Abstract
Climate change will strongly affect the developmental timing of insects, as their development rate depends largely on ambient temperature. However, we know little about the genetic mechanisms underlying the temperature sensitivity of embryonic development in insects. We investigated embryonic development rate in the winter moth (Operophtera brumata), a species with egg dormancy which has been under selection due to climate change. We used RNA sequencing to investigate which genes are involved in the regulation of winter moth embryonic development rate in response to temperature. Over the course of development, we sampled eggs before and after an experimental change in ambient temperature, including two early development weeks when the temperature sensitivity of eggs is low and two late development weeks when temperature sensitivity is high. We found temperature‐responsive genes that responded in a similar way across development, as well as genes with a temperature response specific to a particular development week. Moreover, we identified genes whose temperature effect size changed around the switch in temperature sensitivity of development rate. Interesting candidate genes for regulating the temperature sensitivity of egg development rate included genes involved in histone modification, hormonal signalling, nervous system development and circadian clock genes. The diverse sets of temperature‐responsive genes we found here indicate that there are many potential targets of selection to change the temperature sensitivity of embryonic development rate. Identifying for which of these genes there is genetic variation in wild insect populations will give insight into their adaptive potential in the face of climate change.
Keywords: circadian clock genes, climate change adaptation, diapause, insect embryogenesis, RNAseq, temperature sensitivity
1. INTRODUCTION
Climate change drastically alters the environment that species experience and will in the long run require them to genetically adapt to persist (Visser, 2008). Changes in ambient temperatures can be especially challenging for ectothermic species such as insects, whose development rate is strongly affected by environmental temperature (Van Dyck et al., 2015). Climate change is already altering the developmental timing of many insects, for example driving earlier egg hatching in the winter moth, Operophtera brumata (van Asch et al., 2013), and increasing generation turnover rate in many European butterflies and moths (Altermatt, 2010). Genes involved in the temperature sensitivity of development rate are probably targets of climate change‐induced selection, particularly for herbivorous spring‐feeding insects that need to time their egg hatching to the phenology of their host plant (van Asch & Visser, 2007). However, little is known about the molecular pathways underlying the temperature sensitivity of embryonic developmental timing in insects (Mirth et al., 2021).
The rate of embryonic development is ultimately determined by how metabolic rate scales with temperature (Gillooly et al., 2002). Nevertheless, additional layers of regulation can change the temperature sensitivity of development rate. This is exemplified by insect dormancy: a period of increased environmental stress resistance in which development slows down until environmental conditions become more favourable (Danks, 1987; Wilsterman et al., 2021). Lower rates of development during insect dormancy coincide with important gene expression changes in, for example, cell cycle regulators (Denlinger, 2002; Poupardin et al., 2015). Based on the degree of endogenous programming and the magnitude of developmental suppression, insect dormancy responses can be viewed as a spectrum ranging from quiescence, in which development rate remains responsive to the environment, to diapause: a deep, programmed developmental arrest (Wilsterman et al., 2021).
Insect dormancy responses are often targeted by climate change‐induced selection (Bradshaw & Holzapfel, 2006; Forrest, 2016). An important example comes from the winter moth. Warmer winters due to climate change have caused its eggs to hatch before their food source, young oak leaves, become available (Visser & Holleman, 2001). Winter moths exhibit dormancy during the egg phase, involving slow but progressive embryonic development that takes several months. Egg development rate remains responsive to temperature changes over the course of development, but the magnitude of this direct response to temperature changes over the course of development (Salis et al., 2016; van Dis et al., 2021), suggesting differential regulatory processes. Our previous work showed that the temperature sensitivity of development rate switches from low to high sensitivity once embryos have developed a rudimentary nervous system (van Dis et al., 2021). There is genetic variation for both the baseline temperature sensitivity of embryonic development rate as well as for the change in its temperature sensitivity (van Asch et al., 2007; van Dis et al., 2021), but the genetic mechanisms underlying the temperature sensitivity of winter moth egg development rate remain unknown.
Here, we explore which genes are involved in regulating the temperature sensitivity of winter moth embryonic development rate. Using RNA sequencing (RNAseq), we characterized embryonic gene expression responses to a change in ambient temperature. In total, we sampled four different development weeks, evenly distributed over the course of development, including two development weeks when temperature sensitivity of egg development rate is low and two development weeks when temperature sensitivity is high (van Dis et al., 2021). We first investigate which genes are expressed in each week at a constant control temperature using co‐expression analysis to gain insight into the ongoing developmental processes. Next, we use differential expression analysis to test in each development week which genes change expression in the first 24 h after an increase or decrease in ambient temperature. Using gene ontology (GO) overrepresentation analysis, we highlight groups of temperature‐responsive genes during embryonic development that could be important candidates for regulating the temperature sensitivity of insect development rate.
2. METHODS
2.1. Experimental design
To find temperature‐sensitive genes involved in the regulation of winter moth embryonic development rate, we sampled RNA in a similar split‐brood experiment as previously used (van Dis et al., 2021). The experimental design is visualized in Figure 1. We collected eggs from wild winter moth females caught during the peak of adult emergence in a forest in Doorwerth, the Netherlands (Catch dates: November 26 and 29, and December 3, 2018). Clutches laid in the period after catching (ranging from 190 to 351 eggs) were placed in climate cabinets in constant darkness at a constant 10°C at the start of the experiment (December 14, 2018). Every week, for a period of 8 weeks, a group of six clutches was given a temperature treatment. At the time of treatment, each clutch was divided into eight subclutches of at least 25 eggs. Before the start of the temperature treatment, one subclutch was sampled for RNA extraction and one subclutch was dechorionated with 50% bleach and fixed with 4% formaldehyde to determine the median development stage of the clutch with fluorescence microscopy, as described in van Dis et al. (2021). The remaining six subclutches were divided over three temperature treatments: transferred either to a warm treatment (15°C) or a cold treatment (5°C) or remained at baseline temperature (10°C). To capture early gene expression changes in response to temperature, we sampled RNA for each treatment from one of the subclutches at 3 h and the other subclutch at 24 h after the transfer. RNA samples (0, 3 and 24 h) were always taken at the same time of day in each sampled development week.
FIGURE 1.
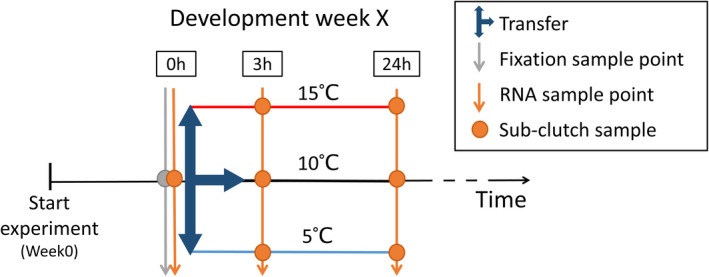
Experimental split‐brood design. From the start of the experiment, clutches of wild winter moth eggs were kept in climate cabinets at a constant 10°C in constant darkness (black timeline). Each clutch was divided into eight subclutches of at least 25 eggs (points). Before the start of the temperature treatment, one subclutch was sampled for RNA extraction (orange arrow at 0 h) and one subclutch was fixed with 4% formaldehyde to determine the median development stage of the clutch with fluorescence microscopy (grey arrow at 0 h). The remaining six subclutches were divided over three temperature treatments (blue arrows), with two transferred to a warm treatment (red line, 15°C), two to a cold treatment (blue line, 5°C) and two remained at baseline temperature (black line, 10°C). For each temperature treatment, we sampled one of the subclutches at 3 h and the other subclutch at 24 h after the transfer for RNA extraction. In total, we gave eggs a temperature treatment in seven development weeks: Every week from 2 weeks after the start of the experiment to week8, six clutches were given a temperature treatment per development week. For developmental weeks 2, 4, 6 and 8, three of these six clutches were used for RNA sequencing, to obtain three biological replicates per sampling point
Winter moth egg development takes ~10 weeks at constant 10°C (van Dis et al., 2021). For RNAseq, we selected four development weeks that were evenly distributed over the course of development (week2, week4, week6 and week8). These included two early development weeks (week2 and week4) with clutches in development stages 7–8 when temperature sensitivity of egg development rate is low (Figure 2); and two later development weeks (week6 and week8) with clutches in development stages 9–11 when temperature sensitivity is high (Figure 2; van Dis et al., 2021). From the available 24 clutches sampled in these development weeks, we selected three clutches per development week for RNA extraction, based on the median development stage of the clutch. A full description of the selection process can be found in Section S1. In total we isolated RNA from three biological replicates per sampled development week (3 × 4 weeks; N = 12 clutches) with seven RNA samples per clutch (1× before transfer, 3× 3 h after, 3× 24 h after transfer; N = 84 samples) and one additional fixed sample to determine the clutch's median developmental stage at the time of the temperature treatment.
FIGURE 2.
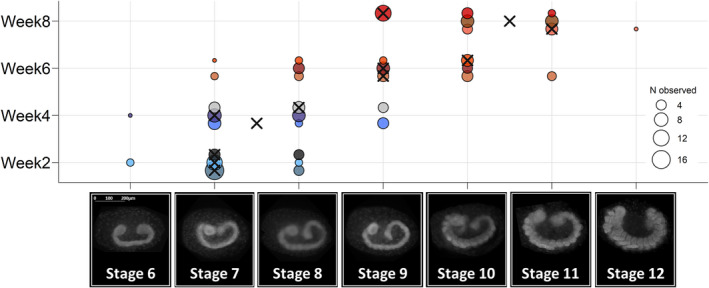
Development stages of sampled clutches at the time of temperature treatment. Before clutches were given a temperature treatment, eggs from each clutch were sampled and fixed for fluorescence microscopy to determine the development stage they were in at the time of temperature treatment. Development stages are indicated with pictures (adopted from van Dis et al., 2021). Each clutch is plotted on a horizontal line and has a unique colour (N = 12), with three clutches per sampled development week. Point size indicates the number of embryos observed in that developmental stage (at least 25 eggs per clutch sampled, 9–21 eggs could be scored), with crosses indicating the median development stage for each of the 12 clutches. Sampled clutches were between development stage 6 and stage 12, covering the period of germband elongation (stage 7) and segmentation (stage 8: Of thorax, stage 9: Of abdomen) to the formation (stage 10) and shaping of the appendages (stages 11–12, for more details see van Dis et al., 2021). This encompasses the switch from low to high temperature sensitivity of egg development rate that occurs after embryos have passed development stage 9 in which they develop a rudimentary nervous system (van Dis et al., 2021)
2.2. RNAseq and processing
Pools of eggs were snap frozen in liquid nitrogen and homogenized in Trizol at the moment of sampling for later RNA extraction with the Zymo Research (ZR) Tissue & Insect RNA MicroPrep kit (R2030; Zymo Research). We extracted total RNA from 84 pools of eggs (12 clutches, each divided into seven subclutches) with at least 25 eggs per pool. Extraction details can be found in Section S2. Library preparation was done with the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB), including a poly‐A selection step. All samples were sequenced in one run, using four lanes on the Illumina NovaSeq6000 platform, aiming for 30 million paired‐end 150‐bp reads per sample.
