Abstract
The Philadelphia 9;22 chromosome translocation has two common isoforms that are preferentially associated with distinct subtypes of leukemia. The p210 variant is the hallmark of chronic myeloid leukemia (CML) whereas p190 is frequently associated with B-cell acute lymphoblastic leukemia. The only sequence difference between the two isoforms is the guanidine exchange factor domain. This guanidine exchange factor is reported to activate RHO family GTPases in response to diverse extracellular stimuli. It is not clear whether and, if so, how RHOA contributes to progression of p210 CML. Here we show that knockout of RHOA in the K562 and KU812, p210-expressing cell lines leads to suppression of leukemogenesis in animal models in vivo. RNA-sequencing analysis of the mock control and null cells demonstrated a distinct change in the gene expression profile as a result of RHOA deletion, with significant downregulation of genes involved in cell activation and cell adhesion. Cellular analysis revealed that RHOA knockout leads to impaired cell adhesion and migration and, most importantly, the homing ability of leukemia cells to the bone marrow, which may be responsible for the attenuated leukemia progression. We also identified IGFBP2 as an important downstream target of RHOA. Further mechanistic investigation showed that RHOA activation leads to relocation of the serum response factor (SRF) into the nucleus, where it directly activates IGFBP2. Knockout of IGFBP2 in CML cells suppressed cell adhesion/invasion, as well as leukemogenesis in vivo. This elevated IGFBP2 expression was confirmed in primary CML samples. Thus, we demonstrate one mechanism whereby the RHOA-SRF-IGFBP2 signaling axis contributes to the development of leukemia in cells expressing the p210 BCR-ABL1 fusion kinase.
Introduction
RHOA is a member of the large family of small GTPases which have diverse functions throughout biology.1 Binding to GDP represents an inactive conformation and activation is achieved through exchange with GTP. The transition between inactive and active forms is facilitated through binding to a guanidine exchange factor (GEF) motif, which is present in an equally diverse set of proteins.1 Activated GTPases serve as signaling intermediates from a variety of external stimuli to downstream effectors.2 RHOA has been most consistently implicated in cell movement through the association with actin cytoskeleton dynamics.3 Because of the central role of actin dynamics in the development and progression of cancer, RHOA has been implicated in invasion and metastasis as well as differentiation of stem cells.4,5 RHOA has also been suggested to play a central role in cancer development, in which it has been implicated as both a tumor suppressor as well as an oncogene.6,7 Early studies involved transfection into normal fibroblasts which led to transformation,8 suggesting an oncogenic role. However, extensive studies involving a wide range of solid tumors failed to demonstrate consistent amplifications or mutations,7 questioning the role of RHOA as an oncogene. In contrast, studies in subtypes of T-cell lymphomas identified consistent mutations, some of which appear to be activating events and hence tumor promoting.9-11 Overall, it appears that the involvement of RHOA in cancer progression may be dependent on the type of cancer cell. Indeed, we recently demonstrated a critical role for RHOA in the development of BCR-FGFR1-driven leukemias and lymphomas through an interaction with the GEF domain in the BCR component of the fusion kinase.12 In this study, deletion of the GEF domain led to a more aggressive development of leukemia, suggesting RHOA activation plays a suppressive role in this model of B-cell acute lymphoblastic leukemia. The same GEF domain is present in the BCR-ABL1 fusion kinase associated with the Philadelphia, 9;22 chromosome translocation (Ph), which is present consistently in a subtype of chronic myelogenous leukemia (CML).13 Both Ph+ CML and the FGFR1-driven stem cell leukemia/lymphoma syndrome are considered to be derived from hematopoietic stem cells and share many similarities but how RHOA contributes to the development of BCR-ABL1-driven CML is not clear.
The Ph 9;22 chromosome translocation results in the creation of a constitutively active chimeric BCR-ABL1 kinase that has two different variants depending on the position of the breakpoint on chromosome 9.14,15 The p210 isoform, which retains the BCR GEF domain and hence activates RHOA, is seen in ~95% cases of CML, which is a relatively benign leukemia.16 The p190 isoform is rarely seen in CML but is present in over 70% of cases of Ph+ B-cell acute lymphoblastic leukemia, which is an acute leukemia and has a far worse prognosis.17 In mouse models, p210 expression gives rise to a slowly progressing disease but when p190 is expressed, there is a short latency and acute development of B-cell leukemia.18,19 Cell line transformation assays revealed that the GEF domain of p210 BCR-ABL1 activates RHOA and contributes to the transformed phenotype.20 Whether and, if so, how this GEF domain-regulated RHOA signaling contributes to CML progression remains elusive.
In this study, we demonstrate that the deletion of RHOA in p210-expressing CML cells leads to suppression of leukemogenesis in vivo, suggesting that RHOA is important for the development and progression of Ph+ CML. RNA-sequencing analysis of mock control (MC) and RHOA knockout (KO) K562 cells identified IGFBP2 as a primary target of RHOA and has been implicated in both cell adhesion and activation. Analysis of gene expression data from human primary CML demonstrated the same high level of IGFBP2 expression as that in normal donors. Loss of RHOA prevents activation and subcellular relocalization of the serum response factor (SRF) which, as a result, leads to downregulation of the IGFBP2 gene. Downregulation of the SRF-IGFBP2 axis signaling cascade leads to suppression of cell adhesion and migration in CML cells and a decreased ability of the cells to home to the bone marrow. These studies suggest an oncogenic role for RHOA in the development of p210 BCRABL1-driven CML.
Methods
Cell and molecular studies
Cell proliferation was assessed using trypan blue exclusion assays and cell viability was measured using the CellTiter Glo assay, western blotting, plasmid transfection, and quantitative reverse transcriptase polymerase chain reaction (RT-PCR), following standard procedures that have been described previously.21,22 A summary of the antibodies used can be found in the Online Supplementary Methods. Luciferase assays were performed as described previously.23
CRISPR/Cas9 knockout
Single-guide RNA used for locus-specific deletion of RHOA in CML cells were designed using CRISPR Targets Track on Genome Browser24 and are described in the Online Supplementary Methods. Virus packing and single clone selection were performed as described previously.25
In vivo studies
All animal experiments were performed under an approved protocol from the Augusta University Institutional Animal Care and Use Committee. Unless specified, 5×106 cells were injected into the tail veins of 6- to 8-week-old female immune deficient NSG mice. Engraftment of luciferase-ex-pressing leukemia cells in the host mice was determined using the Caliper IVIS imaging systems as described previously.26 Homing studies were performed by flow analysis of GFP+ cells in the bone marrow cells from mice implanted 16 hours previously with various cell types.27
Cell adhesion and migration assays
Cell adhesion and migration assays were performed as described previously.27 For the IGFBP2 rescue experiment, 100 ng/mL recombinant human IGFBP2 protein (R&D Systems, #674-B2-025) were added to the growth medium prior to the cell adhesion and migration assays.
