Summary
Cholesterol is a structural component of cell membranes. Most cells are incapable of its catabolism, and intracellular cholesterol accumulation is linked to several disorders including cardiovascular and neurodegenerative diseases. Cholesterol efflux, essential to its metabolism, is dependent on acceptors such as apolipoproteins. Here, we describe an assay to evaluate the capacity of cholesterol acceptors. Cells are treated with an analog of cholesterol tagged with fluorescent BODIPY. Addition of an acceptor leads to BODIPY-cholesterol efflux, measured using a plate reader.
For complete details on the use and execution of this protocol, please refer to Liu et al. (2021).1
Subject areas: Cell-based Assays, Molecular/Chemical Probes
Graphical abstract
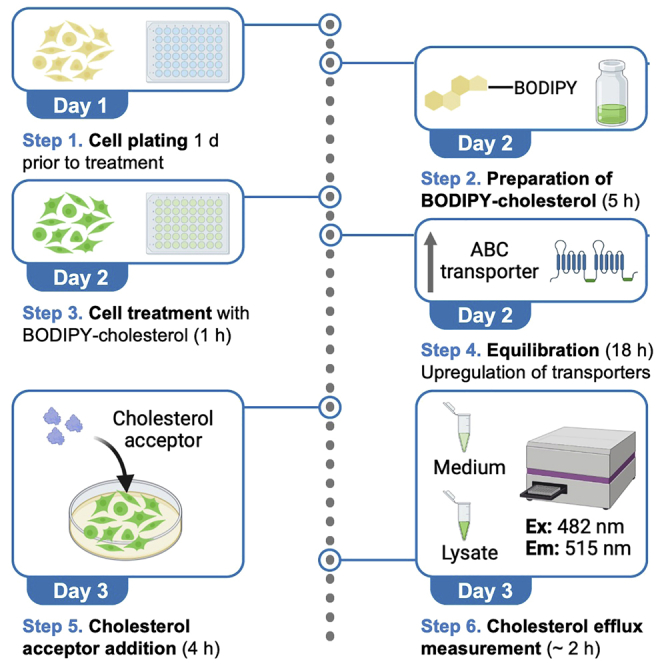
Highlights
-
•
A protocol to characterize cholesterol efflux in vitro
-
•
Characterization of cholesterol acceptors using a fluorescent analog of cholesterol
-
•
Protocol can be optimized to be used in other cell types
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Cholesterol is a structural component of cell membranes. Most cells are incapable of its catabolism, and intracellular cholesterol accumulation is linked to several disorders including cardiovascular and neurodegenerative diseases. Cholesterol efflux, essential to its metabolism, is dependent on acceptors such as apolipoproteins. Here, we describe an assay to evaluate the capacity of cholesterol acceptors. Cells are treated with an analog of cholesterol tagged with fluorescent BODIPY. Addition of an acceptor leads to BODIPY-cholesterol efflux, measured using a plate reader.
Before you begin
The following cholesterol efflux assay is adapted from a previous protocol performed on macrophages, with modifications.2 The protocol below describes a detailed step-by-step procedure for preparing the key reagent for this assay, BODIPY-cholesterol, as well as the specific steps for using primary cortical astrocytes from apolipoprotein E knockout (APOE-KO) mice. We used APOE-KO mice because we were testing the cholesterol accepting capacity of apoE mutants.1 We have also successfully used this protocol in APOE-KO immortalized astrocyte that were generated according to a previously published protocol.3
Primary cortical astrocyte cell culture
Timing: 2 days; cell maintenance over a couple of weeks
-
1.
Coat 1×T-75 tissue culture flask with 25 μg/mL poly-D-lysine one day prior to primary cortical astrocyte culture preparation. On the day of cell plating, rinse flask once with sterile water and let it dry.
-
2.Cortical brain tissue dissection:
-
a.Sacrifice a P1-P4 mouse pup by decapitation.
-
b.Carefully collect the brain and place it in ice-cold Dissection medium (see materials and equipment below).
-
c.Dissect out both hemicortices on a 60 mm petri dish, making sure to carefully peel away the meninges from the cortical tissue.
-
a.
