in vivo by up-regulating the expression of drug efflux-related proteins and activating anti-apoptotic and stem cell-related pathways [7].
Accumulating evidence supports the influence of miRNAs on the maintenance of the CSCs phenotype by modulating the expression of oncogenes and stem cell-related genes. Their roles in the regulation of CSCs characteristics such as asymmetric cell division, tumorigenicity, and treatment resistance have been affirmed [8]. Among these miRNAs, miR-1290 and miR-1825 which have been postulated as CSCs-specific miRNAs with possible roles in the acquisition and maintenance of stem cells features as well as in tumor initiation, progression, invasion, metastasis, chemoresistance, and recurrence [9].
MiR-1290 has been linked to the initiation and progression of several gastrointestinal tumors. It has a tumor-promoting effect and is involved in the occurrence and growth of colorectal and esophageal cancers through regulating the expression of Inositol polyphosphate 4-phosphatase B and nuclear factor I/X genes, respectively [10,11]. It also induces cell proliferation and metastasis of gastric cancer cells by targeting the FOXA1 gene [12]. MiR-1290 has a strong association with TNM staging and the overall survival of colorectal cancer patients [13]. Additionally, its levels are correlated with tumor size, lymphatic and vascular invasion, and tumor differentiation in patients suffering from esophageal, prostate, and colorectal cancers [14,15]. Furthermore, it can influence cell proliferation, invasion, migration, and resistance to chemo-radiation in glioblastoma via targeting the SOCS4 gene [16].
Previous research has shown that miR-1825 plays an important role in the apoptosis, cell proliferation, invasion, and metastasis of various malignancies [17,18]. It has been demonstrated to regulate glioblastoma development by modulating the CDK14 expression and Wnt/β-catenin signaling pathway [19]. Also, it is involved in the progression of prostate cancer via targeting the suppressor of cancer cell invasion gene [20]. Furthermore, it has been found to be an important contributor to the laryngeal carcinogenesis process as a result of its aberrant expression in the CD133+ larynx cancer stem-like cells (CSLCs) [21]. MiR-1825 has been identified as a marker for the pathological grading of glioma and the TNM classification, response to treatment, and prognosis of prostate cancer [17,18].
Deep mechanistic understandings of the CSCs and their regulatory mechanisms in human carcinogenesis may lead to the discovery of a better way for the targeted eradication of CSCs, and thus, can be a future potential tool for cancer management and therapy [22]. Although miR-1290 and miR-1825 and their correlation with the circulating CSCs have been studied in several human malignancies, they have never been investigated in cases of liver cancer. Therefore, the goal of this study was to identify the changes, in circulating LCSCs and their regulatory miRNAs, which take place during the process of hepatitis viruses-induced hepatocarcinogenesis in order to find out potential biomarkers that could be exploited as a tool for early liver cancer detection, management, and therapy.
2. Methodology
2.1. Subjects and sample collection
Among patients who visited outpatients' clinics or were admitted to the Hepato-Gastroenterology Department, Theodor Bilharz Research Institute (TBRI), Giza, Egypt, for the evaluation of their viral hepatitis-related chronic liver disease, 179 patients were chosen to be enrolled in this study after signing informed consent. This study was ethically approved by TBRI Institute's Human Research Ethics Committee in accordance with the guidelines of the 1975 Declaration of Helsinki.
All patients underwent (i) full medical history taking, (ii) thorough clinical examination, (iii) laboratory investigations including CBC, liver functions tests, α-fetoprotein (AFP) measurement, and serological diagnosis of viral hepatitis, (iv) ultrasonography (US), and (v) abdominal CT examination for the confirmation of HCC.
Patients were excluded from the current study if they had history or evidence that pointed to other causes of chronic liver disease, such as Schistosoma infection, biliary disorders, or other malignancies.
Patients were divided into six groups (i) HCV infection group (n = 44), (ii) HCV + liver cirrhosis group (n = 48), (iii) HCV + HCC group (n = 46), (iv) HBV infection group (n = 21), (v) HBV + liver cirrhosis group (n = 12), and (vi) HBV + HCC group (n = 8). Age- and sex-matched individuals (n = 46) were included in this study as healthy controls.
2.2. Flowcytometric analysis
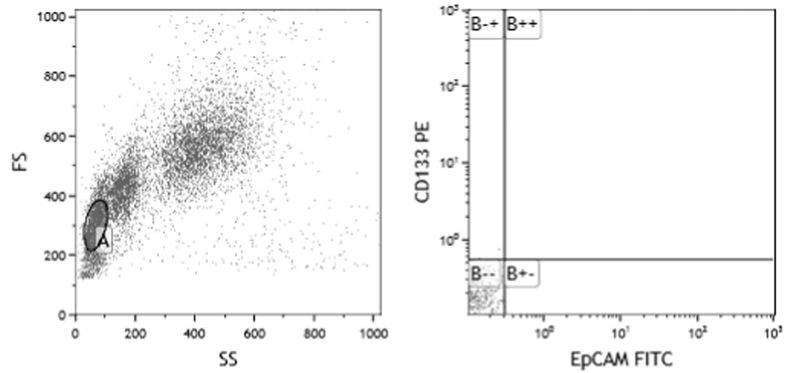
The peripheral blood mononuclear cells were isolated by density gradient centrifugation using Lymphocyte Separation Medium (Lonza, Switzerland). The separated cells were stained using the following fluorescent-labeled antibodies: CD133-phycoerythrin (PE) and EpCAM-fluorescein isothiocyanate (FITC) (Miltenyi Biotec, Germany) and were analyzed by flow cytometry (Beckman Coulter Epics XL-MCL) collecting 10,000 events. As CD133 and EpCAM were proven to be an efficient markers of LCSCs, the percentages of CD133+/EpCAM+ cells were considered as the percentages of LCSCs. The gating strategy used to detect CD133/EpCAM-expressing cells is represented in Fig. 1.
Fig. 1.
Scatter plot of flow cytometry demonstrating the gating strategy used to detect the CD133/EpCAM-expressing cancer stem cells (CSCs).
