Abstract
Cyclin B1 is an essential cyclin-dependent protein that involves in the G2/M transition. Multiple studies report that cyclin B1 is upregulated in cancers and promotes cancer progression. However, the mechanism of cyclin B1 upregulation remains unclear. Here we report that the 5′UTR of cyclin B1 mRNA contains an internal ribosome entry site (IRES) by using a bicistronic fluorescent reporter. We show that IRES can initiate the translation of cyclin B1, and the IRES-mediated translation is further activated under cell stress. Interacting trans-acting factors (ITAFs) are required by most IRES to initiate the translation. We find that PTBP1 promotes the IRES-mediated translation of cyclin B1 by binding to the 5′UTR of cyclin B1. On top of that, PTBP1 promotes the malignancy of ESCC cells. Our data suggest that the IRES-mediated translation of cyclin B1 plays an essential role in the cyclin B1 upregulation in cancers.
Keywords: cyclin B1, internal ribosome entry site, PTBP1, interacting trans-acting factors, esophageal squamous cell carcinoma
Introduction
The cell cycle is a regulated series of molecular events that dictate cellular division and proliferation. The process of cell cycle is well controlled by cyclins and Cdks. The protein levels of cyclins need to be tightly regulated to ensure the cell cycle progresses appropriately. Several different cyclins are active in different parts of the cell cycle, which causes the Cdk to phosphorylate different substrates. Specifically, cyclin B1 is expressed predominantly during the G2/M phase of the cell cycle. In the early stage of mitosis, cyclin B1 and p34 (Cdk1) form the maturation-promoting factor (MPF). The MPF is involved in chromosome condensation, nuclear envelope breakdown, and spindle pole assembly. CyclinB1 is mainly present in the cytoplasm before mitosis and relocates to the nucleus in the late prophase. At the end of mitosis, cyclin B1 is targeted for degradation by APC/C Cdh1.
Cell cycle deregulation is a common feature of human cancer. De-regulation of cyclin B1 will lead to an abnormal cell cycle. Several studies have revealed that cyclin B1 is upregulated in breast cancer, cervical cancer, gastric cancer, colorectal cancer, non-small cell lung cancer, colon cancer, prostate cancer, and esophageal cancer [ 1– 5]. Furthermore, upregulation of cyclin B1 promotes the proliferation of cancer cells and is associated with invasion and metastasis [6]. In addition, the expression level of cyclin B1 is closely related to the prognosis of cancer [ 1, 3, 4].
Translation can be initiated in cap-dependent and cap-independent manner. The cap-dependent translation begins with recognizing the 5′ cap structure by a cap-binding protein eIF4E, thereby scanning the mRNA transcripts. In addition to cap-dependent translation, noncanonical cap-independent translation also plays an essential role in translation. While in stress conditions, such as hypoxia, nutrient limitation, and apoptosis, cap-independent translation will take place due to the inhibition of the cap-dependent translation [ 7, 8]. Internal ribosome entry site (IRES)-mediated translation is a type of cap-independent translation. IRES was first identified in small RNA viruses (picornaviruses) by Nahum Sonnenberg [9] and Eckard Wimmer [10] in 1988, and soon it was found in eukaryotic cells [11]. After that, more IRES have been identified by researchers in eukaryotic cells [12]. IRES is a specific sequence on the 5′UTR of mRNA, which can form a secondary structure to enter the ribosome to initiate translation without cap structure recognition. Under physiological conditions, most of the translations are driven in a cap-dependent manner in eukaryotic cells.
Thousands of human and viral sequences with cap-independent translation activity have been found, which means that ~10% of mammalian mRNAs may be translated by IRES-mediated translation [13]. Cellular IRESs can be classified into two types based on the mechanisms of ribosome recruitment: type I IRESs interact with ribosomes through ITAFs that bind to the RNA-binding motifs and through N-6-methyladenosine (m6A) modification [14]. Type II IRESs, on the other hand, have a short cis-element that pairs with 18S rRNA to recruit ribosomes [15]. Several studies revealed that multiple essential proteins associated with cancer have IRES, such as p53 [12] and c-myc [16].
A bicistronic fluorescent reporter system is a canonical method to measure the activity of IRES. The reporter plasmid contains two cistrons, with the first cistron encoding Renilla luciferase (RLuc) and the second cistron encoding Firefly luciferase (FLuc). Typically, in eukaryotic cells, only the first cistron of the bicistronic plasmid is translated, and the translation will stop due to the presence of the stop codon after the first cistron. However, if the sequence between the two cistrons contains IRES, the translation can directly start from the IRES. The second cistron will be expressed.
In this research, we found that cyclin B1 has an IRES and the IRES can initiate the translation. The IRES-mediated translation is further activated under cell stress. This is one of the reasons that cyclin B1 is upregulated in tumors. We also found that PTBP1 binds to the 5'UTR of cyclin B1 and promotes the IRES-mediated translation.
Materials and Methods
Cell lines and cell culture
Human esophageal squamous cell carcinoma cell lines YES-2, KYSE 150, and KYSE 30 were provided by Professor Shimada of Kyoto University (Kyoto, Japan). The cells were cultured in RPMI 1640 medium (Gibco, Carlsbad, USA) supplemented with 10% fetal bovine serum (FBS), 50 units/mL of penicillin, and 50 μg/mL of streptomycin at 37°C with 5% CO 2.
Plasmids
The plasmids used in this study were all constructed by Jikai Gene Co., Ltd (Shanghai, China). The bicistronic fluorescent reporter plasmid vector is CV094, the elements are CMV-Renilla-MCS-Firefly-SV40-Puromycin, and the cloning site is BamHI/ EcoRI. The 5′UTRs of c-Myc and cyclin B1 were inserted between Renilla luciferase and Firefly luciferase (pR-M-F and pR-B-F). phR-B-F plasmid: a sequence that can form a stable neck ring structure was inserted before the Renilla luciferase, and an additional stop codon was added after the Renilla luciferase.
siRNA and cell transfection
Two micrograms of plasmid or 3 μL of 20 μM siRNA was used for cells in a 6-well plate. Transfections were performed using lipofectamine 2000 (Invitrogen, Carlsbad, USA) according to the manufacturer′s instructions. Sequences are as following: siPTB-1 forward 5′-GCCUCAACGUCAAGUACAATT-3′, reverse 5′-UUGUACUUGACGUUGAGGCTT-3′; siPTB-2 forward 5′-GCGUGAAGAUCCUGUUCAATT-3′, reverse 5′-UUGAACAGGAUCUUCACGCTT-3′; siPTB-3 forward 5′-GCGUCGUCAAAGGAUUCAATT-3′, reverse 5′-UUGAAUCCUUUGACGACGCTT-3′; si-scramble forward 5′-GCAAAUACUCGUGCCUAUACATT-3′, reverse 5′-UGUAUAGGCACGAGUAUUUGCTT-3′.
