Abstract
Objective
The aim of this study was to test the feasibility of PICO (participants, interventions, comparators, outcomes) entity extraction using weak supervision and natural language processing.
Methodology
We re-purpose more than 127 medical and nonmedical ontologies and expert-generated rules to obtain multiple noisy labels for PICO entities in the evidence-based medicine (EBM)-PICO corpus. These noisy labels are aggregated using simple majority voting and generative modeling to get consensus labels. The resulting probabilistic labels are used as weak signals to train a weakly supervised (WS) discriminative model and observe performance changes. We explore mistakes in the EBM-PICO that could have led to inaccurate evaluation of previous automation methods.
Results
In total, 4081 randomized clinical trials were weakly labeled to train the WS models and compared against full supervision. The models were separately trained for PICO entities and evaluated on the EBM-PICO test set. A WS approach combining ontologies and expert-generated rules outperformed full supervision for the participant entity by 1.71% macro-F1. Error analysis on the EBM-PICO subset revealed 18–23% erroneous token classifications.
Discussion
Automatic PICO entity extraction accelerates the writing of clinical systematic reviews that commonly use PICO information to filter health evidence. However, PICO extends to more entities—PICOS (S—study type and design), PICOC (C—context), and PICOT (T—timeframe) for which labelled datasets are unavailable. In such cases, the ability to use weak supervision overcomes the expensive annotation bottleneck.
Conclusions
We show the feasibility of WS PICO entity extraction using freely available ontologies and heuristics without manually annotated data. Weak supervision has encouraging performance compared to full supervision but requires careful design to outperform it.
Keywords: weak supervision, machine learning, information extraction, evidence-based medicine
Lay Summary
Systematic reviews are clinical summaries, incredibly resource-consuming to produce and involve redundant document filtering processes that machine learning could automate. Manual PICO (participant, intervention, comparator, outcome) information analysis aids document filtering but is one of the most resource-intensive stages for writing systematic reviews. Supervised machine learning based PICO information extraction could accelerate document filtering but requires massive hand-labelled datasets for training. We propose a weak supervision approach that uses more than 127 freely available vocabularies and expert-designed rules to label 4081 documents in evidence-based medicine (EBM)-PICO dataset with PICO information. Powerful pretrained transformer models were fine-tuned for PICO extraction using these programmatically labelled documents and compared to the results using expensive hand-labelled EBM-PICO documents. The token-level macro-F1 score was used to compare full supervision and weak supervision. We also examined the errors in the EBM-PICO training data and rectified them in the EBM-PICO test set. The weak supervision approach had promising results overall and had a better F1 score than full supervision for the participant information, albeit it required a careful design. We further rectify the errors made by weak supervision and intend to improve the methodology. Finally, adopting weak supervision for highly compositional PICO information is challenging but feasible and extensible to more clinical entities.
INTRODUCTION
Systematic reviews (SR) are an evidence-based practice of answering clinical questions using a transparent and quantitative approach. The reviewers must collect as many candidate publications as possible, identify the relevant publications, and integrate their results via statistical meta-analysis. A clinical SR question is typically formulated using the PICO (participants, interventions, comparators, outcomes) framework, for example, “Will aerobic exercise (Intervention) improve fatigue (Outcome) in cancer patients (Participant) compared to usual care (Comparator)?.” A publication is only relevant for answering a question if it studies the selected participants, interventions (and their comparators), and outcomes.1 Manually analyzing PICO information from thousands of publications for a single SR often takes 2–8 months of 2 medical experts’ time.2 It can be automated using machine learning (ML) by directly pointing the human reviewers to the PICO descriptions, facilitating quick decision-making for the study’s relevance.
Supervised ML requires hand-labeled data, but hand-labelling data with PICO information require people with combined medical and informatics skills, which is expensive and time-consuming in terms of intensive annotator training and the actual annotation process. Labelling PICO information is tricky because of the high disagreement between human annotators on the exact spans constituting PICO, leading to human errors in hand-labeled corpora.3 Some studies examine the errors in the publicly available evidence-based medicine (EBM)-PICO benchmark.4–6 More importantly, depending upon the SR question, PICO criteria are extended to PICOS (S—study design), PICOC (C—context), PIBOSO (B—background, O—other), and PICOT (T—timeframe).1,7,8 Hand-labeled datasets are static and prohibit quick manual re-labelling in case of human errors or when a downstream task requires new entities. This annotation bottleneck has pivoted attention toward weakly supervised (WS) learning that relies on programmatic labelling sources to obtain training data. Programmatic labelling is quick and allows efficient modifications to the training data labels per the downstream application changes.
