Summary
Here, we present an optimized iDISCO+ protocol combining tissue clearing and light sheet microscopy to map the postnatal development of oxytocin and vasopressin neurons in mouse hypothalamus. We describe tissue preparation, immunostaining, clearing, and imaging. We then detail how to process the 3D cell dataset to analyze cell network using a point-based recording procedure that accurately maps neurons in the Allen brain atlas. This protocol can be applied to any neuronal population, in different brain regions and at different developmental stages.
For complete details on the use and execution of this protocol, please refer to Soumier et al. (2021).1
Subject areas: Bioinformatics, Developmental biology, Microscopy, Neuroscience
Graphical abstract
Highlights
-
•
An optimized iDISCO+ protocol to analyze cell networks in mouse brain
-
•
Intracardiac perfusion procedure and tissue preparation
-
•
Oxytocin and vasopressin cells staining, clearing, and imaging through brain development
-
•
Oxytocin and vasopressin cells 3D mapping in the Allen brain atlas
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we present an optimized iDISCO+ protocol combining tissue clearing and light sheet microscopy to map the postnatal development of oxytocin and vasopressin neurons in mouse hypothalamus. We describe tissue preparation, immunostaining, clearing, and imaging. We then detail how to process the 3D cell dataset to analyze cell network using a point-based recording procedure that accurately maps neurons in the Allen brain atlas. This protocol can be applied to any neuronal population, in different brain regions and at different developmental stages.
Before you begin
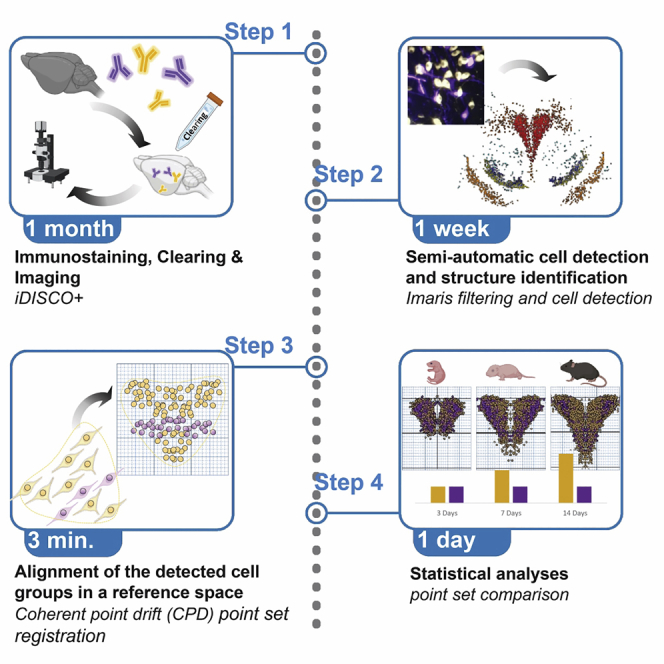
This protocol provides guidelines for the main following steps: 1) Tissue fixation, 2) Immunolabelling, 3) Tissue clearing, 4) Light sheet microscopy, 5) Cell counting, 6) Atlas data registration and mapping.
Before starting the protocol, prepare the fixative solution (4% paraformaldehyde solution) and use it to perform the intracardiac perfusion, then retrieve the clean and fixed brains. Prepare the PBSGT incubation solution in advance because it needs time to reach room temperature.
Institutional permissions
Institutional ethics approval is required prior to the start of the study in order to comply with European rules on animal experimentation.
Animals’ experimentation followed the guidelines of the European legislation (2010/63/EU). This study was approved by the local Ethical Committee (C2EA-UCBL55; project2017112317077317).
Preparation of fixing solution
Timing: 20 min
-
1.Paraformaldehyde solution.Note: Prepare a 4% paraformaldehyde solution (40 mL) up to one week before intracardiac perfusion for an adult mouse of 23–26 gms.
-
a.Use 10 mL 16% PFA bulb and dilute it further in 30 mL PBS-1× to obtain a total of 40 mL 4% PFA (pH = 7.4).
-
b.Store at 4°C for a maximum of 7 days.Note: Multiply the 40 mL aliquots per number of samples.
-
a.
Intracardiac perfusion fixation
Timing: 5 h (2 h for step 2)
To map protein expression in single cells, fixate biological samples via intracardiac perfusion of paraformaldehyde before immunofluorescence labeling and clearing. Intracardiac perfusion is critical for tissue antigenicity and molecular integrity. In addition, this procedure will determine the quality of the three-dimensional mapping of deep brain regions imaged with the light sheet microscope. The experimental setup for intracardiac perfusion is detailed in Figure 1.
-
2.Perfusion fixation.
-
a.Prior to surgery (30 min), inject buprenorphine (0.075 mg/kg) solution subcutaneously (s.c) followed by ketamine/xylasine mixture (300 mg/kg and 15 mg/kg respectively) intraperitoneally (i.p). Additional anesthetic may be delivered to maintain a surgical plane of anesthesia.
-
b.Prior to the first incision, monitor the effect of anesthesia by testing the dorsal foot reflex, breathing pattern, and response to tail pinch. Ensure that the animal is unresponsive (i.e., not moving) to any of the above tests.Then:
-
i.Place the animal on its back and pinch the legs with a needle on a polystyrene plate.
-
ii.Use a blunt scissor to make a 5 cm lateral incision in the abdomen under the rib cage.
-
iii.Cut the diaphragm along the rib cage to expose the lungs.
-
iv.Lift the sternum and clamp the tip of the sternum with a hemostat over the head.
-
v.Make a small incision at the level of the right atrium, and place the perfusion needle into the left ventricle.
-
i.
-
c.Use a peristaltic pump to inject 0.9% of NaCl solution for 1 min (flow rate of 10 mL/min) until lungs turn white.
-
d.Stop the pump and keep the needles inside the heart.
-
e.Switch the tube to a cold 4% PFA solution for 3 min (rate of 10 mL/min).Note: If the perfusion has been successful, fixation tremors should be observed within a few seconds. At the end of the perfusion, the lungs and liver shall be discolored. At the end of the perfusion-fixation, the animal should look rigid.Alternatives: If a peristaltic pump is unavailable, perfusion can be performed manually with a syringe (adapted to the size of the animal until P14). Use these volumes of PFA solution for the following developmental stages: P0 = 10 mL, P7 = 15 mL, P14 = 20 mL.
 CRITICAL: Collect and store paraformaldehyde for later disposal according to your institution’s policy.
CRITICAL: Collect and store paraformaldehyde for later disposal according to your institution’s policy. CRITICAL: Perfuse on the beating heart, to avoid cross-staining with blood vessels as shown in Figure 2.
CRITICAL: Perfuse on the beating heart, to avoid cross-staining with blood vessels as shown in Figure 2.
-
a.
-
3.
Remove the head with scissors and dissect the brain.
CRITICAL: Carefully slide the spatula along the ventral surface of the brain to prevent damage to your brain region of interest (in our study the ventral hypothalamus).
-
4.Post-fixation and storage.
-
a.Maintain the brain in fixative solution (4% PFA) overnight (approximately 15 h) at 4°C.
-
b.Wash the brain 3 times (60 min each wash) with phosphate buffer saline (PBS-1×) and shake at a rate of 30 rotation per minute (rpm). Shaking at room temperature (RT: between 18°C–24°C).
-
a.
