Abstract
To elucidate antigens present on the bacterial surface of Borrelia burgdorferi sensu lato that may be involved in pathogenesis, we characterized a protein, P13, with an apparent molecular mass of 13 kDa. The protein was immunogenic and was expressed in large amounts during in vitro cultivation compared to other known antigens. An immunofluorescence assay, immunoelectron microscopy, and protease sensitivity assays indicated that P13 is surface exposed. The deduced sequence of the P13 peptide revealed a possible signal peptidase type I cleavage site, and computer analysis predicted that P13 is an integral membrane protein with three transmembrane-spanning domains. Mass spectrometry, in vitro translation, and N- and C-terminal amino acid sequencing analyses indicated that P13 was posttranslationally processed at both ends and modified by an unknown mechanism. Furthermore, p13 belongs to a gene family with five additional members in B. burgdorferi sensu stricto. The p13 gene is located on the linear chromosome of the bacterium, in contrast to five paralogous genes, which are located on extrachromosomal plasmids. The size of the p13 transcript was consistent with a monocistronic transcript. This new gene family may be involved in functions that are specific for this spirochete and its pathogenesis.
Borrelia burgdorferi sensu lato is a tick-transmitted spirochete that causes Lyme borreliosis, the most prevalent tick-borne zoonosis in North America and Europe. In recent years, the taxonomy and phylogenetic relationship of different B. burgdorferi sensu lato species have become more extensive and complicated. Besides numerous strains of various origins, three B. burgdorferi taxa with human pathogenic relevance are recognized, B. burgdorferi sensu stricto, B. garinii, and B. afzelii (4, 13). The disease is characterized by dermatological, rheumatological, cardiac, and neurological manifestations (44). Lyme disease and similar infections caused by other Borrelia species are a growing problem worldwide. Therefore, the development of an efficient vaccine against B. burgdorferi sensu lato is being actively pursued.
B. burgdorferi sensu lato contains both an outer membrane and a cytoplasmic membrane analogous to the surface of enteric gram-negative bacteria; however, its membrane composition has several distinct features. For example, the density of transmembrane-spanning proteins in the outer membrane of B. burgdorferi is low compared to that in gram-negative bacteria such as Escherichia coli. In contrast, compared to another spirochete, Treponema pallidum, the transmembrane-spanning proteins are approximately 10-fold more abundant in B. burgdorferi sensu lato, as determined by freeze fracture electron microscopy studies (38, 47). Furthermore, the B. burgdorferi cell includes an extraordinary abundance of lipoproteins (10, 14). A more complete understanding of membrane constituents, their function, and the overall membrane structure of B. burgdorferi sensu lato will provide insights into the strategies that enable the bacterium to establish and maintain chronic infection during Lyme disease.
Infection of a mammalian host by B. burgdorferi sensu lato is generally followed by the production of antibodies directed against a limited number of antigens, several of which have been identified as surface-exposed lipoproteins (8, 51). Among these lipoproteins are the major outer surface proteins, OspA, OspB, OspC, OspD, OspE, and OspF. Lyme disease Borrelia spirochetes may evade phagocytic clearance by varying their recognizable outer surface proteins (32) or by eliminating them entirely from their surface. B. burgdorferi sensu lato strains that no longer express Osp proteins can be isolated by growing Borrelia cell cultures in vitro in the presence of antisera against Osp proteins (40). An immunological characterization of the Osp-less mutant B. burgdorferi strain B313, which lacks OspA, OspB, OspC, and OspD (41), revealed that other antigens were exposed on the surface of the bacteria. The OspA− OspB− OspC− OspD− mutant could not evoke a detectable immune response after intradermal live-cell immunization, but polyclonal mouse serum raised against OspA− OspB− OspC− OspD− cells inhibited in vitro growth of the mutant but not the wild-type cells (41). This indicated that the presence of OspA to OspD in the wild type normally masks alternative antigens from being recognized by the host antibodies. Thus, studies of the proteins recognized by antisera generated after an inoculation with the mutant strain, B313 could lead to the identification of additional surface-exposed B. burgdorferi sensu lato proteins that are important for the pathogenesis of Lyme disease Borrelia (41). Monoclonal antibodies (MAbs) to B313 were produced, and two MAbs with borrelicidal effects were shown to recognize a 13-kDa protein, designated P13 (41).
Previous reports have revealed that the bacterium appears to contain surface-exposed low-molecular-mass proteins that may be required for Borrelia pathogenesis. Many patient sera are known to react to low-molecular-mass antigens, 10 to 13 kDa in size (25, 37, 42). Xu et al. reported that a 14-kDa protein was absent in a noninfectious B31 clone, C-1, although it was present in an infectious clone, C-9 (52). A 14-kDa lipoprotein which induces proliferation and immunoglobulin production in mouse B cells has also been described (22).
In this study, we describe the gene cloning, sequencing, expression, and characterization of another B. burgdorferi sensu lato outer surface-exposed protein, P13, which appears to be distinct from those previously described (22, 25, 37, 42, 52). The deduced sequence of the P13 peptide from three Lyme borreliosis species predicted that P13 is an integral membrane protein with three transmembrane-spanning domains. The protein had no match with predicted proteins of other organisms, but it had similarities to other B. burgdorferi sensu lato proteins harbored on linear plasmids, according to the published B. burgdorferi complete genome sequence (14).
MATERIALS AND METHODS
Bacterial strains, plasmid, and culture conditions.
The Lyme disease Borrelia strains used in this study were the high-passage strain B31 of B. burgdorferi sensu stricto, a tick isolate from North America (ATCC 35210); strain ACAI of B. afzelii, a human skin isolate from Sweden (2); and strain Ip90 of B. garinii, a tick isolate from Asian Russia (27). B. burgdorferi sensu stricto B313 is a mutant strain of B. burgdorferi sensu stricto B31 lacking OspA, OspB, OspC, and OspD (41). The low-passage strain B. burgdorferi sensu stricto N40 was obtained from the strain collection of A. G. Barbour (University of California, Irvine, Calif.) and was originally a gift from S. W. Barthold (Yale University, New Haven, Conn.) The expression plasmid pLF100 (f1+ Ampr ColE1 T7-based pET9 expression vector containing full-length B. burgdorferi OspA sequence) was provided by R. C. Huebner (Pasteur-Mérieux Connaught Laboratories, Swiftwater, Pa.).
The spirochetes were grown in Barbour-Stoenner-Kelly (BSK) II medium supplemented with phosphomycin (100 μg/ml), sulfamethoxazole (250 μg/ml), and rifampin (50 μg/ml). The cells were counted in a Petroff-Hausser chamber under phase-contrast microscopy and harvested as previously described (5, 7). Protein and DNA preparations followed standard protocols. Escherichia coli DH5α and BL21(DE3) were used as hosts for recombinant plasmids and recombinant protein expression. The E. coli strains were grown in Luria broth medium (Gibco BRL, Gaithersburg, Md.) supplemented with 50 μg of carbenicillin (Sigma, St. Louis, Mo.) per ml when required.
SDS-PAGE, immunoblot analysis, and glycoprotein detection.
Bacterial proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (15% polyacrylamide) by the method of Laemmli (28). Protein samples were resuspended in SDS sample buffer and boiled for 5 min before being loaded. The gels were stained with either Coomassie blue R-250 (Sigma) or silver stain (Bio-Rad, Hercules, Calif.). The proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) by electroblotting at 1 mA/cm2 for 45 min to 1 h. The resulting filters were used either for glycoprotein detection using an Immun-Blot apparatus as specified by the manufacturer (Bio-Rad) or for immunodetection.