2.3. MiRNAs expression analysis
MiRNAs were extracted from the isolated mononuclear cells using the miRNeasy Mini Kit (Qiagen, Germany) according to the manufacturer's protocol. Its concentration and purity were assessed using NanoDrop™ 2000 Spectrophotometer (ThermoFischer Scientific, USA). Reverse transcription reactions were performed using TaqMan™ MicroRNA Reverse Transcription Kit (ThermoFischer Scientific, USA) according to the manufacturer's instructions. LCSCs regulatory miRNAs expression analysis was performed using Taqman Universal Master Mix Ii No UNG (ThermoFischer Scientific, USA). The reaction was composed of 10 μL TaqMan® Universal Master Mix II no UNG 2✕, 1 μL TaqMan® miRNA Assay 20✕, 8 μL RNase-free water, and 1 μl cDNA template to reach a final volume of 20 μL. Ready-made TaqMan® miR-1290 and TaqMan® miR-1825 assays were chosen, in the current study, as miR-1290 and miR-1825 were proven to be CSCs-specific miRNAs with potential roles in the acquisition of stem cells features as well as in contributing to tumor initiation, progression, invasion, metastasis, chemoresistance, and recurrence [9,22]. They were provided by ThermoFischer Scientific, USA. TaqMan® RNU6B assay was utilized to normalize the data. The reactions were conducted in triplicates on the StepOnePlus™ Real-Time PCR (Applied Biosystems, USA) under the following conditions: 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing at 60 °C for 1 min. The comparative cycle threshold (CT) method were used to calculate the relative expression of miRNA according to the equation: relative quantification (RQ) = 2-ΔΔCT [23], where ΔΔCT = [ΔCT (unknown sample) ‒ ΔCT (calibrator sample)]
ΔCT (unknown sample) = [CTGI (unknown) ‒ CTGR (unknown)]
ΔCT (calibrator sample) = [CTGI (calibrator) ‒ CTGR (calibrator)]
CT is the number of cycles needed for the fluorescence to reach a specific threshold level of detection, GI is the gene of interest, and GR is the reference gene.
2.4. Statistical analysis
Statistical analyses were performed using SPSS version 25 software (IBM SPSS, Chicago, IL, USA). Means ± standard errors (SE) were computed for the quantitative data. Data means were compared using ANOVA followed by an LSD post hoc test to compare between the groups for the quantitative variables. The receiver operating characteristic curve was constructed by calculating the true- and false-positive fractions of the marker at several cutoff points. The areas under the ROC curve (AUC), sensitivities, and specificities were computed for the levels of CD133/EpCAM-expressing LCSCs and the examined miRNAs. Finally, Spearman correlation was done to identify the correlations between the various biomarkers. The significance level was set as p<0.05.
3. Results
3.1. Participants’ characteristics
In total, 225 participants were recruited for the current study. They were made up of 130 males and 95 females with their ages ranging from 41 to 78 years. They consisted of 44 patients having HCV infection, 48 patients having HCV + liver cirrhosis, 46 patients having HCV + HCC, 21 patients having HBV infection, 12 patients having HBV + liver cirrhosis, 8 patients having HBV + HCC, and 46 age- and gender-matched healthy volunteers. The biochemical parameters, i.e. AST, ALT, total bilirubin, albumin, total protein, prothrombin concentration, platelet count, and AFP were all within the normal range for the control subjects. The demographic and biochemical profiles are presented in Table 1.
Table 1.
The demographic and biochemical profiles of the studied patients’ groups.
| Control |
HCV |
HCV + Cirrhosis |
HCV + HCC |
HBV |
HBV + Cirrhosis |
HBV + HCC |
|
|---|---|---|---|---|---|---|---|
| (n = 46) | (n = 44) | (n = 48) | (n = 46) | (n = 21) | (n = 12) | (n = 8) | |
| Age | 60.11 ± 1.10 | 60.25 ± 1.09 | 60.56 ± 1.22 | 63.26 ± 1.10 | 56.38 ± 2.14 | 57.33 ± 2.44 | 57.25 ± 1.13 |
| AST (IU/L) | 23.30 ± 1.09 | 57.52 ± 3.32* | 59.40 ± 5.75* | 166.04 ± 22.61** | 33.76 ± 11.29 | 101.58 ± 29.41$, ¥ | 218.75 ± 62.79@ |
| ALT (IU/L) | 24.85 ± 2.10 | 52.27 ± 3.33* | 56.27 ± 5.01* | 91.70 ± 15.16** | 24.56 ± 6.65 | 74.58 ± 23.37£ | 97.75 ± 15.72£ |
| Total bilirubin (mg/dL) | 0.62 ± 0.04 | 3.31 ± 0.71* | 2.99 ± 0.59* | 5.46 ± 1.12$,# | 0.65 ± 0.13 | 7.03 ± 2.50£ | 13.68 ± 4.98@ |
| ALB (g/dL) | 4.05 ± 0.06 | 3.43 ± 0.11a | 2.38 ± 0.08b | 2.52 ± 0.09b | 3.81 ± 0.08c | 2.80 ± 0.21d | 2.40 ± 0.13d,e |
| TP (g/dL) | 7.31 ± .076 | 6.75 ± 0.10a | 6.63 ± 0.13a | 6.89 ± 0.14c | 7.03 ± 0.15 | 6.85 ± 0.19c | 6.27 ± 0.15e,f |
| PC (%) | 96.39 ± 0.67 | 77.93 ± 3.16a | 54.49 ± 2.51b | 56.46 ± 2.95b | 87.90 ± 1.43c | 62.18 ± 6.55d | 55.63 ± 9.14d |
| PLT (x103/μL) | 293.33 ± 12.58 | 243.24 ± 14.80a | 152.32 ± 12.62b | 150.01 ± 15.25b | 269.18 ± 18.53 | 127.83 ± 23.74d | 115.63 ± 16.18d |
| AFP (ng/mL) | 4.39 ± 0.26 | 5.31 ± 0.52 | 9.73 ± 1.68 | 3460.29 ± 1008.95** | 7.25 ± 1.14 | 18.05 ± 5.28 | 529.38 ± 341.52@ |
Data are expressed as mean ± standard error (SE).
Hepatitis C virus (HCV); hepatocellular carcinoma (HCC); Hepatitis B virus (HBV); alanine aminotransferase (ALT); aspartate aminotransferase (AST); serum albumin (ALB); total protein (TP); prothrombin concentration (PC); platelet count (PLT); alpha-fetoprotein (AFP).
*Significant increase compared to the control group (p<0.05); **Significant increase compared to the control, HCV, and HCV + cirrhosis groups (p<0.001); $Significant increase compared to the control group (p<0.001); ¥Significant increase compared to the HBV group (p<0.01); @Significant increase compared to the control, HBV, and HBV + cirrhosis groups (p<0.001); #Significant increase compared to the HCV and HCV + cirrhosis groups (p<0.05); £Significant increase compared to the control and HBV groups (p<0.001); aSignificant decrease compared to the control group (p<0.001); bSignificant decrease compared to the control and HCV groups (p<0.001); cSignificant decrease compared to the control group (p<0.05); dSignificant decrease compared to the control and HBV groups (p<0.001); eSignificant decrease compared to the HBV + cirrhosis group (p<0.05).
