Abstract
Background
Drinking motives are robust proximal predictors of alcohol use behaviors and may mediate distinct etiological pathways in the development of alcohol misuse. However, little is known about the genetic and environmental etiology of drinking motives themselves and their potential utility as endophenotypes.
Methods
Here, we leverage a longitudinal study of college students from diverse racial/ethnic backgrounds (phenotypic N = 9889, genotypic N = 4855) to investigate the temporal stability and demographic and environmental predictors of four types of drinking motives (enhancement, social, coping, and conformity). Using genome‐wide association study (GWAS) and in silico tools, we characterize their associated genes and genetic variants (single nucleotide polymorphisms or SNPs).
Results
Drinking motives were stable across four years of college (ICC >0.74). Some robust environmental predictors of alcohol misuse (parental autonomy granting and peer deviance) were broadly associated with multiple types of drinking motives, while others (e.g., trauma exposure) were type specific. Genome‐wide analyses indicated modest SNP‐based heritability (14–22%, n.s.) and several suggestive genomic loci that corroborate findings from previous molecular genetic studies (e.g., PECR and SIRT4 genes), indicating possible differences in the genetic etiology of positive versus negative reinforcement drinking motives that align with an internalizing/externalizing typology of alcohol misuse. Coping motives were significantly genetically correlated with alcohol use disorder diagnoses (r g = 0.71, p = 0.001). However, results from the genetic analyses were largely underpowered to detect significant associations.
Conclusions
Drinking motives show promise as endophenotypes but require further investigation in larger samples to further our understanding of the etiology of alcohol misuse.
Keywords: alcohol misuse, college students, drinking motives, endophenotype, GWAS
We investigated drinking motives as possible endophenotypes for alcohol misuse in a longitudinal sample of college students. Motives were stable across college and robustly associated with environmental factors like trauma, peer deviance, and parenting. Suggestive genomic association signals and a genetic correlation between coping motives and alcohol use disorder indicates their potential for use as endophenotypes. Drinking motives are relatively simple measures to collect and may benefit from inclusion in larger samples and biobanks.
INTRODUCTION
Alcohol misuse, including heavy consumption and alcohol use disorder (AUD), is one of the leading contributors to the global burden of disease (Rehm et al., 2009). It is well established that not all individuals are at equal risk for alcohol misuse, with genetic factors responsible for about 50% of individual differences in liability to developing AUDs (Verhulst et al., 2015). Aside from robust links to alcohol metabolism candidate genes (the ADH and ALDH gene families), early efforts to pinpoint the specific genes underlying this heritability were largely unsuccessful (Hart & Kranzler, 2015). This led researchers to conclude that the genetic etiology of alcohol misuse is characterized by high polygenicity: an aggregate impact of thousands of genetic variants (single nucleotide polymorphisms or SNPs), each with very small effects. Recent work capitalizing on enormous sample sizes through data‐sharing consortia has propelled forward progress in this area, resulting in nearly 100 genomic loci robustly associated with alcohol consumption and/or pathological use (Kranzler et al., 2019; Liu et al., 2019; Sanchez‐Roige et al., 2019; Walters et al., 2018; Zhou et al., 2020). However, significantly associated variants explain less than 10% of the inter‐individual variation in consumption and liability to AUD, leaving much of the heritability to be explained. Further, with the exception of alcohol metabolism genes, the mechanisms linking these genes to differences in alcohol use behaviors are largely still unclear.
An extensive literature indicates a diverse array of interpersonal characteristics and psychiatric disorders associated with alcohol use/misuse (Kessler et al., 2005; Krueger & Markon, 2006; Lynam & Miller, 2004; Whelan et al., 2014). There are also well‐supported divergent etiological pathways to alcohol misuse, most notably the “internalizing” and “externalizing” pathways (Conrod et al., 2006; Mezquita et al., 2014; Zucker, 2008), in which internalizing‐related alcohol misuse is driven by drinking to cope with negative affect (depression, anxiety) and externalizing‐related alcohol misuse stems from a core deficit in impulse control shared with other substance and behavioral addictions, typically with an earlier onset and more severe manifestation (Cloninger et al., 1988). Given the long, winding biological pathways connecting genetic variants in a cell's DNA to the manifestation of a high‐level behavioral outcome, it is likely that genetic variants associated with alcohol misuse have a more direct impact on the intermediate steps of these heterogeneous pathways.
A potential tool to validate and investigate these pathways is the use of “endophenotypes,” factors which sit in the mediational pathway between a genetic predisposition and the distal manifestation of a trait or disorder (Gottesman & Gould, 2003; Hines et al., 2005). There is some debate over the specific definition of an endophenotype, but here we use the recommendations of Lenzenweger (2013) to refer to endophenotypes as measurable constructs located within the causal path between gene(s) and behavior. Fundamental criteria for an endophenotype include heritability, an association with the target phenotype, and shared genetic variance with the target phenotype, which are necessary conditions for mediation of a genetic etiological pathway. Iacono et al. (2017) further recommend that a useful endophenotype should demonstrate strong associations with specific genetic variants that are also linked to the target phenotype, and should be able to illuminate underlying developmental and/or mechanistic processes.
Drinking motives, the reasons why people consume alcohol and what they hope to achieve by drinking, present a clear mechanism by which internalizing and externalizing pathways may lead to alcohol misuse and elucidate the intermediate mechanisms by which such pathways may unfold. As alcohol is a psychoactive drug with both stimulant and sedative effects (Kreusch et al., 2013), motivations for drinking are likely to differ between individuals and, potentially, even between the same individual in different situations. The foremost model of drinking motives, developed by Cooper (1994), proposes four distinct types of drinking motives that fall under two dimensions: valence (negative versus positive reinforcement) and source (internal versus external). Negative reinforcement motives include coping (internal) and conformity (external) motives, which reflect drinking to obtain relief from negative emotions or escape unpleasant affective states. Positive reinforcement motives, on the other hand, underlie drinking to obtain positive affective states or enjoy the pleasurable aspects of alcohol, and are subclassified into enhancement (internal) and social (external) motives. Internal motives are driven by one's own desires or feelings, while external motives are driven by social or environmental influences. Drinking motives, particularly enhancement and coping motives, have repeatedly demonstrated robust, proximal associations with measures of alcohol consumption and alcohol problems (Carpenter & Hasin, 1998; Kuntsche et al., 2005).
Drinking motives are therefore plausible candidate endophenotypes for alcohol misuse, fulfilling the criterion of association with the target phenotype, and the utility of providing mechanistic insight. Despite their strong association with alcohol misuse, there has been little research on the etiology of drinking motives themselves, and even less on their genetic etiology. Moreover, research on the genetic etiology of drinking motives (and of alcohol‐related phenotypes in general) is especially limited among diverse ancestry cohorts. As potential endophenotypes, knowledge of some of the basic epidemiological aspects of drinking motives is needed: how reliable are they, how do they (or do they not) change across time, and what genetic and environmental factors are associated with their development—and might thus be used to modify them or predict individual risk?
Drinking motives have primarily been studied in relation to alcohol, where studies consistently demonstrate that coping and enhancement motives serve as important mediators of the relationship between temperament (neuroticism and impulsivity, respectively) and alcohol use/misuse outcomes (Adams et al., 2012; Mezquita et al., 2010). Coping motives also have strong evidence for mediating the effects of environmental exposures—in particular, trauma—on problematic alcohol use (Hawn et al., 2020). Social drinking motives are the most highly endorsed type but are generally related to normative drinking rather than problematic use or negative consequences of use, while conformity motives have less consistent, and usually transient or weaker associations to alcohol outcomes (Bresin & Mekawi, 2021; Kuntsche et al., 2005). Overall higher motivations are generally associated with higher levels of alcohol use and problems (Bresin & Mekawi, 2021; Cho et al., 2015). The existing research on drinking motives has primarily been carried out in European ancestry individuals, although the existing evidence indicates that similar processes are at play across various ethnic groups and cultures (Ertl et al., 2018; Mezquita et al., 2018; Wicki et al., 2017). Cultural norms surrounding drinking behaviors do, though, play a role in cross‐cultural differences in mean levels of drinking motives (Kuntsche et al., 2015; Wicki et al., 2017).
Drinking motives show moderate stability in young adulthood, with correlations of 0.38–0.64 across a 1‐year period from age 19–20 (Labhart et al., 2016), and standardized loadings of 0.68–0.87 on a latent trait factor across ages 23–33 (Windle & Windle, 2018). However, their stability during the critical period of emerging adulthood in which drinking behaviors are typically established has not yet been well‐characterized.
