Summary
The RNA-binding protein Pcbp2 is widely expressed in the innate and adaptive immune systems and is essential for mouse development. To determine whether Pcbp2 is required for CD4+ T cell development and function, we derived mice with conditional Pcbp2 deletion in CD4+ T cells and assessed their overall phenotype and proliferative responses to activating stimuli. We found that Pcbp2 is essential for T conventional cell (Tconv) proliferation, working through regulation of co-stimulatory signaling. Pcbp2 deficiency in the CD4+ lineage did not impact Treg abundance in vivo or function in vitro. In addition, our data demonstrate a clear association between Pcbp2 control of Runx1 exon 6 splicing in CD4+ T cells and a specific role for Pcbp2 in the maintenance of peripheral CD4+ lymphocyte population size. Last, we show that Pcbp2 function is required for optimal in vivo Tconv cell activation in a T cell adoptive transfer colitis model system.
Subject areas: Molecular biology, Immunology, Cell biology
Graphical abstract

Highlights
-
•
Pcbp2 controls Runx1 exon 6 inclusion in murine CD4+ thymocytes and splenocytes
-
•
Pcbp2 is required for maintaining peripheral CD4+ T lymphocyte population size
-
•
Pcbp2 is required for CD4+ T cell proliferation in vitro
-
•
Pcbp2 deletion in CD4+ Tconv cells attenuates adoptive transfer colitis
Molecular biology; Immunology; Cell biology
Introduction
T lymphocytes (T cells) are critical components of the adaptive immune system and are essential for responses to foreign organisms. T cell development encompasses a highly complex process that starts from thymocyte precursors and involves numerous regulating factors within the thymus and peripheral immune system.1 T cell development and activation have long been understood to depend upon fine-tuned transcriptional events. More recent work has uncovered a critical role for post-transcriptional regulatory mechanisms, controlled by RNA-binding proteins (RBPs) and non-coding RNAs that are essential for immune cell lineage commitment, maintenance, and modulation of immune responses.2,3 RBPs often control sets of genes with related functions (“regulons”) through direct binding of nucleic acid motifs in target mRNAs.4 RBPs have been shown to regulate gene expression in response to T cell activation and differentiation signals through a variety of mechanisms including mRNA stabilization.5,6,7,8 Loss of RBP function in mammalian systems has been shown to cause immune system hyperactivation and autoimmunity,9 uncontrolled inflammation,10 immunodeficiency by impaired viral clearance,11 or result in defective thymopoiesis.12,13 These data suggest that different RBP family proteins may have distinct functions in immune regulation.
Poly(C)-binding proteins (Pcbps; also referred to as hnRNPEs and αCPs) are a widely expressed gene family of RBPs with heterogeneous functions.14,15 The major family members are Pcbp1 and Pcbp2. Pcbp1 is an intronless paralog of Pcbp2 that evolved by retrotransposition of a fully processed Pcbp2 transcript.14,16 Pcbp1 and Pcbp2 have several overlapping functions stemming from their shared structure and binding specificity,14 but also have a subset of isoform-specific functions.17,18 The roles of Pcbp proteins in hematopoiesis, particularly erythroid lineage development and function, have been extensively characterized.18,19 Pcbps integrate nuclear events such as transcript processing (splicing and polyadenylation) with cytoplasmic control over mRNA stabilization and translation.20,21,22 These functions are mediated by direct interactions of Pcbps with cytosine- and pyrimidine-rich motifs in target pre- and mature mRNAs20,21,22,23 and can be controlled by post-translational modifications,17 the intracellular localization of Pcbps,24 or interactions with iron,8 zinc,25 or folate26 across a spectrum of cellular contexts.
We previously have demonstrated that Pcbp1 and Pcbp2 are independently essential for mouse embryonic development and survival.18 Pcbp1-null embryos do not survive the peri-implantation stage of development. Pcbp2-null embryos, in contrast, lose viability at mid-gestation associated with cardiovascular and hematopoietic defects. Splicing analysis of Pcbp2-null embryos showed that inclusion of exon 6 in the Runx1 mRNA required Pcbp2 function.27 Runx1 is an essential regulator of embryonic hematopoiesis28 and exon 6 encodes a protein domain important for Runx1 transcriptional activity.29,30 We have shown that embryos homozygous for a Runx1 mutant allele lacking exon 6 (Runx1ΔE6/ΔE6) had defective definitive erythroid lineage development and reduced embryonic fitness.27 Runx1ΔE6/ΔE6 mice surviving to adulthood demonstrated lymphopenia and thrombocytopenia consistent with the role of Runx1 in the expansion and maintenance of lymphoid and platelet populations.31 Combinatorial inactivation of the Pcbp1 and Pcbp2 loci in murine hematopoietic progenitors (Epo-Cre) demonstrated failed erythroid development and embryonic lethality at mid-gestation.19 This phenotype was more sensitive to Pcbp1 loss and suggested an overall dependency of erythropoiesis on Pcbp gene dose. Notably, analysis of Runx1 exon 6 splicing in this study confirmed that this event is specific to Pcbp2 and not impacted by deletion of Pcbp1 in erythroid progenitors. Using shRNA-mediated knockdown, Pcbp1 has been shown to promote proinflammatory cytokine production in CD4+ T cells through interactions with intracellular iron.8 In T cell lineage conditional knockout mice (CD4-cre and Foxp3-cre), Pcbp1 was shown to act as an intracellular immune checkpoint upregulated in activated T cells to prevent conversion of effector T cells into regulatory T cells, with loss of Pcbp1 resulting in increased Treg differentiation.32 However, Pcbp2 function in the adaptive immune system remains unclear.
The current study was initiated to investigate the impact of Pcbp2 on T cell development and function in an intact mammalian system. We use lineage-specific inactivation of Pcbp2 in murine CD4+ T cells coupled to assays of T cell activation, immunophenotyping, transcriptome analysis, and experimental models of adoptive transfer colitis to identify CD4+ lineage phenotypes associated with loss of Pcbp2 function. We demonstrate that Pcbp2 serves a critical role in T conventional cell (Tconv) proliferation, in part, through regulation of co-stimulatory signaling. We further demonstrate that control of Runx1 exon 6 splicing is associated with a specific role for Pcbp2 in the maintenance of peripheral CD4+ lymphocyte populations. These data establish Pcbp2 as a critical regulator of CD4+ T cell function in a manner that is distinct from its paralog Pcbp1.
Results
Pcbp2 controls Runx1 exon 6 inclusion in murine CD4+ thymocytes and splenocytes
Pcbp2 germline inactivation in mice results in embryonic lethality at E13.518. Therefore, to analyze Pcbp2 function in the CD4+ T cell lineage, we crossed Pcbp2-floxed mice (Pcbp2fl/fl) with CD4cre lines to generate a novel strain with conditional deletion of Pcbp2 in CD4+ T cells (fl-Pcbp2/CD4-cre). Fl-Pcbp2/CD4-cre (CKO) mice were viable and fertile. We next confirmed deletion of the Pcbp2-floxed allele and loss of Pcbp2 expression in fluorescence-activated cell-sorted CD4+CD8+ double-positive (DP) and CD4+ single-positive (SP) thymocytes by qPCR (Figure 1A). We observed no compensatory changes in Pcbp1 mRNA expression, a major Pcbp isoform with overlapping and non-redundant functions to Pcbp2 (Figure 1A).
Figure 1.
Pcbp2 is required for maintaining peripheral CD4+ T cell population size in mice
(A) Verification of conditional Pcbp2 knockout in fl-Pcbp2/CD4-cre (CKO) thymocytes. Total RNA from double-positive (DP, CD4+CD8+) and single-positive (SP, CD4+CD8–) thymocytes was analyzed by qPCR for Pcbp2 mRNA expression from CKO and fl-Pcbp2 (Control) mice. Pcbp1 and Runx 1 were used as expression controls. dCt means and standard errors for each genotype are shown (minimum of three biological replicates per genotype). Statistical significance was determined by Student’s t-test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
(B) Runx1 exon skipping is enhanced in CKO splenic CD4+ T cells. RT-PCR performed across the Runx 1 exon 6 splice junction from bulk splenocytes (Input), CD4+ and CD4– cells purified from CKO and control mice. Amplification diagrams of the Runx1 exon 6 splice junction are illustrated to the right of the gel image. Percentages of exon 6 skipping in each sample are shown. Arrows denote forward and reverse amplification primer positioning.
(C) Runx1 exon skipping is enhanced in CKO DP and SP thymocytes. RT-PCR performed across the Runx 1 exon 6 splice junction from bulk thymocytes (Input), DP and SP cells purified from CKO and control mice.
(D and E) Steady-state thymic T cell populations are not affected by Pcbp2 loss in the CD4 lineage. Flow cytometry analysis of labeled thymocytes from CKO and littermate control mice is shown. Gating illustrates thymic populations as follows: CD4 SP (CD4+CD8–), CD8 SP (CD4–CD8+), DP (CD4+CD8+), DN (CD4–CD8–), and the DN subpopulations [DN1 (CD44+CD25–), DN2 (CD44+CD25+), DN3 (CD44–CD25+), and DN4 (CD44−CD25–)]. Adjacent bar graphs show the percentages and the absolute numbers (mean cell numbers ± standard deviations) of each thymic population. Significance was determined by Student t-test (ns, not significant). Representative data from six independent experiments are shown.
(F and G) Pcbp2 is required for maintaining splenic CD4+ T cell population size. Flow cytometry analysis of labeled splenocytes from CKO and control mice is shown. Gating illustrates splenic populations as follows: CD3+ T cells, CD19+ B cells, CD4+ T cells (CD3+CD4+), CD8+ T cells (CD3+CD8+), and Tregs (CD3+CD4+Foxp3+CD25+). Adjacent bar graphs show the percentages and the absolute numbers (mean cell numbers ± standard deviations) of each splenic population. Significance was determined by Student t-test (ns, ∗p < 0.05). Representative data from at least 5 independent experiments are shown.
We recently demonstrated that Pcbp2 controls Runx1 exon 6 alternative splicing during mammalian embryonic hematopoiesis.27 Conditional Runx1 knockout in CD4+ T cells impairs T cell development and maturation33,34,35 and Runx1 mutant mice homozygous for exon 6 deletion exhibit peripheral lymphopenia.27 For these reasons, we first tested whether adult CKO mice exhibited changes in Runx1 total mRNA levels or in Runx1 mRNA exon 6 inclusion in CD4+ thymocytes and splenocytes. We observed no changes in Runx1 total mRNA between CKO mice and controls (Figure 1A). In contrast, we found higher percentages of Runx1 exon 6 skipping in both DP and SP thymocytes, as well as in CD4+ splenocytes from CKO mice compared to littermate controls (Figures 1B and 1C). These data confirm that Pcbp2 regulates Runx1 exon 6 mRNA inclusion in adult CD4+ lymphoid tissue and suggests that Pcbp2 regulation of Runx1 exon 6 splicing may be a general mechanism of Runx1 mRNA processing in the hematopoietic compartment that is operational from the mammalian fetal period into adulthood.
