Abstract
The prevalence of a high‐energy diet and a sedentary lifestyle has increased the incidence of type 2 diabetes (T2D). T2D is a chronic disease characterized by high blood glucose levels and insulin resistance in peripheral tissues. The pathological mechanism of this disease is not fully clear. Accumulated evidence has shown that noncoding RNAs have an essential regulatory role in the progression of diabetes and its complications. The roles of small noncoding RNAs, such as miRNAs, in T2D, have been extensively investigated, while the function of long noncoding RNAs (lncRNAs) in T2D has been unstudied. It has been reported that lncRNAs in T2D play roles in the regulation of pancreatic function, peripheral glucose homeostasis and vascular inflammation. In addition, lncRNAs carried by small extracellular vesicles (sEV) were shown to mediate communication between organs and participate in diabetes progression. Some sEV lncRNAs derived from stem cells are being developed as potential therapeutic agents for diabetic complications. In this review, we summarize the current knowledge relating to lncRNA biogenesis, the mechanisms of lncRNA sorting into sEV and the regulatory roles of lncRNAs and sEV lncRNAs in diabetes. Knowledge of lncRNAs and sEV lncRNAs in diabetes will aid in the development of new therapeutic drugs for T2D in the future.
Keywords: diabetes, exosomes, long noncoding RNA, small extracellular vesicle, sorting mechanism
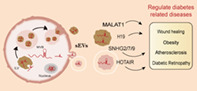
Small extracellular veiscles (sEV) are generated from either plasma membrane budding pathway or endosomal pathway. LncRNAs can be encapsulated in sEV, including MALAT1, H19, SNHGs and HOTAIR. Those sEV lncRNAs mediate communication between organs and participate in diabetic wound healing, obesity, atherosclerosis and diabetic complications.
1. INTRODUCTION
The incidence of type 2 diabetes (T2D) is rapidly increasing with the prevalence of obesity and sedentary lifestyles. Prevention of diabetic complications is the main goal of diabetic treatment, 1 but the current treatment cannot achieve satisfactory effects; for example, half of patients with T2D present with microvascular complications, and 27% present with macrovascular complications in an observational study of 28 countries in Asia, Africa, South America and Europe. 2 Even on the basis of cohort studies from developed countries, the relative risk of complications among patients with diabetes was estimated to be higher than in people without diabetes. 3 Thus, it is important to clarify the pathological mechanism of T2D to carry out more effective therapeutic strategies.
Noncoding RNAs control various levels of gene expression, including chromatin architecture/epigenetic memory, transcription, RNA splicing, editing, translation and turnover. 4 Accumulated evidence has shown that noncoding RNAs have an essential regulatory role in the progression of diabetes and its complications. The roles of small noncoding RNAs, such as miRNAs, in T2D have been extensively investigated, while the function of long noncoding RNAs (lncRNAs) in T2D has been unstudied. Many lncRNAs, such as HOTAIR, MEG3, LET, MALAT1, MIAT, CDKN2BAS1/ANRIL, XIST and GAS5, were shown to change in type 2 diabetic conditions. 5 In addition, lncRNAs carried by small extracellular vesicles (sEV), such as H19, MALAT1, HOTAIR and SNHGs, were shown to mediate communication between organs and participate in diabetes progression. In this review, we summarize the current knowledge relating to lncRNA biogenesis, the mechanisms of lncRNA sorting into sEV, and the regulatory roles of lncRNAs and sEV lncRNAs with a focus on T2D. We hope to aid further therapeutic drug development for T2D.
2. BIOGENESIS AND CHARACTERIZATION OF LONG NONCODING RNAs
Long noncoding RNAs (lncRNAs) are RNA transcripts more than 200 nucleotides (nt) in length that do not encode proteins. LncRNAs are often classified by their gene location relative to nearby protein‐coding genes. 6 As shown in Figure 1A, lncRNA transcripts localized between exons of a protein‐coding gene are defined as intronic lncRNAs. LncRNAs overlapping exons but transcribed in the opposite direction are defined as antisense lncRNAs. Long intervening noncoding RNA (lincRNA, also called long “intergenic” noncoding RNA) refers to lncRNA generated from intergenic genes that do not overlap with exons of either protein‐coding or other non‐lincRNA genes. 7 The biogenesis of lncRNAs is similar to that of mRNAs; for example, both are spliced and modified with 5′‐end m7G caps and 3′‐end poly(A) tails. However, lncRNAs have some unique features that can distinguish them from mRNAs, such as the presence of fewer and longer exons than mRNAs, relatively low expression, the ability to generate circular RNAs or pre‐miRNAs, and strict localization. 8 LncRNAs are transcribed mainly by Pol II, and their expression is highly regulated. SWI/SNF promotes and CAF‐1 inhibits transcriptional initiation and direction, while Dicer and MYC participate in the regulation of elongation 9 (Figure 1A). In the chromatin state, some antisense lncRNA genes are repressed by chromatin remodeling complexes (Swr1, Isw2, Rsc and Ino80), therefore inhibiting the corresponding mRNA expression. 10 It is known that the transcription and splicing efficiency of lncRNAs are low, which causes them to primarily accumulate in the nucleus and form nuclear speckles 11 (Figure 1B), which act as a hub to coordinate all of the nuclear gene expression regulation steps. 12 To date, lncRNAs have been proven to participate in molecular and genomic modulation in various ways, 13 including (1) bringing two distant genes closer (scaffolds, Figure 1C‐I); (2) binding to transcription factors to inhibit gene transcription (decoys, Figure 1C‐II); (3) guiding regulatory proteins to gene sequences, thus affecting gene expression (Figure 1C‐III); and (4) directly binding to gene sequences to affect gene expression (genomic targeting) (Figure 1C‐IV). In pancreatic and diabetic conditions, for example, Hi‐LINC25 binds to transcription factors in islets; H19, LncSHGL and MIST bind to proteins or RNA‐binding proteins (RBPs) to regulate glucose metabolism‐related gene expression; and most of the studied lncRNAs, such as DRAIR, H19 and MALAT1, were shown to target miRNAs to regulate their target gene expression.
FIGURE 1.
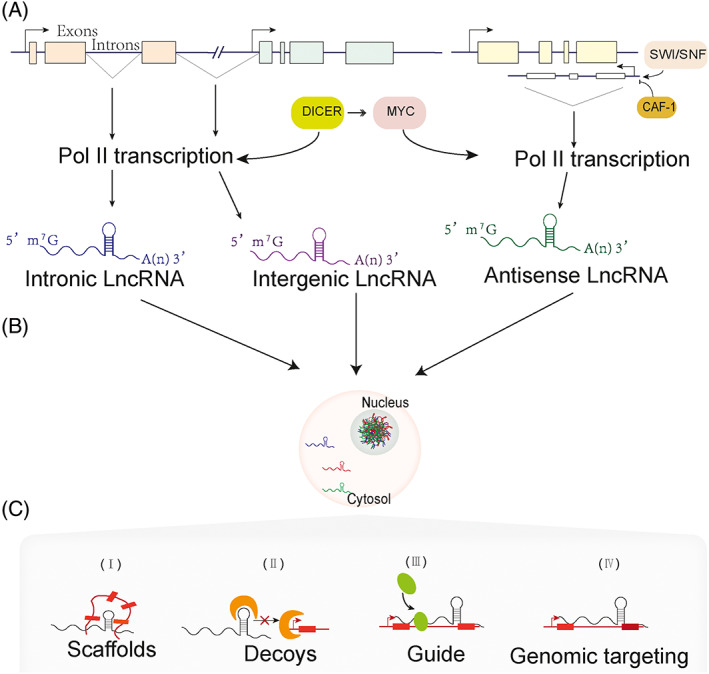
LncRNA biogenesis and functions. (A) lncRNAs classified according to locations of transcripts. Intronic lncRNAs are generated from introns inside a coding‐protein gene. Antisense lncRNAs are transcripts from the opposite direction of a protein‐coding gene. LincRNAs are generated from intergenic genes. (B) The majority of lncRNAs are transcribed by Pol II, which is regulated by MYC and DICER, and lncRNAs are prone to accumulation in the nucleus. (C) LncRNAs function as scaffolds, decoys, genomic targets and guide regulatory proteins
3. ROLES OF lncRNAs IN DIABETES
Although the functional roles of lncRNAs in T2D have only been revealed in recent years, accumulated evidence has demonstrated the biological or pathological roles of lncRNAs in the progression of diabetes and its complications. The mechanisms of lncRNA function in this disease involve pancreatic β cell homeostasis modification, lipid metabolic regulation and inflammatory responses 14 , 15 , 16 (Table 1).
