Abstract
Salmonella enterica serovar Typhimurium infects eukaryotic cells residing within membrane‐bound phagosomes. In this compartment, the pathogen replaces the morphogenetic penicillin‐binding proteins 2 and 3 (PBP2/PBP3) with PBP2SAL/PBP3SAL, two proteins absent in Escherichia coli. The basis for this switch is unknown. Here, we show that PBP3 protein levels drop drastically when S. Typhimurium senses acidity, high osmolarity and nutrient scarcity, cues that activate virulence functions required for intra‐phagosomal survival and proliferation. The protease Prc and the transcriptional regulator OmpR contribute to lower PBP3 levels whereas OmpR stimulates PBP2SAL/PBP3SAL production. Surprisingly, despite being essential for division in E. coli, PBP3 levels also drop in non‐pathogenic and pathogenic E. coli exposed to phagosome cues. Such exposure alters E. coli morphology resulting in very long bent and twisted filaments indicative of failure in the cell division and elongation machineries. None of these aberrant shapes are detected in S. Typhimurium. Expression of PBP3SAL restores cell division in E. coli exposed to phagosome cues although the cells retain elongation defects in the longitudinal axis. By switching the morphogenetic program, OmpR and Prc allow S. Typhimurium to properly divide and elongate inside acidic phagosomes maintaining its cellular dimensions and the rod shape.
Keywords: intracellular, morphogenesis, OmpR‐Prc, peptidoglycan, Salmonella
Salmonella switches morphogenetic peptidoglycan synthases inside eukaryotic cells, a phenomenon triggered by three cues: acid pH, high osmolarity and nutrient scarcity. OmpR responds to acid pH upregulating alternative synthases PBP2SAL and PBP3SAL. Prc fine‐tunes PBP3 levels and prevents escape of PBP2SAL/PBP3SAL production in extracellular bacteria, in which PBP2/PBP3 direct rod shape. Escherichia coli, which lacks PBP2SAL/PBP3SAL, displays gross morphological alterations if exposed to phagosome cues due to decrease in levels of its morphogenetic peptidoglycan synthases.
1. INTRODUCTION
The peptidoglycan (PG), also known as murein sacculus, is the main component of the bacterial cell wall (Egan et al., 2020). Among its unique physicochemical properties are the presence of D‐amino acids and its assembly as a giant covalently bound macromolecule covering the entire cell surface. Because of these properties, the PG determines cell shape and ensures cellular integrity (Egan & Vollmer, 2013; Egan et al., 2017). PG metabolism and morphogenesis have been extensively studied in Escherichia coli and Bacillus subtilis, in which the elongation and division phases are clearly defined in time and space (den Blaauwen et al., 2008; Errington, 2015; Rohs & Bernhardt, 2021). These two phases are orchestrated by PG synthases that crosslink stem peptides in parallel glycan chains (den Blaauwen et al., 2017; Szwedziak & Lowe, 2013). In E. coli, these morphogenetic enzymes are penicillin‐binding proteins 2 and 3 (PBP2 and PBP3) (Egan et al., 2017; Szwedziak & Lowe, 2013), which are components of the elongasome and divisome complexes, respectively (Daitch & Goley, 2020; den Blaauwen & Luirink, 2019; Rohs & Bernhardt, 2021). These complexes are connected to cytoskeletal scaffolds that guide the biosynthetic machineries along the cylindrical or the mid‐cell regions of the cell (Cho, 2015; Daitch & Goley, 2020; Errington, 2015; McQuillen & Xiao, 2020). Based on these roles, PBP2 and PBP3 are essential enzymes not tractable genetically and preferred targets for antimicrobial therapy.
Salmonella enterica serovar Typhimurium (S. Typhimurium) is an intracellular pathogen that encodes two PBPs named PBP2SAL and PBP3SAL that replace PBP2/PBP3 in the host environment (Castanheira et al., 2020). PBP2SAL/PBP3SAL are PBP2/PBP3 paralogues (~67% identity at the protein level) absent in E. coli, active only in acid pH and up‐regulated by S. Typhimurium inside phagosomes (Castanheira et al., 2017, 2020). The exchange of PBP2/PBP3 by PBP2SAL/PBP3SAL taking place in the host is reproduced at some extent in vitro in acidified media with limited amounts of nutrients (Castanheira et al., 2020). Acidity and nutrient limitation trigger the expression of virulence functions like the Salmonella pathogenicity island 2 (SPI‐2), central for survival and proliferation of intracellular bacteria (Deiwick & Hensel, 1999; Garmendia et al., 2003; Kehl et al., 2020; Lee et al., 2000; Pucciarelli & Garcia‐Del Portillo, 2017; Steele‐Mortimer, 2008). Acid pH and nutrient scarcity are therefore cues supposed to be encountered by this pathogen inside phagosomes. Once the pathogen senses acidic pH in this niche, it responds by acidifying its cytoplasm in a process mediated by the transcriptional regulator OmpR (Chakraborty et al., 2015).
Unlike the impact of acid pH and nutrient limitation, the extent to which the osmolarity of the vacuolar compartment signals the pathogen to express virulence factors is still unclear. Early studies concluded that low osmolarity favors SPI‐2 expression (Garmendia et al., 2003; Lee et al., 2000). However, the nutrient‐poor medium developed by Deiwick and Hensel, suitable for inducing expression of SPI‐2 genes, was formulated with 50 mM NaCl (Deiwick & Hensel, 1999). In this medium, SPI‐2 is highly expressed by bacteria exposed up to 2% NaCl, equivalent to ~340 mM of this salt.
To better understand the exchange of essential PG synthases during S. Typhimurium infection, we examined the underlying regulatory mechanisms. Here, we provide evidence for the requirement of a combination of phagosome cues like high osmolarity, acid pH, and nutrient scarcity, to simulate the switch in the morphogenetic machinery, i.e. the substitution of PBP2/PBP3 by PBP2SAL/PBP3SAL. Surprisingly, the combination of these three cues also triggers a significant drop in PBP3 levels in E. coli, a bacterium lacking an alternative enzyme for cell division. The implications of these findings for how the adaptation to the phagosomal environment may require a defined morphogenetic program, are discussed.
2. RESULTS
2.1. OmpR regulates PBP2SAL /PBP3SAL production
S. Typhimurium upregulates in vivo PBP2SAL/PBP3SAL production when colonizing target organs of susceptible mice and inside cultured macrophages and fibroblasts (Castanheira et al., 2017, 2020). The responsible regulator(s) however remain uncharacterized. We targeted as candidates those used by S. Typhimurium to adapt to the intra‐phagosomal lifestyle, including the two‐component systems PhoP‐PhoQ, EnvZ‐OmpR and SsrA‐SsrB (Chen & Groisman, 2013; Dalebroux & Miller, 2014; Feng et al., 2003; Kenney, 2019; Pérez‐Morales et al., 2017) and, the transcriptional regulator SlyA (Buchmeier et al., 1997). Mutants lacking these regulators and bearing 3xFLAG‐tagged alleles of the genes encoding PBP2SAL or PBP3SAL in their native chromosomal locations were monitored for production of these alternative PG synthases. PBP2SAL‐3xFLAG and PBP3SAL‐3xFLAG retain function as they ensure rod shape morphology and cell division in the absence of PBP2 or PBP3, respectively [(Castanheira et al., 2017) and S. Castanheira and F. García‐del Portillo, unpublished data].
PBP2SAL‐3xFLAG and PBP3SAL‐3xFLAG (hereinafter referred as PBP2SAL and PBP3SAL) were almost undetectable in the ompR mutant (Figure 1a,b) with a slight reduction in PBP2SAL levels in the phoP and slyA mutants (Figure 1a). OmpR was also required for PBP2SAL/PBP3SAL production inside eukaryotic cells (Figure 1c). PBP2SAL and PBP3SAL levels decreased up to ~20%, ~50% of wild‐type values, respectively, in intracellular bacteria lacking PhoP (Figure 1c). Immunodetection of PBP2SAL‐3xFLAG and PBP3SAL‐3xFLAG was accompanied in some assays by an unrelated protein that did not affect data interpretation as it showed different electrophoretic mobility (Figure S1). Collectively, these data implicated for the first time OmpR in the regulation of morphogenetic enzymes involved in PG synthesis.
FIGURE 1.
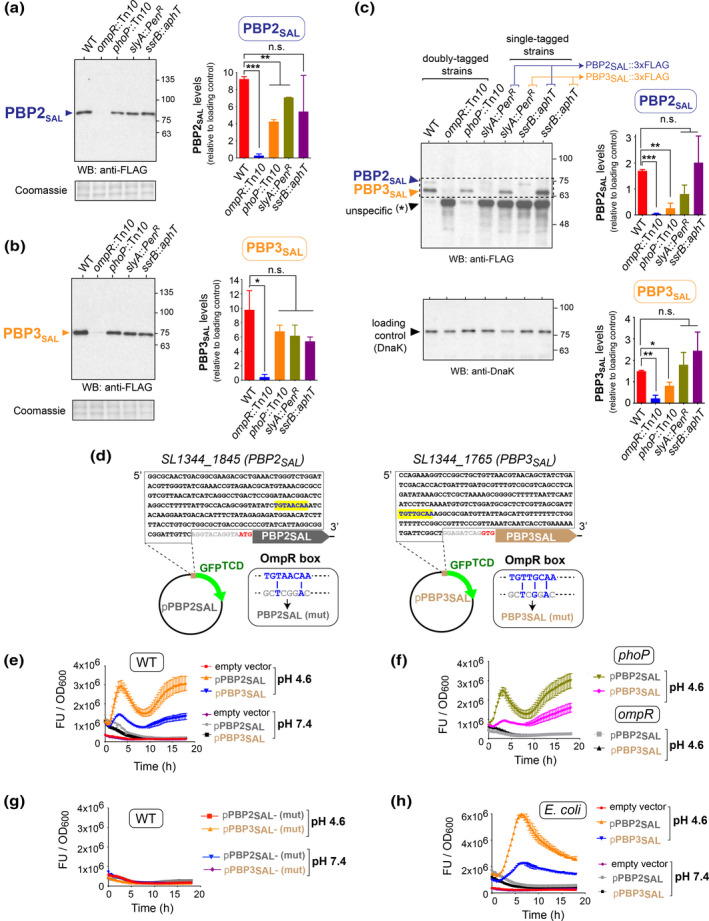
PBP2SAL and PBP3SAL are regulated by OmpR in extracellular and intracellular S. Typhimurium. (a) PBP2SAL protein levels in extracts of isogenic S. Typhimurium strains bearing a PBP2SAL‐3xFLAG allele in its native chromosomal location and mutations in the indicated regulators (OmpR, PhoP, SlyA, and SsrB). Growth conditions were LB medium pH 4.6. The Coomassie staining is shown as loading control. Quantification was performed by densitometry of the bands obtained in the Western. (b) Same as for (a) but for isogenic strains bearing a PBP3SAL‐3xFLAG allele in native chromosomal location. (c) PBP2SAL and PBP3SAL levels produced by intracellular S. Typhimurium at 8 h post‐infection of NRK‐49F rat fibroblasts. Isogenic wild‐type and mutant strains lacking the indicated regulators and either doubly or single‐tagged with the PBP2SAL‐3xFLAG and PBP3SAL‐3xFLAG alleles were used. DnaK was used as loading control. Data are means and standard deviations from a minimum of two independent experiments and are statistically analyzed by unpaired parametric t‐test. *p = 0.01 to 0.05; **p = 0.001 to 0.01; ***p = 0.0001 to 0.001; n.s., not significant. (d) Upstream regions cloned in the promoter‐less reporter plasmid expressing a GFPTCD variant. In yellow are highlighted the OmpR boxes analyzed and the insets depict the mutated versions (mut) generated for each of the regions. (e–h) Activity of the PBP2SAL and PBP3SAL promoters in the indicated strains and growth conditions: (e) wild‐type S. Typhimurium, PCN medium 200 mM NaCl at pH 7.4 and 4.6; (f) S. Typhimurium mutants lacking PhoP or OmpR grown in PCN medium 200 mM NaCl pH 4.6; (g) wild‐type S. Typhimurium bearing reporters with the mutated (mut) versions of each promoter; and (h) wild‐type E. coli expressing the reporter constructs based on the PBP2SAL and PBP3SAL promoters. Data were collected automatically during 18 h in a Tecan plate reader and are represented as mean and standard deviation from three technical replicates. Data are from a representative experiment of a total of three biological replicates. FU, fluorescence units.
