Abstract
RNA interference (RNAi) provides researchers with a versatile means to modulate target gene expression. The major forms of RNAi molecules, genome-derived microRNAs (miRNAs) and exogenous small interfering RNAs (siRNAs), converge into RNA-induced silencing complexes to achieve posttranscriptional gene regulation. RNAi has proven to be an adaptable and powerful therapeutic strategy where advancements in chemistry and pharmaceutics continue to bring RNAi-based drugs into the clinic. With four siRNA medications already approved by the US Food and Drug Administration (FDA), several RNAi-based therapeutics continue to advance to clinical trials with functions that closely resemble their endogenous counterparts. Although intended to enhance stability and improve efficacy, chemical modifications may increase risk of off-target effects by altering RNA structure, folding, and biologic activity away from their natural equivalents. Novel technologies in development today seek to use intact cells to yield true biologic RNAi agents that better represent the structures, stabilities, activities, and safety profiles of natural RNA molecules. In this review, we provide an examination of the mechanisms of action of endogenous miRNAs and exogenous siRNAs, the physiologic and pharmacokinetic barriers to therapeutic RNA delivery, and a summary of the chemical modifications and delivery platforms in use. We overview the pharmacology of the four FDA-approved siRNA medications (patisiran, givosiran, lumasiran, and inclisiran) as well as five siRNAs and several miRNA-based therapeutics currently in clinical trials. Furthermore, we discuss the direct expression and stable carrier-based, in vivo production of novel biologic RNAi agents for research and development.
SIGNIFICANCE STATEMENT
In our review, we summarize the major concepts of RNA interference (RNAi), molecular mechanisms, and current state and challenges of RNAi drug development. We focus our discussion on the pharmacology of US Food and Drug Administration-approved RNAi medications and those siRNAs and miRNA-based therapeutics that entered the clinical investigations. Novel approaches to producing new true biological RNAi molecules for research and development are highlighted.
Introduction
Genome-derived functional microRNA (miRNA) was initially elucidated in Caenorhabditis elegans during the characterization of the lin-4 gene encoding a small RNA (sRNA) with antisense complementarity and the capacity for posttranslational regulation of target gene lin-14 (Lee et al., 1993; Wightman et al., 1993). Later on, miRNAs were identified as a superfamily of conserved and functional noncoding RNAs (ncRNAs) that are present in a wide range of animal species (Pasquinelli et al., 2000; Li et al., 2010), including humans (Friedländer et al., 2014). Since then, the development of RNA interference (RNAi) technology (Napoli et al., 1990; Fire et al., 1998) has offered new routes for the studies of reverse genetics, specific gene expression and regulation, and targeted therapy with the potential to study, manipulate, and achieve control of disease. In particular, RNAi is accomplished through the interactions of sRNA molecules, namely small interfering RNAs (siRNAs) and miRNA, with functions that lie outside the confines of the central dogma of molecular biology (Sen and Blau, 2006; Setten et al., 2019). Aside from miRNAs and siRNAs, a third major class of RNAi molecules exist, termed P-element–induced wimpy testis (PIWI)-interaction RNA (piRNA). Although piRNAs typically function in complex with piwi proteins to posttranscriptionally regulate several pathways important in transposon silencing, genome rearrangement, and germ stem-cell maintenance, the most well studied RNAi molecules are siRNAs and miRNAs that regulate gene expression at the posttranscriptional level by specific or semispecific targeting of messenger RNA (mRNA) (Lai et al., 2013; Catalanotto et al., 2016; Han et al., 2017). Altogether, the discovery and application of RNAi technology provides a unique and adaptable tool for basic genetic and biomedical research and opens doors for its exploitation in the development of novel biotechnologies and therapies.
RNAi therapy uses the natural cellular mechanisms of RNAi to bring gene regulation into clinical practice (Hayes et al., 2014; Mollaei et al., 2019; Yu et al., 2020b; Smith et al., 2022). With four novel RNAi-based therapeutics approval by the US Food and Drug Administration (FDA)—namely patisiran (Onpattro, 2018), givosiran (Givlaari, 2019), lumasiran (Oxlumo, 2020), and inclisiran (Leqvio, 2021)—each siRNA drug selectively acts on a target mRNA transcript to combat a disease. In addition, several siRNA agents (e.g., fitusiran, nedosiran, teprasiran, tivanisiran, and vutrisiran) have entered phase III clinical trials with many other RNAi-based therapeutics progressing through early-stage clinical trials or preclinical development (Zhang et al., 2021a). However, delivery of RNAi therapeutics has long been the root obstacle in the way of clinical success. Once an RNAi drug is administered to the body, there are several physical and pharmacokinetic barriers that limit its actions and desired efficacy (Fan and de Lannoy, 2014; Johannes and Lucchino, 2018; Seth et al., 2019; Smith et al., 2022). Chemical modifications and the development of novel delivery methods compatible with RNAi machinery help researchers overcome these barriers (Maguregui and Abe, 2020; Smith et al., 2022). Although these modifications are intended to enhance stability and improve efficacy, growing evidence suggests that chemical modifications of in vivo synthesized RNAi molecules can increase the risk of off-target effects by altering their structure, folding, biologic activity, and safety away from natural RNAi agents (Morena et al., 2018; Yu et al., 2019). To address this challenge, novel technologies in development today seek to use intact cells to yield true biologic RNA molecules that better recapitulate the structures, stabilities, activities, and safety profiles of endogenous RNAi molecules.
In this review, we provide an examination of the mechanisms of action of endogenous miRNAs and exogenous siRNAs, the pharmacokinetic barriers to therapeutic RNAs, and a summary of the viable chemical modifications and delivery platforms in use. We overview the pharmacology of the four siRNA medications approved by the FDA as well as five therapeutic siRNAs and several RNAi-based therapeutics currently in clinical trials. Furthermore, we discuss novel approaches to in vivo production of RNA agents, including direct expression and utilization of stable carriers, which represent a novel class of real biologic RNAi molecules for research and development.
RNAi Molecules and Mechanisms
Genome-derived miRNAs and exogenous siRNAs are classes of ncRNA molecules with functions in posttranslational gene regulation that make up two of the most well studied RNAi molecules (Carthew and Sontheimer, 2009). Functional genome-derived miRNAs range from 18 to 25 nucleotides in length and are capable of targeting the mRNA transcripts of multiple genes to regulate common or several cellular pathways, whereas exogenous siRNA are generally introduced into the cells to control the expression of a single target gene (Huntzinger and Izaurralde, 2011; Vasudevan, 2012; Catalanotto et al., 2016; Setten et al., 2019; Smith et al., 2022). As such, these ncRNA molecules can play critical roles in modulating essentially all cellular functions, including cell signaling, proliferation, metabolism, immunity, and senescence, among others (Carthew and Sontheimer, 2009; Catalanotto et al., 2016; Piletič and Kunej, 2016).
Genome-Derived miRNAs
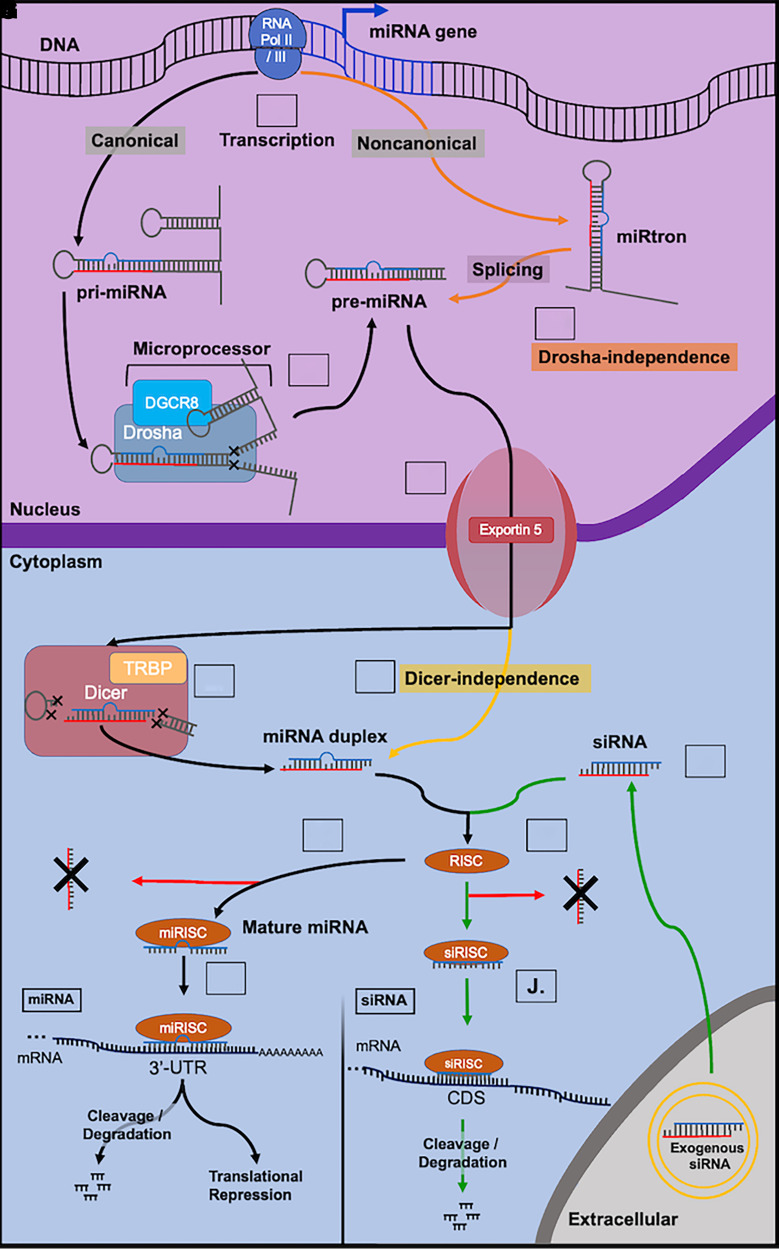
Canonical biogenesis of genome-derived miRNA is categorized into two major phases, nuclear and cytosolic (Fig. 1). Furthermore, depending on the location of the miRNA gene within the genome, miRNAs can be further classified into intergenic- and intragenic-derived miRNAs. Intergenic-derived miRNA come from regions of the genome located in noncoding regions of the DNA between genes, have unique promoter regions, and are transcribed either by RNA polymerase II or III (Ha and Kim, 2014; Valinezhad Orang et al., 2014; Liu et al., 2018). By contrast, the genes of intragenic-derived miRNA are located within exons or introns of protein-coding genes, are coexpressed with their host gene, and are transcribed by RNA polymerase II (Ha and Kim, 2014; Valinezhad Orang et al., 2014). Initiating in the nuclear phase, DNA sequences encoding miRNA genes are transcribed by RNA polymerases into long hairpin transcripts called primary-miRNAs (pri-miRNAs) (Ha and Kim, 2014). A pri-miRNA is typically over 1000 nucleotides in length and consists of three major domains important to miRNA processing: a 33- to 35-nucleotide-long stem, terminal loop, and two single-stranded RNA (ssRNA) segments flaking both ends (Ha and Kim, 2014). Pri-miRNA is further processed in the nucleus by the microprocessor that contains a nuclear RNase III enzyme (Drosha) and its cofactor DiGeorge Syndrome Critical Region 8 (DGCR8) (Denli et al., 2004) to release a smaller hairpin RNA (approximately 65 nucleotides in length) termed precursor microRNA (pre-miRNA). Between the major nuclear and cytosolic phases, pre-miRNA forms a complex with Exportin 5 transporter and GTP-binding nuclear protein (RAN) to exit the nucleus and release the pre-miRNA into the cytosol upon GTP hydrolysis (Okada et al., 2009).
Fig. 1.
Overview of miRNA biogenesis and functions and siRNA mechanisms of action. (A) Intragenic or intergenic miRNA genes are transcribed by RNA polymerases II or III into primary miRNA (pri-miRNA; >1,000 nucleotides) transcripts in canonical pathway (black lines). (B) Pri-miRNAs are subjected to nuclear processing by the microprocessor Drosha-DGCR8 complex to release shorter precursor miRNAs (pre-miRNAs) (e.g., ∼65 nucleotides). (C) Noncanonical miRNA transcripts (e.g., mirtrons) are derived from the genome and subjected to RNA splicing to form pre-miRNAs independent on the microprocessor (orange lines). (D) Pre-miRNAs are exported into the cytoplasm via Exportin 5 transport complex. (E) In the cytoplasm, pre-miRNAs are processed by Dicer/TRBP complex to form miRNA duplexes (18–25 nucleotides). (F) Dicer-independent production of miRNA duplexes (yellow). (G) The guide strand (blue) of the miRNA duplex is selected and loaded into the RNA-induced silencing complex (RISC) to form the miRNA-RISC complex (miRISC) while the passenger strand (red) is degraded. (H) Functional miRNA binds to the 3′-untranslated region (3′UTR) of targeted mRNA to perform posttranscriptional gene regulation, either to accelerate mRNA cleavage or degradation or to repress translation. (I) Exogenous siRNAs can be introduced into cytoplasm through endocytosis or receptor-mediated uptake (green). (J) The antisense strand (blue) is selectively loaded into the RISC to form the siRNA-RISC complex (siRISC) while the sense strand (red) is degraded. (K) Functional siRNA typically acts on the protein coding sequence (CDS) of target transcript to cleave or initiate transcript degradation.
In the cytosolic phase, pre-miRNAs are identified and further processed by RNase III endonuclease (Dicer) and the transactivating response RNA-binding protein (TRBP) (Denli et al., 2004; Okada et al., 2009; Ha and Kim, 2014) at the terminal loops of the hairpins to miRNA duplexes with two 3′ overhangs (Zhang et al., 2004). After a duplex is unwound, the guide (antisense) strand or mature miRNA is loaded into the RNA-induced silencing complex (RISC) to form an miRNA-RISC complex (miRISC) while the passenger (sense) strand is degraded (Yoda et al., 2010). Determining the guide strand from the miRNA duplex is believed to be a result of argonaut 2 endonuclease (AGO2) preference for the 3-prime terminus (3′) or 5-prime terminus (5′) and is typically variable and dependent heavily on both cell type and function (Meijer et al., 2014). AGO2 tends to preferentially select the guide strand (3′ or 5′) of the miRNA duplex with a lower stability or instead the presence of an uracil at its 5′ terminus (Khvorova et al., 2003). After the formation of functional miRISC, the mechanism of regulation is then dependent on the interactions between the miRNA and mRNA sequences (Ha and Kim, 2014; Meijer et al., 2014).
In addition to the canonical pathways, previous studies have identified noncanonical biogenesis of miRNAs. The well studied noncanonical pathways are categorized by the canonical processing steps that they circumvent. The first major noncanonical biogenesis pathway pertains to the nuclear processing phase, independent of Drosha/DGCR8 (Fig. 1). In particular, short hairpin RNA (shRNA)-bearing mirtrons derived from the genome are processed into pre-miRNAs via posttranscriptional splicing to closely resemble Dicer/TRBP recognizable substrates for intracellular translocation and cytoplasmic processing (Westholm and Lai, 2011). After the mirtron-derived pre-miRNAs are exported into the cytosol, the second major noncanonical biogenesis pathway pertains to the cytosolic processing phase independent of the microprocessor (Berezikov et al., 2007; Ruby et al., 2007; Westholm and Lai, 2011) (Fig. 1). However, Dicer/TRBP-independent pre-miRNAs require Ago2 to cleave the 3′ strand and forming a pseudo-miRISC followed by 5′ strand trimming to mature length (Cheloufi et al., 2010; Yang et al., 2010).
Besides canonical and noncanonical biogenesis of miRNAs described above, some miRNAs are directly derived from alternative precursor RNA molecules such as small nucleolar RNAs (snoRNAs) and transfer RNAs (tRNAs) (Ender et al., 2008; Pan et al., 2013; Hasler et al., 2016; Patterson et al., 2017; Li et al., 2018). SnoRNAs have a generalized and well established function of controlling gene expression by modifying ribosomal RNA (rRNA) (Kufel and Grzechnik, 2019). Like canonical miRNAs, some snoRNAs are found to repress mRNAs by incorporation into the RISC complex (Ender et al., 2008; Patterson et al., 2017). miRNAs derived from tRNA precursors constitute a unique source for miRNA maturation where suitable Dicer enzyme substrates are found within the colloquial “clover-leaf” structure of tRNAs (Hasler et al., 2016; Li et al., 2018). Precursor tRNA substrates are cleaved by Dicer into tRNA-derived RNA fragments (tRFs) with the capacity for RISC incorporation and gene regulation (Kumar et al., 2014; Hasler et al., 2016). Further, researchers have also revealed tRFs to guide Ago proteins toward target gene regulation independent of Dicer (Kuscu et al., 2018).
Exogenous siRNAs
Since their discovery following miRNA in 1998, siRNAs have become well established, gene-specific regulatory molecules that function within the RNAi pathway (Fire et al., 1998; Hamilton and Baulcombe, 1999). With several endogenous sources of siRNA identified, a major source for RNAi studies uses exogenous siRNAs that are chemically synthesized and highly selectively to the mRNA transcripts of proteins previously considered to be “undruggable” by small molecule inhibitors (Setten et al., 2019; Smith et al., 2022). Exogenous siRNAs are synthesized as shRNA ranging from 20 to 25 base pairs with 3′ overhangs that can bypass Dicer cleavage for direct incorporation into the RISC (siRISC) to control target gene expression when introduced to the cells (Gaglione and Messere, 2010; Setten et al., 2019) (Fig. 1).
There are two well established approaches to produce siRNAs: chemical (e.g., solid-phase synthesis) and biochemical (e.g., in vitro transcription) syntheses. Solid-phase organic synthesis is a widely used method that can produce large amounts of RNAs and accommodate a wide range of chemical modifications (Amarzguioui et al., 2005). The principle is boiled down to the addition of individual ribonucleosides performed on a solid support or resin, consisting of nucleoside deprotection, coupling, oxidation, and capping. By repeating the cycle for desired number and sequence of nucleosides, the process is ended with oligonucleotide cleavage from the solid support and nucleoside deprotection (Beaucage, 2008; Francis and Resendiz, 2017). This general design uses 2'-hydroxyl protecting groups that provide ribonucleoside phosphoramidites with characteristics key to their synthetic oligomerization (Beaucage, 2008). As such, various protecting group strategies have been developed to allow for site-specific incorporation of chemically modified groups at specific positions within the siRNA (Wilson and Keefe, 2006). In fact, the ability to synthesize siRNA with modify chemistries while retaining their regulatory function leads to the development of the enhanced stability chemistry (ESC) platform used in three out of the four FDA-approved siRNA therapeutics (Foster et al., 2018; Givlaari, 2019; Oxlumo, 2020; Leqvio, 2021).
Another way to produce siRNAs follows the principles of enzymatic reactions that commonly start from in vitro transcription (IVT) (Beckert and Masquida, 2011; Yu et al., 2020a). Divergent from chemical synthesis, IVT requires a DNA template corresponding to target RNAs as well as proper RNA polymerases such as T7 phage RNA polymerase (Donzé and Picard, 2002; Yu et al., 2020a). Specifically, siRNA can be produced in two general steps. Firstly, IVTs are constructed to offer two complementary ssRNAs separately, which can be annealed to offer target double-stranded RNA (dsRNAs) (Wianny and Zernicka-Goetz, 2000; Yang et al., 2002; Yu et al., 2020a). Secondly, recombinant RNases such as Dicer are employed to further process the dsRNAs into desired siRNA agents (Myers et al., 2003; Guiley et al., 2012). Of note, chemically synthesized RNA molecules can also be processed enzymatically to produce target siRNAs in vitro (Yang et al., 2002; Yu et al., 2020a).
