Abstract
Objective:
Pneumonia is an opportunistic infection and it is a major cause of mortality and morbidity among human immunodeficiency virus/acquired immune deficiency syndrome-positive patients. Previous studies have shown the dominant pathogens bacterial isolates were K. pneumoniae 27.0%, S. aureus 20.8%, S. pneumoniae 18.8% and E. coli 8.3%. This study aimed to determine bacteriology of community-acquired pneumonia, antimicrobial susceptibility pattern and associated risk factors among human immunodeficiency virus patients in the Northeast Ethiopia: cross-sectional study.
Methods:
A health facility-based cross-sectional study was conducted from January to April 2021 at six health facilities in Dessie Town. A total of 378 community-acquired pneumonia patients suspected to be human immunodeficiency virus-positive were recruited using a consecutive sampling technique. Sociodemographic and clinical data were collected using a structured questionnaire. A two-milliliter sputum specimen was collected aseptically from each study participant. Samples were cultivated on blood agar, chocolate agar and MacConkey agar to isolate bacterial pathogens. To identify bacteria pathogens Gram stain, colony morphology and biochemical tests were performed. The Kirby-Bauer Disc Diffusion method was used to perform the antimicrobial susceptibility test. Descriptive statistics, logistic regression analysis was carried out using Statistical package for social science version 25 software. p-value < 0.05 with a corresponding 95% confidence interval (CI) was considered for statistical significance.
Result:
The overall prevalence of bacterial pneumonia was 175 (46.3%). Gram-negative bacteria accounted for 119 (68%) and the predominant isolates identified were Streptococcus pneumoniae 49 (28%) followed by Klebsiella pneumoniae 46 (26.3%), Pseudomonas aeruginosa 34 (19.4%). There were 148 (84.6%) multidrug-resistant bacteria overall. Statistically significant factors included viral load, cigarette smoking, cluster of differentiation 4 count, alcohol use, World Health Organization clinical stages III and IV and low white blood cell count.
Conclusion:
The study found that both multidrug resistance and bacterial pneumonia were high. Thus, bacterial culture and antimicrobial susceptibility tests should be routinely performed in health facilities in order to prevent and control the spread of bacterial infection and concurrent drug resistance.
Keywords: pneumonia, bacterial infection, human immunodeficiency virus, Ethiopia
Introduction
Pneumonia is one of the most common pulmonary complications in human immunodeficiency virus (HIV)-positive patients.1 As HIV influences both the humoral and cellular components of innate immunity; these alterations are specific and are evident even with relatively preserved cluster of differentiation 4 (CD4+) T cell counts and undetectable viral loads. Impaired innate immune function, particularly in the setting of CD4+ T cell depletion, may contribute to the pathogenesis of opportunistic lung infections.2
Pneumonia is caused by various groups of micro-organisms. In fact, the most common etiological agents are bacteria.3 Bacterial pneumonia is well known to occur at an elevated incidence in individuals living with HIV.4 Patients with acquired immune deficiency syndrome (AIDS) and HIV infection at the beginning of the HIV outbreak have a documented rise in the rate of bacterial infection.5 HIV patients are more vulnerable to opportunistic and bacterial infections due to alteration of host T-cell function, impaired phagocytic response of neutrophils and macrophages and the inability of B-cells to produce specific and increasing antibodies.6 Infection of pulmonary macrophages and lymphocytes with HIV-1 plays a key role in pulmonary disease pathogenesis in AIDS.7
Pneumonia is the fourth most common cause of death worldwide, after ischemic heart disease, stroke, and chronic obstructive pulmonary disease, and it is the second most common reason for years of life lost.8 It is more frequent in HIV patients, being 5–15 times more common in them than in HIV negative individuals.9
Pulmonary complications are a major cause of morbidity and mortality among HIV-infected patients globally.10 The incidence of bacterial disease among HIV-infected individuals has been substantially reduced by the use of antiretroviral therapy in both developed and developing countries.11 However, even in patients with antiretroviral treatment (ART) and high CD4+ counts, bacterial pneumonia remains high.12 The incidence of bacterial pneumonia among HIV-infected people increased with poor HIV/AIDS control, most commonly manifested by increased viral load and lower CD4 count.13
The most common bacterial pathogens that cause bacterial pneumonia are Streptococcus pneumoniae, Haemophilus influenzae, Klebsiella pneumoniae, Pseudomonas aeruginosa, Escherichia coli and Staphylococcus aureus.14 In Spain, a prospective observational study showed that S. pneumoniae was the leading species in causing bacterial pneumonia, accounted for 31.7%.11 Another study revealed that from 161 HIV-infected individuals, bacterial pneumonia was 17%. In addition, S. Pneumoniae was the most predominant bacteria (30%), followed by H. influenzae and S. aureus, accounted for 2%.15 Moreover, bacterial pneumonia was predominant in HIV-positive patients, accounted for 54%.16 The most common bacterial isolates were K. pneumoniae 27.0%, S. aureus 20.8%, S. pneumoniae 18.8%, E. coli 8.3%, H. influenzae, Klebsiella oxytoca, P. aeruginosa, 4.2% each, Enterobacter spp. 2.1% and unidentified Gram-negative bacteria 10.4%.17
Inappropriate use of antimicrobials provides selective pressure that favors the emergence of resistant strains in pathogenic bacteria. The majority of bacterial isolates found in people infected with HIV have varying levels of antimicrobial resistance.18 Multidrug resistance is defined as a bacterium which is non-susceptible to at least one agent in three or more antimicrobial categories.19,20
In the study area, the most commonly prescribed drugs for pneumonia were amoxicillin, amoxicillin-clavulanic acid, erythromycin, gentamicin and ceftriaxone without antimicrobial susceptibility testing (AST). This empirical method of treatment favors the emergence of drug-resistant bacterial strains. The emergence of drug-resistant bacterial strains poses difficulty in the management of pneumonia among HIV-positive patients. Therefore, this study was aimed to assess bacteriology of community-acquired pneumonia (CAP), antimicrobial susceptibility pattern and associated risk factors among HIV patients in the Northeast Ethiopia: cross-sectional study.
Method and materials
Study setting and design
A health facility-based cross-sectional study was conducted from January to April 2021 at the ART clinics of six health facilities in Dessie town, Northeast Ethiopia. These health facilities were Dessie Comprehensive Specialized Hospital, Selam General Hospital, Borumeda General Hospital, Dessie Health Centre, Ethio General Hospital and Bati General Hospital.
Inclusion and exclusion criteria
All HIV-positive patients from selected health facilities who were enrolled in the ART clinic and showed clinical signs and symptoms of pneumonia, such as shortness of breath, chest pain, fever, chills, fatigue, and cough, during data collection were included. However, patients who were critically ill and unable to expectorate sputum were excluded.
Population
All individuals living with HIV and who were enrolled at the ART clinics of health facilities in Dessie town were the source of population. Whereas HIV patients who were enrolled in the ART clinic of selected health facilities developed clinical evidence of CAP and were available in the ART clinic during the study period.
Sample size and sampling technique
The sample size was determined using a single population proportion formula considering (43.7%) taken from a previous study conducted in patients suspected to have pneumonia in Mekelle, Northern Ethiopia,21 marginal error of 5% and 95% confidence interval (CI) = 1.96 by using the following formula:
where n = the minimum sample size required, Za/2 = the significant value for 95% CI, P = the expected prevalence of pneumonia infection, and d = the margin of error, finally a total of 378 study participants with clinically diagnosed CAP (CD-CAP) were selected using a systematic random sampling. HIV-positive patients who received antibiotics within the past 2 weeks except cotrimoxazole were excluded.
[LE]
Operational definitions
Hand washing habit
Either washing hands with soap and water or killing germs on the hands with an at least 60% alcohol-based hand sanitizer or rub. When you clean your hands, you remove many germs.22
Smoking habit
Smoker consuming between 11–19 cigarettes and above 20 cigarettes or more per day.23
ART failure
ART failure is defined as progression of the disease and high risk of mortality after the beginning of ART.24
Co-infection
Co-infection is the simultaneous infection of a host by multiple pathogen species, for instance multi-parasite infections.25
Body mass index
Body fat based on height and weight that applies to adult men and women.26
The normal range for a white blood cell count is typically between 4000 and 11,000/microliters. Factors like age and sex may determine what a normal range type of WBC: normal percentage of overall lymphocyte: 20% to 40%, neutrophil: 55% to 70%, eosinophil: 1% to 4%, monocyte: 2% to 8% and basophil: 0% to 2%.27
Specimen collection and processing
In total, 2 ml sputum specimens were collected using clean, dry, sterile, wide-necked and leak-proof containers from each study participant. Sputum samples were transported immediately to Wollo University Microbiology Laboratory for analysis. Sputum samples were stored at 4 °C if a specimen processing delay existed.28 Blood samples were also collected for viral load, CD4+ cell and white blood cell count during data collection.
