Abstract
The presence of Salmonella in hatchlings is the single most important risk factor for the introduction of Salmonella into poultry farms, and resistant strains are particularly worrisome, as they could affect treatment outcomes in humans infected through consumption of contaminated poultry products. This study estimated Salmonella prevalence, determined resistance profiles of strains recovered from hatchlings in Nigeria, and determined genetic relatedness between hatchling strains and strains from poultry farms. In this study, 300 fecal samples were collected. Salmonella was isolated by culture and confirmed by PCR, and isolates were tested for susceptibility to antimicrobials by the disk diffusion method. Strains were pair-end sequenced, and genomes were used to obtain serotypes and antibiotic resistance genes. Whole-genome based phylogenetic analysis was used to determine genetic relatedness between these isolates and strains from previously characterized older chicken within the same geographical area. A prevalence of 10.7% was obtained belonging to 13 Salmonella serovars. Resistance to kanamycin (30/32), ciprofloxacin (22/32), nalidixic acid (22/32), and sulfonamides (22/32) were the most commonly observed phenotypic resistances. Twenty-two (68.8%) isolates showed multidrug resistance. In silico predictions identified 36 antimicrobial resistance genes. Four (12.5%) and 22 (68.8%) strains showed point mutations in gyrA and parC. Commonly observed acquired resistance genes included sul1, sul2, sul3, and tet(A) as well as a variety of aminoglycoside-modifying genes. Eleven (34.4%) isolates were predicted to have genes that confer resistance to fosfomycin (fosA7, fosB). A strain of S. Stanleyville was predicted to have optrA, which confers resistance to furazolidone. Strains of S. Kentucky, S. Muenster, and S. Menston obtained from hatchlings showed close genetic relatedness by having less than 30 SNPs difference to strains recovered from chickens at farms previously receiving hatchlings from the same sources.
Key words: prevalence, Salmonella, antibiotic resistance, hatchling, whole genome
INTRODUCTION
Nontyphoidal Salmonella are annually responsible for an estimated 95.1 million human cases of salmonellosis globally, with an estimated 50,771 fatal cases (Hoelzer et al., 2011; Stanaway et al., 2019). Certain Salmonella serovars are causing high economic losses in poultry due to high mortality, morbidity, and impaired production performance. However, the majority of serovars have little impact on poultry health and production, but they may cause human disease through consumption of contaminated poultry products.
Hatcheries occupy a central position between breeders and poultry farms, and they can be key in the transmission of zoonotic pathogens to poultry farms. Ensuring that eggs and hatchlings are free of Salmonella is thus of critical importance for controlling Salmonella in poultry (Bailey et al., 1998). Salmonella can contaminate eggs by both vertical and horizontal transmission, and hatchlings borne from such eggs can be a source of infection, introducing Salmonella into farms (Chao et al., 2007; Osman et al., 2018). Vertical transmission occurs in the ovary or oviduct where Salmonella directly infect the eggs (Wigley et al., 2001). Horizontal transmission occur by fecal contamination of the shell during the passage through the cloaca (Schoeni et al., 1995; Chao et al., 2007).
Next-generating sequencing techniques have become the current standard for profiling of bacterial genomes (Yu et al., 2020). It is increasingly used in public health laboratories for typing and characterizing foodborne pathogens (Kang et al., 2022). This sequence base typing method allow for serotyping, antimicrobial resistance, virulence profiling, and subtyping in a single workflow and provides high resolution and precision (Inns et al., 2017). This technique has been successfully used in Salmonella subtyping for monitoring and traceability (Rounds et al., 2020).
Little is known about the occurrence of Salmonella in hatcheries in Nigeria and to what extent serovars and genotypes found in hatchlings are like those present in poultry farms. Knowledge of such transmission dynamics of Salmonella is essential when establishing effective Salmonella control programs in the poultry industry. In the present study, we aimed to determine the prevalence and serovars of Salmonella in day-old chickens when sold at hatcheries in Nigeria as well as the susceptibility and resistance genes to commonly used antimicrobials. We also assessed the genetic relatedness between Salmonella strains isolated from day-old chickens to those found at commercial poultry farms in Nigeria using whole genome sequence (WGS) analysis.
MATERIALS AND METHODS
Ethical Considerations
Ethical approval for collecting fecal samples from day-old chickens and for conducting the study was obtained from Sokoto State Ministry of Animal Health and Fisheries Developments, Kebbi State Ministry of Animal Health and Husbandry, and Zamfara State Directorate of Animal Health and Livestock Development (Reference numbers MAH&FD/VET/166/11, MAHF/VET/VOL1, and DAHLD/SUB/VET/VOL.1, respectively).
Study Design and Description of Study Area
Cross-sectional sampling was employed to estimate Salmonella prevalence in day-old chicks that arrived from different hatcheries to poultry stores and farms in 3 states of the northwest Nigeria. Poultry stores are registered commercial outlets located across Nigeria that merchant day-old chicks, veterinary medicine, and farm equipment to farmers. Day-old chicks are transported in boxes from hatcheries in the Southwestern Nigeria to the 3 study states with a transport time of up to 48 h. A few of the hatcheries delivering day-old chicks were in the north, and from these locations, transportation time was less than 24 h.
Fecal Sample Collection
Fecal samples were obtained from multiple locations in transport boxes of newly arrived day-old chicks (one box per single hatchery source per farm) into sterile sample bottles. Each transport box measured 49 cm × 29 cm × 13 cm (length × width × depth) and contained a maximum of 51-day-old chicks from the same hatchery source. Three hundred (300) composite fecal samples (approximately 3 g) representing an individual box were collected from January to July 2018. Each sample bottle was labeled and transported on ice packs to the laboratory where microbiological analysis was initiated within 24 h of sample collection.
Salmonella Isolation and Identification
Standard bacteriological methods were used to enrich for, isolate and identify Salmonella strains (Jibril et al., 2020). Briefly, samples were investigated for presence of Salmonella spp. according to the International Standard Organization (2017) method. Presumptive Salmonella isolates were characterized by biochemical tests using commercially available media (Oxoid, Basingstoke, UK), and subjected to serological confirmation by slide agglutination test using polyvalent Salmonella antisera (SSI, Copenhagen, Denmark). As a final confirmation, Salmonella presumptive isolates were subjected to PCR identification using the invA-based method (Rahn et al., 1992; Waghamare et al., 2018).
