Abstract
Recent studies propose that T follicular helper (Tfh) cells possess a high degree of functional plasticity in addition to their well‐defined roles in mediating interleukin‐4‐dependent switching of germinal center B cells to the production of immunoglobulin (Ig)G1 and IgE antibodies. In particular Tfh cells have been proposed to be an essential stage in Th2 effector cell development that are able to contribute to innate type 2 responses. We used CD4‐cre targeted deletion of BCL6 to identify the contribution Tfh cells make to tissue Th2 effector responses in models of atopic skin disease and lung immunity to parasites. Ablation of Tfh cells did not impair the development or recruitment of Th2 effector subsets to the skin and did not alter the transcriptional expression profile or functional activities of the resulting tissue resident Th2 effector cells. However, the accumulation of Th2 effector cells in lung Th2 responses was partially affected by BCL6 deficiency. These data indicate that the development of Th2 effector cells does not require a BCL6 dependent step, implying Tfh and Th2 effector populations follow separate developmental trajectories and Tfh cells do not contribute to type 2 responses in the skin.
Keywords: BCL6, T cell plasticity, Tfh, Th2 differentiation
Tfh cells have been proposed to be an important stage in the development of Th2 effector cells. We show using Bcl6 deficient mice and in vivo cytokine reporter methodology that Th2 effector cells do not require a Tfh step when primed with allergens in the skin.
INTRODUCTION
The cellular and molecular events surrounding the allergen induced differentiation of CD4 T follicular helper (Tfh) cells requires the expression of the transcription factor (TF) BCL6, 1 , 2 the surface expression of CXCR5 enabling the retention Tfh cells in the germinal centers and the production of the cytokines interleukin (IL)‐21 and IL‐4 for driving B cell class switching to immunoglobulin (Ig)G1 and IgE. 3 , 4 , 5 , 6 , 7 Although Tfh cells are classically regarded as a fully differentiated CD4 T cell subset, recent reports indicate that in some experimental models Tfh retain a degree of functional plasticity, are capable of expressing canonical cytokines of all Th lineages and can circulate in the blood and retain the ability to differentiate into effector memory cells. 8 Tfh cells have been shown to differentiate from Th2 committed precursors in the draining lymph node (dLN) after immunization with Schistosoma mansoni eggs. 7 Additionally, Tfh cells have been reported to act as precursors of Th2 effector (Th2eff) cells in lung tissue. 9 In contrast to this, multiple studies report no impact on tissue CD4+ Teff cells in the absence of Tfh. It has been shown that IL‐21+ Tfh cells cannot differentiate into IL‐4/IL‐5/IL‐13 producing Th2eff cells, but can mature into IL‐21+ Teff cells. 10 Using a mixed bone marrow chimera model, it has been shown that BCL6 deficient T cells retain the ability to develop into house dust mite (HDM) specific Teff cells in the lung. 11 This is not limited to Th2 cells as Tfh deficient mice showed equivalent frequency and numbers of RSV‐specific CD4+ Teff cells in the lung. 12 These studies support the hypothesis that BCL6 dependent Tfh cells do not give rise to Teff cells in the tissue. The view that Tfh and Th2 follow different developmental pathways is supported by the observation that Tfh IL‐4 production is regulated by the c‐MAF/NFAT pathway, independently of GATA3, 13 explaining why IL‐4 expression in Tfh is not perturbed by the absence of IL‐4 signaling. 14 In summary, the differentiation potential of Tfh cells following allergen sensitization and their relative contribution to the pool of tissue Th2eff cells after Type 2 sensitization remains unclear and worthy of further investigation.
The Il4/Il13 reporter mouse strain (4C13R) 15 was bred to mice which either retained or had deleted the Tfh subset through CD4cre‐cre/flox specific targeting of BCL6. Local immunization of this novel BCL6fl/fl.4C13R.CD4cre strain allowed the number of allergen‐induced Th2eff cells that accumulated in tissues to be compared in both the presence and absence of the BCL6‐mediated Tfh developmental pathway and to identify what contribution Tfh make to Th2eff responses in the skin and lung. 16
RESULTS
Tfh development is specifically ablated in BCL6fl /fl.CD4cre +/− mice
To investigate the role of Tfh cells in type 2 tissue immune responses, we used an established model of skin Type 2 immunity involving intradermal (i.d.) injection of house dust mite (HDM) allergens into the ear pinna of mice. 16 In this model a significant CD4 T cell response occurs after 7 days in the ear draining auricular lymph node (aLN) and is accompanied by a striking Th2 mediated inflammatory response in the ear tissue. 16 The Tfh cells were identified in the aLN as CD4+CD44hiCXCR5hiPD‐1+ T cells and non‐Tfh were identified as CD4+CD44hiCXCR5− T cells (Supplementary figure 1a). Compared with control PBS injected mice, HDM sensitization of the ear pinna triggered a robust proliferation and expansion in the numbers of non‐Tfh and Tfh cells in the draining aLN of BCL6 sufficient, BCL6fl/fl.CD4cre−/− (BCL6fl/fl) mice. By contrast, HDM‐sensitized BCL6 deficient, BCL6fl/fl.CD4cre+/− (BCL6cKO) mice failed to develop any Tfh cells in the draining aLN, while the expansion of the non‐Tfh cells was unaffected in these BCL6 deficient mice (Figure 1a, b). Tfh cells are essential for providing “help” to B cells via CD40:CD40L signaling to drive B cell activation, maturation and immunoglobulin class switching 3 , 4 and as expected HDM‐sensitized BCL6 sufficient, BCL6fl/fl.CD4cre−/− (BCL6fl/fl) mice had significantly increased levels of serum IgG1 and IgG2a compared with HDM‐sensitized BCL6cKO mice and control PBS injected mice (Figure 1c). These data demonstrate that CD4cre‐mediated deletion of BCL6 abrogates the development of Tfh cells in the aLN after HDM sensitization in the ear pinna and confirms the essential role of BCL6 in Tfh development in this Th2 model.
Figure 1.
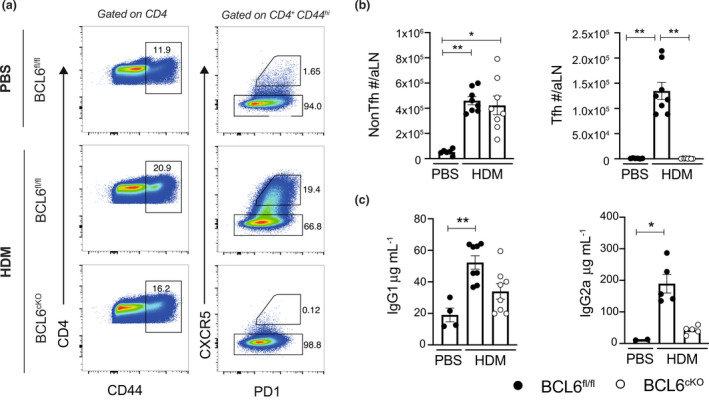
BCL6cKO mice lack Tfh cells, impeding the IgG response. BCL6fl/fl and BCL6cKO mice were sensitized with 200 μg HDM or 30 μL PBS i.d. in the ear pinna. (a) Flow cytometry depicting CD44hi CD4 T cells and Tfh/non‐Tfh cells by CXCR5 and PD‐1 expression day 7 post HDM. (b) Total number of CD4+CD44hi non‐Tfh (CXCR5−) cells and Tfh cells (CXCR5hiPD1+) in aLN day 7 post HDM. (c) IgG1 and IgG2a concentration in serum day 14 post HDM sensitization. Black circles represent BCL6fl/fl, white circles represent BCL6cKO samples. Data are shown as mean ± s.e.m. for six ears per group from one of five repeats that gave similar results except for ELISA data which shows five mice per group from one of two repeats. *P < 0.05, **P < 0.01, not significant is not shown. P‐values were determined using a Kruskal‐Wallis test applying Dunn's multiple comparison post‐test.
