Abstract
The objective of this study was to evaluate the evidence on cost‐effectiveness of pharmacogenetic (PGx)–guided treatment for drugs with Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines. A systematic review was conducted using multiple biomedical literature databases from inception to June 2021. Full articles comparing PGx‐guided with nonguided treatment were included for data extraction. Quality of Health Economic Studies (QHES) was used to assess robustness of each study (0–100). Data are reported using descriptive statistics. Of 108 studies evaluating 39 drugs, 77 (71%) showed PGx testing was cost‐effective (CE) (N = 48) or cost‐saving (CS) (N = 29); 21 (20%) were not CE; 10 (9%) were uncertain. Clopidogrel had the most articles (N = 23), of which 22 demonstrated CE or CS, followed by warfarin (N = 16), of which 7 demonstrated CE or CS. Of 26 studies evaluating human leukocyte antigen (HLA) testing for abacavir (N = 8), allopurinol (N = 10), or carbamazepine/phenytoin (N = 8), 15 demonstrated CE or CS. Nine of 11 antidepressant articles demonstrated CE or CS. The median QHES score reflected high‐quality studies (91; range 48–100). Most studies evaluating cost‐effectiveness favored PGx testing. Limited data exist on cost‐effectiveness of preemptive and multigene testing across disease states.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
There is varying information on the cost‐effectiveness of pharmacogenetic testing for drugs with Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines and no systematic review has collated these data on all drugs with CPIC guidelines to date.
WHAT QUESTION DID THIS STUDY ADDRESS?
What is the evidence on cost‐effectiveness of pharmacogenetic testing for drugs with CPIC guidelines?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
For the first time, we demonstrate that 71% of studies evaluating the cost of pharmacogenetic testing for drugs with CPIC guidelines determined testing was cost‐effective or cost‐saving. The analysis provides details of key study characteristics and differences in cost outcomes.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
These data provide valuable information on the impact of pharmacogenetic testing on cost and may support further coverage determinations by payors to increase accessibility to evidence‐based pharmacogenetic testing.
Pharmacogenomics (PGx) has been studied and applied across many therapeutic areas to evaluate and predict an individual's response to specific drugs based on his or her genetic makeup. 1 PGx‐guided dosing and treatment can be used to normalize systemic drug exposure, reduce side effects, and/or improve clinical response for drugs with established evidence. There are several robust examples where PGx testing can improve drug safety (e.g., abacavir/HLA‐B*5701), 2 and/or enhance clinical effectiveness (e.g., clopidogrel/CYP2C19, 3 psychiatric medications, and CYP2D6/CYP2C19). 4
As of April 2022, the US Food and Drug Administration (FDA) has over 400 biomarkers for roughly 300 distinct medications mentioned in drug labeling, with most related to somatic mutations for cancer targeted therapies. 5 In February 2020, the FDA published a Table of Pharmacogenetic Associations with a focus on germline variations, which was most recently updated in May 2022. 6 These tables outline drug–gene associations for which data support therapeutic management (56 drugs) and have a potential impact on safety or response (19 drugs) or a potential pharmacokinetic impact only (39 drugs). As of July 2022, there are eight drugs with black box warnings related to germline PGx: abacavir/HLA‐B*5701, carbamazepine/HLA‐B*1502, clopidogrel/CYP2C19, codeine and tramadol/CYP2D6, pegloticase and rasburicase/G6PD, and valproic acid/POLG. 5 Greater awareness, improved testing accessibility, and expanding evidence base has increased the adoption of PGx testing at some hospitals.
A key process in implementing PGx testing is the ability to standardize the translation of PGx data into actionable prescribing decisions that are driven by evidence‐based medicine. The Clinical Pharmacogenetics Implementation Consortium (CPIC) is an international group focused on creating, curating, and posting freely available, peer‐reviewed, evidence‐based, updatable, and detailed drug–gene clinical practice guidelines focused on actionability (26 guidelines across roughly 70 drugs, as of April 2022). 7 A key assumption underlying these guidelines is that clinical high‐throughput and preemptive genotyping will become more widespread and that clinicians will be faced with interpreting PGx information even if they have not explicitly ordered a test. Factors considered in guideline development include available evidence supporting PGx intervention, risk of an adverse drug reaction, and available test for a gene or set of genes. The guidelines purposefully do not address cost of testing nor provide guidance on who should be tested, albeit many institutions who have adopted PGx testing utilize CPIC guidelines as a reference for drug–gene inclusion.
Cost is an important consideration to healthcare systems and patients when applying PGx in clinical practice and is often ranked as a major barrier to implementation. 8 Third party payors especially want to understand if PGx test coverage will save or increase healthcare costs downstream. Cost‐effectiveness of PGx testing is changing rapidly as molecular testing platforms are becoming more sophisticated, cheaper, accessible, and are transitioning from single‐gene to multigene tests. 9 While it is understandable that cost is excluded from clinical practice guidelines, it is still critical that cost evaluations are conducted in PGx studies and systematic reviews.
Verbelen et al. conducted an economic evaluation for PGx associations listed in the FDA Table of Pharmacogenomic Biomarkers in Drug Labeling, which included 10 drugs and determined that over half of the 44 evaluations drew conclusions in favor of PGx testing. 10 The evaluation was limited to drugs listed in the FDA website up to September 2015. Given CPIC guidelines have become a universally accepted tool for PGx interpretation among implementers and these guidelines do not include information regarding cost, we proposed an updated systematic review of economic evaluations of drugs with CPIC guidelines. The goal was to evaluate whether the evidence suggested PGx‐guided treatment for CPIC drugs was cost‐effective, cost‐saving, uncertain, or not cost‐effective for all drugs with CPIC level A or B evidence and published guidelines.
MATERIAL AND METHODS
Search strategy
CPIC assigns levels of evidence to gene‐drug pairs based on prescribing actionability ranging from “A” to “D” with “A” meaning genetic information should be used to change prescribing (preponderance of evidence is high or moderate in favor of changing prescribing), “B” meaning genetic information could be used to change prescribing because alternative therapies/dosing are extremely likely to be as effective and as safe as nongenetically based dosing (preponderance of evidence is weak with little conflicting data) and “C” and “D” meaning clinical actionability is unclear. CPIC actionability levels may evolve as more data emerges. As of June 2021, 66 drugs with a published CPIC guideline had an evidence level of “A” or “B,” indicating a prescribing action is recommended for that gene‐drug pair. We conducted a comprehensive literature search from database inception to June 2021 for the 66 drugs with a published CPIC guideline and level “A” or “B” evidence. Pharmacogenomics, genes, and cost terms were searched for in PubMed, Embase, Ovid MEDLINE, and Scopus. Pharmacogenomics and gene terms were also searched in the Tufts CEA registry. Specific search criteria are included in the Supplemental Information . This systematic review was exempt from Institutional Review Board review and was not registered.
Study selection
Primary literature assessing any component of cost within a PGx study for any drug with a published CPIC guideline and evidence level of “A” or “B” was evaluated for inclusion. We excluded studies that were non‐English, review articles without original data, drugs for which there were no CPIC guidelines available, in abstract format only (or full text not available), and if no economic evaluation was presented in the analysis. Only studies comparing a PGx‐guided approach with a non‐PGx‐guided approach were included. Duplicated studies were excluded from the final selection. A.T.A. performed the initial literature search; S.A.M. and C.M. conducted an independent search for additional articles and reviewed the initial list; S.A.M. and J.N.P. reviewed the final list of studies for eligibility.
Data extraction and presentation of results
Data extraction of each study focused on three domains. (i) The gene(s) and drug(s) under review. (ii) Cohort (hypothetical, observational prospective/retrospective, randomized controlled trial) and type of population. (iii) Cost outcomes (cost‐effective, cost‐saving, not cost‐effective, or uncertain), including incremental cost‐effectiveness ratio (ICER) and willingness‐to‐pay thresholds. Studies which demonstrated the PGx intervention resulted in sufficient benefit or value compared with the costs (i.e., below the willingness‐to‐pay threshold) were categorized as cost‐effective. Studies which found the PGx intervention resulted in savings (overall reduction in healthcare costs, as determined by the study investigators) were categorized as cost‐saving. PGx interventions which did not create adequate benefit or value compared with cost (e.g., ICER greater than the willingness‐to‐pay threshold) were categorized as not cost‐effective. Studies which had conflicting results or indicated that cost‐effectiveness was dependent upon one or more parameters in the model (e.g., event rate, sample size, allele frequency, cost of the drug and/or event of interest) were categorized as uncertain.
Other information collected included the publication year, continent and country where the study was performed, perspective of the potential decision maker, time horizon, and funding source. This information was collected or identified as missing for every study and reported in Supplemental Table S1 . No assumptions regarding the data were made. Two reviewers were assigned to extract data independently from each article that passed inclusion criteria. Data, including study characteristics and cost outcomes, are synthesized and reported using descriptive statistics, summary tables, and a narrative method.
