SUMMARY
Infections by root‐feeding nematodes have profound effects on root system architecture and consequently shoot growth of host plants. Plants harbor intraspecific variation in their growth responses to belowground biotic stresses by nematodes, but the underlying mechanisms are not well understood. Here, we show that the transcription factor TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTOR‐9 (TCP9) modulates root system architectural plasticity in Arabidopsis thaliana in response to infections by the endoparasitic cyst nematode Heterodera schachtii. Young seedlings of tcp9 knock‐out mutants display a significantly weaker primary root growth inhibition response to cyst nematodes than wild‐type Arabidopsis. In older plants, tcp9 reduces the impact of nematode infections on the emergence and growth of secondary roots. Importantly, the altered growth responses by tcp9 are most likely not caused by less biotic stress on the root system, because TCP9 does not affect the number of infections, nematode development, and size of the nematode‐induced feeding structures. RNA‐sequencing of nematode‐infected roots of the tcp9 mutants revealed differential regulation of enzymes involved in reactive oxygen species (ROS) homeostasis and responses to oxidative stress. We also found that root and shoot growth of tcp9 mutants is less sensitive to exogenous hydrogen peroxide and that ROS accumulation in nematode infection sites in these mutants is reduced. Altogether, these observations demonstrate that TCP9 modulates the root system architectural plasticity to nematode infections via ROS‐mediated processes. Our study further points at a novel regulatory mechanism contributing to the tolerance of plants to root‐feeding nematodes by mitigating the impact of belowground biotic stresses.
Keywords: root system architecture, root plasticity, TCP transcription factor, cyst nematodes, biotic stress, reactive oxygen species, damage, tolerance, Heterodera schachtii
Significance Statement
Plants utilize root plasticity to cope with adverse environmental conditions in soil, but the molecular and genetic mechanisms regulating the adaptive growth responses to belowground biotic stresses are not well understood. Here, we show that the transcription factor TCP9 modulates plasticity of the root system architecture of Arabidopsis during soil‐borne cyst nematodes infections via reactive oxygen species‐mediated processes, which points at a novel tolerance mechanism based on mitigating the impact rather than the level of biotic stress.
INTRODUCTION
Plant roots are intermittently exposed to various abiotic stresses, such as drought, but also to biotic stresses, such as herbivory by root–parasitic nematodes. Plants utilize root system architectural plasticity to cope with such changing environmental conditions in the soil (Karlova et al., 2021; Koevoets et al., 2016). For instance, secondary roots are formed at the water‐contact side of the roots to adapt root system architecture to the heterogeneous distribution of water in the soil (Karlova et al., 2021). Likewise, root feeding by plant parasitic nematodes results in the formation of secondary roots in the proximity of permanent nematode‐induced feeding structures (Goverse et al., 2000; Lee et al., 2011). Earlier work has shown that plants harbor intraspecific variation in their root growth responses to belowground biotic stress by these cyst nematodes, which correlates with the overall level of tolerance to nematode infections (Miltner et al., 1991; Trudgill & Cotes, 1983). However, the genetic and molecular mechanisms governing root system architectural plasticity in response to biotic stress during nematode infections in plants are not well understood.
The impact of root‐feeding cyst nematodes on plant development and growth leads to large global agricultural losses. This is partly because cyst nematodes can persist in a dormant state in the soil in the absence of host plants for many years (Jones et al., 2013). Hatching of infective juveniles of cyst nematodes primarily occurs in response to root exudates of host plants. These root exudates also provide guidance for the migration of cyst nematodes to the root surface, where they penetrate the root epidermis at the differentiation or maturation zone. Host invasion by cyst nematodes involves piercing of plant cell walls with a needle‐like oral stylet and concomitant secretion of plant cell wall degrading enzymes (Rehman et al., 2009). Once inside the root cortex, infective juveniles migrate intracellularly towards the vascular cylinder, while causing extensive tissue damage. After arriving at the vascular cylinder, they deliver stylet secreted effectors into a host cell to initiate the formation of a permanent feeding site by manipulating plant cell differentiation and growth. Throughout the course of several weeks, this permanent feeding site expands further inside the vascular cylinder by local cell wall degradation and subsequent fusion of neighboring host cells. The permanent feeding site, hereafter named syncytium, provides cyst nematodes access to the flow of assimilates in the plant, which are essential for nematode development and reproduction. Thus, biotic stress on plant root systems during cyst nematode infections is caused by tissue damage during host invasion and loss of assimilates by feeding nematodes.
Members of the TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR1 (TCP) transcription factor family convert environmental signals into adaptive growth responses in plants (Danisman, 2016). In Arabidopsis, the TCP family consists of 24 members and is divided over two groups, named class I and II, based on sequence variation in a conserved TCP domain of about 60 residues (Li, 2015). Class I and II TCPs differ in composition of the nuclear localization signal, the length of the second helix in the basic helix–loop–helix region, and the presence of a positively charged arginine‐rich domain (Cubas et al., 1999). The basic region in the TCP domain is required for binding to GC‐rich motifs in cis‐regulatory elements in DNA sequences upstream of genes that are transcriptionally regulated by TCPs (Kosugi & Ohashi, 2002). Moreover, TCPs are able to form homo‐ and heterodimers, mostly between different members of the same class, which further contributes to functional diversity within the TCP family (Danisman et al., 2012; Viola et al., 2011). Because both class I and II TCP factors can bind to partially overlapping cis‐acting regulatory elements, it is thought that members of the two classes might act antagonistically (Danisman et al., 2012). However, recent reports do not support a strict functional distinction of class I and class II TCP into transcriptional activators and repressors, respectively (Kubota et al., 2017; Lucero et al., 2017; Wang et al., 2020).
Several TCP transcription factors have been shown to drive adaptations in root system architecture in response to abiotic stress. For instance, in Arabidopsis, TCP20 regulates preferential secondary root growth in response to nitrates in a process called root foraging (Guan et al., 2014). To this end, TCP20 interacts with NIN‐like protein (NLP) transcription factors NLP6 and NLP7 to support root meristem growth under nitrogen starvation (Guan et al., 2014). Likewise, TCP13 regulates leaf and root growth in Arabidopsis in response conditions simulating drought (Urano et al., 2022). Furthermore, heterologous expression of PeTCP10 of bamboo (Phyllostachys edulis) in transgenic Arabidopsis induces secondary root growth under treatments simulating drought conditions (Liu et al., 2020) and salt stress (Xu et al., 2021). In a similar experimental design, OsTCP19, a class I TCP from rice, also makes plants more tolerant to salt stress (Mukhopadhyay & Tyagi, 2015). A subset of TCP transcription factors involved in root development and growth is controlled by the microRNA miR319 (Baulies et al., 2022), which together may form a regulatory module tuning plant responses to abiotic stresses (Fang et al., 2021; Zhou et al., 2013).
As members of the TCP family in Arabidopsis regulate root system architectural changes in response to abiotic stresses, we reasoned that they might also be involved in root plasticity under biotic stress. To test this hypothesis, we first analyzed the expression of TCP family members during early stages of infection of the beet cyst nematode Heterodera schachtii in Arabidopsis. As TCP9 was found to be strongly upregulated in association with nematode infections, we further focused our study on this class I TCP transcription factor. Hereto, we investigated the impact of TCP9 on root plasticity during cyst nematode infections by monitoring the primary root growth inhibition response and secondary root formation shortly after inoculation with infective juveniles. Next, we tested if the aberrant root phenotypes observed for tcp9 mutants could be caused by a loss of susceptibility to nematode infections, and therefore lower levels of biotic stress on the root system. To pinpoint possible mechanisms underlying the impact of TCP9 on root system architectural plasticity, we used RNA‐sequencing (RNA‐seq) of nematode‐infected roots of tcp9 mutants at early stages of infection by H. schachtii. The TCP9‐regulated gene expression patterns that we observed herein pointed at changes in reactive oxygen species (ROS)‐mediated processes. We therefore subsequently analyzed if ROS accumulates differently in nematode infection sites in the tcp9 mutants and if root and shoot growth of these mutants responds differently to exogenously applied hydrogen peroxide (H2O2) as compared with wild‐type Arabidopsis plants. Altogether, our data provide insights into a novel mechanism underlying root architecture plasticity in response to biotic stress by endoparasitic cyst nematodes.
RESULTS
TCPs are differentially regulated during Heterodera schachtii infection
To investigate if members of the TCP transcription factor family are differentially regulated during early stages of nematode infection, we assessed their expression levels in whole root systems of Arabidopsis infected with H. schachtii using RNA‐seq. We inoculated 4‐day‐old seedlings with infective H. schachtii juveniles and analyzed the root transcriptome at 6, 12, 24, 48, 72, and 120 h post‐inoculation (hpi). We found that nine TCP family members were differentially expressed in nematode‐infected roots at one or more time points post‐inoculation (Figure 1). Of the class I TCPs, TCP8, TCP9, and TCP20 were upregulated, whereas TCP7, TCP14, and TCP21 were downregulated. Of the class II TCPs, only TCP13 was upregulated throughout this entire time series, whereas both TCP2 and TCP24 were downregulated at the 120 hpi. Our data thus showed that TCP transcription factors might indeed be involved in regulating plant responses during the onset of parasitism by H. schachtii.
Figure 1.
