Key Words: amacrine cell, ganglion cell, horizontal cell, in vivo reprogramming, Müller cell, NeuroD1, photoreceptor, regeneration, retina, retinal degeneration
Abstract
The onset of retinal degenerative disease is often associated with neuronal loss. Therefore, how to regenerate new neurons to restore vision is an important issue. NeuroD1 is a neural transcription factor with the ability to reprogram brain astrocytes into neurons in vivo. Here, we demonstrate that in adult mice, NeuroD1 can reprogram Müller cells, the principal glial cell type in the retina, to become retinal neurons. Most strikingly, ectopic expression of NeuroD1 using two different viral vectors converted Müller cells into different cell types. Specifically, AAV7m8 GFAP681::GFP-ND1 converted Müller cells into inner retinal neurons, including amacrine cells and ganglion cells. In contrast, AAV9 GFAP104::ND1-GFP converted Müller cells into outer retinal neurons such as photoreceptors and horizontal cells, with higher conversion efficiency. Furthermore, we demonstrate that Müller cell conversion induced by AAV9 GFAP104::ND1-GFP displayed clear dose- and time-dependence. These results indicate that Müller cells in adult mice are highly plastic and can be reprogrammed into various subtypes of retinal neurons.
Introduction
Retinal degenerative disease is debilitating, often causes blindness, and effective treatment remains elusive. Regenerating retinal neurons may provide a new way to repair the degenerated retina. Müller glial cells are the major type of glial cells in the retina, and have been reported to regenerate neurons in the retina of zebrafish (Powell et al., 2016). However, this kind of regenerative capacity is rather limited in birds like chickens, and completely lost in mammals like mice (Palazzo et al., 2020). Previous studies have tried various methods of activating the regeneration ability of Müller cells in mammals, including ectopic expression of Ascl1, Math5, Brn3b, or knockout of Ptbp1 (Jorstad et al., 2017; Zhou et al., 2020; Xiao et al., 2021). However, most studies have not achieved high conversion efficiency and often require a combination of more than one transcription factor or other factors to trigger the trans-differentiation. Whether a single transcription factor is capable of reprogramming Müller glia with high efficiency is currently unknown.
NeuroD1 (ND1) is a basic helix-loop-helix transcription factor, which plays an important role during early brain development by inducing neuronal differentiation of neural stem cells (Lee et al., 1995; Goebbels et al., 2005; Pataskar et al., 2016). A series of studies has reported that ND1 alone can reprogram brain astrocytes into neurons in various models of brain disease, including Alzheimer’s disease (Guo et al., 2014), Huntington’s disease (Wu et al., 2020), stroke (Chen et al., 2020), and epilepsy (Zheng et al., 2022), and success has been achieved in both rodents and non-human primate models (Ge et al., 2020). In the retina, ND1 is involved in the development of photoreceptor cells (Ochocinska et al., 2012) and amacrine cells (Cho et al., 2007). Moreover, studies show that loss of ND1 function results in delayed development of amacrine cells and loss of photoreceptor cells. However, it remains unclear whether ND1 can reprogram Müller cells into retinal neurons in adult mice.
In the current study, we investigated whether overexpression of ND1 in Müller cells can reprogram them into retinal neurons. Normal mice or N-methyl-D-aspartic acid (NMDA)-treated mice were intravitreally or subretinally injected with AAV viruses carrying ND1, and immunostaining was carried out at various time points to observe cell morphology and cellular markers of virus-infected cells.
Methods
Animals
Wild-type (C57BL/6J) male mice (weight: approximately 25 g) aged 7–8 weeks were purchased from the Liaoning Changsheng Biotechnology Co., Ltd, China (license No. SYXK (Liao) 2017-0174). The animals were housed in temperature (22°C) and humidity (60%) controlled cages, with a 12/12-hour light/dark cycle and free access to regular food and water. The experimental procedures for animals were conducted following the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals (8th edition) (National Institutes of Health, 2011) and approved by the Qualified Ethics Committee of Jinan University (Approval No. # IACUC-20201118-03) on November 18, 2020. All efforts were made to minimize the number of animals used and their suffering.
For each batch of experiments, the right and left eye of each animal was injected with control virus and ND1 virus, respectively. At least three animals were examined for each time point after virus injection.
Virus construct and injection
Viruses were constructed by OBiO Technology (Shanghai, China) or PackGene Biotech (Guangzhou, China). They included AAV7m8 GFAP::GFP-P2A-ND1 (and the control AAV7m8 GFAP::GFP) and AAV9 GFAP104::ND1-P2A-GFP (and the control AAV9 GFAP104::GFP). For AAV7m8, the GFAP promoter was the synthetic 681-bp gfaABC1D derived from the 2.2-kb gfa2 promoter (Lee et al., 2008). For AAV9, the GFAP104 promoter consists of an EF1α enhancer followed by the gfaABC1D promoter (Perea et al., 2014). The titers used in the experiment were 3 × 1012 GC/mL for AAV7m8 and 1 × 1012 GC/mL for AAV9, except for the titer test where 1011, 1012, and 1013 GC/mL were used for AAV9. All viruses were diluted in phosphate buffered saline (PBS) with 0.001% Pluronic F-68 solution (Poloxamer 188 Solution, PFL01-100ML, Caisson Laboratories, Smithfield, UT, USA) to prevent virus aggregation.
For the injections, animals were anesthetized via intraperitoneal injection of 1.25% tribromoethanol (0.1 mL/10g body weight, Sigma, St. Louis, MO, USA) and pupils were dilated with 0.5% tropicamide solution (Santen Pharmaceutical Co., Osaka, Japan). A glass slide with a diameter of 3 mm was placed on the cornea of the mouse so that the fundus could be clearly seen. After puncturing a hole in the corneal limbus with a sharp 30-gauge needle, 0.8–1.2 μL virus was injected into the vitreous (1.2 μL) or subretinal space (0.8 μL) with a 34-gauge blunt-end needle. If the injection into the subretinal space was successful, a small bleb developed locally in the retina. The needle was kept in the vitreous for 10 extra seconds and was then slowly pulled out, and a drop of antibiotic gel was applied to the eye. Mice were warmed on a safe heating pad until fully awake. Animals that developed cataracts or inflammation were excluded from future experiments. To minimize the variation between animals, the control virus was injected into the right eye and the ND1 virus was injected into the left eye of the same animal.
To induce NMDA injury, 1.2 μL NMDA (ab120052, Abcam, Cambridge, MA, USA) solution (20 mM) was intravitreally injected into the eyes of adult mice one day before the virus injection.
Tissue processing
To examine the effects of virus infection, at 3, 5, 7, 14, 28 days or 8 weeks after virus injection, animals were sacrificed by overdose of anesthesia with tribromoethanol (1.25% solution, 0.14 mL/10 g body weight), and eyes were enucleated and fixed in 4% paraformaldehyde for 30 minutes at room temperature. For retinal slices, eyecups with lenses were then washed with PBS and cryo-protected overnight at 4°C in 0.01 M PBS containing 30% sucrose, and ultimately embedded in the optimal cutting temperature compound (Tissue Tek, Torrance, CA, USA). Retinas were cryo-sectioned on a microtome (Leica Microsystems, Wetzlar, Germany) through the optic disk longitudinally at a thickness of 10 μm. Thereafter, retinal sections were mounted on glass slides for future processing. For whole-mount retinas, after fixation with paraformaldehyde, the retina was removed from other layers, and either flattened on a glass slide and sealed with Vectashield Antifade Mounting Medium (H-1000-NB, Novus Biologicals LLC, Denver, CO, USA), or stored in PBS at 4°C for future immunostaining.