The nonspecific binding was blocked by immersing the membrane for 2 h in 5% nonfat milk powder (Semper, Stockholm, Sweden) in phosphate-buffered saline containing 0.05% Tween 20 (PBS-T). The membranes were then incubated for 1 h with primary antibodies diluted in 2.5% milk powder in PBS-T (1:20 for culture supernatants of MAb 15G6, 1:5 for supernatants of MAb 7D4, and 1:50 for the affinity-purified polyclonal anti P13 antiserum), washed in PBS-T, incubated with the appropriate secondary antibody for 1 h, and finally washed in PBS-T. In the developing reaction, the substrate for the alkaline phosphatase conjugate was 5-bromo-4-chloro-3-indolylphosphate (BCIP) (Sigma). The MAbs were a kind gift from A. G. Barbour and were previously described by Sadziene et al. (41).
Enrichment and purification of P13 antigen.
The subcellular fraction of borrelial outer membrane components (designated fraction B) was prepared as described previously (30). P13 was further purified by SDS-PAGE (15% polyacrylamide) using fraction B obtained from B. burgdorferi B313. The appropriate band was visualized by staining the gel without a fixation step using cold 250 mM KCl. The protein was eluted from the gel in a Schleicher & Schuell Biotrap in 15 mM NH4HCO3 or in SDS running buffer (SDS-glycine) at 200 V for 12 h at 4°C. The purity of the P13 preparation was assessed by two-dimensional gel electrophoresis.
Mass spectrometry (MS) analysis.
Molecular mass determinations of P13 were performed on a VG Platform II mass spectrometer with a range of 2,000 m/z equipped with an electrospray (ES) source (Micromass, Altrincham, United Kingdom). Prior to injection, the purified P13 preparation was precipitated with methanol-chloroform to remove any trace of SDS contamination (50). A P13 solution of 20 pmol/μl in water-acetonitrile (50:50 [vol/vol]) was mixed with 5% formic acid and introduced directly into the ES source at a flow rate of 5 μl/min. Calibration was performed by a separate introduction of horse heart myoglobin (16,951.5 Da). MassLynx software (Fisons Plc, Altrincham, United Kingdom) was used to calculate the molecular mass.
Generation and purification of monospecific polyclonal antiserum.
A 100-μg portion of P13 protein, purified as described above, was used for each of four repeated immunizations of a rabbit. The immunizations were performed at 1- and 2-month intervals. Affinity purification of rabbit antiserum was performed by the method of Sambrook et al. (43).
Protease treatment of spirochetes.
Cell surface proteolysis of intact Borrelia was conducted as previously described by Barbour et al. (6) with minor changes. Briefly, after centrifugation of cultured spirochetes at 2,000 × g for 10 min, washed spirochetes were resuspended in PBS–5 mM MgCl2 (PBS-Mg) to a final concentration of 2 × 109 cells/ml. To 0.5 ml of the cell suspension was added 25 μl of either proteinase K (Boehringer, Mannheim, Germany) in sterile water or trypsin (Sigma) in 1 mM HCl to a final concentration of 12.5 to 200 μg/ml or 6.25 to 400 μg/ml, respectively. As a negative control, sterile water or 1 mM HCl was added to the cell suspension. The mixtures were incubated in 20°C for 1 h, and the proteolytic reactions were stopped by addition of 10 μl of the peptidase inhibitor phenylmethysulfonyl fluoride (50 mg/ml in isopropanol) (Sigma). The suspensions were then centrifuged at 2,000 × g for 10 min and washed twice with PBS-Mg. Pelleted cells were boiled in SDS sample buffer prior to gel loading.
Indirect-immunofluorescence assay and immunogold labeling.
An indirect-immunofluorescence assay was used to study the binding of antibodies to intact or outer membrane-permeabilized spirochetes. The method used was as described by Parveen and Leong (36). Briefly, glass coverslips were placed in 24-well microtiter plates and overlaid with 400 μl of 25 mM HEPES (pH 7.8)–150 mM NaCl–1 mM MgCl2–0.25 mM CaCl2–0.1% glucose–3% bovine serum albumin (BSA). Harvested spirochetes were washed three times with PBS–0.2% BSA, and 100 μl containing 5 × 106 spirochetes was added to each well. After centrifugation of the plates at 200 × g for 10 min, the supernatants were carefully aspirated. The bacteria were fixed to the glass coverslips by adding 500 μl of 3% paraformaldehyde in PBS and subjecting them to gentle rocking at 20°C for 1 h. The cells were washed three times with PBS. To expose periplasmic flagella of fixed cells, 500 μl of cold methanol was added, and then the coverslips were incubated at −20°C for 10 min and washed twice in PBS. Blocking buffer (PBS containing 5% BSA) was added to each coverslip, the coverslips were incubated for 1 h at 20°C, and then primary antibodies diluted in blocking buffer were added. MAbs H9724 (anti-flagellin), H5332 (anti-OspA), both diluted 1:5, and 15G6 (anti-P13), diluted 1:20, were used as primary antibodies. After incubation for 2 h at 20°C, the coverslips were washed three times for 5 min each with PBS. F(ab')2 donkey anti-mouse immunoglobulin G fragments conjugated to Cy3 (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) were used to detect antibody binding. The F(ab')2 fragments diluted 1:200 in PBS–1% BSA were added to each coverslip, and the coverslips were incubated for 1 h at 20°C, washed four times with PBS, and mounted onto glass slides using VECTASHIELD mounting medium (Vector Laboratories, Inc. Burlingame, Calif.). Fluorescence was monitored with a Zeiss Axioplan microscope and Hamamatsu color chilled 3CCD camera.
Immunogold labeling using monoclonal anti-P13 antibody was performed as previously described (23).
Radioactive labeling of lipid precursors.
For labeling of Borrelia cells with [2-3H]glycerol, [9,10(n)-3H]palmitic acid, and [9,10(n)-3H]myristic acid, 1 mCi of radioactive fatty acid was added to 50 ml of culture. To 50 ml of BSK II medium were added 1 ml of isotope in ethanol solution and 0.5 ml of B. burgdorferi B313 culture in the exponential growth phase. For labeling with [1-14C]stearic acid, 625 μl of isotope (125 μCi) in toluene solution was used. Before the isotope was added to the growth medium, the toluene was removed by evaporation and the isotope was resolved in ethanol and added to 15 ml of medium, together with 150 μl of B. burgdorferi B313 culture in the exponential growth phase. The cultures were incubated for 10 days at 32°C. Cells were harvested by centrifugation at 2,000 × g for 10 min, membrane proteins were prepared, and labeled proteins were analyzed by SDS-PAGE and autoradiography.
Amino acid sequencing of P13.
For N-terminal amino acid sequencing, the 13-kDa protein band was isolated from an SDS-PAGE gel and eluted in a Biotrap as described above. The protein was digested with Staphylococcus areus V8 protease endoproteinase Glu-C as specified by the manufacturer (Boehringer). Peptide fragments were transferred to a PVDF membrane by soaking the membrane in protein solution overnight. N-terminal amino acid sequence analysis was performed on a 477A sequencer (Applied Biosystems, Foster City, Calif.) at Umeå University. Electroeluted P13 was further precipitated (50) to remove the added detergent prior to sequencing both the complete protein and the C terminal. The sample was also sent to Eurosequence B. V. (Groningen, The Netherlands) for analysis on an HP 1090 Aminoquant instrument by an automated two-step precolumn derivatization with two reagents, OPA (ophtalaldehyde) for primary and FMOC (9-fluorenylmethylchloroformate) for secondary amino acids. A 477A sequencer (Applied Biosystems) was used for C-terminal amino acid sequencing.
