Abstract
Objective: Long non-coding RNAs (lncRNAs) function as vital regulators in biologic processes and are dysregulated in various tumors; however, little is known about their role in the inflammatory response in asthma. Therefore, this study aimed to investigate the function of antisense HOXA terminal transcriptional RNA (HOTTIP) and its possible mechanism in the ovalbumin (OVA)-induced inflammatory response in asthmatic mice. Methods: Asthma-related data resources from the Gene Expression Omnibus (GEO) database were extracted to explore the relationships between lncRNAs and asthma, and the lncRNA HOTTIP was identified. The probable effect of HOTTIP on airway inflammation was elaborated by ELISA and histopathologic analysis in OVA-sensitized mice. The online database excavation combined with RNA pull-down, RNA immunoprecipitation, luciferase reporter gene assay, and chromatin immunoprecipitation assay were used to analyze the targeted regulation relationship among HOTTIP, CCCTC-binding factor (CTCF), and Ephrin A3 (EFNA3). In addition, in vivo verification of EFNA3’s role in inflammation was conducted in OVA-treated mice. Results: HOTTIP was upregulated in asthmatic mice and downregulating HOTTIP in the mice model of asthma markedly reduced inflammation, and caused less infiltration of inflammatory cells, and secretions of IgE, interleukin (IL)-4, IL-5, and IL-13. Mechanistically, the data indicate that HOTTIP promoted EFNA3 transcription by recruiting CTCF to the EFNA3 promoter. Interestingly, the knockdown of EFNA3 alleviated inflammation in the asthma model. Conclusion: HOTTIP facilitates the airway inflammatory response by regulating EFNA3 transcription, providing a therapeutic target for asthma.
Keywords: Asthma, HOTTIP, CTCF, EFNA3, inflammation
Introduction
Asthma is a chronic respiratory disease that occurs in people of all ages [1,2]. The global incidence of asthma is expected to reach 400 million by 2025 and it is a serious public health threat [3]. Clinically, asthma is characterized by chronic airway inflammation, airflow obstruction, and remodeling [4]. Notably, airway inflammation is the most vital pathologic process in asthma, accompanied by the infiltration of inflammatory cells and the release of various cytokines [5]. In addition, IgE can induce eosinophils to secrete Th2-associated cytokines, like interleukin (IL)-4, interferon-γ, and IL-13 [6], which are involved in the progression of airway inflammation [7]. The prognosis of the current clinical treatments for asthma such as leukotriene modifiers, corticosteroids, and anti-IgE therapy remains unsatisfactory [8,9]. Undoubtedly, the pathogenesis of asthma and effective molecular markers for it remain to be clarified.
Long non-coding RNAs (lncRNAs) are a class of transcripts greater than 200 nucleotides that can interact with DNA, RNA, or proteins to mediate various biologic functions. Dysregulation of lncRNAs is a major culprit in the pathogenesis of asthma. For example, lncRNA five prime to xist functioned as the sponge of miR-590-5p to upregulate janus-activated kinase 2 expression, further facilitating the proliferation and migration of airway smooth muscle cells (ASMCs) [10]. The overexpressed lncRNA lung adenocarcinoma transcript 1 in asthma promotes ASMC proliferation and migration by downregulating microRNA-216a [11]. The knockdown of lncRNA plasmacytoma variant translocation 1 (PVT1) can bring down the levels of inflammatory factors and destroy cell-barrier function [12]. Thus, lncRNAs are involved in asthmatic airway inflammation. Further, a higher expression of IL-4, IL-5, and IL-13 has been observed in asthmatic subjects [13,14], and it appears to be involved in lncRNA PVT1 expression [15].
In this study, we extracted asthma data from the Gene Expression Omnibus (GEO) database. After further evaluation using qPCR, western blotting, and ELISA assay, we identified a lncRNA, antisense HOXA terminal transcriptional RNA (HOTTIP), that was markedly upregulated in asthma. RNA interference-mediated gene silencing was employed to probe the lncRNA HOTTIP’s role in inflammatory responses of asthma and further, its mechanism in asthma was investigated. Our results indicate that lncRNA HOTTIP significantly contributed to asthma pathogenicity and could be a prognostic marker.
Materials and methods
Identification of differently expressed lncRNAs in asthma
The expression dataset in patients with asthma was downloaded from the GEO database at http://www.ncbi.nlm.nih.gov/geo/. All datasets of GSE27876, GSE143192, and GSE64913 were merged and normalized, the differentially expressed genes were screened out, and differently expressed genes and lncRNAs were screened from those datasets by applying the limma package (statistical criterion: |log2FC| > 1 and adjustP < 0.05). HOTTIP-related genes were obtained through Pearson Correlation Coefficient (P < 0.05). Functional enrichment analysis (GO analysis) was performed on HOTTIP-related genes using the clusterProfiler package in R.
Cell culture and transfection
Human bronchial epithelial cells, 16 HBE cells, were obtained from Ningbo Mingzhou Biotechnology Co., Ltd., (MZ-1420, Ningbo, China) and maintained in RPMI 1640 medium (Gibco-Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS and 1% of penicillin and streptomycin in a cell incubator at 37°C and 5% CO2. After confluence, the cells were plated in 6-well plates for shRNA transfection using LipofectamineTM 2000 (Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. Sequence for human HOTTIP: 5’-CAC CGC GAA TTC TTA ATG CAC AAC GCG AAC GTT GTG CAT TAA GAA TTC GC-3’; For mouse HOTTIP: 5’-CAC CGC ATC ACC CTC TAT TTC TTA CCG AAG TAA GAA ATA GAG GGT GAT GC-3’; For CTCF: 5’-CAC CGC CCA TAA ACA TAG GAG AAC TCG AAA GTT CTC CTA TGT TTA TGG GC-3’; For EFNA3: 5’-CAC CGC CAC GAG TAC TAC TAC ATC TCG AAA GAT GTA GTA GTA CTC GTG GC-3’; For shNC: 5’-CAC CCA GCT CCA ACC AGC ACC TGC GAA CAG GTG CTG GTT GGA GCT G-3’.
Development of the animal model
BALB/c mice (female, aged 4-6 weeks) were obtained from the Experimental Animal Centre of East China Normal University (SCXK (Shanghai) 2021-0006) and maintained in pathogen-free animal facilities. All experimental procedures were reviewed and approved by the Animal Care and Use Committee of Kongjiang Hospital (KJ-2020-KY-14). All mice were randomly divided into a control group, asthmatic group, asthma + short hairpin RNA as negative control (shNC) group, and asthma + shRNA-HOTTIP group, with 10 mice in each group. To develop the asthma model in mice, they were challenged by intraperitoneal injection of 20 μg ovalbumin (OVA, Sigma-Aldrich, St. Louis, MO) and 2 mg aluminum hydroxide on day 7, day 14, and day 21. Then, the sensitized mice were placed in a 20 × 20 × 30 cm transparent closed container from day 27 to 30 and treated with atomized 1% OVA solution once per day for 20 min. The control group mice were challenged with an equal amount of normal saline. Additionally, in asthma + shNC and asthma + shHOTTIP groups, 50 μl of AAV6 recombinant vector (OBiO Co., Ltd., Shanghai, China) expressing invalid sequence (shNC) or interference sequence of lncRNA-HOTTIP (shHOTTIP) was infused to the lung of the mice by endotracheal intubation on day 1 and day 7. On day 34, pentobarbital sodium (100 mg/kg, Vetoquinol, Cedex, France) was intraperitoneally injected into all mice, and the bronchoalveolar lavage fluid (BALF) and the lung tissues were gathered for further processing.
