Abstract
Serum glycosylphosphatidylinositol-specific phospholipase D (GPI-PLD) activity is reduced over 75% in systemic inflammatory response syndrome. To investigate the mechanism of this response, expression of the GPI-PLD gene was studied in the mouse monocyte-macrophage cell line RAW 264.7 stimulated with lipopolysaccharide (LPS; 0.5 to 50 ng/ml). GPI-PLD mRNA was reduced approximately 60% in a time- and dose-dependent manner. Oxidative stress induced by 0.5 mM H2O2 or 50 μM menadione also caused a greater than 50% reduction in GPI-PLD mRNA. The antioxidant N-acetyl-l-cysteine attenuated the down-regulatory effect of H2O2 but not of LPS. Cotreatment of the cells with actinomycin D inhibited down-regulation induced by either LPS or H2O2. The half-life of GPI-PLD mRNA was not affected by LPS, or decreased slightly with H2O2, indicating that the reduction in GPI-PLD mRNA is due primarily to transcriptional regulation. Stimulation with tumor necrosis factor alpha (TNF-α) resulted in ∼40% reduction in GPI-PLD mRNA in human A549 alveolar carcinoma cells but not RAW 264.7 cells, suggesting that alternative pathways could exist in different cell types for down-regulating GPI-PLD expression during an inflammatory response and the TNF-α autocrine signaling mechanism alone is not sufficient to recapitulate the LPS-induced reduction of GPI-PLD in macrophages. Sublines of RAW 264.7 cells with reduced GPI-PLD expression exhibited increased cell sensitivity to LPS stimulation and membrane-anchored CD14 expression on the cell surface. Our data suggest that down-regulation of GPI-PLD could play an important role in the control of proinflammatory responses.
Glycosylphosphatidylinositol (GPI)-anchored proteins are numerous on the surface of eukaryotic cells and are involved in a wide variety of physiological functions. In the mammalian immune system, they are involved in the complement cascade, the pro- and anti-inflammatory responses of macrophages, and the activation, development, and proliferation of T cells, as well as the extravasation of leukocytes, tumor invasion, and metastasis (1, 5, 26, 31, 36, 41).
Soluble forms of many GPI-anchored proteins have been observed in body fluids (plasma, urine, cerebrospinal fluid, etc.) and the conditioned medium of cells in culture (reviewed in reference 18). Furthermore, the levels of soluble forms of some GPI-anchored proteins (e.g., CD14, CD16, CD48, CD80, and CD87) increase in a wide range of pathologic conditions (11, 26, 36, 39, 45). Some GPI-anchored cell surface proteins can undergo ectodomain release (also called shedding) by proteolytic cleavage or alternative splicing of primary mRNA transcripts (4). However, there is also substantial evidence for the release of GPI-anchored proteins by intracellular GPI-specific phospholipase D (GPI-PLD) (3, 17, 18, 23, 28, 29, 36, 40, 43, 44, 45).
The GPI-PLD that is predominantly located in plasma is the only well-characterized mammalian phospholipase capable of hydrolyzing the GPI anchor (reviewed in reference 18). GPI-PLD isolated from plasma or serum is unable to release GPI-anchored proteins directly from the surface of intact mammalian cells, probably due to the presence of inhibitory lipid molecules in both the blood plasma and the cell surface (19, 20, 34). However, several studies have indicated that cell-associated GPI-PLD is capable of releasing GPI-anchored proteins, probably at an intracellular site (3, 45). Tsujioka et al. have shown that GPI-PLD expressed in baculovirus-transfected insect cells is more effective at releasing GPI-anchored alkaline phosphatase from CHO cell membranes, implying that posttranslational modification of GPI-PLD may also contribute to the inability of extracellular GPI-PLD to act on cell surfaces (44). Consistent with this concept, GPI-PLD, overexpressed in mammalian cells, was able to cleave GPI anchors early in the secretory pathway, possibly in the endoplasmic reticulum (43).
Although the relative abundance of GPI-PLD in plasma has complicated studies of its cell and tissue distribution, GPI-PLD has been detected in several cell types (e.g., neurons, keratinocytes, bone marrow, leukocytes, pancreatic α and β cells, and mast cells [24, 25, 47]), using a combination of enzyme assay and immunostaining techniques. In many of these studies, the lack of mRNA data makes it difficult to identify the precise source of GPI-PLD: de novo synthesis or uptake from the culture medium. Northern blot analysis of different tissues in humans and mice has revealed some specificity in the tissue expression pattern of GPI-PLD mRNA, with liver and brain having the most (15, 44). Clinical data also showed that patients with liver disease had an altered GPI-PLD activity level in serum, suggesting that the liver is a major source of GPI-PLD in serum (21, 33). However, which cells are responsible for GPI-PLD production in the liver and how this process is regulated remain unclear (34). The change in serum GPI-PLD could reflect altered catabolism of the protein, altered secretion of the enzyme from intracellular compartments, or altered gene expression at both transcriptional and translational levels (6). Although studies focusing specifically on GPI-PLD gene regulation have not been reported, there are indications that GPI-PLD expression may be responsive to extracellular stimuli and different pathophysiological conditions. Furthermore, microarray studies have revealed that GPI-PLD mRNA responds to serum stimulation in human fibroblasts and is actively regulated in some human tumor cell lines (12, 35). GPI-PLD mRNA was also shown to respond to H2O2 stimulation in the murine monocyte-macrophage cell line J774 (30).
Recently, a clinical study showed that the activity of GPI-PLD in sera of patients with systemic inflammatory response syndrome, sepsis, or septic shock was reduced by 75 to 80% compared to a healthy control group (34). In the present study, we have examined the effect of inflammatory stimuli on the expression of GPI-PLD mRNA using the RAW 264.7 monocyte-macrophage cell line as a model system. We have also studied what effect modulation of GPI-PLD has on the cell surface expression of CD14.
MATERIALS AND METHODS
Cell stimulation experiments.
All cells were grown in a humidified incubator at 37°C with 5% CO2. Mouse monocyte-macrophage RAW 264.7 (ATCC [American Type Culture Collection] TIB 71) and human lung carcinoma A549 (ATCC CCL 185) cells were cultured as adherent cells in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml). Mouse monocyte-macrophage J774 (ATCC TIB 67) cells were maintained in DMEM without sodium pyruvate supplemented with 10% heat-inactivated FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml).
Twenty hours before the stimulation experiments, cells were seeded at 50% confluency in 60-mm-diameter dishes. Stimulation was performed in DMEM with 10% FBS unless otherwise indicated. A stock solution (1 mg/ml) of lipopolysaccharide (LPS) from Escherichia coli serotype O127:B8 (Sigma) was freshly prepared by suspending LPS into 0.9% NaCl followed by 30 min of bath sonication at room temperature. The stock was then diluted in medium to achieve the indicated concentrations. A 30% (wt/wt) solution of H2O2 (Sigma) was directly diluted in medium to obtain a concentration of 0.5 mM. Menadione (MQ; Sigma) was freshly prepared in ethanol as a 50 mM stock solution and subsequently diluted 1,000 times in medium. N-Acetyl-l-cysteine (NAC; Sigma) was freshly prepared in ethanol as a 0.5 M stock solution and then diluted to 1 mM in medium. Tumor necrosis factor alpha (TNF-α; Sigma) was diluted in phosphate-buffered saline (PBS) at a concentration of 200 U/μl (2 ng/μl) and stored at −80°C.
RNA isolation and Northern blotting.
Total RNA of cultured cells was purified using TRIZOL reagent (Life Technologies). Concentration and quality of RNA were determined by UV absorbances at 260 and 280 nm. Thirty micrograms of each RNA sample was electrophoresed in 2.2 M formaldehyde–1% agarose gel (some earlier experiments used 0.8%) and blotted to a Nytran SuPerCharge membrane using a TurboBlotter (Schleicher & Schuell). Five micrograms of a 0.24- to 9.5-kb RNA ladder (Life Technologies) mixed with ethidium bromide was run as a marker in parallel. The membrane was UV cross-linked, dried, and stored at 4°C.
