Abstract
Evolution of Bateson‐Dobzhansky‐Muller (BDM) incompatibilities is thought to represent a key step in the formation of separate species. They are incompatible alleles that have evolved in separate populations and are exposed in hybrid offspring as hybrid sterility or lethality. In this study, we reveal a previously unconsidered mechanism promoting the formation of BDM incompatibilities, meiotic drive. Theoretical studies have evaluated the role that meiotic drive, the phenomenon whereby selfish elements bias their transmission to progeny at ratios above 50:50, plays in speciation, and have mostly concluded that drive could not result in speciation on its own. Using the model fungus Neurospora, we demonstrate that the large meiotic drive haplotypes, Sk‐2 and Sk‐3, contain putative sexual incompatibilities. Our experiments revealed that although crosses between Neurospora intermedia and Neurospora metzenbergii produce viable progeny at appreciable rates, when strains of N. intermedia carry Sk‐2 or Sk‐3 the proportion of viable progeny drops substantially. Additionally, it appears that Sk‐2 and Sk‐3 have accumulated different incompatibility phenotypes, consistent with their independent evolutionary history. This research illustrates how meiotic drive can contribute to reproductive isolation between populations, and thereby speciation.
Keywords: Bateson‐Dobzhansky‐Muller incompatibility, meiotic drive, Neurospora, sexual incompatibility, speciation
Throughout the course of the last century, a significant amount of progress has been made in understanding how evolution creates diversity in the natural world, and what factors drive speciation. It is understood that the phenomena controlling phenotypic divergence must coincide with changes at the genotypic level, and in terms of speciation, this divergence is often thought to be precipitated by so called Bateson‐Dobzhansky‐Muller (BDM) incompatibilities. In brief, the BDM model proposes that when two populations become isolated, changes at the genic level occur in such a way as for each population to evolve coadapted alleles. When these two populations meet, the gene products from the different populations are no longer able to perform their function and attempts to mate result in inviable or sterile hybrid offspring. Despite the strong theoretical framework around the BDM model, empirical data on gene interactions that cause reproductive isolation between populations remain relatively rare (Maheshwari and Barbash 2011). As a result, the exact processes that lead to the formation of BMD incompatibilities are also poorly understood.
Meiotic drive is the phenomenon where a selfish genetic element is able to manipulate the sexual cycle of an organism such that it becomes overrepresented in the offspring. Due to the mechanisms through which drivers cheat meiosis, there is often an associated penalty to fertility through the death of meiotic products, such as sperm or spores (Lindholm et al. 2016). For many of the described systems of meiotic drive, multiple genes are required for the drive to function, and these are kept in linkage through genomic inversions to suppress recombination (Presgraves 2009), which effectively isolates the driving haplotype from the nondriving version and can lead to the accumulation of linked deleterious mutations (Dyer et al. 2007). These regions of suppressed recombination are often large and encompass many genes, both related and unrelated to drive, which sets the stage for BDM incompatibilities to form. In agreement with this expectation, many cases have been described where individuals carrying meiotic drive elements show reproductive incompatibilities with nondriving individuals (Wilkinson and Fry 2001; Phadnis and Orr 2009; Zhang et al. 2015). Such incompatibilities may ultimately result in the inability of a driver to sweep to fixation in a population, as for driving alleles to reap the benefits of cheating meiosis, they must reproduce with naïve individuals. Alternatively, the reproductive isolation caused by the drive could effectively split the population and result in speciation. This latter outcome has been under considerable debate, particularly for sex‐biased drives, with opinions regarding the ability and importance of drivers to cause speciation varying wildly (Frank 1991; Coyne and Orr 1993; Zhang et al. 2015; Sweigart et al. 2019). Thus, additional empirical data are required to address the role of meiotic drive in speciation.
The filamentous fungus Neurospora crassa and other closely related Neurospora species have been developed as a model system for comparisons of species recognition criteria and for the study of the evolution of reproductive isolation between species (Dettman et al. 2003a, 2003b, 2006; Menkis et al. 2009; Villalta et al. 2009; Turner et al. 2011). They also harbor meiotic drivers. The three meiotic drivers known in Neurospora are Sk‐1, Sk‐2, and Sk‐3 and these function by means of spore killing, whereby spores that carry the driving element (i.e., the killer gene) will kill their sibling spores that do not (Vogan et al. 2022). Spore killing has a very obvious phenotype in Neurospora. Following meiosis, one round of mitosis occurs to produce eight sexual spores (ascospores). These spores are packaged together in an ascus to form a single row. When spore killing occurs, four viable, black ascospores are observed together with four small aborted white ascospores in each ascus (Turner and Perkins 1979). Sk‐2 and Sk‐3 are multi‐gene drivers that were both discovered in the species Neurospora intermedia. In this species, the killing gene is linked to the resistance gene in a haplotype that extends across a 30‐cM region surrounding the centromere on chromosome 3, resulting in a region of linkage covering roughly 400 genes or 2.5 Mbp (Svedberg et al. 2018). The two spore killers are mutual killers, meaning they produce no viable spores when crossed to each other, which is in contrast to crosses between two Sk‐2 strains (or two Sk‐3 strains), which result in eight viable spores, that is, no spore killing. Sk‐2 and Sk‐3 use different alleles of the same resistance gene and occupy a similar region of chromosome 3 (Hammond et al. 2012), but do not share the same killing gene and appear to have had a long history of evolutionary separation (Svedberg et al. 2018; Rhoades et al. 2019).
Turner (2001) surveyed available Neurospora strains for the presence of spore killing, and during this work discovered that some sensitive strains of N. intermedia were sterile in crosses with Sk‐2 and Sk‐3 spore killer strains. A subset of these strains was later reclassified as N. metzenbergii (Dettman et al. 2003a; Villalta et al. 2009), suggesting the possibility that BDM incompatibilities may have accumulated in the Sk‐2 and/or Sk‐3 spore killing haplotype of N. intermedia. To investigate this hypothesis, we first used molecular markers (Dettman et al. 2003a) to determine the phylogenetic placement of the N. intermedia strains that showed sterility in crosses to Sk‐2 and Sk‐3 and determined that these strains do in fact belong to the sister species N. metzenbergii (Dettman et al. 2003a). Subsequently, by analyzing reproductive success in laboratory crosses, we verified that N. metzenbergii strains display a higher degree of reproductive isolation with N. intermedia strains carrying Sk‐2 or Sk‐3 than with those that do not. Additionally, we found that the mechanism of reproductive isolation appears to differ between Sk‐2 and Sk‐3, suggesting that the separate drivers have captured unique incompatibility factors during their independent evolutionary separation. We conclude that the meiotic drive haplotypes carry genes conferring strong reproductive isolation between strains of Neurospora, either by pleiotropic effects of the killer genes themselves or by the action of other linked genes, and discuss the role that meiotic drive can play in speciation.