Raw reads were quality screened with fastqc version 0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and fastq‐screen version 0.14.1 (Wingett & Andrews, 2018). Using trimmomatic version 0.39 (Bolger et al., 2014), reads were trimmed to remove residual adapter content and leading and trailing bases with poor quality (phred <30). We followed the “new Tuxedo” pipeline for further RNAseq processing (Pertea et al., 2016), using the winter moth reference genome version 1 (Derks et al., 2015) with annotation version 2 (https://www.bioinformatics.nl/wintermoth/portal/data/). Briefly, following read alignment with hisat2 version 2.2.1 (Kim et al., 2019), and alignment quality control with picard tools version 2.23.8 (http://broadinstitute.github.io/picard/), reads marked as optical duplicates by picard markduplicates were filtered out (<1% of reads excluded). We continued with stringtie assembly and quantification version 2.1.2 (Pertea et al., 2015), guided by the winter moth reference genome. As RNAseq studies often find new transcripts, we turned on the stringtie option for predicting novel transcripts and isoforms. Predicted novel transcripts (i.e., transcripts that mapped to intergenic parts of the reference genome as indicated by gffcompare version 0.11.2; Pertea & Pertea, 2020) were blasted against NCBI's nonredundant (nr) protein database (accessed January 28, 2020) using diamond blastx version 2.0.6 (Buchfink et al., 2015) for all six reading frames. We only kept novel transcripts for which at least one homologue was found (25% of novel transcripts excluded). Transcript quantification was done at the gene level, resulting in a data set comprising the expression counts of 29,113 genes.
Raw sequencing reads can be found in the European Nucleotide Archive (ENA) under accession no. PRJEB55675. The transcriptome produced and used for transcript quantification by stringtie , including only transcripts in our data set with for each transcript coordinates to the winter moth reference genome version 1, can be found on Dryad (van Dis et al., 2022).
2.3. Functional annotation transcriptome
Because stringtie found many novel transcripts, we redid the functional annotation of the winter moth reference genome, but only for the transcripts in our data set. Each transcript was translated with emboss version 6.6.0 (Rice et al., 2000) for reading frames 1–3, as strand but not reading frame was specified by stringtie . We then searched for homologues with diamond blastp in three custom databases, only keeping the best hit with all E‐values <0.001: (i) a subset of Swissprot containing all Drosophila melanogaster proteins, (ii) a subset of Swissprot containing all insect proteins, and (iii) a subset of TrEMBL including all insect proteins (UniProtKB: [Bateman et al., 2021], accessed February 10, 2021); supplemented with NCBI's nonredundant (nr) database if no hit was found (4898 genes). To make the GO annotation as complete as possible, UniprotID‐associated GO term annotation (UniprotKB goa_all database, accessed June 16, 2021) was supplemented with interproscan version 5.52–86.0 (Jones et al., 2014) with the Pfam (Punta et al., 2012) and SUPERFAMILY (Wilson et al., 2009) databases; and with pannzer2 web service annotation using default settings and ARGOT_score ≥ 0.7 (Törönen et al., 2018; accessed September 25, 2021). The functionally annotated transcriptome and GO annotation table can be found on Dryad (van Dis et al., 2022).
2.4. Co‐expression analysis
We first characterized embryonic development gene expression at a constant 10°C using weighted gene correlation network analysis ( wgcna r package version 1.70; Langfelder & Horvath, 2008) in r version 4.1.0 (R Core Team, 2021), to gain insight into the ongoing developmental processes in each development week in which eggs received a temperature treatment.
We chose filtering criteria considering all data together (N = 84 samples). We excluded lowly expressed genes with a cut‐off of 20 counts/median library size counts per million (cpm; used cut‐off = 0.17 cpm), following Smyth et al. (2018). Genes were only kept if expressed above this cut‐off at any given time point in at least two of the three biological replicates (at least N = 2 for any given data point, 5152 genes excluded). Counts for the remaining 23,961 genes were normalized using voom (R package limma version 3.50.1; Ritchie et al., 2015) and plotted to validate the used cut‐off (Figure S3; Smyth et al., 2018). No outlier samples were detected using principal components analysis (PCA; Figure S4). Filtered and normalized expression counts for the constant 10°C samples were used for the WGCNA analysis (3 clutches × 4 development weeks × 3 time points; N = 36 samples).
Modules of co‐expressed genes were identified based on similarity in gene expression pattern over the 36 included samples, calculated as pairwise Pearson correlations between each pair of genes to create a signed first‐order network (Langfelder & Horvath, 2008). We chose a soft‐thresholding power of 12 (Figures S5.1 and S5.2), with a module size of at least 30 genes, and a merge cut‐height of 0.40 to prevent extensive overlap between the modules (Figure S5.3). The gene expression pattern of each module was summarized using a PCA, with PC1 showing the gene expression profile of the module. For each module, we tested if the gene expression profile differed significantly between the four development weeks using a linear mixed model with development week as the fixed effect and ClutchID as the random effect. We considered modules with a false discovery rate (FDR) < 0.05 (Table S1.1). For each significant module, only genes with a significant membership to that module were kept (i.e., significant correlation to the module's PC1, FDR <0.05, Table S1.2).
We performed GO overrepresentation analysis (Biological Processes, BP; and Molecular Function, MF) on each module of genes that significantly changed expression over the course of development. We used the R package topgo version 2.44.0 (Alexa et al., 2006) with the complete filtered 23,961 gene set as the background gene list, and only considered GO terms with at least five annotated genes. GO terms were tested with the Fisher exact test using the elim algorithm and considered significant at p < .01 (Alexa et al., 2006). The resulting significant GO terms were hierarchically clustered based on semantic similarity with the R package viseago version 1.6.0 (Brionne et al., 2019), also obtaining the parent term for each cluster to identify major themes. If more than 50 GO terms were significant, we used the categorizer webservice with classification GO_slim (Hu et al., 2008; accessed February 3, 2022) to obtain counts per ancestor term for overview.
2.5. Differential expression analysis
To find temperature‐sensitive genes, we conducted a differential expression analysis including all samples (4 development weeks × 3 clutches × 7 samples; N = 84). Count data were filtered and normalized as described above and were analysed with the R package limma , which uses a linear mixed model with a robust empirical Bayes procedure well suited for the analysis of RNAseq data (Phipson et al., 2016). As we have only three biological replicates for each sampling point, we used two separate models: one for the 3 h response (N = 48 samples) and one for the 24 h response (N = 48 samples) to avoid making the models too complex. In each model, we included temperature treatment, development week and the interaction between the two as fixed effects, and ClutchID as a random effect. Temperature treatment was included as a factor, with the before sample (taken at 0 h) as the reference level. In this way, all temperature treatment estimates are corrected for their common starting point, meaning we can compare the difference in response from 0 to 3 h and 0 to 24 h in each changing temperature treatment (i.e., 5°C increase or decrease) compared to the control constant temperature.
For each gene, we used post hoc tests to assess three different temperature treatment effects. First, we looked for genes with a similar temperature response in each development week (i.e., genes with overall temperature effects). For this, we took the average effect size of the four development weeks for each temperature treatment, comparing the warm and the cold treatment to the control, using estimates from the fixed effect temperature treatment. Second, we looked for genes that responded in a week‐specific manner by comparing the warm and cold treatments to the control for each of the 4 weeks respectively, using estimates from the fixed effects temperature treatment and development week (i.e., genes with within‐week temperature effects). Third, we tested for an interaction effect to find genes whose treatment effect size changed between the developmental weeks, using estimates from the interaction fixed effect between temperature treatment and development week (i.e., genes with between‐week temperature effects). For each post hoc test, p‐value distributions were verified and re‐estimated by correcting the variance of the null model with the R package fdrtool version 1.2.17 (Strimmer, 2008). A gene was considered significant when FDR <0.01.
We tested for overrepresented GO terms in the genes with a significant temperature response, first considering the significant genes for each of the three temperature effect post hoc tests together. We then further explored groups of genes for each temperature effect separately. For each temperature effect, we clustered the 3 h‐responding genes and the 24 h‐responding genes according to their temperature–response expression profile over the four development weeks with Pearson correlation using the R package pheatmap version 1.0.12 (Kolde, 2019). We split each clustered tree into a maximum of five clusters using a cut‐height of 1.75–1.80 and performed GO overrepresentation analysis on each cluster. For the genes with a significant within‐week temperature effect, we also looked for overrepresented GO terms in each week‐specific gene list. For the genes with a significant between‐week temperature effect, we further investigated genes that showed a significant change in temperature effect size around week6 where previous work found evidence for a switch in temperature sensitivity of development rate (van Dis et al., 2021).
3. RESULTS
3.1. Functional annotation
For each of the 84 samples, at least 85% of the reads mapped to the winter moth reference genome version 1 (range 85%–88%). fastq‐screen indicated that samples with lower mapping rates contained a higher percentage of reads that mapped to other genomes (incl. Microbial, up to 4%) indicating minor contamination. Assembling the remaining unmapped reads and performing a blast search did not reveal further contamination (data not shown), indicating that these reads are highly divergent or map to parts of the winter moth reference genome that are not yet well assembled. A large part of the reads mapped to intergenic regions of the reference genome (20%–25%, Figure S6), leading to the discovery of many novel isoforms and transcripts. Of the 29,113 genes in the final data set, transcripts from 15,962 genes matched the reference genome annotation version 2 (54.83%). For 7799 genes, we identified potential novel isoforms (26.79%), and for 11,353 genes novel transcripts of which 10,812 genes did not overlap with the reference genome annotation (39.00%). For 24,971 out of the 29,113 genes (85.77%) we found at least one significant blast hit against one of the functional annotation databases (databases: D. melanogaster Swissprot, Insects Swissprot, Insects TrEMBL, nr); 19,153 genes were also annotated with at least one GO term (65.79%).
3.2. Co‐expression analysis at constant 10°C: Developmental context
To characterize gene expression over the course of development, normalized expression counts from the samples of eggs that were kept at 10°C (i.e., the control treatment) were used in an unsupervised co‐expression analysis (N = 36 samples). The 23,961 genes that passed the low expression cut‐off clustered into 13 gene expression modules and one additional module containing the unclustered genes (N = 3517 genes). Of these modules, the expression profiles of four modules showed significant changes over the course of development (FDR <0.05, Table S1.1), whereas the other modules showed constant low, intermediate or high expression, or more idiosyncratic patterns that differed between clutches (see Figure S5.4).