TABLE 1.
LncRNAs and their effects in T2D
| LncRNA | LncRNA origin | Experimental species | Possible molecular mechanisms | Ref. |
|---|---|---|---|---|
| β cell related | ||||
| HI‐LNC25/HI‐LNC78/HI‐LNC80 | Pancreatic islet tissue | Human | Target islet transcription factor GLIS3 | 17 |
| β‐linc 1 | Pancreatic islet tissue | Human | Affect endocrine cell differentiation | 18 |
| H19 | Pancreatic islet tissue | T2D mice/Rat β cells | Promote β cell proliferation via let‐7/AKT | 19 |
| Adipose development related | ||||
| BATE1/Blnc1 | BAT | Mice | Promote brown and beige adipocyte differentiation | 20 |
| Blnc1 | BAT | Mice | Promote mitochondrial biogenesis | 21 |
| LINC00473 | BAT progenitor cells | Human | Regulate mitochondrial oxidative metabolism | 22 |
| Adi | ADSCs | Rats | Promote adipogenesis via targeting miR‐499 | 23 |
| Dio3os | Fetal BAT from maternal obesity | Mice | Inhibit brown adipose development via maternally imprinted | 24 |
| Glucose homeostasis related | ||||
| H19 | Livers of db/db mice | Mice | Inhibit gluconeogenesis via Foxo1 | 25 |
| Bhmt‐AS | Livers of db/db mice | Mice | Promote gluconeogenesis | 26 |
| lncSHGL | Livers | Mice | Inhibit gluconeogenesis via interacting with hnRNPA1 | 27 |
| Different expressed‐144 lncRNAs | C2C12 skeletal muscle cells | FFA induced insulin resistant cells | Associated with FAO metabolism and insulin signaling | 28 |
| Different expressed‐401 lncRNAs | Skeletal muscles | Rats | Associated with mRNA that regulate glucose metabolism | 29 |
| H19 | Skeletal muscles | Rats | Promote glucose metabolism via DUSP27/AMPK | 30 |
| H19 | Primary satellite cells isolated from muscles | Mice | Promote FAO gene expressions via interacting with hnRNPA1 | 31 |
| Inflammatory related | ||||
| LYPLAL1‐DT | Leukocytes | Human umbilical vein endothelial cells | Inhibit inflammation via miR‐204‐5p/SIRT1 | 32 |
| Linc‐Gm4419 | Renal tissue/Mesangial cells | Mice/HG treated cells | Promote inflammation via NLRP3 | 33 |
| MALAT1 | Brain tissue | Human T2D with OSA | Promote inflammation via miR‐224‐5p/NLRP3 | 34 |
| MALAT1 | Peripheral blood mononuclear cells | NA | Promote inflammation via miR‐1‐3p | 35 |
| SNHG5 | Serum/HK2 cells | Human/HG treated HK2 cells | Promote inflammation via miR‐224‐5p | 36 |
| H19 | Retinal epithelial cells | HG treated retinal epithelial cells | Inhibit inflammation via miR‐93/XBP1 | 37 |
| Lnc uc.48 | Serum/Macrophages | Human/HG and FFA treated cells | Promote inflammation via P2X7R/ERK | 38 |
| Dnm3Os | BMDM | Mice | Promote inflammatory gene expressions via global histone modifications | 39 |
| DRAIR | Monocytes | Human | Inhibit inflammation via epigenetic mechanisms | 40 |
| MIST | Adipose macrophages | Mice | Inhibit inflammation via interacting with PARP‐1 | 41 |
| Tcons_00077866 | Pancreatic β‐TC6 cells | Mice | Promote inflammation via miR‐297b‐5p/SAA3 | 42 |
Abbreviations: ADSCs, adipose derived stromal cells; BAT, brown adipose tissue; BMDM, bone marrow derived macrophages; FFA, fatty acid; HG, high glucose; OSA, obstructive sleep apnea; T2D, type 2 diabetes.
3.1. LncRNAs and β cell homeostasis
Pancreatic β cell dysfunction is a common pathological factor in both type 1 diabetes and T2D. Studies have shown that lncRNAs regulate islet function in various ways. For example, the depletion of lncRNA‐HI‐LNC25, a lncRNA specifically expressed in islet cells, downregulates the expression of the islet transcription factor GLIS3, a mutation that leads to diabetes. 17 This finding indicates a role of lncRNAs in islet development. In addition, the expression of islet lncRNA‐HI‐LNC78 and lncRNA‐HI‐LNC80 is repressed in pancreatic progenitors but is activated in adult islets. Moreover, their expression is also increased in response to high glucose (HG) stimulation, 17 showing the potential regulatory function of lncRNAs in glucose homeostasis. In addition, knockout of β cell lncRNA‐βlinc1 results in defective islet development and disruption of glucose homeostasis in adult mice. 18 LncRNA‐H19 expression promoted the proliferation of β‐cells by targeting the let‐7/AKT pathway in adult mice. 19 All these results demonstrate the essential roles of lncRNAs in the regulation of pancreatic development.
3.2. LncRNAs and adipose development
White adipose tissue (WAT) and brown adipose tissue (BAT) were reported to regulate lipid metabolism and thermogenesis, respectively. 43 Brown and beige adipocytes are known to protect humans and mice from obesity and diabetes, and some adipose‐specific lncRNAs have been shown to affect adipocyte metabolism by regulating mitochondrial function. For example, BATE1 and Blnc1 promote brown and beige adipocyte differentiation and function. 20 Blnc1 protects against diet‐induced obesity by promoting mitochondrial biogenesis in WAT, which accelerates glucose metabolism. 21 LINC00473, a lncRNA specifically expressed in human BAT adipocytes, is decreased in obesity and T2D. LINC00473 was shown to colocalize with mitochondrial and lipid droplet proteins in the cytosol and regulate lipolysis, respiration and transcription of genes associated with mitochondrial oxidative metabolism, thus modifying adipocyte function. 22 In addition, lncRNA‐Adi is highly expressed in adipose tissue‐derived stromal cells (ADSCs) and enhances adipogenesis by interacting with miR‐499, 23 which is a microRNA that has been shown to participate in the regulation of mitochondrial function. 44 Moreover, epigenetic modification of certain lncRNAs affects adipose development. A recent report showed that maternal obesity increases DNA methylation of an antisense lncRNA‐Dio3os promoter in oocytes and offspring brown fat. This is because methylation inhibits Dio3 expression, and as a result, BAT development is inhibited by reduced thyroid hormone synthesis. 24 Interestingly, this maternal repression can be passed to offspring, suggesting that the regulation of lncRNA function in adipose tissue could be transmitted to subsequent generations.
3.3. LncRNAs and glucose homeostasis
Liver gluconeogenesis is the main target for glucose homeostasis regulation. An RNA sequencing analysis in the liver of diabetic mice showed that lncRNA‐H19 depletion increases FOXO1 translocation to the nucleus, which is an essential transcriptional regulator for gluconeogenesis gene expression. 25 LncRNA‐antisense betaine‐homocysteine methyltransferase‐antisense (Bhmt‐AS) is overexpressed in the liver of diabetic mice in response to gluconeogenic hormonal stimuli, and it specifically regulates Bhmt expression and hepatic gluconeogenesis, 26 suggesting that lncRNAs have a role in the regulation of liver gluconeogenesis. Concurrently, lncSHGL enhances the translation efficiency of CALM mRNAs by recruiting hnRNPA1 in the liver of obese mice, which activates the PI3K/AKT pathway in the absence of insulin, 27 revealing an alternative glucose consumption pathway regulated by lncRNAs. In addition, metformin, a first‐line drug for diabetic treatment and an AMPK activator, reduces the glucose effect by accelerating glycolysis and inhibiting gluconeogenesis. Metformin induces different expression levels of lncRNAs. Studies have shown that 456 lncRNAs are upregulated and 409 lncRNAs are downregulated by cAMP stimuli, and nearly half of them are attenuated by metformin treatment. 45 Another study also showed a similar result, in which metformin and resveratrol treatment altered lncRNA profiles in the livers of obese mice, and those lncRNAs regulate insulin signaling pathways. 46 Thus, metformin treatment may exert its antidiabetic effect by altering lncRNA expression.
