Abstract
The rapid development of CRISPR-Cas genome editing tools has greatly changed the way to conduct research and holds tremendous promise for clinical applications. During genome editing, CRISPR-Cas enzymes induce DNA breaks at the target sites and subsequently the DNA repair pathways are recruited to generate diverse editing outcomes. Besides off-target cleavage, unwanted editing outcomes including chromosomal structural variations and exogenous DNA integrations have recently raised concerns for clinical safety. To eliminate these unwanted editing byproducts, we need to explore the underlying mechanisms for the formation of diverse editing outcomes from the perspective of DNA repair. Here, we describe the involved DNA repair pathways in sealing Cas enzyme-induced DNA double-stranded breaks and discuss the origins and effects of unwanted editing byproducts on genome stability. Furthermore, we propose the potential risk of inhibiting DNA repair pathways to enhance gene editing. The recent combined studies of DNA repair and CRISPR-Cas editing provide a framework for further optimizing genome editing to enhance editing safety.
Keywords: genome editing and CRISPR/Cas, DSB repair, chromosomal translocation, large deletion, vector DNA integration, PEM-seq
Introduction
The bacterial clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein (Cas) nucleases have been engineered to achieve efficient gene editing in mammalian cells [ 1– 4] . Among the diverse editing toolboxes, Cas nucleases like Cas9, Cas12a, Cas12e, and Cas12f generate DNA double-stranded breaks (DSBs) to initiate gene disruptions ( Figure 1A) [ 5– 10] , while base editors were developed by fusing Cas nickase with a DNA deaminase enzyme to directly modify DNA nucleotides instead of inducing DSBs ( Figure 1A) [ 11– 13] . Of note, the Cas nickase embedded in base editors can generate single-stranded breaks (SSBs) during the base editing process [ 11– 13] . The more recent prime editors consist of a Cas9 nickase and a reverse transcriptase to induce template-dependent insertions or deletions ( Figure 1A) [14]. Regarding RNA editing, ADAR or nuclease-dead Cas13-fused ADAR can modify RNA nucleotides for gene interference ( Figure 1A) [ 15, 16] . The versatile CRISPR-Cas editing system has been widely used in both scientific research and clinical therapeutics. Many clinical trials employing CRISPR-Cas nucleases are underway and the preliminary results are very promising ( Table 1). These CRISPR-based therapeutic schemes target some very intractable diseases, including T cell, chimeric antigen receptor T cell (CAR T), or T cell receptor-engineered T cell (TCR T) therapy for acquired immunodeficiency syndrome (AIDS) and malignancy [ 17– 19] , modified hematopoietic stem and progenitor cells (HSPCs) for transfusion-dependent β-thalassemia and sickle cell disease [ 20, 21] , correcting CEP290 for Leber Congenital Amaurosis type 10 [22], and Duchenne muscular dystrophy [ 23– 25] .
Figure 1 .
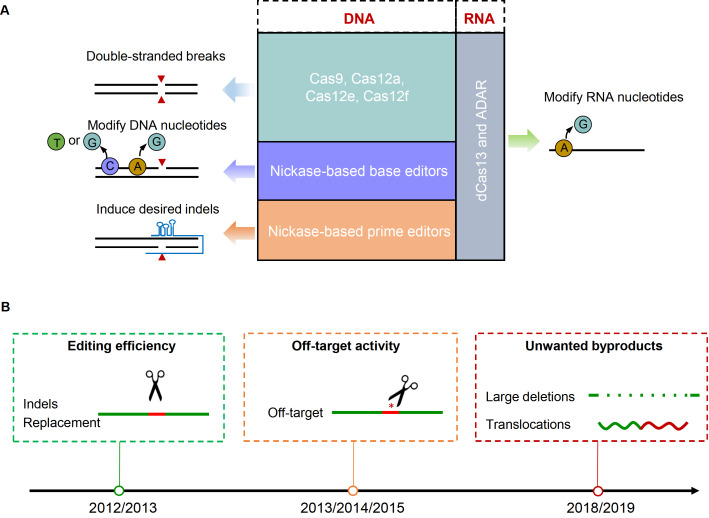
Genome editing tools and the arising concerns
(A) Currently used CRISPR-Cas genome editing tools. CRISPR-Cas editing tools can be subtyped into DSB-dependent nucleases, nickase-based base editors, nickase-based prime editors, and dCas13-based RNA editors. Cas9, Cas12a, Cas12e, and Cas12f are widely-used nucleases for genome editing. Base editors can be classified into CBE (C to T), GBE (C to G), and ABE (A to G) based on the conversion or transversion of the nucleotides. Prime editors induce specific insertions and deletions by using an RNA template in the sgRNA scaffold. RNA base editors are designed by fusing dCas13 and ADAR to convert A to G. (B) Concerns in the genome editing field. Editing efficiencies and off-target activities are early concerns in the field. Until recently, unwanted byproducts like large deletions and translocations are appealed by NIH.
Table 1 Application of Cas9 nuclease in clinical therapy*
|
Target gene |
Clinical ID |
Therapy |
Disease |
Stage |
|
TRAC, B2M |
CAR-T |
B cell leukemia |
Phase 1/2 |
|
|
TRAC, TRBC, B2M |
T cell therapy, CAR-T |
Myeloma, B cell lymphoma |
– |
|
|
TRAC, TRBC, PDCD1 |
TCR-T |
B cell leukemia and solid tumor |
Phase 1 |
|
|
TRAC |
CD19, CD20 or CD22 CAR-T therapy |
Relapsed or refractory hematological malignancies |
Phase 1/2 |
|
|
PDCD1 |
T cell therapy |
Advanced Non-small Cell Lung Cancer |
Phase 1 |
|
|
T cell therapy |
Stage IV bladder cancer |
Phase 1 |
||
|
T cell therapy |
Metastatic renal cell carcinoma |
Phase 1 |
||
|
T cell therapy |
Esophageal cancer |
Phase 1 |
||
|
T cell therapy |
Hormone refractory prostate cancer |
Phase 1 |
||
|
CD7 |
CAR-T |
T cell leukemia |
Phase 1 |
|
|
CD70 |
T cell therapy |
Hematologic malignancies and renal cell carcinoma |
Phase 1 |
|
|
CCR5 |
HSPC therapy |
HIV and leukemia |
– |
|
|
BCL11A |
CRISPR_SCD001 |
HSC therapy |
Transfusion-dependent β-thalassemia (TDT) and sickle cell disease (SCD) |
Phase 1/2 |
|
HBB |
HSC therapy |
TDT |
Phase 1 |
|
|
CEP290 |
AAV therapy |
Leber congenital amaurosis type 10 (LCA10) |
Phase 2 |
|
|
CISH |
NCT03538613, NCT04089891 |
T cell therapy |
Metastatic gastrointestinal epithelial cancer |
Phase 1/2 |
*Because of space limitation, only typical clinical trials are cited here.
Besides the great potential of CRISPR-Cas editing tools, unwanted editing byproducts accompanied with intended editing products have also attracted great attention recently, since they lend additional uncertainty to genome editing [26]. These unwanted editing byproducts include but are not limited to off-target damages, chromosomal structural variations, and exogenous DNA integrations ( Figure 1B). Many efforts have been made to further improve the performance of CRISPR-Cas gene editing tools [ 27– 29] and various methods based on experiments or in silico prediction have been developed to identify or evaluate off-target activity for Cas nucleases ( Table 2, see below for more details) [ 30– 40] . Chromosomal translocations and large deletions have also been routinely observed in different editing scenarios recently [ 31, 37– 39, 41– 45] . For example, chromosomal abnormalities have been discovered to expand in a patient treated by CAR T cells manufactured by Allogene, which leads to the hold on Allogene CAR T therapeutics. An effective method to reduce chromosomal abnormalities during gene editing is still lacking.
Table 2 Methods for the detection of byproducts generated by Cas9*
|
Method |
In vivo/ In vitro/ In silico |
Assay type |
Comment |
|
|
Cas-OFFinder |
In silico |
Off-target |
Sequence alignment |
High false positive |
|
CAST-seq |
In vivo |
Chromosomal structural variations, indels |
Map translocations with induced DSBs |
High-sensitivity; not applicable to limited material |
|
CIRCLE-seq |
In vitro |
Off-target |
Sequence cleaved linear DNA from circularized genomic DNA |
High-sensitivity; Requires in vivo cleavage confirmation |
|
Dig-seq |
In vitro |
Off-target |
Whole-genome sequencing for cleaved chromatin |
High-sensitivity; |
|
Digenome-seq |
In vitro |
Off-target |
Whole-genome sequencing for cleaved naked genomic DNA |
High-sensitivity; Requires in vivo cleavage confirmation |
|
DISCOVER-seq |
In vivo |
Off-target |
Pull down Mre11 binding to broken ends |
Narrow time-window (only maps unjoined ends); low resolution |
|
GUIDE-seq |
In vivo |
Off-target |
Integrate dsODNs into DSB sites |
Unbiased; limited use for blunt-ended DSBs |
|
LAM-HTGTS |
In vivo |
Off-target, chromosomal structural variations |
Map translocations with induced DSBs or recurrent DSBs |
High-sensitivity; not applicable to limited material |
|
PEM-seq |
In vivo |
Off-target, chromosomal structural variations, indels |
Map translocations with induced DSBs or recurrent DSBs |
High-sensitivity; not applicable to limited material |
|
SITE-seq |
In vitro |
Off-target |
Map broken ends with biotinylatedadapters |
High-sensitivity; Requires in vivo cleavage confirmation |
|
UDiTaS |
In vivo |
Chromosomal structural variations, indels |
Map translocations with induced DSBs |
Low sensitivity due to no nested PCR |
*Because of space limitation, only typical methods are cited here.