BALF isolation and cell number assessment
The collected BALF was rinsed thrice with 0.6 ml phosphate-buffered saline (PBS), and the supernatant containing all the cells was collected by centrifugation. The total cells were counted by microscopy using a cell counter. After treatment with red blood lysis buffer, the eosinophils, macrophages, and lymphocytes were stained using Wright’s staining (Jiancheng Technology Co., Ltd., China), and the cell counts were determined using a blood cell analyzer.
RNA isolation and qRT-PCR detection
TRIzolTM reagent (Invitrogen, CA, USA) was used to extract total RNA from the lung tissues or cells. Next, reverse transcription reactions were carried out using the PrimeScript RT reagent kit (Takara Bio, Shiga, Japan). Real-time qPCR reactions were performed on ExicyclerTM 96 system (Bio-Rad Laboratories, USA) following the manufacturer’s protocols. The reaction conditions were as follows: denaturing at 95°C for 10 s, annealing at 60°C for 20 s, and elongation at 70°C for 10 s, for 40 total cycles. The relative gene expression values were calculated by the 2-∆∆Ct method [16].
The primers were as follows: human HOTTIP, 5’-CTT ACG CCC GCA ACA AAA CA-3’ (Forward), 5’-TGG ATG CGC ACA TTC ACT CT-3’ (Reverse); mouse HOTTIP, 5’-TAG CAG CAG GTT TCA GGC TC-3’ (Forward), 5’-ACA GCA CCA GAC TTA GAC GC-3’ (Reverse); EFNA3, 5’-GGG GCC TAC ATC CTT CTT CC-3’ (Forward), 5’-ACC TCC CAT GCT TCG TTT GT-3’ (Reverse); man GAPDH, 5’-GGA TTG TCG ATT GGC-3’ (Forward), 5’-TCC GTT TCA CCA GTA-3’ (Reverse); use GAPDH, 5’-GCC TCG TCC CGT AGA CAA AA-3’ (Forward), 5’-GCA ACA ATC TCC ACT TTG CCA-3’ (Reverse).
Western blotting
Protein extracts were obtained from cells using RIPA lysis buffer (Santa Cruz Biotechnology, Santa Cruz, CA). BCA Kit (Thermo Fisher Scientific, Asheville, NC, USA) was used to quantify the protein concentration in lysates. A similar amount of extracted proteins was separated by 10% SDS-PAGE, and subsequently electrically transferred to polyvinylidene difluoride membranes, then blocked with 5% skim milk, and first probed with indicated primary antibodies including anti-CTCF (1:5,000; ab128873, Abcam, Cambridge, UK), anti-EFNA3 (1:3000; ab153706, Abcam, Cambridge, UK), and anti-GAPDH (1:3000; ab144269, Abcam, Cambridge, UK). Following this the membranes were cultured with an anti-mouse secondary antibody (1:3000; ab97022, Abcam, Cambridge, UK), and visualized with an ECL kit (Solarbio, Beijing, China). GAPDH was used to normalize protein expression.
ELISA
The contents of cytokines IgE, IL-4, IL-5, and IL-13 in BALF were analyzed by ELISA kits (R&D Systems Inc., Minneapolis, MN) following the manufacturer’s instructions.
Histopathologic analysis
The mouse lung tissues were fixed with 10% formalin, dehydrated, paraffin-embedded, and sliced into 5 μm sections. The sliced sections were routinely dewaxed and stained with hematoxylin and eosin (H&E), periodic acid-Schiff stain (PAS), and Masson’s trichrome stain to observe inflammatory cell infiltration, smooth muscle cell hyperplasia, and airway mucus secretion, respectively. All of the sample slides for comparison were assessed under the same magnification with the microscope (Olympus, Tokyo, Japan). The inner perimeter of the bronchial wall (Pi), airway smooth muscle area (WAm), inner area of the bronchial wall (WAi), and the number of bronchial smooth muscle cells (N) were detected.
Immunohistochemistry
For immunohistochemical staining of EFNA3, the lung tissue slides were microwave-heated in antigen retrieval solution, washed with PBS, treated with 3% H2O2 for 20 min in a wet box, and further incubated with goat serum at 37°C for 20 min. Then the samples were treated with the primary antibody to EFNA3 (ab89472, 1:200, Abcam, Cambridge, UK) and further incubated with the secondary antibody (HRP-labeled goat anti-rabbit, 1:200, CST, CA, USA) at room temperature for 1 h. Cell nuclei were stained with hematoxylin and observed under an optical microscope (Olympus CX41, Tokyo, Japan).
RNA pull-down assay
Primers containing EFNA3 promoter were designed, and HOTTIP RNA was obtained by sequence amplification and transcription in vitro, then further treated with RNase-free DNase I and purified with RNeasy Mini Kit (Qiagen, Hilden, Germany). The cells were collected after culture and the total proteins were extracted using the NE-PER nuclear protein extraction kit (Thermo Scientific, Waltham, MA). Biotin-labeled RNA probes were incubated with streptavidin beads to form complexes. After purification, the protein was detected by western blot.
RNA binding protein immunoprecipitation (RIP) assay
The cells were collected and incubated with 0.5-1 ml of pre-cooled RIP lysis buffer at 4°C for 5 min. The cell suspension was centrifuged at 4°C for 10 min, then collected and added to 20 mL Protein A/G magnetic beads, and discarded after stabilizing for 10 s. After that, cells were coimmunoprecipitated with rabbit antibody to CTCF (1:2,000; ab128873, Abcam, Cambridge, UK) or normal rabbit antibody to IgG, and incubated at room temperature for 30 min, then centrifuged and rinsed with RIP wash buffer. TRIzol reagent was appended to immunoprecipitated RNA and the interaction between protein and RNA was determined by RT-qPCR amplification.
Luciferase reporter gene assay
The target promoter fragment of EFNA3 was cloned and inserted into the luciferase reporter gene plasmid vector (Promega, Madison, WI, USA), amplified, and purified for use. Human embryonic kidney 293T cells were cultured and inoculated into 24-well plates. After reaching 80% cell confluency, vectors harboring 800 ng of the reporter and 20 mM/L shCTCF or shNC were co-transfected into the cells. After 24 h, cells were collected and split, and the luciferase substrate was added. The relative luciferase activity was quantified at room temperature in dark conditions following the manufacturer’s instructions. Normalization calculation = RLU firefly luciferase/RLU Renilla luciferase.
Chromatin immunoprecipitation (ChIP) assay
Cells maintained in plates were incubated with 1% formaldehyde solution at 37°C for 10 min, and then supplemented with 0.125 M glycine to terminate the crosslinking. Subsequently, cells were washed twice with cold PBS, lysed using SDS lysis buffer, and 100-900 BP DNA fragments were obtained by ultrasonic fragmentation. Following this, the sample was centrifuged to remove insoluble substances. Anti-CTCF (ab13583, Abcam, Cambridge, UK) or IgG was appended to the ultrasonic broken products and incubated overnight at 4°C. The next day, the sample was treated with NaCl and protease K to remove DNA-protein cross-linking. Further, the DNA was purified, recovered, and analyzed by PCR assay.