Only a few studies have directly examined predictors of drinking motives themselves, but these have provided initial evidence that factors associated with (adolescent) alcohol use, such as stressful life events and peer group alcohol use, are associated with higher levels of all types of drinking motives (Windle & Windle, 2018). Drinking motives, particularly enhancement motives, tend to resemble those of classroom peers (Temmen & Crockett, 2018) and drinking buddies (Kehayes et al., 2021), who seem to set the norms in which one's early drinking motives develop. Van Damme et al. (2015) further demonstrated an effect of parental drinking on enhancement/social motives, although it is unclear whether this is a result of parenting behaviors or an inherited predisposition towards alcohol use that is shared with positive reinforcement motives.
A few twin studies in European ancestry samples have been conducted to estimate the heritability of drinking motives (Agrawal et al., 2008; Kristjansson et al., 2011; Mackie et al., 2011; Prescott et al., 2004; Young‐Wolff et al., 2009), fulfilling a requisite criterion for an endophenotype. These studies have examined Cooper's (1994) Drinking Motives Questionnaire (DMQ) and the related Alcohol Use Inventory (Wanberg & Horn, 1983) subscales and estimated that their heritability ranges from 11% to 40%. There is also some evidence from twin studies that drinking motives mediate the latent genetic overlap between depression and AUD via coping motives (Young‐Wolff et al., 2009). The latent genetic association between the personality traits of neuroticism/impulsivity and AUD are also mediated via coping and enhancement motives, respectively (Littlefield et al., 2011; Prescott et al., 2004), mirroring the relationship observed at the phenotypic level (Adams et al., 2012; Mezquita et al., 2010). However, to date, no robust molecular genetic studies have yet investigated the genetic etiology of drinking motives or their genetic relationship to alcohol misuse.
The aims of this project are thus twofold: (1) to investigate the phenotypic etiology of drinking motives and characterize their stability and associated environmental factors and (2) to investigate the genetic etiology of drinking motives in individuals from diverse ancestry groups using multiple molecular genetic methods. These analyses provide insight into the shared and distinct etiologies of different types of drinking motives and the utility of drinking motives to serve as endophenotypes for alcohol misuse.
MATERIALS AND METHODS
Participants
Data comes from the parent study “Spit for Science” (S4S) (Dick et al., 2014) a longitudinal, prospective study investigating genetic and environmental influences on mental health and substance use in college students at a large, urban, public university. The study enrolled four cohorts of incoming students between 2011 and 2015, and all first‐time freshmen aged 18 years and older were eligible to complete a self‐report survey and provide a saliva sample for DNA collection. Each subsequent spring that participants were enrolled at the university, they were invited to participate in a follow‐up survey. All participants provided informed consent and the S4S study was approved by the university Institutional Review Board.
A total of N = 9889 students from four cohorts were enrolled in the study at the time of analysis. Participation rates have been consistently high across cohorts, with 63–68% of the eligible incoming students enrolling in the study each year and retention rates of 48–75% across follow‐up surveys. The demographic characteristics of the sample are consistent with those of the overall university student population: 61.5% female, with self‐reported race/ethnicity of 0.5% American Indian/Native Alaskan, 16.3% Asian, 18.9% African American, 49.4% Caucasian, 6.0% Hispanic/Latino, 6.2% multiracial, 0.7% Native Hawaiian/Pacific Islander, and 1.9% unknown/unreported. Nearly all participants, 91% of the total sample, also provided a DNA sample, and genotyping was completed for the first three cohorts at the time of analysis, with n = 6325 samples passing quality control.
Measures
All measures were collected via a confidential online self‐report survey. Participants were emailed an individual link to this survey and could complete it at a time and location of their choosing. The surveys assessed a wide range of behaviors, characteristics, and environmental exposures. Data was collected and managed by the secure, web‐based REDCap system of electronic data capture tools (Harris et al., 2009). Measures were largely drawn from psychometrically validated scales administered in an abbreviated version to reduce participant burden. The first survey in the fall of the freshman year (Y1F) was considered a baseline metric and thus assessed lifetime measures of psychopathology and environmental exposures up to the beginning of college, while later assessments in the spring semester of each year (Y1S, Y2S, Y3S, and Y4S) focused on the experience of such factors in the time since the previous survey.
Drinking motives
In each survey, participants who had initiated drinking completed an abbreviated version of the Drinking Motives Questionnaire—Revised (Cooper, 1994). This scale contains four subscales whose items are summed to create scores: Coping (e.g., “because it helps me when I feel depressed or nervous”), Enhancement (e.g., “because it gives me a pleasant feeling”), Conformity (e.g., “to get in with a group I like”), and Social (e.g., “because it makes social gatherings more fun”). Responses are on a Likert‐like scale from 1 = Strongly Agree to 4 = Strongly Disagree (reverse coded). Four items per each subscale were assessed in the Y1F, Y1S, and Y2S surveys, and the one best‐performing item (based on factor loadings) from each subscale was included in the Y3S and Y4S surveys due to space limitations. However, descriptive statistics (see Table 1) and correlations across waves showed that the single‐item scores performed similarly to the multi‐item scale scores.
TABLE 1.
Descriptive statistics for drinking motive scores.
| Measure | Time | N | Min | Max | Mean | SD |
|---|---|---|---|---|---|---|
| Conformity | Y1F | 5852 | 1 | 4 | 1.44 | 0.73 |
| Conformity | Y1S | 4866 | 1 | 4 | 1.42 | 0.72 |
| Conformity | Y2S | 4027 | 1 | 4 | 1.43 | 0.74 |
| Conformity | Y3S | 1726 | 1 | 4 | 1.52 | 0.80 |
| Conformity | Y4S | 1489 | 1 | 4 | 1.53 | 0.82 |
| Coping | Y1F | 5832 | 1 | 4 | 1.84 | 0.96 |
| Coping | Y1S | 4838 | 1 | 4 | 1.86 | 0.96 |
| Coping | Y2S | 4029 | 1 | 4 | 2.04 | 0.95 |
| Coping | Y3S | 1721 | 1 | 4 | 1.94 | 0.99 |
| Coping | Y4S | 1482 | 1 | 4 | 1.97 | 1.00 |
| Enhancement | Y1F | 5849 | 1 | 4 | 2.91 | 0.87 |
| Enhancement | Y1S | 4865 | 1 | 4 | 2.95 | 0.84 |
| Enhancement | Y2S | 4024 | 1 | 4 | 2.86 | 0.84 |
| Enhancement | Y3S | 1733 | 1 | 4 | 2.97 | 0.85 |
| Enhancement | Y4S | 1492 | 1 | 4 | 2.98 | 0.88 |
| Social | Y1F | 5869 | 1 | 4 | 2.94 | 0.84 |
| Social | Y1S | 4894 | 1 | 4 | 3.00 | 0.82 |
| Social | Y2S | 4042 | 1 | 4 | 3.04 | 0.83 |
| Social | Y3S | 1738 | 1 | 4 | 3.07 | 0.82 |
| Social | Y4S | 1495 | 1 | 4 | 3.07 | 0.79 |
Alcohol use
At each wave, participants were asked if they had initiated alcohol use, and, if so, about alcohol use behaviors including typical consumption (gm EtOH/month) and symptoms of DSM‐5 Alcohol Use Disorder (AUDsx), which were measured using the Semi‐Structured Assessment for the Genetics of Alcoholism (SSAGA; Bucholz et al., 1994) and summed.
Environmental risk and protective factors
The S4S surveys assess numerous environmental and psychosocial constructs that have demonstrated associations with alcohol use and psychopathology. Among these are parenting behaviors, specifically the Involvement and Autonomy Granting subscales of Steinberg's Parenting Style scale (Steinberg et al., 1992), peer deviance (the proportion of one's friends who engage in deviant behaviors such as getting drunk and cutting school, as described by Kendler et al., 2008), and exposure to traumatic events such as an assault or natural disaster, measured by the Life Events Checklist (Gray et al., 2004).
Genotyping
Information about genotyping for this sample has been described in detail elsewhere (Dick et al., 2014; Webb et al., 2017). Briefly, samples were genotyped on the Axiom BioBank Array, Catalog Version 2 (Affymetrix Inc., Santa Clara, CA) and imputed to the 1000 Genomes phase 3 all‐ancestries reference panel (The 1000 Genomes Project Consortium, 2015) after removing poor quality SNPs (missingness >5%, Hardy–Weinberg equilibrium [HWE] p values <5 × 10−6) and individual samples (genotyping rate < 98%, heterozygosity outliers, phenotypic/genotypic sex discordance, excess relatedness). Genetic ancestry principal components (PCs) were derived from the 1000 Genomes (phase 3) full reference population and projected onto the S4S samples to identify genetically homogenous ancestral groups for analysis, as described by Peterson et al. (2017). After this procedure, individuals from five continental ancestral super‐populations were available for analysis: Africa (AFR), the Americas (AMR), East Asia (EAS), Europe (EUR), and South Asia (SAS). Within‐group ancestry PCs were then calculated within each of these super‐populations in order to capture fine‐grained differences in allele frequencies that could contribute to residual population stratification. Within groups, additional quality control steps were taken to remove reference population outliers, those with excess relatedness (pi‐hat > 0.1), and SNPs with low ancestry‐specific minor allele frequency (MAF)/HWE.