Whole Genome Sequencing and Quality Control of Sequences by Bioinformatics Analysis
A total of 32 PCR-confirmed Salmonella strains were subjected to whole genome sequence analysis. Colonies were cultured overnight in 5 mL Luria broth (LB) (Difco, Detroit, Michigan) and genomic DNA was extracted using Promega Maxwell DNA automatic extraction robot Maxwell RSC Cultured Cells (Maxwell RSC-16, Madison, Wisconsin) as previously described (Jibril et al., 2020). Sequencing libraries were prepared using Nextera XT kits according to instructions given by the manufacturer (Illumina, San Diego, CA). Genomes were sequenced on an Illumina MiSeq platform using paired-end chemistry (2 × 250-bp) (Illumina). De novo genome assembly of sequences was done using SPAdes version 3.9 (Nurk et al., 2013). The quality of the assembled genome was evaluated using QUAST (Gurevich et al., 2013). Genome characteristics and accession number for each genome are indicated in the Supplementary File 1. The draft genome sequences of day-old chicks are available at the European Nucleotide Archive under study accession numbers PRJEB38609.
Phenotypic and Genotypic Antimicrobial Resistance Analysis
Isolates were phenotypically tested for susceptibility to 11 commonly used antimicrobials representing 9 classes of antimicrobials (ampicillin, cefotaxime, chloramphenicol, gentamicin, ciprofloxacin, sulfonamides, kanamycin, meropenem, nalidixic acid, tetracycline, and trimethoprim) by the Kirby-Bauer disk diffusion test using Clinical and Laboratory Standards Institute (CLSI) protocols (Clinical Laboratory Standards Institute CLSI, 2017) as previously described (Jibril et al., 2021a). A strain was defined as multidrug resistant (MDR) when it was resistant to at least one drug in at least 3 different antimicrobial classes (Magiorakos et al., 18AD). In silico prediction of antimicrobial resistance genes (ARGs) in assemblies of draft genomes was done using Resfinder version 3.2 (Zankari et al., 2013).
In Silico Serotyping and Prediction of Sequence Types
Draft genome sequences were submitted to Salmonella in silico typing resources (SISTR) through the web application programming interface (Yoshida et al., 2016; Robetson et al., 2018). Serotypes were predicted and sequence types (STs) assigned (Achtman et al., 2012).
Phylogenetic Analysis
To determine relatedness between Salmonella isolates from day-old chickens and strains of matching serotypes obtained previously from poultry on the same farm and likewise sequenced (maximum 43 d prior to obtaining isolates from day-old chicken) (Jibril et al., 2020), genetic relatedness was assessed by determining SNP differences. The strains where matching isolates were available (n = 17) belonged to the serovars S. Isangi (ST-216), S. Kentucky (ST-198), S. Larochelle (ST-22), S. Menston (ST-7742), S. Muenster (ST-321), and S. Virchow (ST-6166). Draft assemblies of genomes together with a reference sequence obtained from Salmonella Typhimurium ATCC 14028 (4,889,644 bp) were submitted to CSI Phylogeny 1.4 available at https://cge.cbs.dtu.dk/services/CSIPhylogeny/ to get whole genome SNPs difference (Kaas et al., 9AD). Newick output from CSI phylogeny with strain metadata was imported to iTOL to reconstruct phylogenetic tree (Letunic and Bork, 47AD).
Data and Statistical Analysis
Level of concordance between phenotypic resistance and in silico gene predictions was assessed using Cohen's kappa statistic in SPSS version 28 (IBM, New York, NY). Level of agreement was interpreted as described by McHugh, 22AD. The value of P < 0.05 at 95% confidence interval was considered significant.
RESULTS
Prevalence and Salmonella Serovars
Thirty-two out of the 300 composite fecal samples, each representing one box per shipment to a farm and poultry vendors, were found positive for Salmonella, corresponding to a prevalence of 10.7% in transports of day-old chicks originating from 7 major hatcheries in Nigeria. Samples from hatcheries F (15.0%), G (15.0%), and C (12.7%) had the highest prevalence of Salmonella (Table 1). S. Stanleyville (ST-2562) occurred with the highest occurrence (21.9% of positive boxes) in day-old chicks, followed by S. Isangi (ST-216) (12.5%), S. Virchow (ST-6166) (9.4%), S. Muenster (ST-321) (9.4%), S. Larochelle (ST-22) (9.4%), and S. Menston (ST-7742) (9.4%) (Table 2).
Table 1.
Prevalence of Salmonella in hatchlings in transport boxes immediately after leaving hatcheries in Nigeria.
| Hatchery | Sample site |
Positive |
|||
|---|---|---|---|---|---|
| sources | Number of boxes sampled | Farm | Poultry vendor | Number | (%) |
| Hatchery A | 23 | 7 | 16 | 1 | 4.3 |
| Hatchery B | 31 | 10 | 21 | 2 | 6.5 |
| Hatchery C | 55 | 11 | 44 | 7 | 12.7 |
| Hatchery D | 87 | 29 | 58 | 8 | 9.2 |
| Hatchery E | 24 | 5 | 19 | 2 | 8.3 |
| Hatchery F | 20 | 15 | 5 | 3 | 15.0 |
| Hatchery G | 60 | 14 | 46 | 9 | 15.0 |
| Total | 300 | 91 | 209 | 32 | 10.7 |
Table 2.