Chemical modifications to siRNAs and miRNA mimics can be quite valuable to investigatory and clinical research, such as changes in phosphodiester backbone, ribose, nucleobases, or addition of non-nucleotide molecules (Setten et al., 2019; Smith et al., 2022). The theoretical goals of these optimizations are 3-fold. First, structural optimization may improve RNAi potency and target selectivity besides improved metabolic stability (Bramsen et al., 2009; Janas et al., 2018). Second, optimization may decrease therapeutic immunogenicity and therefore improve overall safety (Robbins et al., 2007; Maguregui and Abe, 2020). Finally, optimization may increase tissue- or organ-targeting specificity by conjugating receptor specific ligands or by changing the physical conformation or chemical properties to increase uptake and endosomal escape into the cytosol (Lönn et al., 2016; Shum and Rossi, 2016; Chakraborty et al., 2017; Foster et al., 2018; Janas et al., 2018; Maguregui and Abe, 2020; Zhang et al., 2021a).
Depending on forms of modifications, a synthetic siRNA can have variable advantages that change properties such as biologic activity, thermodynamic stability, and nuclease resistance and are categorized based on the components modified (Gaglione and Messere, 2010). For example, modifications to the 2’ region of the ribose with 2′-O-methyl, 2′-fluoro, or 2′-O-(2-methoxyethyl) can reduce immunogenicity and improve stability and resistance to degradation by nucleases (Khvorova and Watts, 2017; Maguregui and Abe, 2020). siRNA with a partially phosphorothioated (PS) backbone may increase nonspecific, gymnotic uptake (Lima et al., 2012). It was also reported that replacing the negatively charged phosphodiester backbone with a charge-neutralizing phosphotriester led to an alternative form of siRNAs termed short interfering ribonucleic neutrals that could aid in drug delivery (Meade et al., 2014).
Functions of miRNA and siRNA in Posttranscriptional Gene Regulation
Despite the differences in canonical and noncanonical biogenesis of miRNA as well as the introduction of exogenous RNAi agents, the mechanisms of actions of miRNAs and siRNAs once incorporated within the RISC are largely the same (Fig. 1). However, the type of regulation imposed on the mRNA is dependent on the RNAi agent. SiRISC typically targets and cleaves a single, specific mRNA transcript leading to mRNA cleavage and degradation, whereas miRISC typically targets several specific mRNA transcripts leading to translational repression, mRNA cleavage and degradation, or occasionally translational activation (Ha and Kim, 2014).
Furthermore, both siRNAs and miRNAs are typically selective to specific regions of the mRNAs to regulate gene expression. In general, miRISC acts through imperfect or partial complementarity to the 3′ untranslated region (3′UTR) of its target mRNA transcripts, providing a wider range of target mRNA (Ender et al., 2008; Ha and Kim, 2014; Ipsaro and Joshua-Tor, 2015; Hasler et al., 2016; Patterson et al., 2017; Kuscu et al., 2018). Instead, the typical siRISC will create a perfect complimentary match to the protein coding region or coding DNA sequence (CDS) of a single target mRNA (Lai et al., 2013). Although this is true for most endogenous and synthetic siRNA, three of the FDA-approved siRNA therapeutics actually follow the same mechanism of action as miRISC by binding to the 3′UTR, although they bind with perfect complementarity and are designed to result in transcript cleavage (Coelho et al., 2013; Khvorova, 2017; Liebow et al., 2017) (Fig. 2).
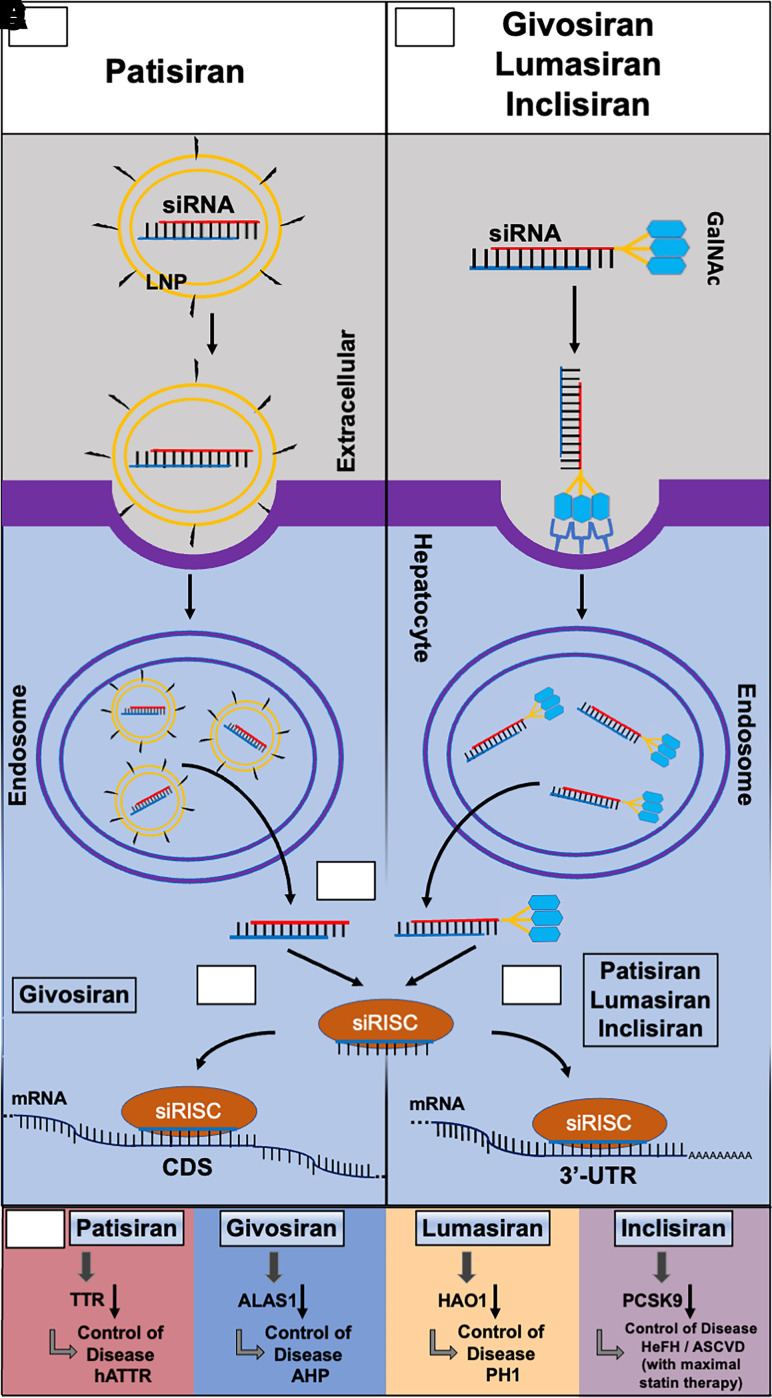
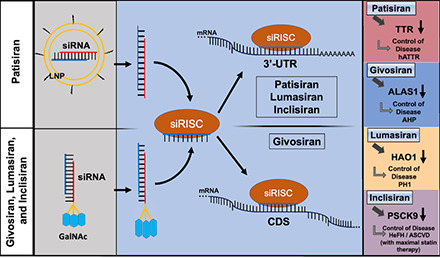
Fig. 2.
Common and specific actions of four FDA-approved siRNA medications in hepatocytes. (A) Patisiran is a double stranded siRNA drug (sense in red and antisense in blue) formulated in a lipid nanoparticle (LNP) decorated with polyethylene glycol (pegylation), and it is administered intravenously for the treatment of hereditary transthyretin-mediated amyloidosis. The LNP induces an opsonization-based immune response and is endocytosed by the hepatocyte prior to endosomal escape. (B) Givosiran, lumasiran, and inclisiran, in which the sense strand (red) is conjugated with three N-acetylgalactosamine (GalNAc) moieties, are administered subcutaneously for the treatments of acute hepatic porphyria, primary hyperoxaluria type 1, and heterozygous familial hypercholesterolemia, respectively. The GalNAc moieties are recognized by asialoglycoprotein receptor 1 (ASGR1), which is highly expressed on the surface of hepatocytes, to facilitate the uptake of siRNAs. (C) The antisense strand (blue) is preferably loaded into the RNA-induced silencing complex (RISC) to form the siRNA-RISC complex (siRISC) while the passenger strand (red) is degraded. (D) Givosiran-derived siRISC binds to the protein coding sequence (CDS) of target mRNA toward cleavage or degradation. (E) Patisiran-, lumasiran-, and inclisiran-derived siRISC interacts with the 3′-untranslated regions (3′UTRs) of target mRNAs to achieve gene silencing. (F) SiRNA drugs (givosiran, patisiran, lumasiran, and inclisiran) target specific mRNAs to achieve the control of their respective diseases. HAO1, hydroxyacid oxidase 1.
Therapeutic siRNAs
Four siRNA medications (patisiran, givosiran, lumasiran, and inclisiran) have been approved by the FDA (in 2018, 2019, 2020, and 2021, respectively), adding to the growing list of oligoribonucleotide drugs (Yu et al., 2019, 2020b; Smith et al., 2022). The designs of these novel siRNA medications are founded on the basic functions and the adaptability of RNAi-based target gene regulation (Onpattro, 2018; Givlaari, 2019; Oxlumo, 2020; Leqvio, 2021) (Table 1). To gain FDA approval, each siRNA-based medication underwent its own series of rigorous clinical trials to determine both the efficacy and safety.
TABLE 1.
FDA-approved siRNA medications
| Medication | Chemistry | Target | Disease | FDA Approval | Reference |
|---|---|---|---|---|---|
| Patisiran (Onpattro) |
Sense: 5′-GUmAACmCmAAGAGUmAUmUmCmCmAUmdTdT-3′ Antisense: 3′-dTdTCAUmUGGUUCUCAUmAAGGUA-5′ |
TTR | hATTR | 2018 | (Onpattro, 2018) |
| Givosiran (Givlaari) |
Sense: 5′-CmsAmsGmAmAmAmGfAmGfUmGfUmCfUmCfAmUmCmUmUmAm-L96-3′ Antisense: 3′-UmsGmsGmUfCmUfUmUfCmUfCmAfCmAfGmAfGmUfAmGfAfsAfsUm-5′ |
ALAS1 | AHP | 2019 | (Givlaari, 2019) |
| Lumasiran (Oxlumo) |
Sense: 5′-GmsAmsCmUmUmCfAmUfCfCfUmGmGmAmAmAmUmAmUmAm-L96-3′ Antisense: 3′-AmsCmsCmUmGmAmAmAfGmUfAmGmGmAmCfCfUmUfUmAmUmsAfsUm-5′ |
HAO1 | PH1 | 2020 | (Oxlumo, 2020) |
| Inclisiran (Leqvio) |
Sense: 5′-CmsUmsAmGmAmCmCfUmGfUmdTUmUmGmCmUmUmUmUmGmUm-L96-3′ Antisense: 3′-AmsAmsGmAmUmCfUmGfGmAfCmAfAmAfAmCfGmAfAfAfAmsCfsAm-5′ |
PCSK9 | HeFH, ASCVD | 2021 | (Leqvio, 2021) |
A, adenosine; Af, adenine 2'-F ribonucleoside; Am, adenine 2'-OMe ribonucleoside; C, cytidine; Cf, cytosine 2'-F ribonucleoside; Cm, cytosine 2'-OMe ribonucleoside; dT, thymidine; G, guanosine; Gf, guanine 2'-F ribonucleoside; Gm, guanine 2'-OMe ribonucleoside; L96, tri-N-acetylgalactosamine; s, phosphorothioate; U, uracil; Uf, uracil 2'-F ribonucleoside; Um, uracil 2'-OMe ribonucleoside.
FDA-Approved siRNA Medications
Patisiran
The development and initial FDA approval of patisiran (brand name: Onpattro) in 2018 for the treatment of hereditary transthyretin-mediated amyloidosis (hATTR) (Table 1) ushered in a first-of-its kind RNAi therapeutic (Onpattro, 2018). Amyloidosis is a rare buildup of amyloid plaques that are not directly produced within the body, formed through the aggregation of several different protein types including transthyretin, a protein encoded by the transthyretin (TTR) mRNA transcript primarily produced in the liver (Gertz, 2016; Kristen et al., 2019). Amyloid formed in hATTR is molecularly characterized as an accumulation of misfolded TTR protein forming amyloid fibrils and clinically manifests as sensorimotor and autonomic neuropathy, cardiomyopathy, arrhythmia, dyspnea, shortness of breath, edema, carpal tunnel syndrome, renal impairment, vitreous opacities, glaucoma, and/or pupillary disorders (Ando et al., 2006; Dungu et al., 2012; Shin and Robinson-Papp, 2012; Kristen et al., 2019).
Patisiran is to exhibit effectiveness in specifically reducing the abundance of both the wild-type and mutant misfolded forms of TTR by RNAi regulation (Onpattro, 2018). Patisiran consists of two 21-nucleotide-long strands with 11 2′-OMe modifications on all pyrimidines present in the sense strand and two uridines in the antisense strand, encapsulated within a lipid nanoparticle (LNP) (Coelho et al., 2013; Onpattro, 2018) (Table 1). This LNP (Fig. 2) includes cholesterol and distearoylphosphatidylcholine (DSPC) [1,2-distearoyl-sn-glycero-3-phosphocholine] with the addition of polyethylene glycol (PEG)-lipids and ionizable amino DLin-MC3-DMA lipids to transport siRNA for circulation stability (Leung et al., 2012; Paunovska et al., 2022). The LNP assists in hepatocyte uptake of patisiran; upon endosomal release, the siRNAs are loaded into the RISC to specifically target and cleave TTR transcripts (Leung et al., 2012; Kristen et al., 2019; Paunovska et al., 2022). Interestingly, the action of patisiran follows that of miRNAs by specifically targeting the 3′UTR (Fig. 2) of TTR instead of the CDS region commonly targeted by an siRNA (Kristen et al., 2019).
Prior to FDA approval, patisiran underwent a series of rigorous clinical trials. Initially, patisiran went through two phase I, placebo-controlled clinical trials (NCT01559077 and NCT02053454), specifically dose-escalation studies ranging from 0.01 to 0.5 mg/kg (Zhang et al., 2019). Phase II placebo-controlled clinical trials were broken into two arms. The first arm (NCT01617967) used a multiple ascending dose study in patients afflicted by hATTR where patients received multiple doses in ascending concentration of 0.01, 0.05, 0.15, or 0.3 mg/kg every 4 weeks or 0.3 mg/kg every 3 weeks (Suhr et al., 2015; Taylor et al., 2018; Kristen et al., 2019). The second arm was a long-term, placebo-controlled, open-label extension (OLE) study (NCT01961921) for individuals who successfully completed the initial phase II study to further characterize the safety and tolerability of long-term patisiran administration (Kristen et al., 2019). Finally, in the pivotal phase III, placebo-controlled clinical trials (APOLLO; NCT1960348), patients received an administration of patisiran (0.3 mg/kg) once every 3 weeks for 18 months (Kristen et al., 2019; González-Duarte et al., 2020; Obici et al., 2020). Conclusion based on the completed phase III study saw a significant improvement in hATTR clinical manifestations as well as in improvement of patient quality of life in patisiran-treated patients (Adams et al., 2018; Obici et al., 2020). In addition, the APOLLO clinical trials assessed the effects of patisiran treatment on the cardiac structure and function on hATTR patients and resulted in a reduction of cardiac wall thickness and global longitudinal strain as well as increased end-diastolic volume and increased cardiac output, suggesting that treatment may also reverse the effects of cardiac associated hATTR (Solomon et al., 2019).
Evidence from clinical trials suggests patisiran to be well tolerated with a consistent and agreeable safety profile in afflicted patients, minimal dispersant of intravenous (i.v.) administered patisiran to off-target organs, and minimal drug accumulation after additional doses, suggesting patisiran siRNA to be cleared via nuclease activity (Coelho et al., 2013; Suhr et al., 2015; Adams et al., 2018). To further evaluate the safety and efficacy of long-term patisiran administration, patients who completed both the phase II OLE and phase III were also enrolled into an ongoing global OLE study (NCT02510261) (Adams et al., 2021). Patisiran is seeing a second ongoing phase III clinical trial (NCT03997383) for hATTR in patients that suffer with a cardiomyopathy comorbidity. Today, patisiran is commercially available with a recommended dose of 0.3 mg/kg every 3 weeks administered through i.v. infusion over 80 minutes (Onpattro, 2018).
Overall, the impact of patisiran provided three major contributions to the areas of RNAi therapy. First, patisiran served as the first RNAi-based therapeutic approved by the FDA that is designed to use endogenous RNAi machinery to control the outcome of disease-causing proteins. This work carved a path of success that other RNAi therapeutics would soon follow. Second, the method of action of patisiran closely mirrors that of endogenous miRNA by targeting the 3′UTR of TTR and supports the potential of future successful miRNA therapeutic development to combat disease. Finally, the development of patisiran is a foundation upon which ongoing research has improved and innovated RNAi technology to be widely applicable and appropriately manipulated to target and remedy molecular disease components.
Givosiran
Givosiran (brand name: Givlaari) was the second siRNA drug to receive its first FDA approval in 2019 for the treatment of acute hepatic porphyria (AHP) (Givlaari, 2019) (Table 1). AHP is a rare genetic disorder derived from the liver and caused by a dysfunction within the heme synthesis pathway that causes an upregulation of aminolevulinate synthase 1 (ALAS1) and an increase ALAS1 protein expression (Bissell et al., 2017; Anderson, 2019; Scott, 2020; Kothadia et al., 2022; Sardh and Harper, 2022). Dysregulated ALAS1 expression is known to increase downstream production of neurotoxic metabolites, aminolevulinic acid, and porphobilinogen, causing AHP (Anderson, 2019; Kothadia et al., 2022). AHP is clinically characterized by severe and debilitating abdominal pain, hypertension, tachycardia, vomiting, seizures, and paralysis and is often associated with neuropathy, chronic kidney disease, and liver disease (Besur et al., 2014; Pallet et al., 2018; Anderson, 2019; Sardh and Harper, 2022).
Givosiran is designed as an effective treatment against AHP by reducing the abundance of ALAS1 by RNAi regulation (Givlaari, 2019). Givosiran consists of a double-stranded, chemically synthesized and fully modified 23-nucleotide-long antisense strand and 21-nucleotide-long sense strand with tri-N-acetylgalactosamine (GalNAc)-conjugation to enhance liver selective delivery (Givlaari, 2019; Debacker et al., 2020; Sardh and Harper, 2022). Further, givosiran also uses 16 2′-F substitutions with the remaining as 2′-OMe substitutions as well as six terminal PS chemical modifications (Givlaari, 2019) (Table 1). Unlike patisiran, givosiran is administered via subcutaneous (s.c.) injection to target and inhibit the translation of ALAS1 mRNA (Givlaari, 2019). The tri-GalNAc delivery platform exploits the biologic interaction between the GalNAc molecules found on damaged glycoproteins and the asialoglycoprotein receptor 1 (ASGR1) found at high levels on hepatocytes to target givosiran to the liver (Debacker et al., 2020; Sardh and Harper, 2022) (Fig. 2). The givosiran siRNA payload is designed to target a specific sequence on the CDS (Fig. 2) of ALAS1, decrease ALAS1 protein abundance and the production of the neurotoxic metabolites, and treat AHP (Agarwal et al., 2020; Sardh and Harper, 2022).