We further designed reporter constructs bearing upstream regions (250 bp from the predicted start codon) of the genes encoding PBP2SAL and PBP3SAL (Figure 1d). The expression assays revealed that these regions are sufficient to drive transcription specifically in acid pH and in an OmpR‐dependent but PhoP‐independent manner (Figure 1e,f). Of interest, these upstream regions bear two sequence motifs, TGTAACAA (PBP2SAL promoter) and TGTTGCAA (PBP3SAL promoter) (Figure 1d) that match the consensus TGT(A/T)ACA(A/T) required for OmpR regulation in S. Typhi and S. Typhimurium (Perkins et al., 2013). Responsiveness of the reporters to acid pH was abrogated by mutations in these motifs (Figure 1g). Interestingly, the reporter constructs were responsive to acid pH in a heterologous system like E. coli (Figure 1h). These latter data showed that OmpR regulates PBP2SAL/PBP3SAL production by increasing transcription of their coding genes without requiring accessory Salmonella‐specific co‐regulator(s).
A recent transcriptomic study reported dacC, a gene encoding the carboxypeptidase PBP6 that trims stem peptides in the PG, as a potentially OmpR‐regulated gene in E. coli and S. Typhimurium (Chakraborty & Kenney, 2018). However, we did not find evidence at the protein level for such regulation in S. Typhimurium using a minimal medium of same composition and even more acidic (pH 4.6) compared to that used in that previous study (Figure S2). We also ruled out the alternative sigma factors RpoS and RpoE, essential for S. Typhimurium to grow and survive inside host cells (Cano et al., 2001; Chen et al., 1996; Osborne & Coombes, 2009), as regulators of PBP2SAL and/or PBP3SAL production (Figure S3). Altogether, these data indicated that OmpR plays a central role in the regulation of PBP2SAL/PBP3SAL in acidic niches and inside eukaryotic cells.
2.2. PBP2SAL /PBP3SAL induction is linked to SPI‐2 expression and parallels a drastic drop in PBP3 levels at high osmolarity
Our previous studies showed that acid pH triggers PBP2SAL/PBP3SAL expression (Castanheira et al., 2017, 2020) and that nutrient limitation causes, in addition, a decrease in PBP2/PBP3 levels (Castanheira et al., 2020). Acidity and nutrient scarcity also signal intravacuolar Salmonella for expression of virulence factors like those of the SPI‐2 regulon (Kehl et al., 2020; Pucciarelli & Garcia‐Del Portillo, 2017; Steele‐Mortimer, 2008). Since acidity and nutrient scarcity stimulate PBP2SAL/PBP3SAL production, we sought to determine the extent at which both SPI‐2 and PBP2SAL/PBP3SAL production responds to a third cue like osmolarity. Low osmolarity was reported to favor expression of the SPI‐2 regulon (Garmendia et al., 2003; Lee et al., 2000) whereas other studies showed high expression in bacteria grown up to ~300 mM NaCl (Deiwick & Hensel, 1999). Based on these contrasting findings, we monitored levels of two representative proteins of the SPI‐2 regulon, the regulator SsrB and the effector protein SseJ, in the nutrient‐poor medium PCN at two pH values (4.6 and 7.4) and two osmolarities, 0 and 200 mM NaCl. This latter NaCl concentration, 200 mM, causes in E. coli K‐12 maximal changes in expression of outer membrane proteins regulated by OmpR (Alphen & Lugtenberg, 1977; Verhoef et al., 1979). Control assays confirmed that addition of 200 mM NaCl to the nutrient‐poor PCN medium at pH 4.6 does not affect S. Typhimurium growth whereas levels in the 300–500 mM range result in progressive reduction in the growth rate (Figure S4).
S. Typhimurium produces SsrB and SseJ at high levels in acidified medium irrespective of the amount of salt added (Figure 2a). This result indicated that the expression of these two proteins of the SPI2 regulon is not affected by osmolarity in the acidified nutrient‐poor PCN medium. Production of PBP2SAL/PBP3SAL in this medium is likewise independent of the level of osmolarity used, although fully dependent on OmpR (Figure 2b).
FIGURE 2.
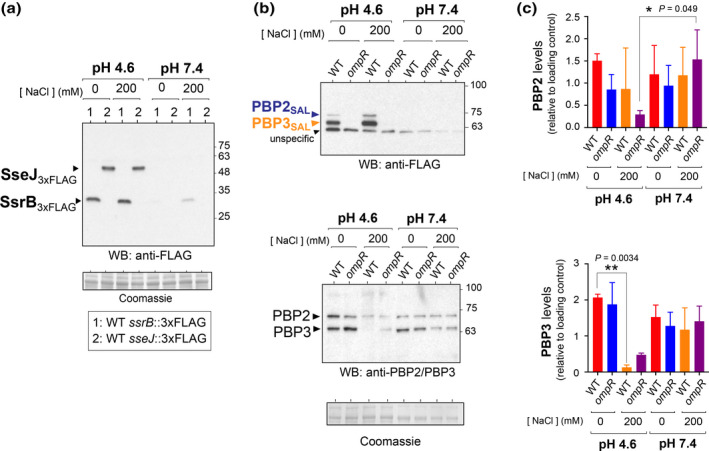
PBP2SAL and PBP3SAL production in S. Typhimurium is linked to that of SPI‐2. (a) Increased levels of the SPI‐2 regulator SsrB and the SPI‐2 effector SseJ in acid pH in the nutrient‐poor medium PCN irrespective of the amount of salt added. (b) PBP2SAL/PBP3SAL and PBP2/PBP3 relative levels detected in wild‐type and ompR strains grown in minimal PCN medium at distinct pH (4.6 and 7.4) and amounts of osmolyte (0, 200 mM NaCl). The Coomassie‐stained membranes are shown as loading controls. (c) Quantification of the relative levels of PBP2 and PBP3 in the indicated conditions by densitometry of the bands obtained in the Western (panel b). Data are means and standard deviations from four independent experiments and statistically analyzed by unpaired parametric t‐test. *p = 0.01 to 0.05; **p = 0.001 to 0.01.
Unlike PBP2SAL/PBP3SAL, osmolarity has a strong effect on the production of PBP2/PBP3. Thus, the levels of these two PG enzymes in acidified medium (pH 4.6) were unaltered at low osmolarity (0 mM NaCl) but diminished drastically at high osmolarity (200 mM NaCl) (Figure 2b). This effect of high osmolarity on lowering PBP3 relative levels was negligible at neutral pH (Figure 2b,c). Therefore, only the combination of three environmental cues associated with the phagosomal niche such as acidity (pH 4.6), nutrient limitation, and high osmolarity (200 mM NaCl) results in reduction of PBP3 levels, a phenomenon observed in vivo when S. Typhimurium infects target organs of mice (Castanheira et al., 2017). This effect was seen in the absence of PhoP (Figure S5a). Intriguingly, low Mg2+ concentrations as of 8 μM, a condition known to activate the PhoP‐PhoQ system (Cunrath & Bumann, 2019; Garcia‐del Portillo et al., 1992; Martin‐Orozco et al., 2006; Röder et al., 2021), caused a decrease in both PBP2 and PBP3 levels but only in acid pH (Figure S5b).
We next asked whether the negative effect of phagosomal cues on PBP3 could involve other essential enzymes of PG metabolism. MurF is an essential cytosolic enzyme required for synthesis of the PG precursor lipid II encoded by a gene mapping downstream of ftsI, the PBP3‐encoding gene. murF and ftsI are part of the d ivision and c ell w all (dcw) gene cluster that functions as a polycistronic transcriptional unit (Figure S6a). Unlike PBP3, MurF levels did not decrease significantly in response to acid pH, nutrient scarcity, or high osmolarity and were not altered in the ompR null mutant (Figure S6b). These data inferred that PBP3 production is post‐transcriptionally regulated in response to a combination of environmental cues occurring in the phagosomal niche. Likewise, PBP3SAL upregulation relies solely on acid pH as activating signal.
2.3. The periplasmic protease Prc targets PBP3 and PBP3SAL in a pH‐dependent manner
In E. coli, the protease Prc processes the C‐terminal region of PBP3 (Hara et al., 1989; Nagasawa et al., 1989), and it has as substrates other enzymes related to PG metabolism like the endopeptidase MepS (Spr) (Singh et al., 2015) and the murein transglycosylase MltG (Hsu et al., 2020). The cleavage site recognized by Prc in E. coli PBP3, of 588 aa, is valine‐577 (V577) (Nagasawa et al., 1989), a residue conserved in PBP3 of S. Typhimurium. PBP3SAL, of 581 aa, has also a valine located 11 residues from the C‐terminus, concretely V570 (Castanheira et al., 2017). Due to this conserved valine residue, we hypothesized a role of Prc in the switch of PBP3 by PBP3SAL occurring in response to phagosome cues. Precursor and processed forms of PBP3 and PBP3SAL were monitored in isogenic S. Typhimurium wild‐type and Δprc strains.
In acid pH (4.6) at low osmolarity (0 mM NaCl), the lack of Prc in S. Typhimurium results in reduced PBP3 levels with wild‐type bacteria unable to process the enzyme under these conditions (Figure 3a). This processing is however detected in E.coli (Figure 3a). As expected, no PBP3 is visualized neither in S. Typhimurium nor in E. coli at pH 4.6, 200 mM NaCl (Figure 3a). At neutral pH 7.4, both PBP3 forms (precursor, mature) are visible in S. Typhimurium wild‐type while only the full‐length PBP3 is detected in the Δprc mutant (Figure 3b). This result indicated that Prc of S. Typhimurium cleaves PBP3, although restricted to the neutral pH condition (compare Figure 3a,b). The capacity of S. Typhimurium Prc to process PBP3 at neutral pH is unrelated to OmpR (Figure 3c).
FIGURE 3.
