Abstract
Prokaryotic Argonaute (pAgo) nucleases with precise DNA cleavage activity show great potential for gene manipulation. Extensive biochemical studies have revealed that recognition of guides with different 5′ groups by Ago is important for biocatalysis. Here, we identified an Ago from the thermophilic Thermus parvatiensis ( TpsAgo) and analyzed the regulatory effect of 5′-modified guides on TpsAgo cleavage activity. Recombinant TpsAgo cleaves single-stranded DNA and RNA targets at 65–90°C, which is mediated by a 5′ hydroxyl or phosphate DNA guide. Notably, TpsAgo can utilize various 5′-modified DNA guides for catalysis, including 5′-NH 2C 6, 5′-Biotin, 5′-FAM and 5′-SHC 6 guides. Moreover, TpsAgo performs programmable cleavage of double-stranded DNA at temperatures over 80°C and strongly tolerates NaCl concentrations up to 3.2 M. These results provide insight into the catalytic performance of Agos by guide regulation, which may facilitate their biotechnological applications.
Keywords: Argonaute, Thermus parvatiensis, endonuclease, 5′-modified guide
Introduction
Argonaute (Ago) proteins are a highly conserved family of nucleic acid-guided proteins that are involved in a wide range of physiological processes in eukaryotes (eukaryotic Agos; eAgos) and prokaryotes (prokaryotic Agos; pAgos) [1]. Structural analyses have revealed that Agos share a conserved domain architecture for the N-terminal (N), PIWI-Argonaute-Zwille (PAZ), Middle (MID), and P element-induced wimpy testis (PIWI) domains [ 1, 2]. The MID domain anchors the 5′-end of the guides to stabilize the binary Ago–guide complex [ 1, 3, 4]. Their interactions are critical for cleavage activity in which guide-mediated cleavage of the complementary target is performed between the 10 th and 11 th nucleotides (nt) of the guide [ 5, 6].
Genomic studies showed that pAgos are more diverse than eAgos [ 1, 7– 9]. Some pAgos derived from thermophilic archaea and mesophilic bacteria have been characterized in detail [ 4, 5, 10– 19]. They generally use guides with a 5′-phosphate for cleavage, but some pAgos use 5′-hydroxyl guides [ 4, 5, 10, 14]. We previously found that Ago from Methanocaldococcus fervens ( MfAgo) can use both 5′-hydroxyl and 5′-phosphate guides [10]. Since the pronounced catalytic activity of Agos is affected by the 5′-end groups, it is interesting to investigate the regulatory effect of the 5′-end group of the guides on the Agos.
Recently, chemically modified guides associated with programmable nucleases have been investigated. Human Ago involved in RNA interference exhibits position-specific chemical modification of small interfering RNAs, which reduces ‘‘off-target’’ transcript silencing [ 20, 21]. As analogs of Agos, CRISPR-Cas nucleases have been demonstrated to use chemically modified guide RNAs (gRNAs) to enhance CRISPR-Cas genome editing [22]. A pAgo from Marinitoga piezophila ( MpAgo) was reported to use 5-bromo-2′-deoxyuridine (5′-BrdU)-modified gRNAs to significantly improve the specificity and affinity of RNA targets [23], which can be programmed as a highly specific RNA-targeting platform to probe RNA biology. Because of the high versatility and potential of pAgos in genetic manipulation [24], chemically modified guides harnessed by pAgos could open new avenues for future biotechnology applications.
In this study, an Ago from the thermophilic prokaryote Thermus parvatiensis ( TpsAgo) was cloned and characterized. As a thermophilic DNA-guided endonuclease, its cleavage activities directed by a variety of 5′-modified guides were systematically analyzed. The data are crucial for improving the understanding of the regulatory effect of guides for Agos and provide insight into guide design for DNA manipulations in the field of biotechnology.
Materials and Methods
Phylogenetic tree and sequence alignment
BLAST was performed based on the PfAgo ( Pyrococcus furiosus) amino acid sequences in the NCBI database. Amino acid sequences with high sequence consistency were selected and analyzed using MEGA 7.0 [25] to construct a phylogenetic tree. Multiple sequence alignment analysis between the TpsAgo sequence and other characterized Ago sequences was performed using ClustalW [26].
Strains and plasmids
Expression strain Escherichia coli BL21(DE3) cells were purchased from Beijing TransGen Biotech (Beijing, China). The cloned strain E. coli TOP10 and plasmid pET-28a (+)- TpsAgo (WP_060384876.1) containing the codon-optimized gene were synthesized by Nanjing GenScript Biotechnology (Nanjing, China).
Protein expression and purification
TpsAgo was codon-optimized and cloned into pET28a(+), which was then transformed into E. coli BL21(DE3). The bacteria were grown at 37°C in LB medium containing 50 μg/mL kanamycin. When the optical density at 600 nm (OD 600) reached 0.6–0.8, isopropyl β-d-1-thiogalactopyranoside (0.5 mM) was added and gene expression was induced at 20°C. The bacteria were harvested by centrifugation and resuspended in 20 mM Tris-HCl containing 1 M NaCl (pH 8.0). The bacteria were disrupted by high pressure, followed by heating at 65°C for 15 min and centrifugation, after which the supernatant was collected. The supernatant was purified using a nickel-nitrilotriacetic acid (Ni–NTA) affinity column, and the eluted proteins were resolved by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The purified protein was concentrated using an ultrafiltration tube and desalted using a PD-10 desalting column (GE Healthcare, Little Chalfont, UK). The protein concentration was determined using a BCA kit (Yeasen, Shanghai, China) according to the manufacturer’s instructions. The purified protein was stored in storage buffer comprised of 20 mM Tris-HCl, 1 M NaCl, and 15% (v/v) glycerol (pH 8.0) at –80°C.