Quality of Health Economic Studies (QHES)
The Quality of Health Economic Studies (QHES) tool was used to assess the robustness of each study, 11 emphasizing appropriate methods, valid and transparent results, and comprehensive reporting. The instrument includes 16 criteria selected by a panel of eight health economics experts. The QHES tool is scored from a scale of 0–100 indicating lowest to highest quality studies. Studies with a QHES score of 75–100 were considered high quality and a score <75 considered not high quality. 12
To standardize the QHES scoring process, a multiple‐step method for inter‐rater reliability was developed: (i) Six raters reviewed and familiarized themselves with the instrument. 13 (ii) All raters individually reviewed and scored five selected papers as a “training set” from the antidepressant studies, which were then discussed among all raters and consensus was formed for each question and the total score. Based on this discussion, a document was created to standardize interpretation of each QHES question. 14 (iii) Raters were arranged into three groups of two. Each pair was assigned articles to score independently then discuss as a pair until consensus was reached to create a single score for each paper. (iv) The original “training set” was re‐scored by two raters so that they would be assessed in the same methodological fashion as the remaining papers. (v) Differences in interpretation or discrepancies in scores between paired raters was brought to the larger group for further discussion and consensus.
RESULTS
Study selection
As shown in Figure 1 , the search strategy identified 2,819 unique articles. We excluded 2,680 articles resulting in 139 full‐text articles assessed for eligibility. Of these, 108 articles were included for data extraction. The most common reasons for exclusion were article(s) did not compare PGx‐guided to non‐PGx‐guided treatment or costs were not evaluated.
Figure 1.
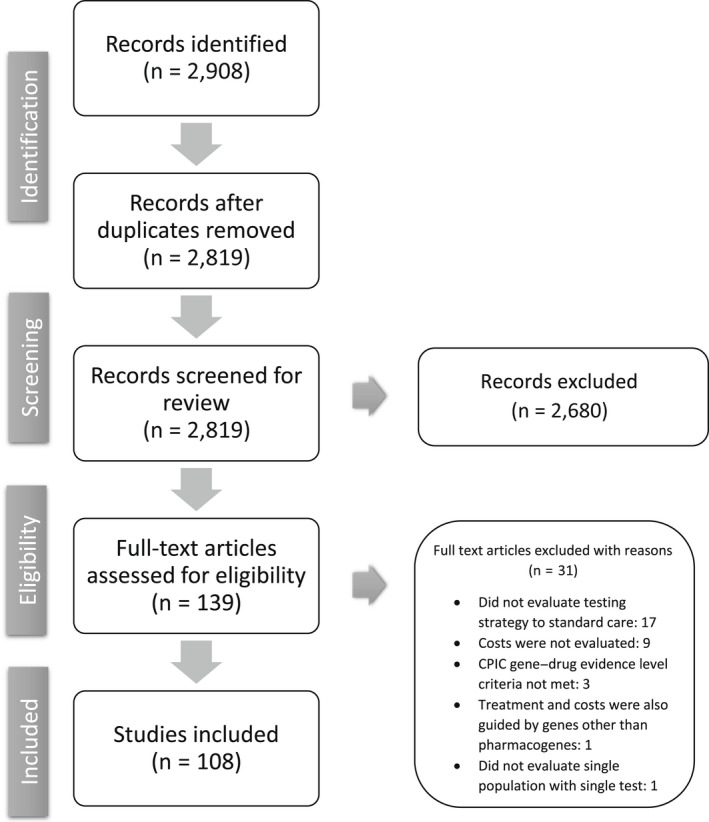
Flowchart of the literature screening. Of the 2,819 unique articles identified, 139 full‐text articles were assessed for eligibility and 108 were included in the final analysis. CPIC, Clinical Pharmacogenetics Implementation Consortium.
Summary of study designs and characteristics
All studies evaluated CPIC level A drug–gene pairs with nine multigene antidepressant studies and one multigene multidrug study also including drug–gene pairs with lower levels of evidence. Of the 108 studies included, 75 (69%) used hypothetical populations with model parameter estimates derived from previously published randomized controlled trials or observational studies (Table 1 ). Most studies were conducted using data from North America (n = 51, 47%), followed by Europe (n = 26, 24%), Asia (n = 28, 26%), and Australia (n = 3, 3%). CYP2C19 had the most single gene studies performed (n = 25, 23%), followed by TPMT (n = 11, 10%). Multigene testing was evaluated in 28 articles (26%), including genotype‐guided trials for depression (e.g., CYP2D6, CYP2C19, HTR2A, and SLC6A4) and warfarin (CYP2C9, VKORC1). Only two studies evaluated cost‐effectiveness of multigene testing across multiple therapeutic areas. 15 , 16 Most studies were published after 2010 (n = 90, 83%). About one‐third of the studies were from the payor perspective (n = 32, 30%) or universal healthcare system perspective (n = 24, 22%). The source of funding primarily came from a grant and/or foundation (n = 46, 43%), whereas 31% (n = 33) did not indicate any funding source or stated that there was no funding used. The entire list of articles reviewed, including drug / drug class, gene, year published, study title, study design, population, continent/country, cost outcome, PubMed ID, QHES score, sample size, perspective, time horizon, funding, willingness‐to‐pay threshold, and ICER are summarized in Supplemental Table S1 .
Table 1.
Summary of study characteristics
| Study characteristics | N = 108 n (%) |
|---|---|
| Year | |
| 2002–2005 | 7 (6%) |
| 2006–2010 | 11 (10%) |
| 2011–2015 | 39 (36%) |
| 2016–2021 | 51 (47%) |
| Continent | |
| North America | 51 (47%) |
| Europe | 26 (24%) |
| Asia | 28 (26%) |
| Australia | 3 (3%) |
| Gene | |
| CYP2B6 | 2 (2%) |
| CYP2C19 | 25 (23%) |
| CYP2C9 | 1 (1%) |
| CYP2D6 | 6 (6%) |
| DPYD | 5 (5%) |
| HLA‐A*31:01 | 1 (1%) |
| HLA‐B*15:02 | 7 (6%) |
| HLA‐B*57:01 | 8 (7%) |
| HLA‐B*58:01 | 10 (9%) |
| IFNL3 (IL‐28B) | 2 (2%) |
| mt‐RNR1 | 1 (1%) |
| TPMT | 11 (10%) |
| UGT1A1 | 1 (1%) |
| Multiple genes | 28 (26%) |
| Antidepressants | 9 (8%) |
| Clopidogrel/tramadol | 1 (1%) |
| Cardiovascular medications | 2 (2%) |
| Multiple drug classes | 1 (1%) |
| Warfarin | 15 (14%) |
| Cohort Type | |
| Hypothetical | 75 (69%) |
| Observational | 19 (18%) |
| Randomized controlled trial | 7 (6%) |
| Multiple | 7 (6%) |
| Perspective | |
| Patient | 1 (1%) |
| Payer | 32 (30%) |
| Societal | 20 (19%) |
| US healthcare system | 13 (12%) |
| Universal healthcare system | 24 (22%) |
| Multiple | 4 (4%) |
| Not stated | 14 (13%) |
| Funding source | |
| Government | 6 (6%) |
| Grant/foundation | 46 (43%) |
| Pharma | 14 (13%) |
| University/institute | 9 (8%) |
| No funding | 16 (15%) |
| Not stated | 17 (16%) |
| QHES scores | |
| High quality (≥75) | 94 (87%) |
| Not high quality (<75) | 14 (13%) |
CYP, cytochrome P450; DPYD, dihydropyrimidine dehydrogenase; HLA, human leukocyte antigen; IFNL3, interferon lambda 3; mt‐RNR1, mitochondrially encoded 12S ribosomal RNA; Pharma, pharmaceutical industry; SLCO1B1, solute carrier organic anion transporter family member 1B1; TPMT, thiopurine‐S‐methyltransferase; UGT1A1, UDP glucuronosyltransferase family 1 member A1.
Quality of studies
The overall median QHES score reflected robust studies (91; range 48–100). Most studies were of high quality with a score >75 (n = 94, 87%). There were 63 studies (58%) in the highest decile (scores between 90 and 100), including 7 studies with a score of 100.
Cost outcomes
Of the 108 articles reviewed, 77 (71%) found PGx‐guided treatment to be cost‐effective (N = 48) or cost‐saving (N = 29), whereas 21 (20%) were found to be not cost‐effective, and 10 (9%) were uncertain (Figure 2 ).
Figure 2.
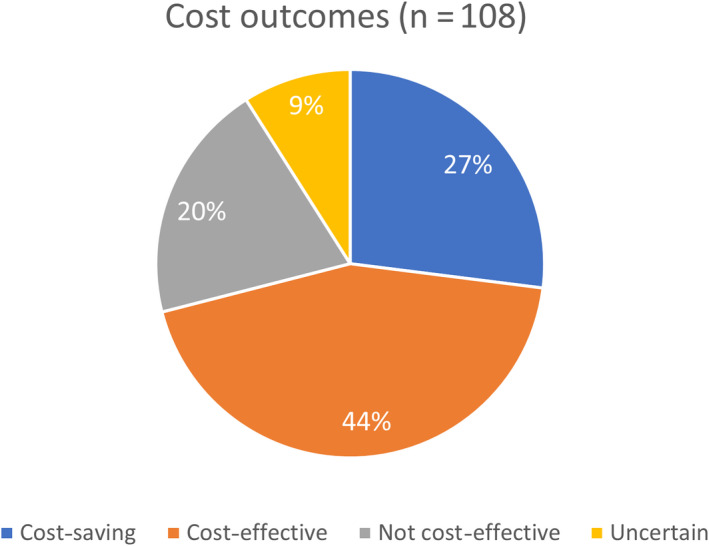
Cost outcomes (N = 108). Of 108 articles included in the systematic review, 44% demonstrated pharmacogenetic testing was cost‐effective, 27% cost‐saving, 20% not cost‐effective, and 9% uncertain.