The 3.3-kb mouse GPI-PLD cDNA probe was prepared by double digestion of plasmid pBluescript SK(+)/GPI-PLD (a gift from Mark Deeg, Indiana University School of Medicine [15]) with EcoRI and XhoI, followed by gel purification using a GeneClean spin kit (BIO-101, Inc.). The 2.9-kb human GPI-PLD cDNA was amplified by PCR from plasmid pSG5/human GPI-PLD (generously provided by Yoshio Misumi, Fukuoka University School of Medicine [44]), followed by gel purification. The 578-bp AvaI/HindIII cDNA fragment encoding amino acids 8 to 157 of human TNF-α was isolated from plasmid pUC-RI-4Large (ATCC). A human glyceraldehyde-3-phosphate dehydrogenase (G3PDH) cDNA control probe was purchased from Clontech Laboratories, Inc. Probes were labeled with [α-32P]dCTP using a Prime-It RmT random primer labeling kit (Stratagene). After prehybridization, blots were hybridized with 32P-labeled probe in QuikHyb solution (Stratagene) at 68°C and washed under conditions recommended by Stratagene before autoradiography. The same blots were stripped of previously hybridized probe and rehybridized with 32P-labeled human G3PDH cDNA as an internal control. Membranes were exposed for different periods. Only mRNA signals within the linear range were used for band volume quantification with a Personal Densitometer SI (Molecular Dynamics). The relative level of GPI-PLD mRNA for each sample was normalized by comparison with the internal G3PDH control. For presentation purposes, the relative level of GPI-PLD mRNA in stimulated cells versus nonstimulated cells was calculated by setting mRNA in nonstimulated cells at 100%. Two-sample t tests were performed using Microsoft Excel data analysis tools.
Determination of mRNA half-life.
The half-life of mouse GPI-PLD mRNA was determined using actinomycin D (Sigma) at a final concentration of 1 μg/ml (104-fold dilution of a stock solution in dimethyl sulfoxide [DMSO]). RAW 264.7 cells were pretreated with 5 ng of LPS per ml or 0.5 mM H2O2 for 2 h. Cells without pretreatment served as controls. Actinomycin D was then added to the cultures, and total RNA was prepared at the times indicated for up to 4 h. The amount of GPI-PLD mRNA as a percentage of the level obtained at 0 h after actinomycin D addition was determined by Northern blot analysis as described above.
Plasmid construction and stable transfection of GPI-PLD.
Mouse GPI-PLD cDNA was PCR amplified from plasmid pBluescript SK(+)/GPI-PLD using primers 5′-dCCCGATATCGAATGACAACATGTCTGC-3′ and 5′-dCCCGAATTCCTTTAGTCTGAGCTGAAG-3′ and cloned into the internal ribosome entry site (IRES) bicistronic expression vector pIRESneo (Clontech) via EcoRV and EcoRI sites. The resultant plasmid construct pIRES/PLD was amplified, purified with EndoFree Maxi kits (Qiagen), and transfected into RAW 264.7 cells seeded in 12-well plates using LipofectAMINE PLUS reagent (Life Tech-nologies). Five hours after transfection, cells were placed in fresh medium. Twenty hours after transfection, cells from each well were split into two 100-mm dishes with medium containing G418 (800 μg/ml; Mediatech, Inc.). Fresh selec-tive medium was fed to cells every 3 to 4 days until G418-resistant colonies appeared. Colonies were then pooled.
GPI-PLD activity assay.
Preparation of detergent cellular extract and analysis of GPI-PLD activity in cells were described previously (47). Protein concentrations of cellular extracts were determined by using bicinchoninic acid reagents (Pierce), and enzyme specific activity (mean ± standard error [SE]) for each extract was calculated.
Detection of mCD14.
Freshly harvested cells (106) were washed twice with ice-cold PBS containing 0.1% FBS and 0.1% NaN3 and resuspended in 100 μl of the same buffer. Phycoerythrin-conjugated rat anti-CD14 monoclonal antibody rmC5-3 (BD Pharmingen) was added for 30 min on ice. Cells were then washed three times with 1 ml of ice-cold PBS containing 0.1% NaN3 and resuspended in 500 μl of the same buffer. The expression of membrane-anchored CD14 (mCD14) on 30,000 cells for each sample was analyzed by flow cytometry on a FACScan (Becton Dickinson).
Cloning of human GPI-PLD promoter and analysis of TNF-α stimulation.
The sequence of the 5′ untranslated region of the human GPI-PLD gene was obtained from 73M23 (GenBank accession no. AL031230). The +1 position was arbitrarily designated the first nucleotide of the available human liver GPI-PLD cDNA (GenBank accession no. L11701). Four forward primers with an XhoI site (5′-dGGGCTCGAGCAGTTCCAGCTGATTACC-3′, 5′-dGGGCTCGAGCACATGGCTGTGTTAAGC-3′, 5′-dGGGCTCGAGATTCTCCCTACTCACTCC-3′, and 5′-dGGTCTCGAGCAAATGCAGGTCGCCATG-3′) and one reverse primer with an EcoRI site (5′-dGGCGAATTCTGGGAATGCTCAGAGCTG) were used to PCR amplify four DNA fragments containing 5′ untranslated regions from -1 to -502, -1 to -1303, -1 to -2615, and -1 to -5407, respectively. These DNA fragments were cloned separately into pSEAP2-Basic (Clontech) through XhoI and EcoRI sites, resulting in constructs p0.5, p1.3, p2.5, and p5.5, respectively (see Fig. 7D).
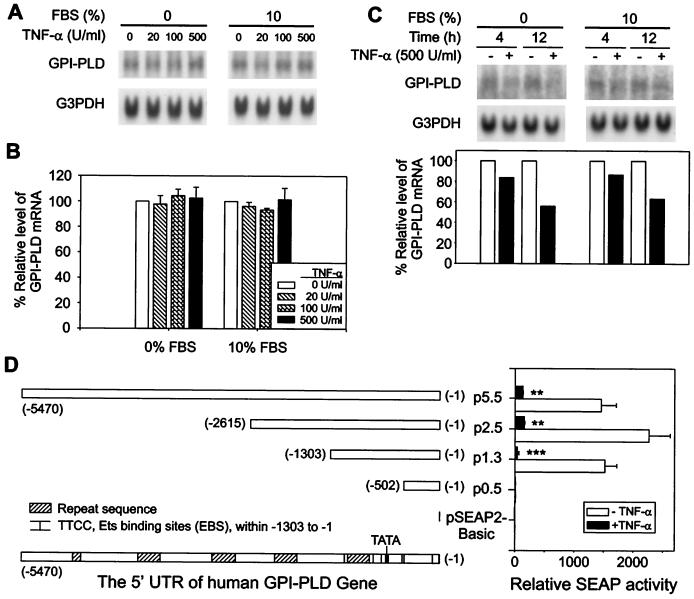
FIG. 7.
TNF-α reduces GPI-PLD expression in human alveolar carcinoma cell line A549 but not in murine macrophage cell line RAW 264.7. (A) The GPI-PLD mRNA level in RAW 264.7 cells was not affected by TNF-α stimulation. RAW 264.7 cells were untreated or treated with TNF-α at the indicated concentrations for 4 h in either serum-free medium or 10% FBS-containing medium. Total RNA was prepared and analyzed by Northern blotting. Top, representative blots hybridized with 32P-labeled GPI-PLD cDNA; bottom, same blot hybridized with 32P-G3PDH. (B) Relative levels of GPI-PLD mRNA in stimulated cells compared to nonstimulated cells with and without the presence of serum. Results are depicted as means ± SE for three independent experiments. (C) Reduction of GPI-PLD mRNA upon TNF-α stimulation in A549 cells is independent of serum. A549 cells were untreated or treated with 500 U of TNF-α per ml in either serum-free medium or 10% FBS-containing medium. Samples were taken at 4 and 12 h, and total RNA was prepared and analyzed by Northern blotting. Shown are representative blots hybridized sequentially with 32P-labeled GPI-PLD cDNA and 32P-G3PDH along with quantification data. (D) TNF-α reduces the expression of SEAP driven by the human GPI-PLD promoter in A549 cells. The four promoter constructs are shown schematically on the left. The positions of five repeat sequences along the 5.5-kb 5′ untranslated region were predicted by using the Repeat Masker server at the University of Washington (http://ftp.genome.washington.edu/) and indicated by gray boxes. The TATA box of the strong promoter located within -502 to -1303 is indicated by a vertical bar. The distribution of 10 potential EBS (TTCC) detected within -1 to -1303 is also denoted by thin vertical bars. A549 cells were transiently transfected with promoter constructs containing a SEAP reporter gene. TNF-α (500 U/ml) treatment and measurement of SEAP activity are described in Materials and Methods. Results are depicted as means ± SE for four independent transfection experiments. ∗∗, P < 0.01; ∗∗∗, P < 0.001.