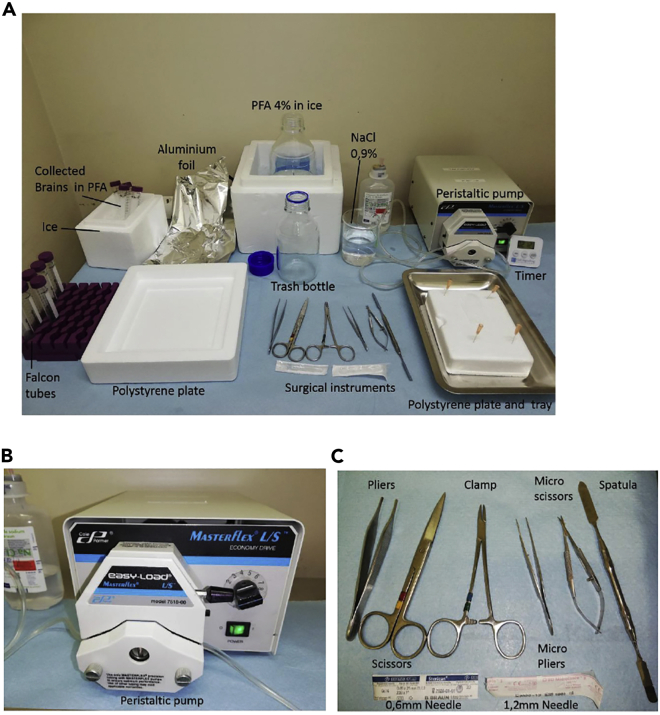
Figure 1.
Surgery equipment and tools for intracardiac perfusion
(A) Left: Falcon tubes filled with 4% PFA for collecting brains to be stored on ice, polystyrene plate for post-mortem dissection, aluminum foil for wrapping the animal’s body. Middle: 4% PFA in ice, garbage bottle to collect blood and PFA, surgical instruments, 0.9% NaCl (or PBS-1×). Right: Peristaltic pump connected with a plastic tube with needle at the end to inject the solution, polystyrene plate with needles to pin the mouse on the back and a tray to collect blood and PFA, timer.
(B) Peristaltic pump to inject 0.9% NaCl followed by 4% PFA solution, with a rate of 10 mL/min (40 mL for an adult animal, resulting in 4 min perfusion).
(C) Surgical instruments: Pliers to hold the skin of the abdomen, scissors to open the abdomen and the chest, clamp to stretch the skin, micro scissors and micro pliers for the heart. Spatula for the dissection. Needles to inject solutions: (0.6 mm diameter until P7 and 1.2 mm since P7).
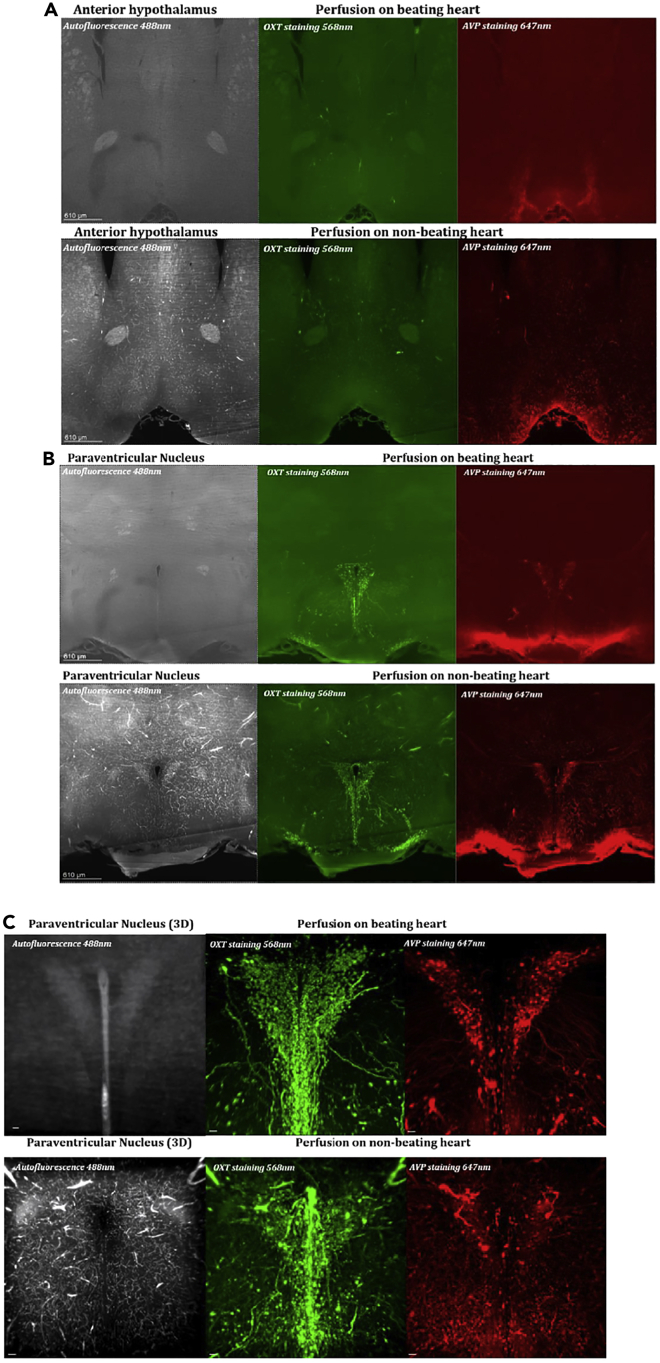
Figure 2.
Staining after perfusion on beating vs non-beating heart
(A) Coronal view of the anterior hypothalamus, scale 610 μm.
(B) Coronal view of the Paraventricular Nucleus, scale 610 μm.
(C) 3D view of the Paraventricular Nucleus, scale 50 μm. Autofluorescence at 488 nm without staining (left panel), oxytocin (OXT) staining at 568 nm (middle panel), vasopressin (AVP) staining at 647 nm (right panel). Perfusion on a deeply anaesthetized mouse with a beating heart (top panel of A, B and C), and a non-beating heart (bottom panel of A, B and C) 1 min (time to open the chest) after death. Staining and clearing was performed following the iDISCO+ protocol. 2D (A, B) and 3D (C) images showing differences in number of blood vessels and vesicles among conditions. Axons are visible in 3D view after the perfusion on beating heart while cells are mixed with blood vessels for the non-beating heart condition. Cell quantification and axon tracing are in the case of a beating heart.
Store brains in phosphate buffer saline with 0.02% sodium azide and store at 4°C. By replacing the storage solution every month, the samples can be conserved for months or even years.
Preparation of phosphate buffer saline gelatin triton (PBSGT) solution
Timing: 1 h
-
5.Procedure for PBSGT solution.
-
a.Dissolve 2 g of Gelatin in 900 mL of distilled water. Heat until complete dissolution (around 15 min). Let it cool down.