WS learning has demonstrated strengths for clinical document classification and relation extraction, but clinical entity extraction tasks have heavily relied on fully supervised (FS) approaches.9–14 Despite the availability of Unified Medical Language System (UMLS), a large compendium of medical ontologies, which can be re-purposed for weak entity labelling, it has not been extensively applied to clinical entity labelling.15 Several legacy clinical applications are also supported by rule-based if-else systems relying on keyword cues that aid weak labelling.16–18 With so many weak labelling sources available, the challenge for weak supervision is efficiently aggregating these sources of varying accuracy. Compare this to crowdsourcing, where an important task is to model the worker's accuracy without the ground truth.19 Though crowdsourcing requires annotator training and quality control, programmatic labelling does not.20
Data programming is a domain-agnostic generative modeling approach combining multiple weak labelling sources and estimating their accuracies. The effectiveness of data programming for biomedical entity recognition has been explored by Fries et al21 in their Trove system. However, Trove only explores well-defined entities like chemical, disease, disorder, and drug. PICO categories are highly compositional spans by definition, fuzzier in comparison and much more intricate in that they can be divided into subclasses. A shortcoming of span extraction is that even after a machine points a human reviewer to the correct PICO span, the reviewer requires to manually read and understand its finer aspects to screen the study for relevance. Span extraction hence leads to semiautomation but hinders full-automation. The entity recognition approach to PICO is not as easy as the entity recognition approach to disease or chemical names, which are more or less standardized. PICO terms are not standard, and even the experts disagree on the exact tokens constituting them.3 WS PICO entity recognition has not garnered as much attention as supervised span recognition. As far as our knowledge goes, only 2 studies exist for WS PICO recognition. One of these approaches only explores distant supervision for intervention extraction using a single labelling source.22 The other approach studies weak supervision for PICO span extraction but still utilizes some supervised annotation signals about whether a sentence includes PICO information.23
The challenges to developing weak supervision approaches to PICO entity recognition are first defining the subclasses within PICO spans and then mapping several available ontologies and terminologies to these. The next challenge is developing WS classifiers by optimally combining ontologies and evaluating their performance compared to full supervision. Another challenge is developing higher-cost expert-generated rules corresponding to these subclasses to aid ontology classifiers and evaluate their combined performance. We also identified limitations in the currently available EBM-PICO training dataset and corrected them in the EBM-PICO test set for reliable evaluation of the WS approaches. Our work demonstrates the feasibility of using weak supervision for PICO entity extraction using the EBM-PICO benchmark and shows how weak supervision overtakes full supervision in certain instances.
METHODOLOGY
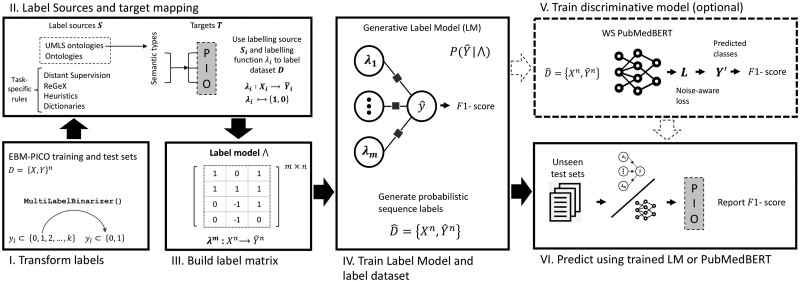
The birds-eye view of our approach is shown in Figure 1.
Figure 1.
Weak PICO entity extraction approach. (I.) Multi-class labels in the EBM-PICO benchmark are binarized. (II.) Low-cost UMLS vocabularies are re-purposed as labelling sources and experts design rules as high-cost labelling sources. (III.) Labelling functions map the training sequences to class labels using labelling sources resulting in an label matrix. (IV. and V.) The label matrix is used to train a generative model that outputs probabilistic labels that a downstream transformer model can use for entity recognition.
Datasets
EBM-PICO is a widely used dataset with multi-level PICO annotations: span-level or coarse-grained and entity-level or fine-grained (refer to Table 1). Span-level annotations encompass the maximum information about each class. Entity-level annotations cover the more fine-grained information at the entity level, with PICO classes further divided into semantic subclasses. The dataset comes predivided into a training set (n = 4933) annotated through crowd-sourcing and an expert annotated gold test set (n = 191) for evaluation.4 The EBM-PICO annotation guidelines caution about variable annotation quality (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6174533/bin/NIHMS988059-supplement-Appendix.pdf). Abaho et al5 developed a framework to post hoc correct EBM-PICO outcomes annotation inconsistencies. Lee and Sun6 studied annotation span disagreements suggesting variability across the annotators. Low annotation quality in the training dataset is excusable, but the errors in the test set can lead to faulty evaluation of the downstream ML methods. We evaluate ∼1% of the EBM-PICO training set tokens to gauge the possible reasons for the fine-grained labelling errors and use this exercise to conduct an error-focused PICO re-annotation for the EBM-PICO gold test set. The dataset is pretokenized and did not require additional preprocessing except the addition of POS tags and token lemma using spaCy (https://spacy.io/). Multi-class fine-grained PICO annotations were binarized, that is, a token label was reset to 1 if the token represented a fine-grained entity.
Table 1.
P (participant), I (intervention), and O (outcome) represent the coarse-grained labels that are further divided into respective fine-grained labels
| P | I/C | O | |
|---|---|---|---|
| 0 | No label | No label | No label |
| 1 | Age | Surgical | Physical |
| 2 | Sex | Physical | Pain |
| 3 | Sample size | Drug | Mortality |
| 4 | Condition | Educational | Side effect |
| 5 | Psychological | Mental | |
| 6 | Other | Other | |
| 7 | Control |
Note: The table is taken from Nye et al4
Binary token labelling
Automatic PICO entity labelling is a classical binary token labelling problem whereby a function maps an input sequence of n text tokens, to output sequence , where is the label for token xi. In weak supervision, is latent and should be estimated by aggregating several weak labelers of variable accuracy. The estimates of are assigned as probabilistic token labels of leading to a weakly labelled dataset that can be used to train discriminative models.