Statistical analysis
Data analysis was performed using SPSS 20.0 software (IBM Corp., Armonk, NY, USA) and all results were presented as mean ± standard deviation (SD). The Student’s t-test was used to analyze the differences between the two groups. One-way ANOVA with Tukey test was employed for multiple comparisons and a value of P < 0.05 was considered significant.
Results
Identification of lncRNA HOTTIP with significant upregulation in asthma
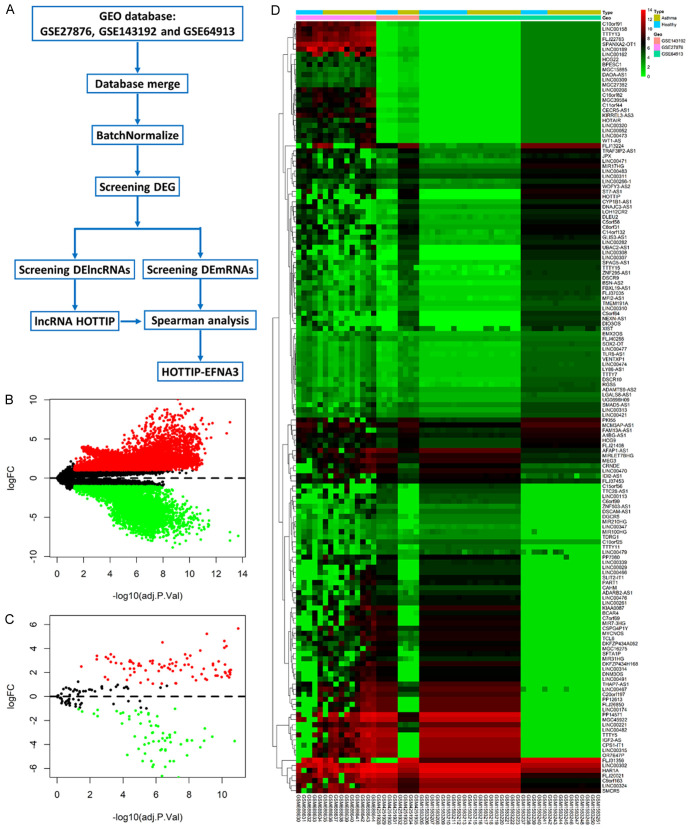
The asthma-related gene expression profiles (GSE27876, GSE143192, and GSE64913) were downloaded from the GEO database, consisting of 15 samples (5 normal and 10 asthma cases), 8 samples (4 normal and 4 asthma cases), and 34 samples (19 normal and 15 asthma cases), respectively (Figure 2A and Table 1). Significant differences in expression patterns between asthmatic and normal specimens were observed using adjustP value < 0.05 and |log2FC| > 1 as cutoffs for expression differences. Compared to normal samples, a total of 8712 genes were specifically dysregulated in asthma samples among the 3 subsets, of which there were 4774 upregulated genes and 3938 downregulated genes (Figure 2B and Table S1). Furthermore, there were 152 specifically dysregulated lncRNAs (85 upregulated lncRNAs and 67 downregulated lncRNAs) in asthma samples (Figure 2C and Table S2). The heat map provided a visual representation of the dysregulated lncRNAs in asthma samples compared to healthy samples (Figure 2D). Based on differential expression analysis, the details of the top 10 lncRNAs, 5 significantly upregulated candidate lncRNAs (ST7-AS1, HOTTIP, UBAC2-AS1, FLJ13224, and LINC00308), and 5 significantly downregulated candidate lncRNAs (LINC00315, MGC45922, CPS1-IT1, PP14571, and TTTY5) are shown in Table 2. Of note, HOTTIP was significantly overexpressed in both human and mouse samples, thus, HOTTIP was selected as the central lncRNA for subsequent analysis.
Figure 2.
Identification of long non-coding RNAs (lncRNA) antisense HOXA terminal transcriptional RNA (HOTTIP) with significant upregulation in asthma. A: Study flow chart of the present study screening lncRNAs in asthma. B: Volcano plots were performed to visualize differently expressed genes between asthma patients and healthy individuals. C: Volcano plots were performed to visualize differently expressed lncRNAs between asthma patients and healthy individuals. D: Heat map and hierarchical clustering demonstrated expression values of differently expressed lncRNAs.
Table 1.
The information of the three microarray datasets (GSE27876, GSE143192, and GSE64913) from gene expression omnibus (GEO) datasets
Table 2.
The details of top 5 upregulated lncRNAs and top 5 downregulated lncRNAs in asthma
| ID | logFC | AveExpr | P.Value | adj.P.Val | B |
|---|---|---|---|---|---|
| ST7-AS1 | 5.669163 | 3.628779 | 4.85E-14 | 1.04E-11 | 21.71473 |
| HOTTIP | 5.222623 | 3.300837 | 9.55E-11 | 8.34E-10 | 14.23162 |
| UBAC2-AS1 | 4.630676 | 3.104768 | 4.85E-12 | 7.26E-11 | 17.17109 |
| FLJ13224 | 4.510021 | 6.295117 | 1.41E-07 | 4.05E-07 | 7.052035 |
| LINC00308 | 4.201737 | 2.655287 | 2.53E-12 | 5.44E-11 | 17.81313 |
| LINC00174 | -5.39639 | 5.609645 | 4.04E-08 | 1.43E-07 | 8.279317 |
| LINC00315 | -5.69119 | 6.744738 | 5.90E-07 | 1.42E-06 | 5.655109 |
| MGC45922 | -5.86867 | 8.199312 | 7.07E-07 | 1.62E-06 | 5.478182 |
ST7-AS1: Suppressor of Tumorigenicity 7 Antisense RNA 1; HOTTIP: Antisense HOXA Terminal Transcriptional RNA; UBAC2-AS1: Ubac2 Antisense RNA 1; FLJ13224: Homo Sapiens Uncharacterized LOC79857; LINC00308: Long Intergenic Non-protein Coding RNA 308; LINC00174: Long Intergenic Non-protein Coding RNA 174; LINC00315: Long Intergenic Non-protein Coding RNA 315; MGC45922: Homo Sapiens Uncharacterized LOC284365; logFC: log2 Fold Change.
Silencing of HOTTIP alleviated inflammation in asthmatic mice
To uncover the role of lncRNA HOTTIP, an asthma model was established by challenging the mice with OVA. The workflow for the induction of asthma is shown in Figure 1. Initially, as tested by qRT-PCR assay, the relative level of HOTTIP in the lung tissues from OVA-induced asthma mice was significantly higher than that of controls, and HOTTIP was successfully silenced using lentivirus infection (Figure 3A). Further data showed that the counts of total cells, eosinophils, macrophages, and lymphocytes in BALF from the asthma model were increased relative to that in the control group. The silencing of HOTTIP led to a markedly reduced number of these inflammatory cells in BALF from OVA-induced asthma mice (Figure 3B-E). The anti-inflammatory effect of HOTTIP was quantified in the supernatant of BALF from asthmatic mice. As observed in the ELISA, the levels of IgE and pro-inflammatory cytokines IL-4, IL-5, and IL-13 in BALF were elevated after OVA treatment; however, treatment with shHOTTIP could significantly suppress these cytokine levels in the BALF supernatants (Figure 3F-I). To evaluate the effect of HOTTIP on airway inflammation, histologic sections of the lung tissues were investigated by H&E staining. As seen in Figure 4A, H&E staining demonstrated that the airways of the OVA-induced mice were extensively affected by cellular infiltration and damaged alveolar structures. These pathological morphology effects in the asthma group were relieved by silencing of HOTTIP. PAS staining was used to further assess the function of HOTTIP on mucus production. OVA treatment caused goblet cell hyperplasia and airway mucus secretions in the mouse model of asthma, whereas these pathologic changes were mitigated in the shRNA-HOTTIP-treated asthma micecompared to the shRNA-NC-treated asthma mice (Figure 4B). Subsequently, to investigate the effect of HOTTIP on smooth muscle hyperplasia, Masson’s staining was carried out. Results showed substantially reduced smooth muscle hyperplasia in the shRNA-HOTTIP-treated asthma mice as compared to shRNA-NC-treated asthma mice (Figure 4C). In addition, the asthma group showed a significant decrease in WAi/WAm levels, and an increase in the levels of N/Pi, WAi/Pi as well as WAm/Pi. By contrast, the treatment with shRNA-HOTTIP significantly reversed the levels of WAi/WAm, N/Pi, WAi/Pi, and WAm/Pi compared to the shRNA-NC group (Figure 4D-G). These data indicated that silencing of HOTTIP alleviated inflammation in OVA-sensitized asthmatic mice.