Data analysis
Data analysis involved two parts. First, we examined drinking motives at a phenotypic level in the full sample. We estimated the Pearson and intraclass correlations between motives across survey waves to estimate their temporal stability, calculated the Pearson correlations between motives and alcohol consumption/AUDsx cross‐sectionally and prospectively (Y1F motives with Y1F‐Y4S alcohol measures), and conducted linear regression analyses to examine how demographic and environmental characteristics predicted mean levels of each type of drinking motive.
Second, we conducted a series of analyses on the genotypic data to investigate the genetic etiology of drinking motives. The genotyped sample (n = 6325) was restricted to include only unrelated individuals with nonmissing phenotype information—i.e., those who initiated alcohol use during the time the measures were collected and chose to answer the relevant survey questions (analytic n = 4855). We used the mean of all available scores across waves (Table 1) for the four drinking motives subscales as the outcome phenotypes. We also utilized relevant covariates to control for possible confounding effects on genetic associations with the phenotypes, which included sex, age (mean across waves where participants had nonmissing drinking motives scores), and within‐ancestry PCs.
We estimated the heritability of drinking motives that could be attributable to measured genetic variants (SNP heritability; h 2 SNP) through the use of genome‐wide complex trait analysis (GCTA; Yang et al., 2011). Genotyped SNPs were filtered for a MAF >0.01 before calculating the genomic relatedness matrix between individuals. GCTA was then run separately in each ancestry using the first 10 within‐ancestry PCs, age, and sex as covariates, and the resulting heritability estimates were meta‐analyzed with a fixed‐effects, inverse variance‐weighted scheme.
Next, we conducted a genome‐wide association (GWAS) of the imputed genetic variants. SNPs were filtered for imputation quality (INFO score >0.5) and ancestry‐specific MAF corresponding to a minor allele count (MAC) > 100. Previous work has established that a MAC > 40 allows for reliable statistical estimation (Bigdeli et al., 2014); we use a more conservative threshold because the somewhat skewed distribution of the phenotypes. Within‐ancestry PCs to include as covariates were decided based on their association with the phenotype in a stepwise linear regression to avoid overfitting (c.f. Webb et al., 2017). The specific PCs used in each analysis are shown in Table S1. GWAS was conducted using SNPTEST (Marchini et al., 2007), again running separately in each ancestry group and meta‐analyzing the association test for each SNP using inverse‐variance weighting in METAL (Abecasis et al., 2012). We present only the results for SNPs that were available in at least 1000 individuals (meaning that the SNP had to pass quality control filters in either the AFR, EUR, or a combination of two or more ancestry sub‐groups), to be certain that spurious results from small samples were not given undue consideration. Multiple testing correction was performed by using a Bonferroni‐corrected threshold for genome‐wide significance of 5 × 10−8 and calculating false discovery rates (FDR) using the qvalue package for R/Bioconductor (Storey et al., 2015). Given the small size of our sample and its corresponding statistical power (see Discussion), we also examine “suggestive” signals (p < 5 × 10−6) and ancestry‐specific results in the largest subgroup (EUR; n = 2537), although these are given lower credibility. Regional association plots for associated loci were visualized using LocusZoom (Pruim et al., 2010). Significant and suggestive signals were uploaded to FUMA (Watanabe et al., 2017) to define candidate genomic loci based on LD (variants with r 2 > 0.6 with the lead SNP, using the 1000 Genomes all‐ancestries reference panel for the LD structure [The 1000 Genomes Project Consortium, 2015]), annotate functional consequences of SNPs in the loci, and map candidate genes to these association signals based on position (within 10 kb), known expression quantitative trait locus (eQTL) effects, or 3‐dimensional chromatin interactions.
After conducting GWAS at the individual variant level, we applied gene‐based and pathway‐based association analyses to test whether the SNP association signal was enriched at these aggregate levels, using MAGMA v.1.08 (de Leeuw et al., 2015). We used the 1000 Genomes as reference panels for the LD structure of each of the five ancestral continental superpopulations. After annotation of the SNPs to genes, genes were also grouped into pathways based on the curated hallmark and canonical pathways gene‐sets from the Molecular Signatures Database (Liberzon et al., 2015). These pathways represent groups of genes whose products are involved in known metabolic and regulatory biochemical processes. Gene‐based analyses were conducted on the ancestry‐specific SNPTEST results and meta‐analyzed with MAGMA, using an inverse variance‐weighted Stouffer's Z test. Pathway analyses were conducted on the meta‐analyzed, transancestral gene‐based results. We also used Popcorn (Brown et al., 2016) to estimate the genetic correlation of the GWAS results across ancestry groups, using the 1000 Genomes as a reference for LD patterns in each ancestry group and calculating the genetic impact correlation (the similarity between SNP effect sizes across ancestry groups, while accounting for differences in allele frequencies).
Finally, we investigated a key endophenotype criterion for drinking motives by estimating their genetic overlap with alcohol‐related phenotypes. Using LD score regression (LDSC; Bulik‐Sullivan et al., 2015), we estimated h 2 SNP from the GWAS summary statistics and, if there was sufficient signal (mean χ2 > 1), we used LDSC to estimate the genome‐wide genetic correlations with three alcohol phenotypes from publicly available GWAS results: consumption (Liu et al., 2019), AUDIT total scores (Sanchez‐Roige et al., 2019), and AUD diagnoses (Walters et al., 2018). As LDSC assumptions rest on ancestry‐specific LD patterns, we used only results of the drinking motives GWAS from the largest group, the EUR ancestry subset (n = 2537), and conducted analyses using EUR LD scores provided with the software and recommended analysis settings (INFO >0.9, MAF >0.01, HapMap3 SNPs). LDSC provides only a global, genome‐wide estimate of the correlation, so we additionally conducted local genetic correlation for a more fine‐grained approach. We curated a list of 98 known alcohol‐related loci based on genome‐wide significant findings from well‐powered GWASs (Liu et al., 2019; Sanchez‐Roige et al., 2019; Walters et al., 2018; Zhou et al., 2020), using a 250 kb window around the lead SNP if a single position rather than a region was provided and merging overlapping loci (within 250 kb) within and across studies. Using LAVA (Werme et al., 2022), we investigated the local genetic correlation between drinking motives (EUR ancestry results) and the same three alcohol‐related phenotypes as for the genome‐wide correlations (consumption, AUDIT‐T, and AUD) within these curated loci. Bivariate correlations were calculated only when there was enrichment (p < 0.0001) in the univariate heritability for both phenotypes at a locus.
RESULTS
Phenotypic etiology
Descriptive statistics for drinking motives across survey waves are shown in Table 1. Mean levels for all four motives increased slightly but remained relatively stable across college. Correlations between each of the motives across time are shown in Table 2. Although there was some decay in the strength of correlation between longer periods of time (e.g., year 1 with year 4), each pair of adjacent time points was moderately correlated (Pearson r = 0.37 to 0.56, p < 5 × 10−20), with excellent overall stability (ICC = 0.75 to 0.81). Conformity motives showed the weakest correlations across time but also had the lowest levels of endorsement. Consistent with previous research, there were significant positive cross‐sectional and longitudinal correlations between motives and alcohol use outcomes, particularly between coping motives and AUDsx and between enhancement/social motives and both consumption and AUDsx (Table 3).
TABLE 2.
Cross‐time correlations for drinking motives across five waves of assessment.
| Wave | Social | Enhancement | Coping | Conformity |
|---|---|---|---|---|
| Y1F‐Y1S | 0.467*** | 0.519*** | 0.460*** | 0.432*** |
| Y1F‐Y2S | 0.408*** | 0.389** | 0.356** | 0.350** |
| Y1F‐Y3S | 0.335** | 0.334** | 0.324** | 0.295** |
| Y1F‐Y4S | 0.313** | 0.347** | 0.255* | 0.279* |
| Y1S‐Y2S | 0.517*** | 0.476*** | 0.403*** | 0.370** |
| Y1S‐Y3S | 0.409** | 0.407** | 0.417** | 0.302** |
| Y1S‐Y4S | 0.383** | 0.416** | 0.377** | 0.260* |
| Y2S‐Y3S | 0.450** | 0.430** | 0.428** | 0.460** |
| Y2S‐Y4S | 0.481** | 0.445** | 0.402** | 0.207* |
| Y3S‐Y4S | 0.550** | 0.558** | 0.557** | 0.431** |
| Intraclass correlation | 0.81*** | 0.81*** | 0.77*** | 0.75*** |
Note: *p < 5e‐10, **p < 5e‐20, ***p < 5e‐100.