Gene Xpert test was performed for mycobacterium analyses.29 Gram staining with more than 25 polymorphic nuclear leukocytes and <10 epithelial cells were considered as good and cultivated. However, specimens with more than 10 epithelial cells and less than 25 polymorphic nuclear leukocytes per high power field (100X) were not good and discarded.28
A sterilized loop of specimens were streaked onto Blood agar (HiMedia™), Chocolate agar and MacConkey agar (HiMedia™). The Chocolate agar was incubated in a candle jar at 37 °C for 24–48 h. Whereas, Blood Agar and MacConkey agar was aerobically incubated for 24 h at 37 °C.30,31 Positive growth on Blood agar and MacConkey agar (HiMedia™) was subculture onto Nutrient agar (HiMedia™) for biochemical and antimicrobial susceptibility test.
Bacterial isolation and identification
The bacteria isolate was characterized using colony morphology, haemolysis pattern, Gram staining, and biochemical tests following the standard microbiological procedure. The significant bacterial count was carried out and the significant bacterial count was reported on the basis of observing an excess of 105 CFU/ml.28 Gram-positive cocci were distinguished and recognized based on Gram stain, blood agar haemolysis patterns, colonial characteristics, catalase test, coagulase test, mannitol fermentation test, and optochin (5 μg) susceptibility. Gram-negative bacteria were identified based on Gram reaction, colony morphology and pigmentation, oxidase test, fermentation of carbohydrates, H2S production, motility, formation of indole, triple sugar iron agar and citrate utilization, lysine decarboxylase or methyl red vogues Proskur utilization, urea hydrolysis and satellite tests.
Antimicrobial susceptibility testing
The isolated organism was tested against antibiotic agents using the Kirby Bauer disc diffusion method on Muller Hilton agar (HMEDIA). Briefly, single pure colonies of isolated species from nutrient agar were picked and transferred to a tube containing 5 ml tryptone-soya broth and mixed to make a homogenous suspension, then incubated at 37 °C until the turbidity of the suspension was matched to a 0.5 McFarland standard. Using sterile swab, the inoculum suspension was inoculated over the entire surface of the Mueller Hinton agar plate.32 After application of the selected antimicrobial disks, the plate was incubated overnight at 37 °C for 16–18 h. Inhibition zone diameter was measured and the degree of susceptibility was interpreted to each antibiotic according to Clinical and laboratory standards institute (CLSI) guideline.33 Antibacterial agents were selected based on local prescription habit and CLSI recommendations.
The standard antibiotic discs (Liofilchem-Italy, HARDY Diagnosis-Santa Maria, USA) and its concentrations used as: penicillin (10 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), clindamycin (30 μg), cefoxitin (30 μg), trimethoprim-sulfamethoxazole (1.25/23.75 µg), cefotaxime (30 µg), ceftriaxone (30 μg), erythromycin (15 μg) and oxacillin (30 μg) are the antimicrobial agents that can be used by Gram-positive bacteria. Chloramphenicol (30 μg), ciprofloxacin (5 μg), tetracycline (30 μg), gentamicin (10 μg), trimethoprim-sulfamethoxazole (1.25/23.75 µg), ceftriaxone (30 μg), piperacillin-tazobactam (100/10 µg), ceftazidime (30 µg), amikacin (30 µg), ampicillin (10 µg), amoxicillin-clavulanic acid (20/10 μg), meropenem (10 μg) and amoxicillin (10 µg) can be used for Gram-negative bacteria.33,34
Diameters of zones of inhibitions were measured using digital caliper. The interpretation of results of antimicrobial susceptibility tests were based on the standardized table supplied by CLSI33 as sensitive, intermediate or resistant. Moreover, an isolate was considered Multidrug resistance (MDR) if it is resistant to at least one agent in three or more antimicrobial categories.35
Quality control
Training was given for data collectors; the completeness of the questionnaires was checked by the principal investigator, and about 5% of the questionnaire was pretested at Meseret specialized clinic. Quality control of culture media was verified for sterility testing by overnight incubation of 5% of uninoculated plates/tubes of the prepared media from each batch. Standard reference strains of S. aureus American type culture collection (ATCC-25,923), E. coli (ATCC-25,922) and P. aeruginosa (ATCC-27,853) and for fastidious organisms, H. influenzae (ATCC 49,247) and S. pneumoniae (ATCC 49,619) were used as control strains.
Statistical analysis
All the data were entered using Epi-data version 4.6.0.4 and exported to Statistical package for social science version 25 for analysis. Descriptive statistics were performed. Bivariate and multivariate analyses were done. Variables with a p-value less or equal to 0.25 in bivariable analysis were subjected to multivariable analysis. Adjusted odd ratio with p value of <0.05 with 95% CI was statistically significant. Results were presented using graphs and tables.
Ethical considerations
Ethical approval was obtained from the ethical review committee of the College of Medicine and Health Sciences, Wollo University with a protocol number of CMHS/HC/354/13. Written informed consent was obtained from the subjects and the legally authorized representatives of the minor subjects prior to study initiation. Confidentiality and any special data security requirements were maintained and assured. All data and sample collected were kept confidential and used only for the purpose of the study. The positive cases were communicated with t physicians in order to initiate their treatment and management accordingly.
Result
Sociodemographic characteristics
The mean age of the study participants was 40.09 (±12.24) years, ranges from 10 to 70 years. Half (50.5%) of the study participants were females and majority of the study participants were urban dwellers (76.5%). In addition, 84% and 35% of participants were married and daily worker, respectively. Two hundred fifty-eight (68.3%) participants were living in a family having less than five members (Table 1).
Table 1.
Sociodemographic characteristics of pneumonia-suspected HIV-positive study participants (n = 378) at Dessie town health facilities, Northeast Ethiopia, 2021.
| Demographic and clinical variables | Frequency | Percentage (%) |
|---|---|---|
| Sex | ||
| Male | 187 | 49.5 |
| Female | 191 | 50.5 |
| Age in years | ||
| 10–16 | 10 | 2.6 |
| 17–30 | 77 | 20.4 |
| 31–45 | 174 | 46 |
| >45 | 117 | 31 |
| Residence | ||
| Rural | 89 | 23.5 |
| Urban | 289 | 76.5 |
| Education status | ||
| Unable to read and write | 104 | 27.5 |
| Only read and write | 37 | 9.8 |
| Primary school completed | 100 | 26.5 |
| Secondary school completed | 64 | 16.9 |
| College and above | 73 | 19.3 |
| Occupation | ||
| Employed | 82 | 21.7 |
| Student | 27 | 6.3 |
| Merchant | 97 | 25.7 |
| House wife | 28 | 7.4 |
| Farmer | 16 | 4.2 |
| Daily laborer | 131 | 34.7 |
| Marital status | ||
| Single | 25 | 6.6 |
| Married | 318 | 84.1 |
| Divorced | 18 | 4.8 |
| Widowed | 17 | 4.5 |
| No of family members | ||
| One | 19 | 5.0 |
| Two | 41 | 10.8 |
| Three | 90 | 23.8 |
| Four | 108 | 28.6 |
| Five | 63 | 16.7 |
| >Six | 57 | 15.1 |
Clinical and behavioural characteristics
Over 378 participants, 258 (68.4%) were non-smokers, 275 (72.8%) were never drink alcohol and 299 (79.1%) were chewed chat. Among 378 participants, 168 (44.4%), 183 (48.4%) and 137 (36.2%) participants were on World Health Organization (WHO) stage-II. viral load and CD4 count of <150 copies/cell and 201–350 cells/mm3, respectively. Moreover, half of the study participants had a white blood cell count of less than 4000 cells/mm3 (Table 2).
Table 2.
Behavioural, clinical and health-related characteristics of pneumonia suspected HIV-positive study participants (n = 378) at Dessie town health facilities, Northeast Ethiopia, 2021.
| Variables | Category | Frequency | Percentage (%) |
|---|---|---|---|
| Hand washing habit | Yes | 366 | (96.8) |
| No | 12 | (3.2) | |
| Cigarette smoking | Yes | 120 | (31.6) |
| No | 258 | (68.4) | |
| Alcohol consumption | Always | 23 | (6.1) |
| Some times | 80 | (21.2) | |
| Never | 275 | (78.7) | |
| Chat chewing | Always | 26 | (6.9) |
| Some times | 53 | (14) | |
| Never | 299 | (79.1) | |
| Current WHO clinical stage | Stage-I | 127 | (33.6) |
| Stage-II | 168 | (44.4) | |
| Stage-III | 67 | (17.7) | |
| Stage-IV | 16 | (4.2) | |
| CD4 count | 0—200 | 51 | (13.5) |
| 201—350 | 137 | (36.2) | |
| 351—500 | 99 | (26.2) | |
| >500 | 91 | (24.1) | |
| White blood cell counts | Low | 185 | (48.9) |
| Normal | 170 | (45.0) | |
| High | 23 | (6.1) | |
| Viral load count | >1000 copies/cell | 180 | (47.6) |
| 150–1000 copies/cell | 15 | (4.0) | |
| <150 copies/cell | 183 | (48.4) | |
| History of hospitalization in the last 6 months | Yes | 43 | (11.4) |
| No | 335 | (88.6) | |
| Prophylaxis TMP.SMX | Yes | 193 | (51.1) |
| No | 185 | (48.9) | |
| Co-infection | Yes | 62 | (16.4) |
| No | 318 | (83.6) | |
| ART failure | Yes | 49 | (13.0) |
| No | 329 | (87.0) | |
| Current BMI | Under weight | 108 | (28.6) |
| Normal | 237 | (62.7) | |
| Over weight | 33 | (8.7) | |
| ART treatment duration | 7–12 months | 3 | (0.8) |
| 1–5 years | 72 | (19) | |
| >5 years | 303 | (80.2) |
WHO = World Health Organization; ART = antiretroviral treatment; BMI = body mass index; CD4 = cluster of differentiation 4.