Non‐Tfh cells show reduced expression of IL‐4 in BCL6cKO . 4C13R mice
To investigate the impact of BCL6 deficiency on the activation of non‐Tfh cells in the aLN, we crossed the BCL6fl/fl.CD4cre+/− strain onto the 4C13R reporter mouse strain to generate BCL6cKO.4C13R mice that lack Tfh cells and report the expression of IL‐4 and IL‐13 by Th2 cells via AmCyan (IL4‐AC) and DsRed (IL13‐DR), respectively. BCL6 sufficient, BCL6fl/fl.4C13R littermates were used as controls. HDM‐sensitized BCL6 sufficient mice significantly increased the frequency and total numbers of non‐Tfh cells expressing IL4‐AC+ in the draining aLN. However, no IL13‐DR+‐expressing non‐Tfh cells could be detected in the in the aLN. Interestingly in HDM‐immunized BCL6 deficient mice, both the frequency and total count of IL4‐AC+ non‐Tfh Th2 cells in the aLN were reduced back to non‐sensitized levels (Figure 2a, b). Of note, we failed to detect expression of IL13‐DR in Tfh cells in our model, in contrast to previous reports of an IL‐13+ high affinity IgE‐promoting Tfh cell. 17 , 18 We hypothesize this is due to the single‐prime sensitization model we employ and suggest that the induction of rare IL‐13+ Tfh cells only occurs in chronic/anaphylactic models of type 2 immunity. When enumerated, these data reveal that non‐Tfh and Tfh cell compartments evenly contribute to the IL‐4+ CD4 T cell pool and that this pool is dramatically reduced in BCL6cKO mice after HDM sensitization (Figure 2d).
Figure 2.
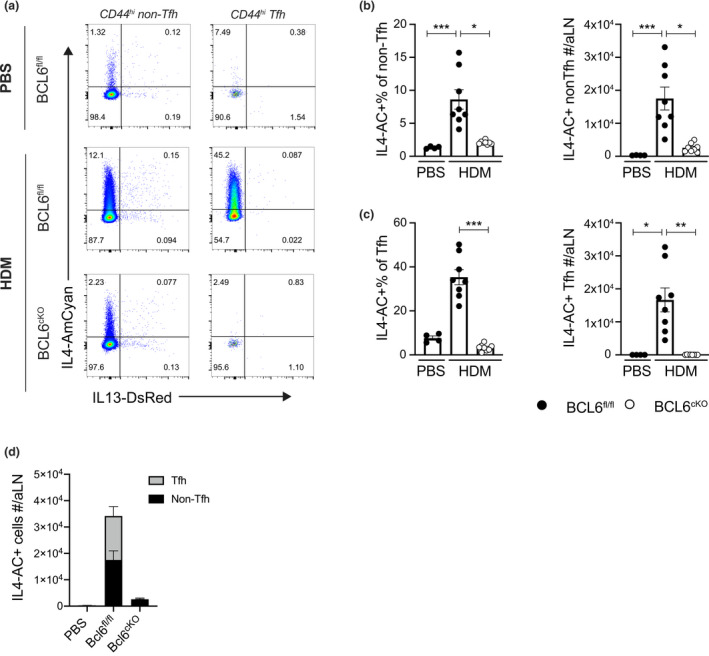
Non‐Tfh cells express reduced levels of IL4‐AC in BCL6cKO.4C13R mice. BCL6fl/fl.4C13R and BCL6cKO.4C13R mice were injected in the ear pinna with 200 μg HDM in 30 μL PBS or PBS alone and cells from the draining aLNs were harvested for analysis on day 7. (a) Flow cytometry plots depicting the expression of IL4‐AC and IL13‐DR in non‐Tfh and Tfh cells. (b, c) Frequency and total number of IL4‐AC+ non‐Tfh (b) and IL4‐AC+ Tfh cells (c). (d) Total number of IL4‐AC+ non‐Tfh and Tfh cells. Black circles represent BCL6fl/fl, white circles represent BCL6cKO samples. Data are shown as mean ± s.e.m. for six aLN per group from one of five repeats that gave similar results. *P < 0.05, **P < 0.01, ***P < 0.001, not significant is not shown. P‐values were determined using a Kruskal‐Wallis test applying Dunn's multiple comparison post‐test.
Taken together our experiments indicate that a clearly defined population of IL‐4+ non‐Tfh Th2 cells appears in the aLN after HDM sensitization, which is in contrast to other studies which observe that 100% of IL‐4+ cells in the dLN are Tfh cells. 9 We also show that BCL6 deficiency in CD4 T cells reduces the number of HDM inducible non‐Tfh cells that express IL‐4.
Skin Th2 effector CD4 cells develop in CD4cre +/− . BCL6cKO .4C13R mice
To assess the impact of Tfh depletion on Th2 cells in tissues, we looked at the effect of BCL6 deletion on CD4 T cell accumulation in the ear pinnae of HDM‐sensitized mice. The CD4 T cells in the ear tissue following allergen sensitization were gated as live, CD45+\CD64−Ly6G−SiglecF−CD11b−Ly6C−TCRγδ−CD3+CD4+ cells (Supplementary figure 1b). HDM sensitization triggered a robust CD4 T cell response in the skin tissue of BCL6fl/fl and BCL6cKO mice, compared with control PBS injected mice. Interestingly, the total number of CD4 T cells trended to being increased compared with BCL6fl/fl controls (Figure 3a). We next used the BCL6cKO.4C13R mouse strain to investigate the effect of BCL6 deletion on the development of Th2eff subsets expressing IL4‐AC+ and IL13‐DR. 14 The Th2eff subsets that can be detected in the skin of these immunized mice include IL4‐AC+ single‐positive (IL4‐AC SP), IL4‐AC+IL13‐DR+ double‐positive (IL4‐AC.IL13‐DR DP), and IL13‐DR+ single positive (IL13‐DR SP) CD4 T cells. The frequency of each of the Th2eff subsets recruited to the ear pinna of HDM‐sensitized mice was unaffected in BCL6cKO.4C13R mice compared with BCL6fl/fl.4C13R (Figure 3b, c). However, the total count of all three Th2eff subsets were significantly increased due to the modest increase in total CD4 T cell accumulation (Figure 3d). This shows that the lack of Tfh cells in BCL6cKO mice does not impede the development of skin Th2eff cell subsets, suggesting that Tfh cells do not contribute to the Th2eff cell pool in the skin in response to HDM sensitization.
Figure 3.