-
3.Trypsin digestion:
-
a.Digest brain cortices with 1.5 mL 0.25% trypsin/EDTA at 37°C for 10 min.
-
b.Neutralize the trypsin with 1.5 mL culture medium.
-
c.Spin the tissue down at 300 × g for 5 min.
-
d.Aspirate the medium and replace with 1.5–2 mL culture medium.
-
e.Triturate the tissue by carefully pipetting up and down until pieces are no longer visible. Avoid creating bubbles.
-
a.
-
4.Cell plating and maintenance:
-
a.Filter the cell suspension through a 100 μm cell strainer.
-
b.Rinse the cell strainer with 10 mL of culture medium.
-
c.Transfer the cell suspension into one PDL-coated T-75 tissue culture flask and place in a 37°C, 5% CO2 incubator.
-
d.Perform culture medium changes every 2–3 days until cells are confluent.
-
e.Remove primary microglia by shaking flask on a tabletop shaker at 350 rpm for 20 min.
-
f.Passage cells with accutase up to 3 times.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Poly-D-lysine | Sigma-Aldrich | A-003-E |
| HBSS (with calcium and magnesium) | Thermo Fisher Scientific | 14025092 |
| 100× Penicillin-Streptomycin (Pen-Strep) | Thermo Fisher Scientific | 15140122 |
| 100× Amphotericin-B (Amp-B) | Gemini Bio-Products | 400-104 |
| DMEM/F-12 with HEPES | Thermo Fisher Scientific | 11330-032 |
| HyClone FBS, heat activated | GE Life sciences | SH30396.03 |
| Recombinant human EGF | Thermo Fisher Scientific | PGH0311 |
| Sodium pyruvate (100 mM) | Thermo Fisher Scientific | 11360070 |
| 0.25% Trypsin/EDTA | Gibco | 25200-056 |
| Trypan blue | Bio-Rad | 1450021 |
| Accutase | Stemcell technologies | 07920 |
| TopFluor Cholesterol (BODIPY-cholesterol) | Avanti Polar Lipids | 810255 |
| Cholesterol (ovine) | Avanti Polar Lipids | 700000 |
| DMSO, molecular biology grade | BioWorld | 40470006 |
| Chloroform | Fisher Chemical | C298-500 |
| Methyl-β-cyclodextrin (MβCD) | Sigma-Aldrich | C4555 |
| DMEM/F-12, no phenol red | Thermo Fisher Scientific | 11039-021 |
| cAMP | Sigma-Aldrich | C8988 |
| Sandoz 58-035 | Sigma-Aldrich | S9318 |
| 10% BSA | MACS Buffer | 130-091-376 |
| ApoA1 | Fitzgerald | 118-30-1047S |
| RIPA lysis buffer | EMD Millipore | 20-188 |
| Other | ||
| Tissue culture flask, T-75 | Corning | 15327 |
| 100 μm cell strainer | Fisher Scientific | 431750 |
| Petri dishes, 60 mm | Corning | 353003 |
| Corning Falcon 15 mL conical centrifuge tubes | Fisher Scientific | 14-959-70C |
| Corning Falcon 50 mL conical centrifuge tubes | Fisher Scientific | 352098 |
| Counting slides | Bio-Rad | 1450015 |
| TC20 cell counter | Bio-Rad | 1450102 |
| Costar® 48-well plate, tissue culture treated | Corning | 3548 |
| Corning™ 96-Well Solid Black Polystyrene Microplates | Corning | 3915 |
| Centrifuge 5424 R | Eppendorf | / |
| Elmasonic S 30 H | Elma | / |
| Orbital incubator shaker Gyromax 703 | Amerex | / |
| Sorvall Legend X1 Centrifuge | Thermo Fisher Scientific | 75004220 |
| Synergy H1 microplate reader | BioTek | / |
Materials and equipment
Dissection medium
| Reagent | Final concentration | Amount |
|---|---|---|
| HBSS | N/A | 98 mL |
| 100× Pen-Strep | 1× | 1 mL |
| 100× Amp-B | 1× | 1 mL |
| Total | N/A | 100 mL |
Stored at 4°C. Prepared on a need basis.