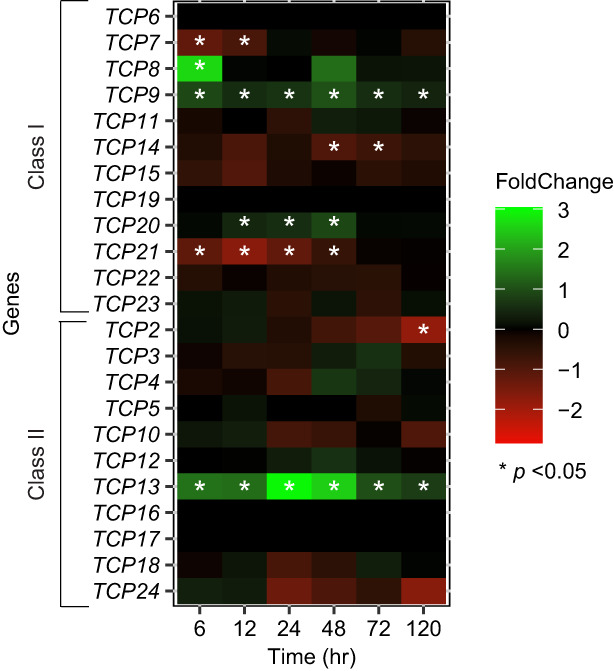
Members of TCP gene family are differentially regulated in whole roots of nematode‐infected Arabidopsis seedlings.
Relative expression level of TCPs as determined by RNA‐sequencing of whole roots of Arabidopsis seedlings either inoculated with 200 infective juveniles of Heterodera schachtii or mock inoculated. Samples were collected at 6, 12, 24, 48, 72, and 120 h post‐inoculation. Heatmap shows the Log2 fold‐change of TCP gene expression of normalized read counts per gene in nematode‐inoculated seedlings versus mock‐inoculated seedlings at different time points. Green indicates upregulation and red indicates downregulation. *Significantly different expression level in nematode‐ versus mock‐inoculated seedlings (*P < 0.05).
TCP9 modulates root system architecture during Heterodera schachtii infection
Both TCP9 and TCP13 were significantly upregulated throughout all early stages of infection by H. schachtii in Arabidopsis. As the function of TCP13 has recently been studied in more detail (Hur et al., 2019; Urano et al., 2022), we decided to focus our research on a possible role for TCP9 in root plasticity responses to biotic stress by H. schachtii. TCP9 can alter root growth in Arabidopsis, but if it does so in response to abiotic and biotic stress is unknown. We also included TCP20 in our study, because it transcriptionally regulates TCP9 expression and forms a functional dimer with TCP9 (Danisman et al., 2012; Wang et al., 2015). First, we challenged 4‐day‐old seedlings of the Arabidopsis loss‐of‐function single mutants tcp9, tcp20, and the double mutant tcp9tcp20 with infective juveniles of H. schachtii to monitor changes in primary root growth at 4 dpi. Notably, at this young age the root system for Arabidopsis seedlings only consists of a primary root. We found that primary root growth of all three tcp mutants and wild‐type Arabidopsis seedlings was inhibited after inoculation with H. schachtii (Figure 2a,b). Because we noticed differences in primary root growth for tcp20 and tcp9tcp20 in mock‐inoculated seedlings (Figure S2), we also calculated the relative growth of the primary root compared with mock‐inoculated plants of the same genotype (Figure 2c). This showed that the relative growth of the primary root of both tcp9 and the tcp9tcp20 mutants was significantly less affected (and thus closer to 1) by cyst nematodes than of the tcp20 mutant and wild‐type Arabidopsis.
Figure 2.
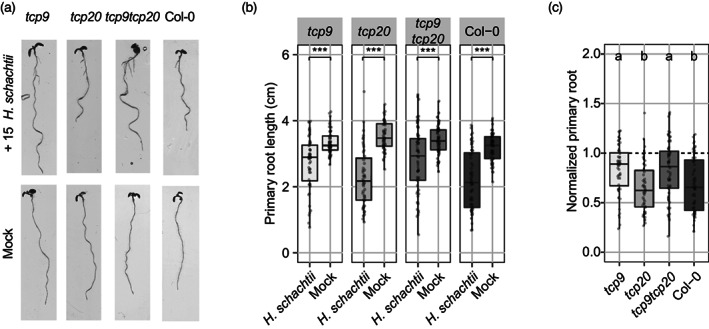
Primary root growth of young tcp9 mutant seedlings is less affected by cyst nematode infections.
Four‐day‐old Arabidopsis seedlings (tcp9, tcp20, tcp9tcp20, and wild‐type Col‐0) were inoculated with 15 juveniles of Heterodera schachtii or a mock solution.
(a) Representative pictures of seedlings either nematode‐ or mock‐inoculated at 4 days post‐inoculation (dpi).
(b) Primary root length (cm) of wild‐type and mutant Arabidopsis seedlings at 4 dpi. Data were analyzed with a two‐way ANOVA with a post‐hoc Tukey HSD test (n = 60). ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
(c) Impact of nematode infection on primary root growth calculated as the primary root length of nematode‐inoculated seedlings divided by the median value of mock‐inoculated seedlings of the same genotype. Data were analyzed with a multiple comparison one‐way ANOVA with a post‐hoc Tukey HSD test. Letters indicate different levels of significance. This experiment was performed three times with similar outcomes and pooled for data analysis.
Next, we tested if TCP9 also affects growth of other root system architecture components in response to H. schachtii. Hereto, we inoculated 9‐day‐old Arabidopsis seedlings having a more elaborate root system with H. schachtii, and measured different root architecture components (i.e., total root length, primary root length, number of secondary roots, total secondary root length) at 7 dpi (Figure 3a). All measurements were transformed to relative values by normalizing the data to the median of the corresponding measurement in mock‐inoculated plants. We found that the total relative root length of only the tcp9 mutant was significantly less affected (and closer to 1) by inoculation with H. schachtii compared with wild‐type Col‐0 plants (Figure 3b). Remarkably, the relative growth of the primary root of the tcp9 mutants was still slightly but no longer significantly different from wild‐type Arabidopsis (Figure 3c). However, the number (Figure 3d) and the total length of the secondary roots (Figure 3e) were less affected by H. schachtii in the tcp9 mutant as compared with wild‐type Arabidopsis Col‐0. We therefore concluded that TCP9 modulates the plasticity of the root system architecture in response to H. schachtii in Arabidopsis.
Figure 3.
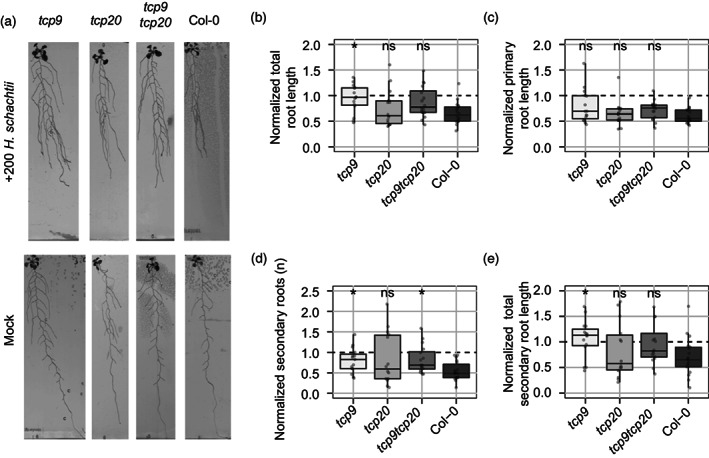
Root system architecture of tcp9 mutants responds less to cyst nematode infections.
Nine‐day‐old Arabidopsis seedlings (tcp9, tcp20, tcp9tcp20, and wild‐type Col‐0) were inoculated with 200 or 0 (Mock) infective juveniles of Heterodera schachtii.
(a) Representative pictures of seedlings at 7 days post‐inoculation (dpi). For data analysis, all values were normalized to the median of the measurements of the corresponding mock‐inoculated plants at 7 dpi.
(b) Total root length of wild‐type and mutant Arabidopsis seedlings.
(c) Primary root length of wild‐type and mutant Arabidopsis seedlings.
(d) Number of secondary roots of wild‐type and mutant Arabidopsis seedlings.
(e) Total secondary root length of wild‐type and mutant Arabidopsis seedlings. This experiment was performed two times with similar outcomes. All data were pooled and analyzed with a pairwise Wilcoxon rank sum test (n = 15–20). ns, not significant, *P < 0.05.
TCP9 does not affect susceptibility of Arabidopsis to Heterodera schachtii
Next, we asked if the root growth response of the tcp9 mutants upon nematode infections was different, because of loss of susceptibility to H. schachtii. In other words, we reasoned that a weaker response in root system architecture of tcp9 and tcp9tcp20 mutants could be due to a smaller number of nematodes inside the roots, and therefore a lower level of biotic stress on the root system. However, we observed no significant differences in the number of nematodes per plant between any of the mutant lines and wild‐type Arabidopsis at 4 dpi (Figure 4a).
Figure 4.
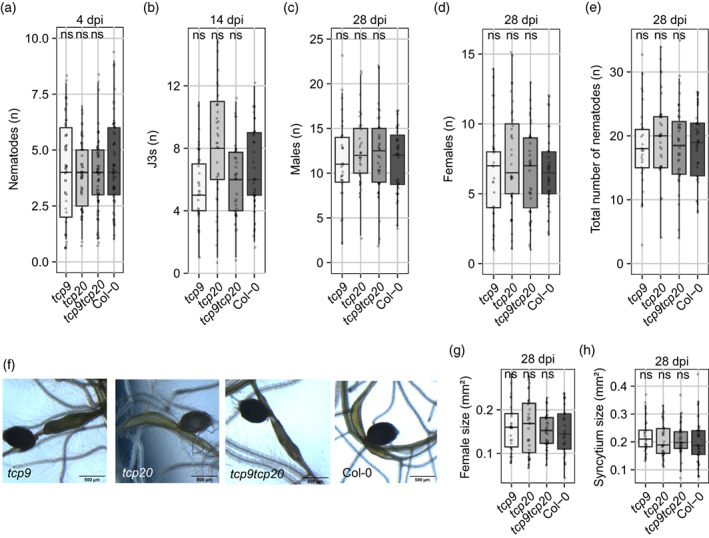
Loss of function mutation in tcp9 does not affect development and growth of Heterodera schachtii in Arabidopsis.