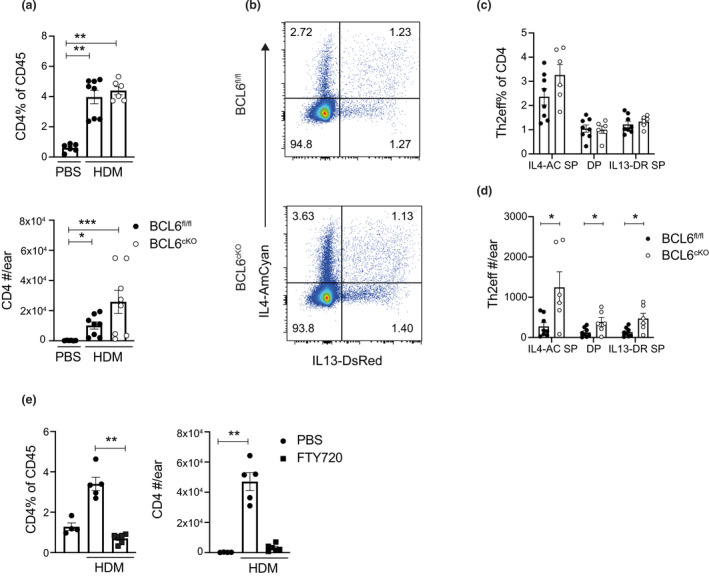
Skin Th2eff develop independently of Tfh cells. Mice were sensitized with injected in the ear pinna with 200 μg HDM in 30 μL PBS or PBS alone. Ear skin tissue was collected for analysis on day 7. (a) Frequency and total number of CD4 T cells in ear tissue from BCL6fl/fl.4C13R and BCL6cKO.4C13R mice. (b) Flow cytometry plots depicting the expression of IL4‐AC and IL13‐DR in CD4 T cells from BCL6fl/fl.4C13R and BCL6cKO.4C13R mice. (c) Enumerated frequencies and (d) total number of Th2eff cell subsets from BCL6fl/fl.4C13R and BCL6cKO.4C13R mice. (e) Frequency and total number of CD4 T cells from C57Bl/6J naïve control mice and mice that were injected with 25 μg FTY720 in PBS or PBS alone on days 2 and 4 post HDM sensitization. Black circles represent BCL6fl/fl, white circles represent BCL6cKO samples. Data are shown as mean ± s.e.m. for six ears per group from one of five repeats that gave similar results. *P < 0.05, **P < 0.01, ***P < 0.001. P‐values for a and e were determined using a Kruskal‐Wallis test applying Dunn's multiple comparison post‐test. P‐values for c and d were determined using a Mann–Whitney U‐test, independently for each Th2eff subset.
To verify that the CD4 T cells accumulating in the skin of HDM allergen‐sensitized mice were derived from the dLN, the egress of lymphocytes was blocked using the sphingosine‐1‐phosphate (S1P) agonist, FTY720. The recruitment of CD4 T cells to the skin tissue was significantly impaired when mice were treated with FTY720 compared with PBS‐treated controls (Figure 3e), indicating that skin CD4 T cells are derived from the aLN.
Taken together, these data show that skin Th2eff cells are primed in the LN, egress and migrate to the skin tissue and that this process is not dependent on a BCL6 mediated step. This suggests that skin Th2eff cells are derived from a non‐Tfh precursor that was primed in the draining aLN.
BCL6 deficiency leads to minimal transcriptional changes in skin Th2eff cells
To ensure that the CD4 Th2eff cells appearing in the skin of HDM‐immunized BCL6cKO.4C13R mice were unaffected by the BCL6 deficiency, we compared the transcriptional profile of each of the defined Th2eff subsets. Ultra‐Low‐Input‐RNA‐seq (ULI‐RNA‐seq) was performed on purified IL4‐AC SP; IL4‐AC.IL13‐DR DP and IL13‐DR SP Th2eff subsets from HDM‐sensitized BCL6cKO.4C13R and BCL6fl/fl.4C13R control mice and gene expression patterns were compared. Ear tissue was harvested and 1000 cells of each skin Th2eff subset were sorted in triplicate for ULI‐RNA‐seq transcriptome analysis. 19 , 20 As CD4 T cells in the ear tissue of naïve mice are rare, naïve CD4 T cells from aLN were sorted and used as a control.
Principal component analysis (PCA) of BCL6cKO.4C13R and BCL6fl/fl.4C13R Th2eff subsets revealed that the PC1 axis accounted for 32.1% of the variance primarily based on the distinct Th2eff subsets. IL4‐AC SP cells clustered on the left of the PCA plot, whilst IL4‐AC.IL13‐DR DP and IL13‐DR SP subsets fell on the right, suggesting that IL4‐AC SP cells are transcriptionally distinct from IL13‐DR+ cells. The PC2 axis accounted for 15.4% of the variance; however, no meaningful separation was noted via PC2. BCL6 sufficiency/deficiency did not influence PCA clustering on the PC1 or PC2 axis; however, samples did segregate based on BCL6 genotype on the PC3 axis that accounted for 9.8% of the variation between samples. BCL6fl/fl samples clustered at the top of the PC3 axis BCL6cKO samples that clustered at the bottom (Figure 4a). This analysis reveals that the primary influence on the transcriptional profile of Th2eff cell subsets was the expression of IL13‐DR, followed by the absence/presence of BCL6.
Figure 4.
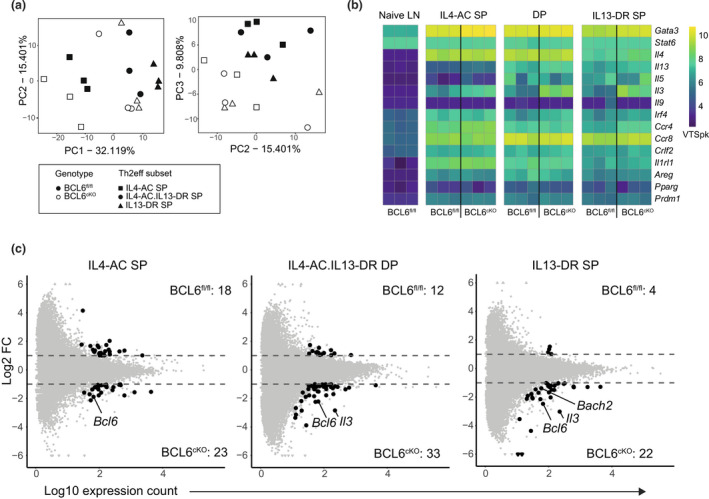
BCL6 deficiency causes minimal transcriptional changes in skin Th2eff cells. Skin Th2eff subsets from BCL6fl/fl.4C13R and BCL6cKO.4C13R mice were harvested from draining aLN and sorted 7 days after HDM sensitization and ULI‐RNAseq was performed. (a) PCA of skin Th2eff subsets, performed on top 1000 highest variance genes for PC1/PC2 and PC2/PC3 axis. BCL6fl/fl samples = black; BCL6cKO samples = white; IL4‐AC SP = square; IL4‐AC.IL13‐DR DP = circle; IL13‐DR SP = triangle. (b) Heatmap showing VSTpk values of canonical Th2 genes in skin Th2eff subsets and naïve LN CD4 T cells. (c) MA plots of skin Th2 subsets comparing BCL6fl/fl.4C13R and BCL6cKO.4C13R samples. Black dots represent DEGs with a log2 FC > 1 or > −1; P‐value < 0.05; and minimum mean read count > 30. Gray dots do not meet differential expression statistical thresholds.
To assess the specific transcriptional regulation of BCL6 in each Th2eff cell subset, the expression of core genes associated with a Th2 phenotype were compared between BCL6cKO.4C13R and BCL6fl/fl.4C13R littermate controls (Figure 4b). Expression values were normalized against gene length and represented as “variance stabilized transcripts per kilobase” (VSTpk). Naïve CD4 T cells isolated from aLN were used as controls. All skin Th2eff cells showed increased expression of key Th2 genes compared with naïve aLN control CD4 T cells. Importantly, Gata3 expression was increased in all Th2eff subsets compared with naïve LN CD4 T cells, indicating that skin Th2eff cells represent the conventional Th2eff subset. Despite the critical role of STAT6 in Th2 cell development, Stat6 was similarly expressed in naïve and Th2eff cells. This is not of concern given that Stat6 mRNA expression does not represent active phosphorylated STAT6. Il4 was expressed highly in both IL4‐AC SP and IL4‐AC.IL13‐DR DP subsets and was expressed at relatively low levels in the IL13‐DR SP subsets and not expressed at all in naïve LN cells. Il13 was expressed highly in IL13‐DR SP and IL4‐AC.IL13‐DR DP subsets and expressed at low to undetectable levels in IL4‐AC SP cells and naïve LN CD4 T cells respectively. Other core Th2 genes including Il5, Il3, Irf4, Ccr4, Ccr8, Crlf2, Il1rl1, Areg, Pparg and Prdm1 were upregulated in all some, if not all, skin Th2eff cell subsets compared with naïve LN controls. Interestingly, Il9 was not detected in any sample, indicating that IL‐9 is not associated with Th2eff cells in our models. Of note, Il5 expression was increased in IL13‐DR+ Th2eff subsets, compared with IL4‐AC SP cells, indicating the linked association between the expression of Il13 and Il5 in skin Th2eff cells. 21 , 22 , 23 These results indicate that IL4‐AC and IL13‐DR reporter expression in 4C13R mice accurately reflects the endogenous Il4 and Il13 gene expression and that all cell subsets sequenced represent bonafide Th2eff cells.