Cloning and sequencing of the p13 gene from Lyme disease Borrelia species.
Plasmid libraries were obtained from borrelial genomic DNA using standard techniques. The amino acid sequence of an internal 25-residue fragment was used to design two degenerate oligonucleotides designated Y5.2 and Y6.2-R (Table 1). These oligonucleotides were used to PCR amplify a 75-bp fragment corresponding to the 25-amino-acid residue. The upstream part of the p13 gene was cloned from a pUC library by using oligonucleotide Y7-R, complementary to the central region of the 75-bp fragment, and primers complementary to the vector in a PCR reaction. The obtained sequence allowed us to design an oligonucleotide, Y9, which was used in combination with oligonucleotide Y7-R to generate a PCR fragment that was used as a probe to screen the B31 and B313 EcoRI-digested genomic DNA libraries by colony hybridization. Full-length p13 clones were thus isolated and designated pLY-100 (B31) and pLY-101 (B313), respectively.
TABLE 1.
Oligonucleotide primers and probes used in this study
| Designation | Sequencea | Purpose | Restriction site | Source; complementary nucleotidesb |
|---|---|---|---|---|
| Y5.2 | 5′-ACNTCNAARCARGAYCCN-3′ | Degenerate primer, probe | 124 to 141 | |
| Y6.2-R | 5′-TGNGCRAARCTNCCDATNCC-3′ | Degenerate primer, probe | 178 to 197 | |
| Y7 | 5′-TGTACCATCTTTATTGAACCTTTTTTTAGGGTTT-3′ | PCR, sequencing primer | 4 to 77 | |
| Y7-R | 5′-AAACCCTAAAAAAAGGTTCAATAAAG-3′ | PCR, sequencing primer | 53 to 77 | |
| Y8-R | 5′-ACCATCAAGCGCTTTGATATC-3′ | Sequencing primer | 276 to 297 | |
| Y9 | 5′-GATTTTTCATTGGATCCCAGAATTTG-3′ | PCR, sequencing primer | BamHI | −217 to −193 |
| Y10-R | 5′-CTATACCAACCGAATTCAAATCCAAG-3′ | PCR, sequencing primer | EcoRI | 226 to 250 |
| Y11 | 5′-GGTTTTTATGGATCCACTTTT-3′ | PCR, sequencing primer | BamHI | −7 to 14 |
| Y12 | 5′-TATGCTACCATGGATCCAGTTTTAA-3′ | PCR, sequencing primer | BamHI | −38 to −14 |
| Y13 | 5′-CGGGATCCGTTTTTTCTAGCTTTGCTCAAGC-3′ | PCR, sequencing primer | BamHI | 32 to 63 |
| Y14 | 5′-GGAATTCCCTGGTTCCGCGTGGATCCATGAATAAACTTTTAATTTTTGTT-3′ | pLY-313 (GST fusion) | EcoRI, BamHI | 1 to 24 |
| Y15-R | 5′-ATAGGATCCTCATCAAATCCAAGA-3′ | Sequencing primer | BamHI | 225 to 248 |
| Y17 | 5′-GGAGTTATGTTAGCAGGTGTGG-3′ | Sequencing primer | 337 to 359 | |
| Y18-R | 5′-TAAAAAAATTTAAAGAAAAGGAGGG-3′ | PCR, sequencing primer | ||
| CMV2-R | 5′-CACCCATTTTCTAGATAAATAAAATTAATAGC-3′ | PCR, sequencing primer | XbaI | 533 to 564 |
| CMV3-R | 5′-ATAAAAGGTACCATAGCTTTTTTTGAAAGACAG-3′ | PCR, sequencing primer | KpnI | 516 to 546 |
| Y30-R | 5′-TTTTAGAATCATTAGCTTGAGC-3′ | p13 primer extension | 55 to 77 | |
| Y33-R | 5′-TAGAATTCAGCAATTGCAATACAG-3′ | pLY-313 (GST fusion) | EcoRI | 574 to 598 |
| Y41-R | 5′-TACCCCAAGTCCATTGAAAAGCAGC-3′ | Northern blot analysis | 310 to 335 |
IUB group codes identifying redundancies in the degenerate primers are as follows: N = A + C + G + T, D = A + G + T, R = A + G, Y = C + T. Restriction endonuclease sites are underlined.
The A in the ATG start codon of the B. burgdorferi B31 p13 sequence was defined as nucleotide position 1.
The full-length p13 genes from B. afzelii ACAI and B. garinii Ip90 were obtained by PCR amplification followed by ligation into pT7 Blue T-vector (Novagen, Madison, Wis.). The following primer pairs were used for PCR amplification of the p13 gene from B. afzelii ACAI: Y9 plus Y18-R, Y9 plus CMV3-R, Y9 plus Y10-R, Y12 plus Y10-R, and Y13 plus CMV3-R. The following primer pairs were used for amplification from B. garinii Ip90: Y9 plus CMV3-R, Y12 plus Y10-R, Y11 plus Y10-R, Y13 plus Y10-R, and Y13 plus CMV2-R.
Plasmid DNAs were sequenced by primer walking using a Pharmacia T7 sequence kit or an ABI Prism Dye Terminator cycle-sequencing ready-reaction kit and an Applied Biosystems 377 A DNA sequencer. Sequences from both strands of plasmid inserts and from at least two different PCR-originated clones were determined. The sequence fragments were assembled using the GCG, Inc., software for the UNIX computer. The EMBL, GenBank, and PIR databases were screened for similar proteins using the National Center for Biotechnology Information Blast electronic mail server (1).
Expression of recombinant P13 and in vitro translation.
Two oligonucleotide primers, Y14 and Y33-R (Table 1), were used to amplify the full-length p13 gene from B. burgdorferi B31. The PCR products were digested with the appropriate restriction enzymes and ligated in frame with glutathione S-transferase (GST) into the tac promoter-based expression vector pGEX-2T (Pharmacia). The GST-P13 fusion protein was induced in E. coli DH5α by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) (Saveen Biotech AB, Malmö, Sweden) (1 mM final concentration). The recombinant protein was subsequently identified by SDS-PAGE and immunoblotting using MAb 15G6.
For in vitro translation experiments, the p13 gene was cloned into pRSETA (Invitrogen, Carlsbad, Calif.) and transformed into BL21(DE3). In vitro translation was performed using TNT Quick Transcription/Translation master mix (Promega Corp., Madison, Wis.) as specified by the manufacturer. The resulting protein labeled with l-[35S]methionine (Amersham Pharmacia Biotech, Solna, Sweden) was separated by SDS-PAGE (15% polyacrylamide) and monitored with Hyperfilm MP (Amersham, Little Chalfont, England).
RNA analysis.