For promoter analysis, A549 cells were seeded into a 48-well plate at a density of 7 × 104 cells/well overnight. Duplicated sets of transient transfection of each construct were performed using LipofectAMINE PLUS reagent. Transfections of promoterless vector pSEAP2-Basic were performed in parallel as negative controls. After 5 h of transfection, cultures were replaced with fresh serum-containing medium and incubated for another 20 h. The medium was removed, and serum-free DMEM with or without TNF-α (500 U/ml) was added to sets of transfected cells for a further 72 h. The activity of the reporter gene, a secreted form of human placental alkaline phosphatase (SEAP), was analyzed in medium supernatant using a SEAP chemiluminescence detection kit (Clontech).
RESULTS
LPS down-regulates GPI-PLD expression in murine macrophage cell lines.
An initial Northern blot screening of several human and murine monocyte-macrophage cell lines demonstrated that two murine cell lines, RAW 264.7 and J774, had a detectable basal level of GPI-PLD mRNA (X. Du and M. G. Low, unpublished observations). The transcript was approximately 7.5 kb, similar to the largest of the three major GPI-PLD transcripts detected in mouse tissues (15). The other two smaller transcripts normally observed in brain and liver tissues (15) were not detectable in mouse monocyte-macrophage cells.
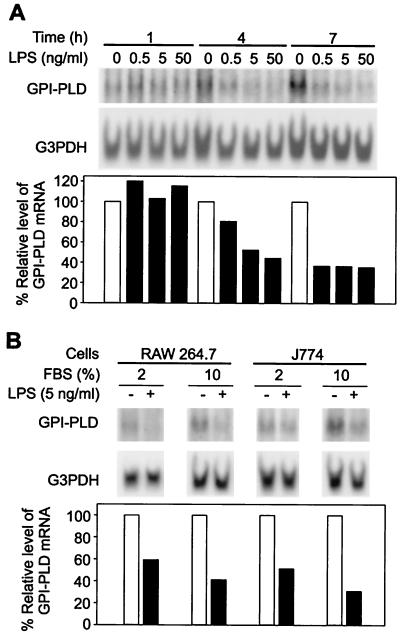
To determine if GPI-PLD mRNA synthesis responded to LPS stimulation, RAW 264.7 cells were treated with LPS at different concentrations (0.5, 5, and 50 ng/ml) for 1, 4, and 7 h, and levels of GPI-PLD mRNA were analyzed by Northern blotting. GPI-PLD mRNA was down-regulated in a time- and dose-dependent manner (Fig. 1A). During the first hour of LPS stimulation, GPI-PLD mRNA remained unchanged in either untreated or LPS-treated cells (Fig. 1A). After 4 h, the mRNA level in LPS-treated cells was reduced in a dose-dependent fashion (Fig. 1A). Down-regulation of GPI-PLD mRNA in RAW 264.7 cells was detectable at LPS concentrations as low as 0.5 ng/ml (Fig. 1A). At the end of the 7-h treatment, GPI-PLD mRNA levels in stimulated cells were reduced by 63 to 65% compared to the nonstimulated cells, even at the lowest LPS concentration (Fig. 1A).
FIG. 1.
Down-regulation of GPI-PLD mRNA by LPS stimulation. (A) Time- and dose-dependent response of GPI-PLD mRNA to LPS stimulation. RAW 264.7 cells were not stimulated (controls) or stimulated with different concentrations of LPS as indicated. Samples were taken at 1, 4, and 7 h for Northern blot analysis using 0.8% RNA denaturing gels. The results are representative of two independent Northern analyses that were hybridized sequentially with 32P-labeled GPI-PLD and G3PDH cDNAs. Intensities of signals, here and in the following figures, were enhanced for presentation purposes; relative levels of GPI-PLD mRNA in stimulated cells in comparison to nonstimulated cells (100%) were calculated as described in Materials and Methods. (B) Reduction of GPI-PLD mRNA by LPS under different serum concentrations. The murine monocyte-macrophage cell lines J774 and RAW 264.7 were stimulated with LPS (5 ng/ml) in medium containing 2 or 10% FBS for 4 h. Total RNA was prepared and analyzed by Northern blotting using 0.8% RNA denaturing gels. Cells without LPS were analyzed in parallel. The results are representative of two independent Northern analyses.
Previous studies of LPS-regulated responses have used serum concentrations ranging from 2 to 10%. A test of the serum dependence of the LPS response revealed that LPS (5 ng/ml for 4 h) effected a greater decrease of mRNA in 10% serum than in 2% serum, using either RAW 264.7 or J774 cells (Fig. 1B). In addition, the basal levels of GPI-PLD mRNA in nonstimulated cells were higher in 10% serum than in 2% serum (Fig. 1B). Medium containing 10% serum was therefore used in all subsequent experiments.
Oxidative stress reduces GPI-PLD expression in RAW 264.7 cells.
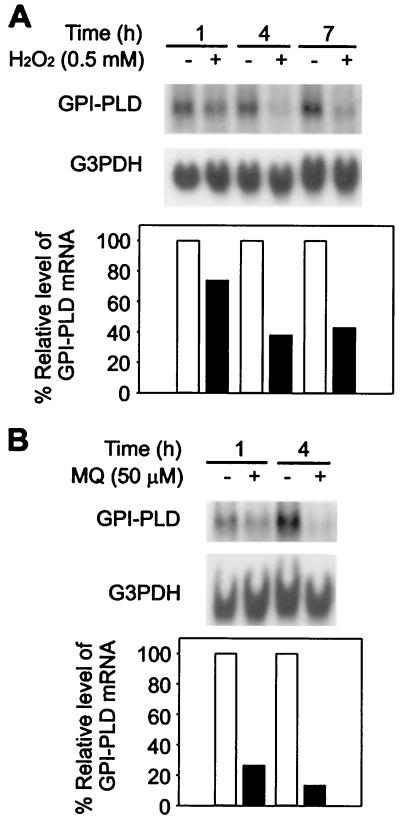
Induction of macrophage inflammatory protein 2 (MIP-2) by LPS can be blocked by antioxidants, and oxidative stress alone can also up-regulate MIP-2 expression (37). To test if H2O2 stimulation changed GPI-PLD mRNA level in macrophage, RAW 264.7 cells were treated with 0.5 mM H2O2 for 1, 4, and 7 h. GPI-PLD mRNA level was reduced within the first 4 h of incubation with H2O2 (Fig. 2A). Compared to nonstimulated cells, the level of GPI-PLD mRNA in H2O2-treated cells decreased by 26% at 1 h, 62% at 4 h, and 57% at 7 h (Fig. 2A).
FIG. 2.
Down-regulation of GPI-PLD mRNA by oxidative stresses. (A) Time course response of GPI-PLD mRNA to H2O2 stimulation. RAW 264.7 cells were untreated (controls) or treated with 0.5 mM H2O2. Samples were taken at 1, 4, and 7 h, and total RNA was prepared and analyzed by Northern blotting. The results are representative of two independent Northern analyses. (B) MQ induces down-regulation of GPI-PLD mRNA. RAW 264.7 cells were stimulated with 50 μM MQ for 1 and 4 h. Cells without MQ were tested in parallel. Total RNA was prepared and analyzed by Northern blotting using 0.8% RNA denaturing gels. The results are representative of two independent Northern analyses.