Givosiran also underwent a series of rigorous clinical trials prior to regulatory approval. In phase I, placebo-controlled clinical trials (NCT02452372) were separated into three dosing regiments based on patient AHP frequency: A (infrequent AHP – single ascending dose s.c. injections of 0.035, 0.10, 0.35, 1.0, or 2.5 mg/kg); B (moderate frequency – once-monthly dose s.c. injections of 0.35 or 1.0 mg/kg); and C (frequent – once-monthly or quarterly dose s.c. injections of 2.5 or 5.0 mg/kg) (Sardh et al., 2019; Sardh and Harper, 2022). Patients who successfully completed phase IC were enrolled in phase I/II, placebo-controlled OLE with the goal to rapidly and sustainably lower hepatic ALAS1 mRNA and urinary neurotoxic metabolite levels as well as reduce the rate of AHP attacks in patients with ongoing attacks through extended givosiran treatment (Bissell et al., 2019; Sardh and Harper, 2022). In the pivotal phase III, placebo-controlled clinical trials (ENVISION; NCT03338816) were performed on symptomatic patients who received once-monthly s.c. injections of givosiran (2.5 mg/kg) for 6 months (Balwani et al., 2020; Sardh and Harper, 2022).
Clinical evidence shows that givosiran-treated patients saw a significant reduction in AHP attacks (74%) and sustained reduction in urinary aminolevulinic acid and porphobilinogen neurotoxic metabolite levels as well as decreased daily pain and an improved quality of life (Balwani et al., 2020; Sardh and Harper, 2022). However, success was accompanied by an increase in adverse hepatic and renal events seen in few givosiran-treated patients (Balwani et al., 2020; Sardh and Harper, 2022). A later study designed to further assess the safety and efficacy of givosiran showed evidence that regular givosiran treatments did indeed reduce the rate of AHP attacks, benefited patients with recurrent AHP attacks, improved quality of life, and supported the safety of givosiran (Sardh and Harper, 2022; Ventura et al., 2022). Today, givosiran is commercially available with a recommended dose of 2.5 mg/kg once a month via s.c. injection (Givlaari, 2019).
The success of givosiran provides three major impacts to the fields of molecular biology and therapeutics. First, as a chemically modified siRNA therapeutic, givosiran serves as evidence that chemical alterations to RNAi technology can be implemented and manipulated to optimize molecular interactions to target and remedy disease. Second, the successful delivery of givosiran demonstrates a major advancement in RNAi therapeutics with a drug delivery platform capable of selective delivery to hepatocytes by the GalNAc delivery platform used in future siRNA therapeutics. Finally, givosiran further supports the use of RNAi-based therapeutics to combat diseases that can be largely at the mercy of ineffective treatment options or deemed undruggable targets by small molecules.
Lumasiran
Lumasiran (brand name: Oxlumo, 2020), was the third siRNA medication to receive its first FDA approval in 2020 for the treatment of primary hyperoxaluria type 1 (PH1) (Oxlumo, 2020) (Table 1). PH1 is a rare autosomal recessive disorder originating in the liver that is caused by a decrease in peroxisomal enzyme alanine-glycolate aminotransferase (AGT), an enzyme responsible for glyoxylate metabolism inhibition, and results in the deposition of calcium oxalate crystals in the kidneys and urinary tract (Cochat and Rumsby, 2013; Garrelfs et al., 2021). PH1 is clinically characterized by nephrolithiasis and nephrocalcinosis of the kidney, leading to kidney disease or failure and systemic oxalosis, the systemic deposition of calcium oxalate crystals (Cochat and Rumsby, 2013).
Lumasiran is designed as an effective treatment of PH1 by reducing the abondance of oxalate deposition by RNAi regulation alternative to dialysis, kidney transplant, or vitamin B6 supplementation (Cochat and Rumsby, 2013; Oxlumo, 2020). Lumasiran consists of a fully modified siRNA with a 23-nucleotide-long antisense strand, 21-nucleotide-long sense strand with tri-GalNAc-conjugation, 10 2′-F, 34 2′-OMe, and six terminal PS chemical modifications (Oxlumo, 2020) (Table 1). Lumasiran is also administered subcutaneously to patients with specific delivery to the liver and targets the 3′UTR (Fig. 2) of the hydroxyacid oxidase 1 (HAO1) mRNA transcript that encodes glycolate oxidase (GO), an upstream enzyme in the oxalate overproduction pathway (Liebow et al., 2017; Debacker et al., 2020; Oxlumo, 2020).
Like that of its predecessors, lumasiran underwent a series of rigorous trials to attain FDA approval. Initial phase I/II, placebo-based clinical trials (NCT02706886) consisted of three different dosing regimens to determine appropriate dosing of either three monthly doses of 1 mg/kg, three monthly doses of 3 mg/kg, or two single doses of 3 mg/kg every 3 months, followed by an OLE (Frishberg et al., 2021). The pivotal phase III, placebo-controlled clinical trials were broken into three unique arms termed ILLUMINATE-A (NCT03681184), ILLUMINATE-B (NCT03905694), and ILLUMINATE-C (NCT03681184). ILLUMINATE-A studied the effects of lumasiran in adults and children over the age of 6 years and received 3 months of lumasiran followed by quarterly maintenance doses of 3 mg/kg that extended into a 54-month dose evaluation OLE period to assess safety and efficacy (Garrelfs et al., 2021). In ILLUMINATE-B, patients under the age of 6 years received similar lumasiran treatments (Sas et al., 2021). ILLUMINATE-C (NCT03681184) is currently ongoing and designed to evaluate the efficacy and safety of lumasiran for patients with advanced PH1 and estimated completion date of July 2025 (Scott and Keam, 2021). After the success of ILLUMINATE-A and ILLUMINATE-B, patients who successfully completed these studies were enrolled into an ongoing OLE period to evaluate the long-term efficacy and safety of lumasiran in adults and children with PH1 (NCT03350451) (Scott and Keam, 2021).
Based on evidence from clinical trials, lumasiran was well tolerated in both children and adult cohorts, demonstrated an acceptable safety, and was shown to beneficially reduce oxalate levels independent of age, sex, race, abnormal kidney function, vitamin B6 use, or history of symptomatic kidney stone events (Garrelfs et al., 2021; Sas et al., 2021). Today, lumasiran is commercially available with a recommended subcutaneous administration dose dependent on body weight (Oxlumo, 2020). For patients less than 10 kg, the recommended administration has a loading dose of lumasiran is 6 mg/kg once monthly for three doses followed by a once-monthly maintenance dose of 3 mg/kg. For patients between 10 kg and 20 kg, the recommended administration has a loading dose of lumasiran is 6 mg/kg once monthly for three doses followed by a once-quarterly maintenance dose of 6 mg/kg. For patients 20 kg and above, the recommended administration has a loading dose of lumasiran is 3 mg/kg once monthly for three doses followed by a once-quarterly maintenance dose of 3 mg/kg.
The success and approval of lumasiran provides two major impacts to the fields of molecular medicine. First, lumasiran serves as an additional RNAi-based therapeutic that further supports the use of RNAi as a means to combat disease, and the method of action of lumasiran once again closely mirrors that of endogenous miRNA and therefore further supports the development of miRNA drugs. Second, successful administration and action of lumasiran further serves as evidence that RNAi technology can be implemented and manipulated to optimize molecular interactions to target and remedy disease.
Inclisiran
The most recent siRNA therapeutics approved by the FDA in 2021, inclisiran (brand name: Leqvio), is a first-in-class siRNA medication designed to treat heterozygous familial hypercholesterolemia (HeFH) and clinical atherosclerotic cardiovascular disease (ASCVD) in combination with maximally tolerated statin therapy (Leqvio, 2021) (Table 1). Both HeFH and ASCVD are similarly characterized by an increase in low-density lipoprotein cholesterol (LDL-C) in circulation (McGowan et al., 2019; Bardolia et al., 2021). HeFH is a familial, autosomal codominant genetic disorder caused by a protein mutation within lipoprotein metabolism that leads to a high accumulation of LDL-C in the blood plasma. Mutations in the LDL receptor (LDL-R) are identified in 85%–90% of cases, and a gain-of-function (GoF) mutation in the proprotein convertase subtilisin/kexin type 9 (PCSK9) gene is identified in roughly 1% of confirmed HeFH cases (Abifadel et al., 2003; McGowan et al., 2019). PCSK9 is a low abundant circulating protein with a critical function in the low-density lipoprotein cholesterol metabolic pathway where a gain-of-function mutation within the PCSK9 gene further enhances its function to decrease natural LDL recycling, untimely resulting in hypercholesterolemia (Abifadel et al., 2003). Chronic elevated LCL-C levels seen in hypercholesterolemia often results in a comorbidity with ASCVD, a thickening and loss of elasticity in the arterial wall (McGowan et al., 2019; Bardolia et al., 2021). Prior to inclisiran, the PCSK9 inhibition was facilitated through the use of two monoclonal antibodies, evolocumab and alirocumab, that function by blocking the LDL receptor; however, their safety and efficacy continue to be under investigation based on data reported by Clinicaltrials.gov (van Bruggen et al., 2020; Santulli et al., 2021).
Unique among siRNA therapeutics, inclisiran is designed as a supplemental, nonstatin therapeutic to improve the effects of current treatments for cholesterol management by reducing the abondance of PCSK9 by RNAi regulation (Bardolia et al., 2021; Leqvio, 2021; Zhang et al., 2021a). Inclisiran consists of a fully modified siRNA with a 23-nucleotide-long antisense strand, 21-nucleotide-long sense strand with tri-GalNAc-conjugation, one 2′-MOE, 11 2′-F, 32 2′-OMe, and six terminal PS chemical modifications designed to target PCSK9 mRNA (Leqvio, 2021) (Table 1). Inclisiran is administered subcutaneously to patients to inhibit PCSK9 protein synthesis, decreasing the influence of PCSK9 on natural LDL recycling by binding to the 3′UTR of PCSK9 mRNA and interfering with its translation (Nair et al., 2014; Khvorova, 2017; Lamb, 2021; Leqvio, 2021) (Fig. 2).
As all approved siRNA therapeutics before it, inclisiran underwent a series of rigorous trials to prior to FDA approval. Initial phase I, placebo-controlled clinical trials (NCT02314442) were broken into two arms to study the effects of inclisiran treatment. Arm one was a single ascending dose study dosed at either 25, 100, 300, 500, or 800 mg and arm two was a multiple ascending dose study with dose regimens of either four weekly doses at 125 mg, two doses every other week at 250 mg, or two monthly doses of either 300 or 500 mg done with or without combination statin therapy (Fitzgerald et al., 2017). Phase II, placebo-controlled clinical trials (NCT02597127) in patients with elevated LDL-C serum levels and high cardiovascular disease risk had two multiple ascending dose trials, receiving either a single dose of either 200, 300, or 500 mg or two doses at days 1 and 90 of either 100, 200, or 300 mg (Ray et al., 2017). The pivotal phase III, placebo-controlled clinical trials were broken into three unique arms termed ORION-9 (NCT03397121), ORION-10 (NCT03399370), and ORION-11 (NCT03400800). ORION-9 was performed on adults with heterozygous familial hypercholesterolemia, receiving 300 mg administered on days 1, 90, 270, and 450 (Raal et al., 2020). ORION-10 and ORION-11 were performed on patients on statin therapy with ASCVD or ASCVD and equivalent ASCVD risk, respectively, receiving 284 mg on days 1 and day 90 followed by a dose every 6 months thereafter for 540 days (Ray et al., 2020).
Clinical results indicate that inclisiran-treated patients saw a reduction of approximately 50% in LDL cholesterol levels of patients treated every 6 months (Raal et al., 2020; Ray et al., 2020). As a newly approved therapeutic, inclisiran is currently under several phase III studies to assess the efficacy, safety, and tolerability of long-term dosing including ORION-8 (NCT03814187), ORION-13 (NCT04659863) and ORION-16 (NCT04652726) (Brandts and Ray, 2021; Reijman et al., 2022). With a recommended initial dose of 284 mg followed by a supplemental dose at 3 months and maintenance doses every 6 months administered via s.c. injection, inclisiran is commercially available and prescribed for combination therapy with the maximally tolerated statins (Leqvio, 2021).
The success after FDA approval of inclisiran in late December 2021 has three major impacts in the fields of molecular biology and therapeutics. First, the approval of inclisiran is further validation that canonical RNAi machinery is adaptable and that modification to RNAi technology can optimize molecular interactions to specifically target and remedy disease. Second, inclisiran is the third siRNA therapeutic to closely mirrors the function of endogenous miRNA and supports the use of miRNA drug development to combat disease. Finally, with the high incidence of hypercholesterolemia, inclisiran offers a unique, first-in-class, nonstatin treatment to help combat hypercholesterolemia, where a 1.0 mmol/l reduction in LDL is suggested to reduce adverse cardiovascular events by roughly 21% (Santulli et al., 2021).
Continued studies into inclisiran explore the pharmacokinetics and safety as well as investigate emerging pharmacodynamic roles for inclisiran on biologic function. In a 2022 study focused on the pharmacokinetics and pharmacodynamics of inclisiran in hepatic-impaired patients, researchers found that patients with impaired hepatic function had approximately a 2-fold increase in systematic exposure to inclisiran with little change in LDL-C compared with patients with no or mild hepatic impairment (Kallend et al., 2022). This suggested that hepatic impairment had little effect on treatment efficacy and showed no issues in the safety or tolerability of inclisiran, which further suggested no need for dose adjustment for patients with mild or moderate hepatic impairment (Kallend et al., 2022). An additional study that was recently published looks into the effects of inclisiran on the formation of oxidized-LDL (ox-LDL)-induced, macrophage-derived foam cells (Wang et al., 2022). In this study, the authors found inclisiran to reduce lipid accumulation and inhibited macrophage-derived foam cell formation through the activation of the peroxisome proliferator-activated receptor gamma (PPARγ) pathway (Wang et al., 2022). Since activation of the PPARγ pathway is important in fatty acid uptake and lipogenesis, the results of this study suggest a possible off-target effect of inclisiran by increasing the expression of increased both the gene and protein expression of PPARγ (Christofides et al., 2021; Wang et al., 2022). Investigations into the pharmacodynamic roles of therapeutics on biologic function are critical not only for understanding how medications interact with the body but also to better understand what pathways or unknown interactions are inadvertently activated that may contribute to the success or potential adverse effects of a therapeutic.
Therapeutic siRNAs in Clinical Trials
In addition to the four FDA-approved siRNA therapeutics, several other novel siRNA therapeutics have entered clinical trials with several in preclinical development (Table 2). Of these, five have reached the pivotal phase III clinical trials, namely fitusiran (Machin and Ragni, 2018), nedosiran (Kletzmayr et al., 2020), teprasiran (Gallagher et al., 2017), tivanisiran (Moreno-Montañés et al., 2018), and vutrisiran (Habtemariam et al., 2021).
TABLE 2.
Therapeutic siRNAs in clinical trials
| Product | Sponsor Company | Target | Disease | Current Development |
|---|---|---|---|---|
| Fitusiran (ALN-AT3SC) | Sanofi Genzyme | Antithrombin | Hemophilia A and B | Phase III – NCT03417245 (ATLAS-A/B) Phase III – NCT03417102 (ATLAS-INH) Phase III – NCT03549871 (ATLAS-PPX) Phase II/III – NCT03974113 (ATLAS-PEDS) Phase III – NCT03754790 (ATLAS-OLE) |
| Nedosiran (DCR-PHXC) | Dicerna Pharmaceuticals | Hepatic LDH | Primary hyperoxaluria | Phase III – NCT04042402 (PHYOX3) Phase II – NCT05001269 (PHYOX8) |
| Teprasiran (QPI-1002) | Quark Pharmaceuticals | p53 | AKI | Phase III – NCT02610296 (ReGIFT) |
| Tivanisiran (SYL-1001) | Sylentis S.A. | Transient receptor potential cation channel subfamily V member 1 | Ocular pain, DED | Phase III – NCT03108664 (HELIX) |
| Vutrisiran (ALN-TTRSC02) | Alnylam Pharmaceuticals | Transthyretin | hATTR | Phase III – NCT03759379 (HELIOS-A) Phase III – NCT04153149 (HELIOS-B) |
| Cosdosiran (QPI-1002) | Quark Pharmaceuticals | Caspase-2 | NAION | Phase II/III (terminated) – NCT02341560 |
| AGN211745(Sirna-027) | Allergan | VEGFR1 | CNV, AMD | Phase I/II – NCT00363714 |
| PF-04523655 | Quark Pharmaceuticals | VEGFR1 | AMD | Phases I – NCT00725686 |
| AMD | Phase II – NCT00713518 | |||
| CNV, RN, DME | Phase II – NCT01445899 | |||
| siG12D LODER | Silenseed Ltd | KRASG12D | PDAC, pancreatic cancer | Phase I – NCT01188785 Phase II – NCT01676259 |
| TKM-080301 | Arbutus Biopharma Corporation | PLK1 | HCC | Phases I/II – NCT02191878 |
| Neuroendocrine tumors, ACC | Phase I/II – NCT01262235 | |||
| Atu027 | Silence Therapeutics GmbH | Protein kinase N3 | Advanced solid tumors | Phase I – NCT00938574 |
| PDC | Phases I/II – NCT01808638 | |||
| TD101 | Pachyonychia Congenita Project | KRT6A | Pachyonychia congenita | Phase I – NCT00716014 |
| ARC-520 | Arrowhead Pharmaceuticals | cccDNA-derived viral mRNA | Safety/tolerability in healthy volunteers | Phase I – NCT01872065 |
| QPI-1002 | Quark Pharmaceuticals | p53 | ARF, AKI | Phase I – NCT00554359 |
| DGF, complications of kidney transplant | Phases I/II – NCT00802347 |
ACC, adrenocortical carcinoma; AMD, age-related macular degeneration; ARF, acute renal failure; CNV, choroidal neovascularization; DGF, delayed graft function; DME, diabetic macular edema; DR, diabetic retinopathy; HCC, hepatocellular carcinoma; LDH, lactate dehydrogenase; NAION, nonarteritic anterior ischemic optic neuropathy; PDAC, pancreatic ductal adenocarcinoma; PDC, pancreatic ductal carcinoma; VEGFR1, vascular endothelial growth factor 1.
Fitusiran
Fitusiran (ALN-AT3SC) is a therapeutic siRNA designed for the treatment of both hemophilia A and B, and fitusiran is currently undergoing phase II and phase III clinical trials (Machin and Ragni, 2018) (Table 2). Hemophilia A and B are X-linked bleeding disorders that are the result of mutations within the genes encoding coagulation factor VIII and IX, respectively, and interfere with normal blood clotting mechanisms of the body (Castaman and Matino, 2019). Fitusiran consists of a fully modified double-stranded siRNA of 21 and 23 nucleotides long with a tri-GalNAc-conjugate and contains 21 2′-F substitutions, 23 2′-OMe substitutions, and six PS modifications at the strand ends (Berk et al., 2021). Fitusiran functions by inhibiting the production of the antithrombin proteins to increase the generation of procoagulation enzyme, thrombin, and improve the blood clotting mechanism (Sehgal et al., 2015; Machin and Ragni, 2018).
