Abstract
Accumulating evidence indicates that ER-phagy serves as a key adaptive regulatory mechanism in response to various stress conditions. However, the exact mechanisms underlying ER-phagy in the pathogenesis of intervertebral disc degeneration remain largely unclear. In the present study, we demonstrated that RETREG1-mediated ER-phagy is induced by glucose deprivation (GD) treatment, along with ER stress activation and cell function decline. Importantly, ER-phagy was shown to be crucial for cell survival under GD conditions. Furthermore, ER stress was suggested as an upstream event of ER-phagy upon GD treatment and upregulation of ER-phagy could counteract the ER stress response. Therefore, our findings indicate that RETREG1-mediated ER-phagy activation protects against GD treatment-induced cell injury via modulating ER stress in human nucleus pulposus cells.
Keywords: intervertebral disc degeneration, senescence, apoptosis, ER-phagy, ER stress
Introduction
Low back pain (LBP) is the most prevalent musculoskeletal condition with a lifetime prevalence of approximately 80%. It is also the leading cause of productivity loss and imposes a substantial socio-economic burden on society [1]. Intervertebral disc degeneration (IDD)-caused pathological changes are recognized as the predominant cause of LBP [2]. Although several factors are involved in the initiation and progress of IDD, including genetics, mechanics, aging, nutrition, etc. [ 3– 5], the specific etiology and pathogenesis of IDD have not yet been fully understood.
The intervertebral disc is composed of three parts: inner gelatinous nucleus pulposus (NP), peripheral laminar annulus fibrosus, and superior and inferior cartilaginous endplates. The central NP tissue is considered to be the key functional executor and is responsible for disc height maintenance and mechanical loading redistribution. NP cells are the main cell type present in NP tissues. These cells produce type II collagens and proteoglycan to maintain extracellular matrix (ECM) homeostasis as well as structural and functional stability [6]. Due to aging and accumulation of unfavorable factors in the intervertebral disc, NP cells are transited from healthy to senescent or apoptotic phenotypes, resulting in the disturbance of ECM metabolic balance. Accordingly, excessive NP cell senescence and apoptosis accumulation can also accelerate IDD progression [7]. However, the precise molecular mechanisms underlying NP cell senescence and apoptosis are still not completely understood.
The endoplasmic reticulum (ER) is a vital intracellular organelle present in all eukaryotic cells and is involved in protein synthesis, folding, and trafficking. To ensure that all nascent proteins in the ER lumen are properly processed, the entire process of protein maturation is tightly controlled by diverse ER quantity control mechanisms, of which the unfolded protein response (UPR) is the most crucial adaptive mechanism [ 8, 9]. When cells are exposed to prolonged or severe stress, the demand for ER function outweighs its capacity, resulting in the accumulation of large amounts of unfolded or misfolded proteins within the ER lumen. This is referred to as ER stress, which subsequently activates the UPR to restore ER homeostasis. UPR classically consists of three main signaling branches: inositol-requiring kinase 1 (IRE1), protein kinase RNA-like ER kinase (PERK), and activating transcription factor 6 (ATF6), which originally binds to the molecular chaperone BiP/GRP78 [8]. However, if UPR fails to restore cellular homeostasis, the ER stress response switches from pro-survival to pro-apoptosis through the activation of C/EBP homologous protein (CHOP) and caspase-12 dependent pathways [ 10, 11]. Previous studies have demonstrated that ER stress-related cell injury plays an important role in the pathogenesis of diseases such as diabetes, immune-related disorders, neurodegeneration, heart diseases, and cancer [ 12, 13]. In addition, upregulated expression of ER stress-related proteins GRP78 and CHOP was detected in degenerated human NP tissues [14], while inhibiting ER stress showed protective effects against cell death and attenuated IDD progression in rats [ 15, 16]. Therefore, an in-depth study of the regulatory mechanisms of ER stress can provide potential therapeutic strategies for rescuing NP cell damage.
ER-phagy, also referred to as reticulophagy, is a recently identified special type of selective autophagy, wherein excessive ER fragments and resident protein aggregates are selectively engulfed by autophagosomes and delivered to lysosomes for degradation and recycling [17]. The highly selective nature of ER-phagy is largely dependent on specific receptors expressed in ER membranes that specifically interact microtubule-associated protein light chain 3 (LC3) with its LC3 interacting region (LIR) [18]. To date, several ER-phagy receptors have been identified, including RETREG1 (reticulophagy regulator 1), CCPG1 (cell-cycle progression gene 1), RTN3L (reticulon 3 long isoform), ATL3 (atlastin 3), SEC62 (SEC62 homolog), and TEX264 (testisexpressed 264), among which RETREG1 was identified first and remains the most studied ER-phagy receptor in mammalian cells [ 19, 20]. Moreover, the elimination of redundant ER by ER-phagy has emerged as a protein quality control mechanism for the maintenance of intracellular homeostasis. ER-phagy was reported to be elicited in response to UPR, and defects in ER-phagy could result in augmented ER stress and even cell damage [ 21– 23]. Functional abnormalities in ER-phagy have been suggested to be involved in the pathogenesis of diverse human disorders such as neuropathy, viral infections, diabetes, and cancer [ 24, 25]. However, the critical role of RETREG1-mediated ER-phagy and its association with ER stress and cell damage have not yet been well defined in the occurrence and progression of IDD.
In this study, we investigated the molecular mechanisms of RETREG1-mediated ER-phagy in modulating ER stress and subsequent cell damage in human NP cells. Our findings indicate that ER stress is involved in GD-induced cellular senescence and apoptosis. Upregulation and downregulation of RETREG1-mediated ER-phagy with genetic manipulation clearly alleviated and exacerbated GD-induced cellular senescence and apoptosis, respectively. In addition, ER-phagy activation triggered by ER stress in turn ameliorated ER stress and related cell damage. Thus, our findings provide novel insights into the regulatory mechanism of RETREG1-mediated ER-phagy and its relationship with ER stress and cell damage in human NP cells.
Materials and Methods
Ethics statement
Studies involving human participants were reviewed and approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China). Written informed consent to participate in this study was provided by the participants or their legal guardian/next of kin.
Reagents and antibodies
Dulbecco’s Modified Eagle Medium (DMEM/F12), glucose-free DMEM, and phosphate-buffered saline (PBS) were obtained from GIBCO (Grand Island, USA). Fetal bovine serum (FBS) was purchased from ScienCell Research Laboratories (Carlsbad, USA). The ER stress-specific inhibitor 4-phenyl butyric acid (4-PBA; 99%) was acquired from MedChemExpress (Monmouth Junction, USA). 4′,6-diamidino-2-phenylindole (DAPI) was obtained from Beyotime (Nantong, China). The specific ER-tracker and Lyso-tracker were purchased from Yeasen Biotechnology (Shanghai, China). Antibodies against p21 (ab109520), Bax (ab104156), Bcl-2 (ab196495), GRP78 (ab21685), caspase-12 (ab62463), IRE1-α (ab37073), and phosphoIRE1-α (ab48417) were obtained from Abcam (Cambridge, UK). LC3B (#83506), p16 (#18769), and cleaved caspase-3 (#9664) were purchased from Cell Signaling Technology (Danvers, USA). Antibodies specific to CHOP (15204-1-AP), RETREG1 (21537-1-AP), p62 (18420-1-AP), p53 (10442-1-AP), β-actin (66009-1-Ig), HRP-conjugated second antibodies (SA00001-1 and SA00001-2), and goat anti-rabbit CoraLite488 and goat anti-mouse CoraLite594 antibodies were acquired from Proteintech (Wuhan, China). RETREG1-siRNA and control-siRNA were constructed by Qijing Biotechnology Co. (Wuhan, China), RETREG1-siRNA1: sense strand 5′-AGCTATCAAAGACCAGTTA-3′, RETREG1-siRNA2: sense strand 5′-CCUCUGAACAGUGACCAAATT-3′. The RETREG1-overexpression lentivirus (NM_001034850) using CV084 (Ubi-MCS-SV40-Neomycin) vector was provided by GeneChem (Shanghai, China).