In skeletal muscle, lncRNAs have a pivotal role in muscle biogenesis and insulin response The skeletal muscle‐specific lncRNAs lincYY1, 47 Dum, 48 and Linc‐RAM 49 were shown to regulate myogenic differentiation and biogenesis. LncRNA profile analysis of insulin‐resistant C2C12 cells treated with palmitic acid showed 70 upregulated and 74 downregulated lncRNAs, which were associated with fatty acid oxidation, lipid oxidation, the PPAR signaling pathway and the insulin signaling pathway. 28 Meanwhile, dysfunction of lncRNA and its highly associated mRNA or prediction of target mRNA (NONRATG017315.2‐Pdk4, NONRATG003318.2‐Stc2, NONRATG011882.2‐Il15, NONRATG013497.2‐Fbxw7, MSTRG.1662‐Ucp3) affects hyperglycemia, glucose intolerance, and increased fatty acid oxidation in diabetic Goto‐Kakizaki rats. 29 In addition, AMPK is an energy sensor that regulates glucose and fatty acid metabolism. LncRNA‐H19 regulates AMPK activation by promoting atypical dual‐specificity phosphatase (DUSP27). 30 Another report showed that H19 interacts with hnRNPA1, thereby increasing fatty acid oxidation‐related transcriptional genes in skeletal muscle cells. 31 These results suggest that lncRNAs have an important role in regulating glucose and fatty acid metabolism in skeletal muscles.
3.4. LncRNAs and inflammatory responses
Inflammation participates in the progression of T2D. 50 Vascular dysfunctions induced by excessive inflammation are major pathological factors that cause diabetic retinopathy, nephropathy and neuropathy. 51 Inflammatory tissue, including adipose tissue, the liver, pancreatic islets, the vasculature and circulating leukocytes, release cytokines, such as TNFα, IL‐6 and IL‐1β. They act in an autocrine and paracrine manner to promote vascular dysfunction by activating the c‐JUN N‐terminal kinase (JNK) and nuclear factor‐kappa B (NF‐κB) pathways. However, the initiation of the inflammatory process remains unclear. On the one hand, a large cytoplasmic multiprotein complex called the inflammasome, especially the nucleotide‐binding oligomerization domain‐like receptor family pyrin domain‐containing 3 (NLRP3) inflammasome, was shown to control the activation of IL‐1β in the progression from obesity to T2D by recognizing microbial products. 52 On the other hand, free fatty acids activate Toll‐like receptor 4 (TLR‐4) in adipocytes, and macrophages also lead to the upregulation of NF‐κB, TNFα and IL‐6. 53 LncRNAs have been found to participate in the inflammatory response in T2D. LncRNA profiles of leukocytes from T2D patients with macrovascular diseases showed that LYPLAL1‐DT has protective effects on endothelial cells under HG and inflammatory conditions. 32 The expression of lncRNA ANRIL was upregulated in T2D patients and in HG‐induced podocytes. LncRNA ANRIL silencing attenuated HG‐induced NLRP3 inflammasome activation and cytokine release. 54 A similar result was observed for lncRNA‐GM4419 in renal tissue from T2D nephropathy mice and HG‐induced mesangial cells. 33 In addition, MALAT1 34 , 35 and SNHG5 36 were shown to regulate inflammation by targeting NLRP3 in T2D brain tissue or neuron cells, and SNHG2 55 , 56 was reported to modulate inflammation by regulating NF‐κB, while H19 37 and lnc uc.48 38 regulate inflammation by activating endoplasmic reticulum stress proteins (XBP1 or ERK), indicating that inflammatory responses are differentially regulated by lncRNAs. In addition, some lncRNAs regulate inflammation by epigenetic modification of inflammation‐related genes. For example, type 2 diabetic conditions induce lncRNA Dnm3os expression via NF‐κB activation, and Dnm3os localizes to the nucleus and alters global histone modifications, thus upregulating inflammation in macrophages. 39 LncRNA DRAIR was downregulated in T2D, and overexpression DRAIR increased anti‐inflammatory but inhibited proinflammatory genes via epigenetic mechanisms. 40 Moreover, lncRNAs participate in the regulation of inflammation in several organs. Except for the renal tissue, brain tissue and retinal tissue mentioned above, macrophage inflammation‐suppressing transcript (Mist) was decreased in adipose tissue macrophages from high‐fat diet‐fed mice. Mist interacts with poly ADP‐ribose polymerase‐1 (PARP1), an activator of inflammatory gene expression and interruption of the Mist‐PARP1 interaction by obesity promotes inflammatory responses. 41 Knockdown of lncRNA TCONS_00077866 (lnc866) inhibited the stearic acid‐induced reduction in insulin secretion and β‐cell inflammation. 42 Therefore, inflammation regulation by lncRNAs has an important role in the incidence and progression of diabetes and makes them possible targets for diabetes therapy.
Based on the functional roles of lncRNAs in diabetes, some lncRNAs have been developed as diagnostic and therapeutic agents for diabetes and its complications. In the clinic, lncRNA ENST00000550337.1 from peripheral blood has high diagnostic value for prediabetes and T2D. 57 LincRNA RP1‐90L14.1 was shown to be related to the pathology of diabetic retinopathy. 58 In addition, two other clinical trials are ongoing (NCT04638556 and NCT04767750). Moreover, researchers found that some lncRNAs are wrapped in extracellular vesicles (EV), which carry them into various organs and function as mediators for cell–cell communications.
4. EV BIOGENESIS AND lncRNA SORTING MECHANISMS
EV are double‐membrane vesicles that are released into extracellular spaces by various types of cells. They cannot replicate and do not have a functional nucleus. EV subtypes are defined by size as large EV (200–2000 nm) or small EVs (sEV, 50–200 nm), by chemical composition as CD63+/CD81+ or Annexin A5 stained, or by the biogenesis pathway as endosomal derived EV (also called exosomes) or plasma membrane derived EV (also called microparticles or microvesicles). sEV are nomenclature by size; they are produced either by direct budding from the plasma membrane or by the fusion of multivesicular bodies (MVBs) with the plasma membrane 59 (Figure 2A). MVBs are generated via a two‐step endocytosis process: an intraluminal vesicle (ILV) is first formed by late endosome invagination, and then ILVs modify the cargos to generate MVBs. Commonly used markers of sEV, such as CD9 and CD63, are vital proteins that participate in endosome‐derived sEV biogenesis. 60 Thus, the widely used term “exosomes” refers explicitly to sEV that are generated via the endosome pathway. However, because of the limitations of purification methods, it is impossible to separate the vesicles based on their biogenesis pathways. According to MISEV 2018, 61 researchers are recommended to categorize EV by size since most studies have not verified the vesicle biogenesis pathway. In this paper, sEV refer to EV less than 150 nm, regardless of the biogenesis pathway. The generation of EV was thought to be a method of quality control to eliminate the “bad” or “useless” proteins. 62 In recent years, many studies have shown that these vesicles carry bioactive cargoes, including lipids, proteins, and nucleic acids, which mediate cell–cell communication and regulate biological functions in recipient cells. 63 , 64
FIGURE 2.