The generation of both intended products and unwanted editing byproducts during genome editing are stimulated by endogenous DSB repair pathways, and understanding how these repair pathways work in depth could help to reduce the side effects of unwanted byproducts during gene editing.
In this review, we begin with the editing mechanism for CRISPR/Cas editing system and then describe the involved DSB repair pathways in the editing process. We next discuss the generation of unwanted genome editing products and propose possible solutions to improve the safety of gene editing.
CRISPR-Cas Induces DNA Breaks to Initiate Gene Editing
The CRISPR-Cas enzyme is an RNA-guided endonuclease that induces DSB at the phage genome. The CRISPR-Cas enzymes have two distinct groups: class I, which applies multi-Cas proteins to achieve DNA cleavage; and class II, which applies a single endonuclease for DNA cleavage [ 46, 47] . Class II is further subtyped into three types: II, V, and VI. The type-II Cas9 recognizing 3′ G rich protospacer adjacent motif (PAM) and type-V Cas12 recognizing 5′ T rich PAM have been engineered for efficient genome editing [48]. Among the engineered Cas9 enzymes, Streptococcus pyogenes Cas9 ( SpCas9) with an NGG (N= “A”, “T”, “C”, or “G”) PAM is the first and most widely used Cas9 for genome editing [ 1– 4, 49] . A smaller size Staphylococcus aureus Cas9 ( SaCas9) was also developed for target sites with NNGRRT (R=“A” or “G”) PAM [50]. Several other Cas9 nucleases including Streptococcus thermophiles Cas9 ( StCas9), Campylobacter jejuni Cas9 ( CjCas9), Francisella novicida Cas9 ( FnCas9), Geobacillus stearothermophilus Cas9 ( GeoCas9), Neisseria Meningitides Cas9 ( NmeCas9), and Streptococcus canis ( ScCas9) were subsequently engineered for genome editing ( Figure 2A) [ 51– 57] . Regarding the Cas12 family, Acidaminococcus sp. Cas12a ( AsCas12a) and Lachnospiraceae bacterium ND2006 Cas12a ( LbCas12a) show great potential in gene editing [6]. Recently, orthologs of small-size Cas12e and Cas12f nucleases have been successfully used for gene editing and show advantages for adeno-associated virus (AAV) package for gene therapy ( Figure 2B) [ 7– 10] .
Figure 2 .
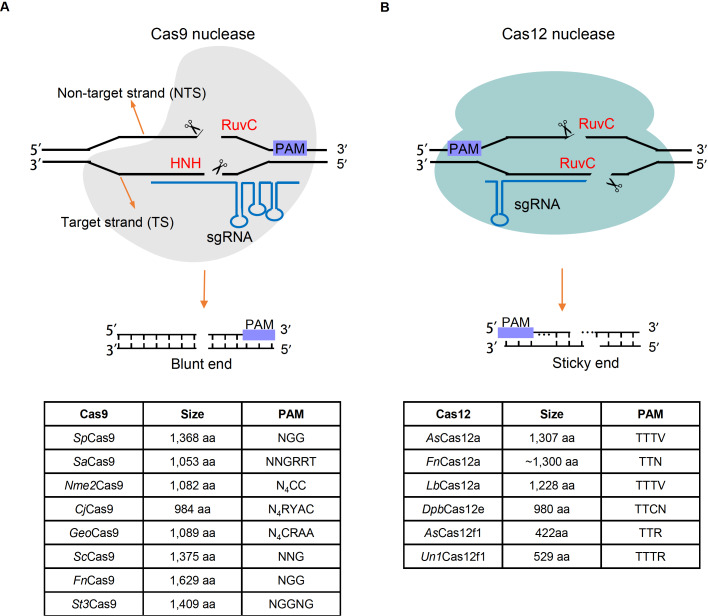
Two main Cas nucleases for genome editing
Cas nucleases are guided by sgRNA to the target site by forming an “R loop”. (A) Cas9 nucleases use the HNH domain to cleave the target strand and RuvC domain to cleave non-target strand both upstream PAM. Cas9 nucleases tend to generate blunt ends. (B) Unlike Cas9, Cas12 nucleases use only the RuvC domain to cleave both target and on-target strand downstream PAM, and Cas12 nucleases generate sticky end. Currently-developed Cas9 and Cas12 nucleases with their size and PAM are displayed on the bottom.
CRISPR-Cas9 system contains a CRISPR RNA (crRNA) for targeting DNA and a trans-activating crRNA (tracrRNA) pairing with crRNA for Cas9 ribonucleoprotein (RNP) package [58]. The crRNA and tracrRNA are further combined into a chimeric single guide RNA (sgRNA), which reserves the high cleavage capacity [5]. Cas9 cleavage begins with the recognition for the PAM sequence located at 3′ of the target DNA, followed by the formation of RNA-DNA hybrid (R loop), Cas9 conformation change, and DNA strand cleavage [ 5, 59, 60] . The target strand (pair with sgRNA) is cleaved by the HNH domain and the non-target strand is cleaved by the RuvC domain and both cleavages occur between the third and fourth nucleotides upstream of PAM, which eventually leads to a blunt-ended DSB ( Figure 2A) [5]. Cas9 can also generate 1-bp staggered ends at some target sequences due to the flexible cleavage position of the RuvC domain, resulting in predictable 1-bp insertions [ 43, 61– 64] . Mutation in either of the two cleavage domains generates Cas9 nickase and mutations in both the cleavage domains generate nuclease-dead Cas9 (dCas9) but reserve DNA-binding activity [ 65, 66] . After cleavage, Cas9 nuclease may stay at the PAM-distal ends until the DNA repair proteins are recruited to seal the broken ends [ 59, 67] . In contrast to Cas9 nucleases, most Cas12 nucleases are guided by a single crRNA and equipped with only a RuvC domain to cleave the DNA strands [ 6, 68] . The RuvC domain of Cas12 nuclease cuts the two DNA strands at varied nucleotides and thus results in sticky-ended DSBs ( Figure 2B).
In addition to CRISPR-Cas nucleases, CRISPR-based base editors and prime editors were mainly developed for mutation corrections. The base editor consists of a Cas9 nickase, a DNA deaminase enzyme, and a uracil-DNA glycosylase inhibitor or uracil-DNA glycosylase, which converts C to T, C to G, or A to G without causing DSBs [ 11– 13] . In this context, AID, APOBEC1, APOBEC3A, and APOBEC3B were used as cytosine base editors (CBEs) for C to T conversion or used as glycosylase base editors (GBEs) for C to G transversion [ 11, 13, 69– 71] . In addition, TadA was engineered as an adenine base editor (ABE) for A to G conversion [12]. ABE was further combined with CBE to generate dual base editors for simultaneous C to T and A to G conversions by several research groups [ 72– 74] . Instead of DNA deaminase enzymes, the prime editors fuse a reverse transcriptase with Cas9 nickase to introduce point mutations not covered by CBEs or ABEs, short insertions, and deletions of several nucleotides by using a template RNA within the sgRNA scaffold [14]. The two base editing systems are complementary to Cas9 nucleases for different editing scenarios but also face several problems. First, the Cas9 nickase-induced SSBs can be converted to DSBs at a frequency of 1 out of 100 [ 75, 76] , which may also lead to chromosomal structural variations as Cas9 nucleases though at a low level. Second, base editors have been reported to have collateral DNA and RNA activities besides off-target activities [ 77– 81] .
DSB Repair Pathways Are Involved in Gene Editing
DSBs are the most deleterious type of DNA lesions, leading to genetic mutations or complex chromosomal rearrangements associated with oncogenesis [ 75, 82– 87] . Each human cell is subjected to 25 endogenous DSBs per day in estimation [75], and thereby robust DSB repair pathways evolve in mammalian cells to recognize and repair emerging DSBs. Typically, the entire process of DSB repair consists of three or four steps: end recognition, end tethering, end processing if necessary, and end joining ( Figure 3A) [82]. The initial end-recognizing and end-binding proteins determine the choice of the DSB pathways, and then other repair proteins are recruited into the DSBs step-by-step until end joining [ 82, 88– 91] . The mammalian cells mainly evolve two types of DSB repair pathways: template-independent end joining repair and template-dependent homology-directed repair ( Figure 3B). These repair pathways compete with each other and are influenced by cell type, cell state, and the nature of the DSBs [92]. The repair of Cas-induced DSBs shares main features with endogenous DSBs except that Cas9 residence at broken ends may have a weak impact on DSB repair [67]. Here we provided a brief overview of these DSB repair pathways involved in gene editing in mammalian cells.