Distribution of Salmonella serovars obtained from boxes with hatchlings in Nigeria.
| Serotypes | Number of strains | Percentage of strains (%) | Antigenic formula | ST |
|---|---|---|---|---|
| S. Chomedey | 1 | 3.1 | C2-C3:z10:e,n,z15 | 3961 |
| S. Haelsingborg | 1 | 3.1 | C1:m,t | 7748 |
| S. Isangi | 4 | 12.5 | C1:d:1,5 | 216 |
| S. Ituri | 2 | 6.3 | B:z10:1,5 | 4498 |
| S. Kentucky | 1 | 3.1 | C2-C3:i:z6 | 198 |
| S. Larochelle | 3 | 9.4 | C1:e,h:1,2 | 22 |
| S. Menston | 3 | 9.4 | C1:g,[s],t:- | 7742 |
| S. Mississipi | 1 | 3.1 | I13,23:d:- | 2194 |
| S. Muenster | 3 | 9.4 | E1:e,h:1,5 | 321 |
| S. Nigeria | 2 | 6.3 | C1:r:1,6 | 4911 |
| S. Stanleyville | 7 | 21.9 | B,z4,z23:1,2 | 2562 |
| S. Takoradi | 1 | 3.1 | C2-C3:i:1,5 | 531 |
| S. Virchow | 3 | 9.4 | C1:r:1,2 | 6166 |
Phenotypic Susceptibility to Antimicrobials and Presence of ARGs
All 32 isolates showed resistance to one or more of the 11 antimicrobials tested. High-level of resistance was observed to kanamycin (30/32; 93.8%), nalidixic acid (22/32; 68.8%), ciprofloxacin (22/32; 68.8%), and sulfonamides (22/32; 68.8%). All strains of S. Isangi were resistant to kanamycin, ciprofloxacin, chloramphenicol, trimethoprim and nalidixic acid. One strain of S. Muenster (ST-321) was resistant to all antimicrobials except meropenem. Twenty-two (68%) of the isolates were MDR; this included strains of the serovars S. Stanleyville (42.9% of strains belonging to this serovar), S. Isangi (100%), S. Muenster (ST-321) (100%), and S. Nigeria (100%) (Table 3).
Table 3.
Phenotypic antimicrobial resistance in Salmonella serovars from boxes of hatchlings in Nigeria.
| Serovar | N | Number of isolates resistant to antimicrobials |
MDR* | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | GEN | KAN | CTX | CIP | SUL | TET | CHL | TMP | NAL | MEM | N | ||
| Chomedey | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 |
| Haelsingborg | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Isangi | 4 | 2 | 3 | 4 | 1 | 3 | 3 | 2 | 4 | 4 | 3 | 0 | 4 |
| Ituri | 2 | 0 | 0 | 2 | 0 | 2 | 1 | 0 | 0 | 0 | 2 | 0 | 0 |
| Kentucky | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 |
| Larochelle | 3 | 0 | 2 | 3 | 1 | 2 | 2 | 0 | 0 | 1 | 3 | 0 | 3 |
| Menston | 3 | 0 | 1 | 3 | 2 | 2 | 1 | 1 | 0 | 0 | 1 | 1 | 1 |
| Mississippi | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 |
| Muenster | 3 | 2 | 2 | 3 | 1 | 2 | 3 | 3 | 1 | 1 | 3 | 0 | 3 |
| Nigeria | 2 | 0 | 0 | 2 | 0 | 1 | 2 | 2 | 0 | 0 | 2 | 0 | 2 |
| Stanleyville | 7 | 1 | 2 | 5 | 5 | 4 | 3 | 0 | 1 | 2 | 3 | 0 | 3 |
| Takoradi | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| Virchow | 3 | 0 | 3 | 3 | 0 | 2 | 2 | 0 | 0 | 0 | 1 | 1 | 1 |
| Total | 32 | 7 | 18 | 30 | 10 | 22 | 22 | 11 | 7 | 9 | 22 | 3 | 22 |
Abbreviations: AMP, ampicillin; CHL, chloramphenicol; CIP, ciprofloxacin; CTX, cefotaxim; GEN, gentamicin; KAN, kanamicin; MEM, meropenem; NAL, nalidixic acid; SUL, sulfonamides; TET, tetracycline; TMP, trimethoprim. *MDR, multidrug resistance.
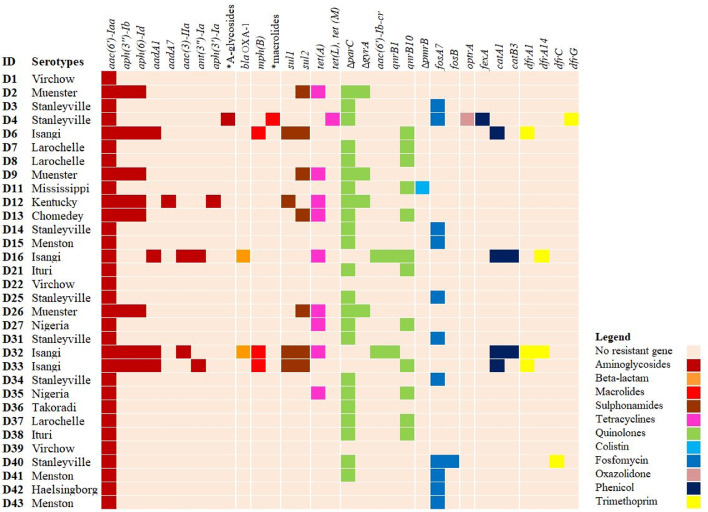
In silico predictions using ResFinder identified 36 ARGs, and it detected point mutations in DNA gyrase (gyrA) and DNA topoisomerase (parC) in 4 (12.5%) and 22 (68.8%) strains, respectively. Point mutations were also observed in pmrB in S. Mississippi, and the specific SNP identified (pmrB; Thr147Pro) is associated with colistin resistance (Jayol et al., 2014). Some isolates (12/32) carried the qnrB10 gene associated with low level ciprofloxacin resistance, or with amplifying resistance mediated by SNPs in quinolone-resistance determining regions of quinolone targets (Veldman et al., 2011). All isolates contained the aac (6′)-Iaa, which confers resistance to kanamycin. Eight isolates carried aph (3") - Ib and aph (6) - Id that both confer resistance to streptomycin. Two S. Isangi strains had blaOXA-1, which confers resistance to ampicillin (Karami et al., 2008). Eleven isolates (34.4%) carried fosfomycin resistance genes (fosA7, fosB), including 7 strains of S. Stanleyville, 3 of S. Menston and one S. Haelsingborg. One S. Stanleyville strain carried multiple ARGs belonging to the classes of furazolidone (optrA), aminoglycosides (str, ant(9)-Ia, ant(6)-Ia), and macrolides (erm(A), erm (B), Inu (B), Isa(A), Isa(E)). Genes conferring resistance to sulfonamides (sul1, sul2), chloramphenicol (fexA, catA1, catB3) and trimethoprim (dfrA, dfrG dfrC) were shown in 25.0%, 15.6%, and 18.8% of the isolates, respectively. One isolate each of S. Virchow, S. Mississippi and S. Menston were phenotypically resistant to meropenem, however, genetic determinants known to encode meropenem resistance were not detected. Genes conferring resistance to more than 3 classes of antimicrobials were observed in S. Kentucky (ST-198), S. Isangi, S. Stanleyville and S. Muenster (ST-321) (Figure 1).