Chromatin immunoprecipitation quantitative polymerase chain reaction
Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP assay kit (Millipore) as described previously.23 In brief, chromatin was cross-linked with 1% formaldehyde for 10 min at room temperature, sheared to an average size of ~500 bp and then immunoprecipitated with an anti-SRF antibody (Cell Signaling, #5147). Details of the ChIP-quantitative polymerase chain reaction (qPCR) primers used are provided in the Online Supplementary Methods. Each immunoprecipitated DNA sample was quantified using qPCR and all ChIP-qPCR signals at the IGFBP2 locus were normalized to an IgG control to calculate relative fold enrichment.
Protein subcellular localization assay
For western blot assays, cytoplasmic and nuclear components were isolated using the NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific). For confocal microscopy, cells were first probed with an anti-SRF antibody (Cell Signaling, #5147), then visualized with Alexa Fluor 488 goat anti-rabbit IgG secondary antibody (Abcam, ab150077).
RNA sequencing and analysis
Two MC clones and the RHOA KO C8 and C9 clones generated from K562 cells were harvested for RNA-sequencing analysis. Preparation of RNA, sequence analysis and gene set enrichment analysis were performed as described previously.28
Statistical analyses
All statistical analyses were performed using the Student t test to determine whether the means of two datasets were significantly different from each other. Error bars represent standard deviations. Unless otherwise stated, in vitro assays were repeated in triplicate and in vivo experiments involved cohorts of five mice. Kaplan-Meier statistical approaches were used to analyze differences in survival between different cohorts of mice.
Results
RHOA knockout in BCR-ABL1-positive chronic myeloid leukemia cells has only a mild effect on cell proliferation/survival
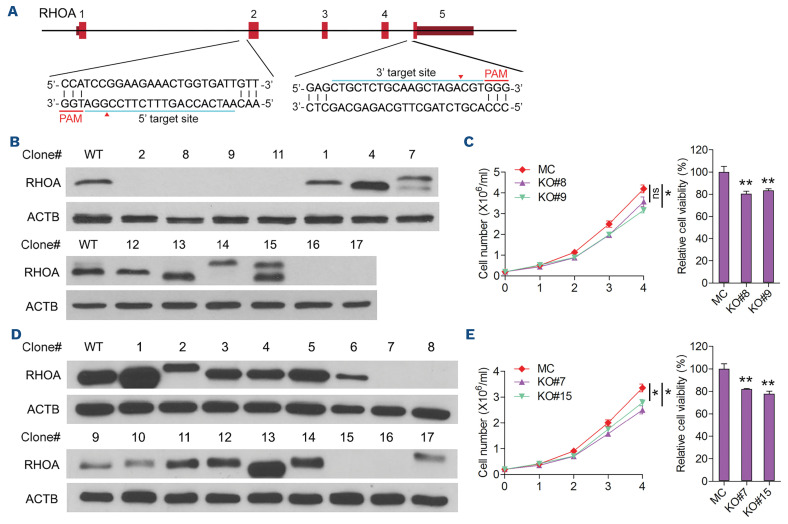
In Ph+ leukemia, there are two major variants of the 9;22 translocation which generate either p190 or p210 variant proteins depending on the position of the translocation breakpoint.13 p210 is the hallmark of CML, whereas p190 occurs in the majority (70%) of cases of Ph+ B-cell acute lymphoblastic leukemia.17 The only sequence difference between p210 and p190 is the inclusion of the tandem double-homology, pleckstrin homology (DH-PH) domains in p210 which provide a GEF domain for activation of the RHOA GTPase, suggesting its potential contribution in the development of CML. Our recent study of the BCR-FGFR1 fusion kinase in stem cell leukemia/lymphoma syndrome demonstrated a role for the GEF domain in the BCR component of the chimeric kinase in activating RHOA and influencing leukemia progression.12 To obtain a better understanding of the role of RHOA in the development of Ph+ CML, we created knockdown clones from two human, p210-expressing cell lines, K562 and KU812,29 using a double targeting CRISPR-Cas9 approach (Figure 1A). Both cell lines were first transduced with pCDH-CMV-NlucP2A-copGFP-T2A-Puro to co-express luciferase and GFP, and then transduced with lentiCRISPR v2 and both the sgRNA and Cas9 to generate the KO cell clones. Using this strategy, exons 2-4 were deleted and a series of RHOA null clones were identified using western blotting (Figure 1B). For further analysis, clones C8 and C9 were randomly selected from K562 and cell proliferation assays showed only small differences between the KO clones and the MC cells (Figure 1C, left). CellTiter Glo assays did not reveal any profound differences in cell survival between the MC and KO cells (Figure 1C, right). Similarly, RHOA was knocked out in KU812 cells and KO clones C7 and C15 also showed only marginal differences in cell proliferation or viability between the KO clones and the MC cell (Figure 1D, E). These observations demonstrate that RHOA has a relatively limited effect on BCR-ABL1-driven CML growth and survival.