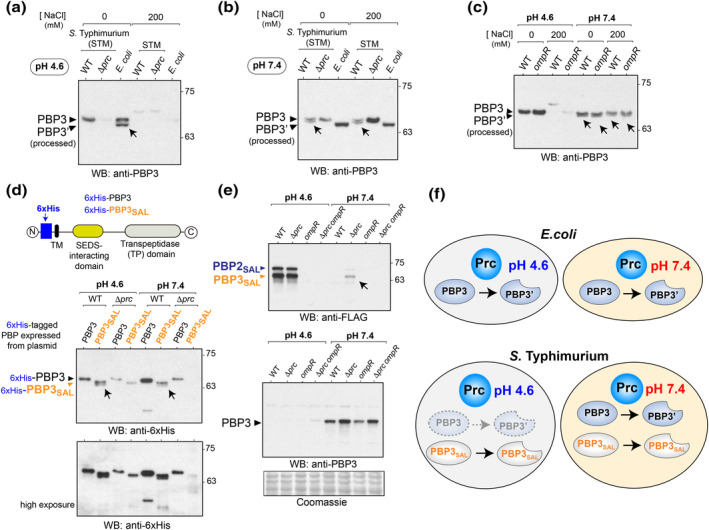
The periplasmic protease Prc selectively targets PBP3 or PBP3SAL at distinct pH. (a) PBP3 processing does not occur in S. Typhimurium at acid pH (4.6) whereas it does in E. coli in the same condition. Arrow indicates the mature processed form of PBP3. (b) S. Typhimurium Prc cleaves PBP3 at neutral pH (7.4) irrespective of the amount of salt added to the medium. The PBP3 form observed for E. coli in these assays is processed as inferred from its higher electrophoretic mobility. Arrows point to the mature form of PBP3. (c) The activity of S. Typhimurium Prc over PBP3 at neutral pH (7.4) occurs independently of OmpR and osmolarity. Arrows point to the mature PBP3 form. (d) Prc cleaves a 6xHis variant of PBP3SAL tagged at the N‐terminus expressed from plasmid at acid (4.6) and neutral (7.4) pH. Arrows point to the processed mature PBP3SAL form. The 6xHis‐PBP3 tagged variant is not cleaved by Prc. (e) Presence of minor amounts of PBP2SAL/PBP3SAL at neutral pH and PBP3 in neutral pH in the absence of Prc. (f) Diagram highlighting the preferred recognition of S. Typhimurium Prc for PBP3SAL at acid pH and for PBP3 at neutral pH. Prc can also cleave PBP3SAL at neutral pH.
With regard to PBP3SAL and, considering its V570 residue, we reasoned it could be recognized by Prc at the C‐terminus. To test this, PBP3SAL and PBP3 variants bearing a 6xHis tag at the N‐terminus (Castanheira et al., 2020) were ectopically expressed in S. Typhimurium wild‐type and Δprc strains at acid (pH 4.6) and neutral (7.4) pH (Figure 3d). Prc cleaves 6xHis‐PBP3SAL at both pH whereas no processing is observed for 6xHis‐PBP3 (Figure 3d). This latter result suggests that tagging PBP3 at its N‐terminal region facing the cytosol could negatively affect its recognition by Prc in the periplasm (Figure 3b).
Given the capacity of Prc to process and eventually destabilize PBP3SAL (Figure 3d), we assessed whether this protease could act as a safeguard preventing the production of the alternative PG synthase PBP3SAL at neutral pH. The lack of Prc results in detection of small amounts of PBP2SAL and PBP3SAL at pH 7.4, which are produced in an OmpR‐dependent manner (Figure 3e). Taken together, these data demonstrated that PBP3 and PBP3SAL are Prc substrates in S. Typhimurium. However, whereas Prc cleaves PBP3 only at neutral pH, this protease processes PBP3SAL at pH 4.6, the physiological condition in which its production is stimulated by OmpR, as well as in neutral pH. Unlike Prc of S. Typhimurium, Prc of E. coli cleaves PBP3 at both acid and neutral pH (Figure 3f).
2.4. The combination of acidity, high osmolarity and nutrient scarcity decreases PBP3 levels in non‐pathogenic and pathogenic E. coli
Considering the drastic drop in PBP3 levels observed in S. Typhimurium exposed to phagosome cues, we sought to analyze this phenomenon in E. coli, a closely related bacterium lacking PBP3SAL. Surprisingly, exposure to acidity, high osmolarity and nutrient scarcity also decreases PBP3 levels in the non‐pathogenic E. coli K‐12 strain MG1655 (Figure 4a).
FIGURE 4.
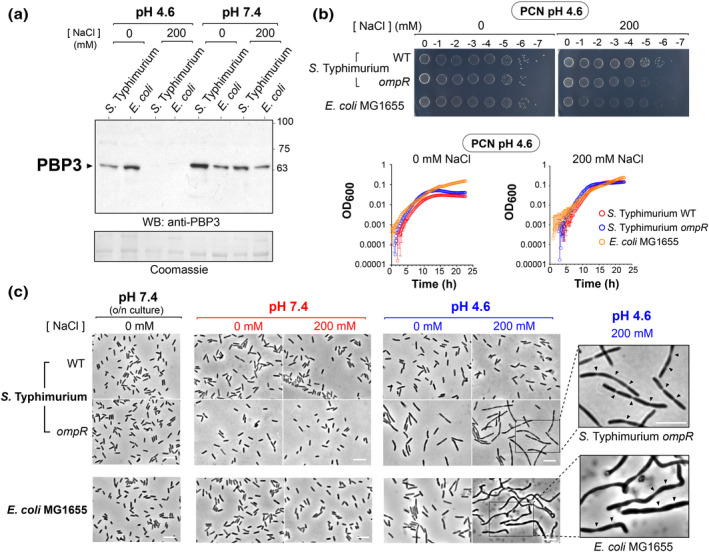
PBP3 levels drop in S. Typhimurium and E. coli in response to environmental cues of the phagosome. (a) PBP3 levels in S. Typhimurium and non‐pathogenic E. coli K‐12 MG1655 grown in minimal PCN medium at the indicated pH and osmolarities. (b) Cultivability of S. Typhimurium (wild‐type and ompR) and E. coli K‐12 MG1655 on PCN medium pH 4.6 plates at the indicated osmolarities (0, 200 mM NaCl). Shown are serial dilutions of an overnight culture. Included are also growth curves of the three strains in liquid PCN medium pH 4.6 with 0 or 200 mM NaCl. (c) Microscopy images of S. Typhimurium (wild‐type and ompR) and E. coli K‐12 strain MG1655 after growing in PCN medium for 4 h at the indicated pH and osmolarities. Shown are also enlargements of S. Typhimurium ompR and E. coli K‐12 strain MG1655 cell in acid pH (4.6) and 200 mM NaCl displaying morphological alterations consistent with arrest in cell division. Arrowheads point to cell division sites. Bar, 5 μm. Data representative of three independent biological replicates.
The reduction in PBP3 levels under these conditions correlated with decreased fitness of the S. Typhimurium ompR mutant, which forms small‐size colonies on solid media containing 200 mM NaCl (Figure 4b). A similar phenotype was observed for E. coli MG1655 (Figure 4b). Both the S. Typhimurium ompR mutant and E. coli MG1655 display similar growth rate in liquid culture compared to wild‐type S. Typhimurium (Figure 4b). This observation in liquid media ruled out a negative effect of the phagosome cues on basal metabolism that could prevent increase in mass. Microscopy analyses showed that, unlike wild‐type S. Typhimurium, its ompR mutant derivate and E. coli MG1655 are impaired for cell division in the acidified nutrient‐poor PCN medium with 200 mM NaCl (Figure 4c). In the case of E. coli MG1655, aberrant shapes including bent and twisted filaments were observed, suggesting that phagosome cues could trigger additional changes in other morphogenetic proteins or PG enzymes besides those affecting PBP3 levels.
To assess whether the morphological alterations detected in E. coli MG1655 were also visible in pathogenic E. coli, we incubated isolates of enteropathogenic (EPEC), enterohemorrhagic (EHEC), and uropathogenic (UPEC) pathotypes in acidified (pH 4.6) nutrient‐poor PCN medium with 200 mM NaCl. To visualize a putative negative effect on cell division, the cultures were started at low optical density values (OD600= 0.005) to allow for at least a minimum of two generations with increase in mass. Final OD600 values were, depending on the E. coli isolate, in the range of ~0.02–0.15. In the PCN pH 4.6, 200 mM NaCl medium, S. Typhimurium reaches final OD600 values of ~0.15–0.20 (Figure S4). Similarly to the non‐pathogenic E.coli MG1655 strain, growth of EPEC, EHEC, and UPEC in PCN pH 4.6, 200 mM NaCl medium results in aberrant morphologies, with cells in most cases adopting shapes of very long irregular twisted filaments (Figure 5a). Such gross morphogenetic alteration was indicative not only of blockage in cell division but also of proper elongation of the PG sacculus along the longitudinal axis. Noteworthy, these morphological changes are not visualized at the same extent in the E. coli isolates grown in acidified PCN pH 4.6 with no salt added, 0 mM NaCl (Figure 5a) or in PCN medium at neutral pH 7.4 irrespective of the amount of salt added (Figure S7a). The morphological alterations of E. coli isolates grown in PCN pH 4.6, 200 mM NaCl are accompanied by a decrease in PBP 3 levels (Figure 5b), consistent with the blockage of cell division observed at the microscope. For UPEC, PBP3 was faintly detected at pH 4.6, irrespective of the amount of salt added (Figure 5b).
FIGURE 5.
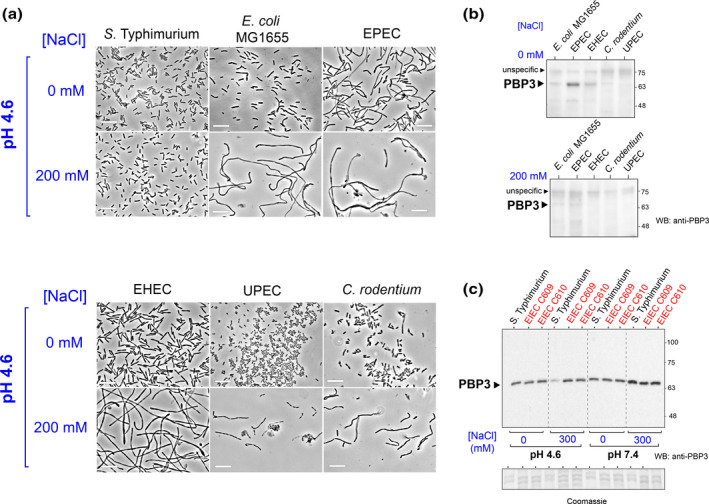
Blockage of cell division and altered morphology in pathogenic and non‐pathogenic E. coli isolates in response to phagosome cues. (a) S. Typhimurium, E. coli isolates, and C. rodentium were incubated overnight in nutrient‐poor medium PCN at pH 4.6 and the indicated osmolarities (0 mM and 200 mM NaCl) and fixed for observation at phase contrast microscopy. The initial OD600 values were adjusted in all cases to 0.005. Note the formation in some cases of atypical morphologies consisting of bent and twisted filaments. Bar, 10 μm. For a representative experiment, final OD600 values reached by the indicated bacteria at pH 4.6 with 0 mM or 200 mM NaCl, respectively, were: S. Typhimurium (0.070, 0.193); E. coli MG1655 (0.038, 0.054); EPEC (0.126, 0.117); EHEC (0.046, 0.057); C. rodentium (0.025, 0.052) and, UPEC (0.024, 0.058). (b) Levels of PBP3 in the E. coli isolates and C. rodentium under the indicated osmolarities in acidified (pH 4.6) nutrient‐poor PCN medium. (c) PBP 3 levels detected in the enteroinvasive E. coli (EIEC) isolates C609 and C610 after growth in nutrient‐rich LB medium with/without additional NaCl (300 mM) added and at two pH values (4.6 and 7.4). Samples in panels (b) and (c) correspond in all cases to total protein extracts. Data shown are from a representative experiment of a total of two independent biological replicates.