Pcbp2 is required for maintaining peripheral CD4+ T lymphocyte population size
Because Runx1 has pleiotropic roles during thymocyte development and peripheral T cell maturation,33,34,35 the hypothesis that Pcbp2 is important for development of the CD4+ T cell lineage was directly tested in CKO mice. The frequency of DP thymocytes, CD4 SP thymocytes, CD8 SP thymocytes, and CD4–CD8– double-negative (DN) thymocytes was comparable between CKO mice and littermate controls (Figures 1D and 1E). We observed no difference in the absolute numbers of these thymic T cell subpopulations (Figure 1E, right panel), or in the relative abundance of DN1-4 subsets (Figure 1E, bottom panel). In contrast, the proportion of CD3+ and CD4+ splenocytes showed a minor, but statistically significant decrease (Figures 1F and 1G) in CKO mice, whereas we observed no difference in the absolute numbers of splenocytes compared to controls (Figure 1G, left panel). In keeping with decreased proportions of CD3+ and CD4+ lineages in the spleen, we observed a corresponding decrease in the absolute number of splenic CD3+ and CD4+ T cells in CKO mice compared to controls (data not shown). There were no significant steady-state proliferation differences in peripheral CD4+ and CD8+ T cells from CKO mice compared to controls as measured by intracellular Ki67 positivity (Figure S1). In contrast to increased thymic and peripheral CD4+Foxp3+CD25+ regulatory T cell (Tregs) abundance occurring in Pcbp1-depleted CD4+ T cells,32 the percentage of Tregs in the CD4+ splenocyte and thymus populations was comparable between CKO mice and controls (Figures 1F and 1G and data not shown). Similar findings as those above also were observed in lymph nodes (Figure S2). Evaluation of the central memory status of splenic and lymph node CD4+FoxP3- and CD8+FoxP3- cells revealed no differences in the distribution of naive (TN, CD44lowCD62L+), central memory (TCM, CD44highCD62L+), or effector memory cells (TEM, CD44highCD62Lneg) between CKO mice and controls (Figure S3). These data reveal that Pcbp2 serves a non-redundant role in the maintenance of steady-state peripheral CD4+ T lymphocyte populations without impacts on the relative abundance of memory cell compartments.
Pcbp2 is required for CD4+ T cell proliferation in vitro
To define the impact of Pcbp2 on CD4+ T cell function, we first assessed the proliferation of purified CD4+CD25− T cells, isolated from CKO mice and controls, using an in vitro proliferation assay. We observed a significant decrease in CKO CD4+ cell proliferation compared to controls (Mean ± SD: 65.2% ± 8.9% vs 92.9% ± 3.9%, p < 0.0001; Figure 2A). To evaluate whether TCRβ or CD3ε expression was altered, we measured TCRβ and CD3ε median fluorescence intensity (MFI) in CD3+CD4+ from lymphoid organs of CKO mice compared to controls. We found a small but statistically significant reduction in TCRβ MFI in CKO CD3+CD4+ T cells across all 3 lymphoid organs tested, while no difference was observed in CD3ε MFI (Figure S4). To determine if CKO CD4+ cells had altered sensitivity to T cell receptor activation by anti-CD3ε antibodies in our system, we performed an anti-CD3ε dose-response analysis. We found that control cells had significantly higher proliferation rates at all tested concentrations of anti-CD3 (0.01–6 μg mL−1) compared to CKO CD4+ cells, with proliferation rates for both mutant and control CD4+ cells reaching a plateau at ∼1 μg mL−1 of anti-CD3ε (Figure 2B). Augmenting T cell receptor (TCR) co-stimulation with anti-CD28 antibodies (1 μg mL−1) resulted in a partial rescue of CKO CD4+ cell proliferation (Figure 2B). In the presence of anti-CD28, the majority of control cells demonstrated 5 or more cycles of division, while the CKO cells had a majority of cells with 3–4 division cycles. These data demonstrated that Pcbp2 function is required for CD4+ T cell in vitro activation and suggest a mechanism whereby co-stimulatory signaling rather than TCR signaling may be impaired in CKO CD4+ cells.
Figure 2.
Loss of Pcbp2 impairs CD4+ T cell proliferation
(A) Anti-CD3ε and irradiated APC-stimulated splenic CKO CD4+ T cells demonstrate decreased proliferation after 72 h. A T cell proliferation assay of Cell Trace Violet (CTV)-labeled CD4+ cells isolated from CKO, and control mice is shown. Adjacent bar graphs illustrate mean percent dividing cells ± standard deviations from Control and CKO CD4+ T cells. Student t-test; ns, ∗∗∗∗p < 0.0001.
(B) CKO CD4+cells have decreased division rates in response to T cell receptor (TCR) activation by anti-CD3ε antibodies. A T cell proliferation assay with anti-CD3ε dose-response (unstimulated, 0.01–6 μg mL−1) analysis is shown. Anti-CD28 costimulation was used to enhance TCR signaling. A plot of mean percent dividing cells (± standard deviations) is illustrated. Multiple t-test with Benjamini and Hochberg correction (FDR<0.05); ∗adj p < 0.05, ∗∗adj p < 0.01.
(C - E) Effect of IL-2 and IL-7 on CKO CD4+ T cell proliferation. (C) T cell proliferation assay histograms of CKO and Control CD4+ cells are shown with the different conditions noted by filled circles (irradiated APC, IL-7, anti-CD3ε, IL-2, anti-CD28; basal stimulation = anti-CD3 and irradiated APC). (D) Bar graphs show mean percent dividing cells (± standard deviations) for CKO and Control CD4+ cells. Multiple t-test with Benjamini and Hochberg correction (FDR<0.05); ns, ∗∗adj p < 0.01, ∗∗∗ adj p < 0.001, ∗∗∗∗adj p < 0.001. (E) Bar graphs of mean percent dividing cells (± standard deviations) for CKO CD4+ T cells illustrated in D. 1-way ANOVA (Kruskal-Wallis test) corrected for multiple comparisons (Benjamini & Hochberg, FDR <0.05), ns, ∗adj p < 0.05.
(F and G) Flow cytometry analysis of Annexin V and fixable viability dye (FVD) stained splenic control and CKO CD4+ T cells at rest and after 24 h of indicated costimulation conditions (basal stimulation = anti-CD3ε and irradiated APC) (F). Adjacent bar graphs show the percentages (mean percentage ± standard deviations) of Annexin V and FVD double-positive T cells occurring under each condition (G); ∗p < 0.05, ∗∗p < 0.01, Student t test.
Because CD28-mediated co-stimulatory signaling promotes both CD4+ cell proliferation and survival,36 we next tested whether other cytokines that regulate these functions in resting and activated CD4+ cells could rescue the proliferation defect we observed in CKO CD4+ cells. Supplementing basal stimulation conditions with IL-2, a cytokine that contributes to clonal expansion, differentiation, and survival of activated T cells, significantly increased proliferation rates of CKO CD4+ cells in a manner comparable to treatment of these cells with anti-CD28 antibodies (Figures 2C–2E). IL-7, a cytokine that supports survival and homeostatic proliferation of resting T cells, increased the 24-h survival of unstimulated CD4+ cells from both controls and CKO mice based on annexin V and fixable viability dye (FVD) staining (Figures 2F and 2G). In contrast, the addition of IL-7 to basal stimulation conditions (irrAPC and anti-CD3 Ab) did not alter proliferation rates of CKO CD4+ cells or controls (Figures 2C–2E). Adding IL-7 to basal stimulation conditions abrogated the modest but significant change in CKO CD4+ cell death at 24 h (Figures 2F and 2G). The addition of IL-7, IL-2, or anti-CD28 antibodies did not change measures of early apoptosis (Annexin V+/FVD−) in unstimulated or stimulated CKO CD4+ cells compared to controls (Figure 2F and data not shown). We then evaluated the cell surface expression of CD28 and CD127 (aka IL7ra) by flow cytometry. We found a statistically significant decrease of CD127 in CKO splenocytes and lymph nodes in both CD4+ and CD8+ cells, while no differences were observed in CD28 expression (Figure S5). Bypassing upstream TCR signaling using PMA/ionomycin stimulation demonstrated that CKO CD4+ T cells had no defects in their capacity to produce inflammatory cytokines but did not reveal any differences in intracellular cytokine production compared to controls (Figure S6). We conclude from these data that Pcbp2 function has a greater impact on proliferative and co-stimulatory signals compared to survival signals and cytokine production. Supplementation with a strong co-stimulation and/or with exogenous IL-2 partially rescued proliferative defects in CD4+ T cells.
CD4+ T cell sensitivity to Treg suppression and Treg suppressive function is not dependent upon functional Pcbp2
We next hypothesized that CKO CD4+ T cells may also be more responsive to suppression by Tregs based on our observation that CKO CD4+ cell in vitro activation was significantly impaired. To test this hypothesis, we performed an in vitro Treg suppression assay with flow-sorted CD4+ cells from CKO mice and littermate controls. We observed no significant difference between CKO and control CD4+ cell suppression in response to co-culture with control Tregs for 72 h (Figures 3A and 3B). To test whether Pcbp2 was required for Tregs to suppress CD4+ cells in vitro, flow-purified Tregs from the spleens of adult CKO mice or littermate controls were co-cultured with activated control CD4+ cells for 72 h. We found that CKO Tregs had comparable suppression ability to control Tregs (Figures 3C and 3D). These data indicate that CD4+ T effector sensitivity to Treg suppression and in vitro Treg suppressive function are not dependent upon functional Pcbp2.
Figure 3.
Conditional Pcbp2 deletion in CD4+ T cells does not affect T conventional sensitivity to Treg suppression and is not required for Treg function
(A and B) (A) Treg suppression assay of CTV-labeled control or CKO CD4+ T cells (Teff) co-cultured with control-T regulatory cells (Treg) in vitro. Serial dilution ratios of control Treg/ CD4+ T cells are noted above histograms. (B) Normalized suppression for each experimental condition in (A) is shown as area under the curve (AUC; see STAR Methods).
(C and D) (C) Treg suppression assay of CTV-labeled control T cells to be suppressed by control or CKO Treg in vitro. Serial dilution ratios of Treg/control CD4+ cells are noted above histograms. (D) Normalized suppression for each experimental condition in (C) is shown as area under the curve (AUC; see STAR Methods).
Representative data from 3 independent experiments are shown. ns, not significant.