Of the four modules that changed expression across development, two relatively small modules (modules 8 and 10, with 208 and 460 genes respectively, FDR <0.05) showed a gene expression pattern where the first sampled development week (week2) was different compared to the other weeks (Figure S5.5). The other two, much larger, modules contained the majority of the genes that changed expression over the course of development (Figure 3, modules 3 and 13 with 3497 and 3280 genes respectively). These genes either gradually went down (module 13, Figure 3a) or up in expression over time (module 3, Figure 3b).
FIGURE 3.
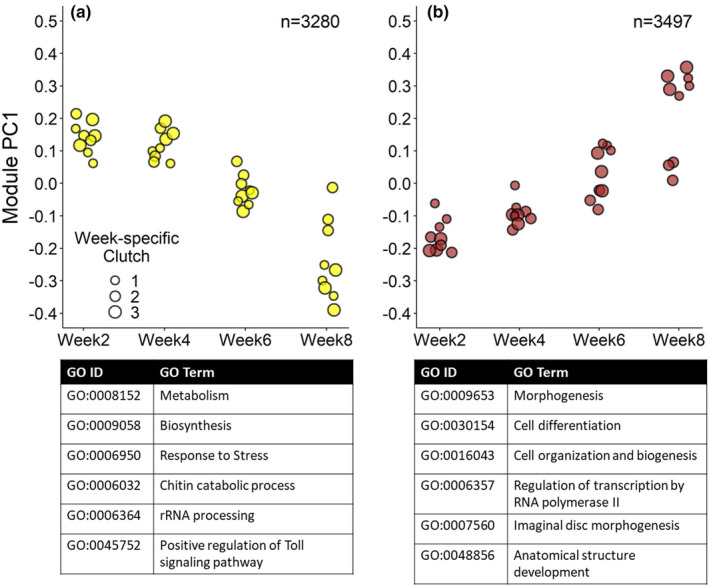
Gene expression profiles of modules involved in developmental progression at constant 10°C. Two major gene modules showed a gene expression profile that changed significantly over the course of embryonic development, either going down in expression over time (a, module 13) or up (b, module 3, FDR <0.05). The expression pattern for each module is summarized by PC1 values for each constant 10°C sample (N = 36) with the number of genes significantly belonging to each module shown in the upper right corner (FDR <0.05). Each point represents a sample where point size indicates the week‐specific clutch it belongs to (three samples at three time points [0, 3 and 24 h] for each of three clutches per development week). Top overrepresented BP GO terms for each module are shown below the figure panels. For each module, the first three GO terms are the most prominent slimmed GO categories (excl. GO:0007275 development) and the last three GO terms are the most significant GO terms (p < .01), for overview showing GO terms that belong to different parent terms and substituting very specific GO terms for the parent term. See Table S2 for a complete list of overrepresented (slimmed) GO terms for each significant module
GO overrepresentation analysis showed that genes with high expression during early development were mainly involved in biosynthesis (Figure 3a; Table S2). Interestingly, overrepresented GO terms included many processes related to environmental stress resistance, such as the immune response, stress response and melanization. A closer look at individual gene annotations showed that expressed immune genes were mostly protease cascade genes that activate the pathway in dorsoventral patterning or immune defences (e.g., blast hits to D. melanogaster immune genes grass, nudel and snake, Table S1.2; Lindsay & Wasserman, 2014), indicating organization of embryonic cell fate positioning or readiness of the immune system rather than an ongoing infection.
For the genes that go up in expression in the latter development weeks (module 3, Figure 3b), overrepresented GO terms were mainly involved in morphogenesis, cell differentiation and cell organization (Table S2). Overrepresented GO terms indicated the development of many anatomical structures, including the eyes, imaginal discs and muscles. Notably, many overrepresented GO terms were involved in nervous system development (Table S2).
3.3. Differential expression analysis: Temperature‐responsive genes
The majority of the 23,961 genes used for analysis were expressed above the low expression threshold in all four development weeks sampled (81.57%), with small sets of genes expressed in only one of the 4 weeks (subsets of 255–582 genes). Similarly, 92.81% of the genes were expressed in all temperature treatments, with small sets of genes expressed in only one of the three treatments (10°C: 389 genes, 5°C: 212 genes, 15°C: 305 genes). In total, we found 837 genes that were significantly differentially expressed (DEGs) between control and temperature change treatments, either at 3 or at 24 h after a 5°C increase and/or decrease in temperature (FDR <0.01, Table S1.3–1.5). There was little overlap in the DEGs between the two sample time points, with only 23 genes differentially expressed both after 3 and 24 h, indicating that we captured two phases of the response to a change in ambient temperature.
There was little overlap between the DEGs from the three temperature‐effect post hoc tests: (i) overall temperature effects (DEGs = 293 genes), (ii) within‐week temperature effects (DEGs = 544 genes) and (iii) between‐week temperature effects (DEGs = 235 genes). DEGs for each temperature effect are discussed in more detail below. Only 98 genes tested significant for more than one temperature effect (11.71%), indicating that each post hoc test captured different types of variation in gene expression. This is reflected in little overlap between overrepresented GO terms found for each temperature effect (Table S3). Interestingly, 56 of the 837 DEGs were only expressed above the low expression threshold either in a specific development week (17 DEGs), or a specific temperature treatment (10 DEGs) or both (29 DEGs, Table S4.1). We highlight a number of these DEGs in Table 1, annotated with a gene function that could potentially be involved in regulating the temperature sensitivity of embryonic development rate. Expression profiles for these genes are visualized in Figure S7.1.
TABLE 1.
Candidate genes for regulating the temperature sensitivity of embryonic development rate
| GeneID | When expressed | Temperature effect compared to the control constant 10°C | D. melanogaster annotation |
|---|---|---|---|
| Within‐week temperature effect | |||
| MSTRG.6256 | week4 at 15°C |
|
pdfr, thought to be an important component of the circadian pathway (FlyBase, accessed April 19, 2022), also thought to be involved in ecdysone production (Iga et al., 2014). |
| MSTRG.19976 | week4 at 15°C |
|
orcokinin a , neuropeptide with diverse regulatory roles in insects, including the regulation of gene transcription and ecdysterodoigenesis (Tanaka, 2021). |
| MSTRG.30412 | week6 at 15°C |
|
asator, a tau‐tubulin kinase involved in ATP binding, protein serine/threonine kinase activity and signal transduction (FlyBase, accessed April 19, 2022) |
| MSTRG.39480 | at 5°C |
|
elys, involved in chromatin binding (FlyBase, accessed April 19, 2022) |
| OBRU01_205030 | week8 after temperature change |
|
JNK interacting protein a , involved in stress‐mediated JNK activation in neurons (Willoughby et al., 2003). |
| Between‐week temperature effect | |||
| MSTRG.37241 | week4 after temperature change |
|
cenG1A, a GTPase that is ecdysone signalling‐dependent (FlyBase, accessed April 19, 2022). |
| OBRU01_214986 | week6 at 15°C |
|
alp4, an alkaline phosphatase involved in nervous system development (FlyBase, accessed April 19, 2022). |
Note: We highlight a number of genes that were only expressed above the low expression threshold in a particular development week and/or temperature treatment, and that showed a significant response to a 5°C increase and/or decrease in temperature compared to a constant 10°C control. These genes were temperature‐responsive either within a specific development week (within‐week temperature effect) or whose temperature effect size changed between development weeks (between‐week temperature effect, see Table S4.1 for a full list including statistics). For each gene, we list when it was expressed in at least two out of three replicates above the low‐expression threshold (see Section 2), the temperature effect that we found (FDR <0.01), and its Drosophila melanogaster annotation with the related gene function obtained from FlyBase (Larkin et al., 2021). Expression profiles for these genes are visualized in Figure S7.1.
Winter moth reference genome annotation instead of D. melanogaster.
3.3.1. Overall temperature effects
From our linear mixed models, we first took the average temperature effect size of the four development weeks for each temperature treatment, and compared the warm and the cold treatment to the control, at 3 and 24 h after a change in temperature (Figure 4a and b respectively). This post hoc test captured genes with a small but consistent response to a change in temperature in each of the four different development weeks and/or genes with a high effect size in one or more weeks (log2FoldChange [log2FC] −2.69 to 3.56, median = ±0.40, mean = ±0.61, FDR <0.01, 293 DEGs in total).
FIGURE 4.
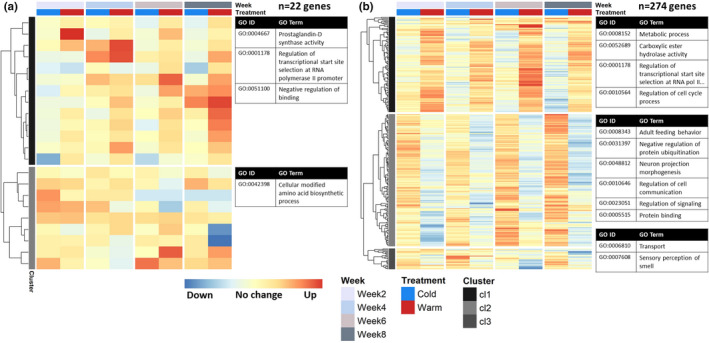
Gene expression clusters of temperature‐responsive genes with a significant overall temperature treatment effect, after 3 h (a) and after 24 h (b). Genes included in the figure on average responded significantly to a 5°C increase and/or decrease in temperature compared to the control constant 10°C in the four developmental weeks (N = 48 samples per panel). These genes showed a small but consistent response to temperature over time and/or had a large effect size in one or more weeks. For each heat map, columns correspond to the four development weeks sampled, and rows correspond to variance‐stabilizing transformation (vst) normalized and z‐transformed expression counts of each included gene, showing the difference in expression from 0 to 3 h (panel a, DEGs = 22 genes) or 0 to 24 h (panel b, DEGs = 274 genes) compared to the control, for the cold (blue column) and warm (red column) treatments. See Table S4.2 for included genes and corresponding statistics. For each identified gene expression profile cluster (indicated by grey scale bands on the left of each heatmap), the most significant GO terms are given (p < .01), with number of terms shown depending on the size of the cluster. For overview, the GO terms shown belong to different parent terms and very specific GO terms have been substituted for their parent term. See Table S5 for a complete list of overrepresented GO terms for each gene expression cluster
Relatively few of the 22 DEGs that responded to a change in temperature after 3 h showed consistent changes across weeks, with most genes having a high effect size in only 1 week (Figure 4a; Table S4.2). After 24 h, we found a relatively large number of genes with a significant overall temperature effect that showed consistent effects across the development weeks (DEGs = 274 genes, Figure 4b; Table S4.2). One large expression cluster (cluster 1) captured genes that were mostly upregulated after 24 h of warmth in each development week, while the second cluster showed a downregulation response after 24 h of warmth and/or upregulation after 24 h of cold. Genes mostly upregulated after 24 h of warmth were involved in processes such as lipid metabolism, regulation of the cell cycle process and mRNA processing (Tables S5.2 and S5.4). Genes mostly downregulated after 24 h of warmth included many processes related to growth such as cellular response to insulin stimulus and G1/S transition of mitotic cell cycle (Tables S5.2 and S5.4). Furthermore, many regulatory processes were overrepresented such as transcriptional regulation, regulation of protein modification, and regulation of cell communication and signalling.