Since S. Typhimurium exhibits normal rod shape in PCN pH 4.6, 200 mM NaCl medium (Figures 4a and 5a) we reasoned that Citrobacter, a genus among several of the Enterobacterales order that encode both a PBP3 and a PBP3 homolog (García‐del Portillo et al., unpublished data), could also exhibit normal rod shape in response to phagosome cues. To our surprise, C. rodentium filaments extensively in PCN pH 4.6, 200 mM NaCl medium (Figure 5a). Considering that the levels of its PBP3 decrease in response to phagosome cues (Figure 5b), this morphological alteration denotes that its PBP3 homolog, if expressed in PCN pH 4.6, 200 mM NaCl, does not promote cell division under these conditions. A key difference between S. Typhimurium and C. rodentium is that only the former evolved an intra‐phagosomal lifestyle.
Taken together, these findings indicate that exposure of enteric bacteria to the combination of acid pH, nutrient scarcity, and high osmolarity, a condition mimicking the intra‐phagosomal niche, causes major changes in the morphogenetic program affecting both cell elongation and cell division. Among the enteric bacteria tested, only S. Typhimurium processes the phagosome cues to preserve proper cellular dimensions and rod shape.
2.5. PBP3SAL restores cell division in E. coli exposed to phagosome cues
The accumulated data inferred that E. coli cannot divide in a phagosomal environment due to the drop in PBP3 levels (Figure 4a), which results in drastic morphological changes (Figure 5a). Since S. Typhimurium responds to phagosome cues upregulating production of PBP3SAL, we assessed whether this alternative PG synthase could promote division in wild‐type E. coli exposed to phagosome cues. We previously showed using an E. coli mutant expressing a thermolabile PBP3 variant that PBP3SAL can promote cell division in acid pH at 42°C (Castanheira et al., 2017); however, these experiments were performed in nutrient‐rich LB medium, a condition that does not cause drop of PBP3 levels in wild‐type E. coli.
Ectopic expression of PBP3SAL in wild‐type E. coli MG1655 exposed to phagosome cues in PCN pH 4.6, 200 mM NaCl partially restores cell division. Although on average the cells producing PBP3SAL are larger than those not exposed to phagosome cues (see Figure 4c), it was evident that some cells in the population undergo cell division (Figure 6a). Noteworthy, most of these PBP3SAL‐expressing cells retain defects in cell elongation showing in some cases curved and twisted shapes (Figure 6a). These morphologies contrasted with those of E. coli MG1655 bearing the empty vector, which filamented at a large extent without any sign of cell division and denoting, in addition, elongation defects (Figure 6a). Immunoassays of protein extracts prepared from these cultures after over‐night incubation in presence of the IPTG inducer confirmed the drop in PBP3 levels in response to the phagosome cues and the ectopic production of PBP3SAL (Figure 6b). Altogether, these data indicated that PBP3SAL can restore cell division in E. coli exposed to phagosome cues although it cannot complement the morphological alterations associated with the elongation machinery.
FIGURE 6.
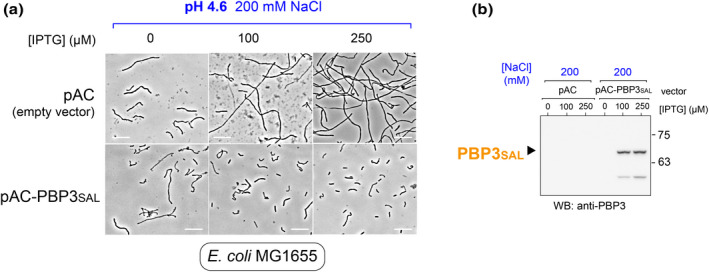
Ectopic expression of PBP3SAL restores cell division in E. coli MG1655 exposed to phagosome cues. (a) Images of E. coli MG1655 harboring the empty vector pAC or a derivate inducible vector expressing the gene encoding PBP3SAL. Induction was carried out by addition of IPTG at the indicated concentrations. Bacteria were maintained overnight in PCN medium at pH 4.6 and 200 mM NaCl. (b) Immunoassay confirming the ectopic production of PBP3SAL in the presence of IPTG. Note that endogenous PBP3 is not visualized since these are conditions mimicking the phagosomal environment. Detection of PBP3SAL with the anti‐PBP3 antibody is explained by the large amount of protein produced from the plasmid and the identity shared between the two proteins which can sustain a small level of cross‐reaction. Data shown are from a representative experiment of a total of two independent biological replicates.
2.6. PBP3 levels decrease in intravacuolar S. Typhimurium persisting inside fibroblasts
The evidence accumulated in nutrient‐poor acidified PCN medium with a high amount of salt, 200 mM NaCl, pointed to phagosomal cues as responsible for impeding the production of PBP3 and upregulating PBP3SAL synthesis (see Figure 2b). To unequivocally demonstrate that this regulation occurs in intra‐phagosomal S. Typhimurium, we infected fibroblasts and obtained protein extracts of intracellular bacteria up to 48 h post‐infection for immunodetection of PBP3, PBP3SAL, and the PG enzyme MurF. Despite the limited proliferation exhibited by these intravacuolar persistent bacteria, PBP3SAL relative levels increase drastically at 8 hpi to further decrease progressively after this post‐infection time (Figure 7a,b). By contrast, and consistently with the data obtained in the nutrient‐poor PCN medium (Figure 2b), PBP3 relative levels decreased from the time bacteria invade the fibroblasts (Figure 7a,b). Interestingly, the relative levels of MurF also decreased from the time of bacterial invasion although remained stable at 24–48 hpi (Figure 7a,b).
FIGURE 7.
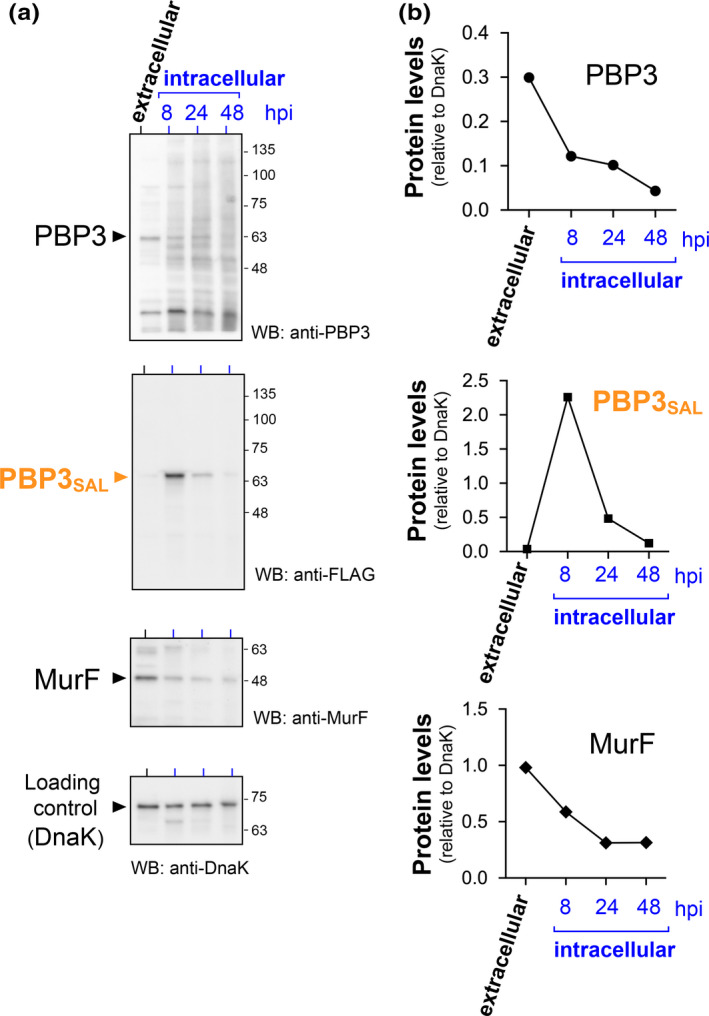
PBP3 levels decrease in intravacuolar S. Typhimurium after invasion of the host cell. NRK‐49F rat fibroblasts were infected with a S. Typhimurium tagged strain bearing the PBP3SAL::3xFLAG allele. Protein extracts were prepared from the inoculum grown overnight that was used for infection (“extracellular” sample) and from infected fibroblasts at the indicated post‐infection times (8, 24, and 48 hpi). (a) Immunodetection of PBP3, PBP3SAL, and MurF. The bacterial chaperone DnaK was also detected as loading control. (b) Levels of PBP3, PBP3SAL, and MurF relativized to those of DnaK, as calculated by densitometry. Data are representative of a total of two independent biological replicates.
Therefore, these data demonstrate that during the initial phase of intracellular infection, PBP3SAL replaces PBP3 in order to support the first cell division events within the phagosome. This phase is followed by the establishment of a perdurable persistence stage. At that stage, only a minor percentage of persistent bacteria could accomplish cell division while maintaining other activities of PG metabolism.
3. DISCUSSION
S. Typhimurium is a unique case of bacterium that uses morphogenetic machineries guided by distinct PG synthases (PBP2/PBP3 versus PBP2SAL/PBP3SAL) to generate a similar (rod) cell shape. Salmonella responds to host signals during the infection by alternating between these two machineries. PBP2SAL/PBP3SAL are functional only in acid pH whereas PBP2/PBP3 are active at both neutral and acid pH (Castanheira et al., 2017, 2020). Such specialization and the acidic environment of phagosomes (Kenney, 2019) prompted us to investigate how the switch of PBP2/PBP3 by PBP2SAL/PBP3SAL modulates S. Typhimurium morphogenesis and its adaptation to the intra‐phagosomal niche.
We identified OmpR as regulator required for PBP2SAL/PBP3SAL production and acid pH as the signal necessary and sufficient for such regulation. OmpR and PhoP control expression of horizontally acquired foreign genes and crosstalk in regulating SPI‐2 (Fass & Groisman, 2009; Kim & Falkow, 2004; Lee et al., 2000; Liew et al., 2019; Worley et al., 2000). However, S. Typhimurium produces PBP2SAL/PBP3SAL in the absence of PhoP (Figure S5) and the SPI‐2 regulator SsrB was also ruled out for controlling PBP2SAL/PBP3SAL expression (Figure 1a,b). PBP2SAL/PBP3SAL and SPI‐2 expression are, therefore, probably not mutually dependent. Furthermore, the reporters bearing the promoter of the PBP2SAL‐ and PBP3SAL‐encoding genes do not respond to PhoP (Figure 1f) indicating that PhoP, if having capacity to modulate PBP2SAL/PBP3SAL production, may act indirectly. A possibility is that PhoP could control regulatory sRNAs capable of binding to the PBP2SAL and/or PBP3SAL transcripts. Future work in this direction is worthy.