Methods
STRAINS
Neurospora strains used in this study were acquired from the Fungal Genetics Stock Center (FGSC; fgsc.net). All strains annotated as N. metzenbergii (n = 17) in the FGSC collection were included in the analysis, as well as all strains annotated as N. intermedia that were collected from New Zealand (n = 11) or Mexico (n = 1). Additional strains of Neurospora (n = 54) were chosen to represent the diversity of the species based on Dettman et al. (2003a). We also included in our study strains of N. intermedia carrying either Sk‐2 or Sk‐3. Note that for strain 7427 and 7428 (which contain the Sk‐2 allele) and strains 3193 and 3194 (containing the Sk‐3 allele), the original strains that carried the spore killer haplotype are not available from the FGSC. Instead, these Sk‐2 strains are F1 progeny of the wild spore killer strain and either of the N. intermedia tester strains 1766 or 1767, and the Sk‐3 strains are the result of three backcrosses from the original wild spore killer strains to the 1767 strain. Strain 7426 represents a wild strain that carries Sk‐2, which was isolated at a later date. All strains used for mating assays are referred to by their FGSC ID number and are listed in Table S1. Strains 8761 and 8762 are single conidial isolates derived from 1766 and 1767, respectively. Locations of strains from the Perkin's collection were taken from the FGSC (www.FGSC.net) and converted to global coordinates using the R package ggmap (Kahle and Wickham 2013).
MOLECULAR MARKERS AND PHYLOGENETIC ANALYSES
As molecular markers, we used four unlinked anonymous nuclear loci (TMI, TML, DMG, and QMA) that were identified in Dettman et al. (2003a) as suitable for species delimitation in Neurospora. We PCR‐amplified and Sanger sequenced the markers according to the protocols given in Dettman et al. (2003a), to assign the investigated strains to species‐level groupings. These markers contain microsatellite sequences, which were removed prior to alignment, and hence only the flanking, nonrepetitive, sequences were aligned and analyzed, as in Dettman et al. (2003a). When comparing our sequencing results of a number of reference strains to those sequences generated by Dettman et al. (2003a) and subsequently used by Villalta et al. (2009), we discovered a large number of cytosines in the Dettman et al. (2003a) sequences that are absent from ours, as well as from sequences from Corcoran et al. (2014) and Svedberg et al. (2018). Due to these discrepancies, we only used marker sequences that we generated ourselves in this study, or that were generated by Corcoran et al. (2014) (Table S2). Sequences with the microsatellite portions removed were concatenated and aligned with the CLC sequence viewer version 7.7 (https://digitalinsights.qiagen.com/), followed by manual curation. The final alignment of 71 sequences had a length of 2093 columns, 915 distinct patterns, 372 parsimony‐informative, 228 singleton sites, and 1493 constant sites. We performed a Maximum Likelihood analysis using IQ‐TREE version 2.0 (Minh et al. 2020) with the default parameters and 100 standard nonparametric bootstraps. The TPM3+F+R2 model was selected based on the Bayesian information criterion. Sequences are deposited in GenBank with accession numbers MW034381–MW034436.
Additionally, we performed a phylogenomic analysis based on SNP data from 40 strains distributed over eight species of Neurospora (Table S2). Genomic data for these strains had been collected in previous studies (Corcoran et al. 2014; Svedberg et al. 2018) and SNPs were called against the N. intermedia strain 8807. For details on SNP calling, see Svedberg et al. (2018). Whole chromosome phylogenies based on a total of 4,680,447 variable sites were inferred using RAxML (Stamatakis 2014) for six of the seven chromosomes separately, using the following parameters: raxmlHPC‐HYBRID‐AVX ‐T 16 ‐m GTRCAT ‐x 45345 ‐p 22455 ‐# 100 ‐f a. Chromosome 3 was excluded to remove the conflicting phylogenetic signal of the nonrecombining spore killer region compared to the rest of the genome, even though that only affects the placement of strains within N. intermedia. The six phylogenetic trees (bipartition trees generated by RAxML) were then merged into a consensus network with 20% pruning, using Splitstree (Huson and Bryant 2006).
CROSSINGS
All crosses were conducted in 10 × 100 mm glass culture tubes containing 1 mL of liquid synthetic crossing media (Westergaard and Mitchell 1947) with no added sucrose. A small strip of filter paper was added to the tubes before autoclaving. Neurospora intermedia strains were used as females for all hybrid crosses: they were inoculated into the tubes and allowed to establish and produce protoperithecia (immature fruiting bodies) over 3 days at room temperature in complete darkness. Conidia (asexual spores that also function as fertilizing agents) from strains used as males in the crosses were then used to fertilize the protoperithecia and after fertilization, the cultures were incubated at 25°C in a 12 h light/dark cycle. After 2–3 weeks, mating had taken place for sexually compatible crosses, perithecia (mature fruiting bodies) were formed, and ascospores had been shot, as evidenced by empty perithecia. To generate offspring from crosses, ascospores were harvested from the walls of the culture tubes using a sterile loop and spread onto the surface of water agar plates. These plates were incubated in a water bath at 60°C for 1 h to induce germination as Neurospora ascospores require a heat treatment to germinate. This treatment kills any contaminating asexual spores (conidia) or mycelia, but will also kill nonpigmented ascospores. Germinated spores were picked from the water agar and placed into sterile glass culture tubes with Vogel's media (Vogel 1956) to establish cultures.