Immunofluorescence
For immunostaining of the retinal glia and neuronal markers retinal sections were blocked with 0.01 M PBS containing 10% normal donkey serum, 3% bovine serum albumin, and 0.3% Triton X-100 for 1 hour, and then blocked in the primary antibodies overnight at 4°C. After thorough washes, retinal sections were incubated with the secondary antibodies for 2 hours at room temperature, and subsequently washed, mounted, and cover-slipped. For 4′,6-diamidino-2-phenylindole (DAPI) staining, sections were incubated with DAPI (1:1000, Electron Microcopy Sciences, Hatfield, PA, USA) for 5 minutes at room temperature before mounting. For whole-mount retinas, retinas were blocked with 0.01 M PBS containing 10% normal donkey serum, 3% bovine serum albumin, and 3% Triton X-100 for 1 hour and incubated in primary antibodies overnight at 4°C, then washed and incubated with secondary antibody for 2 hours at room temperature.
The primary antibodies were: chicken anti-green fluorescent protein (GFP, 1:1000, Aveslabs, Portland, OR, USA, Cat# GFP-1020, RRID: AB_10000240), mouse anti-glutamine synthetase (GS; 1:1000, Millipore, Watford, Herts, UK, Cat# MAB302, RRID: AB_2110656), rabbit anti-Sox9 (1:1000, Millipore, Cat# AB5535, RRID: AB_2239761), rabbit anti-Calretinin (1:1000, SWant, Burgdorf, Bern, Switzerland, Cat# CR7697, RRID: AB_2619710), rabbit anti-RNA binding protein with multiple splicing (RBPMS; 1:1000, Thermo Fisher Scientific, Waltham, MA, USA, Cat# PA5-31231, RRID: AB_2548705), rabbit anti-Recoverin (1:1000, Millipore, Cat# AB5585, RRID: AB_2253622), rabbit anti-Opsin (1:1000, Millipore, Cat# AB5405, RRID: AB_177456), rabbit anti-Calbindin (1:1000, Abcam, Cat# ab49899, RRID: AB_1267903), rabbit anti-ND1 (1:1000, Thermo Fisher Scientific, Cat# PA5-78075, RRID: AB_2736208), and goat anti-Brn3a (1:1000, Santa Cruz, Carlsbad, CA, USA, Cat# sc-31984, RRID: AB_2167511). Secondary antibodies were: Alexa Fluor® 488-donkey anti-chicken 488 (1:1000, Jackson ImmunoResearch Labs, West Grove, PA, USA, Cat# 703-545-155, RRID: AB_2340375), Alexa Fluor™ 647-donkey anti-rabbit 647 (1:1000, Thermo Fisher Scientific, Cat# A-31573, RRID: AB_2536183), and Alexa Fluor™ Plus 594-goat anti-mouse 594 (1:1000, Thermo Fisher Scientific, Cat# A-32744, RRID: AB_2762826).
Image collection, processing, and analyzing
Immunofluorescence-stained tissues were imaged with a confocal microscope (LSM700, Carl Zeiss, Jena, Thuringia, Germany). Because the viruses did not infect the whole retina, only regions with GFP signal were sampled. For the whole-mount retinas, regions with clear and dense GFP+ somas were sampled (examples see Figure 1E). For retinal slices, Z-stacks with 1-μm step were collected to cover the GFP+ regions. For whole-mount retinas, Z-stacks with 1-μm step were collected from the inner nuclei layer (INL; where somas of Müller cells and most amacrine cells are located), the ganglion cell layer (GCL; where ganglion cells and some amacrine cells are located), or the outer nuclei layer (ONL; where photoreceptors are located). GFP+ cells and those double-stained with glial or neuronal markers were counted manually by an independent observer using Zen software (Carl Zeiss). For each retina, values from 3–6 images were averaged to generate one data point, and then all data points from retinas within each group were further averaged to obtain the overall mean value.
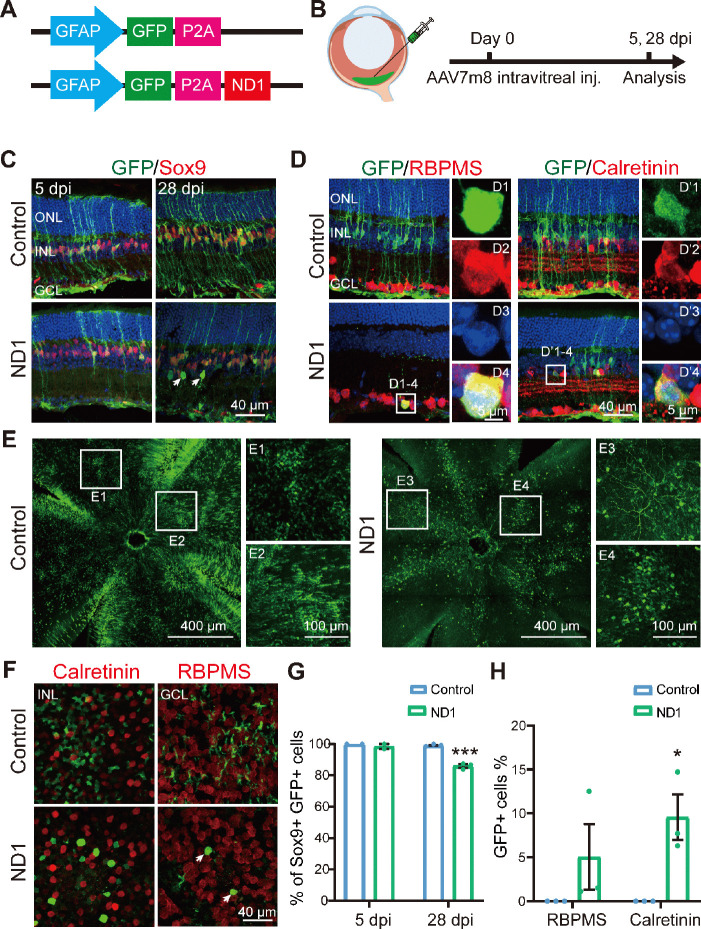
Figure 1.
AAV7m8 GFAP::GFP-ND1 converts a limited number of Müller cells into inner retinal neurons.
(A) Illustration of the virus construct. (B) Illustration of intravitreal injection of the virus (green) and the experimental protocol. (C) Images of retinal slices stained with GFP (green: Alexa Fluor-488) and Müller cell marker Sox9 (red: Alexa Fluor-647) at 5- and 28-days post-injection (dpi) for control and ND1 virus. Nuclei are stained with DAPI (blue). In the ND1 group at 28 dpi, some GFP+ cells were located in the inner retinal layer, away from the Müller cells, and did not express Sox9 (examples indicated by white arrows). In the control group, all GFP+ cells expressed Sox9. (D) Images of retinal slices stained with GFP (green), ganglion cell marker RBPMS (red: Alexa Fluor-647, left panels), or amacrine cell marker Calretinin (red: Alexa Fluor-647, right panels) at 28 dpi. White boxed regions in the ND1 group are enlarged and shown on the right with individual color channels and a merged channel. Nuclei are stained with DAPI (blue). In the ND1 group at 28 dpi, some GFP+ cells were co-labeled with RBPMS or Calretinin, while in the control group, few such cells were observed. (E) Example images of whole-mount retinas showing the virus-infected area, with the boxed region enlarged and shown on the right. GFP signals were widely distributed in the retina in both groups. In the ND1 group, GFP+ branches in ganglion cells were observed, while in the control group, mainly GFP+ processes were observed. (F) Images of whole-mount retinas stained with GFP (green) and Calretinin (red: Alexa Fluor-647, left column) in the inner nuclei layer, or RBPMS (red: Alexa Fluor-647, right column) in the ganglion cell layer. GFP+ cells with oval somas were observed in the ND1 group, while those with polygon shaped somas and end-feet were observed in the control group. (G) Percentage of Sox9+ GFP+ cells among all GFP+ cells at 5 dpi (n = 2 for each group) and 28 dpi (n = 3 for each group) in retinal sections. After infection with ND1, 12.2% of GFP+ cells lost the expression of the Müller cell marker by 28 dpi. (H) Percentage of GFP+ cells with markers RBPMS or Calretinin among all GFP+ cells at 28 dpi. ND1 infection turned 5.0% and 9.6% GFP+ cells into ganglion cells and amacrine cells, respectively. All data are expressed as mean ± SEM. *P < 0.05, ***P < 0.001 (unpaired Student’s t-test). GCL: Ganglion cell layer; GFAP: promoter of glial fibrillary acidic protein; GFP: green fluorescent protein; INL: inner nuclei layer; ND1: NeuroD1; ONL: outer nuclei layer; P2A: porcine teschovirus-1 2A; RBPMS: RNA binding protein with multiple splicing.