TABLE 3.
Cross‐sectional and prospective correlations between drinking motives and alcohol use outcomes.
| Measure | Wave | Motive (Y1F) | |||
|---|---|---|---|---|---|
| Social | Enhancement | Coping | Conformity | ||
| Motive—Social | Y1F | 1 | |||
| Motive—Enhancement | Y1F | 0.63 | 1 | ||
| Motive—Coping | Y1F | 0.27 | 0.30 | 1 | |
| Motive—Conformity | Y1F | 0.11 | 0.04 | 0.26 | 1 |
| AUDsx | Y1F | 0.18 | 0.22 | 0.30 | 0.16 |
| Y1S | 0.26 | 0.26 | 0.24 | 0.10 | |
| Y2S | 0.26 | 0.28 | 0.23 | 0.10 | |
| Y3S | 0.27 | 0.30 | 0.16 | 0.08 | |
| Y4S | 0.28 | 0.31 | 0.17 | 0.09 | |
| Consumption | Y1F | 0.28 | 0.31 | 0.15 | 0.02 |
| Y1S | 0.26 | 0.27 | 0.12 | 0.02 | |
| Y2S | 0.22 | 0.24 | 0.08 | 0.03 | |
| Y3S | 0.24 | 0.26 | 0.06 | −0.01 | |
| Y4S | 0.22 | 0.24 | 0.07 | 0.04 | |
Note: Bolded values are significant after multiple testing correction for 56 tests, adjusted alpha = 0.0009.
Abbreviations: AUDsx, alcohol use disorder symptom count; Y1F‐Y4S, measurement time from Year 1 Fall to Year 4 Spring.
Results from the linear models of predictors of drinking motives are displayed in Table 4. Male gender, older age, and recent trauma exposure were associated with higher levels of conformity motives. Students from racial and ethnic minorities generally had lower or not significantly different levels of all four drinking motives when compared with White students, with the exception that self‐reported Asian ethnicity was associated with higher levels of conformity motives. Parental autonomy granting was associated with lower levels of all drinking motives, while higher parental involvement was associated with higher levels of positive reinforcement (social and enhancement) motives. Peer deviance was associated with higher levels of all motives except conformity. Lifetime (pre‐college) trauma exposure was simultaneously associated with both higher coping motives and lower social motives.
TABLE 4.
Linear regression results of demographic and environmental factors predicting drinking motives.
| Predictor | Social | Enhancement | Coping | Conformity | ||||
|---|---|---|---|---|---|---|---|---|
| Estimate | p | Estimate | p | Estimate | p | Estimate | p | |
| Gender (male) | 0.04 | 0.05 | 0.01 | 0.69 | −0.01 | 0.68 | 0.13 | 1E‐13 |
| Age | 0.02 | 0.16 | 0.00 | 0.97 | 0.03 | 0.04 | 0.04 | 1E‐03 |
| Ethnicity | ||||||||
| American Indian | −0.03 | 0.86 | 0.04 | 0.82 | 0.06 | 0.75 | −0.19 | 0.14 |
| Asian | 0.08 | 6E‐03 | −0.14 | 5E‐07 | 0.04 | 0.21 | 0.21 | 2E‐16 |
| Black | −0.02 | 0.44 | −0.10 | 2E‐04 | −0.10 | 6E‐04 | −0.09 | 2E‐05 |
| Hispanic | 0.01 | 0.72 | −0.03 | 0.44 | −0.13 | 4E‐03 | −0.06 | 0.0 |
| Multiracial | −0.01 | 0.70 | 0.03 | 0.53 | 0.00 | 0.95 | −0.05 | 0.17 |
| Hawaiian/Pacific Islander | 0.04 | 0.73 | −0.15 | 0.19 | −0.01 | 0.92 | −0.06 | 0.51 |
| Unknown | −0.08 | 0.29 | −0.11 | 0.18 | −0.08 | 0.42 | −0.02 | 0.73 |
| Trauma count (during college) | 0.02 | 0.04 | 0.02 | 0.05 | 0.03 | 0.03 | 0.04 | 8E‐06 |
| Trauma count (lifetime) | −0.04 | 3E‐04 | −0.01 | 0.4 | 0.03 | 4E‐03 | −0.02 | 0.03 |
| Parental involvement | 0.02 | 1E‐04 | 0.03 | 6E‐08 | −0.01 | 0.05 | 0.00 | 0.23 |
| Parental autonomy granting | −0.02 | 1E‐06 | −0.02 | 1E‐05 | −0.04 | 3E‐15 | −0.03 | 2E‐14 |
| Mean peer deviance | 0.07 | 2E‐16 | 0.07 | 2E‐16 | 0.04 | 2E‐16 | 0.00 | 0.20 |
Note: Reference category for the Ethnicity measure is White. Bolded values are significant after multiple testing correction for 8 predictor variables, adjusted alpha = 0.0062.
Genetic etiology
SNP Heritability
Heritability estimates from GCTA are displayed in Table S2, with meta‐analysis estimates ranging from 14% (coping) to 22% (enhancement). However, none of the meta‐analytic estimates were significantly differentiable from zero in this sample.
GWAS
Manhattan plots for the SNP‐based GWAS results are shown in Figure 1 (cross‐ancestry meta‐analysis) and Figures S1 and S2 (EUR ancestry subset). There was little evidence of inflation that could indicate bias or population stratification; the median chi‐square statistic (λ) was 0.948–1.021 and the scaled statistic (λ1000) was 1.000 for each meta‐analysis. For social motives, there was no evidence for any locus reaching genome‐wide significance (p < 5 × 10−08), and little evidence of even suggestive association peaks (p < 5 × 10−06). For the other three motive types, at least one genome‐wide significant SNP was identified in addition to several suggestive loci (regional locus plots in Figures S3–S8). The significant and suggestive loci are shown in Table 5. Because true association signals should be found in LD blocks, rather than lone SNPs, Table 5 shows suggestive loci only when three or more SNPs in the same position (±10 kb) pass the suggestive significance threshold. Additional FUMA annotation information on suggestive/significant loci can be found in Table S3.
FIGURE 1.
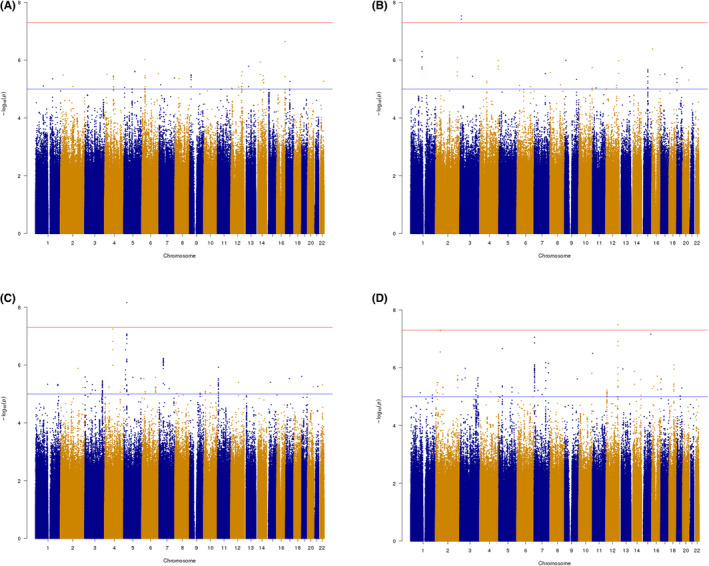
Manhattan plot of genome‐wide association meta‐analysis results for (A) social, (B) enhancement, (C) coping, and (D) conformity drinking motives.
TABLE 5.