Acid pH and high osmolarity signal the sensor EnvZ to activate OmpR (Chakraborty & Kenney, 2018; Kenney, 2019; Quinn et al., 2014). Each signal separately triggers only partly overlapping OmpR regulons (Chakraborty & Kenney, 2018), a view supported by our study. An acid pH of 4.6 alone is sufficient to reach nearly maximal levels of PBP2SAL and PBP3SAL (Figure 1a,b). On the other hand, micromolar levels of magnesium, a signal perceived by the sensor PhoQ to activate PhoP (García Véscovi et al., 1996), are dispensable for PBP2SAL/PBP3SAL production but instead lower PBP2/PBP3 levels (Figure S5). These observations fit with PBP2SAL/PBP3SAL production being stimulated primarily by acidification of the bacterial cytoplasm, known to be a mechanism sustaining the OmpR response to acid pH (Kenney, 2019). Cytoplasm acidification also activates PhoQ, leading to increased expression of PhoP‐regulated genes (Choi & Groisman, 2016). Our observations support an intricate regulatory network that ensures maximal effectiveness in the replacement of PBP2/PBP3 by PBP2SAL/PBP3SAL. Some regulators act to modulate the levels of both pairs of enzymes, such as OmpR, whereas others like PhoP may only modulate levels of PBP2SAL/PBP3SAL in intracellular bacteria. Prc, by preventing production of PBP2SAL/PBP3SAL in neutral pH (Figure 3e) and processing the C‐termini of PBP3 and PBP3SAL, may also act as an additional safeguard mechanism ensuring the correct replacement of the morphogenetic program.
A major unexpected finding of our study was the response of non‐pathogenic and pathogenic E. coli isolates to the same environmental cues that signal S. Typhimurium to change the morphogenetic PG synthases. Under this condition, the E. coli isolates undergo drastic morphological alterations manifested by blockage of cell division and formation of long filaments that, in many occasions, appeared bent and twisted (Figure 5a). Such phenotype denotes failure in the morphogenetic machinery that elongates PG along the longitudinal axis. Intriguingly, B. subtilis mutants deficient in the MreB‐homolog cytoskeletal protein Mdl (Abhayawardhane & Stewart, 1995) were reported to exhibit aberrant morphologies consisting in long bent and twisted filaments that occasionally contained bulges (Jones et al., 2001). Unlike B. subtilis, which has three MreB isoforms, MreB, Mbl, and MreBH, involved in the helical synthesis pattern of the PG along the cylinder region of the cell (Kawai et al., 2009), only one MreB protein is known for S. Typhimurium and E. coli. It is therefore possible that differences exist in the elongasome between both enteric bacteria that when exposed to phagosome cues may not coordinate properly with the division complex in E. coli. Intriguingly, the heterologous expression in E. coli of a protein named Ant, a phage‐encoded putative toxin of unknown function from Streptococcus pneunomiae, results in blockage of cell division and formation of twisted filaments (Chan et al., 2014). This putative toxin may share target(s) among those that respond to phagosome cues.
Phagosomes are acidic and contain large amounts of degradative enzymes and digested material that generate a hyperosmotic environment, only partially alleviated by transporters that excrete material to the cytosol to reduce osmolarity and facilitate vesicular trafficking (Freeman & Grinstein, 2018). Our study links the adaptation to such harsh environment to changes in the morphogenetic machinery of S. Typhimurium, which may not only implicate PBP2SAL/PBP3SAL but also alteration in the activity, interaction and/or amount of other morphogenetic proteins, including cytoskeletal proteins and PG enzymes. S. Typhimurium has a larger repertoire of PG enzymes than E. coli, including a PhoP‐regulated L,D‐endopeptidase named EcgA whose production is activated by intracellular bacteria (Rico‐Pérez et al., 2016) and other proteins related to PG metabolism. In this second group is ScwA, which controls levels of PG hydrolyzing enzymes such as the lytic transglycosylases MltD and Slt, and those of the endo‐ and carboxy‐peptidades, NlpD, PBP4, and AmpH (Cestero et al., 2021). S. Typhimurium also encodes two highly homologous (97% identical) murein lipoproteins, LppA and LppB (Fadl et al., 2005), remaining LppB yet uncharacterized biochemically. These additional proteins linked to PG metabolism are required for virulence, highlighting the central role that morphogenesis has in S. Typhimurium to colonize the phagosomal niche.
Uropathogenic E. coli (UPEC) also undergoes morphogenetic changes in the infection although the underlying mechanism is unknown. Cell division is transiently blocked during urinary tract infections and filamented bacteria exploit this as a strategy to subvert host immune defenses (Horvath et al., 2011; Justice et al., 2006, 2008). This rod‐to‐filament transition, which favors biofilm formation and swarming motility in diverse bacteria (Harshey & Matsuyama, 1994; Yoon et al., 2011), is stimulated in UPEC upon exposure to urine (Khandige et al., 2016), a fluid that in healthy humans is slightly acid (average pH 6.2), harbors a high amount of osmolytes (1025 mOsm/kg) and is poor in nutrients (Rose et al., 2015). The filamented UPEC cells we have observed following exposure to phagosome cues may phenocopy the natural physiological process occurring during infection. At present, we do not have an explanation for why, like UPEC, the other non‐pathogenic and pathogenic E. coli used in our study (MG1655, EPEC, and EHEC isolates) also respond to phagosome cues with gross morphological alterations that involve blockage in cell division – consistent with the drop in PBP3 levels – as well as failure in the elongation machinery. OmpR mediates changes in outer membrane protein composition in response to acid and high osmolarity (Kenney & Anand, 2020) and, based on our findings, it is also probable this has consequences in PG structure. The scheme depicted in Figure 8 underscores the important role that alternative PG enzymes of S. Typhimurium like PBP3SAL may have played for facilitating its adaptation to the intra‐phagosomal lifestyle. Unlike other pathogenic E. coli that even invade eukaryotic cells like EIEC, such additional repertoire of morphogenetic proteins may have been paramount for S. Typhimurium to ensure a normal morphogenetic cycle sustaining the transit from an extra‐ to an intracellular lifestyle.
FIGURE 8.
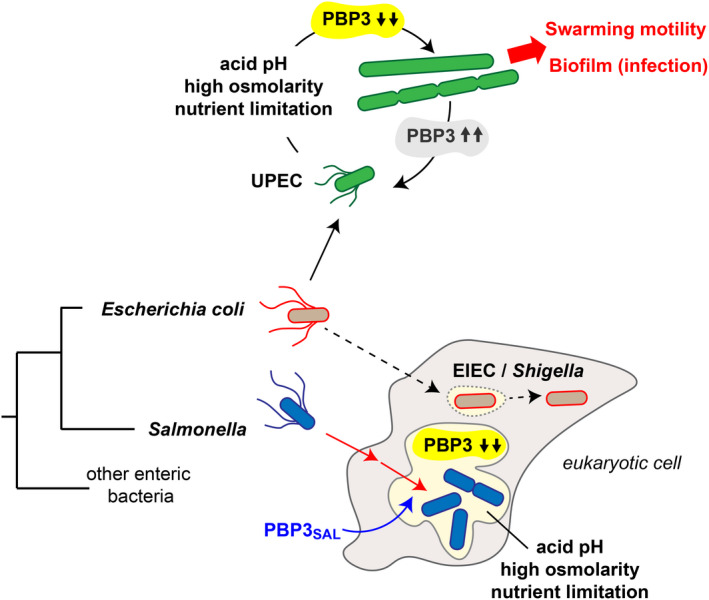
Model integrating the evolutionary impact of the acquisition of PBP3SAL for promoting S. Typhimurium adaptation to an intravacuolar lifestyle. The model highlights how the combination of acidity, high osmolarity, and nutrient scarcity causes transient loss of PBP3 concomitantly to the production of PBP3SAL by intracellular bacteria. Note that other intracellular invasive pathogens like EIEC and Shigella, both lacking PBP3SAL, evolved to lifestyles in the cytosol after the rupture of the initial pathogen‐containing vacuole. The negative regulatory network acting on PBP3 is also proposed to be exploited in extracellular environments by some bacteria, as the representative case of UPEC, to promote biofilm formation during urinary infections or to facilitate swarming motility. EIEC, enteroinvasive E. coli; UPEC, uropathogenic E. coli.
Future studies could also address additional morphogenetic PG enzymes (additional PBP2 and PBP3 homologs encoded by the same bacterium) in genera of the Enterobacteriaceae family that are not professional intra‐phagosomal pathogens. This is the case of Citrobacter spp., Klebsiella spp., and Enterobacter spp., among others. These cases are highly appealing for study when considering the possibility of obtaining mutants lacking their “canonical” PBP2 and PBP3 and to analyze their fitness in acidic niches. In addition, these alternative PBPs can be expressed in heterologous systems to determine their activity at distinct pH. Although C. rodentium filaments when exposed to the combination of phagosome cues (Figure 5a), we have obtained evidence in another Citrobacter species, C. freundii, for a specialization of its PBP3 paralogue for being active only at acid pH (S. Castanheira and F. García‐del Portillo, unpublished data). Consistent with the extracellular lifestyle of Citrobacter species, these findings indicate that C. freundii, and probably also C. rodentium, could use their 91% identical‐97% homologous PBP3 paralogues in acidic environments that, however, may either not be limiting in nutrients or high osmolarity. Other Gammaproteobacteria like Pseudomonas aeruginosa, not belonging to the Enterobacterales order but also not being a professional intra‐phagosomal pathogen, encodes a PBP3 paralog named PBP3X (Liao & Hancock, 1997). While inactivation of PBP3X does not cause morphological alterations, the authors were unable to inactivate the gene encoding PBP3 preventing as a result the characterization of this additional PBP. These cases denote the necessity of eliminating the endogenous canonical morphogenetic proteins to characterize the fascinating alternative programs controlling bacterial shape. Based on our experience with S. Typhimurium, the use of media not normally employed in the laboratory but mimicking conditions encountered in nature by the bacterium of interest is recommended to decipher how alternative morphogenetic programs are regulated.
4. MATERIALS AND METHODS
4.1. Bacterial strains, growth media
Bacterial strains and plasmids used are listed in Table S1. All S. Typhimurium strains used are derivates of virulent wild‐type strain SV5015, an His+ prototroph derivate of virulent strain SL1344 (Vivero et al., 2008). Strains were grown in Luria‐Bertani (LB) broth for conditions with high amount of nutrients. For nutrient‐poor conditions simulating the intra‐phagosomal environment, the phosphate‐carbon‐nitrogen (PCN) (Deiwick & Hensel, 1999) or N (Nelson & Kennedy, 1971) minimal media, were used. The composition of PCN medium is: 4 mM Tricine [N‐[Tris (hydroxymethyl) methyl]glycine], 0.1 mM FeCl3, 376 μM K2SO4, 15 mM NH4Cl, 1 mM MgSO4, 10 μM CaCl2, 0.4% (w/v) glucose, 0.4 mM K2HPO4/KH2PO4, and micronutrients (Deiwick & Hensel, 1999). 200 mM NaCl was added to test responsiveness to high osmolarity. N minimal medium is composed of 5 mM KCl, 7.5 mM (NH4)2S04, 0.5 mM K2S04, and 1 mM KH2PO4 supplemented with 0.4% (w/vol) glucose as carbon source, 0.1% (w/vol) casaminoacids and 10 mM or 8 μM MgCl2 (high or low Mg2+ concentrations, respectively) (Snavely et al., 1991). When necessary, pH was buffered with 80 mM MES [2‐(N‐morpholino) ethanesulfonic acid] adjusted to the desired value with NaOH. To grow strains bearing antibiotic‐resistance genetic elements, media were supplemented with chloramphenicol (10 μg/ml), kanamycin (30 μg/ml), or ampicillin (100 μg/ml). For induction of gene expression from the Plac promoter in pAC‐His plasmids (Table S1), 10 μM IPTG (isopropyl β‐D‐1‐thio‐galactopyranoside) was added. Plasmids expressing N‐terminus tagged 6xHis‐PBP3 and 6xHis‐PBP3SAL variants have been previously described (Castanheira et al., 2020).