Total RNA was extracted from B. burgdorferi B31 and B. burgdorferi B313 cells collected at mid-log phase. The cell pellet was washed once with PBS and then extracted by TRIzol reagent as specified by the manufacturer protocol (Gibco). A 20-μg portion of each RNA preparation were electrophoresed through a 1% agarose–formaldehyde gel at 100 V for 9 h. The gel was neutralized in HEPES (3), and the RNA was transferred to a nylon membrane by capillary blotting (Hybond-N; Amersham). The filter was hybridized at high stringency with a [32P]ATP-end-labeled oligonucleotide (Y41-R). The primer extension reaction was performed with a specific [32P]ATP-end-labeled oligonucleotide primer (Y30-R) and avian myeloblastosis virus reverse transcriptase (Boehringer) as described previously (18). Only a full-length extension product was obtained, and the size of the transcript was compared to that of the DNA sequence of plasmid pLY-100 obtained using the same oligonucleotide primer.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper were submitted to the GenBank database and were assigned the following accession numbers: B. burgdorferi B31 and B313, AF085739; B. afzelii ACA1, AF085740; and B. garinii Ip90, AF085741.
RESULTS
Purification of a 13-kDa protein from B. burgdorferi and production of P13 monospecific polyclonal antibodies.
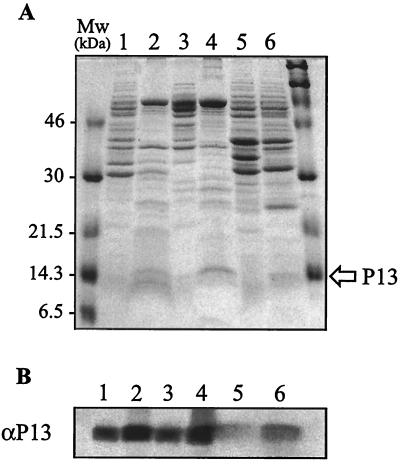
The P13 polypeptide was purified from preparations enriched for membrane proteins in order to facilitate the generation of a P13 antiserum and to identify the p13 gene in B. burgdorferi sensu lato. In the membrane preparation (fraction B) derived from the OspA− OspB− OspC− OspD− mutant, B313, P13 was enriched without contamination by proteins of similar molecular mass to that observed in the B31 preparation (Fig. 1A). The 13-kDa protein was purified by preparative gel electrophoresis of the B313 membrane proteins followed by excision of the 13-kDa protein from the gel. Two-dimensional gel electrophoresis of the resulting protein preparation followed by silver staining or immunoblotting was used to show that there were little or no quantities of contaminating proteins (data not shown).
FIG. 1.
Analysis of P13 expression in different B. burgdorferi sensu lato strains by SDS-PAGE (15% polyacrylamide) and immunoblotting. (A) Coomassie brilliant blue staining (10 μg of protein per lane); (B) immunoblot analysis using polyclonal rabbit sera (αP13) (1 μg of protein is lanes 1 to 4 and 10 μg p in lanes 5 and 6). Lanes: 1, 3, 5, and 6, whole-cell lysates; 2 and 4, membrane fractions (B-fractions). Lanes 1 and 2 correspond to B. burgdorferi B31, lanes 3 and 4 correspond to B. burgdorferi B313, lane 5 correspond to B. afzelii ACA1, and lane 6 corresponds to B. garinii Ip90. Molecular mass (Mw) standards (in kilodaltons) are shown to the left (low molecular mass) and right (high molecular mass). The position of P13 is marked with an arrow.
The purified P13 polypeptide from strain B313 was injected into a rabbit, and antiserum was generated. The polyclonal antiserum raised against purified P13 from B. burgdorferi B313 recognized all Lyme borreliosis species tested (Fig. 1B).
Expression of P13 in different Borrelia species.
The presence and relative levels of P13 in whole-cell extracts were investigated (Fig 1A). P13 was expressed in all whole-cell protein preparations of Lyme disease borreliae regardless of whether it was isolated from a virulent low-passage B. burgdorferi strain, strain N40, or an avirulent high-passage B. burgdorferi sensu lato strain (data not shown). This analysis revealed no major differences among the borrelial strains with respect to either apparent molecular mass or the relative expression levels of P13. In contrast, neither whole-cell protein preparations nor membrane fractions of the relapsing fever species, B. hermsii, B. crocidurae, and B. hispanica, and the avian borreliosis species, B. anserina, revealed any polypeptide of the correct size. Furthermore, Coomassie blue staining and immun-blotting using the monospecific polyclonal antisera showed that P13 was enriched in the membrane fraction (fraction B) in all Lyme borreliosis species. MAbs 15G6 and 7D4, raised against the 13-kDa protein of the B313 strain (41), recognized P13 in B. burgdorferi B31, B313, and N40 and B. afzelii ACAI but did not recognize it in the third Lyme borreliosis species, B. garinii Ip90 (data not shown).
Surface exposure of P13 in B. burgdorferi sensu lato.
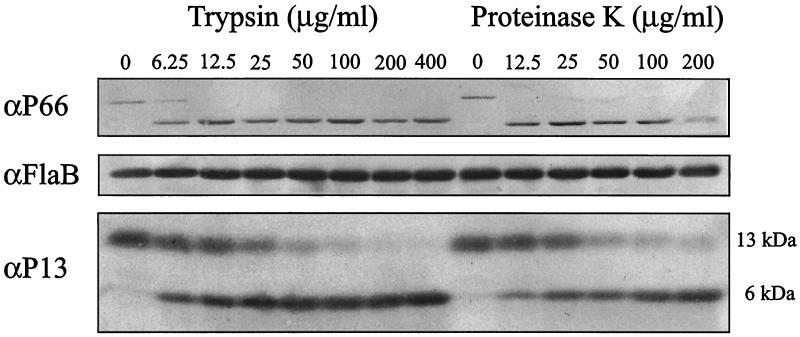
We investigated whether P13 was surface exposed by treatment of intact spirochetes with proteinase K or trypsin (Fig. 2). The protease-treated Borrelia cells were analyzed by SDS-PAGE followed by immunoblot analysis. Our results were in agreement with the results of Sadziene et al. (41), i.e., that a protein of 13 kDa recognized by MAb 15G6 was removed after proteinase K treatment. Immunoblot analysis of protease-treated Borrelia cells using P13-reactive polyclonal rabbit sera showed similar results, but it also labeled a fragment of approximately 6 kDa after protease treatment (Fig. 2). In B. afzelii ACA1 and B. garinii Ip90, the polyclonal rabbit serum also reacted with a 13-kDa protein that was removed after protease treatment (data not shown). To investigate if high concentrations of protease also cleave proteins not exposed on the surface of the spirochete, i.e., proteins situated in the periplasmic space or in the inner membrane, different concentrations of protease were used. The periplasmic flagella were used as a negative control (Fig. 2). When spirochetes were sonicated prior to protease treatment, these internal flagella were susceptible to both proteinase K and trypsin (data not shown). However, no cleavage of FlaB protein from intact B. burgdorferi B31 or B313 was detected and no difference in cleavage pattern was seen for the different protease concentrations used. MAb 9A4, reacting to P66, was used as a positive control, and as previously shown by Bunikis and Barbour (11), the outer membrane-located P66 was cleaved by both proteases. P13 was apparently less sensitive to the protease concentration than was P66. These results indicate that P13 is located in the outer membrane and is surface exposed, since only surface-exposed proteins are cleaved by the proteases used. Similar results were obtained with both B. burgdorferi B31 and B313.
FIG. 2.
Immunoblot analysis of protease-treated B. burgdorferi B31 proteins. Whole intact spirochetes were incubated with different concentrations of either trypsin or proteinase K. The proteins were separated by SDS-PAGE (15% polyacrylamide), and proteins blotted to a PVDF membrane were probed with the MAbs against P66 (9A4) or FlaB (H9724), or the polyclonal rabbit serum against P13. The positions of the P13 protein and 6-kDa cleavage product are shown on the right.