Labelling functions
In a binary token labelling task, a labelling function (LF) is a weak classifier λ that uses domain-specific labelling sources and a logic to emit token labels with labels for a subset of input tokens. An LF designed for a particular target class (here; {participant, intervention, outcome}) should output 1 for the positive token label, 0 for the negative token label, and abstain (−1) on the tokens where it is uncertain . We designed 3 LF types depending on the types of labelling sources. (1) The ontology or dictionary LFs for a target class take a dictionary of terminologies mapped to one of y token labels. Any LF using ontologies or dictionaries used string matching as the labelling heuristic. Relevant bigram word co-occurrences were used to account for fuzzy span matching from the terminologies. A bigram was considered relevant for a vocabulary if it occurred ≤25 times in that vocabulary. (2) A regular expression (ReGeX) LF for a target class takes regex patterns for labels and abstains from the rest. (3) A heuristic LF is personalized for each target class and takes a generic regex pattern and specific part-of-speech (POS) tag signals. Abbreviations in clinical studies are considered using a heuristics abbreviation identifier, and the identified abbreviations were mapped to their respective target classes. Stopwords from Natural Language Toolkit (NLTK) (https://www.nltk.org), spaCy, Gensim (https://radimrehurek.com/gensim/), and scikit learn (https://scikit-learn.org) were used to initialize negative token label templates.
Labelling sources
This section describes the labelling sources used and their mapping to the PICO targets . We used the 2021AB-full release of the UMLS Metathesaurus English subset with 223 vocabularies. After removing non-English and zoonotic vocabularies and the vocabularies containing fewer than 500 terms, we remained with 127 vocabularies.15 Terms in the selected vocabularies were preprocessed by removing stopwords, numbers, and punctuation. Additional vocabularies included disease ontology; human phenotype ontology; ontology of adverse events; chemical entities of biological interest; comparative toxicogenomics database chemical and disease subclasses; clinical trials ontology; gender, sex, and sexual orientation ontology; chemotherapy toxicities ontology; cancer care: treatment outcomes ontology; symptoms ontology; nonpharmacological interventions ontology; and nursing care coordination ontology.24–34 ReGeX and heuristics like POS tag cues were used to capture recurring class-specific patterns otherwise not captured by standardized terminologies. Vocabularies are structured, standardized data sources that do not capture writing variations from clinical literature and custom-built ReGeX are restricted by either task or entity type.35,36 We used distant supervision dictionaries created from the structured fields of clinicaltrials.gov (CTO) as described by Dhrangadhariya and Müller22 Principal investigators of the clinical study manually enter data in CTO, thereby incorporating large-scale writing variations.37
Sources to targets
Along with the source and the logic to map to token labels, an LF needs information about which target label and binary token class to map the source. We report how the LF sources were mapped to PICO targets in this section. UMLS 2021AB-full release contains 16 543 671 concept names, making direct concept to PICO target mapping impractical. These concepts are organized under semantic type categories (eg, disease, sign and symptoms, and age group) (https://www.nlm.nih.gov/research/umls/META3_current_semantic_types.html), which allows mapping these semantic categories to PICO targets, invariably mapping the concepts from the vocabularies to target classes.37 It is a challenging expert-led activity, though decomposing PICO into subclasses greatly helps map sources to target. A semantic category was marked 1 to represent a positive token label for that target class or 0 to represent a negative token label for that target class. Non-UMLS vocabularies were obtained from NCBO bioportal (https://bioportal.bioontology.org/) and were chosen to be PICO target specific and assigned to a single label. Target-specific distant supervision dictionaries were created from the structured fields of clinicaltrials.gov (CTO). The structured field “condition or disease” was mapped to the participant target, and the “intervention/treatment” field was mapped to the intervention target. The semistructured “primary outcome measures” and “secondary outcome measures” fields were mapped to the outcome target. The hand-coded dictionaries were designed using the official websites listing patient-reported outcome (PROMs) questionnaires (https://www.thoracic.org/members/assemblies/assemblies/bshsr/patient-outcome/) and PROMs (https://www.safetyandquality.gov.au/our-work/indicators-measurement-and-reporting/patient-reported-outcomes/proms-lists). Other hand-crafted dictionaries were separately designed for participant gender and sexuality, intervention comparator terms.
LF aggregation
Depending upon the number of sources for each , we had several LFs. Each LF maps a subset of inputs to output sequence with labels yielding a label matrix . The weakly generated labels might have conflicts and overlaps and are generally noisy. The LFs can be ensembled using the majority vote (MV) rule, where a token label is elected only when a majority of vote for it. Ties and abstains lead to the selection of the majority label.
| (1) |
However, MV considers each LF as conditionally independent and does not consider the variable accuracies of different labelling sources weighing them equally. Snorkel implements data programming paradigm into the label model (LM) that re-weights and aggregates LFs into probabilistic labels . To do this, the LM trains a generative model to estimate LF accuracies using stochastic gradient descent to minimize log loss in the absence of labelled data.35,38 Although the ground truth is not observable to estimate accuracies; they can be estimated using observed agreement and disagreement rates between LF pairs , in . Generative modeling ultimately results into token label probabilities for label classes . GridSearch was used to fine-tune the parameters of the LM using the hand-labeled validation set from the EBM-PICO. The parameters are listed in the Experimental details section of the Supplementary Material. Once we have the pseudo-labels generated by majority voting or the LM, these could be used to train a discriminative model.