Differential gene expression and splicing analysis
To investigate the mechanism for impaired CD4+ T cell activation resulting from the loss of Pcbp2, we performed mRNA-seq on unstimulated CKO and control, flow-sorted, CD4+ T cells and cells stimulated with CD3ε/CD28 mouse T-activator beads for 6 h. Overall, in unstimulated CD4+CD25– cells, we found 253 genes in CKO CD4+ T cells with differential expression compared to controls. Of these, 147 were upregulated and 106 were downregulated (Figure 4A and Table S1). In stimulated CKO CD4+ T cells, we found 81 genes with differential expression, of which 70 were upregulated and 11 were downregulated (Figure 4A and Table S2). A set of 42 genes was differentially expressed in both the unstimulated and stimulated conditions (Figures 4A and 4B). Pcbp2 mRNA expression was reduced by greater than 20-fold in unstimulated and stimulated CD4+ T cells, and we observed no significant change in Pcbp1 expression (Figure 4C and Tables S1 and S2), further confirming our Pcbp2-specific gene targeting in the CD4+ lineage. Differentially expressed genes important for immune cell function with a minimum 2-fold change increase in both unstimulated and stimulated cells included myosin 6, NADPH oxidase activator, prostaglandin 12 synthase receptor, and immunoglobulin superfamily member 23. Among transcription factors of interest with differential gene expression, we found ceramide synthase-6 and zinc finger protein 467 (Zfp467) to be upregulated in CKO unstimulated and stimulated CD4+ T cells and RAR-related orphan receptor C (RORC) upregulated in unstimulated cells. Within the set of T cell activation signature genes (GOTERM “T cell activation” (GO:0042110),37 we found 9 genes (Adk, Ager, CD80, CD81, CD83, CD160, IL7r, Rorc, and Zfp683) with differential expression in unstimulated cells (Figure 4B). There were no significant expression differences in the T helper-lineage markers ThPOK, CD40L, and Gata3. A gene ontology pathway analysis was conducted, and no significantly different pathways were identified (data not shown). In summary, these data indicate that Pcbp2 function is essential for normal expression of genes important for immune cell function and CD4+ T cell activation.
Figure 4.
Pcbp2 deletion results in altered gene expression in resting and activated CD4+ T cells
Changes in total gene expression between flow-sorted, splenic, control, and CKO CD4+ T cells were computed by RNA-seq for cells at rest (“unstimulated”) and after 6 h stimulation (“stimulated”) with CD3ε/CD28 mAb-coated T activator beads. Venn diagrams represent gene sets with overlapping differential expression.
(A) Venn diagrams of genes with upregulated or downregulated expression in the absence of Pcbp2 in unstimulated and stimulated CD4+ T cells.
(B) Venn diagram of differentially expressed gene sets that overlap with gene signatures characterizing activated CD4+ T cells. Gene symbols for selected overlaps are illustrated. Differentially expressed genes overlapping all 3 categories are represented in colored font matching the Venn diagram.
(C) Box and whiskers plot of Pcbp1 and Pcbp2 gene expression in resting and activated CD4+ T cells. The box covers the interquartile range and is split by the median. Outliers are shown as an open circle. ∗, adj p-value <0.05; ns, not significant.
To determine which genes had differential alternative splicing in CKO CD4+ T cells, we used MAJIQ, a tool to identify and measure alternative splicing variants from RNA-seq data, defining a significant difference in alternative splicing of a specific splice junction within a local splice variation (LSV) as a change in PSI (ΔѰ) of 0.2 (Probability (|ΔѰ|>=20%) > 95%, see STAR methods). We identified 36 genes with differentially spliced mRNA junctions in unstimulated CKO CD4+ T cells compared to controls (Table S3). In stimulated cells, we found 53 genes with differentially spliced mRNA junctions (Table S4). Among the genes with differential splicing, we found 58 differentially spliced LSVs in unstimulated cells and 74 differentially spliced LSVs in stimulated cells (Figure S7). There were 26 genes that were differentially spliced in both unstimulated and stimulated cells (Tables S3 and S4, and Figure 5A). Two differentially spliced genes, Runx1 and Dlg1, overlapped with the activated CD4+ gene signature (Figure 5B). Two transcription factors, Rfx7 and Runx1, were found to be differentially spliced. Rfx7 has been shown to regulate NK cell survival and function.38 Runx1 alternative splicing identified by this analysis corresponded to exon 6 skipping independently demonstrated by RT-PCR in sorted SP and DP thymocytes and splenocytes (Figures 1B and 1C, and 5C–5E). We also identified Pcbp2 regulated splicing of Ikbkb mRNA (aka, Ikkb) in both stimulated and unstimulated Tconv cells. Ikbkb encodes a kinase in the IKK complex that is activated in response to T cell receptor engagement and is critical for NF-kB activation (Figures 5F–5H).39 The affected junction is complex and includes a retained intron (Figure 5F splicegraph, orange bar), whose relative abundance increases in the absence of Pcbp2 (Figures 5G and 5H). Transcripts with this retained intron would be predicted to result in a premature stop codon and undergo nonsense-mediated decay. Intron retention is a global regulatory mechanism that occurs during CD4+ T cell activation and development of other lymphoid lineages in humans and mice.40,41 We identified several other differentially spliced genes important for immune cell development and function including Dlg1, Map4, Mfge8, Ptprs, Srpk2, and Helz2. We conclude from these data that Pcbp2 serves as a splice factor in murine CD4+ T cells and regulates genes critical for CD4+ cell activation and maintenance.
Figure 5.
Runx1 and Ikbkb mRNA splicing is altered in CKO CD4+ T cells
(A) Venn diagrams of differentially spliced genes (DSGs) from stimulated and unstimulated CD4+ T cell effectors from control and CKO mice.
(B) Venn diagrams of DSGs that overlap with gene signatures characterizing activated CD4+ T cells. Gene symbols for selected overlaps are illustrated. Transcription factors are indicated in bold; differentially expressed genes overlapping all 3 categories are represented in colored font matching the Venn diagram.
(C) The splice graph of the Runx1 transcript in unstimulated control and CKO CD4+ T cells is shown (defined by analysis of RNA-seq using the MAJIQ builder and VOILA visualization tool.42) The numbers above each splice linkage represent the number of junction-spanning raw reads summed across all samples in that group. Runx1 exon 6 skipping is represented by the blue line.
(D and E) Violin plots summarizing the “expected percent selected index” (E(Ψ)) for exon 6 skipping for each genotype (D) and the change in E(Ψ) (E(ΔΨ)) between control and CKO T cells (E). E(Ψ) is a statistical measure of each junction’s inclusion or exclusion level in a given local splice variation (LSV). (See STAR Methods).
(F) Splice graph of the Ikbkb transcript in unstimulated control and CKO CD4+ T cells. Canonical splicing between exons 8 and 9 is illustrated by the blue line. The Ikbkb intron retention event between exons 8 and 9 is represented by the wide orange bar. Other minor splice events in the LSV are captured in red, green, and purple.
(G) E(Ψ) for each splice event in (F) is shown in the violin plot.
(H) Violin plots of E(ΔΨ) between control and CKO CD4+ T cells illustrated in (H).
LSVs were determined to harbor a significantly changing splice junction if the probability that Ψ for any junction in the LSV was >20% changed across different analysis groups (significance threshold shown with dashed lines in (E), (G) and (H)).
We have previously described Pcbp2 as a splice factor in mouse fetal liver hematopoiesis.27 To determine whether Pcbp2 regulates splicing of a common set of genes across hematopoietic lineages in mice, we compared differentially spliced genes identified in Pcbp2 CKO CD4+ T cells to those identified in the fetal liver of Pcbp2 global knockout mice (Pcbp2−/−). We found the following genes to be differentially spliced in both fetal liver and CD4+ T cells lacking Pcbp2: Araf, Armcx2, Dcun1d2, Gemin2, Golga2, Map4, Mfge8, Ptprs, Runx1, and Srpk2 (Figure S8A). Importantly, we found that the Runx1 alternative splicing event involving exon 6 skipping in the fetal liver of Pcbp2 germline knockout mice27 was replicated in CKO CD4+ T cells (Figures 1B and 1C and 5C–5E). We similarly determined the set of differentially expressed genes that overlap between Pcbp2−/− fetal liver and CKO CD4+ T cells (Figure S8B). We found differential expression of Map3k12 to occur in Pcbp2−/− fetal liver and both stimulated and unstimulated CKO CD4+ T cells. Additional genes with differential expression between Pcbp2−/− fetal liver and unstimulated CKO CD4+ T cells included Art4, Cxx1c, Fhdc1, Lmna, March3, Tlr2, and Whrn; between Pcbp2−/− fetal liver and stimulated CKO CD4+ T cells, we identified Il6ra and Tspan32 with overlapping differential expression. Lmna43 and Tlr244 are important for mammalian hematopoiesis. In summary, Pcbp2 function is important for splicing and expression of a common set of genes in murine hematopoietic tissue spanning the fetal period into adulthood.
Pcbp2 deletion attenuates adoptive transfer colitis
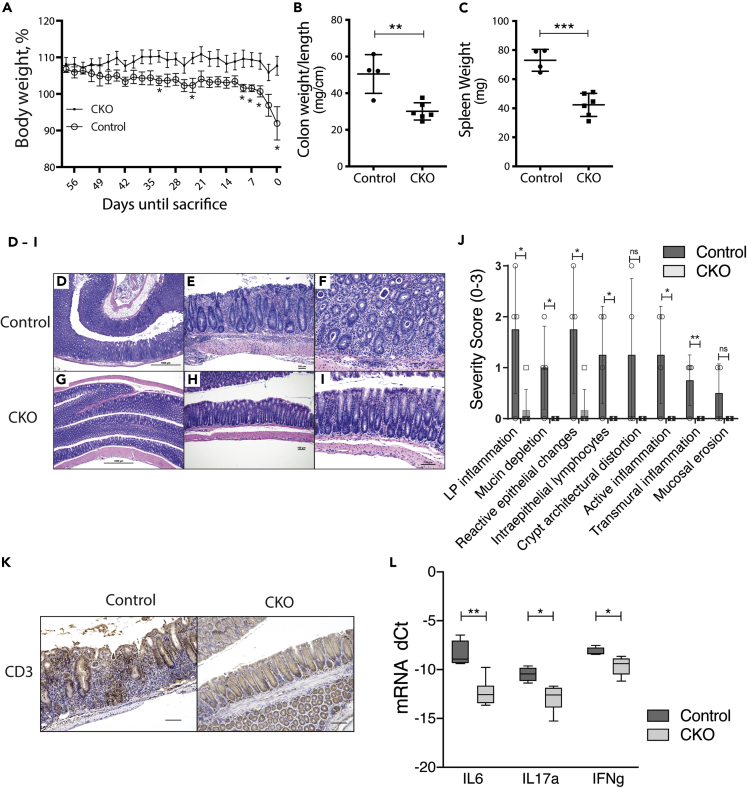
To determine if the impaired CKO CD4+ cell activation phenotype observed in vitro also occurred in an in vivo setting, we utilized a mouse model of autoimmune colitis.45 We injected intravenously 5 × 105 flow-sorted CD4+ T cells isolated from CKO mice or littermate controls into 10-week-old female C57BL6J/Rag1−/− mice (hereafter Rag1−/−) and monitored them biweekly for the development of colitis. Rag1−/− mice adoptively transferred with CKO CD4+ T cells had significantly less weight loss than mice receiving control CD4+ cells (Figure 6A). Similarly, Rag1−/− mice receiving CKO CD4+ T cells also demonstrated significantly lower colon weight/length ratios and developed less splenomegaly (Figures 6B and 6C). Blinded histological assessment was notable for a more severe colitis in control CD4+ cell recipients compared to Rag1−/− mice adoptively transferred with CKO CD4+ T cells (Figures 6D–6I). This observation included increased lamina propria inflammation, mucin depletion, reactive epithelium, intraepithelial lymphocytes, and active and transmural inflammation (Figure 6J). Engraftment and migration of adoptively transferred control and CKO CD4+ T cells to the colonic mucosa were verified by anti-CD3 immunohistochemistry (Figure 6K). We also observed a significant increase in inflammatory cytokine mRNA expression (IL-6, IL-17, and IFN-γ) in Rag1−/− mice adoptively transferred with control CD4+ T cells compared to those receiving CKO CD4+ T cells (Figure 6L). We conclude from these data that Pcbp2 function is required for optimal in vivo CD4+ cell activation in this T cell-mediated colitis model system.