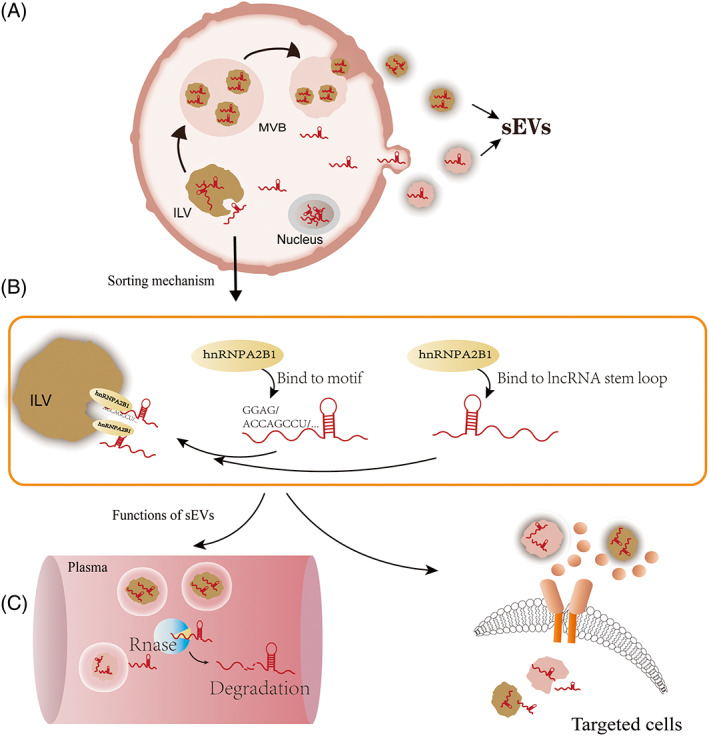
Biogenesis of small extracellular vesicle (sEV) and mechanisms of lncRNA sorting into EVs. (A) sEV generated from multivesicular body fusion with the plasma membrane or direct budding from the plasma membrane; (B) HnRNPA2B1 was shown to guide lncRNA sorting into sEVs by directly binding to stemloops or binding specific sequence motifs. (C) LncRNAs packaging into EV are important to exert their optimal functions. Left, EV protect lncRNAs from degradation by RNase; Right, EV help lncRNAs recognize targeted cells
Recent studies have shown that noncoding RNAs are sorted into EVs, but the molecular mechanism is still unclear. Several proteins or lipids involved in this process have been identified. For example, RBPs, such as heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1), are revealed to guide the sorting of miRNA into EV by interacting with the GGAG motif at the 3′‐end of the miRNAs. 65 The binding specificity of hnRNPA2B1 with the GGAG motif was confirmed by RNA pull‐down analysis, which showed that this interaction was seriously impaired by mutations in the “GGAG” sequence. Overexpression or knockdown of hnRNPA2B1 promotes or suppresses lncRNA‐H19 sorting into sEV in non‐small cell lung cancer (NSCLC) cell lines. 66 In addition, hnRNPA2B1 may also function in the sorting of lncRNAs into sEV. This was evidenced by knockdown or overexpression of hnRNPA2B1, which downregulates or enhances the levels of lncRNA‐AFAP‐AS1 and lncRNA‐AGAP‐AS1 in sEV. 67 , 68 This suggests that the sorting of lncRNAs and miRNAs might share a similar pathway. On the other hand, KRAS mutation, a gene mutation promoting colorectal cancer spread, was shown to affect miRNA sorting into EV in colorectal cancer cells 69 ; however, this mutation has no detectable effect on lncRNA sorting, suggesting that the sorting mechanisms of lncRNAs are at least partially distinct from those of miRNAs. 70 Consistently, a study of the role of sEV lncRNA‐LNMAT2 in lymphatic metastasis of bladder cancer showed that hnRNPA2B1 binds to LNMAT2 via a stem–loop structure located in the 1930–1960 nt region. 71 Coincidentally, a bioinformatic analysis of mRNAs sorted into sEV identified three motifs, ACCAGCCU, CAGUGAGC and UAAUCCCA, which may function as cis‐acting elements guiding RNAs (more than 20% were lncRNAs) to sEV. Strikingly, those motifs from different RNAs were predicted to form similar secondary structures, 72 suggesting that motif specificity and the secondary structures are critical for lncRNA sorting into EV (Figure 2B).
LncRNAs packaging into EV are important to exert their optimal functions (Figure 2C). The double membrane of EV protects them from degradation by cellular RNase, making them have longer than average half‐lives. 73 Proteins or lipids expressed on sEV membranes facilitate their ability to transfer from donor to recipient cells to trigger phenotypic changes in acceptor cells. 64 In particular, some lncRNAs were shown to be abnormally expressed in sEV under disease conditions, showing their considerable promise as novel biomarkers of disease.
5. ROLES OF sEV lncRNAs IN DIABETES
5.1. sEV lncRNA profiling
Profiles of sEV lncRNAs in various diseases have been described. 74 , 75 , 76 Compared with their expression in healthy controls, plasma sEV lncRNAs were found to be expressed differently under distinctive diabetic conditions. For example, lncRNAs are aberrantly expressed in umbilical cord blood sEV from patients with gestational diabetes mellitus. 77 HG‐treated tubular epithelial cell‐derived sEV carried 93 upregulated lncRNAs and 76 downregulated lncRNAs. 78 Similarly, 21 lncRNAs, including SNHG5 and C430049B03Rik, were found to be differentially expressed under high‐glucose conditions in three kinds of progenitor cell lines. 79 In response to proinflammatory cytokine stimuli, human islet cell‐derived sEV contained 31 lncRNAs whose levels were altered. 80 In addition, some lncRNAs were enriched specifically in sEV, and lncRNA‐p3134 levels were found to be four times higher in serum sEV than in controls but remained nearly unchanged in sEV‐free serum. Further experimental results showed that lncRNA‐p3134‐containing sEV promote glucose‐stimulated insulin secretion in β cells, thus regulating pancreatic functions, 81 indicating that the sorting mechanisms of lncRNAs have an important role in lncRNA‐mediated pancreatic function regulation. To date, several sEV lncRNAs have been found to participate in the progression of diabetes and diabetes‐related diseases. 82 Some of them are being developed as promising therapeutic agents for diabetic complications (Table 2).
TABLE 2.
sEV‐LncRNAs and their effects in T2D
| sEV‐LncRNA | sEV‐origin | Target cells | Functions/molecular mechanisms | Ref. |
|---|---|---|---|---|
| p3134 | Human serum | β‐cells | Promote glucose stimulated insulin secretion | 81 |
| H19 | Mice ADSC | Skin fibroblasts | Promote wound healing via miR‐19b/sox9 | 83 |
| H19 | Mice BMMSC | Skin fibroblasts | Promote wound healing via miR‐152‐3p/PTEN | 84 |
| H19 | Mice BMMSC | Osteoblasts | Promote fracture healing via miR‐467/HoxA10 | 85 |
| H19 | Manual nanovesicles | Dermal microvascular endothelial cells | Promote cell proliferation | 86 |
| MALAT1 | Human ADSC | Hippocampal cells | Increase neuro survival via SRSF2/PKCδII | 87 |
| MALAT1 | Mice adipocytes | Hypothalamic neurons | Increase appetite and weight | 88 |
| MALAT1 | Human ADSC | Skin fibroblasts | Accelerate wound healing via miR‐124/wnt/β‐catenin | 89 |
| MALAT1 | HUVEC | Dendritic cells | Prevent atherosclerosis by inhibiting ROS | 90 |
| HOTAIR | Mice Adipose tissue | Intestinal cells | Promote intestinal cell proliferation via NF‐κB | 91 |
| HOTAIR | Mice BMMSC | HUVEC | Promote wound healing via upregulate angiogenic proteins | 92 |
| SNHG2 | Human Macrophages | HUVEC | Inhibit apoptosis of endothelial cells | 93 |
| SNHG7 | Human BMMSC | Human retinal endothelial cells | Repress HG induced endothelial dysfunction | 94 |
| SNHG9 | Human adipocytes | HUVEC | Alleviated inflammation | 95 |
Abbreviations: ADSC, adipose derived stem cells; BMMSC, bone marrow mesenchymal stem cells; HG, high glucose; HUVEC, human umbilical vein endothelial cells; PTEN, phosphatase and tensin homolog; ROS, reactive oxygen species; Sox9, SRY‐Box Transcription Factor 9; SRSF2, Serine and Arginine Rich Splicing Factor 2.
5.2. H19
LncRNA‐H19 is a highly conserved maternally encoded gene. H19 has been shown to be associated with embryonic growth control, tumor growth and T2D. 96 Several clinical studies revealed that serum H19 levels in T2D patients were lower than those in healthy controls. 85 , 97 Patients with diabetes often suffer from slow wound repair and exhibit diabetic foot ulcers. H19 promotes wound healing in diabetic conditions. 98 Experimental evidence showed that sEV H19 derived from adipose mesenchymal stem cells (ADSCs) accelerates wound healing in diabetic mice, and silencing H19 in ADSCs decreased H19 accumulation in sEV, which inhibited skin fibroblast proliferation, migration and invasion by targeting miR‐19b/SOX9. 83 Overexpression of H19 in MSC‐derived sEV promotes wound healing in diabetic foot ulcers by sponging miRNA‐152‐3p, thereafter upregulating phosphatase and tensin homolog (PTEN), a regulator of cell proliferation and growth 84 (Table 2). Furthermore, sEV‐mimicking nanovesicles engineered to increase H19 levels were proven to be an effective nanodrug delivery system. These nanovesicles had a strong ability to rescue cell proliferation signals that were impaired by HG in a diabetic rat model. 86 In addition, H19 carried in sEV was reported to be associated with obesity‐induced retardation of fracture healing. Utilizing normal MSC‐derived sEV can reverse abnormal fracture healing via the miR‐467/HoxA10 axis 85 (Table 2).