Culture medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F-12 with HEPES | N/A | 760 mL |
| HyClone FBS, heat activated | 20% | 200 mL |
| 100× Pen-Strep | 1× | 10 mL |
| 100× Amp-B | 1× | 10 mL |
| 100 mM Sodium pyruvate | 1 mM | 10 mL |
| Recombinant human EGF | 10 ng/mL | 10 μL |
| Total | N/A | 1,000 mL |
Stored at 4°C for up to 6 months.
1× BODIPY-Cholesterol and Equilibration medium stock reagents
| Reagent | Volume | Amount |
|---|---|---|
| 3 mM cAMP | 655 μL DMEM/F-12, no phenol red | 1 mg |
| 2 mg/mL Sandoz 58-035 | 2.5 mL DMSO | 5 mg |
Equilibration medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F-12, no phenol red | N/A | 2.198 mL |
| 10% BSA | 0.2% | 50 μL |
| 3 mM cAMP | 0.3 mM | 250 μL |
| 2 mg/mL Sandoz 58-035 | 2 μg/mL | 2.5 μL |
| Total | N/A | 2.5 mL |
Prepared and used on the day of the experiment.
Note: The amount of BODIPY-cholesterol treatment prepared is dependent on how many cholesterol acceptors are being tested. A volume of 2.5 mL is enough to test 3 different conditions and to include a negative control.
Step-by-step method details
Figure 1 depicts the complete workflow for the procedure.
Figure 1.
Workflow to evaluate the cholesterol accepting capacity of acceptors in mouse primary cortical astrocytes
(A–E) Graphical summary depicting the steps to evaluate the cholesterol accepting capacity of acceptors in mouse primary cortical astrocytes: (A) Primary APOE-knockout (KO) astrocyte culture; (B) Plating astrocytes for experiment; (C) Preparing 10× BODIPY-cholesterol; (D) Cell treatment with BODIPY; (E) Cell treatment with apoA1; data collection.
Cell plating
Timing: 1 h
Cells are plated onto a 48-well plate one day prior to BODIPY-cholesterol treatment. The number of wells prepared depends on how many cholesterol acceptors are being tested (see note below).
-
1.Passage primary astrocytes:
-
a.Aspirate culture medium from cells.
-
b.Rinse cells once with warm DMEM/F-12.
-
c.Cell dissociation:
-
i.Aspirate DMEM/F-12.
-
ii.Add 5 mL of room temperature accutase to the cells.
-
iii.Incubate 10 min in a tissue culture incubator (37°C, 5% CO2).
-
iv.Gently tap the sides of the flask to lift the cells off.
-
v.Add 5 mL of warm culture medium and pipette up and down a couple of times (∼10 times) with a 10-mL serological pipette. Avoid bubbles.
-
vi.Transfer the cell suspension to a 15-mL falcon tube and gently pipette up and down to further dissociate cell clumps.
-
vii.Pellet the cells by centrifugation at 300 × g for 3 min.
-
viii.Resuspend the cell pellet in 1 mL of culture medium and pipette up and down with a 1 mL pipette to ensure the presence of single cells (∼30 times). Avoid bubbles.
-
i.
-
a.
-
2.Cell counting:
-
a.Take 10 μL of the cell suspension and mix thoroughly with 10 μL of trypan blue.
-
b.Load 10 μL of the mixture onto a disposable cell counting slide and count cells (manually or using an automated cell counter).
-
a.
-
3.Cell plating:
-
a.Resuspend cells to a final density of 0.1 × 106 cells/mL in culture medium.
-
b.Plate 25 × 10ˆ3 cells per well in a 48-well plate (250 μL per well).
-
c.Gently shake the plate to distribute the cells evenly across the well.
-
d.Incubate overnight at 37°C, 5% CO2 to allow for the cells to settle and attach to the plate.
-
a.
Note: Alternative counting methods can be performed (step 2). The number of wells prepared (step 3) is dependent on the number of cholesterol acceptor being tested. Typically, we test each condition in triplicate (at least), and we include a baseline control (no acceptor).
CRITICAL: It is important to ensure that the primary astrocytes are not clumped together so cells can be plated as single cells.