(a) Number of juveniles per root system on tcp mutants (tcp9, tcp20, and tcp9tcp20) and wild‐type Arabidopsis plants (Col‐0) at 4 day post‐inoculation (dpi) (n = 60).
(b) The number of third stage juveniles of H. schachtii at 14 dpi.
(c) The number of males of H. schachtii at 28 dpi.
(d) The number of females at 28 dpi.
(e) The total number nematodes at 28 dpi.
(f) Representative images of syncytia and adult females at 28 dpi. Scale bar represents 500 μm.
(g) The maximum two‐dimensional surface area of adult H. schachtii females in a single focal plane of the dissection microscope at 28 dpi.
(h) The maximum two‐dimensional surface area of syncytia at 28 dpi. This experiment was performed three times with similar outcomes. Data were pooled and analyzed with one‐way ANOVA with a post‐hoc Tukey HSD test. ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001 (n = 30–36).
To assess further if TCP9 affects the development of feeding nematodes, we monitored their progression through different development stages inside plants for 28 dpi. We observed that parasitic second stage juveniles did not molt significantly faster into third stage juveniles in any of the mutants compared with wild‐type Arabidopsis plants (Figure 4b). Furthermore, as the supply of nutrients to feeding cyst nematodes can affect the differentiation of juveniles into either males or females, we also examined if TCP9 affects the sex determination of H. schachtii. To this end, we counted the number of males, females, and the total number of individuals at 28 dpi. We observed no significant difference in the number of males or females in any of the mutant lines as compared with wild‐type Arabidopsis (Figure 4c–e). We therefore concluded that TCP9 does not affect normal development of H. schachtii.
Differences in plant susceptibility to parasitic nematodes can also appear in the size of the feeding females. To test if TCP9 indirectly regulates female growth, we measured the maximum surface area of adult females in the two‐dimensional focal plane of a dissecting microscope (Figure 4f). The surface area of adult females in none of the mutants was significantly different compared with wild‐type Arabidopsis plants (Figure 4g). Then, we asked if the size of nematode‐induced syncytium could be affected by TCP9. After measuring the maximum surface area of individual syncytia of feeding adult females, we found no significant difference between the tcp mutants and wild‐type Arabidopsis plants either (Figure 4h). Altogether, we concluded that TCP9 does not affect susceptibility of Arabidopsis to infections by H. schachtii and that lower levels of biotic stress are not the cause of the altered root growth responses in nematode‐infected tcp9 mutants.
TCP9 regulates the expression of genes involved in ROS‐related processes
TCP9 functions as a transcription factor and to understand how it modulates root growth responses, we analyzed changes in the root transcriptome upon infection with H. schachtii in both wild‐type and different mutant Arabidopsis plants. To this end, we performed a time‐series experiment for which we inoculated 4‐day‐old Arabidopsis seedlings of the tcp9, tcp20, and tcp9tcp20 mutants and wild‐type Arabidopsis plants with H. schachtii or a mock solution. Whole roots were sampled at 24, 48, and 72 hpi and subsequently subjected to RNA‐seq. A principal components analysis of the sequence data revealed that most of the overall variance in gene expression was captured by infection with H. schachtii (21.9%) and by time post‐inoculation (9.5%; Figure 5a). The overall variance in gene expression captured by plant genotype was small. Nonetheless, to map specifically the effect of the tcp9, tcp20, and the tcp9tcp20 mutations on gene expression, we used a linear model, including presence of infection, time after inoculation, and plant genotype as factors (Tables S1 and S2). We found a relatively small number of genes in tcp9, tcp20, and tcp9tcp20 mutants with a significantly different expression at different time points post‐inoculation (Figure 5b and Table S1). This showed that the mutations in TCP9 (Figure S3) and TCP20 (Figure S4) most likely have a minor impact on the root transcriptome during H. schachtii infection. Notably, the knock‐out mutation in TCP9, in both tcp9 and tcp9tcp20, had its biggest impact at 48 hpi. Gene Ontology (GO) term enrichment analysis of the set of differentially expressed genes in nematode‐infected tcp9, tcp20, and tcp9tcp20 mutants revealed a significant overrepresentation of reactive oxygen species (ROS)‐related processes associated with absence of TCP9 (Table S3). For instance, 14 Arabidopsis genes annotated as being involved in oxidation–reduction process (GO:0055114) were downregulated in tcp9 at 48 hpi (false discovery rate = 7.57E‐05; Figure 5d; Table S4). Likewise, four of nine Arabidopsis genes annotated as having oxidoreductase activity (GO:0016491) were also downregulated in tcp9 (false discovery rate = 0.0046). Based on these observations, we concluded that the TCP9‐modulated root growth responses during nematode infections most likely involve ROS‐related processes.
Figure 5.
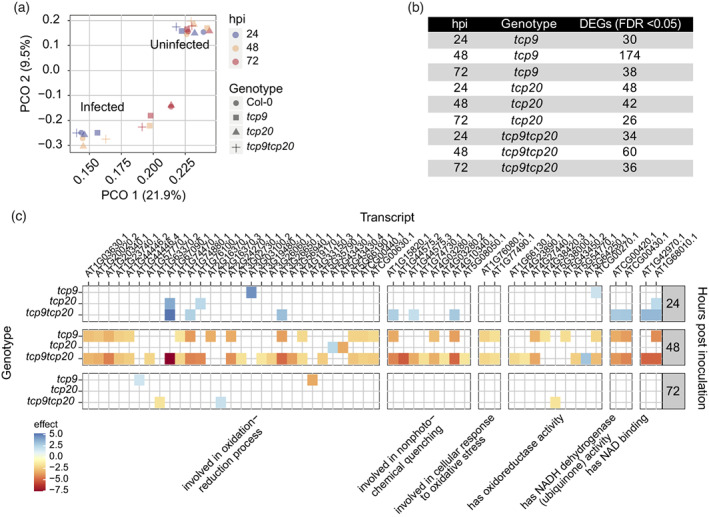
TCP9 regulates gene expression in nematode‐infected roots of Arabidopsis.
Four‐day‐old Arabidopsis seedlings were inoculated with 80 juveniles of Heterodera schachtii or a mock solution. At 24‐, 48‐, and 72‐h post‐inoculation (hpi) whole root samples were collected and subjected to RNA‐sequencing.
(a) Principal components analysis of overall variation in gene expression in wild‐type Arabidopsis Col‐0, and tcp9, tcp20, and tcp9tcp20 mutants at different hpi with H. schachtii (infected) or mock inoculation (uninfected).
(b) Number of genes that were differentially expressed (DEG) in tcp9, tcp20, and tcp9tcp20 at the different hpi when compared with nematode‐infected roots of wild‐type Arabidopsis Col‐0 (false discovery rate, FDR, correction, q < 0.01).
(c) Subset of DEGs (from c) that are classified as being reactive oxygen species‐related in Gene Ontology term enrichment analysis (false discovery rate [FDR], q < 0.01). Colors indicate direction and size of effect of the mutations on gene expression as compared with nematode‐infected wild‐type Arabidopsis Col‐0.
TCP9 alters sensitivity of Arabidopsis to exogenous H2O2
To test if the TCP9‐mediated root growth responses indeed involve ROS‐mediated processes, we first transferred 6‐day‐old seedlings of the tcp9, tcp20, and tcp9tcp20 mutants and wild‐type Arabidopsis to liquid media containing 200 μm H2O2. Eight days after the transfer, we calculated the relative total root length of the seedlings by normalizing the data to the median of the corresponding measurement in mock‐treated plants. This showed that the relative total root length of tcp9 mutant was least affected by H2O2 in the media (and thus closer to 1; Figure 6a,b). Notably, the relative total root length of the tcp20 and tcp9tcp20 mutants was slightly, but not significantly, less affected by H2O2 than wild‐type Arabidopsis seedlings.
Figure 6.
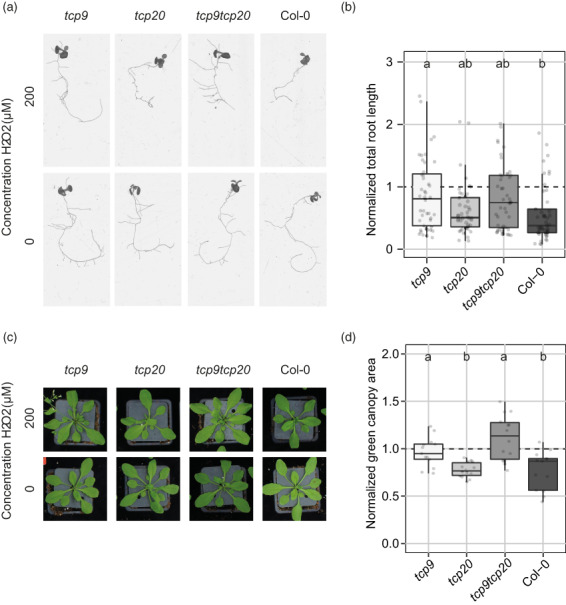
Root and shoot growth of tcp9 mutant responds less to H2O2.