Prevalence of bacterial isolates
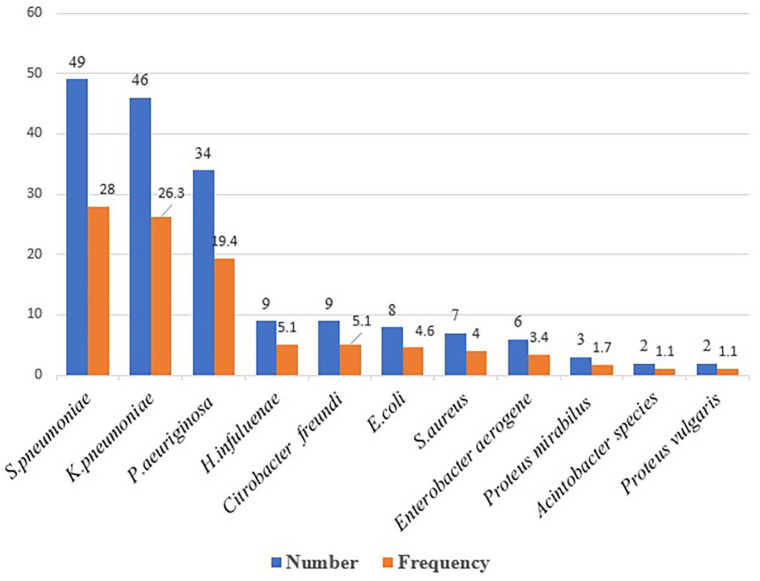
The overall prevalence of bacterial infection was 46.3% (95% CI: 41.3%–51.3%), and almost equal prevalence rate was observed among male and female participants. Of the total 175 isolates, Gram-negative bacteria accounted for 119 (68%). Overall, the predominant isolates identified were S. pneumoniae 49 (28%) followed by K. pneumoniae 46 (26.3%), P. aeruginosa 34 (19.4%). Whereas, the least identified bacteria were Acinetobacter species and Proteus vulugaris 2 (1.1%) each (Figure 1).
Figure 1.
Frequency of bacteria isolated from pneumonia suspected HIV-positive patients at Dessie town health facilities, Northeast Ethiopia, 2021.
Antimicrobial susceptibility profile of bacterial isolates
Gram-positive bacteria showed high levels of resistance to tetracycline 49 (87.5%), penicillin 48 (85.7%), trimethoprim-sulfamethoxazole 34 (78.6%) and chloramphenicol 37 (66.1%). whereas 40 (71.4%) and 42 (75%) of the Gram-positive isolates were sensitive to clindamycin and oxacillin, respectively. Moreover, 57% of S. aureus isolates also showed resistance to methicillin (Table 3).
Table 3.
Antimicrobial resistance and susceptibility pattern of Gram-positive bacteria isolated from pneumonia-suspected HIV-positive patients at Dessie town health facilities, Northeast Ethiopia, 2021.
| Gram-positive bacterial isolates | Antimicrobial agents tested | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E N (%) |
FOX N (%) |
C N (%) |
CXT N (%) |
CRO N (%) |
CIP N (%) |
SXT N (%) |
CL N (%) |
OX N (%) |
TEN (%) | PN (%) | ||
| S. pneumoniae(49) | S | 21 (42.9) | NT | 13 (26.5) | 26 (53.1) | 26 (53.1) | NT | 10 (20.4) | 36 (73.5) | 37 (75.5) | 6 (12.4) | 7 (6.3) |
| R | 28 (57.1) | NT | 36 (73.5) | 23 (46.9) | 23 (46.9) | NT | 39 (79.6) | 13 (27.3) | 12 (24.5) | 43 (87.6) | 41 (83.7) | |
| S. aureus (7) | S | 3 (42.9) | 3 (42.9) | 3 (42.9) | 3 (42.9) | 4 (57.1) | 4 (51.1) | 2 (28.6) | 4 (57.1) | 5 (71.4) | 1 (14.3) | 1 (14.3) |
| R | 4 (57.1) | 4 (57.1) | 4 (57.1) | 4 (57.1) | 3 (42.9) | 3 (42.9) | 5 (71.4) | 3 (42.9) | 2 (28.6) | 6 (85.7) | 6 (85.7) | |
| Total (56) | S | 24 (42.9) | 3 (42.9) | 19 (33.9) | 29 (51.8) | 30 (53.6) | 4 (51.1) | 12 (21.4) | 40 (71.4) | 42 (75) | 7 (12.5) | 8 (14.3) |
| R | 32 (57.1) | 4 (57.1) | 37 (66.1) | 27 (48.2) | 29 (46.3) | 3 (42.9) | 34 (78.6) | 16 (28.6) | 12 (25) | 49 (87.5) | 48 (85.7) | |
CL = Clindamycin; E = Erythromycin; C = Chloramphenicol; CIP = Ciprofloxacin; TE = Tetracycline; SXT = Trimethoprim-Sulfamethoxazole; CRO = Ceftriaxone; P = penicillin; R = Resistant; OX = oxacillin; FOX = Cefoxitin; S = Sensitive; NT = Not tested.
Majority of the Gram-negative isolates showed resistance rate of 67 (88.2%) for tetracycline, 65 (88.2%) for Ampicillin and 65 (87.8%) for amoxacillin-clavulinic acid. Rates of resistance of Gram-negative bacterial isolates against ceftazidime, tetracycline, trimethoprim-sulfamethoxazole, chloramphenicol, amoxacillin-clavulinic acid, cefotaxime, amikacin, ceftriaxone, meropenem and gentamicin ranged from 45 (37.8%)–67 (88.2%). However, Gram-negative bacterial isolates showed relatively low resistance against amikacin 45 (37.8) and meropenem 47 (39.5) (Table 4).
Table 4.
Antimicrobial resistance and susceptibility pattern of Gram-negative bacteria among pneumonia-suspected HIV-positive patients at Dessie town health facilities ART clinic, Northeast Ethiopia, 2021.