The drug classes, types of drugs within each class, gene(s) related to each drug or class, number of articles reviewed, and cost outcomes are summarized in Table 2 . The number of studies ranged between 1 and 23 per drug or drug class, with an average of 7. Clopidogrel had the most publications analyzing cost associated with PGx testing, including 23 articles with 96% concluding CYP2C19 genotyping was cost‐effective or cost‐saving. Warfarin had the second most publications with 16 articles, but only 44% suggested cost‐effectiveness and no article found it to be cost‐saving. Twenty‐six articles evaluated HLA testing for either abacavir, allopurinol, carbamazepine, or phenytoin, 15 of which were found to be cost‐effective or cost‐saving. Nine out of 11 articles evaluating antidepressants were found to be cost‐effective or cost‐saving. Eight articles evaluated fluoropyrimidines (N = 5) or tamoxifen (N = 3), all of which were found to be cost‐effective or cost‐saving except one for tamoxifen, which was uncertain.
Table 2.
Summary of drug–gene pairs and cost outcomes
| Class | Gene | Total articles reviewed | Cost‐saving | Cost‐effective | Not cost‐effective | Uncertain | Drugs |
|---|---|---|---|---|---|---|---|
| Aminoglycosides 23 | mt‐RNR1 | 1 | — | — | 1 | — | Tobramycin |
| Analgesics 24 | CYP2D6 | 1 | — | — | 1 | — | Codeine |
| Anticoagulant 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 | CYP2C9 & VKORC1 | 15 | — | 6 | 5 | 4 | Warfarin |
| CYP2C9 | 1 | — | 1 | — | — | ||
| Antidepressants 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 | CYP2D6 | 2 | — | — | 1 | 1 | Citalopram, escitalopram, fluvoxamine, paroxetine, sertraline, amitriptyline, clomipramine, doxepin, imipramine, nortriptyline, trimipramine, desipramine |
| Multigene panela | 9 | 8 | 1 | — | — | ||
| Antiepileptic 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 | HLA‐A*31:01 | 7 | 1 | 2 | 4 | — | Carbamazepine, phenytoin |
| HLA‐B*15:02 | 1 | — | 1 | — | — | ||
| Antifungal 60 , 61 | CYP2C19 | 2 | 2 | — | — | — | Voriconazole |
| Antigout 62 , 63 , 64 , 65 , 66 , 67 , 68 , 69 , 70 , 71 | HLA‐B*58:01 | 10 | 1 | 4 | 5 | — | Allopurinol |
| Antiplatelet 15 , 72 , 73 , 74 , 75 , 76 , 77 , 78 , 79 , 80 , 81 , 82 , 83 , 84 , 85 , 86 , 87 , 88 , 89 , 90 , 91 , 92 , 93 | CYP2C19 | 22 | 4 | 17 | 1 | — | Clopidogrel |
| CYPC19 & CYP2D6 | 1 | 1 | — | — | — | Clopidogrel, tramadol | |
| Antiretroviral 94 , 95 , 96 , 97 , 98 , 99 , 100 , 101 , 102 , 103 , 104 | HLA‐B*57:01 | 8 | 2 | 4 | 1 | 1 | Abacavir |
| CYP2B6 | 2 | 2 | — | — | — | Efavirenz | |
| UGT1A1 | 1 | — | — | — | 1 | Atazanavir | |
| Cardiovascular medications 18 , 105 | CYP2C9, CYP2C19, SLCO1B1, & VKORC1 | 2 | — | 2 | — | — | Clopidogrel, simvastatin, warfarin |
| Fluoropyrimidines 106 , 107 , 108 , 109 , 110 | DPYD | 5 | 4 | 1 | — | — | Fluorouracil, capecitabine |
| Multidrug 16 | CYP2D6, CYP2C19, CYP2C9, CYP3A4, & VKORC1 | 1 | — | — | — | 1 | Citalopram, escitalopram, fluvoxamine, paroxetine, sertraline, amitriptyline, clomipramine, doxepin, imipramine, nortriptyline, trimipramine, desipramine celecoxib, hydrocodone, flurbiprofen, ibuprofen, meloxicam, tramadol, piroxicam, phenytoin, clopidogrel, codeine, voriconazole |
| Pegylated interferon 111 , 112 | IFNL3 (IL‐28B) | 2 | — | 2 | — | — | Pegylated interferon alpha |
| Proton Pump Inhibitors 113 | CYP2C19 | 1 | 1 | — | — | — | Omeprazole, lansoprazole |
| Selective Estrogen Receptor Modulator 114 , 115 , 116 | CYP2D6 | 3 | — | 2 | — | 1 | Tamoxifen |
| Thiopurines 117 , 118 , 119 , 120 , 121 , 122 , 123 , 124 , 125 , 126 , 127 | TPMT | 11 | 3 | 5 | 2 | 1 | Azathioprine, mercaptopurine |
| Total | All | 108 | 29 | 48 | 21 | 10 | All |
Antidepressant studies contain variable multigene panels ranging from 6 to 13 genes. —, none.
Thirty‐four studies did not calculate an ICER and 36 did not indicate a willingness‐to‐pay threshold. All studies evaluating DPYD and fluoropyrimidines, 10 of 11 thiopurine studies, both voriconazole studies, 2 of 8 abacavir studies, 7 of 11 antidepressant studies, 4 of 23 clopidogrel studies, 1 proton pump inhibitor study, and 2 of 16 warfarin studies did not report an ICER. Of studies classified as either cost‐effective or cost‐saving and reporting both an ICER and willingness‐to‐pay threshold, the ICER was consistently less than the willingness‐to‐pay threshold (Supplemental Table S1 ). Of the studies that did report a willingness‐to‐pay threshold, the currency and value varied drastically (~ US $5,000 to over US $100,000).
Cost outcomes by study characteristics
Figures for cost outcomes stratified by study characteristics, including drug / drug class, gene, publication year, cohort type, perspective, continent, QHES score, and funding are illustrated in Supplemental Figure S1 . Several notable trends existed; however, statistical comparisons were not applied due to small numbers. For example, 13 out of 14 (93%) studies funded by pharmaceutical companies demonstrated PGx testing was cost‐effective or cost‐saving, whereas 64 out of the remaining 94 (68%) studies funded by the government, grants/foundations, universities/institutes, or no funding was used, demonstrated PGx testing was cost‐effective or cost‐saving. Regarding continent, studies performed in Asia (n = 28) had a higher rate of demonstrating PGx testing was not cost‐effective (n = 10; 36%) compared with North America, Europe, and Australia where 11 out of 80 (14%) studies demonstrated PGx testing was not cost‐effective. Nineteen out of 75 studies (25%) based on a hypothetical cohort demonstrated PGx testing was not cost‐effective, whereas 2 out of 33 studies (6%) based on a cohort from observational, retrospective, prospective, or randomized controlled trials demonstrated PGx testing was not cost‐effective. Studies with high QHES scores (75–100) appeared more likely to demonstrate PGx testing was not cost‐effective or uncertain (29 out of 94; 31%) compared with those with QHES scores <75 (2 out of 14; 14%).
DISCUSSION
To aid in the adoption and interpretation of PGx results in clinical practice, CPIC has published many peer‐reviewed guidelines on how to translate PGx results into actionable prescribing decisions, while the FDA has published their standards for which drug–gene pairs have sufficient scientific evidence to suggest PGx impacts drug safety/response and/or metabolism. 6 , 7 Importantly, these resources do not evaluate or reflect on whether PGx testing is cost‐effective, which is a common cited barrier to adoption. 8 In one of the largest systematic reviews evaluating the evidence on cost‐effectiveness of PGx‐guided therapy for drugs with CPIC guidelines, we identified that about three‐quarters of articles determined PGx testing was either cost‐effective or cost‐saving, most of which were based on single‐gene tests.
Although the cost of PGx testing has declined dramatically over the last decade, it continues to be one of the major barriers to widescale adoption in clinical practice. Verbelen et al. provided important insight into which drug–gene pairs with FDA labeling had undergone cost evaluations prior to 2015. 10 The results showed more than half of the 44 articles reviewed found PGx testing was either cost‐effective (30%) or cost‐saving (27%). Warfarin accounted for more than one‐fourth of the studies included. The authors concluded that as genetic testing becomes cheaper, more widely available, and more accessible, further economic evaluations would likely find PGx‐guided treatment to be more cost‐effective compared with standard treatment. Importantly, the focus was on drug–gene pairs with PGx mentioned in the FDA label, whereas our analysis focused on those with CPIC guidelines since these are widely used among implementers in the United States for translating PGx results in clinical practice. Since the Verbelen et al. publication in 2015, 64 articles evaluating cost of a PGx‐guided to non‐PGx‐guided approach for drugs with CPIC guidelines have been published. Of these, 47 (73%) found PGx‐guided treatment to be cost‐effective or cost‐saving.