Preparing 10× BODIPY-cholesterol
Timing: 5 h
The following steps describe the preparation of the stock BODIPY-cholesterol solution. Figure 2 depicts the workflow of this specific procedure.
-
4.Prepare stock solutions of starting materials:
-
a.Dilute cholesterol at 10 mg/mL in chloroform. Store at −20°C for up to one year.
-
b.Dilute TopFluor Cholesterol (BODIPY-cholesterol) at 1 mg/mL in DMSO. Aliquots can be stored at −20°C for several months.
-
c.Prepare MβCD at 20 mM in DMEM/F-12, no phenol red.
-
i.Dissolve 66 mg in 2.5 mL DMEM/F-12, no phenol red.
-
i.
-
a.
Note: 2.5 mL of 10× BODIPY-cholesterol yields 25 mL of 1× BODIPY-cholesterol, which allows for the treatment of 100 wells. The amount of 10× BODIPY-cholesterol should be tailored to the number of wells needed to be treated.
-
5.In a glass test tube, add:
-
a.10 mg/mL cholesterol in chloroform to a final concentration of 0.1 mM (V = 9.75 μL for a final volume of 2.5 mL of 10× BODIPY-cholesterol).
-
b.1 mg/mL TopFluor Cholesterol (BODIPY-cholesterol) in DMSO to a final concentration of 0.05 mM (V = 72.5 μL for a final volume of 2.5 mL of 10× BODIPY-cholesterol).
-
a.
Figure 2.
Preparation of 10× BODIPY-cholesterol stock solution
Pictures depicting the different steps of preparing 10× BODIPY-cholesterol1: BODIPY-cholesterol suspension in organic solvent should look fluorescent green2; Formation of a lipid film is achieved by evaporating solvent under N23; a lipid film has formed on the walls of the glass vial and should appear red; (4) the lipid film is resuspended in DMEM containing 20 Mm MβCD; (5) suspension is sonicated for 30 min at 37°C; (6) and incubated for 3 h at 37°C prior to centrifugation; (7) fluorescence of 10× BODIPY-cholesterol is measured and recorded.
(See Figure 2, step 1).
-
6.
Dry the lipid mixture in a chemical hood in the dark under a flow of N2 until formation of a thin film (see Figure 2, steps 2 and 3).
-
7.
Resuspend the lipid film in 20 mM MβCD in DMEM/F-12, no phenol red (see Figure 2, step 4; 2.5 mL of 20 mM MβCD in DMEM/F-12 for a final volume of 2.5 mL of 10× BODIPY-cholesterol).
-
8.
Sonicate the lipid suspension in a water bath set to 37°C for 30 min, to promote solubilization (Figure 2, step 5).
-
9.
Transfer the sonicated lipid suspension to a capped glass vial and incubate in an orbital incubator shaker at 250 rpm, 37°C for 3 h.
-
10.
Sonicate the lipid suspension again in a water bath set to 37°C for 30 min.
-
11.
Transfer the lipid suspension to a 15-mL falcon tube and centrifuge for 5 min at 4,696 × g to pellet the non-solubilized BODIPY-cholesterol.
-
12.
Carefully transfer the supernatant to a capped glass vial wrapped in foil (Figure 2, step 6). This supernatant is the final 10× BODIPY-cholesterol stock solution.
-
13.Measure the fluorescence of 10× BODIPY cholesterol:
-
a.Add 100 μL to one well of a 96-well black microplate.
-
b.Measure fluorescence in a plate reader:
-
i.Set the excitation wavelength to 482 nm.
-
ii.Set the emission wavelength to 515 nm.
-
iii.Keep a record of the fluorescence measured in RFU.
-
i.
-
a.
Note: Using our plate reader, we typically get a fluorescence measurement over 2,000,000 RFU. The fluorescence of 10× BODIPY-cholesterol will vary with each preparation but we do not recommend using a stock solution with a fluorescence measurement below 1,500,000 RFU. See troubleshooting 1.
-
14.
Store 10× BODIPY-Cholesterol at 4°C for up to one week.