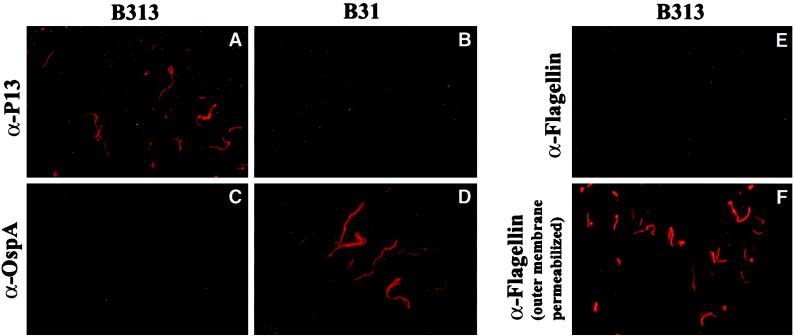
To further show that P13 is surface exposed, we performed an indirect-immunofluorescence assay (Fig. 3). B. burgdorferi B31 and B313 were fixed to glass coverslips with paraformaldehyde, which leaves the outer membrane intact (36). MAb H5332 against OspA was used as a positive control, and as expected, this antibody labeled B. burgdorferi B31 but did not label the OspA− OspB− OspC− OspD− mutant B. burgdorferi B313 (Fig. 3C and D). We confirmed the results, previously shown by Sadziene et al. (41), that the anti-P13 MAb 15G6 binds to intact B. burgdorferi B313 spirochetes but not to cells expressing OspA and OspB (Fig. 3A and B). To show that this labeling of P13 was not the result of a damaged membrane, we used MAb H9724, reactive against the flagellin. This antibody labeled only the periplasmic flagella of B. burgdorferi B313 cells that were methanol permeabilized prior to antibody probing (Fig. 3E and F).
FIG. 3.
Indirect-immunofluorescence assays of B. burgdorferi B31 and B313. Binding of MAbs to intact or membrane-permeabilized spirochetes was detected with Cy3-conjugated F(ab')2 donkey anti-mouse IgG fragments.
To further establish the surface location of P13, we performed an immunogold-labeling experiment. MAb 15G6, directed against the 13-kDa protein of B. burgdorferi B313, was used as the primary antibody for immunogold staining of intact B. burgdorferi B313. The majority of the gold labeling was confined to the outer surface membrane of the Borrelia cells, thus supporting the idea that P13 contained outer surface-exposed domains (data not shown).
Cloning and sequence analysis of the p13 gene from B. burgdorferi sensu lato strains.
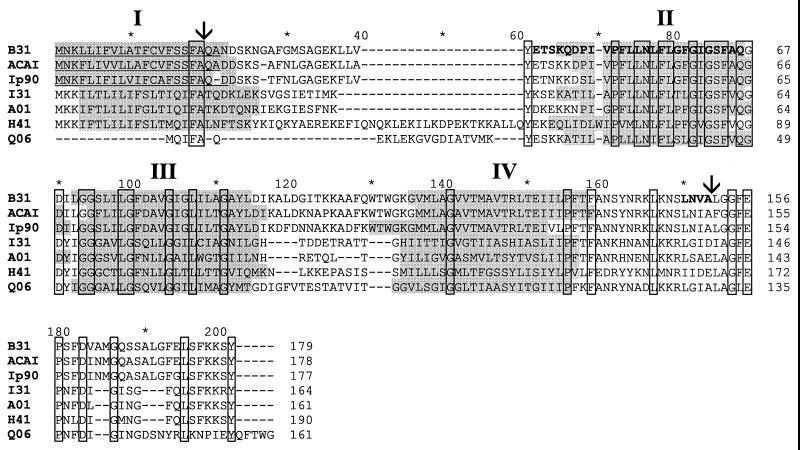
To facilitate the cloning of p13, we undertook an amino acid sequence analysis of the purified P13 polypeptide. An initial attempt to obtain an N-terminal sequence failed due to N-terminal blocking. To circumvent this problem, the P13 protein preparation was digested with S. aureus V8 protease, which resulted in two peptide fragments, as determined by SDS-PAGE analysis. Since one of these fragments was N-terminally blocked, we used this mixture in a second attempt to generate an amino acid sequence. A sequence of 25 consecutive amino acids was obtained (Fig. 4). This sequence was used to design degenerate oligonucleotide primers for PCR amplification of a fragment that defined an internal region of the p13 gene (Table 1). Subsequently, full-length p13 clones were isolated from Borrelia plasmid DNA libraries.
FIG. 4.
Comparison of the deduced amino acid sequences of P13 protein from B. burgdorferi B31 and B313, B. afzelii ACA1, B. garinii Ip90, and the four B. burgdorferi B31 genome paralogues, BBA01, BBI31, BBH41, and BBQ06. Computer-predicted leader peptides with signal peptidase I cleavage sites are underlined. Identical amino acids are shown within open boxes, and transmembrane-spanning domains are gray-shaded and numbered with Roman letters I to IV in bold. The four putative transmembrane-spanning domains were predicted using the SOSUI system available on the Internet access (http://azusa.proteome.bio.tuat.ac.jp/sousi/). Three additional programs, HMMTOP (http://www.enzim.hu/hmmtop/server/submit.html), TMHMM (http://www.cbs.dtu.dk/services), and PSORT (http://psort.nibb.ac.jp/form.html), were also used to predict the secondary structure of the proteins (45). These programs confirmed the location of the transmembrane-spanning domains. The amino acid sequences obtained from both the internal sequencing of S. aureus V8 proteinase (Endo-Glu-C) fragments of B. burgdorferi B313 (ETSKQDPIVPFLLNLFLGFGIGSFAQ) and the C-terminal sequencing, positions 151 to 148 (AVNL), are indicated by boldface letters. Arrows indicate the computer-predicted cleavage site for signal peptidase I and the cleavage site for C-terminal processing by an unknown protease.
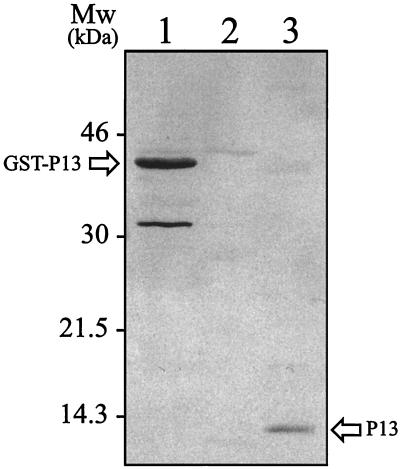
Two recombinant DH5α clones that hybridized to the p13 probe were isolated and designated pLY-100 (from B. burgdorferi strain B31) and pLY-101 (from B. burgdorferi strain B313). Nucleotide sequencing of the inserts of these plasmids revealed identical 537-bp open reading frames (ORFs) initiated by an ATG start codon (Fig. 5). Translation of the DNA sequence also revealed an exact match to the sequence that was previously determined by amino acid sequencing, thus strongly suggesting that the cloned gene indeed encoded the P13 protein. To confirm that the cloned p13 gene encoded the MAb-reactive P13 antigen, we expressed the gene in E. coli. Full-length P13 (19.1 kDa) was fused to GST (26 kDa), and a fusion protein of the expected molecular size (45 kDa) was recognized by the P13 MAb, 15G6, in an immunoblot experiment (Fig. 6). The p13 gene identified did encode the alternative antigen, P13.
FIG. 5.