TpsAgo activity assay
All target and guide oligonucleotide sequences are listed in Supplementary Tables S1 and S2, respectively. For activity assays, 200 nM TpsAgo was mixed with synthetic single-stranded DNA (ssDNA) or RNA guides, and 5′ fluorescently labeled ssDNA or RNA targets in a 1:10:4 ratio of protein: guide: target in reaction buffer containing 15 mM Tris-HCl (pH 8.0), 250 mM NaCl, and 0.5 mM Mn 2+. The reaction mixture was incubated for 0–30 min at 80°C and then rapidly cooled to 4°C. After incubation, loading buffer was added in a 1:1 ratio (v/v), and the samples were resolved on 16% denaturing polyacrylamide gels. The gel was stained with GelRed (Biotium, Fremont, USA) dye for 10–20 min. After staining, the results were observed using a 3500BR gel imaging system (Tanon, Shanghai, China) and quantitatively analyzed using ImageJ software (NIH, Bethesda, USA).
For plasmid cleavage assays, we designed paired guide DNA (gDNA) sets to target 80 bp regions in the pUC19 plasmid with GC contents of 29%, 45%, 53%, and 65%. TpsAgo (750 nM), 2.5 μM synthetic ssDNA guides, and 500 ng pUC19 plasmid were mixed in reaction buffer containing 15 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 0.5 mM Mn 2+ and incubated at 80°C for 2–4 h. The reactions were stopped using Proteinase K (TaKaRa Bio, Shiga, Japan) at 55°C for 1 h. Samples and 5×loading dye (Generay, Shanghai, China) were mixed and then resolved on 1.2% agarose gel. The gels were stained with GelRed dye (Biotium) and visualized using a gel imaging system.
Effects of temperature, metal ions, and NaCl on cleavage activities of TpsAgo
For temperature range assays, 200 nM TpsAgo was mixed with 16 nt 5′-P or 5′-OH ssDNA guides and 60 nt ssDNA targets in a 1:10:4 ratio of protein: guide: target in reaction buffer containing 15 mM Tris-HCl (pH 8.0), 250 mM NaCl, and 0.5 mM Mn 2+. A series of reaction buffers were prepared to ensure a pH of 8.0 at the tested temperature. The reaction mixture was incubated at temperatures of 55–99°C for 15 min.
To determine the cation preference, 200 nM TpsAgo was mixed with 16 nt 5′-P ssDNA guides and 60 nt ssDNA targets in a 1:10:4 ratio of protein:guide:target in reaction buffer containing 15 mM Tris-HCl (pH 8.0) and 250 mM NaCl. Ca 2+, Zn 2+, Co 2+, Mn 2+, Cu 2+, and Mg 2+ (all 0.5 mM) were added and incubated at 80°C for 30 min.
For NaCl concentration assays, 200 nM TpsAgo (without NaCl) was mixed with 16 nt 5′-P ssDNA guides and 60 nt ssDNA targets in a 1:10:4 ratio of protein: guide: target in reaction buffer containing various NaCl concentrations and incubated at 80°C for 30 min.
Effect of guide length on TpsAgo activity
For guide length assays, 200 nM TpsAgo was mixed with 11–21 nt 5′-P or 14–24 nt 5′-OH ssDNA guides and 60 nt ssDNA targets in a 1:10:4 ratio of protein: guide: target in a reaction buffer containing 15 mM Tris-HCl (pH 8.0), 250 mM NaCl, and 0.5 mM Mn 2+ and incubated at 80°C for 15 min.
Effects of 5′-end nucleotides and modification of gDNA on TpsAgo activity
For 5′-end nucleotides preference assays, 200 nM TpsAgo was mixed with 16 nt 5′-P ssDNA guides (5′-A, 5′-T, 5′-G, 5′-C) and 60 nt ssDNA targets in a 1:10:4 ratio of protein: guide: target in reaction buffer containing 15 mM Tris-HCl (pH 8.0), 250 mM NaCl, and 0.5 mM Mn 2+ and incubated at 80°C for 0–30 min. The samples were stained and analyzed as described above, and the data were fitted with Spline using Origin software (OriginLab, Northampton, USA).
To test the preference for the 5′ modification of gDNAs, kinetic analysis of ssDNA cleavage was performed under single-turnover or multiple-turnover conditions. In single-turnover reactions, 800 nM TpsAgo was mixed with 16 nt ssDNA guides (5′-P, 5′-OH, 5′-Biotin, 5′-NH 2C 6, 5′-fluorescein [FAM], 5′-SHC 6), and 60 nt ssDNA targets in a 2:20:1 ratio of protein: guide: target in reaction buffer containing 15 mM Tris-HCl (pH 8.0), 250 mM NaCl, and 0.5 mM Mn 2+, and incubated at 80°C for 0–60 min. The reactions were stopped by treatment with Proteinase K at 55°C for 1 h. In multiple-turnover reactions, 200 nM TpsAgo was mixed with 16 nt ssDNA guides (5′-P, 5′-OH, 5′-Biotin, 5′-NH 2C 6, 5′-FAM, 5′-SHC 6), and 60 nt ssDNA targets in a 1:10:4 ratio of protein: guide: target in reaction buffer containing 15 mM Tris-HCl (pH 8.0), 250 mM NaCl, and 0.5 mM Mn 2+, and incubated at 80°C for 0–30 min.
Effect of the first nucleotides of target DNA on TpsAgo activity
For the first target nucleotides preference assays, 200 nM TpsAgo was mixed with 16 nt 5′-P ssDNA guides and 60 nt ssDNA targets (t1A, t1T, t1G, t1C) in a 1:10:4 ratio of protein:guide:target in reaction buffer containing 15 mM Tris-HCl (pH 8.0), 250 mM NaCl, and 0.5 mM Mn 2+ and incubated at 80°C for 0–30 min. The samples were stained and analyzed as described above, and the data were fitted with Spline using Origin software.
Effect of guide/target mismatch on TpsAgo activity
To test the guide/target mismatch tolerance, 200 nM TpsAgo was mixed with samples of 16 nt 5′-P ssDNA guides harboring a single mismatch at positions 2–16, and 60 nt ssDNA targets in a 1:10:4 ratio of protein: guide: target in reaction buffer containing 15 mM Tris-HCl (pH 8.0), 250 mM NaCl, and 0.5 mM Mn 2+ and incubated at 80°C for 15 min.
