Abstract
MicroRNAs (miRNAs) are involved in the development of human malignancies, and cells have the ability to secrete these molecules into extracellular compartments. Thus, cell-free miRNAs (circulating miRNAs) can potentially be used as biomarkers to evaluate pathophysiological changes. Although circulating miRNAs have been proposed as potential noninvasive tumor biomarkers for diagnosis, prognosis, and response to therapy, their routine application in the clinic is far from being achieved. This review focuses on the recent progress regarding the value of circulating miRNAs as noninvasive biomarkers, with specific consideration of their relevant clinical applications. In addition, we provide an in-depth analysis of the technical challenges that impact the assessment of circulating miRNAs. We also highlight the significance of integrating circulating miRNAs with the standard laboratory biomarkers to boost sensitivity and specificity. The current status of circulating miRNAs in clinical trials as tumor biomarkers is also covered. These insights and general guidelines will assist researchers in experimental practice to ensure quality standards and repeatability, thus improving future studies on circulating miRNAs.
SIGNIFICANCE STATEMENT
Our review will boost the knowledge behind the inconsistencies and contradictory results observed among studies investigating circulating miRNAs. It will also provide a solid platform for better-planned strategies and standardized techniques to optimize the assessment of circulating cell-free miRNAs.
Introduction
Cancer remains a leading cause of morbidity and mortality in men and women. Biomarkers offer a wealth of information with respect to tumor progression and assist in the selection of optimal treatment modalities (Lozano et al., 2012; Ou et al., 2021). Identifying reliable cancer biomarkers that can be used for diagnosis, prognosis, or treatment is a critical need in biomedical research. The National Institutes of Health defines a biomarker as an easily measured and analyzed sign of regular cellular mechanisms, pathogenic processes, or pharmaceutical responsiveness to clinical treatment (FDA-NIH Biomarker Working Group, 2016). A biomarker must meet certain criteria to be regarded as ideal. It must be economical (low-cost), readily available, easily detectable, and can be measured and analyzed using minimally invasive procedures. Another essential metric is the specificity for the disorder under consideration, which must be coupled with sensitivity. Ideally, it should be detectable as early as possible, prior to clinical symptoms, and it should change based on the progression of the disease or therapeutic response. Finally, it is important to establish accurate and reproducible results that can be translated from the laboratory to the clinic (Kingsley and Bhat, 2017).
Recently, microRNAs (miRNA) have gained significant interest as potential biomarkers for cancer and other diseases. MiRNAs are small, single-stranded (18–22 nucleotides) non-coding RNAs that are highly conserved across species. They are generated from precursor molecules referred to as primary miRNAs (pri-miRNAs), which may span hundreds of bases, and are sequentially processed in the nucleus, followed by the cytoplasm by ribonuclease-III complexes (Drosha and Dicer). Mature miRNAs function by coupling to the RNA-induced silencing complex (RISC) and guiding this complex to a complementary sequence known as the seed region, which results in translational suppression (O’Brien et al., 2018). As a result, miRNA expression plays a central role in regulating gene expression at the post-transcriptional level. In recent years, miRNAs have been identified as key molecules in regulating gene expression in humans and other species, with the efficiency to control several molecular pathways (El-Daly, et al., 2019a; Gouhar et al., 2022).
miRNAs are primarily synthesized inside the cell; however, a considerable percentage is exported and can be detected in body fluids. Although cell-free miRNAs may be detected in a variety of bodily fluids (e.g., urine, saliva, spinal fluid, seminal fluid, breast milk), circulating miRNAs are exclusively present in the blood (Zen and Zhang, 2012). Aside from the passive release triggered by apoptotic, necrotic, or inflammatory events, the bulk of circulating miRNAs rely on carriers for active secretion to avoid degradation. Protein complexes, lipoproteins, and extracellular vesicles, such as exosomes and microvesicles, are examples of such carriers (Cheerla and Gevaert, 2017).
Although many studies have demonstrated the value of circulating miRNAs as diagnostic and prognostic markers in cancer, their application remains unrealized in personalized medicine as a reliable diagnostic or therapeutic strategy.
This review focuses on the clinical potential of circulating miRNAs as diagnostic and prognostic markers in cancer and the strategies used to detect and quantify these molecules. Moreover, we discuss previous research efforts to evaluate the diagnostic potential of circulating miRNAs compared with standard diagnostic techniques used in the clinic. The major limitations to circulating miRNAs transitioning from bench to clinic will be discussed in an attempt to offer strategies for overcoming these challenges.
1. Sparkling Stars in the Dark Night: The Potential Use of miRNAs as Biomarkers
Circulating miRNAs have several characteristics that support their potential utility as biomarkers for a wide range of diseases. The extraction of miRNAs from blood, urine, or other bodily fluids is a relatively easy procedure; thus, invasive tissue biopsies may be replaced with a relatively simple analysis of commonly available blood products that may be used for early cancer detection. Circulating miRNAs are distinguished by their high stability, which enables them to remain in bodily fluids after release from cells either as exposed ribonucleoprotein complexes or inside membrane vesicles linked to argonaute proteins (Pritchard et al., 2012b). The ease of detection is another advantage to their potential use in the clinic. Nucleic acid detection techniques are widely available. Of note, the development of new techniques for detecting circulating nucleic acids will save time and money compared with the discovery of new antibodies as protein biomarkers (Li et al., 2016; Ban et al., 2017a). Furthermore, the advent of next-generation techniques, such as microarrays and deep sequencing (Farazi et al., 2011), will facilitate the use of miRNA biomarkers in clinical practice.
Circulating miRNAs exhibit high specificity for both the tissue and cell of origin, and are also associated with disease progression. Thus, they have been used in various studies for categorizing tumor grade and evaluating treatment response (Backes et al., 2016). However, there is a debate regarding this alleged high sensitivity and specificity, as we will discuss later.
In general, the irregular expression patterns of mature or precursor miRNAs have been detected compared with that expressed in normal tissues (Lu et al., 2005; Calin and Croce, 2006; El-Daly et al., 2016). A large number of studies have indicated that human malignancies exhibit abnormal miRNA expression patterns. Hundreds of cases of aberrant miRNAs detected in the plasma and serum of cancer patients compared with healthy participants have been reported over the last decade, whereas other researchers have recognized that circulating miRNAs are potential biomarkers for cancer diagnosis and prognosis (Backes et al., 2016; Kawaguchi et al., 2016; Armand-Labit and Pradines, 2017). A wide range of human tumors, such as lymphoma, glioma, breast, colorectal, and prostate cancers, show abnormal miRNA expression levels that are either decreased or increased (El-Daly, Bayraktar, et al., 2020a; Qasemi Rad et al., 2022; Zou et al., 2022). Alterations in miRNA expression profiles may be a direct reflection of chromosomal or genomic changes in cancer-related genes. Together with the function of cancer-associated miRNAs discovered in a variety of tumor tissue specimens, aberrant miRNA expression likely has major clinical implications (Wu et al., 2012; Schwarzenbach et al., 2014; Grimaldi and Incoronato, 2019).
There are several explanations for the abnormal expression of circulating miRNAs in cancer patients. Approximately half of the genes coding for miRNAs are found in cancer-associated regions of the genome, where they are translocated or activated during carcinogenesis (Calin et al., 2004). Variations in the activities of the enzymes responsible for miRNA biosynthesis, such as Drosha and Dicer 1, are another cause of aberrant miRNA levels (Lin and Gregory, 2015). These enzymes are downregulated in bladder and ovarian cancer but activated in stomach and cervical squamous cell tumors. Finally, alterations in circulating miRNAs in cancer may be induced by pri-miRNA transcriptional errors (Condrat et al., 2020). Considering the variables mentioned above, alterations in miRNA expression in various cancer types result in distinct miRNA fingerprints for each malignancy. Such unique miRNA profiles may be suitable for early cancer diagnosis, prognosis, or therapeutic outcome prediction (Winkle et al., 2021). Given the potential diagnostic value of circulating miRNAs, a comprehensive list of the circulating miRNAs currently recognized as potential biomarkers in various cancers is summarized in Table 1.
TABLE 1.
Clinical correlation of circulating miRNAs as diagnostic/prognostic biomarkers in the most studied solid cancer types (prostate, ovarian, lung, breast, and colorectal cancer)
| Prostate cancer | Clinical Correlation | Sample | References |
|---|---|---|---|
|
Diagnostic miR-221,222 |
Upregulated in metastatic patients | Plasma /serum | (Fendler et al., 2016; Thieu et al., 2014) |
|
Diagnostic miR-21 |
Upregulated in resistant and metastatic patients | Serum | (Zhang et al., 2011) |
|
Diagnostic miR-203a-3p |
Downregulated in Cancer and metastatic patients | Serum | (Qasemi Rad et al., 2022) |
|
Diagnostic miR-221,222 |
Upregulated in metastatic patients | Plasma, serum | (Fendler et al., 2016; Thieu et al., 2014) |
|
Prognostic miR-141 |
Upregulated with good clinical outcome | Plasma | (Gonzales et al., 2011) |
|
Prognostic miR-210 |
Upregulated with poor clinical outcome | Plasma, serum | (H. H. Cheng et al., 2013) |
|
Prognostic miR-132, miR-375, miR-429, miR-200a, miR-200b, miR-200c |
Upregulated with short overall survival | Plasma | (H. H. Cheng et al., 2018; H.-M. Lin et al., 2017a; Souza et al., 2017) |
|
Prognostic miR-141 |
Upregulated with good clinical outcome | Plasma | (Gonzales et al., 2011) |
|
Prognostic miR-210 |
Upregulated with poor clinical outcome | Plasma, serum | (H. H. Cheng et al., 2013) |
| Ovarian cancer | Clinical Correlation | Sample | References |
|
Diagnostic miR-181a, miR-342-3p, and miR-450b-5p, miR-30c-1* |
All miRNAs were downregulated in early stage ovarian cancer patients compared with controls except miR-30c-1* was upregulated | Whole blood | (Häusler et al., 2010) |
|
Diagnostic miR-126, MiR-127, miR-150, miR-155, miR-106b, miR-99b |
Downregulated in newly diagnosed patients compared with controls | Serum | (Resnick et al., 2009; Shapira et al., 2014) |
|
Diagnostic miR-195-5p, miR-451a |
Downregulated in late stage ovarian cancer patients compared with controls | Plasma | (Oliveira et al., 2019) |
|
Diagnostic miR-21, miR-92, miR-93, miR-126, miR-29a, miR-1274a, miR-625-3p, miR-720, miR-375, miR-1307 |
Upregulated in ovarian cancer patients compared with controls, except miR-375, miR-1307 upregulated in metastatic patients | Serum | (Resnick et al., 2009; Shapira et al., 2014; Su et al., 2019) |
|
Diagnostic miR-205-5p, miR-145, miR-10a-5p, miR-346, and miR-328-3p |
Upregulated in patients compared with controls | Plasma | (Wang et al., 2019) |
|
Prognostic miR-34a-5p, miR-93-5p |
Downregulated with better progression free survival | Serum | (Robelin et al., 2020) |
|
Prognostic miR‐135a‐3p |
Upregulated in favorable clinical prognosis | Serum | (Fukagawa et al., 2017) |
|
Prognostic miR-1290 |
Upregulated in patients with short overall survival | Plasma | (Shapira et al., 2014) |
| Lung cancer | Clinical Correlation | Sample | References |
|
Diagnostic hsa-miR-140-5p |
Downregulated in patients of different stages compared with controls | Whole blood | (Fehlmann et al., 2020) |
|
Diagnostic miR-492, miR-590-3p |
Upregulated in early stage patients compared with controls | Serum | (Duan et al., 2021) |
|
Diagnostic miR-631 |
Downregulated in early stage patients compared with controls | Serum | (Duan et al., 2021) |
|
Diagnostic miR-200b-5p, miR-190b, miR-502-5p, miR-629, miR-17, miR-100, miRs-21-5p, miR-103a-3p, miR-126, miR-141-3p, miR-193b-3p, miR-205-5p, miR- 210, miR-301b |
Upregulated in patients with malignant pulmonary nodules compared with benign ones except miR-200b-5p differentiates between nodule lung adenocarcinomas and non-nodule healthy smokers | Plasma | (Cazzoli et al., 2013; Lin et al., 2017b) |
|
Diagnostic miRs-135a-5p, miR-145, mIR-200b-3p |
Downregulated in patients with malignant tumor versus benign tumor | Plasma | (Lin et al., 2017a) |
|
Prognostic miR-98-5p, miR-302e, miR-495-3p, miR-613 |
Upregulated with good clinical outcome | Plasma | (Chen et al., 2016) |
|
Prognostic miR-21, miR-27a, miR-218 |