Comparison of the nucleotide sequences of the p13 genes from B. burgdorferi B31 and B313, B. afzelii ACA1, and B. garinii Ip90. All sequences are aligned with B. burgdorferi B31 (and B313, which was identical to B31). Hyphens indicate homologies between the different species, capital letters indicate differences, and gaps indicate missing or inserted bases. Start and stop codons are indicated by boldface letters and the transcriptional start point is marked by a vertical arrow. The putative −35 and −10 promoter regions and the ribosomal binding sites are overlined. A putative stem-loop structure (→←) for termination was present downstream of p13.
FIG. 6.
Immunoblot analysis of recombinant GST-P13 fusion protein (pGEX-P13) using MAb 15G6. A 10-μg portion of protein was used in SDS-PAGE (15% polyacrylamide), from cultures that were induced with 1 mM IPTG (final concentration) for 3 h. Lanes: 1, pGEX-P13; 2, pGEX alone 3, membrane-fractionated (B-fraction) B. burgdorferi B313.
The sequence obtained from the B. burgdorferi p13 gene enabled us to design oligonucleotide primers that could PCR amplify the p13 gene from B. afzelii ACAI and from B. garinii Ip90. These PCR fragments were cloned, and the DNA sequences of the p13 gene from B. afzelii and from B. garinii were determined (Fig. 5). The ORFs containing putative ATG codons were 537, 540, and 534 nucleotides long for B. burgdorferi B31, B. afzelii ACAI, and B. garinii Ip90, respectively. Putative ribosome binding sites, AGGGG (B. burgdorferi B31 and B313 and B. garinii Ip90) and AGAGG (B. afzelii ACAI) were present 10 bp upstream of the start codons. Further upstream, a ς70-like promoter sequence was found for B. burgdorferi B31 and B313, while the promoter sequences were less highly conserved for Ip90 and ACAI (Fig. 5). The use of these sequences (−10 and −35 regions) as a promoter was supported by the location of the transcriptional start point, which was determined by primer extension analysis (see below). The ORF of p13 terminated at a TAA triplet, which was followed by a putative stem-loop structure for rho-independent transcriptional termination (Fig. 5).
DNA blot analysis of a pulsed-field gel revealed that the p13 gene was located on the linear chromosome (data not shown). This chromosomal location is consistent with the sequenced genome (14).
Analysis of the p13 transcript.
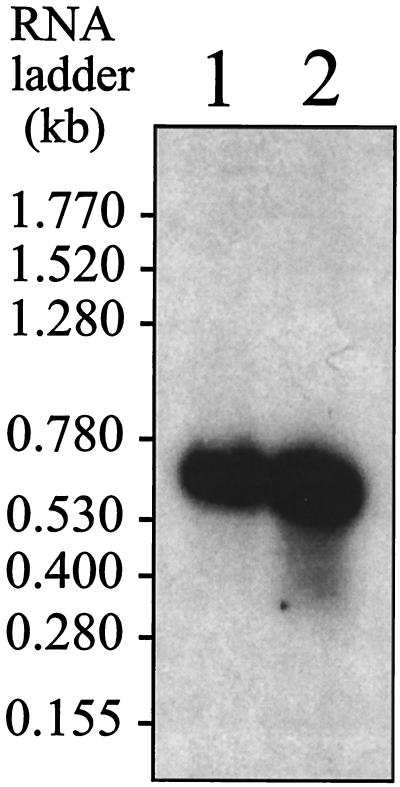
The transcriptional start point of the p13 gene of B. burgdorferi B31 was determined by primer extension analysis (data not shown). The transcriptional start site was identified as an A, 21 bp upstream of the AUG translational start codon (Fig. 5). This analysis identified the possible TTTAAA (−35 box) and TACCAT (−10 box), at positions −32 to −27 and positions −12 to −7, respectively, as the likely promoter signal for the p13 gene. Northern blot analyses were performed to analyze native borrelial mRNA transcripts specific for p13. An oligonucleotide probe, specific for p13 from B. burgdorferi B31 and B313, was used and shown to recognize a transcript approximately 600 bp in size from B31 and B313 (Fig. 7). Thus, the transcriptional start point and the transcript size strongly suggested that p13 was transcribed as a monocistronic mRNA.
FIG. 7.
RNA -blot analysis of mRNA from B. burgdorferi B31 (lane 1) and B313 (lane 2), probed with p13-specific oligonuleotide Y30-R (Table 1). The RNA ladder ranged from 0.16 to 1.77 kb (Gibco).
Leader peptide and transmembrane-spanning domains of P13.
The deduced amino acid sequence of P13 from B. burgdorferi B31, B. afzelii ACAI, and B. garinii Ip90 is presented in Fig. 4. The amino acid sequence of the P13 protein from B. burgdorferi B31 was 85.4 and 80.8% identical to the sequence from B. afzelii ACAI and B. garinii Ip90, respectively. The sequences from the two latter strains had 87.0% identity. Using the SignalP V1.1 WWW prediction server, Center for Biological Sequence Analysis (33) (http://www.cbs.dtu.dk/services/SignalP), the predicted N-terminal end of the P13 protein contained a 21-amino-acid leader peptide sequence in B. burgdorferi B31, and B. afzelii ACAI, whereas in B. garinii Ip90, two possible lengths, 21 or 19 amino acids, were predicted (Fig. 4). These leader peptide sequences were typical of exported proteins with a basic residue followed by a hydrophobic and a potential leader peptidase I cleavage site, according to the criteria established by von Heijne (46). The cleavage of the respective leader sequences would yield a mature P13 protein composed of 158 amino acids with a calculated molecular mass of 16,772 Da for B. burgdorferi B31, 157 amino acids with a calculated molecular mass of 16,864 Da for B. afzelii ACAI, and 156 or 158 amino acids with calculated molecular masses of 16,872 Da or 17,115 Da, respectively for B. garinii Ip90 (http://www.expasy.ch/tools/pi_tool.html).
Homology searches using P13 as query in the protein databases did not reveal a match that had significant sequence similarity, other than borrelial proteins. The published complete genome sequence from the closely related spirochete species T. pallidum revealed that 90 ORFs of unknown function match chromosome-encoded proteins in B. burgdorferi sensu stricto. However, none of the T. pallidum ORFs were homologous to P13 (15). The p13 gene sequence was compared to the recently completed B. burgdorferi B31 genome sequence and was shown to be identical to an ORF called BB0034 (14). Four additional Borrelia proteins with a high degree (34.5 to 40.9%) of sequence identity to P13 were found. Together with these putative peptides, denoted BBA01, BBI31, BBH41, and BBQ06, P13 belongs to a protein family designated paralogous family 48 (14) (http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database-gbb). An additional protein, BBJ03, has been identified as belonging to the same paralogous protein family; this polypeptide was much smaller and had a sequence identity of only 28.6%. Furthermore, no sequence homology to DNA from relapsing-fever Borrelia was revealed in either Southern hybridization or PCR amplification experiments (data not shown).
Computer analysis predicted transmembrane-spanning helices (TM helices) in P13 from all three B. burgdorferi sensu lato species. P13 was suggested to contain four TM helices (three if the signal peptide was excluded), and this feature is also shared among the other members of the paralogous protein family except for the short polypeptide (only 50 amino acids), BBJ03 (Fig. 4). Aligning P13 with the paralogous proteins illustrated that the relative locations of the transmembrane-spanning domains are conserved and that the region of highest similarity between P13 and the paralogues corresponded to the TM helices (Fig. 4).
P13 is posttranslationally processed and modified.