Figure 1.
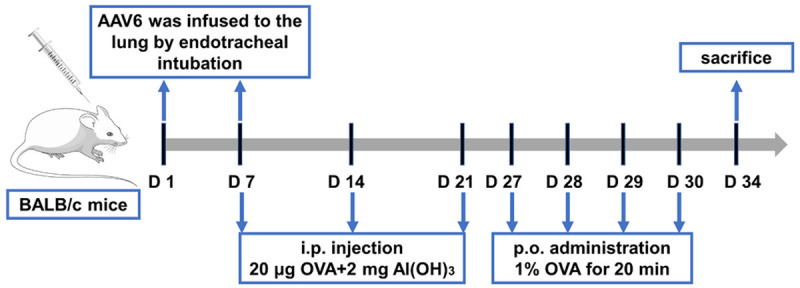
Flow charts showing the development of the ovalbumin (OVA)-induced asthmatic mouse model.
Figure 3.
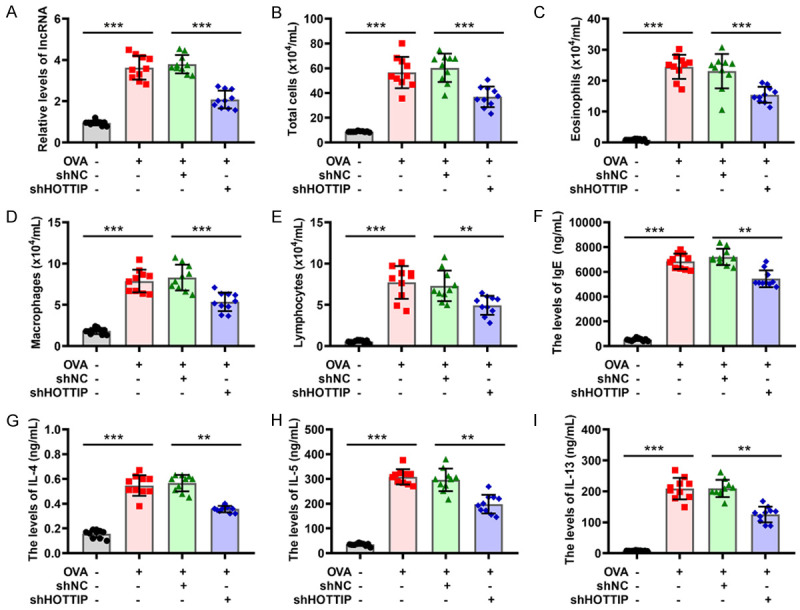
Silencing of HOTTIP alleviates inflammation in asthmatic mice. (A) The expression of HOTTIP was measured in mice treated with OVA or/and HOTTIP knockdown by qRT-PCR. (B-E) The counts of total cells (B), eosinophils (C), macrophages (D), and lymphocytes (E) were detected in bronchoalveolar lavage fluid (BALF) from mice treated with OVA or/and HOTTIP knockdown. (F-I) Levels of IgE (F), Interleukin (IL)-4 (G), IL-5 (H), and IL-13 (I) were determined in BALF from mice treated with OVA or/and HOTTIP knockdown. *P < 0.05, **P < 0.01, ***P < 0.001 of one-way analysis of variance (ANOVA) with Tukey test.
Figure 4.
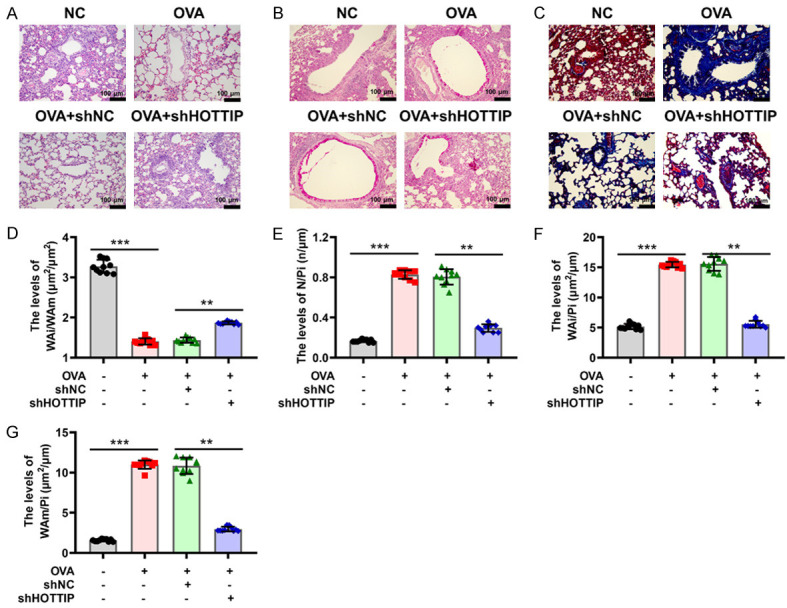
Silencing of HOTTIP attenuates lung tissue damage in asthmatic mice. (A) H&E staining was performed to assess the degree of inflammatory cell infiltration in mice treated with OVA or/and HOTTIP knockdown. Scale bar 100 μm. (B) Periodic acid-Schiff (PAS) staining was performed to detect changes in pathologic morphology in mice treated with OVA or/and HOTTIP knockdown. Scale bar 100 μm. (C) Masson’s staining was conducted to observe changes in mice treated with OVA or/and HOTTIP knockdown. Scale bar 100 μm. (D-G) Pathologic examination cata include inner area of the bronchial wall (WAi)/airway smooth muscle area (WAm) (D), the number of bronchial smooth muscle cells (N)/inner perimeter of the bronchial wall (Pi) (E), WAi/Pi (F), and WAm/Pi (G) of the lung tissues of mice treated with OVA or/and HOTTIP knockdown. *P < 0.05, **P < 0.01, ***P < 0.001 of one-way ANOVA with Tukey test.