Tissue collection, cell culture, and cell treatment
All experimental steps were conducted as previously described [26]. The IDD grade of all participants was determined based on their preoperative T2-phase MRI and graded according to the Pfirrmann classification system: grade I-II as healthy and grade IV-V as degenerated. Healthy NP tissues were obtained from six patients (two male and four female, mean age 15.5 years old, range 11–20 years old) undergoing spinal deformity correction surgery due to adolescent idiopathic scoliosis. Degenerated NP tissues were acquired from six patients (three male and three female, mean age 55.6 years old, range 46–63 years old) who underwent surgery for lumbar spinal stenosis. Detailed demographic information is summarized in Supplementary Table S1. Once separated, the NP tissue was immediately sectioned and frozen in liquid nitrogen or fixed with paraformaldehyde for subsequent western blot analysis and histological analysis.
For isolation and culture of cells, freshly obtained NP tissue specimens were rinsed with PBS, dissected, and enzymatically digested for 8 h. The supernatant was then removed and the precipitate was resuspended in DMEM/F12 containing 10% FBS and 1% penicillin/streptomycin. Human NP cells were identified by flow cytometry, as described previously [26]. All subsequent in vitro experiments were conducted on cells between passage 2 and passage 3.
For cell treatment, the NP cells were exposed to glucose-free DMEM/F12 for different time periods (0, 6, 12, 24, and 48 h), or directly co-cultured with GD in combination with 2.5 mM 4-PBA for 48 h. For RETREG1 overexpression and knockdown experiments, the NP cells were transfected with RETREG1-overexpression lentivirus for 72 h or RETREG1 small interfering RNA (siRNA) for 48 h, followed by GD treatment for 48 h. Transfection efficacies were evaluated by western blot analysis.
Cell viability assay
Cell viability was assessed using the Cell Counting Kit-8 assay kit (CCK-8; Beyotime) according to the manufacturer’s instructions. First, 1 × 10 4 cells/well were seeded in 96-well plates and exposed to GD for different time periods. Thereafter, 10 μL of CCK-8 reaction solution was added to each well, incubated at 37°C for 2 h, and the absorbance value was measured at 450 nm using a spectrophotometer (BioTek, Winooski, USA).
Western blot analysis
Total protein extraction was performed as described previously [ 26, 27]. The protein samples were separated on 8%–12% sodium dodecyl sulfate-polyacrylamide gels, transferred onto polyvinylidene difluoride membranes, blocked with 5% non-fat milk, and finally incubated overnight at 4°C with the antibodies specific to the following proteins: p53 (1:1000), p21 (1:2000), p16 (1:800), Bcl-2 (1:1000), Bax (1:2000), cleaved caspase-3 (1:1000), RETREG1 (1:1000), LC3 (1:600), p62 (1:1000), p-IRE1-α (1:1000), IRE1-α (1:1000), GRP78 (1:1000), caspase-12 (1:1000), CHOP (1:1000), and β-actin (1:5000). After being washed three times with 0.1% TBST, the membranes were incubated with HRP-conjugated secondary antibody (1:2000) for 1 h at room temperature. The protein bands were visualized using an enhanced chemiluminescence imaging system (Bio-Rad) and quantified using ImageJ software.
SA-β-gal staining
SA-β-gal activity was measured using a Senescence β-Galactosidase Staining Kit (Beyotime) to determine cell senescence. SA-β-gal-positive cells were observed under bright-field conditions using a microscope (Olympus, Melville, USA). The percentage of blue-stained cells was quantified to evaluate SA-β-gal activity.
Apoptosis measurement
Apoptosis was detected using the Annexin V-FITC/propidium iodide (PI) double staining assay kit (Beyotime) according to the manufacturer’s instructions. Briefly, cells were collected and resuspended in 300 μL binding buffer. The suspension was incubated with 5 μL Annexin V-FITC and 5 μL PI for 10 min without light. Apoptotic cells were assessed by flow cytometry (BD Biosciences, San Jose, USA). Early (Annexin V+/PI−) and late (Annexin V+/PI+) apoptotic cells were quantified.
Immunofluorescence microscopy
After the indicated treatment, the samples were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Thereafter, 3% bovine serum albumin (BSA) in PBS was used to block non-specific binding sites at room temperature. Human NP cells were then incubated overnight at 4°C with primary antibodies specific to: RETREG1 (1:100), LC3 (1: 100), p16 (1:100), cleaved caspase-3 (1: 100), GRP78 (1:100), and CHOP (1:100). The samples were then incubated with the corresponding fluorescein-labelled secondary antibodies for 1 h at 37°C. The cell nuclei were counterstained with DAPI. Images were captured using a fluorescence microscope (Olympus).
ER and lysosome labeling
Specific ER and lysosome labeling dyes, ER-tracker and Lyso-tracker, were used to determine the morphology and subcellular localization of ER and lysosome, respectively. After being washed with Hanks’ Balanced Salt Solution (HBSS), the cells were incubated with 1 μM ER-tracker (green) and 50 nM Lyso-tracker (red) at 37°C for 30 min, followed by staining with Hoechst 33342 (Beyotime) for 10 min. The cells were then washed with PBS and imaged using a fluorescence microscope (Olympus).
Transmission electron microscopy
Cells were harvested, centrifuged, and fixed with 2.5% glutaraldehyde and immersed in 1% osmium tetroxide. Cells were then dehydrated using a series of graded alcohol solutions. Following permeation and embedding, the samples were cut into 70-nm ultra-thin sections and stained with uranyl acetate and lead citrate. Slides were examined and photographed using a transmission electron microscope (TEM) (Tecnai, FEI, USA).
Immunohistochemical staining
Immunohistochemical staining was performed as described previously [28]. Briefly, human NP samples were fixed with 4% paraformaldehyde, embedded in paraffin, and sliced into 4-μm thin sections. Sections were incubated overnight at 4°C with primary antibodies specific for RETREG1 (1: 150) and LC3 (1: 200) and then incubated for 50 min at room temperature with the indicated secondary antibodies, followed by counterstaining with hematoxylin. The sections were then washed, sealed, and observed under a microscope (Olympus).
Statistical analysis
All experiments were independently conducted at least three times, and the representative results are shown. All data are presented as the mean±SD, and statistical analyses were performed using Student’s t-test or one-way analysis of variance (ANOVA), followed by the Tukey test using GraphPad Prism software (version 8.0). Statistical significance was set at P<0.05.