Homology modeling and structural analysis of MID domain of TpsAgo
Homology modeling was performed using the ternary structure of TtAgo with 5′-P gDNA containing T as the first nucleotide (PDB ID: 4NCA) as a template with YASARA [27]. The quality of the models was evaluated based on the Z-score. The first three nucleotides of 5′-P, 5′-OH, 5′-Biotin, 5′-NH 2C 6, 5′-FAM, and 5′-SHC 6 gDNAs were docked into the TpsAgo model structure using both the AutoDock and AutoDock VINA searching algorithms in the YASARA-Structure program. The interaction between the MID domain of TpsAgo and gDNA was analyzed using PyMol software [28].
K d measurement
The dissociation constants ( K d) for guide DNA binding were determined by bio-layer interferometry (BLI) assay on the Octet RED 96 system (ForteBio, Menlo Park, USA). All steps (equilibrium, loading, association and dissociation step) were performed at 37°C with shaking at 1000 rpm in a black 96-well plate containing 0.2 mL of solution per well for samples or buffer (20 mM Tris-HCl containing 1 M NaCl and 0.5 mM Mn 2+, pH 8.0). Prior to each assay, NTA biosensor tips (Sartorius, Goettingen, Germany)were pre-wetted in 0.2 mL buffer for at least 30 min, followed by equilibration with buffer for 120 s. Afterwards, NTA biosensor tips were loaded with TpsAgo (200 nM), followed by an additional equilibration step (120 s), where a buffer containing 2 mg/mL bovine serum albumin (BSA) and 0.02% (v/v) Tween 20 was used. Subsequently, association of TpsAgo with gDNAs in a concentration range of 400–1000 nM was performed. Association at each studied concentration was carried out for 900 s. Finally, the dissociation was monitored with buffer for 900 s.
Results
TpsAgo uses DNA guides for ssDNA and RNA cleavage
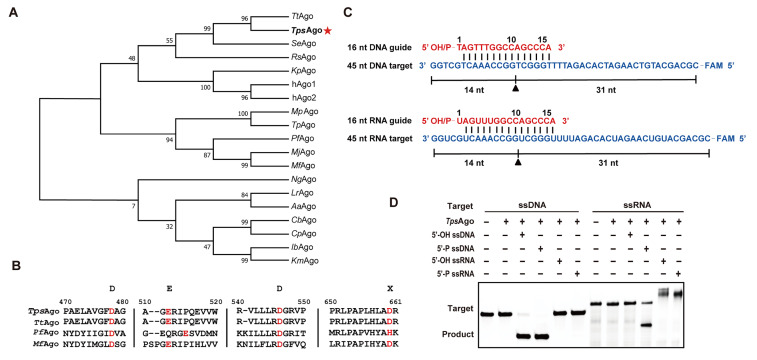
We performed a BLAST search based on the PfAgo amino acid sequences in the NCBI database and constructed a phylogenetic tree ( Figure 1A). Phylogenetic analysis revealed that TpsAgo and TtAgo belong to the same clade, sharing 98.8% identity with 7 amino acids substitution ( Supplementary Figure S1). Multiple sequence alignment showed that TpsAgo contains the conserved DEDD catalytic residues necessary for its cleavage activity, indicating that TpsAgo may be catalytically active ( Figure 1B).
Figure1 .
Phylogenetic analysis and multiple sequence alignment of TpsAgo
(A) Phylogenetic analysis of TpsAgo based on amino acid sequence. TtAgo: Thermus thermophilus Ago. TpsAgo: T. parvatiensis Ago. SeAgo: Synechococcus elongatus Ago. RsAgo: Rhodobacter sphaeroides Ago. KpAgo: Kluyveromyces polysporus Ago. hAgo1 and hAgo2: Homo sapiens Ago. MpAgo: Marinitoga piezophila Ago. TpAgo: Thermotoga profunda Ago. PfAgo: Pyrococcus furiosus Ago. MjAgo: Methanocaldococcus jannaschii Ago. MfAgo: M. fervens Ago. NgAgo: Natronobacterium gregory Ago. LrAgo: Limnothrix rosea Ago. AaAgo: Aquifex aeolicus Ago. CbAgo: Clostridium butyricum Ago. CpAgo: C. perfringens Ago. IbAgo: Intestinibacter bartlettii Ago. KmAgo: Kurthia massiliensis Ago. Numbers at the nodes indicate the bootstrap values for maximum likelihood analysis of 1000 resampled data sets. (B) Multiple sequence alignment of TpsAgo with several other characterized prokaryotic Agos. Red font denotes the key catalytic residues. (C) Schematic diagram of synthesized 45 nt FAM-labeled ssDNA or RNA (blue) as targets and 16 nt DNA or RNA with a 5′-OH or 5′-P group as guides (red). (D) Cleavage activity assay of TpsAgo with FAM-labeled targets.
The recombinant protein was successfully expressed in the soluble form by E. coli BL21(DE3) cells. After purification using a Ni–NTA affinity column, the recombinant proteins were identified by SDS-PAGE. The molecular weight of the purified protein corresponded to the predicted value ( Supplementary Figure S2). To determine the endonuclease activity of the protein, we used synthesized 45 nt FAM labeled ssDNA or RNA as targets and 16 nt DNA or RNA with a 5′-OH or 5′-P group as guides ( Figure 1C). When incubated at 80°C for 30 min, TpsAgo showed DNA nuclease activity with 5′-OH and 5′-P gDNAs, as well as RNA nuclease activity with 5′-P gDNA ( Figure 1D). TpsAgo cleaved the DNA target more efficiently than it cleaved the RNA target ( Supplementary Figure S3).