Upregulated in patients resistant to tyrosine kinase inhibitor therapy | Plasma | (Wang, Su, et al., 2015a) |
|
Prognostic miR-20a, miR-223, miR-145, miR-628-3p miR-29c, miR-210 and miR-1244. |
Upregulated in patients of stage I–II | Serum, plasma | (Moretti et al., 2017) |
| Breast cancer | Clinical Correlation | Sample | References |
|
Diagnostic miR-4270, miR-1225-5p, miR-188-5p, miR-1202, miR-4281, miR-1207-5p, miR-642b-3p, miR-1290, miR-3141 |
Upregulated in patients with stage I, II, and III, compared with stage IV | Serum | (Hamam et al., 2016) |
|
Diagnostic miR-20a, miR-214, miR-21 |
Upregulated in patients with breast cancer and benign disease than in healthy women but only miR-214 discriminate malignant from benign tumors and healthy controls | Serum | (Schwarzenbach et al., 2012) |
|
Diagnostic miR-133a-3p, miR-497-5p, miR-24-3p, and miR-125b-5p, miR-106a, miR-182 |
Upregulated in early stage patients compared with control | Serum | (Wang et al., 2018b; Zou et al., 2022) |
|
Diagnostic miR-127-3p, miR-148b, miR-409-3p, miR-652 and miR-801, miR-1246, miR-206, miR-24, miR-373, miR-505-5p, miR-125b-5p, miR-21-5p, miR-96-5p |
Upregulated in early stage patients compared with control | Plasma | (Cuk et al., 2013; Jang et al., 2021; Matamala et al., 2015) |
|
Diagnostic miR-18a, miR-107, miR-133a, miR-139-5p, miR-143, miR-145, Let-7c, miR-365 and miR-425 |
Downregulated in early stage BC patients | Serum | (Kodahl et al., 2014; Li X-X et al., 2015a) |
|
Prognostic miR-130b-5p, miR-151a-5p, miR-206, miR-222-3p |
Upregulated in patients with shorter survival | Serum | (Wang et al., 2018b) |
|
Prognostic miR-122, miR-148a-3p and miR-374a-5p |
Upregulated as a predictive markers of metastatic recurrence in stages (II-III) | Serum | (X. Wu et al., 2012) |
|
Prognostic miR-148a-3p, miR-374a-5p |
Upregulated in patients with pathologic complete response | Plasma | (Di Cosimo et al., 2020) |
|
Prognostic miR-10b-3p, miR-940, miR-4310 |
Downregulated in metastatic patients sensitive to trastuzumab | Serum | (H. Li et al., 2018) |
| Colorectal cancer | Clinical Correlation | Sample | References |
|
Diagnostic miR-29a, miR-92a, miR-182, miR-409 |
Upregulated in early stage patients compared with control, miR-29a, miR-92a also are upregulated in advanced stage | Plasma | (Huang et al., 2010; Liu et al., 2018; S. Wang, Xiang, et al., 2015b; Yamada et al., 2015) |
|
Diagnostic miR-30a-5p, miR-7, miR-93 |
Downregulated in early stage patients compared with control | Serum, plasma | (Wang, Xiang, et al., 2015b) |
|
Diagnostic miR-506, miR-4316 |
Upregulated in early cancer patients compared with control | Peripheral blood | (Krawczyk et al., 2017) |
|
Diagnostic miR-21, miR-145, miR-203, miR-155, miR-210, miR-31, miR-15b, miR-29a, miR-345, and miR-106a-5p |
Upregulated in advanced stage patients compared with control. | Plasma | (Herreros-Villanueva et al., 2019; Kudelova et al., 2022; Nassar et al., 2021) |
|
Prognostic miR-1290 |
Upregulated with tumor aggressiveness and poor prognosis. | Serum | (Imaoka et al., 2016) |
|
Prognostic miR-203, miR-30a-5p, miR-17-3p, miR-106a, miR-21, miR-92a, miR-1290, miR-210, miR-183, miR-885-5p, miR-592, miR-196b, miR-155 |
Upregulated with lower patient survival rate | Serum | (Hur et al., 2017; Rapado-González et al., 2019; Sun et al., 2019) |
|
Prognostic miR-141 |
Upregulated in patients with lower survival rate and in metastatic patients | Plasma | (H. Cheng et al., 2011) |
|
Prognostic miR-200c |
Upregulated in stage IV patients compared with control | Serum | (Toiyama et al., 2014) |
|
Prognostic miR-21, miR-210 |
Upregulated in Stage IV patients | Plasma | (Nassar et al., 2021) |
|
Prognostic miR-200b, miR-31 |
upregulated in patients with increased recurrence risk | Plasma | (Yuan et al., 2017) |
|
Prognostic miR-19a, miR-19b, miR-15b |
Upregulated in advanced stages | Plasma | (Giráldez et al., 2013) |
|
Prognostic miR-15b, miR-526, miR-96, miR-148a, miR-22 miR-141, miR-628-5p, miR-203 and miR-200b |
Upregulated in advanced stages | Plasma | (Sun et al., 2016) |
2. Pitfalls of Using Circulating miRNAs as Biomarkers
Although previous reports have considered miRNAs as powerful biomarkers, other studies have indicated that the most commonly reported miRNA biomarkers are largely nonspecific because they are associated with a variety of diseases and outcomes. In some circumstances, highly comparable studies with the same aim have shown minimal agreement in their results (Witwer, 2015). For example, Mitchell et al. (2008) reported that miR-141 significantly differentiates prostate cancer patients from normal controls; however, the authors correctly pointed out that abnormal expression has been documented in other epithelial malignancies, such as breast, colon, and lung cancers. Chim et al. (2008) found that miR-141 was upregulated in the blood of pregnant women; thus, the upregulation of miR-141 could be a sign of malignancy or a temporary elevation during pregnancy. Another example of non-specificity is the alterations in levels of circulating miR-16, -155, -21, -126, and -223 that were linked to ten non-neoplastic disorders (Haider et al., 2015).
miR-21 is highly expressed and is considered a reliable prostate cancer biomarker according to several studies (Endzeliņš et al., 2017; Stafford et al., 2022). It has been shown to directly correlate with tumor volume in gastric cancer (Song et al., 2013), and elevated miR-21 expression is associated with aggressive lung cancer and relapse-free survival in diffuse large B-cell lymphoma (Cortez et al., 2011; Wang and Zhang, 2012). Therefore, elevated miR-21 in the blood may be considered an indicator of general illness conditions, such as inflammation, or associated with a variety of cancer types (Egidi et al., 2013). With such low specificity toward specific diseases, miR-21 is considered “the most nonspecific biomarker for all diseases” (Jenike and Halushka, 2021).
Reproducibility is essential for the detection and validation of tumor biomarkers. Misuse of sample methodologies, analytical processes, and statistical methods can all affect reproducibility. This is a significant limitation to the use of circulating miRNAs as a diagnostic tool. Apart from head and neck squamous cell carcinoma, most data generated for circulating miRNAs in cancer has shown a lack of consistency. Many studies have produced results that have later been determined to be non-reproducible (Jarry et al., 2014). The expression pattern of miR-200c is an example of such conflicting findings, with one group reporting that enhanced miR-200c expression is associated with a poor progression and overall survival rate in patients with gastric cancer, whereas others reported a connection with progression-free survival (Valladares-Ayerbes et al., 2012; Williams et al., 2013).
miR-145 represents another example of inconsistency in the expression of circulating miRNAs between studies. miR-145 was reported to be highly overexpressed in plasma from early-stage cancer patients compared with healthy participants, according to several comparable studies conducted in different ethnic groups (Mar-Aguilar et al., 2013; Ashirbekov et al., 2020; Itani et al., 2021). However, other studies have demonstrated that this miRNA was downregulated in tissue and plasma samples of breast cancer patients compared with controls (Visone et al., 2009; Ng et al., 2013).
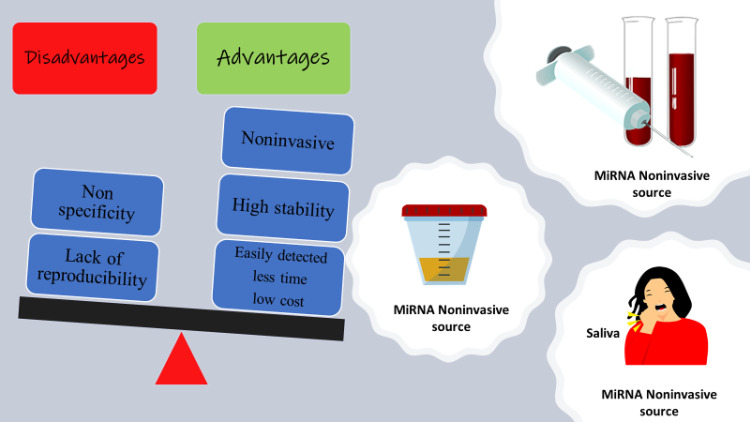
A limited sample size with poor statistical validity, specificity of the detection method, or miRNA degradation may also contribute to the inconsistencies in miRNA results (Alečković & Kang, 2015). Before circulating miRNA profiles can be integrated into tumor progression and clinical outcome, the findings must be verified in large populations. Methodological issues also contribute to this variation (Moldovan et al., 2014). The techniques used by researchers to extract miRNAs from blood samples differ widely, which raises a concern with respect to reproducibility between and within miRNA detection platforms (Cerkovnik et al., 2007; Ban et al., 2017a). In addition, the minimal concentration of circulating miRNAs complicates reliable quantitation and detection (Hengen, 1996). In general, protocols used for the detection of circulating miRNA that require >100 μl of sera could be problematic for some patients (Moret et al., 2013; Niu et al., 2015). Even with the current promise of miRNA biomarkers, the diagnostic specificity and reproducibility of published indicators reveal that fulfilling this promise remains a challenge, as illustrated in Fig. 1. However, this concern is tempered by other studies that have ensured the real power of circulating miRNAs as tumor markers based on their sensitivity and specificity compared with traditional tumor biomarkers.
Fig. 1.
The advantages and disadvantages of circulating miRNA as tumor biomarkers.
3. Clinically Standard Biomarkers Versus Circulating miRNAs
Carcinoembryonic antigen (CEA) and cancer antigen 15.3 (CA15.3) are two standard tumor markers used for the diagnosis and prognosis of breast cancer. The accuracy rates of traditional CA15-3 and CEA for the diagnosis of breast cancer are substantially variable, which may be attributed to an ethnic group, the number of participants, or cancer type (De Cock et al., 2021). A study by Gao et al. (2013) demonstrated that for the diagnosis of breast cancer, miR-21 exhibited much higher sensitivity and specificity (87.6% and 87.3%, respectively) compared with CA153 and CEA, which had sensitivities of 22.47% and 15.73%, respectively. Similarly, two recent studies found that miR-27a, miR-29a, and miR-335 were superior to CEA and CA15.3 for the early diagnosis of breast cancer, and miR-27a exhibited a higher specificity and accuracy for the detection of early-stage and low grade BC, suggesting that miR-27a is a potential biomarker for the early diagnosis of BC, particularly in high-risk patients with early-stage and low-grade malignancies (Ali Ahmed et al., 2020; Swellam et al., 2021).
Bimanual pelvic check and transvaginal ultrasound are widely used diagnostic methods for ovarian cancer (OC). However, they have their own set of limitations, such as being time-consuming, expensive, aggressive, inconvenient, and having poor specificity and sensitivity (Sundar et al., 2015). Some studies have evaluated the efficacy of CA-125 in OC and found an optimal sensitivity and specificity of 0.74 and 0.83, respectively (Wang et al., 2014a; Goff et al., 2017). In a meta-analysis conducted by Zhou et al. (2018), the pooled sensitivity and specificity a panel of circulating miRNAs was 0.76 and 0.81 (95% confidence interval 0.74–0.87), with a high diagnostic accuracy corresponding to an area under the curve (AUC) of up to 0.85. Thus, the accuracy of circulating miRNAs for the diagnosis of OC may be greater compared with traditional biomarkers.
CEA and CA-199 are common diagnostic markers for colorectal cancer (CRC). Despite the fact that these indicators are frequently used in clinical applications, several studies consider CEA and CA 199 as late-stage markers because they are not sufficiently selective to detect early-stage CRC (VukobratBijedic et al., 2013). In a previous study by our group, a receiving operating characteristic curve analysis revealed that serum and tissue miR‐15b, miR‐21, and miR‐29a levels were promising early noninvasive diagnostic markers for CRC as these miRNAs outperformed CEA and CA-199 in terms of diagnosing early transformative changes of the colonic crypt with high AUC values as well as high sensitivity and specificity (El‐Daly et al., 2019b).
Commonly used biomarkers for muscle and liver injury are alanine aminotransferase (ALT) and aspartate aminotransferase (AST), which are increased, especially for drug-induced toxicities viral hepatitis, metastatic tumors, and alcohol consumption (Law and Rudnicka, 2006). In a study by Laterza et al. (2009), miR-122 and miR-133a exhibited high specificity for liver and muscle toxicity, but they were not detected in the plasma of animals with other organ toxicities. This was not the case for ALT and AST, in which both were elevated in response to organ toxicity. Furthermore, when the data were analyzed in conjunction with the histopathology of liver sections, miR-122 demonstrated superior diagnostic sensitivity compared with ALT (Laterza et al., 2009).
The standard indicators used for the diagnosis of gastric cancer (GC) include CEA and CA19-9; however, because these biomarkers are not elevated in early-stage GC, they are not effective for early-stage diagnosis (Zheng et al., 2021). Izumi et al. (2021) evaluated the diagnostic performance of a 10-miRNA signature in over 1900 tissue and serum samples collected from patients with GC at different tumor stages and healthy volunteers. The results indicated that miR-18a, miR-181b, and miR-335 had a high diagnostic precision at all stages with an AUC value of 0.86 (95%CI 0.83–0.90), specifically in stage I patients with an AUC of 0.85 (95%CI, 0.79–0.91). Moreover, this miRNA signature outperformed the commonly used blood biomarkers and surpassed endoscopic screening with respect to cost-effectiveness.
Based on the previously mentioned studies and others as listed in Table 2, circulating miRNAs have a good overall diagnostic accuracy over standard biomarkers used in the clinic.
TABLE 2.