To examine the mechanism of this effect, MQ, which generates reactive oxygen species continuously (37), was used to stimulate RAW 264.7 cells for 1 and 4 h. The relative level of GPI-PLD mRNA was reduced to 26% as early as 1 h following exposure to 50 μM MQ (Fig. 2B). Incubation with MQ for 4 h caused a further decrease of GPI-PLD mRNA in RAW 264.7 cells, to 13% of the level for nonstimulated cells (Fig. 2B).
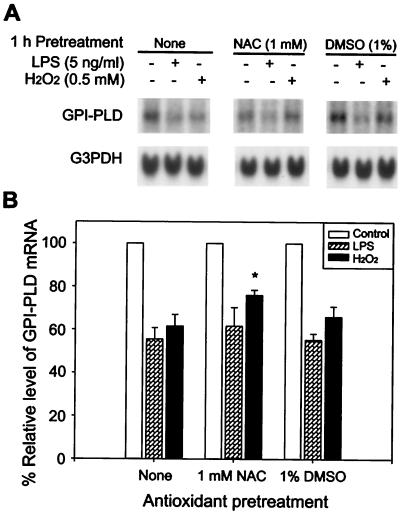
To test if antioxidants could block the stimulatory effect of LPS and H2O2 on GPI-PLD mRNA, RAW 264.7 cells were pretreated with 1 mM NAC or 1% DMSO for 2 h and then challenged with either 5 ng of LPS per ml or 0.5 mM H2O2 for 4 h. Although DMSO at this level had no effect on either LPS or H2O2 stimulation, 1 mM NAC partially blocked the effect of H2O2 but not that of LPS (Fig. 3).
FIG. 3.
Effect of the antioxidant pretreatment on reduction of GPI-PLD mRNA in response to LPS and H2O2. (A) RAW 264.7 cells were untreated (none) or pretreated with 1 mM NAC or 1% DMSO for 1 h and then added with 5 ng of LPS per ml or 0.5 mM H2O2 for another 4 h. Cells without stimulation (control) were tested in parallel. Total RNA for each sample was isolated at the end of the stimulation and analyzed by Northern blotting. The results are representative of three independent experiments. (B) Relative levels of GPI-PLD mRNA in stimulated cells compared to nonstimulated cells under different pretreatments. The level of GPI-PLD mRNA in nonstimulated cells (control) was always set at 100% for either untreated (none) or antioxidant-treated cells. Results are depicted as means ± SE for three independent experiments. ∗, P < 0.05, comparing H2O2-stimulated cells with and without NAC pretreatment.
Down-regulation occurs at the transcriptional level.
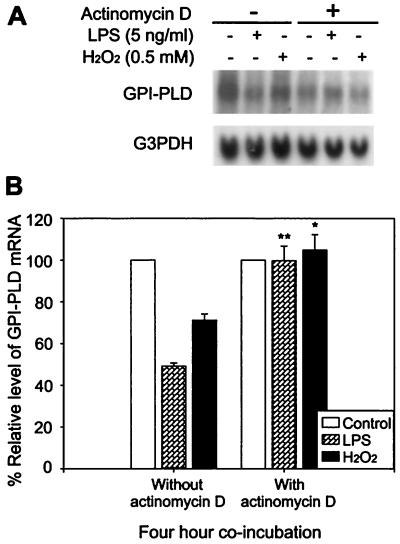
To determine whether the decrease in GPI-PLD mRNA level reflects directly the transcriptional activity of the GPI-PLD gene, actinomycin D, a potent inhibitor of RNA polymerase II-dependent transcription, was included in LPS or H2O2 treatments of RAW 264.7 cells for 4 h. The reduction of GPI-PLD mRNA stimulated by LPS and H2O2 was completely blocked by coincubation with actinomycin D, suggesting that the decrease effected by both of these stimuli involves regulation at the transcriptional level (Fig. 4).
FIG. 4.
Reduction of GPI-PLD mRNA by either LPS or H2O2 is completely inhibited by actinomycin D. (A) RAW 264.7 cells were untreated (control) or treated with 5 ng of LPS per ml or 0.5 mM H2O2 in the presence or absence of 1 μg of actinomycin D per ml for 4 h. Total RNA was extracted and analyzed by Northern blotting. The blots are representative of three independent experiments. (B) Percentages of relative levels of GPI-PLD mRNA in stimulated cells compared to nonstimulated cells with and without the presence of actinomycin D. Results are depicted as means ± SE for three independent experiments. ∗, P < 0.05; ∗∗, P < 0.01.
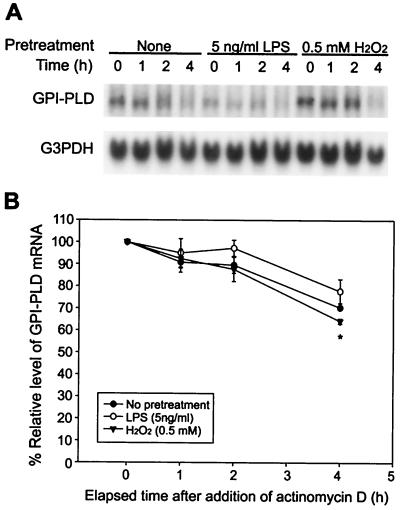
Regulation of posttranscriptional processes might also play a role in the reduction of GPI-PLD mRNA. To examine this possibility, we added actinomycin D to unstimulated, LPS-stimulated, or H2O2-stimulated RAW 264.7 cells and measured the steady-state level of GPI-PLD mRNA over a period of 4 h. The results indicated that GPI-PLD mRNA from untreated cells decreased about 30% after a 4-h treatment with actinomycin D (half-life of ca. 6 h), and its stability was not significantly affected by LPS treatment (half-life of ca. 6 to 7 h [Fig. 5]). H2O2 treatment seems to destabilize GPI-PLD mRNA slightly 4 h after the addition of actinomycin D resulting in a half-life of approximately 5 h (Fig. 5). However, this effect, although statistically significant, was relatively minor. Taken together, our results suggest that the reduction of GPI-PLD mRNA produced by LPS or H2O2 treatment is regulated mainly at the transcriptional level; posttranscriptional regulation appears to play a minor role in H2O2-induced down-regulation.
FIG. 5.
Effect of LPS or H2O2 on the half-life of GPI-PLD mRNA. (A) RAW 264.7 cells were untreated (none) or pretreated with 5 ng of LPS per ml or 0.5 mM H2O2 for 2 h, and then actinomycin D was added to a final concentration of 1 μg/ml. At the times indicated, total RNA was extracted and analyzed by Northern blotting. The results are representative of three independent experiments. (B) Percentage of relative level of GPI-PLD mRNA as a function of time after the addition of actinomycin D. Details of calculation are described in Materials and Methods. Results are depicted as means ± SE for three independent experiments. ∗, P < 0.05, comparing half-lives of mRNA between H2O2-treated and untreated control cells at 4 h.
Decreased GPI-PLD activity increases mCD14 expression on the cell surface and macrophage sensitivity to LPS.
To investigate the possible physiological role of the down-regulation of GPI-PLD during a proinflammatory response induced by LPS, we tried to transfect a mouse GPI-PLD cDNA stably into RAW 264.7 but encountered difficulties. Among the five independent transfection experiments using the cDNA construct pIRES/PLD, only one produced multiple G418-resistant colonies (pooled and labeled IIA), two resulted in no colonies, and the other two resulted in only one colony each (designated IA and IIIA). By comparison, a control transfection of the cloning vector pIRESneo in RAW 264.7 cells readily generated multiple G418-resistant colonies (pooled and designated B). Northern analysis revealed overexpressed mRNA molecules containing GPI-PLD cDNA other than the 7.5-kb genomic transcripts in pIRES/PLD stably transfected lines but not in B and RAW 264.7 cells, suggesting that these lines were not due to neomycin-resistant colonies that had developed spontaneously (data not shown). The GPI-PLD enzyme activity levels in IA, IIA, and IIIA cells were, however, not significantly higher than those in either RAW 264.7 cells or subline B cells. During this process, we also found that continuous culture of the neomycin-resistant RAW 264.7 sublines (B, IA, IIA, and IIIA) in G418 (500 μg/ml) reduced cellular GPI-PLD activity by 40% (Fig. 6C, upper panel).