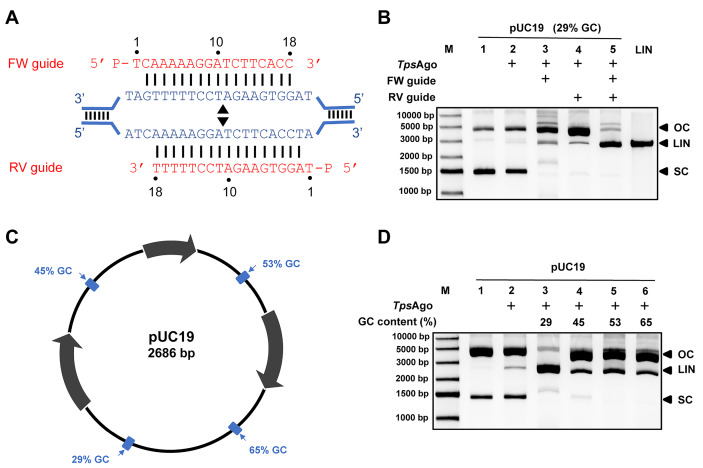
TpsAgo mediates double-stranded DNA cleavage
Previous studies showed that pAgos can successfully generate breaks in plasmid DNA at specific sites defined by paired gDNAs, or in a guide-independent manner [ 4, 11– 13, 16]. Therefore, we first evaluated double-stranded (dsDNA) cleavage in the absence of gDNA. TpsAgo could not cleave the plasmid under the evaluated conditions. We then tested the effect of paired gDNAs on plasmid cleavage of TpsAgo. When a single gDNA was added to the reaction, the supercoiled plasmid disappeared, and the amount of open-circle plasmid greatly increased. Notably, the presence of a pair of gDNAs resulted in large amounts of linear plasmids with fewer open-circle plasmids ( Figure 2A,B). In addition, we designed gDNAs to target regions with different GC contents (29%, 45%, 53%, and 65%). Under the evaluated conditions, the region with a low GC content (29%) was completely cleaved into linear plasmids by TpsAgo. However, regions with higher GC contents were mostly maintained as open circular plasmids ( Figure 2C,D).
Figure2 .
TpsAgo cleaves dsDNA with a pair of complementary guides
(A) Schematic diagram of target regions with 29% GC content (80 bp segments). Black triangles indicate the predicted cleavage sites. (B) Plasmid cleavage in the 29% GC region. (C) Schematic overview of pUC19 target plasmid. The target sites are indicated in blue. Percentages indicate the GC content of the 80 bp segments in which these target sites are located. (D) Plasmid cleavage in different target regions. The reactions were performed with no guides, one guide or one pair of guides at 80°C. Control reactions did not contain Ago proteins. FW, forward guide DNA; RV, reverse gDNA. M, molecular weight marker; Lin, linearized plasmid; SC, supercoiled plasmid; OC, open circular plasmid.
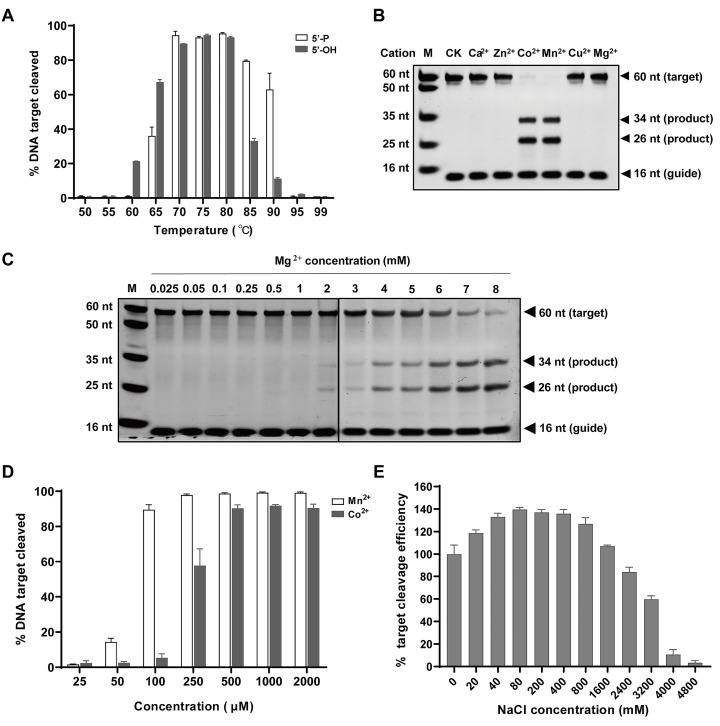
Temperature and chemical factors affect TpsAgo activity
To investigate the temperature range of TpsAgo, we tested the cleavage activity of TpsAgo at temperatures of 50–99°C. The results showed that TpsAgo was most active at 70–80°C using 5′-OH gDNA and 70–85°C using 5′-P gDNA ( Figure 3A).
Figure3 .
Effects of temperature, divalent metal ions, and NaCl concentration on activity of TpsAgo
(A) Effect of temperature on TpsAgo activity. (B) TpsAgo displays Co2+ and Mn2+ mediated ssDNA target cleavage. (C) Mg2+ concentration ranges required for TpsAgo activity. (D) Mn2+ and Co2+ concentration ranges required for TpsAgo activity. (E) Effect of NaCl concentration on TpsAgo activity. In all experiments, TpsAgo: guide: target ratio was 1:10:4. Error bars represent the standard deviation of three independent experiments.
To characterize the effect of metal ions on target cleavage, we investigated the performance of TpsAgo in the presence of various concentrations and types of metal ions. TpsAgo used Co 2+, Mn 2+, and Mg 2+ to mediate DNA-guided DNA target cleavage ( Figure 3B,C). We further investigated the effects of the Co 2+, Mn 2+, and Mg 2+ concentrations on TpsAgo activity. TpsAgo maintained the same cleavage efficiency in the presence of 0.1 to 8 mM Mn 2+, but required a higher concentration of Co 2+ (more than 0.25 mM) and Mg 2+ (more than 4 mM) for active biocatalysis ( Figure 3C,D and Supplementary Figure S4). These findings indicate that Mn 2+ is optimal for TpsAgo cleavage.
NaCl plays an important role in maintaining the stability of pAgos, but high concentrations of NaCl generally inhibit the activity of pAgos: for example, 1.5 M NaCl was shown to almost completely abolish Agos activity in previous studies [ 10, 11, 14]. We used 0–4.8 M NaCl to determine the tolerance of TpsAgo. Unexpectedly, TpsAgo exhibited strong tolerance to 0–2.4 M NaCl. Additionally, approximately 50% of its optimal activity was maintained even at 3.2 M NaCl ( Figure 3E).