3.2. Flowcytometric analysis
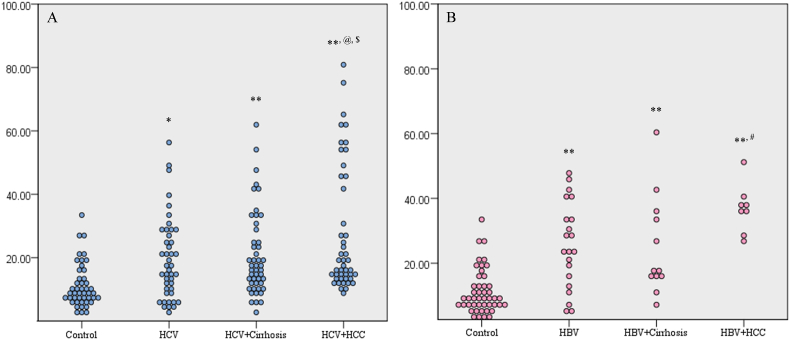
Both the HCV- and HBV-infected patients had a significant increase in the mean percentages of the circulating CD133/EpCAM-expressing LCSCs (19.51 ± 1.94% and 25.74 ± 2.90%, respectively), compared to the control group (11.14 ± 1.01%) (p = 0.005 and p<0.001, respectively). Also, there was a significant elevation of the CD133/EpCAM-expressing cells, in the HCV + and HBV + cirrhosis groups (20.95 ± 1.93 and 25.20 ± 4.46%, respectively), compared to the control group (p<0.001).
Regarding the HCV-associated HCC group, the CD133/EpCAM-expressing cells (28.13 ± 2.97%) increased significantly, compared to the control, HCV, and HCV + cirrhosis groups (p<0.001, p = 0.004, and p = 0.015, respectively). As well, the HBV-related HCC group displayed a significant increase in the CD133/EpCAM-expressing cells (36.81 ± 2.66%) when compared to the control, HBV, and HBV + cirrhosis groups (p<0.001, p = 0.011, and p = 0.015, respectively). However, there was no significant difference between the HCV- and HBV-associated HCC groups, regarding the mean percentage of CD133/EpCAM-expressing cells (Fig. 2).
Fig. 2.
2D dot plot diagrams showing the percentages of CD133/EpCAM-expressing cells in (A) HCV groups and (B) HBV groups.
*significant increase compared to the control group (p<0.01); **significant increase compared to the control group (p<0.001); @significant increase compared to the HCV group (p<0.01); $significant increase compared to the HCV + cirrhosis group (p<0.05); #significant increase compared to the HBV and HBV + cirrhosis groups (p<0.05).
3.3. LCSCs regulatory miRNAs expression profile
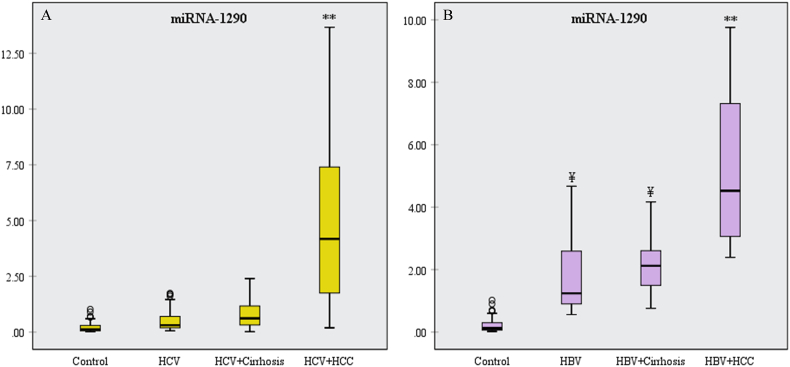
The mean RQ values of miR-1290 were significantly higher in the HCV + HCC group than the control, HCV, and HCV + cirrhosis groups (p<0.001) (Fig. 3A). On the other side, the miR-1290 was significantly elevated in the HBV and HBV + cirrhosis groups, compared to the control group (p<0.001). Also, it showed a significant increase in the HBV + HCC group when compared to the control, HBV, and HBV + cirrhosis groups (p<0.001) (Fig. 3B).
Fig. 3.
Box plot diagrams of miR-1290 expression in (A) HCV groups and (B) HBV groups.
**Significant increase compared to the other groups (p<0.001); ¥significant increase compared to the control group (p<0.001).
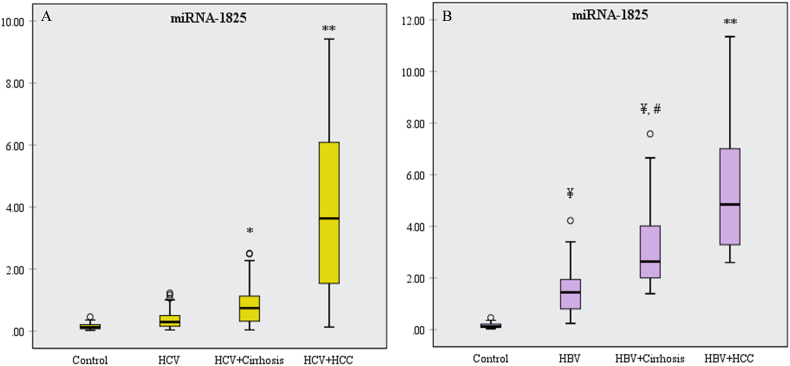
MiR-1825 showed a significant rise in the HCV-associated cirrhosis group, compared to the control (p = 0.023), and in the HCV-associated HCC group, compared to the other groups (p<0.001) (Fig. 4A). Also, it was significantly higher in the HBV group than the control group (p<0.001), in the HBV + cirrhosis group than the control and HBV groups (p<0.001), and in the HBV-associated HCC group than the other groups (p<0.001) (Fig. 4B).
Fig. 4.
Box plot diagrams of miR-1825 expression in (A) HCV groups and (B) HBV groups.
*significant increase compared to the control group (p<0.05); **significant increase compared to the other groups (p<0.001); ¥significant increase compared to the control group (p<0.001); #significant increase compared to the HBV group (p<0.001).