It is important to consider the clinical utility and cost impact of multigene testing vs. single gene. It is recognized that there are minimal cost differences between single‐gene testing and multigene testing platforms for targeted candidate genes. Thus, the cost of one panel test may inform drug/dose selection for many drugs. Hart et al. evaluated the cost‐effectiveness associated with a preemptive multigene test focused on CYP2C19 genotype‐guided antiplatelet therapy and CYP2D6 genotype‐guided pain management, compared with single‐gene test for CYP2C19 with random assignment for pain treatment, and with no testing. 15 As expected, the multigene testing strategy showed a favorable ICER compared with other treatment strategies.
Most articles evaluated in this review addressed PGx testing for one or specific set of genes and one associated drug or drug class in a single therapeutic area, all of which were deemed to be appropriate populations for the drug–gene pair tested. There is likely downstream cost savings if using one multigene panel to estimate the combined cost impact on cancer, cardiology, depression, and other therapeutic areas throughout one's lifetime; however, this has not been quantified. There are many instances when one or two genes can impact dozens of drugs across different drug classes. For example, CYP2C19 genotyping could be informative for antidepressants, clopidogrel, proton pump inhibitors, and voriconazole. The cost of PGx testing is a potentially one‐time fixed cost, while the downstream benefits are multiple and beyond what these studies addressed. Further, in clinical practice patients are often taking more than one drug or are at risk of requiring multiple drugs later in life. Thus, the generalizability of these studies is rather limited by their study design, which is focused on limited gene(s) and drug(s). For example, 9 out of 16 articles did not find genotyping before warfarin to be cost‐effective or cost‐saving, which could potentially change if one considers the lifelong benefit of a one‐time multigene PGx test (i.e., if CYP2C9 and VKORC1 results were readily available). Inevitably, the more drugs the patient is on or may require in the future that are affected by a given gene or set of genes, the more likely a PGx test will yield cost‐effectiveness or cost‐savings.
Preemptive vs. reactive PGx testing is likely to impact cost outcomes. Single‐gene testing is most often performed reactively (e.g., after the patient initiates a specific drug), but as multigene testing becomes more widespread, the results may be used both reactively and preemptively for future medications in various therapeutic areas. Zhu et al. performed a systematic review on cost‐effectiveness of PGx‐guided treatment for cardiovascular disease and identified that two‐thirds of studies were cost‐effective, but only 2 of the 46 articles evaluated panel testing and only one examined preemptive testing. 17 Subsequently, the investigators compared the cost‐effectiveness between preemptive PGx panel testing, reactive testing, and usual care (no testing) in cardiovascular disease management, including CYP2C19‐clopidogrel, CYP2C9/VKORC1‐warfarin, and SLCO1B1‐statins. 18 Using a decision analytic model, the authors demonstrated that preemptive testing was cost‐effective compared with usual care, while reactive testing was not. Those with longer follow up and aged 45 to 64 benefited the most. The PREDICT (Pharmacogenomic Resource for Enhanced Decisions in Care & Treatment) study from Vanderbilt demonstrated that preemptive genotyping of nearly 10,000 patients allowed for almost 15,000 reactive genetic tests to be avoided when data on multiple genes was available beforehand, thereby saving genotyping test costs by reducing the number of single‐gene tests required. 19 The Mayo Clinic RIGHT (Right Drug, Right Dose, Right Time: Using Genomic Data to Individualize Treatment) program reported that among 1,013 patients tested, 99% carried an actionable variant in SLCO1B1, CYP2C19, CYP2C9, or VKORC1. 20 Analysis of the first 5,000 patients tested in the eMERGE‐PGx (partnership of the Electronic Medical Records and Genomics Network and the Pharmacogenomics Research Network) project found that >96% of samples had CPIC‐designated high‐priority actionable variants. 21 It is consistently shown that most patients undergoing multigene panel testing harbor actionable variants. Based on these data and findings from the present systematic review, it is evident preemptive panel testing may yield significant cost savings over the course of a patient's medical care.
Some notable trends emerged when evaluating cost outcomes by study characteristics, such as source of funding, continent where the analysis occurred, and cohort type. While statistical comparisons between these groups was not possible due to small numbers, it is important to note that certain characteristics may influence study findings. Studies funded by pharmaceutical companies overwhelmingly demonstrated that PGx testing was cost‐effective or cost‐saving. Slight differences in methodology, including established willingness‐to‐pay thresholds, may also influence cost outcomes. Country or continent of origin may also impact findings. Studies conducted in Asia or based on hypothetical cohorts were more likely to demonstrate PGx testing was not cost‐effective. Genetic ancestry is also likely to influence allele frequency and number‐needed‐to‐test to avoid an adverse drug reaction.
There are limitations of this analysis, including a limited number of studies for some drug–gene pairs. Of the 108 articles included in this review, 39 focused on warfarin or clopidogrel, accounting for 36% of all articles. The finding that most studies were cost‐effective or cost‐saving cannot be generalized to all drugs with CPIC guidelines given the limited representation of drugs in the literature (e.g., analgesics and proton pump inhibitors only had one study each that evaluated cost). Further, almost half of the drugs with CPIC guidelines have no published studies evaluating cost, which are needed to inform the clinical, research, and economic communities. Publication bias may also impact which studies were available from the systematic literature search. The analysis was meant to be inclusive of any study evaluating cost and PGx testing of a drug with a CPIC guideline with full article available. These comprehensive criteria resulted in the inclusion of some studies which assessed additional drug–gene pairs which are not CPIC level A or B with a guideline (e.g., multigene panels for antidepressant selection). We did not exclude articles based on country of publication, type of journal, study design/cohort, or year. Most articles were based on a hypothetical cohort, which may not translate to real‐world practice. The cost impact could potentially vary between countries due to differences in cost of testing and the frequency of genomic variants based on ancestry. In addition to the range of willingness‐to‐pay thresholds between studies, the currency of the threshold and ICER also varied as most studies reported in the currency of the country of the population evaluated. Thus, it is difficult to generalize cost‐effectiveness or cost‐savings without accounting for potential differences between the populations studied and population of interest. Although cost outcomes were dependent on the respective study investigators' conclusions/findings, the average QHES was overall high, suggesting these studies were mostly robust. Studies with lower QHES scores were more likely to find an intervention cost‐effective, suggesting a more robust evaluation may lend itself to identifying lack of cost‐effectiveness; however, the number of studies with low QHES scores was very low. Lastly, we included drugs with CPIC guidelines as of June 2021. Inevitably, more guidelines have and will be produced as the data evolve and several drugs are considered provisional, which if included, may change the findings of this systematic review.
In conclusion, we identified that most published studies evaluating cost and PGx testing for genes with CPIC guidelines determined that PGx‐guided treatment was cost‐effective or cost‐saving. This is similar to prior publications and systematic reviews, such as the evaluation by Verbelen et al. of drugs with PGx in the FDA label. 10 The novelty in this systematic review is that it is all inclusive of drugs with CPIC guidelines published up to June 2021, which are considered the most widely utilized guidelines to facilitate adoption and interpretation of PGx‐guided therapy in clinical practice in the United States. CPIC guidelines and evidence levels are also now cited in various local coverage determinations such as those from the Centers for Medicare and Medicaid Services. 22 Empey et al. highlights how expanding evidence has led to new PGx payor coverage and proposes mechanisms to overcome implementation obstacles to fully comprehend the value of PGx testing, including harmonization of coverage between states and jurisdictions, greater standardization of testing and interpretation, greater clarity on the regulatory landscape, practitioner education, and point‐of‐care clinical decision support. 22
While some drugs, like clopidogrel, have an abundance of cost data showing there is enough evidence to support the use of PGx testing, others, like anesthesia and NSAIDs, still lack robust cost evaluations. Future studies and cost evaluations should focus on the impact of reactive and preemptive multigene panel testing on downstream clinical outcomes across drugs and drug classes and reduction in healthcare utilization. In addition to data on clinical utility, the data presented herein may support further coverage determinations by payors, thus increasing accessibility to evidence‐based PGx testing.
FUNDING
No funding was received for this work.
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
All authors wrote the manuscript, performed the research, and analyzed the data. S.A.M. and J.N.P. designed the research.