Figure 6.
Pcbp2 deletion attenuates adoptive transfer colitis
(A) Weight curve of Rag1−/− mice adoptively transferred with conventional T cells (CD4+CD25−) from either control or CKO mice. Data in (A) is expressed as mean ± SEM; ∗ adjusted p-value <0.05 (Multiple t-test with Benjamini and Hochberg correction).
(B) Colon weight per unit length from Rag1−/− mice adoptively transferred with T conv cells from either Control or CKO mice.
(C) Spleen weight from Rag1−/− mice adoptively transferred with T conv cells from either control or CKO mice. Data in (B) and (C) is expressed as mean ± SD; ∗∗p < 0.01, ∗∗∗p < 0.001, Student t-test.
(D–I) Hematoxylin and eosin staining of representative specimens from Rag1−/− mice adoptively transferred with control or CKO T conv cells from panels A-C.
(D–F), Control Tconv colon.
(D) Diffuse colitis with hypercellular mucosa and crypt architectural disorder (40× magnification).
(E) Transmural inflammation with focal surface erosion (100×).
(F) Active colitis with lamina propria acute and chronic inflammation, acute cryptitis with intraepithelial neutrophils and lymphocytes, crypt abscesses, and crypt architectural disorder (200×).
(G–I), CKO T cell colon.
(G) Normal colonic mucosa (40× magnification). (H) Regular crypt architecture and lack of muscularis inflammation (100×).
(I) Intact surface epithelium with only rare intraepithelial lymphocytes (200×). Scale bars for (D) and (G) are 1000 μm and 100 μm for (E), (F), (H), and (I).
(J) Blinded histologic analysis of colonic mucosa disease severity. Data expressed as mean ± SD are shown; ns, not significant, ∗adjusted p < 0.05, multiple t-test with Benjamini and Hochberg correction (FDR<0.05).
(K) Anti-CD3 immunohistochemistry of representative colonic mucosa illustrating engraftment of CD4+ T cells in Rag1−/− mice.
(L) mRNA expression analysis of colonic inflammatory cytokines. Total RNA from Rag1−/− colon specimens adoptively transferred with control or CKO T cells was analyzed by qPCR for mRNA expression of IL-6, IL-17, and IFN-γ. dCt (dCt = GAPDH Ct - Cytokine Ct) means and standard errors for each genotype are shown; ∗∗adj p-value <0.01; ∗adjusted p-value <0.05, multiple t-test with Benjamini and Hochberg correction.
Discussion
The experiments presented here show that conditional deletion of the Pcbp2 gene in murine CD4+ T cells results in decreased CD4+ Tconv cell activation with only a marginal effect upon cell viability. The impact of Pcbp2 function on CD4+ Tconv cell activation was demonstrated in two ways. First, we found decreased activation of splenic CD4+ Tconv cells from CKO mice stimulated in vitro by anti-CD3 antibodies and irradiated APCs (Figure 2A). These proliferative defects were almost completely rescued by providing CD28 co-receptor signaling or via IL-2 pathway activation in a manner that could not be achieved with maximal TCR stimulation in our system (Figures 2B–2E). While we did observe a modest decrease (∼10%–15%) in TCRβ cell surface expression in CKO cells (Figure S4), were TCR signaling dramatically impaired, CD28 or IL-2 co-stimulation would not be expected to have such a potent effect on the overall proportion of cells activated to proliferate. Once activated, CKO CD4+ Tconv cells had similar sensitivity to Treg-mediated suppression compared to controls (Figure 3A). Second, we used an in vivo T cell adoptive transfer colitis model and found that CD4+ T cells lacking Pcbp2 caused significantly less clinical, histological, and gene expression manifestations of colitis, all proxy measures of Tconv cell activation in this system (Figure 6). Importantly, we observed no changes in Pcbp1 expression in CD4+ T cells resulting from Pcbp2 genetic deletion (Figures 1A and 4C). Therefore, our studies establish a critical and non-redundant role for Pcbp2 in murine CD4+ Tconv cell activation, which may operate through control of co-stimulatory pathway and/or IL-2 signaling.
We also demonstrate that Pcbp2 is required for maintaining the relative abundance of peripheral CD4+ T lymphocytes (Figures 1F and 1G). In contrast, there were no changes in the abundance or frequency of DP or mature CD4 SP thymocytes in CKO mice (Figures 1D and 1E). Pcbp2 suppression has been associated with cell cycle arrest and the regulation of survival and death in a variety of experimental settings.46 Our data show that CKO CD4+ T cells have only a modest increase in cell death after an activating stimulus compared to controls, whereas unstimulated cells demonstrated no differences (Figures 2F and 2G). IL-2 or IL-7 supplementation does not significantly change cell survival or cell division of control cells (Figures 2C and 2D). Interestingly, the defect in IL-2 signaling in this context does not seem to impact Treg biology (i.e., survival, number, and suppressive function), but affects only Tconv cells. Normal IL-7 signaling is surprising since our RNA-seq data and corresponding CD127 cell surface expression analysis demonstrated that CKO cells have reduced IL-7R expression, but no differences in cell surface CD3ε or CD28 (Figures S4 and S5). It appears that, in our experimental system, the reduction in IL-7R does not have apparent functional consequences on Tconv cells. While we have not identified any gene expression changes in the IL-2 pathway, Runx1 has been shown to be a regulator of IL-2 production in CD4+ T cells47 and impaired Runx1 function could be playing a role in this setting. These data suggest that Pcbp2 is not a critical determinant of cell survival in peripheral CD4+ lymphocytes and led us to consider alternative mechanisms for the decrease in peripheral CD4+ T lymphocytes.
Runx1 is a master hematopoietic transcriptional factor with a well-documented role in the expansion and maintenance of lymphoid and platelet populations.31,33,48,49 Thus, we investigated the possibility that changes in Runx1 expression or isoform abundance could be associated with decreases in peripheral CD4+ CKO lymphocytes. We observed a decrease in Runx1 exon 6 inclusion in processed mRNA from CKO thymocytes and splenocytes compared to controls (Figures 1A–1C, and 5C–5E). The protein domain encoded by exon 6 is located between the DNA-binding domain (Runt homology domain) and C-terminal transactivation and autoinhibitory domains.50 Runx1 by itself is a weak transcriptional activator and commonly heterodimerizes with core binding factor β or other transcription factors to activate or repress transcription.51,52 Notably, the Runx1 exon 6 protein segment interacts with the Sin3a transcriptional repressor complex and contains one of two known Ets-1 transcription factor binding domains within the Runx1 protein.53,54 Both Sin3a and Ets-1 serve an important role in lymphoid development and T cell function.55,56 In vitro and cell line-based overexpression studies have demonstrated that Runx1 proteins lacking exon 6 have reduced transactivation potential.57
In prior work, we demonstrated that Pcbp2 drives development of the definitive erythroid lineage in the mouse fetal liver by operating to promote retention of exon 6 in the Runx1 mRNA.18,27 The observation that both myeloid and lymphoid lineages lacking Pcbp2 show decreased Runx1 exon 6 inclusion suggests that the role of Pcbp2 as a core regulator of Runx1 isoform expression is broadly conserved across hematopoietic lineages. In the current study, peripheral CD4+ T cells from CKO mice have a mRNA ratio of Runx1 full-length to Runx1 lacking exon 6 (Figures 1C and 5D) that is similar to that of heterozygous germline Runx1ΔE6 mutants27 where ∼75%–85% of detectable Runx1 transcripts show skipping of exon 6. Importantly, heterozygous Runx1ΔE6 mice displayed a mild peripheral lymphopenia suggesting a proportional response of this lineage to the relative abundance of Runx1 isoforms lacking exon 6. This may account for the reduction, but not full loss, of peripheral CD4+ T cells in CKO mice since full-length Runx1 transcripts are still expressed, albeit to a much lower degree. Conditional deletion of the full-length Runx1 transcript in CD4cre mouse strains (Runx1 cKO) results in a modest reduction in thymus SP CD4 and CD8 lineages without impacting total thymocyte numbers or DP thymocyte development.34 A larger reduction of SP CD4 cellularity, however, occurs in the peripheral lymphoid organs of Runx1 cKO mice.34,35 Peripheral CD4+ cell loss in Runx1 cKO mice occurs due to a T cell maturation defect in sialylation that has been shown to render Runx1 cKO CD4+ T cells more susceptible to complement-mediated destruction.35 We speculate that a similar T cell maturation defect may occur in CKO CD4+ T cells, rather than defective thymic selection, and may be linked to diminished Runx1 function through isoform switching that results in accumulation of Runx1ΔE6 transcripts. Examination of thymic histologic structure in Pcbp2 CD4+ CKO mice, which was not done in our study, may provide additional clarity to this hypothesis. In summary, the CKO mouse model has demonstrated that Pcbp2 is critical for maintenance of peripheral CD4+ T lymphocyte population size but is dispensable for maintenance of steady-state DP and more mature thymocyte populations in adult mice.
Pcbp1 is a Pcbp2 paralog evolutionarily derived from a Pcbp2 transcript with overlapping and non-redundant functions.14,17,18,27,58 Pcbp1 has been found to control CD4+ T cell proinflammatory cytokine expression in mice through mRNA stabilization (e.g., GM-CSF), but not proliferation or survival in stimulated CD4+ T cells skewed to the Th1 lineage that were depleted of Pcbp1.8,58 Here, in contrast, we demonstrate a significant proliferation defect in CD4+ T cells (Figure 2) without impacts on proinflammatory cytokine mRNA expression, including no impact on GM-CSF, suggesting that Pcbp2 may have a distinct and separable function from Pcbp1 in the CD4+ lineage or its associated CD4+ sub-types. Although we did not observe changes in CD8+ abundance (Figures 1F and 1G) or steady-state CD8+ T cell proliferation in peripheral lymphoid organs (Figure S1), whether Pcbp2 is essential for CD8+ activation following a strong stimulus, such as that demonstrated for CKO CD4+ T cells, was not evaluated in our study and remains an open question. Interestingly, Pcbp2 has been identified through biochemical approaches to be a target protein modulating the antiproliferative effects of 15-deoxyspergualin (DSG), an immunosuppressive agent used in steroid-resistant transplant rejection.59 More recent work by others has shown that CD4+ and CD8+ lineage development in both the thymus and peripheral lymphoid organs are significantly impacted by conditional genetic ablation of Pcbp1 in CD4+ T cells.32 While proliferation was not directly assessed in this study, gene expression analysis revealed decreased expression of genes important for effector T cell activation and function. Conditional Pcbp1 knockout in CD4+ T cells and Foxp3+ cells, respectively, also demonstrated that Pcbp1 is important for regulating T reg differentiation, but not T reg maintenance, and is associated with increased expression of Treg signature genes, including FoxP3. We demonstrate that CD4+ CKO Pcbp2 mice tolerate Pcbp2 loss without detrimental effects upon the abundance of Tregs in the thymus or spleen (Figure 1), or upon Treg function in vitro (Figure 3). We conclude from these data that Pcbp2 has an important and specific role in CD4+ Tconv cell proliferation, but has limited effects upon Treg development and function, while Pcbp1 appears to have a more prominent role in Treg ontogeny.