5.3. MALAT1
Metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) is a long noncoding RNA with a single exon that is predominantly found in nuclear speckles, and it is evolutionarily conserved among mammalian species. 99 MALAT1 was first found to be associated with metastasis in NSCLC patients. 100 Its overexpression increases the risk of metastasis in various cancers. 101 MALAT1 levels were reported to increase in vascular endothelial cells and retinal endothelial cells exposed to HG treatment, 102 , 103 as well as in macrophage‐derived sEV by exposed to HG treatment. 104 However, serum MALAT1 levels were reported to decrease in patients with T2D, as well as sEV MALAT1 levels in the serum of T2D patients. 105 The functional role of sEV MALAT1 in obesity and diabetes‐related diseases varies across tissues and organs. A lncRNA analysis of sEV derived from human ADSCs showed that omental depots of obese donors had increased MALAT1 levels compared with those of omental depots of lean donors. 106 A study also showed that adipocytes of obese mice secreted MALAT1‐containing sEV, which were transferred to hypothalamic anorexigenic neurons by injection into lean mice and increased appetite and weight in lean mice. 88 This result is consistent with a previous study, which showed that human ADSC‐derived sEV containing MALAT1 increase neuronal survival by mediating PKCδII splicing via SRSF2 in the hippocampal cell line (Table 2). Interestingly, it was found that insulin treatment dramatically increased the association of MALAT1 and splicing factors, 87 suggesting that obesity and insulin could affect the crosstalk between organs via MALAT1 in sEV. However, although a high level of MALAT1 could increase the risk of tumorigenesis, ADSC‐derived sEV containing MALAT1 were shown to accelerate wound healing by targeting the miR‐124/Wnt/β‐catenin pathway (Table 2), and ADSC‐sEV subjected to MALAT1 knockdown impaired their ability to protect skin fibroblast proliferation against H2O2 treatment. 89 In addition, human vascular endothelial cell (HUVEC)‐derived sEV exhibited lower MALAT1 expression after oxidized low‐density lipoprotein (ox‐LDL) treatment. MALAT1‐highly enriched sEV from ox‐LDL‐treated HUVECs inhibited ROS accumulation and dendritic cell maturation by interacting with and activating NFR2 (Table 2), thus preventing atherosclerosis development. 90
5.4. HOTAIR
HOX antisense intergenic RNA (HOTAIR) is a 2.2 kb, spliced and polyadenylated transcript that is repressed in the antisense direction at the HOXC locus on chromosome 12 in humans. HOTAIR is highly conserved among species and was the first lncRNA reported to function in trans. 107 Studies have shown that the serum levels of HOTAIR in patients with T2D, diabetic retinopathy and diabetic cardiomyopathy are significantly increased. 108 , 109 , 110 This elevation is also found in different tissues in diabetic states, such as liver tissues, 111 kidney tissues 112 and myocardial tissues. 110 Interestingly, HOTAIR is expressed only in gluteal but not abdominal adipose tissue, and transfecting HOTAIR into abdominal preadipocytes promotes their differentiation into adipocytes. 113 Further study showed that obese subjects with sedentary lifestyles have higher sEV HOTAIR expression in the serum. An experiment showed that gluteal‐femoral fat increases the secretion of sEV HOTAIR, which is taken up by intestinal cells and promotes intestinal cell stemness/proliferation by binding to activated NF‐κB 91 (Table 2), indicating the regulatory role of HOTAIR in metabolic homeostasis. In addition, engineered sEV isolated from HOTAIR‐overexpressing MSCs promoted angiogenesis and wound healing in a diabetic mouse model. Furthermore, MSC‐derived sEV carrying HOTAIR upregulate angiogenic protein expression in endothelial cells. 92
5.5. SNHGs
Small nucleolar RNAs (snoRNAs) are a group of small noncoding RNAs that are localized in the nucleus. snoRNAs are generated from introns; if a full‐length transcript includes introns and exons from a snoRNA gene, it will function as a lan lncRNA, named a small nucleolar RNA host gene (SNHG). 114 , 115 The SNHG family contains various members, from SNHG1 to SNHG13, SNHG15, SNHG17, SNHG20 and SNHG28. 116 Some of these members were recognized as aggressive tumor promoters. For example, SNHG2 was shown to be enriched in sEV compared with their parent cells and to function as an apoptosis marker in tumor cell lines. 117 SNHGs were also shown to be adipogenesis regulators. Overexpression of SNHG2 (also known as GAS5) was shown to reduce lipid accumulation in 3T3‐L1 adipocytes, 118 and SNHG2 was found to be negatively correlated with MSC adipogenic differentiation. 119 Studies of SNHGs contained in sEVs have focused on their regulatory roles in endothelial cell functions. An experimental study demonstrated that SNHG2 was upregulated in a macrophage cell line (TPH‐1) by oxo‐LDL treatment, and sEV shed by SNHG2 knockdown THP‐1 cells inhibited the apoptosis of endothelial cells. 93 Similarly, MSC‐derived sEV containing SNHG7 repress HG‐induced endothelial dysfunction in human retinal microvascular endothelial cells. 94 In addition, sEV derived from SNHG9‐overexpressing adipocytes alleviated inflammation in endothelial cells. 95 Endothelial cell dysfunction is a major contributor to diabetic microvascular‐ or macrovascular‐related complications, such as retinopathy, nephropathy and cardiomyopathy. Thus, targeting sEV‐SNHGs could be a future research direction for diabetic complication therapy.
6. CONCLUSIONS AND FUTURE DIRECTIONS
To date, the relationship between lncRNAs and diabetic pathologies has been investigated extensively. However, the functional roles of lncRNAs carried in sEV in diabetes are still being elucidated. sEV lncRNAs have been shown to regulate diabetes‐related endothelial cell function, lipid metabolism, skin cell proliferation and bone fracture healing. In particular, lncRNAs containing sEV derived from MSCs and ADSCs are promising therapeutic agents for diabetic wound healing and fracture healing. 82 However, using sEV lncRNAs as therapeutic agents in the clinic is still challenging. sEV lncRNA profiles differ according to the disease state and cell conditions; thus, clarifying the origin of sEV is a mandatory step for further functional study. Most studies obtain sEV from the serum of patients, sEV from plasma are a heterogeneous population originating from different cell types and from different sources, which makes it difficult to establish their exact physiological roles and functions. Although various single vesicle technologies have been developed to unravel the heterogeneity of EV, 120 , 121 the current vesicle isolation and enrichment techniques prefer to identify the particular EV subpopulations based on physical properties only or based on compositions and functions. 122 In addition, sEV obtained from a conditioned medium of cells did not always have consistent results in vivo. Additionally, there are still problems with off‐target effects, as the reported sEV lncRNAs all have multiple targets. In addition, although some mechanisms have been revealed for lncRNA sorting into EV, 123 such as RBP binding, miRNA mediating and subcellular localization affecting, further investigation is needed to understand the exact lncRNA sorting mechanisms. Additionally, the lncRNAs secreted by cells to exert paracrine effects or simply for elimination need to be further investigated. Overall, the mechanisms of lncRNAs and sEV lncRNAs in diabetes need more in‐depth research.
FUNDING INFORMATION
This work was supported by National Natural Science Foundation of China (81700704).
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
7.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/tra.12868.
ACKNOWLEDGMENTS
Wenguang Chang, Man Wang, Yuan Zhang and Fei Yu collected all of the data, and Wenguang Chang, Bin Hu, Katarzyna Goljanek‐Whysall and Peifeng Li wrote and revised the manuscript. All authors have read and approved the final version of the manuscript.
Chang W, Wang M, Zhang Y, et al. Roles of long noncoding RNAs and small extracellular vesicle‐long noncoding RNAs in type 2 diabetes. Traffic. 2022;23(11):526‐537. doi: 10.1111/tra.12868
Funding information National Natural Science Foundation of China, Grant/Award Number: 81700704
Contributor Information
Wenguang Chang, Email: changsubmit@126.com.