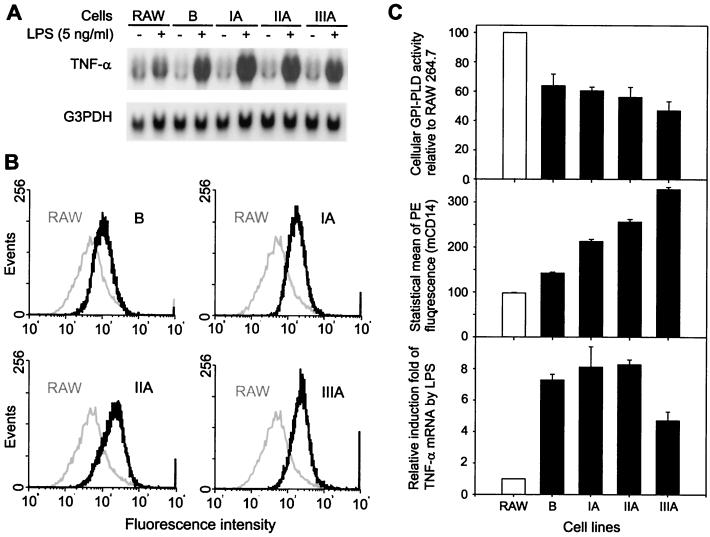
FIG. 6.
Low GPI-PLD activity in RAW 264.7 sublines correlates with an increased stimulatory effect of LPS and increased mCD14 expression on the cell surface. RAW 264.7 sublines B, IA, IIA, and IIIA were obtained by stable transfection as described in Results. (A) RAW 264.6 cells (RAW) and the sublines were unstimulated or stimulated with 5 ng of LPS per ml for 4 h. Total RNA was extracted and analyzed by Northern blotting. Top, representative blot of three independent experiments hybridized with 32P-labeled TNF-α cDNA; bottom, same blot hybridized with 32P-G3PDH cDNA. (B) Comparison of mCD14 expression on the cell surface of RAW 264.7 cells (RAW) and the sublines. mCD14 was detected using phycoerythrin-conjugated rat anti-CD14 monoclonal antibody rmC5-3 as described in Materials and Methods. The histogram of fluorescence (FL2-H) intensity of each subline (black trace) was overlaid with that of RAW 264.7 cells (RAW). The fluorescence background shown by unstained RAW 264.7 cells was below 10, with a mean of 8.85. (C) Decreased GPI-PLD activity in RAW 264.7 sublines is associated with increased mCD14 presentation and induction of TNF-α mRNA by LPS. Three sets of data were aligned for comparison purposes. Top, cellular GPI-PLD specific activities in RAW 264.7 sublines B, IA, IIA, and IIIA (cultured in G418) relative to RAW 264.7 cells (RAW). Results are depicted as means ± SE for three independent experiments. Middle, comparison of statistical means of fluorescence intensities (mCD14) in RAW 264.7 sublines B, IA, IIA, and IIIA with that of RAW 264.7 cells (RAW). Results are depicted as means ± SE for three independent experiments. Bottom, induction of TNF-α mRNA by LPS in RAW 264.7 sublines B, IA, IIA, and IIIA relative to RAW 264.7 cells (RAW). The extent of induction was initially calculated by comparing relative level of TNF mRNA in LPS-stimulated cells to that of nonstimulated cells. For comparison purposes, the induction in RAW 264.7 cells was arbitrarily set at 1. Results are depicted as means ± SE for three independent experiments.
Since LPS reduced GPI-PLD mRNA by about 60%, which is likely to have an effect on GPI-PLD at the protein level, we investigated whether the decreased GPI-PLD activity in these RAW 264.7 sublines (in the presence of G418) had affected cell responsiveness to LPS. RAW 264.7 cells and G418-treated sublines B, IA, IIA, and IIIA were stimulated with LPS (5 ng/ml) for 4 h, using TNF-α mRNA synthesis as the criterion for cell sensitivity to LPS. The magnitude of TNF-α mRNA induction by LPS was five to nine times greater in all four sublines than in the parent cell line (Fig. 6A and C).
It is well established that the GPI-anchored protein mCD14, a component of the LPS receptor complex, plays an essential role in the response of monocytes-macrophages to LPS stimulation. We therefore determined if decreased GPI-PLD activity amplified cell sensitivity to LPS by increasing the expression of mCD14. Cell surface expression of mCD14 was compared on RAW 264.7 and the G418-treated sublines B, IA, IIA, and IIIA. The amount of mCD14 on the cell surface was significantly increased among all sublines that had decreased GPI-PLD activity in comparison to RAW 264.7 cells (Fig. 6B and C).
TNF-α reduces GPI-PLD expression in A549 cells but not in RAW 264.7 cells.
LPS-induced physiological processes such as apoptosis in macrophages are predominantly mediated by the autocrine production of TNF-α (46). To test if TNF-α alone could recapitulate LPS-induced reduction of GPI-PLD mRNA, RAW 264.7 cells were stimulated with TNF-α for 4 h in medium with or without 10% serum (Fig. 7A). No significant change in relative levels of GPI-PLD mRNA was observed between nonstimulated and TNF-α-stimulated cells with either medium (Fig. 7A and B).
During a separate study in which a variety of cell lines were screened, we found that human alveolar carcinoma A549 expressed detectable level of GPI-PLD mRNA. The transcript is 6.5 to 7.0 kb in size, in agreement with the value reported by Tsujioka et al. (44). Since airway epithelium cells respond to the stimulation of proinflammatory cytokines such as TNF-α during airway inflammation, we tested if TNF-α affected the expression of GPI-PLD. A549 cells were stimulated with TNF-α (500 U/ml) during a period of 12 h in the presence of 0 or 10% serum. The levels of GPI-PLD mRNA were reduced in a time-dependent manner, by 12 to 17% at 4 h and 37 to 45% at 12 h, with a slightly greater reduction in serum-free medium (Fig. 7C).
To confirm the TNF-induced down-regulation of GPI-PLD mRNA at the gene transcription level, we cloned the promoter sequence of human GPI-PLD and made four constructs containing different lengths of the 5′ untranslated region in front of a SEAP reporter gene (Fig. 7D). A549 cells were transiently transfected with the promoter constructs and then stimulated with TNF-α. The basal transcriptional activity of the human GPI-PLD gene promoter was located from positions -502 to -1303, as pl.3-transfected cells showed a significantly higher SEAP activity while the p0.5-transfected did not (Fig. 7D). A strong promoter was predicted within this 801-bp region, with a score of 0.91 out of 1, using the bioinformatic tool provided by the Berkeley Drosophila Genome Project (http://www.fruitfly.org/seq_tools/promoter.html). Its TATA box was 724 bp away from the translational start site, with a predicted transcriptional initiation site 694 bp upstream of the first amino acid codon. The stimulation results showed that the promoter region ranging from -1 to -1303 is sufficient for TNF-α-induced down-regulation of GPI-PLD in A549 cells (Fig. 7D). The promoter activity of p1.3 was reduced by over 90% in the presence of TNF-α for 3 days (Fig. 7D), suggesting that the TNF-α regulatory effect on GPI-PLD mRNA occurred at the transcriptional level.
DISCUSSION
It has been suggested that the function of mammalian GPI-PLD is to cleave GPI anchors specifically from GPI-anchored cell surface proteins (reviewed in reference 18). The regulation of GPI-PLD expression and secretion might therefore influence the physiological functions of GPI-anchored proteins and the cells that express them. Previously, the secretion of GPI-PLD was demonstrated to be up-regulated by glucose stimulation in mouse insulinoma βTC3 cells (6), an effect that could be blocked by the addition of cycloheximide. It is likely that newly synthesized protein made a significant contribution to the glucose-stimulated increase in GPI-PLD secretion, but it was not determined whether GPI-PLD gene expression was also up-regulated during the glucose stimulation (6). Our previous study on myeloid cell lines also showed that differentiation of the promyelocytic leukemia cell line HL-60 induced by DMSO or phorbol 12-myristate 13-acetate resulted in a consistent two- to threefold increase in GPI-PLD activity (47). However, the relative levels of GPI-PLD mRNA in HL-60 cells differentiated with either DMSO or phorbol 12-myristate 13-acetate, monitored for 3 days by Northern blotting, revealed no detectable changes (Du and Low, unpublished). This result suggests that the increase in cell-associated GPI-PLD activity previously observed during HL-60 differentiation was due to increased uptake of the enzyme from serum rather than stimulation of its synthesis. The work reported here represents the first systematic study of GPI-PLD mRNA regulation in any cell type.
This study has demonstrated that GPI-PLD mRNA is reduced by either LPS or oxidative stress in mouse RAW 264.7 monocyte-macrophage cells. Down-regulation of GPI-PLD by both LPS and H2O2 was also found in another mouse monocyte-macrophage cell line, J774 (Fig. 1B; Du and Low, unpublished). The results are different from what was previously reported by O'Brien et al.: 1 h of H2O2 (0.5 mM) treatment increased the GPI-PLD mRNA level by 50% in J774 cells (30). The reasons for this difference are unclear.
Our data suggest that the reduction of GPI-PLD mRNA by LPS or H2O2 occurred at the transcriptional level, while posttranslational regulation played a minor role in H2O2-induced reduction. LPS has been shown to regulate genes at the transcriptional, posttranscriptional, or even translational level in macrophages (8, 32). H2O2 has also been demonstrated to induce many genes at both transcriptional and posttranslational levels (37, 38). It was speculated that for some genes such as MIP-2, LPS stimulates gene expression through reactive oxygen species (37). Although we have shown that GPI-PLD mRNA was down-regulated by both LPS and H2O2, it is not clear whether the effects are related. Rather, the fact that NAC can partially block the effect of H2O2 but not that of LPS suggests that LPS and H2O2 may reduce GPI-PLD transcription through different regulatory mechanisms.
It was not surprising to find that TNF-α, an important mediator in the LPS-induced inflammatory response, had no effect on GPI-PLD mRNA in macrophage cells. In fact, the macrophage scavenger receptor A, which is induced fivefold in response to LPS stimulation in RAW 264.7 cells, does not respond to TNF-α treatment either (8). However, we did find that TNF-α reduced GPI-PLD mRNA in pulmonary A549 epithelial cells, suggesting that it might be the mediator to down-regulate GPI-PLD gene expression during an inflammatory response in cells other than macrophages. Whereas reduction of serum GPI-PLD seems to be a general trend during inflammatory responses (34), its mRNA probably is reduced by different paths in macrophages and epithelial cells. Since GPI-PLD was shown to regulate both release and membrane expression of CD87, which is implicated in tumor progression and metastasis of ovarian cancer cell lines (45), the TNF-α inhibitory effect on GPI-PLD expression that we observed in alveolar carcinoma A549 cells might affect cell proliferation and migration by adjusting the balance between membrane-anchored and soluble CD87.
In addition to its capacity for up-regulating the expression of a large number of genes, TNF-α also down-regulates many other genes (reviewed in reference 22). The present work adds a new member to this group of proteins. Although the mechanism by which TNF-α stimulates gene expression has been well characterized, the down-regulation mechanism is not fully understood. Recently, a report showed that Ets-binding sites (EBS; 5′-TTCC) were involved in down-regulating intracellular adhesion molecule 2 (22). Our promoter analysis indicated that the promoter region from -1 to -1303 is sufficient for the response to TNF-α stimulation. A search of this region has detected 10 potential EBS, two of them located, in tandem, between the TATA box and transcriptional start site of the promoter predicted between -502 and -1303 (Fig. 7D). Whether any of these EBS is involved in TNF-α-dependent regulation of the GPI-PLD gene awaits further promoter analysis and mutagenesis experiments. We have also tested the promoter activity in human monocyte THP-1 and promonocyte U-973 cells. Unfortunately, a combination of low transfection efficiency and low level of GPI-PLD expression precluded a promoter analysis (data not shown). Testing of a number of other mouse and rat cells including RAW 264.7 with this human promoter revealed no basal expression (data not shown). The results suggest that the human GPI-PLD promoter might contain cell-specific regulatory elements, in addition to its weak activity.
Although the bicistronic expression system that we used for stable transfection is highly efficient, our attempt to overexpress GPI-PLD stably in RAW 264.7 cells was not successful. The few neomycin-resistant sublines (IA, IIA, and IIIA cells) that were obtained from the transfections did not express higher levels of cellular GPI-PLD activity than the RAW 264.7 or B lines even though they overexpressed mRNA molecules containing GPI-PLD cDNA sequences. These observations suggest that overexpression of GPI-PLD in macrophages might be cytotoxic and only cells with lower GPI-PLD activity survived. In addition, all neomycin-resistant sublines (B, IA, IIA, and IIIA) showed a reduced level of GPI-PLD activity when maintained continuously in G418. This phenomenon could be reversed by 2 weeks of passage without G418, suggesting that the selection procedure itself could have reduced GPI-PLD activity. G418 has previously been shown to influence the metabolism of GPIs and the expression of GPI-anchored proteins in other cell lines, and this might account for our observations (9, 13, 27). However, whatever the mechanism might be, it does not appear to be the result of a direct inhibitory effect on the enzymatic activity of the GPI-PLD protein (Du and Low, unpublished).
Analysis of the RAW 264.7 sublines with decreased GPI-PLD activity showed an inverse correlation between GPI-PLD activity and mCD14 expression or cell sensitivity to LPS. Our data indicate a possible physiological consequence from down-regulation of GPI-PLD mRNA in macrophages during the inflammatory response, which is to decrease GPI-degrading activity, maximize the expression of mCD14 on the cell surface, and thereby optimize the cell response to LPS. Although we tried to detect a change of GPI-PLD at the protein level under different stimulation, the amount of GPI-PLD in cells was below the immunoblotting detection limit (data not shown). We also monitored the enzyme activity of cellular GPI-PLD under different stimulation conditions over a period of 24 h but found no reproducible change in activity (data not shown). Although these results may simply demonstrate the limitations of protein-based detection methods, they could also indicate that the reduction in mRNA level results only in a transient decrease in newly synthesized GPI-PLD in the endoplasmic reticulum, where the degradation of GPI anchors is proposed to occur in vivo (43). This change might not be large enough to affect the overall level of cellular GPI-PLD, the bulk of which may be stored in other cellular compartments (6, 10, 47).
CD14 is the principal LPS-binding protein on the surface of monocytes and macrophages and plays an essential role in the proinflammatory response (reviewed in references 1, 2, 16, and 36). In common with many other GPI-anchored proteins, CD14 has two functionally distinct forms, mCD14 and soluble CD14, (sCD14). The GPI-anchored form (mCD14) is believed to participate directly in LPS signaling via an interaction with toll-like receptor 4. There are also two distinct forms of sCD14 which have no GPI anchor but play an important role in the transfer of LPS between cells (4, 7, 36, 42). LPS stimulation induces a twofold increase in expression of mCD14 and sCD14 as well as a slow increase in CD14 mRNA which enhances the response of macrophages to a second LPS challenge (11, 14). Furthermore, the balance between mCD14 and sCD14 is dramatically altered following LPS stimulation (11). Our data show that GPI-PLD mRNA is rapidly reduced in response to LPS stimulation, and RAW 264.7 sublines with lower GPI-PLD activity have higher levels of mCD14 and a stronger LPS response. These observations suggest that regulation of GPI-PLD synthesis could, in response to a physiological stimulus, modulate the amount of mCD14 available at the macrophage cell surface by altering GPI-degrading activity. The sCD14 formed by anchor degradation would make a relatively small contribution to the total plasma pool of sCD14. However, this process could have a major influence on the balance between the mCD14-LPS and sCD14-LPS complexes encountered at the cell surface by the signaling receptor, toll-like receptor 4.
ACKNOWLEDGMENTS
We are grateful to Jiewei Cai, Hui Liao, and Kim Olson for their excellent suggestions and assistance. We also thank Mark Deeg, Indiana University School of Medicine, and Yoshio Misumi, Fukoka University School of Medicine, for providing GPI-PLD plasmids.
This work was supported by National Institutes of Health grants GM-40083 and GM-35873.
REFERENCES
- 1.Antal-Szalmás P. Evaluation of CD14 in host defence. Eur J Clin Investig. 2000;30:167–179. doi: 10.1046/j.1365-2362.2000.00610.x. [DOI] [PubMed] [Google Scholar]
- 2.Beutler B. Tlr4: central component of the sole mammalian LPS sensor. Curr Opin Immunol. 2000;12:20–26. doi: 10.1016/s0952-7915(99)00046-1. [DOI] [PubMed] [Google Scholar]
- 3.Brunner G, Metz C N, Nguyen H, Gabrilove J, Patel S R, Davitz M A, Rifkin D B, Wilson E L. An endogenous glycosylphosphatidylinositol-specific phospholipase D releases basic fibroblast growth factor-heparan sulfate proteoglycan complexes from human bone marrow cultures. Blood. 1994;83:2115–2125. [PubMed] [Google Scholar]
- 4.Bufler P, Stiegler G, Schuchmann M, Hess S, Krüger C, Stelter F, Eckerskorn C, Schütt C, Engelmann H. Soluble lipopolysaccharide receptor (CD14) is released via two different mechanisms from human monocytes and CD14 transfectants. Eur J Immunol. 1995;25:604–610. doi: 10.1002/eji.1830250244. [DOI] [PubMed] [Google Scholar]
- 5.Deckert M, Ticchioni M, Mari B, Mary D, Bernard A. The glycosylphosphatidylinositol-anchored CD59 protein stimulates both T cell receptor zeta/ZAP-70-dependent and -independent signaling pathways in T cells. Eur J Immunol. 1995;25:1815–1822. doi: 10.1002/eji.1830250704. [DOI] [PubMed] [Google Scholar]
- 6.Deeg M A, Verchere C B. Regulation of glycosylphosphatidylinositol-specific phospholipase D secretion from βTC3 cells. Endocrinology. 1997;138:819–826. doi: 10.1210/endo.138.2.4940. [DOI] [PubMed] [Google Scholar]
- 7.Durieux J-J, Vita N, Popescu O, Guette F, Calzada-Wack J, Munker R, Schmidt R E, Lupker J, Ferrara P, Ziegler-Heitbrock H W L, Labeta M O. The two soluble forms of the lipopolysaccharide receptor, CD14: characterization and release by normal human monocytes. Eur J Immunol. 1994;24:2006–2012. doi: 10.1002/eji.1830240911. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald M L, Moore K J, Freeman M W, Reed G L. Lipopolysaccharide induces scavenger receptor A expression in mouse macrophages: a divergent response relative to human THP-1 monocyte/macrophages. J Immunol. 2000;164:2692–2700. doi: 10.4049/jimmunol.164.5.2692. [DOI] [PubMed] [Google Scholar]
- 9.Gupta D, Tartakoff A, Tisdale E. Metabolic correction of defects in the lipid anchoring of Thy-1 in lymphoma mutants. Science. 1988;242:1446–1448. doi: 10.1126/science.2904699. [DOI] [PubMed] [Google Scholar]
- 10.Hari T, Bütikofer P, Wiesmann U N, Brodbeck U. Uptake and intracellular stability of glycosylphosphatidylinositol-specific phospholipase D in neuroblastoma cells. Biochim Biophys Acta Mol Cell Res. 1997;1355:293–302. doi: 10.1016/s0167-4889(96)00143-7. [DOI] [PubMed] [Google Scholar]
- 11.Hasday J D, Dubin W, Mongovin S, Goldblum S E, Swoveland P, Leturcq D J, Moriarty A M, Bleecker E R, Martin T R. Bronchoalveolar macrophage CD14 expression: shift between membrane-associated and soluble pools. Am J Physiol Lung Cell Mol Physiol. 1997;272:L925–L933. doi: 10.1152/ajplung.1997.272.5.L925. [DOI] [PubMed] [Google Scholar]
- 12.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, Lashkari D, Shalon D, Botstein D, Brown P O. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 13.Küng M, Stadelmann B, Brodbeck U, Bütikofer P. Addition of G418 and other aminoglycoside antibiotics to mammalian cells results in the release of GPI-anchored proteins. FEBS Lett. 1997;409:333–338. doi: 10.1016/s0014-5793(97)00452-3. [DOI] [PubMed] [Google Scholar]
- 14.Landmann R, Knopf H-P, Link S, Sansano S, Schumann R, Zimmerli W. Human monocyte CD14 is upregulated by lipopolysaccharide. Infect Immun. 1996;64:1762–1769. doi: 10.1128/iai.64.5.1762-1769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeBoeuf L C, Caldwell M, Guo Y, Metz C, Davitz M A, Olson L K, Deeg M A. Mouse glycosylphosphatidylinositol-specific phospholipase D (GpldI) characterization. Mamm Genome. 1998;9:710–714. doi: 10.1007/s003359900851. [DOI] [PubMed] [Google Scholar]
- 16.Lien E, Means T K, Heine H, Yoshimura A, Kusumoto S, Fukase K, Fenton M J, Oikawa M, Qureshi N, Monks B, Finberg R W, Ingalls R R, Golenbock D T. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J Clin Investig. 2000;105:497–504. doi: 10.1172/JCI8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lierheimer R, Kunz B, Vogt L, Savoca R, Brodbeck U, Sonderegger P. The neuronal cell-adhesion molecule axonin-1 is specifically released by an endogenous glycosylphosphatidylinositol-specific phospholipase. Eur J Biochem. 1997;243:502–510. doi: 10.1111/j.1432-1033.1997.0502a.x. [DOI] [PubMed] [Google Scholar]
- 18.Low M G. Structure and function of GPI-specific phospholipases. In: Young N S, Moss J, editors. Paroxysmal nocturnal hemoglobinuria and the glycosylphosphatidylinositol-linked proteins. San Diego, Calif: Academic Press; 2000. pp. 239–268. [Google Scholar]
- 19.Low M G, Huang K-S. Factors affecting the ability of glycosylphosphatidylinositol- specific phospholipase D to degrade the membrane anchors of cell surface proteins. Biochem J. 1991;279:483–493. doi: 10.1042/bj2790483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Low M G, Stütz P. Inhibition of the plasma GPI-specific phospholipase D by synthetic analogs of Lipid A and phosphatidic acid. Arch Biochem Biophys. 1999;371:332–339. doi: 10.1006/abbi.1999.1436. [DOI] [PubMed] [Google Scholar]
- 21.Maguire G A, Gossner A. Glycosyl phosphatidylinositol phospholipase D activity in human serum. Ann Clin Biochem. 1995;32:74–78. doi: 10.1177/000456329503200107. [DOI] [PubMed] [Google Scholar]
- 22.McLaughlin F, Ludbrook V J, Kola I, Campbell C J, Randi A M. Characterisation of the tumor necrosis factor (TNF)-α response elements in the human ICAM-2 promoter. J Cell Sci. 1999;112:4695–4703. doi: 10.1242/jcs.112.24.4695. [DOI] [PubMed] [Google Scholar]
- 23.Meri S, Lehto T, Sutton C W, Tyynelä J, Baumann M. Structural composition and functional characterization of soluble CD59: heterogeneity of the oligosaccharide and glycophosphoinositol (GPI) anchor revealed by laser-desorption mass spectrometric analysis. Biochem J. 1996;316:923–935. doi: 10.1042/bj3160923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metz C N, Thomas P, Davitz M A. Immunolocalization of a glycosylphosphatidylinositol-specific phospholipase D in mast cells found in normal tissue and neurofibromatosis lesions. Am J Pathol. 1992;140:1275–1281. [PMC free article] [PubMed] [Google Scholar]
- 25.Metz C N, Zhang Y, Guo Y, Tsang T C, Kochan J P, Altszuler N, Davitz M A. Production of the glycosylphosphatidylinositol-specific phospholipase D by the islets of Langerhans. J Biol Chem. 1991;266:17733–17736. [PubMed] [Google Scholar]
- 26.Morita S, Sato A, Hayakawa H, Ihara H, Urano T, Takada Y, Takada A. Cancer cells overexpress mRNA of urokinase-type plasminogen activator, its receptor and inhibitors in human non-small-cell lung cancer tissue: analysis by northen blotting and in situ hybridization. Int J Cancer. 1998;78:286–292. doi: 10.1002/(SICI)1097-0215(19981029)78:3<286::AID-IJC4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 27.Morris J C, Ping-Sheng L, Zhai H X, Shen T Y, Mensa-Wilmot K. Phosphatidylinositol phospholipase C is activated allosterically by the aminoglycoside G418–2-deoxy-2-fluoro-scyllo-inositol-1-O-dodecylphosphonate and its analogs inhibit glycosylphosphatidylinositol phospholipase C. J Biol Chem. 1996;271:15468–15477. doi: 10.1074/jbc.271.26.15468. [DOI] [PubMed] [Google Scholar]
- 28.Mukasa R, Umeda M, Endo T, Kobata A, Inoue K. Characterization of glycosylphosphatidylinositol (GPI)-anchored NCAM on mouse skeletal muscle cell line C2C12: the structure of the GPI glycan and release during myogenesis. Arch Biochem Biophys. 1995;318:182–190. doi: 10.1006/abbi.1995.1219. [DOI] [PubMed] [Google Scholar]
- 29.Nemoto E, Stohlman S, Dennert G. Release of a glycosylphosphatidylinositol-anchored ADP- ribosyltransferase from cytotoxic T cells upon activation. J Immunol. 1996;156:85–92. [PubMed] [Google Scholar]
- 30.O'Brien K D, Pineda C, Chiu W S, Bowen R, Deeg M A. Glycosylphosphatidylinositol-specific phospholipase D is expressed by macrophages in human atherosclerosis and co-localizes with oxidation epitopes. Circulation. 1999;99:2876–2882. doi: 10.1161/01.cir.99.22.2876. [DOI] [PubMed] [Google Scholar]
- 31.Okazaki I, Moss J. Function of GPI-anchored proteins. In: Young N S, Moss J, editors. Paroxysmal nocturnal hemoglobinuria and the glycosylphosphatidylinositol-linked proteins. San Diego, Calif: Academic Press; 2000. pp. 159–177. [Google Scholar]
- 32.Raabe T, Bukrinsky M, Currie R A. Relative contribution of transcription and translation to the induction of tumor necrosis factor-α by lipopolysaccharide. J Biol Chem. 1998;273:974–980. doi: 10.1074/jbc.273.2.974. [DOI] [PubMed] [Google Scholar]
- 33.Raymond F D, Fortunato G, Moss D W, Castaldo G, Salvatore F, Impallomeni M. Inositol-specific phospholipase D activity in health and disease. Clin Sci. 1994;86:447–451. doi: 10.1042/cs0860447. [DOI] [PubMed] [Google Scholar]
- 34.Rhode H, Lopatta E, Schulze M, Schubert K, Schubert H, Reinhart K, Horn A. Glycosylphosphatidylinositol-specific phospholipase D in blood serum: is the liver the only source of the enzyme? Clin Chim Acta. 1999;281:127–145. doi: 10.1016/s0009-8981(98)00218-6. [DOI] [PubMed] [Google Scholar]
- 35.Ross D T, Scherf U, Eisen M B, Perou C M, Rees C, Spellman P, Iyer V, Jeffrey S S, Van de Rijn M, Waltham M, Pergamenschikov A, Lee J C, Lashkari D, Shalon D, Myers T G, Weinstein J N, Botstein D, Brown P O. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 36.Schütt C. Molecules in focus: CD14. Int J Biochem Cell Biol. 1999;31:545–549. doi: 10.1016/s1357-2725(98)00153-8. [DOI] [PubMed] [Google Scholar]
- 37.Shi M M, Chong I-W, Godleski J J, Paulauskis J D. Regulation of macrophage inflammatory protein-2 gene expression by oxidative stress in rat alveolar macrophages. Immunology. 1999;97:309–315. doi: 10.1046/j.1365-2567.1999.00798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi M M, Godleski J J, Paulauskis J D. Regulation of macrophage inflammatory protein-1α mRNA by oxidative stress. J Biol Chem. 1996;271:5878–5883. doi: 10.1074/jbc.271.10.5878. [DOI] [PubMed] [Google Scholar]
- 39.Smith G M, Biggs J, Norris B, Anderson-Stewart P, Ward R. Detection of a soluble form of the leukocyte surface antigen CD48 in plasma and its elevation in patients with lymphoid leukemias and arthritis. J Clin Immunol. 1997;17:502–509. doi: 10.1023/a:1027327912204. [DOI] [PubMed] [Google Scholar]
- 40.Solter P F, Hoffmann W E. Solubilization of liver alkaline phosphatase isoenzyme during cholestasis in dogs. Am J Vet Res. 1999;60:1010–1015. [PubMed] [Google Scholar]
- 41.Suzuki K, Watanabe T, Sakurai S, Ohtake K, Kinoshita T, Araki A, Fujita T, Takei H, Takeda Y, Sato Y, Yamashita T, Araki Y, Sendo F. A novel glycosylphosphatidyl inositol-anchored protein; a possible role for regulation of neutrophil adherence and migration. J Immunol. 1999;162:4277–4284. [PubMed] [Google Scholar]
- 42.Thieblemont N, Wright S D. Transport of bacterial lipopolysaccharide to the Golgi apparatus. J Exp Med. 1999;190:523–534. doi: 10.1084/jem.190.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsujioka H, Takami N, Misumi Y, Ikehara Y. Intracellular cleavage of glycosylphosphatidylinositol by phospholipase D induces activation of protein kinase Cα. Biochem J. 1999;342:449–455. [PMC free article] [PubMed] [Google Scholar]
- 44.Tsujioka H, Misumi Y, Takami N, Ikehara Y. Posttranslational modifications of glycosylphosphatidylinositol (GPI)-specific phospholipase D and its activity in cleavage of GPI anchors. Biochem Biophys Res Commun. 1998;251:737–743. doi: 10.1006/bbrc.1998.9542. [DOI] [PubMed] [Google Scholar]
- 45.Wilhelm O G, Wilhelm S, Escott G M, Lutz V, Magdolen V, Schmitt M, Rifkin D B, Wilson E L, Graeff H, Brunner G. Cellular glycosylphosphatidylinositol-specific phospholipase D regulates urokinase receptor shedding and cell surface expression. J Cell Physiol. 1999;180:225–235. doi: 10.1002/(SICI)1097-4652(199908)180:2<225::AID-JCP10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 46.Xaus J, Comalada M, Valledor A F, Lloberas J, Lopez-Soriano F, Argiles J M, Bogdan C, Celada A. LPS induces apoptosis in macrophages mostly through the autocrine production of TNF-alpha. Blood. 2000;95:3823–3831. [PubMed] [Google Scholar]
- 47.Xie M, Low M G. Expression and secretion of glycosylphosphatidylinositol-specific phospholipase D by myeloid cells. Biochem J. 1994;297:547–554. doi: 10.1042/bj2970547. [DOI] [PMC free article] [PubMed] [Google Scholar]