Savolitinib plus osimertinib combination therapy has an acceptable safety profile and demonstrated antitumor activity in patients with MET-amplified, EGFR-mutated advanced NSCLC who experienced disease progression on prior EGFR tyrosine kinase inhibitor therapy.
Abstract
MET-inhibitor and EGFR tyrosine kinase inhibitor (EGFR-TKI) combination therapy could overcome acquired MET-mediated osimertinib resistance. We present the final phase Ib TATTON (NCT02143466) analysis (Part B, n = 138/Part D, n = 42) assessing oral savolitinib 600 mg/300 mg once daily (q.d.) + osimertinib 80 mg q.d. in patients with MET-amplified, EGFR-mutated (EGFRm) advanced non–small cell lung cancer (NSCLC) and progression on prior EGFR-TKI. An acceptable safety profile was observed. In Parts B and D, respectively, objective response rates were 33% to 67% and 62%, and median progression-free survival (PFS) was 5.5 to 11.1 months and 9.0 months. Increased antitumor activity may occur with MET copy number ≥10. EGFRm circulating tumor DNA clearance on treatment predicted longer PFS in patients with detectable baseline ctDNA, while acquired resistance mechanisms to osimertinib + savolitinib were mediated by MET, EGFR, or KRAS alterations.
Significance:
The savolitinib + osimertinib combination represents a promising therapy in patients with MET-amplified/overexpressed, EGFRm advanced NSCLC with disease progression on a prior EGFR-TKI. Acquired resistance mechanisms to this combination include those via MET, EGFR, and KRAS. On-treatment ctDNA dynamics can predict clinical outcomes and may provide an opportunity to inform earlier decision-making.
This article is highlighted in the In This Issue feature, p. 1
INTRODUCTION
Osimertinib is a third-generation, irreversible, oral EGFR tyrosine kinase inhibitor (EGFR-TKI) that potently and selectively inhibits both EGFR-TKI–sensitizing and EGFR T790M resistance mutations and has demonstrated efficacy in non–small cell lung cancer (NSCLC), including central nervous system (CNS) metastases (1–5). Osimertinib is considered the preferred first-line option for patients with metastatic NSCLC with EGFR-sensitizing mutations (6, 7) and has recently been approved as an adjuvant treatment in patients with resectable EGFR mutation–positive (EGFRm) NSCLC (Tagrisso US PI, UK SmPC, EU SmPC, and Tagrisso approval in China; ref. 8). Despite osimertinib's proven efficacy in the first-line metastatic setting, many patients with EGFRm NSCLC develop disease progression on osimertinib, with current data suggesting that approximately 25% of these patients develop MET amplification or other MET-based acquired resistance mechanisms, as previously reported based on genetic testing (9–13).
Combination therapy comprising a MET inhibitor and an EGFR-TKI is a potential treatment strategy that could overcome acquired MET-mediated resistance to EGFR-TKIs including osimertinib (14–16). Savolitinib, an oral, potent, and highly selective MET-TKI, has demonstrated preliminary clinical activity in advanced solid tumors (17–20). Preliminary investigation of the combination was studied in Part A of this study: TATTON (NCT02143466; ref. 21).
TATTON was a four-part study (Parts A–D). In the sections of TATTON described here (Parts B and D), patients with EGFRm advanced NSCLC with MET amplification/overexpression following disease progression on a prior EGFR-TKI received savolitinib plus osimertinib (21, 22). Part B was split into three cohorts by prior therapy and T790M status. Part D patients had not received prior third-generation EGFR-TKI and were T790M-negative. Preliminary data from TATTON Parts B and D suggested that savolitinib plus osimertinib may overcome MET-based resistance to EGFR-TKIs in NSCLC (22).
Further data are needed to develop or optimize a MET-amplified/overexpressed biomarker that can identify patients and predict for potential combination benefit. Several challenges exist, as MET expression and MET copy-number changes are continuous variables with no standardized cutoffs (23–25) and different subsets of MET-based resistance are not detected consistently across different testing methods; therefore, using a single assay may lead to patients with tumors sensitive to MET inhibitors being missed. Serial circulating tumor DNA (ctDNA) changes can also provide information on the likelihood of durable benefit with combination therapy, intra/intertumoral heterogeneity that may affect durability, and primary and acquired mediators of resistance (26). In patients with EGFRm NSCLC treated with first- or second-line osimertinib, ctDNA clearance has been shown to correlate with longer progression-free survival (PFS; refs. 27, 28).
Herein, we present the final safety and antitumor activity data from two phase Ib TATTON study expansion cohorts (TATTON Parts B and D). We also report the results of exploratory analyses examining tumor response by the different MET detection methods used in TATTON, acquired resistance mechanisms, and ctDNA clearance of EGFR sensitizing mutations (Ex19del/L858R) and its correlation with PFS at two doses of savolitinib [600 mg once daily (q.d.)/300 mg q.d.] plus osimertinib (80 mg q.d.).
Results
Patient Demographics
Between May 20, 2015, and a data cutoff of March 4, 2020, 138 patients with EGFRm, MET-amplified/overexpressed NSCLC were enrolled into Part B and received savolitinib plus osimertinib second line or later (Supplementary Fig. S1A). Of these patients, 69 were previously treated with a third-generation EGFR-TKI (Part B1 cohort), 51 had never been treated with a third-generation EGFR-TKI and were T790M-negative (Part B2 cohort), and 18 had never been treated with a third-generation EGFR-TKI and were T790M-positive (Part B3 cohort). Median treatment duration was 10.4 months (range, 0.3–40.8) for osimertinib and 8.7 months (range, 0–40.0) for savolitinib (Supplementary Table S1).
Between December 15, 2017, and a data cutoff of March 4, 2020, 42 patients with EGFRm, T790M-negative, MET-amplified/overexpressed NSCLC with no prior third-generation EGFR-TKI treatment were enrolled into Part D and received savolitinib 300 mg q.d. plus osimertinib 80 mg q.d. (Supplementary Fig. S1B). The median treatment duration was 9.8 months (range, 0.4–22.8) for osimertinib and 8.1 months (range, 0.3–23.1) for savolitinib (Supplementary Table S1).
Demographic and baseline disease characteristics (Table 1) were representative of the intended study population and similar across all subcohorts in Part B, with the exception of the number of prior lines of therapy; patients in Part B1 were more likely to have received ≥3 prior lines of anticancer therapy versus patients in Parts B2 and B3 (57% vs. 25% and 6%, respectively). In Part B, all patients assigned to treatment received ≥1 dose of study treatment and were included in the safety analysis set; 137 of 138 patients (99%) had one reportable pharmacokinetic (PK) concentration and were included in the PK analysis set.
Table 1.
Patient demographics and disease characteristics (safety analysis set)
| Part B: osimertinib 80 mg + savolitinib 600/300 mg | Part D: osimertinib 80 mg + savolitinib 300 mg | ||||
|---|---|---|---|---|---|
| B1. Previously received third-generation EGFR-TKI | B2. No prior third-generation EGFR-TKI, T790M-negative | B3. No prior third-generation EGFR-TKI, T790M-positive | All Part B patients | No prior third-generation EGFR-TKI, T790M-negative | |
| n = 69 | n = 51 | n = 18 | N = 138 | N = 42 | |
| Age, years | |||||
| Mean (SD) | 59 (12) | 60 (10) | 60 (9) | 59 (11) | 62 (9) |
| Median (range) | 58 (28–82) | 59 (29–92) | 60 (46–76) | 59 (28–92) | 63 (41–77) |
| Sex | |||||
| Male | 35 (51) | 17 (33) | 5 (28) | 57 (41) | 17 (40) |
| Female | 34 (49) | 34 (67) | 13 (72) | 81 (59) | 25 (60) |
| Race | |||||
| White | 21 (30) | 13 (25) | 4 (22) | 38 (28) | 14 (33) |
| Asian | 48 (70) | 38 (75) | 14 (78) | 100 (72) | 28 (67) |
| ECOG/WHO performance status | |||||
| 0 | 29 (42) | 16 (31) | 6 (33) | 51 (37) | 15 (36) |
| 1 | 40 (58) | 35 (69) | 12 (67) | 87 (63) | 27 (64) |
| Histology | |||||
| Adenocarcinoma | 69 (100) | 48 (94) | 18 (100) | 135 (98) | 40 (95) |
| Squamous and/or squamous features | 0 | 2 (4) | 0 | 2 (2) | 1 (2) |
| Small cell carcinoma | 0 | 1 (2) | 0 | 1 (1) | 1 (2) |
| Overall disease classification | |||||
| Metastatica | 69 (100) | 50 (98) | 18 (100) | 137 (99) | 42 (100) |
| Locally advancedb | 0 | 1 (2) | 0 | 1 (1) | 0 |
| Prior lines of therapyc | |||||
| 1 | 3 (4) | 29 (57) | 12 (67) | 44 (32) | 28 (67) |
| 2 | 27 (39) | 9 (18) | 5 (28) | 41 (30) | 6 (14) |
| ≥3 | 39 (57) | 13 (25) | 1 (6) | 53 (38) | 8 (19) |
NOTE: Data are n (%).
Abbreviations: ECOG, Eastern Cooperative Oncology Group; SD, standard deviation; WHO, World Health Organization.
aPatient could have any metastatic site of disease.
bPatient could have only locally advanced sites of disease.
cOnly anticancer therapies, excluding adjuvant and neoadjuvant, except if they include EGFR-TKI.
In Part D, most (67%) patients had one prior line of anticancer therapy (Table 1), and all patients assigned to treatment received ≥1 dose of study treatment and had 1 reportable PK concentration and were included in the safety analysis set and PK analysis set, respectively.