Results
GD treatment triggers senescence and apoptosis in human NP cells
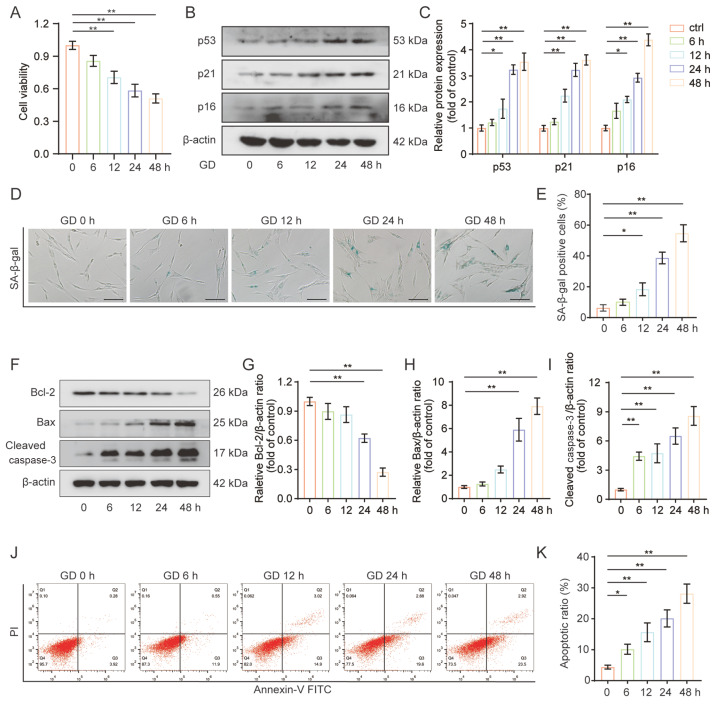
The intervertebral disc is a predominantly avascular organ whose metabolic exchange is largely dependent on diffusion across the cartilage endplate and outer annulus fibrosus. Cartilage endplate calcification and insufficient nutrient availability are believed to be the critical risk factors for NP cell apoptosis and functional damage in degenerated discs [5]. Here, we evaluated the pathological effects of glucose starvation on human NP cells. First, the NP cells were exposed to GD treatment for different time periods (0, 6, 12, 24, and 48 h) and the cell viability was assessed by the CCK-8 assay. Our results showed that GD treatment significantly decreased cell viability in a time-dependent manner ( Figure 1A). Cell senescence was determined by measuring the expressions of senescence-related proteins p53, p21, and p16, and SA-β-gal enzyme activity. Our results indicated that the protein levels of p53, p21, and p16 were significantly increased upon GD treatment ( Figure 1B,C). The number of SA-β-gal-positive cells was also notably increased in response to GD in a time-dependent manner ( Figure 1D,E). Additionally, we investigated the effects of GD on cell apoptosis by detecting the expression levels of apoptosis-related proteins Bcl-2, Bax, and cleaved caspase-3 by western blot analysis and Annexin V/PI double staining followed by flow cytometry. As shown in Figure 1F–I, the apoptosis-related protein Bax and cleaved caspase-3 protein levels were robustly elevated relative to the control group, while Bcl-2 protein level declined under GD treatment in a time-dependent manner. Similarly, the number of early and late apoptotic cells was significantly increased upon GD treatment compared to the control group ( Figure 1J,K). Therefore, our findings demonstrated that GD treatment significantly stimulated human NP cell senescence and apoptosis.
Figure1 .
GD treatment triggered senescence and apoptosis in human NP cells
(A) CCK-8 assay results of NP cells incubated with glucose-free medium for various durations (0, 6, 12, 24, and 48 h). (B,C) Protein expressions of p53, p21, and p16 in NP cells exposed to GD treatment for different durations were measured by western blot analysis. (D,E) SA-β-gal staining assay results from NP cells subjected to GD treatment for different durations, and the quantification of positive SA-β-gal stained NP cells. Scale bar: 100 μm. (F–I) Expressions of Bcl-2, Bax, and cleaved caspase-3 proteins in NP cells subjected to GD treatment for different durations were detected by western blot analysis. (J,K) Flow cytometry analysis of Annexin V/PI double staining was performed to evaluate apoptosis. NP cells were treated as indicated above and both early apoptotic and late apoptotic cells were quantified. *P<0.05, **P<0.01.
ER stress activation is associated with GD treatment-induced senescence and apoptosis in human NP cells
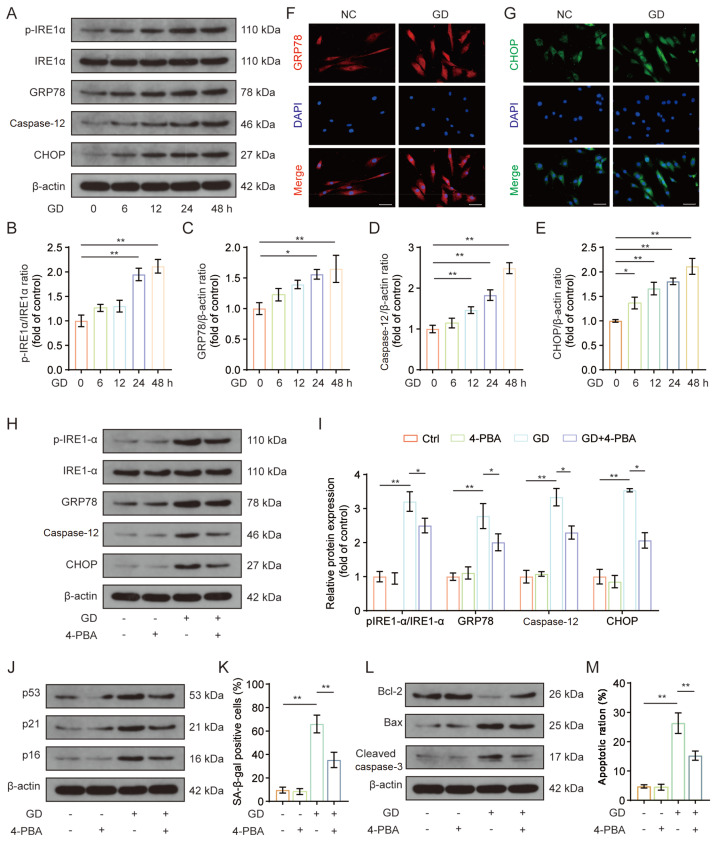
ER stress, a classically conserved cellular stress response, can be initiated in response to a variety of pathological conditions that disturb cellular homeostasis, whereas chronic unabated ER stress could be pathogenic and result in cell damage. Therefore, we explored whether ER stress is involved in GD-induced decline in the functions of human NP cells. Expression levels of ER stress-related markers were determined by western blot analysis. Protein levels of p-IRE1-α/IRE1-α, GRP78, caspase-12, and CHOP were increased in the GD group compared to the control group ( Figure 2A–E). Similarly, immunofluorescence staining results showed that GD treatment significantly elevated GRP78 and CHOP fluorescence intensity ( Figure 2F,G), indicating that ER stress was triggered by GD treatment. To further characterize the role of ER stress in GD-induced cell damage, 4-PBA, a well-known specific inhibitor of ER stress, was administered. Our results showed that the expression levels of ER stress-related proteins were markedly reduced in the GD+4-PBA co-treated group, than the corresponding GD-treated alone group ( Figure 2H,I). Additionally, GD treatment significantly elevated the levels of senescence-related and apoptotic proteins p53, p21, p16, Bax, and cleaved caspase-3, along with decreased Bcl-2 protein levels, while all these alterations induced by GD treatment were partially mitigated by 4-PBA administration ( Figure 2J,L, and Supplementary Figure S1A,B). The flow cytometry and SA-β-gal staining results are consistent with the western blot analysis results ( Figure 2K,M, and Supplementary Figure S1C,D). Consequently, our results suggest that ER stress activation may partially contribute to GD-induced senescence and apoptosis in human NP cells.
Figure2 .
ER stress activation was associated with GD treatment-induced senescence and apoptosis in human NP cells
The NP cells were incubated in glucose-free medium for various durations (0, 6, 12, 24, and 48 h), or NP cells were directly exposed to GD treatment in the presence of 4-PBA for 48 h. (A–E) Western blot analysis of the expressions of ER stress marker proteins p-IRE1-α, IRE1-α, GRP78, caspase-12, and CHOP, and relative quantification results are shown. (F,G) Immunofluorescence staining results of GRP78 and CHOP in NP cells; the nuclei were stained with DAPI (scale bar: 50 μm). (H,I) Western blot analysis of the protein expressions of p-IRE1-α, IRE1-α, GRP78, caspase-12, and CHOP, and the relative quantification results are shown. (J) Western blot analysis of the protein levels of p53, p21, and p16. (K) Quantification analysis of positive SA-β-gal staining NP cells. (L) Western blot analysis of the protein levels of Bcl-2, Bax, and cleaved caspase-3. (M) Quantification analysis of apoptotic NP cell ratio by Annexin V/PI double staining. *P<0.05, **P<0.01.