-
b.Add 100 mL of PBS-10×, 20 mL of 25% Triton-X-100 and 100 mg of thimerosal (pH 7). Store and protect from light.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Oxytocin (Host Rabbit); dilution 1/400 | Phoenix Pharmaceutical | G-051-01 |
| Anti-Vasopressin Neurophysin II (AVP-NPII) (Host Mouse); dilution 1/500 | Merck Millipore | Clone PS41-MABN845 |
| Anti-Oxytocin Neurophysin I (OXT-NPI) (Host Mouse); dilution 1/500 | Merck Millipore | Clone PS38-MABN844 |
| Anti-Vasopressin (Host Rabbit); dilution 1/400 | Phoenix Pharmaceutical | G-065-07 |
| Goat anti-Mouse IgG 647 nm; 1/800 | Abcam | Ab150115 |
| Goat anti-Rabbit IgG 568 nm; 1/800 | Abcam | Ab175471 |
| Chemicals, peptides, and recombinant proteins | ||
| Paraformaldehyde 16% | Thermoscientific | 28908 |
| Phosphate buffer saline 10-X | Roche | 11666789-001 |
| Triton X-100 | Sigma | 9002-93-1 |
| Gelatin | VWR | 9000-70-8 |
| Thimerosal | Sigma | 54-64-8 |
| Saponin | Sigma | 804715-2 |
| Heparin | VWR | EDQMH0200000 |
| Methanol | VWR | 67-56-1 |
| Hydrogen peroxide (H2O2) | Merck | 1085971000 |
| Dichloromethane | Sigma | 75-09-2 |
| Dibenzyl ether | Sigma | 103-50-4 |
| Sodium azide | Merck | S2002-100G |
| Normal goat serum (NGS) | Abcam | Ab7481 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J adult male, 8–9 weeks old | Charles River laboratories | JAX stock number: 000664 |
| Mouse: C57BL6/J male and female, Postnatal Day 0 (PD0) | Offspring of Pregnant females from Charles River lab | JAX stock number: 000664 |
| Mouse: C57BL6/J male and female, Postnatal Day 3 (PD3) | Offspring of Pregnant females from Charles River lab | JAX stock number: 000664 |
| Mouse: C57BL6/J male and female, Postnatal Day 7 (PD7) | Offspring of Pregnant females from Charles River lab | JAX stock number: 000664 |
| Mouse: C57BL6/J male and female, Postnatal Day 14 (PD14) | Offspring of Pregnant females from Charles River lab | JAX stock number: 000664 |
| Software and algorithms | ||
| Imaris | Oxford Instrument | Neuroscientists package- Version 9.5 Imaris Essentials - Microscopy Image Analysis Software - Imaris - Oxford Instruments (oxinst.com) |
| Matlab | MathWorks | R2018-a |
| Coherent Point Drift Algorithm | Myronenko and Song2 | Matlab implementation (Nov. 2020): https://sites.google.com/site/myronenko/research/cpd |
| Light-sheet Microscopy - Point Cloud registration in a Volumetric Atlas. | OSF project: https://doi.org/10.17605/OSF.IO/W5XRA | Matlab implementation: https://osf.io/w5xra/ |
| Oxytocin and Vasopressin Neurodevelopment. | OSF project: https://doi.org/10.17605/OSF.IO/39THQ | Matlab implementation: https://osf.io/39thq/https://doi.org/10.17605/OSF.IO/39THQ |
| Other | ||
| Peristaltic Pump | Cole Parmer | MasterFlex 7518-00 |
| Incubater- Incushaker | Benchmark | 060981 |
| Light Sheet Microscope | Milteniy Biotech | UltraMicroscope II |
| Falcon tube 15 mL | VWR | EPPE0030122.526 |
| Corning tube 15 mL | VWR | 21008-670 |
| Eppendorf tube 5 mL | VWR | 525-0794 |
| Pliers | Fisher Scientific | 16-100-112 |
| Micro pliers | Fisher Scientific | 16-100-127 |
| Scissors | Fisher Scientific | 12-000-148 |
| Micro scissors | Fisher Scientific | 08-953-1B |
| Clamp | Fisher Scientific | 16-100-115 |
| Spatula | Fisher Scientific | 14-373 |
| Needle 1.2 | Euraf | 23750 |
| Needle 0.6 | Euraf | 23758 |
Materials and equipment
PBSGT solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Gelatin | 0.2% (g/100 mL) | 2 g |
| PBS-1× | 9.8% | 100 mL |
| Triton 25% | 2% | 20 mL |
| Thimerosal | 9.8% (g/100 mL) | 100 mg |
| ddH2O | 88% | 900 mL |
| Total | N/A | 1.02L |
Store protected from the light, at RT. Store until one year.
CRITICAL: Thimerosal is a toxic powder, manipulate under a hood.
Primary antibodies solution: combination 1
| Reagent | Final concentration | Amount |
|---|---|---|
| Saponin | 0.1% (g/100 mL) | 10 mg |
| NGS | 1% | 100 μL |
| PBSGT | 99% | 9.9 mL |
| Anti OXT | 1/400 (0.25%) | 25 μL |
| Anti AVP-NPII | 1/500 (0.2%) | 20 μL |
| Total | N/A | 10 mL |
Store at room temperature and protect from light. Do not store the solution more than one week.
Primary antibodies solution: combination 2
| Reagent | Final concentration | Amount |
|---|---|---|
| Saponin | 0.1% (g/100 mL) | 10 mg |
| NGS | 1% | 100 μL |
| PBSGT | 99% | 9.9 mL |
| Anti AVP | 1/400 (0.25%) | 25 μL |
| Anti OXT-NPI | 1/500 (0.2%) | 20 μL |
| Total | N/A | 10 mL |
Store at room temperature and protect from light. Do not store the solution more than one week.
Alternatives: To get a more accurate concentration of antibodies in both combinations, mix the saponin with the NGS and PBSGT, then remove the volume calculated for the antibodies (45 μL total) in order to obtain a perfect total volume of 10 mL. See also below the Step-by-Step immunostaining section.
Step-by-step method details
Samples pretreatment
Timing: 3 days
The removal of lipids is important for effective clearing of tissues and to allow the penetration of external chemicals and molecules. It is mainly achieved by the application of detergents. It is a solvent-based dehydration step.
-
1.Dehydration and decolorization.
-
a.Incubation in increasing series of methanol/water washes: 20%, 40%, 60%, 80%, 100%, 100%, 1 h at each stage, without shaking, at RT and protected from light.
-
b.Incubation in DiChloroMethane (DCM)/Methanol 2:1, overnight at RT with shaking.
-
a.
-
2.Bleaching.
-
a.Do 2× 1 h-rinsing in methanol 100% at RT without shaking and then store in the fridge at 4°C.
-
b.Prepare 5% H2O2 in methanol, and place at 4°C.
-
c.Incubate the brain in the H2O2/methanol solution overnight (about 15 h), at 4°C.
-
a.
Note: H2O2 is utilized in this context for clearing the brain and wash blood vessels. See Figure 3.
Figure 3.
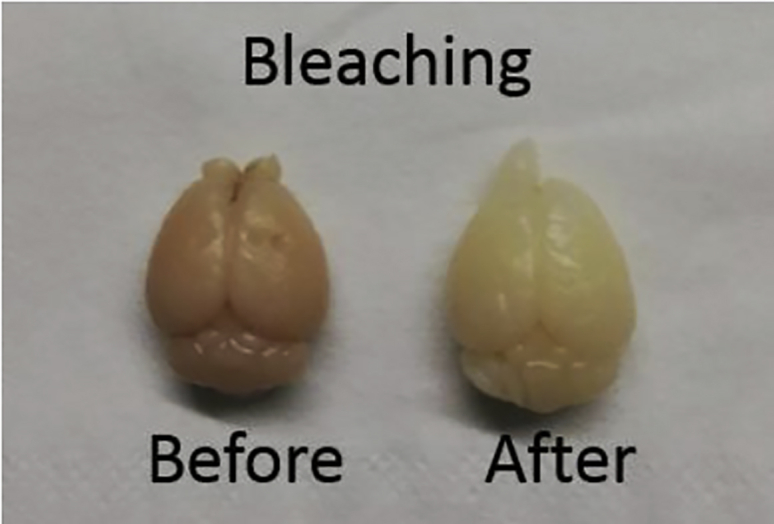
Mouse brain before and after bleaching
-
3.Rehydration.
-
a.Rehydrate your sample by washing in decreasing amounts of methanol: 80%, 60%, 40%, 20% and PBS-1×. One hour for each wash at room temperature.
-
a.
Alternatives: You can skip this phase (samples pretreatment) if the primary antibody is not suited for methanol treatment (for further details see antibodies validation in the troubleshooting section).
Note: 15 mL Falcon tube can be used for all steps 1, 2 and 3.