The circadian clock gene period showed the most significant overall temperature response (Figure 5). This gene showed the expression pattern of cluster 1 at 24 h (Figure 4b), being consistently upregulated after 24 h of warmth (log2FC = 0.85, FDR <0.001), but downregulated after 24 h of cold (log2FC = −0.36, FDR = 0.001, Table S4.2) in each development week. Temperature effect size seemed to increase over time especially for the warm treatment, but this was not significant (FDR >0.01, Table S1.3, GeneID = MSTRG.9278). Interestingly, the GO term circadian rhythm was not overrepresented for gene expression cluster 1, but for cluster 2 (Table S5) it was consistently downregulated after 24 h, with D. melanogaster gene annotations takeout, minibrain, daywake, gawky, and dnc showing the opposite pattern of period.
FIGURE 5.
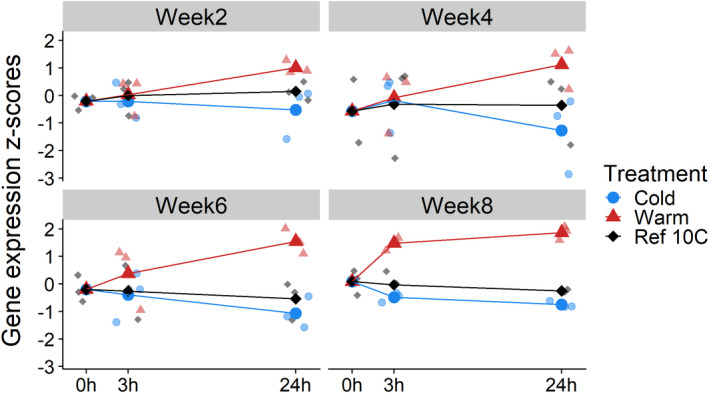
Gene expression profile of circadian clock gene period. After taking an RNA sample at time point 0 h from eggs at constant 10°C (black points), eggs from each clutch (N = 3 clutches per development week) were transferred to a warmer (red points, 15°C) or colder temperature (blue points, 5°C) and RNA was sampled at 3 and 24 h after the transfer (in total, N = 7 samples per clutch). Average, vst normalized and z‐transformed gene expression counts are shown for each development week (bold points and lines), showing that the circadian clock gene period is consistently upregulated after 24 h of warmth (log2FC = 0.85, FDR <0.001), and consistently downregulated after 24 h of cold (log2FC = −0.36, FDR = 0.001, Table S4.2, GeneID = MSTRG.9278)
3.3.2. Within‐week temperature effects
We also tested for each gene whether its expression significantly changed after a 5°C temperature increase or decrease within each development week (544 DEGs in total, FDR <0.01). This second post hoc test captured genes with higher effect sizes (log2FC − 9.91 to 8.47, median = ±2.59, mean = ±2.62), and allowed us to find genes that responded in a week‐specific manner. Indeed, each week had a specific set of genes that responded to an increase or decrease in temperature after 3 and 24 h (Figure 6a and b respectively), with little overlap between week‐specific sets. Of 544 DEGs in total, only 26 genes were significant in more than 1 week, with no genes shared between all weeks.
FIGURE 6.
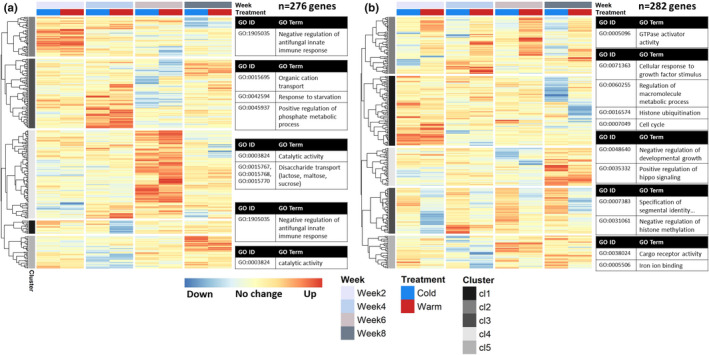
Gene expression clusters of temperature‐responsive genes with a significant within‐week treatment effect, after 3 h (a) and after 24 h (b). Genes included in the figure responded significantly to a 5°C increase and/or decrease in temperature compared to the control constant 10°C in one or more developmental weeks (N = 48 samples per panel), capturing genes that mostly showed week‐specific responses. For each heat map, columns correspond to the four development weeks sampled, and rows correspond to vst normalized and z‐transformed expression counts of each included gene, showing the difference in expression from 0 to 3 h (panel a, DEGs = 276 genes) or 0 to 24 h (panel b, DEGs = 282 genes) compared to the control, for the cold (blue column) and warm (red column) treatments. See Table S4.3 for included genes and corresponding statistics. For each identified gene expression profile cluster (indicated by grey scale bands on the left of each heatmap), the most significant GO terms are given (p < .01), with number of terms shown depending on the size of the cluster. For overview, the shown GO terms belong to different parent terms and very specific GO terms have been substituted for their parent term. See Table S6 for a complete list of overrepresented GO terms for each gene expression cluster
For the response after 3 h (Figure 6a, 276 DEGs), each week had a specific set of genes that was upregulated in both the warm and the cold treatment. For the response after 24 h (Figure 6b, 282 DEGs), the gene expression patterns were more diverse, with clusters of genes that showed similar consistent patterns across development weeks as the overall temperature treatment effects described above (e.g., cluster 2), but also clusters of genes that showed a more week‐specific response. For example, cluster 5 shows a set of genes that was downregulated in response to a temperature change only in development week4 (Table S4.3). When we considered all the genes with a significant within‐week temperature effect together, common processes that were overrepresented included immune response regulation, regulation of GTPase activity, histone modification and zymogen activity (Table S3).
For development week2, significant temperature‐responsive genes were involved in embryonic development processes (e.g., the regulation of cell proliferation), as well as many histone modification processes and gene expression regulation (Tables S6.1 and S6.2). Many of these genes responded after 24 h of temperature change (Figure 6b), with, for example, histone modification genes upregulated (cluster 1, Table S4.3), while most downregulation occurred after 24 h of warmth (cluster 3) of genes involved in, for example, signal transduction (Tables S4.3 and S6.4). Few genes were significantly downregulated 3 h after a change in temperature (Figure 6a, Table S4.3).
Significant temperature‐responsive genes in development week4 were involved in the regulation of GTPase activity, cell homeostasis, and hormone binding (Table S6.1 and S6.2). Interestingly, both at 3 h (Figure 6a) and at 24 h (Figure 6b) after the temperature change, we identified a cluster of genes that was almost exclusively downregulated after both warm and cold treatments in week4 (cluster 5 in both heat maps, Table S4.3). These genes were involved in acyltransferase activity, the regulation of translation and cyclin binding (Tables S6.3–S6.6). Temperature‐responsive genes specific to week4 included pdfr, orcokinin and cenG1A (Table 1).
Overrepresented GO terms for development week6 were neurotransmitter catabolic process, response to cocaine and aromatase activity (Tables S6.1 and S6.2). A large group of genes was upregulated 3 h after a change in temperature in week6, both after cold and warmth (Figure 6a, cluster 4; Table S4.3), which were involved in disaccharide transport and catalytic activity. Gene responses after 24 h were more diverse (Figure 6b), with genes upregulated after 24 h of warmth (e.g., cluster 2) or after 24 h of cold (e.g., cluster 5), while other genes were downregulated after a change in temperature, mostly after cold (e.g., cluster 4, Table S4.3). Temperature‐responsive genes asator and elys were specific to week6 (Table 1).
For development week8, overrepresented GO terms included histone modifications, signal transduction and transcription corepressor activity (Tables S6.1 and S6.2). After 3 h, there was a small group of genes upregulated in both warm and cold treatments in week8 (Figure 6a, cluster 5) involved in acyltransferase activity (Table S6.5). After 24 h (Figure 6b), genes upregulated were involved in, for example, negative regulation of developmental growth (e.g., cluster 4). Genes downregulated at 24 h (e.g., cluster 1) were involved in, for example, histone ubiquitination and Toll signalling (Tables S6.4 and S6.6). A JNK interacting protein was specific to week8 that was significantly downregulated after 24 h of cold (Table 1).
3.3.3. Between‐week temperature effects
With the third and last post hoc test, we tested for an interaction effect between temperature treatment and development week. In this way we compared temperature effects between weeks to find genes whose temperature effect size changed between the weeks. Due to a small sample size, this test was somewhat underpowered and mostly picked up genes with a large change in effect size (log2FC − 13.52 to 13.56, median = ±4.62, mean = ±4.73, 235 DEGs in total). A large proportion of the genes with a significant between‐week effect also showed a significant within‐week effect (137 out of 235 DEGs, 58% overlap). However, there was little overlap between specific week‐to‐week comparisons (only 45 genes significant in more than 1 week‐to‐week comparison). The switch in temperature sensitivity of egg development rate was previously found in development week6 (van Dis et al., 2021). Interestingly, the majority of the significant genes were found around development week6 when comparing gene temperature responses between development week4 and week6 (111 DEGs) and between week6 and week8 (78 DEGs). This is also evident from the gene expression patterns when clustering the between‐week genes (Figure 7), with many clusters showing opposite responses in week4 compared to week6 (e.g., 3 h response in Figure 7a clusters 1 and 2) or week6 compared to week8 (e.g., 24 h response in Figure 7b cluster 3).
FIGURE 7.