RETREG1-mediated ER-phagy is activated in human NP cells in response to GD treatment
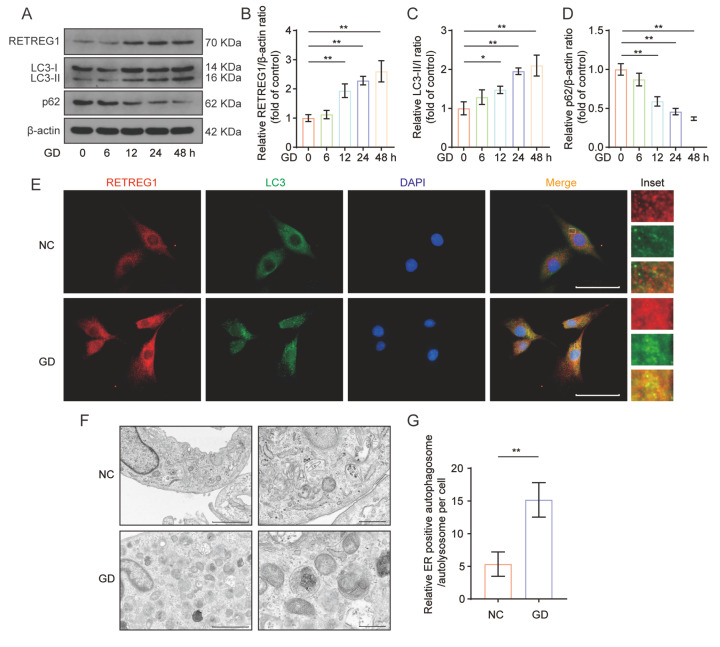
Increasing evidence suggests the crucial role of ER-phagy in maintaining cellular energy and metabolic homeostasis in response to multiple intracellular and extracellular stimulations. To investigate whether RETREG1-mediated ER-phagy is induced under GD conditions, the NP cells were treated as indicated above. The expression of ER-phagy-related proteins, RETREG1, LC3, and p62 were determined by western blot analysis and immunofluorescence assay. As shown in Figure 3A–D, the protein expressions of RETREG1 and LC3 were markedly increased, along with downregulated levels of p62 protein in the GD-treated group, compared to those in the control group. Meanwhile, the RETREG1 and LC3 co-staining results revealed that the fluorescence intensity as well as the colocalization of RETREG1 and LC3 were particularly higher in the GD-treated group than those in the control group ( Figure 3E). Furthermore, the formation and accumulation of autophagosomes/autolysosomes containing ER fragments were determined by TEM for better characterization of the occurrence of ER-phagy. Compared to the control group, GD treatment increased the number of ER-positive autophagosome/autolysosome formation in human NP cells ( Figure 3F,G). Overall, our findings confirmed the initiation of ER-phagy upon GD treatment in human NP cells.
Figure3 .
RETREG1-mediated ER-phagy was activated in human NP cells upon GD treatment
(A–D) Western blot analysis of the protein expressions of RETREG1, LC3, and p62 of NP cells treated with GD for different periods, and relative quantification is shown. (E) Immunofluorescence double staining results of RETREG1 (red) and LC3 (green) in NP cells treated with GD. The nuclei were stained with DAPI and images were observed with a fluorescence microscope. Scale bar: 50 μm. (F) The formation and accumulation of autophagosomes/autolysosomes containing ER fragments or whorls in NP cells were assessed by TEM. Scale bar: 1 μm (left) and 500 nm (right). *P<0.05, **P<0.01.
RETREG1-mediated ER-phagy modulates GD-induced cell senescence and apoptosis in human NP cells
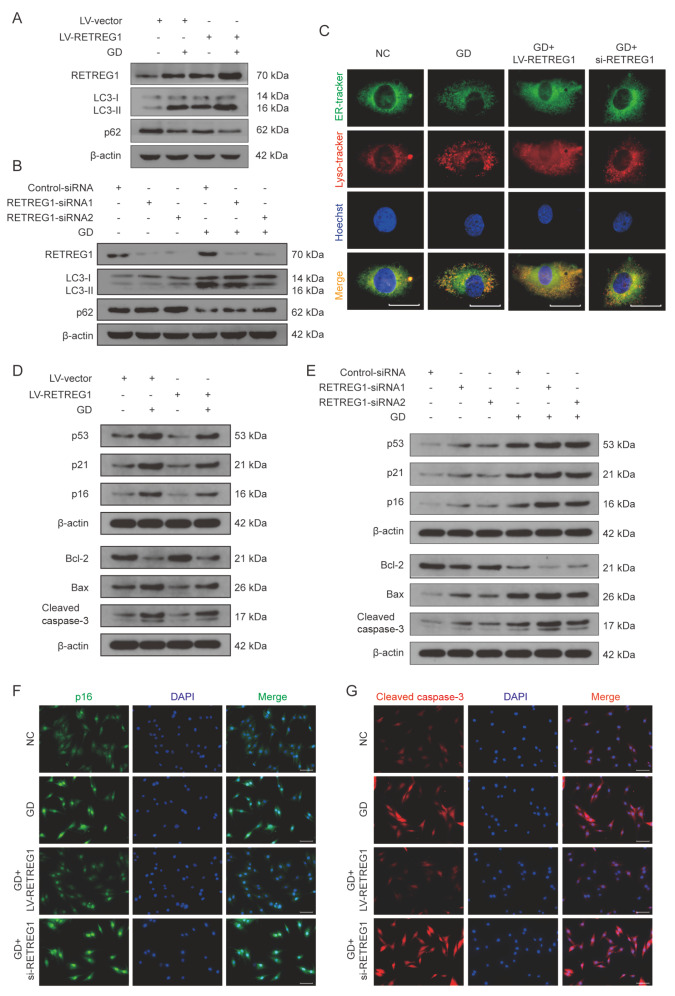
Next, we examined the effects of ER-phagy on senescence and apoptosis by regulating ER-phagy levels via RETREG1-overexpression lentivirus and siRNA transfection. Our results indicated that RETREG1 overexpression further enhanced GD treatment-mediated expressions of ER-phagy related proteins, RETREG1 and LC3, and reduced p62 expression ( Figure 4A and Supplementary Figure S2A). RETREG1 knockdown by RETREG1 siRNA transfection evidently alleviated GD treatment-mediated elevated expression of RETREG1 and LC3 proteins and decreased p62 expression ( Figure 4B and Supplementary Figure S2B). These results suggest that ER-phagy activation cause by GD treatment could be improved by RETREG1-overexpression lentivirus and weakened by siRNA transfection. To further morphologically monitor the alterations of ER-phagy with genetical manipulation of RETREG1, we detected the colocalization of ER and lysosome with specific organelle staining, and the results showed that the co-staining extent of ER puncta with lysosome were obviously amplified with RETREG1 overexpression and declined with RETREG1 knockdown, respectively ( Figure 4C). In addition, the increased expressions of senescence and apoptosis-related proteins p53, p21, p16, Bax, and cleaved caspase-3, and the decreased expression of Bcl-2 following GD treatment were significantly mitigated by RETREG1 overexpression ( Figure 4D and Supplementary Figure S2C,D), while RETREG1 knockdown markedly exacerbated GD-treatment-mediated proapoptotic and prosenescent effects ( Figure 4E and Supplementary Figure S2E,F). Immunofluorescence analysis using anti-p16 and cleaved caspase-3 antibodies showed that the increased fluorescence intensities of p16 and cleaved caspase-3 under GD treatment were decreased by RETREG1 overexpression and knockdown, respectively ( Figure 4F,G). In summary, these results indicated that GD treatment-mediated cell senescence and apoptosis could be relieved by RETREG1-mediated ER-phagy activation.
Figure4 .
RETREG1-mediated ER-phagy modulated GD treatment-induced senescence and apoptosis in human NP cells
(A) After transfection with RETREG1 overexpressing lentivirus for 72 h, the NP cells were exposed to GD treatment for 48 h. The protein levels of RETREG1, LC3, and p62 were determined by western blot analysis. (B) After transfection with RETREG1 siRNA for 48 h, the NP cells were subjected to GD for 48 h. The protein levels of RETREG1, LC3, and p62 were evaluated by western blot analysis. (C) The RETREG1 overexpression and knockdown NP cells were subjected to GD treatment for 48 h, ER and lysosomes were specifically labeled with ER-Tracker Green and LysoTracker Red, respectively. Representative images are shown. Scale bar: 20 μm. (D) The NP cells were treated as indicated in (A), the protein levels of p53, p21, p16, Bcl-2, Bax, and cleaved caspase-3 were detected by western blot analysis. (E) The NP cells were treated as indicated in (B), the protein levels of p53, p21, p16, Bcl-2, Bax, and cleaved caspase-3 were detected by western blot analysis. (F,G) Immunofluorescence staining results of p16 (green) and cleaved caspase-3 (red) in NP cells as treated in (C), the nuclei were stained with DAPI. Scale bar: 100 μm.