| Gram -negative bacterial isolates | Antimicrobials tested | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C N (%) |
TE N (%) |
CIP N (%) |
CXT N (%) |
SXT N (%) |
GN N (%) |
AMP N (%) |
AMC N (%) |
CRO N (%) |
CAZ N (%) |
AMK N (%) |
MEM N (%) |
TZP N (%) |
||
| K. pneumoniae (46) | S | 13 (33.3) | 4 (8.7) | 28 (60.9) | 25 (54.3) | 9 (19.6) | 31 (63.4) | 5 (10.9) | 6 (13) | 25 (54.3) | 21 (45.7) | 32 (69.6) | 32 (69.6) | NT |
| R | 36 (66.7) | 42 (91.3) | 18 (39.1) | 21 (45.7) | 37 (80.4) | 15 (36.6) | 41 (89.1) | 40 (87) | 21 (45.7) | 25 (54.3) | 13 (30.4) | 13 (30.4) | NT | |
| P. aeruginosa (34) | S | NT | NT | 13 (38.2) | 15 (44.1) | NT | 14 (41.2) | NT | NT | 15 (44.1) | 12 (35.3) | 19 (55.9) | 20 (58.8) | 13 (38.2) |
| R | NT | NT | 21 (61.8) | 19 (55.9) | NT | 20 (58.8) | NT | NT | 19 (55.9) | 22 (64.7) | 15 (44.1) | 14 (41.2) | 21 (61.8) | |
| C. freundii (9) | S | 3 (33.6) | 1 (11.1) | 6 (66.7) | 5 (55.6) | 2 (23.3) | 5 (55.6) | 1 (11.1) | NT | 4 (44.4) | 3 (33.3) | 5 (55.6) | 4 (66.7) | NT |
| R | 6 (66.4) | 8 (89.9) | 3 (33.3) | 4 (44.4) | 7 (77.7) | 4 (44.4) | 8 (88.9) | NT | 5 (55.6) | 6 (66.7) | 4 (44.4) | 3 (33.3) | NT | |
| H. influenzae (9) | S | 1 (11.1) | 2 (22.2) | 5 (56.6) | 6 (66.7) | 3 (33.3) | 5 (56.6) | 1 (11.1) | 1 (11.1) | 5 (55.6) | 4 (444) | 5 (56.6) | 5 (55.6) | NT |
| R | 8 (88.9) | 7 (77.8) | 4 (44.4) | 3 (33.3) | 6 (66.7) | 4 (44.4) | 8 (88.9) | 8 (88.9) | 4 (44.4) | 5 (55.6) | 4 (44.4) | 4 (44.4) | NT | |
| E. coli (8) | S | 3 (37.5) | 1 (12.5) | 6 (71.5) | 5 (62.5) | 3 (37.5) | 6 (75) | 1 (12.5) | 1 (12.5) | 5 (62.5) | 4 (50) | 5 (62.5) | 5 (62.5) | NT |
| R | 5 (62.5) | 7 (87.5) | 2 (28.5) | 3 (37.5) | 5 (62.5) | 2 (25) | 7 (87.5) | 7 (87.5) | 3 (37.5) | 4 (50) | 3 (37.5) | 3 (37.5) | NT | |
| Enterobacter aerogene (6) | S | 1 (16.9) | 1 (16.7) | 4 (66.7) | 3 (50) | 2 (33.3) | 4 (66.7) | 1 (16.9) | 1 (16.5) | 4 (66.7) | 3 (50) | 4 (66.7) | 3 (50) | NT |
| R | 5 (83.1) | 5 (83.3) | 2 (33.3) | 3 (50) | 4 (66.7) | 2 (33.3) | 5 (83.1) | 5 (83.5) | 2 (33.3) | 3 (50) | 2 (33.3) | 3 (50) | NT | |
| P. mirabilis (3) | S | 1 (33.3) | NT | 1 (33.3) | 4 (66.7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (66.7) | NT | 2 (66.7) | 1 (33.3) | NT |
| R | 2 (66.7) | NT | 2 (66.7) | 1 (33.3) | 3 (100) | 3 (100) | 3 (100) | 3 (1000) | 1 (33.3) | NT | 1 (33.3) | 2 (66.7) | NT | |
| P. vulgaris (2) | S | 1 (50) | NT | 1 (50) | 1 (100) | 0 (0) | 1 (50) | 0 (0) | 0 (0) | 2 (100) | NT | 1 (50) | 1 (50) | NT |
| R | 1 (50) | NT | 1 (50) | 0 (100) | 2 (100) | 1 (50) | 2 (100) | 2 (100) | 0 (0) | NT | 1 (50) | 1 (50) | NT | |
| Acinetobacter species (2) | S | NT | NT | 0 (0) | NT | NT | 0 (0) | NT | NT | NT | 0 (0) | 1 (50) | 1 (50) | 0 (0) |
| R | NT | NT | 2 (100) | NT | NT | 2 (100) | NT | NT | NT | 2 (100) | 1 (50) | 1 (50) | 2 (100) | |
| Total (119) | S | 23 (27.7) | 9 (11.8) | 64 (53.8) | 64 (54.7) | 19 (22.9) | 66 (55.5) | 9 (11.8) | 9 (12.2) | 62 (53) | 47 (41.2) | 74 (62.2) | 72 (60.5) | 13 (36.1) |
| R | 60 (72.3) | 67 (88.2) | 55 (46.2) | 53 (45.3) | 64 (77.1) | 53 (44.5) | 65 (88.2) | 65 (87.8) | 55 (47) | 67 (58.8) | 45 (37.8) | 47 (39.5) | 23 (63.9) | |
NT = Note tested; AMP = Ampicillin; GN = Gentamicin; AMK = Amikacin; CIP = Ciprofloxacin (5 μg); SXT = trimethoprim-sulfamethoxazole; MEM = Meropenem; AMC = Amoxicillin-clavulanic acid; CTX = cefotaxime; CAZ = Ceftazidime; CRO = ceftriaxone; TE = Tetracycline (30 μg); C = Chloramphenicol; TZP = Piperacillin tazobactam; R = Resistant; S = Sensitive.
MDR patterns of the isolates
Overall, 173 (98.9%) bacterial isolates were resistant to at least one antimicrobial agent and 167 (95.4%) isolates were resistant to ⩾2 antimicrobials. About 25 (143%) isolates had resistance to five and more antimicrobials. The overall prevalence of MDR bacteria was 148 (84.6%). The MDR rate of Gram-positive and Gram-negative isolates was 47 (83.9%) and 101 (84.9%), respectively. About 43 (93.4%) of K. pneumoniae, 7 (87.5%) of E. coli, 42 (85.7%) of S. pneumoniae, 5 (83.3%) of Enterobacter spp. and 67% of H. influenzae isolates developed MDR (Table 5).
Table 5.
Multidrug resistance profile of bacterial isolates (n = 175) from pneumonia-suspected HIV-positive participants at Dessie town health facilities, Northeast Ethiopia, 2021.
| Isolated bacteria | Antimicrobial resistance pattern | MDR; n (%) | |||||
|---|---|---|---|---|---|---|---|
| Ro; n (%) | R1; n (%) | R2; n (%) | R3; n (%) | R4; n (%) | ⩾R5; n (%) | ||
| Gram positive | 0 (0) | 2 (3.6) | 7 (12.5) | 28 (50) | 16 (28.6) | 3 (5.4) | 47 (83.9) |
| S. pneumoniae | 0 (0) | 2 (4.8) | 5 (30.6) | 25 (28.6) | 14 (20.4) | 3 (6.1) | 42 (85.7) |
| S. aureus | 0 (0) | 0 (0) | 2 (28.5) | 3 (43) | 2 (28.5) | 0 (0) | 5 (71.42) |
| Gram negative | 2 (1.7) | 2 (3.4) | 14 (11.8) | 52 (43.7) | 28 (23.5) | 22 (18.5) | 101 (84.9) |
| K. pneumoniae | 0 (0) | 2 (4.3) | 1 (2.2) | 22 (47.8) | 11 (23.9) | 10 (21.7) | 43 (93.4) |
| P. aeruginosa | 1 (2.9) | 0 (0) | 4 (11.8) | 21 (55.9) | 5 (14.7) | 3 (21.4) | 29 (85.3) |
| E. coli | 0 (0) | 0 (0) | 1 (12.5) | 2 (25) | 2 (25) | 3 (37.5) | 7 (87.5) |
| C. freundii | 0 (0) | 0 (0) | 2 (22.2) | 0 (0) | 2 (22.2) | 5 (55.55) | 7 (77.77) |
| Enterobacter spp. | 0 (0) | 0 (0) | 1 (16.7) | 1 (16.7) | 3 (33.3) | 1 (16.7) | 5 (83.3) |
| H. influenzae | 1 (11.1) | 0 (0) | 1 (22.2) | 4 (33.30) | 3 (33.3) | 0 (0) | 7 (77.7) |
| P. mirabilis | 0 (0) | 0 (0) | 1 (33.3) | 0 (0) | 2 (66.7) | 0 (0) | 2 (66.7) |
| P. vulgaris | 0 (0) | 0 (0) | 1 (50) | 1 (50) | 0 (0) | 0 (0) | 1 (50) |
| Acinetobacter speciesa | 0 (0) | 0 (0) | 2 (100) | 2 (100) | 0 (0) | 0 (0) | 2 (100) |
| Overall totalb | 2 (1.1) | 6 (3.4) | 21 (12) | 80 (45.7) | 44 (25.1) | 25 (14.3) | 148 (84.6) |
MDR = multidrug resistance; R0 = No antibiotic resistance; R1 = resistance to one; R2 = resistance to two; R3 = resistance to three; R4 = resistance to four; R4 = resistance to four antibiotics; R5 = resistance to five and more than five antibiotics class.
Percent is computed from the total number of each bacteria species.
Percent is computed from a total number of isolates.
Factors associated with bacterial infection
All variables with a p-value of 0.25 in the bivariate analysis were entered into multivariable logistic regression analysis. Accordingly, alcohol consumption frequently (Adjusted Odds Ratio (AOR) = 3.474, 95% CI: 1.07–11.31, p = 0.039), viral load >1000 copies/ml (AOR = 4.88, 95% CI: 1.88–36.54, p = 0.002), cigarette smoking (AOR = 3.87, 95% CI: 1.56–54.87, p = 0.023), CD4+ cell count less than 200 cells/mm3 (AOR = 6.5, 95% CI: 1.16–11.21, p = 0.027), WHO HIV clinical stage IV (AOR = 9.51, 95% CI: 1.66–54.45, p = 0.011), educational status of participants who only read and write (AOR = 3.415, 95% CI: 1.48–22.88, p = 0.006); and most white blood cell count less than 4000 cells/mm3 (AOR = 3. 5, 95% CI: 1.90–19.57, p = 0.023) were found to have statistically significant association with bacterial infection (Table 6).
Table 6.