Pause point: After rehydration, samples can be stored in a PBS-0.01 M solution (PBS-1×) at 4°C for up to one month. Beyond this time store the sample in PBS 1×- 0.02%. sodium azide solution.
Tissue permeabilization and blocking
Timing: 5 days
The permeabilization step increases porosity and helps in the penetration process of antibodies.
Please note that the blocking step prevent non-specific binding of primary antibodies to the tissue.
-
4.Permeabilization.
-
a.Incubate your sample in a PBSGT + Glycine 0,3 M: 1.13 g of glycine in 50 mL of PBSGT solution, during two days (48 h) (1 day (24 h) for ≤ P7 brains) at 37°C under shaking (30 rpm).
-
a.
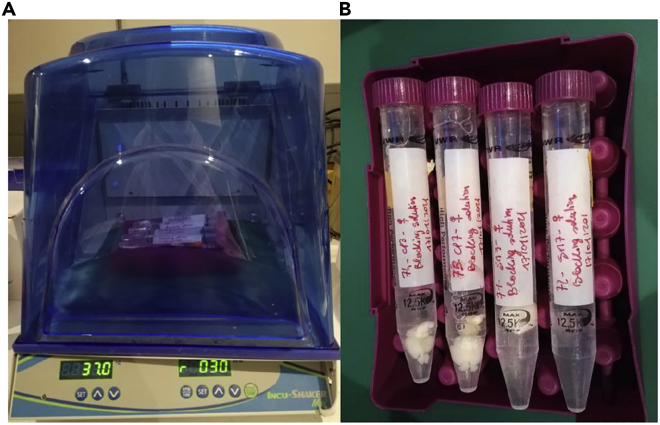
Note: The apparatus used for this step is shown in Figure 4.
Figure 4.
Incushaker and Falcon tubes
(A) Incushaker to incubate sample on shaking at 37°C.
(B) Falcon tubes, (15 mL) elongated in the incushaker.
-
5.Blocking.
-
a.Incubate the sample in PBSGT + 5% NGS (500μL NGS qsp 10 mL PBSGT) during three days (72 h) (1,5 days (36 h) for ≤ P7 brains) at 37°C in a shaking mode (30 rpm).
-
a.
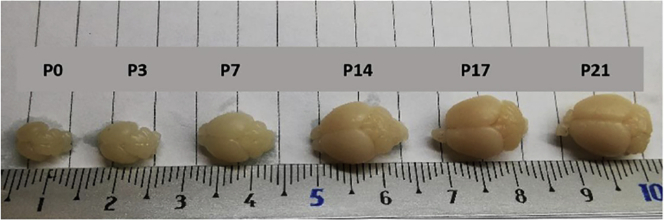
Note: Due to differences in brain size during early postnatal development, reduce the incubation time for blocking and permeabilization by two. See Figure 5.
Figure 5.
Mouse brains at several postnatal ages
The brain at P14, is similar in size to the adult brain and can be handled with the similar procedure (duration, concentration). Ruler from 1 to 10 cm.
Optional: Additionally, wash twice, 60 min each time, in PBSGT, 0.1% saponin with 10 mg/mL of heparin.
Note: For these steps (4, 5), you can use falcon tube 15 mL, and work with a 15 mL volume. See Figure 3.
Immunostaining
Timing: 3.5 weeks
This section describes the labeling process. We have used the oxytocin and vasopressin primary antibodies to target axonal fibers and cell bodies. Then we use secondary antibodies to bind the primary ones. Each secondary antibody contained a fluorochrome in 568 nm or 647 nm. At the end of immunostaining, cell bodies containing oxytocin and vasopressin appear fluorescent when imaged with the light sheet microscope.
-
6.Preparation of the antibody solution.
-
a.Prepare a mixture of PBSGT, 1% NGS and 0.1% saponin (100μL of NGS + 10 mg saponin qsp 10 mL PBSGT).
-
a.
-
7.Primary antibody incubation.
-
a.In the previous solution, incubate the antibodies at the required concentration. We used two different combined antibodies: (see troubleshooting section antibodies double staining):
-
i.anti OXT (Host Rabbit) 1/400 with anti AVP-NPII (Host Mouse) 1/500.
-
ii.anti AVP (Host Rabbit) 1/400 with anti OXT-NPI (Host Mouse) 1/500.Incubate the brains on shaking (30rpm) at 37°C: 3 weeks for adults, 2 weeks for P14 and P7, and only one week for P3 and P0.Note: To save antibodies, place the brains in the incushaker in a 5 mL Eppendorf tube tilted on the side (2 mL Eppendorf tube until P7, fill with 1.5 mL solution) as shown in Figure 6. We obtained 5μL of anti OXT/anti AVP + 4μL of anti OXT-NPI/anti AVP-NPII, qsp 2 mL with antibody solution.Note: Correct antibodies concentration can be assessed by preliminary tests using different concentration. See troubleshooting section antibodies concentration.
-
i.
-
b.In a 15 mL falcon tubes rinse 6 × 1H in PBS-1×, at RT on shaking (30 rpm).
-
a.
-
8.Secondary antibody incubation.
-
a.Prepare a mixture of PBSGT, 1% NGS and 0.1% saponin (100μL of NGS + 10 mg saponin qsp 10 mL PBSGT).
-
b.Prepare the final solution, with the required concentration of antibodies. In our study we used the Goat Anti-Rabbit 568 nm and the Goat anti-Mouse 647 nm at 1/800, for a final volume of 2 mL.
-
i.2.5 μL of Goat Anti-Rabbit 568 nm and 2.5 μL of Goat anti-Mouse 647 nm, qsp 2 mL with antibody solution.Incubate at 37°C, on shaking (30 rpm) during 2 days (48 h).
 CRITICAL: After the staining step as well as during the clearing and storage phase, cover the sample with a foil. This will ensure that the fluorescence labelling is protected from light.
CRITICAL: After the staining step as well as during the clearing and storage phase, cover the sample with a foil. This will ensure that the fluorescence labelling is protected from light.
-
i.
-
c.In 15 mL falcon tubes rinse 6 × 1H in PBS-1×, at RT on shaking (30 rpm).
-
a.
Figure 6.

Antibodies incubation
Brains incubated in 2 mL antibodies solution, in 5 mL Eppendorf tube.
Clearing
Timing: 2 days
This section describes the procedure to obtain transparent sample before imaging with the light sheet. This is obtained by matching the refractive index of the sample with the refractive index of the imaging solution (See also the Renier et al.3).
-
9.Dehydration.
-
a.Incubation in increasing series of methanol in water washes: 20%, 40%, 60%, 80%, 100%, 100% (or overnight in the last washes), 1 h each step, without shaking, at RT and protected from light.
-
a.
-
10.Delipidation.Note: For the delipidation and refractive index matching, use 15 mL corning falcons.
-
a.Incubation in DCM/Methanol 2:1, during 3H at RT on shaking.
-
b.Wash two times (15 min each time) in DCM, on shaking.
-
a.
-
11.Refractive Index Matching.
-
a.Incubate in DiBenzyl Ether solution and wait until samples become transparent. See Figure 7.
-
a.
Figure 7.