Luciferase reporter assay
Cells were harvested at 48 h after transfection, resuspended in luciferase lysis buffer (Promega, Madison, USA), and then incubated on ice for 10 min. Cell debris was removed by centrifugation at 20,000 g at 4°C for 10 min, and the supernatants were used for the luciferase assay. According to the manufacturer’s instructions, Firefly and Renilla luciferase activities were determined using the Dual-Luciferase ® Reporter Assay System (Promega). The ratio of Firefly luciferase to Renilla luciferase activity was defined as IRES activity.
RNA extraction and real-time qPCR
RNA was extracted by using TRIzol Reagent (Invitrogen). RNA concentration was measured with a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, USA). Total RNA (2 μg) was used for cDNA synthesis using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer’s instruction. Real-time qPCR was performed with the ABI 7300 Real-time PCR System (Applied Biosystems, Foster City, USA) using the SYBR ® Premix Ex TaqTM II (TaKaRa, Dalian, China). Primers used in real-time qPCR are: Renilla luciferase forward 5′-GGTAACGCGGCCTCTTCTTA-3′, reverse 5′-AAATGCCAAACAAGCACCCC-3′; Firefly luciferase forward 5′-ACTGGGACGAAGACGAACAC-3′, reverse 5′-GGCGACGTAATCCACGATCT-3′.
Cell migration and invasion assays
Migration of cells was assayed in Transwell cell culture chambers with 6.5-mm diameter polycarbonate membrane filters containing 8 μm pore size (Corning, New York, USA). Briefly, 4×10 4 cells in 100 μL of serum-free medium were added to the upper chamber of the well, and the lower chamber was filled with 600 μL medium containing 20% FBS, and then incubated at 37°C for 10–20 h. The non-migration cells were removed from the upper surface of the membrane with a cotton swab. The filters were then fixed in methanol for 10 min, followed by staining with 10% crystal violet for 15 min. Five random microscopic fields were counted per well, and the mean was determined. For the transwell invasion assay, the membrane of the upper chamber was pre-coated with 50 μL of 2.5 mg/mL matrigel (Falcon/BD, San Diego, USA).
Colony formation assay
One thousand transfected cells were seeded into a 6-well plate and incubated at 37°C with 5% CO 2 for 10–14 days. After being washed with pre-cooled PBS twice, cells were fixed in methanol for 10 min, followed by staining with 10% crystal violet for 15 min. After taking pictures through a microscope, the colonies numbers were counted manually.
Cell proliferation assay
The proliferation ability of different cancer cells was monitored using the xCELLigence Real-Time Cell Analyzer (RTCA)-MP system. (Acea Biosciences/Roche Applied Science, Indianapolis, USA). Culture medium (50 μL) was added to each well of E-Plate 96 (Roche Applied Science) to obtain equilibrium. Transfected cells were incubated in 60 mm culture plates for 24 h, and 2000 cells in 100 μL of culture medium were seeded in E-Plate 96. The E-Plate 96 was locked in the RTCA-MP device at 37°C with 5% CO 2. The cell index directly reflects cellular proliferation on biocompatible microelectrode-coated surfaces. The cell index was read automatically every 15 min, and the recorded curve was shown as the cell index.
Western blot analysis
Cells were lysed in RIPA lysis buffer (Thermo Fisher Scientific) supplemented with protease and phosphatase inhibitor cocktail (Roche, Basel, Switzerland). Protein samples were separated by electrophoresis in a 10%–15% SDS-PAGE gel and transferred to PVDF membranes. The membranes were blocked with 5% BSA. After that, the membranes were incubated with the following primary antibodies: anti-Cyclin B1 antibody (ab32053; Abcam, Cambridge, UK), anti-PTBP1 antibody (12582-1-AP; Proteintech, Rosemont, USA), anti-β-Actin antibody (sc-47778; Santa Cruz, Santa Cruz, USA). After being incubated with HRP-conjugated secondary antibodies (Golden Bridge Biotechnology, Beijing, China), ECL was used to develop the membranes. Membranes were scanned with the ImageQuant LAS (GE Healthcare Life Sciences, Bethesda, USA), and quantification of bands was performed by ImageJ. β-Actin was used as the loading control.
RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) was performed using the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, USA) according to the manufacturer′s instructions. Briefly, 3×10 7 cells were used for one reaction, RIP lysate with the same cell volume was used, mixed with pipetting, and incubated on ice for 5 min, freezed at –80°C overnight, and samples were ready for the subsequent steps.
RNA synthesis in vitro
Linearized plasmids with target sequence were used as templates. Purification was performed by adding 1/20 volumes of 0.5 M EDTA, 1/10 volume of 3 M sodium acetate, 2 volumes of 100% ethanol, and incubated at –80°C for 8 h, followed by centrifugation at 12,000 g at 4°C for 15 min. The plasmid was dissolved in RNase-free water, and the concentration is determined with Nanodrop 2000 spectrophotometer. Biotin-labeled CTP and UTP were used for in vitro transcription. Purification was performed by LiCl precipitation.
RNA pull-down assay
The RNA was incubated at 70°C for 10 min and then put on ice. The beads were washed with 1 mL washing buffer (10 mM HEPES, 50 mM KCl, 1.5 mM MgCl 2, and 0.5% NP-40) three times. Each protein lysate (50 μL) was added and incubated at room temperature for 30 min, followed by centrifugation to remove the supernatant. Biotin-labeled RNA (2 μg) was added to protein lysate with 500 μL hybridization buffer (washing buffer supplemented with 2 mM DTT, 1 mM EDTA, 100 u/mL RNase inhibitor, and 100 μg/mL tRNA) and rotated at room temperature for 1 h. A total of 50 μL beads were added and rotated for 30 min at room temperature, followed by three times wash with washing buffer. The beads were boiled in 1×SDS loading buffer for 5 min, centrifuged, and the supernatant was transferred to a new tube.