Nedosiran
Nedosiran (DCR-PHXC) is another therapeutic siRNA currently in phase III clinical trials that is designed for the treatment of primary hyperoxaluria (Kletzmayr et al., 2020) (Table 2). Nedosiran consists of a nearly fully modified siRNA duplex that forms a tetraloop configuration through interactions between its 22-nucleotide-long antisense strand and 36-nucleotide-long sense strand with tri-GalNAc conjugation as well as 19 2′-F substitution, 35 2′-OMe substitution, and six PS modifications (Lai et al., 2018; Kletzmayr et al., 2020). Nedosiran is an up-and-coming competitor of lumasiran that is being clinically assessed for the treatment of both PH1 and primary hyperoxaluria type 2 (PH2) subtypes of primary hyperoxaluria. However, instead of targeting the upstream glycolate oxidase protein mRNA, nedosiran is designed to target the mRNA of the hepatic enzyme lactate dehydrogenase (LDH), an enzyme that controls the final step in glyoxylate metabolism to oxalate (Cochat and Rumsby, 2013; Kletzmayr et al., 2020).
Teprasiran
Teprasiran (QPI-1002) is a therapeutic siRNA currently in phase III clinical trials that is designed to treat acute kidney injury (AKI), specifically after kidney transplant or cardiovascular surgery (Davidson and McCray, 2011; Gallagher et al., 2017) (Table 2). AKI is not inherently classified as a disease but is instead classified as a clinical syndrome that afflicts many hospitalized patients and is defined as an abrupt decrease in kidney function because of structural damage or impairment (Makris and Spanou, 2016). Teprasiran is unique among current siRNAs in that it only consists of 2′-OMe modifications. Specifically, half of the ribonucleosides within the 19-nucleotide-long siRNA duplex are comprised of 2′-OMe substitutions, and teprasiran is delivered as naked siRNA without another delivery system (Gallagher et al., 2017). Teprasiran functions by targeting the mRNA of the well known tumor suppressor protein p53, which is involved in apoptotic induction during physiologic stress (Molitoris et al., 2009; Thompson et al., 2012; Gallagher et al., 2017).
Tivanisiran
Tivanisiran (SYL-1001) is a therapeutic siRNA designed for the treatment of ocular pain and dry eye disease (DED), and it is currently in phase III clinical trials (Moreno-Montañés et al., 2018) (Table 2). DED is a multifactorial disease of the eye that results in several ocular abnormalities including pain, discomfort, dryness, itching, burning, and photophobia due to a disruption in the healthy tear film, ocular inflammation, or neurosensory abnormalities (Shimazaki, 2018). Tivanisiran stands alone as the only completely unmodified therapeutic siRNA composed of a 19-nucleotide-long duplex in phase III that is also delivered naked and without a delivery platform (Moreno-Montañés et al., 2018). Tivanisiran functions by targeting the mRNA of transient receptor potential cation channel subfamily V member 1 (TRPV1) that plays an important role in several pathways including pain signal transduction, fibrogenesis modulation, the stress response, and the innate inflammatory response (Moreno-Montañés et al., 2018).
Vutrisiran
Vutrisiran (ALN-TTRSC02) is a therapeutic siRNA in phase III clinical trials that is also designed for the treatment of hATTR (Habtemariam et al., 2021) (Table 2). Vutrisiran is an up-and-coming competitor of patisiran since both are designed to treat hATTR. Vutrisiran consists of two fully modified strands of 21 and 23 nucleotides long with the latest version of the tri-GalNAc conjugate delivery platform, and it also contains 35 2′-OMe, nine 2′-F, and six PS modifications at the strand ends (Foster et al., 2018; Weng et al., 2019). With improved delivery to the liver by tri-GalNAc conjugation, vutrisiran is believed to be a more potent than patisiran yet functions similarly in the hepatocyte by targeting a conserved sequence on all TTR variants with similar actions as patisiran to treat hATTR (Foster et al., 2018; Weng et al., 2019; Habtemariam et al., 2021).
miRNA-Based Therapies under Development
Strategies of miRNA-Based Therapies
Alongside the extensive list of siRNAs in clinical trials, several miRNA detection technologies are FDA approved and available today to determine miRNA profiles as potential biomarkers for clinical diagnostic or prognostic purposes (Bonneau et al., 2019; Hanna et al., 2019; Smith et al., 2022). In terms of miRNA-based interventions, there are two major approaches (Bader et al., 2010; Mollaei et al., 2019; Yu et al., 2020b; Yu and Tu, 2022). One strategy, namely miRNA antagonism (anti-miR), is to inhibit or repress the expression or function of a target miRNA; the other approach, namely miRNA replacement therapy, is to restore the expression or function of target miRNA.
Anti-miR therapy relies on sequence complementarity between the single-stranded antagomir (or ASO) and target miRNA (Lima et al., 2018). In particular, antagomirs inhibit miRNA function by complimentary hybridization and/or steric hindrance of miRNA with its target mRNA or for DNA/RNA hybridization degradation by RNase H (Lennox and Behlke, 2011; Lennox et al., 2013; Mie et al., 2018). There is also growing interest in developing small molecule compounds to interfere with miRNA biogenesis or function (Velagapudi et al., 2014; Costales et al., 2017; Meyer et al., 2020; Yu et al., 2020b). Therapeutic antagomirs are typically deployed to target disease causing and overabundant miRNAs in the diseased cells, such as oncogenic miRNAs overexpressed in carcinoma cells (Stenvang et al., 2012; Mollaei et al., 2019). miRNA replacement therapy restores miRNAs downregulated or completely lost in the diseased cells that actually function to suppress the disease, such as tumor suppressor miRNAs (Mollaei et al., 2019). Common methods of replacement typically employ either chemo-engineered miRNA mimics or viral or nonviral vector systems to rebalance the target miRNA profile to combat disease (Yu et al., 2016, 2020b; Mollaei et al., 2019).
Although anti-miR therapy is the prominent miRNA-based intervention currently under clinical trials, the application of miRNA replacement therapy is exemplified in previous study focused on miR-29 mimicry to block pulmonary fibrosis (Montgomery et al., 2014). miR-29 was selected for this study based on its ability to regulate extracellular matrix proteins important in tissue fibrosis and is downregulated in fibrotic diseases (He et al., 2013; Montgomery et al., 2014). In this study, after confirming its functionality in vitro, the chemically modified, synthetic RNA duplex of miR-29 was injected intravenously into a mouse model of pulmonary fibrosis, resulting in a sustained increase miR-29 levels and restored endogenous miR-29 function by decreasing collagen expression and treating the disease (Montgomery et al., 2014).
One major area of study is miRNA-based anticancer treatments to combat various types of cancer (To et al., 2020; Smith et al., 2022). This is because some miRNAs, such as let-7 and miR-34, are master regulators of gene expression with the capability to modulate several critical homeostatic cellular pathways and functions that are often found dysregulated in cancer (Catalanotto et al., 2016; Piletič and Kunej, 2016; To et al., 2020). Specific miRNAs have emerging roles in particular cancer types including human papillomavirus (HPV)-related cancers (Casarotto et al., 2020), colorectal cancers (To et al., 2018), lung cancers (Xue et al., 2017), and acute myeloid leukemias (Lovat et al., 2020), among others. As more and more oncogenic miRNAs are being examined, there has been a large influx of strategies aimed at miRNA inhibition such as miRNA-masks, miRNA-sponges, and miRNA-zippers, among others (To et al., 2020). miRNA-masks are ASOs designed to bind the 3′UTR of target mRNA and shield or protect the mRNA from endogenous miRNA regulation in an inhibitory manner (Wang, 2011; Nguyen and Chang, 2017). miRNA-sponges are often circular RNA (circRNA) molecules that function as a decoy to sequester or “sponge” multiple miRNAs from their target mRNA transcripts and can more easily evade nuclease degradation compared with linearized ASOs (Ebert et al., 2007; Wang, 2011). miRNA-zippers are designed to connect miRNA molecules end-to-end, forming a highly specific, highly stable DNA-RNA duplex to inhibit miRNAs from performing their functions (Meng and Lu, 2017).
Further, miRNA-based therapies are also used in combination with other well established therapies toward optimal outcomes. In fact, combination therapy can come in many flavors and includes either miRNA antagonism in combination miRNA replacement therapy or miRNA-based therapy in combination with existing or new treatment of a given disease. Combination therapies are most prevalent in anticancer therapy to play supportive roles in oncogenic miRNA inhibition or tumor-suppressive miRNA reintroduction. Some common forms of miRNA-based combination therapy include chemotherapy (To, 2013; Zhang and Wang, 2017), immunotherapy (Hand et al., 2010; Ji et al., 2016; Cortez et al., 2019), radiotherapy (Overgaard, 2007; Babar et al., 2011; Moertl et al., 2016; El Bezawy et al., 2019), and photodynamic therapy (El-Daly et al., 2017; Kessel and Oleinick, 2018). Of interest, one form of combination therapy uses multiple tumor-suppressor miRNA reintroduction or anti-miR miRNA inhibition to synergize and bolster the antitumor effects of treatment (Jung et al., 2015; Orellana et al., 2019). In a 2016 article, the expression of miR-621 was demonstrated to be a predictive marker for chemosensitivity to paclitaxel plus carboplatin (PTX/CBP) treatment of breast cancer patients where elevated levels of miR-621 predicted a better response to PTX/CBP treatment (Xue et al., 2016). Researchers further established this correlation in a combination therapy study, demonstrating that increased ectopic expression of miR-621 increased chemosensitivity to PTX/CBP in vitro and in vivo (Xue et al., 2016). This study demonstrates the promise of miRNA therapeutics and supports a potential role in combination therapy.
miRNA-Based Therapies in Clinical Trials
Several miRNAs have been and continue to be tested for remedies of several disease types (Table 3), including anticancer treatment. For example, MRX34, a mimic of miR-34a, was tested as an anticancer therapeutic in two phase II clinical trials (NCT01829971; NCT02862145) for primary liver cancer (PLC), small cell lung cancer (SCLC), lymphoma, melanoma, multiple myeloma (MM), renal cell carcinoma (RCC), and non–small cell lung cancer (NSCLC). miR-34a is a key regulator of tumor suppression controlling the expression of several targets involved in cell cycle (e.g., c-MYC, E2F, CDK4, and CDK6) and apoptosis (e.g., BCL2 and SIRT1) as well as tumor-associated processes such as invasion (e.g., c-MET) (Hermeking, 2010; Misso et al., 2014). miR-34a has also shown antagonistic characteristics to cancer cell viability, stemness, metastasis, and resistance to chemotherapy (Misso et al., 2014). Decreased expression of miR-34a is often associated with several types of cancer, including multiple myeloma (Dimopoulos et al., 2013) and melanoma (Lodygin et al., 2008; Misso et al., 2014). The molecular structure of MRX34 consists of a 23-nucleotide-long, double-stranded miRNA resembling the endogenous miR-34a duplex that is encapsulated in LNP for intravenously administered delivery to tumors located in the liver, bone marrow, spleen, lung, and a variety of other tissues (Beg et al., 2017). Once delivered, MRX34 would interfere with target gene translation by binding to its complementary sequence in the 3′UTR of mRNAs. Although both phase II trials were terminated due to immune-related serious adverse events, MRX34 demonstrated a successful regulation of relevant target genes that provided the necessary proof-of-concept for the efficacy of miRNA-based anticancer therapy (Hong et al., 2020).
TABLE 3.
miRNA-based therapies in clinical and preclinical investigations
| Product | Sponsor Company | miRNA | Disease | Current Development |
|---|---|---|---|---|
| Miravirsen (SPC3649) | Roche/Santaris | Anti-miR-122 | Hepatitis C | Phase II – NCT01727934, NCT01872936, NCT01200420 |
| CDR132L | Cardior Pharmaceuticals GmbH | Anti-miR-132 | MI and AHF (left sided) | Phase II – NCT05350969 |
| Lademirsen (SAR339375, RG-012) | Sanofi Genzyme | Anti-miR-21 | Alport syndrome | Phase I – NCT03373786 |
| AZD4076 (RG-125) | AstraZeneca | Anti-miR-103/107 | NAFLD | Phase I – NCT02826525 |
| NASH | Phase I – NCT02612662 | |||
| RGLS4326 | Regulus Therapeutics | Anti-miR-17 | PKD | Phase I – NCT04536688 |
| TargomiRs | Asbestos Diseases Research Foundation | miR-16 mimic | MPM and NSCLC | Phase I – NCT02369198 |
| MRX34 | Mirna Therapeutics | miR-34 mimic | Melanoma | Phase I/II (withdrawn) – NCT02862145 |
| PLC, SCLC, lymphoma, melanoma, multiple myeloma, RCC, NSCLC | Phase I (terminated) – NCT01829971 | |||
| Remlarsen (MRG-201) | MiRagen Therapeutics/Viridian | miR-29 mimic | Keloid | Phase II – NCT02603224, NCT03601052 |
| MRG-110 | Anti-miR-92a | Angiogenesis/ischemia | Phase I – NCT03603431 | |
| MRG-107 | Anti-miR-155 | Atherosclerosis | Preclinical | |
| MGN-1374 | Anti-miR-15, anti-miR-195 | Post–myocardial infarction | Preclinical | |
| MGN-2677 | Anti-miR-143/145 | Vascular disease | Preclinical | |
| MGN-4220 | Anti-miR-29 | Cardiac fibrosis | Preclinical | |
| MGN-4893 | Anti-miR-451 | ARBCP-like disorders | Preclinical | |
| MGN-5804 | Anti-miR-378 | CMD | Preclinical | |
| MGN-6114 | Anti-miR-92 | PAD | Preclinical | |
| MGN-9103 | Anti-miR-208 | CHF | Preclinical |
AHF, acute heart failure; ARBCP, abnormal red blood cell production; CHF, chronic heart failure; CMD, cardiometabolic disease; MI, myocardial infarction; MPM, malignant pleural mesothelioma; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; PAD, peripheral arterial disease; PKD, polycystic kidney disease; PLC, primary liver cancer; RCC, renal cell carcinoma; SCLC, small cell lung cancer.
Alongside a promising future in miRNA-based anti-cancer therapy, miRNAs also show potential in the treatment of cardiovascular disease. In particular, several miRNAs are under investigation for their roles in cardiomyocyte necrosis, apoptosis, and autophagy as well as cardiac fibroblast proliferation, inflammation, and angiogenesis (Kansakar et al., 2022; Varzideh et al., 2022). One miRNA-based cardiovascular therapeutic, termed CDR132L, is currently starting phase II clinical trials for the treatment of myocardial infarctions and acute heart failure of the left side and functions as an antagomir of miR-132, an miRNA important in maladaptive cardiac remodeling, transformation, and hypertrophy (NCT05350969) (Table 3). A second miRNA-based cardiovascular therapeutic, MRG-110, an antagomir of miR-92a, has currently completed phase I clinical trials and is intended to promote angiogenesis by inhibiting the regulatory function of miR-92a (NCT03603431) (Table 3). Together, these miRNAs demonstrate the critical importance of miRNA regulation to cardiovascular health and further support the potential for miRNA as a therapeutic strategy.
Miravirsen (SPC3649), an antagomir of miR-122 (Table 3), is an example of miRNA antagonism therapy that was evaluated in phase II clinical trials for the treatment of hepatitis C (NCT01727934; NCT01872936; NCT01200420). miR-122 plays a central role in several aspects of liver function and is shown to stimulate hepatitis C virus (HCV) progression by binding the 5′UTR of the HCV genome (Hu et al., 2012). As an anti-miR, miravirsen has sequence complementarity to endogenous mature miR-122 and is composed of several locked nucleic acid (LNAs) ribonucleotides along a DNA PS sequence (Gebert et al., 2014). In its phase 2a clinical trial (NCT01200420), miravirsen-treated patients saw a dose-dependent reduction of HCV RNA levels (Janssen et al., 2013). This early success of antagomirs demonstrated the functionality of anti-miR therapy as the world’s first miRNA-targeted drug and provided a revolutionary drug to treat HCV (Lindow and Kauppinen, 2012).
Following the footsteps of the CDR132L, MRG-110, and miravirsen clinical trials, most current miRNA-based therapies in the clinic investigations are designed as anti-miR therapy discussed previously, whereas restoring miRNAs that are lost or downregulated in diseased cells represents a less tapped means meriting greater attention. As highly potent regulators of gene expression, miRNAs make widely applicable therapeutic candidates since their biologic function is to target several mRNA transcripts to potentially alter several cellular pathways (Catalanotto et al., 2016). This proves an opportunity for one therapeutic to target and alter the effects of several critical pathways found dysregulated in disease. However, it is this same natural property that limits the specificity of an miRNA as treatment to one disease causing pathway and increases the risk of off-target effects, thereby complicating its pharmacology and candidacy as a therapeutics, such as seen with MRX34 (Hong et al., 2020; Segal and Slack, 2020; Zhang et al., 2021b). This characteristic of miRNA has recently been termed “too many targets for miRNA effect” (TMTME), or in other words, the complication that one miRNA therapeutic candidate does indeed have the ability to target many mRNA transcripts and therefore adds another major obstacle unique to miRNA in the path of succuss in clinical trials (Montgomery et al., 2014; Zhang et al., 2021b). Although the probability for a single miRNA therapeutic to achieve its targeted effect is likely, the potency of miRNA toward effecting additional targets that are on-target for the miRNA itself but are off-target for the therapeutics, and can have unprecedented or unpreventable consequences depending on its pharmacological properties (Segal and Slack, 2020; Zhang et al., 2021b). Therefore, much work remains to be done to address toxic off-target effects and enhancing specific or targeted delivery platforms to develop and improve miRNA therapeutics with increased odds of clinical success.
However, in a recently published article researchers sought to tackle the challenge of TMTME by integrating data from multiomic studies with a developed algorithm to identify candidate miRNAs with the potential for miRNA-based therapy for Ewing Sarcoma with minimal effect on essential housekeeping genes (Weaver et al., 2021). Their approach estimates the “network potential” of a tumor based on collective transcriptomic and protein-protein interactions within the tumor, ranks relevant target mRNAs, and identifies prime miRNAs or miRNA combinations to repress those targets (Weaver et al., 2021). This work is a real-time example of current research being done to improve the odds of clinical success and personalize miRNA therapeutics for the greatest effect on cancer progression.
Approaches and Challenges in Delivering RNAi Therapeutics
One obstacle faced by all forms of medications, including RNAi therapeutics discussed in this review, is to ensure an effective and safe level of therapeutic agents to overcome systemic barriers and access the target in specific cells, tissues, or organs (Dowdy, 2017; Paunovska et al., 2022; Smith et al., 2022). Without a viable strategy for protection or tissue-selective delivery, therapeutics RNAs are subject to metabolic enzymes, such as circulating or liver localized hydrolaces and RNases, and rapid clearance by the kidneys after administration, both of which would limit intended target delivery. Nevertheless, hepatic and nonhepatic metabolism as well as renal and biliary clearance are critical components for the body to eliminate xenobiotic agents, including medications and toxins.