ESTIMATION OF REPRODUCTIVE SUCCESS
To measure the reproductive success of crosses between strains of Neurospora, we evaluated the production of perithecia and estimated the proportion of shot black spores to unpigmented white/hyaline spores, following Dettman et al. (2003b). In brief, crosses were allowed to mate until all ascospores had been shot. The sides of the culture tubes were examined under a dissecting microscope at 250× magnification and the proportion of black ascospores was estimated. The results of crosses were scored on a scale from 0 to 6 following Dettman et al. (2003b), where 0 refers to crosses that were entirely sterile (no fruiting bodies produced) and 6 refers to a fully compatible cross (fruiting bodies were produced and shot many black ascospores). The presence of an ostiole was not recorded as a measure of reproductive compatibility; hence, categories 1 and 2 used by Dettman et al. (2003b) are merged in this study. For selected combinations, we set up the crossings in triplicate to determine final values. To verify the accuracy of the method and obtain quantitative values, proportions of black spores were also estimated through perithecial dissections for a limited number of crosses. For this method, we investigated the crosses at an earlier developmental stage, prior to the ejection of ascospores. Mature perithecia were identified based on the presence of long extended ostioles and selected for dissection. Perithecia were dissected on glass slides in a drop of sterile water to obtain rosettes of asci. Only rosettes that contained at least some fully matured black spores were evaluated for the proportion of viable spores. Additionally, perithecia were harvested over a 1‐week period to verify that spore production was evaluated at the optimal time, that is, after most asci had matured, but before most of the ascospores had been shot.
Results
PHYLOGENETIC PLACEMENT OF REPRODUCTIVELY ISOLATED STRAINS
In total, 3038 wild strains of Neurospora have been collected and annotated as N. intermedia based on compatible matings when crossed with reference strains by Dr. David Perkins and others. The details of these crosses are publicly available from the FGSC (http://www.fgsc.net/Neurospora/PerkinsWildCollectionatFGSC.xlsx), and many of the strains are maintained there as well. Strains from New Zealand and Mexico were previously highlighted as mating particularly poorly to spore killers strains (Turner 2001), thus we obtained all available strains from these two locales that are annotated as N. intermedia or N. metzenbergii and determined their phylogenetic placement as per Dettman et al. (2003a). This analysis determined that all strains originally assigned to N. intermedia and showing poor reproduction with spore killers belong to the species N. metzenbergii (Fig. S1), a result that was confirmed with a phylogenomic approach (Fig. S2). This analysis also reveals an interesting biogeographic pattern where it appears that although N. intermedia has a global distribution, it never co‐occurs with N. metzenbergii (Fig. 1). This nonoverlapping distribution is most striking around Mexico, where all isolates were determined as N. metzenbergii, whereas N. intermedia were found immediately to the north, in Texas, and to the south, in Honduras. The one exception to the nonoverlapping distribution is strain 10399 of N. metzenbergii that was isolated from Haiti alongside N. intermedia strains (Fig. 1).
Figure 1.
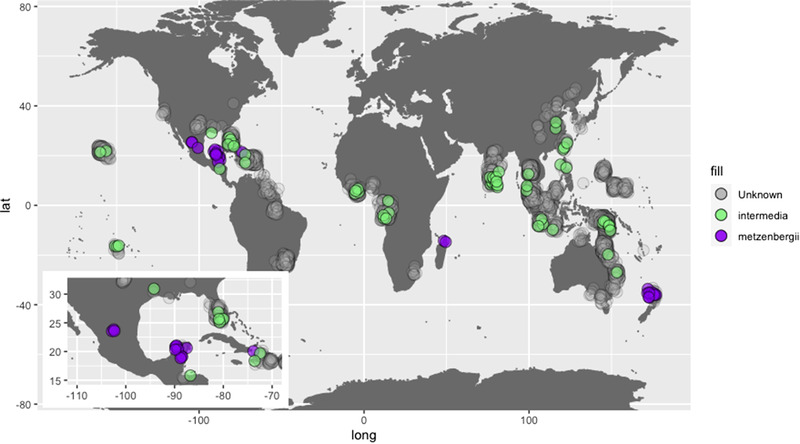
Global distribution of Neurospora intermedia and Neurospora metzenbergii. All strains from the Perkins collection at FGSC (http://www.fgsc.net) that were determined to be N. intermedia through crossing to reference strains are plotted. The geographic origins of those which were confirmed as N. intermedia through molecular evidence are shown in green, and those which were revealed to be N. metzenbergii are shown in purple. The “unknown” strains refer to isolates of N. intermedia with no molecular data. The inset is a magnified view of Mexico and surrounding regions. Note that strains with no precise locale data are visualized as midpoints in their country of origin, including three in Mexico.
THE SPORE KILLERS OF N. intermedia SHOW HIGHER INCOMPATIBILITY WITH N. metzenbergii
Previous work established that N. metzenbergii and N. intermedia are partially reproductively isolated, in that they produce a lower number of viable offspring when crossed to each other as opposed to crosses among strains of their own species (Dettman et al. 2003b; Villalta et al. 2009). We verified that the newly assigned NZ strains of N. metzenbergii show similar levels of reproductive isolation to N. intermedia by crossing representative strains of N. metzenbergii to standard tester strains of N. intermedia and among themselves. We see that, as expected, for both species the within‐species crosses resulted in a high proportion of viable ascospores (>90%; Table S3). Furthermore, most strains of N. metzenbergii were able to mate with the N. intermedia tester strains (1766 and 1767), but produced consistently low proportions of viable spores (60%–80%). Notable exceptions to this range were the N. metzenbergii strain 10399 from Haiti, which produced even fewer viable spores (30%–60%), and 7829 from New Zealand, which did not mate with any N. intermedia strains (Table S3). Previous studies have also shown that some populations of N. intermedia have evolved strong premating barriers with N. crassa (Turner et al. 2011). We investigated two strains from these populations here, 2316 and 6263, which revealed that these barriers may also prevent mating of these N. intermedia strains with strains of N. metzenbergii as successful matings were only achieved with one N. metzenbergii strain, 5120 (Table S3 and S4).
In contrast to crosses with tester strains, crosses of the N. metzenbergii strains to the strains of N. intermedia that carry either Sk‐2 or Sk‐3 showed a dramatic reduction in the proportion of shot black spores. A drop of 50% in the proportion of viable spores is expected due to the spore killing itself; however, many of the crosses resulted in nearly no black spores, or failure to produce perithecia at all, suggesting that there are strong incompatibilities within the spore killer region (Table S3). To confirm that spore killing was occurring in these interspecific crosses, perithecia were dissected to screen for the standard 4:4 white to black spore pattern that should be observed. For all crosses between the spore killer strains and N. metzenbergii strains, the asci mostly contained either small aborted white spores, misshapen black spores, or white spores. In general, few asci were observed that could be properly evaluated for spore killing, but some did appear to have the standard 4:4 pattern, indicating that spore killing is active in the interspecific crosses.