Bivariable and multivariable logistic regression analysis of factors associated with bacterial pneumonia among pneumonia suspected HIV patient’s infection at Dessie town health facilities, Northeast Ethiopia, 2021.
| Variable of study participant | Category | Bacterial growth N (%) | COR (95% CI) | p-Value | AOR (95% CI) | p-Value | |
|---|---|---|---|---|---|---|---|
| Yes | No | ||||||
| Sex | Male | 87 (46.5) | 100 (53.5) | 1.8 (0.68–12.53) | 0.93 | NA | |
| Female | 88 (46) | 103 (54) | Ref | ||||
| Age of study participant | 10–16 | 4 (40) | 6 (60) | Ref | |||
| 17–30 | 27 (35.1) | 50 (64.9) | 1.57 (0.15–2.13) | 0.405 | |||
| 31–45 | 81 (46.6) | 93 (53.4) | 1.46 (0.26–0.84) | 0.34 | |||
| >45 | 63 (53.8) | 54 (46.2) | 1.747 (0.47–1.19) | 0.28 | |||
| Alcohol drinking habit | Always | 13 (56.5) | 10 (43.5) | 9.46 (1.04–5.83) | 0.18 | 3.47 (1.07–11.31) | 0.039* |
| Sometimes | 67 (83.8) | 13 (16.20) | 1.76 (5.13–18.59) | 0.29 | |||
| Never | 95 (34.5) | 180 (65.50) | Ref | ||||
| Occupational status | Employed | 20 (24.4) | 62 (75.6) | Ref | |||
| Student | 9 (37.5) | 15 (62.5) | 1.86 (0.71–4.90) | 0.209 | |||
| Merchant | 52 (53.6) | 45 (46.4) | 1.75 (0.88–16.81) | 0.35 | |||
| House wife | 17 (60.7) | 11 (39.3) | 1.9 (0.92–11.91) | 0.28 | |||
| Farmer | 7 (43.8) | 9 (56.2) | 1.41 (0.80–17.04) | 0.25 | 1.90 (0.06–15.29) | 0.061 | |
| Daily labor | 70 (53.4) | 61 (46.4) | 3.56 (1.93–6.55) | 0.01 | 2.25 (1.09–6.71) | 0.009* | |
| Educational status | Illiterate | 52 (50) | 52 (50) | 1.43 (0.783–2.623) | 0.12 | 3.45 (1.48–22.88) | 0.006* |
| Only read and write | 29 (78.4) | 8 (21.6) | 1.6 (0.89–12.92) | 0.26 | |||
| Primary school | 38 (38) | 62 (62) | 0.88 (0.47–1.63) | 0.681 | |||
| Secondary-school | 26 (40.6) | 38 (59.4) | 0.981 (0.50–1.94) | 0.955 | |||
| College and above | 30 (41.1) | 43 (58.9) | Ref | ||||
| Residence | Rural | 57 (64) | 32 (36) | 2.58 (1.58–4.22) | 0.15 | 2.579 (1.66–18.06) | 0.025* |
| Urban | 118 (40.8) | 171 (59.2) | Ref | ||||
| CD+ count | <200 cells/mm3 | 35 (68.6) | 16 (31.4) | 5.51 (8.04–49.39) | 0.018 | 6.50 (1.16–11.21) | 0.027* |
| 201–350 cells/mm3 | 95 (69.3) | 42 (30.7) | 4.90 (9.46–44.88) | 0.21 | 3.20 (0.72–13.11) | 0.128 | |
| 351–500 cells/mm3 | 36 (36.4) | 63 (63.6) | 0.56 (0.23–11.60) | 0.65 | |||
| >500 cells/mm3 | 9 (9.9) | 82 (90.1) | Ref | ||||
| WBC cells count | Low (<4000) | 134 (72.4) | 51 (27.6) | 11.80 (1.71–19.52) | 0.017 | 3.50 (1.90–19.57) | 0.023* |
| High (>11,000) | 10 (43.5) | 13 (56.5) | 0.74 (0.45–13.76) | 0.35 | |||
| Normal (4000–11,000) | 31 (1) | 139 (82.5) | Ref | ||||
| WHO clinical stage | Stage I | 32 (25.2) | 95 (74.8) | Ref | |||
| Stage II | 88 (52.4) | 80 (47.8) | 0.56 (0.19–11.65) | 0.290 | |||
| Stage III | 49 (73.1) | 18 (26.9) | 1.86 (0.65–5.34) | 0.251 | |||
| Stage IV | 10 (62.5) | 6 (37.5) | 4.54 (1.44–14.29) | 0.010 | 9.51 (1.66–54.47) | 0.011* | |
| Marital status | Single | 5 (20) | 20 (80) | Ref | |||
| Divorced | 7 (38.9) | 11 (61.1) | 1.26 (0.96–0.71) | 0.58 | |||
| Widowed | 7 (41.2) | 10 (58.8) | 1.66 (0.25–1.75) | 0.404 | |||
| Married | 156 (49.1) | 162 (50.9) | 1.73 (0.27–1.96) | 0.528 | |||
| Current smoking habit | Yes | 88 (73.3) | 32 (26.70) | 5.45 (1.35–8.73) | 0.021 | 3.87 (1.56–23.82) | 0.023* |
| No | 87 (33.7) | 171 (66.3) | Ref | ||||
| Chat chewing habit | Always | 20 (76.9) | 6 (23.1) | 3.48 (2.14–14.07) | 0.011 | 8.48 (1.87–30.07) | 0.001* |
| Sometimes | 42 (79.2) | 11 (20.8) | 3.28 (3.11–12.70) | 0.056 | |||
| Never | 113 (37.8) | 186 (62.8) | Ref | ||||
| Viral load | >1000 copies/ml | 147 (81.7) | 33 (18.3) | 6.99 (2.8–58.46) | 0.021 | 4.85 (1.88–36.54) | 0.002* |
| 150–1000 copies/ml | 6 (40) | 9 (60) | 1.88 (0.58–15.03) | 0.21 | |||
| <150 copies/ml | 22 (12.0) | 61 (88) | Ref | ||||
| Underlying chronic diseases | Yes | 22 (84.6) | 4 (15.4) | 2.14 (0.45–11.41) | 0.45 | ||
| No | 153 (43.5) | 199 (56.5) | Ref | ||||
| Coinfection/comorbidity | Yes | 42 (84) | 8 (16) | 1.13 (0.66–25.29) | 0.29 | ||
| No | 133 (40.5) | 195 (59.5) | Ref | ||||
| History of ART failure | Yes | 16 (61.5) | 10 (68.5) | 1.52 (0.23–13.17) | 0.312 | ||
| No | 159 (45.2) | 193 (54.80) | Ref | ||||
| BMI | Under weight | 82 (75.9) | 26 (24.1) | 1.519 (0.39–12.72) | 0.65 | ||
| Over weight | 12 (36.4) | 21 (63.4) | 0.91 (0.43–1.94) | 0.804 | |||
| Normal weight | 81 (36.2) | 156 (65.8) | Ref | ||||
| ART treatment duration | 7–12 months | 2 (66.7) | 1 (33.3) | Ref | |||
| 1–5 years | 28 (38.9) | 44 (61.1) | 0.67 (0.28–1.56) | 0.340 | NA | ||
| >5 years | 145 (47.9) | 158 (52.1) | 0.99 (0.46–2.3) | 0.999 | |||
| Cotrimoxazole prophylaxis | Yes | 143 (74.1) | 50 (25.9) | Ref | |||
| No | 32 (17.3) | 153 (82.7) | 0.65 (0.48–2.10) | 0.35 | NA | ||
ART = antiretroviral therapy; AOR = adjusted odds ratio; BMI = body mass index; CI = confidence interval; COR = crude odds ratio; Ref = reference.
Significant at p < 0.05.
Discussion
Bacterial pneumonia turns into the most common infection in HIV-infected patients and the most joint cause of hospital admission and mortality in high HIV epidemic countries.36
In the present study, the overall prevalence of culture positive sputum among HIV-positive patients was 46.3%. This is comparable with previous studies done in Ethiopia such as Mekelle 43.7%,21 Jimma 45%37 and Arbaminch 42.9%,38 and other parts of the world including Nigeria 42.9%39 and Nepal 46.6%.30 The current finding, however, is higher than previous reports from India (16.6%9 and 17.1%),43 Malawi 29%,40 Nepal 39.7%17 and Ethiopia (32.1% and 40.3%),41,42 but lower than previous prevalence reports from Nigeria (54.07% and 55.6%)43,44 and Spain 53.5%.16 The observed inconsistency might be due to methodological differences, sample size, seasonal variations, or sociodemographic variability of the study participants. Besides, the majority of our study participants had CD4 cell counts of less than 350 cells/mm3 and a higher viral load, which subsequently increases the incidence of bacterial infection and leads to concurrent opportunistic infections.7
Similarly, like most previous study reports in our country and elsewhere, our findings showed that the majority of the isolated etiological agents were Gram-negative bacteria (68%). This finding is compatible with similar studies done in Addis Ababa,41 Tanzania45 and Nepal.17 However, it is different from the study reports in Bahirdar44) and Jimma.43 Etiological differences might be due to environmental contamination and the fact that the majority of Gram-positive bacteria are vulnerable to antibiotics that are widely self-administered, especially the concurrent use of cotrimoxazole drug as a prophylactic agent among HIV patients, which may result in a reduction in the incidence of Gram-positive bacteria.