HOTTIP promotes EFNA3 transcription by recruiting CTCF to the EFNA3 promoter
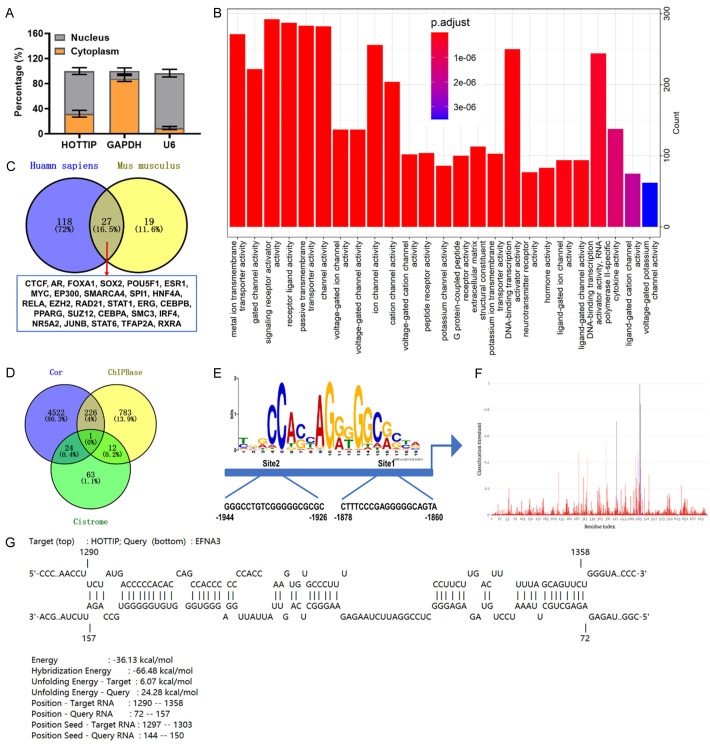
Next, we determined whether HOTTIP was expressed in the nucleus or cytoplasm by conducting a subcellular separation analysis. The data revealed that HOTTIP was localized in the nucleus (Figure 5A), suggesting that this is the main site for HOTTIP to carry out biological functions. Pathway analysis was also applied to confirm the molecular function of HOTTIP. GO analysis showed that HOTTIP was enriched in the following processes: “transporter activity”, “DNA-binding transcription”, “activator activity”, and “neurotransmitter receptor activity” (Figure 5B). Considering the transcriptional regulatory function and cell localization of HOTTIP, the binding transcription factors of HOTTIP were predicted. There were 145 transcription factors in Homo sapiens and 46 transcription factors in Mus musculus from the RNAInter database (http://www.rnainter.org). Among all transcription factors, 27 targets were shared in both Homo and Mus samples, including CCCTC-binding factor (CTCF) (Figure 5C and Table S3), a zinc finger protein that is related to asthma [17]. Subsequently, the ChIPBase databases (https://rna.sysu.edu.cn/chipbase/) and Cistrome database (http://cistrome.org/) were used simultaneously to predict the possible target genes that may be modulated by the transcription factor CTCF. A total of 13 common targets were obtained from the ChIPBase and Cistrome databases. Considering the 4772 related genes of HOTTIP, Venn diagrams illustrated that EFNA3 was the only overlapping gene (Figure 5D and Tables S4, S5, S6). Notably, the two binding sites of CTCF on the EFNA3 promoter were predicted by the JASPAR database (https://jaspar.genereg.net/, Figure 5E). The interaction between HOTTIP and EFNA3 promoter was also predicted and further verified by PRIdictor (http://bclab.inha.ac.kr/pridictor/pridictor.html, Figure 5F). Furthermore, the predicted results from IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp;jsessionid=4570FF5EF08D1A2413A97B6DCF7D05F7) showed an interconnection between HOTTIP and EFNA3 promoter (Figure 5G). Taken together, these results suggest that HOTTIP may regulate EFNA3 transcription by recruiting CTCF.
Figure 5.
Relationship among HOTTIP, CCCTC-binding factor (CTCF) and Ephrin A3 (EFNA3) promoter. A: The percentage of HOTTIP in the cytoplasm and nuclear fractions was determined by nuclear fractionation analyses and qRT-PCR. B: Gene ontology (GO) analysis was performed for HOTTIP-related genes, and the molecular function categories were represented. C: The binding proteins of HOTTIP in Homo sapiens (Blue) and Mus musculus (yellow) were identified by RNAInter database. D: Venn diagram was used to confirm the common targets between HOTTIP-related genes and CTCF targets predicted by ChIPBase database and Cistrome database. E: The JASPAR database was used to predict the CTCF binding motif. F: The binding of HOTTIP to CTCF was predicted by PRIdictor databse. G: The binding site and interaction energy between HOTTIP and EFNA3 promoter were predicted by IntaRNA databse.
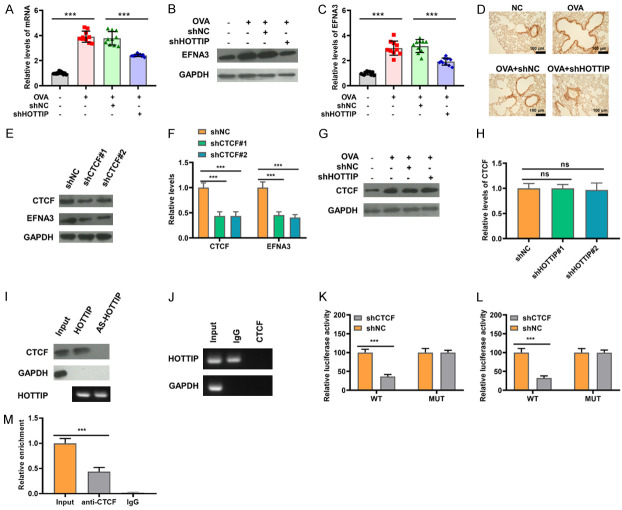
The levels of EFNA3 mRNA and EFNA3 protein were then evaluated by qRT-PCR and western blot to further investigate the underlying molecular mechanism. The levels of EFNA3 mRNA and EFNA3 protein were prominently increased in the asthmatic model group, and the facilitating effect of OVA on the levels of EFNA3 mRNA and EFNA3 protein was significantly reversed by silencing HOTTIP (Figure 6A-C). On the other hand, as seen in Figure 6D of immunohistochemical staining, the lung tissue from asthmatic mice displayed a significant increase in EFNA3 expression, whereas silencing of HOTTIP induced a prominent reduction of EFNA3 expression. To identify the regulation of CTCF on EFNA3, the 16HBE cells stably expressing an shRNA targeting CTCF were built. The western blot results indicated that shRNA sequences (shCTCF#1, #2) could effectively reduce the expression of CTCF and EFNA3 (Figure 6E and 6F). Furthermore, CTCF protein was not decreased in the shRNA-HOTTIP-transduced 16HBE cells when compared with controls (Figure 6G and 6H). To provide direct evidence of the interaction among CTCF, HOTTIP and EFNA3 promoter, an RNA pull-down assay was performed that confirmed that HOTTIP directly interacted with CTCF (Figure 6I). The association between HOTTIP and CTCF was readily confirmed by the RIP assay: the CTCF protein could bind to HOTTIP (Figure 6J). To further determine the specificity of CTCF targeting EFNA3, a luciferase reporter carrying EFNA3 promoter with a putative CTCF binding site was constructed. As shown in Figure 6K and 6L, CTCF knockdown dramatically reduced the luciferase activity of EFNA3 containing a WT promoter but did not decrease the activity of EFNA3 with a MUT promoter. The chip assay further verified the interaction between CTCF and the EFNA3 promoter (Figure 6M). Collectively, our results suggest that HOTTIP promotes EFNA3 transcription by recruiting CTCF to the EFNA3 promoter.
Figure 6.