Genomic annotation for top loci associated with drinking motives.
| CHR | Position | # SNPs | Min. p | Min. q | Max N | Local Genes | eQTL/CI Genes* |
|---|---|---|---|---|---|---|---|
| Social | |||||||
| 4 | 87,532,702 | 7 | 3.54E‐06 | 1.00 | 2533 | MAPK10, PTPN13, SLC10A6 | |
| 6 | 31,310,455 | 4 | 9.63E‐07 | 1.00 | 4820 | HLA‐B | — |
| 9 | 14,733,679 | 5 | 3.24E‐06 | 1.00 | 4820 | CER1, FREM1 | — |
| Enhancement | |||||||
| 1 | 112,418,319 | 5 | 5.08E‐07 | 0.99 | 4509 | KCND3 | — |
| 2 | 216,881,876 | 3 | 8.24E‐07 | 0.99 | 2537 | MREG, PECR | — |
| 3 | 13,613,907 | 2 | 2.97E‐08 | 0.18 | 2537 | FBLN2 | TMEM40 |
| 4 | 183,437,856 | 3 | 1.02E‐06 | 0.99 | 2537 | TENM3 | — |
| 15 | 62,854,341 | 4 | 2.13E‐06 | 1.00 | 4821 | TLN2 | — |
| Coping | |||||||
| 1 | 222,259,677 | 5 | 4.70E‐06 | 0.53 | 4814 | None | — |
| 3 | 174,010,802 | 8 | 3.52E‐06 | 0.53 | 4814 | NLGN1 | — |
| 4 | 81,172,733 | 4 | 5.70E‐08 | 0.11 | 2533 | FGF5 | — |
| 5 | 16,882,772 | 5 | 2.97E‐07 | 0.21 | 3580 | MYO10 | — |
| 5 | 29,577,370 | 17 | 6.90E‐09 | 0.06 | 2533 | None | CDH6 |
| 7 | 42,480,656 | 21 | 5.87E‐07 | 0.24 | 4053 | None | — |
| 11 | 10,529,822 | 10 | 1.19E‐06 | 0.28 | 4814 | AMPD3, MTRNR2L8, RNF141 | — |
| Conformity | |||||||
| 2 | 16,207,569 | 3 | 3.23E‐06 | 0.40 | 4817 | None | — |
| 2 | 46,018,826 | 4 | 5.16E‐08 | 0.21 | 2536 | PRKCE | — |
| 2 | 219,264,333 | 6 | 1.89E‐06 | 0.34 | 3580 | CTDSP1, SLC11A1 | — |
| 3 | 178,679,751 | 6 | 2.22E‐06 | 0.36 | 4817 | ZMAT3 | — |
| 5 | 34,479,237 | 4 | 2.16E‐07 | 0.25 | 3580 | None | — |
| 7 | 4,667,721 | 37 | 8.87E‐08 | 0.21 | 4817 | None | — |
| 7 | 114,732,621 | 3 | 6.69E‐07 | 0.29 | 4817 | None | — |
| 7 | 143,440,695 | 3 | 7.17E‐07 | 0.29 | 4505 | FAM1115C | — |
| 12 | 120,753,429 | 9 | 3.23E‐08 | 0.21 | 2536 | SIRT4, PLA2G1B, PXN | C12orf43, RPLP0, GCN1, POP5, OASL, DYNLL1 |
| 18 | 49,173,330 | 5 | 7.97E‐07 | 0.29 | 4817 | None | — |
Note: Loci are defined by the presence of genome‐wide significant (p < 5e‐08) SNPs or peaks with three or more suggestive (p < 5e‐06) SNPs. Position is the location of the lead SNP (build GRCh37). Local genes are defined by locus position within 10 kb of the gene. *eQTL and chromatin interaction (CI) mapping performed with FUMA only for genome‐wide significant loci.
The genome‐wide significant locus for enhancement motives included 2 SNPs found only in the EUR ancestry group, located on chromosome 3 in the FBLN2 (fibulin 2) gene, which codes for an extracellular matrix protein involved in organ development and differentiation. The regional association plot (Figure S3) showed little association enrichment for SNPs in high LD with the lead SNP, indicating this may be a spurious association. However, FUMA annotation of this region revealed a candidate nonsynonymous exonic variant in FBLN2, rs113265853, which was in LD (r 2 = 0.73) with the lead genome‐wide significant SNP rs149867189. This exonic variant was not directly analyzed but was present in the LD reference panel. Though it may be a spurious signal, an alternate possibility is that the association signal may be tagging this rare (1000 Genomes MAF = 0.01) functional variant. Examining the EUR ancestry‐specific GWAS for enhancement motives, there was also a peak just below genome‐wide significance (p = 6.72 × 10−08), in the PECR (peroxisomal trans‐2‐enoyl‐CoA reductase) gene on chromosome 2 (Figures S1 and S4). This gene has been previously linked to alcohol dependence (Treutlein et al., 2009) and also had a suggestive association in the cross‐ancestry meta‐analysis (p = 8.24 × 10−07).
For the coping motives meta‐analysis, one genome‐wide significant SNP was found in a peak in an intergenic region on chromosome 5 with no nearby genes, although the gene CDH6 was mapped by chromatin interactions with this locus in nine tissues, including stem cells, heart, and liver. The EUR ancestry‐specific GWAS identified additional genome‐wide significant loci in peaks on chromosome 9 (Intergenic, 25 kb from the gene GRIN3A) and 15 (in the pseudogene LOC390617) and a lone significant SNP on chromosome 7 (intergenic), see Figures S5–S7.
For the conformity motives meta‐analysis, one genome‐wide significant SNP alongside a peak of 9 suggestive SNPs was found on chromosome 12 in the SIRT4 (sirtuin 4) gene (Figure S8). No additional significant loci were identified in the EUR ancestry‐specific GWAS.
GWAS summary statistics from the individual ancestry groups were carried forward into gene‐based enrichment testing using MAGMA and meta‐analyzed, and the resulting (meta‐analyzed) gene‐based P values were tested for enrichment in biological pathways. Results from the gene‐based and pathway‐based association analyses are presented in Tables S4 and S5. After Bonferroni correction for the number of tested genes/pathways, there was no evidence for significant enrichment of the SNP association signal in any specific genes or pathways for any of the four motives.
Genetic correlation
LDSC indicated no significant genome‐wide heritability based on the SNP association signals, and a mean χ2 < 1 for social, enhancement, and conformity motives, making them unsuitable for further LDSC analyses. However, there was sufficient SNP heritability for coping motives (h 2 SNP = 0.56, SE = 0.21, mean χ2 = 1.20) to conduct genetic correlation analyses with external GWAS of alcohol‐related phenotypes. After Bonferroni correction for three tests (alpha = 0.05/3 = 0.017), these showed a significant correlation between coping motives and AUD diagnoses (r g = 0.71, p = 0.001), but not alcohol consumption (r g = 0.16, p = 0.069) or AUDIT‐T scores (r g = 0.05, p = 0.644).
Local genetic correlation was carried out to obtain a more fine‐grained view of the genetic relationship between drinking motives and alcohol phenotypes. Examining 98 known alcohol‐related loci, we saw local enrichment (p < 0.0001) for heritability in 13, 20, 44, and 11 of the 98 loci for social, enhancement, coping, and conformity motives, respectively (Table S6). These all exceed the number of loci expected by chance at an alpha of 0.05 (binomial test p = 3.76 × 10−04, 1.43 × 10−04, 3.68 × 10−32 and 0.004, respectively). Local genetic correlations suggested some interesting findings, for example, negative genetic correlations between coping motives and (1) consumption (r g = −0.27, p = 0.004), (2) AUDIT‐T (r g = −0.24, p = 0.041), and (3) AUD (r g = −0.34, p = 0.013) at the ADH locus (chr4:96,764,066‐101,983,024). However, no local genetic correlations survived multiple testing correction.
Trans‐ancestral genetic correlation analysis using Popcorn was limited by the low SNP heritability, so estimates were not calculable for many pairs of GWAS summary statistics (Table S7). For the correlations that could be calculated, there was no evidence that the SNP effects on drinking motives differed between ancestry groups: the correlation between ancestry groups did not significantly differ from r = 1.0 (all p's > 0.19).
DISCUSSION
In this set of analyses, we have uncovered several insights about the nature of drinking motives in college students. First, drinking motives are stable throughout college and represent reliable psychosocial constructs. Second, some robust environmental predictors of alcohol misuse like parental autonomy granting and peer deviance have broad associations with all types of drinking motives, while others, like trauma exposure, have more specific associations. For example, parental autonomy granting predicted lower levels of all drinking motives and peer drinking predicted higher levels of all drinking motives, while lifetime trauma exposure was simultaneously associated with lower social motives and higher coping motives. Third, the investigation into the genetic etiology of drinking motives in college students identified some promising but, as of yet, largely inconclusive results. We found substantial SNP heritability estimates (positive reinforcement motives: 16–22%; negative reinforcement motives: 14–16%). However, these estimates were not significantly different from zero—not surprising given the current sample size—so the ability for inference is limited. In genetic association testing, several loci were identified with suggestive or marginally significant effects, although the results of the gene‐based and pathway‐based analyses showed little evidence of enrichment at the aggregate levels. Genetic correlation analyses indicated substantial sharing of genetic factors between coping motives and AUD that merits follow‐up, though large standard errors again point to the need for replication in more robust samples.