Overall sensitivity and specificity of circulating miRNAs comparing to clinically standard biomarkers
| miRNA | Standard biomarker | Sensitivity and specificity | Cases | Sample source | References |
|---|---|---|---|---|---|
| miR-27a “Upregulated” |
CA15.3 CEA |
miR-27a outperforms both CEA and CA15.3 in terms of early diagnosis of BC, with greater specificity and accuracy in detecting early stages BC patients. | 100 BC patients, 30 benign breast lesions and 20 healthy volunteers |
Human Serum | (Swellam et al., 2021) |
| miR-29a, miR-335 “Downregulated” |
CEA CA15.3 |
The diagnostic fficiency of miR-29a and miR-335 outperformed CEA and CA 15.3, for early identification of BC patients. | 44 BC patients, 25 benign breast lesions and control group served as 19 healthy volunteers | Human serum | (Ali Ahmed et al., 2020) |
| miR‐15b, miR‐21, miR‐29a, miR‐141, miR‐17‐3p “Upregulated” |
CA19.9 CEA |
miR-15b, miR-21, and miR-29a performed best in detecting early-onset colorectal cancer events, while miR-141 and miR-17-3p performed poorly and only upregulated at late stage | Male CD‐1 mice with induced colorectal carcinogenesis | Mice serum | (El-Daly, Morsy, et al., 2019a) |
| miR-21 “Upregulated” |
AFP | miR-21 outperformed α-fetoprotein in distinguishing HCC from chronic hepatitis. |
126 patients with HCC, 30 patients with chronic hepatitis, and 50 healthy volunteers. | Plasma | (Tomimaru et al., 2012). |
| miR-122, miR-133a, and miR-124 “Upregulated” |
ALT AST |
miR-133a was more sensitive to the degree of liver or skeletal muscle damage than AST, while ALT and AST have better specificities for diagnosing and discriminating muscle and liver toxicities. miR-122 is a more diagnostically sensitive marker for identifying liver injury than ALT. | Male and female Sprague Dawley rats treated with liver or muscle toxicants. | Rats plasma |
(Laterza et al., 2009) |
| miR-145, miR-150, miR-223, miR-636, miR-26b, miR-34a, miR-122, miR-126*, miR-145, miR-150, miR-223, miR-505, miR-636, miR-885 “Downregulated” |
CA19-9 | The putative diagnosis efficiency was strengthened by combining both miRNAs panels and serum CA19-9 | 409 pancreatic cancer patients and 25 chronic pancreatitis patients, 312 blood donors as healthy | Human whole blood | (Schultz et al., 2014) |
| miR-18a, miR-181b, and miR-335 “Upregulated” |
CEA CA19-9 |
This miRNA profile outperformed currently used blood markers in terms of diagnostic accuracy for the early diagnosis of gastric cancer, including individuals with stage I tumor. | 598 gastric cancer patient |
Serum | (Izumi et al., 2021). |
| miR-21 “Upregulated” |
CA15.3 CEA |
miR-21 showed higher sensitivity over standard markers in the diagnosis of BC |
89 BC patients and 55 healthy controls | Serum | (Gao et al., 2013) |
| miR-155 “Upregulated” |
CA15.3 CEA | miR-155 showed a higher sensitivity over standard markers in the diagnosis of BC patients | 184 BC patients and 75 control individuals | Serum | (Wang et al., 2014b) |
| miR-182, miR-183, miR-210 “Regulated according to tumor stage” |
CEA | The diagnostic value of CEA was not significantly different from those of miRNAs |
112 Non-small cell lung carcinoma patients and 104 controls | Serum | (Zhu et al., 2016) |
| miR-371a-3p | AFP LDH HCG |
miR-371a-3p showed higher sensitivity than AFP, LDH and HCG | 616 patients with testicular GCTs and 258 male controls | Serum | (Dieckmann et al., 2019) |
| miR-16, miR-195, and miR-199a “Downregulated” |
AFP, DCP AFP-L3 | miR-16 had the highest sensitivity for HCC, followed by miR-199a, AFP, DCP, AFP-L3, and miR-195 | 105 HCC patients, 107 CLD patients, and 71 healthy control | Serum | (Qu et al., 2011) |
| miR-122 and miR-155 “Upregulated” |
ALB AST ALT | miR-122 levels were employed to predict treatment outcome with greater AUC, higher sensitivity, and specificity ratios compared with traditional biomarkers. | 80 HCV patients | Serum | (Fan et al., 2017) |
4. Single Versus Multiple Biomarkers
In most cases, the alteration of biomarker expression in non-malignant disorders is temporary; however, in cancer, dysregulation is either stable or continuously increasing. Because of the issue of non-specificity, diagnosing a patient based on a single tumor biomarker is filled with hazards (McPherson et al., 2021). The use of serial testing and multiple biomarkers can assist in the detection of abnormally increased levels resulting from temporary elevation. Combining traditional tumor markers with highly specific and sensitive miRNAs may be a good option to more accurately reflect the patient’s case and enhance the use of circulating miRNAs as clinical biomarkers (Table 2). A study by Qu et al. (2011) focused on the diagnosis of hepatocellular carcinoma (HCC) using three serum miRNAs, miR-16, miR-195, and miR-199a, alone or in combination with the clinically standard serum indicators, alpha-fetoprotein (AFP), alpha-fetoprotein L3 (AFP-L3), and des-gamma-carboxy prothrombin (DCP). The results indicated that miR-16 was a more sensitive biomarker for HCC, whereas the combination of miR-16, AFP, AFP-L3, and DCP correctly identified approximately 92.4% of the HCC patients with 78.5% specificity. These results indicate that combining circulating miRNAs with standard markers, such as AFP, may improve the accuracy of HCC diagnosis (Qu et al., 2011).
In a lung cancer study, a receiving operating characteristic curve analysis for serum miR-182, miR-183, miR-210, and miR-126 levels revealed their power as a diagnostic biomarker for the early detection of non-small cell lung carcinoma with high sensitivity and specificity. The combination of these four miRNAs with CEA further increased the diagnostic value, which resulted in an AUC of 0.965, a sensitivity of 81.3%, a specificity of 100%, and an accuracy of 90.8% by logistic regression model analysis. The same authors also reported that these miRNAs may be used to differentiate early-stage non-small cell lung carcinoma from cigarette smokers without lung cancer, pneumonia, or GC patients with high sensitivity and specificity (Zhu et al., 2016).
The idea of using miRNAs in combination with other markers was also evaluated in CRC by Cheng et al. (2011). They reported that the combination of miR-141 with CEA increased diagnostic accuracy as the sensitivity was higher compared with applying each marker individually. MiR-141 recognized seven Stage IV colon cancer cases that CEA missed, whereas CEA diagnosed four Stage IV metastatic colon cancer cases that miR-141 did not recognize when the specificity was set to 100%. The combination of multiple miRNAs is also more promising as biomarkers for BC detection compared with individual miRNAs. The supporting evidence demonstrates that a combination of miR-145, miR-155, and miR-382 is a more powerful diagnostic approach for distinguishing BC patients from healthy subjects, with a sensitivity of 0.97 and a specificity of 1.0 compared with individual classifiers (Cui et al., 2015). In a recent work by Jang et al. (2021), a total of 226 plasma samples from patients with BC and 146 healthy non-cancerous controls were evaluated to determine the efficiency of circulating miRNAs as early diagnostic biomarkers. The levels of 9 miRNAs, miR-21, miR-24, miR-202, miR-206, miR-223, miR-246, mi -373, miR-6875, and miR-219B were significantly dysregulated and distinguished healthy subjects from breast cancer patients. Of the nine miRNAs, a combination of four (miR-24, miR-206, miR-373, and miR-1246) exhibited 98% sensitivity, 96% specificity, and an accuracy of 97% for breast cancer diagnosis. This suggests the efficient use of multiple miRNAs as potential biomarkers for the early detection of BC compared with using a single miRNA (Jang et al., 2021).
Overall, these studies clearly demonstrate that using several biomarkers (either a panel of multiple miRNAs or miRNAs plus conventional biomarkers) yields superior diagnostic results compared with the measurement of a single miRNA.
5. Challenges in the Clinical Applications of Circulating miRNAs
Despite the immense progress focused on the role of distinct circulating miRNAs signatures in diverse diseases, a large variety of experimental parameters used for the evaluation process has contributed to the widespread inconsistency in the results. We will discuss the most common (pre-analytical, analytical, and post-analytical) factors contributing to the discrepancies in the circulating miRNAs results.
5.1. Starting Material (Sample Type)
Evaluating the differential expression of miRNAs in circulation may be done using various body fluids. In most cases, whole blood, plasma, or serum samples are used. However, the sample type can have a significant effect on the levels of circulating miRNAs, and it is considered the most important factor responsible for inconsistent results. A large number of studies have revealed that even within the same individual, the expression of circulating miRNAs differs between serum and plasma samples (Table 3). Several studies have conducted a paired comparative analysis to delineate the differences in miRNA expression between serum and plasma of the same patient. Although circulating miRNA yield was comparable in plasma and serum, significantly different miRNA expression profiles between serum and plasma were detected when normalized to an internal reference, and plasma showed more variation than serum (Mompeón et al., 2020). For example, McDonald et al., (2011) reported that the levels of miR-15b, -16, and -24 in plasma were higher compared with that detected in serum. However) observed a different pattern, in which the miRNA levels detected in serum were higher compared with the corresponding plasma samples.
TABLE 3.
Comparing levels of circulating miRNAs in serum and plasma for same individuals
| miRNA | Serum versus Plasma | Condition | Detection Method | References |
|---|---|---|---|---|
| miR-1 | Higher in plasma than in serum | AMI | TaqMan RT-qPCR | (C. Li et al., 2013; Liu et al., 2015) |
| miR-133a, miR-26a, miR-499a | miR-133a and miR-26a levels were significantly upregulated only in serum & miR-499a upregulated only in plasma | AMI | TaqMan RT-qPCR | (Mompeón et al., 2020) |
| miR-1, miR-208b | These miRNAs showed a higher coefficient of variation in plasma samples | AMI | TaqMan RT-qPCR | (Mompeón et al., 2020) |
| miR-21 | Increased in serum, decreased in plasma | AMI | TaqMan RT-qPCR | (Mompeón et al., 2020) |
| miR-15b, miR-16, miR-24, miR-122 | Higher in plasma than in serum | healthy individuals | TaqMan RT-qPCR | (McDonald et al., 2011) |
| Higher in serum than in plasma | healthy individuals | TaqMan RT-qPCR | (Wang et al., 2012a) |
AMI, acute myocardial infarction.
The difference in the miRNA levels between serum and plasma samples may be attributed to the RNA produced by blood cells, leukocytes, and platelets during the coagulation process (Pritchard et al., 2012a). Cell lysis during the coagulation process, particularly red blood cells, may also contribute to the RNA content discrepancy between serum and plasma.
The transfer of miRNAs through extracellular vesicles can control the expression of several genes and reduce the efficiency of miRNAs as disease biomarkers. Therefore, the use of whole blood still faces several limitations for evaluating circulating miRNAs (Tiberio et al., 2015). The discrepancy may also be addressed by the procedures used to separate plasma and serum from whole blood, which results in varying quantities of blood cell contamination in the two fluids. These procedures will be further discussed.
5.2. Sample Processing and Storage
The time interval between sample collection and further processing can influence the miRNA levels, as the cellular components in the blood can contribute irrelevant miRNAs to serum or plasma during storage. Moreover, hemoglobin and lactoferrin may hinder downstream applications of miRNAs (Kim et al., 2012). Therefore, processing a sample within a few hours is highly recommended to reduce the undesired contribution of other components. However, once the sample is processed into serum or plasma, several studies have indicated that the miRNAs are stable from either fresh or frozen specimens. Even after several freeze-thaw cycles, minor to no alterations between fresh and frozen specimens were observed, which confirms that miRNAs are quite stable in circulation (Page et al., 2013b; Glinge et al., 2017; Chorley et al., 2021).
In a study by Grasedieck et al. (2012), the stability of serum miRNAs was detected following short-term (10 days), intermediate (up to 20 months), and long-term (up to 10 years) storage at -80°C and -20°C. Interestingly, storing samples at -20°C had relatively negligible effects on the cycle threshold (ct) values of the measured miRNAs. These results further emphasize the potential use of miRNAs as biomarkers due to their stability during long-term storage. Nevertheless, repeated freeze-thaw cycles should be avoided since minimally represented miRNAs could be affected to some extent if any miRNA degradation occurs (Glinge et al., 2017; Matias-Garcia et al., 2020).
5.3. Centrifugation
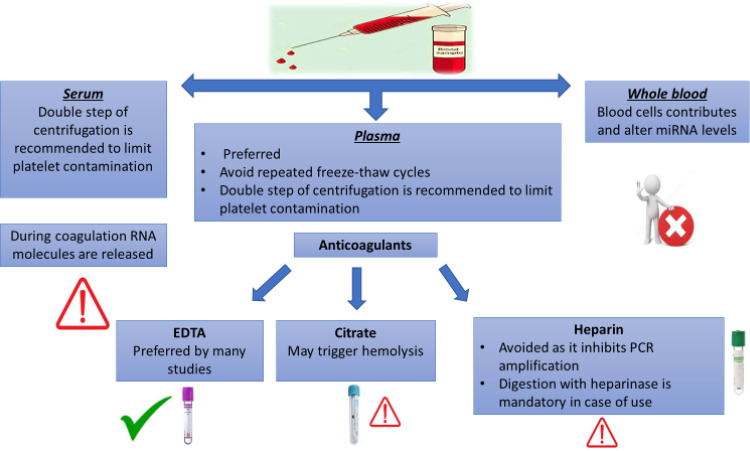
The centrifugation settings, such as speed, duration, and temperature, are critical factors that are often underestimated and may influence the miRNA expression profile of the processed samples. Prolonged high-speed centrifugation may increase sample hemolysis and, as a result, miRNA release from platelets. On the other hand, low-speed and brief centrifugation might result in an inadequate separation of serum or plasma from cellular fractions (Duttagupta et al., 2011). Therefore, standardized centrifugation conditions to separate serum/plasma are necessary for a reliable evaluation. A double step of centrifugation, with a high speed at the second, is usually applied to effectively remove residual platelets from samples and facilitate the assessments of cell-free miRNAs (McDonald et al., 2011; Page et al., 2013a).
5.4. Type of Anticoagulant
The anticoagulant used to collect blood for plasma separation might affect the accuracy of the RNA analysis (Fig. 2). Salts of EDTA, citrate, or heparin are the most common anticoagulants used for plasma collection. Although EDTA and citrate are more likely to be used for miRNA analysis, EDTA collection tubes are preferred over citrate as the chance of hemolysis induction is less if EDTA is used. Heparin is not suggested in blood collection for analysis of circulating miRNAs as it hinders the activity of reverse transcriptase and polymerases in different RNA measurement methodologies (Mitchell et al., 2016; Glinge et al., 2017). However, heparinase treatment of the RNA samples extracted from blood collected with heparin tubes significantly reverses the heparin-induced inhibition of downstream enzymatic assays, enabling a reliable miRNA quantification (Kondratov et al., 2016). This approach comes at a cost, as heparinase concentration and timing should be tuned for each set of samples (Johnson et al., 2003), suggesting the preference of avoiding heparin as an anticoagulant. In a previous study by (Glinge et al., 2017), the expression level of miR-1, miR-21, and miR-29 did not significantly vary in serum or plasma samples collected using either EDTA or citrate for the same individuals. However, in collected lithium-heparin plasma, no expression levels were detected for these miRNAs.