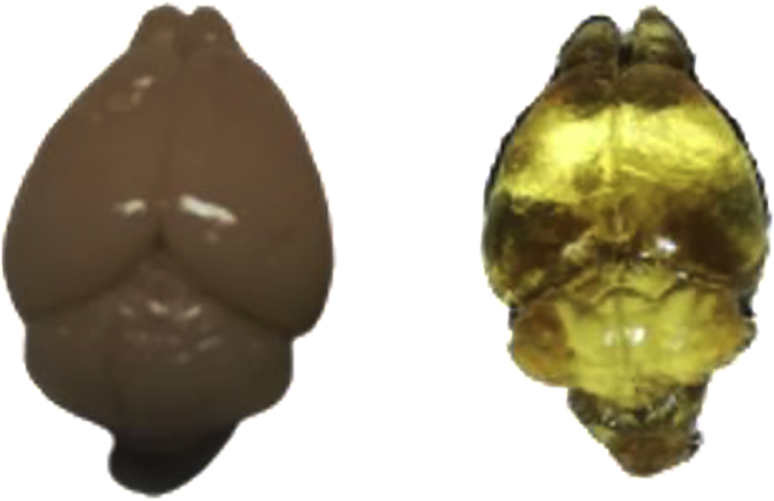
Brains before (left) and after (right) clearing
Imaging
Timing: 3 h
This section describes the procedure for imaging whole brain cell fluorescence with the light sheet microscope. Oxytocin and vasopressin cell bodies were imaged, the data was stored as tiff files and then 3D reconstruction and analysis were performed.
-
12.Imaging.
-
a.Set the laser power (excitation 568 nm for oxytocin and 647 nm for vasopressin) around 20% for the 568 nm and 30% for the 647 nm, with a 150 ms time of exposure.Note: Set source lasers at 25% powered at 16.6 mW. The light sheet numerical aperture is 0.156 (thickness 5 μm), with a width of 70%. Our Ultramicroscope II has 2× objective lenses, with a zoom from 0.63 to 6.3 (1:10). The total magnification is 1.26×-12.6×.
-
b.For the adult brain, scanning hypothalamic regions in coronal plane (see Figure 8) (from the anterior commissure to the median eminence, see Figure 9A); we used a step size of 2 μm and an optical zoom of 1.6.Note: Optical zoom 1.6 correspond to 1.89 μm resolution in x and y. To obtain a cubic voxel, the step size must be set with the same value than the microns size in x and y.Note: Use step size 1,51 μm and optical zoom 2 for P14 and step size 1,21 μm with 2.5 optical zoom for P0, P3 and P7.
-
c.For cell quantification, data acquired in the regions of interest, resulted in voxel size of 0.479 μm3, 0.604 μm3, or 0.75 μm3.
-
a.
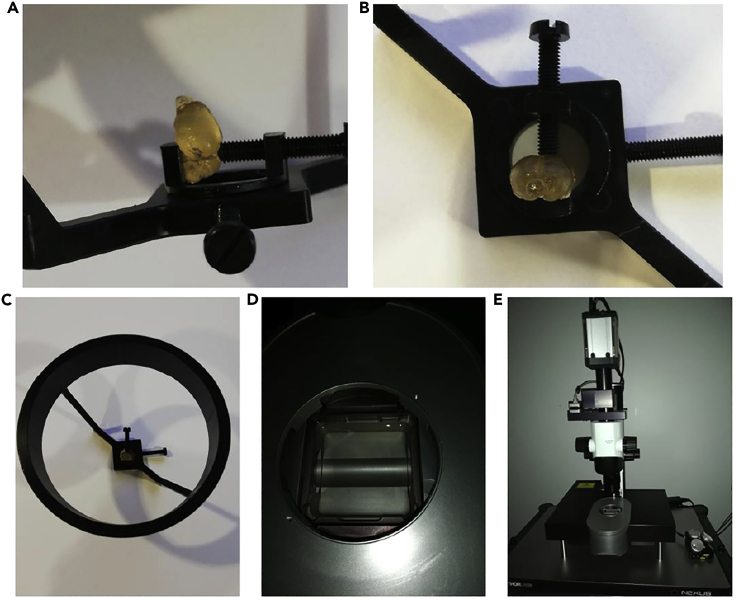
Figure 8.
Light-sheet sample holder
(A–C) For the coronal acquisition, place the brain with the anterior part facing upwards while the dorsal part is in contact with the screw.
(D) The cuve is filled with DBE.
(E) Dip the sample holder into the cuve of the microscope.
Figure 9.
Cell detection and clustering with Imaris
Scale bar 200 μm.
(A) 3D representation of the OXT and AVP staining, in coronal view.
(B) OXT cell detection and region clustering.
(C) AVP cell detection and regions clustering.
Cell counting
Timing: 3 days
In this section, we report how to analyze oxytocin and vasopressin cell network. From light sheet microscope generated tiff files create a 3D file, then perform the quantification as described below. Excel files containing data such as cell size or fluorescence intensity can be obtained for all cells.
-
13.
Conversion to 3D files: With the Imaris converter, you can export 2D tiff images (from the light sheet microscope) to a 3D ims file.
-
14.We performed automatic cell detection with Imaris software as well as through manual checking. For both global and region-by-region cell detection (as illustrated in Figure 10 for ParaVentricular Nucleus) use the Imaris spot detection module.
-
a.Set the cells diameter to 10 microns and start the detection for each channel, OXT and AVP, separately.
-
b.Carefully check each spot to ensure that there is not any missing cells or false positives (i.e., vesicles or axons).
-
c.To process hypothalamic cells, we first performed a global count using the cell counting tool.We then carried out a manual identification of cells in the hypothalamic nuclei.We identified six OXT and AVP cell clusters in the Paraventricular (PVN), Supraoptic (SON), Tuberal (TU) and Medial Preoptic (MPO/MPN) Nuclei as well as in the Bed Nuclei of Stria Terminalis (BNST), and Medial Amygdala (MeA) and in two OXT-specific regions, the Antero Lateral Preoptic (ALPO) and Periventricular (PvPO) areas. Cell coordinates were than mapped into Allen Brain Atlas with a resolution of 1.89 μm3.
-
d.For each region, cell counts were performed using the following size parameters: 0.479 μm3, 0.604 μm3, or 0.75 μm3 voxel. Cells not included in the region of interest were discarded (Figure 10C below). Density analysis within the PVN was conducted by extracting position and fluorescence intensity of each cell.
-
a.
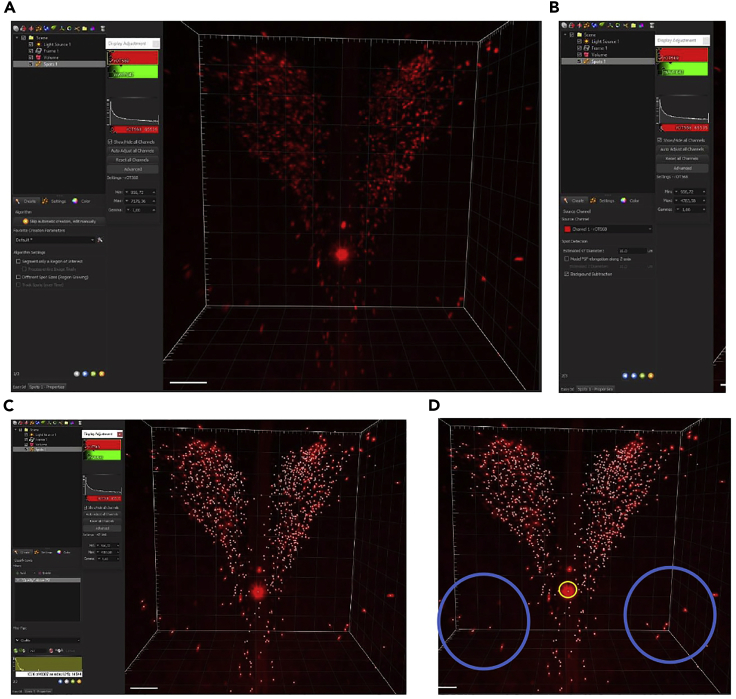
Figure 10.
Oxytocin cells in the paraventricular nucleus detected with the Imaris software
Scale 50 μm.