To estimate the efficiency of local infection, the number of GFP+ cells was counted for each image. To analyze the occurrence of reprogramming, GFP+ cells that no longer expressed the Müller cell marker Sox9 (Sox9-GFP+) were counted and divided by the number of total GFP+ cells. To estimate the reprogramming efficiency for specific types of neurons, GFP+ cells that co-express the marker for that type of neuron were summed and then divided by the total number of GFP+ cells.
Statistical analysis
All data are expressed as the mean ± standard error of the mean (SEM). Statistical significance was assessed with unpaired Student’s t-test using Prism 8.0.2 (GraphPad Software, San Diego, CA, USA, www.graphpad.com). Statistical significance was set at P < 0.05, with P < 0.001 being considered highly significant. Unless otherwise stated, data from one eye of at least three animals were collected for each group. The sample size was determined based on the rule of minimal usage of animals (n ≥ 3).
Results
Our previous studies have demonstrated that overexpressing a single neuronal transcription factor (ND1) in astrocytes can convert them into neurons both in vitro and in vivo when AAV or a retrovirus that only infects dividing glial cells is used (Guo et al., 2014; Chen et al., 2020; Ge et al., 2020; Wu et al., 2020; Zheng et al., 2022). Here, we further investigated whether ND1 can convert Müller glia in the retina into neurons. If possible, this would be a good step toward developing neuroregenerative therapy for treating degenerative diseases of the retina.
ND1 reprograms Müller cells into inner retinal neurons
To test whether ND1 can convert retinal Müller glial cells into neurons, we first intravitreally injected AAV7m8 GFAP::GFP-ND1 or a control virus expressing GFP alone (AAV7m8 GFAP::GFP) into the eyes of the mice (Figure 1A). At 5- or 28-days post-injection (dpi), we stained the retinas for glial and neuronal markers (Figure 1B, experimental protocol illustrated). At 5 dpi, for both control and ND1 groups, essentially all GFP+ cells expressed glial cell marker Sox9 in the Müller cell layer (Figure 1C, left column), confirming the specific infection of Müller glial cells by our AAV. At 28 dpi, all GFP+ cells in the control group still had the typical morphology of Müller cells, with long processes expanding through the whole retina, and cell somas expressing Sox9 (Figure 1C, top right panel). In contrast, although the majority of GFP+ cells in the ND1 group were still Sox9+ (i.e., Müller glia) at 28 dpi, a small percentage of them were not and were located away from the Müller glia layer (Figure 1C, bottom right panel, white arrows). Because all GFP+ cells were initially Sox9+ Müller glia, the fact that some cells stopped expressing Sox9 suggested that they might start to change their identity. To determine whether these observed changes were due to a change in cell type from glia to neuron, we performed immunostaining for markers of inner retinal neurons. In ND1 group at 28 dpi, we identified a few GFP+ cells in the GCL expressing ganglion cell marker RBPMS (Figure 1D, D1 to D4 showing an enlarged cell in ND1 group). We also identified GFP+ cells in the INL expressing amacrine cell marker Calretinin (Figure 1D, D’1 to D’4 showing an enlarged cell in ND1 group).
Examination of whole-mount retinas also revealed differences between the control group and the ND1 group (Figure 1E and F). At 28 dpi, GFP+ cells were spread out and covered a large area of retina, with cells in the control group being more densely infected than those in the ND1 group. Numerous GFP+ Müller cell processes and somas with polygon shapes were seen in the control group (Figure 1E1 and E2), while GFP+ branches of typical RGC dendrites and somas with round or octal shape were observed in ND1 group (Figure 1E3 and E4). Scanning the whole-mount retinas from the INL to the GCL further revealed polygon shaped soma and end-feet processes in the control group (Figure 1F top row), but oval or round shaped somas in the ND1 group, which were similar to the somas of neighboring neurons (Figure 1F bottom row). Quantified data indicated that in the control group, the percentage of Sox9+GFP+ double-positive cells among all the GFP+ cells was nearly 100% from 5 dpi to 28 dpi (blue bars in Figure 1G), indicating that our AAV vectors are highly specific for Müller cells. In contrast, in the ND1 group, the percentage of these double-positive cells dropped from nearly 100% at 5 dpi to 86% at 28 dpi (green bars in Figure 1G), indicating that some GFP+ cells infected by ND1 gradually lost this characteristic of Müller cells. However, the total number of Sox9+ cells was similar between ND1 and control groups, ranging from 27–39 per image at both 5 dpi and 28 dpi.
The drop in the percentage of cells with Müller-cell markers in the ND1 group was similar to the increase in the percentage of markers for inner retinal neurons (Figure 1H, 5.0% GFP+ RBPMS+ double-positive cells, and 9.6% GFP+ Calretinin+ double-positive cells). Similarly, in whole-mount retinas from ND1-group at 28 dpi, we found that 11.6% of cells showed neuron morphology in the INL and 6.8% showed neuron morphology in the GCL layer. Thus, we can conclude that intravitreal injection of AAV7m8 GFAP::GFP-ND1 converted Müller cells into inner retinal neurons, with limited conversion efficiency.
Müller cell conversion in the model of NMDA injury
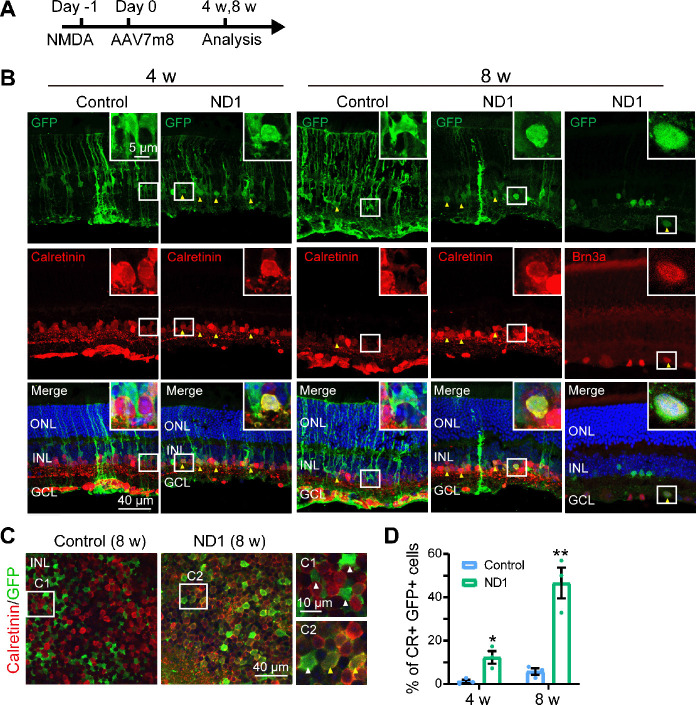
In a previous study, zebrafish Müller cells were transformed into ganglion cells in a model of ganglion cell damage, and into photoreceptors in a model of photoreceptor cell injury (Nagashima et al., 2013), suggesting that injury is a factor for stimulating neural regeneration. Therefore, we investigated whether Müller cells can be converted more efficiently in an injury model. To test this idea, we induced retinal cell death via intravitreal injection of NMDA one day before AAV infection, and then examined the AAV7m8-infected cells 4 and 8 weeks later (protocol shown in Figure 2A). We discovered that in the control group, at both 4 and 8 weeks after virus injection, most GFP+ cells remained Müller cells with typical Müller cell morphology (Figure 2B, left column, typical morphology enlarged in the inset), with very few in the IPL expressing the amacrine cell marker Calretinin (yellow arrowhead in Figure 2B, 8 weeks, left column). However, in the ND1 group, some GFP+ cells appeared as the inner retinal neurons at 4 weeks after virus injection and co-expressed Calretinin (yellow arrowheads in Figure 2B, 4 weeks, right column). By 8 weeks, the majority of ND1-infected cells displayed neuronal morphology and expressed Calretinin (Figure 2B, 8 weeks, middle column) or the ganglion cell marker Brn3a (Figure 2B, 8 weeks, right column). Some GFP+ cells remained typically Müller, with polygon shaped somas and long-extending processes. The reprogramming effect was even more striking when we examined the whole-mount retinas of virally infected regions. In the control group, the somas of GFP+ cells were inlaid nicely between Calretinin+ somas in the INL (Figure 2C left panel, white arrowheads in the enlarged C1 inset). In contrast, in the ND1 group, many GFP+ cell somas were overlaid with Calretinin+ somas, indicating a successful reprogramming by ND1 (Figure 2C, right panel, yellow arrowheads in the enlarged C2 inset). Quantitative analysis revealed that in the retinal section of the NMDA injury model, ND1 converted Müller cells into Calretinin+ amacrine cells with a conversion efficiency of 12.3 ± 2.9% (4 weeks) and 46.6 ± 7.1% (8 weeks) (n = 3 mice each), compared with 1.2 ± 0.8% (4 weeks) and 5.8 ± 1.5% (8 weeks) in the control group (n = 3 mice each) (Figure 2D). Therefore, our data indicate that ND1 can reprogram Müller cells into inner retinal neurons in the NMDA-injured mouse retina, and the efficiency was greater than in non-injured retinas (9.6% at 4 weeks after ND1 injection).