Supporting information
Figure S1
Data S1
Table S1
References
- 1. Wang, L. , McLeod, H.L. & Weinshilboum, R.M. Genomics and drug response. New Engl. J. Med. 364(12), 1144–1153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martin, M.A. , Klein, T.E. , Dong, B.J. , Pirmohamed, M. , Haas, D.W. & Kroetz, D.L. Clinical pharmacogenetics implementation consortium guidelines for HLA‐B genotype and abacavir dosing. Clin. Pharmacol. Therapeut. 91(4), 734–738 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scott, S.A. et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C19 genotype and Clopidogrel therapy: 2013 update. Clin. Pharmacol. Therapeut. 94(3), 317–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hicks, J.K. et al. Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Therapeut. 98(2), 127–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. US Food and Drug Administration Table of pharmacogenomic biomarkers in drug labeling (U.S. Food and Drug Administration; ). https://www.fda.gov/drugs/science‐and‐research‐drugs/table‐pharmacogenomic‐biomarkers‐drug‐labeling Accessed June 24, 2022. [Google Scholar]
- 6. Center for Devices and Radiological Health Table of pharmacogenetic associations (U.S. Food and Drug Administration; ). https://www.fda.gov/medical‐devices/precision‐medicine/table‐pharmacogenetic‐associations Accessed June 24, 2022. [Google Scholar]
- 7. Caudle, K. et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline Development Process. Curr. Drug Metab. 15(2), 209–217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong, W.B. , Carlson, J.J. , Thariani, R. & Veenstra, D.L. Cost effectiveness of pharmacogenomics. Pharmacoeconomics 28(11), 1001–1013 (2010). [DOI] [PubMed] [Google Scholar]
- 9. Berndt, E.R. , Goldman, D.P. & Rowe, J. The value of pharmacogenomic information. In Economic Dimensions of Personalized and Precision Medicine 53–86 (The University of Chicago Press, Chicago, IL, 2019). [Google Scholar]
- 10. Verbelen, M. , Weale, M.E. & Lewis, C.M. Cost‐effectiveness of pharmacogenetic‐guided treatment: are we there yet? Pharmacogenomics J. 17(5), 395–402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chiou, C.‐F. et al. Development and validation of a grading system for the quality of cost‐effectiveness studies. Med. Care 41(1), 32–44 (2003). [DOI] [PubMed] [Google Scholar]
- 12. Spiegel, B.M.R. et al. The quality of published health economic analyses in digestive diseases: a systematic review and quantitative appraisal. Gastroenterology 127(2), 403–411 (2004). [DOI] [PubMed] [Google Scholar]
- 13. Ofman, J.J. et al. Examining the value and quality of health economic analyses: implications of utilizing the QHES. J. Manag. Care Pharm. 9(1), 53–61 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marshall, D.A. et al. Assessing the quality of economic evaluations of clinical nurse specialists and Nurse Practitioners: a systematic review of cost‐effectiveness. NursingPlus Open 1, 11–17 (2015). [Google Scholar]
- 15. Hart, M.R. , Garrison, L.P. , Doyle, D.L. , Jarvik, G.P. , Watkins, J. & Devine, B. Projected cost‐effectiveness for 2 gene‐drug pairs using a multigene panel for patients undergoing percutaneous coronary intervention. Value Health 22(11), 1231–1239 (2019). [DOI] [PubMed] [Google Scholar]
- 16. Brixner, D. et al. The effect of pharmacogenetic profiling with a clinical decision support tool on healthcare resource utilization and estimated costs in the elderly exposed to polypharmacy. J. Med. Econ. 19(3), 213–228 (2015). [DOI] [PubMed] [Google Scholar]
- 17. Zhu, Y. et al. Systematic review of the evidence on the cost‐effectiveness of pharmacogenomics‐guided treatment for cardiovascular diseases. Genet. Med. 22(3), 475–486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu, Y. et al. A model‐based cost‐effectiveness analysis of pharmacogenomic panel testing in Cardiovascular Disease Management: preemptive, reactive, or none? Genet. Med. 23(3), 461–470 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Van Driest, S.L. , Shi, Y. , Bowton, E.A. et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing: a comprehensive analysis of five actionable pharmacogenomic genes using next‐generation DNA sequencing and a customized CYP2D6 genotyping cascade. Clin. Pharmacol. Therapeut. 95(4), 423–431 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ji, Y. et al. Preemptive pharmacogenomic testing for precision medicine: a comprehensive analysis of five actionable pharmacogenomic genes using next‐generation DNA sequencing and a customized CYP2D6 genotyping cascade. J. Mol. Diagn. 18(3), 438–445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bush, W.S. et al. Genetic variation among 82 pharmacogenes: the PGRNSEQ data from the eMERGE network. Clin. Pharmacol. Therapeut. 100(2), 160–169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Empey, P.E. , Pratt, V.M. , Hoffman, J.M. , Caudle, K.E. & Klein, T.E. Expanding evidence leads to new pharmacogenomics payer coverage. Genet. Med. 23(5), 830–832 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Veenstra, D.L. , Harris, J. , Gibson, R.L. , Rosenfeld, M. , Burke, W. & Watts, C. Pharmacogenomic testing to prevent aminoglycoside‐induced hearing loss in cystic fibrosis patients: potential impact on clinical, patient, and economic outcomes. Genet. Med. 9(10), 695–704 (2007). [DOI] [PubMed] [Google Scholar]
- 24. Moretti, M.E. , Lato, D.F. , Berger, H. , Koren, G. , Ito, S. & Ungar, W.J. A cost‐effectiveness analysis of maternal CYP2D6 genetic testing to guide treatment for postpartum pain and avert infant adverse events. Pharmacogenomics J. 18(3), 391–397 (2017). [DOI] [PubMed] [Google Scholar]
- 25. You, J.H.S. , Tsui, K.K. , Wong, R.S. & Cheng, G. Potential clinical and economic outcomes of CYP2C9 and VKORC1 genotype‐guided dosing in patients starting warfarin therapy. Clin. Pharmacol. Therapeut. 86(5), 540–547 (2009). [DOI] [PubMed] [Google Scholar]
- 26. Patrick, A.R. , Avorn, J. & Choudhry, N.K. Cost‐effectiveness of genotype‐guided warfarin dosing for patients with atrial fibrillation. Circ. Cardiovasc. Qual. Outcomes 2(5), 429–436 (2009). [DOI] [PubMed] [Google Scholar]
- 27. Pink, J. , Pirmohamed, M. , Lane, S. & Hughes, D.A. Cost‐effectiveness of pharmacogenetics‐guided warfarin therapy vs. alternative anticoagulation in atrial fibrillation. Clin. Pharmacol. Therapeut. 95(2), 199–207 (2013). [DOI] [PubMed] [Google Scholar]
- 28. Nshimyumukiza, L. et al. Dabigatran versus warfarin under standard or pharmacogenetic‐guided management for the prevention of stroke and systemic thromboembolism in patients with atrial fibrillation: a cost/utility analysis using an analytic decision model. Thrombosis J. 11(1) (2013). 10.1186/1477-9560-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. You, J.H.S. Pharmacogenetic‐guided selection of warfarin versus novel oral anticoagulants for stroke prevention in patients with atrial fibrillation. Pharmacogenet. Genomics 24(1), 6–14 (2014). [DOI] [PubMed] [Google Scholar]
- 30. Eckman, M.H. Cost‐effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann. Intern. Med. 150(2), 73–83 (2009). [DOI] [PubMed] [Google Scholar]
- 31. Leey, J.A. , McCabe, S. , Koch, J.A. & Miles, T.P. Cost‐effectiveness of genotype‐guided warfarin therapy for anticoagulation in elderly patients with atrial fibrillation. Am. J. Geriatr. Pharmacother. 7(4), 197–203 (2009). [DOI] [PubMed] [Google Scholar]
- 32. Meckley, L.M. , Gudgeon, J.M. , Anderson, J.L. , Williams, M.S. & Veenstra, D.L. A policy model to evaluate the benefits, risks and costs of warfarin pharmacogenomic testing. Pharmacoeconomics 28(1), 61–74 (2010). [DOI] [PubMed] [Google Scholar]
- 33. You, J.H. , Tsui, K.K. , Wong, R.S. & Cheng, G. Cost‐effectiveness of dabigatran versus genotype‐guided management of warfarin therapy for stroke prevention in patients with atrial fibrillation. PLoS ONE 7(6), e39640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chong, H.Y. et al. Cost‐effectiveness analysis of pharmacogenetic‐guided warfarin dosing in Thailand. Thromb. Res. 134(6), 1278–1284 (2014). [DOI] [PubMed] [Google Scholar]