Safety
In Part B, all patients (n = 138; 100%) reported ≥1 adverse event (AE; Table 2). The most common AEs in Part B were nausea in 72 (52%) patients, decreased appetite and edema peripheral in 49 (36%) patients, and fatigue and vomiting in 48 (35%) patients (Supplementary Table S2). AEs of grade ≥3 are shown in Supplementary Table S3; of these, those considered possibly causally related to osimertinib or savolitinib are shown in Supplementary Table S4. Osimertinib was discontinued in 15 (11%) patients with AEs considered possibly causally related to study treatment, including 3 (2%) patients with pneumonitis, and 2 (1%) patients with anaphylactic reaction, drug hypersensitivity, or vomiting. Savolitinib was discontinued in 39 (28%) patients due to AEs considered possibly causally related to study treatment, including 5 (4%) patients with anaphylactic reaction and 4 (3%) patients with drug hypersensitivity or vomiting. AEs led to dose reductions or interruptions of osimertinib in 59 (43%) patients and of savolitinib in 61 (44%) patients. Overall, in Part B, serious AEs (SAE) were reported in 67 (49%) patients, the most common of which were pneumonia [7 (5%) patients], anaphylactic reaction and pneumothorax [6 (4%) patients], and dyspnea and pyrexia [5 (4%) patients]. Of interest, AEs included in the grouped term “hypersensitivity” occurred in 67 (49%) patients; these AEs were grade ≥3 in 20 (14%) patients. The only deaths due to AEs considered possibly causally related to study drug occurred in Part B2, in which one case was considered possibly related to savolitinib (acute renal failure) and one to both study treatments (unknown death).
Table 2.
AEs (safety analysis set)
| Part B: osimertinib 80 mg + savolitinib 600/300 mg | Part D: osimertinib 80 mg + savolitinib 300 mg | ||||
|---|---|---|---|---|---|
| B1. Previously received 3G EGFR-TKI | B2. No prior 3G EGFR-TKI, T790M-negative | B3. No prior 3G EGFR-TKI, T790M-positive | All patients | No prior 3G EGFR-TKI, T790M-negative | |
| Patients with an event, n (%) | n = 69 | n = 51 | n = 18 | (N = 138)a | (n = 42) |
| Any AE | 69 (100) | 51 (100) | 18 (100) | 138 (100) | 41 (98) |
| Any AE grade ≥3 | 39 (57) | 35 (69) | 12 (67) | 86 (62) | 21 (50) |
| Possibly causally related to: | |||||
| Osimertinib only | 3 (4) | 3 (6) | 2 (11) | 8 (6) | 2 (5) |
| Savolitinib only | 14 (20) | 13 (25) | 7 (39) | 34 (25) | 5 (12) |
| Osimertinib and savolitinib | 11 (16) | 17 (33) | 9 (50) | 37 (27) | 7 (17) |
| Any AE leading to death | 3 (4) | 3 (6) | 1 (6) | 7 (5) | 2 (5) |
| Any AE leading to discontinuation | |||||
| Osimertinib | 12 (17) | 9 (18) | 3 (17) | 24 (17) | 8 (19) |
| Savolitinib | 19 (28) | 20 (39) | 10 (56) | 49 (36) | 15 (36) |
| AE leading to discontinuation, possibly causally related to any treatment | |||||
| Osimertinib | 8 (12) | 6 (12) | 1 (6) | 15 (11) | 5 (12) |
| Savolitinib | 15 (22) | 17 (33) | 6 (33) | 38 (28) | 12 (29) |
| Any AE leading to interruption or reduction | |||||
| Osimertinib | 22 (32) | 25 (49) | 12 (67) | 59 (43) | 13 (31) |
| Savolitinib | 27 (39) | 25 (49) | 9 (50) | 61 (44) | 16 (38) |
| Any SAEb | 33 (48) | 27 (53) | 7 (39) | 67 (49) | 16 (38) |
| Possibly causally related to: | |||||
| Osimertinib only | 0 | 0 | 0 | 0 | 1 (2) |
| Savolitinib only | 6 (9) | 6 (12) | 3 (17) | 15 (11) | 2 (5) |
| Osimertinib and savolitinib | 5 (7) | 7 (14) | 2 (11) | 14 (10) | 4 (10) |
| Any SAE leading to discontinuation | |||||
| Osimertinib | 9 (13) | 8 (16) | 3 (17) | 20 (14) | 2 (5) |
| Savolitinib | 11 (16) | 11 (22) | 6 (33) | 28 (20) | 6 (14) |
| Any SAE leading to interruption or reduction | |||||
| Osimertinib | 10 (14) | 11 (22) | 6 (33) | 27 (20) | 10 (24) |
| Savolitinib | 9 (13) | 9 (18) | 2 (11) | 20 (14) | 7 (17) |
NOTE: Causality determined by investigator review.
Abbreviations: SAE, serious adverse event; 3G, third-generation.
aMost patients were enrolled to 600 mg savolitinib, prior to weight-based dosing implementation, but, following a protocol amendment, the final 21 patients enrolled in Part B were dosed with savolitinib by body weight as follows: patients who weighed ≤55 kg (n = 7) received 300 mg q.d., and those weighing >55 kg (n = 14) received 600 mg q.d.
bIncluding events leading to death.
Overall, 41 of 42 (98%) patients reported ≥1 AE (Table 2) in Part D. The most common AEs were nausea in 14 (33%) patients, peripheral edema in 13 (31%) patients, rash in 12 (29%) patients, and decreased appetite and paronychia in 9 (21%) patients (Supplementary Table S2). AEs of grade ≥3, including those that were considered causally related to study drug, are reported in Supplementary Tables S3 and S4. Osimertinib was discontinued in 5 (12%) patients due to AEs considered possibly causally related to study treatment, including 2 (5%) patients with interstitial lung disease and 1 patient (2%) with drug hypersensitivity, dyspnea, or pneumonitis. Savolitinib was discontinued in 12 (29%) patients due to AEs considered possibly related to study treatment, including 4 (10%) patients with drug hypersensitivity, 3 (7%) with myalgia, and 2 (5%) each with interstitial lung disease, rash, or pyrexia. The only SAEs reported in more than 1 patient were pneumonia [n = 4 (10%)] and pulmonary embolism [n = 2 (5%)]. The grouped term hypersensitivity occurred in 18 (43%) patients, with these AEs being grade ≥3 in 4 (10%) patients. Two patients died due to AEs in Part D; neither death was considered related to treatment.
Antitumor Activity
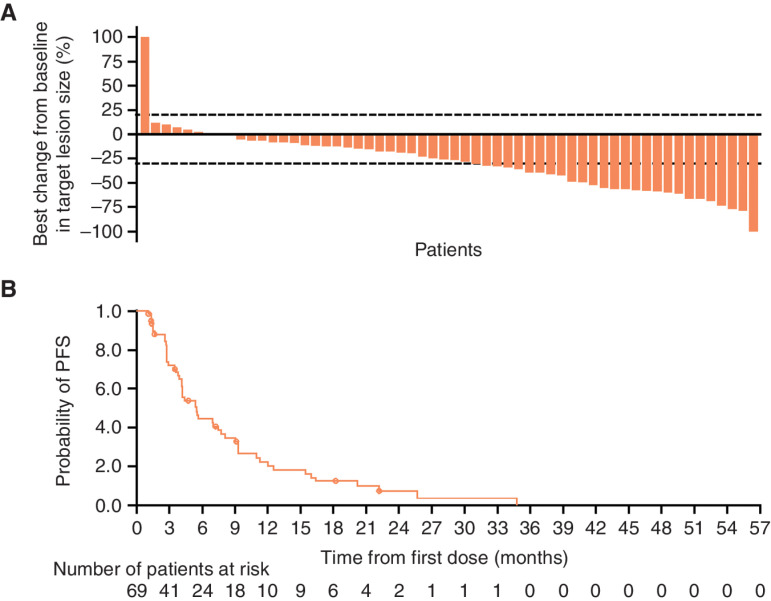
The best percentage change from baseline in target lesion size for Part B1 is shown in Fig. 1A and for Parts B2, B3, and D is shown in Supplementary Fig. S2A. Objective response rates (ORR) were 33% [95% confidence interval (CI), 22–46] in Part B1, 65% (95% CI, 50–78) in Part B2, and 67% (95% CI, 41–87) in Part B3. All confirmed responses were partial responses. The median duration of response (DoR) for patients in Parts B1, B2, and B3 was 9.5 months (95% CI, 4.2–14.7), 10.7 months (95% CI, 6.1–14.8), and 11.0 months [95% CI, 2.8–not calculated (NC)], respectively. The median PFS for patients in Parts B1 (Fig. 1B), B2, and B3 was 5.5 months (95% CI, 4.1–7.7), 9.1 months (95% CI, 5.5–12.8), and 11.1 months (95% CI, 4.1–22.1), respectively (Table 3; Supplementary Fig. S2B).
Figure 1.
A, Waterfall plot of best percentage change from baseline in target lesion size. B, Kaplan–Meier estimates of PFS for Part B subcohort B1 (evaluable response set).
Table 3.