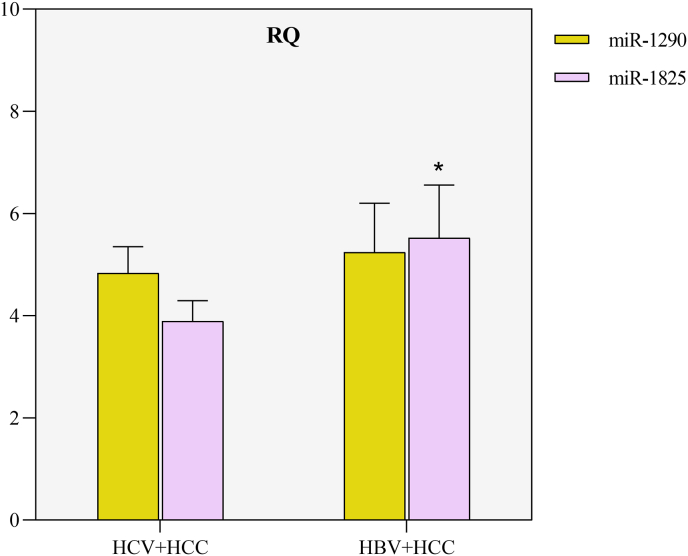
There was a significant difference, in the miR-1825 levels, between the HCV + HCC and HBV + HCC groups (p = 0.004) while no significant difference was detected, regarding the miR-1290 expression, between the HCV + HCC and HBV + HCC groups (Fig. 5).
Fig. 5.
The relative quantification (RQ) values of miR-1290 and miR-1825 expression in the HCV- and HBV-associated HCC groups.
*Significant increase compared to the HCV + HCC group (p<0.01).
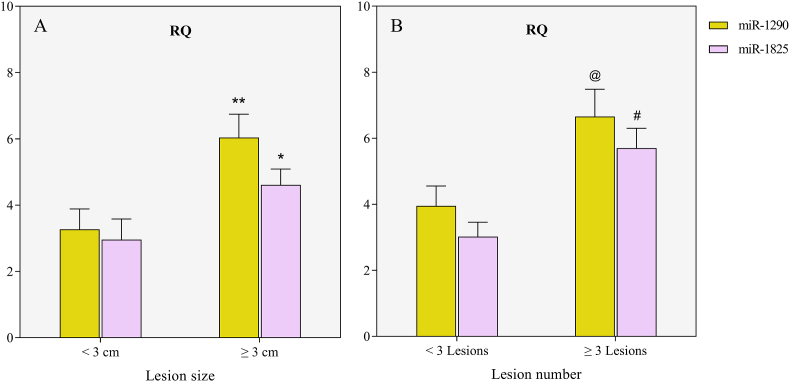
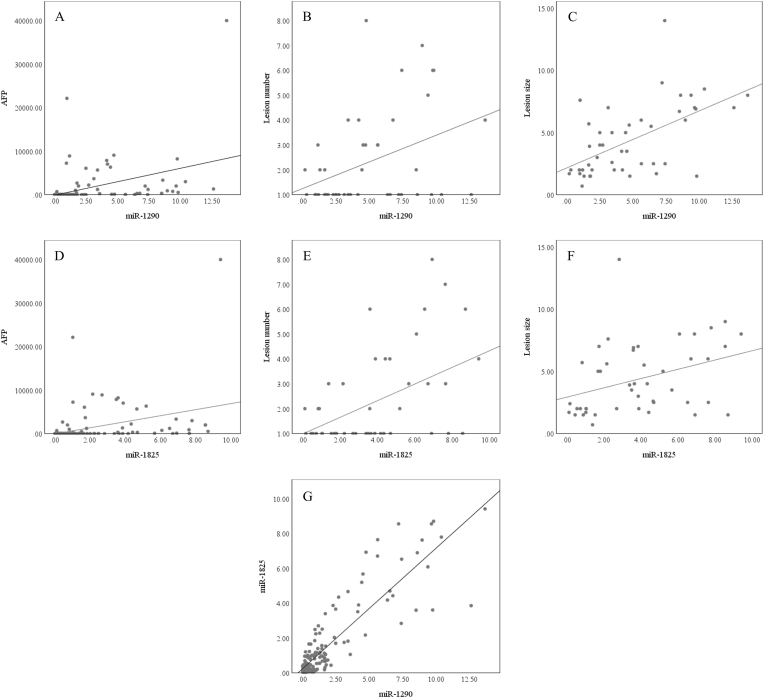
In HCV-, but not in HBV-associated HCC, miR-1290 and miR-1825 expression was significantly higher in patients having focal hepatic lesions measuring 3 cm or more in diameter, compared to those having lesions of less than 3 cm (p = 0.006 and p = 0.037, respectively) (Fig. 6A). Additionally, miR-1290 and miR-1825 showed a significant rise in patients having 3 or more lesions, compared to those having fewer than 3 lesions (p = 0.012 and p = 0.001, respectively) (Fig. 6B).
Fig. 6.
The relative quantification (RQ) values of miR-1290 and miR-1825 expression in the HCV-HCC group according to (A) the lesion size and (B) the focal lesion number.
*Significant increase compared to patients with lesions less than 3 cm in diameter (p<0.05); **significant increase compared to patients with lesions less than 3 cm in diameter (p<0.01); @significant increase compared to patients having fewer than 3 focal lesions (p<0.05); #significant increase compared to patients having fewer than 3 focal lesions (p<0.01).
3.4. Diagnostic performance of the circulating LCSCs and their regulatory miRNAs in predicting HCC
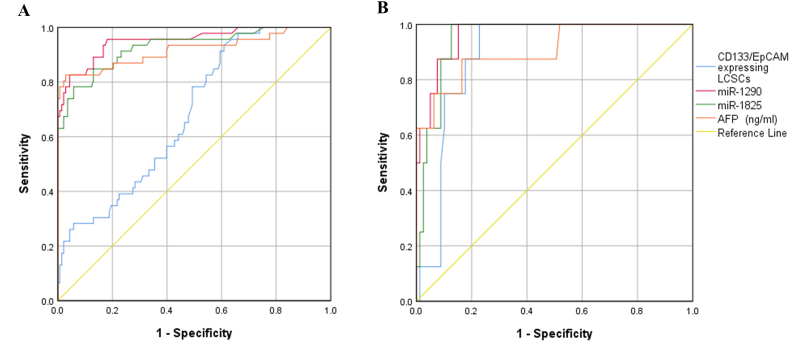
To assess the efficacy of the CD133/EpCAM-expressing cells and miRNAs for predicting HCC, the AUC values were analyzed. It was found that the miR-1290 had the largest AUC, as well as the highest sensitivity and specificity, followed by miR-1825 and CD133/EpCAM-expressing cells (Fig. 7).
Fig. 7.
The receiver operating curve (ROC) of CD133/EpCAM-expressing cells, miR-1290, and miR-1825 for detecting HCC in patients with (A) HCV and (B) HBV.