Figure 3 .
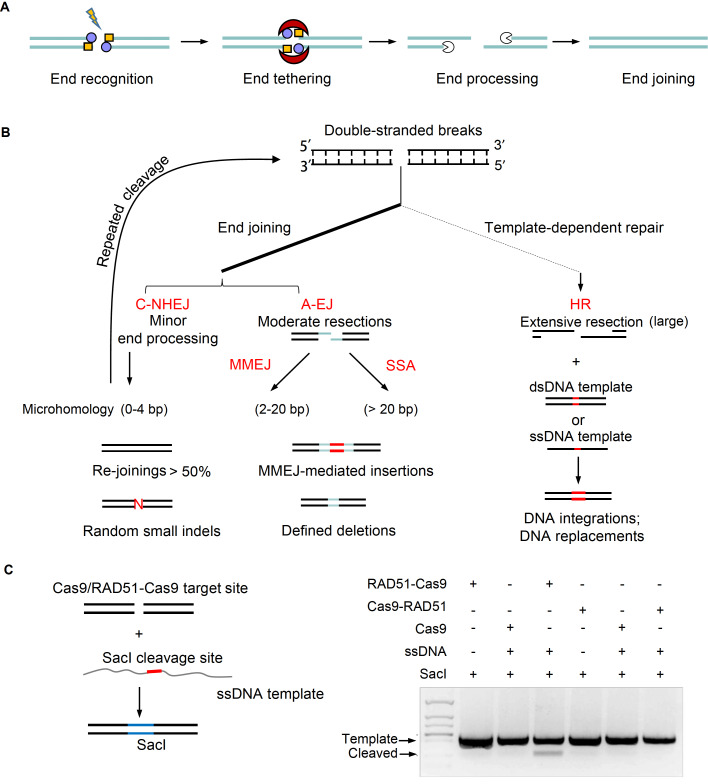
DSB repair pathways involved in genome editing
(A) General steps for DSB repair. End recognition, end tethering, end processing, and end joining. (B) DSB repair pathways for genome editing. DSB repairs are mainly subtyped into end-joining and template-dependent repair. C-NHEJ directly joins two broken ends with small indels in the final products due to limited end processing. Note that more than 50% of products generated by C-NHEJ are re-joinings and will undergo several cycles of repeated cleavage until the formation of indels. The process of end-joining may use homology on the broken ends to generate defined deletion or insertions. Due to the length of the homology, homology length from 2 to 20 bp is recognized as MMEJ and more than 20 bp is recognized as SSA. Template-dependent repair needs extensive resection and uses dsDNA template or ssDNA template. (C) RAD51-Cas9 enhances ssDNA integration. ssDNA template with a SacI cleavage site and homologous arm is co-transfected with Cas9 or RAD51-Cas9. If integration occurs, the DNA bands can be cleaved by the restriction enzyme SacI.
Non-homologous end joining
Classical non-homologous end joining (C-NHEJ) directly re-joins two broken ends and is considered to be the default choice for DSB repair in mammalian cells through cell cycles [93]. In estimation, more than 50% of Cas9-induced DSBs are repaired by NHEJ in human pluripotent stem cells or human cell lines within the first 10 h of DSBs [ 67, 94, 95] . C-NHEJ is an error-prone repair process and usually introduces small nucleotide insertions and deletions (indels). Therefore, the CRISPR-Cas targeting at open reading frames can readily induce gene disruption by C-NHEJ-mediated frameshift. However, it is notable that more than 50% of Cas9-induced breaks are perfectly re-joined without end processing in mouse embryonic stem cells (mESCs) and HEK293T cells [ 96, 97] . In this context, the perfectly re-joined products can be targeted repeatedly by CRISPR-Cas enzymes to accumulate desired editing outcomes.
During C-NHEJ, KU70-KU80 heterodimer immediately binds to the broken ends and recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and/or Artemis endonuclease to mildly process broken ends if needed [ 98– 104] . Next, XRCC4, LIG4, XLF, and recently-identified PAXX proteins are recruited to tether and seal the broken ends [ 105– 112] . Besides Artemis, nucleases such as PALF, MRN complex, and polymerases including the terminal deoxynucleotidyltransferase (TdT), Pol μ, and Pol λ also contribute to the end processing to introduce indels within final products [ 113– 119] . In this context, fusing Cas nucleases with end processing enzymes including T5 and TREX2 facilitates indel formation [ 120, 121] .
Alternative end joining
Alternative end joining (A-EJ) dominates end joining repair when core factors of C-NHEJ are deficient [ 122, 123] . According to the length of microhomology used, A-EJ can be further divided into two subtypes: microhomology-mediated end joining (MMEJ) with homology at approximately 2–20 bp and single-strand annealing (SSA) pathway which requires large homology (>20 bp) ( Figure 3B) [ 43, 124] . In comparison, C-NHEJ only uses microhomology less than 4 bp ( Figure 3B) [ 43, 124, 125] . A-EJ also functions in the presence of C-NHEJ and competes with C-NHEJ to repair Cas9-induced DSBs [ 126, 127] . By examining over 1000 loci cleaved by Cas9, van Steensel and colleagues recently reported that the choice for MMEJ and C-NHEJ may be influenced by chromatin accessibility and MMEJ tends to occur in heterochromatin regions associated with H3K37me3 modification [127]. Moreover, it has been reported that MMEJ displays delayed activity in comparison with C-NHEJ detected by a quantitative time-course study [67].
MMEJ prevalently contributes to the formation of indels during genome editing by generating short deletions between two microhomologous sequences ( Figure 3B). MMEJ-mediated deletions are relatively predictable in the context of embedded microhomology in local sequence [ 61, 62, 64, 127, 128] . MMEJ enhancement by placing two designed microhomologous sequences spanning the CRISPR-Cas9 target site can efficiently induce programmed fragment insertions and deletions during genome editing [ 129– 132] . SSA is useful for large DNA fragment deletion in genome editing and is mainly active in the S/G2 phase for the need of long exposed homology. Zhang and Matlashewski found that up to 90% of editing products in Leishmania were repaired by SSA, and thereby SSA was enhanced to achieve large fragment deletion up to 29 kb [133]. Pol θ, MRN complex and poly (ADP-ribose) polymerase 1 (PARP1) are required in MMEJ [ 134– 137] . SSA shares end resection steps with homologous recombination (HR) to repair DSBs in mammalian cells. For example, CtIP, EXO1, and DNA2 function in both SSA and HR [ 138– 140] .
Homologous recombination
HR requires a homologous template to finish DSB repair and therefore is a relatively precise DNA repair pathway. HR is mainly active in the S and G2 phases in dividing cells, exhibiting a lower utilization rate in comparison with NHEJ in most cells. The deactivation of C-NHEJ makes Cas-induced DSBs prone to be repaired by A-EJ or HR [ 43, 141– 146] . HR is characterized by extended DNA resection and thereby EXO1 and DNA2 responsible for long-distance DNA resection are critical for HR [ 138, 147– 149] . The highly-processed broken ends are then protected by RPA, followed by RAD51-mediated strand invasion and polymerase-mediated fill-in [ 150– 152] . Recently, RNA polymerase III was also reported to function in HR and protect the processed DNA ends [153].
To introduce intended mutations at target sites during gene editing, a double-stranded (ds) or single-stranded (ss) donor DNA is transfected with CRISPR-Cas to activate the HR pathway to induce homology-directed repair (HDR) ( Figure 3B) [ 154, 155] . The homologous sequence for dsDNA donors is usually hundreds in length while the length of the homologous sequence for ssDNA can be as short as dozens of nucleotides [156]. HDR with ssDNA donor is more frequently used for gene editing, due to moderate adverse cellular responses such as avoiding cGAS activation [157]. Given that HDR is at such a low usage rate, inhibitors for C-NHEJ core factors have been used to enhance HDR during gene editing. For example, the small molecule inhibitors 5102 and 5135 were applied to enhance HDR at a 6-fold increase by suppressing the DNA-binding activity of the KU70/KU80 complex [158]. And the inhibitors of DNA-PKcs, NU7026, and KU-0060648 were used to enhance the HDR by 3 folds [143]. Moreover, applying SCR7 to inhibit the LIG4 showed an increase of 5- to 19-fold for HDR usage in mammalian cell lines [ 141, 142] . In addition to C-NHEJ inhibitors, a dominant-negative form of 53BP1 was expressed with CRISPR-Cas9 to enhance HDR frequency up to 86% in various human cell types [159]. Besides suppression of C-NHEJ, stimulating HR can also enhance HDR. In this context, small molecule RS-1, by activating RAD51, could improve HDR usage up to 5 folds in rabbit embryos [160]. Alternatively, fused or co-expressed RAD51 with Cas9 could also improve HDR ( Figure 3C) [ 161– 164] . In addition, a fusion of truncated CtIP and Cas9 also showed at least 2-fold enhancement for HDR in human cell lines, pluripotent stem cells, and rat zygotes [165]. Furthermore, Chin and colleagues fused human GEMININ to the N terminal of Cas9 to specifically express Cas9 in the S/G2/M phase and increased the rate of HDR by up to 87% [166]. Moreover, arresting cells in S or G2/M phase or inhibiting mismatch repair (MMR) has also been reported to enhance single-stranded DNA oligonucleotide-mediated integration for gene editing [ 167– 172] .