Furthermore, since RNAi agents typically interact with intercellular components and on intracellular targets, they need to cross the cellular membrane, which poses another major challenge to delivery due to the large molecular structure and strong, negatively charged nature of RNA and oligonucleotide molecules. Therefore, tailoring “delivery” strategies toward favorable pharmacokinetic properties, including biodistribution of the right levels of therapeutic RNAs to the target tissues and into the target cells to selectively access molecular targets, is critical to achieve the desired efficacy and safety among patients. The introduction of chemical modifications (Micklefield, 2001; Setten et al., 2019; Smith et al., 2022), as discussed previously, is a proven approach that provides an advantage by increasing RNA metabolic stability, ensuring efficacious pharmacological actions, and avoiding nonselective or adverse effects. As such, delivery strategies using chemically modified siRNA such as patisiran, givosiran, lumasiran, and inclisiran, as discussed previously, have attained FDA approval. However, chemical modifications introduce a disadvantage by compromising the initial chemical and physical characteristics and activities of naturally synthesized, modified, and folded RNA (Morena et al., 2018; Yu et al., 2019, 2020a,b). In fact, recent studies on inclisiran discussed previously suggest possible off-target effects of the therapeutic away from its FDA-designated biologic activity, providing the possibility for off-target effects even in RNAi designed with chemically modifications for target specificity (Christofides et al., 2021; Leqvio, 2021; Wang et al., 2022).
Delivery platforms can also be tailored to the unique characteristics of a diseased cell. For example, some delivery tactics connect RNAi agents to tissue- or receptor-specific ligands such as an antibody fragment, an entire antibody, or the FDA-approved tri-GalNAc conjugate to target specific receptors or membrane proteins for internalization (Givlaari, 2019; Seth et al., 2019; Maguregui and Abe, 2020; Oxlumo, 2020; Leqvio, 2021; Paunovska et al., 2022). Although conjugating unique ligands or antibodies may improve tissue specificity or disease targeting, available natural ligands and target receptor density can be limiting factors to an effective delivery and changing the chemical or physical makeup can add additional cost and complexity to both formulation and delivery (Cheng et al., 2012; Paunovska et al., 2022). RNA delivery with nanoparticles (NPs), such as FDA-approved LNPs for RNAi delivery discussed previously and polymer-based systems, is also commonly used to improve blood pharmacokinetics (Pitulle et al., 1995; Karra and Benita, 2012; Leung et al., 2012, 2014; Onpattro, 2018; Boca et al., 2020; Forterre et al., 2020; Paunovska et al., 2022; Smith et al., 2022). For example, in a recently published article, researchers implemented a liposome nanoparticle fitted with polymers consisting of six repeats of aspartate, serine, and serine [(DSS)6] that is shown to target bone formation surfaces to enhance the delivery of a casein kinase-2 interacting protein-1 (Ckip-1) mRNA targeting siRNA to osteogenic lineage cells involved in the progression of osteoporosis (Gao et al., 2022). Both in vitro and in vivo findings supported this novel liposome-based osteoanabolic therapy to treat osteoporosis by the payload siRNA (Gao et al., 2022).
By using NP formulation, an RNAi-based drug like patisiran can remain chemically unmodified or to a lesser degree but protected from degradation by serum RNases (Onpattro, 2018) (Table 4). However, NP-based formulations can be compromised by nonselective tissue distribution barring them from FDA approval for RNAi delivery (Table 4). Therefore, tissue- or receptor-specific ligands are under investigation and may be incorporated into NPs toward an optimal formulation to enhance target tissue delivery and drug internalization (Johannes and Lucchino, 2018; Dasgupta and Chatterjee, 2021; Mitchell et al., 2021; Paunovska et al., 2022; Smith et al., 2022).
TABLE 4.
Nonviral systems and vehicles for the delivery of RNAi therapeutics
| RNAi Delivery Means | System Composition | FDA-Approved RNAi Medications |
|---|---|---|
| Ribonucleoside modifications | Adenine 2'-F ribonucleoside | Yes – givosiran, lumasiran, inclisiran |
| Adenine 2'-OMe ribonucleoside | Yes – givosiran, lumasiran, inclisiran | |
| Cytosine 2'-F ribonucleoside | Yes – givosiran, lumasiran, inclisiran | |
| Cytosine 2'-OMe ribonucleoside | Yes – patisiran, givosiran, lumasiran, inclisiran | |
| Guanine 2'-F ribonucleoside | Yes – givosiran, inclisiran | |
| Guanine 2'-OMe ribonucleoside | Yes – givosiran, lumasiran, inclisiran | |
| Thymidine | Yes – patisiran, inclisiran | |
| Uracil 2'-F ribonucleoside | Yes – givosiran, lumasiran | |
| Uracil 2'-OMe ribonucleoside | Yes – patisiran, givosiran, lumasiran, inclisiran | |
| Phosphate linkage modifications | Phosphorothioate | Yes – givosiran, lumasiran, inclisiran |
| Charge-neutralizing phosphotriester | No | |
| Receptor ligand conjugates | Tri-GalNAc | Yes – givosiran, lumasiran, inclisiran |
| Antibody conjugates | Entire antibody | No |
| Antibody fragment | No | |
| LNP | DSPC | Yes – patisiran |
| PEG-lipids | Yes – patisiran | |
| Ionizable | Yes – patisiran | |
| Polymer-based NP | PEI | No |
| PEI-PEG | No | |
| Polyurethane | No | |
| PACE | No | |
| Micelle | No | |
| Inorganic material-based NP | Gold-mediated | No |
| Mesoporous silicon | No | |
| Graphene oxide mediated | No | |
| Fe3O4 mediated | No | |
| Liposome | Neutral | No |
| Cationic | No | |
| pH-sensitive cationic | No | |
| Ionizable | No | |
| Chemotherapeutic coated | No | |
| Exosome | Exosome | No |
| Exosome-GE11 peptide | No | |
| ELV | No |
DSPC, 1,2-distearoyl-sn-glycero-3-phosphocholine; ELV, exosome-like vesicle; Fe3O4, iron(III) oxide; PACE, poly(amine-co-ester); PEI, polyethylenimine.
Although an adaptable means of delivery, there is the concern about risk of inducing immunogenic effects by non-natural NPs themselves, beyond the RNAi macromolecules, and the barrier for endosomal escape once endocytosed (Kumar et al., 2015; Lu et al., 2018; Xu et al., 2018). To address these concerns, employing exosome-based delivery offers an alternative to NPs as endogenous vehicles under investigation that can load, protect, and deliver therapeutic RNAs (Kumar et al., 2015; Ha et al., 2016; Barile and Vassalli, 2017; Lu et al., 2018). Exosomes are endogenous extracellular vesicles that play important roles in cell-cell communications and can transfer genetic and biochemical information and fuse with cell membranes to directly deliver cargo into the cytoplasm (Lu et al., 2018). Using exosomes over traditional NPs provides a more advantageous natural means of drug delivery that is hoped to increase delivery, membrane permeation efficiency, and biocompatibility to overcome both immunogenicity and endosomal escape (Alvarez-Erviti et al., 2011; van den Boorn et al., 2011; Shtam et al., 2013; Haney et al., 2015; Ha et al., 2016; Lu et al., 2018; Smith et al., 2022). One example is the exosome-GE11 peptide, a modified exosome with a surface GE11 peptide designed to target the epidermal growth factor receptor (EGFR) to deliver RNAi agents to EGFR-expressing cancer tissues (Ohno et al., 2013). However, the use of exosomes comes with some disadvantages, including inefficient or low extraction and isolation yield as well as exosome encapsulation and loading of hydrophilic molecules and the delivery of unwanted, off-target components inherent in exosome composition, providing an opportunity for researchers to investigate and improve exosome extraction methodology, isolation yield, and loading prior to FDA approval for RNAi delivery (Akuma et al., 2019).
Other methods of RNA delivery under investigation include viral systems to express genes holding their RNAi into desired tissues; inorganic material-based NPs such as gold-, mesoporous silicon-, graphene oxide-, or iron(III) oxide (Fe3O4)-mediated NPs; and polymeric vectors, among others (Dowdy, 2017; Fu et al., 2019; Paunovska et al., 2022). Each class of RNA delivery comes with both advantages and disadvantages, increasing the difficulty of choosing the most appropriate delivery vehicle. Viral systems including adenoviral, adeno-associated viral, retroviral, and lentiviral vectors are useful for long-term gene expression by transferring genes into different target tissues (Fu et al., 2019). However, viral delivery is complicated by the induction of immunogenicity and toxicity as well as a low loading capacity, impeding FDA approval for RNAi delivery (Vannucci et al., 2013; Fu et al., 2019) (Table 4). Gold-mediated NPs can interact with thiol and amino functional groups that can be conjugated to RNAi agents to enhance loading and delivery (Chen et al., 2017; Fu et al., 2019; Cai et al., 2020). Mesoporous silica-mediated NPs provide favorable biocompatibility and a large, easily modified, and thermodynamically stable surface area (Mamaeva et al., 2013; Fu et al., 2019). Graphene oxide–mediated NPs contain a unique honeycomb-like network that can absorb a wide array of nucleic acids (Loh et al., 2010; Fu et al., 2019). Fe3O4-mediated NPs can form nanocomplexes with mesoporous magnetic clusters and link with polymeric vectors such as polyethylenimine (PEI) or polyacrylic acid (PAA) to load RNAi agents and increase uptake in vivo (Sun et al., 2017; Fu et al., 2019). However, much like viral based vectors, most nonviral vectors are limited in current clinical research and lack FDA-approved use for RNAi delivery because of their toxicity and potential for off-target delivery (Fu et al., 2019) (Table 4).
In a recent publication, researchers had developed a novel delivery system for miRNA based on in situ self-assembly between gold (Au) salts and tumor suppressor miRNA mimics to form Au-miRNA nanocomplexes (Cai et al., 2020). In this study, in situ self-assembled Au-miRNA nanocomplexes were found not only to be present in cancer cells and to inhibit proliferation in vitro but also demonstrated tumor suppression and enhanced antitumor effects in subcutaneous tumor models in vivo (Cai et al., 2020). Their novel and current work provides supportive evidence for NP delivery strategies to effectively transport large and negatively charged RNA and oligonucleotide molecules cargo. However, the most important aspect in the pursuit of “targeted delivery” is to develop and formulate a drug to improve its pharmacokinetics, pharmacodynamics, and safety profile all while simultaneously maintaining the drugs functionality and, in the case of RNAi, its interactions with endogenous RNAi machinery and intracellular target transcripts at the right level and right time to achieve the right outcomes (Yu et al., 2019; Zhang et al., 2021b).
Novel Biotechnologies To Produce RNAi Agents
Although chemical and biochemical synthesis remains a consistent means of RNA molecule production, there is a growing concern that these methods introduce changes to the physical and chemical properties of RNAi agents that likely exhibit distinct efficacy and safety profiles from those of naturally synthesized and modified RNA equivalents (Morena et al., 2018; Yu et al., 2019, 2020a,b). To address these concerns, alternative methods have emerged to steer RNA production back into live cells for natural products. Two genres of in vivo RNA production are through direct expression using specific host bacteria strains and RNA production using stable carriers (Fig. 3).
Fig. 3.
Novel biotechnologies to produce biologic RNAi agents. (A) Direct expression is one class of novel RNAi agent production. Two forms of direct expression make use of two bacterial strains, Rhodovulum sulfidophilum and an RNase III–deficient Corynebacterium glutamicum. The marine phototropic bacterium R. sulfidophilum has the capability to efflux oligonucleotides such as RNA but not RNases. These characteristics allow R. sulfidophilium to be transformed with an RNA expressing plasmids for direct expression of the target RNA followed by efflux and accumulation in RNase free culture media. Accumulated RNA in culture media can then be isolated and purified to attain the target RNA molecule. A novel strain of C. glutamicum contains a mutation in the RNase III gene (2256LΔrnc) and provides an RNase III ribonuclease–free bacterium for RNA accumulation. Target RNA may be directly expressed and accumulated in C. glutamicum free from RNase degradation for later isolation from cell lysate and purification. (B) The use of stable carriers constitutes the other class of novel RNAi agent production. Transfer RNA (tRNA) and ribosomal RNA (rRNA) can each function as stable RNA scaffolds. By retaining structures and sequences of these highly abundant RNA classes, target RNA is expected to exploit endogenous recognition and accumulate within bacteria. As most recombinant RNAs cannot be overexpressed with tRNA or rRNA scaffold, specific hybrid tRNA/pre-miRNA molecules showing high-level expression in E. coli have been identified, developed, and proven as unique carriers to effectively accommodate a wide variety of target RNAi molecules including RNA aptamers, miRNAs, siRNAs, or sRNAs along with their complimentary sequences for high-yield and large-scale production of biologic RNAi agents.
Direct Expression
In vivo RNA production by direct expression was initiated by the identification of two bacterial strains, Rhodovulum sulfidophilum and an RNase III–deficient Corynebacterium glutamicum (Pereira et al., 2017; Hashiro et al., 2019b) (Fig. 3). R. sulfidophilum is a marine phototrophic bacterium with the capability to efflux nucleic acids but not RNases (Nagao et al., 2015; Pereira et al., 2017; Kikuchi and Umekage, 2018) (Fig. 3). Such characteristics allow R. sulfidophilum to accumulate RNA products in culture medium without fear of nuclease degradation and has shown success in producing a variety of RNA molecules such as tRNAs, rRNAs, aptamers (Ando et al., 2006; Suzuki et al., 2010; Nagao et al., 2014), and human pre-miRNAs (Pereira et al., 2016). RNA accumulated in culture media can be isolated and purified to attain the target RNA molecule (Kikuchi and Umekage, 2018; Yu et al., 2020a) (Fig. 3). However, direct expression by R. sulfidophilum demonstrated a low yield of recombinant RNA production that is likely due to an ineffective diffusion or efflux of RNA into the medium or the presence of intracellular RNases that can readily degrade recombinant RNAs before they can exit the cells and accumulate in medium (Pereira et al., 2017; Kikuchi and Umekage, 2018; Yu et al., 2020a).
Recent exploration into a novel strain of C. glutamicum is being studied to produce RNA molecules in vivo through direct expression (Hashiro et al., 2019a,b) (Fig. 3). This particular strain of C. glutamicum lacks the RNase III ribonuclease because of a disruption in its encoding gene (2256LΔrnc) (Hashiro et al., 2019a,b). Therefore, in combination with a strong promoter driving the overexpression of recombinant RNA, target RNA can be accumulated in RNase III– deficient C. glutamicum toward mass production (Hashiro et al., 2019a,b) (Fig. 3). This novel strain is expected to have direct application to in vivo production of biologic RNAs (Yu et al., 2020a).
Another form of direct expression is coexpression of target RNA molecules with protective RNA-binding proteins. Plant RNA virus tombusvirus-encoded 19-kDa protein (p19) is an RNA-binding protein known to selectively bind and suppress the RNAi function of double-stranded siRNAs with high affinity, and it is being implemented today for the isolation, detection, and stabilization of both siRNA and miRNA (Qiu et al., 2002; Silhavy et al., 2002). Through coexpression, siRNA embedded within an shRNA is designed to be processed by bacterial RNases to target siRNAs and form stable complexes with p19 to avoid degradation (Qiu et al., 2002; Huang et al., 2013). After coexpression, total protein containing siRNA-p19 complexes can be extracted from bacteria and purified, and the complexes can be dissociated with denaturing agents (e.g., SDS) for the fully processed and protected RNAi agents to be purified (Yu et al., 2020a).
Using Stable Carriers
The use of stable carriers offers an alternative to direct expression. There are several stable carriers available to produce recombinant RNA molecules, including viroid-derived circRNA carriers, tRNA or rRNA scaffolds, and chimeric tRNA/pre-miRNA carriers. The viroid-derived circRNA carrier is a relatively new form of stable carrier capable of producing large amounts of stable recombinant RNAs in Escherichia coli (Daròs et al., 2018). Viroids are a special class of infectious agents that contain short, single-stranded circRNAs found in higher plant species (Branch and Robertson, 1984; Branch et al., 1988). The viroid-derived circRNA carrier uses the coexpression of two plasmids to produce and circularize recombinant RNA molecules into viroids (Daròs et al., 2018). The first plasmid offers the recombinant RNA-viroid molecule from the eggplant latent viroid (ELVd) to form the pLELVd-BZB plasmid (Daròs et al., 2018), and the second plasmid produces the tRNA ligase used for viroid circularization into chimeric circRNA (Daròs et al., 2018). The viroid-carrying target RNA can be isolated from bacterial cell lysate and purified by gel electrophoresis or fast protein liquid chromatography (FPLC) methods (Daròs et al., 2018).
Another form of stable carriers uses rRNA as a scaffold to produce recombinant RNA molecules in bacteria (Zhang et al., 2009) (Fig. 3). As the most abundant species of RNA found in living cells by mass, rRNA-based carriers resembling rRNA exploit endogenous recognition and are capable of accommodating and protecting target RNAs (Zhang et al., 2009). The 5S rRNA is used as a scaffold because of its stability to carry and protect a variety of sRNA molecules with DNAzyme-specific sequences within its stem II and stem III site structures (Pitulle et al., 1995; D'Souza et al., 2003; Zhang et al., 2009; Liu et al., 2010) (Fig. 3). By using this stable carrier, a plasmid containing the 5S rRNA-scaffold with an embedded target RNA can be overexpressed in bacteria by rRNA gene promoters rrnB P1 and P2 and rRNA transcription terminators rrnB T1 and T2 prior to recombinant RNA isolation and purification from cell lysate (Zhang et al., 2009; Liu et al., 2010). Target RNAs can then be selectively released from their rRNA-scaffold by DNAzyme-mediated recovery (Zhang et al., 2009; Liu et al., 2010).
Compared with rRNA, the tRNA offers a simple scaffold to produce recombinant RNA molecules in bacteria (Ponchon and Dardel, 2007; Ponchon et al., 2009; Nelissen et al., 2012) (Fig. 3). tRNA is an endogenous molecule found with high abundance in living cells; the tRNA scaffold uses intracellular recognition of the tRNA structure to carry recombinant RNA for overexpression and accumulation in bacteria (Meinnel et al., 1988; Gaudin et al., 2003). In this design, the tRNA anticodon sequence is replaced with an RNA sequence of interest all while leaving the remainder of the tRNA sequence intact to conserve the recognizable cloverleaf structure and overall stability of the tRNA in vivo (Ponchon and Dardel, 2007) (Fig. 3). To implement this stable carrier in vivo, a plasmid containing the recombinant RNA within a tRNA scaffold is overexpressed in E. coli, driven by a murein lipoprotein (lpp) promoter (Ponchon et al., 2009) or T7 promoter (Nelissen et al., 2012), and the overexpressed recombinant RNA is then isolated and purified from cell lysate. The tRNA scaffold has been successfully employed for the design and production of various RNA molecules, including viral RNAs and aptamers (Ponchon and Dardel, 2007), pre-miRNAs (Li et al., 2014, 2015; Wang et al., 2015), and snoRNAs (Peng et al., 2014).
Recently, a novel hybrid tRNA/pre-miRNA carrier platform has been established for large-scale production of bioengineered RNA agents (BioRNA, previously termed BERA) by in vivo fermentation with high yield and purity (Chen et al., 2015; Ho et al., 2018; Li et al., 2021; Petrek et al., 2021) (Fig. 3). The BioRNA consists of an ssRNA molecule with three major components, a tRNA scaffold linked to a pre-miR-34a sequence with an embedded, interchangeable miRNA duplex. As a result, BioRNA can accommodate a wide verity of target RNAi molecules, including RNA aptamers and shRNA as well as miRNA, siRNA, or sRNA along with their complimentary sequence (Ho and Yu, 2016; Yu et al., 2019, 2020a; Yu and Tu, 2022). After introduced into human cells, it is likely that BioRNA enters into endogenous miRNA processing at the pre-miRNA step in the cytoplasm (Dominska and Dykxhoorn, 2010; Yu et al., 2020a) (Fig. 1). However, both canonical processing and selective RNAi agent release (Chen et al., 2015; Ho et al., 2018; Jilek et al., 2019; Tu et al., 2019; Petrek et al., 2021) and Dicer-independent, noncanonical processing (Ho et al., 2018) of BioRNAs have been demonstrated, dependent on the RNAi sequence incorporated into the BioRNA.