Efficacy endpoints (evaluable for response set)
| Part B: osimertinib 80 mg + savolitinib 600/300a mg | Part D: osimertinib 80 mg + savolitinib 300 mg | ||||
|---|---|---|---|---|---|
| Previously treated with a 3G EGFR-TKI | No prior 3G EGFR-TKI, T790M-negative | No prior 3G EGFR-TKI, T790M-positive | No prior 3G EGFR-TKI, T790M-negative | ||
| Endpoint | n = 69 | n = 51 | n = 18 | n = 42 | |
| ORRb, n (%) | 23 (33) | 33 (65) | 12 (67) | 26 (62) | |
| (95% CI) | (22–46) | (50–78) | (41–87) | (46–76) | |
| Complete response | 0 | 0 | 0 | 0 | |
| Partial response | 23 (33) | 33 (65) | 12 (67) | 26 (62) | |
| Stable diseasec | 29 (42) | 12 (24) | 6 (33) | 13 (31) | |
| Progressive disease | 8 (12) | 3 (6) | 0 | 1 (2) | |
| Not evaluable | 9 (13) | 3 (6) | 0 | 2 (5) | |
| Median PFS, months (95% CI) | 5.5 (4.1–7.7) | 9.1 (5.5–12.8) | 11.1 (4.1–22.1) | 9.0 (5.6–12.7) | |
| Total PFS events, n (%) | 51 (74) | 36 (71) | 12 (67) | 29 (69) | |
| PFS rate at 6 months, % (95% CI) | 45 (32–57) | 58 (43–71) | 77 (49–90) | 63 (45–76) | |
| PFS rate at 12 months, % (95% CI) | 21 (11–33) | 38 (24–52) | 47 (23–68) | 38 (23–53) | |
| Median DoR, months (95% CI) | 9.5 (4.2–14.7) | 10.7 (6.1–14.8) | 11.0 (2.8–NC) | 9.7 (4.5–14.3) | |
| Median OS,d months (95% CI) | 30.3 (11.8–NC) | 18.8 (15.1–NC) | NC (24.4–NC) | NC (13–NC) | |
| OS rate at 6 months, % (95% CI) | 86 (74–93) | 90 (77–96) | 94 (65–99) | 93 (79–98) | |
| OS rate at 12 months, % (95% CI) | 62 (47–73) | 69 (52–81) | 94 (65–99) | 78 (61–88) | |
| OS rate at 18 months, % (95% CI) | 53 (38–66) | 52 (36–67) | 87 (58–97) | 66 (49–79) | |
Abbreviation: 3G, third generation
aMost patients were enrolled to 600 mg savolitinib, prior to weight-based dosing implementation, but, following a protocol amendment, the final 21 patients enrolled in Part B were dosed with savolitinib by body weight as follows: patients who weighed ≤55 kg (n = 7) received 300 mg q.d., and those weighing >55 kg (n = 14) received 600 mg q.d.
bAll confirmed responses were partial response.
c≥6 weeks.
dCalculated using the Kaplan–Meier technique. The CI for median OS was derived based on the Brookmeyer–Crowley method.
In Part D, 62% (95% CI, 46–76) of patients had an objective response, all of which were partial responses. The median DoR for patients with an objective response was 9.7 (95% CI, 4.5–14.3) months, with 52% and 39% remaining in response at 9 months and 12 months, respectively. The median PFS was 9.0 months (95% CI, 5.6–12.7), with 29 (69%) events (Table 3; Supplementary Fig. S2B).
Median overall survival (OS), an exploratory outcome, was 30.3 months (95% CI, 11.8–NC) in Part B1, 18.8 months (95% CI, 15.1–NC) in Part B2, NC (95% CI, 24.4–NC) in Part B3, and NC (95% CI, 13.4–NC) in Part D. The survival rates at 18 months for Parts B1, B2, B3, and Part D were 53%, 52%, 87%, and 66%, respectively (Table 3).
Tumor Response by MET Detection Methods in Part B
Valid results were obtained from ≥1 central MET assay of tumors from 120 patients [fluorescent in situ hybridization (FISH) n = 117; next-generation sequencing (NGS) n = 49; immunohistochemistry (IHC) n = 34]. Of these, 105 (88%) were determined as having tumors with MET amplification/overexpression via FISH [n = 100 (95%)], NGS [n = 20 (41%)], or IHC [n = 20 (59%)].
In tumors with both central FISH and IHC data (n = 34), IHC positivity (n = 20) and FISH positivity (n = 27) commonly co-occurred (Supplementary Fig. S3A). IHC positivity was strongly associated with FISH positivity (defined as MET:CEP7 ratio ≥2; n = 16); 13/16 cases of FISH amplification were also IHC-positive (Supplementary Table S5). MET IHC H-scores also tended to be consistently elevated in tumors with MET gene copy number (GCN) ≥10 (Supplementary Fig. S3B). Similarly, in tumors with both central FISH and NGS data (n = 46), NGS (n = 20) and FISH (n = 38) positivity commonly co-occurred (Supplementary Table S5; Supplementary Fig. S3A) and GCN estimates from each assay were positively correlated (Supplementary Fig. S3B and S3C).
In Part B1, ORR by MET detection method was 30% (95% CI, 18–44) for FISH-positive overall; 28% (95% CI, 10–53) for FISH polysomy (GCN ≥5 if MET:CEP7 <2); 31% (95% CI, 17–49) for FISH amplification (MET:CEP7 ratio ≥2); 46% (95% CI, 19–75) for IHC; and 47% (95% CI, 21–73) for NGS-positive. In FISH-positive tumors, ORR trended higher in tumors with MET GCN ≥10 (34%; 95% CI, 18–54) versus tumors with MET GCN 5 to 9 (25%; 95% CI, 10–47; Table 4). A similar pattern of ORR by MET detection method was observed in Part B2, as shown in Table 4.
Table 4.
ORR and DCR by MET biomarker (evaluable for response set)
| N | ORR, n, % (95% CI) | DCR, % (95% CI) | |
|---|---|---|---|
| Part B1: Previously treated with third-generation EGFR-TKI | |||
| All | 69 | 23 | 75 (64–85) |
| 33 (22–46) | |||
| FISH-positive | 53 | 16 | 75 (62–86) |
| 30 (18–44) | |||
| 5–9 copies | 24 | 6 | 67 (45–84) |
| 25 (10–47) | |||
| ≥10 copies | 29 | 10 | 83 (64–94) |
| 34 (18–54) | |||
| Polysomy by FISH | 18 | 5 | 61 (36–83) |
| 28 (10–53) | |||
| Amplification by FISH | 35 | 11 | 83 (66–93) |
| 31 (17–49) | |||
| IHC-positive | 13 | 6 | 85 (55–98) |
| 46 (19–75) | |||
| NGS-positive | 15 | 7 | 73 (45–92) |
| 47 (21–73) | |||
| Part B2: Not previously treated with third-generation EGFR-TKI (T790M-negative) | |||
| All | 51 | 33 | 88 (76–96) |
| 65 (50–78) | |||
| FISH-positive | 34 | 22 | 91 (76–98) |
| 65 (46–80) | |||
| 5–9 copies | 20 | 11 | 85 (62–97) |
| 55 (32–77) | |||
| ≥10 copies | 14 | 11 | 100 (77–100) |
| 79 (49–95) | |||
| Polysomy by FISH | 14 | 8 | 79 (49–95) |
| 57 (29–82) | |||
| Amplification by FISH | 20 | 14 | 100 (83–100) |
| 70 (46–88) | |||
| IHC-positive | 4 | 3 | 100 (40–100) |
| 75 (19–99) | |||
| NGS-positive | 5 | 3 | 100 (48–100) |
| 60 (15–95) | |||
Eight tumors with nonconcordant FISH and IHC results were identified in Part B1: 6 FISH-positive/IHC-negative tumors with 1 partial response [1 partial response, 2 stable disease, 1 progressive disease, 2 not evaluable (NE)] and 2 FISH-negative/IHC-positive tumors with 1 partial response (1 partial response and 1 NE).
ctDNA Analysis
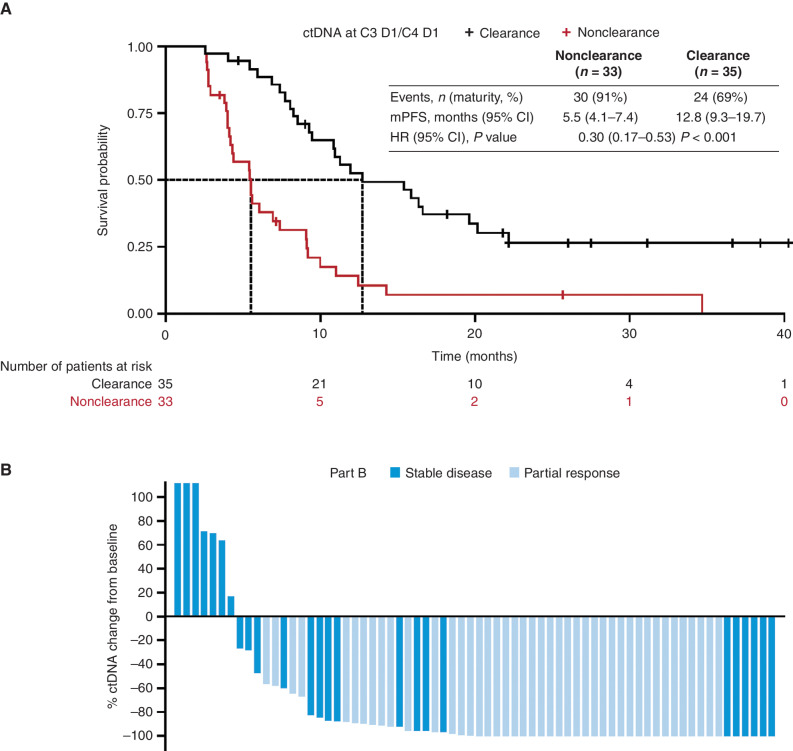
In the ctDNA analysis, 102 patients in Part B (Part B1 n = 52; Part B2 n = 35; Part B3 n = 15) and 37 patients in Part D were eligible for inclusion. EGFRm ctDNA was detectable at baseline in 85 (83%) and 24 (65%) patients tested in Parts B and D, respectively (Supplementary Table S6). ctDNA clearance was comparable at cycle (C) 4 day (D) 1/C5 D1 for Part B2 [n = 24 (69%)] and Part D [n = 22 (59%); Supplementary Fig. S4A and S4B]. Across the two doses of savolitinib, ctDNA clearance was similar (Supplementary Table S6). EGFRm ctDNA clearance at C3 D1/C4 D1 appeared to be predictive of longer PFS for patients with EGFRm, MET-amplified/overexpressed NSCLC with detectable ctDNA at baseline: nonclearance (n = 33) median PFS 5.5 months (95% CI, 4.1–7.4) versus clearance (n = 35) median PFS 12.8 months (95% CI, 9.3–19.7); hazard ratio (HR) 0.30 (95% CI, 0.17–0.53); P < 0.001 (Fig. 2A). Best percentage change from baseline in ctDNA is shown in Fig. 2B.