We demonstrate that Pcbp2 regulates differential expression and splicing of genes important for Tconv cell activation (Figures 4 and 5). Although Pcbp2 CKO CD4+ T cells show apparent defects in co-stimulation (Figure 2), we did not identify expression or splicing differences in any classical T cell resident co-stimulatory molecules (e.g., CD40L, CD28, Tim-1, ICOS, and CD27).60 Similarly, negative co-stimulatory signaling molecules such as CTLA-461 and Tim-362 were neither overexpressed nor differentially spliced in unstimulated or stimulated CKO cells. Validating mRNA expression changes for those genes known to be important for T cell activation (Figure 4B) by testing for differences in cell surface expression, particularly molecules such as CD81 that may impact co-stimulatory pathways,63,64 may increase understanding of the CKO phenotype. At present, co-stimulatory/co-inhibitory pathways are one of the most important targets of anticancer immunotherapy.65 In this regard, future studies that leverage the previously unknown role of Pcbp2 in the regulation of T cell co-stimulation may identify mechanisms that could enhance antitumor responses.
The role of Pcbp2 as a splice factor has been established in both hematopoietic and non-hematopoietic tissues.20,27,66 Regulatory mechanisms of mRNA processing such as alternative splicing, mRNA stabilization, and intron retention have been shown to be important for hematopoietic and lymphoid development and function.40,41,67,68,69 Here, we demonstrate Pcbp2 control of Runx1 cassette exon 6 splicing, as noted above, intron retention of Ikbkb between exons 8 and 9, and differential splicing of several other genes important for T cell activation and maintenance (Figure 5). The kinase encoded by Ikbkb phosphorylates the inhibitor in the inhibitor/NF-kappa-B complex, causing dissociation of the inhibitor and activation of NF-kappa-B. NF-κB signaling is required at multiple stages of T cell development and function.70 Intron retention between Ikbkb exon 8 and 9, which occurred in both unstimulated and stimulated cells, is predicted to result in nonsense-mediated decay and reduce overall Ikbkb mRNA expression. Survey of sequences proximal to the splice donor and acceptor sites in exons 8 and 9, respectively, revealed conserved C-rich tracts consistent with known Pcbp2 and other poly-C-binding protein family protein-binding sites (data not shown),20,25,27,66,71 suggesting that the effect of Pcbp2 on Ikbkb splicing may be through direct interaction with the Ikbkb pre-mRNA. We did not, however, observe differential gene expression of Ikbkb for either condition between CKO’s and controls and speculate that a greater proportion of Ikbkb transcripts with retained intron 8–9 would be needed to detect a significant difference. During both human and murine B cell differentiation, IkB kinase subunits, including Ikbkb, and other NF-κB pathway mediators, have been shown to undergo differential intron retention.41 Notably, intron retention is a global regulatory mechanism that also is prevalent in resting CD4+ T cells that diminishes following activation.40 Our differential splicing data do not support a widespread role for Pcbp2 in global intron retention but rather suggest a more targeted splicing function, including intron retention events, on genes critical for CD4+ T cell differentiation.
In summary, these data highlight the importance of Pcbp2 to the development and function of the CD4+ T cell lineage. The impact of Pcbp2 on CD4+ Tconv cell proliferation, and corresponding rescue of this phenotype with enhanced co-stimulation, supports an essential role for Pcbp2 in regulating CD4 receptor co-stimulatory signaling. While we demonstrate the importance of Pcbp2 in control of gene expression and alternative splicing of genes important for CD4+ Tconv activation, the precise mechanism explaining the co-stimulatory signaling defect remains unclear. Pcbp proteins, including Pcbp2, have been shown to serve as intracellular iron chaperones in a variety of tissue contexts.72,73 Pcbp2 also has been shown to interact with intracellular signaling complexes, such as mTORC2,46 which serve as critical nodes that integrate signals downstream of cytokine and co-stimulatory receptors in CD4+ T cells to control survival, proliferation, and differentiation.74 It is therefore conceivable that mechanisms unrelated to RNA-binding activities, such as regulation of iron metabolism, or direct modulation of PI3K/AKT/mTOR signaling, may account for the defective proliferation phenotype we have observed. In addition, our data demonstrate a clear association between Pcbp2 control of Runx1 exon 6 splicing in CD4+ T cells and a specific role for Pcbp2 in the maintenance of peripheral CD4+ lymphocyte population size, in keeping with our prior observations of lymphopenia in Runx1ΔE6 knockout mice.27 Several well-characterized mechanisms have been shown to control Pcbp protein RNA-binding activity. These include intracellular localization, phosphorylation downstream of TGF-β signaling, and nutritional status.17,24,26 Of clear interest now is to address how Pcbp2 itself is regulated during T cell activation and to understand where Pcbp2 impacts the co-stimulatory signaling cascade. Whether gene expression or splicing changes are direct effects of Pcbp2:RNA interactions on key T cell regulatory gene transcripts or indirect effects of Pcbp2 interacting with splicing machinery complexes also is an open question. Future studies are needed to extend our understanding of Pcbp2 function in the maintenance and activation of CD4+ lymphocytes and to uncover whether and how Pcbp2 may impact human immune-mediated disease.
Limitations of the study
There are several limitations of our study. First, while it was demonstrated that Pcbp2 is essential for activation and proliferation of CD4+ Tconv cells, these proliferation defects were not characterized across all CD4+ T helper subsets or in CD8+ T cells where Pcbp2 genetic deletion also is expected in the CD4-cre system. Therefore, further experimentation is required in these populations to determine whether Pcbp2 serves a more general role regulating T cell proliferation following activating stimuli. Second, the conclusions drawn about Treg function in this study are based on in vitro experimentation. Given the role of Pcbp1 in regulating Treg population size and function, additional in vivo studies, such as adoptive transfer of Pcbp2-null Tregs into mouse models of colitis, or other inflammatory disease models, would further clarify whether Pcbp1 and Pcbp2 have distinct roles in Tregs. Last, the Pcbp2 CD4+ CKO phenotypes described were associated with altered Runx1 exon 6 splicing. Establishing a causal linkage between Runx1 exon 6 splicing changes and impaired CD4+ Tconv proliferation, using full-length Runx1 in rescue experiments for example, is worthy of future investigation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3 (FITC) | BD Pharmingen | clone 145-2C11 |
| CD19 (PE) | BD Pharmingen | clone 1D3 |
| CD4 (BV785) | Biolegend | clone GK 1.5 |
| CD4 (FITC) | eBioscience | clone GK 1.5 |
| CD8 (PE-Cy7) | BD Pharmingen | clone 53–6.7 |
| CD44 (PerCP) | Biolegend | clone IM7 |
| CD62L (APC eFluor 780) | eBioscience | clone MEL-14 |
| Ki67 (eFluor) | eBioscience | clone SolA15 |
| TCR-beta (PE) | eBioscience | clone H57-597 |
| IL-2 (PeCy7) | eBioscience | clone MEL-14 |
| TNF-alpha (FITC) | Biolegend | clone MP6-XT22 |
| IFN-gamma (AF647) | BD Pharmingen | clone XMG1.2 |
| CD25 (BV650) | Biolegend | clone PC61 |
| CD25 (PE) | eBioscience | clone PC61 |
| CD3ε | eBioscience | clone 145-2C11 |
| CD28 | eBioscience | clone 37.51 |
| Foxp3 (APC) | eBioscience | clone FJK6-16s |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Foxp3/Transcription Factor Staining Buffer Set | eBioscience | 00-5523-00 |
| Intracellular staining fixation buffer | eBioscience | 00–8222 |
| Permeabilization buffer | eBioscience | 00–8333 |
| PMA | Sigma | #P8139 |
| Ionomycin | Sigma | #I0634 |
| Brefeldin A Golgi plug | BD Biosciences | #51 230 1KZ |
| Monensin | BioLegend | #420701 |
| IL-2 Recombinant | Roche | 11271164001 |
| IL-7 Recombinant | Biolegend | 57780210 |
| diaminobenzidine tetrahydrochloride (DAB) immunohistochemistry kit | Vector labs | SK-4100 |
| TRIzol Reagent | ThermoFisher | 15596–026 |
| DNase I | Invitrogen | 18068–015 |
| RNasin | Promega | N2511 |
| Annexin V (FITC) | BioLegend | 640906 |
| live/dead fixable aqua dead cell stain kit | Thermofisher Scientific | L34957 |
| CellTrace Violet dye | Invitrogen | C34557 |
| Superscript III | Invitrogen | 18080–044 |
| Fast SYBR green | ThermoFisher | 4385612 |
| oligo(dT)12–18 | Invitrogen | 18418–012 |
| random hexamers | Roche | 11034731001 |
| Critical Commercial Assays | ||
| CD4+CD25+ Regulatory T Cell Isolation Kit, mouse | Miltenyi Biotec, San Diego, CA | 130-091-041 |
| CD3ε/CD28 mouse T-activator beads | Thermofisher | 11456D |
| CD90.2 MicroBeads, mouse | Miltenyi Biotec, San Diego, CA | 130-049-101 |
| Kapa2G Robust HS + DNTPS | Kapa Biosystems | KK5532 |
| Deposited Data | ||
| RNA-seq raw data | Gene Expression Omnibus (GEO) database | GEO: GSE197059 |
| Experimental Models: Organisms/Strains | ||
| CD4-cre transgenic mice (Tg(Cd4-cre)1Cwi) | The Jackson Laboratory | 022071 |
| B6/Rag1−/− | The Jackson Laboratory | 002216 |
| Mouse: Pcbp2fl/fl | Ghanem et al.18 | N.A. |
| Oligonucleotides | ||
| ACACTGCATCTTGGCTTTGC | Integrated DNA Technologies | Ifng-76_F |
| GCTTTTCAATGACTGTGCCGT | Integrated DNA Technologies | Ifng-76_R |
| TGCAAGAGACTTCCATCCAGT | Integrated DNA Technologies | Il6-73_F |
| TTGTGAAGTAGGGAAGGCCG | Integrated DNA Technologies | Il6-73_R |
| CAGCAGCGATCATCCCTCAA | Integrated DNA Technologies | Il17a-184_F |
| TCAGGGTCTTCATTGCGGTG | Integrated DNA Technologies | Il17a-184_R |
| CACTCTGACCATCACCGTCTT | Integrated DNA Technologies | Runx352_F |
| GGATCCCAGGTACTGGTAGGA | Integrated DNA Technologies | Runx352_R |
| Software and Algorithms | ||
| MAJIQ | https://majiq.biociphers.org/ | version 2.0 |
| Prism | Graphpad | version 7 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Louis Ghanem (lghanem@its.jnj.com).
Materials availability
This study did not generate unique reagents.