Length and 5′-end nucleotides of gDNA affect TpsAgo activity
We measured the activity of TpsAgo using 5′-P gDNA ranging from 11 to 21 nt and 5′-OH gDNA ranging from 14 to 24 nt. A minimum of 16 nt gDNA (both for 5′-P and 5′-OH) was required for TpsAgo cleavage activity. The highest cleavage activity was observed for 16–18 nt 5′-P gDNA or 16–19 nt 5′-OH gDNA. Use of longer guides decreased the reaction efficiency. When provided with a 21 nt guide, TpsAgo retained approximately 70% of its optimal activity with 5′-OH gDNA but only 20% of its optimal activity with 5′-P gDNA. The findings indicate that TpsAgo can use longer 5′-OH gDNA than 5′-P gDNA ( Figure 4A,B and Supplementary Figure S5A,B).
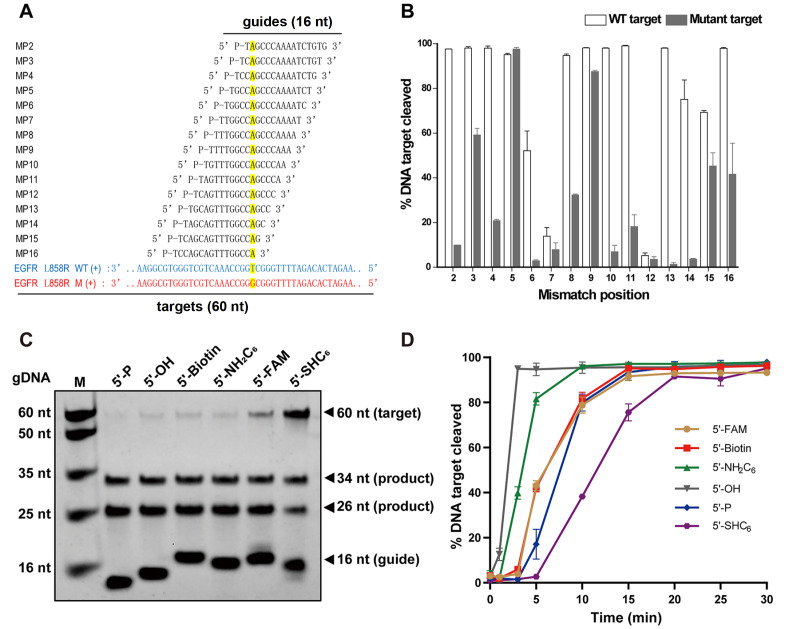
Figure4 .
Effects of length and 5′-end nucleotides of gDNAs on the cleavage activity of TpsAgo
(A) Effect of 5′-P gDNA length on TpsAgo activity. (B) Effect of 5′-OH gDNA length on TpsAgo activity. (C) TpsAgo activity mediated by gDNAs with different 5′-end nucleotides at 80°C for 15 min. (D) Time course experiments for gDNAs with various 5′-end nucleotides. In all experiments, TpsAgo:guide:target ratio was 1:10:4. Error bars represent the standard deviation of three independent experiments.
Preferences for the 5′-end nucleotides of guide sequences have been observed for some Agos [ 29, 30]. To explore the effect of the 5′-end nucleotide of gDNA on the cleavage activity of TpsAgo, we designed 16 nt gDNAs with 5′-A, 5′-T, 5′-G, or 5′-C. After 15 min of incubation, TpsAgo with 5′-T or 5′-G gDNA cleaved 100% of the targets, whereas for 5′-C gDNA and 5′-A gDNA, TpsAgo cleaved 80% and 50% of the targets, respectively. These results show that TpsAgo has a preference for 5′-T and 5′-G gDNA ( Figure 4C,D). We also tested the effect of the first nucleotide of target DNAs on TpsAgo activity and found that TpsAgo showed similar cleavage efficiency for t1A, t1T, and t1C but slightly lower efficiency for t1G ( Supplementary Figure S6).
Single guide/target mismatch curtails TpsAgo activity
Mismatches between the target and guide strands are thought to affect cleavage efficiency [ 4, 5, 17, 31]. To determine the effect of mismatch on the cleavage efficiency of TpsAgo, we designed a series of gDNAs containing single-point mismatch for EGFR (endothelial growth factor receptor) L858R ssDNA, with mismatch sites distributed among positions 2–16 of the guides. We also designed an EGFR wild-type sequence as a control. TpsAgo is sensitive to target/guide mismatch in the seed region (positions 2, 4, 6, 7, 8), and 3′-portion of gDNA (positions 10, 11, 13, 14) ( Figure 5A,B).
Figure5 .
Effects of mismatch in the guide-target duplex and 5′ modification of gDNAs on cleavage activity of TpsAgo
(A) Schematic of gDNAs with different mismatch position. (B) Effect of mismatch in the gDNA-target duplex on TpsAgo activity. (C) TpsAgo activity mediated by different 5′-modified gDNAs at 80°C for 15 min. (D) Time course experiments for various 5′-modified gDNAs. In all experiments, TpsAgo:guide:target ratio was 1:10:4. Error bars represent the standard deviation of three independent experiments.
TpsAgo cleaves with a wide range of 5′-modified guides
To further explore the effect of 5′ chemically modified gDNAs on enzyme activity, we performed the cleavage kinetics assay with 16 nt gDNAs containing a 5′-P, 5′-OH, 5′-Biotin, 5′-NH 2C 6, 5′-FAM, or 5′-SHC 6 at 80°C under either single-turnover conditions or multiple-turnover conditions. In multiple-turnover reactions, TpsAgo used all tested gDNAs to cleave the ssDNA target ( Figure 5C). After 5 min of incubation, TpsAgo with 5′-OH, 5′-NH 2C 6, 5′-Biotin (5′-FAM), 5′-P, and 5′-SHC 6 gDNA cleaved 100%, 80%, 40%, 17%, and 2% of the targets, respectively ( Figure 5D). The highest reaction efficiency was observed for 5′-OH gDNA. Similar cleavage efficiencies were observed for 5′-Biotin and 5′-FAM gDNA, whereas 5′-SHC 6 gDNA showed the lowest efficiency. Furthermore, we measured the equilibrium dissociation constants ( K d) for guide binding by TpsAgo using BLI assay ( Supplementary Table S3). It is seen here that gDNAs with 5′-P, 5′-FAM and 5′-Biotin are bound best, while 5′-SHC 6 modification decreases gDNA affinity, and 5′-OH and 5′-NH 2C 6 guides are bound with dramatically lower affinity than 5′-P guide DNA (>30-fold increase in K d). In single-turnover reactions, the nearly identical reaction efficiencies were observed for 5′-OH, 5′-NH 2C 6 and 5′-Biotin gDNAs. 5′-FAM and 5′-SHC 6 gDNA showed lower cleavage efficiency, whereas the lowest efficiency was observed for 5′-P gDNA ( Supplementary Figure S7).