(a,b) Six‐day‐old Arabidopsis (tcp9, tcp20, tcp9tcp20 and wild‐type Col‐0) seedlings were transferred to liquid KNOP media containing 200 or 0 (mock) μm H2O2. At 8 days post‐transfer, the total root length was measured.
(a) Representative pictures of tcp9, tcp20, tcp9tcp20, and wild‐type Col‐0 roots exposed to 0 and 200 μm H2O2.
(b) Before analysis, the total root length of every seedling treated 200 μm H2O2 was normalized to the median of mock‐inoculated seedlings of the corresponding plant genotypes. This experiment was performed three times with similar outcomes (n = 35).
(c,d) Nine‐day‐old Arabidopsis seedlings (tcp9, tcp20, tcp9tcp20, and wild‐type Col‐0) growth in pots with soil were sprayed with 200 or 0 (Mock) μm H2O2. At 21 days‐post treatment, pictures were made of the green canopy area and the green color was isolated from the pictures by using Photoshop.
(c) Representative pictures of green canopies of plants sprayed with 200 or 0 (Mock) μm H2O2.
(d) Impact of 200 μm H2O2 on green canopy area was calculated as the green canopy area (cm2) of H2O2‐sprayed plants divided by the median value of mock‐sprayed plants of the same genotype. This experiment was performed two times with similar outcomes (n = 14–15). For both experiments, data were pooled and analyzed with a one‐way ANOVA with a post‐hoc Tukey HSD test. Letters indicate different levels of significance (P < 0.05).
Root growth responses to belowground biotic stress by cyst nematodes has been shown to correlate with the aboveground plant growth (Miltner et al., 1991; Trudgill & Cotes, 1983), which could also be mediated by ROS regulation (Labudda et al., 2018). To assess if TCP9 regulates sensitivity to ROS above ground, we sprayed 9‐day‐old seedlings with 200 μm H2O2. Twenty‐one days after spraying the plants, we recorded the effect of H2O2 on the size of the green canopy area. This showed that the green canopy area of tcp9 and tcp9tcp20 mutant plants was less affected by H2O2 than the tcp20 mutant and wild‐type Col‐0 (Figure 6c,d). Based on these findings, we concluded that TCP9 modulates sensitivity to ROS in Arabidopsis both below and above ground.
TCP9 affects ROS‐related processes in nematode infection sites
The tcp9 mutants might be less sensitive to H2O2, because of enhanced ROS scavenging in this mutant. If this would hold true, we expected to find less ROS accumulation in nematode infection sites. To test this hypothesis, we inoculated tcp9, tcp20, and tcp9tcp20 mutants and wild‐type Arabidopsis seedlings with H. schachtii and visualized ROS accumulation in the proximity of the nematode with 3,3′‐diaminobenzidine (DAB) in roots at 72 hpi (Siddique et al., 2014). In wild‐type Arabidopsis plants and the tcp20 and tcp9tcp20 mutants, we could observe clear DAB staining >80% of the infection sites (Figure 7a,b). By contrast, in the tcp9 mutant, only 50% of the nematode infection sites showed DAB staining. These observations indicate that TCP9 modulates ROS homeostasis in response to nematode infections.
Figure 7.
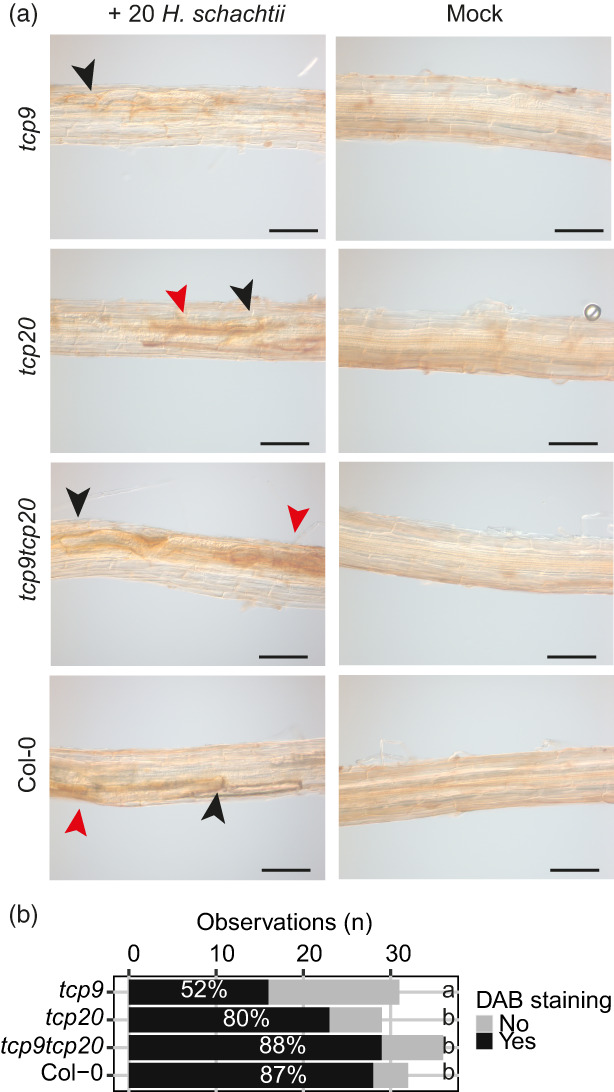
Significant fewer nematode infections showed 3,3′‐diaminobenzidine (DAB) staining in tcp9.
Four‐day‐old Arabidopsis seedlings (tcp9, tcp20, tcp9tcp20, and wild‐type Col‐0) were inoculated with 20 or 0 (Mock) juveniles of Heterodera schachtii. At 3 days post‐inoculation, the accumulation of reactive oxygen species was detected using DAB.
(a) Representative pictures of root segments infected with H. schachtii and non‐infected (Mock). Black arrowhead points at location of nematode head. Red arrowhead points at oxidized DAB staining. Scale bar = 100 μm.
(b) Absolute numbers of individual infections with or without DAB staining. This experiment was done three times with similar outcomes. Data are pooled for statistical analysis in Fisher's exact test. Letters indicate different levels of significance (P < 0.05) (n = 29–33).
DISCUSSION
Plants utilize root system architectural plasticity to cope with adverse environmental conditions in the soil. However, the molecular and genetic underpinnings of adaptive growth responses to belowground biotic stresses are not well understood. As members of the TCP transcription factor family in Arabidopsis are known to regulate root growth responses to abiotic stresses, we reasoned that they might also play a role in root system architecture plasticity under biotic stress by root‐feeding nematodes. In this study, we show that the class I TCP9 transcription factor in Arabidopsis modulates adaptations in the root system architecture in response to endoparasitism by the beet cyst nematode H. schachtii. Our data further suggest that TCP9‐modulated root plasticity in response to H. schachtii involves ROS‐mediated processes, which remarkably do not affect the susceptibility of Arabidopsis seedlings to nematode infections. This contrasts with earlier studies showing that NAPDH oxidase‐generated ROS enhances syncytium growth and nematode development in Arabidopsis infected with H. schachtii (Siddique et al., 2014). Our findings therefore point at a novel tolerance mechanism mitigating the impact of biotic stress rather than targeting the causal agent of this biotic stress in nematode‐infected roots.
Members of the TCP family of transcription factors have been shown to regulate primary root growth and emergence of secondary roots in response to abiotic stress conditions such as high salt levels, drought, and nitrate poor substrates (Li et al., 2020; Ling et al., 2020; Liu et al., 2020; Mukhopadhyay & Tyagi, 2015). We observed that primary root growth in young Arabidopsis seedlings is significantly less inhibited by H. schachtii infections in the tcp9 mutant than in wild‐type Arabidopsis. In older plants and later in the infection cycle, tcp9 reduces the impact of nematode infections on the emergence of secondary roots. It is possible that primary root growth and the emergence of secondary roots are two different outputs of the same stress mitigating process modulated by TCP9. However, these two phenomena could also be independent adaptations driven by different causes of stress on the root system. Here, it should be noted that migratory infective juveniles of H. schachtii induce stress by extensively damaging root tissue during host invasion at the early stages of infection, whereas later in the infection process the loss of assimilates during feeding by sedentary life stages on host cells might be the dominant cause of stress (Wyss & Grundler, 1992). It has been shown that the level and the duration of stress can determine if a condition inhibits or promotes root growth (Julkowska et al., 2014; Zhang et al., 2017). The stress levels during the brute force host invasion by cyst nematodes are probably high but last only a few hours and may induce rapid cessation of primary root growth. Here, TCP9 could modulate damage‐induced stress responses during early stages of the nematode infections, which would also agree with TCP9 being upregulated early in response to mechanical wounding (Figure S5) (Kilian et al., 2007). By contrast, stress by nematode feeding on host cells may be milder but persist for several weeks and could induce the formation of additional secondary roots. In the latter case, TCP9 could modulate these feeding‐induced stress adaptations in root system architecture to mitigate the impact of persistent biotic stress on the root system upon nematode infections.
Previously, it has been reported that TCP9 in conjunction with TCP8 is involved in host defense responses to Pseudomonas syringae pv. maculicola infections by regulating the expression of ICS1 (Wang et al., 2015). We found that TCP9 does not affect host susceptibility to invasions by H. schachtii, which could otherwise alter the level of biotic stress on the root system. A weaker primary root growth inhibition could be caused by a smaller number of nematodes in the tcp9 mutant, but we did not find fewer infections in this mutant. We therefore have no reason to assume that a lower level of biotic stress (i.e., less direct damage by invading infective juveniles) is underlying weaker primary root growth inhibition response in the tcp9 mutant. One could also argue that TCP9 indirectly affects the root growth responses to nematode infections by modulating the re‐allocation of resources from growth to defense. However, we found evidence that TCP9 is not involved in the activation of host defenses, as we observed no difference in the development and growth of nematodes and associated feeding sites between tcp9 mutant and wild‐type Arabidopsis plants. Based on these findings, we believe to have identified a novel mechanism that mitigates the impact of biotic stress independent of susceptibility of Arabidopsis to cyst nematode infections.