ER stress is involved in RETREG1-mediated ER-phagy activation in human NP cells in response to GD treatment
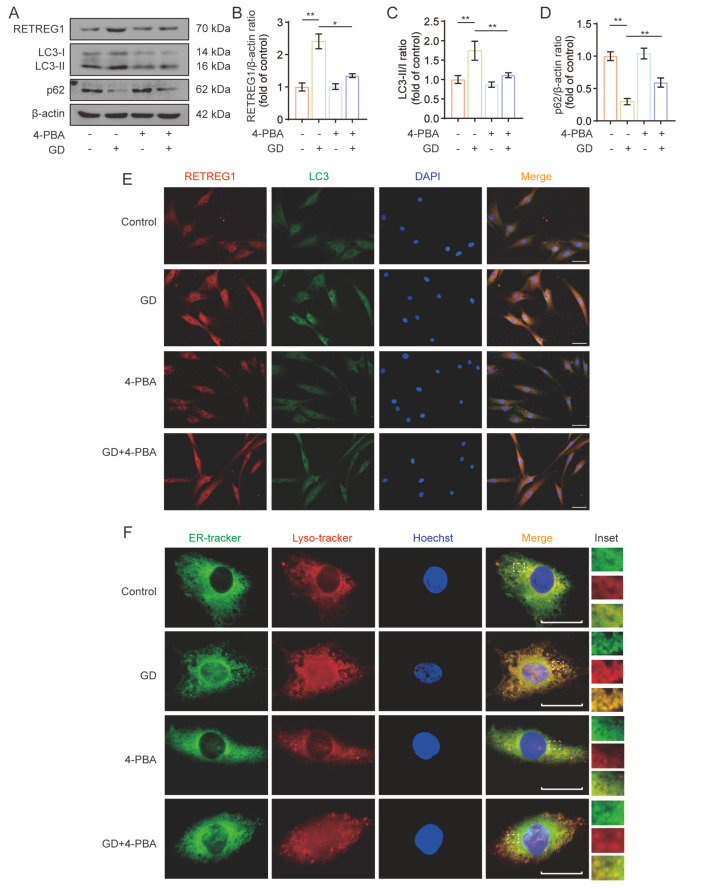
To characterize the critical role of ER stress in inducing ER-phagy, human NP cells were exposed to GD in the presence of the ER stress-specific inhibitor 4-PBA, and the ER-phagy profiles were determined by western blot analysis and immunofluorescence staining. As shown in Figure 5A–D, GD treatment enhanced ER-phagy levels, as indicated by the elevated levels of RETREG1 and LC3 protein expression and the reduced p62 protein expression, which were partially counteracted by 4-PBA administration. Moreover, immunofluorescence double-staining with anti-RETREG1 and anti-LC3 antibodies showed that the fluorescence intensity and colocalization extent of RETREG1 and LC3 in the GD + 4-PBA group were significantly lower than those in the GD alone group ( Figure 5E). Furthermore, we also labeled the ER and lysosomes with ER-tracker (green) and Lyso-tracker (red) respectively, as shown in Figure 5F. Compared to the control group, GD treatment induced a notable increase in the number of ER cluster staining patterns, which mostly colocalized with lysosome cluster staining, suggesting the occurrence of ER lysosomal degradation. In contrast, these effects were suppressed by 4-PBA co-treatment. These findings may indicate that under GD conditions, ER stress activation is associated with the induction of RETREG1-mediated ER-phagy in human NP cells.
Figure5 .
ER stress was involved in RETREG1-mediated ER-phagy activation in human NP cells in response to GD treatment
(A–D) Human NP cells were subjected to GD and treated with 4-PBA for 48 h. The expressions of RETREG1, LC3, and p62 proteins were determined by western blot analysis, and relative quantification results are shown. (E) Immunofluorescence double staining results of RETREG1 (red) and LC3 (green) in human NP cells treated as described above. The nuclei were stained with DAPI and images were observed using a fluorescence microscope. Scale bar: 100 μm. (F) ER-tracker (green) and Lyso-tracker (red) were separately used to specifically label the ER and lysosome, and the representative colocalization results are illustrated. Scale bar: 20 μm. *P<0.05, **P<0.01.
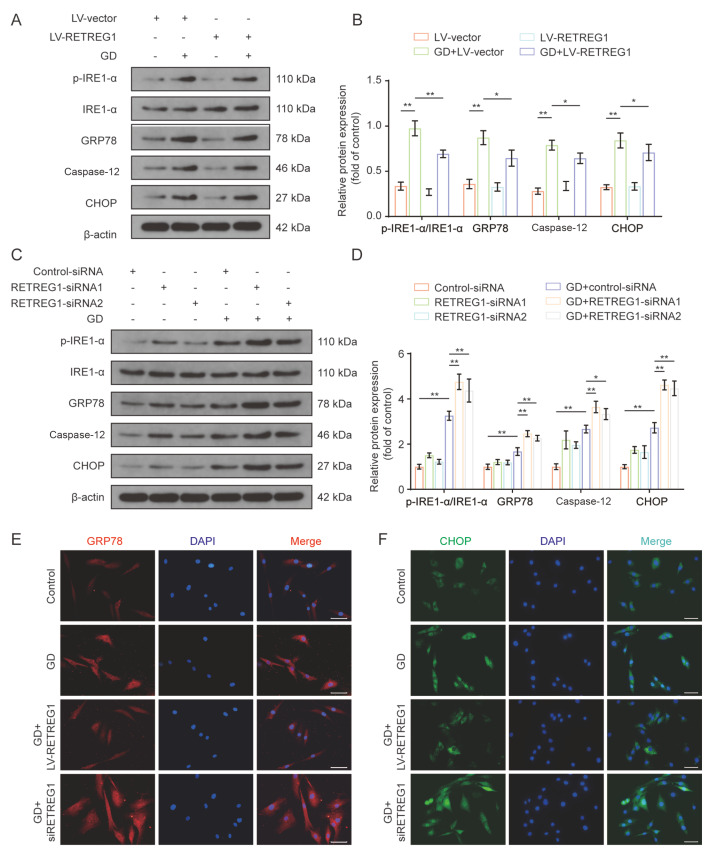
Upregulation of RETREG1-mediated ER-phagy can alleviate ER stress in human NP cells
Having identified that ER-phagy is induced by stress, we further investigated whether ER stress is in turn modulated by ER-phagy based on its fundamental role in restoring ER homeostasis via selective removal of ER fragments. ER-phagy upregulation and downregulation were successfully established with RETREG1-overexpression lentivirus and siRNA transfection. Expression of ER stress marker proteins was detected by western blot analysis and immunofluorescence analysis. Enhanced protein expressions of p-IRE1-α/IRE1-α, GRP78, caspase-12, and CHOP in response to GD treatment were mitigated by RETREG1 overexpression ( Figure 6A,B), while RETREG1 knockdown significantly increased the expressions of ER stress-related proteins ( Figure 6C,D). Consistently, the immunofluorescence staining results showed that GD treatment stimulated GRP78 and CHOP expression, which was partially reversed by RETREG1 overexpression. Conversely, RETREG1 downregulation by siRNA increased GD treatment-induced GRP78 and CHOP expression ( Figure 6E,F). These results confirm that RETREG1-mediated ER-phagy activation can relieve ER stress induced by GD treatment in human NP cells.
Figure6 .