To investigate the impact of BCL6 deficiency on skin Th2eff cells in more depth, differential gene expression analysis was performed on the RNA‐seq data comparing BCL6fl/fl and BCL6cKO samples for each Th2eff subset. Differentially expressed genes (DEGs) were defined as having a log2‐fold change (FC) > 1 or < −1, a P‐value < 0.05 and a minimum mean count > 30. This analysis resulted in less than 50 DEGs between BCL6fl/fl and BCL6cKO samples for each Th2eff subset comparison (Figure 4c, Supplementary tables 1–3), indicating that BCL6 deficiency has a minor effect on the transcriptional profile of Th2eff cells appearing in the skin after HDM sensitization. BCL6 deficiency triggered the upregulation of more genes in Th2eff subsets than downregulation, supporting the role of BCL6 as a transcriptional repressor in Th2eff cells. The expression of Bcl6 was significantly upregulated in all Th2eff subsets of BCL6 deficient BCL6cKO.4C13R mice (Figure 4c). Furthermore, intra‐nuclear transcription factor staining shows that BCL6 is also significantly upregulated at the protein level in BCL6cKO.4C13R mice (Supplementary figure 2a, b). The increased level of Bcl6 reads in BCL6cKO samples has been previously shown to be due to the negative autoregulation of Bcl6. 24 Supplementary figure 2c shows that the increased Bcl6 reads map to exons 1–6 and exon 10, not exons 7–9, which are selectively excised by CD4cre mediated targeting in BCL6cKO mice. 25 This validates the flox/cre system. Bcl6 exons 7–9 encode several critical zinc finger motifs that mediate DNA binding, therefore the deletion of exons 7–9 results in a dysfunctional BCL6 protein that is reflected by the absence of Tfh cells. Therefore, the loss of functional BCL6 protein promotes enhanced transcription of Bcl6, resulting in increased Bcl6 RNA‐seq reads in BCL6cKO samples. Bach2 and Il3 were also significantly upregulated in BCL6cKO IL13‐DR+ Th2eff subsets (Figure 4c), indicating that BCL6 represses the expression of these genes.
Recruitment of type 2 granulocytes is increased in BCL6cKO .4C13R mice
IL‐3 is a canonical type 2 cytokine expressed by effector Th2 cells and is crucial for basophil recruitment and eosinophil survival. 26 , 27 The observation that Il3 mRNA was increased in Th2eff samples from BCL6cKO.4C13R mice led us to investigate whether there was an associated basophil and eosinophil response in our skin HDM model. Strikingly, both basophils and eosinophils in the ear tissue were significantly enriched in BCL6cKO.4C13R mouse ear tissues after HDM sensitization compared with BCL6fl/fl.4C13R counterparts at all time points analyzed (Figure 5a). Furthermore, the MFI of the activation marker CD11b 28 , 29 was not affected in BCL6cKO.4C13R and BCL6fl/fl.4C13R controls (Figure 5b). This demonstrates that allergen activation of BCL6 deficient CD4 T cells triggers an enhanced recruitment of basophils and eosinophils into the skin tissue, but their activation as defined by CD11b upregulation was not affected.
Figure 5.
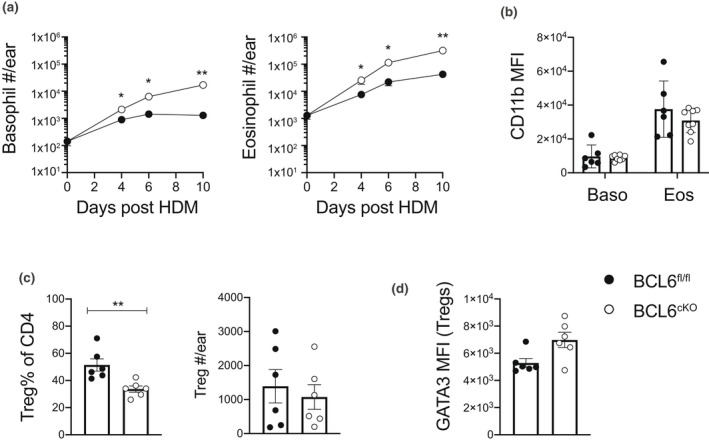
Basophil and eosinophil recruitment to the skin is enhanced in BCL6cKO.4C13R mice and correlates to reduced frequency of Treg cells. BCL6fl/fl.4C13R and BCL6cKO.4C13R mice injected in the ear pinna with 200 μg HDM in 30 μL PBS or PBS alone. Ear skin tissue was collected for analysis on day 7 unless indicated. (a) Kinetics of total number of basophils and eosinophils detected in ear tissue. (b) CD11b MFI in basophils and eosinophils. (c) Frequency and total number of FOXP3+ Treg cells in ear tissue. (d) GATA3 MFI in FOXP3+ Treg cells in ear tissue. Black circles represent BCL6fl/fl, white circles represent BCL6cKO samples. Data are shown as mean ± s.e.m. for six ears per group from one of five repeats that gave similar results. *P < 0.05, **P < 0.01. P‐values were determined using a Mann–Whitney U‐test.
In CD4+ Th2 cells, the transcription factor GATA3 has well‐defined roles in Th2 differentiation and cytokine expression. 30 However, it has also been shown to play a critical role in promoting the expression of FOXP3 and functional suppressive activity in Treg cells. 31 In this regard the Th2 cytokine‐promoting activity of GATA3 must be tightly regulated in Treg cells to prevent the expression of GATA3‐dependent Th2 genes. BCL6 has been shown to be a critical regulator of GATA3 function in Tregs where BCL6 deficient Treg cells show increased expression of GATA3 and Th2 cytokines, resulting in enhanced Th2 inflammation. 32 , 33 This raised the possibility that enhanced eosinophil and basophil infiltration in BCL6cKO.4C13R mice could be due to the indirect effect of BCL6 deficiency on Treg function. To test this, BCL6cKO and BCL6fl/fl mice were sensitized with HDM in the ear pinna and the frequency of Tregs compared in each mouse strain using intranuclear FOXP3 staining and flow cytometry. The frequency of Treg cells in the ear tissue was significantly decreased in BCL6cKO mice compared with BCL6fl/fl littermate controls; however, due to the increased number of CD4 T cells the total number of Treg cells appearing in the ears of BCL6cKO mice was not significantly perturbed (Figure 5c). The MFI of GATA3 was slightly increased in BCL6cKO‐Treg cells, compared with BCL6fl/fl controls (Figure 5d). These data support the finding that BCL6 normally suppresses GATA3 in Treg cells to limit the expression of Th2 genes. In addition to the increased Th2eff cell numbers in the skin of BCL6cKO mice, the Th2‐skewing of Treg cells provides another factor that may contribute to the enhanced recruitment of eosinophils and basophils in the skin tissue of BCL6cKO mice.