RHOA knockout suppresses leukemogenesis in vivo
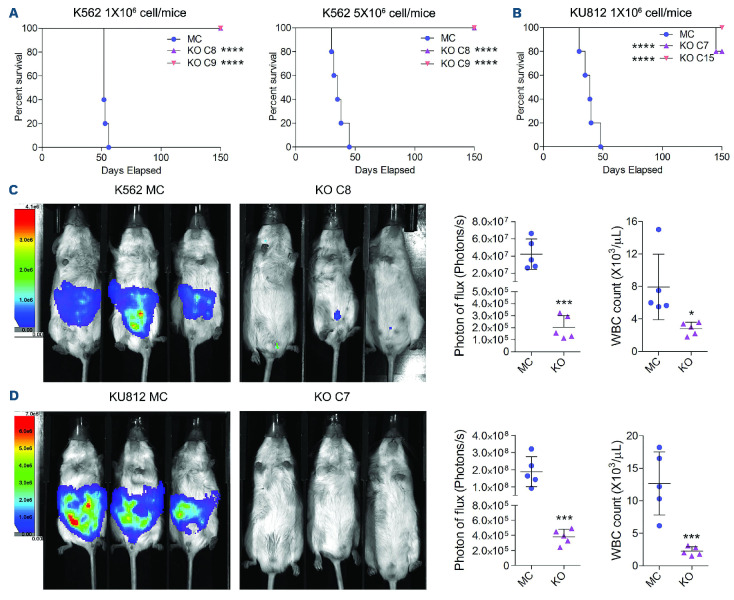
When 1×106 K562 KO C8 or C9 cells were xenografted into immune-compromised NSG mice, Kaplan-Meier analysis showed no mortality over a 150-day observation period, compared with the MC xenografted mice which all died within 56 days (Figure 2A). When the inoculated dose was increased to 5×106 cells/mouse, although the median survival in the MC-cell group decreased from 52 days to 35 days, leukemia-free survival in the RHOA KO cell-engrafted mice was the same. Similarly, RHOA KO clones C7 and C15 from KU812 had a dramatic impact on survival compared with the MC cells. Although one mouse in the KO C7 cellengrafted cohort died late in the observation period, there was no evidences of leukemia at autopsy. Luminescence imaging of the xenografted animals using cells retrofitted to express luciferase showed extensive development of leukemia in the MC-cell-engrafted mice compared to the RHOA KO-cell-engrafted groups from both cell lines (Figure 2C, D). Quantitation of the luminescence signal in the mice as well as white blood cell counts also reflected the distinct disease progression in these mice (Figure 2C, D). Thus, while RHOA does not appear to dramatically affect cell proliferation or survival in this system, it has a profound effect on leukemogenesis in vivo.
RHOA knockout reduces expression levels of genes related to cell adhesion and activation
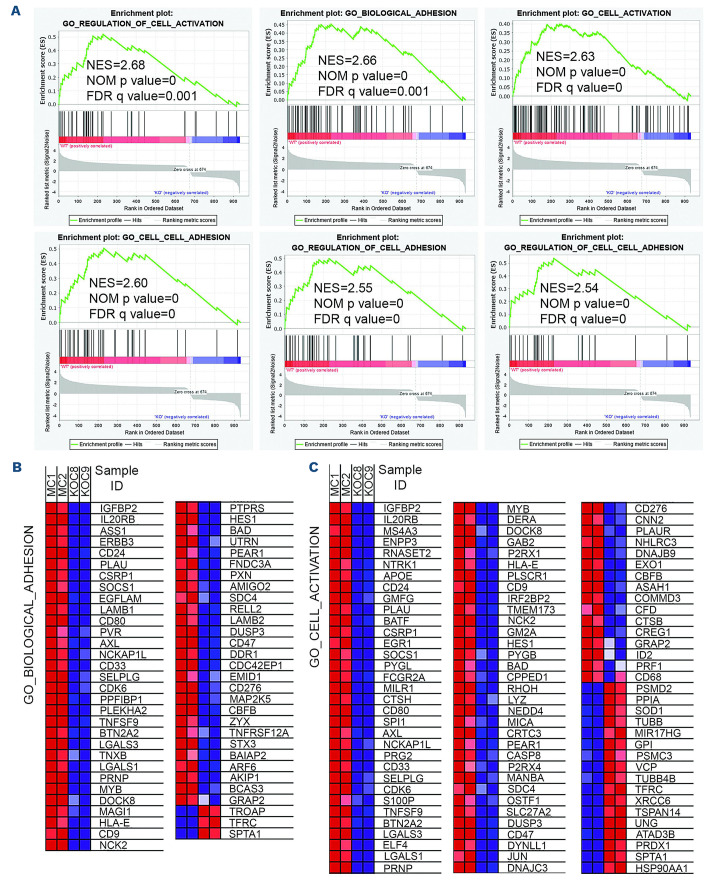
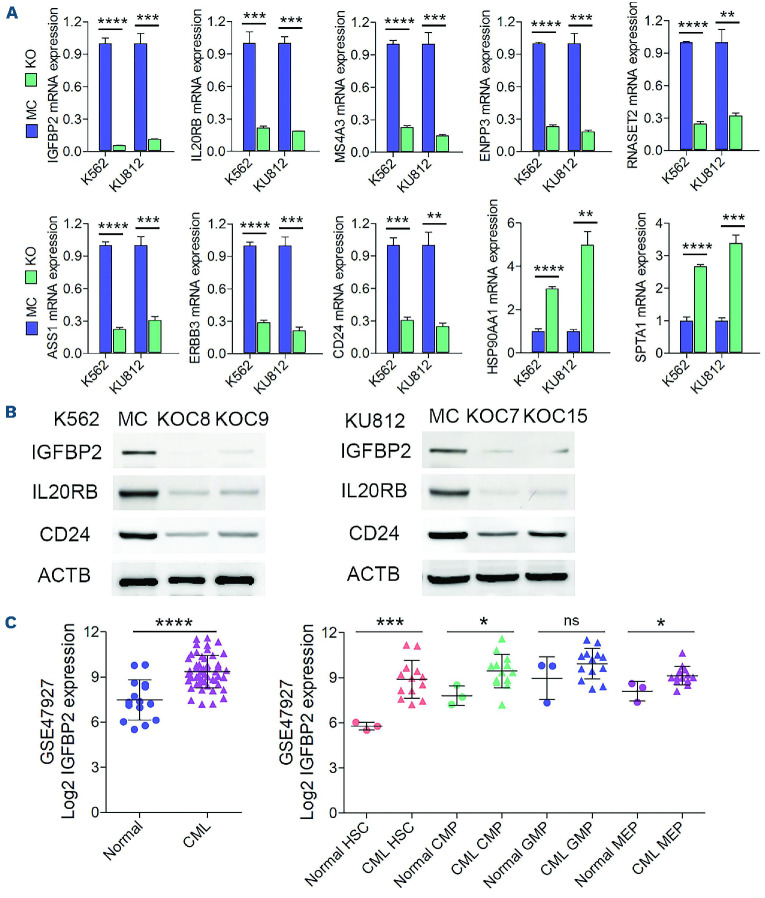
To investigate how RHOA expression might affect CML progression, we analyzed the underlying molecular effects of RHOA using RNA sequencing in a gene expression comparison between K562 MC cells and the two individual KO C8 and C9 clones (Figure 3A). Gene set enrichment analysis of the differentially expressed genes identified changes in major biological functions which included both cell activation and cell adhesion. Heatmap plots clearly show that most of the implicated genes were downregulated as a result of RHOA knockout (Figure 3B). RT-PCR analysis of a subset of genes in these groups (Figure 4A) confirmed the downregulation of IGFBP2, ASS1, IL20RB, ERBB3, MS4A3, CD24 ENPP3 and RNASET2 and upregulation of HSP90AA1 and SPTA1. The same changes in relative gene expression levels seen in the K562 KO cells were also seen in the RHOA KO cells derived from KU812 cells using quantitative RT-PCR. The reduced gene expression levels seen following RHOA depletion were further confirmed at the protein level using western blotting (Figure 4B). In both cell lines, IGFBP2 was the most highly down-regulated gene, suggesting an important role for this gene in the regulation of leukemogenesis.