In the present study, the predominant isolate was S. pneumoniae 49 (26.3%) followed by K. pneumoniae 46 (47.4%) and P. aeruginosa 34 (19.4%). This result is inconsistent with other studies conducted in Ethiopia such as Arbaminch,38 Jimma,43 Bahirdar,44 others with different geographical regions of the world like India,1,46 United Kingdom40 and Spain15 showed that S. pneumoniae was the predominant isolate On the contrary, previous studies conducted in Mekelle,21 Addis Ababa41 Tanzania45 and Bahirdar47; and elsewhere in the world including Nepal,17 Nigeria43,44 and India.14,26,48 This dominance could be attributed to their capsular nature and the emergence of strains from both species with additional genetic traits. The bacteria’s ability to form biofilm, fimbriae and capsular protein to prevent phagocytosis by polymorphonuclear leukocytes and macrophages, as well as the emergence of strains that can acquire additional genetic traits, may explain the bacteria’s dominance49 and capsule that causes severe pneumonia in immunocompromised people.50
According to the international standard for the definition of drug resistance,16 the overall multidrug resistance was found in 84.6% of the total isolated bacteria. This finding was slightly higher as compared to the studies conducted in Bahirdar Ethiopia 76%,42 Nigeria (67.2%)51 and Cameron 79.4%.52 The result was much higher than a study reported from Ethiopia that ranged from 17.9%–56.7%18,29,30; and a systematic review report of 59.7% overall MDR prevalence in Ethiopia.52 The reason for the high MDR prevalence might be due to poor infection control strategies, inappropriate utilization of antimicrobial agents in empirical treatment, extreme antibiotic use, and self-antibiotic prescribing habits.18 MDR was found in 43 (93.4%) of K. pneumoniae isolates, 7 (87.5%) of E. coli isolates, 42 (85.7%) of S. pneumoniae isolates, 5 (83.3%) of Enterobacter spp. isolates and 67% of H. influenzae isolates. This is mainly attributed to the ability of those species to express resistance genes, including external and spread on mobile genetic elements, produce extended-spectrum beta-lactamases and carbapenems, aminoglycoside-modifying enzymes, and porin-efflux mechanisms, which are the main contributing factors of MDR.53,54
In this study, Gram negative bacteria showed the highest resistance rate to 67 (88.2%) for tetracycline, 65 (88.2%) for ampicillin and 65 (87.8%) for amoxacillin-clavulinic acid. Similar findings have been reported in previous studies done in Bahirdar,42 Jimma,41 Mekele,21 Nigeria,43 Nepal17 and India.14 This could be due to the drug’s long-term overuse, the expression of extended-spectrum beta-lactamases, which were developed to resist penicillin, cephalosporins, and monobactams, and the expression of carbapenems, which provides resistance to those -lactams, including carbapenems. The capsule, production of biofilm, efflux pumps and production of polysaccharide matrix that coats the cell can limit the penetration of certain agents.55,56 On the other hand, lower resistance was observed against gentamicin, amikacin, ciprofloxacin, and meropenem, which is indicative of a possible drug of choice for such infections.
In our study, Gram-positive bacteria were resistant to tetracycline 49 (87.5%), penicillin 48 (85.7%), trimethoprim-sulfamethoxazole 34 (78.6%) and chloramphenicol 37 (66.1%). which is in line with previous studies conducted in Arbaminch,38 Jimma,41 India14 and Nepal.57 This might be due to Lack of pump inhibition, efflux proteins, structural modifications.58 Furthermore, long-term usage, easy availability, common prophylaxis and indiscriminate use of commonly used drugs such as trimethoprim-sulfamethoxazole and penicillin could lead to an increase in resistance. On the other hand, Gram-positive isolates showed sensitivity of 70.0% to clindamycin, 62.1% to ceftriaxone, and 72.4% to oxacillin, which was also in agreement with other studies conducted in Mekelle,21 Bahirdar,44 China59 and India.60
Cotrimoxazole has been used as a prophylactic agent against opportunistic infections in HIV/AIDS patients around the world for over 10 years, and it is now used in all HIV/AIDS patients as a prophylactic agent.61 In our study, 193 patients were given cotrimoxazole and 69.5% of bacterial isolates were resistant to cotrimoxazole. This finding was supported by a study report from Tanzania (81.3%).61 This might be attributable to long-term usage of cotrimoxazole as prophylaxis and a common antibacterial agent, which subsequently led to increased development of resistance.61
Patients with CD4+ count less than 200 cells/mm3, viral load 1000 copies/ml and viral load >1000 copies/ml, and white blood cell count less than 4000 cells had a statistically significant association with the occurrence of CAP in the current study, which is comparable to studies from Ethiopia,21 Nepal,30 India,17 Brazil,12 India14 and Nigeria.44 This might be due to the large number of study participants having a low CD4+ count and WHO stages III and IV. When depletion of the CD4+ T cells occurs, bacterial infection may occur. Increasing viral numbers weaken the immune status of the patient, causing impairment of their immunological system (alveolar CD4+ T-cell function is impaired) and is probably related to the impairment of humoral immunity.6,62
Study participants with a viral load of ⩾1000 copies/ml were 4.85 times more likely to develop bacterial infection, respectively, as compared to individuals having <150 copies/ml viral load. This is in agreement with studies conducted in Ethiopia,44 Spain63 and South Africa.64 This might be due to the fact that virus-mediated immunosuppression of the host innate immunity exposes the patient to opportunistic bacteria colonization and viral-induced secondary bacterial infections.62
Cigarette smoking is one of the potential risk factors for the development of pneumonia. In our study, pneumonia patients who smoked frequently were 3.87 times more likely to develop bacterial infection than non-smokers. This was comparable with studies done in Ethiopia,21 Brazil12 and Nepal,17 Kenya65 and Spain63,66 that showed a significant association of cigarette smoking with bacterial infection. This is because cigarette smoking produces structural changes in the respiratory tract that cause peribronchial inflammation and fibrosis, increased mucosal permeability, weakens the muco-ciliary clearance, affects pathogen adherence, and disruption of the respiratory epithelium, which in turn exposes one to lower respiratory tract infections, which may increase the cigarette smoke-induced lung inflammation.67
Alcohol consumption was also found to have a statistically significant association with bacterial infection. In our study, alcohol consumers were 3.474 times more likely to have bacterial pneumonia than non-consumers. This was in line with studies done in Ethiopia,21 South Africa68 and North America.69 This might be because of the sedative effects of alcohol, which can reduce the oropharyngeal tone, leading to an increased risk of aspiration of microbes and altering the pathophysiology by decreasing the phagocytic function of the alveolar macrophages, impairing bacterial clearance and diminishing pulmonary defence against infection. Furthermore, high levels of alcohol blunt mental function and suppress cough and gag reflexes. It also decreases muco-ciliary clearance, impairing both innate and acquired immunity, reduces the production of chemokines, and blunts the chemotaxis of neutrophils.69
Limitation of the study
Due to a lack of resources, we did not attempt to isolate atypical bacterial agents such as Chlamydia, mycoplasma, or Legionella species and respiratory infections. Moreover, molecular characterization of the isolated bacterial agents was not done.
Conclusion
In the current study, relatively higher proportions of K. pneumoniae, P. aeruginosa and S. pneumoniae were the most bacterial isolated identified. In this study, majority of isolated bacteria were Gram-negative. Most of the isolates were found susceptible to gentamicin, cefotaxime and ceftriaxone. However, they were resistant to commonly used antimicrobials like amoxicillin, tetracycline, amoxicillin-clavulanate and co-trimoxazole. However, antimicrobial resistance including MDR was observed to a number of commonly used antibiotics, such as trimethoprim-sulfamethoxazole ampicillin, amoxicillin-clavulanic acid and tetracycline. Detectable recent viral load, low CD4+ count, late WHO clinical stage, cigarette smoking and alcohol consumption were found significant predictors of bacterial infection. Thus, preventive measures to minimize the risk of the disease including life-style factors such as smoking and alcohol consumption; and advocation of proper antimicrobial usage should be done. ART services should strengthen good adherence of ART and monitoring to decrease patients’ viral load and increase CD4+ counts. Moreover, culture screening and antimicrobial susceptibility testing should be practiced routinely; and large-scale population-based surveys are needed to assess effectiveness of cotrimoxazole and molecular-based studies are needed to monitor the epidemiology of MDR bacterial isolates and combat antimicrobial resistance.