Unwanted Editing Byproducts of CRISPR-Cas Increase Genome Instability
Given that the repair of Cas9-induced DSBs is consistent with the repair of endogenous general DSBs, it is inevitable that the sealing of Cas9-induced DSBs results in many diverse outcomes. Besides the intended mutations at the target site, other unwanted byproducts are routinely identified. CRISPR-Cas activities at off-target sites are well explored by developed methods and a dozen of high-fidelity Cas9 variants have been engineered to reduce the off-target activities of CRISPR-Cas9 [ 32, 34– 38, 173– 179] . Chromosomal structural variations such as chromosomal translocations and large deletions have also attracted great attention recently, which may cause genome instability and have pathogenic consequences [ 37, 38, 41– 45] . Furthermore, vector integrations are also frequently detected when AAV or other DNA-based delivery methods are used ( Figure 4) [ 44, 180, 181] . In this section, we will discuss the mechanism underlying the unwanted editing byproducts and summarize the currently used methods for the detection of unwanted editing byproducts.
Figure 4 .
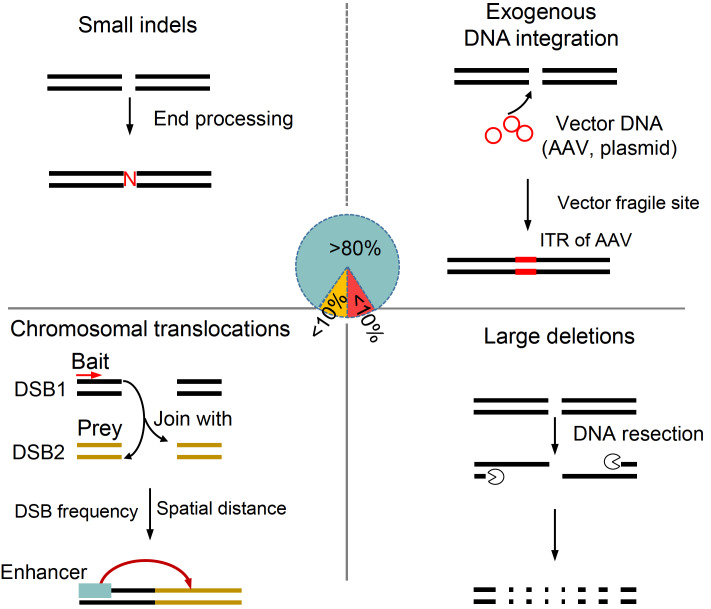
Products and byproducts generated during genome editing
In addition to small indels (top left), vector DNA from plasmid or virus can be integrated into the target site. ITR elements from AAV have been reported to function as enhancers (top right). The juxtaposition of two DSBs forms chromosomal translocations. Both DSBs frequency and spatial distance contribute to the translocation formation (bottom left). Large deletions are generated by end-joining after extensive DNA resection at the target site (bottom right).
Off-target activities
Off-target sites of CRISPR-Cas9 are highly homologous to the target sites with higher mutation tolerance at the PAM-distal region [182]. The seed sequence in the target DNA (10–12 nucleotides located in the 3 prime ends of the 20-nt sgRNA) is vital for Cas9 cleavage, and mutations in the seed region nearly block Cas9 cleavage, yet mutations in other regions cause off-target cleavage [ 183– 185] . To improve the specificity of Cas proteins, a dozen of high-fidelity variants have been developed to obtain lower off-target activities. e SpCas9(1.1), SpCas9-HF1, and HypaCas9 were developed based on Cas9-DNA structures and Cas9 conformation change before cleavage [ 177– 179] . Sniper-Cas9, evo-Cas9, and xCas9 were developed by high-throughput screening methods [ 174, 176, 186] . These high-fidelity variants perform well at some target loci, however, the sacrifice of the editing efficiency was also detected at certain loci for some variants [ 44, 173] . Moreover, because chromosomal structural variations are byproducts during the process of DSB repair and mainly occur at the target sites, high-fidelity variants could not reduce chromosomal translocations and large deletions caused by Cas nuclease [44].
Chromosomal translocations
The juxtaposition of two DSBs can form translocation at a very low frequency. A single DSB generated by meganuclease I-SceI or CRISPR-Cas9 could join to any DSB induced by ion irradiation, implying that any two escaped DSBs can form translocation [ 37, 43, 187] . High levels of chromosomal translocations were identified in the presence or absence of C-NHEJ, indicating that both C-NHEJ and MMEJ are involved in translocation formation [43]. Chromosomal translocations may generate new fusion oncogenes or significantly change the expression levels of genes related to cancer, which possibly results in oncogenesis ( Figure 4, bottom left).
During genome editing, DSBs at the target sites are the dominant DSBs and thereby the vast majority of editing outcomes are re-joinings of the two broken ends of the target DSBs. However, other broken ends that occur simultaneously within the edited cells may also have a chance to join with target DSBs to form translocations. These involved DSBs can be categorized into three types: other target DSBs, off-target DSBs, and general DSBs. Correspondingly, the translocations involving these DSBs are referred to as target translocations, off-target translocations, and general translocations, respectively. The target translocation mainly occurs in the multiplex gene editing system and multiple CRISPR-Cas-induced target DSBs join together to induce a high level of chromosomal translocations. The off-target translocations involving DSBs at off-target sites are also dependent on CRISPR-Cas enzymes. As for general translocations, general DSBs induced by various DNA metabolism activities arise randomly in the genome and can also be captured by CRISPR-Cas-induced target DSBs to form chromosomal translocations. These general DSBs may occur in certain physiological processes including V(D)J recombination or class switch recombination in lymphocytes [ 187– 192] , or are triggered by genomic transcription or DNA replication [ 37, 38, 43, 44, 193– 197] . General translocations are distributed widely over the genome with an obvious accumulation at the transcription start site (TSS) [ 38, 43, 44, 187] . Generally, the frequencies of these translocations are in an order of target translocations > off-target translocations >> general translocations.
Several previous reports showed that target chromosomal translocations frequently arose during multiplex genome editing in CAR T manufacturing [ 18, 198– 200] . Chromosomal translocations are also occasionally captured during single-gene editing by many laboratories [ 201, 202] . Using the high-throughput primer-extension-mediated sequencing (PEM-seq), we found that chromosomal translocations occur at a frequency of 1.0%–2.4% in embryonic stem cells (ESCs) and up to 10% in HEK293T during genome editing [ 43, 44] . Cathomen and colleagues also found that chromosomal rearrangements occurred at a ratio of up to 1.6% in edited stem cells [31]. Off-target translocations can be largely suppressed by using high-fidelity Cas9 variants to reduce the break frequency at off-target sites, but the solution to reduce general translocations or translocations among multiple editing loci is still lacking [44]. A recent clinical trial on TCR T therapy indicated that engineered T cells containing translocations among TRAC-TRBC-PDCD1 remained in the blood at even hundreds of days post-infusion into the patients [18], raising a great concern for these chromosomal abnormalities.
Chromosomal large deletions
Chromosomal large deletions induced by CRISPR-Cas routinely occur at the target site and range from several hundred bases to megabases, resulting in the loss of a large chromosome fragment around the target site or even the entire chromosome [ 43, 44, 193] . Large deletions arise at a frequency of up to 10% in various human and mouse cell lines based on the sequencing data of PEM-seq or Nanopore DNA sequencing [ 38, 43, 44, 203] ; Bradley and colleagues found that more than 20% of edited mESCs contained deletions more than 250 bp, extending up to 6 kb [41]; Thomas and colleagues found that about 57% edited mouse zygotes contained large deletions up to 2.3 kb [45]. A 3.5 Mb deletion was also identified at the UROS loci in HEK293T cells [42]. The mechanism underlying the generation of chromosomal large deletions is not fully understood. We and others found that MMEJ contributes to the formation of large deletions (up to 76.7%) in mESCs and other cells [ 43, 204] . C-NHEJ deficiency could increase the large deletions up to 3 folds [43]. Yet no solutions have been proposed to reduce large deletions during CRISPR-Cas-mediated genome editing ( Figure 4, bottom right).
Exogenous vector DNA integration
Integration of exogenous DNA originating from vectors or viruses into the genome was another concern of genome editing ( Figure 4, top right). Specifically, the target site is the most frequent integration site [ 44, 180] . György and colleagues found high level of AAV integration (up to 47%) in murine neurons, mouse brain ( APP SW , Mecp2, and Dnmt3b), and moused muscle ( Dmd) [180]. As in HEK293T cells, up to 41.3% of edited cells contain vector integration at RAG1, DNMT1, EMX1, VEGFA, and C-MYC loci, for both Cas9 and its variants [ 43, 44] . Microhomology is widely detected between the integration sites and the integrated fragments, indicating the involvement of MMEJ in vector integration [43]. Additionally, some fragile elements at vectors such as the AAV inverted terminal repeat (ITR) regions can greatly elevate the vector integration level [ 43, 180] .