Figure 2.
ctDNA clearance at C3 D1/C4 D1 in patients with baseline detectable ctDNA (Part B; n = 68). A, Kaplan–Meier estimates of PFS in patients with ctDNA clearance and nonclearance. B, Waterfall plot of best percentage change from baseline in ctDNA. mPFS, median progression-free survival.
Baseline Genomics and Acquired Resistance
In total, 68 patients were eligible for the baseline genomic analysis and 45 patients were eligible for the analysis evaluating mechanisms of acquired resistance.
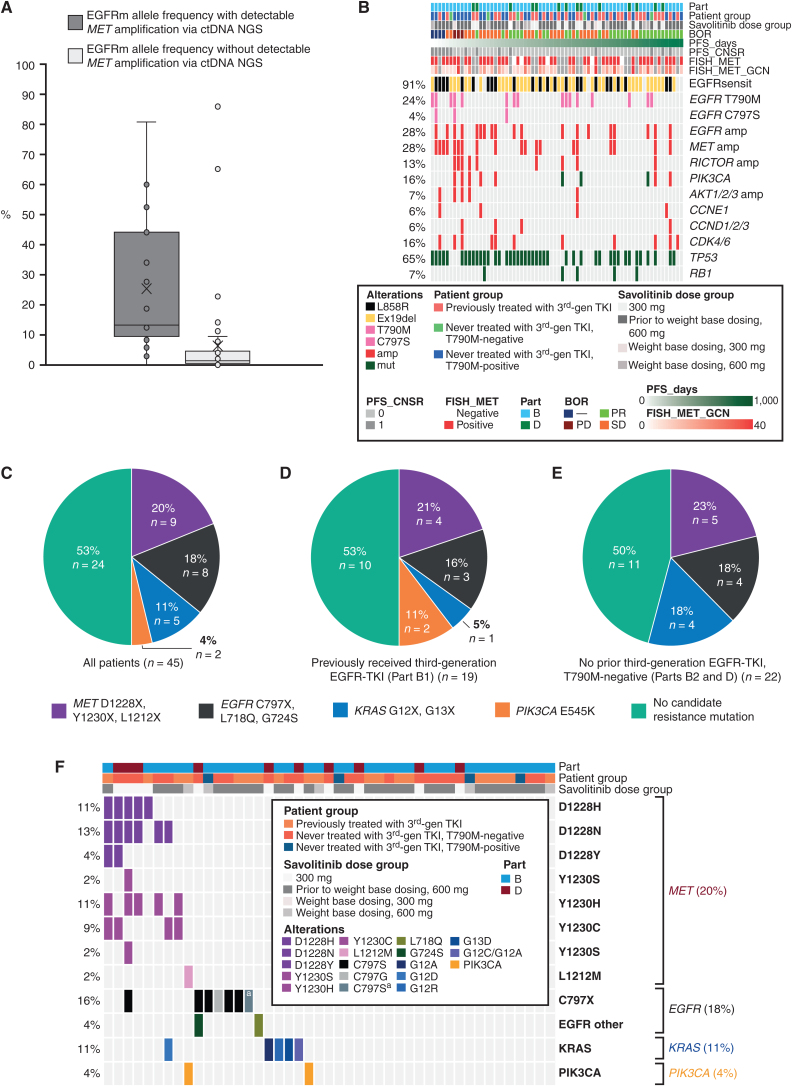
For the baseline genomics analysis, 63 patients had valid central MET FISH results and of these 55 (87%) were MET FISH-positive; only 15 (27%) MET FISH-positive tumors had detectable MET amplification in ctDNA at baseline. Detection of MET amplification via ctDNA was strongly associated with ctDNA content, as estimated by the EGFRm allele frequency: (n = 68) median EGFRm allele frequency with and without detectable MET amplification via ctDNA NGS was 13.1 versus 1.4, respectively (Welch t test P < 0.01; Fig. 3A). In Part B1 patients, detection of baseline EGFR C797S (n = 1/31) and RICTOR amplification (n = 4/31; Fig. 3B) appeared to correlate with poor response, although this preliminary observation should be treated with caution due to the small sample size.
Figure 3.
EGFRm allele frequency with and without detectable MET amplification via ctDNA NGS (A; n = 68); baseline genomics analysis (B); and an overview of baseline genomics in all patients with samples assessed in acquired resistance analysis (C), patients who had previously received third-generation EGFR-TKI (Part B1; D), and patients who had not previously received third-generation EGFR-TKI and were T790M-negative (Parts B2 and D combined; E). amp, amplification; BOR, best objective response; CNSR, censor; ex19del, exon 19 deletion; mut, mutant; sensit, sensitizing.F, Detailed breakdown of acquired resistance mutations in all patients. Patients who had not previously received third-generation EGFR-TKI and were T790M-positive (Part B3): no candidate resistance mechanism (n = 3), EGFR C797X, L718Q, G724S (n = 1). Due to small numbers, these data were not included as a figure. aTwo independent genotypes resulting in C797S.
For acquired resistance analysis (n = 45), in 53% (n = 24) of the samples compared between baseline and disease progression, no candidate resistance mutations were found. In the remaining 47% of samples, the following acquired mutations were recorded (exclusivity between genes in most patients): MET D1228X, Y1230X, and L1212X (20%, 9/45); EGFR C797X, L718Q, and G724S (18%, n = 8); KRAS G12X and G13X (11%, n = 5); and PIK3CA E545K (4%, n = 2; Fig. 3C, D and E). Across the 5 patients with KRAS G12X or G13X alterations, 6 mutants were detected: G12A (n = 2), G12C (n = 1), G12D (n = 1), G12R (n = 1), and G13D (n = 1); 1 patient had both G12C and G12A (Fig. 3F). Most patients who developed MET-based resistance (7/9) developed more than 1 MET mutation, suggestive of polyclonal resistance. The resistance profiles appeared similar by prior EGFR-TKI status and by savolitinib dose across both Parts B and D (Supplementary Table S7). For patients with MET-mediated resistance (n = 9), EGFR-, KRAS-, or PIK3CA-mediated resistance (n = 12), or no known resistance mechanism detected (n = 24), the ORR was 67% (6/9; 95% CI, 30–93), 42% (5/12; 95% CI, 15–72), and 46% (11/24; 95% CI, 26–67), respectively. At data cutoff, 89% (8/9), 100% (12/12), and 92% (22/24) had progressed or died, respectively. Median PFS (95% CI) tended to be slightly longer in patients with MET-mediated resistance [10.9 months (3.0–16.4)] or EGFR-, KRAS-, or PIK3CA-mediated resistance [9.1 months (5.5–12.5)] versus those with no known resistance mechanisms detected [4.2 months (4.0–6.9); Supplementary Fig. S5A and S5B). However, these data should be interpreted with caution, as they cut across different cohorts and the patient values are very low.
PK
Following oral administration of 600 mg/300 mg q.d. doses of savolitinib in combination with 80 mg q.d. osimertinib in Parts B and D, savolitinib was rapidly absorbed, with median time to maximum plasma concentration (tmax) and time to maximum plasma concentration at steady-state (tssmax) values ranging from 2 to 4 hours after dose, and rapidly eliminated with mean terminal elimination half-life (t1/2λz) of ∼2 to 4 hours following single and multiple dosing, respectively (Supplementary Fig. S6A–S6D and Supplementary Table S8). Metabolites M2 and M3 were rapidly formed, with median tmax and tssmax values similar to savolitinib at 2 to 4 hours after dose, and rapidly eliminated with mean t1/2λz of 4 to 6 hours from single and multiple dosing. Overall, steady-state savolitinib PK was consistent with single-dose observations, suggesting neither apparent accumulation nor time-dependent PK changes upon multiple dosing. There was no apparent impact of prior osimertinib administration on the PK of savolitinib and corresponding metabolites on C1 D1. The PK of savolitinib and osimertinib was consistent with other patient populations in TATTON Part A and previous studies (21).
DISCUSSION
These final results of TATTON Parts B/D confirm our earlier findings that the combination of savolitinib plus osimertinib demonstrates antitumor activity across several subcohorts of patients with EGFRm, MET-amplified/overexpressed NSCLC with a manageable safety profile that is broadly in line with other oral MET-TKIs. Our results suggest that savolitinib plus osimertinib may overcome MET-based resistance in patients with NSCLC, whose disease has progressed on prior EGFR-TKI.