HOTTIP promotes EFNA3 transcription by recruiting CTCF to the EFNA3 promoter. A: mRNA levels of EFNA3 in mice treated with OVA or/and HOTTIP knockdown were measured by qRT-PCR. B, C: The expression of EFNA3 in mice treated with OVA or/and HOTTIP knockdown was measured by western blot. D: EFNA3 expression in lung tissues of mice treated with OVA or/and HOTTIP knockdown was detected with immunohistochemical staining. E, F: The expression of CTCF and EFNA3 in 16HBE cells with CTCF knockdown were determined by western blot. G, H: The expression of CTCF in 16HBE cells transfected with HOTTIP knockdown were measured by western blot assay. I: RNA pull-down assay was performed to test the binding relationship between HOTTIP and CTCF. J: The interaction between HOTTIP and CTCF was confirmed by RNA binding protein immunoprecipitation (RIP) assay. K: Dual luciferase assay was performed to confirm the interaction between CTCF on site 1 of EFNA3 promoter. L: Dual luciferase assay was performed to confirm the interaction between CTCF on site 2 of EFNA3 promoter. M: The interaction between CTCF and EFNA3 promoter was confirmed by chromatin immunoprecipitation (ChIP) assay. *P < 0.05, **P < 0.01, ***P < 0.001 of one-way ANOVA with Tukey test or Student’s t-test.
Knockdown of EFNA3 alleviated inflammation in asthmatic mice
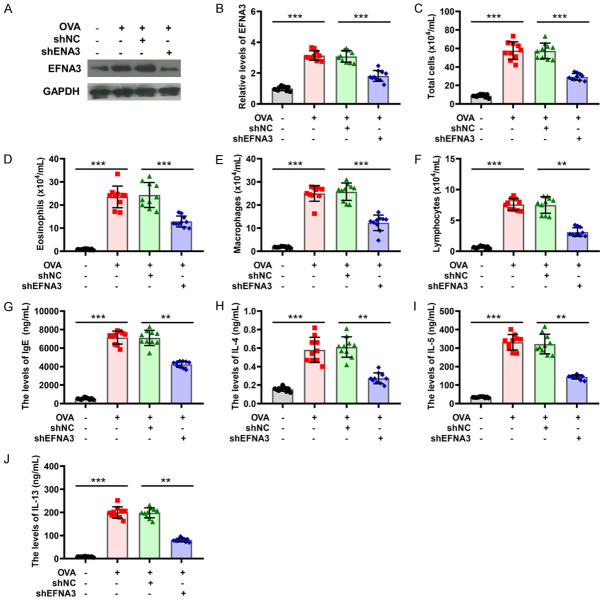
To probe the role of EFNA3 in inflammatory responses of asthma, the changes in EFNA3 expression in BALF were detected using a western blot assay. As seen in Figure 7A, EFNA3 was found to be upregulated with the most significant change in asthmatic mice. EFNA3 expression was inhibited by transfection of shEFNA3, and the transfection efficiency was verified by western blot analysis (Figure 7A and 7B). Rescue experiments were performed and showed that EFNA3 knockdown significantly rescued the OVA-mediated promotion of total cells in BALF (Figure 7C). Similar observations were presented for the percentages of eosinophils, macrophages, and lymphocytes in BALF of asthma model mice (Figure 7D-F). Moreover, ELISA analysis revealed that the increased number of IgE responses to OVA stimuli in asthmatic mice was inhibited by the silencing of EFNA3 (Figure 7G). As for the secretion of IL-4, IL-5, and IL-13, the knockdown of EFNA3 also induced a marked decrease in their levels in BALF supernatant from asthmatic mice compared to asthmatic mice with shNC treatment (Figure 7H-J). H&E staining was conducted to evaluate inflammation influence in the lung tissues with the elimination of EFNA3 by pretreatment with shRNA. As shown in Figure 8A, H&E staining demonstrated that the knockdown of EFNA3 weakened the effect of increased infiltrating inflammatory cells and thickened airway walls induced by OVA administration. Using PAS staining, the function of EFNA3 on mucus production was further assessed, and a tendency for decreased inflammatory cells was observed in the lungs of asthmatic mice following the application of EFNA3 knockdown, as shown in Figure 8B. Masson’s staining revealed the role of EFNA3 in smooth muscle hyperplasia and showed that the knockdown of EFNA3 led to a decrease in smooth muscle hyperplasia induced by OVA (Figure 8C). In addition, the downregulation of EFNA3 exacerbated a marked decrease in WAi/WAm levels, and a marked increase of N/Pi, WAi/Pi, and WAm/Pi levels in asthmatic mice compared to asthmatic mice with shNC treatment (Figure 8D-G). Thus, the knockdown of EFNA3 may be responsible for suppressing inflammation in the mouse model of asthma.
Figure 7.
Knockdown of EFNA3 alleviates inflammation in asthmatic mice. (A, B) The expression of EFNA3 was measured in mice treated with OVA or/and EFNA3 knockdown by western blot. (C-F) The number of total cells (C), eosinophils (D), macrophages (E), and lymphocytes (F) were detected in BALF from mice treated with OVA or/and EFNA3 knockdown. (G-J) Levels of IgE (G), IL-4 (H), IL-5 (I), and IL-13 (J) were determined in BALF from mice treated with OVA or/and EFNA3 knockdown. *P < 0.05, **P < 0.01, ***P < 0.001 of one-way ANOVA with Tukey test.
Figure 8.
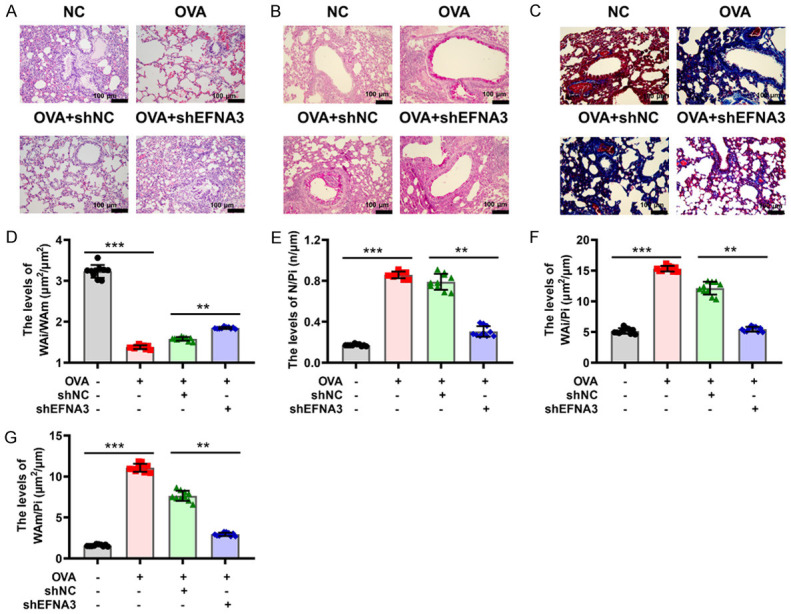
Knockdown of EFNA3 attenuates lung tissue damage in asthmatic mice. (A) H&E staining was performed to assess the degree of inflammatory cell infiltration in mice treated with OVA or/and EFNA3 knockdown. Scale bar 100 μm. (B) PAS staining was performed to detect changes in pathologic morphology in mice treated with OVA or/and EFNA3 knockdown. Scale bar 100 μm. (C) Masson’s staining was conducted to determine changes in mice treated with OVA or/and EFNA3 knockdown. Scale bar 100 μm. (D-G) Pathologic features including WAi/WAm (D), N/Pi (E), WAi/Pi (F), and WAm/Pi (G) of the lung tissues of mice treated with OVA and/or EFNA3 knockdown. *P < 0.05, **P < 0.01, ***P < 0.001 of one-way ANOVA with Tukey test.