Figure 2.
Müller glia are converted into amacrine cells by ND1 in NMDA injured retina.
(A) Illustration of the experimental protocol. (B) Images of retinal sections stained with GFP (green: Alexa Fluor-488) and amacrine cell marker Calretinin (red: Alexa Fluor-647, lane 1–4) or ganglion cell marker Brn3a (red: Alexa Fluor-647, lane 5) at 4 and 8 weeks after virus injection. In the ND1 group, some GFP+ cells co-expressed Calretinin and Brn3a, while there were few such cells in the control group. Yellow arrows point to GFP+ cells that co-expressed neuronal markers. Boxed regions of typical cells are enlarged in the top-right corners of each panel. (C) Images of whole-mount retinas stained with GFP (green) and Calretinin (red: Alexa Fluor-647) in the INL, with white-boxed regions enlarged on the right of the panels. Yellow arrows point to GFP+ cells that co-expressed Calretinin, and white arrows point to GFP+ cells that did not express Calretinin. In the ND1 group, numerous GFP+ cells co-expressed Calretinin, while in the control group few such cells were observed. (D) Percentage of GFP+ cells with Calretinin (CR) among all GFP+ cells at 4 and 8 weeks after virus injection. All data are expressed as mean ± SEM (n = 3 for each group at each time points). *P < 0.05, **P < 0.01 (unpaired Student’s t-test). CR: Calretinin; GCL: ganglion cell layer; GFP: green fluorescent protein; INL: inner nuclei layer; ND1: NeuroD1; ONL: outer nuclei layer.
Enhanced ND1 expression in normal mice converts Müller cells into outer retinal neurons in a time-dependent manner
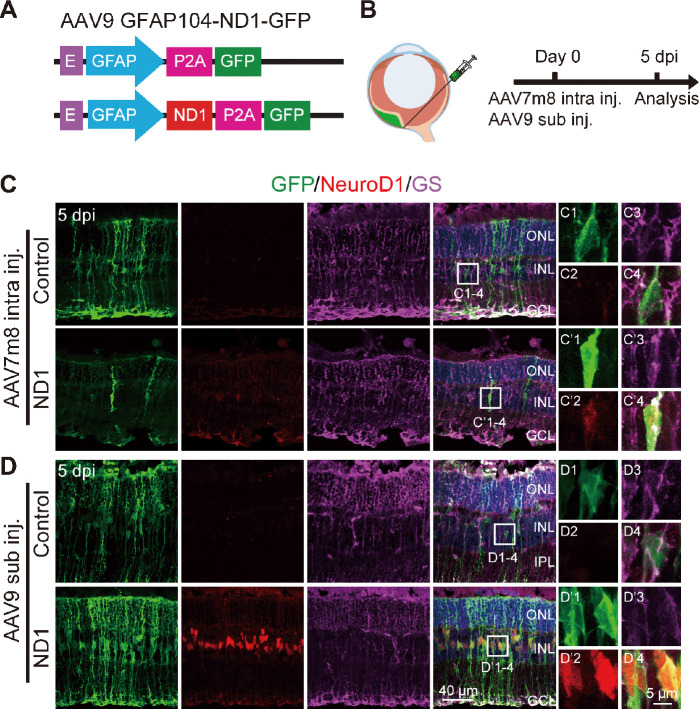
Because conversion efficiency among viral infected Müller cells after intravitreal injection of AAV7m8 GFP-ND1 was low, we examined ND1 expression level via immunostaining. Surprisingly, we could barely detect ND1 signals in the ND1-infected cells, even though GFP signals were obvious. This might have occurred for several reasons. For instance, maybe it was because the AAV serotype was not ideal, or some factor related to the GFP-P2A-ND1 sequence, or because of the intravitreal injection. To find the answer, based on our studies of the mouse brain (Chen et al., 2020; Xiang et al., 2021; Zheng et al., 2022), we engineered a new vector AAV9 GFAP104::ND1-P2A-GFP that changed into a different serotype (AAV9), added the EF1α enhancer to increase ND1 expression, and switched the sequence to ND1-P2A-GFP (Figure 3A). We also changed the injection site from intravitreal to subretinal (Figure 3B). Interestingly, after these changes, we found that subretinal injection of AAV9 ND1-GFP resulted in efficient expression of ND1 (red) in infected Müller cells (green), which was confirmed by immunostaining with Müller glia marker GS (purple) (Figure 3D bottom row). In contrast, only weak expression of ND1 was observed in AAV7m8 GFP-ND1 infected retinas (Figure 3C, bottom row) and no ND1 expression was observed in either control group (top rows in Figure 3C, and D). Therefore, we employed this new AAV9 vector that produced high ND1 expression levels for further experimental analyses.
Figure 3.
AAV9 GFAP104::ND1-GFP enhances the expression of ND1.
(A) Construct of the AAV9 virus. (B) Illustration of the subretinal injection (sub inj.) and the experimental protocol to detect ND1 expression. (C) Images of retinal slices after intravitreal injection (intra inj.) of AAV7m8 GFAP::GFP-ND1 or control virus at 5 dpi. Slices were stained with GFP (green: Alexa Fluor-488), NeuroD1 antibody (red: Alexa Fluor-647), and Müller cell marker GS (purple: Alexa Fluor-594). White boxed region in each group is enlarged and shown on the right with individual color channels and a merged channel. In the ND1 group, the soma of GFP+ cells expressed weak ND1 and GS. (D) Images of retinal slices after subretinal injection of AAV9 GFAP104::ND1-GFP or control virus at 5 dpi. In the ND1 group, the soma of GFP+ cells strongly expressed ND1. For the control virus, all GFP+ cells expressed GS, but none expressed ND1. E: EF1α enhancer; GFAP: promoter of glial fibrillary acidic protein; P2A: porcine teschovirus-1 2A; ND1: NeuroD1; GFP: green fluorescent protein; GS: glutamine synthetase; ONL: outer nuclei layer; INL: inner nuclei layer; GCL: ganglion cell layer; Intra inj.: intravitreal injection; Sub inj.: subretinal injection.