Figure 1.
Generation of RHOA knockout clones and the effect of knockout on cell proliferation in vitro. (A) Overview of the CRISPR strategy to generate RHOA knockout shows the location of target regions within the gene flanking exons 2 and 4 and the protospacer adjacent motif. (B) Western blot analysis of clones recovered following RHOA targeting in K562 cells identified six clones that are null for RHOA. (C) Trypan blue exclusion assays (n=3) over a 4-day proliferation period showed minor differences in cell growth between the mock control (MC) cells and cells from knockout (KO) clones C8 and C9. CellTiter Glo viability assays at day 3 also showed only a marginal difference in cell survival between the MC and KO cells (n=3). (D, E) In the same analysis of KO clones identified from the KU812 cells (D), there is only a minor difference in both cell proliferation and survival (E). Differences between the KO and MC cells were evaluated using the Student t test. *P<0.01, **P≤0.001, ns: not significant. PAM: protospacer adjacent motif.
While a role for RHOA in leukemogenesis was suggested from the findings of the knockout studies, these were performed in highly evolved cell lines. The question therefore arises whether RHOA activation influences primary human CML development. To address this issue, we analyzed the GSE 47927 gene expression dataset that was derived from human CML patients’ samples (n=52) compared with cells from normal donor samples (n=15). Within the CML cases the samples were further analyzed by isolating various stem/progenitor cells from these leukemias. These included hematopoietic stem cells, common myeloid progenitors, granulocyte monocytic progenitors and megakaryocyte erythroid progenitors. Since we have shown that functional RHOA activates IGFBP2 expression, we further analyzed the expression of IGFBP2 in the various subpopulations of cells as a surrogate for RHOA activation. There was a significant increase in expression levels in the overall leukemic cell population (Figure 4C). Within the subgroups, highly significant increases were seen in the hematopoietic stem cells with less significant increases in the common myeloid progenitors and megakaryocyte erythroid progenitors. There was no difference in the granulocyte monocytic progenitor population. Although cytogenetic findings were not reported for the CML cases, since 95% of CML carry the p210 rearrangement, these data suggest that in human CML, RHOA activated by p210 upregulates expression of IGFBP2 and contributes to CML progression, as observed in our cell line models.
Figure 2.
RHOA knockout suppresses chronic myeloid leukemia progression in vivo. (A) Kaplan-Meier survival analysis of mice xenografted with 1x106 mock control (MC) cells or knockout (KO) K562 cells shows that those that received MC cells had a median survival of 52 days; none of the mice xenografted with the KO cells developed disease over the 150-day observation period. When the inoculum was increased to 5x106 cells, while there was a reduction in the mean survival time to 35 days for recipients of the MC cells, there was no difference in survival for those that received the KO cells. (B) Similarly, for the KU812 cells, while the mean survival of mice in the MC group was 39 days, none of the mice injected with the KO cells developed of leukemia over the observation period. (C, D) This significant difference in disease development for K562 cells (C) and KU812 cells (D) could be visualized in mice using luminescence tracking which was mirrored by the relative levels of white blood cells in the peripheral blood at the time of sacrifice (N=5). Differences between the KO and MC cells were evaluated using the Student t test. *P<0.01, ***P≤0.0001, ****P=0.00001.
Cell adhesion and homing are affected by RHOA
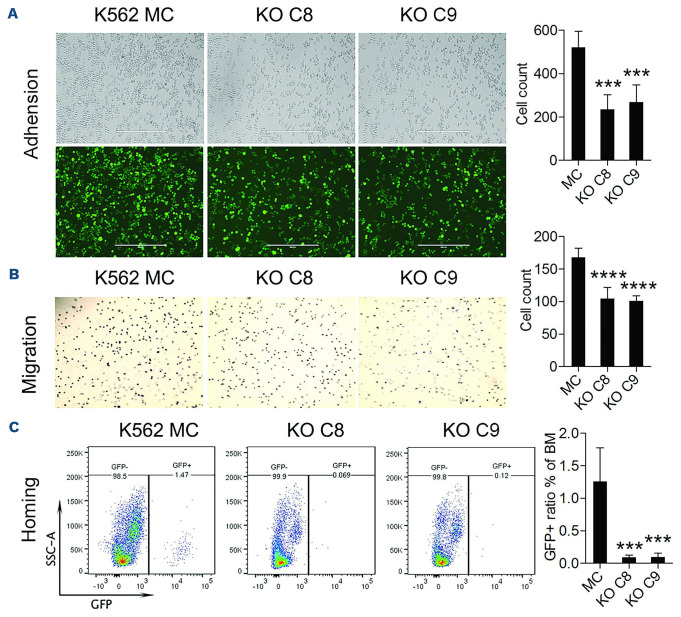
Since the gene set enrichment analysis defined cell activation and adhesion as the most significantly affected biological processes following deletion of RHOA, we analyzed these phenotypes in more depth. Adhesion was evaluated by plating 1x105 GFP+ cells onto fibronectin fragment, CH-296-coated plates as we have described previously.30 The number of cells was determined by counting GFP+ cells per unit area using ImageJ software (Figure 5A). Compared to the K562 MC cells, the RHOA KO clones, C8 and C9, showed significantly reduced adhesion properties.
In a similar comparison of migration through extracellular matrix, the RHOA KO cells showed a significantly reduced migration potential compared with the K562 MC cells (Figure 5B). Since it is possible that reduced cell invasion and migration may lead to a reduced ability of leukemic cells to home to the bone marrow, we used homing assays to compare the homing ability of MC and RHOA KO cells. When 5x106 K562 MC cells were injected into the tail veins of NSG mice, the presence of GFP+ leukemic cells in the bone marrow 16 h after injection was significantly increased compared to the number of GFP+ cells following injection of KO C8 or C9 cells, demonstrating a reduced capacity for homing following RHOA KO (Figure 5C). These empirical observations provide support for the gene expression studies implicating RHOA in these two biological processes in CML leukemia cells and their contribution to leukemia progression.
Figure 3.