Lung Th2eff cells are reduced in BCL6cKO .4C13R mice after primary Nippostrongylus brasiliensis infection
Given previous findings that IL‐4+ Tfh cells can give rise to Th2eff cells in the lung, 9 we next employed a lung model of Type 2 immunity and tested the dependency of lung Th2eff cells on Tfh cells. BCL6cKO.4C13R and BCL6fl/fl.4C13R littermate controls were infected subcutaneously with 550 live Nb L3 larvae or PBS, and the lung draining mediastinal lymph node (medLN) and lung tissue were analyzed to determine the frequency of Th2eff in the lung. Cellular responses were analyzed on day 10 post‐Nb infection, at the peak of the Th2 CD4 response to this stimulus. 34 , 35 As seen in the HDM skin sensitization model, the development of Tfh cells in the medLN was completely inhibited in the BCL6cKO.4C13R mice, whilst the number of non‐Tfh cells was similar to the BCL6fl/fl.4C13R controls (Figure 6a, b). Live Nb infection triggered a robust increase in the frequency and total number of IL4‐AC+ non‐Tfh cells compared with the PBS controls and this trended to being decreased in BCL6cKO.4C13R mice (Figure 6c, d). Compared with the PBS controls, Tfh cells in Nb infected mice significantly upregulated the expression of IL4‐AC and due to the complete ablation of Tfh cells, this population was absent in BCL6cKO.4C13R mice (Figure 6c, e). These results indicate that as observed in the skin HDM sensitization model, a significant population of IL‐4 producing non‐Tfh appears in the medLN following Nb infection. Furthermore these data agree with the aLN/skin data that the loss of Tfh cells and/or the loss of intrinsic BCL6 regulation reduces the frequency of IL‐4 producing non‐Tfh cells.
Figure 6.
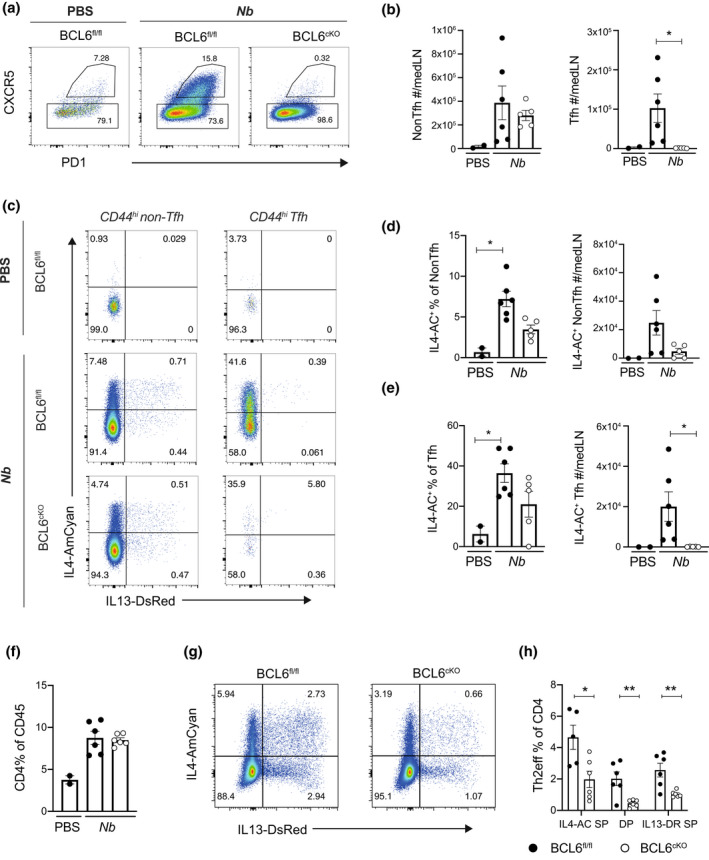
BCL6cKO.4C13R mice show a partial dependence on Tfh cells for Th2eff cell development in the lung. BCL6fl/fl.4C13R and BCL6cKO.4C13R mice were infected with 550 live Nb L3 subcutaneously in neck scruff. Lung and draining LN tissues were collected and cells harvested for analysis on day 10. (a) Flow cytometry plots gated on CD4+ CD44hi depicting expression of CXCR5 and PD1 to identify Tfh and non‐Tfh cells. (b) Total number of non‐Tfh and Tfh cells in medLN. (c) Flow cytometry plots depicting the expression of IL4‐AC and IL13‐DR in non‐Tfh and Tfh cells in the medLN. (d, e) Frequency and total number of IL4‐AC+ non‐Tfh (d) and IL4‐AC+ Tfh cells (e). (f) Frequency of CD4 T cells in the lung. (g) Flow cytometry plots depicting IL4‐AC and IL13‐DR expression in lung CD4 T cells. (h) Frequencies of Th2eff subsets in the lung tissue. Black circles represent BCL6fl/fl, white circles represent BCL6cKO samples. Data are shown as mean ± s.e.m. for 2–6 mice per group from one of two repeats that gave similar results. *P < 0.05, **P < 0.01. P‐values were determined for b an d–f using a Kruskal‐Wallis test applying Dunn's multiple comparison post‐test. P‐values for h were determined using a Mann–Whitney U‐test, independently for each Th2eff subset.
The frequency of lung infiltrating CD4 T cells was increased approximately 2‐fold in Nb infected mice compared with PBS controls and this was not affected by BCL6 deficiency in BCL6cKO.4C13R mice compared with BCL6fl/fl.4C13R controls (Figure 6f). The frequencies of all three Th2eff subsets were significantly decreased in BCL6cKO.4C13R mice (Figure 6g, h), indicating that BCL6 may have a role in the development of Th2eff in the lung. This contrasts with the HDM skin sensitization data which showed no role for Tfh cells in skin Th2eff development (Figure 3b–d), indicating that tissue‐specific regulation may distinguish the role of Tfh cells in skin and lung type 2 effector responses.
DISCUSSION
We describe the use of a conditional CD4, BCL6 deficient Il4/Il13 reporter mouse strain to examine the contribution that the Tfh subset makes to the development of Th2eff cells in the skin and lung. We report that Tfh cells do not contribute to Th2eff cell development in the skin as comparable numbers of all three Il4 and Il13 expressing Th2eff subsets develop in the skin of HDM sensitized BCL6 sufficient and BCL6/Tfh‐deficient mice. This conclusion is consistent with Lonnberg et al. 36 who showed an early bifurcation of Th1 and Tfh cells from a common precursor in spleen CD4 T cells in malaria‐infected mice and reported no evidence of functional plasticity once the commitment to a T helper cell fate had been established.