Figure 1.
Presence of predicted antimicrobial resistance genes in Salmonella isolates from boxes of hatchlings in Nigeria. The row represents isolates ID, while columns represent predicted antimicrobial resistant genes. *A-glycosides; other aminoglycoside resistant genes predicted (str, ant (9)-Ia, ant (6)-Ia). *Macrolides; other macrolide resistant genes predicted (erm(A), erm(B), Inu(B), Isa(A), Isa(E)).
Comparison between phenotypic and genotypic resistances revealed a substantial agreement with statistical significance between resistance genes identified and phenotypic resistance to tetracycline (k = 0.78, P = 0.001), moderate agreement for chloramphenicol (k = 0.59, P = 0.001), and fair agreement for ampicillin (k = 0.38, P = 0.006) and trimethoprim (k = 0.39, P = 0.02) (Supplementary Table 1).
Phylogenetic Comparison
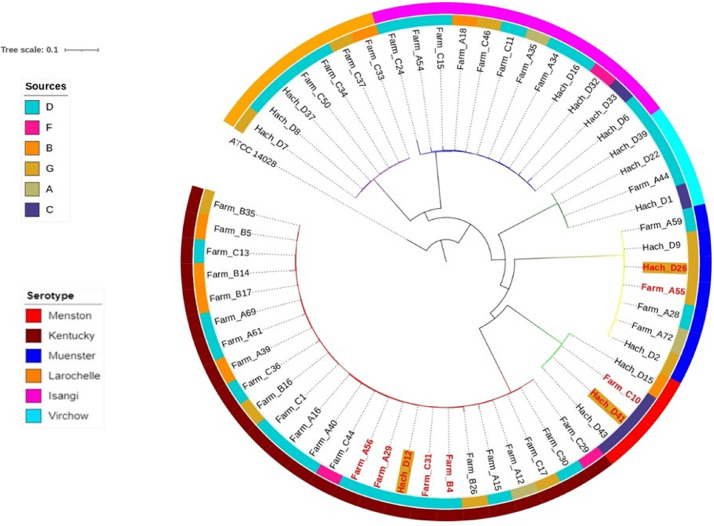
Seventeen isolates of 6 serovars obtained from day-old chicks (S. Kentucky, S. Muenster, S. Larochelle, S. Virchow, S. Isangi, S. Menston) matches according to serovar with strains previously obtained from chickens raised at the same farms (Jibril et al.‚ 2020). Comparison was done to these isolates by SNP analysis. The percentage of reference genome covered by all isolates was 81.0%, with 3,946,763 SNP positions. Strains clustered according to serovar to form 6 clades supported with a bootstrap value of 100%, confirming the phylogenetic relevance of serovars (Figure 2). Result from the SNP matrix analysis (Supplementary File 2) showed that strains of S. Kentucky had SNPs differences ranging from 2 to 2,889. One S. Kentucky strain from day-old chicks aligned closely with a strain from poultry at the same farm (22 SNPs apart). Strains of S. Menston had SNPs difference ranging from 27 to 2,475. A strain from day-old chicks of this serovar differed in only 27 SNPs from a strain isolated at a poultry farm that previously had sourced chicken from the same hatchery. SNP difference for S. Muenster ranged from 6 to 2,396 and notably 2 strains from day-old chickens showed less than 15 SNP differences with strains from poultry on a farm that had previously sourced day-old chickens from the same hatchery (Figure 2). Within the S. Kentucky, S. Menston and S. Muenster clades, 4 isolates from hatcheries differed ≤30 SNPs from a farm isolate.
Figure 2.
Whole genome SNP rooted phylogenetic tree of matching Salmonella serovars obtained from boxes of hatchlings and farms in Nigeria. Each branch in separate colors represent a serovar. The strain used as reference genome is Salmonella Typhimurium ATCC 14028. Inner ring represents hatchery sources, where farmers obtained their chicks and each color bar represent hatchery as indicated in the legend located in the top left. The outer ring represents matching serovars of Salmonella from hatchlings and older chickens. Red colored node labels are strains with low number of SNPs between strains from hatchling and older chickens. Hatchling strains are with gray background.
DISCUSSION
The Salmonella prevalence of 10.7% found in our day-old chicks are somewhat lower than estimates from previous studies, which reported 27.0% of newly arrived day-old chicks in the United States (Habing et al., 2015) and 18.6% of imported day-old chicks in Egypt (Osman et al., 2010). A lower prevalence (7.5%) of Salmonella was however estimated in a more recent study of day-old chicks in Egypt (Sedeik et al., 2019). In any case, the presence of Salmonella in day-old chicks can jeopardize any control measures to prevent Salmonella at the farms (Bailey et al., 1998). We have recently reported presence of S. Kentucky (ST-198), S. Isangi, S. Larochelle and S. Muenster (ST-321) in commercial poultry farms in the same geographical area investigated in the current study. While detection of the same serovars in day-old chicks and in poultry farms suggests that hatcheries could be sources and may play an important role in the transmission of Salmonella, genome comparison or other detailed typing of isolates is required to firmly conclude on the transmission dynamics.