4.2. Construction of chromosomal epitope‐tagged genes
Strains bearing chromosomal 3 × FLAG‐epitope‐tagged genes were constructed as described (Uzzau et al., 2001). The phage P22 HT 105/1 int201 was used for transductional crosses to mobilize mutant alleles for strain construction
4.3. Construction of S. Typhimurium Δprc derivates expressing PBP2SAL‐3xFLAG and PBP3SAL‐3xFLAG tagged variants
A P22 HT 105/1 int201 lysate was obtained from S. Typhimurium strain SV6246 (Δprc::KmR) (Hernández et al., 2013), which lacks the entire prc coding sequence (Hernández et al., 2013). The phage lysate was used to transduce recipient strain MD5064 (PBP2SAL‐3xFLAG PBP3SAL‐3xFLAG) to obtain MD5416 (Δprc::KmR PBP2SAL‐3xFLAG PBP3SAL‐3xFLAG) (Table S1).
4.4. DNA techniques
All primers used in this study are listed in Table S2. PCR was performed using Q5 polymerase (New England Biolabs) according to the manufacturer's instructions. PCR fragments were purified using the NucleoSpin Gel and PCR Clean‐up kit (Macherey‐Nagel, ref. 740609.50). Plasmids were purified using the NZYMiniprep kit (NZYTech, ref. MB01001).
4.5. Reporter plasmids bearing PBP2SAL and PBP3SAL promoter regions
To monitor activity of the predicted promoter regions for the genes encoding PBP2SAL and PBP3SAL a vector with the backbone of plasmid pGEN222 (Galen et al., 1999) bearing an ovoalbumin (OVA)‐encoding gene under the PompC promoter and a gene expressing GFPTCD variant (Corcoran et al., 2010), was used. This vector is named pGEN222‐ova‐gfp. Regions encompassing 250 pb upstream the start codon of PBP2SAL‐ and PBP3SAL‐encoding genes (SL1344_1845 and SL1344_1765, respectively) were amplified from S. Typhimurium SV5015 genomic DNA (gDNA) using the primers Ppbp2sal_EcoRI_NotI_Fw/Ppbp2sal_EcoRI_NotI_Rv and Ppbp3sal_EcoRI_NotI_Fw/ Ppbp3sal_EcoRI_NotI_Rv (Table S2). The PCR products containing the PBP2SAL and PBP3SAL promoter regions were digested with EcoRI and NotI and ligated in an EcoRI/NotI‐digested pGEN222‐ova‐gfp replacing, as a result, the pompC‐ova region. As negative control, this pompC‐ova region was excised with EcoRI/NotI from pGEN222‐ova‐gfp and the plasmid was relegated. To this aim, pGEN222‐ova‐gfp digested with EcoRI and NotI was incubated with T4 DNA polymerase (Roche) for 15 min at 12°C, then 2.5 μl of EDTA‐Na+ 0.1 M was added. The mixture was re‐incubated for 20 min at 75°C and the purified DNA product further incubated overnight at 16°C with a T4 DNA ligase (Roche)
4.6. Mutated versions of the OmpR boxes
An annealing extension procedure was used for generating PBP2SAL and PBP3SAL promoters with altered OmpR boxes. These regions were amplified in two fragments from plasmids pGEN222(ΔompC‐ova)‐PPBP2 SAL ‐gfp TCD and pGEN222(ΔompC‐ova)‐PPBP3 SAL ‐gfp TCD, respectively. Fragments with mutations at putative OmpR‐binding sites were amplified using the following primers: (i) pPBP2sal_mut_Fw/pPBP2sal_EcoRI_NotI_Rv and pPBP2sal_mut_Rv/ pPBP2sal_EcoRI_NotI_Fw for the PBP2SAL promoter; and, (ii) pPBP3sal_mut_Fw/ pPBP3sal_EcoRI_NotI_Rv and pPBP3sal_mut_Rv/ pPBP3sal_EcoRI_NotI_Fw, for the PBP3SAL promoter (Table S2). Then, the fragments for each construction were co‐incubated for 5 cycles of annealing extension and the resulting full‐length fragment were amplified using external primers Ppbp2sal_EcoRI_NotI_Fw/ Ppbp2sal_EcoRI_NotI_Rv and Ppbp3sal_EcoRI_NotI_Fw/ Ppbp3sal_EcoRI_NotI_Rv for 30 cycles. The resulting fragments were digested with EcoRI and NotI restriction enzymes, purified and ligated in a EcoRI‐NotI digested pGEN222‐derivative vector. All constructs were sequenced with primer pGEN222_Fw (Table S2) to rule out the presence of undesired mutations. The reporter plasmids vectors harboring the mutated versions of the PBP2SAL and PBP3SAL promoters were electroporated into the desired S. Typhimurium and E. coli strains.
4.7. Promoter activity with reporter plasmids expressing GFPTCD
The S. Typhimurium and E. coli strains harboring the reporter plasmids with the PBP2SAL and PBP3SAL promoter regions were grown overnight in minimal PCN medium pH 7.4, 200 mM NaCl at 37°C, then washed once (4300 × g, 2 min, RT) with PCN 200 mM NaCl and pH of either 4.6 or 7.4 and, finally resuspended in these respective media at an initial OD600 of 0.02. These cultures were transferred with volumes of 180 μl per well in triplicate into 96‐well flat clear bottom black polystyrene TC‐treated microplates (Corning, ref. 3904). Bacteria were incubated at 37°C for 18 h with 20 sec of orbital agitation every 20 min, followed by OD600 and fluorescence measurements in a Spark microplate multimode reader (Tecan). Fluorescence units (FU) were measured with wavelength excitation at 475/15 nm and emission at 510/15 nm. The background values of both optical density and fluorescence were subtracted in the values of the bacterial samples. Fluorescence units were finally corrected by optical density for graphical representation.
4.8. Ectopic expression of PBP3SAL in wild‐type E. coli MG1655
Cultures of E. coli strain K‐12 MG1655 harboring plasmids pAC‐lac or pAC‐lac‐PBP3SAL (Table S1) were grown overnight at 37°C in PCN minimal medium 0 mM NaCl pH 7.4 with chloramphenicol (10 μg/ml). Cultures were then washed in PCN 0 mM NaCl pH 4.6 and diluted to an initial OD600 of 0.005 in PCN 200 mM NaCl pH 4.6 with chloramphenicol (10 μg/ml) and 38 mM glycerol as carbon source. For induction of PBP3SAL expression, isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) was used at 0, 100, or 250 μM from the time of dilution in the pH 4.6 medium. Bacteria were grown for 15 h at 37°C and harvested and processed for microscopy and western blot for detection of PBP3 levels.
4.9. Large‐scale infection of fibroblasts
Large‐scale infections were performed to obtain protein from intracellular bacteria, essentially as previously described (Núñez‐Hernández et al., 2013). NRK‐49F normal rat kidney fibroblasts (ATCC CRL‐1570) were seeded in Nunc Square BioAssay Dishes (Thermo Scientific, ref. 166508) with 50 ml Dulbecco's Modified Eagle Medium (DMEM) culture medium supplemented with 5% (v/v) fetal bovine serum (FBS) and 4 mM L‐glutamine to a confluence of 80% (~5.0 × 107 cells). The fibroblast culture was infected with the different S. Typhimurium strains, previously grown overnight at 37°C in static LB culture, for 40 min at an MOI of 10:1 (bacteria: fibroblast). At this post‐infection time, cells were washed two times with prewarmed complete PBS solution (PBS pH 7.4 with 0.9 mM CaCl2 0.5 mM MgCl2) and then incubated in fresh DMEM‐5% FBS culture medium containing 100 μg/ml of gentamicin until 2 h post‐infection. The culture medium was then replaced with fresh DMEM‐5% FBS medium containing 10 μg/ml gentamicin until 8 h post‐infection. At that time, the infected fibroblasts were washed five times with 20 ml cold complete PBS and lysed in 17 ml of a solution containing 1% (v/v) pH 6.6–7.9 basic phenol, 19% (v/v) ethanol and 0.4% (w/v) SDS in water. A volume of 1.2 μl DNase (10 mg/ml) was added to each plate. After 30 min of incubation at 4°C, the lysate was collected in 40 ml polypropylene tubes (Sorvall) and centrifuged (27,500 × g, 4°C, 30 min). The resultant pellet was washed twice with 1 ml of a 1% basic phenol, 19% ethanol solution (29,400 × g, 15 min, 4°C). Intracellular bacteria were resuspended in 40 μl Laemmli lysis buffer.
4.10. Phase‐contrast microscopy
Bacteria in overnight cultures were centrifuged (4300 × g, 2 min, RT), washed in PCN medium with the corresponding pH (4.6 or 7.4) and salt concentration (0 mM or 200 mM NaCl), and used to inoculate the respective media to an initial optical OD600 of 0.02. After 4 h growing with agitation (150 rpm) at 37°C, bacteria were harvested (6800 × g, 4 min, 4°C), washed twice in PBS buffer, fixed with 3% paraformaldehyde (PFA) for 10 min, and adjusted to a final paraformaldehyde (PFA) concentration of 1%. For microscopy, fixed bacteria were centrifuged (4300 × g, 4 min, RT) and resuspended in an equal volume of PBS. A volume of 30 μl was dropped on poly‐L‐Lys precoated coverslips and incubated for 10 min at RT. Attached bacteria were washed four times with PBS and the coverslip was mounted on slides using ProLong Gold Antifade (Molecular Probes). Images were acquired on an inverted Leica DMI 6000B microscope with an automated CTR/7000 HS controller (Leica Microsystems) and an Orca‐R2 charge‐coupled‐device (CCD) camera (Hamamatsu Photonics).
4.11. Immunofluorescence microscopy
NRK‐49F fibroblasts were seeded on coverslips to a confluence of 40–50% in 24‐well plates and infected as described before, with SV5015 and MD5460 strains. Infected cells were fixed at 24 h post‐infection in 3% paraformaldehyde (15 min, RT), and then processed for immunofluorescence microscopy as described previously (López‐Montero et al., 2016). For localization of intracellular Salmonella and E. coli, rabbit polyclonal anti‐S. Typhimurium LPS (ref. 2948‐47‐6, 1:1000, Difco Laboratories) and rabbit polyclonal anti‐E. coli surface antigens (ref. B65001R, 1:1000, Life Science) were used as primary antibody, respectively. Goat polyclonal anti‐rabbit IgG conjugated to Alexa 488 (1:1000, Molecular Probes) was used as secondary antibody. Nuclei were stained with Dapi (5 μg/ml). Images were acquired on an inverted Leica DMI 6000B fluorescence microscope with an automated CTR/7000 HS controller (Leica Microsystems) and an Orca‐R2 charge‐coupled‐device (CCD) camera (Hamamatsu Photonics).