One potential limitation of the BCL6cKO mice used in our study is the observation that BCL6 could potentially regulate gene expression in non‐Tfh CD4 T cell compartments such as Th2 cells and other non‐Th2 cells including Tregs. By using RNA‐seq we showed that despite its prominent role as a transcriptional repressor, BCL6 does not play a significant role in the intrinsic regulation of allergen‐primed skin Th2eff cells. This was evident from differential gene analysis that resulted in less than 50 DEGs between BCL6 sufficient and deficient cells, for each Th2eff cell subset. Of note, Bach2 was one of the 22 genes upregulated in BCL6cKO IL13‐DR SP samples. BACH2 is a transcriptional repressor that has been shown to inhibit Th2 differentiation. 37 BACH2 inhibits BLIMP‐1, 38 an antagonistic partner of BCL6. Therefore, the upregulation of Bach2 in BCL6cKO IL13‐DR SP cells may represent a feedback loop whereby BACH2 indirectly promotes the expression of BCL6 through repression of BLIMP‐1 in BCL6 deficient cells. However, despite altered Bach2 expression, the limited transcriptional differences suggests the functional expression of BACH2 was not substantially altered in the absence of BCL6. It is possible that the lack of BCL6 transcriptional repression in BCL6cKO Th2eff cells may lead to enhanced IL‐4 and IL‐13 expression, concealing the loss of Tfh‐derived Th2eff cells in our experiments. However, if this were the case, we would expect to see increased expression of Il4 and Il13 in BCL6cKO Th2eff cells. RNA‐seq analysis did reveal that Il13 was significantly upregulated in BCL6cKO IL4‐AC SP cells; however, given that IL4‐AC SP cells do not express IL13‐DR, this upregulation of Il13 was not substantial enough to induce DsRed expression from the 4C13R transgene. Furthermore, the expression of Il13 in IL4‐AC SP BCL6cKO cells remained substantially lower than IL13‐DR+ counterparts. In summary, the overall lack of intrinsic BCL6 regulation in skin Th2eff cells negates the possibility that Th2 deficiency in the skin is masked. Instead these results give confidence to our conclusion that Th2eff cells are derived from non‐Tfh cells in the aLN and not Tfh cells.
Interestingly, we did observe that BCL6 regulation could occur in the Treg compartment as seen by a reduction in the frequency of Treg cells in the skin accompanied by increased expression of GATA3 in Treg cells from BCL6cKO mice. We hypothesize this contributes to the enhanced infiltration of eosinophils and basophils observed in the skin tissue. Increased eosinophil and basophil recruitment has been reported in germline BCL6 KO mice, although it was attributed to BCL6‐dependent suppression of chemokine expression in macrophages. 39 , 40 Here, we provide evidence that BCL6 acts to suppress the GATA3‐dependent Th2 program in Treg cells, which we postulate aids the regulatory function of Treg cells and limits the recruitment of eosinophils and basophils to the skin tissue. In addition to Tregs, BCL6 deficient IL4‐AC.IL13‐DR DP and IL13‐DR SP Th2eff cells upregulated Il3 compared with BCL6 sufficient counterparts. This enhanced Il3 expression, along with the general increase in total numbers of Th2eff cell subsets, due to increased CD4 T cell counts, may also contribute to the increased recruitment of eosinophils and basophils to the tissue.
In the two models of type 2 immunity employed in this study, we report that approximately half of IL‐4+ CD4 T cells in the aLN are of the Tfh lineage and that the frequency of IL‐4+ non‐Tfh cells is reduced in BCL6cKO mice. Therefore, the loss of Tfh cells results in a substantial decrease of IL‐4 availability in the aLN, which may explain the reduced frequency of IL‐4+ non‐Tfh “classical” Th2 cells, given the critical role of IL‐4 in Th2 differentiation. T helper cell differentiation is thought to take place in the draining lymph node, alongside MHC‐II/TCR mediated activation of naïve CD4 T cells by dendritic cells, and is dependent on the cytokines present in the environmental milieu of the lymph node at the time of activation. 41 The allergen induced Th2eff cells appearing in the tissues are thought to be derived from IL‐4+ non‐Tfh Th2 cells in the aLN. However, our analysis of skin Th2eff cells indicates that the reduction of Th2 cells in aLN of BCL6cKO mice does not result in reduced numbers of skin Th2eff cells. This is most probably related to LN output of Th2 cells capable of migrating to tissues and becoming Th2eff, which is more than able to fill the available inflammatory niches at the sensitized skin site in the ear pinna.
The observation that Th2eff cells were reduced in the lungs of Tfh‐deficient BCL6cKO.4C13R mice by approximately 2‐fold in response to a live Nb infection leaves open the possibility that Th2eff cells in the lung may be partially derived from Tfh cells in the medLN. This supports a previous finding by Ballesteros et al. 9 who reported that 100% of IL‐4+ cells in the medLN were Tfh cells and that inhibition of Tfh cells using a BCL6 inhibitor reduced the development of both IL‐4+ and IL‐13+ Th2eff cells in the lung tissue by approximately 3‐fold. Alternatively, the lung may not hold a maximum niche for Th2eff cells like the skin, and thus if medLN non‐Tfh Th2 cells are the precursors to Th2eff cells in the lung, the reduction of IL‐4+ non‐Tfh Th2 cells would directly translate to the reduced Th2eff cells observed in the lung. Given that Ballesteros et al. did not detect IL‐4+ non‐Tfh cells in the medLN, this alternate hypothesis of non‐Tfh derived Th2eff cells in the lung does not apply to their chronic, recall challenge model of HDM‐induced lung inflammation. Given the complex nature of the lung microenvironment, the difficulty of accurately enumerating cell numbers, and model specific differences, further studies will be required to determine whether Tfh cells play a role in lung Th2eff cell development.
In summary, our data show that Tfh are not necessary for the differentiation of Th2eff cells in a primary HDM sensitization model in the skin; however, in a chronic lung type 2 model a partial Tfh‐dependent Th2eff response may occur, but not as significantly as reported elsewhere. This work supports the hypothesis that allergen activated Tfh cells diverge early in the CD4 T cell differentiation pathway and represent a distinct subset of CD4 T cells in the aLN that do not contribute to the pool of Th2eff cells that are activated and migrate to the skin. Additionally, we report that BCL6 normally acts to suppress CD4 T cell mediated type 2 inflammatory response in the skin, with increased eosinophil and basophil infiltration in BCL6cKO mice. We suggest that this is due to a number of contributing factors, including the regulatory role of BCL6 in maintaining Treg function through inhibition of the GATA3 induced Th2 genes. We support the notion that BCL6 does not influence Th2eff cell differentiation despite its obligatory role in Tfh development, and indicate that primary skin Th2eff cells are derived from a non‐Tfh precursor in the lymph node.
METHODS
Mice
All mice were bred and maintained on a C57BL/6 background in a specific pathogen‐free barrier conditions in the Malaghan Institute of Medical Research Biomedical Research Unit. The 4C13R 15 reporter mice bacterial artificial chromosome does not interfere with normal immune function with similar levels of T cells, B cells, immunoglobulin and inflammatory cells being induced by immunization compared with wild type C57BL/6 mice. Bcl6fl/fl and CD4cre strains were imported from the John Curtin School of Medical Research, ANU, Canberra. All animal procedures were approved by the Victoria University of Wellington Animal Ethics committee and performed in accordance with institutional guidelines.
Immunizations and treatments
Mice were anesthetized using xylazine and ketamine (Phoenix, Auckland, New Zealand). 30 μL of a solution containing 200 μg HDM (Greer Laboratories, Lenoir, NC, USA) was injected intradermally into the ear pinna and mice were killed 7 days later. For live Nippostrongylus brasiliensis infections, the mice were injected with 550 L3 larvae subcutaneously in the scruff of the neck and killed 10 days later. To block egress of cells from lymph nodes the mice were treated with 25 μg FTY720 in PBS intra‐peritoneally at day 2 and again on day 4 after HDM immunization.