Supplemental Material
Supplemental material, sj-docx-1-smo-10.1177_20503121221145569 for Bacteriology of community-acquired pneumonia, antimicrobial susceptibility pattern and associated risk factors among HIV patients, Northeast Ethiopia: cross-sectional study by Mihret Tilahun, Daniel Gebretsadik, Abdurahaman Seid, Alemu Gedefie, Melaku Ashagrie Belete, Melkam Tesfaye, Edosa Kebede and Agumas Shibabaw in SAGE Open Medicine
Acknowledgments
The authors would like to acknowledge Department of Medical Laboratory Science, College of Medicine and Health Sciences, Wollo University for providing the laboratory set up and facilities to conduct the experiments. Health facilities in Dessie town and all study participants are gratefully acknowledged for their kind cooperation during data collection.
Footnotes
Authors’ contributions: Conceived and designed the experiments: Mihret Tilahun, Abdurrahman Seid, Daniel Gebretsadik; performed the experiments: Mihret Tilahun, Abdurrahman Seid, Agumas Shibabaw; analyzed the data: Mihret Tilahun, Abdurrahman Seid, Daniel Gebretsadik, Alemu Gedefie, Melaku Ashagrie Belete; and wrote and edited the manuscript: Mihret Tilahun, Abdurrahman Seid, Alemu Gedefie, Agumas Shibabaw, Melaku Ashagrie Belete. All authors read and approved the final manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics approval: The study was done from HIV-positive CAP suspected patients from antiretroviral clinics in the health facilities of the Dessie town. Supportive letter to conduct this study was obtained from college of medicine and health science community service, research and post graduate office, Wollo University with a protocol number of CMHS/HC/354/13. A permission letter was obtained from the Dessie town health office and then in each health institutions.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Informed consent: Written informed consent was obtained from the subjects and the legally authorized representatives of the minor subjects prior to study initiation.
ORCID iDs: Mihret Tilahun  https://orcid.org/0000-0002-2343-2119
https://orcid.org/0000-0002-2343-2119
Daniel Gebretsadik  https://orcid.org/0000-0001-5032-5143
https://orcid.org/0000-0001-5032-5143
Alemu Gedefie  https://orcid.org/0000-0002-9678-5513
https://orcid.org/0000-0002-9678-5513
Edosa Kebede  https://orcid.org/0000-0001-7006-303X
https://orcid.org/0000-0001-7006-303X
Data availability statement: Data supporting the conclusions of this article are within the manuscript and are available on reasonable request from the principal investigators due to ethical reasons.
Supplemental material: Supplemental material for this article is available online.
References
- 1. Shah RR. Microbiological profile of respiratory tract infections among HIV-infected and HIV-non infected patients: a comparative study. Int Arch Integr Med 2016; 3(2): 51–59. [Google Scholar]
- 2. Morris A, Crothers K, Beck J, et al. An official ATS workshop report: emerging issues and current controversies in HIV-associated pulmonary diseases. Proc Am Thorac Soc 2011; 8(1): 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tural Önür S, Dalar L, İliaz S, et al. Pneumonia in HIV-infected patients. Eurasian J Pulmonol 2016; 18(1): 11–17. [Google Scholar]
- 4. Ethiopian Federal Ministry of Health. Federal HIV/AIDS prevention and control office federal ministry of health of Ethiopia. Guidelines for management of opportunistic infections and anti retroviral treatment in adolescents and adults in Ethiopia. 2008. [Google Scholar]
- 5. Gordin FM, Roediger MP, Girard P-M, et al. Pneumonia in HIV-infected persons: increased risk with cigarette smoking and treatment interruption. Am J Respir Crit Care Med 2008; 178(6): 630–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crothers K, Thompson BW, Burkhardt K, et al. HIV-associated lung infections and complications in the era of combination antiretroviral therapy. Proc Am Thorac Soc 2011; 8(3): 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lazarous DG, O’Donnell AE. Pulmonary infections in the HIV-infected patient in the era of highly active antiretroviral therapy: an update. Curr Infect Dis Rep 2007; 9(3): 228–232. [DOI] [PubMed] [Google Scholar]
- 8. Ferkol T, Schraufnagel D. The global burden of respiratory disease. Ann Am Thorac Soc 2014; 11(3): 404–406. [DOI] [PubMed] [Google Scholar]
- 9. Kandati J, Boorsu SK, Lakshmi Ponugoti M, et al. Bacterial and fungal agents causing lower respiratory tract infections in patients with human immunodeficiency virus infection. Int J Res Med Sci 2016; 4(8): 3595–3600. [Google Scholar]
- 10. Koss CA, Jarlsberg LG, den Boon S, et al. A clinical predictor score for 30-day mortality among HIV-infected adults hospitalized with pneumonia in Uganda. PLoS One 2015; 10(5): e0126591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Curran A, Falco V, Crespo M, et al. Bacterial pneumonia in HIV-infected patients: use of the pneumonia severity index and impact of current management on incidence, aetiology and outcome. HIV Med 2008; 9(8): 609–615. [DOI] [PubMed] [Google Scholar]
- 12. Lamas CC, Coelho LE, Grinsztejn BJ, et al. Community-acquired lower respiratory tract infections in HIV-infected patients on antiretroviral therapy: predictors in a contemporary cohort study. Infection 2017; 45(6): 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gingo MR, Morris A. Pathogenesis of HIV and the lung. Curr HIV/AIDS Rep 2013; 10(1): 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shilpa A, Anuradha K, Venkatesha D. Drug susceptibility pattern of aerobic bacterial isolates from pulmonary infection in HIV seropositives and their correlation with CD4 Count. IOSR J Dent Med Sci 2014; 13(3): 37–41. [Google Scholar]
- 15. Cilloniz C, Torres A, Polverino E, et al. Community-acquired lung respiratory infections in HIV-infected patients: microbial aetiology and outcome. Eur Respir J 2014; 43(6): 1698–1708. [DOI] [PubMed] [Google Scholar]
- 16. Conklin LM, Bigogo G, Jagero G, et al. High Streptococcus pneumoniae colonization prevalence among HIV-infected Kenyan parents in the year before pneumococcal conjugate vaccine introduction. BMC Infect Dis 2015; 16(1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khushbu Y, Satyam P. Bacteriological Profile of Lower Respiratory Tract Infection (LRTI) among HIV Seropositive Cases in Central Terai of Nepal. Int J Curr Microbiol App Sci 2015; 4(11): 431–442. [Google Scholar]
- 18. Mulu W, Yizengaw E, Alemu M, et al. Pharyngeal colonization and drug resistance profiles of Morraxella catarrrhalis, Streptococcus pneumoniae, Staphylococcus aureus, and Haemophilus influenzae among HIV infected children attending ART Clinic of Felegehiwot Referral Hospital, Ethiopia. PLoS One 2018; 13(5): e0196722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Magiorakos A-P, Srinivasan A, Carey Rt, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18(3): 268–281. [DOI] [PubMed] [Google Scholar]
- 20. Sweeney MT, Lubbers BV, Schwarz S, et al. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J Antimicrob Chemother 2018; 73(6): 1460–1463. [DOI] [PubMed] [Google Scholar]
- 21. Adhanom G, Gebreegziabiher D, Weldu Y, et al. Species, risk factors, and antimicrobial susceptibility profiles of bacterial isolates from HIV-infected patients suspected to have pneumonia in Mekelle zone, Tigray, northern Ethiopia. BioMed Res Int 2019; 2019: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dastider D, Jyoti Sen D, Kumar Mandal S, et al. Hand sanitizers bid farewell to germs on surface area of hands. Eur J Pharm Med Res 2020; 7(4): 648–656. [Google Scholar]
- 23. Webb MS, Carey MP. Tobacco smoking among low-income Black women: demographic and psychosocial correlates in a community sample. Nicotine Tob Res 2008; 10(1): 219–229. [DOI] [PubMed] [Google Scholar]
- 24. Saracino A, Lorenzini P, Caputo SL, et al. Increased risk of virologic failure to the first antiretroviral regimen in HIV-infected migrants compared to natives: data from the ICONA cohort. Clin Microbiol Infect 2016; 22(3): 288.e1–288.e8. [DOI] [PubMed] [Google Scholar]
- 25. McShane H. Co-infection with HIV and TB: double trouble. Int J STD AIDS 2005; 16(2): 95–101. [DOI] [PubMed] [Google Scholar]
- 26. Sperrin M, Marshall AD, Higgins V, et al. Body mass index relates weight to height differently in women and older adults: serial cross-sectional surveys in England (1992–2011). J Public Health 2016; 38(3): 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Enawgaw B, Birhan W, Abebe M, et al. Haematological and immunological reference intervals for adult population in the state of Amhara, Ethiopia. Trop Med Int Health 2018; 23(7): 765–773. [DOI] [PubMed] [Google Scholar]