P13 was named after its apparent molecular mass in SDS-PAGE (15% polyacylamide), but the sequence data suggested that it is larger. To investigate this discrepancy, we studied the migration of P13 in both 18% polyacrylamide gradient gels and 16.5% polyacrylamide-Tris-Tricine gels (BioRad), relative to molecular mass standards. The apparent molecular mass in both systems was still 13 to 14 kDa (data not shown), although Tris-Tricine gels are specifically designed to accurately determine the molecular mass of small proteins that can otherwise comigrate with SDS and give rise to false apparent masses. However, the theoretical mass for this protein is 19,104 Da, and using an in vitro translational system, it was shown in practice to encode a protein corresponding to the estimated theoretical mass (data not shown).
Many surface-exposed proteins in Borrelia are posttranslationally modified by fatty acids, and some examples of carbohydrate-modified proteins exist (10, 16). Since the P13 signal sequence contains a cysteine at position 13, which, together with the surrounding amino acids (LATFC), constitutes a potential leader peptidase II cleavage-lipid modification site, we wanted to investigate if P13 might be posttranslationally modified in vivo. Labeling with [3H]palmitic acid, [3H]myristic acid, [1-14C]stearic acid, or [3H]glycerol revealed no lipidation of P13 (19). Since known lipoproteins such as OspA and OspC were labeled with [3H]palmitic acid, this analysis suggested that P13 is not lipidated. We also tested whether P13 is glycosylated by using an Immun-Blot kit for glycoprotein detection. This analysis revealed that P13 was not glycosylated whereas the positive control, ovalbumin, was easily detected as glycosylated (data not shown).
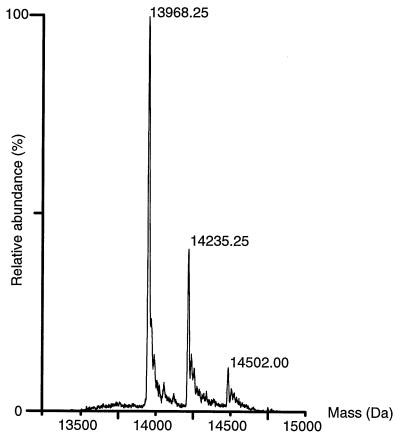
As an independent measure of the P13 molecular mass, we subjected purified P13 from B. burgdorferi B313 to electrospray mass spectrometry (ESMS) analysis and obtained a molecular mass of 13,968 ± 1 Da (Fig. 8). We also performed N- and C-terminal amino acid sequencing, which revealed a blocked N terminus and a processed C terminus that lacked the last 28 amino acids. Computer predictions of the theoretical mass with N-terminal processing by signal peptidase I after position 21 and C-terminal processing for B. burgdorferi sensu stricto B31 resulted in a theoretical mass of 13,789 Da. This apparent mass calculation is different from the ESMS result, 13,968 Da. If the mass calculation were instead made using a possible cleavage after position 19, the native P13 protein would start with a glutamine and the theoretical mass for B. burgdorferi sensu stricto B31 P13 would be 13,988 Da. This calculation is based on the finding of an additional computer-predicted cleavage site after position 19 for P13 of B. garinii Ip90.
FIG. 8.
ESMS results for purified P13 protein. The molecular masses are indicated above the peaks.
Since P13 is found to be blocked for Edman degradation, one possible modification that would fit to the theoretical mass and the experimentally obtained mass is a pyroglutamic acid modification of the possible N-terminal glutamine. Pyroglutamic acid modification reduces the molecular mass by 17.3 Da, which would give a theoretical mass of 13,971 Da, only 3 Da different from the mass measured by MS.
DISCUSSION
The present study was initiated to clarify previous observations suggesting that in the absence of the major Osp proteins, i.e., OspA, OspB, OspC, OspD, OspE, and OspF, other antigens become accessible for the host immune response. Using an Osp-less mutant (OspA− OspB− OspC− OspD−) Sadziene et al. (41) showed that growth-inhibiting antibodies were produced against non-Osp components, and further characterization identified MAbs directed against a protein with an apparent molecular mass of 13kDa. In this study we identified and characterized the novel P13 antigen, which appears to be nonlipidated and present in both infectious and noninfectious strains and thus to be distinct from other low-molecular-mass proteins described in earlier studies (22, 25, 37, 42, 52).
Previous results indicate that a 13-kDa protein of B. burgdorferi sensu stricto is surface exposed (41). MAb 15G6 labels this 13-kDa protein from whole-cell B. burgdorferi sensu stricto cells in Western blot analyses, but after proteinase K treatment of intact Borrelia cells this band disappears. Previously, in a growth inhibition assay using whole immunoglobulin and the Fab fragment, the growth of the OspA− OspB− OspC− OspD− B313 mutant was inhibited (41). Here we provide more evidence to prove that the P13 protein is surface exposed. Immunoflourescence assay results show that anti-P13 MAb 15G6 binds to intact B. burgdorferi B313 (Fig. 3). However, this antibody does not label B. burgdorferi B31, which can be explained by the hindrance phenomenon similar to that shown for another surface-exposed protein, P66, by Bunikis and Barbour (11). These results are also in agreement with results showing that this antibody inhibits the growth of B. burgdorferi B313 but not cells expressing OspA and OspB in a growth inhibition assay (41).
To further show that P13 is surface exposed and to rule out the possibility that high concentrations of protease do not cleave proteins present in the periplasmic space or inner membrane, we used different concentrations of protease to treat intact Borrelia cells. Since the surface-exposed P66 protein is known to be cleaved by both proteinase K and trypsin at different concentrations (11), we used P66 as a positive control and the periplasmic flagella as a negative control. Consistent with the results of Bunikis and Barbour (11), no differences in proteolysis of the flagella were detected between the different concentrations of protease, indicating that the proteins not exposed on the surface of the bacteria were not affected by the proteases. The P13 protein was cleaved by all concentrations of protease tested, although in a more concentration-dependent manner than P66, which indicates that P13 is less accessible for proteases than is P66. The discrepancy in MAb 15G6 binding and surface proteolysis of P13 can be explained by different accessibilities of antibodies and proteases. Hence, the accessibility of the P13 epitope to MAbs is masked by other proteins while proteases remove other surface-exposed proteins, thereby accessing the P13 protein.
The purification of P13 facilitated the generation of an antiserum that recognized a 13-kDa protein from all Lyme disease Borrelia species investigated. An amino acid sequence obtained from the purified peptide enabled us to clone the p13 gene from B. burgdorferi B31 and B313 and subsequently from B. afzelii ACAI and B. garinii Ip90.
Given the large number of bacterial genomes already sequenced, it is surprising that sequence homology searches using P13 as a query revealed no homologues in other genera. Interestingly, five similar sequences were found in B. burgdorferi sensu stricto. Thus, P13 belongs to a protein family that appears to be unique to B. burgdorferi sensu stricto. Whether this protein family is also present in other Lyme disease Borrelia species is currently under investigation. The importance of these paralogous proteins is not known, but all of them, except BBJ03, contain an amino terminus likely to encode a signal sequence. Possibly this P13 protein family is involved in functions that are specific for the B. burgdorferi sensu lato pathogenesis, such as the interaction between the bacteria and the arthropod and vertebrate hosts. The finding that P13 appears not to be present in relapsing fever Borrelia species and that no sequence similarity to P13 was found in the T. pallidum genome sequence makes it a very good tool for both molecular and serological diagnosis of Lyme disease. Thus, its uniqueness for Lyme disease Borrelia species and its location in the outer surface mean that the P13 protein could also be a good candidate for inclusion in a second generation vaccine.