The public health importance of the trade of Salmonella-positive day-old chicks cannot be underscored as nontyphoidal Salmonella illness in humans have been linked to zoonotic transmission from day-old chicks via poultry products to humans (Behravesh et al., 2014), and day-old chicks have previously been associated with multistate outbreaks of salmonellosis (Habing et al., 2015). Some of the serovars we detected (S. Kentucky, S. Isangi and S. Muenster) in the day-old chicks are of public health importance as they have been implicated in human salmonellosis (van Cauteren et al., 2009; Suleyman et al., 2016). Furthermore, isolation of S. Stanleyville and S. Virchow, which have been associated with zoonotic transmission of invasive nontyphoidal Salmonella (iNTS), is indeed worrisome. S. Stanleyville is mostly detected in poultry and cattle (European Food Safety Authority and European Center for Disease Prevention and Control ECDC, 2015; Cui et al., 2016; Fagbamila et al., 2017 ), but reports have associated this serovar with invasive human infections in West and Central Africa (Tennant et al., 2010; Mossoro-Kpinde et al., 2015), and several outbreaks in Europe (Cibin et al., 2019). S. Virchow seems associated with poultry (Andoh et al., 2016; Bertrand et al., 2006) and accounted for 25% of NTS human infections in Australia (Parisi et al., 2018).
Phenotypically, all isolates showed resistance to at least one antimicrobial with 68% of these isolates being MDR. Multiple ARGs were identified with good agreement to phenotypic resistance. Resistance to aminoglycosides, sulfonamides, ciprofloxacin, chloramphenicol, and trimethoprim were commonly observed and genes and point mutations encoding these resistances were found in the isolates. Resistances to third generation cephalosporins and ciprofloxacin were combined in 6 isolates (18.8%) representing 5 different serovars. Empiric treatment of human salmonellosis often uses these antimicrobials. Notably, 34.4% of isolates belonging to S. Stanleyville, S. Menston and S. Haelsingborg were shown to contain fosfomycin resistance genes, however we did not test for phenotypic resistance against this drug. Fosfomycin is a broad spectrum antimicrobial that is often used as drug of last resort in the treatment of chronic bacterial infections in human and is presently being promoted as a treatment of choice for life-threatening sepsis in African settings (Falagas et al., 2016; Kane et al., 2021; Obiero et al., 2022). Detection of fosfomycin resistance in day-old chicks is worrisome, because it indicates possible administration of this antimicrobial to day-old chicks or grandparent stock. Reports have documented that antimicrobials are sometime administered in hatcheries to control early mortality rate associated with colibacillosis. The antimicrobials are administered in ovo or by subcutaneous injection to hatchlings, together with Marek's disease vaccination (McReynolds et al., 79AD; Heinrich et al., 2013; Baron et al., 2014). In some instances, this include the use of third generation cephalosporins, and this has been strongly linked to the increase in resistance to this class of antimicrobials in poultry production (Dutil et al., 2010; Persoons et al., 2011; Baron et al., 2014). However, information on the use of third generation cephalosporins and fosfomycin in the Nigerian hatchery industry is unavailable and needs investigation. In addition, a strain of S. Stanleyville contained multiple ARGs including optrA (furazolidone). Detection of furazolidone resistance genes in day-old chicks is worrisome, as this drug is banned for use in Nigeria (National Agency for Food and Drug Administration and Control NAFDAC, 2017). Jibril et al., 2021b demonstrated in a recent study that 5% of poultry farms used products containing nitrofurantoin, a furazolidone antibiotic.
We observed that strains from day-old chicks were of the same serovars as strains isolated from poultry at the same farm. SNPs differences of less than 30, typically used to infer isolate point source for enterobacteriales (den Bakker et al., 2011; Taylor et al., 2015; Gymoese et al., 2017; Kudirkiene et al., 86AD) was observed in S. Kentucky, S. Menston and S. Muenster between strains from day-old chicks and poultry, while strains of the same serovars from different farms had higher number of SNPs. This could be a strong indication that hatchling was the source of Salmonella in the farms; however, the comparison of strains obtained in 2 cross sectional studies (the current study and the study of Jibril et al., 2020) was not ideal. For firm conclusions, a longitudinal study in the farms after receiving the hatchlings would have been optimal. Different SNP threshold have been used to establish genetic relatedness between strains of Salmonella strains (den Bakker et al., 2011; Taylor et al., 2015; Gymoese et al., 2017; Kudirkiene et al., 86AD). With a strict cut-off of 30 SNPs, isolates obtained from hatchlings and poultry on the same farm often showed less than 30 SNPs difference, which was far less than the diversity observed in strain, which were not from the same hatchery. This suggests clonal transmission between hatcheries and farms is likely, therefore underscoring the important of hatcheries in the transmission dynamics of Salmonella infection in poultry farms (Martelli et al., 45AD; Sharma et al., 2018).
CONCLUSIONS
In conclusion, moderate prevalence of Salmonella was reported in hatchlings leaving the hatcheries. Notably, strains were often MDR, and resistant genes to fosfomycin was detected in S. Stanleyville (ST-2562), S. Menston (ST-321), and S. Haelsingborg (ST-7748). Furthermore, this study demonstrated S. Kentucky (ST-198), S. Muenster (ST-321), and S. Menston (ST-321) obtained in hatchlings were closely related strains obtained from poultry in the farms which sourced hatchlings from same hatchery.
Acknowledgments
This work was part-supported by an African Research Leader Award to INO from the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement that is also part of the EDCTP2 program supported by the European Union. Authors express their profound appreciation to farm managers and poultry store merchants for their support during sample collection.
Disclosures
Authors declared there is no conflicting interest that could influence the publication of this study.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.102427.