- 35. Mitropoulou, C. et al. Economic evaluation of pharmacogenomic‐guided warfarin treatment for elderly Croatian atrial fibrillation patients with ischemic stroke. Pharmacogenomics 16(2), 137–148 (2015). [DOI] [PubMed] [Google Scholar]
- 36. Kim, D.J. , Kim, H.S. , Oh, M. , Kim, E.Y. & Shin, J.G. Cost effectiveness of genotype‐guided warfarin dosing in patients with mechanical heart valve replacement under the fee‐for‐service system. Appl. Health Econ. Health Policy 15(5), 657–667 (2017). [DOI] [PubMed] [Google Scholar]
- 37. Martes‐Martinez, C. et al. Cost‐utility study of warfarin genotyping in the VACHS affiliated anticoagulation clinic of Puerto Rico. P. R. Health Sci. J. 36(3), 165–172 (2017). [PMC free article] [PubMed] [Google Scholar]
- 38. You, J.H.S. Universal versus genotype‐guided use of direct oral anticoagulants in atrial fibrillation patients: A decision analysis. Pharmacogenomics 16(10), 1089–1100 (2015). [DOI] [PubMed] [Google Scholar]
- 39. Verhoef, T.I. et al. Cost‐effectiveness of pharmacogenetic‐guided dosing of warfarin in the United Kingdom and Sweden. Pharmacogenomics J. 16(5), 478–484 (2016). [DOI] [PubMed] [Google Scholar]
- 40. Chan, F. , Wong, R. , Cheng, G. & You, J. The potential clinical and economic outcomes of pharmacogenetics‐oriented management of warfarin therapy – A decision analysis. Thromb. Haemost. 92(9), 590–597 (2004). [DOI] [PubMed] [Google Scholar]
- 41. Sluiter, R.L. , Janzing, J.G. , van der Wilt, G.J. , Kievit, W. & Teichert, M. An economic model of the cost‐utility of pre‐emptive genetic testing to support pharmacotherapy in patients with major depression in primary care. Pharmacogenomics J. 19(5), 480–489 (2019). [DOI] [PubMed] [Google Scholar]
- 42. Berm, E.J. , Gout‐Zwart, J.J. , Luttjeboer, J. , Wilffert, B. & Postma, M.J. A model based cost‐effectiveness analysis of routine genotyping for CYP2D6 among older, depressed inpatients starting nortriptyline pharmacotherapy. PLOS ONE 11(12), e0169065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hornberger, J. , Li, Q. & Quinn, B. Cost‐effectiveness of combinatorial pharmacogenomic testing for treatment‐resistant major depressive disorder patients. Am. J. Manag. Care 21(6), e357–e365 (2015) PMID: 26247576. [PubMed] [Google Scholar]
- 44. Groessl, E.J. , Tally, S.R. , Hillery, N. , Maciel, A. & Garces, J.A. Cost‐effectiveness of a pharmacogenetic test to guide treatment for major depressive disorder. J. Manag. Care Spec. Pharm. 24(8), 726–734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perlis, R.H. , Mehta, R. , Edwards, A.M. , Tiwari, A. & Imbens, G.W. Pharmacogenetic testing among patients with mood and anxiety disorders is associated with decreased utilization and cost: a propensity‐score matched study. Depress. Anxiety 35(10), 946–952 (2018). [DOI] [PubMed] [Google Scholar]
- 46. Najafzadeh, M. , Garces, J.A. & Maciel, A. Economic evaluation of implementing a novel Pharmacogenomic Test (IDgenetix®) to guide treatment of patients with depression åand/or anxiety. Pharmacoeconomics 35(12), 1297–1310 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tanner, J.A. , Davies, P.E. , Overall, C.C. , Grima, D. , Nam, J. & Dechairo, B.M. Cost–effectiveness of combinatorial pharmacogenomic testing for depression from the Canadian Public Payer Perspective. Pharmacogenomics 21(8), 521–531 (2020). [DOI] [PubMed] [Google Scholar]
- 48. Winner, J.G. et al. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a 1 year prospective evaluation. Curr. Med. Res. Opin. 31(9), 1633–1643 (2015). [DOI] [PubMed] [Google Scholar]
- 49. Fagerness, J. et al. Pharmacogenetic‐guided psychiatric intervention associated with increased adherence and cost savings. Am. J. Manag. Care 20(5), e146–e156 (2014). [PubMed] [Google Scholar]
- 50. Olson, M.C. et al. Clinical impact of pharmacogenetic‐guided treatment for patients exhibiting neuropsychiatric disorders. Prim. Care Companion CNS Disord. 19(2) (2017). 10.4088/pcc.16m02036. [DOI] [PubMed] [Google Scholar]
- 51. Maciel, A. , Cullors, A. , Lukowiak, A.A. & Garces, J. Estimating cost savings of pharmacogenetic testing for depression in real‐world clinical settings. Neuropsychiatr. Dis. Treat. 14, 225–230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Plumpton, C.O. , Yip, V.L. , Alfirevic, A. , Marson, A.G. , Pirmohamed, M. & Hughes, D.A. Cost‐effectiveness of screening for HLA‐A*31:01 prior to initiation of carbamazepine in epilepsy. Epilepsia 56(4), 556–563 (2015). [DOI] [PubMed] [Google Scholar]
- 53. Dong, D. , Sung, C. & Finkelstein, E.A. Cost‐effectiveness of HLA‐B*1502 genotyping in adult patients with newly diagnosed epilepsy in Singapore. Neurology 79(12), 1259–1267 (2012). [DOI] [PubMed] [Google Scholar]
- 54. Tiamkao, S. , Jitpimolmard, J. , Sawanyawisuth, K. & Jitpimolmard, S. Cost minimization of HLA‐B*1502 screening before prescribing carbamazepine in Thailand. Int. J. Clin. Pharm. 35(4), 608–612 (2013). [DOI] [PubMed] [Google Scholar]
- 55. Chong, H.Y. et al. Is universal HLA‐b*15:02 screening a cost‐effective option in an ethnically diverse population? A case study of Malaysia. Br. J. Dermatol. 177(4), 1102–1112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Choi, H. & Mohit, B. Cost‐effectiveness of screening for HLA‐B*1502 prior to initiation of carbamazepine in epilepsy patients of Asian ancestry in the United States. Epilepsia 60(7), 1472–1481 (2019). [DOI] [PubMed] [Google Scholar]
- 57. Chen, Z. , Liew, D. & Kwan, P. Real‐world cost‐effectiveness of pharmacogenetic screening for epilepsy treatment. Neurology 86(12), 1086–1094 (2016). [DOI] [PubMed] [Google Scholar]
- 58. Yuliwulandari, R. et al. Cost‐effectiveness analysis of genotyping for HLA‐B*15:02 in Indonesian patients with epilepsy using a generic model. Pharmacogenomics J. 21(4), 476–483 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rattanavipapong, W. , Koopitakkajorn, T. , Praditsitthikorn, N. , Mahasirimongkol, S. & Teerawattananon, Y. Economic evaluation of HLA‐B*15:02 screening for carbamazepine‐induced severe adverse drug reactions in Thailand. Epilepsia 54(9), 1628–1638 (2013). [DOI] [PubMed] [Google Scholar]
- 60. Mason, N.T. , Bell, G.C. , Quilitz, R.E. , Greene, J.N. & McLeod, H.L. Budget impact analysis of CYP2C19‐guided voriconazole prophylaxis in AML. J. Antimicrob. Chemother. 70(11), 3124–3126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Patel, J.N. et al. Evaluation of CYP2C19 genotype‐guided voriconazole prophylaxis after allogeneic hematopoietic cell transplant. Clin. Pharmacol. Therapeut. 107(3), 571–579 (2019). [DOI] [PubMed] [Google Scholar]
- 62. Dong, D. , Tan‐Koi, W.C. , Teng, G.G. , Finkelstein, E. & Sung, C. Cost–effectiveness analysis of genotyping for HLA‐B*5801 and an enhanced safety program in gout patients starting allopurinol in Singapore. Pharmacogenomics 16(16), 1781–1793 (2015). [DOI] [PubMed] [Google Scholar]
- 63. Ke, C.H. et al. Cost‐effectiveness analysis for genotyping before allopurinol treatment to prevent severe cutaneous adverse drug reactions. J. Rheumatol. 44(6), 835–843 (2017). [DOI] [PubMed] [Google Scholar]
- 64. Saokaew, S. , Tassaneeyakul, W. , Maenthaisong, R. & Chaiyakunapruk, N. Cost‐effectiveness analysis of HLA‐B*5801 testing in preventing allopurinol‐induced SJS/ten in Thai population. PLoS One 9(4), e94294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jutkowitz, E. , Dubreuil, M. , Lu, N. , Kuntz, K.M. & Choi, H.K. The cost‐effectiveness of HLA‐B*5801 screening to guide initial urate‐lowering therapy for gout in the United States. Semin. Arthritis Rheum. 46(5), 594–600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chong, H.Y. , Lim, Y.H. , Prawjaeng, J. , Tassaneeyakul, W. , Mohamed, Z. & Chaiyakunapruk, N. Cost‐effectiveness analysis of HLA‐B*58:01 genetic testing before initiation of allopurinol therapy to prevent allopurinol‐induced Stevens–Johnson syndrome/toxic epidermal necrolysis in a Malaysian population. Pharmacogenet. Genomics 28(2), 56–67 (2018). [DOI] [PubMed] [Google Scholar]