Additionally, we used perithecial dissections of a subset of strains to quantify the degree of reproductive incompatibility (Table S4). For all interspecific crosses, few asci were observed to have a full component of eight black ascospores. Nearly all asci contained at least one small aborted ascospore, and some asci appeared to be entirely aborted. Large proportions of partially pigmented ascospores or brown spores were also observed. Categorizing brown spores within asci is difficult, as these spores may have still developed into black spores prior to shooting. Additionally, some number of brown spores are viable and survive the heat treatment to germinate (Ho 1986). Due to these caveats, brown spores were considered viable in the data presented here. The perithecial dissections confirmed that some of the crosses of N. metzenbergii strains to Sk‐2 and Sk‐3 strains show a dramatic reduction in black spore production as compared to crosses with non‐spore killer strains. Strains 10395 and 7830 did not exhibit a decrease in the proportion of black spores that was greater than the 50% expected from spore killing alone when crossed to the lab strains of Sk‐2 and Sk‐3; however, the methods used here may ignore completely aborted asci and so only quantify within‐ascus incompatibility. Strains 5119 and 8881 both show a considerable reduction in the proportion of black spores produced, with 8881 being the most severe. Crosses to the wild Sk‐2 strain 7426 were similar to the lab‐derived strains for 10395, but strains from NZ showed a large decrease in black spore production, suggesting that there may be additional incompatibilities in the non‐spore killing region that have been lost in the lab‐derived strains (Fig. 2).
Figure 2.
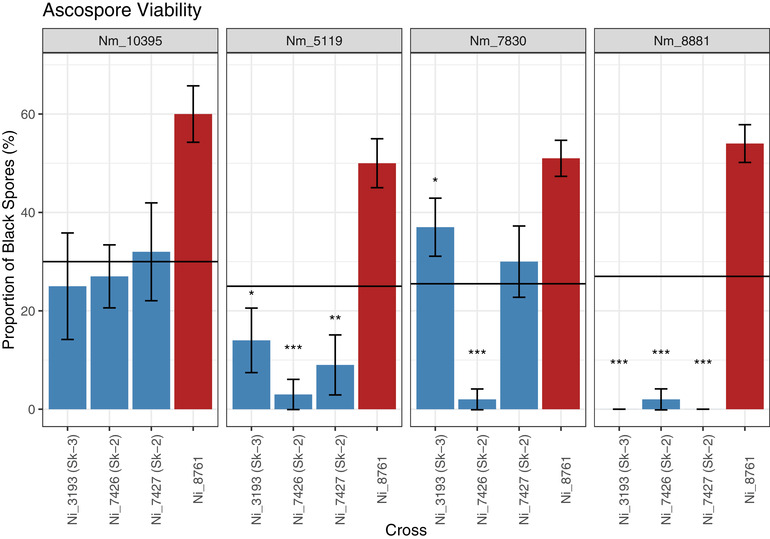
Proportion of black spores produced by crosses between four Neurospora metzenbergii strains (10395 [Mexico], 5119 [New Zealand], 7830 [New Zealand], and 8881 [Madagascar]) and four N. intermedia strains (3193 [Sk‐3], 7426 [Sk‐2], 7427 [Sk‐2], and 8761 [sensitive]). Horizontal lines represent half the value of the cross to 8761 to the given N. metzenbergii strain as an expectation for a decrease in germination due to spore killing alone. Asterisks represent significant deviations from this expectation according to a chi square test (* <0.05, ** <0.01, *** <0.001); whiskers denote one standard error.
DIFFERENT INCOMPATIBILITY PHENOTYPES OF Sk‐2 AND Sk‐3 WITH N. metzenbergii
A number of N. metzenbergii strains displayed different incompatibility phenotypes when crossed to Sk‐2 than when crossed to Sk‐3 despite the fact that the genomic background of the spore killer strains should be highly similar outside of the nonrecombining spore killer region. With the Madagascar strains, 8880 and 8881, and strain 8846 (Mexico), no spores are shot when crossed to the Sk‐2 strains, but many small aborted white spores are shot when crossed to the Sk‐3 strains, suggesting that there are different incompatibilities present within the different spore killer regions. To confirm that the spore killing region is specifically associated with the reproductive isolation observed between the Madagascar strains and the Sk‐2 and Sk‐3 strains, F1 progeny from crosses between 1767 and either 3193 (Sk‐3) or 7427 (Sk‐2) was backcrossed four times to 1766 to generate spore killer strains with a more isogenic background to 1766. Crosses to these 5× backcrossed strains showed identical phenotypes as to the parental spore killer strains, confirming the association of the phenomenon to the spore killer region. To verify these phenotypes, perithecial dissections were conducted. These confirmed that perithecia appear to be formed normally (ostioles are present) in crosses to both spore killers, but that in the Sk‐2 crosses, most asci abort at the spore‐forming stage (Fig. 3a), whereas in the Sk‐3 crosses, spores are formed, but nearly all are abortive (small and white) (Fig. 3b). With crosses to Sk‐3, occasionally viable spores can be found (Fig. 3b), and these can germinate after heat treatment, showing they are fully viable. Given this low rate of viable spore production, it is likely due to rare recombination events within the spore killer region, which is hypothesized to occur occasionally (Svedberg et al. 2018).
Figure 3.