Peifeng Li, Email: peifli@hotmail.com.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Pfeiffer AF, Klein HH. The treatment of type 2 diabetes. Dtsch Arztebl Int. 2014;111(5):69‐81. quiz 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zheng Y, Ley SH, Hu FB. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol. 2018;14(2):88‐98. [DOI] [PubMed] [Google Scholar]
- 3. Gregg EW, Sattar N, Ali MK. The changing face of diabetes complications. Lancet Diabetes Endocrinol. 2016;4(6):537‐547. [DOI] [PubMed] [Google Scholar]
- 4. Deogharia M, Gurha P. The “guiding” principles of noncoding RNA function. Wiley Interdiscip Rev RNA. 2022;13(4):e1704. [DOI] [PubMed] [Google Scholar]
- 5. Sathishkumar C, Prabu P, Mohan V, Balasubramanyam M. Linking a role of lncRNAs (long non‐coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum Genomics. 2018;12(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quinn JJ, Chang HY. Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47‐62. [DOI] [PubMed] [Google Scholar]
- 9. Zheng GX, Do BT, Webster DE, Khavari PA, Chang HY. Dicer‐microRNA‐Myc circuit promotes transcription of hundreds of long noncoding RNAs. Nat Struct Mol Biol. 2014;21(7):585‐590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alcid EA, Tsukiyama T. ATP‐dependent chromatin remodeling shapes the long noncoding RNA landscape. Genes Dev. 2014;28(21):2348‐2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ip JY, Nakagawa S. Long non‐coding RNAs in nuclear bodies. Dev Growth Differ. 2012;54(1):44‐54. [DOI] [PubMed] [Google Scholar]
- 12. Galganski L, Urbanek MO, Krzyzosiak WJ. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 2017;45(18):10350‐10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904‐914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hua F. New insights into diabetes mellitus and its complications: a narrative review. Ann Transl Med. 2020;8(24):1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang W, Wang J. Exosomes and their noncoding RNA cargo are emerging as new modulators for diabetes mellitus. Cell. 2019;8(8):853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang W. Non‐coding RNAs and Berberine: a new mechanism of its anti‐diabetic activities. Eur J Pharmacol. 2017;795:8‐12. [DOI] [PubMed] [Google Scholar]
- 17. Moran I, Akerman I, van de Bunt M, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue‐specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16(4):435‐448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arnes L, Akerman I, Balderes DA, Ferrer J, Sussel L. betalinc1 encodes a long noncoding RNA that regulates islet beta‐cell formation and function. Genes Dev. 2016;30(5):502‐507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanchez‐Parra C, Jacovetti C, Dumortier O, et al. Contribution of the long noncoding RNA H19 to beta‐cell mass expansion in neonatal and adult rodents. Diabetes. 2018;67(11):2254‐2267. [DOI] [PubMed] [Google Scholar]
- 20. Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell. 2014;55(3):372‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang S, Zhu W, Zheng F, et al. The long noncoding RNA Blnc1 protects against diet‐induced obesity by promoting mitochondrial function in white fat. Diabetes Metab Syndr Obes. 2020;13:1189‐1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tran KV, Brown EL, DeSouza T, et al. Human thermogenic adipocyte regulation by the long noncoding RNA LINC00473. Nat Metab. 2020;2(5):397‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Y, Li K, Zhang X, Chen J, Li M, Liu L. The novel long noncoding RNA lncRNA‐Adi regulates adipogenesis. Stem Cells Transl Med. 2020;9(9):1053‐1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen YT, Yang QY, Hu Y, et al. Imprinted lncRNA Dio3os preprograms intergenerational brown fat development and obesity resistance. Nat Commun. 2021;12(1):6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goyal N, Sivadas A, Shamsudheen KV, et al. RNA sequencing of db/db mice liver identifies lncRNA H19 as a key regulator of gluconeogenesis and hepatic glucose output. Sci Rep. 2017;7(1):8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen X, Zhang Y, Zhang X, et al. Long non‐coding RNA Bhmt‐AS attenuates hepatic gluconeogenesis via modulation of Bhmt expression. Biochem Biophys Res Commun. 2019;516(1):215‐221. [DOI] [PubMed] [Google Scholar]
- 27. Wang J, Yang W, Chen Z, et al. Long noncoding RNA lncSHGL recruits hnRNPA1 to suppress hepatic gluconeogenesis and lipogenesis. Diabetes. 2018;67(4):581‐593. [DOI] [PubMed] [Google Scholar]
- 28. Han M, You L, Wu Y, et al. RNA‐sequencing analysis reveals the potential contribution of lncRNAs in palmitic acid‐induced insulin resistance of skeletal muscle cells. Biosci Rep. 2020;40(1):BSR2019252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang W, Bai Y, Chen Z, et al. Comprehensive analysis of long non‐coding RNAs and mRNAs in skeletal muscle of diabetic Goto‐Kakizaki rats during the early stage of type 2 diabetes. PeerJ. 2020;8:e8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geng T, Liu Y, Xu Y, et al. H19 lncRNA promotes skeletal muscle insulin sensitivity in part by targeting AMPK. Diabetes. 2018;67(11):2183‐2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gui W, Zhu WF, Zhu Y, et al. LncRNAH19 improves insulin resistance in skeletal muscle by regulating heterogeneous nuclear ribonucleoprotein A1. Cell Commun Signal. 2020;18(1):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu X, Liu Y, Cui J, et al. LncRNA LYPLAL1‐DT screening from type 2 diabetes with macrovascular complication contributes protective effects on human umbilical vein endothelial cells via regulating the miR‐204‐5p/SIRT1 axis. Cell Death Discov. 2022;8(1):245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yi H, Peng R, Zhang LY, et al. LincRNA‐Gm4419 knockdown ameliorates NF‐kappaB/NLRP3 inflammasome‐mediated inflammation in diabetic nephropathy. Cell Death Dis. 2017;8(2):e2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Du P, Wang J, Han Y, Feng J. Blocking the LncRNA MALAT1/miR‐224‐5p/NLRP3 Axis inhibits the hippocampal inflammatory response in T2DM with OSA. Front Cell Neurosci. 2020;14:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ashjari D, Karamali N, Rajabinejad M, et al. The axis of long non‐coding RNA MALAT1/miR‐1‐3p/CXCR4 is dysregulated in patients with diabetic neuropathy. Heliyon. 2022;8(3):e09178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cai Q, Wang C, Huang L, et al. Long non‐coding RNA small nucleolar RNA host gene 5 (SNHG5) regulates renal tubular damage in diabetic nephropathy via targeting MiR‐26a‐5p. Horm Metab Res. 2021;53(12):818‐824. [DOI] [PubMed] [Google Scholar]
- 37. Luo R, Xiao F, Wang P, Hu YX. lncRNA H19 sponging miR‐93 to regulate inflammation in retinal epithelial cells under hyperglycemia via XBP1s. Inflamm Res. 2020;69(3):255‐265. [DOI] [PubMed] [Google Scholar]
- 38. Wu H, Wen F, Jiang M, Liu Q, Nie Y. LncRNA uc.48+ is involved in the diabetic immune and inflammatory responses mediated by P2X7 receptor in RAW264.7 macrophages. Int J Mol Med. 2018;42(2):1152‐1160. [DOI] [PubMed] [Google Scholar]
- 39. Das S, Reddy MA, Senapati P, et al. Diabetes mellitus‐induced long noncoding RNA Dnm3os regulates macrophage functions and inflammation via nuclear mechanisms. Arterioscler Thromb Vasc Biol. 2018;38(8):1806‐1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reddy MA, Amaram V, Das S, et al. lncRNA DRAIR is downregulated in diabetic monocytes and modulates the inflammatory phenotype via epigenetic mechanisms. JCI Insight. 2021;6(11):e143289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stapleton K, das S, Reddy MA, et al. Novel long noncoding RNA, macrophage inflammation‐suppressing transcript (MIST), regulates macrophage activation during obesity. Arterioscler Thromb Vasc Biol. 2020;40(4):914‐928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu H, Guo R, Zhang Y, et al. Inhibition of lncRNA TCONS_00077866 ameliorates the high stearic acid diet‐induced mouse pancreatic beta‐cell inflammatory response by increasing miR‐297b‐5p to downregulate SAA3 expression. Diabetes. 2021;70(10):2275‐2288. [DOI] [PubMed] [Google Scholar]