Methods for the Detection of Unwanted Editing Byproducts
Many methods have been developed to detect off-target activities of CRISPR-Cas enzymes, both in vivo and in vitro. The in vivo or ex vivo methods include LAM-HTGTS, GUIDE-seq, DISCOVER-seq, and PEM-seq, while the in vitro methods include but are not limited to Digenome-seq, Dig-seq, CIRCLE-seq, and SITE-seq ( Table 2). These methods have been summarized very well in previous literature [ 28, 35, 38] . Here we focus on the methods to detect other unwanted editing byproducts including chromosomal translocations and large deletions. Quantitative RT-PCR has been widely used to detect chromosomal translocations between two target sites [ 18, 198, 205] , but the resolution is very limited. Whole-genome sequencing or exon sequencing have also been used to identify chromosomal structural variations [ 78, 206] , but these methods are costly and difficult to analyze. Recently, enrichment of target chromatin fragments before sequencing has been introduced to develop several new methods including PEM-seq, LAM-HTGTS, UDiTaS, and CAST-seq (see below for more details). Better enrichment assay or the third-generation sequencing may further facilitate the development of new assays to detect chromosomal translocations or large deletions.
PEM-seq and LAM-HTGTS
Based on chromosomal translocation capture, both PEM-seq and LAM-HTGTS rely on a Cas enzyme-generated “bait” DSB to capture genome-wide “prey” DSBs in vivo [ 37, 38, 207, 208] . The prey-bait junctions are cloned using 1-cycle primer extension for PEM-seq and 80-cycle linear amplification for LAM-HTGTS, followed by ligation with bridge adapters. Subsequent PCR further amplifies the products for next-generation sequencing. Both methods can be used to detect off-target sites that form chromosomal translocations with the bait DSBs as well as large deletions and genome-wide translocations. The LAM-HTGTS was further improved as iHTGTS after optimization of the experimental procedures and the introduction of the random molecular barcode [ 193, 209] . In comparison to LAM-HTGTS and iHTGTS, PEM-seq is a quantitative method which can be used to calculate the frequency of different editing outcomes including vector integrations [ 38, 43] . These methods have been widely applied in mESCs, hESCs, human and mouse primary T cells, various tumor cell lines, and mouse tissues to evaluate the fidelity of Cas9 and Cas12a and their orthologs [ 38, 43, 44, 193, 194] .
UDiTaS and CAST-seq
UDiTaS, which is based on Tn5 shearing, employs primers on bait and Tn5-introduced adapters to amplify target DSB-involved junctions to identify both chromosomal structural variations and on-target indels [39]. UDiTas was used to identify complex chromosomal rearrangements for CEP290 and TCR loci in HEK293T cells. A recently developed method CAST-seq employs decoy primers to amplify bait-prey junctions and can be used to detect chromosomal structural variations [31].
Perspectives
The great improvement of CRISPR-Cas nucleases in clinics shows great potential in the treatment of intractable diseases. Yet DSB is a double-edged sword: off-target damages, chromosomal translocations, large deletions are other non-negligible unwanted editing byproducts that consist of up to 10% of total editing events. The high-fidelity Cas9 variants, especially e SpCas9(1.1), SpCas9-HF1, FeCas9, and HypaCas9, are indeed able to effectively reduce off-target activities [44]. However, the solution for other unwanted editing byproducts is still lacking. The decrease of chromosomal translocations or large deletions is usually accompanied by the decline of editing efficiency in previous reports [38]. Given that more than 50% of Cas-induced DSBs are perfect re-joinings and can be cleaved again by CRISPR-Cas until the formation for final indels or degradation of CRISPR-Cas, a Cas enzyme prefers to generate indels rather than perfect re-joinings may narrow the time windows of free DSBs and restrict the generation of various unwanted editing byproducts. On this basis, Cas9TX has been recently developed by our group to greatly reduce chromosomal structural variations by fusing an optimized TREX2 with Cas9. We applied Cas9TX to the next-generation chimeric antigen receptor T (CAR T) engineering and found the levels of deleterious translocations were decreased by tens of folds among multiple targeting sites [210].
Many methods employ inhibitors for DNA repair proteins to change the choice of DNA repair pathways in editing cells [ 141– 143, 158, 159, 211] . However, the perturbation of DNA repair pathways may bring unpredicted editing byproducts that greatly affect genome integrity. For example, the inhibition of C-NHEJ often leads to elevated levels of chromosomal translocations, large deletions, and vector integrations [43]. Moreover, deactivation of p53 in editing cells can also cause genome stability and lead to cancers [ 212– 215] .
Supporting information
Acknowledgments
We thank Yaxuan Zhou for her contribution to the work on RAD51-Cas9. We thank members of the Hu lab, especially Dr. Yang Liu for their helpful comments.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Key R&D Program of China (No. 2017YFA0506700), the National Natural Science Fundition of China (Nos. 32122018 and 31771485), the SLS-Qidong Innovation Fund, and the PKU-TSU Center for Life Sciences.
References
- 1.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. . 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 2.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. . 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. . 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, et al. RNA-guided human genome engineering via Cas9. Science. . 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. . 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. . 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu JJ, Orlova N, Oakes BL, Ma E, Spinner HB, Baney KLM, Chuck J, et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. . 2019;566:218–223. doi: 10.1038/s41586-019-0908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DY, Lee JM, Moon SB, Chin HJ, Park S, Lim Y, Kim D, et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat Biotechnol. . 2021;40:94–102. doi: 10.1038/s41587-021-01009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z, Zhang Y, Yu H, Pan D, Wang Y, Wang Y, Li F, et al. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat Chem Biol. . 2021;17:1132–1138. doi: 10.1038/s41589-021-00868-6. [DOI] [PubMed] [Google Scholar]
- 10.Xu X, Chemparathy A, Zeng L, Kempton HR, Shang S, Nakamura M, Qi LS. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol Cell. . 2021;81:4333–4345. doi: 10.1016/j.molcel.2021.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. . 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. . 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao D, Li J, Li S, Xin X, Hu M, Price MA, Rosser SJ, et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat Biotechnol. . 2021;39:35–40. doi: 10.1038/s41587-020-0592-2. [DOI] [PubMed] [Google Scholar]
- 14.Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. . 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. RNA editing with CRISPR-cas13. Science. . 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu L, Yi Z, Zhu S, Wang C, Cao Z, Zhou Z, Yuan P, et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat Biotechnol. . 2019;37:1059–1069. doi: 10.1038/s41587-019-0178-z. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Wang J, Liu Y, Xie L, Su B, Mou D, Wang L, et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N Engl J Med. . 2019;381:1240–1247. doi: 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- 18.Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E, Mangan PA, et al. CRISPR-engineered T cells in patients with refractory cancer. Science. . 2020;367:eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y, Xue J, Deng T, Zhou X, Yu K, Deng L, Huang M, et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med. . 2020;26:732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 20.Frangoul H, Ho TW, Corbacioglu S. CRISPR-cas9 gene editing for sickle cell disease and β-thalassemia. N Engl J Med. . 2021;384:e91. doi: 10.1056/NEJMc2103481. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Zeng J, Roscoe BP, Liu P, Yao Q, Lazzarotto CR, Clement K, et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med. . 2019;25:776–783. doi: 10.1038/s41591-019-0401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS, Bumcrot D, et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med. . 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 23.Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, Caballero D, Harron R, et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. . 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moretti A, Fonteyne L, Giesert F, Hoppmann P, Meier AB, Bozoglu T, Baehr A, et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat Med. . 2020;26:207–214. doi: 10.1038/s41591-019-0738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells . Science. . 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha K, Sontheimer EJ, Brooks PJ, Dwinell MR, Gersbach CA, Liu DR, Murray SA, et al. The NIH somatic cell genome editing program. Nature. . 2021;592:195–204. doi: 10.1038/s41586-021-03191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. . 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 28.Kim D, Luk K, Wolfe SA, Kim JS. Evaluating and enhancing target specificity of gene-editing nucleases and deaminases. Annu Rev Biochem. . 2019;88:191–220. doi: 10.1146/annurev-biochem-013118-111730. [DOI] [PubMed] [Google Scholar]
- 29.Zhang F. Development of CRISPR-Cas systems for genome editing and beyond. Quart Rev Biophys. . 2019;52:e6. doi: 10.1017/S0033583519000052. [DOI] [Google Scholar]