Appendix. Supplementary materials
REFERENCES
- Achtman M., Wain J., Weill F.X., Nair S., Zhou Z., Sangal V., Krauland M.G., Hale J.L., Harbottle H., Uesbeck A., Dougan G., Harrison L.H., Brisse S. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh L.A., Dalsgaard A., Obiri-Danso K., Newman M.J., Barco L., Olsen J.E. Prevalence and antimicrobial resistance of Salmonella serovars isolated from poultry in Ghana. Epidemiol. Infect. 2016;144:3288–3299. doi: 10.1017/S0950268816001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S., Jouy E., Larvor E., Eono F., Bougeard S., Kempf I. Impact of third-generation-cephalosporin administration in hatcheries on fecal Escherichia coli antimicrobial resistance in broilers and layers. Antimicrob. Agents Chemother. 2014;58:5428–5434. doi: 10.1128/AAC.03106-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behravesh C.B., Brinson D., Hopkins B.A., Gomez T.M. Backyard poultry flocks and salmonellosis: A recurring, yet preventable public health challenge. Clin. Infect. Dis. 2014;58:1432–1438. doi: 10.1093/cid/ciu067. [DOI] [PubMed] [Google Scholar]
- Bertrand S., Weill F.X., Cloeckaert A., Vrints M., Mairiaux E., Fraud K., Dierick K., Wildemauve C., Godard C., Butaye P., Imberechts H., Grimont P.A.D., Collard J.M. Clonal emergence of extended-spectrum β-lactamase (CTX-M-2)-producing Salmonella enterica serovar virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003) J. Clin. Microbiol. 2006;44:2897–2903. doi: 10.1128/JCM.02549-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M.R., Hsien C.H., Yeh C.M., Chou S.J., Chu C., Su Y.C., Yu C.Y. Assessing the prevalence of Salmonella enterica in poultry hatcheries by using hatched eggshell membranes. Poult. Sci. 2007;86:1651–1655. doi: 10.1093/ps/86.8.1651. [DOI] [PubMed] [Google Scholar]
- Cibin V., Busetti M., Longo A., Petrin S., Knezevich A., Ricci A., Barco L., Losasso C. Whole genome sequencing of Salmonella Serovar Stanleyville from two Italian outbreaks resulted in unexpected genomic diversity within and between outbreaks. Foodborne Pathog. Dis. 2019;16:307–308. doi: 10.1089/fpd.2018.2564. [DOI] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. Wayne; Pennsylvania USA: 2017. [Google Scholar]
- Cui M., Xie M., Qu Z., Zhao S., Wang J., Wang Y., He T., Wang H., Zuo Z., Wu C. Prevalence and antimicrobial resistance of Salmonella isolated from an integrated broiler chicken supply chain in Qingdao, China. Food Control. 2016;62:270–276. [Google Scholar]
- Bailey J.S., Cason J.A., Cox N.A. Effect of Salmonella in young chicks on competitive exclusion treatment. Poult. Sci. 1998;77:394–399. doi: 10.1093/ps/77.3.394. [DOI] [PubMed] [Google Scholar]
- den Bakker H.C., Swit A.I.M., Cummings C.A., Hoelzer K., Degoricija L., Rodriguez-Rivera L.D., Wright E.M., Fang R., Davis M., Root T., Schoonmaker-Bopp D., Musser K.A., Villamil E., Waechter H.N., Kornstein L., Furtado M.R., Wiedmann M. A whole-genome single nucleotide polymorphism-based approach to trace and identify outbreaks linked to a common Salmonella enterica subsp. enterica serovar montevideo pulsed-field gel electrophoresis type. Appl. Environ. Microbiol. 2011;77:8648–8655. doi: 10.1128/AEM.06538-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutil L., Irwin R., Finley R., Ng L.K., Avery B., Boerlin P., Bourgault A.M., Cole L., Daignault D., Desruisseau A., Demczuk W., Hoang L., Horsman G.B., Ismail J., Jamieson F., Maki A., Pacagnella A., Pillai D.R. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 2010;16:48–54. doi: 10.3201/eid1601.090729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagbamila I.O., Barco L., Mancin M., Kwaga J., Ngulukun S.S., Zavagnin P., Lettini A.A., Lorenzetto M., Abdu P.A., Kabir J., Umoh J., Ricci A. and Muhammad M., Salmonella serovars and their distribution in Nigerian commercial chicken layer farms, PLoS One, 12, 2017, 9;12:e0173097. [DOI] [PMC free article] [PubMed]
- Falagas M.E., Vouloumanou E.K., Samonis G., Vardakas K.Z. Fosfomycin fgfd. Clin. Microbiol. Rev. 2016;29:321–347. doi: 10.1128/CMR.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. F1000Prime recommendation of: QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymoese P., Sørensen G., Litrup E., Olsen J.E., Nielsen E.M., Torpdahl M. Investigation of outbreaks of Salmonella enterica serovar typhimurium and its monophasic variants using whole-genome sequencing, Denmark. Emerg. Infect. Dis. 2017;23:1631–1639. doi: 10.3201/eid2310.161248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habing G.G., Kessler S.E., Mollenkopf D.F., Wittum T.E., Anderson T.C., Barton Behravesh C., Joseph L.A., Erdman M.M. Distribution and diversity of Salmonella strains in shipments of hatchling poultry, United States, 2013. Zoonoses Public Health. 2015;62:375–380. doi: 10.1111/zph.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich K., Chan D., Fussell R.J., Kay J.F., Sharman M. Food Additives and Contaminants - Part A Chemistry, Analysis, Control, Exposure and Risk Assessment. 2013. Can the unauthorised use of ceftiofur be detected in poultry? pp. 1733–1738. [DOI] [PubMed] [Google Scholar]
- Hoelzer K., Switt A.I.M., Wiedmann M. Animal contact as a source of human non-typhoidal salmonellosis. Vet. Res. 2011;42:34. doi: 10.1186/1297-9716-42-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inns T., Ashton P.M., Herrera-Leon S., Lighthill J., Foulkes S., Jombart T., Rehman Y., Fox A., Dallman T., De Pinna E., Browning L., Coia J.E., Edeghere O., Vivancos R. Prospective use of whole genome sequencing (WGS) detected a multi-country outbreak of Salmonella Enteritidis. Epidemiol. Infect. 2017;145:289–298. doi: 10.1017/S0950268816001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority and European Center for Disease Prevention and Control (ECDC), 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014, EFSA J., 2015, 13:4329.
- International Standard Organization. (2017) Microbiology of the Food Chain-Horizontal method for the detection, enumeration and serotyping of Salmonella-Part 1: Detection of Salmonella. 2017.