Fig. 2.
General concerns and suggestions regarding sample collection.
5.5. Sample Volume
The initial volume of the sample used for RNA extraction is another factor to be considered which has a significant influence on the quality of miRNA quantification. Although miRNA concentrations in the liquid biopsies are relatively low, this does not justify the use of a higher amount of the startup material as it can enhance RNA contamination with the higher number of contaminants carried over with the starting sample. Moreover, a saturation of the purification columns would commonly occur (Androvic et al., 2019). Most RNA isolation kits recommended a starting input sample of 200 µl. However, this can vary based on the sample type. In the study conducted by Androvic et al. (2019), the authors evaluated the impact of the sample volume and the type of the startup sample using an miRNeasy Serum/Plasma Advanced Kit (Qiagen Corp). According to this study, the optimal input volume varies depending on the sample type, human plasma or serum, or rat serum. Higher input volumes of >300 µl for human samples and >150 µl for rat serum samples resulted in poor RNA isolation efficiency and less reproducible results when compared with moderate input volumes of 200–300 µl for human serum or plasma and 100–150 µl for rat serum.
5.6. Hemolysis Effect
Since blood cells significantly contribute to circulating miRNAs, any perturbations in blood cell counts or hemolysis can alter the levels of circulating miRNAs, reflecting a blood cell-based feature instead of a disease-related cause (Kirschner et al., 2013). Hemolysis can usually happen throughout sampling and handling procedures, and the released cellular miRNAs can alter the measured miRNA levels up to 50-fold, according to Pritchard et al. (2012b), making it difficult to understand the findings biologically. Moreover, hemolysis can alter the normalization step. For example, miR-16-5p, the most often miRNA used as an internal reference, is an abundant miRNA in erythrocytes. Thus, its level is altered even at a low level of hemolysis (Kirschner et al., 2011). Hemolysis can be limited by following the standard operating procedures of blood collection. This includes using proper diameter needles (not too small to prevent blood cell damage), avoiding improper tube mixing, and prolonged tourniquet in addition to other standard procedures (Khan et al., 2017).
Unless the investigated cell-free miRNAs are not impacted by hemolysis, excluding hemolyzed samples from further procedures is a critical step. Usually, hemolysis can be evaluated through visual inspection or measuring the sample's absorption spectroscopically through scanning at a wavelength of 350 to 650 nm (Khan et al., 2017). The optical density at 414 nm (free oxyhemoglobin absorption peak) is used to estimate the degree of hemolysis, with secondary peaks at 541 and 576 nm reflecting elevated levels of hemolysis. If the absorption at 414 nm is more than 0.2, the sample is considered hemolyzed (Kirschner et al., 2013). However, this approach is not optimal since lipemic plasma samples create an invalid rise in absorbance values at 414 nm even though no hemoglobin-related peak is present (Heckl et al., 2021). A hemolysis biomarker based on miRNA expression is suggested if the original serum or plasma sample is no longer accessible. It was reported that miRNAs are abundant in erythrocytes, such as miR-144, miR-451, or miR-23a, which could be used to detect potential hemolyzed samples (Rasmussen et al., 2010; Blondal et al., 2013). Blondal et al. (2013) suggested that a ΔCq of more than 7 for the ratio of miR-23a/miR-451 is an indication of high hemolysis. miR-23a is a prevalent miRNA in plasma and serum and is unaffected by hemolysis. However, miR-451a is exceptionally abundant in erythrocytes and rises extremely in response to hemolysis (Pritchard et al., 2012b; Blondal et al., 2013). A two-tailed quality control technique, suggested by Androvic et al. (2019), combines two sets of exogenous synthetic spike-in molecules and three endogenous miRNAs; let-7a, miR-23a, and miR-451a, all quantified with the two-tailed RT-qPCR method, have been offered as a cost-effective and accurate technique to assess the quality of isolated miRNAs and potential erythrocyte contamination.
To conclude, data from studies using various blood fractions should not be directly compared, and only miRNAs that are not slightly up- or downregulated will be useful as clinical biomarkers.
5.7. RNA Extraction Protocols
Biofluids have a marginal amount of RNA. As a result, RNA isolation protocols impact the outcomes of circulating miRNA measurement. Although there is no consensus regarding the optimum extraction protocol, nucleic acid extraction protocols are generally divided into three main categories; 1) guanidine-phenol-chloroform-based protocols, 2) commercial kits utilizing columns or beads, and 3) a combination of the two previously mentioned protocols (Tiberio et al., 2015). The ideal RNA extraction technique is one that is uncomplicated, fast, inexpensive, and, most importantly, reproducible, with minimal variability across samples, and capable of preserving RNA purity and integrity. In the past few years, several studies focused on comparing the efficiency of the various RNA extraction protocols and commercial kits. Comparative studies developed by researchers used several RNA extraction methods (Moret et al., 2013; Page et al., 2013a; Sourvinou et al., 2013; Monleau et al., 2014; Li et al., 2015b; Tan et al., 2015), including manual protocol, such as TRIzol (Invitrogen), or the following commercial kits:
Phenol and column methods, such as miRNeasy Serum/Plasma kit (Qiagen) and mirVana PARIS kit (Life Technologies).
Protein precipitation and column methods, such as miRCURY RNA Isolation Kit from Qiagen and HigherPurity Total RNA Extraction Kit from Canvax.
Proteinase K and column methods, such as Quick-RNA Miniprep Kit (Zymo Research) and Monarch RNA Purification Kit (New England Biolabs).
Magnetic beads method, such as Agencourt RNAdvance Blood Kit (Beckman Coulter Life Sciences).
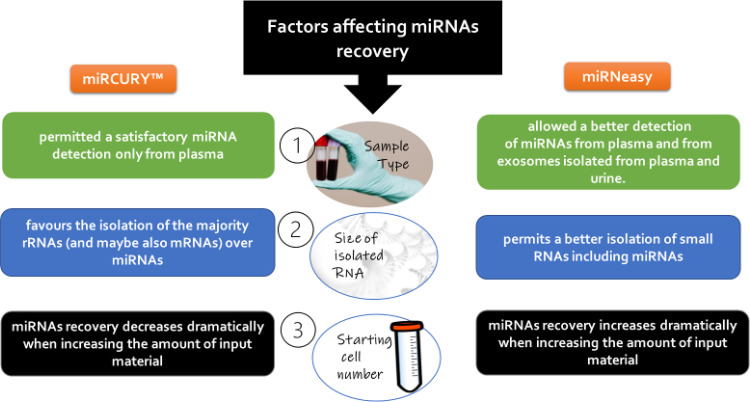
Although there is no consensus on which approach is the best (considering that some studies recommended miRNeasy Serum/Plasma kit to produce an enriched miRNA fraction from plasma samples), most of the comparative publications highlighted the merits of column-based extraction techniques over Trizol (Moret et al., 2013; Felekkis and Papaneophytou, 2020). In addition, increasing the volume of starting material produces a less efficient recovery of miRNAs (McAlexander et al., 2013b; El-Khoury et al., 2016), and adding modest doses of an RNA carrier et (Moret et al., 2013). There is an agreement that the different isolation methods provide different yields and/or quality of miRNAs (Felekkis abd Papaneophytou, 2020). Even different kits using the same isolation method provide different miRNA yields and quality. El-Khoury et al, (2016) compared the results from the most commercially available isolation kits miRCURY and miRNeasy, and their recommendations are summarized in Fig. 3.
Fig. 3.
Factors affecting miRNA recovery during the process of extraction.
In the extraction protocol, precipitation is the most important step that may affect the final yield. As a result, optimal precipitation conditions are required to enhance miRNA recovery from biofluids. Carriers such as glycogen, RNA bacteriophage carrier (MS2), linear acrylamide, and yeast transfer RNA (tRNA) are often used to enhance extraction efficiency and improve reproducibility (McAlexander et al., 2013a). These carriers function by trapping RNA, and since they are insoluble, they form a visible pellet which facilitates the RNA extraction (Wang et al., 2002; Cuk et al., 2013; McAlexander et al., 2013b; Moret et al., 2013). Glycogen is the most frequent carrier used; however, coupling yeast transfer RNA with glycogen has been found to considerably enhance RNA yield, as well as boost miRNA extraction rather than either using glycogen or yeast transfer RNA alone (Ban et al., 2017b). In this context, it is also suggested that the use of glycogen alone is preferred in some experiments since other carriers, that include nucleic acid components, may interfere with downstream analysis such as next-generation sequencing, and the exogenous RNAs of the added carriers could consume sequencing reads (Androvic et al., 2019; Bryzgunova et al., 2021).
5.8. miRNA Quantification
Developing an accurate miRNA quantification approach is a critical factor to obtaining valid results, especially using array hybridization platforms that require a precise quantity of starting material (Lee et al., 2014). Spectrophotometry quantitation platforms, such as Nanodrop or NanoQuant, used to measure RNA quantity and quality could be unreliable when applied to serum and plasma samples that contain low concentrations of RNA. Spectrophotometry quantitation platforms function by measuring the absorbance at 260 nm, which could also detect free nucleotides and DNA (El-Khoury et al., 2016). Contaminants from freeze-thaw cycles or isolation reagents, such as phenols (absorbs at 270 nm) or EDTA, could also interfere with the absorbance (Garcia-Elias et al., 2017). Moreover, RNA measurements of samples that are extracted using RNA carriers will be invalid because the contribution of signal from the RNA carrier is much stronger compared with that of the sample itself and consequently masks the signal of the RNA of interest (Ramón-Núñez et al., 2017). Other quantitation techniques, such as fluorometric analysis, may be more suitable than spectrophotometry. Unlike the NanoQuant and Nanodrop assays, the Qubit fluorometer (Life Technologies, Thermo Fisher Scientific, Inc.) incorporates fluorescent dyes that are selective for short RNA sequences, even at low concentrations, over other forms of RNA. As a result, the values produced using this platform correspond primarily to the miRNA component of the sample (Mauger et al., 2015). Garcia-Elias et al. (2017) compared the optimal methodology for miRNA quantitation in plasma samples using different platforms, including the NanoQuant spectrophotometer (Tecan Infinite 200 PRO), Nanodrop 2000 spectrophotometer (Thermo Scientific TM) and Qubit 2.0 fluorometer (Life Technologies). Based on the data, both spectrophotometer-based platforms overestimated the level of miRNAs compared with the fluorometric approach by detecting nucleotides and other contaminants. In this context, using an RNA isolation protocol that is more specific to miRNA will decrease contaminating RNA species and reduce the quantitative discrepancies between the Qubit and spectrophotometric techniques. The Qubit 2.0 Fluorometer also enabled a lower detection range for miRNAs (0.05 ng/µl) compared with the common spectrophotometry platforms (2 ng/µl for Nanodrop, and 3 ng/µl for Nano- quant), which is essential for biofluids. These findings suggest that the Qubit-based miRNA technique produces a more accurate estimate of miRNA levels, particularly for plasma samples. Of note, Qubit was more suitable compared with spectrophotometer-based platforms for miRNA quantification, even from tissue samples in a comparison of different quantitation platforms Deben et al., (2013).
5.9. miRNA Detection Platforms
Because circulating miRNAs are promising biomarkers for various human disorders, the development of a reliable and relatively simple detection method remains a challenging because of the intrinsic characteristics of miRNAs, which include small size, low quantitation in biofluids, and a high sequence similarity among miRNA family members (Tiberio et al., 2015). With the abundance of techniques available for miRNA detection, such as northern blot, quantitative polymerase chain reaction (RT-qPCR), microarray, and next-generation sequencing, choosing the appropriate detection technique depends upon the nature of the samples and the study target. Each detection platform has advantages and disadvantages that must be evaluated before developing an application (Table 4).
Table 4.
Common detection platform currently applied for circulating miRNAs
| Detection platform | Advantages/strengths | Pitfalls/weaknesses | References |
|---|---|---|---|
| Amplification-based methods | - Reasonable specificity, sensitivity, reproducibility, low cost, and easily conducted technique |
- Detect only annotated miRNAs - Inner reference gene is necessary for data normalization |
(Mestdagh et al., 2008; Schmittgen et al., 2008) |
| Hybridization-based arrays | - Profiling a large number of miRNAs in a high-throughput technique - Require a relatively small volume of RNA as input (100 ng or less) - Reasonable reproducibility |
- Difficulty in distinguishing the difference between mature and unprocessed miRNA - Cross-hybridization within miRNA family members due to sequence homology - Lower specificity than RT-qPCR and sequencing - Detect only annotated miRNAs - Hard to detect low abundant miRNAs |
(Reid et al., 2011; Schwarzenbach et al., 2014) |
| Sequencing (next-generation sequencing and deep sequencing) | - Higher specificity and more precise than other platforms - Identifying novel miRNAs with unknown sequences - Discriminate between isomiRs |
- Data processing needs bioinformatics analysis - A relatively high-cost technique - Time-consuming |
(Johansen et al., 2011) |
5.9.1. Amplification-based technique
The quantitative reverse transcription polymerase chain reaction (RT-qPCR) platform is the most applied method, referred to as the “gold standard” to quantify specific miRNA due to its sensitivity, simplicity of use, and relatively low cost. RT-qPCR is mostly the validation technique that is applied following miRNA profiling obtained from platforms, such as arrays and sequencing (Mestdagh et al., 2008; Schmittgen et al., 2008).