- 30.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. . 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turchiano G, Andrieux G, Klermund J, Blattner G, Pennucci V, El Gaz M, Monaco G, et al. Quantitative evaluation of chromosomal rearrangements in gene-edited human stem cells by CAST-Seq. Cell Stem Cell. . 2021;28:1136–1147.e5. doi: 10.1016/j.stem.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets . Nat Methods. . 2017;14:607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D, Kim JS. DIG-seq: a genome-wide CRISPR off-target profiling method using chromatin DNA. Genome Res. . 2018;28:1894–1900. doi: 10.1101/gr.236620.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. . 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 35.Wienert B, Wyman SK, Richardson CD, Yeh CD, Akcakaya P, Porritt MJ, Morlock M, et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq . Science. . 2019;364:286–289. doi: 10.1126/science.aav9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. . 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. . 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin J, Liu M, Liu Y, Wu J, Gan T, Zhang W, Li Y, et al. Optimizing genome editing strategy by primer-extension-mediated sequencing. Cell Discov. . 2019;5:18. doi: 10.1038/s41421-019-0088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giannoukos G, Ciulla DM, Marco E, Abdulkerim HS, Barrera LA, Bothmer A, Dhanapal V, et al. UDiTaS™, a genome editing detection method for indels and genome rearrangements. BMC Genomics. . 2018;19:212. doi: 10.1186/s12864-018-4561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cameron P, Fuller CK, Donohoue PD, Jones BN, Thompson MS, Carter MM, Gradia S, et al. Mapping the genomic landscape of CRISPR–Cas9 cleavage. Nat Methods. . 2017;14:600–606. doi: 10.1038/nmeth.4284. [DOI] [PubMed] [Google Scholar]
- 41.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. . 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cullot G, Boutin J, Toutain J, Prat F, Pennamen P, Rooryck C, Teichmann M, et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun. . 2019;10:1136. doi: 10.1038/s41467-019-09006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu M, Zhang W, Xin C, Yin J, Shang Y, Ai C, Li J, et al. Global detection of DNA repair outcomes induced by CRISPR–Cas9. Nucleic Acids Res. . 2021;49:8732–8742. doi: 10.1093/nar/gkab686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W, Yin J, Zhang-Ding Z, Xin C, Liu M, Wang Y, Ai C, et al. In-depth assessment of the PAM compatibility and editing activities of Cas9 variants. Nucleic Acids Res. . 2021;49:8785–8795. doi: 10.1093/nar/gkab507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, et al. Large deletions induced by Cas9 cleavage. Nature. . 2018;560:E8–E9. doi: 10.1038/s41586-018-0380-z. [DOI] [PubMed] [Google Scholar]
- 46.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol. . 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. . 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. . 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. . 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, et al. In vivo genome editing using Staphylococcus aureus Cas9 . Nature. . 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edraki A, Mir A, Ibraheim R, Gainetdinov I, Yoon Y, Song CQ, Cao Y, et al. A compact, high-accuracy cas9 with a dinucleotide PAM for in vivo genome editing . Mol Cell. . 2019;73:714–726.e4. doi: 10.1016/j.molcel.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni . Nat Commun. . 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatterjee P, Jakimo N, Jacobson JM. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci Adv. . 2018;4:eaau0766. doi: 10.1126/sciadv.aau0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, Nakane T, et al. Structure and engineering of Francisella novicida Cas9 . Cell. . 2016;164:950–961. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harrington LB, Paez-Espino D, Staahl BT, Chen JS, Ma E, Kyrpides NC, Doudna JA. A thermostable Cas9 with increased lifetime in human plasma. Nat Commun. . 2017;8:1424. doi: 10.1038/s41467-017-01408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Müller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, et al. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome . Mol Ther. . 2016;24:636–644. doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis . Proc Natl Acad Sci USA. . 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. . 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. . 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. . 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allen F, Crepaldi L, Alsinet C, Strong AJ, Kleshchevnikov V, De Angeli P, Páleníková P, et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat Biotechnol. . 2018;37:64–72. doi: 10.1038/nbt.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O, Cassa CA, et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. . 2018;563:646–651. doi: 10.1038/s41586-018-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shou J, Li J, Liu Y, Wu Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell. . 2018;71:498–509.e4. doi: 10.1016/j.molcel.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 64.Chakrabarti AM, Henser-Brownhill T, Monserrat J, Poetsch AR, Luscombe NM, Scaffidi P. Target-specific precision of CRISPR-mediated genome editing. Mol Cell. . 2019;73:699–713.e6. doi: 10.1016/j.molcel.2018.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. . 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. . 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brinkman EK, Chen T, de Haas M, Holland HA, Akhtar W, van Steensel B. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Mol Cell. . 2018;70:801–813.e6. doi: 10.1016/j.molcel.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, Fedorova I, et al. Crystal structure of cpf1 in complex with guide RNA and target DNA. Cell. . 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gehrke JM, Cervantes O, Clement MK, Wu Y, Zeng J, Bauer DE, Pinello L, et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat Biotechnol. . 2018;36:977–982. doi: 10.1038/nbt.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. . 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 71.Jin S, Fei H, Zhu Z, Luo Y, Liu J, Gao S, Zhang F, et al. Rationally designed APOBEC3B cytosine base editors with improved specificity. Mol Cell. . 2020;79:728–740.e6. doi: 10.1016/j.molcel.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 72.Li C, Zhang R, Meng X, Chen S, Zong Y, Lu C, Qiu JL, et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat Biotechnol. . 2020;38:875–882. doi: 10.1038/s41587-019-0393-7. [DOI] [PubMed] [Google Scholar]
- 73.Grünewald J, Zhou R, Lareau CA, Garcia SP, Iyer S, Miller BR, Langner LM, et al. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat Biotechnol. . 2020;38:861–864. doi: 10.1038/s41587-020-0535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang X, Zhu B, Chen L, Xie L, Yu W, Wang Y, Li L, et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat Biotechnol. . 2020;38:856–860. doi: 10.1038/s41587-020-0527-y. [DOI] [PubMed] [Google Scholar]
- 75.Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. . 2017;168:644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci USA. . 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin S, Zong Y, Gao Q, Zhu Z, Wang Y, Qin P, Liang C, et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. . 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 78.Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H, Yuan L, et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. . 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lei Z, Meng H, Lv Z, Liu M, Zhao H, Wu H, Zhang X, et al. Detect-seq reveals out-of-protospacer editing and target-strand editing by cytosine base editors. Nat Methods. . 2021;18:643–651. doi: 10.1038/s41592-021-01172-w. [DOI] [PubMed] [Google Scholar]
- 80.Zhou C, Sun Y, Yan R, Liu Y, Zuo E, Gu C, Han L, et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. . 2019;571:275–278. doi: 10.1038/s41586-019-1314-0. [DOI] [PubMed] [Google Scholar]
- 81.Grünewald J, Zhou R, Garcia SP, Iyer S, Lareau CA, Aryee MJ, Joung JK. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. . 2019;569:433–437. doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. . 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. . 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lieber MR. Mechanisms of human lymphoid chromosomal translocations. Nat Rev Cancer. . 2016;16:387–398. doi: 10.1038/nrc.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Casellas R, Basu U, Yewdell WT, Chaudhuri J, Robbiani DF, Di Noia JM. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat Rev Immunol. . 2016;16:164–176. doi: 10.1038/nri.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. . 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annu Rev Genet. . 2012;46:455–473. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 88.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. . 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 89.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. . 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 90.Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. . 2019;20:698–714. doi: 10.1038/s41580-019-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu T, Huang J. Quality control of homologous recombination. Cell Mol Life Sci. . 2014;71:3779–3797. doi: 10.1007/s00018-014-1649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chapman JR, Taylor MRG, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. . 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 93.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. . 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, Aach J, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. . 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hollywood JA, Lee CM, Scallan MF, Harrison PT. Analysis of gene repair tracts from Cas9/gRNA double-stranded breaks in the human CFTR gene. Sci Rep. . 2016;6:32230. doi: 10.1038/srep32230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guo T, Feng YL, Xiao JJ, Liu Q, Sun XN, Xiang JF, Kong N, et al. Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing. Genome Biol. . 2018;19:170. doi: 10.1186/s13059-018-1518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Song B, Yang S, Hwang GH, Yu J, Bae S. Analysis of NHEJ-based DNA repair after CRISPR-mediated DNA cleavage. Int J Mol Sci. . 2021;22:6397. doi: 10.3390/ijms22126397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. . 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 99.Griffith AJ, Blier PR, Mimori T, Hardin JA. Ku polypeptides synthesized in vitro assemble into complexes which recognize ends of double-stranded DNA. J Biol Chem. . 1992;267:331–338. doi: 10.1016/S0021-9258(18)48498-0. [DOI] [PubMed] [Google Scholar]
- 100.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. . 2002;108:781–794. doi: 10.1016/S0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 101.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. . 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 102.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. . 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-W. [DOI] [PubMed] [Google Scholar]