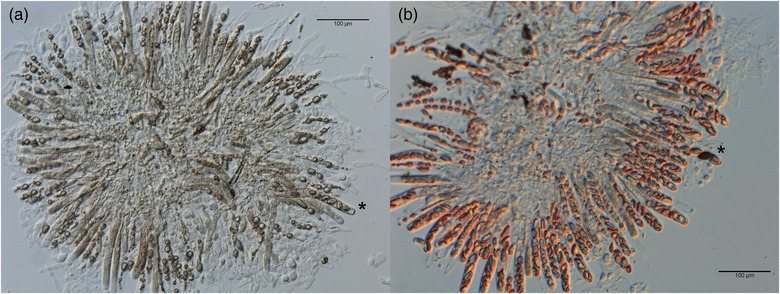
Rosettes of asci from crosses between Neurospora metzenbergii 8881 and (a) the Sk‐2 backcross strains or (b) the Sk‐3 backcross strain. For crosses to Sk‐2, nearly all asci are aborted without producing spores of any kind, but occasionally will produce an ascus with small aborted spores (*). For Sk‐3 crosses, asci contain only small aborted spores most of the time, but occasionally viable spores are found (*).
To investigate the incompatibilities further, hybrids were collected from crosses between the Madagascar strain 8880 and the N. intermedia strains 8761 and 8795, and crossed to the Sk‐2 and Sk‐3 strains. Most of the hybrids maintain the specific incompatibilities when crossed to Sk‐2 and Sk‐3 tester strains; however eight out of 36 recover some fertility. A few strains show differential recovery of fertility between Sk‐2 and Sk‐3, in that some black spores were observed in crosses to the Sk‐2 strain, but not the Sk‐3 strain, or vice versa, supporting the hypothesis that the different phenotypes observed between Sk‐2 and Sk‐3 crosses represent different underlying incompatibilities.
Discussion
Here, we provide evidence that meiotic drive in Neurospora contributes to reproductive isolation, both directly through killing half of all spores and through the accumulation of additional incompatibilities, a process that is often overlooked in discussions of drive systems and speciation (Sweigart et al. 2019). Although the low frequency of both drive and full incompatibility suggests that the precise incompatibilities at play in Sk‐2 and Sk‐3 are unlikely to have been major contributors in the divergence of N. metzenbergii and N. intermedia, it nevertheless provides an example of a scenario where meiotic drive could lead to speciation in other systems.
To understand what conditions may have led to the evolution of reproductive incompatibilities between N. intermedia and N. metzenbergii, it is important to determine what evolutionary forces were likely most dominant during their divergence. The delimitation between N. intermedia and N. metzenbergii aligns well with the recognition criteria for both the biological species concept and the phylogenetic species concept, in that molecular markers demonstrate their monophyly and mating assays show them to reproduce better with conspecifics than interspecifics (Dettman et al. 2003a, 2003b). Here, we have demonstrated that strains of Neurospora that were isolated from New Zealand, and previously designated as N. intermedia, in fact belong to N. metzenbergii, nearly doubling the number of known N. metzenbergii samples in collections and highlighting the unusual geographical distribution of the two species. This distribution is consistent with them competing to the end of competitive exclusion, but as strains were collected at different times, temporal factors cannot be eliminated. Together, these facts all point to the possibility that the N. intermedia and N. metzenbergii originally diverged allopatrically (species with overlapping ranges tend to compete less [Letcher et al. 1994]), thereby fulfilling the main requirement for the formation of BDM incompatibilities.
We have provided evidence that additional reproductive barriers exist within the spore killer region of Sk‐2 and Sk‐3 strains of N. intermedia that inhibit the ability of said strains to hybridize with N. metzenbergii. The reduced fertility is partially driven by the 50% loss of spores due to spore killing, but the strains carrying Sk‐2 and Sk‐3 also exhibit a strong reduction in fertility with some strains of N. metzenbergii that go beyond what can be explained by the activity of the spore killer elements alone. It could be expected that the reason crosses between the spore killers and N. metzenbergii strains show a decrease in viable progeny is the production of unbalanced gametes as a result of the mismatched inversions, but as a reduced fertility is not observed between crosses of spore killer and sensitive strains within N. intermedia, it is unlikely to be a significant issue. Additionally, the fact that the strongest incompatibilities with N. metzenbergii strains appear to be polymorphic further argues against a significant role of the inversions. The occurrence of polymorphic incompatibility factors between populations has been discussed as variable reproductive isolation (VRI), and has been observed in a variety of systems (Cutter 2012). Under a VRI scenario, these polymorphic alleles could originate either from ancestral, incomplete lineage sorting or as derived alleles within N. metzenbergii, but derived alleles are more likely to contribute to reproductive isolation (Orr 1995). Moreover, as only the killer haplotype is incompatible with these N. metzenbergii strains, the incompatible locus of N. intermedia must be located within the Sk region. Additionally, the differential phenotypes exhibited between Sk‐2 and Sk‐3 crosses suggest that discrete incompatible loci of large effect exist within the Sk‐2 and Sk‐3 regions. Conversely, as Sk‐2 and Sk‐3 do not appear to share the same killer gene (Rhoades et al. 2019) it could be possible that the spore killing genes themselves are responsible for the incompatibility through pleiotropic effects on reproduction, as has been observed with one meiotic driver from Drosophila (Phadnis and Orr 2009). Whether the incompatibility is caused by the killer gene or another gene confined within the haplotype needs further investigation.
The fact that incompatibilities are observed to accumulate in the nonrecombining killer haplotype potentially provides deeper insights into the evolution of BDM incompatibilities. Although the per base pair rate of divergent substitutions between N. intermedia and N. metzenbergii is higher in the Sk region than the rest of the genome, these still represent the minority of total divergent sites across the genome (10,345/191,911 substitutions) and the spore killer strains, in fact, have slightly fewer nonsynonymous substitutions in total than sensitive strains (59,077 vs. 63,279) (Svedberg et al. 2018). It is known that selection plays an important role in the accumulation of BDM incompatibilities and although a lot of attention has been paid to directional selection and stabilizing selection (Welch 2004; Fierst and Hansen 2010; Nosil and Schluter 2011), less has been given to decreased purifying selection. Additionally, a considerable amount of theory and modeling have discussed the role of deleterious recessive mutations linked to drivers or regions of reduced recombination in speciation (Navarro and Barton 2003; Welch 2004; Kirkpatrick and Barton 2006). We can be confident that deleterious recessive mutations are not a factor in Sk‐2 or Sk‐3 as crosses among killer strains of the same type show no decrease in the production of viable offspring. Nonetheless, the spore killer regions may be accumulating nonfunctional mutations due to the relaxed selection in the Sk haplotype. Importantly, only nonfunctional mutations that can be compensated for by the genomic background of N. intermedia will persist, as others will be highly deleterious or lethal. However, in the hybrid crosses to N. metzenbergii, alleles allowing for such compensation may not be present due to recombination and/or independent assortment of chromosomes. So, although phylogenomic analyses suggest that most sites in the Sk haplotype are diverging under neutral expectation, the region may be a hotspot for the formation of BDM incompatibilities (Svedberg et al. 2018). These ideas should hold true for any genomic region experiencing suppressed recombination, such as inversions, and, for this reason, have broader implications beyond meiotic drive.