- 43. Heeren J, Scheja L. Brown adipose tissue and lipid metabolism. Curr Opin Lipidol. 2018;29(3):180‐185. [DOI] [PubMed] [Google Scholar]
- 44. Liu J, Liang X, Zhou D, et al. Coupling of mitochondrial function and skeletal muscle fiber type by a miR‐499/Fnip1/AMPK circuit. EMBO Mol Med. 2016;8(10):1212‐1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y, Tang H, Ji X, et al. Expression profile analysis of long non‐coding RNAs involved in the metformin‐inhibited gluconeogenesis of primary mouse hepatocytes. Int J Mol Med. 2018;41(1):302‐310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shu L, Hou X, Song G, Wang C, Ma H. Comparative analysis of long noncoding RNA expression profiles induced by resveratrol and metformin treatment for hepatic insulin resistance. Int J Mol Med. 2021;48(5):206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou L, Sun K, Zhao Y, et al. Linc‐YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun. 2015;6:10026. [DOI] [PubMed] [Google Scholar]
- 48. Wang L, Zhao Y, Bao X, et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015;25(3):335‐350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu X, Zhang Y, Li T, et al. Long non‐coding RNA Linc‐RAM enhances myogenic differentiation by interacting with MyoD. Nat Commun. 2017;8:14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol. 2011;11(2):98‐107. [DOI] [PubMed] [Google Scholar]
- 51. Feng Y, Ge Y, Wu M, et al. Long noncoding RNAs regulate inflammation in diabetic peripheral neuropathy by acting as ceRNAs targeting miR‐146a‐5p. Diabetes Metab Syndr Obes. 2020;13:413‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang Y, Xu W, Zhou R. NLRP3 inflammasome activation and cell death. Cell Mol Immunol. 2021;18(9):2114‐2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mitchell S, Vargas J, Hoffmann A. Signaling via the NFkappaB system. Wiley Interdiscip Rev Syst Biol Med. 2016;8(3):227‐241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cai R, Jiang J. LncRNA ANRIL silencing alleviates high glucose‐induced inflammation, oxidative stress, and apoptosis via upregulation of MME in podocytes. Inflammation. 2020;43(6):2147‐2155. [DOI] [PubMed] [Google Scholar]
- 55. Zhao J, Liu B, Li C. Knockdown of long noncoding RNA GAS5 protects human cardiomyocyte‐like AC16 cells against high glucose‐induced inflammation by inhibiting miR‐21‐5p‐mediated TLR4/NF‐kappaB signaling. Naunyn Schmiedebergs Arch Pharmacol. 2020;393(8):1541‐1547. [DOI] [PubMed] [Google Scholar]
- 56. Jiang L, Wang C, Shen X. LncRNA GAS5 suppresses ER stressinduced apoptosis and inflammation by regulating SERCA2b in HGtreated retinal epithelial cell. Mol Med Rep. 2020;22(2):1072‐1080. [DOI] [PubMed] [Google Scholar]
- 57. Li X, Zhao Z, Gao C, et al. The diagnostic value of whole blood lncRNA ENST00000550337.1 for pre‐diabetes and type 2 diabetes mellitus. Exp Clin Endocrinol Diabetes. 2017;125(6):377‐383. [DOI] [PubMed] [Google Scholar]
- 58. Awata T, Yamashita H, Kurihara S, et al. Correction: a genome‐wide association study for diabetic retinopathy in a Japanese population: potential association with a long intergenic non‐coding RNA. PLoS One. 2015;10(4):e0126789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chang W, Xiao D, Fang X, Wang J. Phospholipids in small extracellular vesicles: emerging regulators of neurodegenerative diseases and cancer. Cytotherapy. 2022;24(2):93‐100. [DOI] [PubMed] [Google Scholar]
- 60. Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cell. 2019;8(7):727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487‐514. [DOI] [PubMed] [Google Scholar]
- 63. Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226‐1232. [DOI] [PubMed] [Google Scholar]
- 64. Mathieu M, Martin‐Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell‐to‐cell communication. Nat Cell Biol. 2019;21(1):9‐17. [DOI] [PubMed] [Google Scholar]
- 65. Villarroya‐Beltri C, Gutiérrez‐Vázquez C, Sánchez‐Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lei Y, Guo W, Chen B, Chen L, Gong J, Li W. Tumorreleased lncRNA H19 promotes gefitinib resistance via packaging into exosomes in nonsmall cell lung cancer. Oncol Rep. 2018;40(6):3438‐3446. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67. Han M, Gu Y, Lu P, et al. Exosome‐mediated lncRNA AFAP1‐AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol Cancer. 2020;19(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68. Zheng Z, Chen M, Xing P, Yan X, Xie B. Increased expression of Exosomal AGAP2‐AS1 (AGAP2 antisense RNA 1) in breast cancer cells inhibits trastuzumab‐induced cell cytotoxicity. Med Sci Monit. 2019;25:2211‐2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cha DJ, Franklin JL, Dou Y, et al. KRAS‐dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hinger SA, Cha DJ, Franklin JL, et al. Diverse long RNAs are differentially sorted into extracellular vesicles secreted by colorectal cancer cells. Cell Rep. 2018;25(3):715‐725.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen C, Luo Y, He W, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. 2020;130(1):404‐421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Batagov AO, Kuznetsov VA, Kurochkin IV. Identification of nucleotide patterns enriched in secreted RNAs as putative cis‐acting elements targeting them to exosome nano‐vesicles. BMC Genomics. 2011;12(Suppl 3):S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. O'Grady T, Njock MS, Lion M, et al. Sorting and packaging of RNA into extracellular vesicles shape intracellular transcript levels. BMC Biol. 2022;20(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sun Z, Yang S, Zhou Q, et al. Emerging role of exosome‐derived long non‐coding RNAs in tumor microenvironment. Mol Cancer. 2018;17(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xie F, Liu YL, Chen XY, et al. Role of MicroRNA, LncRNA, and exosomes in the progression of osteoarthritis: a review of recent literature. Orthop Surg. 2020;12(3):708‐716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cheng J, Meng J, Zhu L, Peng Y. Exosomal noncoding RNAs in glioma: biological functions and potential clinical applications. Mol Cancer. 2020;19(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cao M, Zhang L, Lin Y, et al. Differential mRNA and long noncoding RNA expression profiles in umbilical cord blood exosomes from gestational diabetes mellitus patients. DNA Cell Biol. 2020;39(11):2005‐2016. [DOI] [PubMed] [Google Scholar]
- 78. Zhou S, Fang J, Hu M, et al. Determining the influence of high glucose on exosomal lncRNAs, mRNAs, circRNAs and miRNAs derived from human renal tubular epithelial cells. Aging (Albany NY). 2021;13(6):8467‐8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Huang G, Garikipati VNS, Zhou Y, et al. Identification and comparison of hyperglycemia‐induced extracellular vesicle transcriptome in different mouse stem cells. Cells. 2020;9(9):2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Krishnan P, Syed F, Jiyun Kang N, G. Mirmira R, Evans‐Molina C. Profiling of RNAs from human islet‐derived exosomes in a model of type 1 diabetes. Int J Mol Sci. 2019;20(23):5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ruan Y, Lin N, Ma Q, et al. Circulating LncRNAs analysis in patients with type 2 diabetes reveals novel genes influencing glucose metabolism and islet beta‐cell function. Cell Physiol Biochem. 2018;46(1):335‐350. [DOI] [PubMed] [Google Scholar]
- 82. Chang W, Li M, Song L, Miao S, Yu W, Wang J. Noncoding RNAs from tissue‐derived small extracellular vesicles: roles in diabetes and diabetic complications. Mol Metab. 2022;58:101453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Qian L, Pi L, Fang BR, Meng XX. Adipose mesenchymal stem cell‐derived exosomes accelerate skin wound healing via the lncRNA H19/miR‐19b/SOX9 axis. Lab Invest. 2021;101(9):1254‐1266. [DOI] [PubMed] [Google Scholar]