In its initial design, BioRNA used a chimeric bacterial methionyl tRNA (btRNAMet) fused with human hsa-pre-miR-34a (btRNAMet/hsa-pre-miR-34a) to determine its capability of overexpression in bacteria (Chen et al., 2015). What separates hybrid BioRNA from tRNA stable carriers is the inclusion of a pre-miR-34a as a critical component designed for effective and natural cleavage, guidance, and incorporation into the RISC complex by endogenous machinery (Li et al., 2021). To improve compatibility in human cells, the BioRNA design was adapted by substituting the bacterial tRNA with human versions (Li et al., 2021). Several human tRNA (htRNA) were screened and identified to effetely couple with hsa-pre-miR-34a, among them seryl (htRNASer) and leucyl (htRNALeu) htRNAs, which led to an improved overexpression in E. coli, making up over 40% of the total bacterial RNA (Li et al., 2021).
The BioRNA carrier enables recombinant RNA production in live culture to yield more natural RNA molecules (Chen et al., 2015; Ho et al., 2018; Li et al., 2021). To do so, the BioRNA coding sequence is cloned into a target vector with a lipoprotein promoter, and the BioRNA-expressing plasmid is transformed into E. coli for overexpression of the recombinant RNA (Li et al., 2021). Total RNA is then extracted from cell lysate and purified by anion exchange fast protein liquid chromatography to yield an BioRNA product with preserved structure, stability, activity, and safety like that of endogenous RNAi molecules (Wang et al., 2015; Ho et al., 2018; Li et al., 2021). The resulting products are high-quality, biologically active RNA molecules that are well tolerated in animal models and resemble the functions of endogenous cellular RNAi mechanisms (Chen et al., 2015; Li et al., 2015; Jian et al., 2017; Jilek et al., 2017, 2019; Alegre et al., 2018; Ho et al., 2018; Zhang et al., 2018; Li et al., 2019; Tu et al., 2019, 2020; Xu et al., 2019; Umeh-Garcia et al., 2020; Yi et al., 2020; Deng et al., 2021; Li et al., 2021).
Conclusions and Perspectives
Since its discovery, evidence suggests that RNAi is an adaptable and versatile tool that can be modified by researchers to study reverse genetics and specific gene repression and to develop targeted therapies. More importantly, the components that make up the RNAi pathway are not a static set of sequential steps but instead an expanding pool of adaptable components that function to deliver gene-specific regulation (RNAi Molecules and Mechanisms) (Fig. 1). This is evident in new and developing technologies that use the foundations of RNAi to develop novel synthesis; improve functionality, potency, stability, and pharmacology (Fig. 3); establish new methods of delivery (Table 4); and form groundbreaking therapeutic strategies against diseases by acting on previously considered un- or nondruggable targets. This notion is supported in the wake of the fourth therapeutic siRNA to gain approval by the FDA (inclisiran) and is further supported by the numerous RNAi-based therapeutics advancing through clinical trials (Tables 1–3). However, with most RNAi therapeutics functioning as siRNAs (Tables 1 and 2) or anti-miRs, there are few miRNA mimics making it into clinical trials (Table 3). This leaves a vastly untapped market for miRNA replacement therapy. miRNA has the unique property of targeting several mRNA transcripts and creates an opportunity for a single drug to regulate multiple targets or biologic pathways that are often dysregulated in a disease. To achieve this, factors such as dosing, cross-reactivity, on-target efficacy, and unwanted effects need to be addressed through comprehensive basic and clinical research before they can attain regulatory approval.
Efforts continue to be made to bring the production of various RNAi molecules back into the cells and away from chemical modifications by using direct expression in bacteria and by using stable carriers to preserve these natural properties while continuing to maintain stability, biologic activity, and safety as discussed in Novel Biotechnologies To Produce RNAi Agents. Among them, the tRNA/pre-miRNA carrier-based platform technology has proven to be a robust and versatile means of harnessing in vivo RNA production of biologically active miRNAs, siRNAs, aptamers, and sRNAs that are designed to recapitulate several of the physical and chemical properties of natural RNA molecules needed for RNAi-based therapy. Future research exploring the use of bioengineered or recombinant RNAi molecules against chemo-engineered RNA analogs will provide insight and define their efficacy as biochemical candidates for both biologic research and as clinical treatments for disease.
Abbreviations
- AGO2
argonaut 2 endonuclease
- AHP
acute hepatic porphyria
- AKI
acute kidney injury
- ALAS1
aminolevulinate synthase 1
- anti-miR
miRNA antagonism
- ASCVD
atherosclerotic cardiovascular disease
- ASO
antisense oligonucleotide
- BioRNA
bioengineered RNA agent
- CDS
coding DNA sequence or protein coding region
- circRNA
circular RNA
- DED
dry eye disease
- DGCR8
DiGeorge syndrome critical region 8
- EGFR
epidermal growth factor receptor
- FDA
US Food and Drug Administration
- GalNAc
N-acetylgalactosamine
- HAO1
hydroxyacid oxidase 1
- hATTR
hereditary transthyretin-mediated amyloidosis
- HCV
hepatitis C virus
- HeFH
heterozygous familial hypercholesterolemia
- HhtRNA
humanized tRNA
- IVT
in vitro transcription
- LDL-C
low-density lipoprotein cholesterol
- LNP
lipid nanoparticle
- miR or miRNA
microRNA
- miRISC
microRNA in RNA-induced silencing complex
- ncRNA
noncoding RNA
- NP
nanoparticle
- NSCLC
non–small cell lung cancer
- OLE
open-label extension
- p19
plant tombusvirus-encoded 19-kDa protein
- PCSK9
proprotein convertase subtilisin/kexin type 9
- PEG
polyethylene glycol
- PH1
primary hyperoxaluria type 1
- PPARγ
peroxisome proliferator-activated receptor gamma
- pre-miRNA
precursor microRNA
- pri-miRNA
primary microRNA
- PS
phosphorothioate
- PTX/CBP
paclitaxel plus carboplatin
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- rRNA
ribosomal RNA
- shRNA
short hairpin RNA
- siRISC
small interfering RNA in RNA-induced silencing complex
- siRNA
small interfering RNA
- snoRNA
small nucleolar RNA
- sRNA
small RNA
- ssRNA
single-stranded RNA
- TRBP
transactivating response RNA-binding protein
- tRNA
transfer RNA
- TTR
transthyretin
- 3′UTR
3′ untranslated region
Authorship Contributions
Performed data analysis: Traber, Yu.
Wrote or contributed to the writing of the manuscript: Traber, Yu.
Footnotes
This work was supported by National Institutes of Health National Institute of General Medical Sciences [Grant R35-GM140835], National Institute of General Medical Sciences-funded pharmacology training program [Grant T32-GM099608] (to G.M.T.), and National Cancer Institute [Grant R01-CA225958] and [Grant R01-CA253230].
No author has an actual or perceived conflict of interest with the contents of this article.
References
- Abifadel MVarret MRabès JPAllard DOuguerram KDevillers MCruaud CBenjannet SWickham LErlich D, et al. (2003) Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 34:154–156. [DOI] [PubMed] [Google Scholar]
- Adams DGonzalez-Duarte AO’Riordan WDYang CCUeda MKristen AVTournev ISchmidt HHCoelho TBerk JL, et al. (2018) Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med 379:11–21. [DOI] [PubMed] [Google Scholar]
- Adams DPolydefkis MGonzález-Duarte AWixner JKristen AVSchmidt HHBerk JLLosada López IADispenzieri AQuan D, et al. ; patisiran global OLE study group (2021) Long-term safety and efficacy of patisiran for hereditary transthyretin-mediated amyloidosis with polyneuropathy: 12-month results of an open-label extension study. Lancet Neurol 20:49–59. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Simon AR, Goel V, Habtemariam BA, Clausen VA, Kim JB, Robbie GJ (2020) Pharmacokinetics and pharmacodynamics of the small interfering ribonucleic acid, givosiran, in patients with acute hepatic porphyria. Clin Pharmacol Ther 108:63–72. [DOI] [PubMed] [Google Scholar]
- Akuma P, Okagu OD, Udenigwe CC (2019) Naturally occurring exosome vesicles as potential delivery vehicle for bioactive compounds. Front Sustain Food Syst DOI: 10.3389/fsufs.2019.00023. [Google Scholar]
- Alegre F, Ormonde AR, Snider KM, Woolard K, Yu AM, Wittenburg LA (2018) A genetically engineered microRNA-34a prodrug demonstrates anti-tumor activity in a canine model of osteosarcoma. PLoS One 13:e0209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ (2011) Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29:341–345. [DOI] [PubMed] [Google Scholar]
- Amarzguioui M, Rossi JJ, Kim D (2005) Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett 579:5974–5981. [DOI] [PubMed] [Google Scholar]
- Anderson KE (2019) Acute hepatic porphyrias: current diagnosis & management. Mol Genet Metab 128:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Suzuki H, Nishimura S, Tanaka T, Hiraishi A, Kikuchi Y (2006) Characterization of extracellular RNAs produced by the marine photosynthetic bacterium Rhodovulum sulfidophilum. J Biochem 139:805–811. [DOI] [PubMed] [Google Scholar]
- Babar IA, Czochor J, Steinmetz A, Weidhaas JB, Glazer PM, Slack FJ (2011) Inhibition of hypoxia-induced miR-155 radiosensitizes hypoxic lung cancer cells. Cancer Biol Ther 12:908–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Brown D, Winkler M (2010) The promise of microRNA replacement therapy. Cancer Res 70:7027–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwani MSardh EVentura PPeiró PARees DCStölzel UBissell DMBonkovsky HLWindyga JAnderson KE, et al. ; ENVISION Investigators (2020) Phase 3 trial of RNAi therapeutic givosiran for acute intermittent porphyria. N Engl J Med 382:2289–2301. [DOI] [PubMed] [Google Scholar]
- Bardolia C, Amin NS, Turgeon J (2021) Emerging non-statin treatment options for lowering low-density lipoprotein cholesterol. Front Cardiovasc Med 8:789931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile L, Vassalli G (2017) Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther 174:63–78. [DOI] [PubMed] [Google Scholar]
- Beaucage SL (2008) Solid-phase synthesis of siRNA oligonucleotides. Curr Opin Drug Discov Devel 11:203–216. [PubMed] [Google Scholar]
- Beckert B, Masquida B (2011) Synthesis of RNA by in vitro transcription. Methods Mol Biol 703:29–41. [DOI] [PubMed] [Google Scholar]
- Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS (2017) Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs 35:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC (2007) Mammalian mirtron genes. Mol Cell 28:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk C, Civenni G, Wang Y, Steuer C, Catapano CV, Hall J (2021) Pharmacodynamic and pharmacokinetic properties of full phosphorothioate small interfering RNAs for gene silencing in vivo. Nucleic Acid Ther 31:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besur S, Hou W, Schmeltzer P, Bonkovsky HL (2014) Clinically important features of porphyrin and heme metabolism and the porphyrias. Metabolites 4:977–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell DM, Anderson KE, Bonkovsky HL (2017) Porphyria. N Engl J Med 377:862–872. [DOI] [PubMed] [Google Scholar]
- Bissell MAnderson KEBonkovsky DHLSardh EHarper PBalwani MRees DStein PBloomer JRParker C, et al. (2019) A phase 1/2 open-label extension study of givosiran, an investigational RNAi therapeutic, in patients with acute intermittent porphyria. Eur J Neurol 73:S553–S554 DOI: 10.21037/tcr.2016.11.37. [Google Scholar]
- Boca S, Gulei D, Zimta AA, Onaciu A, Magdo L, Tigu AB, Ionescu C, Irimie A, Buiga R, Berindan-Neagoe I (2020) Nanoscale delivery systems for microRNAs in cancer therapy. Cell Mol Life Sci 77:1059–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau E, Neveu B, Kostantin E, Tsongalis GJ, De Guire V (2019) How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC 30:114–127. [PMC free article] [PubMed] [Google Scholar]
- Bramsen JBLaursen MBNielsen AFHansen TBBus CLangkjaer NBabu BRHøjland TAbramov MVan Aerschot A, et al. (2009) A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res 37:2867–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch AD, Benenfeld BJ, Robertson HD (1988) Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc Natl Acad Sci USA 85:9128–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch AD, Robertson HD (1984) A replication cycle for viroids and other small infectious RNA’s. Science 223:450–455. [DOI] [PubMed] [Google Scholar]
- Brandts J, Ray KK (2021) Clinical implications and outcomes of the ORION phase III trials. Future Cardiol 17:769–777. [DOI] [PubMed] [Google Scholar]
- Cai WFeng HYin LWang MJiang XQin ZLiu WLi CJiang HWeizmann Y, et al. (2020) Bio responsive self-assembly of Au-miRNAs for targeted cancer theranostics. EBioMedicine 54:102740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarotto M, Fanetti G, Guerrieri R, Palazzari E, Lupato V, Steffan A, Polesel J, Boscolo-Rizzo P, Fratta E (2020) Beyond microRNAs: emerging role of other non-coding RNAs in HPV-driven cancers. Cancers (Basel) 12:1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaman G, Matino D (2019) Hemophilia A and B: molecular and clinical similarities and differences. Haematologica 104:1702–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C, Cogoni C, Zardo G (2016) MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci 17:1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS (2017) Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol Ther Nucleic Acids 8:132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ (2010) A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QX, Wang WP, Zeng S, Urayama S, Yu AM (2015) A general approach to high-yield biosynthesis of chimeric RNAs bearing various types of functional small RNAs for broad applications. Nucleic Acids Res 43:3857–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Xianyu Y, Jiang X (2017) Surface modification of gold nanoparticles with small molecules for biochemical analysis. Acc Chem Res 50:310–319. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Al Zaki A, Hui JZ, Muzykantov VR, Tsourkas A (2012) Multifunctional nanoparticles: cost versus benefit of adding targeting and imaging capabilities. Science 338:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofides A, Konstantinidou E, Jani C, Boussiotis VA (2021) The role of peroxisome proliferator-activated receptors (PPAR) in immune responses. Metabolism 114:154338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochat P, Rumsby G (2013) Primary hyperoxaluria. N Engl J Med 369:649–658. [DOI] [PubMed] [Google Scholar]
- Coelho TAdams DSilva ALozeron PHawkins PNMant TPerez JChiesa JWarrington STranter E, et al. (2013) Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med 369:819–829. [DOI] [PubMed] [Google Scholar]
- Cortez MA, Anfossi S, Ramapriyan R, Menon H, Atalar SC, Aliru M, Welsh J, Calin GA (2019) Role of miRNAs in immune responses and immunotherapy in cancer. Genes Chromosomes Cancer 58:244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costales MG, Haga CL, Velagapudi SP, Childs-Disney JL, Phinney DG, Disney MD (2017) Small molecule inhibition of microRNA-210 reprograms an oncogenic hypoxic circuit. J Am Chem Soc 139:3446–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza LM, Larios-Sanz M, Setterquist RA, Willson RC, Fox GE (2003) Small RNA sequences are readily stabilized by inclusion in a carrier rRNA. Biotechnol Prog 19:734–738. [DOI] [PubMed] [Google Scholar]
- Daròs JA, Aragonés V, Cordero T (2018) A viroid-derived system to produce large amounts of recombinant RNA in Escherichia coli. Sci Rep 8:1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta I, Chatterjee A (2021) Recent advances in miRNA delivery systems. Methods Protoc 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson BL, McCray PB Jr (2011) Current prospects for RNA interference-based therapies. Nat Rev Genet 12:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debacker AJ, Voutila J, Catley M, Blakey D, Habib N (2020) Delivery of oligonucleotides to the liver with GalNAc: from research to registered therapeutic drug. Mol Ther 28:1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Petrek H, Tu MJ, Batra N, Yu AX, Yu AM (2021) Bioengineered miR-124-3p prodrug selectively alters the proteome of human carcinoma cells to control multiple cellular components and lung metastasis in vivo. Acta Pharm Sin B 11:3950–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432:231–235. [DOI] [PubMed] [Google Scholar]
- Dimopoulos K, Gimsing P, Grønbæk K (2013) Aberrant microRNA expression in multiple myeloma. Eur J Haematol 91:95–105. [DOI] [PubMed] [Google Scholar]
- Dominska M, Dykxhoorn DM (2010) Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci 123:1183–1189. [DOI] [PubMed] [Google Scholar]
- Donzé O, Picard D (2002) RNA interference in mammalian cells using siRNAs synthesized with T7 RNA polymerase. Nucleic Acids Res 30:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdy SF (2017) Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol 35:222–229. [DOI] [PubMed] [Google Scholar]
- Dungu JN, Anderson LJ, Whelan CJ, Hawkins PN (2012) Cardiac transthyretin amyloidosis. Heart 98:1546–1554. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4:721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bezawy RTinelli STortoreto MDoldi VZuco VFolini MStucchi CRancati TValdagni RGandellini P, et al. (2019) miR-205 enhances radiation sensitivity of prostate cancer cells by impairing DNA damage repair through PKCε and ZEB1 inhibition. J Exp Clin Cancer Res 38:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Daly SM, Abba ML, Gamal-Eldeen AM (2017) The role of microRNAs in photodynamic therapy of cancer. Eur J Med Chem 142:550–555. [DOI] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G (2008) A human snoRNA with microRNA-like functions. Mol Cell 32:519–528. [DOI] [PubMed] [Google Scholar]
- Fan J, de Lannoy IA (2014) Pharmacokinetics. Biochem Pharmacol 87:93–120. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KWhite SBorodovsky ABettencourt BRStrahs AClausen VWijngaard PHorton JDTaubel JBrooks A, et al. (2017) A highly durable RNAi therapeutic inhibitor of PCSK9. N Engl J Med 376:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre A, Komuro H, Aminova S, Harada M (2020) A comprehensive review of cancer microRNA therapeutic delivery strategies. Cancers (Basel) 12:1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster DJBrown CRShaikh STrapp CSchlegel MKQian KSehgal ARajeev KGJadhav VManoharan M, et al. (2018) Advanced siRNA designs further improve in vivo performance of GalNAc-siRNA conjugates. Mol Ther 26:708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis AJ, Resendiz MJE (2017) Protocol for the solid-phase synthesis of oligomers of RNA containing a 2'-O-thiophenylmethyl modification and characterization via circular dichroism. J Vis Exp 125:56189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MRLizano EHouben AJBezdan DBáñez-Coronel MKudla GMateu-Huertas EKagerbauer BGonzález JChen KC, et al. (2014) Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol 15:R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishberg YDeschênes GGroothoff JWHulton SAMagen DHarambat JVan’t Hoff WGLorch UMilliner DSLieske JC, et al. ; study collaborators (2021) Phase 1/2 study of lumasiran for treatment of primary hyperoxaluria type 1: a placebo-controlled randomized clinical trial. Clin J Am Soc Nephrol 16:1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Chen J, Huang Z (2019) Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA 1:24 DOI: 10.1186/s41544-019-0024-y. [Google Scholar]