Under the alternative scenario where the killer genes themselves are responsible for the sexual incompatibilities, a different model may explain the observations. Instead of BDMs being more likely to form in the nonrecombining haplotype, it is possible that spore killer genes are involved in pathways related to sexual reproduction, and that genes in these pathways are more prone to become BDM incompatibilities. This appears to be the case with Overdrive in Drosophila pseudoobscura, where Overdrive is a meiotic drive gene, but also has many downstream trans targets that it interacts with. It has accumulated more nonsynonymous mutations than any of its targets, suggesting it may be diverging faster than the background rate (Phadnis and Orr 2009). The molecular mechanisms through which Sk‐2 and Sk‐3 operate are largely unknown, although in the case of Sk‐2 it does not appear to be directly acting through a disrupted sexual cycle, as transformants can be constructed that kill during vegetative growth (Rhoades and Hammond 2021). Whatever the cause of the observed sterility in crosses to N. metzenbergii, it must be specific to the N. metzenbergii background, as both Sk‐2 and Sk‐3 do not show similar patterns when crossed to the more distantly related N. crassa (Turner and Perkins 1979).
Hybrid incompatibilities have been linked to meiotic drivers in numerous systems (Braidotti and Barlow 1997; Tao et al. 2001; Wilkinson et al. 2003; McDermott and Noor 2010), but the discussion of how they could contribute to speciation has largely centered around the co‐evolution of complex suppressor systems, and the divergence of those systems in disparate populations (Courret et al. 2019). This discussion may be largely irrelevant here as no suppressors are known to be involved in Sk‐2 or Sk‐3, nor is there any evidence of other meiotic drivers within N. metzenbergii. With spore killing (and mechanistically similar meiotic drivers), if mutual killers, such as Sk‐2 and Sk‐3, fix in different populations, then crosses between individuals of those populations will be entirely incompatible. Such a scenario appears to have arisen in the fission yeast, Schizosaccharomyces pombe, where the diverse group of wtf genes show unique distributions in individual lineages (Bravo Núñez et al. 2020). Mutual killing may be a common phenomenon as it is observed in three well‐described systems: S. pombe, N. intermedia, and for the Spok genes of Podospora anserina (Vogan et al. 2019). However, such a mechanism will only remain in place as long as the drive is active, if the driver reaches fixation in a given population, it may be prone to decay (as observed in both S. pombe and P. anserina) and the reproductive barrier that it caused would disappear. Therefore, cases where the driving mechanism and/or co‐evolved suppressor genes implicitly cause reproductive isolation may not be expected to persist through time to impact speciation. However, if BDM incompatibilities have time to form during this phase or, as discussed above, are even more prone to formation in these regions, then drive could still contribute significantly to the reproductive isolation between populations, even if said drive then subsequently vanishes from the populations.
It should be noted that both the Spok and wtf genes are single gene drivers, which should be much less prone to the accumulation of linked incompatibilities illustrated here. Additionally, these small loci could be prone to cross species boundaries, as is similarly observed in Drosophila (Meiklejohn et al. 2018). In fact, the third known spore killer in Neurospora, Sk‐1 from N. sitophila, is also a single gene driver and has also been observed to jump species boundaries (Svedberg et al. 2021). In a scenario where reproductive isolation is caused by mutual killers, if an individual were able to acquire both drivers, it would be resistant to both drives and therefore viable with both species/populations. The selective advantage for such an individual could be quite strong, arguing against single gene drives as speciation genes. This implies that meiotic drivers that constitute large haplotypes may have greater impacts on population structure than single gene drives, and hence be observed more often during interspecies/population crossings. Therefore, although until recently, meiotic drive systems were thought to require multiple linked loci, it may be the case that single gene drivers are much more common but are observed less due to minimal linked phenotypes.
AUTHOR CONTRIBUTIONS
AV designed and conducted experiments, ran analyses, made figures, and wrote the first draft. JS performed project inception, ran analyses, and edited drafts. MGS conducted experiments and edited drafts. HJ performed project inception, edited drafts, and provided funding.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA ARCHIVING
Sequences generated in this study are deposited in GenBank with accession numbers MW034381–MW034436.
Associate Editor: T. Bataillon
Handling Editor: P. T. Chapman
Supporting information
Supplementary Figure 1. Maximum likelihood tree of 69 Neurospora strains, based on four genetic markers, TMI, TML, DMG, and QMA. N. discreta was set as the outgroup and the branch delineating the outgroup was shortened for illustration purposes. The spore killer strains are marked in red and blue for Sk‐2 and Sk‐3, respectively. Sequences that were generated in this study are marked with an asterisk (*), all of which were formerly annotated as N. intermedia. The Nx prefixes prior to the FGSC numbers denote the species of the given strain.
Supplementary Figure 2. Network of the terminal Neurospora clade based on maximum likelihood trees inferred from whole chromosome SNP data from six of the seven chromosomes; Chromosome 3 carries the Sk locus and so was excluded from the analysis. Colored ovals denote species of Neurospora. Note that strain 4723 falls outside all currently delimited species and may represent an undescribed species. Strains carrying Sk‐2 are marked with red text and, the one Sk‐3 strain is marked in blue. Network pruning level was set to 20%.
Supplementary Table 1. Strains used in this study
Supplementary Table 2. Strains and data used for phylogenetic analyses
Supplementary Table 3. Proportion of shot black ascospores from laboratory crosses.
Supplementary Table 4. Proportion of black ascospores of laboratory crosses from dissected perithecia.
ACKNOWLEDGMENTS
This work was supported by the European Research Council (ERC) grant ERC‐2014‐CoG (project 648143, SpoKiGen) and The Swedish Research Council (to HJ). We thank the support given by the National Genomics Infrastructure (NGI)/Uppsala Genome center on massive parallel DNA sequencing. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under the project SNIC 2017/1‐567. We would also like to thank D. Jacobson for the invaluable advice and data on the Perkin's collection.