- 28. Walia K, Madhumathi J, Veeraraghavan B, et al. Establishing antimicrobial resistance surveillance & research network in India: journey so far. Indian J Med Res 2019; 149(2): 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ioannidis P, Papaventsis D, Karabela S, et al. Cepheid GeneXpert MTB/RIF assay for Mycobacterium tuberculosis detection and rifampin resistance identification in patients with substantial clinical indications of tuberculosis and smear-negative microscopy results. J Clin Microbiol 2011; 49(8): 3068–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ojha CR, Rijal N, Khagendra K, et al. Lower respiratory tract infections among HIV positive and control group in Nepal. VirusDisease 2015; 26(1): 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murray PR, Rosenthal KS, Pfaller MA. Medical microbiology E-Book. Philadelphia, PA: Elsevier Health Sciences, 2020. [Google Scholar]
- 32. Zapata A, Ramirez-Arcos S. A comparative study of McFarland turbidity standards and the Densimat photometer to determine bacterial cell density. Curr Microbiol 2015; 70(6): 907–909. [DOI] [PubMed] [Google Scholar]
- 33. Weinstein MP, Lewis JS. The clinical and laboratory standards institute subcommittee on antimicrobial susceptibility testing: background, organization, functions, and processes. J Clin Microbiol 2020; 58(3): e01864-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abbey TC, Deak E. What’s new from the CLSI subcommittee on antimicrobial susceptibility testing M100 29th edition. Clin Microbiol Newsl 2020; 41(23): 203–209. [Google Scholar]
- 35. Magiorakos A-P, Srinivasan A, Carey R, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18(3): 268–281. [DOI] [PubMed] [Google Scholar]
- 36. Kengne M, Lebogo MBB, Nwobegahay JM, et al. Antibiotics susceptibility pattern of Streptococcus pneumoniae isolated from sputum cultures of human immunodeficiency virus infected patients in Yaoundé, Cameroon. Pan African Med J 2018; 31: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Regasa B, Yilma D, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from community-acquired pneumonia patients in Jimma University Specialized Hospital, Jimma, Ethiopia. 2015. [Google Scholar]
- 38. Regasa B. Drug resistance patterns of bacterial pathogens from adult patients with pneumonia in Arba Minch hospital, South Ethiopia. Glob J Med Res 2014; 14(5): 1–4. [Google Scholar]
- 39. Salami A, Oluboyo P, Akambi A, et al. Bacterial pneumonia in the AIDS patients. West African J Med 2006; 25(1): 1–5. [DOI] [PubMed] [Google Scholar]
- 40. Hartung TK, Chimbayo D, van Oosterhout JJ, et al. Etiology of suspected pneumonia in adults admitted to a high-dependency unit in Blantyre, Malawi. Am J Trop Med Hyg 2011; 85(1): 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nurahmed N, Kedir S, Fantahun S, et al. Bacterial profile and antimicrobial susceptibility patterns of lower respiratory tract infection among patients attending selected health centers of Addis Ababa, Ethiopia. Egypt J Chest Dis Tuberc 2020; 69(2): 399. [Google Scholar]
- 42. Temesgen D, Bereded F, Derbie A, et al. Bacteriology of community acquired pneumonia in adult patients at Felege Hiwot Referral Hospital, Northwest Ethiopia: a cross-sectional study. Antimicrob Resist Infect Control 2019; 8(1): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ojo-Bola O, Oluyege A. Antibiotics resistance of bacteria associated with pneumonia in HIV/AIDS patients in Nigeria. Am J Infect Dis Microbiol 2014; 2(6): 138–144. [Google Scholar]
- 44. Urama EU, Enweani IB, Oshim IO, et al. Microbiological profile of respiratory tract infections among HIV sero-positive subjects attending Nnamdi Azikiwe University Teaching Hospital Nnewi, Nigeria. Amer J Med and Med Sci 2018; 8: 37–42. [Google Scholar]
- 45. Kishimbo P, Sogone NM, Kalokola F, et al. Prevalence of gram negative bacteria causing community acquired pneumonia among adults in Mwanza City, Tanzania. Pneumonia 2020; 12(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mane A, Gujar P, Gaikwad S, et al. Aetiological spectrum of severe community-acquired pneumonia in HIV-positive patients from Pune, India. Indian J Med Res 2018; 147(2): 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Genetu DE, Zenebe Y. Bacterial profile and their antibiotic resistance pattern among HIV patients diagnosed with pneumonia in Felege-Hiwot referral hospital, Bahir Dar, Northwest Ethiopia. 2020. [Unpublished manuscript]. [Google Scholar]
- 48. Hidron AI, Kempker R, Moanna A, et al. Methicillin-resistant Staphylococcus aureus in HIV-infected patients. Infect Drug Resist 2010; 3: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Effah CY, Sun T, Liu S, et al. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob 2020; 19(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee C-R, Lee JH, Park KS, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol 2017; 7: 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Adeyemi FM, Adejuyigbe E, Ebhodaghe BI, et al. Molecular characterizationand antibiotic resistance pro. Arch Clin Microbiol 2018; 6(1): 269–276. [Google Scholar]
- 52. Marbou WJT, Kuete V. Bacterial resistance and immunological pro. J Infect Public Heal 2017; 10(3): 269–276. [DOI] [PubMed] [Google Scholar]
- 53. Muhie OA. Antibiotic use and resistance pattern in Ethiopia: systematic review and meta-analysis. Int J Microbiol 2019; 2019: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cag Y, Caskurlu H, Fan Y, et al. Resistance mechanisms. Ann Transl Med 2016; 4(17): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gudata D, Begna F. Antimicrobial resistance. Int J Res Granthaalayah 2018; 6(11): 77–93. [Google Scholar]
- 56. Liwa AC, Jaka H. Antimicrobial resistance: mechanisms of action of antimicrobial agents In: Méndez-Vilas A. (ed.) The battle against microbial pathogens: basic science, technological advances and educational programs. Madrid: Formatex, 2015, vol. 5, pp. 876–885. [Google Scholar]
- 57. Shrestha S, Acharya A, Gautam A, et al. Lower respiratory tract pathogens and their antimicrobial susceptibility pattern in a medical hospital of central Nepal. Int J Biomed Adv Res 2013; 4(5): 335–340. [Google Scholar]
- 58. Schindler BD, Kaatz GW. Multidrug efflux pumps of gram-positive bacteria. Drug Resist Updat 2016; 27: 1–13. [DOI] [PubMed] [Google Scholar]
- 59. Jinghua M, Gaizhuang L, Qiaoli C. Pathogens and antibiotic resistance of children with community-acquired pneumonia. 2017. [Google Scholar]
- 60. Menon RU, George AP, Menon UK. Etiology and anti-microbial sensitivity of organisms causing community acquired pneumonia: a single hospital study. J Family Med Prim Care 2013; 2(3): 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marwa KJ, Mushi MF, Konje E, et al. Resistance to cotrimoxazole and other antimicrobials among isolates from HIV/AIDS and non-HIV/AIDS patients at Bugando Medical Centre, Mwanza, Tanzania. AIDS Res Treat 2015; 2015: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cribbs SK, Crothers K, Morris A. Pathogenesis of HIV-related lung disease: immunity, infection, and inflammation. Physiol Rev 2020; 100(2): 603–632. [DOI] [PubMed] [Google Scholar]
- 63. Benito N, Moreno A, Miro J, et al. Pulmonary infections in HIV-infected patients: an update in the 21st century. Eur Respir J 2012; 39(3): 730–745. [DOI] [PubMed] [Google Scholar]
- 64. Feldman C, Anderson R. HIV-associated bacterial pneumonia. Clin Chest Med 2013; 34(2): 205–216. [DOI] [PubMed] [Google Scholar]
- 65. Muthumbi E, Lowe BS, Muyodi C, et al. Risk factors for community-acquired pneumonia among adults in Kenya: a case–control study. Pneumonia 2017; 9(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rivero-Calle I, Pardo-Seco J, Aldaz P, et al. Incidence and risk factor prevalence of community-acquired pneumonia in adults in primary care in Spain (NEUMO-ES-RISK project). BMC Infect Dis 2016; 16(1): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Voss M, Wonnenberg B, Honecker A, et al. Cigarette smoke-promoted acquisition of bacterial pathogens in the upper respiratory tract leads to enhanced inflammation in mice. Respir Res 2015; 16(1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Segal LN, Methé BA, Nolan A, et al. HIV-1 and bacterial pneumonia in the era of antiretroviral therapy. Proc Am Thorac Soc 2011; 8(3): 282–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Simou E, Britton J, Leonardi-Bee J. Alcohol and the risk of pneumonia: a systematic review and meta-analysis. BMJ Open 2018; 8(8): e022344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-smo-10.1177_20503121221145569 for Bacteriology of community-acquired pneumonia, antimicrobial susceptibility pattern and associated risk factors among HIV patients, Northeast Ethiopia: cross-sectional study by Mihret Tilahun, Daniel Gebretsadik, Abdurahaman Seid, Alemu Gedefie, Melaku Ashagrie Belete, Melkam Tesfaye, Edosa Kebede and Agumas Shibabaw in SAGE Open Medicine