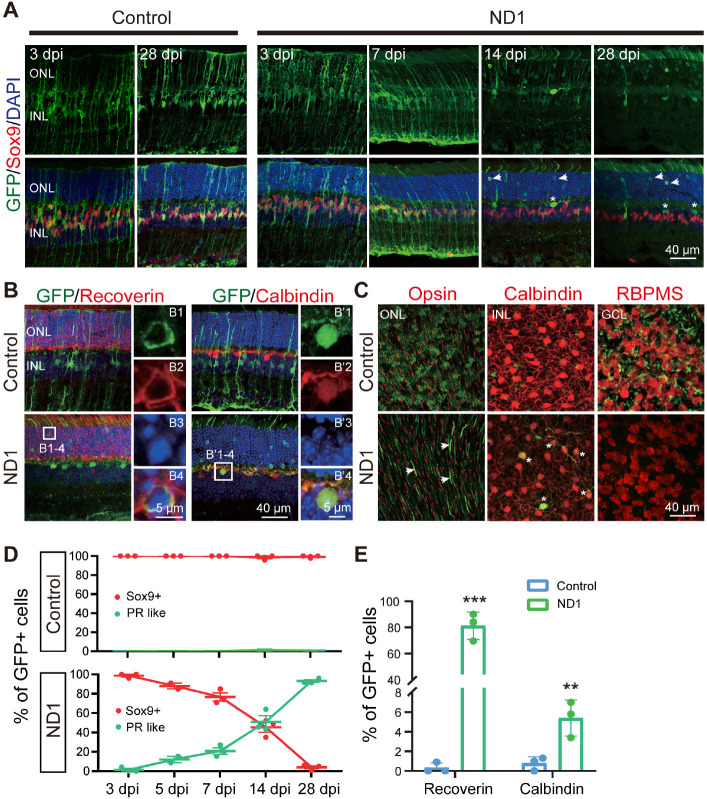
After confirming ND1 expression in Müller cells, we continued to investigate whether this new AAV9 ND1 vector can convert Müller cells into neurons. To understand how ND1 might cause changes in Müller cells, we looked at more time points (3, 5, 7, 14 and 28 dpi). In the control group, GFP+ cells stayed as Müller cells with Sox9 expression until the last measurement at 28 dpi (Figure 4A, left two columns). In the ND1 group, the AAV9 ND1-GFP-infected cells displayed typical Müller cell morphology from 3 dpi until 7 dpi, and the majority of GFP+ cells expressed Sox9 (Figure 4A, ND1 group, 3 and 7 dpi, 5 dpi not shown). However, by 14 dpi, Müller cell morphology disappeared among the ND1-GFP-infected cells; instead, the majority of GFP+ cells appeared in the ONL, where photoreceptor cell somas were typically identified (Figure 4A, ND1 group, 14 dpi). We also observed a number of GFP+ filaments protruding outside the ONL, reminiscent of the filament from photoreceptor cells (Figure 4A, ND1 group, 14 dpi). By 28 dpi, many GFP+ cells could be found in the ONL, and the number of filament-like protrusions outside the ONL had increased (Figure 4A, ND1 group, 28 dpi). Immunostaining for Recoverin indicated that the majority of the converted cells were photoreceptors (Figure 4B, left column, squared region enlarged in B1–4, quantification shown in Figure 4E). Additionally, we also found a small number of converted cells expressing Calbindin, a marker for horizontal cells (Figure 4B, right column, squared region enlarged in B’1–4). ND1-induced cell conversion was also clear from the whole-mount retina images (Figure 4C). In the control group, GFP+ cells showed no overlap with Opsin, Calbindin, or RBPMS (Figure 4C, top row) in the ONL, INL, or GCL respectively. In contrast, in the ND1 group, there were many GFP+ filaments in ONL, and a few GFP+ cell somas co-localized with Calbindin in INL, but few co-localized with RBPMS in GCL (Figure 4C, bottom row). Quantitative analyses at different time points revealed that in the control group, essentially all the viral infected cells were Sox9+ Müller cells, and were rarely located in the ONL (Figure 4D, top panel, red line). In the ND1 group, the ND1-GFP-infected cells showed a gradual loss of Sox9, accompanied by a gradual increase in photoreceptors (green line) in the ONL (Figure 4D, bottom panel). Quantified immunostaining results (28 dpi) also showed that 81.3% of ND1-GFP-infected cells expressed Recoverin, and 5.4% of ND1-GFP-infected cells expressed Calbindin (Figure 4E). These results indicate an ND1-induced time-dependent gradual conversion of Müller cells into outer retinal neurons in the adult mouse retina.
Figure 4.
Enhanced ND1 converts Müller cells into outer retinal neurons overtime.
(A) Images of retinal slices stained with GFP (green) and Müller cell marker Sox9 (red: Alexa Fluor-647) at different time points after injection of control or ND1 virus. Top row shows the GFP channel only, bottom row shows the merged channels. Nuclei are stained with DAPI (blue). Numerous GFP+ photoreceptor-like cells (arrows) and a few horizontal-like cells (asterisk) were observed in the ND1 group over time. (B) Images of retinal slices stained with GFP (green), photoreceptor marker Recoverin (red: Alexa Fluor-647, left panels), or horizontal cell marker Calbindin (red: Alexa Fluor-647, right panels) at 28 dpi. White boxed regions in the ND1 group are enlarged and shown on the right with individual color channels and a merged channel. Nuclei are stained with DAPI (blue). In the ND1 group, most GFP+ cells co-labeled with Recoverin or Calbindin, while in the control group, few such cells were observed. (C) Images of whole-mount retinas stained with GFP (green: Alexa Fluor-488) and Opsin above the ONL (red: Alexa Fluor-647, left panels), with Calbindin at the top of the INL (red: Alexa Fluor-647, middle panels), or with RBPMS in the GCL (red: Alexa Fluor-647, right panels). In the ND1 group, GFP+ filaments in the shape of photoreceptor outer segments (arrows) were observed, and all GFP+ cells in the INL were co-labelled with Calbindin (asterisks), but no GFP+ cells were observed in the GCL. In the control group, GFP+ outer processes, inner processes, and end-feet of Müller cells were observed in each of their appropriate layer. (D) Percentage of Sox9+ GFP+ cells (red lines) or GFP+ photoreceptor-like cells (green lines) among all GFP+ cells at various time points. After infection with ND1, 23.3%, 54.7%, and 96.0% of GFP+ cells stopped expressing the Müller cell marker at 7, 14, and 28 days respectively. (E) Percentage of GFP+ cells expressing Recoverin or Calbindin among all GFP+ cells at 28 dpi. ND1 infection turned 81.3% of GFP+ cells into photoreceptors and 5.4% into horizontal cells. All data are expressed as mean ± SEM (n = 3 for each group). **P < 0.01, ***P < 0.001 (unpaired Student’s t-test). GCL: Ganglion cell layer; GFP: green fluorescent protein; INL: inner nuclei layer; ND1: NeuroD1; ONL: outer nuclei layer; PR like: photoreceptor like cells; RBPMS: RNA binding protein with multiple splicing.