Our study provides several lines of evidence that TCP9‐modulated root plasticity under biotic stress by root‐feeding nematodes involves ROS‐mediated regulation of plant growth. We show that TCP9 is required for transcriptional activation of enzymes involved in ROS‐related processes in nematode‐infected roots. We also demonstrate that the lack of TCP9 in the tcp9 mutant makes the root and shoot system of Arabidopsis seedlings less sensitive to exogenous ROS (i.e., H2O2). Finally, we found that TCP9 modulates the local accumulation of ROS in response to nematode infections without affecting plant susceptibility (Hewezi et al., 2010; Jin et al., 2011). Several studies have demonstrated that perturbations in local ROS homeostasis contribute to stress‐induced root growth adaptations (reviewed by Considine & Foyer, 2021). For instance, local ROS homeostasis plays a key role in balancing cell differentiation and elongation in root apical meristems thereby determining the rate at which roots grow (Tsukagoshi et al., 2010). ROS accumulation is shown to be required for the emergence and development of secondary roots in Arabidopsis (Huang et al., 2020; Orman‐Ligeza et al., 2016). Our finding that TCP9‐modulated root plasticity involves ROS‐mediated processes also agrees with earlier observations on mechanisms underlying stress‐induced adaptive plant growth responses by other TCPs. For instance, PeTCP10 of Phyllostachys edulis is thought to increase drought tolerance in transgenic Arabidopsis via ROS‐regulated root growth (Liu et al., 2020; Xu et al., 2021). McTCP1 is shown to control ROS‐regulated vegetative growth of the liverwort Marchantia polymorpha (Busch et al., 2019). The salt stress responses mediated by OsTCP19 in rice results in less ROS accumulation in detached leaf explants (Mukhopadhyay & Tyagi, 2015). Nevertheless, further research is needed to link causally the transcriptional regulation of ROS‐related processes, ROS sensitivity, and ROS accumulation to the stress‐induced growth response modulated by TCP9 in nematode‐infected roots.
Our study also shows the complex interplay between different members of the TCP transcription factor family. Although in earlier work, TCP20 has been shown to transcriptionally regulate TCP9 during leaf development (Danisman et al., 2012), we have found no evidence that such an interaction is underlying the different phenotypes observed in our assays. We found that the tcp20 single mutant does not phenocopy the tcp9 mutant, and instead it behaved like wild‐type Arabidopsis in our experiments. TCP20 is therefore not required for the ROS‐related processes mediated by TCP9 in nematode‐infected Arabidopsis roots. Nonetheless, at the transcriptome level we identified multiple genes being uniquely regulated in the tcp9tcp20 double mutant that were not regulated in either of the single mutants. Moreover, the impact of tcp9 on some of the ROS‐related phenotypes seems to be partially mitigated in the tcp9tcp20 double mutant (e.g., Figure 6b), which suggests that at least some degree of interplay occurs between the networks activated by TCP9 and TCP20.
The TCP9‐regulated plasticity of the root system architecture points at a novel mechanism underlying tolerance to root‐feeding nematodes. Infections by cyst nematodes induce alterations in root system architecture, which correlate with the level of tolerance to root‐feeding nematodes (Miltner et al., 1991). However, the molecular and genetic mechanisms underlying tolerance to cyst nematode infections based on root system architecture plasticity are not clear. An important conceptual difficulty here is the discrimination of resistance and tolerance phenotypes of nematode‐infected plants. While resistance is aimed at reducing the cause of biotic stress (or stressor), tolerance concerns the mitigation of the impact of biotic stress. Our data show that TCP9 does not affect susceptibility (and resistance) of Arabidopsis to cyst nematode infections, and therefore does not regulate the level of stress by reducing the parasite load in the plant. Instead, we show that TCP9 mitigates the impact of cyst nematode infections on primary root growth and formation of novel secondary roots. This is reminiscent to earlier observations with other TCP transcription factors contributing to tolerance to abiotic stress by regulating compensatory adaptations in root system architecture (Mukhopadhyay & Tyagi, 2015). In this context, tolerance to biotic stresses such as root‐feeding nematode could be mechanistically related to tolerance to abiotic stress. The involvement of ROS‐related processes in the TCP‐mediated mitigation of both biotic and abiotic stresses points to this as well. Further parallels could be tested by investigating if intraspecific variation in tolerance to root‐feeding nematodes correlates with variation in tolerance to abiotic stresses, such as salt and drought.
To conclude, tolerance to biotic stress, such as root‐feeding by cyst nematodes, based on root system architectural plasticity is an important agronomic trait of crops as it ultimately determines damage thresholds for soil‐borne diseases. It has been shown that some resistant crop varieties do not tolerate low levels of nematode infections well, resulting in significantly lower damage thresholds. In contrast, more susceptible but tolerant varieties display a higher damage threshold for cyst nematodes, leading to lower yield losses despite high infection rates. Our work on the root growth responses meditated by TCP9 provides novel insights into the genetic underpinnings of tolerance to root feeding nematodes, which can provide an additional layer of protection for securing global food production besides disease resistance.
EXPERIMENTAL PROCEDURE
Plant culturing
For seed sterilization, Arabidopsis seeds (Col‐0, tcp9, tcp20, and tcp9tcp20; Danisman et al., 2012) were placed in Eppendorf tubes in a desiccator. The seeds were vapor sterilized for 3–4 h using a mixture of hydrochloric acid (25%) and sodium hypochlorite (50 g/L). Finally, the sterile seeds were stratified for 4 days. For in vitro assays, seeds were sown on square petri dishes (120 × 120 mm) or 12‐well plates containing modified Knop medium (Sijmons et al., 1991). Seedlings were grown at 21°C and 16‐h light/8‐hdark conditions. For in vivo pot experiments, Arabidopsis seeds (Col‐0, tcp9, tcp20, tcp9tcp20) were placed in Eppendorf tubes and stratified for 4 days and sown in 200‐ml pots containing Lentse potgrond (Lensi B.V. Bleiswijk).
Nematode hatching and sterilization
Heterodera schachtii cysts (Woensdrecht population from IRS, the Netherlands) were collected from sand of Brassica oleracea infected plants as previously described (Baum et al., 2000). Heterodera schachtii cysts were rinsed and transferred into a clean Erlenmeyer. Water was added to a maximum volume of 100 ml containing 0.02% sodium azide. This mixture was stirred for 20 min. Later, sodium azide was vigorously removed by washing with tap water. Cysts were then placed on a hatching sieve in a glass petri dish. An antibiotic solution was added containing 1.5 mg ml−1 gentamycin, 0.05 mg ml−1 nystatin and 3 mm zinc chloride. The cysts were incubated in the dark for 4–7 days. Eventually, nematode juveniles were collected in a 2‐ml Eppendorf tube. The J2s were surface sterilized with HgCl2 solution (0.002% Triton X‐100 w/v, 0.004% NaN3 w/v, 0.004% HgCl2 w/v) for 20 min. After incubation, the nematodes were spun down and the supernatant was removed. Nematodes were washed with sterile water and spun down again. This was repeated three times. Finally, the nematodes were resuspended in 0.7% gelrite (Duchefa Biochemie, Haarlem, the Netherlands).
Root architecture assay
Four‐ or nine‐day‐old seedlings were inoculated with either 0 (negative control; mock), 15 or 200 sterile second stage H. schachtii juveniles (J2) in 5 μl gelrite (Baum et al., 2000). At 4‐ or 7 dpi, seedlings were scanned and analyzed using WhinRHIZO (WinRhizo pro2015; Regent Instruments Inc., Ville de Québec, Canada). To compare the effect of nematode infection on the root system component between genotypes we normalized the values of nematode infected seedlings to the median of the corresponding root system component of mock‐treated seedlings.
Fuchsin staining
To determine how many H. schachtii juveniles entered the root at 4 dpi, Arabidopsis seedlings were transferred to a 2.5% bleach solution for 5 min and afterwards washed and incubated in water for 10 min. The roots were placed in fuchsine solution (15 ml:500 μl fuchsine stock; 0.35 g acid fuchsin, 25 ml glacial acetic acid, 75 ml water). The solution was boiled for 1 min in a microwave and cooled down to room temperature. Subsequently, the roots were washed with water and stored in 40% glycerol. Using a stereo microscope, the number of juveniles were counted.
Nematode infection assay
Fourteen‐day‐old Arabidopsis seedlings in a 12‐well plate were inoculated with approximately 250 sterile H. schachtii juveniles in 5 μl gelrite (Baum et al., 2000). At 14 and 28 dpi, the number of J3 stage nematodes, male and female nematodes were counted. The counting and imaging were done under a light microscope (ZEISS microscopes and cameras, ZEN 3.2 [Blue edition]). ImageJ (1.53c) was used to measure the size of the females and the feeding sites.
RNA‐seq analysis
Experimental setup and RNA isolation
To check the expression of TCPs during cyst nematode infection, 4‐day‐old A. thaliana Col‐0 seedlings were inoculated with 200 sterile H. schachtii juveniles in modified Knop's medium. To understand how TCP9 transcriptionally regulates root growth responses during cyst nematode infections, 4‐day‐old A. thaliana seedlings of Col‐0, tcp9, tcp20, and the double mutant tcp9tcp20 were inoculated with 80 sterile H. schachtii juveniles in modified Knop's medium. Root tissue was harvested and snap frozen in liquid nitrogen. Root tissue was ground in liquid nitrogen and total RNA was extracted with the Maxwell® 16 LEV plant RNA kit (Promega, Madison, WI, USA) in the Maxwell 16 AS2000 instrument (Promega), following the manufacturer's instructions. Three biological replicates of approximately 80 plants/sample per condition were generated.