Note: Other protocols filter their lipid suspension through a membrane filter to discard any undissolved BODIPY-cholesterol. We find that 10× BODIPY-cholesterol binds to PVDF membrane filters, resulting in a dramatic decrease in fluorescence post-filtering.
Pause point: 10× BODIPY-cholesterol can be kept at 4°C for up to a week in the dark. We would not recommend using 10× BODIPY-cholesterol after one week as there could be both a decrease in fluorescence, and the risk of formation of cholesteryl esters, which would not efflux from cells.
CRITICAL: Cyclodextrins can form complexes with cholesterol and fluorescent cholesterol analogs, facilitating their solubilization into aqueous solutions. It is thus important not to omit it from the preparation. One way to ensure that the 10× BODIPY-cholesterol solution was adequately prepared is by checking its fluorescence with a plate reader (ex: 482 nm; em: 515 nm). We find that a 10× BODIPY-cholesterol with low fluorescence (below 1,000,000 RFU) yields poor results.
Cholesterol efflux assay
Timing: 2 days
The steps below describe the loading of cells with BODIPY-cholesterol. This is followed by an equilibration step with cAMP to up-regulate ABCA1, essential to facilitate ABCA1-mediated cholesterol efflux. Finally, cholesterol acceptors are added to the cells. Their cholesterol accepting capacity is defined by the associated baseline corrected fractional efflux of BODIPY-cholesterol (see quantification and statistical analysis below). All following experimental steps are performed in the presence of an acetyl-choline esterase inhibitor (Sandoz 58-035) to inhibit the formation of cholesteryl esters. ApoA1 at 10 μg/mL was used as cholesterol acceptors to illustrate this assay.
-
15.Loading the cells with 1× BODIPY-cholesterol:
-
a.Dilute 10× BODIPY-cholesterol to 1× BODIPY-cholesterol in DMEM/F-12, no phenol red supplemented with Sandoz (an acetyl-choline esterase inhibitor) at 2 μg/mL (see materials and equipment above).
-
b.Aspirate culture medium from cells.
-
c.Add 250 μL of 1× BODIPY-cholesterol in each experimental well.
-
d.Incubate for 1 h in an incubator at 37°C, 5% CO2.
-
a.
-
16.Equilibration with cAMP:
-
a.Prepare Equilibration medium (see materials and equipment above).
-
b.Remove 1× BODIPY-cholesterol from cells.
-
c.Rinse cells twice with DMEM/F-12, no phenol red supplemented with 2 μg/mL Sandoz 58-035.
-
d.Add 250 μL Equilibration medium per well.
-
e.Incubate overnight (∼16 h) in an incubator at 37°C, 5% CO2.
-
a.
-
17.Addition of ApoA1:
-
a.ApoA1 was diluted to 10 μg/mL in DMEM/F-12, no phenol red, supplemented with 2 μg/mL Sandoz.
-
b.The equilibration medium was aspirated and 250 μL of 10 μg/mL apoA1 were added to each well. ApoA1 was tested in in triplicate.
-
c.250 μL of DMEM/F-12, no phenol red, supplemented with 2 μg/mL Sandoz, were added in triplicate to measure the baseline cholesterol efflux (in the absence of apoA1).
-
d.Incubate for 4 h in an incubator at 37°C, 5% CO2.
-
a.
-
18.Measurement of BODIPY-cholesterol:
-
a.Transfer medium from each well to a 96-well plate.
-
b.Rinse cells once with PBS.
-
c.Add 250 μL of RIPA lysis buffer containing protease inhibitor to cells and incubate at 4°C on a shaker for 20 min.
-
d.Transfer 95 μL of culture medium to a 96-black fluorescence plate in duplicate.
-
e.Measure BODIPY-cholesterol in the medium using a plate reader (ex: 482 nm; em: 515 nm).
-
f.Transfer RIPA cell lysate from each well to a 96-well plate.
-
g.Centrifuge plate for 20 min at 2,000 × g.
-
h.Transfer 95 μL of culture medium to a 96-well black fluorescence plate in duplicate.
-
i.Measure BODIPY-cholesterol in the lysate using a plate reader (ex: 482 nm; em: 515 nm).
-
a.