4.12. Growth conditions for preparation of protein extracts
When using minimal PCN medium, S. Typhimurium and E. coli strains were first grown overnight with agitation at 37°C, pH 7.4 without NaCl supplementation. To monitor the effect of osmolarity on PBPs production, the bacteria from overnight cultures were washed [4300 × g, 2 min, room temperature (RT)] with PCN medium at the conditions of interest: pH 4.6 or 7.4, and NaCl concentrations of 0 mM or 200 mM. The initial optical density at 600 nm (OD600) of the culture was established at 0.02 in 20 ml cultures. When using N minimal medium, S. Typhimurium strains were grown overnight with agitation at 37°C, pH 7.4 with 10 mM MgCl2. Overnight cultures were then washed (4300 × g, 2 min, RT) with N minimal medium pH 4.6 or 7.4, and MgCl2 concentrations of 8 μM or 10 mM, according to culture growth conditions, and diluted to an initial OD600 of 0.02 in 20 ml cultures. In minimal media, bacteria were collected after 4 h of growth at 37°C in agitation (150 rpm). When using LB, bacteria were first grown overnight in agitation at 37°C in non‐buffered neutral pH LB. Overnight cultures were diluted to an initial OD600 of 0.02 in 20 ml cultures with LB or LB pH 4.6 buffered with MES and bacteria collected at 3 h post‐inoculation. To trigger production of PBP3 or PBP3SAL from pAC‐His plasmids (Table S1), strains were grown overnight with agitation (150 rpm) at 37°C in LB and diluted to an OD600 of 0.02 in fresh LB or LB pH 4.6 buffered with MES. Strains were then grown at 37°C for 1.5 h, induced with 10 μM IPTG for gene expression and grown for further 1.5 h before being collected for total protein extraction. In all growth conditions, bacteria were harvested by centrifugation at 18,000 × g, 5 min, 4°C, washed twice in phosphate‐buffered saline (PBS), and resuspended in 100 μl of Laemmli lysis buffer per optical density unit.
4.13. Immunoblot assays
For SDS/PAGE, samples were incubated at 100°C for 5–10 min and centrifuged to remove cell debris (6800 × g, 5 min, RT) for SDS/PAGE. Proteins resolved by gels were transferred to polyvinylidene difluoride (PVDF) membranes and incubated with the corresponding antibodies. Detection was performed by Clarity Western ECL Substrate chemiluminescence kit (Bio Rad, ref. 1705061). Once used for the Western blot, the PVDF membranes were stained with Coomassie solution to confirm proper adjustment of samples except samples from intracellular bacteria, in which immunodetection of DnaK chaperone was performed. Proteins were routinely resolved in 6% polyacrylamide gels. To increase the separation of mature and processed PBP3 forms, 11% gels for the Tricine‐electrophoresis system (Schägger & von Jagow, 1987), were used. Similarly, to increase separation between PBP2SAL and PBP3 SAL, 4%–20% Mini‐Protean TGX Precast Protein gels (BioRad, ref. 4561096) were used. The following antibodies were used as primary antibodies: mouse monoclonal anti‐Flag (M2 clone, 1:5000; Sigma), mouse monoclonal anti‐6xHis (1:2500; R&D Systems), rabbit polyclonal anti‐PBP2 (1:1000; lab collection) (Castanheira et al., 2020), rabbit polyclonal anti‐PBP3 (1:1000; lab collection) (Castanheira et al., 2017), and mouse monoclonal anti‐DnaK (clone 8E2/2, 1:10000; Enzo Life Sciences). Goat polyclonal anti‐mouse (ref. 1706516, 1:20,000, BioRad) and anti‐rabbit IgG (ref. 1706515, 1:30,000, BioRad) conjugated to horseradish peroxidase (Bio‐Rad) were used as secondary antibodies.
4.14. Statistical analyses
Protein bands were analyzed with Fiji distribution of ImageJ2 (version 1.52i). For densitometry, the films of the immunoassays showing similar intensity for PBP2SAL (or PBP3SAL) in independent experiments in wild‐type bacterial samples were used for comparison to the Coomassie‐stained gel. The integration value of the PBP2SAL or PBP3SAL bands obtained in wild‐type and mutants was further used to calculate the ratio versus the two major Coomassie‐stained bands (extracellular samples, bacteria grown in nutrient media) or versus levels of the chaperone DnaK (intracellular samples, bacterial extracts prepared from infected fibroblasts). Ratios from independent experiments for each sample (wild‐type and mutants) were used to calculate means and standard deviation. Data were analyzed with GraphPad Prism software v8.0 (GraphPad Inc. San Diego, CA) using unpaired two‐tailed Student's t test. Significance was established at p‐values <0.05.
AUTHOR CONTRIBUTIONS
David López‐Escarpa: Conceptualization; data curation; formal analysis; investigation; methodology; writing – review and editing. Sónia Castanheira: Formal analysis; investigation; methodology; resources; writing – review and editing. Francisco García‐del Portillo: Conceptualization; funding acquisition; project administration; resources; supervision; writing – original draft; writing – review and editing.
CONFLICT OF INTEREST
The authors report there are no competing interests to declare.
ETHICS STATEMENT
No animal experimentation was performed in this study.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
D.L.‐E. is supported by a PhD fellowship from the “Programa de Formación de Personal Investigador (FPI)” of the Spanish Ministry of Science and Innovation (ref. BES‐2017‐080709). This work was supported by the Spanish Ministry of Science and Innovation under grant PID2020‐112971GB‐I00/10.13039/501100011033 to F.G‐dP. We thank Josep Casadesús (University of Seville, Spain), M. Hensel (University of Osnabrück, Germany), Luis Ángel Fernández and Álvaro San Millán (CNB‐CSIC, Madrid, Spain) for the gift of S. Typhimurium and E. coli strains. We also thank Gadea Rico‐Pérez for the construction of some tagged S. Typhimurium strains and Henar González for the technical support.
López‐Escarpa, D. , Castanheira, S. & García‐del Portillo, F. (2022). OmpR and Prc contribute to switch the Salmonella morphogenetic program in response to phagosome cues. Molecular Microbiology, 118, 477–493. 10.1111/mmi.14982
This paper is dedicated to the memory of Josep Casadesús, Professor of Genetics at the University of Seville in Spain, who was a reference in epigenetics, phenotypic heterogeneity in microbial communities and Salmonella pathogenesis. J. Casadesús unexpectedly passed away on August 2, 2022.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Abhayawardhane, Y. & Stewart, G.C. (1995) Bacillus subtilis possesses a second determinant with extensive sequence similarity to the Escherichia coli mreB morphogene. Journal of Bacteriology, 177, 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alphen, W.V. & Lugtenberg, B. (1977) Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli . Journal of Bacteriology, 131, 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen, T. , Hamoen, L.W. & Levin, P.A. (2017) The divisome at 25: the road ahead. Current Opinion in Microbiology, 36, 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen, T. & Luirink, J. (2019) Checks and balances in bacterial cell division. mBio, 10, e00149‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen, T. , de Pedro, M.A. , Nguyen‐Disteche, M. & Ayala, J.A. (2008) Morphogenesis of rod‐shaped sacculi. FEMS Microbiology Reviews, 32, 321–344. [DOI] [PubMed] [Google Scholar]
- Buchmeier, N. , Bossie, S. , Chen, C.Y. , Fang, F.C. , Guiney, D.G. & Libby, S.J. (1997) SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infection and Immunity, 65, 3725–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano, D.A. , Martínez‐Moya, M. , Pucciarelli, M.G. , Groisman, E.A. , Casadesús, J. & García‐Del Portillo, F. (2001) Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infection and Immunity, 69, 6463–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira, S. , Cestero, J.J. , Rico‐Pérez, G. , García, P. , Cava, F. , Ayala, J.A. et al. (2017) A specialized peptidoglycan synthase promotes Salmonella cell division inside host cells. mBio, 8, e01685‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira, S. , Lopez‐Escarpa, D. , Pucciarelli, M.G. , Cestero, J.J. , Baquero, F. & Garcia‐Del Portillo, F. (2020) An alternative penicillin‐binding protein involved in Salmonella relapses following ceftriaxone therapy. eBioMedicine, 55, 102771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestero, J.J. , Castanheira, S. , Pucciarelli, M.G. & García‐Del Portillo, F. (2021) A novel Salmonella periplasmic protein controlling cell wall homeostasis and virulence. Frontiers in Microbiology, 12, 633701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, S. & Kenney, L.J. (2018) A new role of OmpR in acid and osmotic stress in Salmonella and E. coli . Frontiers in Microbiology, 9, 2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, S. , Mizusaki, H. & Kenney, L.J. (2015) A FRET‐based DNA biosensor tracks OmpR‐dependent acidification of Salmonella during macrophage infection. PLoS Biology, 13, e1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, W.T. , Yeo, C.C. , Sadowy, E. & Espinosa, M. (2014) Functional validation of putative toxin‐antitoxin genes from the Gram‐positive pathogen Streptococcus pneumoniae: phd‐doc is the fourth bona‐fide operon. Frontiers in Microbiology, 5, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Y. , Eckmann, L. , Libby, S.J. , Fang, F.C. , Okamoto, S. , Kagnoff, M.F. et al. (1996) Expression of Salmonella typhimurium rpoS and rpoS‐dependent genes in the intracellular environment of eukaryotic cells. Infection and Immunity, 64, 4739–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.D. & Groisman, E.A. (2013) The biology of the PmrA/PmrB two‐component system: the major regulator of lipopolysaccharide modifications. Annual Review of Microbiology, 67, 83–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H. (2015) The role of cytoskeletal elements in shaping bacterial cells. Journal of Microbiology and Biotechnology, 25, 307–316. [DOI] [PubMed] [Google Scholar]
- Choi, J. & Groisman, E.A. (2016) Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Molecular Microbiology, 101, 1024–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran, C.P. , Cameron, A.D.S. & Dorman, C.J. (2010) H‐NS silences gfp, the green fluorescent protein gene: gfpTCD is a genetically remastered gfp gene with reduced susceptibility to H‐NS‐mediated transcription silencing and with enhanced translation. Journal of Bacteriology, 192, 4790–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunrath, O. & Bumann, D. (2019) Host resistance factor SLC11A1 restricts Salmonella growth through magnesium deprivation. Science, 366, 995–999. [DOI] [PubMed] [Google Scholar]
- Daitch, A.K. & Goley, E.D. (2020) Uncovering unappreciated activities and niche functions of bacterial cell wall enzymes. Current Biology, 30, R1170–R1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux, Z.D. & Miller, S.I. (2014) Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Current Opinion in Microbiology, 17, 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiwick, J. & Hensel, M. (1999) Regulation of virulence genes by environmental signals in Salmonella typhimurium . Electrophoresis, 20, 813–817. [DOI] [PubMed] [Google Scholar]
- Egan, A.J. , Cleverley, R.M. , Peters, K. , Lewis, R.J. & Vollmer, W. (2017) Regulation of bacterial cell wall growth. The FEBS Journal, 284, 851–867. [DOI] [PubMed] [Google Scholar]
- Egan, A.J.F. , Errington, J. & Vollmer, W. (2020) Regulation of peptidoglycan synthesis and remodelling. Nature Reviews Microbiology, 18, 446–460. [DOI] [PubMed] [Google Scholar]
- Egan, A.J. & Vollmer, W. (2013) The physiology of bacterial cell division. Annals of the New York Academy of Sciences, 1277, 8–28. [DOI] [PubMed] [Google Scholar]
- Errington, J. (2015) Bacterial morphogenesis and the enigmatic MreB helix. Nature Reviews. Microbiology, 13, 241–248. [DOI] [PubMed] [Google Scholar]