These findings have encouraging implications for the use of drinking motives as endophenotypes of internalizing and externalizing pathways to alcohol misuse. Their stability across time is important for establishing that these are trait‐like outcomes linked to enduring temperamental dimensions (e.g., personality), and thus are viable for aiding gene identification efforts (Gottesman & Gould, 2003). These results were consistent with estimates in a slightly older sample across 10 years (Windle & Windle, 2018) and somewhat higher than those of a similar aged sample that looked across a shorter (1 year) time frame (Labhart et al., 2016), indicating high stability when motives are considered in the long‐term. However, their moderate correlations across shorter intervals also point to their potential to be modified via environmental interventions.
A few promising results from the genetic analyses are also worth considering. First, we found a suggestive association with enhancement motives in the PECR gene in Europeans. Although not quite reaching the threshold of genome‐wide significance, this signal showed a clear peak with enrichment of association signal in a large number of SNPs within a single locus, which bolsters confidence that it is a true effect. This gene is highly expressed in the liver and has been previously implicated in a GWAS of early onset alcohol dependence (Treutlein et al., 2009). Such evidence is consistent with the hypothesized connection between enhancement motives and an externalizing pathway/subtype of alcohol dependence characterized by early age of onset and stronger genetic influences (e.g., Cloninger et al., 1988). The PECR region (±10 kb, chr2:216893111‐216956539) has not been implicated in any larger GWAS of alcohol phenotypes since the Treutlein et al. (2009) study, although no such study has focused specifically on a severe/early‐onset subtype. This region had minimal association with alcohol consumption (p = 0.0015), AUDIT total scores (p = 0.0071), or AUD (p = 0.0061) in recent large‐scale GWAS analyses (Liu et al., 2019; Sanchez‐Roige et al., 2019; Walters et al., 2018). There was also evidence for a genome‐wide significant association in the SIRT4 gene with conformity motives. This gene is a close relative of the SIRT1 gene that was implicated in the genetic etiology of major depression (CONVERGE Consortium, 2015) and may suggest a common predisposition shared between internalizing psychopathology and this negative reinforcement motive. However, there is also a large number of other genes linked to this locus by positional, eQTL, and chromatin interaction mapping, any of which could be driving the identified association effect. The SIRT4 region (±10 kb, chr12:120730124‐120761045) had minimal association with alcohol consumption (p = 0.0026), AUDIT total scores (p = 0.0004), or AUD (p = 0.0370) in recent large‐scale GWAS analyses (Liu et al., 2019; Sanchez‐Roige et al., 2019; Walters et al., 2018).
An unusual result from the local genetic correlation analysis was the finding of a negative correlation (r g = −0.34) between coping motives and AUD at the ADH locus, despite an overall positive genome‐wide correlation (r g = 0.71). Although this result did not survive multiple testing correction, the strong biological evidence for ADH and the consistency of the negative genetic correlation across related measures (r g = −0.27 with consumption and r g = −0.24 with AUDIT scores) indicates that this may be worth further, cautious, consideration. We speculate that this may be a consequence of the relatively specific association of coping motives with alcohol problems/AUD, rather than with heaviness of consumption (Bresin & Mekawi, 2021; Kuntsche et al., 2005). Variants in the ADH genes are, of course, associated with multiple domains of alcohol use/misuse, but their foremost effect is on heaviness of consumption due to lowered sensitivity to the intoxicating effects of EtOH (Hart & Kranzler, 2015). Therefore while the aggregate genetic effects on AUD are positively correlated with coping motives, the specific effects of ADH variants are not. However, it's still surprising to see a negative genetic correlation rather than simply a null or weaker positive one. Another possible explanation is that coping motives primarily reflect a negative reinforcement mechanism while those with lower sensitivity to intoxication due to ADH variants are pursuing positive reinforcement, (i.e., chasing a high), and drinking more to achieve that reward state rather than to escape from a negative affective state.
We conclude that our findings at this stage thus provide only modest insight into the biology underlying drinking motives and their potential genetic pathways to alcohol misuse. This is perhaps unsurprising given trends in gene identification efforts for complex, and particularly psychiatric/behavioral traits. The emerging landscape of the field indicates that tens if not hundreds of thousands of samples may be needed for credible GWAS results, even for “less genetically complex” endophenotypes (Flint & Munafo, 2007). Power calculations with the Genetic Power Calculator (Purcell et al., 2003) indicated that we had 0.5–22% power to detect individual SNP associations with a typical effect size for a complex trait (MAF = 0.10, explaining 0.2–0.5% of the trait variance), and that a sample size of at least 8500–21,500 would be needed to detect SNP effects in that range. Our sample had >80% power to detect larger effect sizes (e.g., 1% of trait variance explained), which might be expected either from larger effects on endophenotypes that lie closer together in biological pathways, or from the aggregation of individual SNP effects into genes and pathways. Whether or not they prove to be simpler genetic targets, endophenotypes retain uniquely valuable roles, such as their ability to provide insight into the etiological mechanisms of a distal target phenotype and allow for prospective identification of individuals at risk (Iacono et al., 2017).
In the first reported GWAS of drinking motives, we already show promising results for an endophenotype—plausible candidate genes and robust genetic correlation with alcohol misuse measures—in a small sample of a few thousand participants, suggesting that GWAS samples for drinking motives may not need to be as large as for alcohol misuse measures to achieve similar gains in gene identification progress. Further, unlike many endophenotypes (e.g., brain imaging, neurophysiological measures) whose main drawback is their costliness to collect, drinking motives are reliably measured by a simple survey and could be collected at scale for GWAS in much the same way as alcohol misuse measures are now, possibly with a much greater return on investment. Recently developed statistical methods, such as genomic structural equation modeling (Grotzinger et al., 2019) might provide a means to investigate the overlapping and/or mediational role of drinking motives in the genetic etiology of alcohol misuse. The potential insights that drinking motives can provide into the mechanisms underlying alcohol misuse underscores their value for study with larger samples and more powerful study designs in the future.
CONFLICT OF INTEREST
The authors report no financial disclosures or potential conflicts of interest.
Supporting information
Figures S1–S8
Tables S1–S7
ACKNOWLEDGMENTS
Spit for Science has been supported by Virginia Commonwealth University, P20 AA017828, R37AA011408, K02AA018755, P50 AA022537, and K01AA024152 from the National Institute on Alcohol Abuse and Alcoholism, and UL1RR031990 from the National Center for Research Resources and National Institutes of Health Roadmap for Medical Research. This research was also supported by the National Institute on Drug Abuse of the National Institutes of Health under Award Number U54DA036105 and the Center for Tobacco Products of the U.S. Food and Drug Administration. JES was supported by F31AA024378 from the NIAAA. RE Peterson is supported by NIMH K01MH113848, The Brain & Behavior Research Foundation NARSAD grant 28632 P&S Fund, and NIAAA P50AA022537. The content is solely the responsibility of the authors and does not necessarily represent the views of the NIH or the FDA. The funding agencies had no role in the study design, data analysis, manuscript preparation, or decision to submit for publication. Data from this study are available to qualified researchers via dbGaP (phs001754.v2.p1). We would like to thank the Spit for Science participants for making this study a success, as well as the many University faculty, students, and staff who contributed to the design and implementation of the project. Portions of this work were conducted as part of a PhD dissertation project and have been made available online via ProQuest.
The Spit for Science Working Group
Director: Karen Chartier Co‐Director: Ananda Amstadter. Past Founding Director: Danielle M. Dick. Registry management: Emily Lilley, Renolda Gelzinis, Anne Morris. Data cleaning and management: Katie Bountress, Amy E. Adkins, Nathaniel Thomas, Zoe Neale, Kimberly Pedersen, Thomas Bannard & Seung B. Cho. Data collection: Amy E. Adkins, Kimberly Pedersen, Peter Barr, Holly Byers, Erin C. Berenz, Erin Caraway, Seung B. Cho, James S. Clifford, Megan Cooke, Elizabeth Do, Alexis C. Edwards, Neeru Goyal, Laura M. Hack, Lisa J. Halberstadt, Sage Hawn, Sally Kuo, Emily Lasko, Jennifer Lend, Mackenzie Lind, Elizabeth Long, Alexandra Martelli, Jacquelyn L. Meyers, Kerry Mitchell, Ashlee Moore, Arden Moscati, Aashir Nasim, Zoe Neale, Jill Opalesky, Cassie Overstreet, A. Christian Pais, Kimberly Pedersen, Tarah Raldiris, Jessica Salvatore, Jeanne Savage, Rebecca Smith, David Sosnowski, Jinni Su, Nathaniel Thomas, Chloe Walker, Marcie Walsh, Teresa Willoughby, Madison Woodroof & Jia Yan. Genotypic data processing and cleaning: Cuie Sun, Brandon Wormley, Brien Riley, Fazil Aliev, Roseann Peterson & Bradley T. Webb.