Cell cycle assay
Cells were fixed in 70% ethanol at 4°C overnight, washed with PBS twice and resuspended in 300–500 μL PI (Roche)/Triton X-100 staining solution consisting of 10 mL of 0.1% (v/v) Triton X-100 in PBS, 2 mg DNase-free RNase A (Sigma), and 0.4 mL of 500 μg/mL PI. Cells were incubated at 37°C for 15 min, and then analyzed by flow cytometry within 2 h.
In vivo tumor assay
Nude mice were obtained from Vital River Laboratory Animal Technology Co., Ltd (Beijing, China), and all animal care and procedures were in accordance with national and institutional policies for animal health and well-being and approved by Cancer Institute and Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China). KYSE 30 cells were collected by trypsinization when the confluence reached 70%–80%. The cell suspension was transferred to a sterile centrifuge tube and centrifuged at 200 g for 5 min. The cells were washed twice with PBS, and the cell concentration was adjusted to 1×10 7 cells/ml. Cells were re-suspended in FBS-free medium. About 1×10 6 cells were injected into the flank of nude mice by intraperitoneal injection. Thirty days after injection, tumors were harvested and the tumor weight was measured.
Statistical analysis
Data were presented as the mean±standard deviation (SD). Unpaired t-test was used for comparison between two groups and one-way or two-way analysis of variance (ANOVA) for comparison among multiple groups. All analyses were performed using GraphPad Prism 8.0 Software. P<0.05 was considered to be statistically significant.
Results
The 5′UTR of cyclin B1 contains an IRES
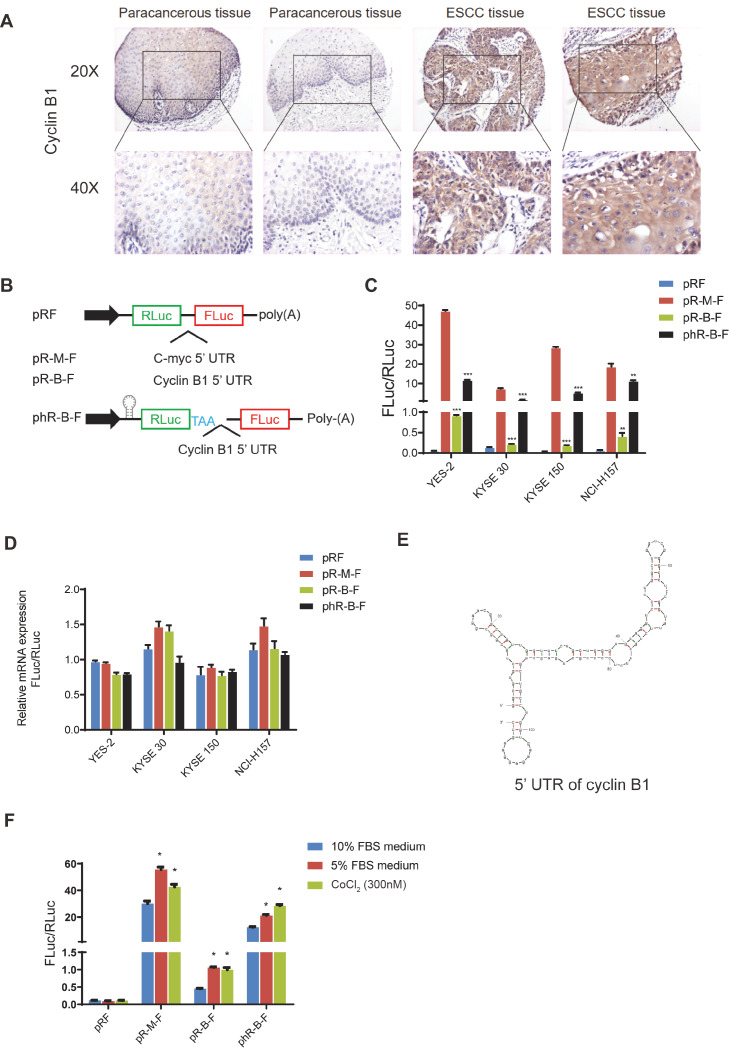
The upregulation of cyclinB1 generally results in tumorigenesis. The previous research in our lab demonstrated that cyclin B1 is aberrantly upregulated in esophageal squamous carcinoma (ESCC) and cyclin B1 promotes the motility and migration of tumors [6]. Overexpression of cyclin B1 antagonizes chemotherapeutic-induced apoptosis through the PTEN/Akt pathway in human ESCC cells [17]. Here we showed the expression of cyclin B1 in ESCC by immunohistochemistry (IHC). The IHC result showed that cyclin B1 expression in ESCC tissue is higher than that in paracancerous tissue ( Figure 1A). The RNA-sequencing data showed that the RNA expression level of cyclin B1 in ESCC tissue was higher than that in normal tissue, but not as dramatic as its protein level (data not shown). These data indicated that the translation of cyclin B1 may be instrumental. In the cyclins family, cyclin D1 plays a crucial role in the G1/S phase. Multiple studies showed that the upregulation of cyclin D1 in cancers is caused by IRES-mediated translation [ 18, 19]. Based on these findings, we hypothesize that the IRES-mediated translation may contribute to the high expression level of cyclin B1 in cancer.
Figure1 .
The 5′UTR of cyclin B1 contains an IRES
(A) The immunohistochemistry of cyclin B1 in esophageal squamous cell carcinoma (ESCC) and paracancerous tissue. (B) The structure of bicistronic fluorescent reporter plasmid. There are two cistrons in the plasmid. The first cistron is Renilla luciferase (RLuc), and the second cistron is Firefly luciferase (FLuc). Sequences of 5′UTR of cyclin B1 and c-myc are inserted into the plasmid between the two luciferases. A hairpin and an additional stop code are inserted flank the first cistron to prevent the read-through translation. (C) Four cell lines were transfected with the reporter plasmids. Dual-Luciferase® Reporter Assay System was used to measure the activity of the Luciferase. The ratio of FLuc and RLuc indicates the activity of the IRES. Data are presented as the mean±SD of five independent experiments. *P<0.05, **P<0.01, ***P<0.001. (D) The mRNA levels of the two luciferases. RNA was extracted from the four cell lines at 48 h after transfection. Data are presented as the mean±SD of three independent experiments of real-time PCR. (E) The secondary structure of 5’UTR of cyclin B1 mRNA predicted by the RNA Folding Form V2.3. (F) KYSE 30 cells were treated with medium contaning 5% FBS or CoCl2 (300 nM) for 48 h. The ratio of FLuc and RLuc indicates the activity of the IRES. Data are presented as the mean±SD of three independent experiments. *P<0.05.