REFERENCES
- Braidotti, G. & Barlow, D.P. (1997) Identification of a male meiosis‐specific gene, Tcte2, which is differentially spliced in species that form sterile hybrids with laboratory mice and deleted in t chromosomes showing meiotic drive. Developmental Biology, 186, 85–99. [DOI] [PubMed] [Google Scholar]
- Bravo Núñez, M.A. , Sabbarini, I.M. , Eickbush, M.T. , Liang, Y. , Lange, J.J. , Kent, A.M. & Zanders, S.E. (2020) Dramatically diverse Schizosaccharomyces pombe wtf meiotic drivers all display high gamete‐killing efficiency. PLOS Genetics, 16, e1008350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran, P. , Dettman, J.R. , Sun, Y. , Luque, E.M. , Corrochano, L.M. , Taylor, J.W. , Lascoux, M. & Johannesson, H. (2014) A global multilocus analysis of the model fungus Neurospora reveals a single recent origin of a novel genetic system. Molecular Phylogenetics and Evolution, 78, 136–147. [DOI] [PubMed] [Google Scholar]
- Courret, C. , Chang, C.‐H. , Wei, K.H.‐C. , Montchamp‐Moreau, C. & Larracuente, A.M. (2019) Meiotic drive mechanisms: lessons from Drosophila . Proceedings. Biological sciences / The Royal Society, 286, 20191430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J.A. & Orr, H.A. (1993) Further evidence against meiotic‐drive models of hybrid sterility. Evolution; Internation Journal of Organic Evolution, 47, 685–687. [DOI] [PubMed] [Google Scholar]
- Cutter, A.D. (2012) The polymorphic prelude to Bateson–Dobzhansky–Muller incompatibilities. Trends in Ecology & Evolution, 27, 209–218. [DOI] [PubMed] [Google Scholar]
- Dettman, J.R. , Jacobson, D.J. & Taylor, J.W. (2003a) A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution; Internation Journal of Organic Evolution, 57, 2703–2720. [DOI] [PubMed] [Google Scholar]
- Dettman, J.R. , Jacobson, D.J. , Turner, E. , Pringle, A. & Taylor, J.W. (2003b) Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution; Internation Journal of Organic Evolution, 57, 2721–2741. [DOI] [PubMed] [Google Scholar]
- Dettman, J.R. , Jacobson, D.J. & Taylor, J.W. (2006) Multilocus sequence data reveal extensive phylogenetic species diversity within the Neurospora discreta complex. Mycologia, 98, 436–446. [DOI] [PubMed] [Google Scholar]
- Dyer, K.A. , Charlesworth, B. & Jaenike, J. (2007) Chromosome‐wide linkage disequilibrium as a consequence of meiotic drive. PNAS, 104, 1587–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierst, J.L. & Hansen, T.F. (2010) Genetic architecture and postzygotic reproductive isolation: evolution of Bateson‐Dobzhansky‐Muller incompatibilities in a polygenic model. Evolution; Internation Journal of Organic Evolution, 64, 675–693. [DOI] [PubMed] [Google Scholar]
- Frank, S.A. (1991) Divergence of meiotic drive‐suppression systems as an explanation for sex‐ biased hybrid sterility and inviability. Evolution; Internation Journal of Organic Evolution, 45, 262–267. [DOI] [PubMed] [Google Scholar]
- Hammond, T.M. , Rehard, D.G. , Xiao, H. & Shiu, P.K.T. (2012) Molecular dissection of Neurospora Spore killer meiotic drive elements. PNAS, 109, 12093–12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, C.C. (1986) Identity and characteristics of Neurospora intermedia responsible for oncom fermentation in Indonesia. Food microbiology, 3, 115–132. [Google Scholar]
- Huson, D.H. & Bryant, D. (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution, 23, 254–267. [DOI] [PubMed] [Google Scholar]
- Kahle, D. & Wickham, H. (2013) ggmap: spatial visualization with ggplot2. The R journal, 5, 144‐161. [Google Scholar]
- Kirkpatrick, M. & Barton, N. (2006) Chromosome inversions, local adaptation and speciation. Genetics, 173, 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letcher, A.J. , Purvis, A. , Nee, S. & Harvey, P.H. (1994) Patterns of overlap in the geographic ranges of palearctic and British mammals. Journal of Animal Ecology, 63, 871–879. [Google Scholar]
- Lindholm, A.K. , Dyer, K.A. , Firman, R.C. , Fishman, L. , Forstmeier, W. , Holman, L. , Johannesson, H. , Knief, U. , Kokko, H. , Larracuente, A.M. , et al. (2016) The ecology and evolutionary dynamics of meiotic drive. Trends in Ecology & Evolution, 31, 315‐326. [DOI] [PubMed] [Google Scholar]
- Maheshwari, S. & Barbash, D.A. (2011) The genetics of hybrid incompatibilities. Annual Review of Genetics, 45, 331–355. [DOI] [PubMed] [Google Scholar]
- McDermott, S.R. & Noor, M.A.F. (2010) The role of meiotic drive in hybrid male sterility. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 365, 1265–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn, C.D. , Landeen, E.L. , Gordon, K.E. , Rzatkiewicz, T. , Kingan, S.B. , Geneva, A.J. , Vedanayagam, J.P. , Muirhead, C.A. , Garrigan, D. , Stern, D.L. , et al. (2018) Gene flow mediates the role of sex chromosome meiotic drive during complex speciation. eLife, 7, e35468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkis, A. , Bastiaans, E. , Jacobson, D.J. & Johannesson, H. (2009) Phylogenetic and biological species diversity within the Neurospora tetrasperma complex. Journal of Evolutionary Biology, 22, 1923–1936. [DOI] [PubMed] [Google Scholar]
- Minh, B.Q. , Schmidt, H.A. , Chernomor, O. , Schrempf, D. , Woodhams, M.D. , von Haeseler, A. & Lanfear, R. (2020) IQ‐TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular biology and evolution, 37, 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, A. & Barton, N.H. (2003) Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution; Internation Journal of Organic Evolution, 57, 447–459. [DOI] [PubMed] [Google Scholar]
- Nosil, P. & Schluter, D. (2011) The genes underlying the process of speciation. Trends in Ecology & Evolution, 26, 160–167. [DOI] [PubMed] [Google Scholar]