5.9.2. Hybridization-based microarrays
Array-based platforms are a common approach for miRNA profiling available from several suppliers. Among the most popular hybridization-based arrays is Affymetrix Gene Chip miRNA Array, Agilent Human miRNA Microarray, and miRCURY LNA miRNA Arrays (Li and Ruan, 2009). The microarray-based approaches are especially appealing because of their high-throughput profiling ability, allowing for simultaneous detection of a broad number of diverse miRNAs in several samples processed in parallel in a single test. The arrays are designed based on miRNA sequences deposited in the miRBase database (Reid et al., 2011; Schwarzenbach et al., 2014).
5.9.3. Sequencing
Detection of miRNAs through sequencing is a promising approach that has recently become the preferred method as it avoids several challenges faced by the other detection platforms. The identification of novel miRNAs has been considerably boosted with the use of sequencing as a detection platform (Fox et al., 2009). Over the last years, Illumina's Genome Analyzer (GA), HiSeq, Roche 454 Genome sequencing system, and Applied Biosystems SOLiD technologies have been commercially accessible. However, next-generation sequencing-based methods suffer from drawbacks, including requiring bioinformatician support for data analysis, tedious sequencing library construction processes, and a potential sequence bias resulting from the sequencing library construction process (Johansen et al., 2011).
5.9.4. Nanopore sensing
Several research studies have described a unique approach for electrically detecting miRNAs in tissue or biofluids that do not involve labeling, enzyme reactions, or amplification (Chen et al., 2009; Garcia-Elias et al., 2017; Asano et al., 2019). Wang et al. (2011) demonstrated a nanopore sensor capable of detecting subpicomolar levels of cancer-associated miRNAs and differentiating single nucleotide variants across miRNA family members by employing a programmable oligonucleotide probe.
5.10. Lack of Standard miRNA as an Internal Reference
Technical variability between samples is more prevalent as a result of several factors, such as differences in starting material, RNA extraction method, or reaction efficiency during the labeling or hybridization techniques. To obtain an accurate determination of miRNA levels in a specific tissue, body fluid, or cell type and to ensure that the data are reliable, it is important to normalize the data once it is generated (Witwer, 2015; Wang et al., 2018a).
Currently, there is no housekeeping circulating miRNA that could be accurately used as an internal control. Reference miRNAs used for tissue miRNA analysis, such as RNU6 and RNU48, cannot be continuously recognized in the circulation because of their significant RNAse-mediated degradation (Wang et al., 2012b). miR-16 has been considered a miRNA housekeeping, and it is the most popular in the literature. However, it is also among the most severely affected by hemolysis (Pritchard et al., 2012b) and thus should not be used as an miRNA reference for data normalization. Several miRNAs have been proposed as possible internal references in multiple studies (Chen et al., 2013). For example, Faraldi et al. (2019) tested a panel of 179 miRNAs following RNA extraction from plasma using RT-qPCR and used multiple normalization procedures based on endogenous miRNAs and exogenous oligonucleotides for quantitation. Based on the results, miR-320d was identified as the most suitable reference miRNA for eliminating technical variability across replicates.
A collection of non-human spike-in RNAs is often used for normalization in miRNA measurements, particularly for qPCR-based profiling (Vigneron et al., 2016). However, a global agreement for a reliable internal reference is lacking. Correcting the plasma/serum volume has also been suggested as an option for normalization, as volume is a clinical standard for other biomarkers (Faraldi et al., 2019). Although defining a normalizing miRNA with broad applicability would be more accurate, shifting focus from normalizers to normalizing the techniques is more realistic. The introduction of consistent methodological instructions, rather than a universal set of normalizers, will improve the accurate quantitation and comparison of circulating miRNAs (Marabita et al., 2016).
6. Other Individual Factors (External Factors)
Other important variables that may significantly affect the appropriate interpretation of circulating miRNAs in disease biomarker studies are linked to inter-individual variability and the impact of disease-independent factors. Individual factors, such as race (Wang et al., 2018a), gender, ethnicity, age, physical activity, drug use (de Boer et al., 2013), smoking (Badrnya et al., 2014), and lifestyle may all have an impact on the level of circulating miRNAs (XenomiRs, 2012; Becker and Lockwood, 2013).
- Gender variations have a significant impact on the biologic composition of bodily fluids (Wang et al., 2013). In addition, sex-biased expression of circulating miRNAs has been reported. A clear example was reported by Wang et al. (2013) in which circulating levels of let-7g and miR-221 exhibited a female-specific increase in patients with metabolic syndrome, but this increase was not observed in male patients. Several studies have reported an effect of gonadal steroids on miRNA expression (Morgan and Bale, 2012). Menstrual cycles and pregnancy are among the events that also have an effect on total circulating miRNA profiles (Rekker et al., 2013; Luizon et al., 2021).
- The type of diet consumed prior to sampling may also influence circulating miRNA expression. Because of the remarkable sequence conservation of several miRNAs across species, it is possible that some circulating miRNAs are not produced in cells of the body, but from external sources. Many miRNAs are present in dietary supplements, such as catechins, indoles, curcumin, and resveratrol, that could be indistinguishable from endogenous miRNAs at the sequence and/or functional level in the circulation (Witwer and Hirschi, 2014; Witwer, 2015; Lee et al., 2017; El-Daly et al., 2020b; Salem et al., 2021).
- Circadian cycles represent an external factor that has an impact on the release of extracellular vesicles and miRNAs in the circulation (Heegaard et al., 2016; Koritzinsky et al., 2019). Shende et al. (2011) evaluated a panel of miRNAs targeting the clock gene, Bmal1, in the serum of mice exposed to a standard 12 hour light/12 hour dark cycle. Of these, miR-152 and miR-494 were expressed in diurnal oscillations with expression peaks detected near the middle of the day and 8 h or 12 h later during the night. This study and others connect miRNAs with the control of the circadian timekeeping system (Cheng et al., 2007; Heegaard et al., 2016; Khalyfa et al., 2020) and highlights the necessity of considering biooscillations in circulating miRNA studies to reduce any potential variability.
- Gravitational change is also considered a variant factor that affects the expression of circulating miRNAs. In the study by Jirak et al., (2020), profiling 213 miRNAs by next-generation sequencing following exposure of healthy volunteers to gravitational changes revealed a significant alteration in the expression of four miRNAs, miR-24-3p, miR-941, miR-486-5p, and miR-223-3p, suggesting that the secretion pattern of some miRNAs are affected by changes in gravity.
- Physical activity has also been found to affect circulating miRNA levels as diagnostic markers. Activities ranging from a marathon run as well as acute aerobic or endurance exercises have been reported to induce significant changes in the expression of different miRNA panels (Gomes et al., 2014; Nielsen et al., 2014; Horak et al., 2018). Horak et al. (2018) demonstrated that the expression of plasma miR-93, miR-222, and miR-16 were significantly changed (down- or upregulated) in young athletes that performed high-intensity interval training and hypertrophic or explosive strength training for eight weeks compared with baseline expression. In a recent study by Eyileten et al. (2022), engaging in extreme physical exercises, such as an Eileen ultramarathon, was coupled to alterations in the expression of a panel of circulating miRNAs associated with fibrosis, inflammation, and cardiac muscle function. From the deregulated list of miRNAs, the expression of miR-1-3p was altered by race duration. miR-1-3p was significantly higher in participants who completed the event in under 10 hours compared with runners that required more than 10 hours.
The type of exercise also affects the expression of circulating miRNAs (Uhlemann et al., 2014; Horak et al., 2018). For example, running a marathon resulted in a significant elevation of plasma miR-126 and miR-133 (Uhlemann et al., 2014). In contrast, performing a maximal symptom-limited exercise or bicycling for four hours under an anaerobic threshold raised the level of plasma miR-126 with no effect on plasma levels of miR-133. In contrast, eccentric resistance exercise caused an isolated elevation in plasma miR-133 with no effect on miR-126 levels (Uhlemann et al., 2014).
- Disease conditions and pharmacological treatment can affect the circulating miRNA expression profiles. For example, miR-122 is known to be elevated in most hepatocellular cancer cases, but it is also elevated during hepatitis B or C infection or liver damage. Moreover, miR-122 levels may change in response to hepatitis medications. The administration of some drugs that target platelets can modulate the expression pattern of platelet-derived miRNAs. Aspirin has been reported to dysregulate the expression levels of platelet miRNAs, such as miR-126, miR-191, miR-223, miR-126, and miR-150 (de Boer et al., 2013; Willeit et al., 2013).
Although some of the above-mentioned individual aspects may be addressed and taken into account during the evaluation of circulating miRNAs as biomarkers, some external factors may be difficult to control or adequately take into consideration. Thus, while evaluating circulating miRNA data, their expression levels may be affected by the sum of individual behavior in addition to disease factors.
7. Circulating miRNAs in Clinical Trials
The necessary requirements for the clinical validation of circulating miRNAs include specificity, consistent expression across blood and tissue samples, and reproducibility (Grimaldi and Incoronato, 2019). Because these requirements have not been verified, translation to the clinic has not yet been realized. To present an overview of the current advances in the clinical translation of circulating miRNAs as cancer diagnostic biomarkers, we searched the database, ClinicalTrials.gov, for the terms “tumor,” “cancer,” and “neoplasm” in the disease field and “miRNA,” “circulating,” “blood” in the other terms field. Our search yielded 36 studies, which included 13 interventional studies, 21 observational studies, and 5 studies listed as observational, Patient Registry Studies. Most of these studies focused on assessing the role of circulating miRNAs in predicting response to therapeutic interventions. The majority were done on female participants, with the main focus on breast cancer. The study phase was not applicable for most of these studies. However, one study was in phase I, two studies in phase II, and one study was in phase IV. For 34 clinical trials, no results have been uploaded or introduced, whereas 10 studies were listed as completed. The most recent clinical study started on May 12, 2021, by Fondazione IRCCS Istituto Nazionale dei Tumori, Milano with the aim of collecting genetic data from primary tissues and blood to determine whether certain mutations and/or miRNA overexpression were linked to limited iodine absorption or radioresistance in patients with metastatic differentiated thyroid cancer.
Conclusion and Future Directions
Based on the challenges described above, there is a need to create standard operating protocols for the evaluation of miRNAs in circulation, including the selection of a starting material, sample isolation protocols, detection, and normalization methods. In this review, we attempted to summarize the standard operating procedures that have been suggested by multiple studies as optimal procedures, which may help to improve the detection of circulating miRNAs as tumor biomarkers in the clinic. However, there are currently no optimal strategies for analyzing circulating miRNAs, and research into developing and optimizing novel methods is ongoing. Efforts to improve and standardize existing technologies and the discovery of novel strategies have the potential to shift this approach from the laboratory to clinical practice (bench to bedside).
Finally, the scientific community has the burden of developing standardized procedures through which the reproducibility of miRNA results can be improved. Acknowledging the experimental limitations of any study will alert other researchers to conduct their studies using standard guidelines set by the scientific community. A final important point to be considered is the transparency and reporting of the exact experimental procedures followed by each study.
Abbreviations
- AFP
alpha-fetoprotein
- AFP-L3
alpha-fetoprotein L3
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- AUC
area under the curve
- BA
breast cancer
- CA15.3
cancer antigen 15.3
- CEA
carcinoembryonic antigen
- CRC
colorectal cancer
- DCP
des-gamma-carboxy prothrombin
- GC
gastric cancer
- HCC
hepatocellular carcinoma
- miRNA
microRNA
- OC
ovarian cancer
- RT-qPCR
reverse transcription quantitative polymerase chain reaction
Authorship Contributions
Participated in research design: El-Daly, Gouhar.
Contributed with the analytic tools: El-Daly, Gouhar, Abd Elmageed.
Wrote or contributed to the writing of the manuscript: El-Daly, Gouhar, Abd Elmageed.
Footnotes
The research work of Dr. Sherien M. El-Daly is supported by the National Research Centre and the Science and Technology Development Funds (STDF, Grant #26388). The research work of Dr. Zakaria Y. Abd Elmageed is partially supported by National Institutes of Health National Cancer Institute [Grant R21-CA194750].
All authors declare no conflict of interest or competing interests that could influence the current study.
S.M.E-D. and S.A.G. contributed equally to this work.