| (2) |
Experiments
The LFs were used to label the EBM-PICO training set and obtain . We tested MV and LM to aggregate LFs. LM output probabilistic labels for the training set were used as weak supervision signals to train downstream PubMedBERT to minimize noise-aware cross-entropy loss. PubMedBERT was trained on PubMed literature and was chosen because of its domain similarity to our training data (PubMed abstracts) and task.39 It was tuned on fixed parameters listed in the experimental details section in the Supplementary Material.
| (3) |
UMLS ontologies are readily available sources of weak supervision, while searching the non-UMLS ontologies requires an additional effort and understanding of the target class and domain. On the contrary, designing the rules requires understanding the idiosyncratic clinical patterns for the target classes. Therefore, we experiment and report results on 3 “expense” tiers to gauge the performance changes: (1) UMLS labelling sources, (2) UMLS and non-UMLS labelling sources, and (3) UMLS, non-UMLS and expert-generated rules. We test label aggregation via MV and LM along with WS PubMedBERT for the above tiers. The WS experiments were compared against a competitive FS PubMedBERT trained using the hand-labeled EBM-PICO training set. For all the experiments, 80% of the EBM-PICO dataset was used for training and 20% for validation.
UMLS ontologies were ranked based on the total number of n-gram overlaps between the respective terminology and the EBM-PICO validation set. These were then partitioned into 127 partitions , where the first partition combined the entire UMLS into a single LF and was used as the baseline. The last partition kept all the terminologies as separate LFs.
Partition-wise performance over the validation set was tracked.
Evaluation
We report the classical macro-averaged F1 and recall for MV, LM, and WS transformer models and the FS PubMedBERT models. Token-level macro-F1 was chosen to make it comparable to the PICO extraction literature. Mean macro-averaged scores are reported over 3 runs of each model, with the top 3 random seeds (0, 1, and 42) used in Python. The models were separately trained for each target class recognition task using the raw (inside, out) tagging scheme. We used Student’s t-test with an alpha α threshold of 0.05 to measure the statistical significance.
RESULTS
PICO decomposition
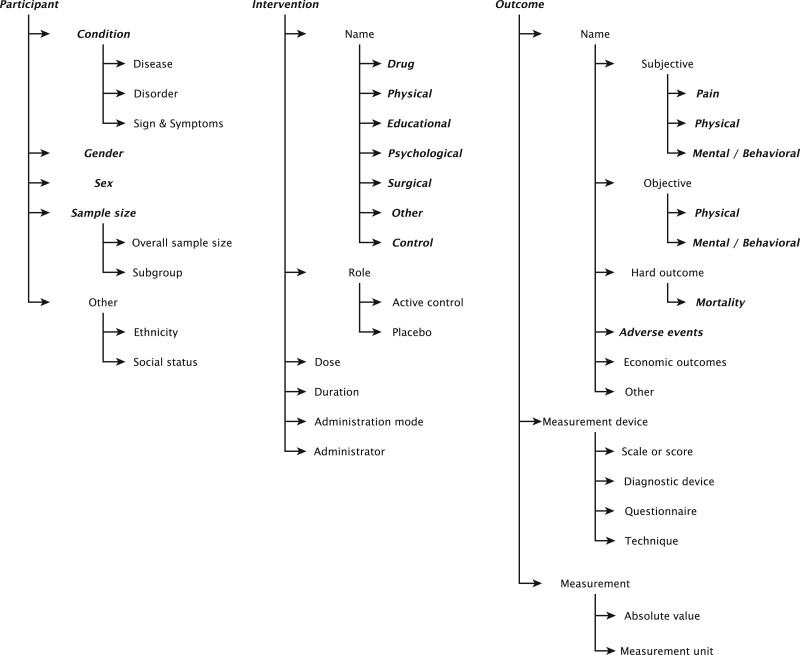
We extended the EBM-PICO subclasses (refer to Table 1) to better query the labelling sources and design LFs (see Figure 2). For a more comprehensive subgrouping, we propose developing a PICO ontology.40 It is more straightforward to search for ontologies representing adverse events or diseases rather than fending for an ontology describing the entire participant or outcome spans. It is easier to grasp cues separately for outcome terms and instruments of outcome measurement to develop heuristics. The intervention span can include the intervention name, role (primary intervention or comparator), dosage, frequency, mode of administration, and administrator. The outcome span can include the outcome names, the scales, techniques or instruments used to measure them, and the absolute outcome measurement values. The EBM-natural language processing guidelines restrict annotating the outcome name, how it was measured, and the intervention’s name and role (control, placebo), leaving out the other subclasses.
Figure 2.
Hierarchical representation of PICO subclasses. The categories marked in bold–italic are the same as the fine-grained categories in the EBM-PICO corpus.
Error rectification
We rectified the errors in the EBM-PICO validation set and categorized them for each PIO class, as shown in Table 2. Of 12 960 (∼1% of 1 303 169) training tokens evaluated to gauge the errors, 18.30% of the intervention class tokens, 23.39% of the participant class tokens, and 20.21% of the outcome class tokens were errors. These error categories are elaborated in the Supplementary Material. The error analysis was used to correct fine-grained annotation errors in the EBM-PICO test set, and both the EBM-PICO and its updated version were used for evaluation. We were constrained with obtaining multiple annotators for the re-annotation to calculate inter-annotator agreement. Therefore, we calculated Cohen's between the original EBM-PICO gold set and our re-annotation over 200 documents and compared it to Cohen's (see Figure 3) provided by the authors of the original corpus.4
Table 2.