- 103.Blier PR, Griffith AJ, Craft J, Hardin JA. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. . 1993;268:7594–7601. doi: 10.1016/S0021-9258(18)53216-6. [DOI] [PubMed] [Google Scholar]
- 104.Mimori T, Hardin JA. Mechanism of interaction between Ku protein and DNA. J Biol Chem. . 1986;261:10375–10379. doi: 10.1016/S0021-9258(18)67534-9. [DOI] [PubMed] [Google Scholar]
- 105.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. . 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 106.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. . 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 107.Xing M, Yang M, Huo W, Feng F, Wei L, Jiang W, Ning S, et al. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat Commun. . 2015;6:6233. doi: 10.1038/ncomms7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, Tamura N, Travers J, et al. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. . 2015;347:185–188. doi: 10.1126/science.1261971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buck D, Malivert L, de Chasseval R, Barraud A, Fondanèche MC, Sanal O, Plebani A, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. . 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 110.Dai Y, Kysela B, Hanakahi LA, Manolis K, Riballo E, Stumm M, Harville TO, et al. Nonhomologous end joining and V(D)J recombination require an additional factor. Proc Natl Acad Sci USA. . 2003;100:2462–2467. doi: 10.1073/pnas.0437964100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, Stamato TD, Taccioli GE, et al. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. . 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 112.Wei YF, Robins P, Carter K, Caldecott K, Pappin DJ, Yu GL, Wang RP, et al. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol Cell Biol. . 1995;15:3206–3216. doi: 10.1128/MCB.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li S, Kanno S, Watanabe R, Ogiwara H, Kohno T, Watanabe G, Yasui A, et al. Polynucleotide kinase and aprataxin-like forkhead-associated protein (PALF) acts as both a single-stranded DNA endonuclease and a single-stranded DNA 3′ exonuclease and can participate in DNA end joining in a biochemical system. J Biol Chem. . 2011;286:36368–36377. doi: 10.1074/jbc.M111.287797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kanno S, Kuzuoka H, Sasao S, Hong Z, Lan L, Nakajima S, Yasui A. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. . 2007;26:2094–2103. doi: 10.1038/sj.emboj.7601663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bebenek K, Pedersen LC, Kunkel TA. Structure–function studies of DNA polymerase λ. Biochemistry. . 2014;53:2781–2792. doi: 10.1021/bi4017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Aoufouchi S, Flatter E, Dahan A, Faili A, Bertocci B, Storck S, Delbos F, et al. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. . 2000;28:3684–3693. doi: 10.1093/nar/28.18.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo . Immunity. . 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 118.Quennet V, Beucher A, Barton O, Takeda S, Löbrich M. CtIP and MRN promote non-homologous end-joining of etoposide-induced DNA double-strand breaks in G1. Nucleic Acids Res. . 2011;39:2144–2152. doi: 10.1093/nar/gkq1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juárez R, Bebenek K, Kee BL, Blanco L, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. . 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 120.Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJY, Konečná E, Belanto JJ, et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell. . 2017;29:1196–1217. doi: 10.1105/tpc.16.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu Y, Yuan Q, Zhu Y, Gao X, Song J, Yin Z. Improving FnCas12a genome editing by exonuclease fusion. CRISPR J. . 2020;3:503–511. doi: 10.1089/crispr.2020.0073. [DOI] [PubMed] [Google Scholar]
- 122.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. . 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. . 1996;15:5093–5103. doi: 10.1002/j.1460-2075.1996.tb00890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. . 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. . 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bae S, Kweon J, Kim HS, Kim JS. Microhomology-based choice of Cas9 nuclease target sites. Nat Methods. . 2014;11:705–706. doi: 10.1038/nmeth.3015. [DOI] [PubMed] [Google Scholar]
- 127.Schep R, Brinkman EK, Leemans C, Vergara X, van der Weide RH, Morris B, van Schaik T, et al. Impact of chromatin context on Cas9-induced DNA double-strand break repair pathway balance. Mol Cell. . 2021;81:2216–2230.e10. doi: 10.1016/j.molcel.2021.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen W, McKenna A, Schreiber J, Haeussler M, Yin Y, Agarwal V, Noble WS, et al. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Res. . 2019;47:7989–8003. doi: 10.1093/nar/gkz487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nakade S, Tsubota T, Sakane Y, Kume S, Sakamoto N, Obara M, Daimon T, et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun. . 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sakuma T, Nakade S, Sakane Y, Suzuki KIT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc. . 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 131.Kim SI, Matsumoto T, Kagawa H, Nakamura M, Hirohata R, Ueno A, Ohishi M, et al. Microhomology-assisted scarless genome editing in human iPSCs. Nat Commun. . 2018;9:939. doi: 10.1038/s41467-018-03044-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Grajcarek J, Monlong J, Nishinaka-Arai Y, Nakamura M, Nagai M, Matsuo S, Lougheed D, et al. Genome-wide microhomologies enable precise template-free editing of biologically relevant deletion mutations. Nat Commun. . 2019;10:4856. doi: 10.1038/s41467-019-12829-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang WW, Matlashewski G. Single-strand annealing plays a major role in double-strand DNA break repair following CRISPR-cas9 cleavage in leishmania . mSphere. . 2019;4:e00408. doi: 10.1128/mSphere.00408-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Truong LN, Li Y, Shi LZ, Hwang PYH, He J, Wang H, Razavian N, et al. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci USA. . 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat Struct Mol Biol. . 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. . 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. . 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 138.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. . 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro . Mol Cell Biol. . 1997;17:2764–2773. doi: 10.1128/MCB.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. . 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. . 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 142.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. . 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Robert F, Barbeau M, Éthier S, Dostie J, Pelletier J. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med. . 2015;7:93. doi: 10.1186/s13073-015-0215-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, et al. In vivogenome editing via CRISPR/Cas9 mediated homology-independent targeted integration . Nature. . 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zelensky AN, Schimmel J, Kool H, Kanaar R, Tijsterman M. Inactivation of Pol θ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat Commun. . 2017;8:66. doi: 10.1038/s41467-017-00124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Riesenberg S, Chintalapati M, Macak D, Kanis P, Maricic T, Pääbo S. Simultaneous precise editing of multiple genes in human cells. Nucleic Acids Res. . 2019;47:e116. doi: 10.1093/nar/gkz669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. . 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2–Sgs1–RPA and its stimulation by Top3–Rmi1 and Mre11–Rad50–Xrs2. Nature. . 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. . 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. . 1998;26:5388–5393. doi: 10.1093/nar/26.23.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang X, Haber JE. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair . PLoS Biol. . 2004;2:e21. doi: 10.1371/journal.pbio.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McVey M, Khodaverdian VY, Meyer D, Cerqueira PG, Heyer WD. Eukaryotic DNA polymerases in homologous recombination. Annu Rev Genet. . 2016;50:393–421. doi: 10.1146/annurev-genet-120215-035243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu S, Hua Y, Wang J, Li L, Yuan J, Zhang B, Wang Z, et al. RNA polymerase III is required for the repair of DNA double-strand breaks by homologous recombination. Cell. . 2021;184:1314–1329.e10. doi: 10.1016/j.cell.2021.01.048. [DOI] [PubMed] [Google Scholar]
- 154.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. . 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. . 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 156.Zhang X, Li T, Ou J, Huang J, Liang P. Homology-based repair induced by CRISPR-Cas nucleases in mammalian embryo genome editing. Protein Cell. . 2021;13:316–335. doi: 10.1007/s13238-021-00838-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat Immunol. . 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 158.Pawelczak KS, Gavande NS, VanderVere-Carozza PS, Turchi JJ. Modulating DNA repair pathways to improve precision genome engineering. ACS Chem Biol. . 2018;13:389–396. doi: 10.1021/acschembio.7b00777. [DOI] [PubMed] [Google Scholar]
- 159.Jayavaradhan R, Pillis DM, Goodman M, Zhang F, Zhang Y, Andreassen PR, Malik P. CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat Commun. . 2019;10:2866. doi: 10.1038/s41467-019-10735-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun. . 2016;7:10548. doi: 10.1038/ncomms10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pinder J, Salsman J, Dellaire G. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. . 2015;43:9379–9392. doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rees HA, Yeh WH, Liu DR. Development of hRad51–Cas9 nickase fusions that mediate HDR without double-stranded breaks. Nat Commun. . 2019;10:2212. doi: 10.1038/s41467-019-09983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kurihara T, Kouyama-Suzuki E, Satoga M, Li X, Badawi M, Thiha M, Baig DN, et al. DNA repair protein RAD51 enhances the CRISPR/Cas9-mediated knock-in efficiency in brain neurons. Biochem Biophysl Res Commun. . 2020;524:621–628. doi: 10.1016/j.bbrc.2020.01.132. [DOI] [PubMed] [Google Scholar]