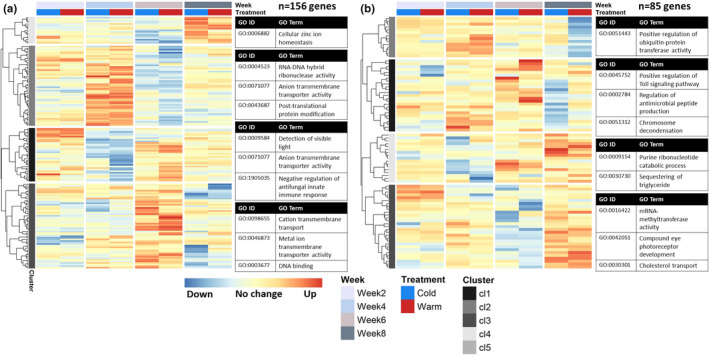
Gene expression clusters of temperature‐responsive genes whose temperature treatment effect significantly changed between development weeks, after 3 h (a) and after 24 h (b). Genes included in the figure showed a significantly different temperature effect size when comparing development weeks (N = 48 samples per panel, FDR <0.01), capturing genes whose response to a 5°C increase and/or decrease in ambient temperature changed during the course of development. For each heat map, columns correspond to the four development weeks sampled, and rows correspond to vst normalized and z‐transformed expression counts of each included gene, showing the difference in expression from 0 to 3 h (panel a, DEGs = 156 genes) or 0 to 24 h (panel b, DEGs = 85 genes) compared to the control, for the cold (blue column) and warm (red column) treatments. See Table S4.4 for included genes and corresponding statistics. For each identified gene expression profile cluster (indicated by grey scale bands on the left of each heatmap), the most significant GO terms are given (p < .01), with number of terms shown depending on the size of the cluster. For overview, the shown GO terms belong to different parent terms and very specific GO terms have been substituted for their parent term. See Table S7 for a complete list of overrepresented GO terms for each gene expression cluster
Overrepresented GO terms for the 235 between‐week DEGs together included immune response regulation terms and oxidoreductase activity (Table S3). Genes related to immune response regulation were, for example, significantly downregulated after 24 h of cold in week8 compared to the other weeks (Figure 7b, cluster 1; Table S4.4), while genes involved in oxidoreductase activity differed in the 3 h response to cold in week8 compared to week6 (Figure 7a, cluster 4; Table S4.4). Overall, temperature effect size changes around week6 were involved in carbohydrate catabolic process and RNA–DNA hybrid ribonuclease activity (week4 vs. week6), and zinc ion transport, binding and enzyme activity (week6 vs. week8, Tables S7.1 and S7.2). Genes with development week and temperature‐specific expression patterns whose temperature response changed between week4 and week6 included cenG1A and alp4 (Table 1).
Interestingly, the temperature response of transmembrane transporter activity seemed to change over the course of development. This process was significantly overrepresented in all four clusters of genes whose temperature response after 3 h differed significantly between development weeks (Figure 7a; Tables S7.3 and S7.5). This mostly encompassed genes whose temperature response changed significantly between development week4 and week6. For example, genes involved in anion and cation transmembrane transport were upregulated 3 h after a temperature change in week6, while they did not respond or were downregulated in week4 (clusters 1 and 3, Table S4.4).
3.4. Overlap between differential expression and co‐expression analysis
We investigated how many of the temperature‐responsive genes were involved in week‐specific development processes by looking at the overlap between the differential expression analysis results (837 DEGs across all three temperature effect post hoc tests) and the co‐expression analysis results (i.e., the four WGCNA gene modules that changed in expression over the course of development at constant 10°C). Fewer than half of the genes that responded significantly to a change in temperature overlapped with the genes in the four modules that changed expression over the course of development at 10°C (Figure 8; 363 genes overlapped out of 837 DEGs). This indicates that the 474 genes that did not overlap with these gene modules were constantly expressed throughout development. Indeed, only 37 of these 474 genes showed a temperature‐specific and/or week‐specific expression pattern (see section 3.3 and Table S4.1).
FIGURE 8.
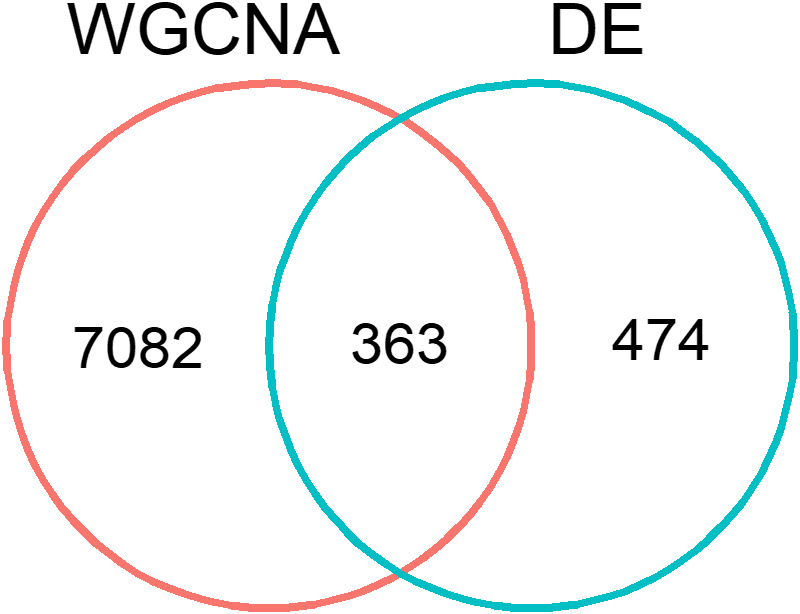
Overlap between co‐expression analysis (WGCNA) and differential expression analysis (DE). Of the 837 genes that were significantly differentially expressed after a 5°C temperature increase and/or decrease (FDR <0.01), 363 genes overlapped with the four modules of genes that showed a significant change between development weeks at the control constant 10°C (FDR <0.05, table S1.3). The expression patterns of the other 7082 genes that changed expression between development weeks at 10°C were not affected by a change in temperature, while 474 genes that did not significantly change expression over the course of development were differentially expressed after a change in temperature
The 363 genes that were both temperature‐responsive and changed in expression over the course of development at constant 10°C (Figure 8) showed diverse responses to a 5°C increase and/or decrease in temperature, including a similar proportion of DEGs from each temperature effect post hoc test (overall temperature effects: 55%, within‐week effects: 40%, between‐week effects: 39%). More than half of the overlap was with WGCNA module 3 (Figure 3a; 224 out of 363 DEGs), which encompassed genes that were lowly expressed in the first two development weeks and went up in expression from development week6 onwards. Overrepresented GO terms for these 363 overlapping genes included many regulatory processes such as negative regulation of protein ubiquitination and regulation of transcription by RNA polymerase II, as well as oxidoreductase activity and more general binding GO terms (e.g., transcription coactivator binding, Tables S8.1 and S8.2).
By splitting up the overlap for week‐specific DEGs over the four WGCNA modules, we found small groups of genes that showed an unexpected overlap. For each development week, we found a small group of genes (7–46 genes) that overlapped with a WGCNA module that showed low expression in that particular week at constant 10°C (Table S8.3). For example, 42 and 56 genes that were temperature‐responsive in development week6 and week8, respectively, overlapped with WGCNA module 13 or module 10 (Figure 3a; Figure S5.5a). At constant 10°C, these genes were high in expression in development week2 and/or week4, but low in expression in week6 and week8. Similarly, we found genes that were temperature‐responsive in week2 and week4 (20 and 12 genes respectively) that overlapped with genes that only became high in expression in week6 and week8 at a constant 10°C (WGCNA module 3, Figure 3b). We highlight a number of these DEGs in Table 2, annotated with a gene function that could potentially be involved in the regulation of the temperature sensitivity of embryonic development rate. Expression profiles for these genes are visualized in the Figure S7.2.
TABLE 2.
Candidate genes for regulating the temperature sensitivity of embryonic development rate – Continued
| GeneID | When highly expressed at constant 10°C | Temperature effect compared to the control constant 10°C | D. melanogaster annotation |
|---|---|---|---|
| Within‐week temperature effect | |||
| MSTRG.20613 | Module 13: week2 and week4 |
|
tao, a Ser/Thr kinase involved in Hippo‐signalling (FlyBase, accessed May 24, 2022) |
| MSTRG.12645 | Module 13: week2 and week4 |
|
fdl, involved in restructuring the brain via hormonal control during metamorphosis (FlyBase, accessed May 24, 2022). |
| MSTRG.3087 | Module 13: week2 and week4 |
|
tey, a protein that regulates neuromuscular target specificity, involved in the negative regulation of transcription and protein ubiquitination (FlyBase, accessed May 24, 2022). |
| MSTRG.22817 | Module 13: week2 and week4 |
|
gld, a glucose dehydrogenase involved in oxidoreductase activity (FlyBase, accessed May 24, 2022). |
| MSTRG.8284 | Module 13: week2 and week4 |
|
cyp6a13, a cytochrome P450 involved in oxidoreductase activity that may be involved in the metabolism of insect hormones (FlyBase, accessed May 24, 2022). |
| MSTRG.32147 | Module 13: week2 and week4 |
|
sp7, a serine protease involved in the activation of the melanization cascade (FlyBase, accessed May 24, 2022) |
| MSTRG.13770 | Module 13: week2 and week4 |
|
atlastin, encodes a membrane‐bound GTPase that enables protein binding (FlyBase, accessed May 24, 2022) |
| MSTRG.11979 | Module 13: week2 and week4 |
|
nanos, which codes for an RNA‐binding protein that forms part of a translational repressor complex (FlyBase, accessed April 13, 2022). |
| MSTRG.25766 | Module 13: week2 and week4 |
|
cycC a , coactivator involved in regulating gene transcription of nearly all RNA polymerase II‐dependent genes (FlyBase, accessed May 24, 2022). |
| MSTRG.11006 | Module 3: week6 and week8 |
|
Psc, member of the polycomb gene group that regulates gene expression through epigenetic marks. Involved in chromatin binding, DNA binding, and cell cycle control (FlyBase, accessed May 24, 2022). |
| MSTRG.10930 | Module 3: week6 and week8 |
|
polo, a Ser/Thr kinase thought to be a cell cycle regulator (FlyBase, accessed May 24, 2022) |
Note: We highlight a number of genes with a significant response to a 5°C increase and/or decrease in temperature within a specific development week (within‐week temperature effect), which at the control constant 10°C were lowly expressed in that same development week, but highly expressed in another development week (see Table S8.3 for a full list including statistics). For each gene, we list when it was highly expressed during development at constant 10°C (belonging to one of four significant WCGNA gene modules, see Section 2), the temperature effect that was found (FDR <0.01), and its Drosophila melanogaster annotation with the related gene function obtained from FlyBase (Larkin et al., 2021). Expression profiles for these genes are visualized in Figure S7.2.
D. pseudoobscura annotation instead of D. melanogaster.
4. DISCUSSION
Climate change will strongly impact the developmental timing of insects, but we know little about the molecular pathways underlying the temperature sensitivity of insect embryonic development rate (Mirth et al., 2021). Here we used RNAseq to find candidate genes underlying the regulation of temperature sensitivity in winter moth embryos, a species whose egg development rate has been under selection due to climate change (van Asch et al., 2007). We sampled eggs over the course of development, including two early development weeks and two late development weeks, and identified a diverse set of temperature‐responsive genes. A total of 837 genes significantly responded to a 5°C increase or decrease in temperature compared to a constant 10°C control. These genes either responded in a consistent way in each development week sampled, or showed a week‐specific response. Moreover, we found genes whose temperature effect size changed between early and late development weeks. This diversity suggests that the temperature sensitivity of insect embryonic development rate is a polygenic trait, with many potential gene targets that climate change selection could act upon.