A bicistronic fluorescent reporter plasmid containing the cyclin B1 5′UTR (pR-B-F) was constructed for the IRES activity ( Figure 1B). The Renilla luciferase (RLuc) sequence is the first cistron and the second cistron encodes the Firefly luciferase (FLuc). The ratio of firefly luciferase to Renilla luciferase (FLuc/RLuc) reflects the IRES activity. Based on previous studies [ 20, 21], we constructed a bicistronic fluorescent reporter enzyme plasmid containing c-myc 5′UTR (pR-M-F) as a positive control ( Figure 1B).
In specific scenarios, the bicistronic fluorescent reporter system will have false positives. The second cistron can be translated if the inserted sequence contains hidden promoters or splicing sites. Another scenario is that if read-through occurs in the translation, it can also result in the expression of the second cistron [22]. To exclude the hidden promoter, the mRNA levels of Renilla luciferase and firefly luciferase were measured by real-time qPCR. To exclude the false positives caused by abnormal splicing and read-through, a sequence that could form a stable hairpin structure was inserted before the first cistron and an additional stop codon was added after the first cistron ( Figure 1B).
Three human ESCC cell lines YES-2, KYSE 30, and KYSE 150, as well as one human non-small cell lung adenocarcinoma cell NCI-H157 were transfected with the reporter plasmids. The cells were collected at 48 h after transfection, and the ratio of Firefly luciferase to Renilla luciferase (FLuc/RLuc) was measured by luciferase activity assay ( Figure 1C). The data showed that the reporter plasmid containing cyclin B1 5′UTR has a significantly higher ratio than the negative control (pRF). The phR-B-F plasmid, which has a hairpin structure before the first cistron, has an even higher FLuc/RLuc ratio. This result suggests that there is no false positive caused by variable splicing and read-through ( Figure 1C). To exclude the effect of the hidden promoters, we measured the mRNA level of the two luciferases by real-time qPCR. No significant difference was found between the RNA levels of these two luciferases ( Figure 1D). Typically, for IRES-mediated translation, a cloverleaf-like secondary structure is needed to initiate the process [23]. We generated a predicted secondary structure of the 5′UTR RNA of cyclin B1 by using the RNA Folding Form V2.3 [24]. The cyclin B1 5′UTR can form a cloverleaf-like structure, which is a typical structure of IRES ( Figure 1E). These results demonstrate that the cyclin B1 5′UTR has an IRES.
The IRES activity is further induced under stress conditions such as nutrient deficiency, DNA damage, and hypoxia [25]. To determine the IRES activity of the cyclin B1 under stress conditions, YES-2 cells were transfected with reporter plasmids and cultured in a complete medium containing 10% FBS (as negative control), medium supplemented with 5% FBS, and medium supplemented with 10% FBS and 300 nM CoCl 2. The 5% FBS is a nutrient-deficient condition, and CoCl 2 can induce hypoxia in cells. The cells were harvested after 48 h, and the luciferase activity assay was performed to measure the activity of IRES ( Figure 1F). The results showed that the ratio of Firefly luciferase activity to Renilla luciferase activity (FLuc/RLuc) was significantly increased in cells cultured in medium containing 5% FBS and in cells cultured in medium containing CoCl 2, compared with that in cells cultured in medium containing 10% FBS. These results indicate that the IRES of cyclin B1 is further activated under nutrient deficiency or hypoxia.
PTBP1 promotes IRES-mediated translation of cyclin B1
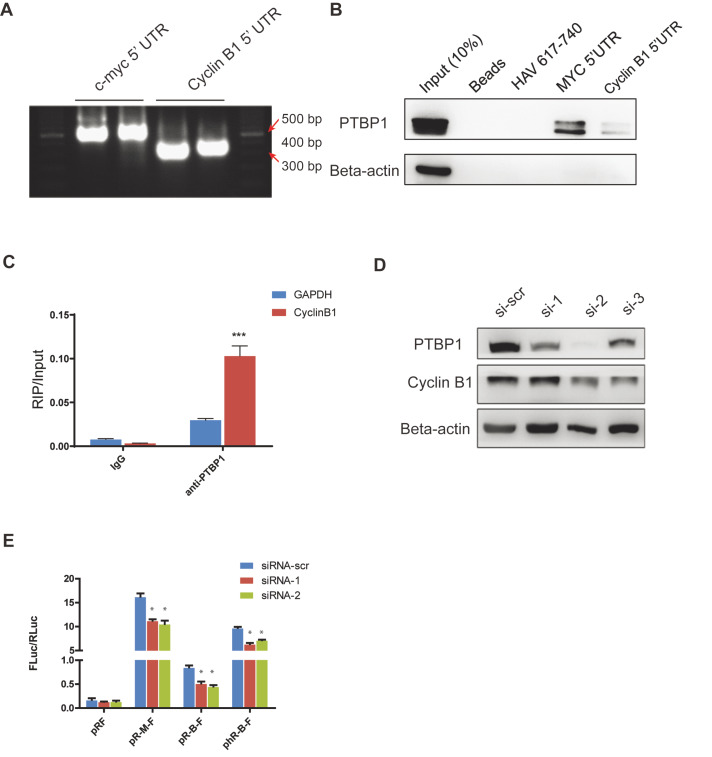
Interacting trans-acting factors (ITAFs) are required by most IRES to initiate the translation. Typically, ITAFs can directly bind to IRES and promote translation. For example, ITAFs such as PTBP1, hnRNPC2/C2, HuR, Unr, PCBP1, and La have been reported to be involved in IRES-mediated translation [ 10, 26]. An RNA pull-down assay was performed to find ITAFs involved in cyclin B1 IRES-mediated translation. The 5′UTR sequence of cyclin B1 was synthesized with biotin-labeled UTP and CTP by in vitro transcription assay ( Figure 2A). After screening several classical ITAFs involved in the IRES initiation (data not shown), RNA pull-down assay showed that PTBP1 binds to the 5′UTR of cyclin B1. Meanwhile, the 5′UTR sequence of c-myc was used as a positive control and the sequence of HAV 617-740 was used as a negative control [ 27, 28] ( Figure 2B). These data were confirmed by RIP assay. PTBP1 was precipitated by a specific antibody, and the bound RNAs were measured by real-time PCR. There was significantly more cyclin B1 bound to the PTBP1 than bound to the control ( Figure 2C). All these results support the conclusion that PTBP1 binds to cyclin B1 5′UTR.