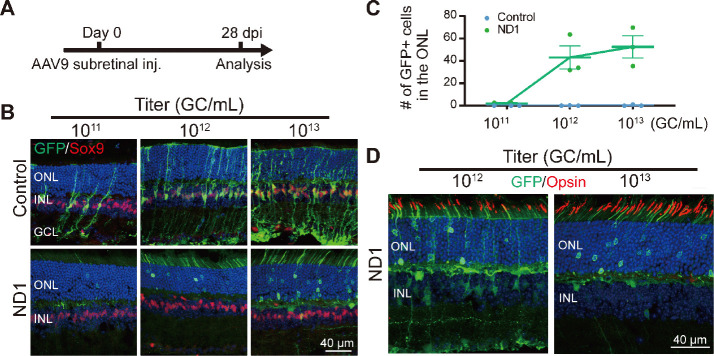
ND1-induced Müller cell conversion is dose-dependent
Next, we investigated different AAV doses to find the best one for converting Müller cells to neurons. Subretinal injection of the control AAV9 GFAP104::GFP and the experimental AAV9 GFAP104::ND1-P2A-GFP at 1011, 1012, or 1013 genome copies per milliliter (GC/mL) (with the same volume of 0.8 μL) was performed, and retinas were collected at 28 dpi for immunostaining (Figure 5A and B). In the control group (Figure 5B, top row), we observed a small number of GFP-infected Müller cells expressing Sox9 at the lowest dose of 1011 GC/mL, but the number of GFP+ cells were significantly higher for the medium dose of 1012 GC/mL, and even higher at the largest dose of 1013 GC/mL. No obvious GFP+ cells were identified in the ONL of the control group at any of the three doses (Figure 5B, top row). In the ND1 group (Figure 5B, bottom row), we rarely detected GFP+ Müller cells at the lowest dose, except for a few GFP+ cells in the ONL. However, at the medium dose we observed more ND1-GFP-converted cells in the ONL, with many GFP+ filaments in the outer segment of the ONL (Figure 5B, bottom row). At the high dose, we detected even more ND1-GFP-converted cells in the ONL. The dose-dependent conversion curve is shown in Figure 5C. Immunostaining with Opsin at the medium and high doses revealed that except for a few instances, the majority of GFP+ filaments in the ND1 group did not express Opsin (Figure 5D), suggesting that these cells were likely rod photoreceptor cells rather than cone cells. On the basis of these dose-dependent results, we recommend 1012 GC/mL for future retinal conversion studies, because the conversion efficiency was close to what the higher dose achieved. Together, our results suggest that expression of ND1 can convert Müller cells into rod photoreceptors in a dose-dependent manner.
Figure 5.
Dose-dependent conversion of Müller cells by ND1.
(A) The experimental protocol. (B) Images of retinal slices after subretinal injection (inj) of AAV9 GFAP104::ND1-GFP or control virus with increasing titers from 1011 to 1013 GC/mL. Slices were stained with GFP (green: Alexa Fluor-488) and Sox9 (red: Alexa Fluor-647). The number of GFP+ cells increased with the virus titer. In the control group, GFP+ cells remained as Müller cells, while in the ND1 group, numerous GFP+ somas and outer-segment structures were observed in the ONL where photoreceptors are located. A few GFP+ cells with horizontal cell morphology were also observed at high titer. (C) Number of GFP+ cells in the ONL increased with ND1 virus titer. In contrast, hardly any GFP+ cells were observed in ONL for the control virus even at high titer. (D) Retinal sections stained with GFP (green: Alexa Fluor-488) and cone outer segment marker Opsin (red: Alexa Fluor-647) under two titers. Most GFP filaments did not express Opsin. GC/mL: Genome copies per milliliter; GCL: ganglion cell layer; GFP: green fluorescent protein; INL: inner nuclei layer; ND1: NeuroD1; ONL: outer nuclei layer.
Discussion
We demonstrate here that specific overexpression of ND1 in Müller cells can convert them into retinal neurons in a time- and dose-dependent manner. Interestingly, weak expression of ND1 through AAV7m8 resulted in conversion of Müller cells into amacrine cells, but high expression of ND1 through AAV9 primarily resulted in conversion of Müller cells into photoreceptors. Such striking differences in the reprogrammed cell fate after infection by different viral vectors suggest that Müller cells are highly plastic and may have the potential to be reprogrammed into diversified neuronal subtypes depending on the transcription factors and their expression level.
Different cell fates induced by different viral vectors
One of the most striking results of this study is the discovery that Müller cells can be converted into different types of neurons by the same neural transcription factor (ND1), depending on which AAV vector was used to deliver it. We tested two different AAV serotypes (AAV7m8 and AAV9) with two different GFAP promoters (GFAP681 vs. GFAP104) and different orders of ND1 and GFP (GFP-NeuroD1 vs. NeuroD1-GFP). Previous studies have reported that AAV7m8 is good for infecting the retina (Hickey et al., 2017). Our study confirmed that AAV7m8 indeed infected Müller cells very well, but the expression level of ND1 was relatively low. We cannot exclude the possibility that this was because of a weak promoter or the sequence of GFP in front of ND1 (AAV7m8 GFAP681::GFP-P2A-ND1). In our own previous studies, we reported strong infection and high expression of ND1 in astroglial cells using AAV9 in both brain and spinal cord (Chen et al., 2020; Puls et al., 2020; Wu et al., 2020; Zheng et al., 2022). Therefore, we tested AAV9 in the retina as well in this study. Interestingly, we found that subretinal injection of AAV9 GFAP104::NeuroD1-P2A-GFP not only infected Müller cells very well but also led to greater ND1 expression compared with injection of AAV7m8 GFAP681::GFP-P2A-ND1. Even more interestingly, despite expressing the same neural transcription factor (ND1), these two different AAV vectors induced different Müller cell conversion: AAV7m8 GFAP681::GFP-P2A-ND1 converted Müller cells into amacrine cells in the INL, whereas AAV9 GFAP104::NeuroD1-P2A-GFP converted Müller cells into photoreceptors in the ONL.
Such different cell fates induced by different viral vectors, both expressing the same ND1 in Müller cells, is rather surprising and several explanations are possible. Our first speculation is that the different cell fates are associated with the ND1 expression level, which differed between the two different viral vectors. Another factor might be the location of the injection, which we changed for the AAV9 injections. Multiple factors, including AAV serotypes, promoters, enhancers, injection site, virus titer, retinal injury or not, all could have some impact on the cell fate of these converted cells. For example, our studies together with previous studies (Jorstad et al., 2017) found that NMDA injury appears to help or initiate Müller glia conversion, possibly through activating Müller glia and altering their epigenetic landscape. More extensive studies are needed to determine the precise mechanisms underlying the differential cell fates triggered by different AAV vectors. We are currently designing a new set of experiments using a stronger promoter and enhancer to increase ND1 expression in lineage-traced Müller glia in retinal injury and disease models.
Comparison between different neural transcription factors
Previous studies have reported that mouse retinal Müller cells can be reprogrammed into multiple subtypes of retinal neurons using different transcription factors. For example, Jorstad et al. (2017) used Ascl1 and the histone deacetylase inhibitor TSA to transdifferentiate Müller cells into bipolar cells and amacrine cells, respectively. In mice with NMDA injury, Xiao et al. (2021) overexpressed Math5 in Müller cells and transdifferentiated them into ganglion cells and amacrine cells. In this study, we used a single neuronal transcription factor (ND1) to transdifferentiate retinal Müller cells into rod cells, amacrine cells, and a few ganglion neurons and horizontal cells. Comparing our study with other studies, we find that a developmental relationship might exist between the types of converted neurons and the type of neuronal transcription factors that is used. It might be that Müller cells in mice still have some neurogenic potential, like that shown in zebrafish where Müller cells can differentiate into ganglion cells and photoreceptor cells when the retina is damaged (Nagashima et al., 2013).
Neuronal transcription factors might convert Müller cells into the same types of neurons that are made when they act on neural stem cells during early retinal development. According to previous developmental studies, Math5 (Xiao et al., 2021) is involved in both ganglion cell and amacrine cell development (Yang et al., 2003), Ascl1 (Jorstad et al., 2017) is involved in both bipolar cell and amacrine cell development (Brzezinski et al., 2011), and ND1 plays an important role in the development of photoreceptor cells and amacrine cells (Inoue et al., 2002; Cherry et al., 2011). Moreover, it is possible to combine different neural transcription factors together to generate more specific types of neurons. For example, combining Math5 and Brn3b together induced ganglion cells more specifically than did Math5 alone (Xiao et al., 2021). In another study, three neural transcription factors (Otx2, Crx, Nrl) that are critical during rod cell development can transdifferentiate Müller cells into rod cells with high specificity (Yao et al., 2018). Together, these studies suggest that different transcription factors can reprogram Müller cells into different subtypes of neurons.