Upregulation of RETREG1-mediated ER-phagy alleviated ER stress in human NP cells
(A,B) Human NP cells were transfected with RETREG1 overexpressing lentivirus for 72 h, followed with 48 h GD treatment. The expressions of p-IRE1-α, IRE1-α, GRP78, caspase-12, and CHOP proteins were evaluated by western blot analysis, and relative quantification results are shown. (C,D) The NP cells transfected with RETREG1 siRNA were subjected to 48 h of GD treatment. The expressions of p-IRE1-α, IRE1-α, GRP78, caspase-12, and CHOP proteins were evaluated using western blot analysis, and relative quantification results are shown. (E,F) Results of immunofluorescence staining of p16 (green) and cleaved caspase-3 (red) proteins in human NP cells treated as mentioned above. The nuclei were stained with DAPI, scale bar: 50 μm. *P<0.05, **P<0.01.
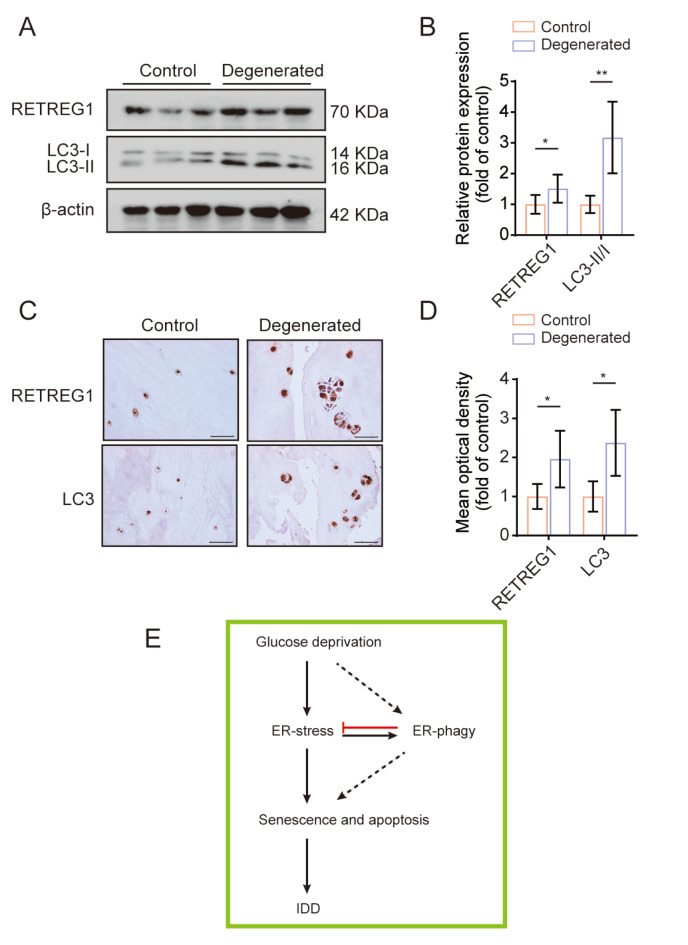
Enhanced expression of RETREG1 and LC3 in degenerated human NP tissues
To further investigate the potential involvement of RETREG1-mediated ER-phagy in IDD, we examined the protein expressions of RETREG1 and LC3 in NP tissues obtained from six relatively healthy and six patients with degenerated human NP tissues by western blot analysis and immunohistochemical staining. As shown in Figure 7A,B, compared to the relatively healthy NP tissues, degenerated NP tissues had significantly higher expressions of RETREG1 and LC3 proteins. Similarly, as detected by immunohistochemical staining ( Figure 7C,D), an elevated expression of RETREG1 and LC3 was observed in degenerated human NP tissues compared to those in the relatively healthy NP tissues. Thus, we concluded that RETREG1-mediated ER-phagy abnormality may be clinically relevant for the initiation and progression of IDD.
Figure7 .
Enhanced expression of RETREG1 and LC3 in degenerated human NP tissues
(A,B) The expressions of RETREG1 and LC3 proteins in relatively healthy and degenerated human NP tissues were measured by western blot analysis, and relative quantification results are shown. n=12. (C,D) Immunohistochemical staining images for RETREG1 and LC3 in human NP tissues were obtained and relative mean optical density was quantified. n=12. (E) Schematic illustration of the possible mechanisms by which RETREG1-mediated ER-phagy relieves ER stress response and subsequent cell damage under GD conditions. *P<0.05, **P<0.01.
Degeneration of the intervertebral disc is typically featured by poor nutrient availability, excessive cell apoptosis, and impaired cellular functional resistance to stress conditions. In this study, we sought to establish the potential role of RETREG1-mediated ER-phagy in relieving ER stress and subsequent senescence and apoptosis in human NP cells induced by GD. We found that GD treatment resulted in time-dependent increase in apoptosis and senescence in human NP cells, which was mediated by the ER stress pathway. Alteration of RETREG1-mediated ER-phagy with genetic manipulation showed its potent influence on GD-mediated apoptosis and senescence. Moreover, ER stress was identified as a trigger of ER-phagy under GD conditions, while ER-phagy activation relieved ER stress in human NP cells ( Figure 7E).
Discussion
The NP tissues of the disc are anatomically avascular, and their primary nutrient supply is achieved via diffusion from the capillaries of the outer annulus fibrosus and cartilage endplates. With ageing and degeneration, altered structure and composition of the endplate compromise proper nutrient diffusion, metabolic products elimination, and even NP cell viability and survival [ 29, 30]. DeLucca et al. [31] quantified human endplates mechanics and composition and demonstrated that the permeability of human endplates decreased by 50%–60% during degeneration. Wills et al. [32] constructed a novel, composition-based permeability model for cartilage endplates and revealed that cartilage endplate degeneration might locally affect oxygen and lactate levels, reduce glucose concentration, and ultimately induce cell starvation and death. In serum starvation conditions, Li et al. [33] established that NP stem cells underwent quiescence and their proliferation was suppressed. Moreover, Wang et al. [34] demonstrated that glucose and serum deficiency could result in mitochondrial dysfunction, apoptosis, and matrix degradation in rat NP cells. In the present study, we found that the expression of senescence- and apoptosis-related proteins was visibly enhanced after GD, combined with an increased proportion ratio of Annexin V positive and SA-β-gal-positive human NP cells, which further consolidated the pathological role of GD in driving human NP cell damage and even cell death.
ER stress has been documented as a highly conserved stress response in eukaryotic cells, and is activated when cells are unable to balance the protein-folding demands and capabilities. Generally, ER stress acts as a double-edged sword in cell function and fate; moderate ER stress is beneficial to cells as it resolves protein folding and restores ER homeostasis, while irremediable ER stress leads to cell death [13]. Over the past decade, chronic ER stress-related cell damage has been implicated in a wide range of human pathologies, such as diabetes, neurodegeneration, heart diseases, and osteoarthritis [ 35– 37]. Concurrently, an accumulation of ER stress-related proteins, including GRP78, CHOP, IRE1, and PERK, is observed in degenerated NP tissues [ 14, 38]. Several in vitro studies have confirmed that irremediable ER stress is generated, resulting in advanced glycation end product accumulation, mechanical stress, oxidative stress, and compression, which eventually leads to NP cell damage [ 15, 28, 39, 40]. In addition, pharmacological targeting of ER stress showed favorable effects on NP cells and delayed IDD progression in rats [ 16, 41]. However, the involvement of ER stress in GD-mediated apoptosis and senescence in human NP cells has not been well defined. In this study, an elevated expression of ER stress-related proteins was observed in response to GD treatment of human NP cells, which was potently mitigated by 4-PBA administration. Notably, GD-mediated senescence and apoptosis were also weakened in the presence of 4-PBA, indicating that ER stress activation plays a vital role in GD treatment-induced senescence and apoptosis of human NP cells.