Error distribution and error categories in the analyzed tokens (∼1%) of the EBM-PICO corpus
| Error category | Participant | Intervention | Outcome |
|---|---|---|---|
| Repeat mention unmarked | 213 | 227 | 207 |
| Remain un-annotated | 47 | 59 | 71 |
| Inconsistency | 46 | 18 | 85 |
| Punctuation/article | 15 | 23 | 48 |
| Conjunction connector | 30 | 36 | 57 |
| Junk | 53 | 79 | 30 |
| Extra information | 80 | 146 | 58 |
| Generic mention | 70 | 120 | 85 |
| Total errors | 554 | 708 | 641 |
Figure 3.
Cohen's between the expert annotated EBM-PICO gold test set and EBM-PICO compared to the Cohen's for EBM-PICO gold test set annotations.
Table 3 reports macro-averaged F1 for the experiments detailed in the experiments section compared to the FS approach. Error rectification leads to an overall average F1 improvement of 4.88% across the experiments using a weakly labeled training set with the highest average improvement of 8.25% (7.15–9.52%) for participants and 2.68% (−0.11% to 4.28%) for outcomes. For the participant class, both the LM and the WS F1 scores increase the full supervision score by 0.90–1.71%. It has to be noticed that weak supervision outperforms full supervision on the rectified benchmark only for the participant entity.
Table 3.
Macro-averaged F1 scores for UMLS, UMLS + other, and rule-based weak supervision
| Target | LF source | #LF | MV |
LM |
WS |
FS |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fine | Corr | Fine | Corr | Fine | Corr | Fine | Corr | |||
| P | UMLS | 3–4 | 62.13 | 69.28 | 64.28 | 72.22 | 65.32 | 73.49 | 72.99 | 74.41 |
| +Ontology | 4 | 61.72 | 69.32 | 64.23 | 72.18 | 64.76 | 72.31 | |||
| +Rules | 19–119 | 63.08 | 72.06 | 65.79 | 75.31 | 66.73 | 76.12 | |||
| I/C | UMLS | 8–95 | 59.7 | 63.94 | 60.11 | 64.28 | 59.17 | 61.72 | 83.37 | 81.06 |
| +Ontology | 5–101 | 62.14 | 66.92 | 62.83 | 67.09 | 67.06 | 69.76 | |||
| +Ru1es | 4–35 | 58.51 | 63.45 | 64.34 | 68.17 | 70.27 | 72.39 | |||
| O | UMLS | 5–6 | 55.79 | 59.85 | 58.76 | 62.36 | 60.83 | 63.55 | 81.2 | 80.53 |
| +Ontology | 4–5 | 56.006 | 59.64 | 59.27 | 62.34 | 59.55 | 60.46 | |||
| +Rules | 3–5 | 55.08 | 59.36 | 60.9 | 62.87 | 60.5 | 60.39 | |||
Note: Underlined values show the best score without manually labelled training data. Bold values show the best overall F1 score in any category. Fine: EBM-PICO fine-grained annotations; Corr: EBM-PICO fine-grained annotations (EBM-PICO updated). I/C: intervention/comparator; O: outcome; P: participant.
MV versus LM versus WS
The LM improved the average performance by 2.74% (0.17–5.83%) compared to majority voting. However, PubMedBERT did not guarantee improved performance across the targets leading to performance drops between 0.4 and 2.56%. Though the WS PubMedBERT models did not always improve the performance compared to their LM counterparts, they had the best F1 score for each target class. The majority voting had higher recall across experiments compared to precision, while LM focused on precision (see Figure 4), making it a possible choice for recall-oriented PICO extraction tasks.
Figure 4.
Precision and recall across the experiments for the (A) majority vote models (left) and (B) label models (right).
LF tiers
Adding non-UMLS LFs to the UMLS tier increases performance for the intervention target by an average of 4.48% but leads to performance drops for the participants and outcomes targets by 0.36% and 0.64%, respectively. Adding task-specific LFs increased the overall F1 by a negligible 0.98%. Heuristics improved performance for the interventions LM by 11.1%.
UMLS partitions
To investigate the optimal number of UMLS LFs required, we used the same methodology as in Trove, holding all non-UMLS and heuristics LFs fixed across all ablation tiers and computed performance across partitions of the UMLS terminology. We noticed an increased performance for the first few partitions. However, we did not see the performance drop with a further increase in the participant and intervention target partition number. Partitions with more than 100 LFs performed better. This situation contrasts with Trove, where an increase in partitions leads to a drop in performance across targets (see Figure 5). For the outcomes target, an increase in the number of partitions leads to an increased performance initially but a drop with a further increase in the partition numbers. LM outperforms MV on training performance across the 2 targets and experiments except for the intervention target, where the MV model combining UMLS and additional ontologies outperforms LM. The simple baseline collapsing UMLS into a single LF usually did not perform better than the others in UMLS partitions for any of the 3 experiment tiers (refer to the #LF columns in Table 3).
Figure 5.
The relationship between the number of UMLS partitions and the macro-averaged F1 score for (A) participants target, (B) outcomes target, and (C) interventions target.