Figure2 .
PTBP1 promotes IRES-mediated translation of cyclin B1
(A) The RNA bands of cyclin B1 and c-myc 5′UTR. RNA sequence of cyclin B1 and c-myc 5′UTR were transcriptions from DNA template using the MEGAscript® kits. CTP and UTP were labeled with biotin. (B) RNA pull-down assay of cyclin B1 5′UTR. The RNA sequences were synthesized by in vitro transcription. Biotin UTPs and CTPs were added to the reaction. The MYC 5′UTR sequence was used as a positive control and the HAV 617-740 sequence as a negative control. (C) RNA IP assay of PTBP1. The experiment was performed by using the Magna RIPTM RNA-Binding Protein Immunoprecipitation kit. The cell lysate was from KYSE 30 cells. Data are shown as the mean±SD of three independent experiments of real-time qPCR. GAPDH is a negative control. ***P<0.001. (D) Knockdown of PTBP1 in KYSE 30 cells. Three siRNAs were used to knock down the PTBP1. (E) Dual-luciferase reporter assay after knockdown of PTBP1. The activity of IRES was measured after the knockdown of PTBP1. The ratio of FLuc and RLuc indicates the activity of the IRES. Data are presented as the mean±SD of five independent experiments. *P<0.05.
PTBP1 belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). PTBP1 is associated with pre-mRNAs in the nucleus and influences pre-mRNA processing and other mRNA metabolism and transport aspects. To further investigate the function of PTBP1 in cyclin B1 translation, we transfected KYSE 30 cells with siRNA of PTBP1. The result showed that the expression level of cyclin B1 was decreased after PTBP1 knockdown ( Figure 2D). To determine whether the decrease of cyclin B1 expression is caused by inhibition of the IRES-mediated translation, we performed the bicistronic fluorescent reporter assay in the KYSE 30 cells after PTBP1 knockdown. As expected, the IRES activity of cyclin B1 was impaired after the knockdown of PTBP1 ( Figure 2E). These results suggest that PTBP1 promotes the IRES-mediated translation of cyclin B1.
PTBP1 is upregulated in tumors and correlated with cyclinB1
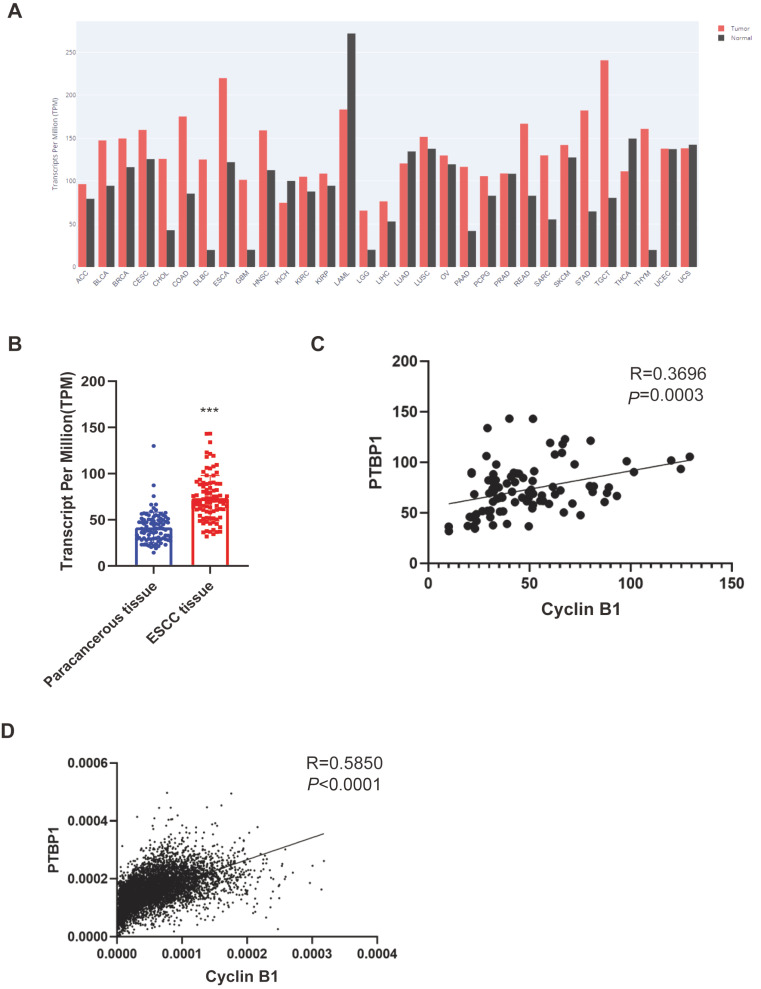
To further investigate the function of PTBP1 and the relationship between PTBP1 and cyclin B1, we used the expression data from the pan-cancer analysis database. The GEPIA website provides a user-friendly analysis and visualization of the results based on the TCGA database. Data showed that PTBP1 is upregulated in most kinds of cancers ( Figure 3A). Several research groups have reported that PTBP1 promotes cancer cell growth, as well as malignant and chemotherapy resistance in breast cancer, pancreatic cancer, and bladder cancer [ 29– 31]. However, the function of PTBP1 in ESCC remains unclear, which made us focus on the function of PTBP1 in ESCC. The RNA-seq data of 91 ESCC patients showed that the expression of PTPB1 was significantly higher in ESCC tissue than in the paracancerous tissue ( Figure 3B). The correlation analysis of PTBP1 and cyclin B1 in 91 ESCC tissue and TCGA databases showed that PTBP1 and cyclin B1 had a positive correlation ( Figure 3C,D). All these findings suggest that PTBP1 plays an important role in ESCC.
Figure3 .
PTBP1 is upregulated in tumors and correlated with cyclin B1
(A) RNA expression of PTBP1 from TCGA and the visualization chart from the GEPIA. (B) RNA-seq data of 91 ESCC patients. Values are tested for statistical difference by paired t-test. ***P<0.0001. (C) Correlation of PTBP1 and cyclin B1 in 91 ESCC tissues analyzed by the Pearson correlation coefficient analysis. R=0.3696, P=0.0003, N=91. (D) Correlation of PTBP1 and cyclin B1 analyzed by Pearson correlation coefficient analysis of pan-cancer database from TCGA. R=0.5850, P<0.0001, N=8386.