4.1. Dormancy phenotype in winter moth eggs
Dormancy during winter moth embryonic development involves slow but progressive development rather than a period of developmental arrest (van Dis et al., 2021). At constant 10°C, changes in gene expression over the course of development were reflected in four gene expression profiles encompassing a total of 7445 genes (Figure 3). In the early two development weeks, there was an overrepresentation of genes involved in biosynthesis, switching to morphogenesis in the later two development weeks. Despite the differences in ongoing developmental processes between weeks, many genes consistently responded to a change in ambient temperature in each development week. These temperature‐responsive genes might be behind the direct response to temperature of winter moth embryonic development rate: dormancy in which embryos remain responsive to the environment (Wilsterman et al., 2021). These genes included many regulatory processes that could be involved in the regulation of development rate such as cell cycle regulation and transcriptional regulation.
Interestingly, we found that early in development, when egg development rate has low temperature sensitivity, many biological processes involved in environmental stress resistance were overrepresented. This environmental stress resistance included the upregulation of genes involved in chaperone‐mediated protein folding and immune genes, which are also upregulated during dormancy in other insects, specifically during deep‐programmed diapause (Denlinger, 2002; Kubrak et al., 2014). Upregulation of stress‐related genes, such as chaperone proteins, can suppress growth (Feder et al., 1992; López‐Maury et al., 2008). This transcriptional pattern thus suggests that early in development winter moth eggs have a form of diapause, in which low temperature sensitivity of development rate is achieved through the upregulation of genes that suppress the magnitude of the direct response to temperature. For example, genes from the Toll pathway were upregulated after a change in temperature in development week2, which is an important innate immune pathway in insects, as well as an important determinant of dorsoventral patterning (Lindsay & Wasserman, 2014). This response changed significantly over the course of development, with Toll pathway genes downregulated after cold in week8.
4.2. Candidate gene groups
In our analysis, we found temperature‐responsive genes at different levels of gene expression regulation, including genes involved in transcriptional, post‐transcriptional, translational and epigenetic regulation. In this section, we highlight three groups of genes from our results that represent interesting candidates for regulating the temperature sensitivity of insect embryonic development rate: (1) circadian clock genes, (2) nervous system development genes and (3) genes involved in histone modifications. Other interesting candidates included (4) hormonal control genes, (5) cell cycle regulators and (6) protein binding genes, which are discussed in more detail in Section S7.
From the 837 genes that significantly changed their gene expression in response to an increase and/or decrease in temperature, we expect only a small subset to be regulator genes of development rate. We hypothesize that the six gene groups we highlight are such regulators based on what is currently known about their biological functions. For the other temperature‐responsive genes, we expect them to be either responding to the regulator genes of developmental rate or to be directly responding to temperature. For example, week‐specific genes that responded at 3 h after a change in temperature tended to respond in the same direction for the cold and warm temperature treatment, which could indicate a week‐specific stress response (Figure 6a). In the experiment, eggs received quite an abrupt change in temperature, so these genes might only represent a short‐term response to temperature.
4.2.1. Circadian clock genes
Diapause and developmental timing in insects have been linked to circadian clock genes in many species (Dolezel, 2015). For example, in the European corn borer moth, genetic variation in the circadian clock gene period results in different timing of diapause termination (Kozak et al., 2019). Period was the most significant temperature‐responsive gene in our analysis, being upregulated after warmth and downregulated after cold in each development week (Figure 5). The effect of temperature on period furthermore seemed to increase over the course of development, and we found many other circadian clock genes that responded significantly to a change in temperature, such as pdfr, takeout and daywake, often showing the opposite response of period. This transcriptional pattern suggests that period might be a regulator gene of development rate with the other circadian clock genes responding to this regulation. In Drosophila, a latitudinal length polymorphism in the period gene is thought to determine the extent of temperature compensation of the circadian clock (Costa et al., 1992; Sawyer et al., 1997), making period an intriguing candidate for regulating the temperature sensitivity of embryonic developmental timing in the winter moth.
4.2.2. Nervous system development
In the winter moth, a major embryonic development event that coincides with the switch from low to high temperature sensitivity in development week6 is nervous system development (van Dis et al., 2021). This is reflected in our results: at a constant 10°C, many processes involved in nervous system development became highly expressed from development week6 onwards (Figure 3). Interestingly, many of the genes involved in nervous system development also responded to a change in ambient temperature, including processes such as neuron projection morphogenesis and differentiation (e.g., gene alp4, Table 1; and gene tao, Table 2). We previously hypothesized that nervous system development is involved in the increased temperature sensitivity of development rate from development week6 onwards, as this opens up the possibility for the integration of internal and environmental stimuli to actively regulate important developmental processes (van Dis et al., 2021). Indeed, an important component of regulating developmental transitions in response to the environment is through the development of sensory neurons (Faunes & Larraín, 2016). In our analysis, two important genes involved in the development of dendrites were temperature responsive: nanos and pumilio (Table 2 and Table S4.1). In D. melanogaster, nanos and pumilio jointly control the elaboration of class IV neurons (Ye et al., 2004), which are associated with temperature sensing (Terada et al., 2016).
Further indications for the involvement of neuronal signalling in the regulation of temperature sensitivity came from temperature‐responsive genes involved in processes such as the negative regulation of neurotransmitter secretion, response to cocaine and the positive regulation of voltage‐gated potassium channel activity. Most striking was the large group of genes involved in ion transmembrane transporter activity whose temperature effect size changed drastically from development week4 to week6 (Figure 7a). This might indicate the activation of neuronal regulation from development week6 onwards, since ion channels play a central role in neuronal signalling and membrane potential regulation (Hodgkin & Huxley, 1952). Whereas early in development lower temperature sensitivity might be modulated by repressor genes (see discussion above on the winter moth's dormancy phenotype), we hypothesize that genes involved in neuronal regulation can enhance the response of development rate to temperature changes late in development.
4.2.3. Histone modifications
Epigenetic regulation at the molecular level includes mechanisms such as DNA methylation and histone modifications that determine how accessible genes are for transcription (Jaenisch & Bird, 2003). Our results point to histone modifications as an important mechanism to regulate the temperature sensitivity of embryonic development rate. Histone modifications alter the packaging structure of DNA, called chromatin, which regulates transcription (Kouzarides, 2007) and can suppress gene expression for example during hibernation (Storey, 2015). In our results, histone modifications, such as histone methylation, responded to temperature changes in a consistent way across development weeks, as well as showing week‐specific responses. We hypothesize that early in development transcriptional control through histone modifications increases, while late in development this control decreases in response to temperature, which could modulate the switch from low to high temperature sensitivity in development week6. For example, early in development the psc gene was upregulated after cold in development week2, which regulates gene expression through epigenetic marks in D. melanogaster, influencing chromatin binding, DNA binding and cell cycle control (Table 2). Late in development, genes involved in the negative regulation of histone methylation and histone ubiquitination were temperature‐responsive, with in development week6 another chromatin binding gene elys being downregulated after a change in temperature (Table 1).
Genes involved in histone modifications during embryonic development represent an intriguing target for climate change selection. Such chromatin regulation seems to play an important role in the variation and evolution of gene expression during insect embryonic development (Liu et al., 2020). In D. melanogaster, chromatin regulators are prime candidates for local temperature adaptation. For example, natural variation in the chromatin regulator gene cramped, involved in the modulation of signalling pathways Hedgehog and Wingless during embryonic development, shifts the reaction norm of many temperature‐sensitive traits such as abdominal pigmentation (Gibert et al., 2011).
4.3. Potential paths to climate change adaptation
Regulatory genes are a probable target for rapid adaptation, because genetic variation in regulatory genes can increase variation in gene expression in response to the environment while maintaining functional robustness (Wagner, 2011). We have used RNAseq to highlight groups of regulatory genes that represent interesting candidates for the regulation of the temperature sensitivity of development rate, including histone modifications, nervous system development, hormonal control (Section S7) and circadian clock genes. Future research will need to confirm the role of these genes in regulating development rate through functional assessment. Moreover, we need to identify for which of these genes there is genetic variation present in wild populations to gain insight into their adaptive potential in the face of climate change (Waldvogel et al., 2020).
Climate change has been exerting a strong selection pressure on the temperature sensitivity of winter moth egg development rate (van Asch et al., 2007). Warmer winters have advanced the timing of its egg hatching more than the timing of its food source, oak budburst, resulting in mistiming of more than 10 days. Within the span of a decade, the temperature sensitivity of winter moth embryonic development rate has genetically adapted such that eggs are now better timed to the timing of oak budburst (van Asch et al., 2013). Our long‐term field data suggest this genetic adaptation is ongoing, as there is still a timing mismatch between winter moth egg hatching and oak budburst of on average 5 days, which has been progressively improving in the period beyond the years analysed in van Asch et al. (2013). One path adaptation could take to change the temperature sensitivity of egg development rate in the winter moth could be through targeting the overall temperature effect genes we identified, such as the circadian clock gene period (Figure 5). Changing the transcriptional response to temperature of such genes could change the baseline temperature sensitivity of development rate. In D. melanogaster, there is extensive genetic variation for the duration of embryogenesis at a constant temperature (Horváth et al., 2016), while the relative timing of development stages remains constant (Kuntz & Eisen, 2014), indicating the involvement of such overall temperature effect genes. Alternatively, adaptation could focus on temperature‐responsive genes that are involved in developmental progression and/or genes that are only expressed or temperature‐responsive at a specific point during development. For example, selection could change the temperature sensitivity of winter moth development rate at a specific development stage by changing the magnitude of the neuronal signalling response to temperature in development week6, or by changing the extent of transcriptional control through histone modifications in development week2 (discussed above).
In addition to the genes we found here, other regulatory mechanisms might similarly play a role in the regulation of development rate in response to temperature, such as alternative splicing (Anduaga et al., 2019) and microRNAs (Faunes & Larraín, 2016), but these were beyond the scope of this paper. We have focused on regulatory genes, but nonsynonymous mutations that change the enzymatic reactions of proteins to temperature could similarly play a role in the adaptation to climate change. In D. melanogaster, long‐term artificial selection on the duration of total development (from embryo to adult fly) at a constant temperature involved a number of nonsynonymous mutations in developmental genes (Burke et al., 2010). In our analysis, many genes involved in developmental progression were also temperature‐responsive (Figure 8), such as genes involved in imaginal disc morphogenesis and regionalization. If genetic variation exists for such development genes in wild populations of the winter moth, its rapid genetic adaptation to climate change could involve regulatory and/or nonregulatory genes.