Cell isolation and flow cytometry
Draining lymph nodes were pressed through a 70‐μm cell strainer into complete media [IMDM (Gibco, ThermoFisher Scientific, MA, USA) supplemented with 5% FBS (Sigma‐Aldrich, MO, USA), 100 U mL−1 penicillin per 100 μg mL−1 streptomycin (Invitrogen™, ThermoFisher Scientific, Waltham, MA, USA) and 55 μm β‐mercaptoethanol (Invitrogen, Waltham, MA, USA) to make a single cell suspension. Dorsal and ventral ear sheets were separated and minced into very fine pieces using scissors. Each ear was digested in 3 mL digestion media [IMDM (Gibco), containing 1.2 mg collagenase IV (Sigma) and 120 μg mL−1 DNAse I (Roche, Basel, Switzerland)] and incubated in a 37°C shaking incubator at 200 rpm for 30 min. Ear wash buffer [PBS containing 1% BSA (Sigma, St Louis, MO, USA) and 5 mm EDTA (Invitrogen)] was added immediately to stop the digestion. The solution was pipetted up and down to further dissociate tissue and passed through a 70‐μm cell strainer. The ears were washed once more in ear wash buffer then red blood cells were lysed using ammonium‐chloride‐potassium buffer [1 mm EDTA (Invitrogen), 150 mm NH4Cl (Sigma), 10 mm NaHCO3 (Sigma) in ddH2O] before being resuspended in complete media. Half the lung was taken and used for digestion. Briefly, the lungs were finely minced and digested in 2 mL digestion media [DMEM (Gibco), containing 2.4 mg mL−1 Collagenase I (Sigma) and 120 μg mL−1 DNAse I (Roche)] for 1 h in a 37°C shaking incubator at 200 rpm. The solution was pipetted up and down to further dissociate tissue and passed through a 70‐μm cell strainer. Lungs were washed in complete media before being counted and stained for flow cytometry. Live cell counts were performed using a hemocytometer and trypan blue (Invitrogen) exclusion. For FACS analysis, the cells were resuspended in FACS buffer [2% FBS (Sigma), 1 mm EDTA (Invitrogen), 0.01% sodium azide (Sigma) in PBS] and 2 × 106 cells were plated for staining. The cells were first incubated with anti‐CD16/32 antibody (clone 2.4G2) before staining with fluorophore‐conjugated antibodies. Lymph node samples were stained for CXCR5 (2G8, Biotin) at 37°C for 30 min separately from full antibody cocktail mix. Cell surface antibody cocktails were made up using combinations of: SiglecF‐BV421, CD11b‐BV570, CD90.2‐BV605, CD90.2‐PECy7, NK1.1‐BV650, Ly6G‐BV711, CD3‐PECy5, CD3‐BV786, CD8‐BV786, CD8‐PECy7, CD8‐AF700, CD8a‐FITC, CD4‐PerCPCy5.5, CD4‐APCH7, CD4‐BV605, CD4‐APCCy7, cKit‐BB700, CD64‐PE dazzle‐594, CD49b‐PECy7, CD49b‐FITC, CD200R3‐APC, CD45‐APCCy5.5, Ly6C‐AF700, PD1‐PerCPef710, CD44‐AF700, CD44‐PECy5, TCRgd‐PECy7, CD19‐APCH7 and Streptavidin‐BV605 (BioLegend, San Diego, CA, USA). The cells were either stained for viability with Zombie‐NIR fixable dye prior to 24G2 blocking in PBS, or after cell surface staining with DAPI. Data were acquired on a Cytek Aurora Spectral flow cytometer (Cytek Biosciences, Fremont, CA, USA). Flow data were analyzed using FlowJo software (Treestar).
ELISA and Legendplex
IgG1 and IgG2 serum ELISAs were performed using Invitrogen kits Cat # 88–50 410 and 88–50 420 respectively.
RNA sequencing
Wild type 4C13R or BCL6cKO.4C13R reporter mice were immunized with HDM as above, killed on day 7 and the Th2 effector subsets isolated from ear tissue were sorted for using a BD Influx cell sorter. Th2eff cell subsets were gated as CD45+, CD3int, TCRγd−, CD4+, CD8− and subsequently either IL4‐AC SP, IL4‐AC.IL13‐DR DP or IL13‐DR SP. CD4+ cells from naïve aLN were also sequenced as a control. 1000 cells were sorted directly into 5 μL lysis buffer [TCL Buffer (Qiagen, Hilden, Germany)] with 1% 2‐mercaptoethanol), Smart‐seq2 libraries were prepared as described previously 19 , 20 with slight modifications. Briefly, total RNA was captured and purified on RNAClean XP beads (Beckman Coulter, Brea, CA, USA). Polyadenylated mRNA was then selected using an anchored oligo(dT) primer (5′–AAGCAGTGGTATCAACGCAGAGTACT30VN‐3′) and converted to cDNA via reverse transcription. First strand cDNA was subjected to limited PCR amplification followed by Tn5 transposon based fragmentation using the Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA, USA). The samples were then PCR amplified for 18 cycles using barcoded primers such that each sample carries a specific combination of eight base Illumina P5 and P7 barcodes and pooled together prior to Smart sequencing. Smart‐seq paired‐end sequencing was performed on an Illumina NextSeq500 using 2 × 38 bp reads. Trimmomatic 42 (v0.36) was used to remove Illumina adapters, base pairs lower than 30 Phred quality score, and reads shorter than 20 bases. STAR 43 (v2.7.1a) was used to align reads to the GENCODE GRCm38/mm10 M16 genome primary assembly. 44 Gene level counts were calculated using feature Counts 45 (v1.28.1) in R 46 (v3.4.4). Gene level counts were calculated as variance stabilized transcripts per kilobase (VSTpk). VSTpk expression represents how many transcripts matched to a gene per 1000 base pairs of gene length, with extra normalization to control for differences in the varying sequencing depth per sample. Replicated samples were checked for similarity by Pearson correlation scores (≥ 0.90) on transcripts with higher than an average of 5 reads and below the 99th percentile of reads counted. Heatmap and PCA plots (R packages: heatmap, 47 ggbiplot 48 , 49 were created from log2‐transformed raw gene counts from feature Counts. Differential gene expression analysis was performed using DESeq2 50 and genes with a log2‐fold‐change > 1 or < 1; P‐value < 0.05 and a minimum mean read expression count > 30 were determined as DEGs.
Statistical analysis
Statistical analysis was performed using Prism 8 (GraphPad Software, San Diego, CA, USA). Differences between two groups were compared using a non‐parametric Mann–Whitney U‐test. Differences between three or more groups were calculated using a non‐parametric Kruskal‐Wallis test. Not significant, not displayed; (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). All figures show the mean ± s.e.m. Differences are not significant unless stated otherwise.
AUTHOR CONTRIBUTIONS
Jodie Chandler: Conceptualization; data curation; formal analysis; methodology; visualization; writing – original draft; writing – review and editing. Melanie Prout: Data curation; formal analysis. Sam Old: Formal analysis; visualization. Cynthia Morgan: Formal analysis. Franca Ronchese: Conceptualization; investigation; writing – review and editing. Christophe Benoist: Methodology; resources. Graham Le Gros: Conceptualization; formal analysis; funding acquisition; project administration; supervision; writing – review and editing.
CONFLICT OF INTEREST
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting information
ACKNOWLEDGMENTS
We thank the Hugh Green Cytometry Centre for technical assistance with cell sorting and the Biomedical Research Unit at the Malaghan Institute of Medical Research for animal husbandry to generate the BCL6fl/fl.4C13R.CD4cre mouse line. This work was funded by Independent Research Organization funding from the Health Research Council of New Zealand and the Marjorie Barclay Trust. The authors declare no competing financial interests.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Zenodo at http://doi.org/10.5281/zenodo.7048910.