Savage, J.E. , Peterson, R.E. , Aliev, F. , Spit for Science Working Group & Dick, D.M. (2022) Genetic and environmental etiology of drinking motives in college students. Alcoholism: Clinical and Experimental Research, 46, 1783–1796. Available from: 10.1111/acer.14930
The Spit for Science Working Group members are given in Appendix.
REFERENCES
- Abecasis, G.R. , Auton, A. , Brooks, L.D. , Depristo, M.A. , Durbin, R.M. , Handsaker, R.E. et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, Z.W. , Kaiser, A.J. , Lynam, D.R. , Charnigo, R.J. & Milich, R. (2012) Drinking motives as mediators of the impulsivity‐substance use relation: pathways for negative urgency, lack of premeditation, and sensation seeking. Addictive Behaviors, 37, 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal, A. , Dick, D.M. , Bucholz, K.K. , Madden, P.A. , Cooper, M.L. , Sher, K.J. et al. (2008) Drinking expectancies and motives: a genetic study of young adult women. Addiction, 103, 194–204. [DOI] [PubMed] [Google Scholar]
- Bigdeli, T.B. , Neale, B.M. & Neale, M.C. (2014) Statistical properties of single‐marker tests for rare variants. Twin Research and Human Genetics, 17, 143–150. [DOI] [PubMed] [Google Scholar]
- Bresin, K. & Mekawi, Y. (2021) The “Why” of drinking matters: a meta‐analysis of the association between drinking motives and drinking outcomes. Alcoholism, Clinical and Experimental Research, 45, 38–50. [DOI] [PubMed] [Google Scholar]
- Brown, B.C. , Ye, C.J. , Price, A.L. & Zaitlen, N. (2016) Transethnic genetic‐correlation estimates from summary statistics. The American Journal of Human Genetics, 99, 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz, K.K. , Cadoret, R. , Cloninger, C.R. , Dinwiddie, S.H. , Hesselbrock, V.M. , Nurnberger, J.L. et al. (1994) A new, semi‐structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. Journal of Studies on Alcohol, 55, 149–158. [DOI] [PubMed] [Google Scholar]
- Bulik‐Sullivan, B.K. , Loh, P.R. , Finucane, H.K. , Ripke, S. , Yang, J. , Patterson, N. et al. (2015) LD Score regression distinguishes confounding from polygenicity in genome‐wide association studies. Nature Genetics, 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, K.M. & Hasin, D.S. (1998) Reasons for drinking alcohol: relationships with DSM‐IV alcohol diagnoses and alcohol consumption in a community sample. Psychology of Addictive Behaviors, 12, 168–184. [Google Scholar]
- Cho, S.B. , Llaneza, D.C. , Adkins, A.E. , Cooke, M. , Kendler, K.S. , Clark, S.L. et al. (2015) Patterns of substance use across the first year of college and associated risk factors. Frontiers in Psychiatry, 6, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloninger, C.R. , Sigvardsson, S. , Gilligan, S.B. , Von Knorring, A.L. , Reich, T. & Bohman, M. (1988) Genetic heterogeneity and the classification of alcoholism. Advances in Alcohol & Substance Abuse, 7, 3–16. [DOI] [PubMed] [Google Scholar]
- Conrod, P.J. , Stewart, S.H. , Comeau, N. & Maclean, A.M. (2006) Efficacy of cognitive‐behavioral interventions targeting personality risk factors for youth alcohol misuse. Journal of Clinical Child and Adolescent Psychology, 35, 550–563. [DOI] [PubMed] [Google Scholar]
- CONVERGE consortium . (2015) Sparse whole‐genome sequencing identifies two loci for major depressive disorder. Nature, 523, 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, M.L. (1994) Motivations for alcohol use among adolescents: development and validation of a four‐factor model. Psychological Assessment, 6, 117–128. [Google Scholar]
- de Leeuw, C.A. , Mooij, J.M. , Heskes, T. & Posthuma, D. (2015) MAGMA: generalized gene‐set analysis of GWAS data. PLoS Computational Biology, 11, e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick, D.M. , Nasim, A. , Edwards, A.C. , Salvatore, J.E. , Cho, S.B. , Adkins, A. et al. (2014) Spit for Science: launching a longitudinal study of genetic and environmental influences on substance use and emotional health at a large US university. Frontiers in Genetics, 5, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl, V. , Preuss, M. & Neuner, F. (2018) Are drinking motives universal? Characteristics of motive types in alcohol‐dependent men from two diverse populations. Frontiers in Psychiatry, 9, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint, J. & Munafo, M.R. (2007) The endophenotype concept in psychiatric genetics. Psychological Medicine, 37, 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman, I.I. & Gould, T.D. (2003) The endophenotype concept in psychiatry: etymology and strategic intentions. The American Journal of Psychiatry, 160, 636–645. [DOI] [PubMed] [Google Scholar]
- Gray, M.J. , Litz, B.T. , Hsu, J.L. & Lombardo, T.W. (2004) Psychometric properties of the life events checklist. Assessment, 11, 330–341. [DOI] [PubMed] [Google Scholar]
- Grotzinger, A.D. , Rhemtulla, M. , de Vlaming, R. , Ritchie, S.J. , Mallard, T.T. , Hill, W.D. et al. (2019) Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nature Human Behaviour, 3, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, P.A. , Taylor, R. , Thielke, R. , Payne, J. , Gonzalez, N. & Conde, J.G. (2009) Research electronic data capture (REDCap)—A metadata‐driven methodology and workflow process for providing translational research informatics support. Journal of Biomedical Informatics, 42, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, A.B. & Kranzler, H.R. (2015) Alcohol dependence genetics: lessons learned from Genome‐Wide Association Studies (GWAS) and post‐GWAS analyses. Alcoholism, Clinical and Experimental Research, 39, 1312–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawn, S.E. , Bountress, K.E. , Sheerin, C.M. , Dick, D.M. & Amstadter, A.B. (2020) Trauma‐related drinking to cope: a novel approach to the self‐medication model. Psychology of Addictive Behaviors, 34, 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines, L.M. , Ray, L. , Hutchison, K. & Tabakoff, B. (2005) Alcoholism: the dissection for endophenotypes. Dialogues in Clinical Neuroscience, 7, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono, W.G. , Malone, S.M. & Vrieze, S.I. (2017) Endophenotype best practices. International Journal of Psychophysiology, 111, 115–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehayes, I.L. , Mackinnon, S.P. , Sherry, S.B. , Leonard, K.E. & Stewart, S.H. (2021) The influence of drinking buddies: a longitudinal investigation of drinking motivations and drinking behaviors in emerging adults. Substance Use & Misuse, 56, 286–296. [DOI] [PubMed] [Google Scholar]
- Kendler, K.S. , Jacobson, K. , Myers, J.M. & Eaves, L.J. (2008) A genetically informative developmental study of the relationship between conduct disorder and peer deviance in males. Psychological Medicine, 38, 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, R.C. , Berglund, P. , Demler, O. , Jin, R. , Merikangas, K.R. & Walters, E.E. (2005) Lifetime prevalence and age‐of‐onset distributions of dsm‐iv disorders in the national comorbidity survey replication. Archives of General Psychiatry, 62, 593–602. [DOI] [PubMed] [Google Scholar]
- Kranzler, H.R. , Zhou, H. , Kember, R.L. , Vickers Smith, R. , Justice, A.C. , Damrauer, S. et al. (2019) Genome‐wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nature Communications, 10, 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreusch, F. , Vilenne, A. & Quertemont, E. (2013) Assessing the stimulant and sedative effects of alcohol with explicit and implicit measures in a balanced placebo design. Journal of Studies on Alcohol and Drugs, 74, 923–930. [DOI] [PubMed] [Google Scholar]
- Kristjansson, S.D. , Agrawal, A. , Littlefield, A.K. , Pergadia, M.L. , Lessov‐Schlaggar, C.N. , Sartor, C.E. et al. (2011) Drinking motives in female smokers: factor structure, alcohol dependence, and genetic influences. Alcoholism: Clinical and Experimental Research, 35, 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger, R.F. & Markon, K.E. (2006) Reinterpreting comorbidity: a model‐based approach to understanding and classifying psychopathology. Annual Review of Clinical Psychology, 2, 111–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntsche, E. , Knibbe, R. , Gmel, G. & Engels, R. (2005) Why do young people drink? A review of drinking motives. Clinical Psychology Review, 25, 841–861. [DOI] [PubMed] [Google Scholar]
- Kuntsche, E. , Wicki, M. , Windlin, B. , Roberts, C. , Gabhainn, S.N. , van der Sluijs, W. et al. (2015) Drinking motives mediate cultural differences but not gender differences in adolescent alcohol use. The Journal of Adolescent Health, 56, 323–329. [DOI] [PubMed] [Google Scholar]