RHOA knockout changes the gene expression profiles in K562 chronic myeloid leukemia cells. (A) Gene set enrichment analysis of RNA-sequencing data from K562 mock control (MC) or RHOA knockout (KO) clones (N=2) showed significant enrichment of genes involved in cell adhesion and activation biological processes in the RHOA-expressing MC cells. Gene set enrichment analysis of RNA-sequencing data from K562 mock control (MC) or RHOA knockout (KO) clones (N=2) showed significant enrich ment of genes involved in cell adhesion and activation biological processes in the RHOA-expressing MC cells. (B) A comparison in expression changes of genes associated with cell adhesion is illustrated and predominantly shows significant downregulation in the KO cells. (C) Similarly genes involved in cell activation biological processes predominantly show significant downregulation in the KO cells. NES: normalized enrichment score; NOM p-values: nominal P values; FDR q-values: false discovery rate q values.
RHOA regulates IGFBP2 expression in a serum response factor-dependent manner
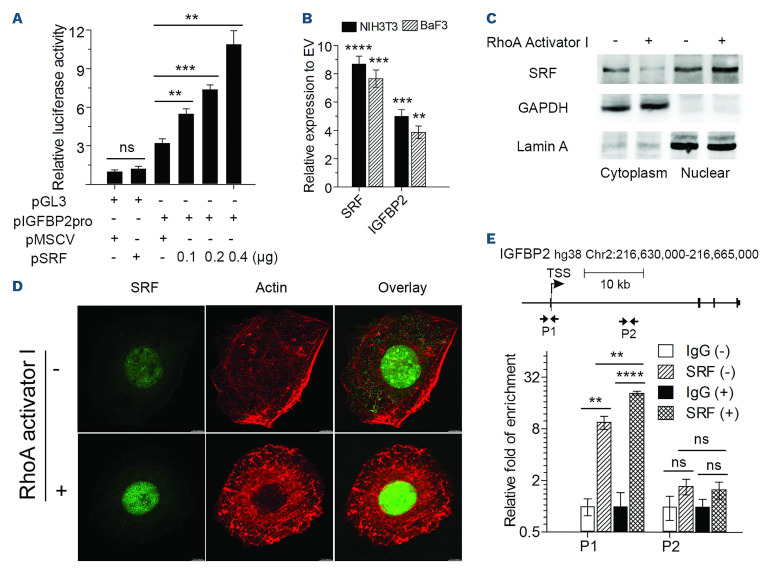
The SRF regulates many genes involved in cell migration and adhesion,31 which are also the biological processes most significantly affected as a result of RHOA deletion. In addition, RHOA has been shown to regulate gene expression, in part, through the regulation of the SRF transcription factor.32 Both SRF and IGFBP2 have been shown to orchestrate cell motility and invasion phenotypes in different cancers.33,34 To determine whether the downregulation of IGFBP2 in RHOA KO cells is dependent on SRF, we generated reporter constructs in the pGL3 vector containing the 1 kb fragment spanning the IGFBP2 promoter. In the absence of the promoter, SRF was unable to induce expression of luciferase. However, when the promoter region was present, luciferase activity increased in the presence of SRF, in a dose-dependent manner (Figure 6A). Thus, it appears that SRF can regulate IGFBP2 gene expression directly.
Figure 4.
Detection of the expression levels of potential RHOA downstream targets in chronic myeloid leukemia cells and primary patients’ samples. (A) Quantitative reverse transcriptase polymerase chain reaction (RT-qPCR) analysis was used to verify differential expression of several of the most highly downregulated and upregulated genes in K562 RHOA knockout (KO) cells compared with mock control (MC) cells (N=3). Analysis of the same genes in the KU812 RHOA KO cells using RT-qPCR showed the same differential expression patterns. (B) The reduced expression levels for IGFBP2, IL20RB and CD24 were further confirmed using western blotting. (C) Analysis of the GSE47927 chronic myeloid leukemia (CML) dataset revealed a highly significant difference in IGFBP2 expression in CML samples (n=52) compared with normal controls (n=15). When cells from these CML cases were sub-fractionated into different stem/progenitor subpopulations, there was a highly significant increase in IGFBP2 expression in CML hematopoietic stem cells compared with normal counterparts. In similar comparisons, a significant increase in IGFBP2 expression was seen in common myeloid progenitors and megakaryocyte-erythroid progenitors but no difference was seen in granulocyte-monocytic progenitors. Statistical significance was established using the Student t test. **P≤0.001, ***P≤0.0001, ****P≤0.00001. HSC: hematopoietic stem cells; CMP: common myeloid progenitors; GMP: granulocyte-monocytic progenitors; MEP: megakaryocyte-erythroid progenitors.
In addition, when SRF was overexpressed in BaF3 hematopoietic cells or NIH 3T3 cells, there was a proportional increase in IGFBP2 expression compared with the expression of cells transduced with the empty vector (Figure 6B). To further investigate how RHOA regulates SRF activity in leukemia, we treated both K562 and NIH3T3 cells with RHOA activator I. Following activation of RHOA, western blotting demonstrated an increased localization of SRF to the nucleus in K562 cells (Figure 6C), which could be visualized directly in adherent NIH 3T3 cells (Figure 6D). ChIP-qPCR analysis (Figure 6E) demonstrated direct binding of SRF to the IGFBP2 promoter region in K562 cells and its promoter occupancy was further enhanced when the cells were exposed to RHOA activator I in the culture medium. Thus, loss of RHOA activity in p210 BCRABL1-expressing CML cells leads to reduced IGFBP2 expression as a result of a reduced ability to promote SRF-mediated transcription activation.