The CD133/EpCAM-expressing cells, miR-1290, and miR-1825 demonstrated sensitivities of 78%, 96%, and 91%, respectively, and specificities of 51%, 82%, and 77%, respectively. On the other side, AFP values of more than 8.4 ng/mL had a sensitivity and specificity of 87% and 80%, respectively (Table 2).
Table 2.
The areas under the curves, sensitivities, and specificities of the CD133/EpCAM-expressing cells and the examined miRNAs.
| AUC | 95% CI | p value | Sensitivity | Specificity | |
|---|---|---|---|---|---|
| HCV | |||||
| Total CD133/EpCAM | 0.681 | 0.598–0.763 | p<0.001 | 78% | 51% |
| miR-1290 | 0.952 | 0.914–0.990 | p<0.001 | 96% | 82% |
| miR-1825 | 0.928 | 0.881–0.976 | p<0.001 | 91% | 77% |
| AFP (ng/mL) | 0.917 | 0.858–0.977 | p<0.001 | 87% | 80% |
| HBV | |||||
| Total CD133/EpCAM | 0.890 | 0.82–0.962 | p<0.001 | 87.5% | 82.3% |
| miR-1290 | 0.964 | 0.920–1.000 | p<0.001 | 87.5% | 92.4% |
| miR-1825 | 0.949 | 0.902–0.996 | p<0.001 | 87.5% | 91.1% |
| AFP (ng/mL) | 0.907 | 0.788–1.000 | p<0.001 | 87.5% | 83.5% |
3.5. Correlations
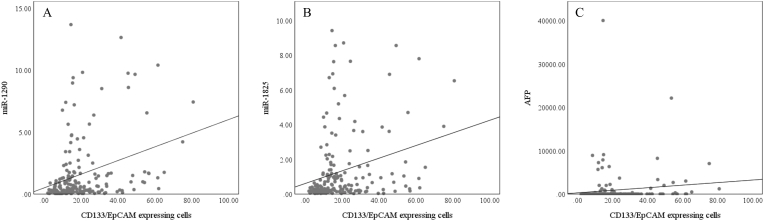
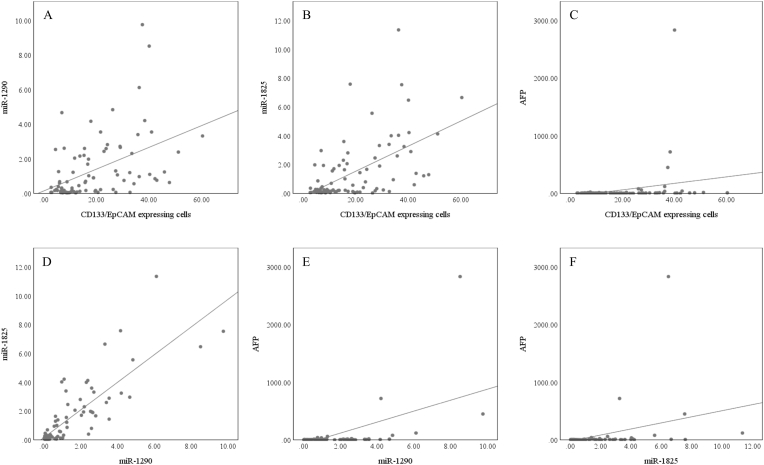
In HCV infection, the CD133/EpCAM-expressing cells significantly correlated with miR-1290 (r = 0.38, p<0.001), miR-1825 (r = 0.35, p<0.001), and AFP (r = 0.18, p = 0.016) but not with the number or size of tumor lesion (Fig. 8). Also, miR-1290 and miR-1825 were positively correlated with each other (r = 0.79, p<0.001), AFP (r = 0.49, p<0.001 and r = 0.48, p<0.001, respectively), number of focal lesions (r = 0.38, p = 0.01 and r = 0.41, p = 0.004, respectively), and focal lesion size (r = 0.55, p<0.001 and r = 0.42, p = 0.004, respectively) (Fig. 9). On the other side, AFP didn't show any correlation with the focal lesion number or size.
Fig. 8.
Scatter plot diagrams of the correlations between the CD133/EpCAM-expressing cells and (A) miR-1290, (B) miR-1825, and (C) AFP, in the HCV groups.
Fig. 9.
Scatter plot diagrams of the correlations between the miR-1290 and (A) AFP, (B) focal lesion number , and (C) lesion size, and between the miR-1825 and (D) AFP, (E) focal lesion number, (F) lesion size, and (G) miR-1290, in the HCV groups.
In HBV infection, the CD133/EpCAM-expressing cells exhibited significant correlations with miR-1290 (r = 0.51, p<0.001), miR-1825 (r = 0.55, p<0.001), and AFP (r = 0.32, p = 0.002). Also, the miR-1290 and miR-1825 were shown to be positively correlated with each other (r = 0.81, p<0.001), as well as with AFP (r = 0.44, p<0.001 and r = 0.42, p<0.001, respectively) (Fig. 10). However, none of them, in addition to the AFP, showed any correlation with the number or size of the malignant lesion.
Fig. 10.
Scatter plot diagrams of the correlations between the CD133/EpCAM-expressing cells and (A) miR-1290, (B) miR-1825, and (C) AFP, between the miR-1290 and (D) miR-1825 and (E) AFP, and between the miR-1825 and (F) AFP, in the HBV groups.
4. Discussion
In the present study, individuals with viral hepatitis alone or in conjunction with cirrhosis showed a significant increase in the mean percentages of the CD133/EpCAM-expressing cells when compared to the control group. Also, the viral hepatitis-associated hepatocellular carcinoma (HCC) patients had higher percentages of CD133/EpCAM-expressing cells than the other corresponding groups. MiR-1290 showed a significant increase in the HBV and HBV-associated cirrhosis groups, compared to the control group. Additionally, miR-1825 expression was significantly up-regulated in the HCV-associated cirrhosis group. Besides, it demonstrated a significant progressive escalation in the HBV, HBV-associated cirrhosis, and HBV-associated HCC which had the highest expression level. The mean relative quantification values of miR-1290 and miR-1825 were considerably higher in the HCV- and HBV-associated HCC groups than in the other equivalent groups, and they were positively correlated with the tumor lesion size and number in the HCV-associated HCC patients. Moreover, only miR-1825 was able to differentiate between HCV- and HBV-associated HCC groups. These findings indicate the relevance of CD133/EpCAM expressing cells and their regulatory miRNAs in the pathophysiology of liver cirrhosis and hepatocellular carcinoma developed as a consequence of either chronic hepatitis C (HCV) or hepatitis B viruses (HBV) infection.