References
- Alečković M, Kang Y (2015) Regulation of cancer metastasis by cell-free miRNAs. Biochim Biophys Acta 1855:24–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Ahmed E, Abd El-Basit SA, Mohamed MA, Swellam M (2020) Clinical role of MiRNA 29a and MiRNA 335 on breast cancer management: their relevance to MMP2 protein level. Arch Physiol Biochem 1–8. [DOI] [PubMed] [Google Scholar]
- Androvic P, Romanyuk N, Urdzikova-Machova L, Rohlova E, Kubista M, Valihrach L (2019) Two-tailed RT-qPCR panel for quality control of circulating microRNA studies. Sci Rep 9:4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armand-Labit V, Pradines A (2017) Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts 8:61–81. [DOI] [PubMed] [Google Scholar]
- Asano N, Matsuzaki J, Ichikawa M, Kawauchi J, Takizawa S, Aoki Y, Sakamoto H, Yoshida A, Kobayashi E, Tanzawa Y, et al. (2019) A serum microRNA classifier for the diagnosis of sarcomas of various histological subtypes. Nat Commun 10:1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashirbekov Y, Abaildayev A, Omarbayeva N, Botbayev D, Belkozhayev A, Askandirova A, Neupokoyeva A, Utegenova G, Sharipov K, Aitkhozhina N (2020) Combination of circulating miR-145-5p/miR-191-5p as biomarker for breast cancer detection. PeerJ 8:e10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes C, Meese E, Keller A (2016) Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol Diagn Ther 20:509–518. [DOI] [PubMed] [Google Scholar]
- Badrnya S, Baumgartner R, Assinger A (2014) Smoking alters circulating plasma microvesicle pattern and microRNA signatures. Thromb Haemost 112:128–136. [DOI] [PubMed] [Google Scholar]
- Ban E, Chae D-K, Yoo YS, Song EJ (2017a) An improvement of miRNA extraction efficiency in human plasma. Anal Bioanal Chem 409:6397–6404. [DOI] [PubMed] [Google Scholar]
- Ban E, Chae D-K, Yoo YS, Song EJ (2017b) An improvement of miRNA extraction efficiency in human plasma. Anal Bioanal Chem 409:6397–6404. [DOI] [PubMed] [Google Scholar]
- Becker N, Lockwood CM (2013) Pre-analytical variables in miRNA analysis. Clin Biochem 46:861–868. [DOI] [PubMed] [Google Scholar]
- Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, Dahlsveen IK (2013) Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59:S1–S6. [DOI] [PubMed] [Google Scholar]
- Bryzgunova O, Konoshenko M, Zaporozhchenko I, Yakovlev A, Laktionov P (2021) Isolation of Cell-Free miRNA from Biological Fluids: Influencing Factors and Methods. Diagnostics (Basel) 11:865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6:857–866. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN, Pass HI (2013) microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol 8:1156–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerkovnik P, Perhavec A, Zgajnar J, Novakovic S (2007) Optimization of an RNA isolation procedure from plasma samples. Int J Mol Med 20:293–300. [PubMed] [Google Scholar]
- Cheerla N, Gevaert O (2017) MicroRNA based pan-cancer diagnosis and treatment recommendation. BMC Bioinformatics 18:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liang H, Guan D, Wang C, Hu X, Cui L, Chen S, Zhang C, Zhang J, Zen K, et al. (2013) A combination of Let-7d, Let-7g and Let-7i serves as a stable reference for normalization of serum microRNAs. PLoS One 8:e79652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu Y, Liao X, Liao R, Zhang L, Niu K, Li T, Li D, Chen Z, Duan Y, et al. (2016) Plasma miRNAs in predicting radiosensitivity in non-small cell lung cancer. Tumour Biol 37:11927–11936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Gelfond JAL, McManus LM, Shireman PK (2009) Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics 10:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H-YM, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, et al. (2007) microRNA modulation of circadian-clock period and entrainment. Neuron 54:813–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HH, Mitchell PS, Kroh EM, Dowell AE, Chéry L, Siddiqui J, Nelson PS, Vessella RL, Knudsen BS, Chinnaiyan AM, et al. (2013) Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer-associated hypoxia. PLoS One 8:e69239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HH, Plets M, Li H, Higano CS, Tangen CM, Agarwal N, Vogelzang NJ, Hussain M, Thompson IM Jr, Tewari M, et al. (2018) Circulating microRNAs and treatment response in the Phase II SWOG S0925 study for patients with new metastatic hormone-sensitive prostate cancer. Prostate 78:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W (2011) Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One 6:e17745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim SSC, Shing TKF, Hung ECW, Leung TY, Lau TK, Chiu RWK, Lo YM (2008) Detection and characterization of placental microRNAs in maternal plasma. Clin Chem 54:482–490. [DOI] [PubMed] [Google Scholar]
- Chorley BN, Atabakhsh E, Doran G, Gautier J-C, Ellinger-Ziegelbauer H, Jackson D, Sharapova T, Yuen PST, Church RJ, Couttet P, et al. (2021) Methodological considerations for measuring biofluid-based microRNA biomarkers. Crit Rev Toxicol 51:264–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, Suciu N, Cretoiu SM, Voinea SC (2020) miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 9:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA (2011) MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol 8:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z, Lin D, Song W, Chen M, Li D (2015) Diagnostic value of circulating microRNAs as biomarkers for breast cancer: a meta-analysis study. Tumour Biol 36:829–839. [DOI] [PubMed] [Google Scholar]
- Cuk K, Zucknick M, Madhavan D, Schott S, Golatta M, Heil J, Marmé F, Turchinovich A, Sinn P, Sohn C, et al. (2013) Plasma microRNA panel for minimally invasive detection of breast cancer. PLoS One 8:e76729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer HC, van Solingen C, Prins J, Duijs JMGJ, Huisman MV, Rabelink TJ, van Zonneveld AJ (2013) Aspirin treatment hampers the use of plasma microRNA-126 as a biomarker for the progression of vascular disease. Eur Heart J 34:3451–3457. [DOI] [PubMed] [Google Scholar]
- De Cock L, Heylen J, Wildiers A, Punie K, Smeets A, Weltens C, Neven P, Billen J, Laenen A, Wildiers H (2021) Detection of secondary metastatic breast cancer by measurement of plasma CA 15.3. ESMO Open 6:100203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deben C, Zwaenepoel K, Boeckx C, Wouters A, Pauwels P, Peeters M, Lardon F, Baay M, Deschoolmeester V (2013) Expression analysis on archival material revisited: isolation and quantification of RNA extracted from FFPE samples. Diagn Mol Pathol 22:59–64. [DOI] [PubMed] [Google Scholar]
- Di Cosimo S, Appierto V, Pizzamiglio S, Silvestri M, Baselga J, Piccart M, Huober J, Izquierdo M, de la Pena L, Hilbers FS, et al. (2020) Early Modulation of Circulating MicroRNAs Levels in HER2-Positive Breast Cancer Patients Treated with Trastuzumab-Based Neoadjuvant Therapy. Int J Mol Sci 21:1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann K-P, Radtke A, Geczi L, Matthies C, Anheuser P, Eckardt U, Sommer J, Zengerling F, Trenti E, Pichler R, et al. (2019) Serum Levels of MicroRNA-371a-3p (M371 Test) as a New Biomarker of Testicular Germ Cell Tumors: Results of a Prospective Multicentric Study. J Clin Oncol 37:1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Qiao S, Li D, Li S, Zheng Z, Wang Q, Zhu X (2021) Circulating miRNAs in Serum as Biomarkers for Early Diagnosis of Non-small Cell Lung Cancer. Front Genet 12:673926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour DR, Lott JA, Nolte FS, Gretch DR, Koff RS, Seeff LB (2000) Diagnosis and monitoring of hepatic injury. II. Recommendations for use of laboratory tests in screening, diagnosis, and monitoring. Clin Chem 46:2050–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW (2011) Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One 6:e20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egidi MG, Cochetti G, Serva MR, Guelfi G, Zampini D, Mechelli L, Mearini E (2013) Circulating microRNAs and kallikreins before and after radical prostatectomy: are they really prostate cancer markers? BioMed Res Int 2013:241780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Daly SM, Abba ML, Patil N, Allgayer H (2016) miRs-134 and -370 function as tumor suppressors in colorectal cancer by independently suppressing EGFR and PI3K signalling. Sci Rep 6:24720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Daly SM, Bayraktar R, Anfossi S, Calin GA (2020a) The Interplay between MicroRNAs and the Components of the Tumor Microenvironment in B-Cell Malignancies. Int J Mol Sci 21:3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Daly SM, Gamal-Eldeen AM, Gouhar SA, Abo-Elfadl MT, El-Saeed G (2020b) Modulatory Effect of Indoles on the Expression of miRNAs Regulating G1/S Cell Cycle Phase in Breast Cancer Cells. Appl Biochem Biotechnol 192:1208–1223. [DOI] [PubMed] [Google Scholar]
- El-Daly SM, Morsy SM, Medhat D, El-Bana MA, Latif YA, Omara EA, Awadallah JR, Gamal-Eldeen AM (2019a) The diagnostic efficacy of circulating miRNAs in monitoring the early development of colitis-induced colorectal cancer. J Cell Biochem 120:16668–16680. [DOI] [PubMed] [Google Scholar]
- El-Daly SM, Omara EA, Hussein J, Youness ER, El-Khayat Z (2019b) Differential expression of miRNAs regulating NF-κB and STAT3 crosstalk during colitis-associated tumorigenesis. Mol Cell Probes 47:101442. [DOI] [PubMed] [Google Scholar]
- El-Khoury V, Pierson S, Kaoma T, Bernardin F, Berchem G (2016) Assessing cellular and circulating miRNA recovery: the impact of the RNA isolation method and the quantity of input material. Sci Rep 6:19529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endzeliņš E, Berger A, Melne V, Bajo-Santos C, Soboļevska K, Ābols A, Rodriguez M, Šantare D, Rudņickiha A, Lietuvietis V, et al. (2017) Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 17:730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyileten C, Wicik Z, Fitas A, Marszalek M, Simon JE, De Rosa S, Wiecha S, Palatini J, Postula M, Malek LA (2022) Altered Circulating MicroRNA Profiles After Endurance Training: A Cohort Study of Ultramarathon Runners. Front Physiol 12:792931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Zhang Q, Chen H, He P, Li Y, Si M, Jiao X (2017) Circulating microRNAs as a biomarker to predict therapy efficacy in hepatitis C patients with different genotypes. Microb Pathog 112:320–326. [DOI] [PubMed] [Google Scholar]
- Faraldi M, Gomarasca M, Sansoni V, Perego S, Banfi G, Lombardi G (2019) Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci Rep 9:1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Horlings HM, Ten Hoeve JJ, Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F, van Kouwenhove M, et al. (2011) MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res 71:4443–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehlmann TKahraman MLudwig NBackes CGalata VKeller VGeffers LMercaldo NHornung DWeis Tet al. (2020) Evaluating the Use of Circulating MicroRNA Profiles for Lung Cancer Detection in Symptomatic Patients. JAMA Oncol 6:714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felekkis K, Papaneophytou C (2020) Challenges in Using Circulating Micro-RNAs as Biomarkers for Cardiovascular Diseases. Int J Mol Sci 21:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendler A, Stephan C, Yousef GM, Kristiansen G, Jung K (2016) The translational potential of microRNAs as biofluid markers of urological tumours. Nat Rev Urol 13:734–752. [DOI] [PubMed] [Google Scholar]
- Fox, S, Filichkin, S, Mockler, TC (2009). Applications of ultra-high-throughput sequencing. Methods Mol Biol 553:79–108. [DOI] [PubMed] [Google Scholar]
- Fukagawa S, Miyata K, Yotsumoto F, Kiyoshima C, Nam SO, Anan H, Katsuda T, Miyahara D, Murata M, Yagi H, et al. (2017) MicroRNA-135a-3p as a promising biomarker and nucleic acid therapeutic agent for ovarian cancer. Cancer Sci 108:886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zhang Q, Xu J, Guo L, Li X (2013) Clinical significance of serum miR-21 in breast cancer compared with CA153 and CEA. Chin J Cancer Res 25:743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Elias A, Alloza L, Puigdecanet E, Nonell L, Tajes M, Curado J, Enjuanes C, Díaz O, Bruguera J, Martí-Almor J, et al. (2017) Defining quantification methods and optimizing protocols for microarray hybridization of circulating microRNAs. Sci Rep 7:7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giráldez MD, Lozano JJ, Ramírez G, Hijona E, Bujanda L, Castells A, Gironella M(2013) Circulating microRNAs as biomarkers of colorectal cancer: results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol 11:681–688.e3. [DOI] [PubMed] [Google Scholar]
- Glinge C, Clauss S, Boddum K, Jabbari R, Jabbari J, Risgaard B, Tomsits P, Hildebrand B, Kääb S, Wakili R, et al. (2017) Stability of Circulating Blood-Based MicroRNAs - Pre-Analytic Methodological Considerations. PLoS One 12:e0167969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff BA, Agnew K, Neradilek MB, Gray HJ, Liao JB, Urban RR (2017) Combining a symptom index, CA125 and HE4 (triple screen) to detect ovarian cancer in women with a pelvic mass. Gynecol Oncol 147:291–295. [DOI] [PubMed] [Google Scholar]
- Gomes CPC, Oliveira GP Jr, Madrid B, Almeida JA, Franco OL, Pereira RW (2014) Circulating miR-1, miR-133a, and miR-206 levels are increased after a half-marathon run. Biomarkers 19:585–589. [DOI] [PubMed] [Google Scholar]
- Gonzales JC, Fink LM, Goodman OB Jr, Symanowski JT, Vogelzang NJ, Ward DC (2011) Comparison of circulating MicroRNA 141 to circulating tumor cells, lactate dehydrogenase, and prostate-specific antigen for determining treatment response in patients with metastatic prostate cancer. Clin Genitourin Cancer 9:39–45. [DOI] [PubMed] [Google Scholar]
- Gouhar SA, Abo-Elfadl MT, Gamal-Eldeen AM, El-Daly SM (2022) Involvement of miRNAs in response to oxidative stress induced by the steroidal glycoalkaloid α-solanine in hepatocellular carcinoma cells. Environ Toxicol 37:212–223. [DOI] [PubMed] [Google Scholar]
- Grasedieck S, Schöler N, Bommer M, Niess JH, Tumani H, Rouhi A, Bloehdorn J, Liebisch P, Mertens D, Döhner H, et al. (2012) Impact of serum storage conditions on microRNA stability. Leukemia 26:2414–2416. [DOI] [PubMed] [Google Scholar]
- Grimaldi AM, Incoronato M. (2019). Clinical Translatability of “Identified” Circulating miRNAs for Diagnosing Breast Cancer: Overview and Update. Cancers 11:901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA-NIH Biomarker Working Group. (2016). BEST ( Biomarkers, EndpointS, and other Tools). Food and Drug Administration and National Institutes of Health, Maryland. [PubMed] [Google Scholar]