- Fadl, A.A. , Sha, J. , Klimpel, G.R. , Olano, J.P. , Galindo, C.L. & Chopra, A.K. (2005) Attenuation of Salmonella enterica Serovar Typhimurium by altering biological functions of murein lipoprotein and lipopolysaccharide. Infection and Immunity, 73, 8433–8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass, E. & Groisman, E.A. (2009) Control of Salmonella pathogenicity Island‐2 gene expression. Current Opinion in Microbiology, 12, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, X. , Oropeza, R. & Kenney, L.J. (2003) Dual regulation by phospho‐OmpR of ssrA/B gene expression in Salmonella pathogenicity Island 2. Molecular Microbiology, 48, 1131–1143. [DOI] [PubMed] [Google Scholar]
- Freeman, S.A. & Grinstein, S. (2018) Resolution of macropinosomes, phagosomes and autolysosomes: osmotically driven shrinkage enables tubulation and vesiculation. Traffic, 19, 965–974. [DOI] [PubMed] [Google Scholar]
- Galen, J.E. , Nair, J. , Wang, J.Y. , Wasserman, S.S. , Tanner, M.K. , Sztein, M.B. et al. (1999) Optimization of plasmid maintenance in the attenuated live vector vaccine strain Salmonella typhi CVD 908‐htrA. Infection and Immunity, 67, 6424–6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Véscovi, E. , Soncini, F.C. & Groisman, E.A. (1996) Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell, 84, 165–174. [DOI] [PubMed] [Google Scholar]
- Garcia‐del Portillo, F. , Foster, J.W. , Maguire, M.E. & Finlay, B.B. (1992) Characterization of the micro‐environment of Salmonella typhimurium‐containing vacuoles within MDCK epithelial cells. Molecular Microbiology, 6, 3289–3297. [DOI] [PubMed] [Google Scholar]
- Garmendia, J. , Beuzón, C.R. , Ruiz‐Albert, J. & Holden, D.W. (2003) The roles of SsrA‐SsrB and OmpR‐EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI‐2 type III secretion system. Microbiology (Reading), 149, 2385–2396. [DOI] [PubMed] [Google Scholar]
- Hara, H. , Nishimura, Y. , Kato, J. , Suzuki, H. , Nagasawa, H. , Suzuki, A. et al. (1989) Genetic analyses of processing involving C‐terminal cleavage in penicillin‐binding protein 3 of Escherichia coli . Journal of Bacteriology, 171, 5882–5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey, R.M. & Matsuyama, T. (1994) Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface‐induced differentiation into hyperflagellate swarmer cells. Proceedings of the National Academy of Sciences, 91, 8631–8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández, S.B. , Ayala, J.A. , Rico‐Pérez, G. , García‐del Portillo, F. & Casadesús, J. (2013) Increased bile resistance in Salmonella enterica mutants lacking Prc periplasmic protease. International Microbiology, 16, 87–92. [DOI] [PubMed] [Google Scholar]
- Horvath, D.J. , Li, B. , Casper, T. , Partida‐Sanchez, S. , Hunstad, D.A. , Hultgren, S.J. et al. (2011) Morphological plasticity promotes resistance to phagocyte killing of uropathogenic Escherichia coli . Microbes and Infection, 13, 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P.‐C. , Chen, C.‐S. , Wang, S. , Hashimoto, M. , Huang, W.‐C. & Teng, C.‐H. (2020) Identification of MltG as a Prc protease substrate whose dysregulation contributes to the conditional growth defect of Prc‐deficient Escherichia coli . Frontiers in Microbiology, 11, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L.J. , Carballido‐López, R. & Errington, J. (2001) Control of cell shape in bacteria: helical, Actin‐like filaments in Bacillus subtilis . Cell, 104, 913–922. [DOI] [PubMed] [Google Scholar]
- Justice, S.S. , Hunstad, D.A. , Cegelski, L. & Hultgren, S.J. (2008) Morphological plasticity as a bacterial survival strategy. Nature Reviews. Microbiology, 6, 162–168. [DOI] [PubMed] [Google Scholar]
- Justice, S.S. , Hunstad, D.A. , Seed, P.C. & Hultgren, S.J. (2006) Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proceedings of the National Academy of Sciences of the United States of America, 103, 19884–19889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, Y. , Asai, K. & Errington, J. (2009) Partial functional redundancy of MreB isoforms, MreB, Mbl and MreBH, in cell morphogenesis of Bacillus subtilis . Molecular Microbiology, 73, 719–731. [DOI] [PubMed] [Google Scholar]
- Kehl, A. , Noster, J. & Hensel, M. (2020) Eat in or take out? Metabolism of intracellular Salmonella enterica . Trends in Microbiology, 28, 644–654. [DOI] [PubMed] [Google Scholar]
- Kenney, L.J. (2019) The role of acid stress in Salmonella pathogenesis. Current Opinion in Microbiology, 47, 45–51. [DOI] [PubMed] [Google Scholar]
- Kenney, L.J. & Anand, G.S. (2020) EnvZ/OmpR two‐component signaling: an archetype system that can function noncanonically. EcoSal Plus, 9. 10.1128/ecosalplus.ESP-0001-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandige, S. , Asferg, C.A. , Rasmussen, K.J. , Larsen, M.J. , Overgaard, M. , Andersen, T.E. et al. (2016) DamX controls reversible cell morphology switching in uropathogenic Escherichia coli . mBio, 7, e00642‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.C. & Falkow, S. (2004) Delineation of upstream signaling events in the Salmonella pathogenicity Island 2 transcriptional activation pathway. Journal of Bacteriology, 186, 4694–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, A.K. , Detweiler, C.S. & Falkow, S. (2000) OmpR regulates the two‐component system SsrA‐ssrB in Salmonella pathogenicity Island 2. Journal of Bacteriology, 182, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, X. & Hancock, R.E. (1997) Identification of a penicillin‐binding protein 3 homolog, PBP3x, in Pseudomonas aeruginosa: gene cloning and growth phase‐dependent expression. Journal of Bacteriology, 179, 1490–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew, A.T.F. , Foo, Y.H. , Gao, Y. , Zangoui, P. , Singh, M.K. , Gulvady, R. et al. (2019) Single cell, super‐resolution imaging reveals an acid pH‐dependent conformational switch in SsrB regulates SPI‐2. eLife, 8, e45311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Montero, N. , Ramos‐Marquès, E. , Risco, C. & García‐Del Portillo, F. (2016) Intracellular Salmonella induces aggrephagy of host endomembranes in persistent infections. Autophagy, 12, 1886–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Orozco, N. , Touret, N. , Zaharik, M.L. , Park, E. , Kopelman, R. , Miller, S. et al. (2006) Visualization of vacuolar acidification‐induced transcription of genes of pathogens inside macrophages. Molecular Biology of the Cell, 17, 498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillen, R. & Xiao, J. (2020) Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ‐ring. Annual Review of Biophysics, 49, 309–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa, H. , Sakagami, Y. , Suzuki, A. , Suzuki, H. , Hara, H. & Hirota, Y. (1989) Determination of the cleavage site involved in C‐terminal processing of penicillin‐binding protein 3 of Escherichia coli . Journal of Bacteriology, 171, 5890–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.L. & Kennedy, E.P. (1971) Magnesium transport in Escherichia coli. Inhibition by cobaltous ion. The Journal of Biological Chemistry, 246, 3042–3049. [PubMed] [Google Scholar]
- Núñez‐Hernández, C. , Tierrez, A. , Ortega, A.D. , Pucciarelli, M.G. , Godoy, M. , Eisman, B. et al. (2013) Genome expression analysis of nonproliferating intracellular Salmonella enterica serovar Typhimurium unravels an acid pH‐dependent PhoP‐PhoQ response essential for dormancy. Infection and Immunity, 81, 154–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, S.E. & Coombes, B.K. (2009) RpoE fine tunes expression of a subset of SsrB‐regulated virulence factors in Salmonella enterica serovar Typhimurium. BMC Microbiology, 9, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Morales, D. , Banda, M.M. , Chau, N.Y.E. , Salgado, H. , Martínez‐Flores, I. , Ibarra, J.A. et al. (2017) The transcriptional regulator SsrB is involved in a molecular switch controlling virulence lifestyles of Salmonella . PLoS Pathogens, 13, e1006497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, T.T. , Davies, M.R. , Klemm, E.J. , Rowley, G. , Wileman, T. , James, K. et al. (2013) ChIP‐seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization. Molecular Microbiology, 87, 526–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucciarelli, M.G. & Garcia‐Del Portillo, F. (2017) Salmonella intracellular lifestyles and their impact on host‐to‐host transmission. Microbiology Spectrum, 5. 10.1128/microbiolspec.MTBP-0009-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn, H.J. , Cameron, A.D.S. & Dorman, C.J. (2014) Bacterial regulon evolution: distinct responses and roles for the identical OmpR proteins of Salmonella Typhimurium and Escherichia coli in the acid stress response. PLoS Genetics, 10, e1004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico‐Pérez, G. , Pezza, A. , Pucciarelli, M.G. , de Pedro, M.A. , Soncini, F.C. & García‐del Portillo, F. (2016) A novel peptidoglycan D,L‐endopeptidase induced by Salmonella inside eukaryotic cells contributes to virulence. Molecular Microbiology, 99, 546–556. [DOI] [PubMed] [Google Scholar]
- Röder, J. , Felgner, P. & Hensel, M. (2021) Comprehensive single cell analyses of the nutritional environment of intracellular Salmonella enterica . Frontiers in Cellular and Infection Microbiology, 11, 624650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs, P.D.A. & Bernhardt, T.G. (2021) Growth and division of the peptidoglycan matrix. Annual Review of Microbiology, 75, 315–336. [DOI] [PubMed] [Google Scholar]
- Rose, C. , Parker, A. , Jefferson, B. & Cartmell, E. (2015) The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Critical Reviews in Environmental Science and Technology, 45, 1827–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger, H. & von Jagow, G. (1987) Tricine‐sodium dodecyl sulfate‐polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Analytical Biochemistry, 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Singh, S.K. , Parveen, S. , SaiSree, L. & Reddy, M. (2015) Regulated proteolysis of a cross‐link‐specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proceedings of the National Academy of Sciences of the United States of America, 112, 10956–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snavely, M.D. , Gravina, S.A. , Cheung, T.T. , Miller, C.G. & Maguire, M.E. (1991) Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression. The Journal of Biological Chemistry, 266, 824–829. [PubMed] [Google Scholar]
- Steele‐Mortimer, O. (2008) The Salmonella‐containing vacuole: moving with the times. Current Opinion in Microbiology, 11, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak, P. & Lowe, J. (2013) Do the divisome and elongasome share a common evolutionary past? Current Opinion in Microbiology, 16, 745–751. [DOI] [PubMed] [Google Scholar]
- Uzzau, S. , Figueroa‐Bossi, N. , Rubino, S. & Bossi, L. (2001) Epitope tagging of chromosomal genes in Salmonella . Proceedings of the National Academy of Sciences of the United States of America, 98, 15264–15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef, C. , Lugtenberg, B. , van Boxtel, R. , de Graaff, P. & Verheij, H. (1979) Genetics and biochemistry of the peptidoglycan‐associated proteins b and c of Escherichia coli K12. Molecular & General Genetics, 169, 137–146. [DOI] [PubMed] [Google Scholar]
- Vivero, A. , Baños, R.C. , Mariscotti, J.F. , Oliveros, J.C. , García‐del Portillo, F. , Juárez, A. et al. (2008) Modulation of horizontally acquired genes by the Hha‐YdgT proteins in Salmonella enterica serovar Typhimurium. Journal of Bacteriology, 190, 1152–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley, M.J. , Ching, K.H. & Heffron, F. (2000) Salmonella SsrB activates a global regulon of horizontally acquired genes. Molecular Microbiology, 36, 749–761. [DOI] [PubMed] [Google Scholar]
- Yoon, M.Y. , Lee, K.‐M. , Park, Y. & Yoon, S.S. (2011) Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS One, 6, e16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