ORR was consistent between patients in Part B2 (65%; 95% CI, 50–78), Part B3 (67%; 95% CI, 41–87), and Part D (62%; 95% CI, 46–76). However, ORR was considerably lower in patients in Part B1 (33%; 95% CI, 22–46), which is a population of particular interest given the importance of acquired MET amplification in patients with EGFRm NSCLC treated with osimertinib (9, 11–13). Similar preliminary activity (ORR 41%; stable disease, 41%) of osimertinib plus savolitinib in patients with EGFRm advanced NSCLC and MET alterations following first-line osimertinib treatment was reported in an interim analysis of ORCHARD (ref. 29; NCT03944772; n = 20). Dual MET and EGFR inhibition with tepotinib plus gefitinib also demonstrated antitumor activity in the INSIGHT study (NCT01982955) in patients with EGFRm, T790M-negative NSCLC with MET overexpression (by IHC 2+ or 3+) or MET amplification (by FISH) and acquired resistance to previous EGFR-TKIs, with MET amplification potentially being a predictive marker for this treatment combination (30). Our results also suggest that there is no apparent difference in antitumor activity between savolitinib doses, which is also supported by the PFS data.
We further demonstrate that ORR was largely comparable irrespective of the MET assay used, although some preliminary signals of increased efficacy were observed in tumors with MET amplification/overexpression by GCN ≥10, NGS, and IHC. This supports the continued use of FISH, IHC, and NGS for MET detection in patients following disease progression on third-generation EGFR-TKIs. However, it should be noted there was lower co-occurrence for detecting MET positivity between NGS and FISH versus IHC and FISH, and there may have been potential bias in the patient selection, as a larger proportion were included in the analysis based on a FISH result versus NGS or IHC.
Our data, along with preliminary findings from the phase II SAVANNAH study (NCT03778229; ref. 31), highlight the challenges and importance of monitoring MET amplification during treatment, particularly considering the observed association between tumor content and MET amplification detection and the degree of ctDNA decrease during treatment, resulting in a high level of false negatives. This affects the potential to distinguish between true loss of MET amplification versus ctDNA content just decreasing to an undetectable level.
We also found that EGFRm ctDNA clearance may be predictive of longer PFS for patients with EGFRm, MET-amplified/overexpressed NSCLC with detectable ctDNA at baseline, which is in line with previous studies (27, 32, 33). The proportion of patients with EGFRm ctDNA clearance was comparable at C4 D1/C5 D1 for Part B2 and Part D, consistent with ORR and PFS. Serial ctDNA testing also provided insights into the longitudinal development of acquired resistance mutations. Approximately half of all evaluable patients had an identifiable acquired resistance mechanism to the savolitinib plus osimertinib combination. Resistance in the remaining half of evaluable patients may be identified with broader genomic analyses or via nongenomic mechanisms, as analysis of resistance mechanisms using plasma ctDNA does have limitations, including detection of copy-number variations. Our analysis may be missing these forms of acquired resistance. It is also possible that these tumors were never fully sensitive to the savolitinib plus osimertinib combination and thus never developed resistance. This is supported by the shorter PFS for patients without a detectable mechanism of resistance (Supplementary Fig. S5). Detected acquired resistance mechanisms appeared to be predominantly mediated by additional aberrations in either MET, EGFR, or KRAS and were representative of on- and off-target acquired resistance mechanisms previously associated with osimertinib or savolitinib monotherapy in NSCLC (9, 34, 35). Consistent with our observations, acquired EGFR mutations (L718Q, C797S; refs. 36, 37), MET mutations (D1228N/H, Y1230C; refs. 36–39), and KRAS mutations (40) have previously been reported following combination therapy with osimertinib (or EGFR-TKI therapy including osimertinib) plus savolitinib or crizotinib. Most of these reports are from small patient samples or case studies, but it is notable that they suggest that acquired resistance to crizotinib may differ from that with savolitinib. While co-occurring mutations across multiple genes were rarely detected, multiple acquired mutations were often detected in a specific gene, particularly MET, suggesting that individual tumors showed inherent resistance pathway dependencies. Currently, there is little evidence to predict what predetermines escape mutations and whether factors such as PK or pharmacology play a part in this process. However, a retrospective small analysis (n = 38) suggested that the type of EGFR mutation prior to osimertinib treatment may play a role in which resistance pattern emerges (41). A recent study proposed that the relative proportion of EGFR:MET in a tumor predicts dependence on the signaling from either protein (42). It is possible that the EGFR:MET ratio may also predict the bifurcated EGFR- versus MET-mediated resistance observed in this study. Our acquired resistance findings may inform treatment options to overcome subsequent resistance to savolitinib and osimertinib combination therapy; for example, tumors with acquired EGFR mutations may respond to different treatment combinations. The ORCHARD (43) trial is testing osimertinib plus gefitinib in second-line patients following disease progression on osimertinib, who acquired EGFR C797S mutations (43).
Consistent with the known safety profiles of osimertinib and savolitinib (2, 20, 44), and in line with the AE profile for this combination reported in Part A of the study (21), savolitinib increased the proportion of patients who had nausea, diarrhea, and fatigue expected with osimertinib alone and also of peripheral edema, a known side effect of MET-TKIs.
Overall, the safety profile of the savolitinib plus osimertinib combination was manageable, although the data suggest that the lower 300 mg savolitinib dose may be better tolerated than the 600 mg dose. The AE profile of patients in Part D was slightly improved versus that of the Part B subcohorts, particularly the proportion of patients experiencing AEs grade ≥3 or SAEs. The incidence of individual AEs was generally consistent between Parts B and D; however, numerically, more cases of some AEs were reported in the Part B subcohorts versus Part D. The grouped term hypersensitivity was similar across groups.
To address the emerging safety profile of the savolitinib plus osimertinib combination, which saw the incidence of drug hypersensitivity–related AEs including pyrexia and anaphylactic reaction in several patients, toxicity management guidelines were updated several times throughout the prolonged duration of the study. This included the implementation of weight-based dosing in the savolitinib plus osimertinib, second line or later, MET-amplified/overexpressed cohorts (the last group of patients enrolled in Part B) and management by proactive use of corticosteroids, antihistamines, and antipyretics, with restart only being permitted in an institution prepared for management of anaphylaxis and resuscitation. It is possible that the implementation of these toxicity management guidelines for hypersensitivity-related AEs may have affected awareness, identification, and early management of these toxicities as the study progressed. Similar toxicity management is being utilized in the SAVANNAH study in which no grade ≥3 drug hypersensitivity–related AEs were reported in a recent preliminary analysis (31). The mechanism underlying drug hypersensitivity–related AEs observed in TATTON is not yet known but is being explored. For example, examples of drug hypersensitivity have been linked with specific HLA genotypes (45). HLA profiling has been implemented in subsequent savolitinib studies (including SAVANNAH) to explore whether any observed drug hypersensitivity–related AEs are associated with specific HLA genotypes.
We identified several limitations with the study. First, TATTON was a single-arm, phase I study with a relatively small, heterogeneous prior EGFR-TKI treatment population. This includes the possibility that some patients may have received a non–EGFR-TKI therapy as their most recent anticancer therapy, potentially resulting in some clinical activity seen due to EGFR-TKI retreatment. Second, CNS scans were not mandatory in all patients, and therefore it was not possible to robustly assess antitumor activity in the CNS. Third, the protocol did not require third-generation EGFR-TKIs previously received by patients in Part B1 to be named or if they were the most recent line of anticancer therapy; thus, a breakdown of previously received third-generation EGFR-TKIs or their line of use was not available. Finally, as mentioned above, bias may have been introduced into the analysis of tumor response by the different MET-detection methods, as most patients were included in the analysis based on a FISH result. The COVID-19 pandemic was judged to have no meaningful impact on the quality of the study.
In conclusion, savolitinib plus osimertinib combination therapy has an acceptable risk–benefit profile and encouraging antitumor activity in patients with MET-amplified/overexpressed, EGFRm advanced NSCLC who experienced disease progression on a previous first-, second-, or third-generation EGFR-TKI and represents an approach worthy of further investigation to improve outcomes in EGFRm advanced NSCLC. Serial ctDNA analyses showed that EGFRm ctDNA clearance on treatment was predictive of longer PFS in patients with detectable ctDNA at baseline, which may provide an opportunity to inform earlier decision-making. Acquired resistance mechanisms to this combination were found to include those via MET, EGFR, and KRAS. For patients with MET-mediated resistance to osimertinib, savolitinib plus osimertinib is currently being assessed prospectively in the ongoing SAVANNAH (31), ORCHARD (43), and SAFFRON (NCT05261399) studies, and tepotinib plus osimertinib is being assessed in the INSIGHT2 study (NCT03940703; ref. 46).
METHODS
Study Design
TATTON was a multiarm, multicenter, open-label phase Ib trial conducted in eight countries (Canada, Japan, Ukraine, Poland, Russia, South Korea, Taiwan, USA) to assess the safety, tolerability, and antitumor activity of osimertinib 80 mg orally q.d. in combination with selumetinib, savolitinib, or durvalumab at escalating doses in patients with EGFRm advanced NSCLC who had progressed after EGFR-TKI therapy.
The study methodology has been previously published (21, 22). In brief, the study consisted of four parts, A through D (Supplementary Fig. S7). In Part A, osimertinib plus investigational agent combination regimens were identified in initial dose-finding cohorts under assessment by the Safety Review Committee (21); these doses were selected for further assessment in dose-expansion cohorts. In Part B, patients received either savolitinib plus osimertinib, osimertinib plus intermittent doses of selumetinib, or osimertinib plus durvalumab. We report the results of the final clinical analysis of the Part B initial expansion savolitinib plus osimertinib cohorts and the Part D subsequent expansion cohort for the same combination. Part C is a Japan-specific savolitinib dose-finding substudy, which is not reported here (47).
Part B included three savolitinib cohorts of patients with MET-amplified/overexpressed, EGFRm NSCLC: B1 (prior third-generation EGFR-TKI received), B2 [no prior third-generation EGFR-TKI received (T790M-negative)], and B3 [no prior third-generation EGFR-TKI received (T790M-positive)]. Part D comprised patients with MET-amplified/overexpressed, EGFRm NSCLC who had received no prior third-generation EGFR-TKI, but had received ≥1 prior line of therapy with a first-/second-generation EGFR-TKI, and whose tumors were T790M-negative. Inclusion and exclusion criteria have been previously described (ref. 22; Supplementary Methods).