- 67. Park, D.J. et al. Cost‐effectiveness analysis of HLA‐B5801 genotyping in the treatment of gout patients with chronic renal insufficiency in Korea. Arthritis Care Res. 67(2), 280–287 (2015). [DOI] [PubMed] [Google Scholar]
- 68. Plumpton, C.O. , Alfirevic, A. , Pirmohamed, M. & Hughes, D.A. Cost effectiveness analysis of HLA‐B*58:01 genotyping prior to initiation of allopurinol for gout. Rheumatology 56(10), 1729–1739 (2017). [DOI] [PubMed] [Google Scholar]
- 69. Cheng, H. , Yan, D. , Zuo, X. , Liu, J. , Liu, W. & Zhang, Y. A retrospective investigation of HLA‐B*5801 in hyperuricemia patients in a Han population of China. Pharmacogenet. Genomics 28(5), 117–124 (2018). [DOI] [PubMed] [Google Scholar]
- 70. Teng, G.G. , Tan‐Koi, W.‐C. , Dong, D. & Sung, C. Is HLA‐B*58:01 genotyping cost effective in guiding allopurinol use in gout patients with chronic kidney disease? Pharmacogenomics 21(4), 279–291 (2020). [DOI] [PubMed] [Google Scholar]
- 71. S‐ling, P. , Jeon, Y. , Pearce, F. , Thong, B. & Aziz, M. Cost‐effectiveness of sequential urate lowering therapies for the management of gout in Singapore. J. Med. Econ. 23(8), 838–847 (2020). [DOI] [PubMed] [Google Scholar]
- 72. Reese, E.S. , Daniel Mullins, C. , Beitelshees, A.L. & Onukwugha, E. Cost‐effectiveness of cytochrome P450 2C19 genotype screening for selection of antiplatelet therapy with clopidogrel or prasugrel. Pharmacotherapy 32(4), 323–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sorich, M.J. , Horowitz, J.D. , Sorich, W. , Wiese, M.D. , Pekarsky, B. & Karnon, J.D. Cost–effectiveness of using CYP2C19 genotype to guide selection of clopidogrel or ticagrelor in Australia. Pharmacogenomics 14(16), 2013–2021 (2013). [DOI] [PubMed] [Google Scholar]
- 74. Lala, A. , Berger, J.S. , Sharma, G. , Hochman, J.S. , Scott Braithwaite, R. & Ladapo, J.A. Genetic testing in patients with acute coronary syndrome undergoing percutaneous coronary intervention: A cost‐effectiveness analysis. J. Thromb. Haemost. 11(1), 81–91 (2013). [DOI] [PubMed] [Google Scholar]
- 75. Crespin, D.J. , Federspiel, J.J. , Biddle, A.K. , Jonas, D.E. & Rossi, J.S. Ticagrelor versus genotype‐driven antiplatelet therapy for secondary prevention after acute coronary syndrome: a cost‐effectiveness analysis. Value Health 14(4), 483–491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Panattoni, L. , Brown, P.M. , Te Ao, B. , Webster, M. & Gladding, P. The cost effectiveness of genetic testing for CYP2C19 variants to guide thienopyridine treatment in patients with acute coronary syndrome: a New Zealand evaluation. Pharmacoeconomics 30(11), 1067–1084 (2012). [DOI] [PubMed] [Google Scholar]
- 77. Kazi, D.S. et al. Cost‐effectiveness of genotype‐guided and dual antiplatelet therapies in acute coronary syndrome. Ann. Intern. Med. 160(4), 221–232 (2014). [DOI] [PubMed] [Google Scholar]
- 78. Wang, Y. , Yan, B.P. , Liew, D. & Lee, V.W. Cost‐effectiveness of cytochrome P450 2C19 *2 genotype‐guided selection of clopidogrel or ticagrelor in Chinese patients with acute coronary syndrome. Pharmacogenomics J. 18(1), 113–120 (2017). [DOI] [PubMed] [Google Scholar]
- 79. Fragoulakis, V. et al. Cost‐effectiveness analysis of pharmacogenomics‐guided clopidogrel treatment in Spanish patients undergoing percutaneous coronary intervention. Pharmacogenomics J. 19(5), 438–445 (2019). [DOI] [PubMed] [Google Scholar]
- 80. Mitropoulou, C. et al. Economic analysis of pharmacogenomic‐guided clopidogrel treatment in Serbian patients with myocardial infarction undergoing primary percutaneous coronary intervention. Pharmacogenomics 17(16), 1775–1784 (2016). [DOI] [PubMed] [Google Scholar]
- 81. Borse, M.S. , Dong, O.M. , Polasek, M.J. , Farley, J.F. , Stouffer, G.A. & Lee, C.R. CYP2C19‐guided antiplatelet therapy: a cost–effectiveness analysis of 30‐day and 1‐year outcomes following percutaneous coronary intervention. Pharmacogenomics 18(12), 1155–1166 (2017). [DOI] [PubMed] [Google Scholar]
- 82. Kim, K. , Touchette, D.R. , Cavallari, L.H. , Ardati, A.K. & DiDomenico, R.J. Cost‐effectiveness of strategies to personalize the selection of P2Y12 inhibitors in patients with acute coronary syndrome. Cardiovasc. Drugs Ther. 33(5), 533–546 (2019). [DOI] [PubMed] [Google Scholar]
- 83. Jiang, M. & You, J.H.S. CYP2C19 genotype plus platelet reactivity‐guided antiplatelet therapy in acute coronary syndrome patients. Pharmacogenet. Genomics 25(12), 609–617 (2015). [DOI] [PubMed] [Google Scholar]
- 84. Jiang, M. & You, J.H.S. CYP2C19 LOF and GOF‐guided antiplatelet therapy in patients with acute coronary syndrome: a cost‐effectiveness analysis. Cardiovasc. Drugs Ther. 31(1), 39–49 (2017). [DOI] [PubMed] [Google Scholar]
- 85. Jiang, M. & You, J.H.S. Cost–effectiveness analysis of personalized antiplatelet therapy in patients with acute coronary syndrome. Pharmacogenomics 17(7), 701–713 (2016). [DOI] [PubMed] [Google Scholar]
- 86. Fu, Y. , X‐yi, Z. , S‐bei, Q. et al. Cost–effectiveness of CYP2C19 LOF‐guided antiplatelet therapy in Chinese patients with acute coronary syndrome. Pharmacogenomics 21(1), 33–42 (2020). [DOI] [PubMed] [Google Scholar]
- 87. Limdi, N.A. et al. Cost‐effectiveness of CYP2C19‐guided antiplatelet therapy in patients with acute coronary syndrome and percutaneous coronary intervention informed by real‐world data. Pharmacogenomics J. 20(5), 724–735 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Narasimhalu, K. , Ang, Y.K. , Tan, D.S. , De Silva, D.A. & Tan, K.B. Cost effectiveness of genotype‐guided antiplatelet therapy in Asian ischemic stroke patients: ticagrelor as an alternative to clopidogrel in patients with CYP2C19 loss of function mutations. Clin. Drug Investig. 40(11), 1063–1070 (2020). [DOI] [PubMed] [Google Scholar]
- 89. Cai, Z. et al. Cost‐effectiveness of CYP2C19 genotyping to guide antiplatelet therapy for acute minor stroke and high‐risk transient ischemic attack. Sci. Rep. 11(1), 7383 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. AlMukdad, S. , Elewa, H. , Arafa, S. & Al‐Badriyeh, D. Short‐ and long‐term cost‐effectiveness analysis of CYP2C19 genotype‐guided therapy, universal clopidogrel, versus universal ticagrelor in post‐percutaneous coronary intervention patients in Qatar. Int. J. Cardiol. 331, 27–34 (2021). [DOI] [PubMed] [Google Scholar]
- 91. Kim, J.H. , Tan, D.S.‐Y. & Chan, M.Y. Cost‐effectiveness of CYP2C19‐guided antiplatelet therapy for acute coronary syndromes in Singapore. Pharmacogenomics J. 21(2), 243–250 (2021). [DOI] [PubMed] [Google Scholar]
- 92. Johnson, S.G. , Gruntowicz, D. , Chua, T. & Morlock, R.J. Financial Analysis of CYP2C19 genotyping in patients receiving dual antiplatelet therapy following acute coronary syndrome and percutaneous coronary intervention. J. Manag. Care Spec. Pharm. 21(7), 552–557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Patel, V. et al. Cost‐utility analysis of genotype‐guided antiplatelet therapy in patients with moderate‐to‐high risk acute coronary syndrome and planned percutaneous coronary intervention. Pharm. Pract. (Internet) 12(3), 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kauf, T.L. , Farkouh, R.A. , Earnshaw, S.R. , Watson, M.E. , Maroudas, P. & Chambers, M.G. Economic efficiency of genetic screening to inform the use of abacavir sulfate in the treatment of HIV. Pharmacoeconomics 28(11), 1025–1039 (2010). [DOI] [PubMed] [Google Scholar]
- 95. Nieves Calatrava, D. , Óde, C.‐M. , Iribarren‐Loyarte, J.A. et al. Cost‐effectiveness analysis of HLA‐B*5701 typing in the prevention of hypersensitivity to abacavir in HIV+ patients in Spain. Enferm. Infecc. Microbiol. Clin. 28(9), 590–595 (2010). [DOI] [PubMed] [Google Scholar]
- 96. Kapoor, R. et al. Reducing hypersensitivity reactions with HLA‐B*5701 genotyping before abacavir prescription: clinically useful but is it cost‐effective in Singapore? Pharmacogenet. Genomics 25(2), 60–72 (2015). [DOI] [PubMed] [Google Scholar]