RNA‐seq and quantification
The RNA was sequenced at BGI using the DNBseq platform. Approximately 50 million paired reads of 150 bp were generated per sample. Raw reads were deposited at ArrayExpress under E‐MTAB‐11649. Code used for the analysis has been reposited under https://git.wur.nl/published_papers/willig_2022_tcp.
Reads were aligned to the TAIR10 genome of A. thaliana (Berardini et al., 2015) using hisat2 v2.1.0 with option –dta enabled (Kim et al., 2015). Transcripts were predicted and quantified as transcripts per kilobase million (TPM) using stringtie v2.2.0 based on the TAIR10 reference gene annotations using the –e option (Pertea et al., 2016). To check the expression of TCPs at different time points in Col‐0 inoculated with H. schachtii, we calculated gene expression levels with RSEM (v1.2.12) using default parameters (#Add Ref https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471‐2105‐12‐323) and determined differentially expressed genes using DEseq2 (Add Ref: https://genomebiology.biomedcentral.com/articles/10.1186/s13059‐014‐0550‐8) with a fold‐change ≥2.00 and adjusted P ≤ 0.05.
Presence of TCP9 and TCP20 mutations
To visually confirm the tcp9‐, tcp20‐ and double knockout mutants, samples of RNA‐seq reads mapped to the TAIR10 genome were visualized in the JBrowse genome browser (Buels et al., 2016), along with the TAIR10 reference gene annotations. We used Col‐0_72_‐_2 (S2) as control, tcp9_72_‐_3 (S5) for the tcp9 mutant, tcp20_72_‐1 (S38) for the tcp20 mutant, and tcp9/20_72_‐_1 (S42) for the tcp9tcp20 double mutant. Screenshots were taken at the genomic loci of the TCP9 (AT2G45685) and TCP20 (A0A178VAT1) genes on the TAIR10 genome.
RNA‐seq analysis
Data were analyzed using “R” (v. 3.5.3 x64) in Rstudio (R Core Team, 2019). For analysis, the dplyr and tidyr packages were used for data organization (Wickham et al., 2018a; Wickham et al., 2018b) and plots were generated using ggplot2 (Hadley Wickham, 2009). Before analysis, the TPM values were filtered and transformed (as described in Diaz‐Granados et al., 2019). First, the A. thaliana gene‐expression was filtered for reads detected in all samples, resulting in 28 600 detected genes (of 48 359 protein coding genes in the TAIR11 assembly). We did not notice a batch‐effect over the biological replicates. In subsequent analyses either log‐transformed or the raw TPM values were used.
Principal components analysis
To understand the sources of variance in the gene expression data principal components analysis was used.
To ensure equal weight on the differences in expression, the gene expression data E were transformed by:
where R is the log2 ratio with the mean of gene i and sample j, E TPM is the expression measured as TPM and is the mean expression of gene i over all samples. This procedure was followed for both species (for H. schachtii only on the infected samples). The expression data were analyzed using the prcomp function. The first four axes of both species were examined.
Linear model on Arabidopsis thaliana expression
To understand the role of time, treatment, presence of the tcp9 mutation, and presence of the tcp20 mutation on gene expression in A. thaliana, we used various linear models, where differential expression of the genes was determined by comparison with the wild‐type reference genotype Col‐0. For each combination of genotype (tcp9, tcp20, tcp9tcp20), time point (24, 48, or 72 h), and treatment (mock or inoculated with H. schachtii) the following model was fitted
where E TPM,log2 is the log2‐normalized expression and G is genotype (Col‐0 versus the selected mutant genotype). This led to 18 comparisons. To determine the amount of differentially expressed genes per comparison, we first selected for effect‐size (¦effect¦ > 1.5) and set the threshold for the P < 0.01, to determine the false‐positive rate, we used the p.adjust function with the fdr method (Benjamini & Hochberg, 1995). We found that this selection had a median FDR = 0.066 (see Table S1). This led to the identification of 21–244 differentially expressed genes per comparison (see Tables S1 and S2).
Enrichment analysis
Gene enrichment analyses were conducted using the TAIR10 GO annotations. Groups were selected on minimal three genes representing one group and with minimal two genes in overlap (FDR <0.01) (see Table S3).
DAB staining
Sterile H. schachtii juveniles were inoculated onto 4‐day‐old Arabidopsis seedlings that were grown in square petri dishes. Three days after inoculation, Arabidopsis roots were stained with DAB staining solution in the dark for 3 h (Siddique et al., 2014). Later the DAB staining solution was replaced by a bleaching solution (ethanol/acetic acid/glycerol = 3:1:1). Finally, the DAB staining on the roots was visualized with a Zeiss, Axio Imager.A2 microscope via 10× and 20× objectives. Significance of the differences between proportions was calculated by a in Fisher's exact test.
Root architecture assay of H2O2 ‐treated seedlings
We first tested which concentration of H2O2 is the most informative. To this end, 6‐day‐old wild‐type Col‐0 seedlings that were grown in a square petri dish were transferred to a 24‐well plate containing sterile liquid Knop media, which contained 0, 100, 200, or 300 μm H2O2. Eight days after growth, the roots of seedlings were scanned and analyzed using WhinRHIZO. Statistical significance of the pairwise differences between mock and nematode infection groups were tested using a two‐way ANOVA with Tukey correction (Figure S1). Seedlings treated with 200 μm H2O2 gave the most informative result. Therefore, we decided to continue with only the treatments 0 (mock) and 200 μm H2O2. To determine if there are differences between tcp9, tcp20, tcp9tcp20, and wild‐type Col‐0 seedlings we followed the same procedure. To compare if genotypes were less affected by H2O2 treatment, we normalized the values of H2O2‐treated plants to the median of mock‐treated plants.
Shoot growth assay of H2O2 ‐treated plants
Nine‐day‐old Arabidopsis seedlings that were grown in Lentse potgrond were exposed to 200 μm H2O2 that was applied by spraying (Sewelam et al., 2013). Mock‐treated plants were both sprayed with water. At 21 dpi, the green canopy area was imaged using a Sony α33 camera equipped with a 28 mm lens. By using the “Color Threshold” and “Analyze Particles” tools in ImageJ, we calculated the green canopy area. To compare the effect of spraying H2O2 on the shoot system, we normalized the values of H2O2‐treated plants to the median of the mock‐sprayed plants.
Statistical analysis
Statistical analyses were performed using the R software version 3.6.3 (Windows, x64). The R packages used are tidyverse (https://CRAN.R‐project.org/package=tidyverse), artool (https://CRAN.R‐project.org/package=ARTool) and multcompview (https://CRAN.R‐project.org/package=multcompView). For normally distributed data, significance of the differences among means was calculated by a one‐way or two‐way ANOVA followed by Tukey's HSD test for multiple comparisons. A non‐parametric pairwise Wilcoxon test followed by false discovery rate correction for multiple comparisons was used for non‐normally distributed data with one grouping factor.
AUTHOR CONTRIBUTIONS
GS, JJW, and JB conceived the project. JJW, JJLT, WJ, and JC designed the experiments and performed data collection. Data were analyzed and interpreted by JJW and MGS. JJW and GS wrote the article with inputs from all co‐authors.
CONFLICT OF INTERESTS
The authors declare that they have no competing interests.
OPEN RESEARCH BADGES
This article has earned an Open Data badge for making publicly available the digitally‐shareable data necessary to reproduce the reported results. The data is available at https://www.ebi.ac.uk/arrayexpress/experiments/E‐MTAB‐11649/.
Supporting information
Figure S1. Effect of increasing concentrations of H2O2 on the root growth of Col‐0. Six‐day‐old Arabidopsis (tcp9, tcp20, tcp9tcp20, and wild‐type Col‐0) seedlings were transferred to liquid KNOP media containing 0‐300 μm H2O2. At 8 days post‐transfer the total root length was measured. (a) Representative pictures of wild‐type Col‐0 roots exposed to 0–300 μm H2O2. (b) The root architecture of wild‐type Col‐0 seedlings was quantified by total root length (cm). Data were analyzed with a one‐way ANOVA with a post‐hoc Tukey HSD test. ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001 (n = 35). Letters indicate different levels of significance (P < 0.05).
Figure S2. Primary roots of tcp20 and tcp9tcp20 are significantly longer compared with wild‐type Col‐0. Primary root length (cm) of wild‐type and mutant Arabidopsis seedlings at 8 days post‐germination. Data were analyzed with a one‐way ANOVA with a post‐hoc Tukey HSD test (n = 60). ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001. This experiment was performed three times with similar outcomes and pooled for data analysis.
Figure S3. TCP9 mutation in tcp9 single and tcp9tcp20 double mutants. RNA‐seq reads were mapped to the TAIR10 Arabidopsis genome. Normal length of TCP9 transcript indicated in yellow.
Figure S4. TCP20 mutation in tcp20 single and tcp9tcp20 double mutants. RNA‐seq reads were mapped to the TAIR10 Arabidopsis genome. Normal length of TCP20 transcript indicated in yellow and blue.
Figure S5. TCP9 is upregulated in response to wounding, oxidate stress, drought, and genotoxic stress. Figure and data were retrieved from http://bar.utoronto.ca/eplant/ and Kilian et al. (2007) respectively. Image was generated by Waese et al. (2017).