Over the past years, the early diagnosis of HCC has relied on surveillance with the serological assessments of AFP and ultrasonograhy (US). However, neither the sensitivity nor the specificity of either is adequate for detecting early-onset HCC. For instance, at a cut-off value of 20 ng/mL, the reported sensitivity of AFP for diagnosing HCC at any stage ranges between 41% and 65%, with specificity ranging from 80% to 94% [24]. Also, the US has a sensitivity of 84% for HCC detection at any stage and a sensitivity of 47% for the identification of early‐stage HCC. Moreover, the US imaging analysis is operator-dependent [25]. In hepatic lesions measuring ≥ 1 cm, the CT and MRI imaging techniques provide a definitive diagnosis with high accuracy allowing for the early detection of HCC. Yet, these modalities are still too expensive for widespread applications. As a result, new imaging techniques are being developed to improve the HCC detectability and characterization of the HCC nodules. In addition, significant HCC protein, RNA, and DNA biomarkers with potential clinical value have emerged to aid in HCC surveillance [24].
The identification of cancer stem cells (CSCs), also known as cancer-initiating cells, within a tumor signifies a breakthrough in cancer research and management. To effectively and completely treat the malignant disorder and prevent tumor recurrence, diagnostic and therapeutic techniques that target these cells and their regulators, in addition to the differentiated tumor cells, are required [26]. This necessitates unraveling the underlying molecular, genetic, and epigenetic mechanisms responsible for regulating the development of CSCs.
Similar to the results of the current study, it has been shown that CD133+EpCAM+ cells exhibit tumor-initiating properties such as self-renewal, drug resistance, and tumorigenicity [27], and tumorigenic CSCs can be obtained from HBV- or HCV-related HCC [28]. Also, sustained HCV expression has been demonstrated to promote stem cell-like features with enhanced expression of CD133, AFP, CK19, and c-Myc [29]. Furthermore, it has been found that HBV boosts the production of CSCs-related markers, e.g. CD133, and the expression of CSCs-related genes in tumor cells [30]. Additionally, the X protein of HBV contributes to the stem-like properties of CSCs in HCC, including self-renewal and chemoresistance [31].
Measuring the levels of circulating miRNAs shows a lot of promise to be utilized as biomarkers for malignancies because they are highly stable in human formalin-fixed tissue, serum, and various body fluids, are not obviously influenced by age, gender, smoking status, body mass index, or other basic characteristics, and have a tissue-specific expression pattern [32]. In the present study, miR-1290 exhibited the highest sensitivity and specificity for detecting HCC associated with chronic HCV infection (96% and 82%, respectively), followed by the miR-1825 (91% and 77%, respectively) . Meanwhile, AFP, at a value of >8.4 ng/mL, had a sensitivity and specificity of 87% and 80%, respectively. Analogous to this finding, it has been shown that the miR-1290 has sensitivities of 75%, 80%, 26%, and 98.96% and specificities of 97.5%, 96.7%, 90.0%, and 100% for identifying pancreatic cancer [33], lung adenocarcinoma [34], prostate cancer [35], and gastric cancer [14], respectively.
MiR-1290 was initially discovered in human embryonic stem cells (ESCs). It is encoded by the first intron of the aldehyde dehydrogenase 4 family member A1 gene [36]. It has been revealed that miR-1290 raises the percentage of cells in the G2/M phase while decreasing their percentage in the G0/G1 peak, meaning that miR-1290 facilitates cell cycle progression through mitosis which is crucial in directing cell proliferation [37]. Moreover, miR-1290 activates the Akt and NF-κB pathways to overcome the cytokinesis-induced proliferation delay and is implicated in cancer cell reprogramming by up-regulating the expression of Nanog and c-Myc or down-regulating the expression of Oct4 and Sox2 [38].
Several lines of evidence have indicated that miR-1290 promotes epithelial-mesenchymal transition (EMT) and the subsequent acquisition of CSC phenotypes in malignancies. Additionally, It has been presented that miR-1290 has a considerably higher expression in the CSLCs, and down-regulating its expression in the CD133+ cells by antagomir-1290 reduces tumor growth, clonogenicity, proliferation, migration, and invasion [39].
In line with the findings of the present study, the onco-miR-1290 expression has been observed to be significantly increased in non-small cell lung cancer (NSCLC) and colorectal cancer (CRC) tissues when compared to the adjacent non-tumor normal tissues [40,41]. Also, it has been demonstrated that miR-1290 overexpression is positively correlated with advanced tumor stage and positive lymph node metastasis, predicts poor overall survival, and is an independent prognostic factor for NSCLC, esophageal squamous cell carcinoma, pancreatic cancer, and CRC [15,33,40,41]. Likewise, there has been a positive correlation between the serum levels of miR-1290 and the tumor size of pancreatic cancer and lung adenocarcinoma [33,34]. However, there has been no evidence of a significant association between the miR-1290 expression and NSCLC tumor size [41].
MiR-1825 is another stemness-associated miRNA investigated in the present study. It resides within the 20q11.21 chromosomal region which has been linked to a recurrent gain of function abnormality in the human ESCs and induced pluripotent stem cells [42]. Interestingly, in human ESCs, 20q11.21 amplification has resulted in the acquisition of a gene-expression signature enriched for cancer-related genes, improvement of the colony-forming potential, and reduction of apoptosis [43].
In agreement with this study, miR-1825 overexpression has been detected in larynx cancer and has been identified as a key contributor to carcinogenesis as a consequence of its dysregulation in the laryngeal CSLCs [21]. Also, it has been proposed that it is involved in the acquisition and maintenance of stem cells-associated characteristics as well as in tumor development, progression, metastasis, chemoresistance, and recurrence [22]. For instance, it has been found to be highly up-regulated in prostate cancer tissues and to function as a biomarker for prostate cancer [18]. Dissimilarly, miR-1825 down-regulation has been associated with glioma carcinogenesis. Too, a reduced miR-1825 expression has been reported in glioblastoma and has been utilized to predict poor prognosis, whereas its re-expression has impeded glioblastoma cell survival, invasiveness, migration, and metastasis [19].
Despite the promising results, the current study has certain limitations. First, the sample size was relatively small, and a larger sample size from different ethnic populations is needed to further confirm the findings. Second, the levels of LCSCs and the examined miRNAs were not validated in HCC tissue specimens. Third, the correlation between the LCSCs, the miRNAs expression, and patient prognosis and survival was not investigated. Therefore, a long-term follow-up study will be planned to determine whether they can serve as predictive indicators for patients’ survival.