- 97. Hughes, D.A. , Vilar, F.J. , Ward, C.C. , Alfirevic, A. , Park, B.K. & Pirmohamed, M. Cost‐effectiveness analysis of HLA B*5701 genotyping in preventing abacavir hypersensitivity. Pharmacogenetics 14(6), 335–342 (2004). [DOI] [PubMed] [Google Scholar]
- 98. Schackman, B.R. , Scott, C.A. , Walensky, R.P. , Losina, E. , Freedberg, K.A. & Sax, P.E. The cost‐effectiveness of HLA‐B*5701 genetic screening to guide initial antiretroviral therapy for HIV. AIDS 22(15), 2025–2033 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ruiz‐Iruela, C. et al. HLA‐B*57:01 genotyping in the prevention of hypersensitivity to abacavir: 5 years of experience. Pharmacogenet. Genomics 26(8), 390–396 (2016). [DOI] [PubMed] [Google Scholar]
- 100. Goh, K.S. , Kapoor, R. , Lee, C.C. , Ng, C.Y.L. & Leong, K.P. HLA‐B*5701 genotyping for abacavir prescription: Re‐examination of its cost‐effectiveness in Singapore. Ann. Acad. Med. Singapore 48(4), 133–138 (2019). [PubMed] [Google Scholar]
- 101. Wolf, E. et al. Cost impact of prospective HLA‐B*5701‐screening prior to abacavir/lamivudine fixed dose combination use in Germany. Eur. J. Med. Res. 15(4), 145–151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schackman, B.R. , Haas, D.W. , Park, S.S. , Li, X.C. & Freedberg, K.A. Cost–effectiveness of CYP2B6 genotyping to optimize Efavirenz dosing in HIV clinical practice. Pharmacogenomics 16(18), 2007–2018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Martín, A.S. et al. Dose reduction of Efavirenz: an observational study describing cost–effectiveness, pharmacokinetics and pharmacogenetics. Pharmacogenomics 15(7), 997–1006 (2014). [DOI] [PubMed] [Google Scholar]
- 104. Schackman, B.R. et al. Cost‐effectiveness analysis of UGT1A1 genetic testing to inform antiretroviral prescribing in HIV disease. Antivir. Ther. 18(3), 399–408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Dong, O.M. et al. Cost‐effectiveness of Multigene pharmacogenetic testing in patients with acute coronary syndrome after percutaneous coronary intervention. Value Health 23(1), 61–73 (2020). [DOI] [PubMed] [Google Scholar]
- 106. Murphy, C. et al. Cost implications of reactive versus prospective testing for dihydropyrimidine dehydrogenase deficiency in patients with colorectal cancer: a single‐institution experience. Dose‐Response 16(4), 155932581880304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Fragoulakis, V. et al. Estimating the effectiveness of DPYD genotyping in Italian individuals suffering from cancer based on the cost of chemotherapy‐induced toxicity. Am. J. Human Genet. 104(6), 1158–1168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Deenen, M.J. et al. Upfront genotyping of DPYD*2A to individualize fluoropyrimidine therapy: a safety and cost analysis. J. Clin. Oncol. 34(3), 227–234 (2016). [DOI] [PubMed] [Google Scholar]
- 109. Cortejoso, L. , García‐González, X. , García, M.I. , García‐Alfonso, P. , Sanjurjo, M. & López‐Fernández, L.A. Cost–effectiveness of screening for DPYD polymorphisms to prevent neutropenia in cancer patients treated with fluoropyrimidines. Pharmacogenomics 17(9), 979–984 (2016). [DOI] [PubMed] [Google Scholar]
- 110. Henricks, L.M. et al. A cost analysis of upfront DPYD genotype–guided dose individualization in fluoropyrimidine‐based anticancer therapy. Eur. J. Cancer 107, 60–67 (2019). [DOI] [PubMed] [Google Scholar]
- 111. Liu, S. , Cipriano, L.E. , Holodniy, M. , Owens, D.K. & Goldhaber‐Fiebert, J.D. New protease inhibitors for the treatment of chronic hepatitis C. Ann. Intern. Med. 156(4), 279–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Blázquez‐Pérez, A. , San Miguel, R. & Mar, J. Cost‐effectiveness analysis of triple therapy with protease inhibitors in treatment‐naive hepatitis C patients. Pharmacoeconomics 31(10), 919–931 (2013). [DOI] [PubMed] [Google Scholar]
- 113. Lehmann, D.F. , Medicis, J.J. & Franklin, P.D. Polymorphisms and the pocketbook: The cost‐effectiveness of cytochrome P450 2C19 genotyping in the eradication of Helicobacter pylori infection associated with duodenal ulcer. J. Clin. Pharmacol. 43(12), 1316–1323 (2003). [DOI] [PubMed] [Google Scholar]
- 114. Wei, X. et al. Cost‐effectiveness analysis of CYP2D6*10 pharmacogenetic testing to guide the adjuvant endocrine therapy for postmenopausal women with estrogen receptor positive early breast cancer in China. Clin. Drug Investig. 40(1), 25–32 (2019). [DOI] [PubMed] [Google Scholar]
- 115. Woods, B. , Veenstra, D. & Hawkins, N. Prioritizing pharmacogenetic research: a value of information analysis of CYP2D6 testing to guide breast cancer treatment. Value Health 14(8), 989–1001 (2011). [DOI] [PubMed] [Google Scholar]
- 116. Wei, X. et al. CYP2D6*10 pharmacogenetic‐guided SERM could be a cost‐effective strategy in Chinese patients with hormone receptor‐positive breast cancer. Pharmacogenomics 21(1), 43–53 (2020). [DOI] [PubMed] [Google Scholar]
- 117. Thompson, A.J. , Newman, W.G. , Elliott, R.A. , Roberts, S.A. , Tricker, K. & Payne, K. The cost‐effectiveness of a pharmacogenetic test: a trial‐based evaluation of TPMT genotyping for azathioprine. Value Health 17(1), 22–33 (2014). [DOI] [PubMed] [Google Scholar]
- 118. Marra, C.A. , Esdaile, J.M. & Anis, A.H. Practical pharmacogenetics: the cost effectiveness of screening for thiopurine s‐methyltransferase polymorphisms in patients with rheumatological conditions treated with azathioprine. J. Rheumatol. 29, 2507–2512 (2002). [PubMed] [Google Scholar]
- 119. Oh, K.T. Pharmacoeconomic analysis of thiopurine methyltransferase polymorphism screening by polymerase chain reaction for treatment with azathioprine in Korea. Rheumatology 43(2), 156–163 (2003). [DOI] [PubMed] [Google Scholar]
- 120. Priest, V.L. et al. Pharmacoeconomic analyses of azathioprine, methotrexate and prospective pharmacogenetic testing for the management of inflammatory bowel disease. Pharmacoeconomics 24(8), 767–781 (2006). [DOI] [PubMed] [Google Scholar]
- 121. Winter, J. , Walker, A. , Shapiro, D. , Gaffney, D. , Spooner, R.J. & Mills, P.R. Cost‐effectiveness of thiopurine methyltransferase genotype screening in patients about to commence azathioprine therapy for treatment of inflammatory bowel disease. Aliment. Pharmacol. Ther. 20(6), 593–599 (2004). [DOI] [PubMed] [Google Scholar]
- 122. Dubinsky, M.C. , Reyes, E. , Ofman, J. , Chiou, C.‐F. , Wade, S. & Sandborn, W.J. A cost‐effectiveness analysis of alternative disease management strategies in patients with crohn's disease treated with azathioprine or 6‐Mercaptopurine. Am. J. Gastroenterol. 100(10), 2239–2247 (2005). [DOI] [PubMed] [Google Scholar]
- 123. Hagaman, J. , Kinder, B. & Eckman, M. Thiopurine S‐methyltranferase testing in idiopathic pulmonary fibrosis: a pharmacogenetic cost‐effectiveness analysis. Lung 188(2), 125–132 (2010). [DOI] [PubMed] [Google Scholar]
- 124. van den Akker‐van Marle, M.E. , Gurwitz, D. , Detmar, S.B. et al. Cost‐effectiveness of pharmacogenomics in clinical practice: a case study of thiopurine methyltransferase genotyping in acute lymphoblastic leukemia in Europe. Pharmacogenomics 7(5), 783–792 (2006). [DOI] [PubMed] [Google Scholar]
- 125. Zarca, K. , Durand‐Zaleski, I. , Loriot, M.‐A. , Chatellier, G. & Pallet, N. Modeling the outcome of systematic TPMT genotyping or phenotyping before azathioprine prescription: a cost‐effectiveness analysis. Mol. Diagn. Ther. 23(3), 429–438 (2019). [DOI] [PubMed] [Google Scholar]
- 126. Donnan, J.R. , Ungar, W.J. , Mathews, M. , Hancock‐Howard, R.L. & Rahman, P. A cost effectiveness analysis of thiopurine methyltransferase testing for guiding 6‐mercaptopurine dosing in children with acute lymphoblastic leukemia. Pediatr. Blood Cancer 57(2), 231–239 (2011). [DOI] [PubMed] [Google Scholar]
- 127. Sluiter, R.L. et al. Genotype‐guided thiopurine dosing does not lead to additional costs in patients with inflammatory bowel disease. J. Crohn's Colitis 13(7), 838–845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Data S1
Table S1