ND1 reprogramming in the central nervous system
In our current study, we overexpressed ND1 in the Müller cells, a major type of glial cell in the retina, and converted them into neurons. This is consistent with our previous work on other parts of the central nervous system, including the brain and spinal cord. In rodent models, overexpression of ND1 converts reactive astrocytes into functional neurons in various brain disease or injury models, including ischemic injury (Chen et al., 2020), stab injury-induced glial scar (Zhang et al., 2020), temporal lobe epilepsy (Zheng et al., 2022), Huntington’s disease (plus Dlx2) (Wu et al., 2020), Alzheimer’s disease (Guo et al., 2014), and spinal cord injury (Puls et al., 2020). ND1-mediated reprogramming was also reported by several other labs (Chen et al., 2017; Jiang et al., 2021; Tang et al., 2021). In addition to the promising reparative effect of ND1 reprogramming in rodent models, ND1 led to a marked increase in local neuronal density in a non-human primate model of ischemic stroke, and protected interneurons in the converted areas (Ge et al., 2020).
Unlike in the brain and spinal cord where AAV9 vectors infect astrocytes efficiently, we were surprised that they do not infect astrocytes in the retina (Figure 6). Instead, our AAV9 vectors mainly targeted Müller cells with high specificity and infection efficiency. Because retinal astrocytes are also GFAP-positive, one possibility may be that retinal astrocytes lack the appropriate AAV receptors to endocytose AAV particles. Another possibility is that the lineage of Müller cells and retinal neurons is very close, both derived from retinal progenitor cells, while retinal astrocytes come from optic nerves (Hamon et al., 2016; Reemst et al., 2016). Therefore, Müller cells may be more likely to trans-differentiate into their neighboring neurons than retinal astrocytes that come from a different lineage. The precise mechanism needs further investigation.
Figure 6.
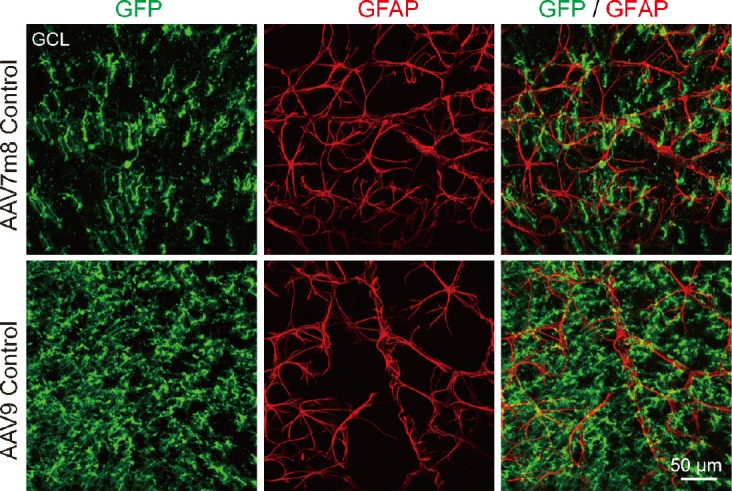
Neither virus with GFAP promoter infects retinal astrocytes.
Images of whole-mount retinas stained with GFP (green: Alexa Fluor-488) and GFAP (red: Alexa Fluor-647) in the GCL, where astrocytes are located. Images were collected 7 weeks after injection of AAV7m8 GFAP-GFP or AAV9 GFAP104-GFP. For both viruses, no GFAP+ astrocytes expressed GFP. GCL: Ganglion cell layer; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein.
Limitations and potential application
As a promising technology, in vivo cell trans-differentiation is a hot topic in the field of gene therapy, but recently a debate has arisen regarding the origin of cell trans-differentiation. To confirm our findings, we recommend the use of different viral systems for a series of dose- and time-dependent experiments to ensure that the induced neurons are derived from Müller cells. Lineage tracing is one way to track the origin of induced neurons, but we have found that while lineage-traced astrocytes in the mouse brain can be converted into neurons (Xiang et al., 2021), these Cre-LoxP-Mediated recombinant astrocytes have a high conversion barrier and are more difficult to convert (Chen, 2021). We are currently designing new experiments to test Müller conversion in the retina using lineage-traced mice. Another way is to use single-cell sequencing at various time points after virus injection to check the intermediate state between Müller cells and retinal neurons during the reprogramming process.
One concern is viral leakage into preexisting neurons. While no one can completely exclude this possibility when injecting viruses into healthy retinas, our current data suggest that our AAV system is relatively specific in targeting Müller cells through GFAP promoter restriction. First, few GFP+ neurons were observed in the control GFAP::GFP group, even after applying a high titer of AAVs (1013 GC/mL). Second, expression of ND1 was restricted to Müller cells at the early stage after being infected by GFAP::NeuroD1-GFP (5 dpi). If ND1 does not convert Müller cells at all, one would expect to see Müller cells being labeled by both ND1 and GFP forever (as we saw in the control group at 28 dpi). The actual outcome was the disappearance of GFP-labeled Müller cells in the experimental group, accompanied by the appearance of GFP-labeled photoreceptor cells at the same time, a clear indication of conversion. The ultimate goal of our research is to regenerate retinal neurons from Müller cells to restore vision in degenerated retinas. In this study, we identified AAV9 GFAP104::NeuroD1-GFP as a potential tool for reprograming Müller cells into photoreceptor cells after subretinal injection. In the next step, we will investigate how well ND1 reprogramming can repair degenerated photoreceptors in a mouse model. Meanwhile, we are also testing various combinations of transcription factors beyond ND1 for their ability to regenerate of RGCs and other subtypes of retinal neurons. Ultimately, our findings could provide a means to treat the millions of patients suffering from retinopathy.
Additional file: Open peer review reports 1 (91.2KB, pdf) and 2 (89.9KB, pdf) .
Footnotes
Conflicts of interest: GC is a co-founder of NeuExcell Therapeutics Inc. The other authors have no conflict of interest.
Availability of data and materials: All data generated or analyzed during this study are included in this published article and its supplementary information files.
Open peer reviewers: Han van der Want, Norwegian University of Science and Technology, Norway; Herbert M Geller, National Heart, Lung, and Blood Institute, USA; Javier Francisco-Morcillo, University of Extremadura, Spain.
P-Reviewers: van der Want H, Geller HM, Francisco-Morcillo J; C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Yu J, Song LP; T-Editor: Jia Y
Funding: This study was supported by the Guangdong Grant Key Technologies for Treatment of Brain Disorders, China, No. 2018B030332001 (to GC); the Guangzhou Key Projects of Brain Science and Brain-Like Intelligence Technology, No. 20200730009 (to YX); and the Guangdong Basic and Applied Basic Research Foundation, No. 2020A1515110898 (to WYC).