Table S1. Number of genes that are differentially regulated in tcp9, tcp20, and tcp9tcp20 compared with Col‐0.
Table S2. Differentially expressed genes.
Table S3. Enrichment analysis of differentially expressed genes.
Table S4. Reactive oxygen species‐related genes that are differentially expressed.
ACKNOWLEDGEMENTS
We thank Richard Immink for sharing the tcp9, tcp20 and tcp9tcp20 Arabidopsis mutants. We thank Stefan van de Ruitenbeek for helping with depositing the RNA‐seq data. This work was supported by the Graduate School Experimental Plant Sciences (EPS). JJW is funded by Dutch Top Sector Horticulture & Starting Materials (TU18152). MGS was supported by NWO domain Applied and Engineering Sciences VENI grant (17282). JLLT was supported by NWO domain Applied and Engineering Sciences VENI (14250) and VIDI (18389) grants.
DATA AVAILABILITY STATEMENT
All relevant data can be found within the manuscript, its supporting materials and E‐MTAB‐11649 (https://www.ebi.ac.uk/arrayexpress/experiments/E‐MTAB‐11649/).
References
- Baulies, J.L. , Bresso, E.G. , Goldy, C. , Palatnik, J.F. & Schommer, C. (2022) Potent inhibition of TCP transcription factors by miR319 ensures proper root growth in Arabidopsis. Plant Mol. Biol., 108, 93–103. 10.1007/s11103-021-01227-8 [DOI] [PubMed] [Google Scholar]
- Baum, T.J. , Wubben, M.J. , Hardyy, K.A. , Su, H. & Rodermel, S.R. (2000) A screen for Arabidopsis thaliana mutants with altered susceptibility to Heterodera schachtii. Journal of Nematology, 32, 166–173. [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. & Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing author ( s ): Yoav Benjamini and Yosef Hochberg source. Journal of the Royal Statistical Society. Series B ( Methodological ), 57(1), 289–300. [Google Scholar]
- Berardini, T.Z. , Reiser, L. , Li, D. , Mezheritsky, Y. , Muller, R. , Strait, E. et al. (2015) The arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis, 53, 474–485. 10.1002/dvg.22877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buels, R. , Yao, E. , Diesh, C.M. , Hayes, R.D. , Munoz‐Torres, M. , Helt, G. et al. (2016) JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biology, 17, 1–12. Available at:. 10.1186/s13059-016-0924-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, A. , Deckena, M. , Almeida‐Trapp, M. , Kopischke, S. , Kock, C. , Schüssler, E. et al. (2019) Mp TCP 1 controls cell proliferation and redox processes in Marchantia polymorpha. The New Phytologist, 224, 1627–1641. 10.1111/nph.16132 [DOI] [PubMed] [Google Scholar]
- Considine, M.J. & Foyer, C.H. (2021) Stress effects on the reactive oxygen species‐dependent regulation of plant growth and development F. Van Breusegem, ed. Journal of Experimental Botany, 72, 5795–5806. https://academic.oup.com/jxb/article/72/16/5795/6295483 [DOI] [PubMed] [Google Scholar]
- Cubas, P. , Lauter, N. , Doebley, J. & Coen, E. (1999) The TCP domain: a motif found in proteins regulating plant growth and development. The Plant Journal, 18, 215–222. 10.1046/j.1365-313X.1999.00444.x [DOI] [PubMed] [Google Scholar]
- Danisman, S. (2016) TCP transcription factors at the Interface between environmental challenges and the Plant's growth responses. Frontiers in Plant Science, 7, 1–13. 10.3389/fpls.2016.01930/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman, S. , Wal, F. , van der Dhondt, S. et al. (2012) Arabidopsis class I and class II TCP transcription factors regulate Jasmonic acid metabolism and leaf development antagonistically. Plant Physiology, 159, 1511–1523. https://academic.oup.com/plphys/article/159/4/1511/6109445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz‐Granados, A. , Sterken, M.G. , Persoon, J. et al. (2019) SIZ1 is a nuclear host target of the nematode effector GpRbp1 from Globodera pallida that acts as a negative regulator of basal plant defense to cyst nematodes. bioRxiv, 725697. 10.1101/725697v1 [DOI] [Google Scholar]
- Fang, Y. , Zheng, Y. , Lu, W. , Li, J. , Duan, Y. , Zhang, S. et al. (2021) Roles of miR319‐regulated TCPs in plant development and response to abiotic stress. Crop Journa, 9, 17–28. 10.1016/j.cj.2020.07.007 [DOI] [Google Scholar]
- Goverse, A. , Overmars, H. , Engelbertink, J. , Schots, A. , Bakker, J. & Helder, J. (2000) Both induction and morphogenesis of cyst nematode feeding cells are mediated by auxin. Molecular Plant‐Microbe Interactions, 13, 1121–1129. 10.1094/MPMI.2000.13.10.1121 [DOI] [PubMed] [Google Scholar]
- Guan, P. , Wang, R. , Nacry, P. , Breton, G. , Kay, S.A. , Pruneda‐Paz, J.L. et al. (2014) Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proceedings of the National Academy of Sciences, 111, 15267–15272. 10.1073/pnas.1411375111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi, T. , Howe, P.J. , Maier, T.R. , Hussey, R.S. , Mitchum, M.G. , Davis, E.L. et al. (2010) Arabidopsis spermidine synthase is targeted by an effector protein of the cyst nematode Heterodera schachtii. Plant Physiology, 152, 968–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A. , Wang, Y. , Liu, Y. , Wang, G. & She, X. (2020) Reactive oxygen species regulate auxin levels to mediate adventitious root induction in Arabidopsis hypocotyl cuttings. Journal of Integrative Plant Biology, 62, 912–926. 10.1111/jipb.12870 [DOI] [PubMed] [Google Scholar]
- Hur, Y.S. , Kim, J. , Kim, S. , Son, O. , Kim, W.Y. , Kim, G.T. et al. (2019) Identification of TCP13 as an upstream regulator of ATHB12 during leaf development. Genes (Basel), 10, 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J. , Hewezi, T. & Baum, T.J. (2011) Arabidopsis peroxidase AtPRX53 influences cell elongation and susceptibility to heterodera schachtii. Plant Signaling & Behavior, 6, 1778–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.T. , Haegeman, A. , Danchin, E.G.J. , Gaur, H.S. , Helder, J. , Jones, M.G.K. et al. (2013) Top 10 plant‐parasitic nematodes in molecular plant pathology. Molecular Plant Pathology, 14, 946–961. 10.1111/mpp.12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska, M.M. , Hoefsloot, H.C.J. , Mol, S. , Feron, R. , Boer, G.‐J. de, Haring, M.A. and Testerink, C. (2014) Capturing Arabidopsis root architecture dynamics with <scp>root‐fit</scp> reveals diversity in responses to salinity. Plant Physiology, 166, 1387–1402. Available at: https://academic.oup.com/plphys/article/166/3/1387/6111199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova, R. , Boer, D. , Hayes, S. & Testerink, C. (2021) Root plasticity under abiotic stress. Plant Physiology, 187, 1057–1070. https://academic.oup.com/plphys/article/187/3/1057/6359830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian, J. , Whitehead, D. , Horak, J. , Wanke, D. , Weinl, S. , Batistic, O. et al. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV‐B light, drought and cold stress responses. The Plant Journal, 50, 347–363. 10.1111/j.1365-313X.2007.03052.x [DOI] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. & Salzberg, S.L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nature Methods, 12, 357–360. http://www.nature.com/articles/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koevoets, I.T. , Venema, J.H. , Elzenga, J.T.M. & Testerink, C. (2016) Roots withstanding their environment: exploiting root system architecture responses to abiotic stress to improve crop tolerance. Frontiers in Plant Science, 7, 1–19. 10.3389/fpls.2016.01335/abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S. & Ohashi, Y. (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. The Plant Journal, 30, 337–348. 10.1046/j.1365-313X.2002.01294.x [DOI] [PubMed] [Google Scholar]
- Kubota, A. , Ito, S. , Shim, J.S. , Johnson, R.S. , Song, Y.H. , Breton, G. et al. (2017) TCP4‐dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis S. Hake, ed. PLOS Genetics, 13, e1006856. Available at:. 10.1371/journal.pgen.1006856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labudda, M. , Różańska, E. , Czarnocka, W. , Sobczak, M. & Dzik, J.M. (2018) Systemic changes in photosynthesis and reactive oxygen species homeostasis in shoots of Arabidopsis thaliana infected with the beet cyst nematode Heterodera schachtii. Molecular Plant Pathology, 19, 1690–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. , Chronis, D. , Kenning, C. , Peret, B. , Hewezi, T. , Davis, E.L. et al. (2011) The novel cyst nematode effector protein 19C07 interacts with the Arabidopsis auxin influx transporter LAX3 to control feeding site development. Plant Physiology, 155, 866–880. https://academic.oup.com/plphys/article/155/2/866/6111577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Yuan, H. , Liu, F. , Luan, J. , Yang, Y. , Ren, L. et al. (2020) BpTCP7 gene from Betula platyphylla regulates tolerance to salt and drought stress through multiple hormone pathways. Plant Cell, Tissue and Organ Culture, 141, 17–30. 10.1007/s11240-019-01748-7 [DOI] [Google Scholar]