- Labhart, F. , Kuntsche, E. , Wicki, M. & Gmel, G. (2016) Reciprocal influences of drinking motives on alcohol use and related consequences: a full cross‐lagged panel study among young adult men. Behavioral Medicine, 43, 277–284. [DOI] [PubMed] [Google Scholar]
- Lenzenweger, M.F. (2013) Thinking clearly about the endophenotype–intermediate phenotype–biomarker distinctions in developmental psychopathology research. Development and Psychopathology, 25, 1347–1357. [DOI] [PubMed] [Google Scholar]
- Liberzon, A. , Birger, C. , Thorvaldsdóttir, H. , Ghandi, M. , Mesirov, J.I.L.L.P. & Tamayo, P. (2015) The molecular signatures database hallmark gene set collection. Cell Systems, 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield, A.K. , Agrawal, A. , Ellingson, J.M. , Kristjansson, S. , Madden, P.A.F. , Bucholz, K.K. et al. (2011) Does variance in drinking motives explain the genetic overlap between personality and alcohol use disorder symptoms? A twin study of young women. Alcoholism: Clinical and Experimental Research, 35, 2242–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M. , Jiang, Y. , Wedow, R. , Li, Y. , Brazel, D.M. , Chen, F. et al. (2019) Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nature Genetics, 51, 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynam, D.R. & Miller, J.D. (2004) Personality pathways to impulsive behavior and their relations to deviance: results from three samples. Journal of Quantitative Criminology, 20, 319–341. [Google Scholar]
- Mackie, C.J. , Conrod, P.J. , Rijsdijk, F. & Eley, T.C. (2011) A systematic evaluation and validation of subtypes of adolescent alcohol use motives: genetic and environmental contributions. Alcoholism: Clinical and Experimental Research, 35, 420–430. [DOI] [PubMed] [Google Scholar]
- Marchini, J. , Howie, B. , Myers, S. , Mcvean, G. & Donnelly, P. (2007) A new multipoint method for genome‐wide association studies by imputation of genotypes. Nature Genetics, 39, 906–913. [DOI] [PubMed] [Google Scholar]
- Mezquita, L. , Stewart, S.H. & Ruipérez, M.Á. (2010) Big‐five personality domains predict internal drinking motives in young adults. Personality and Individual Differences, 49, 240–245. [Google Scholar]
- Mezquita, L. , Ibáñez, M.I. , Moya, J. , Villa, H. & Ortet, G. (2014) A longitudinal examination of different etiological pathways to alcohol use and misuse. Alcoholism: Clinical and Experimental Research, 38, 1770–1779. [DOI] [PubMed] [Google Scholar]
- Mezquita, L. , Bravo, A.J. , Ortet, G. , Pilatti, A. , Pearson, M.R. & Ibáñez, M.I. (2018) Cross‐cultural examination of different personality pathways to alcohol use and misuse in emerging adulthood. Drug and Alcohol Dependence, 192, 193–200. [DOI] [PubMed] [Google Scholar]
- Peterson, R.E. , Edwards, A.C. , Bacanu, S.A. , Dick, D.M. , Kendler, K.S. & Webb, B.T. (2017) The utility of empirically assigning ancestry groups in cross‐population genetic studies of addiction. The American Journal on Addictions, 26, 494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, C.A. , Cross, R.J. , Kuhn, J.W. , Horn, J.L. & Kendler, K.S. (2004) Is risk for alcoholism mediated by individual differences in drinking motivations? Alcoholism, Clinical and Experimental Research, 28, 29–39. [DOI] [PubMed] [Google Scholar]
- Pruim, R.J. , Welch, R.P. , Sanna, S. , Teslovich, T.M. , Chines, P.S. , Gliedt, T.P. et al. (2010) LocusZoom: regional visualization of genome‐wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S. , Cherny, S.S. & Sham, P.C. (2003) Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics, 19, 149–150. [DOI] [PubMed] [Google Scholar]
- Rehm, J. , Mathers, C. , Popova, S. , Thavorncharoensap, M. , Teerawattananon, Y. & Patra, J. (2009) Global burden of disease and injury and economic cost attributable to alcohol use and alcohol‐use disorders. The Lancet, 373, 2223–2233. [DOI] [PubMed] [Google Scholar]
- Sanchez‐Roige, S. , Palmer, A.A. , Fontanillas, P. , Elson, S.L. , Adams, M.J. , Howard, D.M. et al. (2019) Genome‐wide association study meta‐analysis of the Alcohol Use Disorders Identification Test (AUDIT) in two population‐based cohorts. American Journal of Psychiatry, 176, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, L. , Lamborn, S.D. , Dornbusch, S.M. & Darling, N. (1992) Impact of parenting practices on adolescent achievement: authoritative parenting, school involvement, and encouragement to succeed. Child Development, 63, 1266–1281. [DOI] [PubMed] [Google Scholar]
- Storey, J.D. , Bass, A.J. , Dabney, A. & Robinson, D. (2015) qvalue: Q‐value estimation for false discovery rate control. R package version 2.2.2 . Available from: http://github.com/jdstorey/qvalue
- Temmen, C.D. & Crockett, L.J. (2018) Adolescent predictors of social and coping drinking motives in early adulthood. Journal of Adolescence, 66, 1–8. [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium . (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein, J. , Cichon, S. , Ridinger, M. , Wodarz, N. , Soyka, M. , Zill, P. et al. (2009) Genome‐wide association study of alcohol dependence. Archives of General Psychiatry, 66, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Damme, J. , Maes, L. , Kuntsche, E. , Crutzen, R. , de Clercq, B. , van Lippevelde, W. et al. (2015) The influence of parental drinking on offspring's drinking motives and drinking: a mediation analysis on 9 year follow‐up data. Drug and Alcohol Dependence, 149, 63–70. [DOI] [PubMed] [Google Scholar]
- Verhulst, B. , Neale, M.C. & Kendler, K.S. (2015) The heritability of alcohol use disorders: a meta‐analysis of twin and adoption studies. Psychological Medicine, 45, 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, R.K. , Polimanti, R. , Johnson, E.C. , Mcclintick, J.N. , Adams, M.J. , Adkins, A.E. et al. (2018) Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nature Neuroscience, 21, 1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanberg, K.W. & Horn, J.L. (1983) Assessment of alcohol use with multidimensional concepts and measures. The American Psychologist, 38, 1055–1069. [DOI] [PubMed] [Google Scholar]
- Watanabe, K. , Taskesen, E. , van Bochoven, A. & Posthuma, D. (2017) FUMA: functional mapping and annotation of genetic associations. bioRxiv. 10.1101/110023 [DOI] [PMC free article] [PubMed]
- Webb, B.T. , Edwards, A.C. , Wolen, A.R. , Salvatore, J.E. , Aliev, F. , Riley, B.P. et al. (2017) Molecular genetic influences on normative and problematic alcohol use in a population‐based sample of college students. Frontiers in Genetics, 8, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werme, J. , van der Sluis, S. , Posthuma, D. & de Leeuw, C.A. (2022) An integrated framework for local genetic correlation analysis. Nature Genetics, 54, 274–282. [DOI] [PubMed] [Google Scholar]
- Whelan, R. , Watts, R. , Orr, C.A. , Althoff, R.R. , Artiges, E. , Banaschewski, T. et al. (2014) Neuropsychosocial profiles of current and future adolescent alcohol misusers. Nature, 512, 185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicki, M. , Kuntsche, E. , Eichenberger, Y. , Aasvee, K. , Bendtsen, P. , Dankulincová Veselská, Z. et al. (2017) Different drinking motives, different adverse consequences? Evidence among adolescents from 10 European countries. Drug and Alcohol Review, 36, 731–741. [DOI] [PubMed] [Google Scholar]
- Windle, R.C. & Windle, M. (2018) Adolescent precursors of young adult drinking motives. Addictive Behaviors, 82, 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Lee, S.H. , Goddard, M.E. & Visscher, P.M. (2011) GCTA: a tool for genome‐wide complex trait analysis. American Journal of Human Genetics, 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young‐Wolff, K.C. , Kendler, K.S. , Sintov, N.D. & Prescott, C.A. (2009) Mood‐related drinking motives mediate the familial association between major depression and alcohol dependence. Alcoholism: Clinical and Experimental Research, 33, 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Sealock, J.M. , Sanchez‐Roige, S. , Clarke, T.‐K. , Levey, D.F. , Cheng, Z. et al. (2020) Genome‐wide meta‐analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nature Neuroscience, 23, 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker, R.A. (2008) Anticipating problem alcohol use developmentally from childhood into middle adulthood: what have we learned? Addiction, 103(Suppl 1), 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S8
Tables S1–S7