Discussion
Asthma is a complex, highly prevalent chronic respiratory disease with an increasing incidence worldwide [18]. Recently, the function of lncRNA on asthma progression has been re-recognized. In this study, the expression datasets of patients with asthma were downloaded from the GEO database and there were 85 upregulated lncRNAs and 67 downregulated lncRNAs in asthma samples. HOTTIP, a non-coding RNA molecule derived from the 5’-terminal of HOXA, was significantly overexpressed in both human and mouse asthma samples. We built an OVA-induced asthma mouse model to explore the underlying role of HOTTIP.
HOTTIP is a critical oncogenic factor, and its dysregulation was reported to promote the occurrence and development of ovarian, liver, esophageal, and lung cancers, and other tumors [19-22]. Moreover, it has previously been shown that lncRNA-HOTTIP contributed to the regulation of inflammation. For example, it was reported that HOTTIP silencing reversed the inflammation caused by high-glucose treatment in diabetic nephropathy SV40-MES13 cells [23]. Inhibition of HOTTIP lessened the production of IL-6 and TNF-α in oral submucous fibrosis [24]. In addition, the previous work of Xumin et al. suggested that HOTTIP induces inflammatory responses in a rheumatoid arthritis rat model [25]. The inhibition of HOTTIP has also been implicated to alleviate proinflammatory cytokine expression in in vitro and in vivo models of Parkinson’s disease [26]. In line with the data reported in the above-mentioned diseases [24,27], our study revealed clearly that the inhibition of HOTTIP significantly decreased the numbers of inflammatory cells, and the secretion of IgE, IL-4, IL-5, and IL-13 in the BALF, thereby attenuating inflammation in the asthmatic model mice. Further, the lung tissues from challenged mice showed airway structural changes consistent with cellular infiltration in goblet cell hyperplasia, airway mucus secretions, and smooth muscle hyperplasia as evidenced by H&E, Masson’s, and PAS staining. These effects of OVA sensitization were recovered by inhibition of HOTTIP. These findings might provide potential therapeutic candidates for the treatment of asthma. Nonetheless, the limitations of the study should be acknowledged; other lncRNAs excavated by bioinformatics analysis have not been verified through in vitro and in vivo experiments.
Based on the online database results, we predicted that CTCF was the transcription factor of HOTTIP and EFNA3 was the target gene of CTCF. Therefore, we hypothesized that the silencing of HOTTIP would have an anti-inflammatory effect in OVA-challenged mice, probably through the CTCF/EFNA3 signaling pathway. CTCF, an evolutionarily conserved zinc-finger protein, is ubiquitously expressed in eukaryotes. In a previous study, RNA was isolated from airway tissues of 12 pairs of asthmatic and non-asthmatic samples to test the differences in gene expression. As a result, differentially expressed genes were discovered to comprise CTCF motifs in their upstream promoters in asthmatic airways, indicating that CTCF is closely associated with the pathogenesis of asthma [28]. Consistent with these findings, our results indicated that CTCF expression was markedly elevated in the asthma model. Using RNA pull-down, RIP assay, and luciferase reporter gene assays, we proved that HOTTIP could interact with CTCF. Further data indicated the upregulation of CTCF at the protein levels in 16HBE cells, which was inhibited following the silencing of HOTTIP. As a crucial member of the ephrin family, EFNA3 was speculated to be a key modulator of diverse clinical diseases such as oral tumors, and lung adenocarcinoma [29,30]. Our results of chip assay authenticated the interaction between CTCF and EFNA3 promoter, indicating that HOTTIP resulted in consequent facilitation of EFNA3 transcription by recruiting CTCF to EFNA3 promoter. Information obtained from these findings may offer a better understanding of the regulatory mechanisms of HOTTIP in asthma. Nonetheless, we must consider the following limitations of this study: (1) Specific to transcription factors, only CTCF was verified in this study. Thus, validation in other transcription factors is needed, and (2) The mechanism was only studied at the cellular level; hence, further in vivo experiments are warranted.
To date, only a few studies have evaluated the interplay between EFNA3 and asthma. Interestingly, several studies have shown that EFNA3 is correlated with inflammation. Hypoxia induces EFNA3 in an HIF isoform-dependent manner, participating in inflammation in rheumatoid arthritis [31]. Intravitreal injection of nanoceria effectively triggered EFNA3 suppression, which is involved in inflammatory pathological mechanisms of age-related macular degeneration [32]. Upregulation of EFNA3 was detected in gastric cancer, hepatocellular carcinoma, and other diseases [33], which is consistent with our western blot results in asthmatic mice. In our in vivo asthma mice model, we found that knocking out EFNA3 can suppress inflammation. Hence, EFNA3 may be a promising biomarker in asthma patients and might provide more evidence for the regulatory role of HOTTIP in asthma.
Conclusions
In conclusion, our study showed that silencing of HOTTIP inhibits the inflammatory responses in asthma by recruiting CTCF to the EFNA3 promoter. HOTTIP may be a novel therapeutic target in asthma.
Disclosure of conflict of interest
None.