- Orr, H.A. (1995) The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics, 139, 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis, N. & Orr, H.A. (2009) A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science, 323, 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. (2009) Drive and sperm. Pp. 471–506 in Birkhead, T.R. , Hosken, D. & Pitnick, S. , eds. Sperm biology. Academic Press, Lond. [Google Scholar]
- Rhoades, N.A. & Hammond, T.M. (2021) RNA editing controls meiotic drive by a Neurospora Spore killer. bioRxi, 10.1101/2020.12.30.424869. [DOI] [Google Scholar]
- Rhoades, N.A. , Harvey, A.M. , Samarajeewa, D.A. , Svedberg, J. , Yusifov, A. , Abusharekh, A. , Manitchotpisit, P. , Brown, D.W. , Sharp, K.J. , Rehard, D.G. , et al. (2019) Identification of rfk‐1, a meiotic driver undergoing RNA editing in Neurospora . Genetics, 212, 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedberg, J. , Hosseini, S. , Chen, J. , Vogan, A.A. , Mozgova, I. , Hennig, L. , Manitchotpisit, P. , Abusharekh, A. , Hammond, T.M. , Lascoux, M. , et al. (2018) Convergent evolution of complex genomic rearrangements in two fungal meiotic drive elements. Nature Communication, 9, 4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedberg, J. , Vogan, A.A. , Rhoades, N.A. , Sarmarajeewa, D. , Jacobson, D.J. , Lascoux, M. , Hammond, T.M. & Johannesson, H. (2021) An introgressed gene causes meiotic drive in Neurospora sitophila . PNAS, 118, e2026605118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart, A.L. , Brandvain, Y. & Fishman, L. (2019) Making a murderer: the evolutionary framing of hybrid gamete‐killers. Trends in Genetics, 35, 245–252. [DOI] [PubMed] [Google Scholar]
- Tao, Y. , Hartl, D.L. & Laurie, C.C. (2001) Sex‐ratio segregation distortion associated with reproductive isolation in Drosophila . PNAS, 98, 13183–13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, B.C. (2001) Geographic distribution of neurospora spore killer strains and strains resistant to killing. Fungal Genetics and Biology, 32, 93–104. [DOI] [PubMed] [Google Scholar]
- Turner, B.C. & Perkins, D.D. (1979) Spore killer, a chromosomal factor in neurospora that kills meiotic products not containing it. Genetics, 93, 587–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, E. , Jacobson, D.J. & Taylor, J.W. (2011) Genetic architecture of a reinforced, postmating, reproductive isolation barrier between Neurospora species indicates evolution via natural selection. Plos Genetics, 7, e1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalta, C.F. , Jacobson, D.J. & Taylor, J.W. (2009) Three new phylogenetic and biological Neurospora species: N. hispaniola, N. metzenbergii and N. perkinsii . Mycologia, 101, 777–789. [DOI] [PubMed] [Google Scholar]
- Vogan, A.A. , Ament‐Velásquez, S.L. , Granger‐Farbos, A. , Svedberg, J. , Bastiaans, E. , Debets, A.J. , Coustou, V. , Yvanne, H. , Clavé, C. , Saupe, S.J. , et al. (2019) Combinations of Spok genes create multiple meiotic drivers in Podospora . eLife, 8, e46454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogan, A.A. , Martinossi‐Allibert, I. , Ament‐Velásquez, S.L. , Svedberg, J. & Johannesson, H. (2022) The spore killers, fungal meiotic driver elements. Mycologia, 114, 1–23. [DOI] [PubMed] [Google Scholar]
- Vogel, H.J. (1956) A convenient growth medium for Neurospora crassa . Microbial Genetics Bulletin, 13, 42–47. [Google Scholar]
- Welch, J.J. (2004) Accumulating Dobzhansky‐Muller incompatibilities: reconciling theory and data. Evolution; Internation Journal of Organic Evolution, 58, 1145–1156. [DOI] [PubMed] [Google Scholar]
- Westergaard, M. & Mitchell, H.K. (1947) Neurospora V. A synthetic medium favoring sexual reproduction. American Journal of Botany, 34, 573–577. [Google Scholar]
- Wilkinson, G.S. & Fry, C.L. (2001) Meiotic drive alters sperm competitive ability in stalk‐eyed flies. Proceedings. Biological sciences / The Royal Society, 268, 2559–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, G.S. , Swallow, J.G. , Christensen, S.J. & Madden, K. (2003) Phylogeography of sex ratio and multiple mating in stalk‐eyed flies from southeast Asia. Genetica, 117, 37–46. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Sun, T. , Woldesellassie, F. , Xiao, H. & Tao, Y. (2015) Sex ratio meiotic drive as a plausible evolutionary mechanism for hybrid male sterility. Plos Genetics, 11, e1005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Maximum likelihood tree of 69 Neurospora strains, based on four genetic markers, TMI, TML, DMG, and QMA. N. discreta was set as the outgroup and the branch delineating the outgroup was shortened for illustration purposes. The spore killer strains are marked in red and blue for Sk‐2 and Sk‐3, respectively. Sequences that were generated in this study are marked with an asterisk (*), all of which were formerly annotated as N. intermedia. The Nx prefixes prior to the FGSC numbers denote the species of the given strain.
Supplementary Figure 2. Network of the terminal Neurospora clade based on maximum likelihood trees inferred from whole chromosome SNP data from six of the seven chromosomes; Chromosome 3 carries the Sk locus and so was excluded from the analysis. Colored ovals denote species of Neurospora. Note that strain 4723 falls outside all currently delimited species and may represent an undescribed species. Strains carrying Sk‐2 are marked with red text and, the one Sk‐3 strain is marked in blue. Network pruning level was set to 20%.
Supplementary Table 1. Strains used in this study
Supplementary Table 2. Strains and data used for phylogenetic analyses
Supplementary Table 3. Proportion of shot black ascospores from laboratory crosses.
Supplementary Table 4. Proportion of black ascospores of laboratory crosses from dissected perithecia.
Data Availability Statement
Sequences generated in this study are deposited in GenBank with accession numbers MW034381–MW034436.