(A) Unbox the spot size option (to set the cell size in step 2), and unbox the region of interest (or keep this option and design your ROI).
(B) Choose the correct channel (here OXT 568), set the diameter size and check the box background subtraction.
(C) Automatic detection obtained with the Imaris tool counting.
(D) Example of false detection: in blue outside the region (i.e., Medial Preoptic Area), in yellow false positive.
Expected outcomes
PVN and SON identification
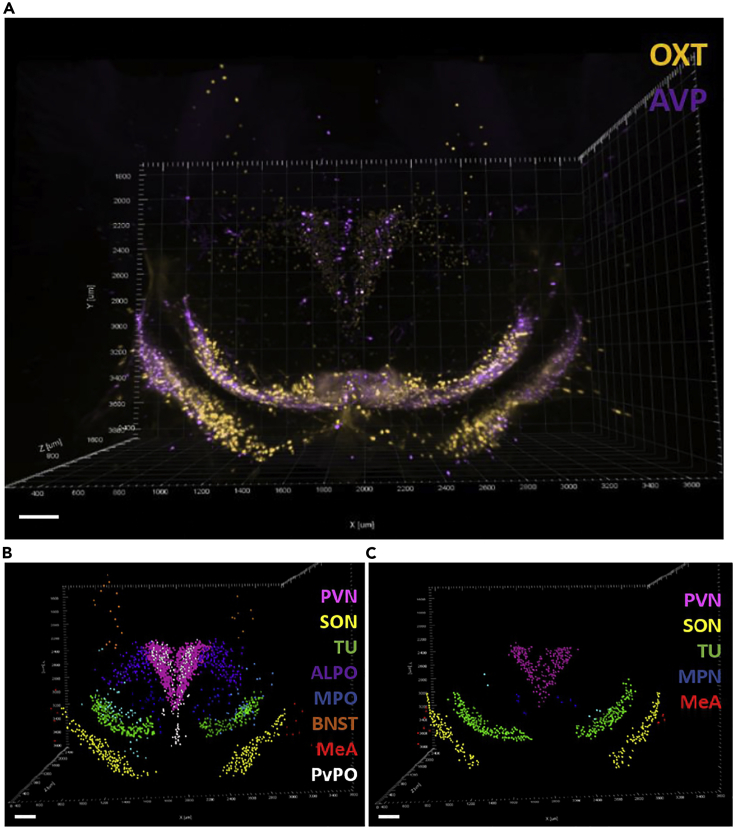
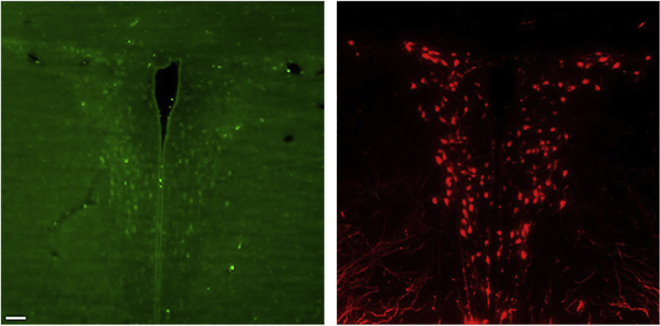
To use a common frame of reference, the spatial position of individual cells in the para-ventricular (PVN) and supra-optic (SON) nuclei of the adult animal was processed together. The two regions can be easily identified because of the excellent signal-to-noise ratio of the labeled OXT and AVP cells and because they form an easily recognizable spatial triad in the Allen brain atlas: the PVN is surrounded in the center by two elongated shapes forming the left and right SON.
Specifically, on the medio-lateral axis, the PVN cells are located around the boundary of the third ventricle, whereas on a coronal view, they form the known butterfly shape, as shown in Figure 2C. On the ventral axis, the SON is identified by its elongate shape above the optic tract, and is symmetrically located on the left and right sides. Staining of hypothalamic cells in the PVN and SON is shown in Figure 9.
An example of the 3D pattern of OXT and AVP cells identified in one adult animal is shown in Figures 9B and 9C.
Quantification and statistical analysis
Atlas registration
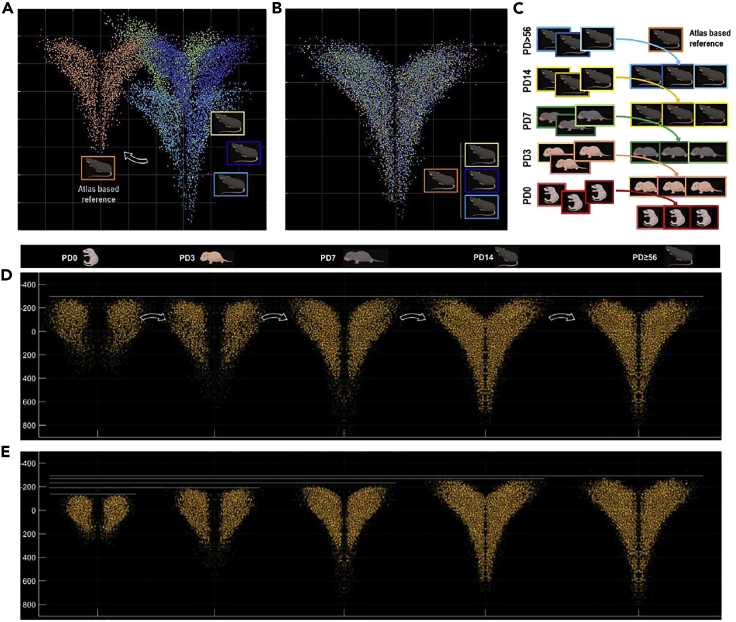
We estimated spatial transformations to map the cell data into a common spatial reference frame in the Allen brain mouse atlas (https://atlas.brain-map.org/). This step involves aligning the cell coordinates (Figure 11A) into a spatial matrix of annotations to extract the volume in the theoretical location. The PVN and SON were the reference areas for cells mapping in the atlas (Figure 11B).
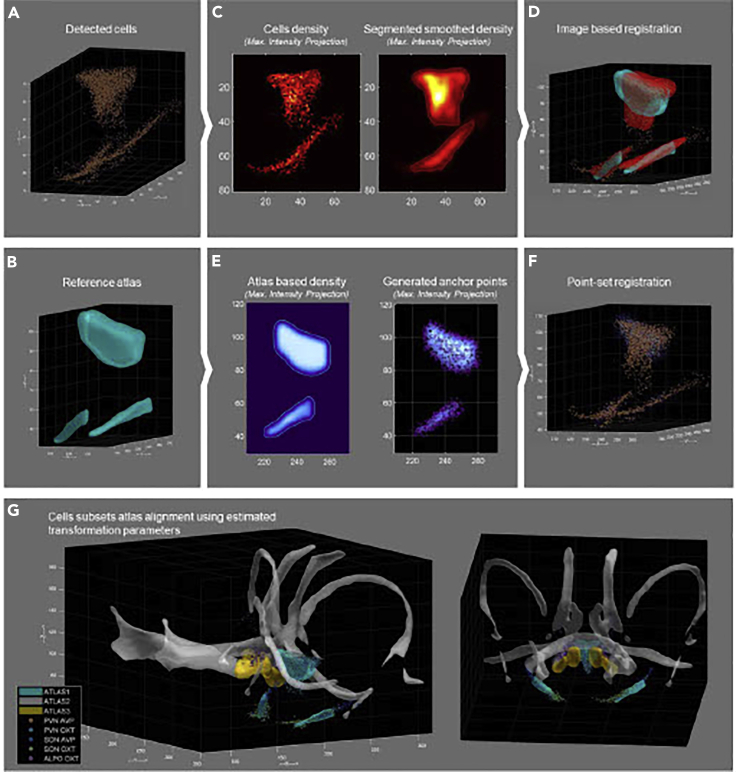
Figure 11.