Taken together, the results of the present study demonstrate that the CD133/EpCAM expressing LCSCs play an important role in the pathogenesis of liver cirrhosis and HCC development as a consequence of chronic HCV or HBV infection, and this is in part orchestrated by dysregulated miRNAs profiles. This suggests that CD133/EpCAM and stemness-associated miRNAs, miR-1290 and miR-1825, could serve as promising diagnostic and prognostic biomarkers as well as therapeutic targets in patients suffering from liver cirrhosis or HCC.
Research involving human participants
All procedures performed in this study were in accordance with the ethical standards of the Institutional Research Committee and Review Board (FWA00010609) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all participants included in this study.
Data availability statement
All data generated or analyzed during this study are included in this published article.
CRediT authorship contribution statement
Marwa Hassan: Conceptualization, Funding acquisition, Data curation, Formal analysis, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. Sami Mohamed Nasr: Conceptualization, Project administration, Data curation, Formal analysis, Methodology, Validation. Noha Abdelaal Amin: Data curation, Formal analysis, Methodology, Visualization, Validation. Eman El-Ahwany: Conceptualization, Investigation, Project administration, Supervision, Validation. Mona Zoheiry: Conceptualization, Funding acquisition, Formal analysis, Supervision, Validation. Mohamed Elzallat: Conceptualization, Project administration, Data curation, Formal analysis, Methodology, Visualization.
Declaration of competing interest
The authors have no conflicts of interest to declare.
Acknowledgement
This work was kindly supported by the internal project No. 107 Diagnosis, Theodor Bilharz Research Institute, Giza, Egypt.
References
- 1.Ehsan N., Sweed D., Elsabaawy M. Evaluation of HCV-related liver fibrosis post-successful DAA therapy, Egypt. Liver J. 2021;11:1–7. doi: 10.1186/s43066-021-00129-0. [DOI] [Google Scholar]
- 2.El-Ahwany E.G.E., Mourad L., Zoheiry M.M.K., Abu-Taleb H., Hassan M., Atta R., Hassanien M., Zada S. MicroRNA-122a as a non-invasive biomarker for HCV genotype 4-related hepatocellular carcinoma in Egyptian patients. Arch. Med. Sci. 2019;15:1454–1461. doi: 10.5114/aoms.2019.86621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ringelhan M., Pfister D., O'Connor T., Pikarsky E., Heikenwalder M. The immunology of hepatocellular carcinoma. Nat. Immunol. 2018;193:222–232. doi: 10.1038/s41590-018-0044-z. 19 (2018. [DOI] [PubMed] [Google Scholar]
- 4.Okasha H., Hassan M., Aboushousha T., Samir S. Effect of Interferon-Beta (IFN-β) on tumor suppressor and apoptotic markers in hepatocellular carcinoma cell line. Int. J. Res. Pharm. Sci. 2019;10:2936–2943. doi: 10.26452/ijrps.v10i4.1574. [DOI] [Google Scholar]
- 5.Yang J.D., Hainaut P., Gores G.J., Amadou A., Plymoth A., Roberts L.R. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang L., Cheng Q., Zhang B.H., Zhang M.Z. Circulating microRNAs as biomarkers in hepatocellular carcinoma screening a validation set from China. Medicine (Baltim.) 2015;94 doi: 10.1097/MD.0000000000000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lv D., Chen L., Du L., Zhou L., Tang H. Emerging regulatory mechanisms involved in liver cancer stem cell properties in hepatocellular carcinoma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.691410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nassar D., Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu. Rev. Pathol. 2016;11:47–76. doi: 10.1146/annurev-pathol-012615-044438. [DOI] [PubMed] [Google Scholar]
- 9.Kim G., An H.J., Lee M.J., Song J.Y., Jeong J.Y., Lee J.H., Jeong H.C. Hsa-miR-1246 and hsa-miR-1290 are associated with stemness and invasiveness of non-small cell lung cancer. Lung Cancer. 2016;91:15–22. doi: 10.1016/j.lungcan.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Ma Q., Wang Y., Zhang H., Wang F. miR-1290 contributes to colorectal cancer cell proliferation by targeting INPP4B. Oncol. Res. 2018;26:1167–1174. doi: 10.3727/096504017X15051741798389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao Y., Liu J., Zhang D., Li B. MiR-1290 promotes cancer progression by targeting nuclear factor I/X(NFIX) in esophageal squamous cell carcinoma (ESCC) Biomed. Pharmacother. 2015;76:82–93. doi: 10.1016/j.biopha.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Lin M., Shi C., Lin X., Pan J., Shen S., Xu Z., Chen Q. SMicroRNA-1290 inhibits cells proliferation and migration by targeting FOXA1 in gastric cancer cells. Gene. 2016;582:137–142. doi: 10.1016/j.gene.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Kang E., Jung S.C., Nam S.K., Park Y., Seo S.H., Park K.U., Oh H.K., Kim D.W., Kang S.B., Lee H.S. Tissue miR-200c-3p and circulating miR-1290 as potential prognostic biomarkers for colorectal cancer. Sci. Rep. 2022;12:1–11. doi: 10.1038/s41598-022-06192-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L., Cai Y., Chen X., Zhu Y., Cai J. Circulating MiR-1290 as a potential diagnostic and disease monitoring biomarker of human gastrointestinal tumors. BMC Cancer. 2021;21:989. doi: 10.1186/s12885-021-08729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun H., Wang L., Zhao Q., Dai J. Diagnostic and prognostic value of serum miRNA-1290 in human esophageal squamous cell carcinoma. Cancer Biomarkers. 2019;25:381–387. doi: 10.3233/CBM-190007. [DOI] [PubMed] [Google Scholar]
- 16.Khalighfard S., Kalhori M.R., Haddad P., Khori V., Alizadeh A.M. Enhancement of resistance to chemo-radiation by hsa-miR-1290 expression in glioblastoma cells. Eur. J. Pharmacol. 2020;880 doi: 10.1016/j.ejphar.2020.173144. [DOI] [PubMed] [Google Scholar]
- 17.Xing W., Zeng C. A novel serum microRNA-based identification and classification biomarker of human glioma. Tumor Biol. 2017;39 doi: 10.1177/1010428317705339. [DOI] [PubMed] [Google Scholar]