IGFBP2 knockout can partially recapitulate the effects of RHOA knockout on chronic myeloid leukemia progression
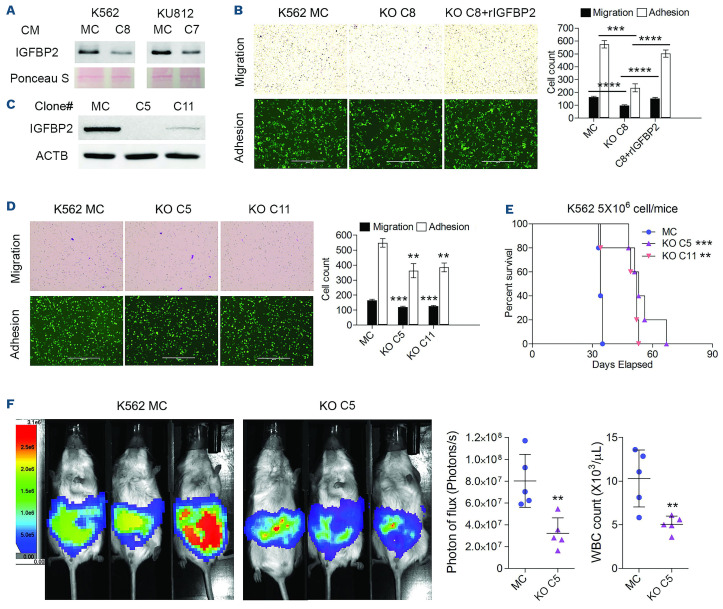
IGFBP2 is a secreted protein and exerts its effect through controlling the distribution, function, and activity of insulin-like growth factors (IGF) in the pericellular space, although it also has IGF-independent mechanisms of action.35 Analysis of the culture medium recovered from MC K562 and KU812 cells expressing RHOA showed relatively high levels of IGFBP2 but these levels were dramatically reduced in KO cells derived from these two cell lines (Figure 7A). To determine whether the level of secretion of IGFBP2 from these cells affects the adhesion and migration phenotypes, we introduced exogenous IGFBP2 into the culture medium of K562 KO C8 cells. While the KO cells showed a reduction in migration and adhesion compared with the MC cells, there was a significant increase in the adhesion and migration potential of these cells after adding IGFBP2 to the culture medium (Figure 7B). To explore this effect further, we generated IGFBP2 KO clones (C5 and C11) from K562 cells using CRISPR (Figure 7C). When these clones were subjected to migration and adhesion assays (Figure 7D), both phenotypes were significantly impaired, demonstrating the importance of IGFBP2 in the regulation of these phenotypes in CML cells. In vivo engraftment of these cells (Figure 6E) demonstrated that, as a result of the IGFBP2 loss, there was a highly significant increase in survival in mice xenografted with the KO cells compared with the MC cells. These results were confirmed using luminescence imaging (Figure 6F) and white blood cell counts in the peripheral blood of the animals. Thus, IGFBP2 is clearly an important target of RHOA in the regulation of the oncogenic phenotype of BCR-ABL1-expressing leukemic cells.
Discussion
RHOA has been associated with diverse aspects of cancer development and progression but whether it can be described as an oncogene or a tumor suppressor gene has been an open question.1,6 In fibroblast transformation assays RHOA acted as an oncogene but activation deficient mutations did not.8 In murine studies, specific inactivation of RHOA did not support an oncogenic function in vivo. Analysis of mutations in human cancer showed that RHOA is infrequently amplified and is often predicted to be deleted, suggesting a tumor suppressor function.7,31 Many of the mutations used to study RHOA function in vitro are not observed in human cancers and so their direct role in cancer development has been difficult to interpret.7 Despite the variable presence of RHOA mutations and amplifications in human solid tumors, consistent RHOA mutations predicted to be activating events have been found in certain T-cell and B-cell leukemias and lymphomas.9-11 In the specific example of Ph+ CML, we show that deletion of RHOA leads to suppression of leukemogenesis, favoring an oncogenic role in cells expressing the p210 BCR-ABL chimeric kinase harboring the GEF domain that activates RHOA.
The role of RHOA in cancer development appears to be due both to actin cytoskeleton-initiated molecular signaling and a direct role in regulating various aspects of cell movement and invasion through interactions with the actin cytoskeleton machinery.1,4 Overall, however, the role of RHOA in cancer development appears to be cell context-dependent, which may be further complicated by the differential availability of the various binding partners in different cell types. Our studies here define a clear role for RHOA in promoting leukemogenesis of Ph+ CML, since its knockout completely suppressed leukemogenesis in vivo. However, in our studies in stem cell leukemia/lymphoma syndrome driven by the BCR-FGFR1 chimeric kinase, prevention of RHOA activation as a result of deletion of the GEF domain led to a more aggressive disease,12 further highlighting the contextual role of RHOA in different cancer cell types.
Figure 5.
RHOA regulates the cell mobility of chronic myeloid leukemia cells. (A) K562 mock control (MC) and knockout (KO) C8 and C9 cells were plated on CH-296-coated plates (N=3) and after 24 hours randomly selected fields were photographed (N=5) and the number of attached cells per field was counted using ImageJ software, which showed a significant reduction in cell adhesion for the KO cells. (B) Using transwell assays, there was a significant reduction in the number of cells migrating through the membrane for the KO cells compared with the MC cells. (C) For the homing assay, mice were xenografted with 5x106 cells and, after 16 hours, the ratio of GFP+ cells in the bone marrow (BM) was determined using flow cytometry. There was a significant reduction in the number of KO cells homing to the BM compared with the number of MC cells. The scale bar in (A) represents 400 µm, with the same magnification for images in (A) and (B). Differences between the KO and MC cells were evaluated using the Student t test. ***P≤0.0001, ****P≤0.00001.
While the mechanism by which RHOA influences the malignant phenotype is still emerging, in the CML cell model we describe, RHOA clearly has a profound influence on gene expression profiles and appears to activate many genes involved in cell migration and actin cytoskeleton interactions. It was shown some time ago that activated RHOA is required for transcriptional activation by SRF32 and that this is facilitated by RHOA-dependent relocation of SRF into the nucleus. We now show that migration and invasion of CML cells are regulated by the SRF transcription factor, which is well known to promote expression of genes involved in these phenotypes.31 IGFBP2 was the most highly affected gene resulting from RHOA knockout, which we now show is regulated directly by SRF in a RHOA-dependent manner. IGFBP2 also directly influences cell migration and adhesion as well as homing of cells to the bone marrow.
Knockout of IGFBP2 led to impaired cell migration and adhesion and improved survival in the CML animal model, implying a significant direct role in leukemogenesis. IGFBP2 is highly expressed in a broad range of cancers, including leukemias, and has become a valuable biomarker for poor outcome. In some cancer types IGFBP2 also promotes cell migration and invasion36 and the fact that it is regulated by SRF further demonstrates the close interaction to promote RHOA-mediated signaling. As a secreted protein, it serves its main function through binding to IGF receptors to initiate signal transduction involved in oncogenesis and fetal development, although it has also been shown to signal through IGF-independent mechanisms.31 In cancers such as gliomas, it is recognized as an oncogene and potential target for therapy.35 In particular, IGFBP2 binds to integrins,37 which are involved in tumor progression, migration and metastasis. IGFBP2 is a pivotal regulator of the process of epithelial-mesenchymal transition through the PI3K/AKT axis and the NFKB pathway by activating PI3K/ATK and suppressing PTEN.36-38 The observations presented here confirm a direct role for IGFBP2 in the regulation of leukemia cell mobility which contributes to leukemia progression. In addition, tumorigenic cell-derived IGFBP2 has also been shown to support in vitro and in vivo expansion of hematopoietic stem cells.39-41 It appears therefore that IGFBP2 expression in the leukemia cells, through RHOA-SRF signaling, may also contribute to leukemia progression by influencing the bone marrow microenvironment and modulating hematopoietic cells, concepts that are worthy of further investigation.