- 164.Wilde JJ, Aida T, Del Rosario RCH, Kaiser T, Qi P, Wienisch M, Zhang Q, et al. Efficient embryonic homozygous gene conversion via RAD51-enhanced interhomolog repair. Cell. . 2021;184:3267–3280.e18. doi: 10.1016/j.cell.2021.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Charpentier M, Khedher AHY, Menoret S, Brion A, Lamribet K, Dardillac E, Boix C, et al. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat Commun. . 2018;9:1133. doi: 10.1038/s41467-018-03475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gutschner T, Haemmerle M, Genovese G, Draetta GF, Chin L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. . 2016;14:1555–1566. doi: 10.1016/j.celrep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 167.Bialk P, Rivera-Torres N, Strouse B, Kmiec EB. Regulation of gene editing activity directed by single-stranded oligonucleotides and CRISPR/Cas9 systems. PLoS ONE. . 2015;10:e0129308. doi: 10.1371/journal.pone.0129308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9–mediated editing of germline DNA. Science. . 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Olsen PA, McKeen C, Krauss S. Branched oligonucleotides induce in vivo gene conversion of a mutated EGFP reporter . Gene Ther. . 2003;10:1830–1840. doi: 10.1038/sj.gt.3302079. [DOI] [PubMed] [Google Scholar]
- 170.van de Vrugt HJ, Harmsen T, Riepsaame J, Alexantya G, van Mil SE, de Vries Y, Bin Ali R, et al. Effective CRISPR/Cas9-mediated correction of a Fanconi anemia defect by error-prone end joining or templated repair. Sci Rep. . 2019;9:768. doi: 10.1038/s41598-018-36506-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dekker M, Brouwers C, te Riele H. Targeted gene modification in mismatch-repair-deficient embryonic stem cells by single-stranded DNA oligonucleotides. Nucleic Acids Res. . 2003;31:27e–27. doi: 10.1093/nar/gng027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Olsen PA, Randol M, Krauss S. Implications of cell cycle progression on functional sequence correction by short single-stranded DNA oligonucleotides. Gene Ther. . 2005;12:546–551. doi: 10.1038/sj.gt.3302454. [DOI] [PubMed] [Google Scholar]
- 173.Schmid-Burgk JL, Gao L, Li D, Gardner Z, Strecker J, Lash B, Zhang F. Highly parallel profiling of Cas9 variant specificity. Mol Cell. . 2020;78:794–800.e8. doi: 10.1016/j.molcel.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, Lee K, et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun. . 2018;9:3048. doi: 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, Bode NM, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. . 2018;24:1216–1224. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Casini A, Olivieri M, Petris G, Montagna C, Reginato G, Maule G, Lorenzin F, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol. . 2018;36:265–271. doi: 10.1038/nbt.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, et al. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature. . 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. . 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. . 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hanlon KS, Kleinstiver BP, Garcia SP, Zaborowski MP, Volak A, Spirig SE, Muller A, et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat Commun. . 2019;10:4439. doi: 10.1038/s41467-019-12449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Norris AL, Lee SS, Greenlees KJ, Tadesse DA, Miller MF, Lombardi HA. Template plasmid integration in germline genome-edited cattle. Nat Biotechnol. . 2020;38:163–164. doi: 10.1038/s41587-019-0394-6. [DOI] [PubMed] [Google Scholar]
- 182.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. . 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 183.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. . 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. . 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. . 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. . 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. . 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hu J, Tepsuporn S, Meyers RM, Gostissa M, Alt FW. Developmental propagation of V(D)J recombination-associated DNA breaks and translocations in mature B cells via dicentric chromosomes. Proc Natl Acad Sci USA. . 2014;111:10269–10274. doi: 10.1073/pnas.1410112111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hu J, Zhang Y, Zhao L, Frock RL, Du Z, Meyers RM, Meng F, et al. Chromosomal loop domains direct the recombination of antigen receptor genes. Cell. . 2015;163:947–959. doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Meng FL, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon KR, Meyers RM, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. . 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu X, Liu T, Shang Y, Dai P, Zhang W, Lee BJ, Huang M, et al. ERCC6L2 promotes DNA orientation-specific recombination in mammalian cells. Cell Res. . 2020;30:732–744. doi: 10.1038/s41422-020-0328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bai W, Zhu G, Xu J, Chen P, Meng F, Xue H, Chen C, et al. The 3′-flap endonuclease XPF-ERCC1 promotes alternative end joining and chromosomal translocation during B cell class switching. Cell Rep. . 2021;36:109756. doi: 10.1016/j.celrep.2021.109756. [DOI] [PubMed] [Google Scholar]
- 193.Zuo E, Huo X, Yao X, Hu X, Sun Y, Yin J, He B, et al. CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol. . 2017;18:224. doi: 10.1186/s13059-017-1354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ling X, Chang L, Chen H, Gao X, Yin J, Zuo Y, Huang Y, et al. Improving the efficiency of CRISPR-Cas12a-based genome editing with site-specific covalent Cas12a-crRNA conjugates. Mol Cell. . 2021;81:4747–4756.e7. doi: 10.1016/j.molcel.2021.09.021. [DOI] [PubMed] [Google Scholar]
- 195.Liu Y, Ai C, Gan T, Wu J, Jiang Y, Liu X, Lu R, et al. Transcription shapes DNA replication initiation to preserve genome integrity. Genome Biol. . 2021;22:176. doi: 10.1186/s13059-021-02390-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wei PC, Chang AN, Kao J, Du Z, Meyers RM, Alt FW, Schwer B. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell. . 2016;164:644–655. doi: 10.1016/j.cell.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Canela A, Maman Y, Huang SYN, Wutz G, Tang W, Zagnoli-Vieira G, Callen E, et al. Topoisomerase II-induced chromosome breakage and translocation is determined by chromosome architecture and transcriptional activity. Mol Cell. . 2019;75:252–266.e8. doi: 10.1016/j.molcel.2019.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Poirot L, Philip B, Schiffer-Mannioui C, Le Clerre D, Chion-Sotinel I, Derniame S, Potrel P, et al. Multiplex genome-edited t-cell manufacturing platform for “off-the-shelf” adoptive t-cell immunotherapies. Cancer Res. . 2015;75:3853–3864. doi: 10.1158/0008-5472.CAN-14-3321. [DOI] [PubMed] [Google Scholar]
- 199.Liu X, Zhang Y, Cheng C, Cheng AW, Zhang X, Li N, Xia C, et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. . 2017;27:154–157. doi: 10.1038/cr.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ren J, Liu X, Fang C, Jiang S, June CH, Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin Cancer Res. . 2017;23:2255–2266. doi: 10.1158/1078-0432.CCR-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. . 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Amit I, Iancu O, Levy-Jurgenson A, Kurgan G, McNeill MS, Rettig GR, Allen D, et al. CRISPECTOR provides accurate estimation of genome editing translocation and off-target activity from comparative NGS data. Nat Commun. . 2021;12:3042. doi: 10.1038/s41467-021-22417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wen W, Quan ZJ, Li SA, Yang ZX, Fu YW, Zhang F, Li GH, et al. Effective control of large deletions after double-strand breaks by homology-directed repair and dsODN insertion. Genome Biol. . 2021;22:236. doi: 10.1186/s13059-021-02462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Owens DDG, Caulder A, Frontera V, Harman JR, Allan AJ, Bucakci A, Greder L, et al. Microhomologies are prevalent at Cas9-induced larger deletions. Nucleic Acids Res. . 2019;47:7402–7417. doi: 10.1093/nar/gkz459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bothmer A, Gareau KW, Abdulkerim HS, Buquicchio F, Cohen L, Viswanathan R, Zuris JA, et al. Detection and modulation of DNA translocations during multi-gene genome editing in T cells. CRISPR J. . 2020;3:177–187. doi: 10.1089/crispr.2019.0074. [DOI] [PubMed] [Google Scholar]
- 206.Karakoc E, Alkan C, O′Roak BJ, Dennis MY, Vives L, Mark K, Rieder MJ, et al. Detection of structural variants and indels within exome data. Nat Methods. . 2011;9:176–178. doi: 10.1038/nmeth.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, Frock RL. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification–mediated high-throughput genome-wide translocation sequencing. Nat Protoc. . 2016;11:853–871. doi: 10.1038/nprot.2016.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu Y, Yin J, Gan T, Liu M, Xin C, Zhang W, Hu J. PEM-seq comprehensively quantifies DNA repair outcomes during gene-editing and DSB repair. STAR Protocols. . 2022;3:101088. doi: 10.1016/j.xpro.2021.101088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yin J, Liu M, Liu Y, Hu J. Improved HTGTS for CRISPR/Cas9 off-target detection. BIO-PROTOCOL. . 2019;9:e3229. doi: 10.21769/BioProtoc.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Yin J, Lu R, Xin C, Wang Y, Ling X, Li D, Zhang W, et al. Cas9 exo-endonuclease eliminates chromosomal translocations during genome editing. Nat Commun. . 2022;13:1204. doi: 10.1038/s41467-022-28900-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Paulsen BS, Mandal PK, Frock RL, Boyraz B, Yadav R, Upadhyayula S, Gutierrez-Martinez P, et al. Ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR–Cas9 genome editing. Nat Biomed Eng. . 2017;1:878–888. doi: 10.1038/s41551-017-0145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. . 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ferguson DO, Sekiguchi JAM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci USA. . 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Difilippantonio MJ, Petersen S, Chen HT, Johnson R, Jasin M, Kanaar R, Ried T, et al. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J Exp Med. . 2002;196:469–480. doi: 10.1084/jem.20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yan CT, Kaushal D, Murphy M, Zhang Y, Datta A, Chen C, Monroe B, et al. XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proc Natl Acad Sci USA. . 2006;103:7378–7383. doi: 10.1073/pnas.0601938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.