Homology modeling and structural analysis of MID domain of TpsAgo
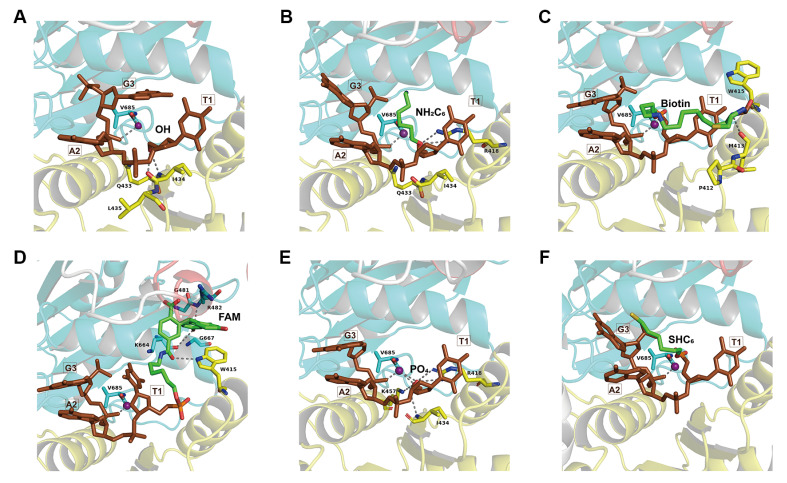
To better understand the specific guide recognition mechanism of TpsAgo, the structure of TpsAgo was built with the structure of TtAgo with 5′-P gDNA (PDB ID: 4NCA) as a template ( Supplementary Figure S8), and the first three nucleotides of 5′-P, 5′-OH, 5′-Biotin, 5′-NH 2C 6, 5′-FAM, and 5′-SHC 6 gDNAs were individually docked into the TpsAgo model structure ( Figure 6A–F). As the 5′ ends of the guides are anchored in the MID domains of Agos [5], we examined the potential interaction between the MID domain of TpsAgo and different 5′-modified gDNAs. Structural analysis of TpsAgo revealed multiple hydrophobic residues and hydrogen bonds around the 5′-OH, 5′-NH 2C 6, 5′-Biotin, and 5′-FAM groups, whereas more hydrogen bonds but fewer hydrophobic residues were observed around the 5′-P group ( Table 1 and Figure 6A–E). For the 5′-SHC 6 group, weaker interactions derived from the hydrophobic residues and hydrogen bonds were formed ( Figure 6F).
Figure6 .
Structural analysis of MID domain of TpsAgo
Structural analysis of MID domain of TpsAgo with 5′-OH (A), 5′-NH2C6 (B), 5′-Biotin (C), 5′-FAM (D), 5′-P (E), and 5′-SHC6 (F) gDNAs were shown. It should be noted that 5′-NH2C6, 5′-Biotin, 5′-FAM, and 5′-SHC6 gDNAs still contain a 5′-phosphate group but not at the specific 5′-terminal. The amino acid residues of the MID domain of TpsAgo involved in interactions with the 5′-end group are highlighted in yellow; the Mn2+ involved in interactions with both gDNA and PIWI domain (cyan) is shown as purple sphere; the first three nucleotides of guide are colored in brown; the abovementioned interactions of hydrogen bond or ionic bond are indicated as gray dotted lines.
Table1 Hydrophobic interaction and hydrogen bonds formed by TpsAgo and the 5′-end of the guides
|
Guides |
Hydrophobic interaction-related residues |
Hydrogen bond-related residues (groups) |
Hydrogen bonds ( n) |
|
5′-OH gDNA |
Gln 433, Ile 434, Leu 435, Val 685 |
Ile 434 (COOH, NH 2) |
2 |
|
5′-NH 2C 6 gDNA |
Gln 433, Ile 434, Val 685 |
Arg 418 (NH 2) |
2 |
|
5′-Biotin gDNA |
Pro 412, Met 413, Trp 415 |
Met 413 (COOH), Trp 415 (NH 2) |
2 |
|
5′-FAM gDNA |
Trp 415, Gly 481, Gly 667 |
Trp 415 (C 6H 6), Arg 482 (NH 2), Lys 664 (COOH) |
3 |
|
5′-P gDNA |
Val 685 |
Arg 418 (NH 2), Ile 434 (NH 2), Lys 457 (NH 2) |
4 |
|
5′-SHC 6 gDNA |
Val 685 |
none |
0 |
Discussion
Compared with eAgos, which exclusively use RNA guides to cleave RNAs, pAgos use DNA guides or RNA guides to cleave complementary nucleic acid targets. Most pAgos use 5′-P guides, while few have a preference for 5′-OH guides [5]. pAgos that can use other 5′-modified guides remain to be explored. We characterized thermophilic pAgo from T. parvatiensis as a programmable endonuclease with DNA-guided cleavage of DNA and RNA targets. Most characterized pAgos have an exclusive target cleavage activity, except for TtAgo, MpAgo, and KmAgo, which can simultaneously act on DNA and RNA targets [ 5, 12, 16, 32]. The ability of TpsAgo to cleave multiple substrates (DNA and RNA) can expand the ability to perform genetic manipulation.