- Jayol A., Poirel L., Brink A., Villegas M.V., Yilmaz M., Nordmann P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 2014;58:4762–4766. doi: 10.1128/AAC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jibril A.H., Okeke I.N., Dalsgaard A., Kudirkiene E., Akinlabi O.C., Bello M.B., Olsen J.E. Prevalence and risk factors of Salmonella in commercial poultry farms in Nigeria. PLoS One. 2020;15 doi: 10.1371/journal.pone.0238190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jibril A.H., Okeke I.N., Dalsgaard A., Menéndez V.G., Olsen J.E. Genomic analysis of antimicrobial resistance and resistance plasmids in Salmonella serovars from poultry in Nigeria. Antibiotics. 2021;10:1–22. doi: 10.3390/antibiotics10020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jibril A.H., Okeke I.N., Dalsgaard A., Olsen J.E. Association between antimicrobial usage and resistance in Salmonella from poultry farms in Nigeria. BMC Vet. Res. 2021;17:1–10. doi: 10.1186/s12917-021-02938-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas R.S., Leekitcharoenphon P., Aarestrup F.M., Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane Z., Gastine S., Obiero C., Williams P., Murunga S., Thitiri J., Ellis S., Correia E., Nyaoke B., Kipper K., Van Den Anker J., Sharland M., Berkley J.A., Standing J.F. IV and oral fosfomycin pharmacokinetics in neonates with suspected clinical sepsis. J. Antimicrob. Chemother. 2021;76:1855–1864. doi: 10.1093/jac/dkab083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Wang M., Meng C., Li A., Jiao X., Pan Z. Prevalence and whole-genome sequencing analysis of Salmonella reveal its spread along the duck production chain. Poult. Sci. 2022;101 doi: 10.1016/j.psj.2022.101993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karami N., Hannoun C., Adlerberth I., Wold A.E. Colonization dynamics of ampicillin-resistant Escherichia coli in the infantile colonic microbiota. J. Antimicrob. Chemother. 2008;62:703–708. doi: 10.1093/jac/dkn263. [DOI] [PubMed] [Google Scholar]
- Kudirkiene E., Sørensen G., Torpdahl M., de Knegt L.V., Nielsen L.R., Rattenborg E., Ahmed S., Olsen J.E. Epidemiology of Salmonella enterica serovar Dublin in Cattle and humans in Denmark, 1996 to 2016: a retrospective whole-genome-based study. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.01894-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., Paterson D.L., Rice L.B., Stelling J., Struelens M.J., Vatopoulos A., Weber J.T., Monnet D.L. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- Martelli F., Birch C., Davies R.H. Observations on the distribution and control of Salmonella in commercial duck hatcheries in the UK. Avian Pathol. 2016;45:261–266. doi: 10.1080/03079457.2016.1146820. [DOI] [PubMed] [Google Scholar]
- McHugh M.L. Interrater reliability: the kappa statistic. Biochem. Med. 2012;22:276–282. [PMC free article] [PubMed] [Google Scholar]
- McReynolds J.L., Caldwell D.Y., Barnhart E.T., Deloach J.R., McElroy A.P., Moore R.W., Hargis B.M., Caldwell D.J. The effect of in ovo or day-of-match subcutaneous antibiotic administration on competitive exclusion culture (PREEMPTTM) establishment in neonatal chickens. Poult. Sci. 2000;79:1524–1530. doi: 10.1093/ps/79.11.1524. [DOI] [PubMed] [Google Scholar]
- Mossoro-Kpinde C.D., Manirakiza A., Mbecko J.R., Misatou P., Le Faou A., Frank T. Antimicrobial resistance of enteric Salmonella in Bangui, Central African Republic. J. Trop. Med. 2015 doi: 10.1155/2015/483974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S., Bankevich A., Antipov D., Gurevich A., Korobeynikov A., Lapidus A., Prjibelsky A., Pyshkin A., Sirotkin A., Sirotkin Y., Stepanauskas R., McLean J., Lasken R., Clingenpeel S.R., Woyke T., Tesler G., Alekseyev M.A., Pevzner P.A. Assembling genomes and mini-metagenomes from highly chimeric reads. J Comput Biol. 2013,;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obiero C.W., Williams P., Murunga S., Thitiri J., Omollo R., Walker A.S., Egondi T., Nyaoke B., Correia E., Kane Z., Gastine S., Kipper K., Standing J.F., Ellis S., Sharland M., Berkley J.A. Randomised controlled trial of fosfomycin in neonatal sepsis: pharmacokinetics and safety in relation to sodium overload. Arch. Dis. Child. 2022;107:802–810. doi: 10.1136/archdischild-2021-322483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman K.M., Kappell A.D., Elhadidy M., Elmougy F., El-Ghany W.A.A., Orabi A., Mubarak A.S., Dawoud T.M., Hemeg H.A., Moussa I.M.I., Hessain A.M., Yousef H.M.Y. Poultry hatcheries as potential reservoirs for antimicrobial-resistant Escherichia coli: a risk to public health and food safety. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-23962-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman K.M., Yousef A.M.M., Aly M.M., Radwan M.I. Salmonella spp. infection in imported 1-day-old chicks, ducklings, and Turkey poults: A public health risk. Foodborne Pathog. Dis. 2010;7:383–390. doi: 10.1089/fpd.2009.0358. [DOI] [PubMed] [Google Scholar]
- Parisi A., Crump J.A., Stafford R., Glass K., Howden B.P., Kirk M.D. Increasing incidence of invasive nontyphoidal Salmonella infections in Queensland, Australia, 2007-2016. PLoS Negl. Trop. Dis. 2018;13:2007–2016. doi: 10.1371/journal.pntd.0007187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Agency for Food and Drug Administration and Control (NAFDAC), Veterinary Medicine and Allied Products (VMAP), 2017. Accessed Dec. 25, 2022. https://www.nafdac.gov.ng/wp-content/uploads/Files/Resources/Directorate_Resources/VMAP/LIST-OF-BANNED-VETERINARY-DRUGS.pdf.