Modeling and mapping oxytocin and vasopressin cell networks
(A) PVN and SON cell coordinates of an adult animal.
(B) PVN and SON volumes extracted from the Allen Brain Mouse Atlas (25 μm).
(C) Maximum intensity projection representation of the cellular density estimated on a grid of 25 μm (Left), smoothed using a gaussian kernel size of σ = 50 μm, scaled between 0 and 1, and segmented with a threshold of 0.1 to estimates the volume of PVN and SON in the recorded dataset (Right).
(D) Illustration of the atlas volumes (blue) and the recording volumes (red) aligned using image base registration.
(E) Maximum intensity projection of the estimated cellular density using a smoothed version (gaussian filter of σ = 50 μm) of the Atlas (Left) and anchor points generated from this density (Right).
(F) Illustration of the original cell coordinates (orange) aligned on the atlas reference space using anchor points (blue) with point-set registration.
(G) Illustration of other recorded cell groups and other Allen Brain Atlas derived volumes aligned in the same reference space using the previously estimated spatial transformation parameters. Such alignments can then be used for a precise identification of the recorded and identified cell groups.
Because the cell data are in two different formats, two strategies were considered for performing the alignment. The first is to convert the cell coordinates to a raster space with the same resolution as the regional resolution of the atlas and to estimate the occupancy volume of this cell network (Figure 11C). Then, the classical 3D image registration procedure is used to perform the alignment between the atlas and the data (Figure 11D). The second technique is to estimate a theoretical cell density probability from the reference atlas and generate a set of anchor points from it with a random number generator (Figure 11E). This set of anchor points can then be used to align the coordinates of the cells in the reference space using a point-based registration method.
In this study, we chose the second method because it retains the original spatial resolution of the recorded structure (the atlas being of lower resolution), is robust in estimating the shape/contour of the atlas regions, and is much less expensive in terms of computational load (alignments can be performed in seconds on a standard workstation).
Thus, the alignment procedure in the atlas was performed as follows:
-
1.
PVN and SON cell coordinates of adult animals were collected with high quality recordings (acquisition at 1.89 μm3).
-
2.
A hypothetical cell density was estimated by extracting SON and PVN volumes from the Allen mouse brain atlas with 25 μm resolution smoothed with a Gaussian filter of size σ = 50 μm.
-
3.
Considering N, the number of cell coordinates to align, N anchor points were generated from the estimated densities.
-
4.
The translation T, rotation R, and scale S parameters enabling the alignment of the data in the atlas were estimated by applying a rigid point-based registration with the CPD (Coherent Point Drift) algorithm.
-
5.
The estimated parameters (T, R and S) were used for alignment and visualization of other cell groups identified in the atlas during the same recording (Figure 11G).
-
6.Since the PVN and SON definitions are of higher resolution in our recordings as compared to those in the atlas (5 μm thick virtual slices every two microns vs. 25 μm every 100 μm for the Allen atlas), alignments of the remaining recordings in the reference space are performed using the same point-based registration method, but with the original and recorded cell coordinate data as anchor points.
- A Matlab demo showing our methods is available at: https://osf.io/w5xra/.
Intra-group and inter-group point set-registration of PVN cells
The final analysis focuses on the quantification of cells in the PVN region and their spatial changes during the developmental stages investigated in our study. To achieve this goal, we aligned the recorded cells from several animals. First, we merged the cells of individuals (N = 4) of the same age with different spatial coordinates but with a priori similar layouts and densities. Second, we aligned data from different age groups (N = 5) for which we expected slight differences in cell density and spatial pattern. Point-based registration methods, such as CPD, align a cloud of points (i.e., cells) while minimizing the distance between them. These methods are therefore well suited to this type of analysis as they do not require a perfect correspondence of cell coordinates and can align incomplete data.
The procedure for identifying changes during PVN cells development was designed as follows:
-
7.
Using the CPD method, we aligned the recorded adult PVN data set onto the spatial coordinates of previously identified adult PVN cells mapped in a reference space based on Allen brain atlas (Figures 12A and 12B).
-
8.
The CPD method aligns all recorded cells of an experimental group to the coordinates of the nearest experimental group. Thus, 14-day data are aligned with adult data, 7-day data are aligned with 14-day data, and so forth (Figure 12C).
-
9.
With all the datasets aligned and scaled in the same space, it is then possible to use signal processing methods (e.g., based on K-nearest neighbor methods) and statistical analyses to highlight experimental effects while controlling structure growth during development.
-
10.
From the scaling parameters estimated by the method, it is possible to recover the original cells sizes and to quantify the growth of the region during development.
Figure 12.
Modeling the distribution of oxytocin cells in the paraventricular region as a function of age
(A) Coordinates of PVN cells of an adult animal in a reference space and cells PVN coordinates of three adult animals before alignment.
(B) PVN cell coordinates of all adult animals aligned in the reference space after rigid CPD registration.
(C) Illustration of the procedure used to align many animals with different ages.
(D) Illustration of the aligned PVN cells for the different experimental group. Although the overall variation in structure size is corrected, the evolution of the PVN shape over the course of development is distinctively visible. Scale bar in Y axis: microns.
(E) PVN cells for the different experimental groups aligned but with the original size preserved using only estimated translation and rotation parameters after CPD registration. The global expansion of the structure is then visible. Scale bar in Y axis: microns.
A demo Matlab script illustrating the methods is available here: https://osf.io/39thq/.
Limitations
Due to the use of several reagents, if the brain tissue is weak and damaged after dissection, it may not be resistant to the rough treatment of the different solvents. Similarly, a methanol-resistant antibody is essential with this method.
For example, the Thermofisher TOPRO-3 iodide nuclear dye (Invitrogen T3605) stains very well, whereas the neuronal marker NeuN (Merck MAB377) is not suitable for the iDISCO+ protocol (Figure 13).
Figure 13.
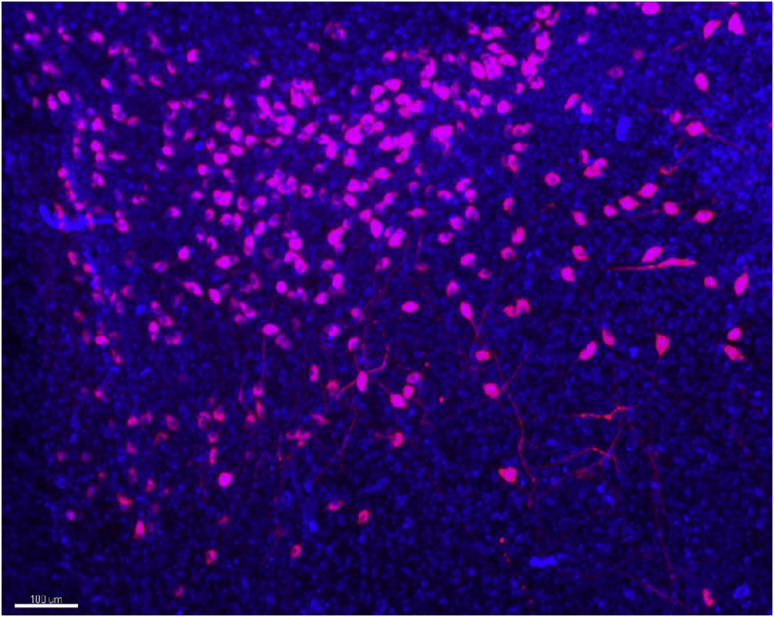
TOPRO-3 and Oxytocin in the ParaVentricular Nucleus (PVN)
TOPRO-3, nuclear dye, in blue and Oxytocin cells in red. Scale bar 100 microns.