TpsAgo functions over a wide temperature range (65–90°C), whereas different temperature spectra were observed when using 5′-P gDNA and 5′-OH gDNA. TpsAgo cleaved targets at temperatures greater than 85°C with 5′-P gDNA but not with 5′-OH gDNA. This phenomenon has also been reported for CbAgo and MfAgo [ 4, 10]. The interactions between the 5′-phosphate and MID binding pocket can stabilize Ago–guide complexes at elevated temperatures [4]. When the temperature exceeds 85°C, the binding affinity between the phosphate group and the MID domain may be stronger than that of the hydroxyl group.
For thermophilic Agos, complementary base pairing of approximately 15 nt between the guide and target is required to form a relatively stable double helix structure at high temperatures, which is conducive to subsequent cleavage. For TpsAgo activity, a minimum of 16 nt gDNA is required, which is similar to other thermophilic Agos [ 5, 11, 31]. The bias for the 5′-end nucleotides varies for different Agos [ 12, 16, 29, 30, 33]. TpsAgo showed a preference for 5′-T and 5′-G of gDNA, possibly because of the higher binding affinity between 5′-T or 5′-G and TpsAgo than that of 5′-A or 5′-C.
Previous studies showed that mismatch between the target and guide in eAgos and some pAgos significantly affects cleavage efficiency. For RsAgo and MpAgo, a mismatch in the seed region (2–8 nt) greatly reduces target recognition and cleavage; for CbAgo, LrAgo, and SeAgo, a mismatch in the seed region has little or no effect on cleavage efficiency, whereas mismatch at the 3′-portion of gDNA significantly reduces cleavage efficiency [ 4, 5, 17, 34]. In this study, we found that TpsAgo has a low tolerance for guide/target mismatches both in the seed region and 3′-portion of gDNA, and evaluation of TpsAgo cleavage at single-base resolution may facilitate its application in precise DNA manipulation.
Previous studies suggested that pAgo can successfully generate dsDNA breaks in plasmids. Similar to reported mesophilic Agos [4], guide-dependent dsDNA cleavage by TpsAgo depends on the presence of AT-rich regions. Interestingly, guide-independent plasmid processing activity has been found for some Agos but was suppressed at elevated temperatures [ 4, 16, 35] or in the presence of excess guides [31]. Here, no guide-independent activity for TpsAgo at 80°C was observed, which may be related to the high temperature and saturated gDNA (gDNA: TpsAgo=10:1) .
Most Agos reported to date rely on 5′-P guides, but some Ago variants can accept 5′-OH guides [ 4, 5, 10, 14, 16]. The ability of various Agos to use other 5′-modified guides remains unclear. We showed that pAgo can use many 5′-modified guides, including 5′-P, 5′-OH, 5′-Biotin, 5′-NH 2C 6, 5′-FAM, and 5′-SHC 6 but with different cleavage efficiencies in multiple-turnover conditions. TpsAgo displayed the highest cleavage efficiency when using 5′-OH gDNA, indicating its preference for 5′-OH. We also found that 5′-NH 2C 6, 5′-Biotin, and 5′-FAM had higher cleavage activity than 5′-P. Subsequently, homologous modeling was performed to analyze the impact of these modified guides on the changes of interactions with MID domain. Through in silico analysis, we described the multiple interactions that may respond to TpsAgo conformation changes and alter their activity mediated by different modified guides. It should be noted that the third nucleotide (G3) and its phosphate group of 5′-OH gDNA showed a different orientation, which could impair the base-paring with target or the gDNA binding. The reason may be that only three first nucleotides were used for modeling without the downstream guide DNA sequence. On the other hand, it is possible that the G3 of 5′-OH guide does not form a base-pair with the target, because recent studies showed that a mismatch in the seed region stimulated target cleavage for some Agos [ 4, 36]. In fact, TpsAgo associated with 5′-OH and 5′-NH 2C 6 guides displaying higher cleavage efficiency in multiple-turnover reactions showed a much lower binding affinity, while high binding affinity guides (5′-P, 5′-Biotin and 5′-FAM modifications) resulted in lower cleavage efficiency. We propose that the difference in the kinetics of cleavage may be resulted from the different turnover rates for different 5′-end modifications, since previous studies indicated that the rate-limiting step in the reaction is the dissociation of products from the complex after cleavage. To test this hypothesis, we measured the kinetics of ssDNA cleavage under the single-turnover conditions (the binary complex of TpsAgo with guide DNA was present in 2-fold over target). The nearly identical reaction rates observed for 5′-OH, 5′-NH 2C 6 and 5′-Biotin guides seem to support the proposal that the rate-limiting step in the reaction is the dissociation of products from the complex after cleavage. Interestingly, TpsAgo has a much lower cleavage efficiency towards to 5′-SHC 6 and 5′-P guides. The lower cleavage efficiency observed for 5′-SHC 6 guide may be explained by its oxidation due to its instability [35], while 5′-P guide results the lowest cleavage efficiency, suggesting that a fraction of complexes formed by TpsAgo is catalytically inactive. Although previous studies have shown that the rate-limiting step in the reaction is the dissociation of products from the complex after cleavage [ 4, 37], our results suggested that there may be other factors which can affect the cleavage efficiency, such as target recognition and duplex propagation [3].
The cleavage site of LrAgo is shifted 1–2 nt downstream from the 5′-end in the absence of the 5′-phosphate group in the guide molecule. Changes in the slicing site were also observed in hAgo2 with non-phosphorylated guides. Therefore, the phosphate group can help determine the precise cleavage site [ 4, 38]. However, changes in the slicing site were not observed for TpsAgo, indicating that TpsAgo can perform precise cleavage with non-phosphorylated guides. Thus, further structural research is required to understand the specific guide recognition mechanism of TpsAgo. Our findings expand the understanding of the catalytic diversity of pAgos and may facilitate widespread use of pAgos in genetic manipulations.
Supplementary Data
Supplementary Data is available at Acta Biochimica et Biphysica Sinica online.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the Natural Science Foundation of China (No. 31770078) and the Ministry of Science and Technology (No. 2020YFA0907700).