All patients in the Part B savolitinib cohorts and Part D were required to have locally confirmed MET amplification prior to study entry. MET testing could be performed on tumor tissue prospectively during screening using central laboratory FISH (MET GCN ≥5 or MET:CEP7 ratio ≥2 was required), IHC (MET +3 expression in ≥50% of tumor cells was required), or NGS (≥20% tumor cells, coverage of ≥200× sequencing depth and ≥4 copies of MET over tumor ploidy were required); local test results were allowed. Where samples were available from patients enrolled based on a local test, retrospective confirmation of MET status was performed using central tissue FISH, NGS, and IHC per the above criteria; samples were available for most patients. In Part B, MET amplification was defined as a patient who harbored either a MET gene amplification and/or a MET exon 14 genomic alteration but did not harbor a MET 1228 or 1230 resistance mutation. Additionally, in Part D, a patient could exhibit MET overexpression.
The study was conducted in accordance with the principles of the Declaration of Helsinki and Good Clinical Practice guidelines of the International Conference on Harmonisation. The protocol was approved by the local institutional review board at each site and complied with local country regulations; patients provided written informed consent before participation.
Outcomes
The primary objective was to investigate the safety and tolerability of osimertinib plus savolitinib. The secondary objectives were the assessment of antitumor activity of savolitinib plus osimertinib [ORR, DoR, change in target lesion size from baseline, and PFS using Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1] and the characterization of the PK of osimertinib and savolitinib and their metabolites (after single dosing, and at steady state after multiple dosing, when given in combination). Exploratory objectives included the preliminary assessment of the effect of osimertinib plus savolitinib on OS; the assessment of whether longitudinal clearance (longitudinal sample with EGFRm <0.5%) of baseline detectable EGFRm ctDNA could be a predictor of PFS in patients with EGFRm, MET-amplified/overexpressed NSCLC; and the comparison of ORR in patients with MET amplification/overexpression detected using FISH, IHC, or NGS. Definitions for these outcomes are provided in the Supplementary Methods.
Procedures
The procedures for TATTON Parts B and D have been previously described (21, 22). Briefly, all patients in the Part B savolitinib cohorts were initially administered osimertinib 80 mg q.d. plus savolitinib 600 mg q.d. Following the emergence of hypersensitivity and anaphylaxis events, a protocol amendment on March 12, 2018, led to a weight-based savolitinib dosing regimen for the last group of patients enrolled in Part B in parallel to the lower dose of savolitinib 300 mg for all patients enrolled in Part D to improve the safety and tolerability profile of the combination. The weight-based savolitinib dosing regimen in Part B was as follows: Patients weighing ≤55 kg received 300 mg q.d., and those weighing more than 55 kg received 600 mg q.d. In Part D, patients received osimertinib 80 mg q.d. and savolitinib 300 mg q.d.
Physical examinations were conducted on D1 of each 28-day cycle; liver function tests, clinical chemistry, and electrocardiograms were assessed at screening, C1 (D1, 8, 15, and 22), at D1 of C2 through C7, and every 8 weeks until treatment discontinuation. RECIST assessments were performed at screening and on D1 of C1 and every 6 weeks (relative to first dose) thereafter until progression. Blood samples for biomarker analysis were taken at screening, predose on D1 and D15/22 of C1, every 6 weeks (relative to the first dose) in C2 through C6, and every 8 weeks (relative to the first dose) thereafter until progression. Blood samples for PK analysis were taken at D1 of C1, C2, C3, C6, and C11. AEs were collected from informed consent to 28 days (± 7 days) after study treatment was discontinued (end of the follow-up period) and graded according to the NCI Common Terminology Criteria for Adverse Events version 4.0.
Patients continued to receive osimertinib plus savolitinib beyond disease progression if they continued to show clinical benefit, as judged by the investigator, and in the absence of discontinuation criteria. Safety assessments were continued while patients were receiving any study treatment beyond disease progression. Beyond disease progression, patients were followed up every 12 weeks until withdrawal from the study or death. Information on dose interruptions and modifications has been previously described (ref. 22; details are also included in the Supplementary Methods).
MET Detection Methods
A biomarker analysis was performed on Part B of the study to report tumor response rates (ORR) across different MET detection methods (FISH, IHC, and NGS); here, only centrally confirmed data are reported. During patient screening for the study, MET testing was performed on tumor tissue collected after the most recent disease progression. MET testing was performed in a central laboratory using FISH [Kreatech MET FISH LDT MET/SE 7 (D7Z1) DNA probe (Kreatech, Inc.) or Vysis MET FISH CDx IUO (Abbott Molecular)], NGS (FoundationOne Clinical Laboratory Improvement Amendments assay, which is based on the FDA-approved F1CDx; P170019), and/or central IHC testing (Ventana SP44 antibody). MET GCN by FISH was determined as amplification (MET:CEP7 ratio ≥2) or polysomy (MET GCN ≥5 if MET:CEP7 <2). For NGS, standard Foundation Medicine MET focal (<20 MB) amplification calls (20% tumor cells, coverage of ≥200× sequencing depth, ≥5 copies of MET over tumor ploidy) were used. The MET IHC assay used the rabbit monoclonal antibody SP44, directed at the c-terminus of the MET protein, with the UltraView DAB Detection Kit on Ventana Benchmark XT Instrument. IHC positivity was defined as 3+ MET staining in ≥50% of tumor cells.
ctDNA Analysis
NGS-based analysis (Resolution Bioscience) and droplet digital polymerase chain reaction (ddPCR; BioDesix) was used to assess EGFRm (Ex19del/L858R) ctDNA clearance in Parts B and D of the study. A subset of samples had duplicate plasma aliquots run on both the NGS and ddPCR platforms, which showed high concordance. ctDNA samples were collected as per the schedule described above for the biomarker analysis. The analysis included Part B patients evaluable for efficacy and with a baseline plus ≥1 longitudinal ctDNA sample at/before C6 D1. Data from ctDNA evaluable Part B patients (n = 49) identified that ctDNA clearance correlates with longer PFS and that C4 D1/C5 D1 was the optimal time point for PFS prediction; thus, the first 20 patients from Part D with available plasma samples at C1 D1 and C4 D1 were included in the analysis. All patients in the ctDNA analysis received savolitinib 600 mg flat dose (Part B) and savolitinib 300 mg flat dose (Part D).
Acquired Resistance Analysis
For the acquired resistance analysis, paired plasma samples collected at baseline and following disease progression and/or treatment discontinuation up to the data cutoff date (March 4, 2020) were assessed. Plasma ctDNA samples were analyzed using NGS (Guardant Health; Guardant360 73-gene panel or Omni 500-gene panel). The Omni 500-gene panel included all 73 genes from the Guardant360 panel, and analyses from each patient were reported only for genes included in both panels with genomic alterations identified using Guardant Health's pipeline, which included mutations and amplifications of EGFR and MET. Disease progression was assessed by the investigator, according to RECIST 1.1. Assessments were completed for patients with PFS of over 2 months. ORR and PFS were assessed in samples according to whether resistance was via a known detectable mechanism (MET-driven or non–MET-driven) or an undetected mechanism.
PK
Where possible, the PK analysis assessed the following parameters for savolitinib and osimertinib and their respective metabolites after single dosing: maximum plasma concentration (Cmax), tmax, area under the plasma concentration–time curve (AUC), AUC from zero to 24 hours (AUC0–24), AUC from zero to last sample time (AUC0-t), terminal half-life (t1/2), apparent clearance (CL/F), and volume of distribution (Vz/F). After multiple dosing, the following PK parameters were assessed: tssmax, maximum plasma concentration at steady state (Cssmax), minimum plasma concentration at steady state (Cssmin), AUC at steady state (AUCss), CL/F at steady state (CLss/F), the extent of accumulation on multiple dosing (RAC), and time dependency of the PK (TCP).
Statistical Methods
Final study data cutoff was March 4, 2020. For the ctDNA analysis, two different interim data cutoffs were used: February 28, 2018, for Part B and March 20, 2019, for Part D.
Approximately 40 evaluable patients per cohort with centrally confirmed MET amplification were planned to be recruited in Part B. In Part D, 40 patients were planned to be enrolled to have approximately 25 centrally confirmed MET-amplified/overexpressed patients. It was expected that this number of patients would enable adequate tolerability, safety, PK, and pharmacodynamic data to be obtained while exposing as few patients as possible to the investigational product and procedures. PFS, OS, ORR, DoR, tumor response, and change in tumor size were analyzed using evaluable patients, who were patients with centrally confirmed MET-amplified/overexpressed status and ≥1 post–baseline RECIST assessment (evaluable for response set). Patients who were enrolled based on local MET testing results who were not subsequently confirmed by central testing were not counted as evaluable and were not included in the efficacy summaries for that cohort. ORR was presented along with 95% exact (Clopper–Pearson) CIs. PFS and OS were summarized using time-to-event methods, including Kaplan–Meier plots.
Safety data were assessed in the safety analysis set, which comprised all dosed patients. Safety and biomarker data were not formally statistically analyzed.
The PK analysis set comprised all patients who received ≥1 dose of osimertinib or savolitinib for whom any post-dose PK data were available. For the PK analysis, actual sampling times were used in the parameter calculations and PK parameters were derived using standard noncompartmental methods.
Statistical analyses were performed with SAS (version 9.1.3). This study was registered at ClinicalTrials.gov under ID NCT02143466.
Data Availability
Data underlying the findings described in this article may be obtained in accordance with AstraZeneca's data sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure.