- Gaglione M, Messere A (2010) Recent progress in chemically modified siRNAs. Mini Rev Med Chem 10:578–595. [DOI] [PubMed] [Google Scholar]
- Gallagher KM, O’neill S, Harrison EM, Ross JA, Wigmore SJ, Hughes J (2017) Recent early clinical drug development for acute kidney injury. Expert Opin Investig Drugs 26:141–154. [DOI] [PubMed] [Google Scholar]
- Gao Y, Xin H, Cai B, Wang L, Lv Q, Hou Y, Liu F, Dai T, Kong L (2022) RNA interference-based osteoanabolic therapy for osteoporosis by a bone-formation surface targeting delivery system. Biochem Biophys Res Commun 601:86–92. [DOI] [PubMed] [Google Scholar]
- Garrelfs SFFrishberg YHulton SAKoren MJO’Riordan WDCochat PDeschênes GShasha-Lavsky HSaland JMVan’t Hoff WG, et al. ; ILLUMINATE-A Collaborators (2021) Lumasiran, an RNAi therapeutic for primary hyperoxaluria type 1. N Engl J Med 384:1216–1226. [DOI] [PubMed] [Google Scholar]
- Gaudin C, Nonin-Lecomte S, Tisné C, Corvaisier S, Bordeau V, Dardel F, Felden B (2003) The tRNA-like domains of E. coli and A. aeolicus transfer-messenger RNA: structural and functional studies. J Mol Biol 331:457–471. [DOI] [PubMed] [Google Scholar]
- Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J (2014) Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res 42:609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz MA (2016) Amyloidosis, in Goldman-Cecil Medicine, 25th ed (Goldman L, Schafer AI, eds) vol 1, Elsevier/Saunders, Philadelphia, PA. [Google Scholar]
- Givlaari. (2019) Package insert. Alnylam Pharmaceuticals, Cambridge, MA. [Google Scholar]
- González-Duarte ABerk JLQuan DMauermann MLSchmidt HHPolydefkis MWaddington-Cruz MUeda MConceição IMKristen AV, et al. (2020) Analysis of autonomic outcomes in APOLLO, a phase III trial of the RNAi therapeutic patisiran in patients with hereditary transthyretin-mediated amyloidosis. J Neurol 267:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiley KZ, Pratt AJ, MacRae IJ (2012) Single-pot enzymatic synthesis of Dicer-substrate siRNAs. Nucleic Acids Res 40:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha D, Yang N, Nadithe V (2016) Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B 6:287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524. [DOI] [PubMed] [Google Scholar]
- Habtemariam BAKarsten VAttarwala HGoel VMelch MClausen VAGarg PVaishnaw AKSweetser MTRobbie GJ, et al. (2021) Single-dose pharmacokinetics and pharmacodynamics of transthyretin targeting N-acetylgalactosamine-small interfering ribonucleic acid conjugate, vutrisiran, in healthy subjects. Clin Pharmacol Ther 109:372–382. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950–952. [DOI] [PubMed] [Google Scholar]
- Han YN, Li Y, Xia SQ, Zhang YY, Zheng JH, Li W (2017) PIWI proteins and PIWI-interacting RNA: emerging roles in cancer. Cell Physiol Biochem 44:1–20. [DOI] [PubMed] [Google Scholar]
- Hand TW, Cui W, Jung YW, Sefik E, Joshi NS, Chandele A, Liu Y, Kaech SM (2010) Differential effects of STAT5 and PI3K/AKT signaling on effector and memory CD8 T-cell survival. Proc Natl Acad Sci USA 107:16601–16606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney MJKlyachko NLZhao YGupta RPlotnikova EGHe ZPatel TPiroyan ASokolsky MKabanov AV, et al. (2015) Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J Control Release 207:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Hossain GS, Kocerha J (2019) The potential for microRNA therapeutics and clinical research. Front Genet 10:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiro S, Mitsuhashi M, Chikami Y, Kawaguchi H, Niimi T, Yasueda H (2019a) Construction of Corynebacterium glutamicum cells as containers encapsulating dsRNA overexpressed for agricultural pest control. Appl Microbiol Biotechnol 103:8485–8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiro S, Mitsuhashi M, Yasueda H (2019b) Overexpression system for recombinant RNA in Corynebacterium glutamicum using a strong promoter derived from corynephage BFK20. J Biosci Bioeng 128:255–263. [DOI] [PubMed] [Google Scholar]
- Hasler D, Lehmann G, Murakawa Y, Klironomos F, Jakob L, Grässer FA, Rajewsky N, Landthaler M, Meister G (2016) The lupus autoantigen La prevents mis-channeling of tRNA fragments into the human microRNA pathway. Mol Cell 63:110–124. [DOI] [PubMed] [Google Scholar]
- Hayes J, Peruzzi PP, Lawler S (2014) MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 20:460–469. [DOI] [PubMed] [Google Scholar]
- He Y, Huang C, Lin X, Li J (2013) MicroRNA-29 family, a crucial therapeutic target for fibrosis diseases. Biochimie 95:1355–1359. [DOI] [PubMed] [Google Scholar]
- Hermeking H (2010) The miR-34 family in cancer and apoptosis. Cell Death Differ 17:193–199. [DOI] [PubMed] [Google Scholar]
- Ho PYDuan ZBatra NJilek JLTu MJQiu JXHu ZWun TLara PNDeVere White RW, et al. (2018) Bioengineered noncoding RNAs selectively change cellular miRNome profiles for cancer therapy. J Pharmacol Exp Ther 365:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PY, Yu AM (2016) Bioengineering of noncoding RNAs for research agents and therapeutics. Wiley Interdiscip Rev RNA 7:186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong DSKang YKBorad MSachdev JEjadi SLim HYBrenner AJPark KLee JLKim TY, et al. (2020) Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer 122:1630–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Xu Y, Hao J, Wang S, Li C, Meng S (2012) MiR-122 in hepatic function and liver diseases. Protein Cell 3:364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Jin J, Deighan P, Kiner E, McReynolds L, Lieberman J (2013) Efficient and specific gene knockdown by small interfering RNAs produced in bacteria. Nat Biotechnol 31:350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12:99–110. [DOI] [PubMed] [Google Scholar]
- Ipsaro JJ, Joshua-Tor L (2015) From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat Struct Mol Biol 22:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas MMSchlegel MKHarbison CEYilmaz VOJiang YParmar RZlatev ICastoreno AXu HShulga-Morskaya S, et al. (2018) Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat Commun 9:723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen HLReesink HWLawitz EJZeuzem SRodriguez-Torres MPatel Kvan der Meer AJPatick AKChen AZhou Y, et al. (2013) Treatment of HCV infection by targeting microRNA. N Engl J Med 368:1685–1694. [DOI] [PubMed] [Google Scholar]
- Ji Y, Hocker JD, Gattinoni L (2016) Enhancing adoptive T cell immunotherapy with microRNA therapeutics. Semin Immunol 28:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian CTu MJHo PYDuan ZZhang QQiu JXDeVere White RWWun TLara PNLam KS, et al. (2017) Co-targeting of DNA, RNA, and protein molecules provides optimal outcomes for treating osteosarcoma and pulmonary metastasis in spontaneous and experimental metastasis mouse models. Oncotarget 8:30742–30755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilek JL, Tian Y, Yu AM (2017) Effects of microRNA-34a on the pharmacokinetics of cytochrome P450 probe drugs in mice. Drug Metab Dispos 45:512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilek JL, Zhang QY, Tu MJ, Ho PY, Duan Z, Qiu JX, Yu AM (2019) Bioengineered Let-7c inhibits orthotopic hepatocellular carcinoma and improves overall survival with minimal immunogenicity. Mol Ther Nucleic Acids 14:498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L, Lucchino M (2018) Current challenges in delivery and cytosolic translocation of therapeutic RNAs. Nucleic Acid Ther 28:178–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Yeom C, Choi YS, Kim S, Lee E, Park MJ, Kang SW, Kim SB, Chang S (2015) Simultaneous inhibition of multiple oncogenic miRNAs by a multi-potent microRNA sponge. Oncotarget 6:20370–20387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallend D, Stoekenbroek R, He Y, Smith PF, Wijngaard P (2022) Pharmacokinetics and pharmacodynamics of inclisiran, a small interfering RNA therapy, in patients with hepatic impairment. J Clin Lipidol 16:208–219. [DOI] [PubMed] [Google Scholar]
- Kansakar U, Varzideh F, Mone P, Jankauskas SS, Santulli G (2022) Functional role of microRNAs in regulating cardiomyocyte death. Cells 11:983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karra N, Benita S (2012) The ligand nanoparticle conjugation approach for targeted cancer therapy. Curr Drug Metab 13:22–41. [DOI] [PubMed] [Google Scholar]
- Kessel D, Oleinick NL (2018) Cell death pathways associated with photodynamic therapy: an update. Photochem Photobiol 94:213–218. [DOI] [PubMed] [Google Scholar]
- Khvorova A (2017) Oligonucleotide therapeutics – a new class of cholesterol-lowering drugs. N Engl J Med 376:4–7. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209–216. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Watts JK (2017) The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 35:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Umekage S (2018) Extracellular nucleic acids of the marine bacterium Rhodovulum sulfidophilum and recombinant RNA production technology using bacteria. FEMS Microbiol Lett 365:fnx268. [DOI] [PubMed] [Google Scholar]
- Kletzmayr A, Ivarsson ME, Leroux JC (2020) Investigational therapies for primary hyperoxaluria. Bioconjug Chem 31:1696–1707. [DOI] [PubMed] [Google Scholar]
- Kothadia JP, LaFreniere K, Shah JM (2022) Acute hepatic porphyria, in StatPearls, StatPearls Publishing, Treasure Island, FL. [PubMed] [Google Scholar]
- Kristen AV, Ajroud-Driss S, Conceição I, Gorevic P, Kyriakides T, Obici L (2019) Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener Dis Manag 9:5–23. [DOI] [PubMed] [Google Scholar]
- Kufel J, Grzechnik P (2019) Small nucleolar RNAs tell a different tale. Trends Genet 35:104–117. [DOI] [PubMed] [Google Scholar]
- Kumar L, Verma S, Vaidya B, Gupta V (2015) Exosomes: natural carriers for siRNA delivery. Curr Pharm Des 21:4556–4565. [DOI] [PubMed] [Google Scholar]
- Kumar P, Anaya J, Mudunuri SB, Dutta A (2014) Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A (2018) tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24:1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CPursell NGierut JSaxena UZhou WDills MDiwanji RDutta CKoser MNazef N, et al. (2018) Specific inhibition of hepatic lactate dehydrogenase reduces oxalate production in mouse models of primary hyperoxaluria. Mol Ther 26:1983–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CF, Chen CY, Au LC (2013) Comparison between the repression potency of siRNA targeting the coding region and the 3′-untranslated region of mRNA. BioMed Res Int 2013:637850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb YN (2021) Inclisiran: first approval. Drugs 81:389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854. [DOI] [PubMed] [Google Scholar]
- Lennox KA, Behlke MA (2011) Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther 18:1111–1120. [DOI] [PubMed] [Google Scholar]
- Lennox KA, Owczarzy R, Thomas DM, Walder JA, Behlke MA (2013) Improved performance of anti-miRNA oligonucleotides using a novel non-nucleotide modifier. Mol Ther Nucleic Acids 2:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leqvio. (2021) Package insert. Novartis Pharmaceuticals, East Hanover, NJ. [Google Scholar]
- Leung AK, Hafez IM, Baoukina S, Belliveau NM, Zhigaltsev IV, Afshinmanesh E, Tieleman DP, Hansen CL, Hope MJ, Cullis PR (2012) Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J Phys Chem C Nanomater Interfaces 116:18440–18450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Tam YY, Cullis PR (2014) Lipid nanoparticles for short interfering RNA delivery. Adv Genet 88:71–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Addepalli B, Tu MJ, Chen QX, Wang WP, Limbach PA, LaSalle JM, Zeng S, Huang M, Yu AM (2015) Chimeric microRNA-1291 biosynthesized efficiently in Escherichia coli is effective to reduce target gene expression in human carcinoma cells and improve chemosensitivity. Drug Metab Dispos 43:1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Wang WP, Wu WJ, Huang M, Yu AM (2014) Rapid production of novel pre-microRNA agent hsa-mir-27b in Escherichia coli using recombinant RNA technology for functional studies in mammalian cells. Drug Metab Dispos 42:1791–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PCTu MJHo PYBatra NTran MMLQiu JXWun TLara PNHu XYu AX, et al. (2021) In vivo fermentation production of humanized noncoding RNAs carrying payload miRNAs for targeted anticancer therapy. Theranostics 11:4858–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xu Z, Sheng J (2018) tRNA-derived small RNA: a novel regulatory small non-coding RNA. Genes (Basel) 9:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SC, Chan WC, Hu LY, Lai CH, Hsu CN, Lin WC (2010) Identification of homologous microRNAs in 56 animal genomes. Genomics 96:1–9. [DOI] [PubMed] [Google Scholar]
- Li X, Tian Y, Tu MJ, Ho PY, Batra N, Yu AM (2019) Bioengineered miR-27b-3p and miR-328-3p modulate drug metabolism and disposition via the regulation of target ADME gene expression. Acta Pharm Sin B 9:639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebow ALi XRacie THettinger JBettencourt BRNajafian NHaslett PFitzgerald KHolmes RPErbe D, et al. (2017) An investigational RNAi therapeutic targeting glycolate oxidase reduces oxalate production in models of primary hyperoxaluria. J Am Soc Nephrol 28:494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima JF, Cerqueira L, Figueiredo C, Oliveira C, Azevedo NF (2018) Anti-miRNA oligonucleotides: a comprehensive guide for design. RNA Biol 15:338–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima WFPrakash TPMurray HMKinberger GALi WChappell AELi CSMurray SFGaus HSeth PP, et al. (2012) Single-stranded siRNAs activate RNAi in animals. Cell 150:883–894. [DOI] [PubMed] [Google Scholar]
- Lindow M, Kauppinen S (2012) Discovering the first microRNA-targeted drug. J Cell Biol 199:407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Shyr Y, Cai J, Liu Q (2018) Interplay between miRNAs and host genes and their role in cancer. Brief Funct Genomics 18:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Stepanov VG, Strych U, Willson RC, Jackson GW, Fox GE (2010) DNAzyme-mediated recovery of small recombinant RNAs from a 5S rRNA-derived chimera expressed in Escherichia coli. BMC Biotechnol 10:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J, Hermeking H (2008) Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7:2591–2600. [DOI] [PubMed] [Google Scholar]
- Loh KP, Bao Q, Eda G, Chhowalla M (2010) Graphene oxide as a chemically tunable platform for optical applications. Nat Chem 2:1015–1024. [DOI] [PubMed] [Google Scholar]
- Lönn P, Kacsinta AD, Cui XS, Hamil AS, Kaulich M, Gogoi K, Dowdy SF (2016) Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci Rep 6:32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovat FNigita GDistefano RNakamura TGasparini PTomasello LFadda PIbrahimova NCatricalà SPalamarchuk A, et al. (2020) Combined loss of function of two different loci of miR-15/16 drives the pathogenesis of acute myeloid leukemia. Proc Natl Acad Sci USA 117:12332–12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Xing H, Xun Z, Yang T, Ding P, Cai C, Wang D, Zhao X (2018) Exosome-based small RNA delivery: progress and prospects. Asian J Pharm Sci 13:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin N, Ragni MV (2018) An investigational RNAi therapeutic targeting antithrombin for the treatment of hemophilia A and B. J Blood Med 9:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vMaguregui A, Abe H (2020) Developments in siRNA modification and ligand conjugated delivery to enhance RNA interference ability. ChemBioChem 21:1808–1815. [DOI] [PubMed] [Google Scholar]
- Makris K, Spanou L (2016) Acute kidney injury: definition, pathophysiology and clinical phenotypes. Clin Biochem Rev 37:85–98. [PMC free article] [PubMed] [Google Scholar]
- Mamaeva V, Sahlgren C, Lindén M (2013) Mesoporous silica nanoparticles in medicine--recent advances. Adv Drug Deliv Rev 65:689–702. [DOI] [PubMed] [Google Scholar]
- McGowan MP, Hosseini Dehkordi SH, Moriarty PM, Duell PB (2019) Diagnosis and treatment of heterozygous familial hypercholesterolemia. J Am Heart Assoc 8:e013225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade BRGogoi KHamil ASPalm-Apergi Cvan den Berg AHagopian JCSpringer ADEguchi AKacsinta ADDowdy CF, et al. (2014) Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat Biotechnol 32:1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Smith EM, Bushell M (2014) Regulation of miRNA strand selection: follow the leader? Biochem Soc Trans 42:1135–1140. [DOI] [PubMed] [Google Scholar]
- Meinnel T, Mechulam Y, Fayat G (1988) Fast purification of a functional elongator tRNAmet expressed from a synthetic gene in vivo. Nucleic Acids Res 16:8095–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Lu M (2017) RNA interference-induced innate immunity, off-target effect, or immune adjuvant? Front Immunol 8:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SM, Williams CC, Akahori Y, Tanaka T, Aikawa H, Tong Y, Childs-Disney JL, Disney MD (2020) Small molecule recognition of disease-relevant RNA structures. Chem Soc Rev 49:7167–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklefield J (2001) Backbone modification of nucleic acids: synthesis, structure and therapeutic applications. Curr Med Chem 8:1157–1179. [DOI] [PubMed] [Google Scholar]
- Mie Y, Hirano Y, Kowata K, Nakamura A, Yasunaga M, Nakajima Y, Komatsu Y (2018) Function control of anti-microRNA oligonucleotides using interstrand cross-linked duplexes. Mol Ther Nucleic Acids 10:64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misso GDi Martino MTDe Rosa GFarooqi AALombardi ACampani VZarone MRGullà ATagliaferri PTassone P, et al. (2014) Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids 3:e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R (2021) Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov 20:101–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moertl S, Mutschelknaus L, Heider T, Atkinson MJ (2016) MicroRNAs as novel elements in personalized radiotherapy. Transl Cancer Res 5 (Suppl 6):S1262–S1269 DOI: 10.21037/tcr.2016.11.37. [Google Scholar]
- Molitoris BADagher PCSandoval RMCampos SBAshush HFridman EBrafman AFaerman AAtkinson SJThompson JD, et al. (2009) siRNA targeted to p53 attenuates ischemic and cisplatin-induced acute kidney injury. J Am Soc Nephrol 20:1754–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollaei H, Safaralizadeh R, Rostami Z (2019) MicroRNA replacement therapy in cancer. J Cell Physiol 234:12369–12384. [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Yu G, Latimer PA, Stack C, Robinson K, Dalby CM, Kaminski N, van Rooij E (2014) MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol Med 6:1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morena F, Argentati C, Bazzucchi M, Emiliani C, Martino S (2018) Above the epitranscriptome: RNA modifications and stem cell identity. Genes (Basel) 9:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Montañés J, Bleau AM, Jimenez AI (2018) Tivanisiran, a novel siRNA for the treatment of dry eye disease. Expert Opin Investig Drugs 27:421–426. [DOI] [PubMed] [Google Scholar]
- Myers JW, Jones JT, Meyer T, Ferrell JE Jr (2003) Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat Biotechnol 21:324–328. [DOI] [PubMed] [Google Scholar]
- Nagao N, Hirose Y, Misawa N, Ohtsubo Y, Umekage S, Kikuchi Y (2015) Complete genome sequence of Rhodovulum sulfidophilum DSM 2351, an extracellular nucleic acid-producing bacterium. Genome Announc 3:e00388-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao N, Suzuki H, Numano R, Umekage S, Kikuchi Y (2014) Short hairpin RNAs of designed sequences can be extracellularly produced by the marine bacterium Rhodovulum sulfidophilum. J Gen Appl Microbiol 60:222–226. [DOI] [PubMed] [Google Scholar]