- Rounds JM, Taylor AJ, Eikmeier D, Nichols MM, Lappi V, Wirth SE, Boxrud DJ, Smith KE, Medus C. Prospective Salmonella Enteritidis surveillance and outbreak detection using whole genome sequencing, Minnesota 2015-2017. Epidemiol Infect. 2020:148:e254. [DOI] [PMC free article] [PubMed]
- Persoons D., Haesebrouck F., Smet A., Herman L., Heyndrickx M., Martel A., Catry B., Berge A.C., Butaye P. and Dewulf J., Risk factors for ceftiofur resistance in Escherichia coli from Belgian broilers, Epidemiol Infect, 2011. 139:765–771. [DOI] [PubMed]
- Rahn K., De Grandis S.A., Clarke R.C., McEwen S.A., Galin J.E., Ginocchio C., Curtiss R., Gyles C.L. Amplification of invA gene of Salmonella by polymerase chain reaction (PCR) as a specific method for detection of Salmonellae. Mol. Cell. Probes. 1992;6:271–279. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- Robertson J., Yoshida C., Kruczkiewicz P., Nadon C., Nichani A., Taboada E.N., Nash J.H.E. Comprehensive assessment of the quality of Salmonella whole genome sequence data available in public sequence databases using the Salmonella in silico typing resource (SISTR) Microb. Genom. 2018;4 doi: 10.1099/mgen.0.000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeni J.L., Glass K.A., McDermott J.L., Wong A.C.L. Growth and penetration of Salmonella Enteritidis, Salmonella Heidelberg and Salmonella Typhimurium in eggs. Int. J. Food Microbiol. 1995;24:385–396. doi: 10.1016/0168-1605(94)00042-5. [DOI] [PubMed] [Google Scholar]
- Sedeik M.E., El-shall N.A., Awad A.M., Elfeky S.M., Abd El-Hack M.E., Hussein E.O.S., Alowaimer A.N., Swelum A.A. Isolation, conventional and molecular characterization of Salmonella spp. from newly hatched broiler chicks. AMB Express. 2019;9(1):136. doi: 10.1186/s13568-019-0821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Erdman M.M., Muñoz-Vargas L., Mollenkopf D.F., Habing G.G. Changes in the prevalence, genotypes and antimicrobial resistance phenotypes of non-typhoidal Salmonella recovered from mail-order hatchling poultry sold at US feed stores, 2013–2015. Zoonoses Public Health. 2018;65:e102–e112. doi: 10.1111/zph.12416. [DOI] [PubMed] [Google Scholar]
- Stanaway J.D., Parisi A., Sarkar K., Blacker B.F., Reiner R.C., Hay S.I., Nixon M.R., Dolecek C., James S.L., Mokdad A.H., Abebe G., Ahmadian E., Alahdab F., Alemnew B.T.T., Alipour V., Allah Bakeshei F., Animut M.D., Ansari F., Arabloo J.…Crump J.A. The global burden of non-typhoidal Salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019;19:1312–1324. doi: 10.1016/S1473-3099(19)30418-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suleyman G., Perri M., Vager D., Samuel L., Zervos M.J., Alangaden G., Tibbetts R.J. Characterization of Salmonella Isangi possessing a CTX-M15 ESBL associated with an outbreak in a US Hospital. Diagn. Microbiol. Infect. Dis. 2016;85:386–390. doi: 10.1016/j.diagmicrobio.2016.03.016. [DOI] [PubMed] [Google Scholar]
- Taylor A.J., Lappi V., Wolfgang W.J., Lapierre P., Palumbo M.J., Medus C., Boxrud D. Characterization of foodborne outbreaks of Salmonella enterica serovar enteritidis with whole-genome sequencing single nucleotide polymorphism-based analysis for surveillance and outbreak detection. J. Clin. Microbiol. 2015;53:3334–3340. doi: 10.1128/JCM.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant S.M., Diallo S., Levy H., Livio S., Sow S.O., Tapia M., Fields P.I., Mikoleit M., Tamboura B., Kotloff K.L., Nataro J.P., Galen J.E., Levine M.M. Identification by PCR of non-typhoidal Salmonella enterica serovars associated with invasive infections among febrile patients in Mali. PLoS Negl.Trop. Dis. 2010;4 doi: 10.1371/journal.pntd.0000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Cauteren D., Jourdan-da Silva N., Weill F.X., King L., Brisabois A., Delmas G., Vaillant V., de Valk H. Outbreak of Salmonella enterica serotype Muenster infections associated with goat’s cheese, France, March 2008. Euro Surveill. 2009;14 doi: 10.2807/ese.14.31.19290-en. [DOI] [PubMed] [Google Scholar]
- Veldman K., Cavaco L.M., Mevius D., Battisti A., Franco A., Botteldoorn N., Bruneau M., Perrin-Guyomard A., Cerny T., de Frutos Escobar C., Guerra B., Schroeter A., Gutierrez M., Hopkins K., Myllyniemi A.L., Sunde M., Wasyl D., Aarestrup F.M. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J. Antimicrob. Chemother. 2011;66:1278–1286. doi: 10.1093/jac/dkr084. [DOI] [PubMed] [Google Scholar]
- Waghamare R.N., Paturkar A.M., Vaidya V.M., Zende R.J., Dubal Z.N., Dwivedi A., Gaikwad R.V. Phenotypic and genotypic drug resistance profile of Salmonella serovars isolated from poultry farm and processing units located in and around Mumbai city, India. Vet. World. 2018;11:1682–1688. doi: 10.14202/vetworld.2018.1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley P., Berchieri A., J., Page K.L., Smith A.L., Barrow P.A. Salmonella enterica serovar pullorum persists in splenic macrophages and in the reproductive tract during persistent, disease-free carriage in chickens. Infect. Immun. 2001;69:7873–7879. doi: 10.1128/IAI.69.12.7873-7879.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C.E., Kruczkiewicz P., Laing C.R., Lingohr E.J., Gannon V.P.J., Nash J.H.E., Taboada E.N. The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One. 2016;11:1–17. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Du F., Ban R., Zhang Y. SimuSCoP: reliably simulate Illumina sequencing data based on position and context dependent profiles. BMC Bioinf. 2020;21:331. doi: 10.1186/s12859-020-03665-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Kaas R.S., Seyfarth A.M., Agersø Y., Lund O., Larsen M.V., Aarestrup F.M. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013;68:771–777. doi: 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.