DISCUSSION
Our study results show the promising performance of weak supervision compared to full supervision, surpassing it for participant extraction. It has to be noticed that weak supervision requires careful LF design consideration to surpass full supervision, primarily due to the compositional nature of PICO classes. In another study, we use this weak supervision approach to successfully extend PICO to PICOS extraction (S—study type) without needing additional annotated “study type” data to quickly power applications.41
Although it is easy to re-purpose the vocabularies for labelling, it is challenging to map them to the correct PICO targets. A decreased or stagnant F1 after adding non-UMLS LFs to the UMLS tiers indicates this. Task-specific ReGeX and heuristics were developed upon inspection of the most frequent terms in the EBM-PICO validation set. The same procedure was followed across the targets. However, the performance boost using rule-based LFs was only observed in the participants and interventions and was detrimental to the outcomes.
Even though LM improves performance compared to MV, MV has a higher recall across experiments indicating a good corpus coverage of the LFs (refer to Figure 4). While some studies press on PICO extraction being a recall-oriented task, this is debated in practice. In practice, high recall might lead to an accompanying high false-positive (FP) rate, leading to the reviewers spending more time to manually weed out FP noise than reading and annotating the abstract with the entities.23
LM only considers the information encoded in the weak sources to label phrases from the training text but does not consider the contextual information around the phrases. Transformers consider the contextual information and should generalize beyond the LMs in theory. It is empirically confirmed by the performance boost that PubMedBERT brings this on top of the LM for some instances, but the WS outcomes extraction results refute it.
Error analysis
We conducted an error analysis on 18 (n = 5291 tokens) out of 200 EBM-PICO gold test set documents to contextualize the weak supervision models. Table 4 shows token-level errors divided into either of the 4 classes: (1) false negative (FN)—if the entire entity that the token was part of was missed out by the LM, (2) FP—if the entire entity that the token was part of was falsely recognized as an entity, (3) boundary error (BE)—if the boundary tokens were missed out, but otherwise the entity was identified, and (4) overlapping error—if LM made an error in the nonperipheral tokens of an otherwise identified entity mention. Nonperipheral tokens are all tokens except the first and last of the multi-token entity.
Table 4.
Distribution of the token-level errors made by the best label models on EBM-PICO gold
| FP | FN | BE | OE | |
|---|---|---|---|---|
| Participants | 160 | 76 | 80 | 10 |
| Interventions | 308 | 119 | 60 | 0 |
| Outcomes | 233 | 306 | 139 | 7 |
In future, we aim to reduce FNs and dig deeper into this category. Besides participant disease, tokens representing participant sample size, age group, gender, and symptoms subclasses went unrecognized. The LM labeled these FNs with low confidence, meaning the LFs did encode this information, but the signals were not strong enough for correct classification. Such FNs could be mitigated by weighting LFs for these subclasses. Considerable standard and unusual abbreviation terms were missed out, especially the ones encompassed by brackets, for example, metabolic syndrome (MetS), testicular cancer + testicular self-examination, and left ventricular hypertrophy (LVH). The model did not pick some of the standard abbreviations, for example, LVH and MetS, due to a faulty mapping of these abbreviations to the incorrect PICO target.
A similar pattern was observed for the intervention (eg, inference-based approach and radiotherapy) and outcomes class abbreviations too. The mismapping is now amended. LM did not capture the abbreviations enclosed in a bracket (eg, “(COPD)”) as the LFs were not designed to tag these brackets.
Intervention LMs did not recognize common drug names, for example, Fenofibrate and CP-529414.
In addition, many nonstandardized treatment names went unrecognized, for example, substance abuse prevention program, inference-based approach, high-concentration contrast agents, and epigastric impedance. Such terms are absent from UMLS and non-UMLS vocabularies leading to FNs, so the LFs do not encode them. Similarly, intervention BEs were the nonidiosyncratic tokens partially misrecognized because the vocabulary did not encode this partial information.
For example, the term “internal stenting” is partially recognized because “stenting” is a UMLS concept but not “internal.” Similarly, in the term “endopyelotomy stent placement,” only the UMLS concept token “stent” was identified. Participant BE FNs were usually the extra information that described more about the participant's disease, for example, the information about disease staging went unrecognized in the participant's disease entity (in “advanced carcinoma,” the word “advanced” was a BE FN). Such entities not encoded by the LFs contribute to the FNs and could be mitigated by adding relevant vocabulary and rules.42 It is straightforward to add vocabulary but challenging to map a semantic group or a vocabulary to PICO categories, especially for the outcomes class. Our current source-to-target mapping approach is manual and based on subjective expert judgment, which an objective algorithm can improve. This mapping could have led to several unexplainable errors, especially for the outcomes and, to a less extent, for the intervention class. In addition, it took time to identify semantic categories and UMLS vocabularies corresponding to the study outcomes pointing toward the gap in developing one.
CONCLUSION
We adapted weak supervision for PICO spans and developed models for predicting PICO entities without a hand-labeled corpus. We also identified errors pertinent to the current PICO benchmark, updated the dataset, and used both datasets to evaluate the recognition models. The approach achieves promising performance compared to full supervision and warrants further research into weak supervision for compositional entities like PICO. The approach can be extended to more clinical SR entities without a manually labeled corpus, thereby being a starting point to overcome the annotation bottleneck. In the future, we will work on extending the data programming approach to inspect strategies for objectively mapping ontologies to PICO subclasses and experiment using external models like MetaMap as LFs.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr Jason A. Fries for his valuable comments and insights on the labelling function design and LM execution phase of the study. The authors thank in advance the anonymous reviewers for their time and feedback.
CONFLICT OF INTEREST STATEMENT
None declared.