Figure 6.
RHOA activates IFGBP2 expression through serum response factor. (A) Luciferase expression analysis shows that in the presence of the IGFBP2 promoter there was an increase in activity that was proportional to the levels of serum response factor (SRF) transfected in the same cells. (B) When SRF was overexpressed in either NIH3T3 or BaF3 cells, there was a proportional increase in IGFBP2 expression. (C) When K562 cells were treated with RHOA activator I, levels of SRF declined in the cytoplasm but increased in the nucleus. The relative purity of the cytoplasmic and nuclear protein enrichment was demonstrated by the almost exclusive presence of proteins GAPDH (cytoplasm) and Lamin A (nucleus). (D) The same increased levels of SRF in the nucleus of adherent 3T3 cells following RHOA activation was seen using confocal microscopy. (E) Chromatin immunoprecipitation quantitative polymerase chain reaction analysis from K562 cells using primers P1, which target the IGFBP2 promoter region, showed occupancy of SRF on the IGFBP2 promoter (SRF-), which was increased when the cells were treated with the RHOA activator (SRF+). In the same experimental model, no significant changes were seen in the downstream intron region defined by the P2 primers. The scale bar in (D) represents 50 µm. Differences between the knockout and matched control cells were evaluated using the Student t test. Cells transduced with empty vector were used as the control for comparison in (B). *P<0.01, **P≤0.001, ***P≤0.0001, ****P≤0.00001, ns: not significant. MSCV: murine stem cell virus; EV: empty vector.
In our studies we focused on the role of RHOA in the development of p210-expressing CML cells. While the structural loss of the GEF domain in p190-expressing leukemias precludes RHOA activation, these leukemias show a more aggressive disease course. Here, we clearly demonstrate a critical disease-promoting role for RHOA in p210-expressing CML. With regard to leukemogenesis in p190-expressing cells, it appears that these cells have evolved other molecular oncogenic mechanisms that are independent of RHOA function. Indeed, in gene expression studies comparing p190- and p210-expressing leukemias there was a significant difference in gene expression patterns.42 p190-expressing CML samples exhibited an upregulation of interferon, IL1R and p53 and hyperactivation of STAT1/JAK1, SRC and PAK1 signaling, all of which are related to aggressive cell growth. p190-expressing CML, although rare, showed gene expression patterns similar to the p190-expressing acute lymphoblastic leukemia samples. In addition, using quantitative comparative proteomics, strong differences in the interaction and cellular phosphoproteome were identified between p190- and p210-expressing cells in two different studies.43,44 Thus, it appears that the course of disease progression in cells expressing the variant Ph chromosome translocations is likely due to their different overall genetic signatures and that RHOA activation is specifically important on the background of the p210-directed genetic changes driving leukemogenesis.
Figure 7.
Knockout of IFGBP2 partially recapitulates the phenotype of RHOA loss. (A) Analysis of the supernatant from K562 C8 and KU812 C7 knockout (KO) cells when compared with the respective mock control (MC) cells, showed a dramatic reduction in IGFBP2 protein levels in the KO cell culture medium. (B) In migration and adhesion analysis, K562 KO C8 cells showed a reduction in both phenotypes compared with MC cells. When exogenous, recombinant IGFBP2 (rIGFBP2) was added to the culture medium, there was a significant increase in both phenotypes in the KO C8 cells. (C, D) CRISPR knockout of IGFBP2 in K562 cells generated two clones, C5 and C11 (C), which when subjected to migration and adhesion assays (D) showed reduced levels compared with MC cells. (E) When these cells were xenografted into NSG hosts, mice receiving KO clones C5 and C11 showed an increased survival compared with MC-engrafted mice. (F) This result was consistent with luminescence intensity in the mice after 28 days, which showed that the tumor burden was significantly reduced in the mice grafted with the KO C5 cells compared to that of mice engrafted with MC cells. White blood cell count at the time of sacrifice was also reduced in the mice grafted with KO C5 cells. The scale bars in (B) and (D) represent 400 µm, with the magnification for the images in (B) and (D) being the same. Differences between the KO and MC cells were evaluated using the Student t test. Pairwise comparisons are indicated by the horizontal lines. **P≤0.001, ***P≤0.0001, ****P ≤0.00001. CM: culture medium; WBC: white blood cells.
Supplementary Material
Acknowledgments
This research was conducted in part in the Georgia Cancer Center Shared Resources (Cores).
Funding Statement
Funding: This work was supported by grant CA076167 (to JKC) from the National Institutes of Health, USA.