5. CONCLUSIONS
Our RNAseq results indicate potential candidates for regulating the temperature sensitivity of embryonic development rate in the winter moth. These were temperature‐responsive genes involved in a diverse range of biological processes, regulating gene expression at different layers, from transcription to signal transduction. This diversity suggests that the temperature sensitivity of embryonic development rate is a polygenic trait with many potential gene targets that climate change‐induced selection could act upon. The winter moth is one of the few species for which we have evidence that it has been genetically adapting to climate change (Scheffers et al., 2016; van Asch et al., 2013). To identify which paths to adaptation are possible and even likely, we need to determine for which of the candidate genes identified here there is genetic variation present in wild insect populations, to gain insight into their adaptive potential in the face of climate change.
AUTHOR CONTRIBUTIONS
N.E.v.D., M.E.V., B.W. and R.A.H. designed the experiment; N.E.v.D. performed the experiment; A.S.P optimized the RNA extraction protocol and did the RNA extractions; N.E.v.D. conducted the sequence processing and statistical analysis with the help of J.E.R.; N.E.v.D. wrote the manuscript with input from all authors.
6. CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
OPEN RESEARCH BADGES
This article has earned an Open Data Badge for making publicly available the digitally‐shareable data necessary to reproduce the reported results. The data is available at [provided https://doi.org/10.5061/dryad.hx3ffbghd].
Supporting information
Appendix S1:
ACKNOWLEDGEMENTS
We thank Bart van Lith for field collection; Christa Mateman for laboratory assistance; Martijn Derks for the version 2 annotation of the winter moth reference genome and helpful suggestions on functional annotation; Maurijn van der Zee for valuable discussions on the results; Ruth Fawthrop for help with English grammar; and Melanie Lindner for making her WGCNA script publicly available, which was very helpful for doing our analysis. We would also like to thank three anonymous reviewers for their constructive and insightful comments that helped to improve the manuscript. This research was supported by an Adaptive Life Program grant [IVA AL 3.2C DIS to N.E.v.D., B.W., R.A.H. and M.E.V.] made possible by the Board of the University of Groningen, the Faculty of Science and Engineering, and the Groningen Institute for Evolutionary Life Science (GELIFES).
van Dis, N. E. , Risse, J. E. , Pijl, A. S. , Hut, R. A. , Visser, M. E. , & Wertheim, B. (2022). Transcriptional regulation underlying the temperature response of embryonic development rate in the winter moth. Molecular Ecology, 31, 5795–5812. 10.1111/mec.16705
Handling Editor: Sean D Schoville
DATA AVAILABILITY STATEMENT
Raw RNAseq reads (incl. metadata) used in this article can be found at the European Nucleotide Archive (ENA) under accession no. PRJEB55675. The processed data used for analysis, including the annotated transcriptome, GO annotation table, gene count matrix and supporting data can be found on Dryad https://doi.org/10.5061/dryad.hx3ffbghd (van Dis et al., 2022). Bioinformatic and statistical analysis scripts to reproduce the results can be found on GitHub at https://github.com/NEvanDis/WM_RNAseq.
REFERENCES
- Alexa, A. , Rahnenfuhrer, J. , & Lengauer, T. (2006). Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics, 22(13), 1600–1607. 10.1093/bioinformatics/btl140 [DOI] [PubMed] [Google Scholar]
- Altermatt, F. (2010). Climatic warming increases voltinism in european butterflies and moths. Proceedings of the Royal Society B: Biological Sciences, 277(1685), 1281–1287. 10.1098/rspb.2009.1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anduaga, A. M. , Evanta, N. , Patop, I. L. , Bartok, O. , Weiss, R. , & Kadener, S. (2019). Thermosensitive alternative splicing senses and mediates 2 temperature adaptation in Drosophila . eLife, 8, 1–31. 10.7554/eLife.44642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A. , Martin, M.‐J. , Orchard, S. , Magrane, M. , Agivetova, R. , Ahmad, S. , Ahmad, S. , Alpi, E. , Bowler‐Barnett, E. H. , Britto, R. , Bursteinas, B. , Bye‐A‐Jee, H. , Coetzee, R. , Cukura, A. , Da Silva, A. , Denny, P. , Dogan, T. , Ebenezer, T. G. , Fan, J. , … Teodoro, D. (2021). UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Research, 49(D1), D480–D489. 10.1093/nar/gkaa1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30(15), 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw, W. E. , & Holzapfel, C. M. (2006). Evolutionary response to rapid climate change. Science, 312(5779), 1477–1478. 10.1126/science.1127000 [DOI] [PubMed] [Google Scholar]
- Brionne, A. , Juanchich, A. , & Hennequet‐Antier, C. (2019). ViSEAGO: A Bioconductor package for clustering biological functions using gene ontology and semantic similarity. BioData Mining, 12(1), 16. 10.1186/s13040-019-0204-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink, B. , Xie, C. , & Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nature Methods, 12(1), 59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- Burke, M. K. , Dunham, J. P. , Shahrestani, P. , Thornton, K. R. , Rose, M. R. , & Long, A. D. (2010). Genome‐wide analysis of a long‐term evolution experiment with Drosophila . Nature, 467, 587–590. 10.1038/nature09352 [DOI] [PubMed] [Google Scholar]
- Costa, R. , Peixoto, A. A. , Barbujani, G. , & Kyriacou, C. P. (1992). A latitudinal cline in a Drosophila clock gene. Proceedings of the Royal Society of London. Series B: Biological Sciences, 250(1327), 43–49. 10.1098/rspb.1992.0128 [DOI] [PubMed] [Google Scholar]
- Danks, H. V. (1987). Insect dormancy: An ecological perspective. Biological Survey of Canada (Terres‐ trial Arthropods). [Google Scholar]
- Denlinger, D. L. (2002). Regulation of diapause. Annual Review of Entomology, 47, 93–122. [DOI] [PubMed] [Google Scholar]
- Derks, M. F. L. , Smit, S. , Salis, L. , Schijlen, E. , Bossers, A. , Mateman, C. , Pijl, A. S. , de Ridder, D. , Groenen, M. A. M. , Visser, M. E. , & Megens, H.‐J. (2015). The genome of winter moth (Operophtera brumata) provides a genomic perspective on sexual dimorphism and phenology. Genome Biology and Evolution, 7(8), 2321–2332. 10.1093/gbe/evv145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel, D. (2015). Photoperiodic time measurement in insects. Current Opinion in Insect Science, 7, 98–103. 10.1016/j.cois.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Faunes, F. , & Larraín, J. (2016). Conservation in the involvement of heterochronic genes and hormones during developmental transitions. Developmental Biology, 416(1), 3–17. 10.1016/j.ydbio.2016.06.013 [DOI] [PubMed] [Google Scholar]
- Feder, J. H. , Rossi, J. M. , Solomon, J. , Solomon, N. , & Lindquist, S. (1992). The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes and Development, 6(8), 1402–1413. [DOI] [PubMed] [Google Scholar]
- Forrest, J. R. (2016). Complex responses of insect phenology to climate change. Current Opinion in Insect Science, 17, 49–54. 10.1016/j.cois.2016.07.002 [DOI] [PubMed] [Google Scholar]
- Gibert, J.‐M. , Karch, F. , & Schlötterer, C. (2011). Segregating variation in the Polycomb group gene cramped alters the effect of temperature on multiple traits. PLoS Genetics, 7(1), e1001280. 10.1371/journal.pgen.1001280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly, J. , Charnov, E. , West, G. , Savage, V. , & Brown, J. (2002). Effects of size and temperature on developmental time. Nature, 417, 70–73. [DOI] [PubMed] [Google Scholar]
- Hodgkin, A. L. , & Huxley, A. F. (1952). A quantitative description of membrane current and its application to conduction and excitation in nerve. The Journal of Physiology, 117, 500–544. 10.1113/jphysiol.1952.sp004764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth, B. , Betancourt, A. J. , & Kalinka, A. T. (2016). A novel method for quantifying the rate of embryogenesis uncovers considerable genetic variation for the duration of embryonic development in Drosophila melanogaster. BMC Evolutionary Biology, 16(1), 200. 10.1186/s12862-016-0776-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z.‐L. , Bao, J. , & Reecy, J. (2008). CateGOrizer: A web‐based program to batch analyze gene ontology classification categories. Online Journal of Bioinformatics, 9, 108–112. [Google Scholar]
- Iga, M. , Nakaoka, T. , Suzuki, Y. , & Kataoka, H. (2014). Pigment dispersing factor regulates ecdysone biosynthesis via Bombyx neuropeptide G protein coupled receptor‐B2 in the prothoracic glands of Bombyx mori. PLoS One, 9(7), e103239. 10.1371/journal.pone.0103239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch, R. , & Bird, A. (2003). Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nature Genetics, 33(3S), 245–254. 10.1038/ng1089 [DOI] [PubMed] [Google Scholar]
- Jones, P. , Binns, D. , Chang, H.‐Y. , Fraser, M. , Li, W. , McAnulla, C. , McWilliam, H. , Maslen, J. , Mitchell, A. , Nuka, G. , Pesseat, S. , Quinn, A. F. , Sangrador‐Vegas, A. , Scheremetjew, M. , Yong, S.‐Y. , Lopez, R. , & Hunter, S. (2014). InterProScan 5: Genome‐scale protein function classification. Bioinformatics, 30(9), 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Paggi, J. M. , Park, C. , Bennett, C. , & Salzberg, S. L. (2019). Graph‐based genome alignment and genotyping with HISAT2 and HISAT‐genotype. Nature Biotechnology, 37(8), 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde, R. (2019). Pheatmap: Pretty Heatmaps. R package version 1.0.12 .