- 84. Li B, Luan S, Chen J, et al. The MSC‐derived exosomal lncRNA H19 promotes wound healing in diabetic foot ulcers by upregulating PTEN via MicroRNA‐152‐3p. Mol Ther Nucleic Acids. 2020;19:814‐826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang Y, Chen W, Zhao L, et al. Obesity regulates miR‐467/HoxA10 axis on osteogenic differentiation and fracture healing by BMSC‐derived exosome LncRNA H19. J Cell Mol Med. 2021;25(3):1712‐1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tao SC, Rui BY, Wang QY, Zhou D, Zhang Y, Guo SC. Extracellular vesicle‐mimetic nanovesicles transport LncRNA‐H19 as competing endogenous RNA for the treatment of diabetic wounds. Drug Deliv. 2018;25(1):241‐255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. El Bassit G, Patel RS, Carter G, et al. MALAT1 in human adipose stem cells modulates survival and alternative splicing of PKCdeltaII in HT22 cells. Endocrinology. 2017;158(1):183‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gao J, Li X, Wang Y, et al. Adipocyte‐derived extracellular vesicles modulate appetite and weight through mTOR signalling in the hypothalamus. Acta Physiol (Oxf). 2020;228(2):e13339. [DOI] [PubMed] [Google Scholar]
- 89. He L, Zhu C, Jia J, et al. ADSC‐exos containing MALAT1 promotes wound healing by targeting miR‐124 through activating Wnt/beta‐catenin pathway. Biosci Rep. 2020;40(5):BSR20192549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Li H, Zhu X, Hu L, Li Q, Ma J, Yan J. Loss of exosomal MALAT1 from ox‐LDL‐treated vascular endothelial cells induces maturation of dendritic cells in atherosclerosis development. Cell Cycle. 2019;18(18):2255‐2267. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91. Lu X, Bai D, Liu X, Zhou C, Yang G. Sedentary lifestyle related exosomal release of Hotair from gluteal‐femoral fat promotes intestinal cell proliferation. Sci Rep. 2017;7:45648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Born LJ, Chang K‐H, Shoureshi P, et al. HOTAIR‐loaded mesenchymal stem/stromal cell extracellular vesicles enhance angiogenesis and wound healing. Adv Healthc Mater. 2021;11:e2002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen L, Yang W, Guo Y, et al. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS One. 2017;12(9):e0185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cao X, Xue LD, di Y, Li T, Tian YJ, Song Y. MSC‐derived exosomal lncRNA SNHG7 suppresses endothelial‐mesenchymal transition and tube formation in diabetic retinopathy via miR‐34a‐5p/XBP1 axis. Life Sci. 2021;272:119232. [DOI] [PubMed] [Google Scholar]
- 95. Song Y, Li H, Ren X, Li H, Feng C. SNHG9, delivered by adipocyte‐derived exosomes, alleviates inflammation and apoptosis of endothelial cells through suppressing TRADD expression. Eur J Pharmacol. 2020;872:172977. [DOI] [PubMed] [Google Scholar]
- 96. Zhu L, Li Y, Xia F, et al. H19: a vital long noncoding RNA in the treatment of diabetes and diabetic complications. Curr Pharm des. 2022;28(12):1011‐1018. [DOI] [PubMed] [Google Scholar]
- 97. Alfaifi M, Verma AK, Alshahrani MY, et al. Assessment of cell‐free long non‐coding RNA‐H19 and miRNA‐29a, miRNA‐29b expression and severity of diabetes. Diabetes Metab Syndr Obes. 2020;13:3727‐3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhou Y, Yi Y, Wang J, et al. Discovery of novel pleuromutilin derivatives as potent antibacterial agents. Eur J Med Chem. 2022;237:114403. [DOI] [PubMed] [Google Scholar]
- 99. Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 2017;14(12):1705‐1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ji P, Diederichs S, Wang W, et al. MALAT‐1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early‐stage non‐small cell lung cancer. Oncogene. 2003;22(39):8031‐8041. [DOI] [PubMed] [Google Scholar]
- 101. Goyal B, Yadav SRM, Awasthee N, Gupta S, Kunnumakkara AB, Gupta SC. Diagnostic, prognostic, and therapeutic significance of long non‐coding RNA MALAT1 in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875(2):188502. [DOI] [PubMed] [Google Scholar]
- 102. Gong YP, Zhang YW, Su XQ, Gao HB. Inhibition of long noncoding RNA MALAT1 suppresses high glucose‐induced apoptosis and inflammation in human umbilical vein endothelial cells by suppressing the NF‐kappaB signaling pathway. Biochem Cell Biol. 2020;98(6):669‐675. [DOI] [PubMed] [Google Scholar]
- 103. Radhakrishnan R, Kowluru RA. Long noncoding RNA MALAT1 and regulation of the antioxidant defense system in diabetic retinopathy. Diabetes. 2021;70(1):227‐239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Shyu KG, Wang BW, Fang WJ, Pan CM, Lin CM. Exosomal MALAT1 derived from high glucose‐treated macrophages up‐regulates resistin expression via miR‐150‐5p downregulation. Int J Mol Sci. 2022;23(3):1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Tello‐Flores VA, Valladares‐Salgado A, Ramírez‐Vargas MA, et al. Altered levels of MALAT1 and H19 derived from serum or serum exosomes associated with type‐2 diabetes. Noncoding RNA Res. 2020;5(2):71‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Patel RS, Carter G, El Bassit G. Adipose‐derived stem cells from lean and obese humans show depot specific differences in their stem cell markers, exosome contents and senescence: role of protein kinase C delta (PKCdelta) in adipose stem cell niche. Stem Cell Investig. 2016;3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311‐1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang H, Xia Y, Zhang Y. Diagnostic significance of serum lncRNA HOTAIR and its predictive value for the development of chronic complications in patients with type 2 diabetes mellitus. Diabetol Metab Syndr. 2021;13(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shaker OG, Abdelaleem OO, Mahmoud RH, et al. Diagnostic and prognostic role of serum miR‐20b, miR‐17‐3p, HOTAIR, and MALAT1 in diabetic retinopathy. IUBMB Life. 2019;71(3):310‐320. [DOI] [PubMed] [Google Scholar]
- 110. Qi K, Zhong J. LncRNA HOTAIR improves diabetic cardiomyopathy by increasing viability of cardiomyocytes through activation of the PI3K/Akt pathway. Exp Ther Med. 2018;16(6):4817‐4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Li M, Guo Y, Wang XJ, Duan BH, Li L. HOTAIR participates in hepatic insulin resistance via regulating SIRT1. Eur Rev Med Pharmacol Sci. 2018;22(22):7883‐7890. [DOI] [PubMed] [Google Scholar]
- 112. Majumder S, Hadden MJ, Thieme K, et al. Dysregulated expression but redundant function of the long non‐coding RNA HOTAIR in diabetic kidney disease. Diabetologia. 2019;62(11):2129‐2142. [DOI] [PubMed] [Google Scholar]
- 113. Divoux A, Karastergiou K, Xie H, et al. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity (Silver Spring). 2014;22(8):1781‐1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12(2):84‐88. [DOI] [PubMed] [Google Scholar]
- 115. Zimta AA, Tigu AB, Braicu C, Stefan C, Ionescu C, Berindan‐Neagoe I. An emerging class of long non‐coding RNA with oncogenic role arises from the snoRNA host genes. Front Oncol. 2020;10:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shi J, Ding W, Lu H. Identification of long non‐coding RNA SNHG family as promising prognostic biomarkers in acute myeloid leukemia. Onco Targets Ther. 2020;13:8441‐8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Dolan KL, Peña J, Allison SD, Martiny JBH. Phylogenetic conservation of substrate use specialization in leaf litter bacteria. PLoS One. 2017;12(3):e0174472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Liu H, Li H, Jin L, et al. Long noncoding RNA GAS5 suppresses 3T3‐L1 cells adipogenesis through miR‐21a‐5p/PTEN signal pathway. DNA Cell Biol. 2018;37(9):767‐777. [DOI] [PubMed] [Google Scholar]
- 119. Xu L, Gong ZR, Tao MJ, Ai Q. Artificial light harvesting by dimerized Mobius ring. Phys Rev E. 2018;97(4–1):042124. [DOI] [PubMed] [Google Scholar]
- 120. Nolan JP, Duggan E. Analysis of individual extracellular vesicles by flow cytometry. Methods Mol Biol. 2018;1678:79‐92. [DOI] [PubMed] [Google Scholar]
- 121. Han C, Kang H, Yi J, et al. Single‐vesicle imaging and co‐localization analysis for tetraspanin profiling of individual extracellular vesicles. J Extracell Vesicles. 2021;10(3):e12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Bordanaba‐Florit G, Royo F, Kruglik SG, Falcón‐Pérez JM. Using single‐vesicle technologies to unravel the heterogeneity of extracellular vesicles. Nat Protoc. 2021;16(7):3163‐3185. [DOI] [PubMed] [Google Scholar]
- 123. He L, Lin M, Shen J, Qi H. Emerging role of exosomal long non‐coding RNAs in lung cancer. Mol Biol Rep. 2022;49(6):4989‐4997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