REFERENCES
- 1. Johnston RJ, Poholek AC, DiToro D. et al. Bcl6 and Blimp‐1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 2009; 325: 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yu D, Rao S, Tsai LM. et al. The transcriptional repressor Bcl‐6 directs T follicular helper cell lineage commitment. Immunity 2009; 31: 457–468. [DOI] [PubMed] [Google Scholar]
- 3. Breitfeld D, Ohl L, Kremmer E. et al. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med 2000; 192: 1545–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schaerli P, Willimann K, Lang AB. et al. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J Exp Med 2000; 192: 1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reinhardt RL, Liang HE, Locksley RM. Cytokine‐secreting follicular T cells shape the antibody repertoire. Nat Immunol 2009; 10: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. King IL, Mohrs M. IL‐4‐producing CD4+ T cells in reactive lymph nodes during helminth infection are T follicular helper cells. J Exp Med 2009; 206: 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Glatman Zaretsky A, Taylor JJ, King IL, Marshall FA, Mohrs M, Pearce EJ. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. J Exp Med 2009; 206: 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 2011; 29: 621–663. [DOI] [PubMed] [Google Scholar]
- 9. Ballesteros‐Tato A, Randall TD, Lund FE, Spolski R, Leonard WJ, Leon B. T follicular helper cell plasticity shapes pathogenic T helper 2 cell‐mediated immunity to inhaled house dust mite. Immunity 2016; 44: 259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coquet JM, Schuijs MJ, Smyth MJ. et al. Interleukin‐21‐producing CD4+ T cells promote type 2 immunity to house dust mites. Immunity 2015; 43: 318–330. [DOI] [PubMed] [Google Scholar]
- 11. Hondowicz BD, An D, Schenkel JM. et al. Interleukin‐2‐dependent allergen‐specific tissue‐resident memory cells drive asthma. Immunity 2016; 44: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pyle CJ, Labeur‐Iurman L, Groves HT. et al. Enhanced IL‐2 in early life limits the development of TFH and protective antiviral immunity. J Exp Med 2021; 218: e20201555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yusuf I, Kageyama R, Monticelli L. et al. Germinal center T follicular helper cell IL‐4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J Immunol 2010; 185: 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prout MS, Kyle RL, Ronchese F, Le Gros G. IL‐4 is a key requirement for IL‐4 and IL‐4/IL‐13 expressing CD4 Th2 subsets in lung and skin. Front Immunol 2018; 9: 1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roediger B, Kyle R, Yip KH. et al. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat Immunol 2013; 14: 564–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Camberis M, Prout M, Tang SC. et al. Evaluating the in vivo Th2 priming potential among common allergens. J Immunol Methods 2013; 394: 62–72. [DOI] [PubMed] [Google Scholar]
- 17. Gowthaman U, Chen JS, Zhang B. et al. Identification of a T follicular helper cell subset that drives anaphylactic IgE. Science 2019; 365: 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clement RL, Daccache J, Mohammed MT. et al. Follicular regulatory T cells control humoral and allergic immunity by restraining early B cell responses. Nat Immunol 2019; 20: 1360–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart‐seq2 for sensitive full‐length transcriptome profiling in single cells. Nat Methods 2013; 10: 1096–1098. [DOI] [PubMed] [Google Scholar]
- 20. Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full‐length RNA‐seq from single cells using smart‐seq2. Nat Protoc 2014; 9: 171–181. [DOI] [PubMed] [Google Scholar]
- 21. Tibbitt CA, Stark JM, Martens L. et al. Single‐cell RNA sequencing of the T helper cell response to house dust mites defines a distinct gene expression signature in airway Th2 cells. Immunity 2019; 51: 169–184. [DOI] [PubMed] [Google Scholar]
- 22. Morimoto Y, Hirahara K, Kiuchi M. et al. Amphiregulin‐producing pathogenic memory T helper 2 cells instruct eosinophils to secrete osteopontin and facilitate airway fibrosis. Immunity 2018; 49: 134–150. [DOI] [PubMed] [Google Scholar]
- 23. Wambre E, Bajzik V, DeLong JH. et al. A phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci Transl Med 2017; 9: eaam9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X, Li Z, Naganuma A, Ye BH. Negative autoregulation of BCL‐6 is bypassed by genetic alterations in diffuse large B cell lymphomas. Proc Natl Acad Sci USA 2002; 99: 15018–15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hollister K, Kusam S, Wu H. et al. Insights into the role of Bcl6 in follicular Th cells using a new conditional mutant mouse model. J Immunol 2013; 191: 3705–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tai PC, Sun L, Spry CJ. Effects of IL‐5, granulocyte/macrophage colony‐stimulating factor (GM‐CSF) and IL‐3 on the survival of human blood eosinophils in vitro . Clin Exp Immunol 1991; 85: 312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shen T, Kim S, Do JS. et al. T cell‐derived IL‐3 plays key role in parasite infection‐induced basophil production but is dispensable for in vivo basophil survival. Int Immunol 2008; 20: 1201–1209. [DOI] [PubMed] [Google Scholar]
- 28. Johansson MW. Activation states of blood eosinophils in asthma. Clin Exp Allergy 2014; 44: 482–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. MacGlashan D Jr. Marked differences in the signaling requirements for expression of CD203c and CD11b versus CD63 expression and histamine release in human basophils. Int Arch Allergy Immunol 2012; 159: 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wan YY. GATA3: a master of many trades in immune regulation. Trends Immunol 2014; 35: 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA‐3 for the function of regulatory T cells. Immunity 2011; 35: 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sawant DV, Sehra S, Nguyen ET. et al. Bcl6 controls the Th2 inflammatory activity of regulatory T cells by repressing Gata3 function. J Immunol 2012; 189: 4759–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sawant DV, Wu H, Yao W, Sehra S, Kaplan MH, Dent AL. The transcriptional repressor Bcl6 controls the stability of regulatory T cells by intrinsic and extrinsic pathways. Immunology 2015; 145: 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Harvie M, Camberis M, Tang SC, Delahunt B, Paul W, Le Gros G. The lung is an important site for priming CD4 T‐cell‐mediated protective immunity against gastrointestinal helminth parasites. Infect Immun 2010; 78: 3753–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Panhuys N, Tang SC, Prout M. et al. In vivo studies fail to reveal a role for IL‐4 or STAT6 signaling in Th2 lymphocyte differentiation. Proc Natl Acad Sci USA 2008; 105: 12423–12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lonnberg T, Svensson V, James KR. et al. Single‐cell RNA‐seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Sci Immunol 2017; 2: eaal2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsukumo S, Unno M, Muto A. et al. Bach2 maintains T cells in a naive state by suppressing effector memory‐related genes. Proc Natl Acad Sci USA 2013; 110: 10735–10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ochiai K, Katoh Y, Ikura T. et al. Plasmacytic transcription factor Blimp‐1 is repressed by Bach2 in B cells. J Biol Chem 2006; 281: 38226–38234. [DOI] [PubMed] [Google Scholar]
- 39. Mondal A, Sawant D, Dent AL. Transcriptional repressor BCL6 controls Th17 responses by controlling gene expression in both T cells and macrophages. J Immunol 2010; 184: 4123–4132. [DOI] [PubMed] [Google Scholar]
- 40. Toney LM, Cattoretti G, Graf JA. et al. BCL‐6 regulates chemokine gene transcription in macrophages. Nat Immunol 2000; 1: 214–220. [DOI] [PubMed] [Google Scholar]
- 41. Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol 2010; 28: 445–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dobin A, Davis CA, Schlesinger F. et al. STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frankish A, Diekhans M, Ferreira AM. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019; 47: D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 2019; 47: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. RCoreTeam . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 47. Kolde R. pheatmap: Pretty Heatmaps. R package version 1.0.12 ed: R; 2019.
- 48. Vu VQ. ggbiplot: A ggplot2 based biplot. R package version 0.55 ed: R; 2011.
- 49. Wickham H. Ggplot2: Elegant Graphics for Data Analysis. New York: Springer; 2009. [Google Scholar]
- 50. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in Zenodo at http://doi.org/10.5281/zenodo.7048910.