Note: We recommend using a 96-well plate to store the medium and lysate to avoid pipetting variation between each sample.
CRITICAL: It is important to ensure the presence of Sandoz, the acetyl-choline esterase inhibitor, at all steps to avoid esterification of cholesterol and BODIPY-cholesterol. Up-regulation of ABCA1 with 0.3 mM cAMP is also essential to facilitate a measurable cholesterol efflux.
Expected outcomes
This protocol is intended for the characterization of the cholesterol accepting capacity of cholesterol acceptors such as apolipoproteins. This is achieved by loading cells with BODIPY-cholesterol, a fluorescence analog of cholesterol. The addition of cholesterol acceptors extracellularly leads to ABCA1-mediated cholesterol efflux, which can be quantified by measuring fluorescence using a plate reader. This assay has been successfully used to compare the cholesterol accepting capacity of different apoE isoforms, with the Jacksonville mutation at position 236 in apoE3 leading to enhanced BODIPY-cholesterol efflux.1
Here we used apoA1 at 10 μg/mL as cholesterol acceptor. The full experiment was conducted on 3 separate occasions (3 biological replicates). Treatment with apoA1 led to an additional 10% BODIPY-cholesterol efflux compared to baseline (Figure 3A). This increase was statistically significant (paired t-test; p<0.05) When comparing different cholesterol acceptors, BODIPY-cholesterol efflux is baseline corrected to subtract the efflux due to diffusion (see quantification and statistical analysis below and Figure 3B)(paired t-test; p<0.05).
Figure 3.
Cholesterol accepting capacity of apoA1
(A) ApoA1 added to the cells at 10 μg/mL leads to approximately 20% efflux of cholesterol from primary mouse astrocytes (paired t-test; p = 0.0128).
(B) ApoA1 at 10 μg/mL leads to approximately a 10% increase in cholesterol efflux compared to baseline (no acceptor added to cells) (paired t-test; p = 0.0133).
Quantification and statistical analysis
BODIPY-cholesterol fluorescence is measured in duplicate in the medium and in the cell lysate using a plate reader (em: 482 nm; ex: 515 nm; Tables 1 and 2).
Table 1.
Example of raw fluorescence data measured in the medium on a plate reader
| Fluorescence in the medium (FM) | ||||||
|---|---|---|---|---|---|---|
| ApoA1 technical repeat 1 | ApoA1 technical repeat 2 | ApoA1 technical repeat 3 | No acceptor technical repeat 1 | No acceptor technical repeat 2 | No acceptor technical repeat 3 | |
| A | 2162 | 2271 | 2127 | 1058 | 1049 | 1005 |
| B | 2207 | 2287 | 2035 | 1147 | 1006 | 984 |
| Average | 2184.5 | 2279 | 2081 | 1102.5 | 1027.5 | 994.5 |
Table 2.
Example of raw fluorescence data measured in the RIPA lysate on a plate reader
| Fluorescence in the RIPA lysate (FL) | ||||||
|---|---|---|---|---|---|---|
| ApoA1 technical repeat 1 | ApoA1 technical repeat 2 | ApoA1 technical repeat 3 | No acceptor technical repeat 1 | No acceptor technical repeat 2 | No acceptor technical repeat 3 | |
| A | 7828 | 7896 | 7258 | 8056 | 8667 | 8990 |
| B | 8034 | 7693 | 7005 | 8304 | 9045 | 8250 |
| Average | 7931 | 7794.5 | 7131.5 | 8180 | 8856 | 8620 |
Total BODIPY-cholesterol fluorescence intensity is defined by the sum of the fluorescence in the medium and the fluorescence remaining inside the cells. Fractional efflux of BODIPY-cholesterol is defined as the ratio of the fluorescence intensity in the medium to the total fluorescence intensity. BODIPY-cholesterol can efflux from cells by diffusion. Background BODIPY-cholesterol efflux, which occurs at baseline in the absence of acceptors, is subtracted from the fractional BODIPY-cholesterol efflux to yield a baseline corrected fractional BODIPY-cholesterol efflux for each acceptor (%BODIPY-cholesterol efflux, Equation 1).