Experimental model and subject details
Animal husbandry
All mouse studies were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee at the Perelman School of Medicine at the University of Pennsylvania (Protocol number 804434). All mice were housed in standard cages within a barrier facility under 12 hour on/off light cycling conditions and were given ad libitum access to standard mouse chow and water. The Pcpb2 floxed mouse line used in this study was previously described.18 CD4-cre transgenic mice (Tg(Cd4-cre)1Cwi; strain #022071) and B6/Rag1−/− (strain #002216) were obtained from The Jackson Laboratory (Bar Harbor, ME). All experiments were performed in female mice between 6-12 weeks of age.
Adoptive transfer colitis model
Ten-week-old C57BL6/Rag1−/− mice were adoptively transferred by tail vein injection with 5 × 105 CD4+CD25− Tconv cells from either control or CKO mice isolated by cell sorting (purity >98%) (Mo Flo Astrios EQ, Beckman Coulter). Colitis was assessed by the daily monitoring of body weight.75 Gut and lymphoid tissues were collected for histologic and flow cytometric analysis.
Method details
Flow cytometry
Cells were analyzed on a LSR II flow cytometer (BD Biosciences) in the Flow Cytometry Core Laboratory of The Children’s Hospital of Philadelphia Research Institute, and data were analyzed using FlowJo (version 10.1). Antibodies used for staining murine antigens were CD3 (FITC, BD Pharmingen, clone 145-2C11), CD19 (PE, BD Pharmingen, clone 1D3), CD4 (Brilliant Violet 785, BioLegend and FITC, eBioscience, clone GK 1.5), CD8 (PE-Cy7, BD Pharmingen, clone 53–6.7), CD25 (Brilliant Violet 650, BioLegend and PE, eBioscience, clone PC61), CD44 (PerCp, BioLegend, clone IM7), CD62L (APC eFluor 780, eBioscience, clone MEL-14), Foxp3 (APC, eBioscience, clone FJK-16s), Ki67 (eFluor 450, eBioscience, clone SolA15), TCR-β (PE, eBioscience, clone H57-597), IFN-γ (Alexa Fluor 647, BD Pharmingen, Clone XMG1.2), IL-2 (PeCy7, eBioscience, Clone MEL-14) and TNF-α (FITC, BioLegend, Clone MP6-XT22). Intranuclear staining was performed using a Foxp3/Transcription Factor Staining Buffer Set (cat. #00-5523-00, eBioscience) per the manufacturer’s instructions. The populations of immune cells in the thymus were defined as follows: double positive (CD4+CD8+), single CD4+, single CD8+ and double negative (DN1: CD4–CD8–CD25–CD44+; DN2: CD4–CD8–CD25+CD44+; DN3: CD4–CD8–CD25+ CD44–; and DN4: CD4–CD8–CD25–CD44–). The populations of immune cells in the spleen and lymph nodes were defined as follows: CD4+ T cells (CD3+CD4+CD8–CD19–), CD8+ T cells (CD3+CD8+CD4–CD19–), B cells (forward and side scatter intensities corresponding to lymphocytes; and CD3–CD19+). Flow cytometric detection of apoptosis and necrosis was performed with FITC–labeled Annexin V (cat. #640906, BioLegend) and the live/dead fixable aqua dead cell stain kit (cat. #L34957; Thermofisher Scientific).
T cell isolation
We isolated lymphocytes from the spleen. After preparing single cell suspensions, red blood cells lysis and cell counting, we used magnetic beads (Miltenyi Biotec, San Diego, CA) to isolate Tconv cells (CD4+CD25–), T regulatory cells (Treg) (CD4+CD25+) and antigen-presenting cells (APC) (CD90.2–). To achieve high purity, Tconv single cell suspensions were incubated with 1.5 times the recommended amount of antibodies (biotin conjugated antibodies against non-CD4 cells and anti-biotin microbeads), as previously reported.76 Isolation was performed in the dark and the total amount of cells for the LD column was restricted to 1 × 108 cells. Purified Tconv cells were used in downstream proliferation assays. Proliferation assays were replicated with Tconv cells purified by cell sorting (Mo Flo Astrios EQ, Beckman Coulter, >98% purity).
T cell activation and proliferation
After isolation, control and CKO Tconv were labelled with 5 μM of Cell Trace Violet (CTV) (Invitrogen), as previously reported77 and were cultured in T cell media [Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U mL−1), streptomycin (100 mg mL−1), and 55 nM β2-mercaptoethanol)] in the presence of irradiated APC (100 Gy) purified from control mice, and stimulated with CD3ε monoclonal antibody (eBioscience, clone 145-2C11) at a concentration of 1 μg mL−1, in 96-well round-bottom plates (5 × 104 cells per well) for 72 hours (37° Celsius and 5% CO2). Cells were then harvested, labeled with anti-CD4-FITC (eBioscience, clone GK 1.5), and analyzed by dye dilution flow cytometry.
In vitro assays to investigate Treg suppressive function
Isolated CD4+CD25+ Tregs from CKO or control littermates were co-cultured in 96-well round-bottom plates with CTV labelled Tconv cells (5 × 104 cells per well) from control mice in the presence of irradiated (100 Gy) APCs (5 × 104 cells per well) and 1 μg mL−1 anti-CD3ε. In order to precisely establish the suppression capacity, Tregs were added in culture at 5 different dilutions with respect to effector T cells (TE) (TE:Treg - 1:1, 1:2, 1:4, 1:8, 1:16). Cells were harvested after 72 hours, labeled with CD4-FITC and CD25 Brilliant Violet 650 and analyzed by flow cytometry. Treg suppressive function was determined using a modified area-under-curve (AUC) method.77 The AUC approach allows for reliable measures of Treg suppressive function when TE cells have different cell division rates, as we observed between control and CKO TE cells (Figure 2).77
Proliferation rescue
Isolated and CTV labeled control and CKO CD4+ T cells were co-cultured (5 × 104 cells per well) for 72 hours in the presence of irradiated APC (100 Gy), 1 μg mL−1 CD3ε monoclonal antibody and either: 1) 1 μg mL−1 CD28 mAb (eBioscience, clone 37.51); 2) 20 IU mL−1 recombinant IL-2 Recombinant (Roche); 3) 1 ng mL−1 IL-7 Recombinant (BioLegend); or 4) both IL-2 (20 IU mL−1) and IL-7 (1 ng mL−1). Cells were harvested after 72 hours, labeled with CD4-FITC and analyzed by flow cytometry.
Intracellular cytokine staining
For assessment of in vitro cytokine production, we incubated freshly isolated splenocytes from CKO and control mice (37°C, 5% CO2) in 24-well plates. PMA (Sigma, #P8139), ionomycin (Sigma, #I0634), Brefeldin A Golgi plug (BD Biosciences #51 230 1KZ) and Monensin (BioLegend #420701) were added to reach final concentrations of 6 ng mL−1 PMA, 1 μg mL−1 ionomycin, 5 μg mL−1 Brefeldin A and 5 μg mL−1 Monensin. Cells were incubated for 4 hours and then harvested for flow cytometry. Intracellular staining fixation buffer (eBioscience, Cat. 00–8222) and permeabilization buffer (eBioscience, cat. 00–8333) were used for intracellular staining.
Histology
Colons were fixed in 10% neutral buffered formalin overnight, washed and paraffin-embedded. Hematoxylin and eosin stained sections were prepared at the Molecular Pathology and Imaging core in the Department of Gastroenterology at the University of Pennsylvania. Sections were scored by a pathologist (B.J.W.) blinded to the treatment conditions. We used a modified version of the scoring system previously reported,76 using the following parameters: 1) degree of lamina propria infiltration (0–3); 2) degree of mucin depletion as evidenced by loss of goblet cells (0–2); 3) reactive epithelial changes (0–3); 4) number of intraepithelial lymphocytes per high-power field within crypts (0–3); 5) degree of crypt architectural distortion (0–3); 6) degree of inflammatory activity (0–2); 7) degree of transmural inflammation (0–2); and 8) degree of mucosal surface erosion up to total surface ulceration (0–2). Qualitative assessment of Tconv cell engraftment in Rag1−/− mice was done by assessing CD3ε expression (Cd3e, eBioscience, Clone 145-2C11) within colon sections using a diaminobenzidine tetrahydrochloride (DAB) immunohistochemistry kit (Vector labs, SK-4100) per the manufacturer’s instructions.
RNA isolation and quantitative PCR
Spleen, thymus and colon samples were placed in Eppendorf tubes containing 300 μL TRIzol Reagent (ThermoFisher, 15596–026). Samples were homogenized by pestle and volumes adjusted to 1 mL with additional TRIzol Reagent. RNA extraction was carried out according to the manufacturer’s instructions. RNA was resuspended in nuclease-free diethyl pyrocarbonate (DEPC)-treated water and quantified by NanoDrop (NanoDrop 1000, ThermoFisher). One microgram of total RNA was DNase I treated (18068–015; Invitrogen) per the manufacturer’s instructions and used for cDNA synthesis. DNase-treated RNA was primed with oligo(dT)12–18 (18418–012; Invitrogen) and random hexamers (11034731001; Roche) in the presence of 40 units of RNasin (N2511; Promega) and reverse transcribed for 45 min at 50°C per the vendor’s protocol (Superscript III, 200 units/20 μL reaction mixture [18080–044; Invitrogen]). Complementary DNA (cDNA) was diluted 2.5-fold, and 2 μL of diluted cDNA was used for each 20 μL quantitative PCR (qPCR) (Fast SYBR Green mix, catalog number 4385612; ThermoFisher). Synthesized primers for mouse Pcbp1, Pcbp2, Runx1, and GAPDH were previously described.27 Primers for the mouse cytokines Ifng, Il-6, and Il-17a were as follows: Ifng-76_F (5′-ACACTGCATCTTGGCTTTGC-3′), Ifng-76_R (5′-GCTTTTCAATGACTGTGCCGT-3′), Il6-73_F (5′-TGCAAGAGACTTCCATCCAGT-3′), Il6-73_R (5′-TTGTGAAGTAGGGAAGGCCG-3′), Il17a-184_F (5′-CAGCAGCGATCATCCCTCAA-3′), and Il17a-184_R (5′-TCAGGGTCTTCATTGCGGTG-3′). Thermocycling was performed on an Applied Biosystems QuantStudio 5 for each primer pair as follows: 95°C for 20 s and then 40 cycles of 95°C for 3 s and 60°C for 30 s. A melt curve analysis was performed at the end of each qPCR run, and single dissociation peaks were verified for all primer pairs. As additional verification, all primer pairs were noted to generate single amplicons of the expected sizes by agarose gel electrophoresis.
RT-PCR
Complementary DNA (cDNA) prepared from spleen and thymus was amplified using a standard KAPA2G reaction (Kapa Biosystems, KK5532) with 1 μL cDNA input. Primers to amplify mouse Runx1 exon 6 were Runx352_F (5′-CACTCTGACCATCACCGTCTT-3′) and Runx352_R (5′-GGATCCCAGGTACTGGTAGGA-3′). Thermocycling was performed using an Applied Biosystems ProFlex machine: 95°C for 2 minutes; 32 cycles of 95°C for 20 seconds, 60°C for 20 seconds, and 72°C for 20 seconds; 72°C for 2 minutes; and a 4°C hold. Amplified cDNAs were run on a 10% TBE-acrylamide gel, stained with EtBr, and quantified using a BioRad ChemiDoc MP imaging platform.