Supplementary Material
Supplementary methods, 8 tables, 7 figures
Acknowledgments
The authors thank all the patients and their families. The study (NCT02143466) was funded by AstraZeneca, the manufacturer of the drugs savolitinib and osimertinib. The authors acknowledge Bernadette Tynan, MSc, of Ashfield MedComms, an Inizio Company, for medical writing support that was funded by AstraZeneca in accordance with Good Publications Practice (GPP3) guidelines (http://www.ismpp.org/gpp3).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
R.J. Hartmaier reports personal fees from AstraZeneca during the conduct of the study, as well as a patent for US11066709B2 issued to Genentech and Foundation Medicine. A.A. Markovets reports other support from AstraZeneca during the conduct of the study, as well as other support from AstraZeneca outside the submitted work. M.J. Ahn reports personal fees from Yuhan Pharmaceuticals, Lilly, Takeda, Merck, AstraZeneca, MSD, Daiichi Sankyo, and Alpha Pharmaceuticals outside the submitted work. L.V. Sequist reports grants from AstraZeneca during the conduct of the study; personal fees from AstraZeneca, Janssen, Pfizer, and Genentech, and grants from Novartis and Boehringer Ingelheim outside the submitted work; and a patent for the combination use of osimertinib and BLU-667 pending to Blueprint Medicines. J.-Y. Han reports grants from Pfizer and Takeda outside the submitted work. B.C. Cho reports nonfinancial support from the American Society of Clinical Oncology, AstraZeneca, Guardant, Roche, the European Society for Medical Oncology, the International Association for the Study of Lung Cancer, the Korean Cancer Association, the Korean Society of Medical Onoclogy, the Korean Society of Thyroid-Head and Neck Surgery, the Korean Cancer Study Group, Novartis, MSD, the Chinese Thoracic Oncology Society, and Pfizer, personal fees (advisory board) from KANAPH Therapeutics, Bridgebio therapeutics, Cyrus therapeutics, Guardant Health, and Oscotec, personal fees (member of the board of directors) from Interpark Bio Convergence Corp. and J INTS BIO, personal fees (full or part-time employment) from Yonsei University Health System, personal fees (stocks/shares) from TheraCanVac, Gencurix, Bridgebio therapeutics, KANAPH Therapeutics, Cyrus therapeutics, Interpark Bio Convergence Corp., and J INTS BIO, personal fees (royalties) from Champions Oncology, grants from MOGAM Institute, LG Chem, Oscotec, Interpark Bio Convergence Corp., GIInnovation, GI-Cell, Abion, AbbVie, AstraZeneca, Bayer, Blueprint Medicines, Boehringer Ingelheim, Champions Oncology, CJ Bioscience, CJ Blossom Park, Cyrus Therapeutics, Dizal Pharma, Genexine, Janssen, Eli Lilly, MSD, Novartis, Nuvalent, Oncternal, Ono Pharmaceuticals, Regeneron, Dong-A ST, Bridgebio therapeutics, Yuhan Pharmaceuticals, ImmuneOncia, Illumina, KANAPH Therapeutics, Therapex, J INTS BIO, and Hanmi, personal fees (advisory role) from Abion, BeiGene, Novartis, AstraZeneca, Boehringer Ingelheim, Roche, Bristol Myers Squibb, CJ, CureLogen, Cyrus therapeutics, Ono Pharmaceuticals, Onegene Biotechnology, Yuhan, Pfizer, Eli Lilly, GI-Cell, Guardant, HK Inno-N, Imnewrun Biosciences, Janssen, Takeda, MSD, Medpacto, Blueprint medicines, RandBio, and Hanmi, and other support from DAAN Biotherapeutics outside the submitted work. H.A. Yu reports grants and personal fees from AstraZeneca during the conduct of the study, as well as grants and personal fees from Cullinan, Blueprint Medicines, Black Diamond, and Daiichi Sankyo and grants from ERASCA, Novartis, and Pfizer outside the submitted work. J.C.-H. Yang reports personal fees and other support from Amgen, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Daiichi Sankyo, Merck KGaA, Marmstadt Germany, MSD, and Novartis, grants, personal fees, and other support from AstraZeneca, other support from Eli Lilly, Janssen, Puma Oncology, Gilead, and GSK, and personal fees from Ono Pharmaceuticals, Pfizer, Roche/Genentech, Takeda, and Yuhan Pharmaceuticals outside the submitted work. D.M. Kowalski reports advisory boards for AstraZeneca, Roche, Bristol Myers Squibb, Merck, MSD, Amgen, Boehringer Ingelheim, Takeda, Novartis, Sanofi-Aventis, and Pfizer. S. Ren reports other support from AstraZeneca outside the submitted work. P. Frewer reports personal fees and other support from AstraZeneca during the conduct of the study, as well as personal fees and other support from AstraZeneca outside the submitted work. D.A.E. Cross reports personal fees from AstraZeneca outside the submitted work. N. Kurian reports employment with and holds stock in AstraZeneca at the time of submission of the manuscript. M. Cantarini reports other support from AstraZeneca during the conduct of the study. P.A. Jänne reports grants and personal fees from AstraZeneca during the conduct of the study; grants and personal fees from Boehringer Ingelheim, Eli Lilly, Daiichi Sankyo, and Takeda, personal fees from Pfizer, Roche/Genentech, Chugai, SFJ Pharmaceuticals, Voronoi, Biocartis, Novartis, Sanofi Oncology, Mirati Therapeutics, Transcenta, Silicon Therapeutics, Syndax, Nuvalent, Bayer, Eisai, Allorion Therapeutics, Accutar Biotech, and AbbVie, and grants from PUMA, Revolution Medicines, and Astellas Pharmaceuticals outside the submitted work; and a patent for EGFR mutations issued, licensed, and with royalties paid from Labcorp. No disclosures were reported by the other authors.
Authors’ Contributions
R.J. Hartmaier: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–review and editing. A.A. Markovets: Formal analysis, investigation, visualization, writing–review and editing. M.J. Ahn: Supervision, writing–review and editing. L.V. Sequist: Investigation, writing–review and editing. J.-Y. Han: Resources, formal analysis, investigation, writing–review and editing. B.C. Cho: Resources, data curation, investigation, writing–original draft, writing–review and editing. H.A. Yu: Conceptualization, resources, data curation, formal analysis, supervision, investigation, methodology, writing–review and editing. S.-W. Kim: Resources, data curation, investigation, writing–review and editing. J.C.-H. Yang: Conceptualization, supervision, validation, investigation, methodology, project administration, writing–review and editing. J.-S. Lee: Investigation, writing–review and editing. W.-C. Su: Investigation, writing–review and editing. D.M. Kowalski: Formal analysis, supervision, investigation, writing–review and editing. S. Orlov: Data curation. S. Ren: Conceptualization, formal analysis, writing–review and editing. P. Frewer: Formal analysis, methodology. X. Ou: Validation, writing–original draft, writing–review and editing. D.A.E. Cross: Writing–review and editing. N. Kurian: Data curation, methodology, writing–original draft, writing–review and editing. M. Cantarini: Conceptualization, formal analysis, supervision, investigation, writing–review and editing. P.A. Jänne: Conceptualization, resources, writing–review and editing.