We also tried several antibodies for oxytocin and vasopressin labeling from different suppliers and species:
Validated antibodies for iDISCO+ clearing:
Oxytocin (Mouse), Merck Millipore MABN844.
Oxytocin (Rabbit), Immunostar 20068.
Oxytocin (Rabbit), Phoenix pharma G-051-01.
Vasopressin (Mouse), Merck MABN845.
Vasopressin (Rabbit), Immunostar 20069.
Vasopressin (Rabbit), Phoenix pharma G-065-07.
Low signal antibody:
Oxytocin (Rabbit), Merck AB911.
Troubleshooting
Problem 1
Antibodies validation.
As described in the original protocol, the antibodies are not all resistant to methanol pre-treatment. Considering the time required by the 3D protocol, the use of the wrong antibodies with the iDISCO method may represent a considerable loss of time (Figure 14).
Figure 14.
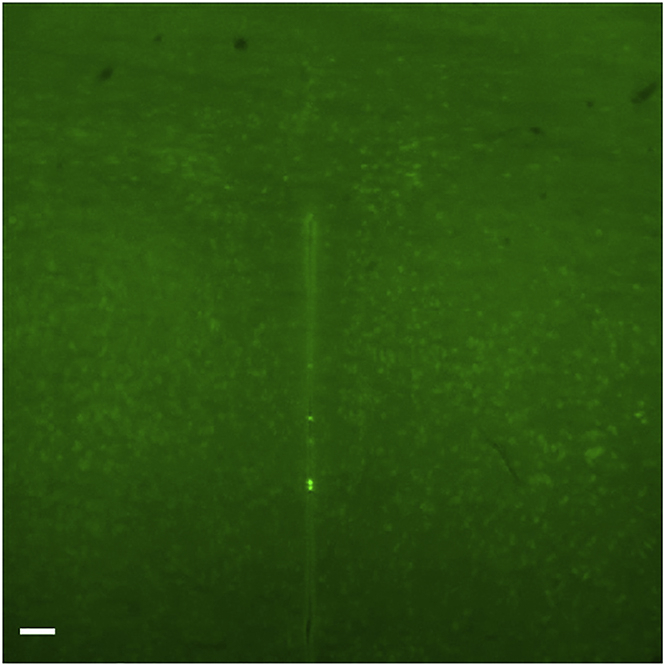
PVN without staining
This antibody (NeuN Merck MAB377) is not compatible with the methanol treatment. Scale 50 μm.
This should be performed before the samples pretreatment in the step-by-step section.
Potential solution
To make sure that your antibody works with the methanol treatment, you can first test its effect on brain slices.
-
•
Perform a 3H incubation of slices in methanol 100%.
-
•
Perform your classical 2D immunohistochemistry with the chosen antibody.
-
•
Image your slice and check if you obtain a staining.
-
•
If you have a staining: your antibody will resist to methanol and then you can perform your 3D analysis.
-
•
If you do not observe any staining, try the iDISCO+ protocol by skipping the sample pretreament setp, or try another antibody.
Problem 2
Antibodies concentration.
Whole brain staining requires a high concentration of primary and secondary antibodies.
Poor staining due to low antibody concentration compromises the analysis of cells (see Figure 15). You can test different concentrations from 1/100 to 1/1000 for 3D whole brain staining to find the most appropriate one. The concentration used in our protocol is indicated in the immunostaining part in the step-by-step section.
Figure 15.
Effect of antibodies concentration on staining
PVN with low staining (green) vs high staining (red). Scale bar 50 μm.
Potential solution
You can increase the concentration of the antibodies, for example by using the primary at about 1:500, but you can also adjust to 1:100 or 1:200 if necessary.
Problem 3
Antibodies double staining.
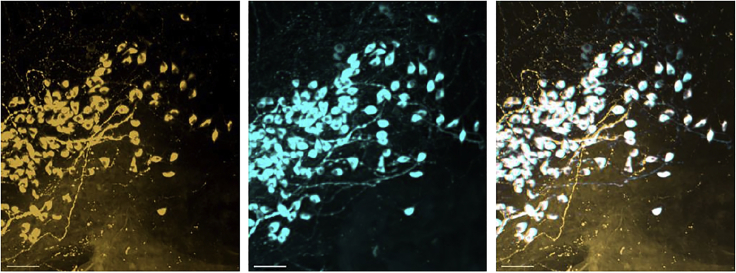
To perform double staining, we used antibodies from two different species (i.e., rabbit and mouse). The mouse antibodies are OXT-NPI and AVP-NPII and the rabbit antibodies AVP and OXT. We used two different combinations, AVP (rabbit) with OXT-NPI (mouse) and OXT (rabbit) with AVP-NPII (mouse) in order to be certain that the OXT (rabbit) antibody targets the same cells as the OXT-NPI (mouse) and this also holds for AVP and AVP-NPII. More details about staining in the immunostaining part of the step-by-step section (see Figure 16).
Figure 16.
Staining in the SON
OXT (rabbit) in gold, OXT-NPI in cyan, and merge in white. Scale 50 μm.
Potential solution
Before starting our cell counting, we did a pilot on young brains as control to make sure that the two OXT and the two AVP antibodies are targeting the same cell population. We performed a co-staining for OXT mouse and rabbit antibodies (i.e., OXT and OXT NPI) and also for AVP.
Resource availability
Lead contact
Further information and requests for resources and reagents should be sent to the lead contact: Angela Sirigu; sirigu@isc.cnrs.fr.
Materials availability
This study did not generate new reagents or materials.
Acknowledgments
This research is funded by Centre National de la Recherche Scientifique, le Vinatier Hospital and Labex Cortex (ANR-11–LABEX-0042) to A.S. We thank the genotyping facility (SFR Biosciences-ENS Lyon) and the animal facility (iXplora-CarMen Laboratory U1060) for allowing access to the animal facility.
Author contributions
M.H. performed experiments and contributed to analyses and writing. G.L. contributed to editing codes, performing analyses, and writing. A. Soumier contributed to review. C.D. contributed to review and funding. A. Sirigu contributed to supervision, writing, review, and funding.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Marie Habart, Email: marie.habart@isc.cnrs.fr.
Guillaume Lio, Email: guillaume.lio@isc.cnrs.fr.
Angela Sirigu, Email: sirigu@isc.cnrs.fr.
Data and code availability
All original Matlab codes for processing immunolabelled cells detected with Imaris software have been posted on the Open Science Framework website. The accession number for the data reported in this paper is: https://doi.org/10.17605/OSF.IO/39THQ. These data are part of the Oxytocin and Vasopressin Neurodevelopment project, and are publicly available as of the date of publication. A Matlab demo showing our methods is available at: https://osf.io/w5xra/. DOIs are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Soumier A., Habart M., Lio G., Demily C., Sirigu A. Differential fate between oxytocin and vasopressin cells in the developing mouse brain. iScience. 2021;25:103655. doi: 10.1016/j.isci.2021.103655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Myronenko A., Song X. Point-set registration: coherent point Drift. IEEE Trans. Pattern Anal. Mach. Intell. 2010;32:2262–2275. doi: 10.1109/TPAMI.2010.46. [DOI] [PubMed] [Google Scholar]
- 3.Renier N., Wu Z., Simon D.J., Yang J., Ariel P., Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All original Matlab codes for processing immunolabelled cells detected with Imaris software have been posted on the Open Science Framework website. The accession number for the data reported in this paper is: https://doi.org/10.17605/OSF.IO/39THQ. These data are part of the Oxytocin and Vasopressin Neurodevelopment project, and are publicly available as of the date of publication. A Matlab demo showing our methods is available at: https://osf.io/w5xra/. DOIs are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.