Recent research has focused on the mechanisms underlying ER-phagy that selectively delivers ER fragments to the lysosomes via the autophagy pathway, counterbalancing ER expansion and restoring intracellular homeostasis [42]. ER-phagy serves as an adaptive mechanism that is rapidly upregulated and exerts a cytoprotective function under various external or internal stress conditions, including nutrient deprivation, ER stress, and pathogen attack [ 43– 45]. RETREG1 was the first identified ER-phagy mediator in eukaryotic cells and promotes ER scission via its membrane-bending capacity and interacts with LC3 or GABARAP to facilitate ER degradation [46]. ER network integrity and turnover are tightly regulated by RETREG1, and the downregulation of RETREG1 protein in human cells may result in dilation of ER volume and accumulation of misfolded proteins [47]. In addition, mutated RETREG1 protein that cannot induce ER-phagy, could lead to the degeneration of sensory neurons and result in sensory neuropathy in humans. Intriguingly, excessive ER-phagy has been reported to impair ER homeostasis and trigger cell death in HeLa cells [48]. In the present study, we observed that the expression levels of ER-phagy related proteins were dramatically upregulated in human NP cells in response to GD treatment, and attenuated GD-induced senescence and apoptosis, providing evidence for the fundamental role played by ER-phagy in conferring cells resistance to adverse stimuli.
Selective ER sequestration into autophagosomes was first noticed during the investigation of UPR-induced ER remodeling and its importance was realized in overcoming folding stress and maintaining homeostasis control [ 49, 50]. ER-phagy is triggered or substantially enhanced after ER stress is elicited in response to treatment with drugs or starvation stress [ 51, 52]. ER-phagy impairment is related to augmented ER stress, resulting in cell death in certain cell types [18]. ATF4, an ER stress-related transcription factor, was recently established as a novel mechanistic link between ER stress and ER-phagy in glioblastoma cells [53]. Strikingly, excessive ER-phagy by RETREG1 gain-of-function mutation or drug administration could facilitate inappropriate ER degradation, aggravate ER stress, and ultimately promote cell death [ 48, 54]. The relationship between ER stress and ER-phagy seems complicated, and may be associated with cell type and the extent of stimulation. In the present study, the suppression of GD-induced ER stress with 4-PBA administration partially reduced the extent of RETREG1-mediated ER-phagy, whereas the reinforcement of RETREG1-mediated ER-phagy could counteract GD treatment-induced ER stress. This result strongly suggests the existence of a close link between RETREG1-mediated ER-phagy and ER homeostasis in human NP cells.
However, there are some limitations in our research. Firstly, although our study strongly points toward a close connection between ER stress and RETREG1-mediated ER-phagy, the direct molecular mechanisms responsible for communication between the two processes remain unclear. Secondly, apart from RETREG1, we did not investigate whether other ER-phagy receptors are involved in the activation of ER stress-mediated ER-phagy. Lastly, enhanced RETREG1-mediated ER phagocytosis as a potential therapeutic strategy needs to be verified in vivo in future studies.
In conclusion, this study demonstrated that ER stress is an upstream event of RETREG1-mediated ER-phagy activation in response to GD, and the upregulation of RETREG1-mediated ER-phagy could relieve GD-induced ER stress and subsequent cell injury in human NP cells. These findings advance our understanding of the pathophysiology of IDD and provide insights into the development of novel therapeutic strategies.
Supporting information
Supplementary Data
Supplementary Data is available at Acta Biochimica et Biphysica Sinica online.
COMPETING INTERESTS
The authors declared that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Natural Science Foundation of China (Nos. 81902261, 81772401, and 82002366), the Application Foundation and Advanced Program of Wuhan Science and Technology Bureau (No. 2019020701011457), and the Fundamental Research Funds for the Central Universities (No. 2019kfyXMBZ063).
References
- 1.Cieza A, Causey K, Kamenov K, Hanson SW, Chatterji S, Vos T. Global estimates of the need for rehabilitation based on the Global Burden of Disease study 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. . 2020;396:2006–2017. doi: 10.1016/S0140-6736(20)32340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knezevic NN, Candido KD, Vlaeyen JWS, Van Zundert J, Cohen SP. Low back pain. Lancet. . 2021;398:78–92. doi: 10.1016/S0140-6736(21)00733-9. [DOI] [PubMed] [Google Scholar]
- 3.Vergroesen PPA, Kingma I, Emanuel KS, Hoogendoorn RJW, Welting TJ, van Royen BJ, van Dieën JH, et al. Mechanics and biology in intervertebral disc degeneration: a vicious circle. Osteoarthritis Cartilage. . 2015;23:1057–1070. doi: 10.1016/j.joca.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 4.Kadow T, Sowa G, Vo N, Kang JD. Molecular basis of intervertebral disc degeneration and herniations: what are the important translational questions? Clin Orthopaedics Relat Res. . 2015;473:1903–1912. doi: 10.1007/s11999-014-3774-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang YC, Urban JPG, Luk KD. Intervertebral disc regeneration: do nutrients lead the way? Nat Rev Rheumatol. . 2014;10:561–566. doi: 10.1038/nrrheum.2014.91. [DOI] [PubMed] [Google Scholar]
- 6.Colombier P, Clouet J, Hamel O, Lescaudron L, Guicheux J. The lumbar intervertebral disc: from embryonic development to degeneration. Joint Bone Spine. . 2014;81:125–129. doi: 10.1016/j.jbspin.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Liao Z, Li S, Lu S, Liu H, Li G, Ma L, Luo R, et al. Metformin facilitates mesenchymal stem cell-derived extracellular nanovesicles release and optimizes therapeutic efficacy in intervertebral disc degeneration. Biomaterials. . 2021;274:120850. doi: 10.1016/j.biomaterials.2021.120850. [DOI] [PubMed] [Google Scholar]
- 8.Korennykh A, Walter P. Structural basis of the unfolded protein response. Annu Rev Cell Dev Biol. . 2012;28:251–277. doi: 10.1146/annurev-cellbio-101011-155826. [DOI] [PubMed] [Google Scholar]
- 9.Yu M, Lun J, Zhang H, Wang L, Zhang G, Zhang H, Fang J. Targeting UPR branches, a potential strategy for enhancing efficacy of cancer chemotherapy. Acta Biochim Biophys Sin. . 2021;53:1417–1427. doi: 10.1093/abbs/gmab131. [DOI] [PubMed] [Google Scholar]
- 10.Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta (BBA) - Mol Cell Res. . 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iurlaro R, Muñoz-Pinedo C. Cell death induced by endoplasmic reticulum stress. FEBS J. . 2016;283:2640–2652. doi: 10.1111/febs.13598. [DOI] [PubMed] [Google Scholar]
- 12.Grootjans J, Kaser A, Kaufman RJ, Blumberg RS. The unfolded protein response in immunity and inflammation. Nat Rev Immunol. . 2016;16:469–484. doi: 10.1038/nri.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oakes SA, Papa FR. The role of endoplasmic reticulum stress in human pathology. Annu Rev Pathol Mech Dis. . 2015;10:173–194. doi: 10.1146/annurev-pathol-012513-104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao Z, Luo R, Li G, Song Y, Zhan S, Zhao K, Hua W, et al. Exosomes from mesenchymal stem cells modulate endoplasmic reticulum stress to protect against nucleus pulposus cell death and ameliorate intervertebral disc degeneration in vivo . Theranostics. . 2019;9:4084–4100. doi: 10.7150/thno.33638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo R, Song Y, Liao Z, Yin H, Zhan S, Wang K, Li S, et al. Impaired calcium homeostasis via advanced glycation end products promotes apoptosis through endoplasmic reticulum stress in human nucleus pulposus cells and exacerbates intervertebral disc degeneration in rats. FEBS J. . 2019;286:4356–4373. doi: 10.1111/febs.14972. [DOI] [PubMed] [Google Scholar]
- 16.Luo R, Liao Z, Song Y, Yin H, Zhan S, Li G, Ma L, et al. Berberine ameliorates oxidative stress-induced apoptosis by modulating ER stress and autophagy in human nucleus pulposus cells. Life Sci. . 2019;228:85–97. doi: 10.1016/j.lfs.2019.04.064. [DOI] [PubMed] [Google Scholar]