Contributor Information
Anjani Dhrangadhariya, Institute of Informatics, University of Applied Sciences Western Switzerland (HES-SO), Sierre, Switzerland; University of Geneva (UNIGE), Geneva, Switzerland.
Henning Müller, Institute of Informatics, University of Applied Sciences Western Switzerland (HES-SO), Sierre, Switzerland; University of Geneva (UNIGE), Geneva, Switzerland.
FUNDING
HES-SO Valais-Wallis, Sierre, Switzerland, funded this work.
AUTHOR CONTRIBUTIONS
AD developed and adapted the work plan for WS PICO information extraction from experimental design to the python implementations of LFs, LM development, and training the WS transformers to data analysis. HM oversaw the work. AD drafted the article, and HM reviewed and provided input on preliminary versions. AD and HM approved the final version of the paper to be published. AD agrees to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
ETHICS STATEMENT
Our study focuses on the applicability of weak supervision for information extraction using publicly available datasets. Our investigation neither introduces any social or ethical concerns. We do not foresee any direct social consequences or ethical issues.
SUPPLEMENTARY MATERIAL
Supplementary material is available at JAMIA Open online.
DATA AVAILABILITY
The code is available on GitHub (https://github.com/anjani-dhrangadhariya/distant-PICO) and will be made public upon publication. The EBM-PICO error analysis file for the validation set, the error-rectified EBM-PICO gold test set, and other resources like distant supervision CTO dictionaries and hand-crafted dictionaries are available on DRYAD (https://doi.org/10.5061/dryad.ncjsxkszr). More information can be found in the supplementary file.
REFERENCES
- 1. Uman LS. Systematic reviews and meta-analyses. J Can Acad Child Adolesc Psychiatry 2011; 20 (1): 57–9. [PMC free article] [PubMed] [Google Scholar]
- 2. Borah R, Brown AW, Capers PL, Kaiser KA.. Analysis of the time and workers needed to conduct systematic reviews of medical interventions using data from the PROSPERO registry. BMJ Open 2017; 7 (2): e012545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brockmeier AJ, Ju M, Przybyła P, Ananiadou S.. Improving reference prioritisation with PICO recognition. BMC Medical Inform Decis Mak 2019; 19 (1): 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nye B, Li JJ, Patel R, et al. A corpus with multi-level annotations of patients, interventions and outcomes to support language processing for medical literature. In: proceedings of the conference. Association for Computational Linguistics (ACL) Meeting, Vol. 2018, Melbourne, Australia; July 2018: 197. NIH Public Access. [PMC free article] [PubMed]
- 5. Abaho MBD, Williamson P, Dodd S. Correcting crowdsourced annotations to improve detection of outcome types in evidence based medicine. In: CEUR workshop proceedings, Vol. 2429; January 1, 2019: 1–5.
- 6. Lee GE, Sun A. A study on agreement in PICO span annotations. In: proceedings of the 42nd international ACM SIGIR conference on research and development in information retrieval, Paris, France; ACM; July 18, 2019: 1149–52.
- 7. Riva JJ, Malik KM, Burnie SJ, Endicott AR, Busse JW.. What is your research question? An introduction to the PICOT format for clinicians. J Can Chiropr Assoc 2012; 56 (3): 167. [PMC free article] [PubMed] [Google Scholar]
- 8. Methley AM, Campbell S, Chew-Graham C, McNally R, Cheraghi-Sohi S.. PICO, PICOS and SPIDER: a comparison study of specificity and sensitivity in three search tools for qualitative systematic reviews. BMC Health Serv Res 2014; 14 (1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meng Y, Shen J, Zhang C, Han J. Weakly-supervised neural text classification. In: proceedings of the 27th ACM international conference on information and knowledge management, Torino, Italy; ACM; October 17, 2018:983–92.
- 10. Wang Y, Sohn S, Liu S, et al. A clinical text classification paradigm using weak supervision and deep representation. BMC Med Inform Decis Mak 2019; 19 (1): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mintz M, Bills S, Snow R, Jurafsky D. Distant supervision for relation extraction without labeled data. In: proceedings of the joint conference of the 47th annual meeting of the ACL and the 4th international joint conference on natural language processing of the AFNLP, Suntec, Singapore; August 2009:1003–11.
- 12. Elangovan A, Davis M, Verspoor K, Assigning function to protein-protein interactions: a weakly supervised BioBERT based approach using PubMed abstracts. arXiv:2008.08727, 2020.
- 13. Weber L, Thobe K, Migueles Lozano OA, Wolf J, Leser U.. PEDL: extracting protein--protein associations using deep language models and distant supervision. Bioinformatics 2020; 36 (Suppl_1): i490–i498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mallory EK, de Rochemonteix M, Ratner A, et al. Extracting chemical reactions from text using Snorkel. BMC Bioinform 2020; 21 (1): 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Humphreys BL, Lindberg DA, Schoolman HM, Barnett GO.. The unified medical language system: an informatics research collaboration. J Am Med Inform Assoc 1998; 5 (1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Friedlin FJ, McDonald CJ.. A software tool for removing patient identifying information from clinical documents. J Am Med Inform Assoc 2008; 15 (5): 601–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim YS, Yoon D, Byun J, et al. Extracting information from free-text electronic patient records to identify practice-based evidence of the performance of coronary stents. PLoS One 2017; 12 (8): e0182889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang H, Garibaldi JM.. Automatic detection of protected health information from clinic narratives. J Biomed Inform 2015; 58 (Suppl): S30–S38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Imamura H, Sato I, Sugiyama M. Analysis of minimax error rate for crowdsourcing and its application to worker clustering model. In: proceedings of 35th international conference on machine learning (ICML), Stockholm, Sweden; July 3, 2018:2147–56.