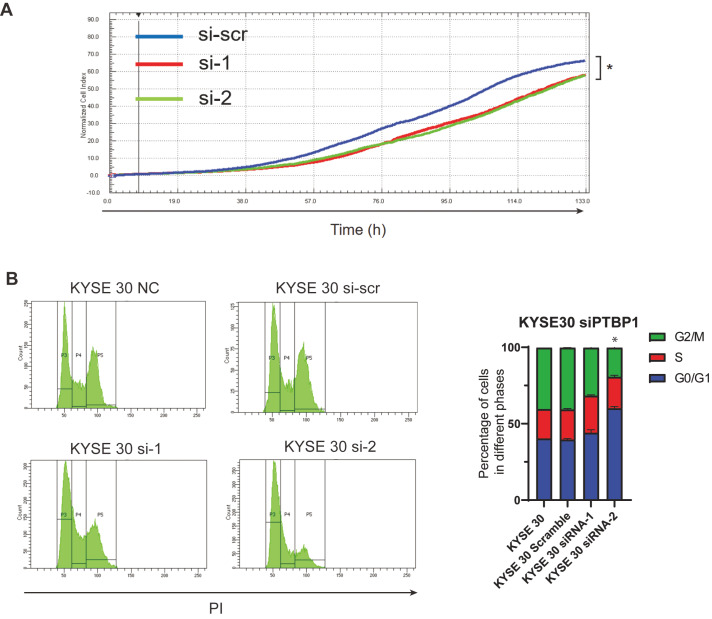
To investigate the function of PTBP1 in ESCC cells, KYSE 30 cells were transfected with PTBP1 siRNA and cell proliferation assay was performed. The cell growth rate was monitored by the xCELLigence RTCA MP system. Results showed that cells with PTBP1 knockdown grew more slowly than cells transfected with the si-scramble ( Figure 4A). Typically, the inhibition of proliferation is caused by abnormal cell cycles of the cell. To determine the cell cycle of PTBP1-knockdown cells, the DNA of cells was stained by PI and analyzed by flow cytometry. The cell cycle assay showed that PTBP1-knockdown cells had more cells in the G0/G1 phase and fewer cells in the G2/M phase than the NC cells and si-scramble cells ( Figure 4B). This result indicates that PTBP1 is required to maintain the accelerated cell cycle and proliferation rate in cancer cells.
Figure4 .
PTBP1 promotes the proliferation of ESCC cells
(A) Cell proliferation assay. KYSE 30 cells were transfected with PTBP1 siRNA. The cell growth rate was monitored by the xCELLigence RTCA MP system. The cell index was normalized by the index of 6 h after plating. *P<0.05, n=5. (B) Cell cycle analysis. KYSE 30 cells were transfected with PTBP1 siRNA. After fixation with 70% ethanol, the nuclei were stained with Propidium Iodide (PI), cells were analyzed by flow cytometry. *P<0.05, n=3. NC: untransfected cells.
PTBP1 promotes malignancy of ESCC cells
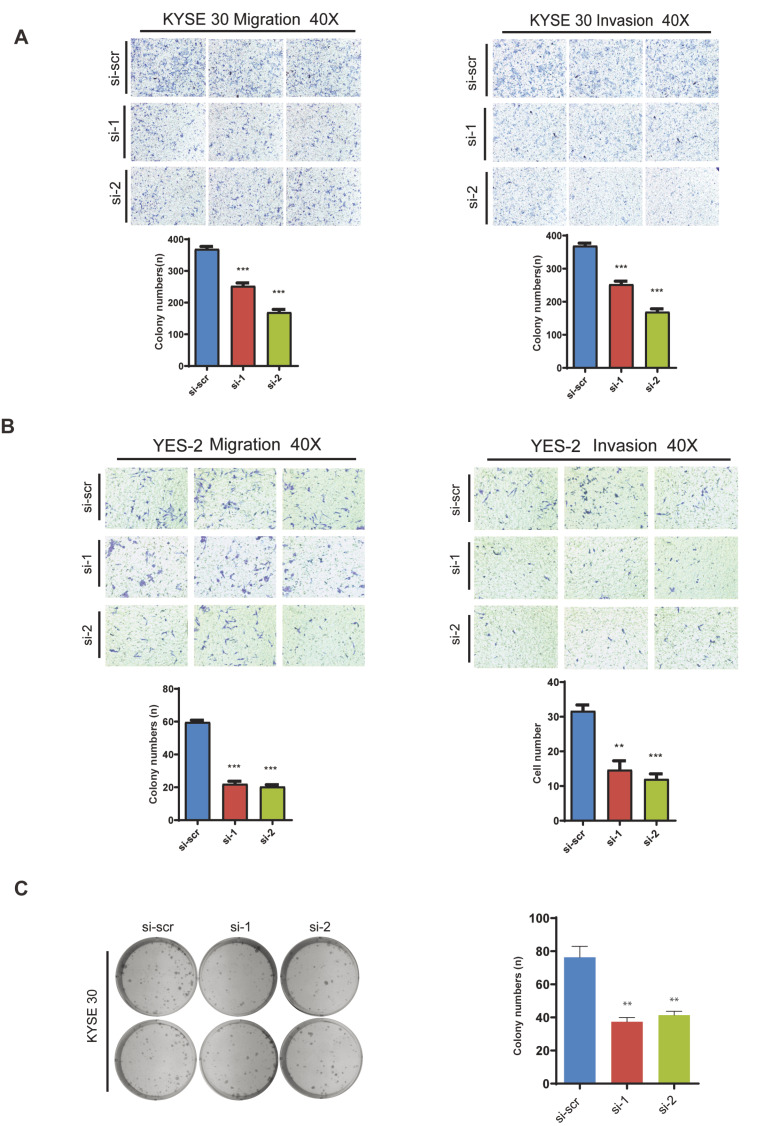
Invasion and migration are two central factors of tumor cells. Most deaths from cancer are caused by metastases. To determine if PTBP1 promotes invasion and migration in ESCC, transwell invasion and migration assays were performed. The data showed that knockdown of PTBP1 resulted in the inhibition of migration and invasion in ESCC cells ( Figure 5A,B). In addition to the increased invasion and migration, unlimited proliferation and increased colony formation abilities are critical for cancer cells to survive and progress. Therefore, colony formation assay was further performed after PTBP1 knockdown, and the result showed that knockdown of PTBP1 impaired the colony formation ability of ESCC cells ( Figure 5C).