Gene expression analysis
For gene expression profiling, bulk splenic Tconv cells from control and CKO littermates were flow sorted to obtain a high purity (>99%) CD4+CD25– population. CD4+ cell activation was achieved by incubating 1 million sorted Tconv cells with 3 μL CD3ε/CD28 mouse T-activator beads (Thermofisher, 11456D) per 1 mL T cell media for 6 hours followed immediately by pelleting then disruption and freezing in TRIzol. TruSeq stranded mRNA sequencing libraries were generated from 100 nanograms of total RNA (RIN range 5.0–8.7, median 7.4, average 7.0) (GeneWiz, South Plainfield, NJ). Libraries were indexed, pooled and sequenced using Illumina HiSeq 2 × 150 bp paired end sequencing. Datasets are available at the Gene Expression Omnibus (GEO) database under accession number (GEO: GSE197059). RNA-sequencing reads were trimmed using Trimmomatic v.0.36. The trimmed reads were mapped to the Mus musculus GRCm38 reference genome using STAR v.2.5.2b. For differential gene expression analysis, uniquely aligned RNA-seq reads were calculated using featureCounts from the Subread package v.1.5.2. Differentially expressed genes across different conditions were determined using the limma voom pipeline in edgeR. Genes quantified at < 1 count per million (CPM) were removed for being lowly expressed and then read counts were log2 transformed and normalized by the TMM procedure. Significantly differentially expressed genes were identified using the moderated t-statistic and adjusted for multiple hypothesis testing using the Benjamini-Hochberg procedure. A false discovery rate threshold of <0.05 was used to determine significance. Gene ontology analysis was performed using the PantherDB resource.78 Specifically, the set of significantly differentially expressed genes with FDR <0.05 was compared against a reference list of all genes in the Mus musculus genome for evidence of overrepresentation using gene groupings as defined in the “PANTHER GO-Slim Biological Process” set. Statistical significance was determined using Fishers Exact Test with FDR correction. MAJIQ version 2.0 was used for the analysis of alternative splicing. RNA sequencing reads overlapping annotated and de novo exon junctions were quantified using the MAJIQ Builder. For each Local Splicing Variation (LSV), percent selected index (PSI, denoted Ψ) and changes in Ψ between each junction across analysis groups were calculated using the MAJIQ deltapsi function. LSVs were determined to harbor a significantly changing splice junction if the probability that Ψ for any junction in the LSV was >20% changed across different analysis groups.42
Quantification and statistical analysis
Data were analyzed using Graphpad software version 7 (La Jolla, CA). All data were tested for normal distribution of variables, and normally distributed data were displayed as means ± SD. Comparisons between two groups were assessed with Student’s t-test, or unpaired t-test corrected for multiple comparisons using the Benjamini and Hochberg false discovery rate methodology (FDR<0.05) in all other cases.79
Acknowledgments
We are grateful for productive discussions with Stephen Liebhaber, Edward Behrens, Andrew Wells, Florin Tuluc, David Piccoli, and Robert Baldassano.
This work was supported by the Children’s Hospital of Philadelphia Research Institute (L.R.G.) and the Children's Hospital of Philadelphia Inflammatory Bowel Disease Center (M.M.).
Author contributions
Conceptualization, M.M., U.H.B., and L.R.G.; methodology, M.M., T.A., U.H.B., and L.R.G.; formal analysis, M.M., D.S.M.L., T.A., and L.R.G.; investigation, M.M., G.A., A.K., N.N., and B.J.W.; writing (original draft), M.M., D.S.M.L., and L.R.G.; writing (review and editing), T.A., U.H.B., and L.R.G.; supervision, L.R.G.; funding acquisition, L.R.G.
Declaration of interests
We have no conflicts of interest to declare.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105860.
Supplemental information
Data and code availability
RNA-seq data was generated in this study. Datasets are available at the Gene Expression Omnibus (GEO) database under accession number GSE197059. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. This paper does not report original code.
References
- 1.Zhu J., Yamane H., Paul W.E. Differentiation of effector CD4 T cell populations (∗) Annu. Rev. Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivanov P., Anderson P. Post-transcriptional regulatory networks in immunity. Immunol. Rev. 2013;253:253–272. doi: 10.1111/imr.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kafasla P., Skliris A., Kontoyiannis D.L. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat. Immunol. 2014;15:492–502. doi: 10.1038/ni.2884. [DOI] [PubMed] [Google Scholar]
- 4.Keene J.D. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 5.Vogel K.U., Bell L.S., Galloway A., Ahlfors H., Turner M. The RNA-binding proteins Zfp36l1 and Zfp36l2 enforce the thymic β-selection checkpoint by limiting DNA damage response signaling and cell cycle progression. J. Immunol. 2016;197:2673–2685. doi: 10.4049/jimmunol.1600854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner M., Díaz-Muñoz M.D. RNA-binding proteins control gene expression and cell fate in the immune system. Nat. Immunol. 2018;19:120–129. doi: 10.1038/s41590-017-0028-4. [DOI] [PubMed] [Google Scholar]
- 7.Díaz-Muñoz M.D., Turner M. Uncovering the role of RNA-binding proteins in gene expression in the immune system. Front. Immunol. 2018;9:1094. doi: 10.3389/fimmu.2018.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z., Yin W., Zhu L., Li J., Yao Y., Chen F., Sun M., Zhang J., Shen N., Song Y., Chang X. Iron drives T helper cell pathogenicity by promoting RNA-binding protein PCBP1-mediated proinflammatory cytokine production. Immunity. 2018;49:80–92.e7. doi: 10.1016/j.immuni.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Pratama A., Ramiscal R.R., Silva D.G., Das S.K., Athanasopoulos V., Fitch J., Botelho N.K., Chang P.P., Hu X., Hogan J.J., et al. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity. 2013;38:669–680. doi: 10.1016/j.immuni.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Yiakouvaki A., Dimitriou M., Karakasiliotis I., Eftychi C., Theocharis S., Kontoyiannis D.L. Myeloid cell expression of the RNA-binding protein HuR protects mice from pathologic inflammation and colorectal carcinogenesis. J. Clin. Invest. 2012;122:48–61. doi: 10.1172/JCI45021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George C.X., Samuel C.E. Host response to polyomavirus infection is modulated by RNA adenosine deaminase ADAR1 but not by ADAR2. J. Virol. 2011;85:8338–8347. doi: 10.1128/JVI.02666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodson D.J., Janas M.L., Galloway A., Bell S.E., Andrews S., Li C.M., Pannell R., Siebel C.W., MacDonald H.R., De Keersmaecker K., et al. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat. Immunol. 2010;11:717–724. doi: 10.1038/ni.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frischmeyer-Guerrerio P.A., Montgomery R.A., Warren D.S., Cooke S.K., Lutz J., Sonnenday C.J., Guerrerio A.L., Dietz H.C. Perturbation of thymocyte development in nonsense-mediated decay (NMD)-deficient mice. Proc. Natl. Acad. Sci. USA. 2011;108:10638–10643. doi: 10.1073/pnas.1019352108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makeyev A.V., Liebhaber S.A. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philpott C.C. Coming into view: eukaryotic iron chaperones and intracellular iron delivery. J. Biol. Chem. 2012;287:13518–13523. doi: 10.1074/jbc.R111.326876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makeyev A.V., Liebhaber S.A. Identification of two novel mammalian genes establishes a subfamily of KH-domain RNA-binding proteins. Genomics. 2000;67:301–316. doi: 10.1006/geno.2000.6244. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhury A., Hussey G.S., Ray P.S., Jin G., Fox P.L., Howe P.H. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghanem L.R., Kromer A., Silverman I.M., Chatterji P., Traxler E., Penzo-Mendez A., Weiss M.J., Stanger B.Z., Liebhaber S.A. The poly(C) binding protein Pcbp2 and its retrotransposed derivative Pcbp1 are independently essential to mouse development. Mol. Cell Biol. 2016;36:304–319. doi: 10.1128/MCB.00936-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji X., Jha A., Humenik J., Ghanem L.R., Kromer A., Duncan-Lewis C., Traxler E., Weiss M.J., Barash Y., Liebhaber S.A. RNA-binding proteins PCBP1 and PCBP2 are critical determinants of murine erythropoiesis. Mol. Cell Biol. 2021;41 doi: 10.1128/MCB.00668-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji X., Park J.W., Bahrami-Samani E., Lin L., Duncan-Lewis C., Pherribo G., Xing Y., Liebhaber S.A. αCP binding to a cytosine-rich subset of polypyrimidine tracts drives a novel pathway of cassette exon splicing in the mammalian transcriptome. Nucleic Acids Res. 2016;44:2283–2297. doi: 10.1093/nar/gkw088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji X., Wan J., Vishnu M., Xing Y., Liebhaber S.A. αCP Poly(C) binding proteins act as global regulators of alternative polyadenylation. Mol. Cell Biol. 2013;33:2560–2573. doi: 10.1128/MCB.01380-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji X., Kong J., Liebhaber S.A. An RNA-protein complex links enhanced nuclear 3' processing with cytoplasmic mRNA stabilization. EMBO J. 2011;30:2622–2633. doi: 10.1038/emboj.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji X., Kong J., Carstens R.P., Liebhaber S.A. The 3' untranslated region complex involved in stabilization of human alpha-globin mRNA assembles in the nucleus and serves an independent role as a splice enhancer. Mol. Cell Biol. 2007;27:3290–3302. doi: 10.1128/MCB.02289-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chkheidze A.N., Liebhaber S.A. A novel set of nuclear localization signals determine distributions of the alphaCP RNA-binding proteins. Mol. Cell Biol. 2003;23:8405–8415. doi: 10.1128/MCB.23.23.8405-8415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yabe-Wada T., Philpott C.C., Onai N. PCBP2 post-transcriptionally regulates sortilin expression by binding to a C-rich element in its 3' UTR. FEBS open bio. 2020;10:407–413. doi: 10.1002/2211-5463.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y.S., Khan R.A., Zhang Y., Xiao S., Wang M., Hansen D.K., Jayaram H.N., Antony A.C. Incrimination of heterogeneous nuclear ribonucleoprotein E1 (hnRNP-E1) as a candidate sensor of physiological folate deficiency. J. Biol. Chem. 2011;286:39100–39115. doi: 10.1074/jbc.M111.230938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghanem L.R., Kromer A., Silverman I.M., Ji X., Gazzara M., Nguyen N., et al. Poly(C)-Binding protein Pcbp2 enables differentiation of definitive erythropoiesis by directing functional splicing of the Runx1 transcript. Mol. Cell Biol. 2018;38 doi: 10.1128/MCB.00175-18. e001755–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A.H., Speck N.A. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komeno Y., Yan M., Matsuura S., Lam K., Lo M.C., Huang Y.J., Tenen D.G., Downing J.R., Zhang D.E. Runx1 exon 6-related alternative splicing isoforms differentially regulate hematopoiesis in mice. Blood. 2014;123:3760–3769. doi: 10.1182/blood-2013-08-521252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka Y., Joshi A., Wilson N.K., Kinston S., Nishikawa S., Göttgens B. The transcriptional programme controlled by Runx1 during early embryonic blood development. Dev. Biol. 2012;366:404–419. doi: 10.1016/j.ydbio.