- 18.Guo X., Han T., Hu P., Guo X., Zhu C., Wang Y., Chang S. Five microRNAs in serum as potential biomarkers for prostate cancer risk assessment and therapeutic intervention. Int. Urol. Nephrol. 2018;50:2193–2200. doi: 10.1007/s11255-018-2009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu F., Li C., Sun Y., Jia T., Li N., Li H. Upregulation of miR-1825 inhibits the progression of glioblastoma by suppressing CDK14 though Wnt/β-catenin signaling pathway. World J. Surg. Oncol. 2020;18:147. doi: 10.1186/s12957-020-01927-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai J.-C., Huang G.-Y. miR-1825 accelerates cell proliferation and inhibits cell apoptosis of prostate cancer via targeting suppressor of cancer cell invasion. J. Biomater. Tissue Eng. 2021;11:820–831. doi: 10.1166/jbt.2021.2681. [DOI] [Google Scholar]
- 21.Karatas O.F., Suer I., Yuceturk B., Yilmaz M., Oz B., Guven G., Cansiz H., Creighton C.J., Ittmann M., Ozen M. Identification of microRNA profile specific to cancer stem-like cells directly isolated from human larynx cancer specimens. BMC Cancer. 2016;16:853. doi: 10.1186/s12885-016-2863-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan A., Ahmed E., Elareer N., Junejo K., Steinhoff M., Uddin S. Role of miRNA-regulated cancer stem cells in the pathogenesis of human malignancies. Cells. 2019;8:840. doi: 10.3390/cells8080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Wang W., Wei C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020;7:308–319. doi: 10.1016/j.gendis.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adeniji N., Dhanasekaran R. Current and emerging tools for hepatocellular carcinoma surveillance. Hepatol. Commun. 2021;5:1972–1986. doi: 10.1002/hep4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun J.H., Luo Q., Liu L.L., Bin Song G. Liver cancer stem cell markers: progression and therapeutic implications. World J. Gastroenterol. 2016;22:3547–3557. doi: 10.3748/wjg.v22.i13.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Yu D., Zhang H., He H., Zhang C., Zhao W., guang Shao R. CD133+EpCAM+ phenotype possesses more characteristics of tumor initiating cells in hepatocellular carcinoma Huh7 Cells. Int. J. Biol. Sci. 2012;8:992–1004. doi: 10.7150/ijbs.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita T., Honda M., Nakamoto Y., Baba M., Nio K., Hara Y., Zeng S.S., Hayashi T., Kondo M., Takatori H., Yamashita T., Mizukoshi E., Ikeda H., Zen Y., Takamura H., Wang X.W., Kaneko S. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology. 2013;57:1484–1497. doi: 10.1002/hep.26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali N., Allam H., May R., Sureban S.M., Bronze M.S., Bader T., Umar S., Anant S., Houchen C.W. Hepatitis C virus-induced cancer stem cell-like signatures in cell culture and murine tumor xenografts. J. Virol. 2011;85:12292–12303. doi: 10.1128/jvi.05920-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z., Dai X., Wang T., Zhang C., Zhang W., Zhang W., Zhang Q., Wu K., Liu F., Liu Y., Wu J. Hepatitis B virus PreS1 facilitates hepatocellular carcinoma development by promoting appearance and self-renewal of liver cancer stem cells. Cancer Lett. 2017;400:149–160. doi: 10.1016/j.canlet.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 31.Wang C., Da Wang M., Cheng P., Huang H., Dong W., Zhang W.W., Li P.P., Lin C., Pan Z.Y., Wu M.C., Zhou W.P. Hepatitis B virus X protein promotes the stem-like properties of OV6+ cancer cells in hepatocellular carcinoma. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2016.493. e2560–e2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., Peng R., Wang J., Qin Z., Xue L. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin. Epigenet. 2018;10:1–10. doi: 10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei J., Yang L., Wu Y.N., Xu J. Serum miR-1290 and miR-1246 as potential diagnostic biomarkers of human pancreatic cancer. J. Cancer. 2020;11:1325–1333. doi: 10.7150/jca.38048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y., Wei J., Zhang W., Xie M., Wang X., Xu J. Serum exosomal miR-1290 is a potential biomarker for lung adenocarcinoma. OncoTargets Ther. 2020;13:7809–7818. doi: 10.2147/OTT.S263934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H.P., Lai H.M., Guo Z. Prostate cancer early diagnosis: circulating microRNA pairs potentially beyond single microRNAs upon 1231 serum samples. Briefings Bioinf. 2021;22 doi: 10.1093/bib/bbaa111. bbaa111. [DOI] [PubMed] [Google Scholar]
- 36.Morin R.D., O'Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., Eaves C.J., Marra M.A. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yelamanchili S.V., Morsey B., Harrison E.B., Rennard D.A., Emanuel K., Thapa I., Bastola D.R., Fox H.S. The evolutionary young miR-1290 favors mitotic exit and differentiation of human neural progenitors through altering the cell cycle proteins. Cell Death Dis. 2014;5:e982. doi: 10.1038/cddis.2013.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J., Ji X., Zhu L., Jiang Q., Wen Z., Xu S., Shao W., Cai J., Du Q., Zhu Y., Mao J. Up-regulation of microRNA-1290 impairs cytokinesis and affects the reprogramming of colon cancer cells. Cancer Lett. 2013;329:155–163. doi: 10.1016/J.CANLET.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 39.Sun B., Yang N., Jiang Y., Zhang H., Hou C., Ji C., Liu Y., Zuo P. Antagomir-1290 suppresses CD133+ cells in non-small cell lung cancer by targeting fyn-related Src family tyrosine kinase. Tumor Biol. 2015;36:6223–6230. doi: 10.1007/s13277-015-3307-4. [DOI] [PubMed] [Google Scholar]
- 40.Soheilifar M.H., Pornour M., Saidijam M., Najafi R., Azizi Jalilian F., Keshmiri Neghab H., Amini R. miR-1290 contributes to oncogenesis and angiogenesis via targeting of THBS1, DKK3 and, SCAI. Bioimpacts. 2021;11 doi: 10.34172/BI.2021.23571. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mo D., Gu B., Gong X., Wu L., Wang H., Jiang Y., Zhang B., Zhang M., Zhang Y., Xu J., Pan S. miR-1290 is a potential prognostic biomarker in non-small cell lung cancer. J. Thorac. Dis. 2015;7:1570–1579. doi: 10.3978/J.ISSN.2072-1439.2015.09.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen H.T., Geens M., Spits C. Genetic and epigenetic instability in human pluripotent stem cells. Hum. Reprod. Update. 2013;19:187–205. doi: 10.1093/humupd/dms048. [DOI] [PubMed] [Google Scholar]
- 43.Werbowetski-Ogilvie T.E., Bossé M., Stewart M., Schnerch A., Ramos-Mejia V., Rouleau A., Wynder T., Smith M.J., Dingwall S., Carter T., Williams C., Harris C., Dolling J., Wynder C., Boreham D., Bhatia M. Characterization of human embryonic stem cells with features of neoplastic progression. Nat. Biotechnol. 2009;27:91–97. doi: 10.1038/nbt.1516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.