- 17.Molinari M. ER-phagy: eating the factory. Mol Cell. . 2020;78:811–813. doi: 10.1016/j.molcel.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson S. Emerging principles of selective ER autophagy. J Mol Biol. . 2020;432:185–205. doi: 10.1016/j.jmb.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grumati P, Dikic I, Stolz A. ER-phagy at a glance. J Cell Sci. . 2018;131:jcs217364. doi: 10.1242/jcs.217364. [DOI] [PubMed] [Google Scholar]
- 20.Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. . 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Wang S, Wang Z, Ding M, Li X, Guo J, Han G, et al. Dexmedetomidine alleviated endoplasmic reticulum stress via inducing ER-phagy in the spinal cord of neuropathic pain model. Front Neurosci. . 2020;14:90. doi: 10.3389/fnins.2020.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng Y, Li B, Zhang W, Jiang L. ER-phagy and ER stress response (ERSR) in plants. Front Plant Sci. . 2019;10:1192. doi: 10.3389/fpls.2019.01192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song S, Tan J, Miao Y, Zhang Q. Crosstalk of ER stress‐mediated autophagy and ER‐phagy: Involvement of UPR and the core autophagy machinery. J Cell Physiol. . 2018;233:3867–3874. doi: 10.1002/jcp.26137. [DOI] [PubMed] [Google Scholar]
- 24.Mo J, Chen J, Zhang B. Critical roles of FAM134B in ER-phagy and diseases. Cell Death Dis. . 2020;11:983. doi: 10.1038/s41419-020-03195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hübner CA, Dikic I. ER-phagy and human diseases. Cell Death Differ. . 2020;27:833–842. doi: 10.1038/s41418-019-0444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y, Li S, Geng W, Luo R, Liu W, Tu J, Wang K, et al. Sirtuin 3-dependent mitochondrial redox homeostasis protects against AGEs-induced intervertebral disc degeneration. Redox Biol. . 2018;19:339–353. doi: 10.1016/j.redox.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang L, Hu J, Weng Y, Jia J, Zhang Y. Sirtuin 6 prevents matrix degradation through inhibition of the NF-κB pathway in intervertebral disc degeneration. Exp Cell Res. . 2017;352:322–332. doi: 10.1016/j.yexcr.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 28.Lu S, Song Y, Luo R, Li S, Li G, Wang K, Liao Z, et al. Ferroportin-dependent Iron homeostasis protects against oxidative stress-induced nucleus pulposus cell ferroptosis and ameliorates intervertebral disc degeneration in vivo . Oxid Med Cell Longev. . 2021;2021:1–18. doi: 10.1155/2021/6670497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freemont AJ. The cellular pathobiology of the degenerate intervertebral disc and discogenic back pain. Rheumatology. . 2008;48:5–10. doi: 10.1093/rheumatology/ken396. [DOI] [PubMed] [Google Scholar]
- 30.Fields AJ, Ballatori A, Liebenberg EC, Lotz JC. Contribution of the endplates to disc degeneration. Curr Mol Bio Rep. . 2018;4:151–160. doi: 10.1007/s40610-018-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLucca JF, Cortes DH, Jacobs NT, Vresilovic EJ, Duncan RL, Elliott DM. Human cartilage endplate permeability varies with degeneration and intervertebral disc site. J Biomech. . 2016;49:550–557. doi: 10.1016/j.jbiomech.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz Wills C, Foata B, González Ballester MÁ, Karppinen J, Noailly J. Theoretical explorations generate new hypotheses about the role of the cartilage endplate in early intervertebral disk degeneration. Front Physiol. . 2018;9:1210. doi: 10.3389/fphys.2018.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Sun C, Sun J, Yang MH, Zuo R, Liu C, Lan WR, et al. Autophagy mediates serum starvation-induced quiescence in nucleus pulposus stem cells by the regulation of P27. Stem Cell Res Ther. . 2019;10:118. doi: 10.1186/s13287-019-1219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Yang Y, Zuo R, Wu J, Zhang C, Li C, Liu M, et al. FOXO3 protects nucleus pulposus cells against apoptosis under nutrient deficiency via autophagy. Biochem Biophys Res Commun. . 2020;524:756–763. doi: 10.1016/j.bbrc.2020.01.168. [DOI] [PubMed] [Google Scholar]
- 35.Uehara Y, Hirose J, Yamabe S, Okamoto N, Okada T, Oyadomari S, Mizuta H. Endoplasmic reticulum stress-induced apoptosis contributes to articular cartilage degeneration via C/EBP homologous protein. Osteoarthritis Cartilage. . 2014;22:1007–1017. doi: 10.1016/j.joca.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 36.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. . 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 37.Scheper W, Hoozemans JJM. The unfolded protein response in neurodegenerative diseases: a neuropathological perspective. Acta Neuropathol. . 2015;130:315–331. doi: 10.1007/s00401-015-1462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen T, Xue P, Ying J, Cheng S, Liu Y, Ruan D. The role of unfolded protein response in human intervertebral disc degeneration: perk and IRE1-α as two potential therapeutic targets. Oxid Med Cell Longev. . 2021;2021:1–9. doi: 10.1155/2021/6492879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang B, Ke W, Wang K, Li G, Ma L, Lu S, Xiang Q, et al. Mechanosensitive ion channel Piezo1 activated by matrix stiffness regulates oxidative stress-induced senescence and apoptosis in human intervertebral disc degeneration. Oxid Med Cell Longev. . 2021;2021:1–13. doi: 10.1155/2021/8884922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin H, Peng Y, Li J, Wang Z, Chen S, Qing X, Pu F, et al. Reactive oxygen species regulate endoplasmic reticulum stress and er-mitochondrial Ca2+ crosstalk to promote programmed necrosis of rat nucleus pulposus cells under compression. Oxid Med Cell Longev. . 2021;2021:1–20. doi: 10.1155/2021/8810698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W, Qing X, Wang B, Ma K, Wei Y, Shao Z. Tauroursodeoxycholic acid protects nucleus pulposus cells from compression-induced apoptosis and necroptosis via inhibiting endoplasmic reticulum Stress. Evid-Based Complement Alternat Med. . 2018;2018:1–11. doi: 10.1155/2018/6719460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anding AL, Baehrecke EH. Cleaning house: selective autophagy of organelles. Dev Cell. . 2017;41:10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loi M, Fregno I, Guerra C, Molinari M. Eat it right: ER-phagy and recovER-phagy. Biochem Soc Trans. . 2018;46:699–706. doi: 10.1042/BST20170354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cebollero E, Reggiori F, Kraft C. Reticulophagy and ribophagy: regulated degradation of protein production factories. Int J Cell Biol. . 2012;2012:1–9. doi: 10.1155/2012/182834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakatogawa H, Mochida K. Reticulophagy and nucleophagy: New findings and unsolved issues. Autophagy. . 2015;11:2377–2378. doi: 10.1080/15548627.2015.1106665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. . 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 47.Chino H, Mizushima N. ER-Phagy: Quality control and turnover of endoplasmic reticulum. Trends Cell Biol. . 2020;30:384–398. doi: 10.1016/j.tcb.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Liao Y, Duan B, Zhang Y, Zhang X, Xia B. Excessive ER-phagy mediated by the autophagy receptor FAM134B results in ER stress, the unfolded protein response, and cell death in HeLa cells. J Biol Chem. . 2019;294:20009–20023. doi: 10.1074/jbc.RA119.008709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. . 2014;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- 50.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. . 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deegan S, Saveljeva S, Gorman AM, Samali A. Stress-induced self-cannibalism: on the regulation of autophagy by endoplasmic reticulum stress. Cell Mol Life Sci. . 2013;70:2425–2441. doi: 10.1007/s00018-012-1173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lipatova Z, Segev N. A role for macro-ER-phagy in ER quality control. PLoS Genet. . 2015;11:e1005390. doi: 10.1371/journal.pgen.1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zielke S, Kardo S, Zein L, Mari M, Covarrubias-Pinto A, Kinzler MN, Meyer N, et al. ATF4 links ER stress with reticulophagy in glioblastoma cells. Autophagy. . 2021;17:2432–2448. doi: 10.1080/15548627.2020.1827780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Leonibus C, Cinque L, Settembre C. Beating the ER: novel insights into FAM134B function and regulation. EMBO J. . 2020;39:e104546. doi: 10.15252/embj.2020104546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.