References
- 1.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Ann Rev Cell Dev Biol. 2005;21:247-269. [DOI] [PubMed] [Google Scholar]
- 2.Thumkeo D, Watanabe S, Narumiya S. Physiological roles of Rho and Rho effectors in mammals. Eur J Cell Biol. 2013;92(10-11):303-315. [DOI] [PubMed] [Google Scholar]
- 3.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70(3):389-399. [DOI] [PubMed] [Google Scholar]
- 4.Lawson CD, Ridley AJ. Rho GTPase signaling complexes in cell migration and invasion. J Cell Biol. 2018;217(2):447-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen E, Brakebusch C. Rho GTPase function in development: how in vivo models change our view. Exp Cell Res. 2012;318(14):1779-1787. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert-Ross M, Marcus AI, Zhou W. RhoA, a novel tumor suppressor or oncogene as a therapeutic target? Genes Dis. 2015;2(1):2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svensmark JH, Brakebusch C. Rho GTPases in cancer: friend or foe? Oncogene. 2019;38(50):7447-7456. [DOI] [PubMed] [Google Scholar]
- 8.Prendergast GC, Khosravi-Far R, Solski PA, Kurzawa H, Lebowitz PF, Der CJ. Critical role of Rho in cell transformation by oncogenic Ras. Oncogene. 1995;10(12):2289-2296. [PubMed] [Google Scholar]
- 9.Yoo HY, Sung MK, Lee SH, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(4):371-375. [DOI] [PubMed] [Google Scholar]
- 10.Sakata-Yanagimoto M, Enami T, Yoshida K, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(2):171-175. [DOI] [PubMed] [Google Scholar]
- 11.Palomero T, Couronné L, Khiabanian H, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46(2):166-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu T, Chong Y, Lu S, et al. Loss of the BCR-FGFR1 GEF domain suppresses RHOA activation and enhances B-lymphomagenesis in mice. Cancer Res. 2019;79(1):114-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quintás-Cardama A, Cortes J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 2009;113(8):1619-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pane F, Intrieri M, Quintarelli C, Izzo B, Muccioli GC, Salvatore F. BCR/ABL genes and leukemic phenotype: from molecular mechanisms to clinical correlations. Oncogene. 2002;21(56):8652-8667. [DOI] [PubMed] [Google Scholar]
- 15.Wong S, Witte ON. The BCR-ABL story: bench to bedside and back. Annu Rev Immunol. 2004;22:247-306. [DOI] [PubMed] [Google Scholar]
- 16.Baccarani M, Iacobucci I, Chiaretti S, et al. In Ph+BCR-ABL1(P210+) acute lymphoblastic leukemia the e13a2 (B2A2) transcript is prevalent. Leukemia. 2020;34(3):929-931. [DOI] [PubMed] [Google Scholar]
- 17.Burmeister T, Schwartz S, Bartram CR, Gökbuget N, Hoelzer D, Thiel E; GMALL study group. Patients' age and BCR-ABL frequency in adult B-precursor ALL: a retrospective analysis from the GMALL study group. Blood. 2008;112(3):918-919. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Ilaria RL Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189(9):1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovacic B, Hoelbl A, Litos G, et al. Diverging fates of cells of origin in acute and chronic leukaemia. EMBO Mol Med. 2012;4(4):283-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahay S, Pannucci NL, Mahon GM, et al. The RhoGEF domain of p210 Bcr-Abl activates RhoA and is required for transformation. Oncogene. 2008;27(14):2064-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Kim J, Teng Y, et al. Loss of ATF3 promotes hormone-induced prostate carcinogenesis and the emergence of CK5+CK8+ epithelial cells. Oncogene. 2016;35(21):3555-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowell JK, Qin H, Hu T, Wu Q, Bhole A, Ren M. Mutation in the FGFR1 tyrosine kinase domain or inactivation of PTEN is associated with acquired resistance to FGFR inhibitors in FGFR1-driven leukemia/lymphomas. Int J Cancer. 2017;141(9):1822-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu T, Wu Q, Chong Y, et al. FGFR1 fusion kinase regulation of MYC expression drives development of stem cell leukemia/lymphoma syndrome. Leukemia. 2018;32(11):2363-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu T, Pi W, Zhu X, et al. Long non-coding RNAs transcribed by ERV-9 LTR retrotransposon act in cis to modulate long-range LTR enhancer function. Nucleic Acids Res. 2017;45(8):4479-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piranlioglu R, Lee E, Ouzounova M, et al. Primary tumor-induced immunity eradicates disseminated tumor cells in syngeneic mouse model. Nat Commun. 2019;10:1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taniguchi Ishikawa E, Chang KH, Nayak R, et al. Klf5 controls bone marrow homing of stem cells and progenitors through Rba5-mediated B1/b2-integrin trafficking. Nat Commun. 2013;4:1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren M, Qin H, Kitamura E, Cowell JK. Dysregulated signaling pathways in the development of CNTRL-FGFR1-induced myeloid and lymphoid malignancies associated with FGFR1 in human and mouse models. Blood. 2013;122(6):1007-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deregowska A, Pepek M, Pruszczyk K, Machnicki MM, Wnuk M, Stoklosa T. Differential regulation of telomeric complex by BCR-ABL1 kinase in human cellular models of chronic myeloid leukemia - from single cell analysis to next-generation sequencing. Genes. 2020;11(10):1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu T, Chong T, Lu S, McGuinness M, Williams DA, Cowell JK. RAC1/2 activation promotes FGFR1 driven leukemogenesis in stem cell leukemia/lymphoma syndrome. Haematologica. 2020;105(2):e68-e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onuh JO, Qiu H. Serum response factor-cofactor interactions and their implications in disease. FEBS J. 2021;288(10):3120-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81(7):1159-1170. [DOI] [PubMed] [Google Scholar]
- 33.Ikeda T, Hikichi T, Miura H, et al. Srf destabilizes cellular identity by suppressing cell-type-specific gene expression programs. Nat Commun. 2018;9(1):1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, Zheng J, Zou Y, Song C, Hu X, Zhang CC. IGF binding protein 2 is a cell-autonomous factor supporting survival and migration of acute leukemia cells. J Hematol Oncol. 2013;6(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickard A, McCance DJ. IGF-binding protein 2 - oncogene or tumor suppressor? Front Endocrinol. 2015;6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao S, Sun Y, Zhang X, et al. IGFBP2 activates the NF-κB pathway to drive epithelial-mesenchymal transition and invasive character in pancreatic ductal adenocarcinoma. Cancer Res. 2016;76(22):6543-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang GK, Hu L, Fuller GN, Zhang W. An interaction between insulin-like growth factor-binding protein 2 (IGFBP2) and integrin alpha5 is essential for IGFBP2-induced cell mobility. J Biol Chem. 2006;281(20):14085-14091. [DOI] [PubMed] [Google Scholar]
- 38.Li T, Forbes ME, Fuller GN, Li J, Yang X, Zhang W. IGFBP2: integrative hub of developmental and oncogenic signaling network. Oncogene. 2020;39(11):2243-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huynh H, Iizuka S, Kaba M, et al. Insulin-like growth factorbinding protein 2 secreted by a tumorigenic cell line supports ex vivo expansion of mouse hematopoietic stem cells. Stem Cells. 2008;26(6):1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang CC, Kaba M, Iizuka S, Huynh H, Lodish HF. Angiopoietin-like 5 and IGFBP2 stimulate ex vivo expansion of human cord blood hematopoietic stem cells as assayed by NOD/SCID transplantation. Blood. 2008;111(7):3415-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huynh H, Zheng J, Umikawa M, et al. IGF binding protein 2 supports the survival and cycling of hematopoietic stem cells. Blood. 2011;118(12):3236-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adnan-Awad S, Kim D, Hohtari H, et al. Characterization of p190-Bcr-Abl chronic myeloid leukemia reveals specific signaling pathways and therapeutic targets. Leukemia. 2021;35(7):1964-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cutler JA, Tahir R, Sreenivasamurthy SK, et al. Differential signaling through p190 and p210 BCRABL fusion proteins revealed by interactome and phosphoproteome analysis. Leukemia. 2017;31(7):1513-1524. [DOI] [PubMed] [Google Scholar]
- 44.Reckel S, Hamelin R, Georgeon S, et al. Differential signaling networks of Bcr–Abl p210 and p190 kinases in leukemia cells defined by functional proteomics. Leukemia. 2017;31(7):1502-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.