Formula to calculate the baseline corrected BODIPY-cholesterol efflux:
| (Equation 1) |
Limitations
The success of this protocol is highly dependent on the quality of the BODIPY-cholesterol treatment, as well as dependent on the quality of the primary astrocyte culture. We do not recommend proceeding with the experiment if the fluorescence of 10× BODIPY-cholesterol is below 1,500,000 RFU. We also find that there are batch to batch differences in the quality of 10× BODIPY-cholesterol. Conducting biological replicates with the same stock of 10× BODIPY-cholesterol may improve reproducibility.
We do not recommend proceeding with the experiment if the cells plated for the experiment do not look healthy as we have incidentally notice that poor cultures lead to no difference in BODIPY-cholesterol efflux between baseline and the cholesterol acceptor treated cells.
Troubleshooting
Problem 1
Low fluorescence of 10× BODIPY cholesterol.
We do not recommend using 10× BODIPY-cholesterol preparations that have low fluorescence (below 1,500,000 RFU using the Synergy H1 microplate reader).
Potential solution
-
•
Get a different lot of BODIPY-cholesterol reagent: we find that different lots result in a difference in the final fluorescence of the 10× BODIPY-cholesterol stock solution.
-
•
Replace cholesterol.
-
•
Use glass containers and glass syringes to transfer reagents.
Problem 2
No BODIPY-cholesterol efflux.
The 10× BODIPY-cholesterol and 1×BODIPY-cholesterol display good fluorescence values but no BODIPY-cholesterol efflux can be measured.
Potential solution
-
•
Make sure that all steps include the presence of an acetylcholine esterase inhibitor (Sandoz 58-035 used here) to avoid esterification of cholesterol. Esterified cholesterol will accumulate in the cells and not efflux.
-
•
Replace Sandoz 58-035 and prepare fresh.
-
•
Increase the concentration of the cholesterol acceptor. This step may need to be optimized for each acceptor. Include a positive control (such as apoA1) to make sure that the assay is working.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Chia-Chen Liu (Liu.chiachen@mayo.edu).
Materials availability
This protocol did not generate new unique reagents.
Acknowledgments
The authors thank Dr. Na Zhao and Dr. Yuka Martens for technical support. This research was supported by the National Institute of Health (NIH) (grants U19AG069701 to C.-C.L. and G.B.; R01AG062110 to C.-C.L.), a Cure Alzheimer’s Fund (G.B.), and a grant from Brightfocus (C.-C.L.), and A-C.R. is supported by a BrightFocus Fellowship.
Author contributions
A.-C.R. performed the experiments; A-C.R. and C.-C.L. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Ana-Caroline Raulin, Email: raulin.ana-caroline@mayo.edu.
Chia-Chen Liu, Email: liu.chiachen@mayo.edu.
Guojun Bu, Email: guojun.bu@molecularneurodegeneration.org.
Data and code availability
No custom code is necessary to perform this protocol.
References
- 1.Liu C.C., Murray M.E., Li X., Zhao N., Wang N., Heckman M.G., Shue F., Martens Y., Li Y., Raulin A.C., et al. APOE3-Jacksonville (V236E) variant reduces self-aggregation and risk of dementia. Sci. Transl. Med. 2021;13:eabc9375. doi: 10.1126/scitranslmed.abc9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sankaranarayanan S., Kellner-Weibel G., de la Llera-Moya M., Phillips M.C., Asztalos B.F., Bittman R., Rothblat G.H. A sensitive assay for ABCA1-mediated cholesterol efflux using BODIPY-cholesterol. J. Lipid Res. 2011;52:2332–2340. doi: 10.1194/jlr.D018051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morikawa M., Fryer J.D., Sullivan P.M., Christopher E.A., Wahrle S.E., DeMattos R.B., O'Dell M.A., Fagan A.M., Lashuel H.A., Walz T., et al. Production and characterization of astrocyte-derived human apolipoprotein E isoforms from immortalized astrocytes and their interactions with amyloid-beta. Neurobiol. Dis. 2005;19:66–76. doi: 10.1016/j.nbd.2004.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No custom code is necessary to perform this protocol.