- Kouzarides, T. (2007). Chromatin modifications and their function. Cell, 128(4), 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kozak, G. M. , Wadsworth, C. B. , Kahne, S. C. , Bogdanowicz, S. M. , Harrison, R. G. , Coates, B. S. , & Dopman, E. B. (2019). Genomic basis of circannual rhythm in the European corn borer moth. Current Biology, 29, 3501–3509. 10.1101/633362 [DOI] [PubMed] [Google Scholar]
- Kubrak, O. I. , Kučerová, L. , Theopold, U. , & Nässel, D. R. (2014). The sleeping beauty: How reproductive diapause affects hormone signaling, metabolism, immune response and somatic maintenance in Drosophila melanogaster. PLoS One, 9(11), e113051. 10.1371/journal.pone.0113051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz, S. G. , & Eisen, M. B. (2014). Drosophila embryogenesis scales uniformly across temperature in developmentally diverse species. PLoS Genetics, 10(4), e1004293. 10.1371/journal.pgen.1004293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder, P. , & Horvath, S. (2008). WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics, 9(1), 559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, A. , Marygold, S. J. , Antonazzo, G. , Attrill, H. , dos Santos, G. , Garapati, P. V. , Goodman, J. L. , Gramates, L. S. , Millburn, G. , Strelets, V. B. , Tabone, C. J. , Thurmond, J. , & FlyBase Consortium . (2021). FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Research, 49(D1), D899–D907. 10.1093/nar/gkaa1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, S. A. , & Wasserman, S. A. (2014). Conventional and non‐conventional Drosophila toll signaling. Developmental & Comparative Immunology, 42(1), 16–24. 10.1016/j.dci.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Frochaux, M. , Gardeux, V. , Deplancke, B. , & Robinson‐Rechavi, M. (2020). Inter‐embryo gene expression variability recapitulates the hourglass pattern of evo‐devo. BMC Biology, 18(1), 1–12. 10.1186/s12915-020-00842-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Maury, L. , Marguerat, S. , & Bähler, J. (2008). Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nature Reviews Genetics, 9(8), 583–593. 10.1038/nrg2398 [DOI] [PubMed] [Google Scholar]
- Mirth, C. K. , Saunders, T. E. , & Amourda, C. (2021). Growing up in a changing world: Environmental regulation of development in insects. Annual Review of Entomology, 66, 81–99. 10.1146/annurev-ento-041620-083838 [DOI] [PubMed] [Google Scholar]
- Pertea, G. , & Pertea, M. (2020). GFF utilities: GffRead and GffCompare. F1000Research, 9(304), 304. 10.12688/f1000research.23297.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, M. , Kim, D. , Pertea, G. M. , Leek, J. T. , & Salzberg, S. L. (2016). Transcript‐level expression analysis of RNA‐seq experiments with HISAT, StringTie and ballgown. Nature Protocols, 11(9), 1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, M. , Pertea, G. M. , Antonescu, C. M. , Chang, T. C. , Mendell, J. T. , & Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA‐seq reads. Nature Biotechnology, 33(3), 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson, B. , Lee, S. , Majewski, I. J. , Alexander, W. S. , & Smyth, G. K. (2016). Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. The Annals of Applied Statistics, 10(2), 946–963. 10.1214/16-AOAS920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupardin, R. , Schöttner, K. , Korbelová, J. , Provazník, J. , Doležel, D. , Pavlinic, D. , Beneš, V. , & Koštál, V. (2015). Early transcriptional events linked to induction of diapause revealed by RNAseq in larvae of drosophilid fly, Chymomyza costata. BMC Genomics, 16(720), 720. 10.1186/s12864-015-1907-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta, M. , Coggill, P. C. , Eberhardt, R. Y. , Mistry, J. , Tate, J. , Boursnell, C. , Pang, N. , Forslund, K. , Ceric, G. , Clements, J. , Heger, A. , Holm, L. , Sonnhammer, E. L. , Eddy, S. R. , Bateman, A. , & Finn, R. D. (2012). The Pfam protein families database. Nucleic Acids Research, 40(Database issue), D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing. [Google Scholar]
- Rice, P. , Longden, I. , & Bleasby, A. (2000). EMBOSS: The European molecular biology open software suite. Trends in Genetics, 16(6), 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Ritchie, M. E. , Phipson, B. , Wu, D. , Hu, Y. , Law, C. W. , Shi, W. , & Smyth, G. K. (2015). Limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Research, 43(7), e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis, L. , Lof, M. , van Asch, M. , & Visser, M. E. (2016). Modeling winter moth Operophtera brumata egg phenology: Nonlinear effects of temperature and developmental stage on developmental rate. Oikos, 125(12), 1772–1781. 10.1111/oik.03257 [DOI] [Google Scholar]
- Sawyer, L. A. , Hennessy, J. M. , Peixoto, A. A. , Rosato, E. , Parkinson, H. , Costa, R. , & Kyriacou, C. P. (1997). Natural variation in a Drosophila clock gene and Temperature compensation. Science, 278(5346), 2117–2120. 10.1126/science.278.5346.2117 [DOI] [PubMed] [Google Scholar]
- Scheffers, B. R. , De Meester, L. , Bridge, T. C. L. , Hoffmann, A. A. , Pandolfi, J. M. , Corlett, R. T. , Butchart, S. H. M. , Pearce‐Kelly, P. , Kovacs, K. M. , Dudgeon, D. , Pacifici, M. , Rondinini, C. , Foden, W. B. , Martin, T. G. , Mora, C. , Bickford, D. , & Watson, J. E. M. (2016). The broad footprint of climate change from genes to biomes to people. Science, 354(6313), aaf7671. 10.1126/science.aaf7671 [DOI] [PubMed] [Google Scholar]
- Smyth, G. K. , Ritchie, M. E. , Law, C. W. , Alhamdoosh, M. , Su, S. , Dong, X. , & Tian, L. (2018). RNA‐seq analysis is easy as 1‐2‐3 with limma, Glimma and edgeR. F1000Research, 5, 1–30. 10.12688/f1000research.9005.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, K. B. (2015). Regulation of hypometabolism: Insights into epigenetic controls. Journal of Experimental Biology, 218(1), 150–159. 10.1242/jeb.106369 [DOI] [PubMed] [Google Scholar]
- Strimmer, K. (2008). fdrtool: A versatile R package for estimating local and tail area‐based false discovery rates. Bioinformatics, 24(12), 1461–1462. 10.1093/bioinformatics/btn209 [DOI] [PubMed] [Google Scholar]
- Tanaka, Y. (2021). Orcokinins. In Ando H., Ukena K., & Nagata S. (Eds.), Handbook of hormones (2nd ed., pp. 791–793). Academic Press. [Google Scholar]
- Terada, S.‐I. , Matsubara, D. , Onodera, K. , Matsuzaki, M. , Uemura, T. , & Usui, T. (2016). Neuronal processing of noxious thermal stimuli mediated by dendritic Ca2+ influx in Drosophila somatosensory neurons. eLife, 5, 1–26. 10.7554/eLife.12959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törönen, P. , Medlar, A. , & Holm, L. (2018). PANNZER2: A rapid functional annotation web server. Nucleic Acids Research, 46(W1), W84–W88. 10.1093/nar/gky350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Asch, M. , Salis, L. , Holleman, L. J. M. , Van Lith, B. , & Visser, M. E. (2013). Evolutionary response of the egg hatching date of a herbivorous insect under climate change. Nature Climate Change, 3, 244–248. 10.1038/NCLIMATE1717 [DOI] [Google Scholar]
- van Asch, M. , van Tienderen, P. H. , Holleman, L. J. M. , & Visser, M. E. (2007). Predicting adaptation of phenology in response to climate change, an insect herbivore example. Global Change Biology, 13(8), 1596–1604. 10.1111/j.1365-2486.2007.01400.x [DOI] [Google Scholar]
- van Asch, M. , & Visser, M. E. (2007). Phenology of Forest caterpillars and their host trees: The importance of synchrony. Annual Review of Entomology, 52, 37–55. 10.1146/annurev.ento.52.110405.091418 [DOI] [PubMed] [Google Scholar]
- van Dis, N. E. , Risse, J. E. , Pijl, A. S. , Hut, R. A. , Visser, M. E. , & Wertheim, B. (2022). Transcriptional regulation underlying the temperature response of embryonic development rate in the winter moth. Dryad, Dataset. 10.5061/dryad.hx3ffbghd [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dis, N. E. , van der Zee, M. , Hut, R. A. , Wertheim, B. , & Visser, M. E. (2021). Timing of increased temperature sensitivity coincides with nervous system development in winter moth embryos. Journal of Experimental Biology, 224(17), 1–10. 10.1242/jeb.242554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck, H. , Bonte, D. , Puls, R. , Gotthard, K. , & Maes, D. (2015). The lost generation hypothesis: Could climate change drive ectotherms into a developmental trap? Oikos, 124(1), 54–61. 10.1111/oik.02066 [DOI] [Google Scholar]
- Visser, M. E. (2008). Keeping up with a warming world; assessing the rate of adaptation to climate change. Proceedings of the Royal Society B, 275, 649–659. 10.1098/rspb.2007.0997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser, M. E. , & Holleman, J. M. (2001). Warmer springs disrupt the synchrony of oak and winter moth phenology. Proceedings of the Royal Society B: Biological Sciences, 268, 289–294. 10.1098/rspb.2000.1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, A. (2011). Innovation through regulation. In The Origins of Evolutionary Innovations (pp. 28–29). Oxford Univ. Press. 10.1093/acprof:oso/9780199692590.003.0050 [DOI] [Google Scholar]
- Waldvogel, A. , Feldmeyer, B. , Rolshausen, G. , Exposito‐Alonso, M. , Rellstab, C. , Kofler, R. , Mock, T. , Schmid, K. , Schmitt, I. , Bataillon, T. , Savolainen, O. , Bergland, A. , Flatt, T. , Guillaume, F. , & Pfenninger, M. (2020). Evolutionary genomics can improve prediction of species' responses to climate change. Evolution Letters, 4(1), 4–18. 10.1002/evl3.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby, E. A. , Perkins, G. R. , Collins, M. K. , & Whitmarsh, A. J. (2003). The JNK‐interacting Protein‐1 scaffold protein targets MAPK Phosphatase‐7 to dephosphorylate JNK. Journal of Biological Chemistry, 278(12), 10731–10736. 10.1074/jbc.M207324200 [DOI] [PubMed] [Google Scholar]
- Wilson, D. , Pethica, R. , Zhou, Y. , Talbot, C. , Vogel, C. , Madera, M. , Chothia, C. , & Gough, J. (2009). SUPERFAMILY—Sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Research, 37(Database issue), D380–D386. 10.1093/nar/gkn762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsterman, K. , Ballinger, M. A. , & Williams, C. M. (2021). A unifying, eco‐physiological framework for animal dormancy. Functional Ecology, 35(1), 11–31. 10.1111/1365-2435.13718 [DOI] [Google Scholar]
- Wingett, S. W. , & Andrews, S. (2018). FastQ screen: A tool for multi‐genome mapping and quality control. F1000Research, 7, 1338. 10.12688/f1000research.15931.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, B. , Petritsch, C. , Clark, I. E. , Gavis, E. R. , Jan, L. Y. , & Jan, Y. N. (2004). Nanos and pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Current Biology, 14(4), 314–321. 10.1016/j.cub.2004.01.052 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1:
Data Availability Statement
Raw RNAseq reads (incl. metadata) used in this article can be found at the European Nucleotide Archive (ENA) under accession no. PRJEB55675. The processed data used for analysis, including the annotated transcriptome, GO annotation table, gene count matrix and supporting data can be found on Dryad https://doi.org/10.5061/dryad.hx3ffbghd (van Dis et al., 2022). Bioinformatic and statistical analysis scripts to reproduce the results can be found on GitHub at https://github.com/NEvanDis/WM_RNAseq.