- Haider S, Liaquat L, Shahzad S, Sadir S, Madiha S, Batool Z, Tabassum S, Saleem S, Naqvi F, Perveen T (2015) A high dose of short term exogenous D-galactose administration in young male rats produces symptoms simulating the natural aging process. Life Sci 124:110–119. [DOI] [PubMed] [Google Scholar]
- Hamam R, Ali AM, Alsaleh KA, Kassem M, Alfayez M, Aldahmash A, Alajez NM (2016) microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci Rep 6:25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler SFM, Keller A, Chandran PA, Ziegler K, Zipp K, Heuer S, Krockenberger M, Engel JB, Hönig A, Scheffler M, et al. (2010) Whole blood-derived miRNA profiles as potential new tools for ovarian cancer screening. Br J Cancer 103:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl C, Lang A, Rühm A, Sroka R, Duffield T, Vogeser M, Paal M(2021) Spectrophotometric evaluation of hemolysis in plasma by quantification of free oxyhemoglobin, methemoglobin, and methemalbumin in presence of bilirubin. J Biophotonics 14:e202000461. [DOI] [PubMed] [Google Scholar]
- Heegaard NHH, Carlsen AL, Lilje B, Ng KL, Rønne ME, Jørgensen HL, Sennels H, Fahrenkrug J (2016) Diurnal Variations of Human Circulating Cell-Free Micro-RNA. PLoS One 11:e0160577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengen PN (1996) Carriers for precipitating nucleic acids. Trends Biochem Sci 21:224–225. [DOI] [PubMed] [Google Scholar]
- Herreros-Villanueva M, Duran-Sanchon S, Martín AC, Pérez-Palacios R, Vila-Navarro E, Marcuello M, Diaz-Centeno M, Cubiella J, Diez MS, Bujanda L, et al. (2019) Plasma MicroRNA Signature Validation for Early Detection of Colorectal Cancer. Clin Transl Gastroenterol 10:e00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak M, Zlamal F, Iliev R, Kucera J, Cacek J, Svobodova L, Hlavonova Z, Kalina T, Slaby O, Bienertova-Vasku J (2018) Exercise-induced circulating microRNA changes in athletes in various training scenarios. PLoS One 13:e0191060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X (2010) Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 127:118–126. [DOI] [PubMed] [Google Scholar]
- Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, Goel A (2017) Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 66:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaoka H, Toiyama Y, Fujikawa H, Hiro J, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, et al. (2016) Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol 27:1879–1886. [DOI] [PubMed] [Google Scholar]
- Itani MM, Nassar FJ, Tfayli AH, Talhouk RS, Chamandi GK, Itani ARS, Makoukji J, Boustany RN, Hou L, Zgheib NK, et al. (2021) A Signature of Four Circulating microRNAs as Potential Biomarkers for Diagnosing Early-Stage Breast Cancer. Int J Mol Sci 22:6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi D, Zhu Z, Chen Y, Toden S, Huo X, Kanda M, Ishimoto T, Gu D, Tan M, Kodera Y, et al. (2021) Assessment of the Diagnostic Efficiency of a Liquid Biopsy Assay for Early Detection of Gastric Cancer. JAMA Netw Open 4:e2121129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JY, Kim YS, Kang KN, Kim KH, Park YJ, Kim CW (2021) Multiple microRNAs as biomarkers for early breast cancer diagnosis. Mol Clin Oncol 14:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarry J, Schadendorf D, Greenwood C, Spatz A, van Kempen LC (2014) The validity of circulating microRNAs in oncology: five years of challenges and contradictions. Mol Oncol 8:819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenike AE, Halushka MK (2021) miR-21: a non-specific biomarker of all maladies. Biomark Res 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirak P, Wernly B, Lichtenauer M, Franz M, Knost T, Abusamrah T, Kelm M, Bimpong-Buta N-Y, Jung C (2020) Next-generation sequencing analysis of circulating micro-RNA expression in response to parabolic flight as a spaceflight analogue. NPJ Microgravity 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen SD, Karlsen BO, Furmanek T, Andreassen M, Jørgensen TE, Bizuayehu TT, Breines R, Emblem A, Kettunen P, Luukko K, et al. (2011) RNA deep sequencing of the Atlantic cod transcriptome. Comp Biochem Physiol Part D Genomics Proteomics 6:18–22. [DOI] [PubMed] [Google Scholar]
- Johnson ML, Navanukraw C, Grazul-Bilska AT, Reynolds LP, Redmer DA (2003) Heparinase treatment of RNA before quantitative real-time RT-PCR. Biotechniques 35:1140–1142, 1144. [DOI] [PubMed] [Google Scholar]
- Kawaguchi T, Komatsu S, Ichikawa D, Tsujiura M, Takeshita H, Hirajima S, Miyamae M, Okajima W, Ohashi T, Imamura T, et al. (2016) Circulating MicroRNAs: A Next-Generation Clinical Biomarker for Digestive System Cancers. Int J Mol Sci 17:1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalyfa A, Gaddameedhi S, Crooks E, Zhang C, Li Y, Qiao Z, Trzepizur W, Kay SA, Andrade J, Satterfield BC, et al. (2020) Circulating Exosomal miRNAs Signal Circadian Misalignment to Peripheral Metabolic Tissues. Int J Mol Sci 21:6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J, Lieberman JA, Lockwood CM (2017) Variability in, variability out: best practice recommendations to standardize pre-analytical variables in the detection of circulating and tissue microRNAs. Clin Chem Lab Med 55:608–621. [DOI] [PubMed] [Google Scholar]
- Kim D-J, Linnstaedt S, Palma J, Park JC, Ntrivalas E, Kwak-Kim JYH, Gilman-Sachs A, Beaman K, Hastings ML, Martin JN, et al. (2012) Plasma components affect accuracy of circulating cancer-related microRNA quantitation. J Mol Diagn 14:71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley SMK, Bhat BV (2017) Role of microRNAs in sepsis. Inflamm Res 66:553–569. [DOI] [PubMed] [Google Scholar]
- Kirschner MB, Edelman JJB, Kao SC-H, Vallely MP, van Zandwijk N, Reid G (2013) The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front Genet 4:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, Reid G (2011) Haemolysis during sample preparation alters microRNA content of plasma. PLoS One 6:e24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodahl AR, Lyng MB, Binder H, Cold S, Gravgaard K, Knoop AS, Ditzel HJ (2014) Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: a case control study. Mol Oncol 8:874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov K, Kurapeev D, Popov M, Sidorova M, Minasian S, Galagudza M, Kostareva A, Fedorov A (2016) Heparinase treatment of heparin-contaminated plasma from coronary artery bypass grafting patients enables reliable quantification of microRNAs. Biomol Detect Quantif 8:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koritzinsky EH, Street JM, Chari RR, Glispie DM, Bellomo TR, Aponte AM, Star RA, Yuen PST (2019) Circadian variation in the release of small extracellular vesicles can be normalized by vesicle number or TSG101. Am J Physiol Renal Physiol 317:F1098–F1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk P, Powrózek T, Olesiński T, Dmitruk A, Dziwota J, Kowalski D, Milanowski J (2017) Evaluation of miR-506 and miR-4316 expression in early and non-invasive diagnosis of colorectal cancer. Int J Colorectal Dis 32:1057–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudelova E, Holubekova V, Grendar M, Kolkova Z, Samec M, Vanova B, Mikolajcik P, Smolar M, Kudela E, Laca L, et al. (2022) Circulating miRNA expression over the course of colorectal cancer treatment. Oncol Lett 23:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, Johnson JM, Sina JF, Fare TL, Sistare FD, et al. (2009) Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem 55:1977–1983. [DOI] [PubMed] [Google Scholar]
- Law M, Rudnicka AR (2006) Statin safety: a systematic review. Am J Cardiol 97 (8A):52C–60C. [DOI] [PubMed] [Google Scholar]
- Lee I, Baxter D, Lee MY, Scherler K, Wang K (2017). The importance of standardization on analyzing circulating RNA. Mol Diagn Ther 21:259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JTY, Cheung KMC, Leung VYL (2014) Correction for concentration overestimation of nucleic acids with phenol. Anal Biochem 465:179–186. [DOI] [PubMed] [Google Scholar]
- Li C, Fang Z, Jiang T, Zhang Q, Liu C, Zhang C, Xiang Y (2013) Serum microRNAs profile from genome-wide serves as a fingerprint for diagnosis of acute myocardial infarction and angina pectoris. BMC Med Genomics 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liu J, Chen J, Wang H, Yang L, Chen F, Fan S, Wang J, Shao B, Yin D, et al. (2018) A serum microRNA signature predicts trastuzumab benefit in HER2-positive metastatic breast cancer patients. Nat Commun 9:1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ruan K (2009) MicroRNA detection by microarray. Anal Bioanal Chem 394:1117–1124. [DOI] [PubMed] [Google Scholar]
- Li X-X, Gao S-Y, Wang P-Y, Zhou X, Li Y-J, Yu Y, Yan Y-F, Zhang H-H, Lv C-J, Zhou H-H, et al. (2015a) Reduced expression levels of let-7c in human breast cancer patients. Oncol Lett 9:1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kong D, Chen H, Liu S, Hu H, Wu T, Wang J, Chen W, Ning Y, Li Y, et al. (2016) miR-155 acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1. Sci Rep 6:21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mauro M, Williams Z (2015b) Comparison of plasma extracellular RNA isolation kits reveals kit-dependent biases. Biotechniques 59:13–17. [DOI] [PubMed] [Google Scholar]
- Lin H-M, Mahon KL, Spielman C, Gurney H, Mallesara G, Stockler MR, Bastick P, Briscoe K, Marx G, Swarbrick A, et al. (2017a) Phase 2 study of circulating microRNA biomarkers in castration-resistant prostate cancer. Br J Cancer 116:1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Gregory RI (2015) MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Leng Q, Jiang Z, Guarnera MA, Zhou Y, Chen X, Wang H, Zhou W, Cai L, Fang H, et al. (2017b) A classifier integrating plasma biomarkers and radiological characteristics for distinguishing malignant from benign pulmonary nodules. Int J Cancer 141:1240–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fan Z, Zhao T, Cao W, Zhang L, Li H, Xie Q, Tian Y, Wang B (2015) Plasma miR-1, miR-208, miR-499 as potential predictive biomarkers for acute myocardial infarction: An independent study of Han population. Exp Gerontol 72:230–238. [DOI] [PubMed] [Google Scholar]
- Liu X, Xu T, Hu X, Chen X, Zeng K, Sun L, Wang S (2018) Elevated circulating miR-182 acts as a diagnostic biomarker for early colorectal cancer. Cancer Manag Res 10:857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et al. (2012) Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838. [DOI] [PubMed] [Google Scholar]
- Luizon MR, Conceição IMCA, Viana-Mattioli S, Caldeira-Dias M, Cavalli RC, Sandrim VC (2021) Circulating MicroRNAs in the Second Trimester From Pregnant Women Who Subsequently Developed Preeclampsia: Potential Candidates as Predictive Biomarkers and Pathway Analysis for Target Genes of miR-204-5p. Front Physiol 12:678184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar-Aguilar F, Mendoza-Ramírez JA, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, Rodríguez-Padilla C, Reséndez-Pérez D (2013) Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers 34:163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabita F, de Candia P, Torri A, Tegnér J, Abrignani S, Rossi RL (2016) Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform 17:204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamala N, Vargas MT, González-Cámpora R, Miñambres R, Arias JI, Menéndez P, Andrés-León E, Gómez-López G, Yanowsky K, Calvete-Candenas J, et al. (2015) Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem 61:1098–1106. [DOI] [PubMed] [Google Scholar]
- Matias-Garcia PR, Wilson R, Mussack V, Reischl E, Waldenberger M, Gieger C, Anton G, Peters A, Kuehn-Steven A (2020) Impact of long-term storage and freeze-thawing on eight circulating microRNAs in plasma samples. PLoS One 15:e0227648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger F, Dulary C, Daviaud C, Deleuze J-F, Tost J (2015) Comprehensive evaluation of methods to isolate, quantify, and characterize circulating cell-free DNA from small volumes of plasma. Anal Bioanal Chem 407:6873–6878. [DOI] [PubMed] [Google Scholar]
- McAlexander MA, Phillips MJ, Witwer KW (2013a) Comparison of Methods for miRNA Extraction from Plasma and Quantitative Recovery of RNA from Cerebrospinal Fluid. Front Genet 4:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A (2011) Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem 57:833–840. [DOI] [PubMed] [Google Scholar]
- McPherson RA, Msc MD, Pincus MR (2021) Henry’s Clinical Diagnosis and Management by Laboratory Methods E-book, Elsevier Health Sciences. [Google Scholar]
- Mestdagh P, Feys T, Bernard N, Guenther S, Chen C, Speleman F, Vandesompele J (2008) High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res 36:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AJ, Gray WD, Hayek SS, Ko Y-A, Thomas S, Rooney K, Awad M, Roback JD, Quyyumi A, Searles CD (2016) Platelets confound the measurement of extracellular miRNA in archived plasma. Sci Rep 6:32651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M (2014) Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med 18:371–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mompeón A, Ortega-Paz L, Vidal-Gómez X, Costa TJ, Pérez-Cremades D, Garcia-Blas S, Brugaletta S, Sanchis J, Sabate M, Novella S, et al. (2020) Disparate miRNA expression in serum and plasma of patients with acute myocardial infarction: a systematic and paired comparative analysis. Sci Rep 10:5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monleau M, Bonnel S, Gostan T, Blanchard D, Courgnaud V, Lecellier C-H (2014) Comparison of different extraction techniques to profile microRNAs from human sera and peripheral blood mononuclear cells. BMC Genomics 15:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moret I, Sánchez-Izquierdo D, Iborra M, Tortosa L, Navarro-Puche A, Nos P, Cervera J, Beltrán B (2013) Assessing an improved protocol for plasma microRNA extraction. PLoS One 8:e82753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, D’Antona P, Finardi E, Barbetta M, Dominioni L, Poli A, Gini E, Noonan DM, Imperatori A, Rotolo N, et al. (2017) Systematic review and critique of circulating miRNAs as biomarkers of stage I-II non-small cell lung cancer. Oncotarget 8:94980–94996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan CP, Bale TL (2012) Sex differences in microRNA regulation of gene expression: no smoke, just miRs. Biol Sex Differ 3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar FJ, Msheik ZS, Itani MM, Helou RE, Hadla R, Kreidieh F, Bejjany R, Mukherji D, Shamseddine A, Nasr RR, et al. (2021) Circulating miRNA as Biomarkers for Colorectal Cancer Diagnosis and Liver Metastasis. Diagnostics (Basel) 11:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EKO, Li R, Shin VY, Jin HC, Leung CPH, Ma ESK, Pang R, Chua D, Chu K-M, Law WL, et al. (2013) Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One 8:e53141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S, Åkerström T, Rinnov A, Yfanti C, Scheele C, Pedersen BK, Laye MJ (2014) The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS One 9:e87308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Zhang L, Qiu H, Wu Y, Wang Z, Zai Y, Liu L, Qu J, Kang K, Gou D (2015) An improved method for detecting circulating microRNAs with S-Poly(T) Plus real-time PCR. Sci Rep 5:15100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J, Hayder H, Zayed Y, Peng C (2018) Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DNP, Carlsen AL, Heegaard NHH, Prahm KP, Christensen IJ, Høgdall CK, Høgdall EV (2019) Diagnostic plasma miRNA-profiles for ovarian cancer in patients with pelvic mass. PLoS One 14:e0225249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou F-S, Michiels S, Shyr Y, Adjei AA, Oberg AL (2021) Biomarker Discovery and Validation: Statistical Considerations. J Thorac Oncol 16:537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page K, Guttery DS, Zahra N, Primrose L, Elshaw SR, Pringle JH, Blighe K, Marchese SD, Hills A, Woodley L, et al. (2013a) Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One 8:e77963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page K, Guttery DS, Zahra N, Primrose L, Elshaw SR, Pringle JH, Blighe K, Marchese SD, Hills A, Woodley L, et al. (2013b) Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One 8:e77963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M (2012a) Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 5:492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M (2012b) Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 5:492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M (2011) Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol 45:355–360. [DOI] [PubMed] [Google Scholar]