References
- 1.Swarts DC, Makarova K, Wang Y, Nakanishi K, Ketting RF, Koonin EV, Patel DJ, et al. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol. . 2014;21:743–753. doi: 10.1038/nsmb.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J, Yang J, Cho WC, Zheng Y. Argonaute proteins: Structural features, functions and emerging roles. J Adv Res. . 2020;24:317–324. doi: 10.1016/j.jare.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lisitskaya L, Aravin AA, Kulbachinskiy A. DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins. Nat Commun. . 2018;9:5165. doi: 10.1038/s41467-018-07449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuzmenko A, Yudin D, Ryazansky S, Kulbachinskiy A, Aravin AA. Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea . Nucleic Acids Res. . 2019;47:5822–5836. doi: 10.1093/nar/gkz379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaya E, Doxzen KW, Knoll KR, Wilson RC, Strutt SC, Kranzusch PJ, Doudna JA. A bacterial Argonaute with noncanonical guide RNA specificity. Proc Natl Acad Sci USA. . 2016;113:4057–4062. doi: 10.1073/pnas.1524385113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng G, Zhao H, Wang J, Rao Y, Tian W, Swarts DC, van der Oost J, et al. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage . Proc Natl Acad Sci USA. . 2014;111:652–657. doi: 10.1073/pnas.1321032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin EV. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence. Biol Direct. . 2017;12:5. doi: 10.1186/s13062-017-0177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarova KS, Wolf YI, van der Oost J, Koonin EV. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct. . 2009;4:29. doi: 10.1186/1745-6150-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hegge JW, Swarts DC, van der Oost J. Prokaryotic argonaute proteins: novel genome-editing tools? Nat Rev Microbiol. . 2018;16:5–11. doi: 10.1038/nrmicro.2017.73. [DOI] [PubMed] [Google Scholar]
- 10.Chong Y, Liu Q, Huang F, Song D, Feng Y. Characterization of a recombinant thermotolerant argonaute protein as an endonuclease by broad guide utilization. Bioresour Bioprocess. . 2019;6:1–10. doi: 10.1186/s40643-019-0254-8. [DOI] [Google Scholar]
- 11.Swarts DC, Hegge JW, Hinojo I, Shiimori M, Ellis MA, Dumrongkulraksa J, Terns RM, et al. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA . Nucleic Acids Res. . 2015;43:5120–5129. doi: 10.1093/nar/gkv415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, Wang Y, et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. . 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zander A, Willkomm S, Ofer S, van Wolferen M, Egert L, Buchmeier S, Stöckl S, et al. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii . Nat Microbiol. . 2017;2:17034. doi: 10.1038/nmicrobiol.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Y, Sun W, Wang J, Sheng G, Xiang G, Zhang T, Shi W, et al. Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single- and double-stranded DNA at 37°C. Cell Discov. . 2019;5:38. doi: 10.1038/s41421-019-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegge JW, Swarts DC, Chandradoss SD, Cui TJ, Kneppers J, Jinek M, Joo C, et al. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute . Nucleic Acids Res. . 2019;47:5809–5821. doi: 10.1093/nar/gkz306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Li W, Jiang X, Wang Y, Zhang Z, Liu Q, He R, et al. A programmable omnipotent Argonaute nuclease from mesophilic bacteria Kurthia massiliensis . Nucleic Acids Res. . 2021;49:1597–1608. doi: 10.1093/nar/gkaa1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olina A, Kuzmenko A, Ninova M, Aravin AA, Kulbachinskiy A, Esyunina D. Genome-wide DNA sampling by Ago nuclease from the cyanobacterium Synechococcus elongatus . RNA Biol. . 2020;17:677–688. doi: 10.1080/15476286.2020.1724716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage . Mol Cell. . 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kropocheva E, Kuzmenko A, Aravin AA, Esyunina D, Kulbachinskiy A. A programmable pAgo nuclease with universal guide and target specificity from the mesophilic bacterium Kurthia massiliensis . Nucleic Acids Res. . 2021;49:4054–4065. doi: 10.1093/nar/gkab182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. . 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. . 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 22.Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. . 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapinaite A, Doudna JA, Cate JHD. Programmable RNA recognition using a CRISPR-associated Argonaute. Proc Natl Acad Sci USA. . 2018;115:3368–3373. doi: 10.1073/pnas.1717725115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryazansky S, Kulbachinskiy A, Aravin AA. The expanded universe of prokaryotic argonaute proteins. mBio. . 2018;9:e01935–18. doi: 10.1128/mBio.01935-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. . 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. . 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krieger E, Vriend G. Models@Home: distributed computing in bioinformatics using a screensaver based approach. Bioinformatics. . 2002;18:315–318. doi: 10.1093/bioinformatics/18.2.315. [DOI] [PubMed] [Google Scholar]
- 28.Delano WL. The PyMOL molecular graphics system. 2002, http://www.pymol.org
- 29.Willkomm S, Oellig CA, Zander A, Restle T, Keegan R, Grohmann D, Schneider S. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat Microbiol. . 2017;2:17035. doi: 10.1038/nmicrobiol.2017.35. [DOI] [PubMed] [Google Scholar]
- 30.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. . 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 31.Song J, Hegge JW, Mauk MG, Chen J, Till JE, Bhagwat N, Azink LT, et al. Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics . Nucleic Acids Res. . 2020;48:e19. doi: 10.1093/nar/gkz1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. . 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olovnikov I, Chan K, Sachidanandam R, Newman DK, Aravin AA. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol Cell. . 2013;51:594–605. doi: 10.1016/j.molcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Esyunina D, Olovnikov I, Teplova M, Kulbachinskiy A, Aravin AA, Patel DJ. Accommodation of helical imperfections in Rhodobacter sphaeroides argonaute ternary complexes with guide RNA and target DNA . Cell Rep. . 2018;24:453–462. doi: 10.1016/j.celrep.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X, Sun Y, Chen L, Huang F, Liu Q, Feng Y. A hyperthermophilic argonaute from Ferroglobus placidus with specificity on guide binding pattern . Front Microbiol. . 2021;12:654345. doi: 10.3389/fmicb.2021.654345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen GR, Sive H, Bartel DP. A seed mismatch enhances argonaute2-catalyzed cleavage and partially rescues severely impaired cleavage found in fish. Mol Cell. . 2017;68:1095–1107.e5. doi: 10.1016/j.molcel.2017.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. . 2012;151:1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. . 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]