References
- 1.Brzezinski JAt, Kim EJ, Johnson JE, Reh TA. Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development. 2011;138:3519–3531. doi: 10.1242/dev.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen G. In vivo confusion over in vivo conversion. Mol Ther. 2021;29:3097–3098. doi: 10.1016/j.ymthe.2021.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zhang B, Xu S, Lin R, Wang W. Lentivirus carrying the NeuroD1 gene promotes the conversion from glial cells into neurons in a spinal cord injury model. Brain Res Bull. 2017;135:143–148. doi: 10.1016/j.brainresbull.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Chen YC, Ma NX, Pei ZF, Wu Z, Do-Monte FH, Keefe S, Yellin E, Chen MS, Yin JC, Lee G, Minier-Toribio A, Hu Y, Bai YT, Lee K, Quirk GJ, Chen G. A NeuroD1 AAV-based gene therapy for functional brain repair after ischemic injury through in vivo astrocyte-to-neuron conversion. Mol Ther. 2020;28:217–234. doi: 10.1016/j.ymthe.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherry TJ, Wang S, Bormuth I, Schwab M, Olson J, Cepko CL. NeuroD factors regulate cell fate and neurite stratification in the developing retina. J Neurosci. 2011;31:7365–7379. doi: 10.1523/JNEUROSCI.2555-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho JH, Klein WH, Tsai MJ. Compensational regulation of bHLH transcription factors in the postnatal development of BETA2/NeuroD1-null retina. Mech Dev. 2007;124:543–550. doi: 10.1016/j.mod.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge LJ, Yang FH, Li W, Wang T, Lin Y, Feng J, Chen NH, Jiang M, Wang JH, Hu XT, Chen G. In vivo neuroregeneration to treat ischemic stroke through NeuroD1 AAV-based gene therapy in adult non-human primates. Front Cell Dev Biol. 2020;8:590008. doi: 10.3389/fcell.2020.590008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goebbels S, Bode U, Pieper A, Funfschilling U, Schwab MH, Nave KA. Cre/loxP-mediated inactivation of the bHLH transcription factor gene NeuroD/BETA2. Genesis. 2005;42:247–252. doi: 10.1002/gene.20138. [DOI] [PubMed] [Google Scholar]
- 9.Guo Z, Zhang L, Wu Z, Chen Y, Wang F, Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer's disease model. Cell Stem Cell. 2014;14:188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamon A, Roger JE, Yang XJ, Perron M. Müller glial cell-dependent regeneration of the neural retina: An overview across vertebrate model systems. Dev Dyn. 2016;245:727–738. doi: 10.1002/dvdy.24375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Institutes of Health. Washington, DC, USA: National Academies Press; 2011. Guide for the Care and Use of Laboratory Animals, 8th edition. [Google Scholar]
- 12.Hickey DG, Edwards TL, Barnard AR, Singh MS, de Silva SR, McClements ME, Flannery JG, Hankins MW, MacLaren RE. Tropism of engineered and evolved recombinant AAV serotypes in the rd1 mouse and ex vivo primate retina. Gene Ther. 2017;24:787–800. doi: 10.1038/gt.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, Kageyama R. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–842. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- 14.Jiang MQ, Yu SP, Wei ZZ, Zhong W, Cao W, Gu X, Wu A, McCrary MR, Berglund K, Wei L. Conversion of reactive astrocytes to induced neurons enhances neuronal repair and functional recovery after ischemic stroke. Front Aging Neurosci. 2021;13:612856. doi: 10.3389/fnagi.2021.612856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jorstad NL, Wilken MS, Grimes WN, Wohl SG, VandenBosch LS, Yoshimatsu T, Wong RO, Rieke F, Reh TA. Stimulation of functional neuronal regeneration from Müller glia in adult mice. Nature. 2017;548:103–107. doi: 10.1038/nature23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y, Messing A, Su M, Brenner M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia. 2008;56:481–493. doi: 10.1002/glia.20622. [DOI] [PubMed] [Google Scholar]
- 18.Nagashima M, Barthel LK, Raymond PA. A self-renewing division of zebrafish Müller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development. 2013;140:4510–4521. doi: 10.1242/dev.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochocinska MJ, Muñoz EM, Veleri S, Weller JL, Coon SL, Pozdeyev N, Iuvone PM, Goebbels S, Furukawa T, Klein DC. NeuroD1 is required for survival of photoreceptors but not pinealocytes: results from targeted gene deletion studies. J Neurochem. 2012;123:44–59. doi: 10.1111/j.1471-4159.2012.07870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palazzo I, Deistler K, Hoang TV, Blackshaw S, Fischer AJ. NF-κB signaling regulates the formation of proliferating Müller glia-derived progenitor cells in the avian retina. Development. 2020;147:dev183418. doi: 10.1242/dev.183418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pataskar A, Jung J, Smialowski P, Noack F, Calegari F, Straub T, Tiwari VK. NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J. 2016;35:24–45. doi: 10.15252/embj.201591206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perea G, Yang A, Boyden ES, Sur M. Optogenetic astrocyte activation modulates response selectivity of visual cortex neurons in vivo. Nat Commun. 2014;5:3262. doi: 10.1038/ncomms4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell C, Cornblath E, Elsaeidi F, Wan J, Goldman D. Zebrafish Müller glia-derived progenitors are multipotent, exhibit proliferative biases and regenerate excess neurons. Sci Rep. 2016;6:24851. doi: 10.1038/srep24851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puls B, Ding Y, Zhang F, Pan M, Lei Z, Pei Z, Jiang M, Bai Y, Forsyth C, Metzger M, Rana T, Zhang L, Ding X, Keefe M, Cai A, Redilla A, Lai M, He K, Li H, Chen G. Regeneration of functional neurons after spinal cord injury via in situ NeuroD1-mediated astrocyte-to-neuron conversion. Front Cell Dev Biol. 2020;8:591883. doi: 10.3389/fcell.2020.591883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reemst K, Noctor SC, Lucassen PJ, Hol EM. The indispensable roles of microglia and astrocytes during brain development. Front Hum Neurosci. 2016;10:566. doi: 10.3389/fnhum.2016.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y, Wu Q, Gao M, Ryu E, Pei Z, Kissinger ST, Chen Y, Rao AK, Xiang Z, Wang T, Li W, Chen G, Chubykin AA. Restoration of visual function and cortical connectivity after ischemic injury through neurod1-mediated gene therapy. Front Cell Dev Biol. 2021;9:720078. doi: 10.3389/fcell.2021.720078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z, Parry M, Hou XY, Liu MH, Wang H, Cain R, Pei ZF, Chen YC, Guo ZY, Abhijeet S, Chen G. Gene therapy conversion of striatal astrocytes into GABAergic neurons in mouse models of Huntington's disease. Nat Commun. 2020;11:1105. doi: 10.1038/s41467-020-14855-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang Z, Xu L, Liu M, Wang Q, Li W, Lei W, Chen G. Lineage tracing of direct astrocyte-to-neuron conversion in the mouse cortex. Neural Regen Res. 2021;16:750–756. doi: 10.4103/1673-5374.295925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao D, Jin K, Qiu S, Lei Q, Huang W, Chen H, Su J, Xu Q, Xu Z, Gou B, Tie X, Liu F, Liu S, Liu Y, Xiang M. In vivo regeneration of ganglion cells for vision restoration in mammalian retinas. Front Cell Dev Biol. 2021;9:755544. doi: 10.3389/fcell.2021.755544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Yao K, Qiu S, Wang YV, Park SJH, Mohns EJ, Mehta B, Liu X, Chang B, Zenisek D, Crair MC, Demb JB, Chen B. Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas. Nature. 2018;560:484–488. doi: 10.1038/s41586-018-0425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Lei Z, Guo Z, Pei Z, Chen Y, Zhang F, Cai A, Mok G, Lee G, Swaminathan V, Wang F, Bai Y, Chen G. Development of neuroregenerative gene therapy to reverse glial scar tissue back to neuron-enriched tissue. Front Cell Neurosci. 2020;14:594170. doi: 10.3389/fncel.2020.594170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng J, Li T, Qi S, Qin B, Yu J, Chen G. Neuroregenerative gene therapy to treat temporal lobe epilepsy in a rat model. Prog Neurobiol. 2022;208:102198. doi: 10.1016/j.pneurobio.2021.102198. [DOI] [PubMed] [Google Scholar]
- 34.Zhou H, Su J, Hu X, Zhou C, Li H, Chen Z, Xiao Q, Wang B, Wu W, Sun Y, Zhou Y, Tang C, Liu F, Wang L, Feng C, Liu M, Li S, Zhang Y, Xu H, Yao H, et al. Glia-to-neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice. Cell. 2020;181:590–603.e16. doi: 10.1016/j.cell.2020.03.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.