- 20. Foncubierta Rodríguez A, Müller H. Ground truth generation in medical imaging: a crowdsourcing-based iterative approach. In: proceedings of the ACM multimedia 2012 workshop on crowdsourcing for multimedia, New York, United States; October 29, 2012:9–14.
- 21. Fries JA, Steinberg E, Khattar S, et al. Ontology-driven weak supervision for clinical entity classification in electronic health records. Nat Commun 2021; 12 (1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dhrangadhariya A, Müller H. DISTANT-CTO: a zero cost, distantly supervised approach to improve low-resource entity extraction using clinical trials literature. In: proceedings of the 21st workshop on biomedical language processing, Dublin, Ireland; Association for Computational Linguistics; May 2022:345–58.
- 23. Liu S, Sun Y, Li B, Wang W, Bourgeois FT, Dunn AG. Sent2Span: span detection for PICO extraction in the biomedical text without span annotations. In: findings of the association for computational linguistics: EMNLP 2021, Punta Cana, Dominican Republic; Association for Computational Linguistics; 2021:1705–15.
- 24. Schriml LM, Arze C, Nadendla S, et al. Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res 2012; 40 (Database issue): D940–D946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S.. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet 2008; 83 (5): 610–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He Y, Sarntivijai S, Lin Y, et al. OAE: the ontology of adverse events. J Biomed Sem 2014; 5 (1): 29–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Matos P, Alcántara R, Dekker A, et al. Chemical entities of biological interest: an update. Nucleic Acids Res 2010; 38 (Database issue): D249–D254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin AY, Gebel S, Li QL, et al. CTO: a community-based clinical trial ontology and its applications in PubChemRDF and SCAIView. In: CEUR workshop proceedings, Vol. 2807; 2020. NIH Public Access. [PMC free article] [PubMed]
- 29. Kronk CA, Dexheimer JW.. Development of the gender, sex, and sexual orientation ontology: evaluation and workflow. J Am Med Inform Assoc 2020; 27 (7): 1110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geifman N, Rubin E.. Towards an age-phenome knowledge-base. BMC Bioinform 2011; 12 (1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rogier A, Coulet A, Rance B. Using an ontological representation of chemotherapy toxicities for guiding information extraction and integration from EHRs. In: Medinfo 2021—18th World Congress on Medical and Health Informatics, Vol. 290, Virtual; IOS Press; 2021:. 91. [DOI] [PubMed]
- 32. Lin FP, Groza T, Kocbek S, Antezana E, Epstein RJ.. Cancer care treatment outcome ontology: a novel computable ontology for profiling treatment outcomes in patients with solid tumors. JCO Clin Cancer Inform 2018; 2: 1–14. [DOI] [PubMed] [Google Scholar]
- 33. Mohammed O, Benlamri R, Fong S. Building a diseases symptoms ontology for medical diagnosis: an integrative approach. In: the first international conference on future generation communication technologies, London, United Kingdom; IEEE; December 12–14, 2012:104–8.
- 34. Ninot G, Boulze-Launay I, Bourrel G, et al. De la définition des Interventions Non Médicamenteuses (INM) à leur ontologie. Hegel 2018; 11: 21–7. [Google Scholar]
- 35. Ratner A, Bach SH, Ehrenberg H, Fries J, Wu S, Ré C. Snorkel: rapid training data creation with weak supervision. In: proceedings of the VLDB endowment. International conference on very large data bases, Munich, Germany; 2017:269–82 [DOI] [PMC free article] [PubMed]
- 36. Safranchik E, Luo S, Bach S.. Weakly supervised sequence tagging from noisy rules. Proc AAAI Conf Artif Intell 2020; 34 (04): 5570–8. [Google Scholar]
- 37. Califf RM, Zarin DA, Kramer JM, Sherman RE, Aberle LH, Tasneem A.. Characteristics of clinical trials registered in ClinicalTrials. gov, 2007-2010. JAMA 2012; 307 (17): 1838–47. [DOI] [PubMed] [Google Scholar]
- 38. Dunnmon JA, Ratner A, Saab eJK, et al. Cross-modal data programming enables rapid medical machine learning. Patterns 2020; 1 (2): 100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gu Y, Tinn R, Cheng H, et al. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans Comput Healthc 2022; 3 (1): 1–23. [Google Scholar]
- 40. Sanchez-Graillet O, Witte C, Grimm F, Cimiano P.. An annotated corpus of clinical trial publications supporting schema-based relational information extraction. J Biomed Semant 2022; 13 (1): 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dhrangadhariya A, Müller H, Manzo G.. PICO to PICOS: Extracting Study Type and Design Information without Labelled Data; 2023.
- 42. Augenstein I, Derczynski L, Bontcheva K.. Generalisation in named entity recognition: a quantitative analysis. Comput Speech Lang 2017; 44: 61–83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code is available on GitHub (https://github.com/anjani-dhrangadhariya/distant-PICO) and will be made public upon publication. The EBM-PICO error analysis file for the validation set, the error-rectified EBM-PICO gold test set, and other resources like distant supervision CTO dictionaries and hand-crafted dictionaries are available on DRYAD (https://doi.org/10.5061/dryad.ncjsxkszr). More information can be found in the supplementary file.