References
- 1. Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 2014;4:1046–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non–small-cell lung cancer. N Engl J Med 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 3. Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017;376:629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu YL, Ahn MJ, Garassino MC, Han JY, Katakami N, Kim HR, et al. CNS efficacy of osimertinib in patients with T790M-positive advanced non-small-cell lung cancer: data from a randomized phase III trial (AURA3). J Clin Oncol 2018;36:2702–9. [DOI] [PubMed] [Google Scholar]
- 5. Reungwetwattana T, Nakagawa K, Cho BC, Cobo M, Cho EK, Bertolini A, et al. CNS response to osimertinib versus standard epidermal growth factor receptor tyrosine kinase inhibitors in patients with untreated EGFR-mutated advanced non–small-cell lung cancer. J Clin Oncol 2018;36:3290–7. [DOI] [PubMed] [Google Scholar]
- 6. Hanna NH, Robinson AG, Temin S, Baker S Jr, Brahmer JR, Ellis PM, et al. Therapy for stage IV non-small-cell lung cancer with driver alterations: ASCO and OH (CCO) joint guideline update. J Clin Oncol 2021;39:1040–91. [DOI] [PubMed] [Google Scholar]
- 7. Planchard D, Popat S, Kerr K, Novello S, Smit EF, Faivre-Finn C, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29 Suppl 4:iv192–iv237. (Updated version published 2020 Sep 15.) Available from:https://www.esmo.org/guidelines/guidelines-by-topic/lung-and-chest-tumours/clinical-practice-living-guidelines-metastatic-non-small-cell-lung-cancer. [DOI] [PubMed]
- 8. Wu YL, Tsuboi M, He J, John T, Grohe C, Majem M, et al. Osimertinib in resected EGFR-mutated non–small-cell lung cancer. N Engl J Med 2020;383:1711–23. [DOI] [PubMed] [Google Scholar]
- 9. Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non–small cell lung cancer. Br J Cancer 2019;121:725–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Piotrowska Z, Isozaki H, Lennerz JK, Gainor JF, Lennes IT, Zhu VW, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov 2018;8:1529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol 2018;4:1527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ramalingam SS, Cheng Y, Zhou C, Ohe Y, Imamura F, Cho BC, et al. Mechanisms of acquired resistance to first-line osimertinib: preliminary data from the phase III FLAURA study. Ann Oncol 2018;29Suppl 8:viii740. [Google Scholar]
- 14. York ER, Varella-Garcia M, Bang TJ, Aisner DL, Camidge DR. Tolerable and effective combination of full-dose crizotinib and osimertinib targeting MET amplification sequentially emerging after T790M positivity in EGFR-mutant non–small cell lung cancer. J Thorac Oncol 2017;12:e85–e8. [DOI] [PubMed] [Google Scholar]
- 15. Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039–43. [DOI] [PubMed] [Google Scholar]
- 16. Gainor JF, Niederst MJ, Lennerz JK, Dagogo-Jack I, Stevens S, Shaw AT, et al. Dramatic response to combination erlotinib and crizotinib in a patient with advanced, EGFR-mutant lung cancer harboring de novo MET amplification. J Thorac Oncol 2016;11:e83–e5. [DOI] [PubMed] [Google Scholar]
- 17. Hua Y, Shen L, Gan H, Lickliter J, Millward M, Xu J, et al. Phase I studies of a selective cMet inhibitor AZD6094 (HMPL504/volitinib) in patients with advanced solid tumors [abstract]. In: Proceedings of the 106th Annual Meeting of the American Association for Cancer Research; 2015 Apr 18–22; Philadelphia, PA. Philadelphia (PA): AACR; Cancer Res 2015;75(15 Suppl):Abstract nr CT305. [Google Scholar]
- 18. Jia H, Dai G, Weng J, Zhang Z, Wang Q, Zhou F, et al. Discovery of (S)-1-(1-(Imidazo[1,2-a]pyridin-6-yl)ethyl)-6-(1-methyl-1H-pyrazol-4-yl)-1H-[1,2, 3]triazolo[4,5-b]pyrazine (volitinib) as a highly potent and selective mesenchymal-epithelial transition factor (c-Met) inhibitor in clinical development for treatment of cancer. J Med Chem 2014;57:7577–89. [DOI] [PubMed] [Google Scholar]
- 19. Gavine PR, Ren Y, Han L, Lv J, Fan S, Zhang W, et al. Volitinib, a potent and highly selective c-Met inhibitor, effectively blocks c-Met signaling and growth in c-MET amplified gastric cancer patient-derived tumor xenograft models. Mol Oncol 2015;9:323–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gan HK, Millward M, Hua Y, Qi C, Sai Y, Su W, et al. First-in-human phase I study of the selective MET inhibitor, savolitinib, in patients with advanced solid tumors: safety, pharmacokinetics, and antitumor activity. Clin Cancer Res 2019;25:4924–32. [DOI] [PubMed] [Google Scholar]
- 21. Oxnard GR, Yang JC, Yu H, Kim SW, Saka H, Horn L, et al. TATTON: a multi-arm, phase Ib trial of osimertinib combined with selumetinib, savolitinib, or durvalumab in EGFR-mutant lung cancer. Ann Oncol 2020;31:507–16. [DOI] [PubMed] [Google Scholar]
- 22. Sequist LV, Han JY, Ahn MJ, Cho BC, Yu H, Kim SW, et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non–small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol 2020;21:373–86. [DOI] [PubMed] [Google Scholar]
- 23. Noonan SA, Sachs PB, Camidge DR. Transient asymptomatic pulmonary opacities occurring during osimertinib treatment. J Thorac Oncol 2016;11:2253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Finocchiaro G, Toschi L, Gianoncelli L, Baretti M, Santoro A. Prognostic and predictive value of MET deregulation in non–small cell lung cancer. Ann Transl Med 2015;3:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Drilon A, Cappuzzo F, Ou SI, Camidge DR. Targeting MET in lung cancer: will expectations finally be MET? J Thorac Oncol 2017;12:15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu C, Cao H, Shi C, Feng J. The role of circulating tumor DNA in therapeutic resistance. Onco Targets Ther 2019;12:9459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou C, Imamura F, Cheng Y, Okamoto I, Cho BC, Lin MC, et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib and comparator EGFR-TKIs in the FLAURA trial. J Clin Oncol 37, 2019(suppl; abstr 9020). [Google Scholar]
- 28. Shepherd FA, Papadimitrakopoulou V, Mok T, Wu YL, Han JY, Ahn MJ, et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib in the AURA3 trial. J Clin Oncol 36,2018(suppl; abstr 9027). [Google Scholar]
- 29. Yu HA, Ambrose H, Baik C, Cho BCC, Cocco E, Goldberg SB, et al. ORCHARD osimertinib + savolitinib interim analysis: a biomarker-directed phase II platform study in patients (pts) with advanced non-small cell lung cancer (NSCLC) whose disease has progressed on first-line (1L). Ann Oncol 2021;32Suppl 5:S978–S9. Abstract nr 1239P. [Google Scholar]
- 30. Wu YL, Cheng Y, Zhou J, Lu S, Zhang Y, Zhao J, et al. Tepotinib plus gefitinib in patients with EGFR-mutant non-small-cell lung cancer with MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitor (INSIGHT study): an open-label, phase 1b/2, multicentre, randomised trial. Lancet Respir Med 2020;8:1132–43. [DOI] [PubMed] [Google Scholar]
- 31. Ahn MJDe Marinis D, Bonanno L, Cho BC, Kim TM, Cheng S, et al. , MET biomarker-based preliminary efficacy analysis in SAVANNAH: savolitinib + osimertinib in EGFRm NSCLC post-osimertinib. J Thorac Oncol 2022;17(9 Suppl 4):S469–70. Abstract nr EP08.02-140.
- 32. Chae YK, Davis AA, Agte S, Pan A, Simon NI, Iams WT, et al. Clinical implications of circulating tumor DNA tumor mutational burden (ctDNA TMB) in non–small cell lung cancer. Oncologist 2019;24:820–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ebert EBF, McCulloch T, Hansen KH, Linnet H, Sorensen B, Meldgaard P. Clearing of circulating tumour DNA predicts clinical response to first line tyrosine kinase inhibitors in advanced epidermal growth factor receptor mutated non–small cell lung cancer. Lung Cancer 2020;141:37–43. [DOI] [PubMed] [Google Scholar]
- 34. Mu Y, Hao X, Xing P, Hu X, Wang Y, Li T, et al. Acquired resistance to osimertinib in patients with non-small-cell lung cancer: mechanisms and clinical outcomes. J Cancer Res Clin Oncol 2020;146:2427–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Recondo G, Bahcall M, Spurr LF, Che J, Ricciuti B, Leonardi GC, et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14-mutant NSCLC. Clin Cancer Res 2020;26:2615–25. [DOI] [PubMed] [Google Scholar]
- 36. Piper-Vallillo AJ, Halbert BT, Rangachari D, Kobayashi SS, Costa DB. Acquired resistance to osimertinib plus savolitinib is mediated by MET-D1228 and MET-Y1230 mutations in EGFR-mutated MET-amplified lung cancer. JTO Clin Res Rep 2020;1:100071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu L, Qu J, Heng J, Zhou C, Xiong Y, Yang H, et al. A large real-world study on the effectiveness of the combined inhibition of EGFR and MET in EGFR-mutant non-small-cell lung cancer after development of EGFR-TKI resistance. Front Oncol 2021;11:722039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kang J, Chen HJ, Wang Z, Liu J, Li B, Zhang T, et al. Osimertinib and cabozantinib combinatorial therapy in an EGFR-mutant lung adenocarcinoma patient with multiple MET secondary-site mutations after resistance to crizotinib. J Thorac Oncol 2018;13:e49–53. [DOI] [PubMed] [Google Scholar]
- 39. Lim SM, Yang SD, Lim S, Shim HS, Cho BC. Brief Report: heterogeneity of acquired resistance mechanisms to osimertinib and savolitinib. JTO Clin Res Rep 2021;2:100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deng L, Kiedrowski LA, Ravera E, Cheng H, Halmos B. Response to dual crizotinib and osimertinib treatment in a lung cancer patient with MET amplification detected by liquid biopsy who acquired secondary resistance to EGFR tyrosine kinase inhibition. J Thorac Oncol 2018;13:e169–e72. [DOI] [PubMed] [Google Scholar]
- 41. Wang C, Zhao K, Hu S, Li M, Song Y. Patterns and treatment strategies of osimertinib resistance in T790M-positive non-small cell lung cancer: a pooled analysis. Front Oncol 2021;11:600844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eser PÖ, Paranal RM, Son J, Ivanova E, Kuang Y, Haikala HM, et al. Oncogenic switch and single-agent MET inhibitor sensitivity in a subset of EGFR-mutant lung cancer. Sci Transl Med 2021;13:eabb3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu HA, Goldberg SB, Le X, Piotrowska Z, Goldman JW, De Langen AJ, et al. Biomarker-directed phase II platform study in patients with EGFR sensitizing mutation-positive advanced/metastatic non–small cell lung cancer whose disease has progressed on first-line osimertinib therapy (ORCHARD). Clin Lung Cancer 2021;22:601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jänne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689–99. [DOI] [PubMed] [Google Scholar]
- 45. Alfirevic A, Pirmohamed M. Drug induced hypersensitivity and the HLA complex. Pharmaceuticals (Basel) 2011;4:69–90. [Google Scholar]
- 46. Smit EF, Dooms C, Raskin J, Nadal E, Tho LM, Le X, et al. INSIGHT 2: a phase II study of tepotinib plus osimertinib in MET-amplified NSCLC and first-line osimertinib resistance. Future Oncol 2022;18:1039–54. [DOI] [PubMed] [Google Scholar]
- 47. Yoh K, Hirashima T, Saka H, Kurata T, Ohe Y, Hida T, et al. Savolitinib ± osimertinib in japanese patients with advanced solid malignancies or EGFRm NSCLC: Ph1b TATTON Part C. Target Oncol 2021;16:339–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods, 8 tables, 7 figures
Data Availability Statement
Data underlying the findings described in this article may be obtained in accordance with AstraZeneca's data sharing policy described at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure.