Abstract
Mesenchymal stem cells have contributed to the continuous progress of tissue engineering and regenerative medicine. Adipose‐derived stem cells (ADSC) possess many advantages compared to other origins including easy tissue harvesting, self‐renewal potential, and fast population doubling time. As multipotent cells, they can differentiate into osteoblastic cell linages. In vitro bone models are needed to carry out an initial safety assessment in the study of novel bone regeneration therapies. We hypothesized that 3D bone‐on‐a‐chip models containing ADSC could closely recreate the physiological bone microenvironment and promote differentiation. They represent an intermedium step between traditional 2D–in vitro and in vivo experiments facilitating the screening of therapeutic molecules while saving resources. Herein, we have differentiated ADSC for 7 and 14 days and used them to fabricate in vitro bone models by embedding the pre‐differentiated cells in a 3D collagen matrix placed in a microfluidic chip. Osteogenic markers such as alkaline phosphatase activity, calcium mineralization, changes on cell morphology, and expression of specific proteins (bone sialoprotein 2, dentin matrix acidic phosphoprotein‐1, and osteocalcin) were evaluated to determine cell differentiation potential and evolution. This is the first miniaturized 3D‐in vitro bone model created from pre‐differentiated ADSC embedded in a hydrogel collagen matrix which could be used for personalized bone tissue engineering.
Keywords: ADSC differentiation, bone‐on‐a‐chip, in vitro bone model, microfluidics
Abbreviations
- ADSC
Adipose‐derived stem cells
- ALP
Alkaline phosphatase
- ARS
Alizarin Red Staining
- BSP‐2
Bone sialoprotein (2)
- ECM
Extracellular Matrix
- D‐ADSC
Differentiated ADSC
- DMP‐1
Dentin matrix production (1)
- HOB‐c
Human primary osteoblasts
- MSC
Mesemchymal stem cells
- OCM
Osteogenic culture medium
- OCN
Osteocalcein
- PBS
Phosphate buffered saline
- PDMS
Polydimethylsiloxane
- PFA
Paraformaldehyde
- pNPP
p‐Nitrophenyl Phosphate
- U‐ADSC
Undifferentiated ADSC
- 7D‐ADSC
Differentiated ADSC for 7 days
- 14D‐ADSC
Differentiated ADSC for 14 days
1. INTRODUCTION
Bone remodeling is a dynamic and continuous process required for the maintenance of the skeleton architecture which responds to mechanical stimuli. 1 The success of this process requires an equilibrium between bone resorption and formation. 2 , 3 When this equilibrium is disturbed, some pathologies arise such as osteoporosis. Moreover, this imbalance could also be enhanced after the placement of an implant due to the changes in the mechanical conditions, which may end up in an aseptic loosening of the implant. 4
The absence of bone matrix production inside the scaffolds is one of the main limitations in the field of bone tissue engineering. Current bone substitutes do not succeed in recapitulating the unique remodeling capacity of bone tissue yet. Thus, a deeper understanding of cellular behavior is clearly necessary for the main types of bone cells. 5 A wider comprehension of the events that occur during cell‐microenvironment interaction and crosstalk are crucial to improve natural bone formation and final bone remodeling after placing an implant. 3
The development of in vitro systems that recreate bone‐like tissue has two main purposes: understanding the mechanisms of bone formation to improve the bone graft outcome, and replicating in vivo conditions to study specific diseases and potential therapies. 6 In vitro cell studies have demonstrated to be a cost‐effective tool to understand bone regeneration. In general, reliable in vitro models of bone tissue have become a prerequisite for a wider comprehension of the biological mechanisms taking place during bone tissue formation and remodeling. 7 However, the widely used 2D cell culture models have the great disadvantage of not providing a physiological microenvironment for the cells. One of the most relevant technologies adapted in the past years to perform in vitro studies is microfluidics, which offers superior complexity over 2D models and has the advantage of being able to incorporate elements of the natural cellular microenvironment. 8 Furthermore, this technology allows to work with volumes at small scale, 9 which implies a large reduction in dimensions. This involves saving chemical reagents, space, and waste. Furthermore, working with microscale systems allows greater pH and temperature control and versatility that is not possible for macroscale cell studies. 10 , 11 Microfluidic‐based devices are a good approach to evaluate the regulatory effects of single mechanical, chemical, and biochemical cues on cell behavior, which are uncontrollable in 2D in vitro studies. They also have shown potential for reproducing bone tissue‐like models by combining multiple stimuli and, hence, closely mimicking an in vivo‐like environment. 12 Besides, microfluidic platforms allow co‐culturing different cell lines simultaneously and also the long‐term culture of bone cells in a 3D environment (essential for bone tissue culture) using a limited number of cells and reagents that can even be coupled with high‐resolution real time imaging and feedback control. 13 , 14
So far, the literature shows that different bone‐on‐a‐chip models have been created to study various phenomena like bone cells functions, 15 bone regeneration, 16 bone innervation, 17 bone healing, 18 and bone metastases. 19 In addition, in order to recreate physiological cellular microenvironments, bio‐mimetic hydrogels can be introduced inside those microfluidic devices creating 3D natural scaffolds. Importantly, hydrogels allow generating controlled chemical gradients and carrying out long term microscopy tracking and different biochemical assays. Yin et al. showed that compared to 3D cultures larger growth rate inhibition is observed when culturing in the traditional 2D cell monolayers. 20 This fact combined with the aforementioned advantages of microfluidics implies a closer approach to a physiological environment.
Stem cells have unlimited potential in regenerative medicine, tissue engineering and cell therapies because they are able to self‐renew and differentiate into multiple cell lineages providing therapeutic solutions for numerous diseases. 21 Mesenchymal stem cells (MSCs), originally identified in bone marrow, are adult stem cells which can be isolated from almost every connective tissue in the body including adipose tissue, trabecular bone, skin, skeletal muscle, or umbilical cord. 22 , 23
It has been reported that in order to isolate MSCs from the body, large amounts of tissue need to be harvested. 24 Thus, tissue availability and donor site morbidity are key factors to select MSCs sources. Adipose‐derived stem cells (ADSC) fulfill previous preferential selecting factors. Additionally, they show many advantages over other sources used in cell therapy research such as bone marrow‐derived stem cells. Previous studies have demonstrated a multipotent fate of ADSC, hence they can differentiate into, for instance, adipocytes, chondrocytes, myoblasts, and osteoblasts in vitro. 25 , 26 Osteogenic, adipogenic, and chondrogenic differentiation of ADSC can be achieved by the addition of specific supplemented culture media. 21 Although bone marrow derived stem cells have shown the largest osteogenic potential, previous in vitro research on bone tissue demonstrated that osteogenic differentiation of ADSC by the addition of osteogenic culture media (OCM) was achieved in only 14–21 days when using 2D‐cultures. 27 , 28 Even though more information about the properties of ADSC is needed to standardize their clinical applications, all of the aforementioned makes ADSC a valuable stem cell source to study potential therapies in the treatment of bone‐associated pathologies. Moreover, ADSC offer an accessible and plentiful tissue source, easy isolation and culture, and higher proliferation rates than bone marrow derived stem cells. Furthermore, donor site morbidity is considerably lower.
Other authors have also reported bone tissue on a chip by using decellularized bone and bone marrow stem cells (BMSC). 29 , 30 However, unlike the previously mentioned bone‐on‐a‐chip models, here we have used easily harvested human cells together with an easy‐to‐handle and simply manufactured device, which includes an off‐the‐shelf collagen‐based hydrogel being one of the main components of the extracellular matrix (ECM). This would potentially allow the development and translation of a personalized, facile, and cost‐effective bone model. In addition, by using highly proliferative cells like ADSC, incubation time is reduced. We previously studied osteoblast‐osteocyte transition in a cell density dependent experiment and we concluded that high‐cell density is needed to succeed when creating a mature bone network. 31 Manssorifar et al. 17 thoroughly reviewed the current state of the art of bone‐on‐a‐chip models for different applications, but none of them achieve a fully osteogenic differentiation inside a 3D culture starting from MSCs.
Here we hypothesized that ADSC are able to mimic the bone microenvironment and they carry an MSC‐bone cells transition on a bone‐on‐a‐chip collagen hydrogel‐based model. Osteogenic markers as alkaline phosphatase (ALP) activity, calcium mineralization, cell morphology changes and specific proteins expression (Osteocalcein [OCN], bone sialoprotein‐2 (BSP‐2), and dentin matrix acidic phosphoprotein‐1 (DMP‐1)) were evaluated inside the developed microfluidic devices to determine cell differentiation and the evolution of the culture.
In this work, hydrogels made of type‐I collagen were selected because it constitutes the major protein component of natural bone and the matrix for direct bone repair. 12 ADSC at different stages of osteogenic differentiation (D‐ADSC) were used to fabricate in vitro bone models by embedding the culture in a 3D collagen matrix inside microfluidic devices. Although osteogenic differentiation of ADSC has been widely reported, the possibility of using them to introduce an in vitro bone‐on‐a‐chip collagen hydrogel‐based model allows a more physiological scenario than other traditional 3D cultures such as the hanging drop method. Our model would bring all the advantages of microfluidics, specially cell number reduction, control over the microenvironment and promotion of nutrients and biochemicals diffusion through the hydrogel. Besides, the possibility of easily obtaining these cells from a patient's fat deposits makes the use of ADSCs very attractive for personalized bone‐on‐a‐chip models.
The use of this in vitro bone model could constitute an intermediate step between traditional in vitro and in vivo experiments, which would consequently reduce animal testing. Besides, it could be also possible to rapidly test in vitro the effects of different chemical stimuli (e.g., growth factor and high throughput drug testing) over stem cells osteogenesis for individual patients. As far as we know, this is the first 3D‐bone‐on‐a‐chip collagen hydrogel‐based model fabricated from human adipose derived stem cells.
2. MATERIALS AND METHODS
2.1. Study design
Human ADSCs were cultured in osteogenic medium (OCM) for different periods of time (7 and 14 days) in 2D flasks (Figure 1‐left). ADSCs cultured in their regular expansion media were used as comparison. Flow cytometry was used to characterized the 2D cultured ADSCs in terms of cell cycle, viability, apoptosis, necrosis, and osteogenic differentiation (α‐BSP‐2, α‐OCN, and α‐DMP‐1). Additionally, an Alizarin Red staining (ARS) was used to assess mineralization, being a late marker of osteogenic differentiation.
FIGURE 1.
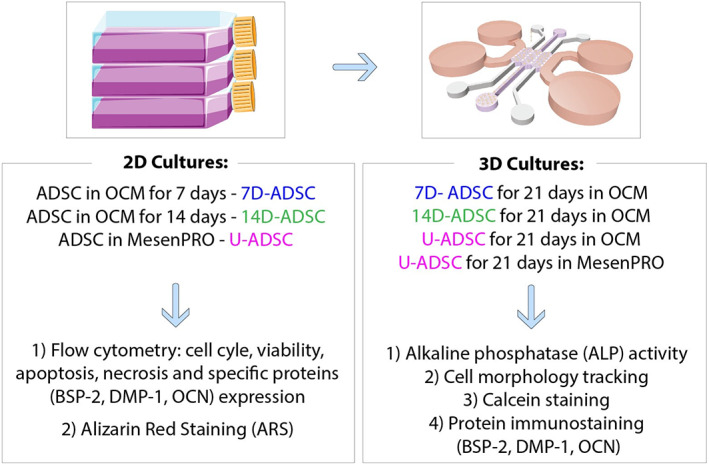
Study design. Cells were incubated and differentiated in osteogenic culture medium (OCM) in 2D flasks: Adipose‐derived stem cells (ADSC) differentiated for 7 days (7D‐ADSC) or 14 days (14D‐ADSC). ADSC in expansion medium (MesenPRO) were incubated as control, undifferentiated cells (U‐ADSC). Results from flow cytometry and ARS were referred to these 2D cultures. D‐ADSC or U‐ADSC were seeded in collagen hydrogels deposited inside the microfluidic device and the rest of the assays were referred to 3D cultures. ARS, Alizarin red staining
ADSCs at different stages of differentiation were cultured in microfluidic devices (see Figure 1‐right and Figure 2), embedded in a collagen type I matrix. Assessment of osteogenic differentiation inside the chip was carried out by: ALP quantification, cell morphology tracking, calcein staining, and specific proteins (α‐BSP‐2, α‐OCN, and α‐DMP‐1) immunostaining. Figure 1 summarizes the study design.
FIGURE 2.
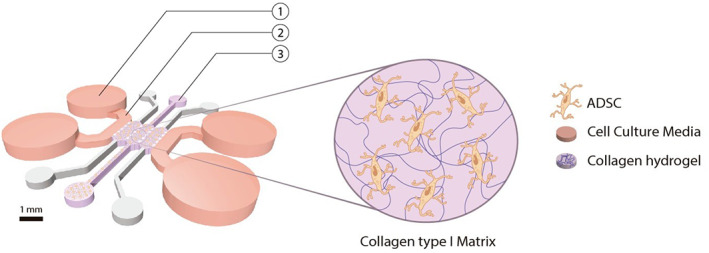
Scheme of the microfluidic device geometry composed of three micro channels. Collagen type I matrix embedding ADSC was enclosed inside the channels through the loading ports (3). The hydrogel was attached to the channels and the culture was hydrated with osteogenic medium through the medium entry ports (1) by diffusion between the chamber and the media channel (2). Cultures were incubated for 21 days inside the device where all the biochemical assays were performed. ADSC, adipose‐derived stem cells
2.2. ADSC differentiation
StemPRO ADSC were purchased from Invitrogen (R7788‐110, US, from lipoaspirate of a 43 years old female) and cultured (2D) under standard conditions (5% CO 2, 37°C) up to 70%–80% confluence in their regular expansion medium MesenPRO RS Medium (Invitrogen, 12,746–012). For cell expansion, cultures were washed with phosphate buffered saline (PBS; Lonza), detached with TrypLE Express (Invitrogen) and plated in T25 cell culture flasks (Thermo Fisher Scientific) at a density of 15,000 cells/cm2. To differentiate the cells, expansion medium was exchanged with OCM, prepared according to the previous bibliography using Dulbecco's modified eagle medium (DMEM) supplemented with: 10% fetal bovine serum (Thermo Fisher Scientific, Gibco, 31,885–023), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM L‐glutamine (all from Lonza); and with osteogenic factors: β‐glycerol phosphate 10 mM, ascorbic acid 50 μM. 32
Differentiation was conducted in T25 culture flasks (Thermo Fisher Scientific) for 7 or 14 days before cell seeding in the microfluidic devices' channels. Differentiated (D‐ADSC) and undifferentiated cells (U‐ADSC) as negative control were embedded at a final concentration of 106 cells/ml in a high density type I collagen hydrogel (6 mg/ml) and seeded inside polydimethylsiloxane (PDMS)‐based microfluidic devices (Section 2.4). 31 This bone‐on‐a‐chip platform enabled a static 3D culture, which was subsequently incubated up to 21 days (Section 2.5 details the 3D cell seeding protocol).
2.3. Flow cytometry
Distribution of cell cycle, viability, apoptosis, necrosis, and specific protein markers (DMP‐1, BSP‐2, and OCN) related to osteogenic differentiation were analyzed by flow cytometry in a FACSARIA BD equipment using the FACSDIVA BD software (Cell Separation and Cytometry Unit, CIBA, IIS Aragon, Spain). D‐ADSC, U‐ADSC (as control) and primary human osteoblasts (HOB‐c, PromoCell, C‐12720, Germany) were studied. HOB‐c is the cell line that we used in our previous bone model 31 and it was also assayed as control to check its closeness with D‐ADSC. HOB‐c are fully differentiated osteoblasts isolated from the femoral trabecular bone tissue of the knee or the hip joint region of a healthy single donor.
Mouse anti‐human primary antibodies (α‐BSP‐2, α‐OCN, and α‐DMP‐1) were purchased from Sigma‐Aldrich and Alexa Fluor 405‐labeled antibodies (Thermofisher Scientific) were used as secondary ones. Cell cycle, viability, apoptosis, and necrosis measurements were also tested by flow cytometry for every cell line to check their loss in the proliferation rate over the differentiation time.
2.4. Microfluidic device fabrication
Bone‐on‐a‐chip devices were in‐house designed and fabricated in poly(dimethlysiloxane) (PDMS) by soft lithography, following the methodology previously described. 33 A commercial product was used to produce the silicone elastomer (Sylgard 184 Silicone Elastomer Kit, Dow Chemical, Germany), which comprises a polymeric base and a silicone resin solution as curing agent. The two liquid parts were mixed in a 10:1 ratio (base: curing agent) and poured in a master made of SU‐8 where the microengineered geometry was patterned by photolithography. Those PDMS‐based devices were bonded to a 35 mm glass (Ibidi, Germany) by plasma treatment (PDC‐32G Basic Plasma Cleaner, Harrick Plasma, NY, USA) under vacuum. They were then coated with PDL (poly‐D‐lysine; 1 mg/ml in PBS; Sigma‐Aldrich, Germany) to enhance collagen matrix adhesion.
A previously reported geometry, developed by our group was selected for this study. 34 The geometry consists on a central culturing chamber divided into three compartments Briefly, the length of each single compartment is 2.5 mm, the width is 1.145 mm, and the average height is 290 μm. The chip uses equally spaced (175 μm) trapezoidal columns to separate the compartments and support the hydrogel via surface tension. A plasma treatment and poly‐D‐lysine (Sigma‐Aldrich, P7886) coating was also needed to ensure collagen attachment to the channels due to PDMS hydrophobicity. Figure 2 offers a sketch of a seeded microdevice.
The features of the device and its geometry allowed us to maintain the 3D cultures for 21 days in their collagen matrix. Meanwhile, daily microscopy tracking is possible and also the cell culture fixation and subsequent staining in the same platform (immunostaining and calcein staining). Thus, we present here a valuable and versatile tool, that at the same time it is easy to manufacture and handle, which allows a complex bone‐on‐a‐chip creation.
2.5. ADSC seeding–bone‐on‐a‐chip
U‐ADSC and D‐ADSC were used at passages 4 to 7 and mixed with the collagen‐based hydrogel solution. The final cell density of 106 cells/ml was based on recent findings on osteogenic differentiation. 31 , 35 Cell laden collagen hydrogels, having a final collagen concentration of 6 mg/ml, were prepared by mixing in an ice bath collagen type I solution (Rat Tail; 10.80 mg/ml; Corning, NY), 10x Dulbecco's phosphate buffered saline (DPBS; Sigma‐Aldrich), 0.5 M NaOH (Sigma‐Aldrich) to adjust the pH to 7.4–7.6, and cells suspended in their expansion medium. In the present work, this collagen concentration of 6 mg/ml was selected due to the similarity of the resulting scaffolds with the natural bone matrix mechanical properties as previously reported. 36 The prior solution was polymerized for 20 min in a humid chamber at 37°C. During the polymerization process, collagen fibers create an interpenetrating polymeric network in the presence of living cells. 37
Approximately 15 μl of hydrogel is introduced inside each device, containing 15.000 cells initially. Once loaded in the bone‐on‐a‐chip devices, cells were 3D cultured in OCM for 21 days. Four timepoints over this period were selected to study culture evolution: at 3, 7, 14, and 21 days from the beginning of the 3D seeding. At each timepoint, three independent samples (n = 3) were studied and the rest of them continued their incubation. A total of four batches were studied: 7D‐ADSC, 14D‐ADSC, U‐ADSC cultured in OCM and U‐ADSC was also cultured in stem cell expansion media (MesenPRO). Every culture was repeated three times to get statistical significance. Figure 1 shows an outline of the experiments performed.
2.6. Hydrogel microstructure visualization: SEM inspection
Scanning electron microscopy (SEM) images were acquired using an Inspect SEM F50 (FEI Company, US) in an energy range between 0 and 30 keV. 6 mg/ml collagen hydrogel was fabricated and polymerized in wells according the aforementioned protocol with no cellular component. Once the polymerization was completed, sample preparation procedure started by a drying stage using different ethanol concentrations in water. Then, samples were frozen separately in liquid nitrogen. Subsequently, samples were submitted to a critical point drying stage (Leica EM CPD300 Critical Point Dryer). Finally, the samples were coated with a carbon film before they were examined by SEM.
The mechanical properties of the collagen fibrous hydrogels used in this study were characterized in previous studies by our group. 36 The collagen hydrogel of 6 mg/ml presented a 0.69 μm ± 0.05 pore size, 90.53% ± 0.93 porosity and 254.05 Pa ± 29.06 storage shear modulus. Here, we imaged the micro‐architecture of the hydrogel by SEM. Finally, Image J specific tools were used to measure the collagen fibers thickness.
2.7. Cell morphology tracking
A Nikon D‐Eclipse C1 microscope was used to image the evolution of cell cultures over time. At each timepoint, microfluidic devices from every experiment were placed on the microscope and several pictures were taken at different magnifications. Changes in morphology were analyzed qualitatively in terms of: protrusion length, number of protrusions, cell body size and connections between cells. All of the mentioned features are commonly used as morphological markers of osteogenic differentiation.
2.8. ALP activity quantification
2.8.1. ALP enzyme presents in the culture
The metalloenzyme ALP initiates the calcification process by providing inorganic phosphates. 38 It is used as a marker for osteoblastic activity, since ALP expression changes over time when osteoblast differentiation occurs. 39 Extracellular ALP activity was measured using a colorimetric assay of enzymatic activity (SIGMAFAST p‐NPP Kit, Sigma Aldrich). It uses pnitrophenyl phosphate (pNPP) as colorimetric substrate that changes its absorbance when dephosphorylated by the presence of ALP.
Cell culture medium (OCM or MesenPRO depending on the batch) was exchanged and sampled after 2 h of incubation after 3, 7, 14, and 21 days of culture. Samples were stored at −80°C and, once the experiment was finished, they were thawed. 40 μl of medium were added to a 96‐well plate in triplicate with 50 μl of pNPP solution. 40 Samples were incubated at room temperature in the dark for 1 h and absorbance was read at 405 nm by spectrophotometry (Synergy HX, Biotek, Instruments, USA). Readings were converted to ALP production using a standard curve, with samples containing no ALP subtracted as background. ALP production was normalized by the DNA content of each sample in order to get comparable estimations of ALP activity between samples with different initial cell seeding density. Thus, results are expressed as mU (ALP‐pNPP dephosphorylated) per mg of DNA in the sample.
2.8.2. DNA content measurement
Proliferation over time was monitored by measuring DNA content via the Hoechst 33–258 kit assay (Sigma‐Aldrich), according to the manufacturer's protocol. Briefly: after medium extraction, cells were isolated by digesting the collagen matrix overnight in a solution of collagenase from Clostridium histolyticum (Sigma‐Aldrich, 2 mg/ml, ≥125 CDU/mg) and centrifuging the cell suspension at 13.000 rpm for 15 min. 50 μl of Hoechst buffer (1 mM EDTA, 10 mM Tris [hydroxymethyl] aminomethane and 0.1 M Sodium Chloride at pH 7.4, all reagents from Sigma‐Aldrich) were added to the pellet. Cells were lysed by applying three cycles of freezing–thawing (−80°C) before running the biochemical assay. Later, 20 μl of cells lysate or DNA standards were suspended in 200 μl of Hoechst dye solution (0.1% vol/vol, Sigma‐Aldrich) and added in a full black 96‐well plate in triplicate. Fluorescence was then measured (excitation/emission: 380/440 nm) using a fluorescence spectrophotometer (Synergy HT Multi‐mode microplate reader, BioTek Instruments, USA). Readings were converted to DNA content using a standard curve, according to the manufacturer's protocol, with samples containing no cells subtracted as background.
2.9. Calcium deposition
2.9.1. Alizarin red staining
Alizarin red is a dye which binds to Ca2+ ions forming a strong red complex. ARS was used as an indicator of calcium deposition in 2D mineralizing cells (in culture flasks, Figure 1 left). 7D‐ADSC, 14D‐ADSC, and U‐ADSC as control were also stained according to the manufacturer's protocol. Briefly, the medium was removed from the culture flasks and cells were fixed in 4% paraformaldehyde (PFA) in PBS for 15 min. Then, fixed cells were washed with deionized water three times for 5 min. ARS staining 1% solution (pH adjusted to 4.2) was added to the cells and incubated at room temperature for 10 min. The stain was removed and the cells were rinsed five times in PBS for 5 min. Cells morphological changes and formation of osteogenic indicative nodules were examined under an inverted phase microscope (Olympus IX‐81).
2.9.2. Calcein staining
Calcein green staining (Sigma Aldrich) was performed to analyze calcium deposition inside the 3D culture (Figure 1 right). As it has been studied before, 41 the staining was conducted at 7, 14, and 21 days of culture without affecting cell viability. Briefly, calcein was dissolved in 0.5 M NaOH solution at 0.25% (wt/vol) and 3D cultures were incubated with the calcein solution (25 mg/ml of calcein solution in OCM 1:100) for 5 days, washed three times in PBS and fixed using 4% (wt/vol) PFA for 20 min. Samples were observed with a Zeiss LSM880 confocal scanning microscope (ex/em 493/515 nm, Microscopy and Imaging Core Facility, Instituto Aragonés de Ciencias de la Salud [IACS]) and maximum intensity images were generated from z‐stacks using ZEN Blue orthogonal projection and NIH ImageJ software.
2.10. Protein immunostaining
Protein staining was conducted inside the microdevice (Figure 1 right). First, medium was removed from the 3D culture after 3, 7, 14, and 21 days and rinsed twice with PBS. All the buffers and solutions were poured inside the device through access medium ports (Figures 1 and 2) and they reached the culture by diffusion through the hydrogel.
First, cells were fixed using 4% PFA for 20 min and, then, rinsed three times in PBS. Samples were subsequently permeabilized for 15 min with 0.1% Triton‐X and blocked overnight with 5% BSA. Later, samples were separately incubated with α‐BSP‐2, α‐DMP‐1, or α‐OCN (all from Sigma Aldrich) monoclonal antibodies at a dilution of 1:100 at 4°C overnight. After washing three times with PBS, samples were incubated for 6 h with Alexa Fluor R 633 goat anti‐mouse secondary antibody at a dilution of 1:200 (Thermo‐Fisher). Cell nuclei and cytoskeleton were then counterstained with DAPI (Invitrogen) and Phalloidin–Tetramethylrhodamine (TRICT, Santa Cruz Biotechnology). Finally, samples were observed with a Zeiss LSM880 confocal scanning microscope. Images were collected using the microscope in sequential mode with a 40× oil immersion lens (lens specification, Plan‐Apochromat 40×, NA 1.3), a bit depth of 16 and a format of 1024 × 1024 pixels. The confocal pinhole was 1 Airy unit. We hypothesized that results from confocal microscopy would support flow cytometry results considering that the diffusion and protein release mechanisms are quite different in 2D (cell growth and differentiation before cytometry testing) and 3D cultures (samples stained in the chip).
2.11. Statistics
All experiments were conducted in triplicates. Matlab programming language was used to run all statistical analyses. We used the ANOVA test to assess significant differences between time points and the pair‐wise multiple comparison procedure using the Turkey's HSD test. p‐value <.05 was considered as significant.
3. RESULTS AND DISCUSSION
Countless previous works have highlighted the relevance of MSCs in bone tissue engineering. 42 Adult stem cells include BMSC, ADSC, and dental pulp stem cells. BMSCs are most frequently used in bone tissue engineering for their high‐osteogenic differentiation potentials. Nevertheless, the harvesting of BMSCs is an unpleasant procedure and their proliferation rates are generally low. 24 For an alternative, adipose tissue is particularly attractive because of its minimal extraction risk from lipoaspirates. ADSC have high‐proliferation rates, immunosuppressive properties and secrete numerous polypeptides, hormones, and potent growth factors to stimulate angiogenesis and osteogenesis. 43 In view of all these properties, ASCs are used for bone tissue regeneration in clinical trials. However, more information about the properties of ASCs is needed to standardize their clinical applications.
Here we hypothesized that pre‐differentiated ADSC, which can maintain self‐renewal, are abundant and accessible stem cell sources and may create an in vitro bone‐on‐a‐chip model which closely mimics bone tissue matrix. This is achieved thanks to the hydrogel matrix, composed of type‐I collagen, which is the major protein component of bone. Moreover, this bone‐on‐a‐chip model shows the ability to use a low number of cells. For this study we chose commercially available primary human cells (details in Section 2.2) as a model in our proof of concept to validate our hypothesis.
The selected geometry has been designed in our group and previously reported to quantify chemotaxis of human dermal fibroblasts in 3D 34 and to study osteoblast‐osteocyte transition in a cell density‐dependent manner. 31 Stability of the hydrogel, which needs to be attached to the micro‐columns, requested this geometry having three hydrogel‐channels instead of only a single one. This allows a static 3D culture in a controlled environment, making possible a differentiation process of the culture. In addition, this kind of devices allow easy microscopy tracking in real time without the need of complex external structures while maintaining the sterility in the sample. Finally, despite the simplicity of the design, it is possible to conduct in situ cell culture fixation and the subsequent specific staining.
In order to create the bone‐on‐a‐chip model, osteogenic differentiation of the ADSC was conducted through incubation in OCM for 7 and 14 days. These cells were then embedded in collagen to create the 3D culture of differentiated ADSC inside the microfluidic devices. To determine the development of mature bone formation from ADSC at different stages of differentiation, we investigated the major aspects underlying osteogenic differentiation: cell morphology variation, matrix mineralization, and protein synthesis.
MSCs osteogenic differentiation is a multi‐step process. In the first stage (5–14 days), the expression of ALP increases. Then, the expression of other proteins such as OCN and BSP‐2, is accompanied by a downregulation of ALP. At the final stages of osteogenic differentiation into osteocytes, the cell culture expresses proteins such as DMP‐1 and osteopontin, 43 , 44 together with some changes in cell morphology such as dendrites creation, inducing changes in cytoskeleton as well as a reduction in the number of organelles. 45 All of those aforementioned markers were selected to determine the state and evolution of the culture.
3.1. Stem cells differentiation and maturation
Figure 3 describes a comparison between the four cell batches analyzed (HOB‐c, U‐ADSC, 7D‐ADSC, 14D‐ADSC) in terms of cell viability (A), cell cycle (B) using 2D incubated cells in T‐25 flasks and selected bone markers expression of the same cellular batches (C). Numerical data from a single flow cytometry experiment were compiled into Tables 1, 2, 3.
FIGURE 3.
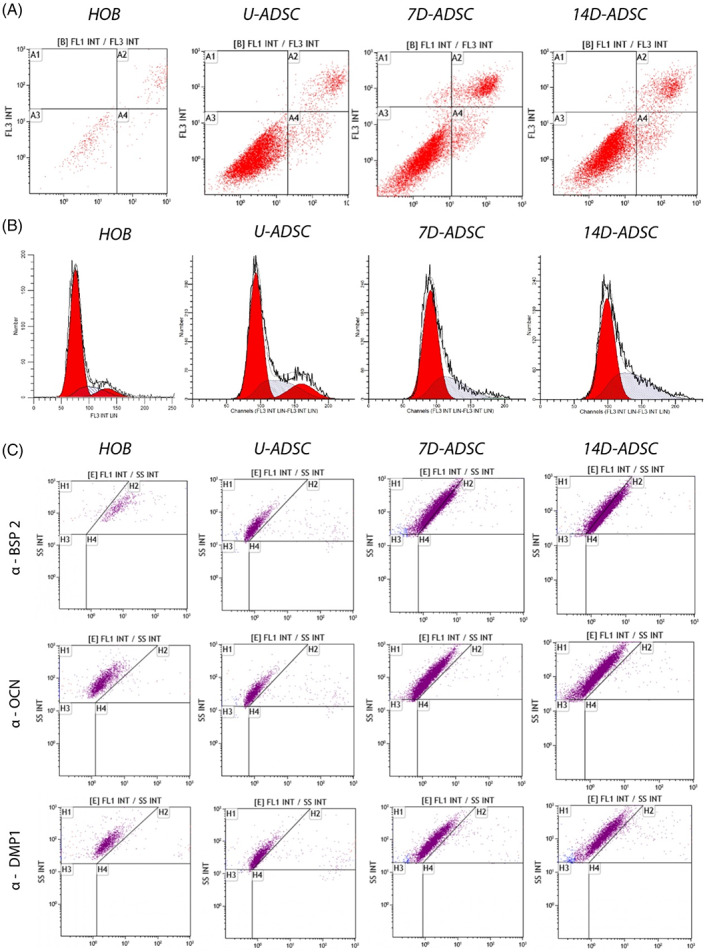
Flow cytometry of different cell lines: Human primary osteoblasts (HOB‐c), undifferentiated ADSC (U‐ADSC), and ADSC differentiated for 7 and 14 days (7D‐ADSC and 14D‐ADSC, respectively). (A) cell viability, apoptosis, and necrosis and (B) cell cycle and (C) osteogenic markers expression (BSP‐2, OCN, and DMP‐1)
TABLE 1.
Quantitative data from the cell viability experiment conducted on the different cell lines: Human primary osteoblasts (HOB‐c), undifferentiated ADSC (U‐ADSC), and ADSC differentiated for 7 and 14 days (7D‐ADSC and 14D‐ADSC, respectively)
| Parameter | HOB‐c | U‐ADSC | 7D‐ADSC | 14D‐ADSC |
|---|---|---|---|---|
| % Cell viability | 67.28 | 90.88 | 75.88 | 71.02 |
| % Necrosis | 0.13 | 0.07 | 1.33 | 3.24 |
| % Early apoptosis | 6.60 | 3.45 | 6.53 | 3.75 |
| % Late apoptosis | 25.99 | 5.61 | 16.27 | 21.99 |
TABLE 2.
Quantitative data from the cell cycle experiment conducted on the different cell lines: Human primary osteoblasts (HOB‐c), undifferentiated ADSC (U‐ADSC), and ADSC differentiated for 7 and 14 days (7D‐ADSC and 14D‐ADSC, respectively)
| Parameter | HOB‐c | U‐ADSC | 7D‐ADSC | 14D‐ADSC |
|---|---|---|---|---|
| % G1 | 75.13 | 76.70 | 80.44 | 84.78 |
| % S | 4.04 | 10.6 | 9.03 | 4.88 |
| % G2 | 20.83 | 12.70 | 10.53 | 10.34 |
TABLE 3.
Quantitative data from the osteogenic markers expression conducted on the different cell lines: Human primary osteoblasts (HOB‐c), undifferentiated ADSC (U‐ADSC), and ADSC differentiated for 7 and 14 days (7D‐ADSC and 14D‐ADSC, respectively)
| Parameter | HOB‐c | U‐ADSC | 7D‐ADSC | 14D‐ADSC |
|---|---|---|---|---|
| % α BSP − 2 | 25.17 | 5.29 | 6.13 | 29.51 |
| % α OCN | 15.14 | 1.29 | 1.83 | 14.53 |
| % α DM P − 1 | 8.99 | 1.37 | 2.23 | 7.59 |
ADSC presented higher viability than HOB‐c, with a small percentage of apoptotic cells and almost no necrotic cells. Meanwhile, incubation of differentiated cells and HOB‐c in a 2D culture showed an increased number of necrotic and apoptotic cells and a lower percentage of viable cells. As ADSC were differentiated, viability decreased mainly due to apoptosis, but in all cases percentage viability of ADSC was above 70% (Table 1 and Figure 3A).
Cell cycle analysis also confirmed that U‐ADSC and D‐ADSC showed higher proliferation rates (i.e., enhanced S‐phase) than HOB‐c (Table 2 and Figure 3B). Slight differences were also observed during differentiation where an increase in the number of cells in G1 phase was seen which is indicative of an increased protein and mRNA synthesis needed for the subsequent DNA replication and cell division with differentiation time, while the percentage of cells in S‐phase decreased, indicating a delay in G1/S transition. Yin et al. 20 also showed that during differentiation in 2D cultures ADSCs underwent replicative senescence, and cell growth was reduced, but also they showed that the decline in 2D culture was greater than when using 3D cultures based on hydrogel matrices. They confirmed that 3D culturing exerted positive effects in delaying senescence in ADSCs. Also, we observed that the S‐phase decreased over differentiation time (Figure 3B) which is in agreement with previously reported data where undifferentiated stem cells own a high‐proliferation rate that decreases when differentiation takes place. 46 , 47
BSP‐2 is a non‐collagenous protein upregulated by osteoblasts during the tissue mineralization phase and downregulated when osteocyte differentiation occurs. DMP‐1 is an ECM protein associated to osteocytes. OCN is a bone‐specific protein synthesized by osteoblasts that represents a good marker for late osteogenic maturation. 45 , 48 Thus, OCN, BSP‐2 and DMP‐1 markers expression using both flow cytometry and immunofluorescent staining can determine the osteoblastic and osteocytic phenotypes.
It was observed that ADSC incubated for 14 days in OCM showed specific osteogenic marker proteins in the same manner that primary osteoblasts did (Table 3 and Figure 3C). 7D‐ADSC also showed markers expression, which was higher than U‐ADSC in OCM. However, as it is detailed later, the expression of those markers was not so clear when using 3D cultures. ADSC culture evolution differences in 2D and 3D have been previously studied. 20 Unlike the aforementioned work, our 3D culture presented a fibrous collagen matrix inside a thoroughly controlled environment thanks to the use of microfluidics. This model might replicate the physiological matrix closer than traditional 3D cultures, as the ones performed in well‐plates or the hanging‐drop models.
Finally, the last outcome that can be noticed from Figure 3 is the increased number of viable cells (see also Table 1). Despite both HOB‐c and ADSC lines being primary cells and used at the same passages, it is clear that there were much fewer cellular events measured in the flow cytometry tests when assaying osteoblasts than stem cells; independently of whether they were differentiated or not. This can be attributed to the differences in the proliferation rates between osteoblastic human cells and stem cells. This conclusion also highlights one of the main strengths of using stem cells in tissue engineering research and regenerative medicine.
3.2. Collagen network mimics bone extracellular matrix
Bone ECM consists mainly of collagen type I (90%) and non‐collagenous proteins (10%). 49 In our model, we mimic the bone ECM thanks to the fabrication of a high‐density collagen type I hydrogel and the specifically supplemented culture medium (OCM). Samples of the hydrogel with no cells were imaged by SEM to observe the hydrogel micro‐ and nano‐structure.
Figure 4 shows representative SEM images of the non‐seeded collagen hydrogel used in our bone‐on‐a‐chip model. As it can be seen, a porous matrix is achieved, with a wide range of pores sizes. It can be seen a heterogeneous and fibrous network with a large superficial area for cells to attach. Image J specific tools allowed the measurement of the diameter of the collagen fibers (Figure 4B), and the values varied between 60 and 90 nm. The presence of nano and micro patterns in our collagen matrix is very important as these appear in the natural ECM and cells in tissues are naturally in contact with nano and micro features. 50
FIGURE 4.
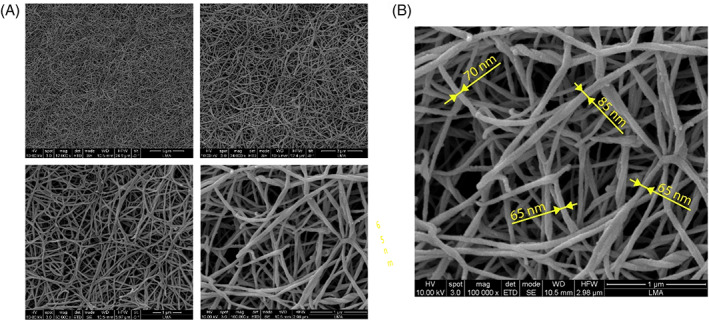
Micro‐ and nano‐structure of collagen hydrogel: (A) Representative area of 6 mg/ml collagen hydrogel at different magnifications (12.000×, 24.000×, 50.000×, and 100.000×), (B) 100.000× magnification image highlighting collagen nano‐structure
The concentration of the hydrogel used here was 6 mg/ml of collagen. However, collagen density is a tunable value: it can be diluted or concentrated depending on the final mechanical and rheological properties required. 36 For instance, for in vitro creation of endothelial vessels, Pérez et al. 51 compared two different collagen hydrogel concentrations (2.5 and 6 mg/ml) and showed that this directly affects the integrity of endothelial vessels, which decreases when high‐concentration hydrogels were used. This fact makes collagen hydrogels a versatile tool for different fields of tissue engineering. This collagenous matrix has been used to study, among others, osteoblasts migration, 12 or cancer dissemination, 52 but this is the first time they act as an osteogenic differentiation substrate for MSC.
3.3. Changes in morphology by 3D network creation
The literature shows that cell morphology is a prime property of cells because changes in cellular function, including cell phenotype, cause morphological variations, and vice versa. 53 , 54 When osteogenic differentiation of stem cells occurs, the cellular morphology changes from the characteristic fibroblastic monolayer to a multilayered fibroblastic culture. As cells differentiate, the organization of cytoskeletal elements leads to these morphological changes. A marked change in the organization of the cytoskeleton was previously observed in ADSCs during osteogenic differentiation which is attributed to the assembly and disassembly kinetics of actin microfilaments. 55
We use brightfield images to show the culture morphology evolution over time. Figure 5 pictures cell culture morphological changes (at a 10× magnification) in all cell lines along the different incubation days. Individual images belong to the central picture of a brightfield full z‐stack taken through the whole depth of the gel. On the other hand, Figure 6 shows the cultured collagen hydrogel attached to the three micro channels and cellular connections at different magnifications. Cells three‐dimensionality appreciated in the pictures also revealed the success in the 3D seeding protocol and the long‐time culture feasibility. Apart from that, confocal images obtained to target calcium deposition (Figure 7) also show changes in single‐cell morphology.
FIGURE 5.
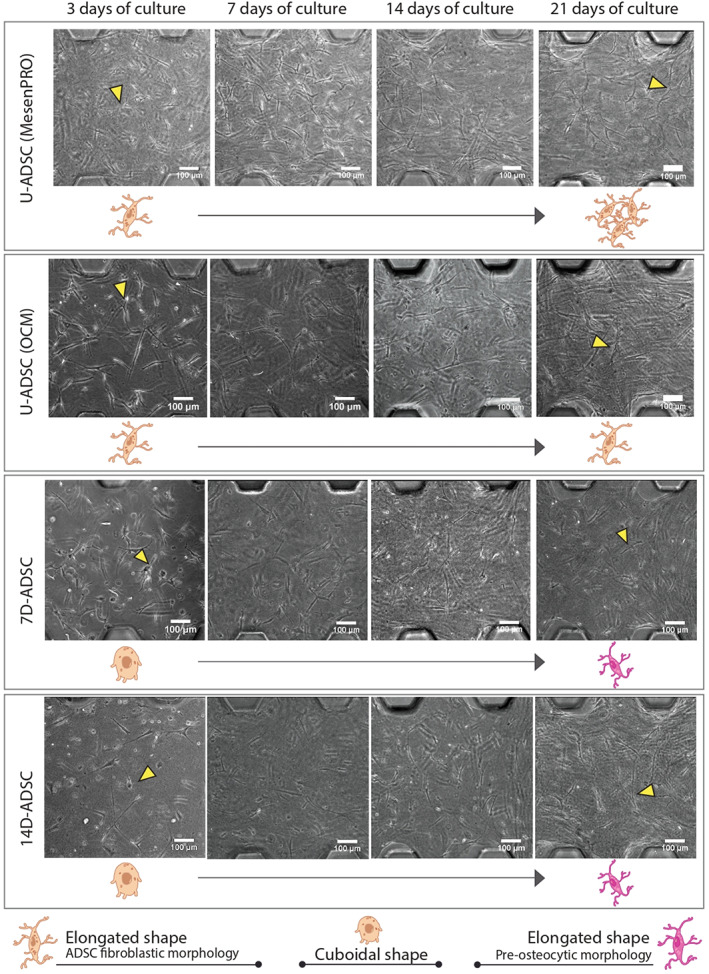
Comparison of ADSCs morphology at four timepoints and different states of differentiation (7D‐ADSC, 14D‐ADSC, and U‐ADSC as control incubated with both culture mediums: osteogenic (OCM) and regular for ADSC incubation (MesenPRO) inside the bone‐on‐a‐chip models using brightfield microscopy. Schemes of cell morphology evolution are shown underneath the microscopy images and yellow arrows point out an individual example. Individual scale bars: 100 μm. ADSC, adipose‐derived stem cells
FIGURE 6.
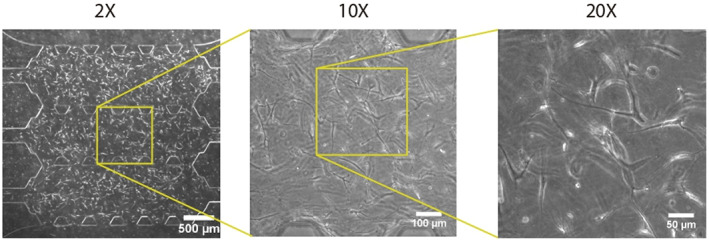
Brightfield microscopy image of cell‐embedded collagen hydrogel attached to the three micro channels at different magnifications: 2×, 10×, and 20×
FIGURE 7.
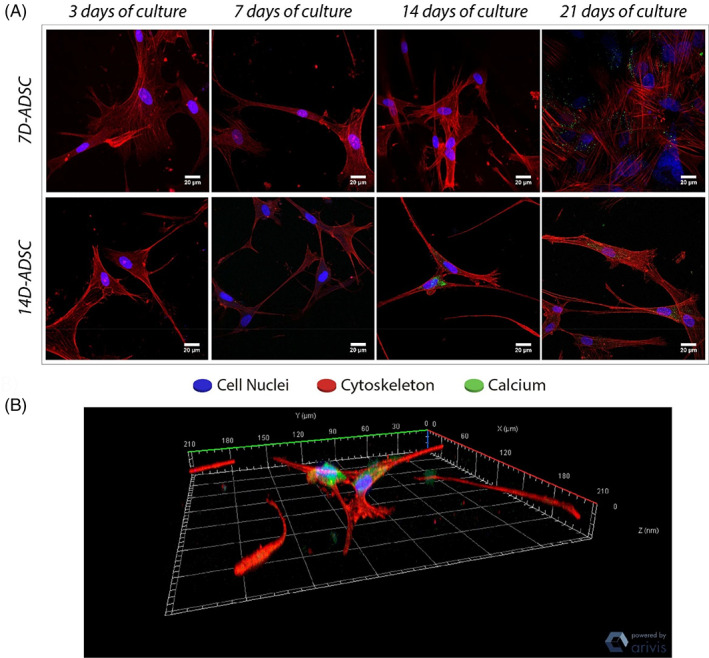
(A) Calcium deposition on collagen type I matrix. Individual scale bars: 20 μm. (B) Cell–cell connection 3D reconstruction was done automatically from a confocal z‐stack image thanks to the Zen‐Blue software
It can be observed (Figure 5) that U‐ADSC incubated in MesenPRO (control) maintained their elongated fibroblastic shape over the culturing time when using 3D arrangements. Besides, we could observe an increase in the number of cells and cell connections. Similarly, U‐ADSC incubated in OCM also maintained their shape over the culture period. This is due to the fact that osteogenic differentiation was not fully achieved inside the 3D culture, as it is confirmed along the results section. If ADSC became osteoblasts in these conditions, a morphological change from elongated toward a rounded shape would be expected. On the other hand, D‐ADSC showed in the 3D culture a rounded morphology, as it was expected due to the previous differentiation period. Over the culturing time, cells changed their morphology toward an elongated configuration, exhibiting dendrites and protrusions; typical osteocytic feature. Pictures also revealed that 14D‐ADSC (which presented major osteogenic phenotype) are more elongated and dendritic than 7D‐ADSC at the end of the culturing time.
In agreement with our results, Mcbeath et al. 56 demonstrated that spread cells tend to differentiate into osteoblasts whereas round cells tended to differentiate into adipocytes by using the micro‐patterning technique. This fact was attributed to the activation of the RhoA‐ROCK signaling, which was triggered by the actin‐myosin‐generated tension. Regarding the number of protrusions and length, no qualitative changes were observed due to the fibroblast like shape of D‐ADSC. 55 However, in our previous study in a 3D HOB‐c culture, a significant increment in both protrusion length and number of protrusions was quantified when high‐cell density was seeded. 31 However, several qualitative conclusions were obtained from morphology tracking microscopy in this work: every cell line assayed here shows more connections and a larger number of elongated cells over time. Differentiated cells (7D‐ADSC and 14D‐ADSC) held a wider morphology, which is an osteoblastic feature, 28 at the beginning of the experiment. This initial cubical shape changed over time and cells turned elongated. Going through U‐ADSC (both in OCM and MesenPRO) pictures we can appreciate an increased number of cells over time, which means they were still proliferating. Other authors also tracked cell protrusions as osteogenic indicator. Ochiai‐Shino et al. 57 studied human‐induced pluripotent stem cells differentiation in monolayers and one of the selected markers was cell morphology. They observed a change from cuboidal to dendritic shape in the cells by SEM. Other works studied the effect of the microarchitecture of the scaffold on cell morphology and its implications in human MSCs osteogenic differentiation. 58
Morphology changes are easily detected thanks to different microscopy techniques. Besides, microfluidic devices as the ones presented here allow a live culture follow up with no need of culture fixations. Researchers are able to distinguish different cell phenotypes depending on cell morphology in both 2D and 3D cultures. All of the above thanks to the close relationships between cytoskeleton shape and cell function. 56
3.4. Calcium deposition and matrix mineralization
Calcein is a fluorochrome binding calcium at the bone mineralization front, 59 thus identifying newly mineralized ECM. Figure 7 show confocal images of calcein stained samples fixed at 7, 14, and 21 days of culture. Calcein staining at 3 days of incubation was not feasible because calcein solutions needed 5 days of incubation previous to calcium fixation. However, we hypothesize that no calcium ions were deposited at that timepoint.
Matrix mineralization can be observed by confocal microscopy as stained in green color (Figure 7A), which is observed in 7D‐ADSC images at 21 days of culture and in 14D‐ADSC at 14 and 21 days of culture. Individual pictures belong to orthogonal projections to a full z‐stack confocal image. The figure also shows a 3D reconstruction of a cell–cell connection of the z‐stack which exhibits the cells' union through their dendrites (Figure 7B). This calcium staining showed that D‐ADSC were able to mineralize the ECM. 7D‐ADSC began mineralization at the end of the culture period (picture at 21 days) meanwhile 14D‐ADSC started matrix calcification after 14 days of culture.
ARS has also been widely used to confirm osteogenic differentiation in ADSC in the past. 60 , 61 , 62 ARS performed in 2D culture flasks on U‐ADSC, 7D‐ADSC, and 14D‐ADSC (Figure 8) revealed similar conclusions than calcein staining did: 14D‐ADSC culture released more calcium deposits than 7D‐ADSC cultures did. The same staining procedure was performed on U‐ADSC which did not show any staining. Cells growing superficially release calcium ions, which are in charge of future collagen matrix mineralization. As it was expected, after 14 days of differentiation, there was a superior calcium ion release. Thus, 14D‐ADSC line had more matrix mineralization potential once cultured in 3D than 7D‐ADSC. This is why, calcein stained samples were observed sooner.
FIGURE 8.
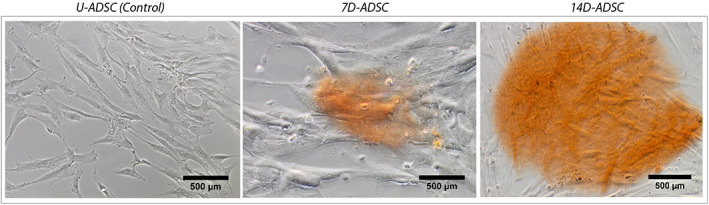
ARS T‐25 culture flask of cell cultures (2D): U‐ADSC (as control), 7D‐ADSC and 14D‐ADSC. Olympus IX‐81 inverted phase microscope was used at 20× magnification. ADSC, adipose‐derived stem cells; ARS, Alizarin red staining
Other authors have also studied calcium evolution in 3D cultures to evaluate matrix mineralization. McGarrigle et al. 35 quantified calcium content in mouse‐like cell lines cultured in 24 well plates and saw that calcium content rapidly increased accompanied by an ALP down‐regulation and DMP‐1 release to the matrix. Similarly to our study, microfluidic bone models using human osteoblasts at high cellular density also showed collagen matrix calcification at the end of the culture, 31 although no ARS analysis was conducted to observe initial ions. Tenstad et al. 63 used ARS and Oil Red staining inside microfluidic devices to determine whether iMSC3 (a human telomerised bone marrow mesenchymal cells) differentiate towards osteogenic or adipogenic linage, respectively showing that an adequate long‐term microfluidic cultivation is possible in those devices.
3.5. ALP activity reveals ADSC osteogenic differentiation
The transition from a proliferative to a quiescent phase during osteogenic differentiation is linked to the upregulation of ALP and its later reduction. 26 , 64 In bone tissue engineering, up‐regulation of ALP expression is related to osteogenesis being a precursor of tissue mineralization. 38 , 65 To assess ALP cells secretion during the experiments, we quantified the evolution of the enzymatic activity in the culture medium.
Figure 9 A compares ALP expression over time for the different cell cultures. 14D‐ADSC had the highest expression of ALP enzyme showing a maximum at the beginning of the culturing time (p = .0003 comparing day 3 and 7 of culture) and continued downregulating while bone cells were getting mature. Once a fully osteogenic differentiation was achieved, ALP expression did not present significant differences (p = .9228 and p = .7622 comparing 7–14 and 21 days of culture). 7D‐ADSC presents the same pattern but the highest production occurred later owing to the initial maturation state of the culture (p = .0134 between days 7 and 14 of culture). After the maximum, enzymatic activity was reduced and kept stable (p = .7547 between 14 and 21 days of culture). This ALP expression pattern matched with the expected one from previous studies performed in primary human osteoblasts, 31 where they showed this maximum expression after 7 days of culture. The peak value in the expression was comparable with the one expressed by 14D‐ADSC at 3 days of culture (±26 mU/mg DNA for primary human osteoblasts at day 7 and ± 20 mU/mg DNA for 14D‐ADSC at day 3). However, 7D‐ADSC showed exactly the same pattern of expression, but the maximum of enzymatic activity value presented with differentiated stem cells was lower (±17 mU/mg DNA). The activity of ALP depends on the maturation state during new bone formation. In a first stage proliferation and extracellular matrix formation take place, then the extracellular matrix maturation is accompanied by an increase in ALP activity. Finally, after mineralization ALP activity decreases which is in agreement with our results.
FIGURE 9.
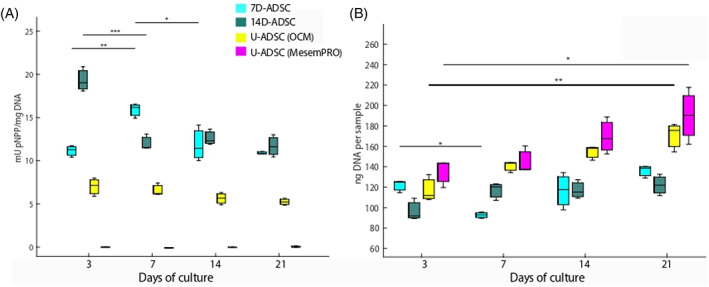
ALP activity (A) and DNA content evolution over time (B) at different timepoints. Three samples for each group were collected at each timepoint. Then, every experiment was conducted three times (n = 9). ANOVA + post hoc. Turkey‐Kramer test type with statistical significance: *p < .05, **p < .01, ***p < .005. ALP, alkaline phosphatase
As it was expected, U‐ADSC incubated in MesenPRO did not express enzymatic activity. ALP expression value was almost non‐existent and did not present any significant change over time (p = .4476). U‐ADSC incubated in OCM showed slight ALP expression. Despite of the slight marker expression, enzyme release pattern did not match with the expected one for differentiating cells and did not show significant changes over time (p = .0834). Thus, in order to fabricate an in vitro bone model from human stem cells, a previous differentiation period was required. Here, we measured the temporal evolution of ALP activity for different cell lines during the whole culturing time. So, we suggest that differentiated ADSC cultured in a 3D natural collagen scaffold had a phase of increased mineralization at the beginning of the culture (day 3 for 14D‐ADSC and day 7 for 7D‐ADSC), which later continued toward their total differentiation process. Thus, it can be seen that 14D‐ADSC exhibit the highest differentiation potential. In addition, it was observed that a pre‐differentiation period was necessary to obtain the enzymatic activity.
ALP is a widely used marker for osteogenesis which helps to study whether some chemical (e.g., growth factors application) or mechanical stimuli (e.g., flow or substrate stiffness) may affect osteogenic differentiation. Previous MSCs models have been developed on monolayers (2D cultures). For instance, Angle et al. 66 monitored ALP activity of rat bone marrow stromal cells monolayers to study the effects of low‐intensity pulsed ultrasound exposure on stem cell osteogenesis. The cellular monolayer showed an ALP maximum expression when cells started to differentiate and afterwards, enzyme expression declined. This tendency agreed with our results for 7D‐ADSC and 14D‐ADSC. Other studies in microfluidic devices compared ALP values of a monolayer under dynamic hydraulic pressure. 67 Those authors determined that mechanical stimulation increased the production of osteogenic matrix components. Wang et al. 68 also proved that bone morphogenetic proteins enhance a faster and more efficient osteogenesis because ALP activity was five times higher than the one of control cells after 14 days of monolayer incubation.
Others have studied reinforced natural hydrogels to observe the effect of the matrix nature on bone marrow stem cell differentiation. Hasany et al. 69 worked on alginate and hyaluronic acid hydrogels supplemented, for instance, with nanoclays. They conducted ALP activity evaluation of different gel combinations composed of alginate, hyaluronic acid, and two‐dimensional nanoclays, and expression curves after 3 weeks also showed a maximum at 14 or 21 days (depending on the hydrogel), matching with the curves presented in this work.
Increasing the scale of the experiment, MSCs have been cultured in 3D scaffolds. Garcia‐Gareta et al. used bone marrow MSCs on titanium scaffolds and they measured their differentiation by ALP quantification. 70 ALP expression of 3D constructs followed the same pattern than the one in this work. Former in vivo experiments carried out with MSCs have been performed and they can be distinguished into one‐step processes or multi‐step processes depending on whether they have and in vitro differentiation pre‐treatment or not. 71
Yang et al. used rat D‐ADSC with 12 days of previous incubation is OCM. 72 They measured OCN production and matrix mineralization, confirming osteoblastic differentiation and induced bone formation in vitro. Besides, those authors also revealed that implantation of D‐ADSC could induce new bone formation in vivo. They also found direct U‐ADSC applications in in vivo experiments for bone‐defect treatments. Calabrese et al. compared the osteogenesis around collagen‐hydroxyapatite scaffolds with and without human‐ADSC in mice for 2, 4, and 8 weeks. 73 Several markers were evaluated, including ALP activity, OCN production and scaffold mineralization among others. At the end, those authors determined that the ADSC addition to the scaffolds previous to mice implantation enhanced mineralization, osteogenesis, and angiogenesis. In our 3D microfluidic model, concordant results were obtained with no need of animal experimentation. Thus, those microfluidic platforms may be used as an intermediary step between in vitro and in vivo research.
3.6. DNA content in the samples
As it has been reported in the literature, undifferentiated stem cells own a high‐proliferation rate; which decreases when differentiation takes place. 74 , 75 Our results also support this finding as it was observed in both cell cycle analysis and DNA content evaluation in the microdevices over time.
At the beginning of every culture, the same amount of DNA would be expected considering that the same cell density was seeded. However, an exact number of cells cannot be established at the beginning owing to DNA amount differences in every cell because it depends on its origin and status. Figure 9 (right) compares the amount of genetic material in ng of DNA per sample over the four timepoints studied during the course of the experiment. DNA content of U‐ADSC incubated in MesenPRO increased over the timepoints (p = .0181). These outcomes match with cell cycle conclusions from flow cytometry and confirm that cell proliferation rate decreases when stem cells acquire their final phenotype. U‐ADSC incubated in OCM presented also DNA increase (p = .0013) because they were not getting a fully osteogenic phenotype. On the other side, differentiated cells showed no significant differences in DNA content at days 3 and 21 of culture (p = .3773 for 7D‐ADSC batch and p = .1047 for 14D‐ADSC batch).
Other authors have studied osteogenic markers normalized according to the DNA content in 2D monolayer cultures 76 , 77 and 3D cultures. 31 , 35 McGarrigle et al. (2016) studied osteogenic differentiation depending on the substrate stiffness in a 3D mouse cell line culture. 35 They found that culture DNA content grew fast and constantly during 56 days. However, this fact occurred due to the non‐apoptotic behavior of MC3T3‐E1 cells. Similar to our work, DNA content measured in HOB‐c was slightly reduced over time along the experiment when seeding with high cell density but remained constant when seeding low cellular density. 31 The proliferation rate of osteogenic mature cells was low, especially in 3D cultures as the one just mentioned. In this work, the same density as HOB‐c was seeded at the beginning of the culture (106 cells/ml). Results indicated that the tendency observed in DNA content in the D‐ADSC batches studied here (both 7D‐ADSC and 14D‐ADSC) followed the expected trend due to the mature osteogenic cellular behavior.
3.7. Immunostaining reveals osteogenic markers in 3D
As flow cytometry confirmed, we succeeded in differentiating ADSC into an osteoblastic phenotype after 14 days of culture in OCM. Our experiments consisted on 21 days of subsequent incubation (in 3D) in microchannels hydrated with OCM. Thus, at the end of the culture time, osteogenic markers expression from all experiments (but negative control) could be expected. However, U‐ADSC incubated in OCM did not show the markers expected in osteogenic differentiation medium. This fact may be attributed due to the differences between the diffusion coefficient in 2D culture flasks and inside a high‐density 3D matrix. 20
Confocal images showed the expression of bone specific markers for both differentiation periods. DMP‐1, as a marker for osteocytes, was only observed in 3D for 14D‐ADSC at 14 and 21 days of culture (Figure 10). BSP‐2 was detected for 14D‐ADSC (first stages of the culture: 3 days) and 7D‐ADSC (by the end of the culture: 21 days); determining osteoblastic features. OCN was slightly detected for both batches; which also matches with flow cytometry results given that OCN presented less expression than BSP‐2 as osteoblastic marker. Negative control ensured that the specific antibody bindings and stained samples cultured with U‐ADSC did not show any of the bone markers. Figure 10 shows orthogonal projections of the most revealing single pictures. To observe the full evolution of protein release, please see Figures S1 and S2 in the supplemental information section.
FIGURE 10.
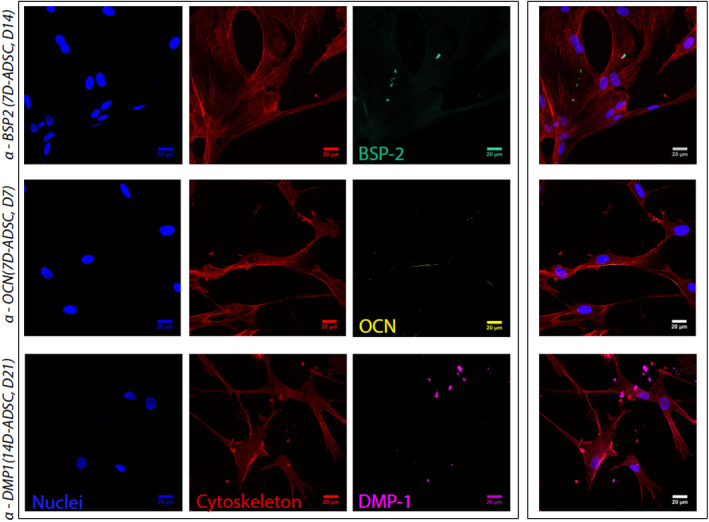
Orthogonal projection of Z‐stack confocal pictures. 3D cell culture was stained with DAPI and Phalloidin to observe cell nuclei and cytoskeleton, respectively. Specific protein secretion was targeted by immunostaining. BSP‐2 stained sample from 7D‐ADSC at 14 days of culture, OCN stained sample from 14D‐ADSC at 7 days of culture, DMP‐1 stained sample from 14D‐ADSC at 21 days of culture. Individual channels are shown together with the merged image, all of them at 40× magnification (scale bar 20 μm). ADSC, adipose‐derived stem cells; OCN, osteocalcein
According to the bone markers expression assayed with growing cells in 2D, ADSC 2D culture expressed osteogenic markers at 14 days of differentiation, especially BSP‐2 (immature bone cells, Figure 3). This batch of 14D‐ADSC cultured in microdevices expressed BSP‐2 at the beginning of the 3D culture, but they finally expressed DMP‐1 as a sign of complete osteogenic differentiation. In addition, 2D culture of ADSC did not express bone markers after 7 days of differentiation (Figure 3). After seeding 7D‐ADSC batches in 3D, no markers were detected at the beginning of the culture, but finally BSP‐2 and OCN were observed, probing the osteoblastic behavior of these cells. Consequently, it was anew observed that a pre‐differentiation period for the ADSC is needed to obtain osteogenic phenotype in the 3D culture. Besides, only 14D‐ADSC expressed DMP1, a late osteogenic marker.
Many authors usually performed this kind of staining using 2D cultures. For instance, Oh et al. stained a monolayer of MSCs to track osteopontin secretion and determined osteogenic differentiation under the presence or absence of other markers (OCN, BSP‐2, and ALP) by real time PCR. 78 However, when immunofluorescence needs to be performed in a physiological scenario, 3D staining protocol is required.
Previous works have used immunostaining to track specific proteins in a 3D matrix. Bernhardt et al. studied fully osteoblast‐osteocyte differentiation inside 3D modified‐collagen hydrogels seeded in 48 well plates and they stained DMP‐1 to determine which hydrogel modification could enhance osteogenesis. 79 Microfluidics also allows a controlled diffusion and devices features make possible a high‐quality real time microscopy. Nasello et al. stained DMP‐1 and BSP‐2 in microfluidic devices to observe osteoblasts maturation and determined that high‐cell density inside the chip created a 3D network able to carry a fully osteogenic differentiation. 31 Here, BSP‐2, DMP‐1 and OCN were selected to determine osteogenic differentiation of a 3D‐ADSC culture. As detailed above, our results are in agreement with both preliminary results from flow cytometry and previous findings from the literature commented above.
Throughout the study, analysis of osteogenic markers showed that pre‐differentiated cells for 14 days produced mineralized bone matrix earlier than the ones differentiated for 7 days or the undifferentiated ones. When inducing pre‐differentiation stage, cells were already committed to the osteogenic lineage, therefore they started producing mineralized bone matrix earlier. 80 Other studies reported similar outcomes both in vitro and in vivo. 81 , 82 From the clinical point of view, we consider this may be an advantage since cells can be pre‐differentiated while expanding them. Thus, it is possible to shorten the time necessary to create the bone model.
3.8. Relevance of the study and potential future applications
Traditional in vitro systems already showed that ADSC undergo osteogenic differentiation under specific conditions. Here, we translated the findings of the traditional macro‐models on stem cells differentiation to the organ‐on‐a‐chip scale. We created a minimal functional unit that captures several aspects of the osteogenic differentiation of ADSCs. As mentioned before, this was a proof of concept study and therefore, commercial cells from only one patient were used. Thus, future work would involve increasing the number of biological subjects.
The use of the microengineered technology to study this process in a 3D fibrous extracellular matrix takes advantage of the limited number of cells required by these systems, meanwhile the physiological environment is closely maintained. Moreover, microfluidics has been widely reported as an innovative and powerful tool to study physiological events in vitro. Park et al. 67 compared osteogenesis of a monolayer of human bone marrow MSCs and ADSCs under dynamic hydraulic compression. They concluded that stimulated stem cells showed increased osteogenic gene expression. However, this model lacks the benefits of a 3D culture. Another potential use of the model presented here is the introduction of the gel in previously‐fabricated constructs. For instance, Marturano‐Kruik et al. 29 created a co‐culture on a scaffold to study metastatic colonization. Thus, this multi‐channel geometry could also allow a co‐culture osteoblasts‐osteoclasts, or their mesenchymal and hematopoietic precursors respectively, to study bone remodeling‐on‐a‐chip. In addition, our collagen type‐I‐based hydrogel loaded with differentiated ADSC may be introduced in potential bone‐grafts to study tissue‐biomaterial interactions and scaffold degradation rates among other variables.
Another potential field of application of this ADSC bone‐on‐a‐chip model is drug delivery. Mancera‐Andrade et al. 83 thoroughly reviewed different tissue micro–models for drug delivery studies comparing different carriers, drugs and the final envisaged applications. Giving all the advantages provided by our model in terms of collagen hydrogel diffusion and ease to daily culture follow‐up, cultures responses to growth factors as BMP2 and BMP7 68 could be studied. In addition, as it was commented before, collagen hydrogel density is a tunable variable. Its mechanical properties can be adjusted depending on the final application by modifying collagen concentration. Thus, our approach may not be limited to bone tissue engineering or stem cell differentiation. The versatility of this technology allows the use of different cell lines to study a wide variety of physiological processes.
Discussing clinical translation, it is important to consider that the technique presented here could allow creating a bone‐on‐a‐chip model out of a specific patient or group of patients (e.g., different models depending demographic population features). All of that would be created in a highly controlled miniaturized environment and conducted in a relatively short time. Moreover, the highest benefit of the use of this model would be the large reduction in the number of cells needed. Currently, after stem cell harvesting, long periods of time are required to grow extensive cell cultures. Taking into consideration the number of cells reduction, standby periods could be reduced. Microfluidic platforms may serve as an in vitro bone model and allow the study of the different behaviors of stem cells derived from different sources.
4. CONCLUSIONS
We translated the findings of traditional macro‐models on stem cells osteogenic differentiation to the organ‐on‐a‐chip scale. We created a minimal functional unit that captures the major aspects underlying the differentiation of ADSC into osteoblasts‐osteocytes. The ability of undergoing osteogenic differentiation without any external stimulation when seeded on an osteoconductive environment makes ADSC promising candidates for bone tissue engineering. Different aspects were assessed to determine whether differentiated and undifferentiated ADSC exhibit osteogenic markers. Finally, it was proven that differentiated cells during 14 days of incubation, created a mature bone model inside the microfluidic platforms. Cells differentiated during 7 days also created an incipient bone model, in terms of presenting a cellular coordinated network, but the expression of osteogenic markers did not indicate a fully differentiated phenotype. The fact that no dynamic fluid flow was used to create the model also offers an important advantage in terms of experimental set‐up simplicity, which is important to potentially use the model in the clinic. From a clinical perspective, our in vitro bone model proof of concept may provide a miniaturized patient‐specific tool to study individual osteogenic potential and the effects of different therapies for the treatment of bone‐associated pathologies.
CONFLICT OF INTEREST
There are no conflicts to declare.
Supporting information
Figure S1 α‐BSP2, α‐OCN, and α‐DMPI immunostaining of 7D‐ADSC. Evolution over time at different timepoints. Individual pictures are orthogonal projections of a ≂80 μm Z‐stack. Scale bar is 20 μm and pictures are magnified 40 times.
Figure S2 α‐BSP2, α‐OCN, and α‐DMPI immunostaining of 14D‐ADSC. Evolution over time at different timepoints. Individual pictures are orthogonal projections of a ≂80 μm Z‐stack. Scale bar is 20 μm and pictures are magnified 40 times.
Table S1 List of acronyms used along the full main text
ACKNOWLEDGMENTS
Authors would like to acknowledge the use of Servicio General de Apoyo a la Investigación‐SAI (Universidad de Zaragoza), the use of Servicios Científico Técnicos del CIBA (IACS‐Universidad de Zaragoza), Spanish Ministry of Economy and Competitiveness through Projects DPI2017‐84780‐C2‐1‐R and PID2020‐113819RB‐I00 and the Government of Aragon in the form of grant awarded to Pilar Alamán‐Díez (Grant No.2018‐22). Elena García‐Gareta is funded by a “Maria Zambrano Fellowship” from the Spanish Ministry of Universities (2021 Funding Program for attraction of international talent to the Spanish academic system).
Alamán‐Díez P, García‐Gareta E, Arruebo M, Pérez María Ángeles. A bone‐on‐a‐chip collagen hydrogel‐based model using pre‐differentiated adipose‐derived stem cells for personalized bone tissue engineering. J Biomed Mater Res. 2023;111(1):88‐105. doi: 10.1002/jbm.a.37448
Funding information Gobierno de Aragón, Grant/Award Number: 2018‐22; Maria Zambrano Fellowship from the Spanish Ministry of Universities (2021 Funding Program for attraction of international talent to the Spanish academic system); Ministerio de Ciencia e Innovación, Grant/Award Number: DPI2017‐84780‐C2‐1‐R;PID2020‐113819RB‐I00
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request
REFERENCES
- 1. Burr DB, Robling AG, Turner CH. Effects of biomechanical stress on bones in animals. Bone. 2002;30(5):781‐786. [DOI] [PubMed] [Google Scholar]
- 2. Burger EH, Klein‐Nulend J. Mechanotransduction in bone ‐ role of the lacuno‐canalicular network. FASEB J. 1999;13(9001):S101‐S112. [PubMed] [Google Scholar]
- 3. Kohli N, Ho S, Brown SJ, et al. Bone remodelling in vitro: where are we headed?: a review on the current understanding of physiological bone remodelling and inflammation and the strategies for testing biomaterials in vitro. Bone. 2018;110:38‐46. doi: 10.1016/j.bone.2018.01.015 [DOI] [PubMed] [Google Scholar]
- 4. Deng Z, Wang Z, Jin J, et al. SIRT1 protects osteoblasts against particle‐induced inflammatory responses and apoptosis in aseptic prosthesis loosening. Acta Biomater. 2017;49:541‐554. [DOI] [PubMed] [Google Scholar]
- 5. Stevens MM, George JH. Exploring and engineering the cell surface interface. Science. 2005;310(5751):1135‐1138. [DOI] [PubMed] [Google Scholar]
- 6. Pirosa A, Gottardi R, Alexander PG, Tuan RS. Engineering in‐vitro stem cell‐based vascularized bone models for drug screening and predictive toxicology. Stem Cell Res Therapy. 2018;9(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. George EL, Truesdell SL, York SL, Saunders MM. Lab‐on‐a‐chip platforms for quantification of multicellular interactions in bone remodeling. Exp Cell Res. 2018;365(1):106‐118. doi: 10.1016/j.yexcr.2018.02.027 [DOI] [PubMed] [Google Scholar]
- 8. Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368‐373. [DOI] [PubMed] [Google Scholar]
- 9. Abgrall P, Gué AM. Lab‐on‐chip technologies: making a microfluidic network and coupling it into a complete microsystem ‐ a review. J Micromech Microeng. 2007;17(5):R15‐R49. [Google Scholar]
- 10. Moreno‐Arotzena O, Borau C, Movilla N, Vicente‐Manzanares M, García‐Aznar JM. Fibroblast migration in 3D is controlled by haptotaxis in a non‐muscle myosin II‐dependent manner. Ann Biomed Eng. 2015;43(12):3025‐3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Polacheck WJ, Li R, Uzel SGM, Kamm RD. Microfluidic platforms for mechanobiology. Lab Chip. 2013;13(12):2252‐2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Del Amo C, Olivares V, Cóndor M, et al. Matrix architecture plays a pivotal role in 3D osteoblast migration: the effect of interstitial fluid flow. J Mech Behav Biomed Mater. 2018;83(January):52‐62. [DOI] [PubMed] [Google Scholar]
- 13. Bersini S, Jeon JS, Dubini G, et al. A microfluidic 3D invitro model for specificity of breast cancer metastasis to bone. Biomaterials. 2014;35(8):2454‐2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huh D, Ys T, Hamilton GA, Kim HJ, Ingber DE. Microengineered physiological biomimicry: organs‐on‐chips. Lab Chip. 2012;12(12):2156‐2164. [DOI] [PubMed] [Google Scholar]
- 15. Babaliari E, Petekidis G, Chatzinikolaidou M. A precisely flow‐controlled microfluidic system for enhanced preosteoblastic cell response for bone tissue engineering. Bioengineering. 2018;5(3):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Z, Li Z, Li EN, et al. Osteochondral tissue chip derived from iPSCs: modeling OA pathologies and testing drugs. Front Bioeng Biotechnol. 2019;7:411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mansoorifar A, Gordon R, Bergan RC, Bertassoni LE. Bone‐on‐a‐chip: microfluidic technologies and microphysiologic models of bone tissue. Adv Funct Mater. 2021;31(6):1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee JH, Gu Y, Wang H, Lee WY. Microfluidic 3D bone tissue model for high‐throughput evaluation of wound‐healing and infection‐preventing biomaterials. Biomaterials. 2012;33(4):999‐1006. [DOI] [PubMed] [Google Scholar]
- 19. Beyer S, Blocki A, Cheung MCY, Wan ZHY, Mehrjou B, Kamm RD. Lectin staining of microvascular glycocalyx in microfluidic cancer cell extravasation assays. Life. 2021;11(3):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yin Q, Xu N, sheng Xu D, xin Dong M, min Shi X, Wang Y, et al. Comparison ofsenescence‐related changesbetween three‐ and two‐dimensional cultured adipose‐derived mesenchymal stem cells 2020;p. 1–12. [DOI] [PMC free article] [PubMed]
- 21. Si Z, Wang X, Sun C, et al. Adipose‐derived stem cells: sources, potency, and implications for regenerative therapies. Biomed Pharmacother. 2019;114(March):108765. doi: 10.1016/j.biopha.2019.108765 [DOI] [PubMed] [Google Scholar]
- 22. Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143‐147. [DOI] [PubMed] [Google Scholar]
- 23. Bianco P. “Mesenchymal” stem cells. Ann Rev Cell Dev Biol. 2014;30:677‐704. [DOI] [PubMed] [Google Scholar]
- 24. Kargozar S, Mozafari M, Hamzehlou S, Milan PB, Kim HW, Baino F. Bone tissue engineering using human cells: a comprehensive review on recent trends, current prospects, and recommendations. 2019;9. [Google Scholar]
- 25. Strem BM, Hicok KC, Zhu M, et al. Multipotential differentiation of adipose tissue‐derived stem cells. Keio J Med. 2005;54(3):132‐141. [DOI] [PubMed] [Google Scholar]
- 26. Zuk PA, Zhu M, Ashjian P, et al. Human adipose tissue is a source of multipotent. Stem Cells. 2002;13(December):4279‐4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fomby P, Cherlin AJ, Hadjizadeh A, et al. Stem cells and cell therapies in lung biology and diseases: conference report. Ann Am Thorac Soc. 2010;12(3):181‐204. doi: 10.1016/j.trsl.2010.06.007 [DOI] [Google Scholar]
- 28. Bunnell BA, Estes BT, Guilak F, Gimble JM. Differentiation of adipose stem cells. Methods Mol Biol. 2008;456:155‐171. [DOI] [PubMed] [Google Scholar]
- 29. Marturano‐Kruik A, Nava MM, Yeager K, et al. Human bone perivascular niche‐on‐a‐chip for studying metastatic colonization. Proc Natl Acad Sci U S A. 2018;115(6):1256‐1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Truesdell SL, George EL, Van Vranken CC, Saunders MM. A lab‐on‐a‐chip platform for stimulating osteocyte mechanotransduction and analyzing functional outcomes of bone remodeling. J Vis Exp. 2020;2020(159):1‐11. [DOI] [PubMed] [Google Scholar]
- 31. Nasello G, Alamán‐Díez P, Schiavi J, Pérez MÁ, McNamara L, García‐Aznar JM. Primary human osteoblasts cultured in a 3D microenvironment create a unique representative model of their differentiation into osteocytes. Front Bioeng Biotechnol. 2020;8(April):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langenbach F, Handschel J. Effects of dexamethasone, ascorbic acid and β ‐glycerophosphate on the osteogenic differ‐ entiation of stem cells in vitro. Stem Cell Res Therapy. 2013;4(5):1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shin Y, Han S, Jeon JS, et al. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat Protoc. 2012;7:1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Del Amo C, Borau C, Movilla N, Asín J, García‐Aznar JM. Quantifying 3D chemotaxis in microfluidic‐based chips with step gradients of collagen hydrogel concentrations. Integr Biol. 2017;9(4):339‐349. [DOI] [PubMed] [Google Scholar]
- 35. McGarrigle MJ, Mullen CA, Haugh MG, Voisin MC, McNamara LM. Osteocyte differentiation and the formation of an interconnected cellular network in vitro. Eur Cells Mater. 2016;31(353):323‐340. [DOI] [PubMed] [Google Scholar]
- 36. Valero C, Amaveda H, Mora M, García‐Aznar JM. Combined experimental and computational characterization of crosslinked collagen‐based hydrogels. PLoS ONE. 2018;13(4):1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rowe SL, Stegemann JP. Interpenetrating collagen‐fibrin composite matrices with varying protein contents and ratios. Biomacromolecules. 2006;7(11):2942‐2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Golub EE, Boesze‐Battaglia K. The role of alkaline phosphatase in mineralization. Curr Opin Orthop. 2007;18(5):444‐448. [Google Scholar]
- 39. Marom R, Shur I, Solomon R, Benayahu D. Characterization of adhesion and differentiation markers of osteogenic marrow stromal cells. J Cell Physiol. 2005;202(1):41‐48. [DOI] [PubMed] [Google Scholar]
- 40. Birmingham E, Niebur GL, Mchugh PE, et al. Osteogenic differentiation of mesenchymal stem cells is regulated by osteocyte and osteoblast cells in a simplified bone niche. Eur Cell Mater. 2012;23(353):13‐27. [DOI] [PubMed] [Google Scholar]
- 41. Schiavi J, Keller L, Morand DN, et al. Active implant combining human stem cell microtissues and growth factors for bone‐regenerative nanomedicine. Nanomedicine. 2015;10(5):753‐763. [DOI] [PubMed] [Google Scholar]
- 42. Jiang Y, Jahagirdar BN, Reinhardt RL, et al. Pluripotency of mes‐ enchymal stem cells derived from adult marrow. Nature. 2007;447(7146):879‐880. [Google Scholar]
- 43. Shafaei H, Kalarestaghi H. Adipose‐derived stem cells: an appropriate selection for osteogenic differentiation. J Cell Physiol. 2020;235(11):8371‐8386. [DOI] [PubMed] [Google Scholar]
- 44. Grottkau BE, Lin Y. Osteogenesis of adipose‐derived stem cells. Bone Res. 2013;1:133‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dallas SL, Bonewald LF. Dynamics of the transition from osteoblast to osteocyte. Ann NY Acad Sci. 2010;1192:437‐443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pattappa G, Heywood HK, de Bruijn JD, Lee DA. The metabolism of human mesenchymal stem cells during proliferation and differentiation. J Cell Physiol. 2011;226(10):2562‐2570. [DOI] [PubMed] [Google Scholar]
- 47. Rodríguez JP, Rios S, Gonzalez M. Modulation of the proliferation and differentiation of human mesenchymal stem cells by copper. J Cell Biochem. 2002;85(1):92‐100. [PubMed] [Google Scholar]
- 48. Nakamura A, Dohi Y, Akahane M, et al. Osteocalcin secretion as an early marker of osteogenic differentiation of rat mesenchymal stem cells. Tissue Eng ‐ Part C: Methods. 2009;15(2):169‐180. [DOI] [PubMed] [Google Scholar]
- 49. Lin X, Patil S, Gao YG, Qian A. The bone extracellular matrix in bone formation and regeneration. Front Pharmacol. 2020;11:757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Salmasi S, Kalaskar DM, Yoon WW, Blunn GW, Seifalian AM. Role of nanotopography in the development of tissue engineered 3D organs and tissues using mesenchymal stem cells. World J Stem Cells. 2015;7(2):266‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pérez‐Rodríguez S, Huang SA, Borau C, García‐Aznar JM, Polacheck WJ. Microfluidic model of monocyte extravasation reveals the role of hemodynamics and subendothelial matrix mechanics in regulating endothelial integrity. Biomicrofluidics. 2021;15(5):054102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Plou J, Juste‐Lanas Y, Olivares V, del Amo C, Borau C, García‐Aznar JM. From individual to collective 3D cancer dissemination: roles of collagen concentration and TGF‐β . Sci Rep. 2018;8(1):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Prasad A, Alizadeh E. Cell form and function: interpreting and controlling the shape of adherent cells. Trends Biotechnol. 2019;37(4):347‐357. doi: 10.1016/j.tibtech.2018.09.007 [DOI] [PubMed] [Google Scholar]
- 54. Frost OG, Owji N, Thorogate R, et al. Cell morphology as a design parameter in the bioengineering of cell‐biomaterial surface interactions. Biomater Sci. 2021;9(23):8032‐8050. [DOI] [PubMed] [Google Scholar]
- 55. Pablo Rodríguez J, González M, Ríos S, Cambiazo V. Cytoskeletal organization of human mesenchymal stem cells (MSC) changes during their osteogenic differentiation. J Cell Biochem. 2004;93(4):721‐731. [DOI] [PubMed] [Google Scholar]
- 56. McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483‐495. [DOI] [PubMed] [Google Scholar]
- 57. Ochiai‐Shino H, Kato H, Sawada T, et al. A novel strategy for enrichment and isolation of osteoprogenitor cells from induced pluripotent stem cells based on surface marker combination. PLoS ONE. 2014;9(6): e99534. doi: 10.1371/journal.pone.0099534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Phadke A, Hwang Y, Kim SH, et al. Effect of scaffold microarchitecture on osteogenic differentiation of human mesenchymal stem cells. Eur Cell Mater. 2013;25:114‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yeh SCA, Wilk K, Lin CP, Intini G. In vivo 3D histomorphometry quantifies bone apposition and skeletal progenitor cell differentiation. Sci Rep. 2018;8(1):2‐11. doi: 10.1038/s41598-018-23785-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Uddin SMZ, Qin YX. Enhancement of osteogenic differentiation and proliferation in human mesenchymal stem cells by a modified low intensity ultrasound stimulation under simulated microgravity. PLoS ONE. 2013;8(9):1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ye X, Zhang P, Xue S, Xu Y, Tan J, Liu G. Adipose‐derived stem cells alleviate osteoporosis by enchancing osteogenesis and inhibiting adipogenesis in a rabbit model. Cytotherapy. 2014;16(12):1643‐1655. [DOI] [PubMed] [Google Scholar]
- 62. Carelli S, Colli M, Vinci V, Caviggioli F, Klinger M, Gorio A. Mechanical activation of adipose tissue and derived mesenchymal stem cells: novel anti‐inflammatory properties. Int J Mol Sci. 2018;19(1):267. doi: 10.3390/ijms19010267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tenstad E, Tourovskaia A, Folch A, Myklebost O, Rian E. Extensive adipogenic and osteogenic differentiation of patterned human mesenchymal stem cells in a microfluidic device. Lab Chip. 2010;10(11):1401‐1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boukhechba F, Balaguer T, Michiels JF, et al. Human primary osteocyte differentiation in a 3D culture system. J Bone Miner Res. 2009;24(11):1927‐1935. [DOI] [PubMed] [Google Scholar]
- 65. Halling Linder C, Ek‐Rylander B, Krumpel M, et al. Bone alkaline phosphatase and tartrate‐resistant acid phosphatase: potential co‐regulators of bone mineralization. Calcif Tissue Int. 2017;101(1):92‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Angle SR, Sena K, Sumner DR, Virdi AS. Osteogenic differentiation of rat bone marrow stromal cells by various intensities of low‐intensity pulsed ultrasound. Ultrasonics. 2011;51(3):281‐288. [DOI] [PubMed] [Google Scholar]
- 67. Park SH, Sim WY, Min BH, Yang SS, Khademhosseini A, Kaplan DL. Chip‐based comparison of the osteogenesis of human bone marrow‐ and adipose tissue‐derived mesenchymal stem cells under mechanical stimulation. PLoS ONE. 2012;7(9):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Q, Chen GX, Guo L, Yang L. The osteogenic study of tissue engineering bone with BMP2 and BMP7 gene‐modified rat adipose‐derived stem cell. J Biomed Biotechnol. 2012;2012:410879. doi: 10.1155/2012/410879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hasany M, Thakur A, Taebnia N, et al. Combinatorial screening of nanoclay‐reinforced hydrogels: a glimpse of the "holy grail" in orthopedic stem cell therapy? ACS Appl Mater Interfaces. 2018;10(41):34924‐34941. [DOI] [PubMed] [Google Scholar]
- 70. García‐Gareta E, Hua J, Rayan F, Blunn GW. Stem cell engineered bone with calcium‐phosphate coated porous titanium scaffold or silicon hydroxyapatite granules for revision total joint arthroplasty. J Mater Sci Mater Med. 2014;25(6):1553‐1562. [DOI] [PubMed] [Google Scholar]
- 71. Najman S, Najdanović J, Cvetković V. Application of adipose‐derived stem cells in treatment of bone tissue defects. Clinical Implementation of Bone Regeneration and Maintenance. IntechOpen; 2020. [Google Scholar]
- 72. Yang M, Ma QJ, Dang GT, Ma KT, Chen P, Zhou CY. In vitro and in vivo induction of bone formation based on ex vivo gene therapy using rat adipose‐derived adult stem cells expressing BMP‐7. Cytotherapy. 2005;7(3):273‐281. [DOI] [PubMed] [Google Scholar]
- 73. Calabrese G, Giuffrida R, Forte S, et al. Human adipose‐derived mesenchymal stem cells seeded into a collagen‐hydroxyapatite scaffold promote bone augmentation after implantation in the mouse. Sci Rep. 2017;7(1):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Peister A, Mellad JA, Larson BL, Hall BM, Gibson LF, Prockop DJ. Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood. 2004;103(5):1662‐1668. [DOI] [PubMed] [Google Scholar]
- 75. Shahdadfar A, Frønsdal K, Haug T, Reinholt FP, Brinchmann JE. In vitro expansion of human mesenchymal stem cells: choice of serum is a determinant of cell proliferation, differentiation, gene expression, and transcriptome stability. Stem Cells. 2005;23(9):1357‐1366. [DOI] [PubMed] [Google Scholar]
- 76. Mullen CA, Haugh MG, Schaffler MB, Majeska RJ, McNamara LM. Osteocyte differentiation is regulated by extracellular matrix stiffness and intercellular separation. J Mech Behav Biomed Mater. 2013;28:183‐194. doi: 10.1016/j.jmbbm.2013.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Puwanun S, Delaine‐Smith RM, Colley HE, Yates JM, MacNeil S, Reilly GC. A simple rocker‐induced mechanical stimulus upregulates mineralization by human osteoprogenitor cells in fibrous scaffolds. J Tissue Eng Regen Med. 2018;12(2):370‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Oh SA, Lee HY, Lee JH, et al. Collagen three‐dimensional hydrogel matrix carrying basic fibroblast growth factor for the cultivation of mesenchymal stem cells and osteogenic differentiation. Tissue Eng Part A. 2012;18(9–10):1087‐1100. [DOI] [PubMed] [Google Scholar]
- 79. Bernhardt A, Weiser E, Wolf S, Vater C, Gelinsky M. Primary human osteocyte networks in pure and modified collagen gels. Tissue Eng ‐ Part A. 2019;25(19–20):1347‐1355. [DOI] [PubMed] [Google Scholar]
- 80. Wankhade UD, Shen M, Kolhe R, Fulzele S. Advances in adipose‐derived stem cells isolation, characterization, and application in regenerative tissue engineering. Stem Cells Int. 2016;2016:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Peters A, Toben D, Lienau J, et al. Locally applied osteogenic predifferentiated progenitor cells are more effective than undifferentiated mesenchymal stem cells In the treatment of delayed bone healing. Tissue Eng Part A. 2009;15(10):2947‐2954. [DOI] [PubMed] [Google Scholar]
- 82. Kandalam U, Kawai T, Ravindran G, et al. Predifferentiated gingival stem cell‐induced bone regeneration in rat alveolar bone defect model. Tissue Eng Part A. 2021;27(5‐6):424‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mancera‐Andrade EI, Parsaeimehr A, Arevalo‐Gallegos A, Ascencio‐Favela G, Parra‐Saldivar R. Microfluidics technology for drug delivery: a review. Front Biosci ‐ Elite. 2018;10(1):74‐91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 α‐BSP2, α‐OCN, and α‐DMPI immunostaining of 7D‐ADSC. Evolution over time at different timepoints. Individual pictures are orthogonal projections of a ≂80 μm Z‐stack. Scale bar is 20 μm and pictures are magnified 40 times.
Figure S2 α‐BSP2, α‐OCN, and α‐DMPI immunostaining of 14D‐ADSC. Evolution over time at different timepoints. Individual pictures are orthogonal projections of a ≂80 μm Z‐stack. Scale bar is 20 μm and pictures are magnified 40 times.
Table S1 List of acronyms used along the full main text
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request