Figure5 .
PTBP1 promotes migration, invasion, and colony formation of ESCC cells
(A,B) KYSE 30 and YES 2 cells were transfected with the siRNA of PTBP1. Cell invasion and migration were measured using transwell plates coated with and without matrix gel respectively. Data are presented as the mean±SD of three independent experiments. **P<0.01; ***P<0.001. (C) KYSE 30 cells were transfected with PTBP1 siRNA. Cells were plated into 6-well plates at the density of 1000 cells per well at 24 h after the transfection and cultured for 14 days. The number of colonies was counted. Data are presented as the mean±SD of three independent experiments **P<0.01.
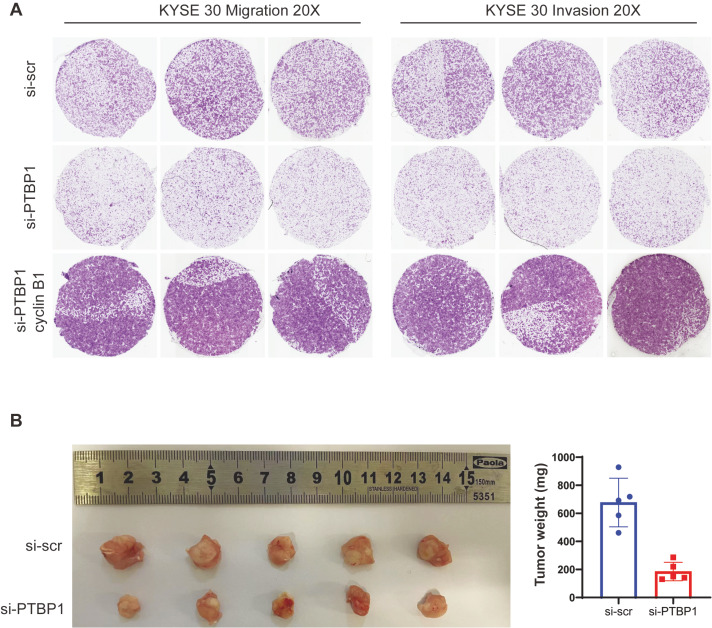
As PTBP1 was found to promote the IRES-mediated translation of cyclin B1, we therefore infer that PTBP1 may act as a cancer-promoter by regulating the expression of cyclin B1. A rescue experiment was performed to verify this hypothesis. After the knockdown of PTBP1, cyclin B1 was overexpressed in the ESCC cells. The invasion and migration assay showed that overexpression of cyclin B1 can reverse the inhibition of invasion and migration in PTBP1-knockdown ESCC cells ( Figure 6A). To confirm the function of PTBP1 in ESCC cancer, the PTBP1-knockdown cells were injected into the nude mice. Data showed that knockdown of PTBP1 impaired the ESCC cell’s tumor growth in vivo ( Figure 6B). These data indicate that PTBP1 has a cancer-promoting function in ESCC cells.
Figure6 .
Overexpression of cyclin B1 can rescue the PTBP1-knockdown phenotype in ESCC cells
(A) Transwell migration and invasion assays. After knockdown of PTBP1, cyclin B1 was overexpressed in the ESCC cells. KYSE 30 cells were used for the transwell migration and invasion assays as previously described. (B) In vivo tumorigenesis assay. A total of 1×106 KYSE 30 cells were injected into the flank of nude mice by intraperitoneal injection. Tumors were harvested 30 days after injection. n=5.
Discussion
The viral IRESs are well understood, but the function and mechanisms of cellular IRES remain to be further explored. Recently, with the improvement of next-generation sequencing and bioinformatics algorithms, a new type of endogenous RNAs has become the focus of the transcriptome research. Circular RNAs (circRNAs) are non-coding RNAs formed by covalently closed loops through back splicing. CircRNAs play essential roles in various biological processes as transcriptional regulators and microRNA sponges which bind with RNA-binding proteins [ 32– 34]. Recent studies have indicated that some cytoplasmic circRNAs can be effectively translated into detectable peptides, and IRES has been suggested as the potential mechanism for circRNA translation [ 35, 36]. These new findings imply that IRES translates not only the canonical protein but also unique peptides. However, the new challenge is that we need better methods to identify the new peptides translated from the circRNA, and we also need more research to understand the functions of these unique peptides. Therefore, IRES-mediated translation is more critical than expected, which may be a potential therapeutic target for cancer treatment.
Sustaining proliferative signaling is one of the hall markers of cancer cells. The abnormal cell cycle is a critical reason for sustaining proliferation. Cyclin B1, as an essential cyclin-dependent protein, plays a crucial role in the G2/M transition. In this study, we reported that the 5′UTR of cyclin B1 contains an IRES. The IRES activity of cyclin B1 is further activated under stress conditions. We propose that the IRES-mediated translation of cyclin B1 is one of the reasons that cyclin B1 is overexpressed in cancer cells.
Furthermore, we demonstrated that PTBP1 can promote cyclin B1 IRES-mediated translation by binding to the 5′UTR of cyclin B1 transcript. Our work also revealed that PTBP1 is upregulated, which promotes cancer malignancy in ESCC cells. Our results suggest that PTBP1 may be a cancer-promoting gene in ESCC cells by promoting the IRES-mediated translation of cyclin B.
The cap-dependent translation is generally suppressed in cancer cells because most cancer cells are under stress conditions. In addition, IRES-mediated translation is activated in cancer cells as a supplementary of cap-dependent translation. Cyclin B1 has been found to be overexpressed in multiple cancers and promote cancer progression. However, due to the important role of cyclin B1 in cell cycle, it is hard to use cyclin B1 as a therapeutic target, because the cell cycle of a normal cell will also be affected, and this will cause unpredictable side effects. In contrast, we can use IRES-mediated translation as a therapeutic target, which can suppress cyclin B1 expression in cancer cells and keep normal cells unaffected.