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichikawa M., Asai T., Saito T., Seo S., Yamazaki I., Yamagata T., Mitani K., Chiba S., Ogawa S., Kurokawa M., Hirai H. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 32.Ansa-Addo E.A., Huang H.C., Riesenberg B., Iamsawat S., Borucki D., Nelson M.H., Nam J.H., Chung D., Paulos C.M., Liu B., et al. RNA binding protein PCBP1 is an intracellular immune checkpoint for shaping T cell responses in cancer immunity. Sci. Adv. 2020;6:eaaz3865. doi: 10.1126/sciadv.aaz3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniuchi I., Osato M., Egawa T., Sunshine M.J., Bae S.C., Komori T., Ito Y., Littman D.R. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 34.Egawa T., Tillman R.E., Naoe Y., Taniuchi I., Littman D.R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu F.C., Shapiro M.J., Dash B., Chen C.C., Constans M.M., Chung J.Y., Romero Arocha S.R., Belmonte P.J., Chen M.W., McWilliams D.C., Shapiro V.S. An essential role for the transcription factor Runx1 in T cell maturation. Sci. Rep. 2016;6 doi: 10.1038/srep23533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boise L.H., Minn A.J., Noel P.J., June C.H., Accavitti M.A., Lindsten T., Thompson C.B. CD28 costimulation can promote T cell survival by enhancing the expression of Bcl-XL. Immunity. 1995;3:87–98. doi: 10.1016/1074-7613(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 37.Smith C.L., Eppig J.T. The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009;1:390–399. doi: 10.1002/wsbm.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castro W., Chelbi S.T., Niogret C., Ramon-Barros C., Welten S.P.M., Osterheld K., Wang H., Rota G., Morgado L., Vivier E., et al. The transcription factor Rfx7 limits metabolism of NK cells and promotes their maintenance and immunity. Nat. Immunol. 2018;19:809–820. doi: 10.1038/s41590-018-0144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh H., Ghosh S. NF-κB: roles and regulation in different CD4(+) T-cell subsets. Immunol. Rev. 2013;252:41–51. doi: 10.1111/imr.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni T., Yang W., Han M., Zhang Y., Shen T., Nie H., Zhou Z., Dai Y., Yang Y., Liu P., et al. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res. 2016;44:6817–6829. doi: 10.1093/nar/gkw591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ullrich S., Guigó R. Dynamic changes in intron retention are tightly associated with regulation of splicing factors and proliferative activity during B-cell development. Nucleic Acids Res. 2020;48:1327–1340. doi: 10.1093/nar/gkz1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaquero-Garcia J., Barrera A., Gazzara M.R., González-Vallinas J., Lahens N.F., Hogenesch J.B., Lynch K.W., Barash Y. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife. 2016;5 doi: 10.7554/eLife.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin J.W., Spinler K.R., Swift J., Chasis J.A., Mohandas N., Discher D.E. Lamins regulate cell trafficking and lineage maturation of adult human hematopoietic cells. Proc. Natl. Acad. Sci. USA. 2013;110:18892–18897. doi: 10.1073/pnas.1304996110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagai Y., Garrett K.P., Ohta S., Bahrun U., Kouro T., Akira S., Takatsu K., Kincade P.W. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eden K. Adoptive transfer colitis. Methods Mol. Biol. 2019;1960:207–214. doi: 10.1007/978-1-4939-9167-9_18. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh D., Srivastava G.P., Xu D., Schulz L.C., Roberts R.M. A link between SIN1 (MAPKAP1) and poly(rC) binding protein 2 (PCBP2) in counteracting environmental stress. Proc. Natl. Acad. Sci. USA. 2008;105:11673–11678. doi: 10.1073/pnas.0803182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong W.F., Kurokawa M., Satake M., Kohu K. Down-regulation of Runx1 expression by TCR signal involves an autoregulatory mechanism and contributes to IL-2 production. J. Biol. Chem. 2011;286:11110–11118. doi: 10.1074/jbc.M110.166694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen M.J., Yokomizo T., Zeigler B.M., Dzierzak E., Speck N.A. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voon D.C.C., Hor Y.T., Ito Y. The RUNX complex: reaching beyond haematopoiesis into immunity. Immunology. 2015;146:523–536. doi: 10.1111/imm.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levanon D., Groner Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- 51.Ito Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv. Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- 52.Durst K.L., Hiebert S.W. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene. 2004;23:4220–4224. doi: 10.1038/sj.onc.1207122. [DOI] [PubMed] [Google Scholar]
- 53.Lutterbach B., Westendorf J.J., Linggi B., Isaac S., Seto E., Hiebert S.W. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 2000;275:651–656. doi: 10.1074/jbc.275.1.651. [DOI] [PubMed] [Google Scholar]
- 54.Kim W.Y., Sieweke M., Ogawa E., Wee H.J., Englmeier U., Graf T., Ito Y. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 1999;18:1609–1620. doi: 10.1093/emboj/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cowley S.M., Iritani B.M., Mendrysa S.M., Xu T., Cheng P.F., Yada J., et al. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol. Cell Biol. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. https://journals.asm.org/doi/10.1128/MCB.25.16.6990-7004.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zamisch M., Tian L., Grenningloh R., Xiong Y., Wildt K.F., Ehlers M., Ho I.C., Bosselut R. The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J. Exp. Med. 2009;206:2685–2699. doi: 10.1084/jem.20092024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bae S.C., Ogawa E., Maruyama M., Oka H., Satake M., Shigesada K., Jenkins N.A., Gilbert D.J., Copeland N.G., Ito Y. PEBP2 alpha B/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol. Cell Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242-3252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho J.J.D., Robb G.B., Tai S.C., Turgeon P.J., Mawji I.A., Man H.S.J., Marsden P.A. Active stabilization of human endothelial nitric oxide synthase mRNA by hnRNP E1 protects against antisense RNA and microRNAs. Mol. Cell Biol. 2013;33:2029–2046. doi: 10.1128/MCB.01257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murahashi M., Simizu S., Morioka M., Umezawa K. Identification of poly(rC) binding protein 2 (PCBP2) as a target protein of immunosuppressive agent 15-deoxyspergualin. Biochem. Biophys. Res. Commun. 2016;476:445–449. doi: 10.1016/j.bbrc.2016.05.142. [DOI] [PubMed] [Google Scholar]
- 60.Magee C.N., Boenisch O., Najafian N. The role of costimulatory molecules in directing the functional differentiation of alloreactive T helper cells. Am. J. Transplant. 2012;12:2588–2600. doi: 10.1111/j.1600-6143.2012.04180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chambers C.A., Kuhns M.S., Egen J.G., Allison J.P. CTLA-4-mediated inhibition in regulation of T cell responses: mechanisms and manipulation in tumor immunotherapy. Annu. Rev. Immunol. 2001;19:565–594. doi: 10.1146/annurev.immunol.19.1.565. [DOI] [PubMed] [Google Scholar]
- 62.Wolf Y., Anderson A.C., Kuchroo V.K. TIM3 comes of age as an inhibitory receptor. Nat. Rev. Immunol. 2020;20:173–185. doi: 10.1038/s41577-019-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Witherden D.A., Boismenu R., Havran W.L. CD81 and CD28 costimulate T cells through distinct pathways. J. Immunol. 2000;165:1902–1909. doi: 10.4049/jimmunol.165.4.1902. [DOI] [PubMed] [Google Scholar]
- 64.Cevik S.I., Keskin N., Belkaya S., Ozlu M.I., Deniz E., Tazebay U.H., Erman B. CD81 interacts with the T cell receptor to suppress signaling. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waldman A.D., Fritz J.M., Lenardo M.J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Georgiadou D., Boussata S., Keijser R., Janssen D.A.M., Afink G.B., van Dijk M. Knockdown of splicing complex protein PCBP2 reduces extravillous trophoblast differentiation through transcript switching. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.671806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mallory M.J., Allon S.J., Qiu J., Gazzara M.R., Tapescu I., Martinez N.M., Fu X.D., Lynch K.W. Induced transcription and stability of CELF2 mRNA drives widespread alternative splicing during T-cell signaling. Proc. Natl. Acad. Sci. USA. 2015;112:E2139–E2148. doi: 10.1073/pnas.1423695112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diaz-Muñoz M.D., Osma-Garcia I.C. The RNA regulatory programs that govern lymphocyte development and function. Wiley Interdiscip. Rev. RNA. 2022;13:e1683. doi: 10.1002/wrna.1683. [DOI] [PubMed] [Google Scholar]
- 69.Nicolet B.P., Zandhuis N.D., Lattanzio V.M., Wolkers M.C. Sequence determinants as key regulators in gene expression of T cells. Immunol. Rev. 2021;304:10–29. doi: 10.1111/imr.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blanchett S., Boal-Carvalho I., Layzell S., Seddon B. NF-κB and extrinsic cell death pathways - entwined do-or-die decisions for T cells. Trends Immunol. 2021;42:76–88. doi: 10.1016/j.it.2020.10.013. [DOI] [PubMed] [Google Scholar]
- 71.Van Nostrand E.L., Freese P., Pratt G.A., Wang X., Wei X., Xiao R., Blue S.M., Chen J.Y., Cody N.A.L., Dominguez D., et al. A large-scale binding and functional map of human RNA-binding proteins. Nature. 2020;583:711–719. doi: 10.1038/s41586-020-2077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frey A.G., Nandal A., Park J.H., Smith P.M., Yabe T., Ryu M.S., Ghosh M.C., Lee J., Rouault T.A., Park M.H., Philpott C.C. Iron chaperones PCBP1 and PCBP2 mediate the metallation of the dinuclear iron enzyme deoxyhypusine hydroxylase. Proc. Natl. Acad. Sci. USA. 2014;111:8031–8036. doi: 10.1073/pnas.1402732111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ryu M.S., Zhang D., Protchenko O., Shakoury-Elizeh M., Philpott C.C. PCBP1 and NCOA4 regulate erythroid iron storage and heme biosynthesis. J. Clin. Invest. 2017;127:1786–1797. doi: 10.1172/JCI90519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eri R., McGuckin M.A., Wadley R. T cell transfer model of colitis: a great tool to assess the contribution of T cells in chronic intestinal inflammation. Methods Mol. Biol. 2012;844:261–275. doi: 10.1007/978-1-61779-527-5_19. [DOI] [PubMed] [Google Scholar]
- 76.Akimova T., Xiao H., Liu Y., Bhatti T.R., Jiao J., Eruslanov E., Singhal S., Wang L., Han R., Zacharia K., et al. Targeting sirtuin-1 alleviates experimental autoimmune colitis by induction of Foxp3+ T-regulatory cells. Mucosal Immunol. 2014;7:1209–1220. doi: 10.1038/mi.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Akimova T., Levine M.H., Beier U.H., Hancock W.W. Standardization, evaluation, and area-under-curve analysis of human and murine Treg suppressive function. Methods Mol. Biol. 2016;1371:43–78. doi: 10.1007/978-1-4939-3139-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mi H., Thomas P. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 2009;563:123–140. doi: 10.1007/978-1-60761-175-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data was generated in this study. Datasets are available at the Gene Expression Omnibus (GEO) database under accession number GSE197059. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. This paper does not report original code.