- Nair JKWilloughby JLChan ACharisse KAlam MRWang QHoekstra MKandasamy PKel’in AVMilstein S, et al. (2014) Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc 136:16958–16961. [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen FH, Leunissen EH, van de Laar L, Tessari M, Heus HA, Wijmenga SS (2012) Fast production of homogeneous recombinant RNA--towards large-scale production of RNA. Nucleic Acids Res 40:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D-D, Chang S (2017) Development of novel therapeutic agents by inhibition of oncogenic microRNAs. Int J Mol Sci 19:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obici LBerk JLGonzález-Duarte ACoelho TGillmore JSchmidt HHSchilling MYamashita TLabeyrie CBrannagan TH 3rd, et al. (2020) Quality of life outcomes in APOLLO, the phase 3 trial of the RNAi therapeutic patisiran in patients with hereditary transthyretin-mediated amyloidosis. Amyloid 27:153–162. [DOI] [PubMed] [Google Scholar]
- Ohno STakanashi MSudo KUeda SIshikawa AMatsuyama NFujita KMizutani TOhgi TOchiya T, et al. (2013) Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther 21:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T (2009) A high-resolution structure of the pre-microRNA nuclear export machinery. Science 326:1275–1279. [DOI] [PubMed] [Google Scholar]
- Onpattro (2018) Package insert. Alnylam Pharmaceuticals, Cambridge, MA. [Google Scholar]
- Orellana EA, Li C, Lisevick A, Kasinski AL (2019) Identification and validation of microRNAs that synergize with miR-34a - a basis for combinatorial microRNA therapeutics. Cell Cycle 18:1798–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard J (2007) Hypoxic radiosensitization: adored and ignored. J Clin Oncol 25:4066–4074. [DOI] [PubMed] [Google Scholar]
- Oxlumo (2020) Package insert. Alnylam Pharmaceuticals, Cambridge, MA. [Google Scholar]
- Pallet N, Karras A, Thervet E, Gouya L, Karim Z, Puy H (2018) Porphyria and kidney diseases. Clin Kidney J 11:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YZ, Zhou A, Hu Z, Yu AM (2013) Small nucleolar RNA-derived microRNA hsa-miR-1291 modulates cellular drug disposition through direct targeting of ABC transporter ABCC1. Drug Metab Dispos 41:1744–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AEReinhart BJSlack FMartindale MQKuroda MIMaller BHayward DCBall EEDegnan BMüller P, et al. (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408:86–89. [DOI] [PubMed] [Google Scholar]
- Patterson DGRoberts JTKing VMHouserova DBarnhill ECCrucello APolska CJBrantley LWKaufman GCNguyen M, et al. (2017) Human snoRNA-93 is processed into a microRNA-like RNA that promotes breast cancer cell invasion. NPJ Breast Cancer 3:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunovska K, Loughrey D, Dahlman JE (2022) Drug delivery systems for RNA therapeutics. Nat Rev Genet 23:265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Yu G, Tian S, Li H (2014) Co-expression and co-purification of archaeal and eukaryal box C/D RNPs. PLoS One 9:e103096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira P, Pedro AQ, Queiroz JA, Figueiras AR, Sousa F (2017) New insights for therapeutic recombinant human miRNAs heterologous production: Rhodovolum sulfidophilum vs Escherichia coli. Bioengineered 8:670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira P, Pedro AQ, Tomás J, Maia CJ, Queiroz JA, Figueiras A, Sousa F (2016) Advances in time course extracellular production of human pre-miR-29b from Rhodovulum sulfidophilum. Appl Microbiol Biotechnol 100:3723–3734. [DOI] [PubMed] [Google Scholar]
- Petrek H, Yan Ho P, Batra N, Tu MJ, Zhang Q, Qiu JX, Yu AM (2021) Single bioengineered ncRNA molecule for dual-targeting toward the control of non-small cell lung cancer patient-derived xenograft tumor growth. Biochem Pharmacol 189:114392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piletič K, Kunej T (2016) MicroRNA epigenetic signatures in human disease. Arch Toxicol 90:2405–2419. [DOI] [PubMed] [Google Scholar]
- Pitulle C, Hedenstierna KO, Fox GE (1995) A novel approach for monitoring genetically engineered microorganisms by using artificial, stable RNAs. Appl Environ Microbiol 61:3661–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponchon L, Beauvais G, Nonin-Lecomte S, Dardel F (2009) A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat Protoc 4:947–959. [DOI] [PubMed] [Google Scholar]
- Ponchon L, Dardel F (2007) Recombinant RNA technology: the tRNA scaffold. Nat Methods 4:571–576. [DOI] [PubMed] [Google Scholar]
- Qiu W, Park JW, Scholthof HB (2002) Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol Plant Microbe Interact 15:269–280. [DOI] [PubMed] [Google Scholar]
- Raal FJKallend DRay KKTurner TKoenig WWright RSWijngaard PLJCurcio DJaros MJLeiter LA, et al. ; ORION-9 Investigators (2020) Inclisiran for the treatment of heterozygous familial hypercholesterolemia. N Engl J Med 382:1520–1530. [DOI] [PubMed] [Google Scholar]
- Ray KKLandmesser ULeiter LAKallend DDufour RKarakas MHall TTroquay RPTurner TVisseren FL, et al. (2017) Inclisiran in patients at high cardiovascular risk with elevated LDL cholesterol. N Engl J Med 376:1430–1440. [DOI] [PubMed] [Google Scholar]
- Ray KKWright RSKallend DKoenig WLeiter LARaal FJBisch JARichardson TJaros MWijngaard PLJ, et al. ; ORION-10 and ORION-11 Investigators (2020) Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N Engl J Med 382:1507–1519. [DOI] [PubMed] [Google Scholar]
- Reijman MD, Schweizer A, Peterson ALH, Bruckert E, Stratz C, Defesche JC, Hegele RA, Wiegman A (2022) Rationale and design of two trials assessing the efficacy, safety, and tolerability of inclisiran in adolescents with homozygous and heterozygous familial hypercholesterolaemia. Eur J Prev Cardiol DOI: 10.1093/eurjpc/zwac025 [published ahead of print]. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I (2007) 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther 15:1663–1669. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Jankauskas SS, Gambardella J (2021) Inclisiran: a new milestone on the PCSK9 road to tackle cardiovascular risk. Eur Heart J Cardiovasc Pharmacother 7:e11–e12. [DOI] [PubMed] [Google Scholar]
- Sardh E, Harper P (2022) RNAi therapy with givosiran significantly reduces attack rates in acute intermittent porphyria. J Intern Med 291:593–610. [DOI] [PubMed] [Google Scholar]
- Sardh EHarper PBalwani MStein PRees DBissell DMDesnick RParker CPhillips JBonkovsky HL, et al. (2019) Phase 1 trial of an RNA interference therapy for acute intermittent porphyria. N Engl J Med 380:549–558. [DOI] [PubMed] [Google Scholar]
- Sas DJMagen DHayes WShasha-Lavsky HMichael MSchulte ISellier-Leclerc ALLu JSeddighzadeh AHabtemariam B, et al. ; ILLUMINATE-B Workgroup (2021) Phase 3 trial of lumasiran for primary hyperoxaluria type 1: a new RNAi therapeutic in infants and young children. Genet Med 24:654–662. [DOI] [PubMed] [Google Scholar]
- Scott LJ (2020) Givosiran: first approval. Drugs 80:335–339. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Keam SJ (2021) Lumasiran: first approval. Drugs 81:277–282. [DOI] [PubMed] [Google Scholar]
- Segal M, Slack FJ (2020) Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin Drug Discov 15:987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal ABarros SIvanciu LCooley BQin JRacie THettinger JCarioto MJiang YBrodsky J, et al. (2015) An RNAi therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia. Nat Med 21:492–497. [DOI] [PubMed] [Google Scholar]
- Sen GL, Blau HM (2006) A brief history of RNAi: the silence of the genes. FASEB J 20:1293–1299. [DOI] [PubMed] [Google Scholar]
- Seth PP, Tanowitz M, Bennett CF (2019) Selective tissue targeting of synthetic nucleic acid drugs. J Clin Invest 129:915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setten RL, Rossi JJ, Han SP (2019) The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov 18:421–446. [DOI] [PubMed] [Google Scholar]
- Shimazaki J (2018) Definition and diagnostic criteria of dry eye disease: historical overview and future directions. Invest Ophthalmol Vis Sci 59:DES7–DES12. [DOI] [PubMed] [Google Scholar]
- Shin SC, Robinson-Papp J (2012) Amyloid neuropathies. Mt Sinai J Med 79:733–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtam TA, Kovalev RA, Varfolomeeva EY, Makarov EM, Kil YV, Filatov MV (2013) Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun Signal 11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum K, Rossi J (2016) Preface. Methods Mol Biol 1364:v–vi. [DOI] [PubMed] [Google Scholar]
- Silhavy D, Molnár A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyán J (2002) A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J 21:3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ES, Whitty E, Yoo B, Moore A, Sempere LF, Medarova Z (2022) Clinical Applications of short non-coding RNA-based therapies in the era of precision medicine. Cancers (Basel) 14:1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon SDAdams DKristen AGrogan MGonzález-Duarte AMaurer MSMerlini GDamy TSlama MSBrannagan TH 3rd, et al. (2019) Effects of patisiran, an RNA interference therapeutic, on cardiac parameters in patients with hereditary transthyretin-mediated amyloidosis. Circulation 139:431–443. [DOI] [PubMed] [Google Scholar]
- Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S (2012) Inhibition of microRNA function by antimiR oligonucleotides. Silence 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhr OBCoelho TBuades JPouget JConceicao IBerk JSchmidt HWaddington-Cruz MCampistol JMBettencourt BR, et al. (2015) Efficacy and safety of patisiran for familial amyloidotic polyneuropathy: a phase II multi-dose study. Orphanet J Rare Dis 10:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SWang YZhou RDeng ZHan YHan XTao WYang ZShi CHong D, et al. (2017) Targeting and regulating of an oncogene via nanovector delivery of microRNA using patient-derived xenografts. Theranostics 7:677–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Ando T, Umekage S, Tanaka T, Kikuchi Y (2010) Extracellular production of an RNA aptamer by ribonuclease-free marine bacteria harboring engineered plasmids: a proposal for industrial RNA drug production. Appl Environ Microbiol 76:786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L, Gidal B, Blakey G, Tayo B, Morrison G (2018) A phase I, randomized, double-blind, placebo-controlled, single ascending dose, multiple dose, and food effect trial of the safety, tolerability and pharmacokinetics of highly purified cannabidiol in healthy subjects. CNS Drugs 32:1053–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Kornbrust DJ, Foy JW, Solano EC, Schneider DJ, Feinstein E, Molitoris BA, Erlich S (2012) Toxicological and pharmacokinetic properties of chemically modified siRNAs targeting p53 RNA following intravenous administration. Nucleic Acid Ther 22:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KK (2013) MicroRNA: a prognostic biomarker and a possible druggable target for circumventing multidrug resistance in cancer chemotherapy. J Biomed Sci 20:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KK, Tong CW, Wu M, Cho WC (2018) MicroRNAs in the prognosis and therapy of colorectal cancer: from bench to bedside. World J Gastroenterol 24:2949–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KKW, Fong W, Tong CWS, Wu M, Yan W, Cho WCS (2020) Advances in the discovery of microRNA-based anticancer therapeutics: latest tools and developments. Expert Opin Drug Discov 15:63–83. [DOI] [PubMed] [Google Scholar]
- Tu MJ, Duan Z, Liu Z, Zhang C, Bold RJ, Gonzalez FJ, Kim EJ, Yu AM (2020) MicroRNA-1291-5p sensitizes pancreatic carcinoma cells to arginine deprivation and chemotherapy through the regulation of arginolysis and glycolysis. Mol Pharmacol 98:686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu MJ, Ho PY, Zhang QY, Jian C, Qiu JX, Kim EJ, Bold RJ, Gonzalez FJ, Bi H, Yu AM (2019) Bioengineered miRNA-1291 prodrug therapy in pancreatic cancer cells and patient-derived xenograft mouse models. Cancer Lett 442:82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeh-Garcia M, Simion C, Ho PY, Batra N, Berg AL, Carraway KL, Yu A, Sweeney C (2020) A novel bioengineered miR-127 prodrug suppresses the growth and metastatic potential of triple-negative breast cancer cells. Cancer Res 80:418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M (2014) Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics 2014:970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bruggen FH, Nijhuis GBJ, Zuidema SU, Luijendijk H (2020) Serious adverse events and deaths in PCSK9 inhibitor trials reported on ClinicalTrials.gov: a systematic review. Expert Rev Clin Pharmacol 13:787–796. [DOI] [PubMed] [Google Scholar]
- van den Boorn JG, Schlee M, Coch C, Hartmann G (2011) siRNA delivery with exosome nanoparticles. Nat Biotechnol 29:325–326. [DOI] [PubMed] [Google Scholar]
- Vannucci L, Lai M, Chiuppesi F, Ceccherini-Nelli L, Pistello M (2013) Viral vectors: a look back and ahead on gene transfer technology. New Microbiol 36:1–22. [PubMed] [Google Scholar]
- Varzideh F, Kansakar U, Donkor K, Wilson S, Jankauskas SS, Mone P, Wang X, Lombardi A, Santulli G (2022) Cardiac remodeling after myocardial infarction: functional contribution of microRNAs to inflammation and fibrosis. Front Cardiovasc Med 9:863238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S (2012) Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA 3:311–330. [DOI] [PubMed] [Google Scholar]
- Velagapudi SP, Gallo SM, Disney MD (2014) Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat Chem Biol 10:291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura PBonkovsky HLGouya LAguilera-Peiró PMontgomery Bissell DStein PEBalwani MAnderson DKEParker CKuter DJ, et al. ; ENVISION Investigators (2022) Efficacy and safety of givosiran for acute hepatic porphyria: 24-month interim analysis of the randomized phase 3 ENVISION study. Liver Int 42:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WPHo PYChen QXAddepalli BLimbach PALi MMWu WJJilek JLQiu JXZhang HJ, et al. (2015) Bioengineering novel chimeric microRNA-34a for prodrug cancer therapy: high-yield expression and purification, and structural and functional characterization. J Pharmacol Exp Ther 354:131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z (2011) The principles of MiRNA-masking antisense oligonucleotides technology. Methods Mol Biol 676:43–49. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen X, Liu J, Wang Y, Zhang S (2022) Inclisiran inhibits oxidized low-density lipoprotein-induced foam cell formation in Raw264.7 macrophages via activating the PPARγ pathway. Autoimmunity 55:223–232. [DOI] [PubMed] [Google Scholar]
- Weaver DT, Pishas KI, Williamson D, Scarborough J, Lessnick SL, Dhawan A, Scott JG (2021) Network potential identifies therapeutic miRNA cocktails in Ewing sarcoma. PLOS Comput Biol 17:e1008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Xiao H, Zhang J, Liang XJ, Huang Y (2019) RNAi therapeutic and its innovative biotechnological evolution. Biotechnol Adv 37:801–825. [DOI] [PubMed] [Google Scholar]
- Westholm JO, Lai EC (2011) Mirtrons: microRNA biogenesis via splicing. Biochimie 93:1897–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M (2000) Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol 2:70–75. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75:855–862. [DOI] [PubMed] [Google Scholar]
- Wilson C, Keefe AD (2006) Building oligonucleotide therapeutics using non-natural chemistries. Curr Opin Chem Biol 10:607–614. [DOI] [PubMed] [Google Scholar]
- Xu C, Haque F, Jasinski DL, Binzel DW, Shu D, Guo P (2018) Favorable biodistribution, specific targeting and conditional endosomal escape of RNA nanoparticles in cancer therapy. Cancer Lett 414:57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JSun JHo PYLuo ZMa WZhao WRathod SBFernandez CAVenkataramanan RXie W, et al. (2019) Creatine based polymer for codelivery of bioengineered microRNA and chemodrugs against breast cancer lung metastasis. Biomaterials 210:25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue JChi YChen YHuang SYe XNiu JWang WPfeffer LMShao ZMWu ZH, et al. (2016) MiRNA-621 sensitizes breast cancer to chemotherapy by suppressing FBXO11 and enhancing p53 activity. Oncogene 35:448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Yang J, Luo M, Cho WC, Liu X (2017) MicroRNA-targeted therapeutics for lung cancer treatment. Expert Opin Drug Discov 12:141–157. [DOI] [PubMed] [Google Scholar]
- Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, Bishop JM (2002) Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci USA 99:9942–9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC (2010) Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci USA 107:15163–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Tu MJ, Liu Z, Zhang C, Batra N, Yu AX, Yu AM (2020) Bioengineered miR-328-3p modulates GLUT1-mediated glucose uptake and metabolism to exert synergistic antiproliferative effects with chemotherapeutics. Acta Pharm Sin B 10:159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y (2010) ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol 17:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, Batra N, Tu MJ, Sweeney C (2020a) Novel approaches for efficient in vivo fermentation production of noncoding RNAs. Appl Microbiol Biotechnol 104:1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, Choi YH, Tu MJ (2020b) RNA drugs and RNA targets for small molecules: principles, progress, and challenges. Pharmacol Rev 72:862–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, Jian C, Yu AH, Tu MJ (2019) RNA therapy: are we using the right molecules? Pharmacol Ther 196:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, Tian Y, Tu MJ, Ho PY, Jilek JL (2016) MicroRNA pharmacoepigenetics: posttranscriptional regulation mechanisms behind variable drug disposition and strategy to develop more effective therapy. Drug Metab Dispos 44:308–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, Tu MJ (2022) Deliver the promise: RNAs as a new class of molecular entities for therapy and vaccination. Pharmacol Ther 230:107967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W (2004) Single processing center models for human Dicer and bacterial RNase III. Cell 118:57–68. [DOI] [PubMed] [Google Scholar]
- Zhang MM, Bahal R, Rasmussen TP, Manautou JE, Zhong XB (2021a) The growth of siRNA-based therapeutics: updated clinical studies. Biochem Pharmacol 189:114432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QY, Ho PY, Tu MJ, Jilek JL, Chen QX, Zeng S, Yu AM (2018) Lipidation of polyethylenimine-based polyplex increases serum stability of bioengineered RNAi agents and offers more consistent tumoral gene knockdown in vivo. Int J Pharm 547:537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Cheng Z, Wang Y, Han T (2021b) The risks of miRNA therapeutics: in a drug target perspective. Drug Des Devel Ther 15:721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Goel V, Robbie GJ (2019) Pharmacokinetics of patisiran, the first approved RNA interference therapy in patients with hereditary transthyretin-mediated amyloidosis. J Clin Pharmacol 60:573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Potty AS, Jackson GW, Stepanov V, Tang A, Liu Y, Kourentzi K, Strych U, Fox GE, Willson RC (2009) Engineered 5S ribosomal RNAs displaying aptamers recognizing vascular endothelial growth factor and malachite green. J Mol Recognit 22:154–161. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang J (2017) MicroRNAs are important regulators of drug resistance in colorectal cancer. Biol Chem 398:929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]