In conclusion, we provide new insights into the mechanism of cyclin B1 overexpressed in cancer cells. IRES-mediated translation may be a new and better potential therapeutic target which can inhibit cyclin B1 expression with less toxicity and side effects.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Natural Science Foundation of China (Nos. 81472661 and 81872398).
References
- 1.Begnami MD, Fregnani JHTG, Nonogaki S, Soares FA. Evaluation of cell cycle protein expression in gastric cancer: cyclin B1 expression and its prognostic implication. Hum Pathol. . 2010;41:1120–1127. doi: 10.1016/j.humpath.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Androic I, Krämer A, Yan R, Rödel F, Gätje R, Kaufmann M, Strebhardt K, et al. Targeting cyclin B1 inhibits proliferation and sensitizes breast cancer cells to taxol. BMC Cancer. . 2008;8:1. doi: 10.1186/1471-2407-8-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou L, Li J, Zhao YP, Cui QC, Zhou WX, Guo JC, You L, et al. The prognostic value of Cyclin B1 in pancreatic cancer. Med Oncol. . 2014;31:107. doi: 10.1007/s12032-014-0107-4. [DOI] [PubMed] [Google Scholar]
- 4.Aaltonen K, Amini RM, Heikkilä P, Aittomäki K, Tamminen A, Nevanlinna H, Blomqvist C. High cyclin B1 expression is associated with poor survival in breast cancer. Br J Cancer. . 2009;100:1055–1060. doi: 10.1038/sj.bjc.6604874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang A, Yoshimi N, Ino N, Tanaka T, Mori H. Overexpression of cyclin B1 in human colorectal cancers. J Cancer Res Clin Oncol. . 1997;123:124–127. doi: 10.1007/BF01269891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song Y, Zhao C, Dong L, Fu M, Xue L, Huang Z, Tong T, et al. Overexpression of cyclin B1 in human esophageal squamous cell carcinoma cells induces tumor cell invasive growth and metastasis. Carcinogenesis. . 2008;29:307–315. doi: 10.1093/carcin/bgm269. [DOI] [PubMed] [Google Scholar]
- 7.Komar AA, Hatzoglou M. Cellular IRES-mediated translation. Cell Cycle. . 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. . 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 9.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. . 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 10.Jang SK, Kräusslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation . J Virol. . 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macejak DG, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. . 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 12.Mokrejš M, Masek T, Vopálensky V, Hlubucek P, Delbos P, Pospísek M. IRESite—a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res. . 2010;38:D131–D136. doi: 10.1093/nar/gkp981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, et al. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. . 2016;351:aad4939. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 14.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, et al. 5′ UTR m(6)A promotes cap-independent translation. Cell. . 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dresios J, Chappell SA, Zhou W, Mauro VP. An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat Struct Mol Biol. . 2006;13:30–34. doi: 10.1038/nsmb1031. [DOI] [PubMed] [Google Scholar]
- 16.Stoneley M, Chappell SA, Jopling CL, Dickens M, MacFarlane M, Willis AE. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol. . 2000;20:1162–1169. doi: 10.1128/MCB.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou Y, Ma L, Ma L, Huang Z, Zhou W, Zhao C, Zhang B, et al. Overexpression of cyclin B1 antagonizes chemotherapeutic-induced apoptosis through PTEN/Akt pathway in human esophageal squamous cell carcinoma cells. Cancer Biol Ther. . 2012;14:45–55. doi: 10.4161/cbt.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jo OD, Martin J, Bernath A, Masri J, Lichtenstein A, Gera J. Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J Biol Chem. . 2008;283:23274–23287. doi: 10.1074/jbc.M801185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi YJ, Sharma A, Wu H, Lichtenstein A, Gera J. Cyclin D1 and c-myc internal ribosome entry site (IRES)-dependent translation is regulated by AKT activity and enhanced by rapamycin through a p38 MAPK- and ERK-dependent pathway. J Biol Chem. . 2005;280:10964–10973. doi: 10.1074/jbc.M407874200. [DOI] [PubMed] [Google Scholar]
- 20.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. . 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 21.Chappell SA, LeQuesne JP, Paulin FE, deSchoolmeester ML, Stoneley M, Soutar RL, Ralston SH, et al. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: A novel mechanism of oncogene de-regulation. Oncogene. . 2000;19:4437–4440. doi: 10.1038/sj.onc.1203791. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Wang Z. IRES-mediated cap-independent translation, a path leading to hidden proteome. J Mol Cell Biol. . 2019;11:911–919. doi: 10.1093/jmcb/mjz091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez-Salas E, Quinto SL, Ramos R, Fernández-Miragall O. IRES elements: features of the RNA structure contributing to their activity. Biochimie. . 2002;84:755–763. doi: 10.1016/s0300-9084(02)01408-6. [DOI] [PubMed] [Google Scholar]
- 24.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. . 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godet AC, David F, Hantelys F, Tatin F, Lacazette E, Garmy-Susini B, Prats AC. IRES trans-acting factors, key actors of the stress response. Int J Mol Sci. . 2019;20:924. doi: 10.3390/ijms20040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoneley M, Subkhankulova T, Le Quesne JP, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. . 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol Cell. . 2003;11:757–771. doi: 10.1016/S1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell SA, Brown EC, Coldwell MJ, Jackson RJ, Willis AE. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol Cell Biol. . 2001;21:3364–3374. doi: 10.1128/MCB.21.10.3364-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calabretta S, Bielli P, Passacantilli I, Pilozzi E, Fendrich V, Capurso G, Fave GD, et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. . 2016;35:2031–2039. doi: 10.1038/onc.2015.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He X, Arslan AD, Ho TT, Yuan C, Stampfer MR, Beck WT. Involvement of polypyrimidine tract-binding protein (PTBP1) in maintaining breast cancer cell growth and malignant properties. Oncogenesis. . 2014;3:e84. doi: 10.1038/oncsis.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bielli P, Panzeri V, Lattanzio R, Mutascio S, Pieraccioli M, Volpe E, Pagliarulo V, et al. The splicing factor PTBP1 promotes expression of oncogenic splice variants and predicts poor prognosis in patients with non–muscle-invasive bladder cancer. Clin Cancer Res. . 2018;24:5422–5432. doi: 10.1158/1078-0432.CCR-17-3850. [DOI] [PubMed] [Google Scholar]
- 32.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. . 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. . 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. . 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. . 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petkovic S, Müller S. RNA circularization strategies i n vivo and in vitro . Nucleic Acids Res. . 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]