- Li, S. (2015) The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signaling & Behavior, 10, e1044192. 10.1080/15592324.2015.1044192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, L. , Zhang, W. , An, Y. , Du, B. , Wang, D. & Guo, C. (2020) Genome‐wide analysis of the TCP transcription factor genes in five legume genomes and their response to salt and drought stresses. Functional & Integrative Genomics, 20, 537–550. 10.1007/s10142-020-00733-0 [DOI] [PubMed] [Google Scholar]
- Liu, H. , Gao, Y. , Wu, M. , Shi, Y. , Wang, H. , Wu, L. et al. (2020) TCP10, a TCP transcription factor in moso bamboo (Phyllostachys edulis), confers drought tolerance to transgenic plants. Environmental and Experimental Botany, 172, 104002. 10.1016/j.envexpbot.2020.104002 [DOI] [Google Scholar]
- Lucero, L.E. , Manavella, P.A. , Gras, D.E. , Ariel, F.D. & Gonzalez, D.H. (2017) Class I and class II TCP transcription factors modulate SOC1‐dependent flowering at multiple levels. Molecular Plant, 10, 1571–1574. https://linkinghub.elsevier.com/retrieve/pii/S1674205217302447 [DOI] [PubMed] [Google Scholar]
- Miltner, E.D. , Karnok, K.J. & Hussey, R.S. (1991) Root response of tolerant and intolerant soybean cultivars to soybean cyst nematode. Agronomy Journal, 83, 571–576. 10.2134/agronj1991.00021962008300030014x [DOI] [Google Scholar]
- Mukhopadhyay, P. & Tyagi, A.K. (2015) OsTCP19 influences developmental and abiotic stress signaling by modulatingABI4‐mediated pathways. Scientific Reports, 5, 9998. http://www.nature.com/articles/srep09998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman‐Ligeza, B. , Parizot, B. , Rycke, R. , de Rycke, R. , Fernandez, A. , Himschoot, E. et al. (2016) RBOH‐mediated ROS production facilitates lateral root emergence in Arabidopsis. Development, 143, 3328–3339. 10.1242/dev.136465/258571/RBOH-mediated-ROS-production-facilitates-lateral [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, M. , Kim, D. , Pertea, G.M. , Leek, J.T. & Salzberg, S.L. (2016) Transcript‐level expression analysis of RNA‐seq experiments with HISAT, StringTie and ballgown. Nature Protocols, 11, 1650–1667. Available at:. 10.1038/nprot.2016-095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2019) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.r‐project.org/ [Google Scholar]
- Rehman, S. , Butterbach, P. , Popeijus, H. , Overmars, H. , Davis, E.L. , Jones, J.T. et al. (2009) Identification and characterization of the Most abundant cellulases in stylet secretions from Globodera rostochiensis. Phytopathology, 99, 194–202. 10.1094/PHYTO-99-2-0194 [DOI] [PubMed] [Google Scholar]
- Sewelam, N. , Kazan, K. , Thomas‐Hall, S.R. , Kidd, B.N. , Manners, J.M. & Schenk, P.M. (2013) Ethylene response factor 6 is a regulator of reactive oxygen species signaling in Arabidopsis. PLoS One, 8, e70289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique, S. , Matera, C. , Radakovic, Z.S. , Shamim Hasan, M. , Gutbrod, P. , Rozanska, E. et al. (2014) Parasitic Worms stimulate host NADPH oxidases to produce reactive oxygen species that limit plant cell death and promote infection. Science Signaling, 7, 1–11. 10.1126/scisignal.2004777 [DOI] [PubMed] [Google Scholar]
- Sijmons, P.C. , Grundler, F.M.W. , Mende, N. , Burrows, P.R. & Wyss, U. (1991) Arabidopsis thaliana as a new model host for plant‐parasitic nematodes. The Plant Journal, 1, 245–254. 10.1111/j.1365-313X.1991.00245.x [DOI] [Google Scholar]
- Trudgill, D.L. & Cotes, L.M. (1983) Tolerance of potato to potato cyst nematodes (Globodera rostochiensis and G. pallida) in relation to the growth and efficiency of the root system. The Annals of Applied Biology, 102, 385–397. 10.1111/j.1744-7348.1983.tb02708.x [DOI] [Google Scholar]
- Tsukagoshi, H. , Busch, W. & Benfey, P.N. (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell, 143, 606–616. Available at:. 10.1016/j.cell.2010.10.020 [DOI] [PubMed] [Google Scholar]
- Urano, K. , Maruyama, K. , Koyama, T. , Gonzalez, N. , Inzé, D. , Yamaguchi‐Shinozaki, K. et al. (2022) CIN‐like TCP13 is essential for plant growth regulation under dehydration stress. Plant Mol. Biol., 108, 257–275. 10.1007/s11103-021-01238-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola, I.L. , Uberti Manassero, N.G. , Ripoll, R. & Gonzalez, D.H. (2011) The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA‐binding properties due to the presence of a threonine residue at position 15 of the TCP domain. The Biochemical Journal, 435, 143–155. https://portlandpress.com/biochemj/article/435/1/143/88336/The‐Arabidopsis‐class‐I‐TCP‐transcription‐factor [DOI] [PubMed] [Google Scholar]
- Waese, J. , Fan, J. , Pasha, A. , Yu, H. , Fucile, G. , Shi, R. et al. (2017) ePlant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell, 29, 1806–1821. https://academic.oup.com/plcell/article/29/8/1806‐1821/6100398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Gao, J. , Zhu, Z. , Dong, X. , Wang, X. , Ren, G. et al. (2015) TCP transcription factors are critical for the coordinated regulation of ISOCHORISMATE SYNTHASE 1 expression in Arabidopsis thaliana. The Plant Journal, 82, 151–162. 10.1111/tpj.12803 [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Yu, Y. , Wang, J. , Chen, Q. & Ni, Z. (2020) Heterologous overexpression of the GbTCP5 gene increased root hair length, root hair and stem trichome density, and lignin content in transgenic Arabidopsis. Gene, 758, 144954. 10.1016/j.gene.2020.144954 [DOI] [PubMed] [Google Scholar]
- Wickham, H. (2009) ggplot2: Elegant Graphics for Data Analysis. New York: Springer US. [Google Scholar]
- Wickham, H. F., R. ; Henry, L ; Müller, K. (2018a) dplyr: A Grammar of Data Manipulation.
- Wickham, H. F., R. ; Henry, L ; Müller, K. (2018b) tidyr: Easily Tidy Data with ‘spread()’ and ‘gather()’ Functions, pp.
- Wyss, U. & Grundler, F.M.W. (1992) Feeding behavior of sedentary plant parasitic nematodes. Netherlands Journal of Plant Pathology, 98, 165–173. 10.1007/BF01974483 [DOI] [Google Scholar]
- Xu, Y. , Liu, H. , Gao, Y. , Xiong, R. , Wu, M. , Zhang, K. et al. (2021) The TCP transcription factor PeTCP10 modulates salt tolerance in transgenic Arabidopsis. Plant Cell Reports, 40, 1971–1987. Available. at, 10.1007/s00299-021-02765-7 [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Huber, H. , Beljaars, S.J.M. , Birnbaum, D. , Best, S. de, Kroon, H. de and Visser, E.J.W. (2017) Benefits of flooding‐induced aquatic adventitious roots depend on the duration of submergence: linking plant performance to root functioning. Annals of Botany, 120, 171–180. Available at: http://academic.oup.com/aob/article/120/1/171/3861691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, M. , Li, D. , Li, Z. , Hu, Q. , Yang, C. , Zhu, L. et al. (2013) Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping Bentgrass. Plant Physiology, 161, 1375–1391. https://academic.oup.com/plphys/article/161/3/1375‐1391/6110813 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Effect of increasing concentrations of H2O2 on the root growth of Col‐0. Six‐day‐old Arabidopsis (tcp9, tcp20, tcp9tcp20, and wild‐type Col‐0) seedlings were transferred to liquid KNOP media containing 0‐300 μm H2O2. At 8 days post‐transfer the total root length was measured. (a) Representative pictures of wild‐type Col‐0 roots exposed to 0–300 μm H2O2. (b) The root architecture of wild‐type Col‐0 seedlings was quantified by total root length (cm). Data were analyzed with a one‐way ANOVA with a post‐hoc Tukey HSD test. ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001 (n = 35). Letters indicate different levels of significance (P < 0.05).
Figure S2. Primary roots of tcp20 and tcp9tcp20 are significantly longer compared with wild‐type Col‐0. Primary root length (cm) of wild‐type and mutant Arabidopsis seedlings at 8 days post‐germination. Data were analyzed with a one‐way ANOVA with a post‐hoc Tukey HSD test (n = 60). ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001. This experiment was performed three times with similar outcomes and pooled for data analysis.
Figure S3. TCP9 mutation in tcp9 single and tcp9tcp20 double mutants. RNA‐seq reads were mapped to the TAIR10 Arabidopsis genome. Normal length of TCP9 transcript indicated in yellow.
Figure S4. TCP20 mutation in tcp20 single and tcp9tcp20 double mutants. RNA‐seq reads were mapped to the TAIR10 Arabidopsis genome. Normal length of TCP20 transcript indicated in yellow and blue.
Figure S5. TCP9 is upregulated in response to wounding, oxidate stress, drought, and genotoxic stress. Figure and data were retrieved from http://bar.utoronto.ca/eplant/ and Kilian et al. (2007) respectively. Image was generated by Waese et al. (2017).
Table S1. Number of genes that are differentially regulated in tcp9, tcp20, and tcp9tcp20 compared with Col‐0.
Table S2. Differentially expressed genes.
Table S3. Enrichment analysis of differentially expressed genes.
Table S4. Reactive oxygen species‐related genes that are differentially expressed.
Data Availability Statement
All relevant data can be found within the manuscript, its supporting materials and E‐MTAB‐11649 (https://www.ebi.ac.uk/arrayexpress/experiments/E‐MTAB‐11649/).