- Qasemi Rad M, Pouresmaeil V, Hosseini Mojahed F, Amirabadi A, Aalami AH (2022) Clinicopathological utility of miR-203a-3p in diagnosing colorectal cancer. Mol Biol Rep 49:6975–6985. [DOI] [PubMed] [Google Scholar]
- Ramón-Núñez LA, Martos L, Fernández-Pardo Á, Oto J, Medina P, España F, Navarro S (2017) Comparison of protocols and RNA carriers for plasma miRNA isolation. Unraveling RNA carrier influence on miRNA isolation. PLoS One 12:e0187005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapado-González Ó, Álvarez-Castro A, López-López R, Iglesias-Canle J, Suárez-Cunqueiro MM, Muinelo-Romay L (2019) Circulating microRNAs as Promising Biomarkers in Colorectal Cancer. Cancers (Basel) 11:898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, Horos R, Von Lindern M, Enright AJ, O’Carroll D (2010) The miR-144/451 locus is required for erythroid homeostasis. J Exp Med 207:1351–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Kirschner MB, van Zandwijk N (2011) Circulating microRNAs: Association with disease and potential use as biomarkers. Crit Rev Oncol Hematol 80:193–208. [DOI] [PubMed] [Google Scholar]
- Rekker K, Saare M, Roost AM, Salumets A, Peters M (2013) Circulating microRNA Profile throughout the menstrual cycle. PLoS One 8:e81166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE (2009) The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol 112:55–59. [DOI] [PubMed] [Google Scholar]
- Robelin P, Tod M, Colomban O, Lachuer J, Ray-Coquard I, Rauglaudre G, Joly F, Chevalier-Place A, Combe P, Lortholary A, et al. (2020) Comparative analysis of predictive values of the kinetics of 11 circulating miRNAs and of CA125 in ovarian cancer during first line treatment (a GINECO study). Gynecol Oncol 159:256–263. [DOI] [PubMed] [Google Scholar]
- Salem AZ, Medhat D, Fathy SA, Mohamed MR, El-Khayat Z, El-Daly SM (2021) Indole glucosinolates exhibit anti-inflammatory effects on Ehrlich ascites carcinoma cells through modulation of inflammatory markers and miRNAs. Mol Biol Rep 48:6845–6855. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C (2008) Real-time PCR quantification of precursor and mature microRNA. Methods 44:31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, Calatayud D, Nielsen SE, Yilmaz M, Holländer NH, et al. (2014) MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA 311:392–404. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H, Milde-Langosch K, Steinbach B, Müller V, Pantel K (2012) Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res Treat 134:933–941. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H, Nishida N, Calin GA, Pantel K (2014) Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 11:145–156. [DOI] [PubMed] [Google Scholar]
- Shapira I, Oswald M, Lovecchio J, Khalili H, Menzin A, Whyte J, Dos Santos L, Liang S, Bhuiya T, Keogh M, et al. (2014) Circulating biomarkers for detection of ovarian cancer and predicting cancer outcomes. Br J Cancer 110:976–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shende VR, Goldrick MM, Ramani S, Earnest DJ (2011) Expression and rhythmic modulation of circulating microRNAs targeting the clock gene Bmal1 in mice. PLoS One 6:e22586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Bai Z, Zhang J, Meng H, Cai J, Deng W, Bi J, Ma X, Zhang Z (2013) Serum microRNA-21 levels are related to tumor size in gastric cancer patients but cannot predict prognosis. Oncol Lett 6:1733–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourvinou IS, Markou A, Lianidou ES (2013) Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability. J Mol Diagn 15:827–834. [DOI] [PubMed] [Google Scholar]
- Souza MF, Kuasne H, Barros-Filho MC, Cilião HL, Marchi FA, Fuganti PE, Paschoal AR, Rogatto SR, Cólus IMS (2017) Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS One 12:e0184094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford MYC, Willoughby CE, Walsh CP, McKenna DJ (2022) Prognostic value of miR-21 for prostate cancer: a systematic review and meta-analysis. Biosci Rep 42:BSR20211972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YY, Sun L, Guo ZR, Li JC, Bai TT, Cai XX, Li WH, Zhu YF (2019) Upregulated expression of serum exosomal miR-375 and miR-1307 enhance the diagnostic power of CA125 for ovarian cancer. J Ovarian Res 12:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liu Y, Cogdell D, Calin GA, Sun B, Kopetz S, Hamilton SR, Zhang W (2016) Examining plasma microRNA markers for colorectal cancer at different stages. Oncotarget 7:11434–11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Yang B, Lin M, Yu H, Chen H, Zhang Z (2019) Identification of serum miR-30a-5p as a diagnostic and prognostic biomarker in colorectal cancer. Cancer Biomark 24:299–305. [DOI] [PubMed] [Google Scholar]
- Sundar S, Neal RD, Kehoe S (2015) Diagnosis of ovarian cancer. BMJ 351:h4443. [DOI] [PubMed] [Google Scholar]
- Swellam M, Zahran RFK, Ghonem SA, Abdel-Malak C (2021) Serum MiRNA-27a as potential diagnostic nucleic marker for breast cancer. Arch Physiol Biochem 127:90–96. [DOI] [PubMed] [Google Scholar]
- Tan GW, Khoo ASB, Tan LP (2015) Evaluation of extraction kits and RT-qPCR systems adapted to high-throughput platform for circulating miRNAs. Sci Rep 5:9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieu W, Tilki D, de Vere White R, Evans CP (2014) The role of microRNA in castration-resistant prostate cancer. Urol Oncol 32:517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiberio P, Callari M, Angeloni V, Daidone MG, Appierto V (2015) Challenges in using circulating miRNAs as cancer biomarkers. BioMed Res Int 2015:731479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A(2014) Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg 259:735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I, Umeshita K, et al. (2012) Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol 56:167–175. [DOI] [PubMed] [Google Scholar]
- Uhlemann M, Möbius-Winkler S, Fikenzer S, Adam J, Redlich M, Möhlenkamp S, Hilberg T, Schuler GC, Adams V (2014) Circulating microRNA-126 increases after different forms of endurance exercise in healthy adults. Eur J Prev Cardiol 21:484–491. [DOI] [PubMed] [Google Scholar]
- Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, Iglesias-Díaz P, Lorenzo-Patiño MJ, Haz M, Santamarina I, Blanco M, Fernández-Tajes J, Quindós M, et al. (2012) Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med 10:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron N, Meryet-Figuière M, Guttin A, Issartel J-P, Lambert B, Briand M, Louis M-H, Vernon M, Lebailly P, Lecluse Y, et al. (2016) Towards a new standardized method for circulating miRNAs profiling in clinical studies: Interest of the exogenous normalization to improve miRNA signature accuracy. Mol Oncol 10:981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Rassenti LZ, Veronese A, Taccioli C, Costinean S, Aguda BD, Volinia S, Ferracin M, Palatini J, Balatti V, et al. (2009) Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood 114:3872–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukobrat-Bijedic Z, Husic-Selimovic A, Sofic A, Bijedic N, Bjelogrlic I, Gogov B, Mehmedovic A (2013) Cancer Antigens (CEA and CA 19-9) as Markers of Advanced Stage of Colorectal Carcinoma. Med Arch 67:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhang Q (2012) The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol 138:1659–1666. [DOI] [PubMed] [Google Scholar]
- Wang F, Hou J, Jin W, Li J, Yue Y, Jin H, Wang X (2014b) Increased circulating microRNA-155 as a potential biomarker for breast cancer screening: a meta-analysis. Molecules 19:6282–6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Peng R, Wang J, Qin Z, Xue L (2018a) Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenetics 10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gao J, Yao H, Wu Z, Wang M, Qi J (2014a) Diagnostic accuracy of serum HE4, CA125 and ROMA in patients with ovarian cancer: a meta-analysis. Tumour Biol 35:6127–6138. [DOI] [PubMed] [Google Scholar]
- Wang K, Yuan Y, Cho J-H, McClarty S, Baxter D, Galas DJ (2012b) Comparing the MicroRNA spectrum between serum and plasma. PLoS One 7:e41561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QT, Xiao W, Mindrinos M, Davis RW (2002) Yeast tRNA as carrier in the isolation of microscale RNA for global amplification and expression profiling. Biotechniques 33:788, 790, 792 passim. [DOI] [PubMed] [Google Scholar]
- Wang S, Su X, Bai H, Zhao J, Duan J, An T, Zhuo M, Wang Z, Wu M, Li Z, et al. (2015a) Identification of plasma microRNA profiles for primary resistance to EGFR-TKIs in advanced non-small cell lung cancer (NSCLC) patients with EGFR activating mutation. J Hematol Oncol 8:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X, Yu L, Wang L, Wang J, Wu Y, et al. (2015b) A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer 136:152–161. [DOI] [PubMed] [Google Scholar]
- Wang, W, Wu, L, Li, C, Zhou, X, Liu, P, Jia, X, Chen, Y, Zhu, W (2019). Five serum microRNAs for detection and predicting of ovarian cancer. Eur J Obstet Gynecol Reprod Biol X 3:100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-T, Tsai P-C, Liao Y-C, Hsu C-Y, Juo S-HH (2013) Circulating microRNAs have a sex-specific association with metabolic syndrome. J Biomed Sci 20:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yin W, Lin Y, Yin K, Zhou L, Du Y, Yan T, Lu J (2018b) Downregulated circulating microRNAs after surgery: potential noninvasive biomarkers for diagnosis and prognosis of early breast cancer. Cell Death Discov 4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zheng D, Tan Q, Wang MX, Gu L-Q (2011) Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat Nanotechnol 6:668–674 DOI: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willeit P, Zampetaki A, Dudek K, Kaudewitz D, King A, Kirkby NS, Crosby-Nwaobi R, Prokopi M, Drozdov I, Langley SR, et al. (2013) Circulating microRNAs as novel biomarkers for platelet activation. Circ Res 112:595–600. [DOI] [PubMed] [Google Scholar]
- Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, Tuschl T (2013) Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci USA 110:4255–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkle M, El-Daly SM, Fabbri M, Calin GA (2021) Noncoding RNA therapeutics challenges and potential solutions. Nat Rev Drug Discov 20:629–651 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer KW (2015) Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem 61:56–63 10.1373/clinchem.2014.221341. [DOI] [PubMed] [Google Scholar]
- Witwer KW, Hirschi KD (2014) Transfer and functional consequences of dietary microRNAs in vertebrates: concepts in search of corroboration: negative results challenge the hypothesis that dietary xenomiRs cross the gut and regulate genes in ingesting vertebrates, but important questions persist. BioEssays 36:394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Somlo G, Yu Y, Palomares MR, Li AX, Zhou W, Chow A, Yen Y, Rossi JJ, Gao H, et al. (2012) De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med 10:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer K (2012) XenomiRs and miRNA homeostasis in health and disease: Evidence that diet and dietary miRNAs directly and indirectly influence circulating miRNA profiles. RNA Biol 9:1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, et al. (2015) Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia. Clin Cancer Res 21:4234–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Baker K, Redman MW, Wang L, Adams SV, Yu M, Dickinson B, Makar K, Ulrich N, Böhm J, et al. (2017) Dynamic plasma microRNAs are biomarkers for prognosis and early detection of recurrence in colorectal cancer. Br J Cancer 117:1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zen K, Zhang C-Y (2012) Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev 32:326–348. [DOI] [PubMed] [Google Scholar]
- Zhang H-L, Yang L-F, Zhu Y, Yao X-D, Zhang S-L, Dai B, Zhu Y-P, Shen Y-J, Shi G-H, Ye D-W (2011) Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 71:326–331. [DOI] [PubMed] [Google Scholar]
- Zheng M, Lou A, Zhang H, Zhu S, Yang M, Lai W (2021) Serum KL-6, CA19-9, CA125 and CEA are Diagnostic Biomarkers for Rheumatoid Arthritis-Associated Interstitial Lung Disease in the Chinese Population. Rheumatol Ther 8:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zuo M-Z, He Z, Li H-R, Li W (2018) Identification of circulating microRNAs as diagnostic biomarkers for ovarian cancer:A pooled analysis of individual studies. Int J Biol Markers 33:1724600818766500. [DOI] [PubMed] [Google Scholar]
- Zhu W, Zhou K, Zha Y, Chen D, He J, Ma H, Liu X, Le H, Zhang Y (2016) Diagnostic value of serum miR-182, miR-183, miR-210, and miR-126 levels in patients with early-stage non-small cell lung cancer. PLoS One 11:e0153046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou R, Loke SY, Tang YC, Too H-P, Zhou L, Lee ASG, Hartman M (2022) Development and validation of a circulating microRNA panel for the early detection of breast cancer. Br J Cancer 126:472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]