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
References
- 1.Li H, Wang K, Huang H, Cheng W, Liu X. A meta-analysis of anti-interleukin-13 monoclonal antibodies for uncontrolled asthma. PLoS One. 2019;14:e0211790. doi: 10.1371/journal.pone.0211790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaczmarek KA, Clifford RL, Knox AJ. Epigenetic changes in airway smooth muscle as a driver of airway inflammation and remodeling in asthma. Chest. 2019;155:816–824. doi: 10.1016/j.chest.2018.10.038. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Fei H, Lin Q, Liang F, You Y, Li M, Wu M, Qu Y, Li P, Yuan Y, Chen T, Jiang H. ZEB2 facilitates peritoneal metastasis by regulating the invasiveness and tumorigenesis of cancer stem-like cells in high-grade serous ovarian cancers. Oncogene. 2021;40:5131–5141. doi: 10.1038/s41388-021-01913-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Xu J, Meng Y, Adcock IM, Yao X. Role of inflammatory cells in airway remodeling in COPD. Int J Chron Obstruct Pulmon Dis. 2018;13:3341–3348. doi: 10.2147/COPD.S176122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahy JV. Type 2 inflammation in asthma-present in most, absent in many. Nat Rev Immunol. 2015;15:57–65. doi: 10.1038/nri3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samitas K, Delimpoura V, Zervas E, Gaga M. Anti-IgE treatment, airway inflammation and remodelling in severe allergic asthma: current knowledge and future perspectives. Eur Respir Rev. 2015;24:594–601. doi: 10.1183/16000617.00001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chupp GL, Kaur R, Mainardi A. New therapies for emerging endotypes of asthma. Annu Rev Med. 2020;71:289–302. doi: 10.1146/annurev-med-041818-020630. [DOI] [PubMed] [Google Scholar]
- 8.Nannini LJ. Treat to target approach for asthma. J Asthma. 2020;57:687–690. doi: 10.1080/02770903.2019.1591443. [DOI] [PubMed] [Google Scholar]
- 9.Donohue JF, Herje N, Crater G, Rickard K. Characterization of airway inflammation in patients with COPD using fractional exhaled nitric oxide levels: a pilot study. Int J Chron Obstruct Pulmon Dis. 2014;9:745–51. doi: 10.2147/COPD.S44552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen Y, Yang G, Zhuo S, Zhuang H, Chen S. lncRNA FTX promotes asthma progression by sponging miR-590-5p and upregulating JAK2. Am J Transl Res. 2021;13:8833–8846. [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Wang FH, Wang L, Li Y, Lu J, Chen J. LncRNA MALAT1 promotes proliferation and migration of airway smooth muscle cells in asthma by downregulating microRNA-216a. Saudi J Biol Sci. 2021;28:4124–4131. doi: 10.1016/j.sjbs.2021.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, Zhang Q, Hao J, Wang J, Wang C. LncRNA PVT1 exacerbates the inflammation and cell-barrier injury during asthma by regulating miR-149. J Biochem Mol Toxicol. 2020;34:e22563. doi: 10.1002/jbt.22563. [DOI] [PubMed] [Google Scholar]
- 13.Huang Z, Luo Q, Yao F, Qing C, Ye J, Deng Y, Li J. Identification of differentially expressed long non-coding RNAs in polarized macrophages. Sci Rep. 2016;6:19705. doi: 10.1038/srep19705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin H, Liu MH, Gao F, Shang HM. Pro-inflammatory and pro-fibrotic role of long non-coding RNA RMRP in pediatric asthma through targeting microRNA-206/CCL2 axis. J Biol Regul Homeost Agents. 2021;35:71–83. doi: 10.23812/20-505-A. [DOI] [PubMed] [Google Scholar]
- 15.Wei Y, Han B, Dai W, Guo S, Zhang C, Zhao L, Gao Y, Jiang Y, Kong X. Exposure to ozone impacted Th1/Th2 imbalance of CD(4+) T cells and apoptosis of ASMCs underlying asthmatic progression by activating lncRNA PVT1-miR-15a-5p/miR-29c-3p signaling. Aging (Albany NY) 2020;12:25229–25255. doi: 10.18632/aging.104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang J, Chen X, Liu H, Shao J, Xie R, Gu P, Duan C. Polypyrimidine tract-binding protein 1 promotes proliferation, migration and invasion in clear-cell renal cell carcinoma by regulating alternative splicing of PKM. Am J Cancer Res. 2017;7:245–259. [PMC free article] [PubMed] [Google Scholar]
- 17.Sun X, Shen W, Li Z, Zhang W. CCCTC-binding factor transcriptionally regulates Galectin-7 and activates the JNK/STAT3 axis to aggravate bronchial epithelial cell injury. Pediatr Pulmonol. 2022;57:90–99. doi: 10.1002/ppul.25726. [DOI] [PubMed] [Google Scholar]
- 18.Jassal MS. Special considerations-asthma in children. Int Forum Allergy Rhinol. 2015;5(Suppl 1):S61–7. doi: 10.1002/alr.21577. [DOI] [PubMed] [Google Scholar]
- 19.Dong YJ, Feng W, Li Y. HOTTIP-miR-205-ZEB2 axis confers cisplatin resistance to ovarian cancer cells. Front Cell Dev Biol. 2021;9:707424. doi: 10.3389/fcell.2021.707424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge Y, Yan X, Jin Y, Yang X, Yu X, Zhou L, Han S, Yuan Q, Yang M. MiRNA-192 [corrected] and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet. 2015;11:e1005726. doi: 10.1371/journal.pgen.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Liu Y, Chen G, Liu H, Wu Y, Liu J, Zhang Z. HOTTIP is upregulated in esophageal cancer and triggers the drug resistance. J BUON. 2021;26:1056–1061. [PubMed] [Google Scholar]
- 22.Sun Y, Zhou Y, Bai Y, Wang Q, Bao J, Luo Y, Guo Y, Guo L. A long non-coding RNA HOTTIP expression is associated with disease progression and predicts outcome in small cell lung cancer patients. Mol Cancer. 2017;16:162. doi: 10.1186/s12943-017-0729-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu XJ, Gong Z, Li SJ, Jia HP, Li DL. Long non-coding RNA Hottip modulates high-glucose-induced inflammation and ECM accumulation through miR-455-3p/WNT2B in mouse mesangial cells. Int J Clin Exp Pathol. 2019;12:2435–2445. [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YH, Yu CC, Hsieh PL, Liao YW, Yu CH, Su TR. Inhibition of lncRNA HOTTIP ameliorated myofibroblast activities and inflammatory cytokines in oral submucous fibrosis. J Formos Med Assoc. 2021;120:1188–1193. doi: 10.1016/j.jfma.2020.11.013. [DOI] [PubMed] [Google Scholar]
- 25.Hu X, Tang J, Hu X, Bao P, Deng W, Wu J, Liang Y, Chen Z, Gao L, Tang Y. Silencing of long non-coding RNA HOTTIP reduces inflammation in rheumatoid arthritis by demethylation of SFRP1. Mol Ther Nucleic Acids. 2020;19:468–481. doi: 10.1016/j.omtn.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Lun P, Ji T, Wan DH, Liu X, Chen XD, Yu S, Sun P. HOTTIP downregulation reduces neuronal damage and microglial activation in Parkinson’s disease cell and mouse models. Neural Regen Res. 2022;17:887–897. doi: 10.4103/1673-5374.322475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao P, Liu H, Xue Y, Xiang T, Sun Z. LncRNA HOTTIP promotes inflammatory response in acute gouty arthritis via miR-101-3p/BRD4 axis. Int J Rheum Dis. 2022;00:1–11. doi: 10.1111/1756-185X.14514. [DOI] [PubMed] [Google Scholar]
- 28.Pascoe CD, Obeidat M, Arsenault BA, Nie Y, Warner S, Stefanowicz D, Wadsworth SJ, Hirota JA, Jasemine Yang S, Dorscheid DR, Carlsten C, Hackett TL, Seow CY, Paré PD. Gene expression analysis in asthma using a targeted multiplex array. BMC Pulm Med. 2017;17:189. doi: 10.1186/s12890-017-0545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Wang L, Zhou X, Luo X, Liu K, Jiang E. OSCC exosomes regulate miR-210-3p targeting EFNA3 to promote oral cancer angiogenesis through the PI3K/AKT pathway. Biomed Res Int. 2020;2020:2125656. doi: 10.1155/2020/2125656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng M, Tong R, Zhang Z, Wang T, Liang C, Zhou X, Hou G. EFNA3 as a predictor of clinical prognosis and immune checkpoint therapy efficacy in patients with lung adenocarcinoma. Cancer Cell Int. 2021;21:535. doi: 10.1186/s12935-021-02226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen H, Muz B, Khong TL, Feldmann M, Paleolog EM. Differential effects of Th1 versus Th2 cytokines in combination with hypoxia on HIFs and angiogenesis in RA. Arthritis Res Ther. 2012;14:R180. doi: 10.1186/ar3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kyosseva SV, Chen L, Seal S, McGinnis JF. Nanoceria inhibit expression of genes associated with inflammation and angiogenesis in the retina of Vldlr null mice. Exp Eye Res. 2013;116:63–74. doi: 10.1016/j.exer.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng P, Liu X, Li H, Gao L, Yu Y, Wang N, Chen H. EFNA3 is a prognostic biomarker correlated with immune cell infiltration and immune checkpoints in gastric cancer. Front Genet. 2021;12:796592. doi: 10.3389/fgene.2021.796592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.