A closer sequence examination of the P13 gene family revealed that all members contain putative transmembrane domains, which are conserved. The region of highest similarity between P13 and its paralogues also corresponded to two TM helices and a possible surface-exposed intervening sequence (Fig. 4). Given the conservation of the TM helices between P13 and the paralogous proteins, we suggest that all members of the P13 genome paralogues are localized to the membrane, which is further supported by the finding of a predicted N-terminal signal peptide. However, the P13 signal peptide was predicted to be cleaved by signal peptidase, but the corresponding sequence of the paralogues predicted an uncleavable signal, suggesting that the paralogues are not posttranslationally processed at the N terminus (46). Alternatively, they are processed by a currently unknown borrelial protease, which recognizes a different cleavage site from that is other prokaryotes.
P13 was blocked for Edman degradation. Therefore, further experiments were performed to elucidate the nature of a possible modification, such as lipidation or glycosylation. Since P13 contained a cysteine at position 13, which, together with the surrounding amino acids (LATFC), constitutes a potential leader peptidase II cleavage-lipid modification site, experiments were performed to find possible lipidation. However, our results indicated that P13 is not lipidated or glycosylated, and the absence of fatty acid modification might be explained by the relatively early location of the cysteine, i.e., position 13, which would result in an unusually short putative signal peptide in P13. The lipoprotein acylation sites are usually located within the first 16 to 28 amino acids of borrelial lipoproteins (9, 26, 29, 35, 48, 49) and within the first 16 to 30 amino acids of gram-negative and gram-positive bacteria (21). Therefore, it would appear that fatty acids or carbohydrates do not posttranslationally modify P13.
The deduced molecular mass of the N-terminally signal peptidase I-cleaved P13 protein was 16.8 kDa, but the observed molecular mass as determined by SDS-PAGE was approximately 13 kDa. To address this discrepancy, C-terminal amino acid sequencing and MS analysis were employed. This analysis confirmed a mass of about 13.9 kDa. Given the molecular mass obtained by the ESMS analysis, in vitro translation analysis, and N- and C-terminal amino acid sequencing, we propose that P13 is posttranslationally processed. Further analyses are required to determine the nature of the N- and C-terminal processing. C-terminal processing is a phenomenon found in several bacterial species, such as E. coli, Azotobacter vinelandii, Synechocystis spp., Bartonella bacilliformis, and Desulfovibrio desulfuricans, but it is much more common in eukaryotes (17, 20, 31, 39). Interestingly, the Borrelia genome project identified a carboxyl-terminal protease (Ctp) (BB0359) based on sequence identity (30%) to Synechocystis sp. strain PCC6803. This putative borrelial Ctp had sequence identity (32%) to CtpA from B. bacilliformis. It is noteworthy that in D. desulfuricans ATCC 7757 the C-terminal processing of [Fe] hydrogenase was suggested to be involved in its export to the periplasm (20).
The genomic localization of p13 and the paralogous genes were investigated by computer analysis of the B. burgdorferi complete genome sequence. The p13 gene is localized to the chromosome, which was confirmed by pulsed-field electrophoresis, but the paralogous genes were carried on linear plasmids (lp54:BBA01, lp28-4:BBI31, lp38:J03, lp28-3:H41, and lp56:BBQ06). Furthermore, RNA blot analysis, using an internal p13-specific oligonucleotide as probe, revealed a transcript of about 600 bp. This size is consistent with a monocistronic P13 transcript encoded by the gene localized on the chromosome.
The p13 gene was compared between different genospecies of Lyme disease borrelia. The genes showed a very high degree of sequence conservation (87 to 90% sequence identity), which is typical among genes carried on the chromosome. Such a high degree of homology has also been found among the chromosomal p66 and the flaB genes (12, 34). In contrast, the plasmid-encoded major outer surface proteins OspA, OspB, and OspC exhibit a significant species- and strain-dependent genetic and antigenic polymorphism (6, 24, 51).
In conclusion, the p13 gene is chromosomal and the P13 protein is surface exposed. This is perhaps important for the pathogenesis and virulence of the bacteria. P13 appears to be surface exposed based on its sensitivity to proteinase treatments of intact cells, immunofluorescense, and immunogold examination using a P13-specific antiserum. As the sequence of P13 revealed TM helices, these results further strengthen the notion that this protein has one or several surface-exposed domains and that P13 is most probably associated with the outer membrane of the Lyme disease borreliae.
ACKNOWLEDGMENTS
This study was supported by Swedish Medical Research Council grant 07922, Swedish Council for Forestry and Agricultural Research grant 23.0161, Symbicom AB, Medical Science Council, the Royal Academy of Science (Olof Ahlöfs Foundation), and the J. C. Kempe Foundation.
We thank Alan Barbour for providing MAbs 15G6 and 7D4 and B. burgdorferi strains N40 and B313 and for discussions throughout the study, Robert Huebner for kindly providing plasmid pLF100, Per Ingvar Ohlsson for performing amino acid sequencing, Kerstin Jacobsson for performing the two-dimensional gel electrophoresis, and Gunilla Jäger for performing ESMS analysis. Åsa Schääf-Olsson, Linda Öberg, and Ingela Nilsson are gratefully acknowledged for skillful technical assistance, A. M. Cockshutt, J. Jass, and A. G. Barbour are acknowledged for critical reading of the manuscript.
Laila Noppa and Yngve Östberg contributed equally to this work.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Åsbrink E, Hovmark A, Hederstedt B. The spirochetal etiology of acrodermatitis chronica atrophicans Herxheimer. Acta Dermatol Venereol. 1984;64:506–512. [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G S, A. J, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1992. [Google Scholar]
- 4.Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J C, Assous M, Grimont P A. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- 5.Barbour A G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour A G, Tessier S L, Hayes S F. Variation in a major surface protein of Lyme disease spirochetes. Infect Immun. 1984;45:94–100. doi: 10.1128/iai.45.1.94-100.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbour A G, Tessier S L, Todd W J. Lyme disease spirochetes and ixodid tick spirochetes share a common surface antigenic determinant defined by a monoclonal antibody. Infect Immun. 1983;41:795–804. doi: 10.1128/iai.41.2.795-804.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barthold S W, Fikrig E, Bockenstedt L K, Persing D H. Circumvention of outer surface protein A immunity by host-adapted Borrelia burgdorferi. Infect Immun. 1995;63:2255–2261. doi: 10.1128/iai.63.6.2255-2261.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergström S, Bundoc V G, Barbour A G. Molecular analysis of linear plasmid-encoded major surface proteins, OspA and OspB, of the Lyme disease spirochaete Borrelia burgdorferi. Mol Microbiol. 1989;3:479–486. doi: 10.1111/j.1365-2958.1989.tb00194.x. [DOI] [PubMed] [Google Scholar]
- 10.Brandt M E, Riley B S, Radolf J D, Norgard M V. Immunogenic integral membrane proteins of Borrelia burgdorferi are lipoproteins. Infect Immun. 1990;58:983–991. doi: 10.1128/iai.58.4.983-991.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunikis J, Barbour A G. Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect Immun. 1999;67:2874–2883. doi: 10.1128/iai.67.6.2874-2883.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunikis J, Noppa L, Bergström S. Molecular analysis of a 66-kDa protein associated with the outer membrane of Lyme disease Borrelia. FEMS Microbiol Lett. 1995;131:139–145. doi: 10.1111/j.1574-6968.1995.tb07768.x. [DOI] [PubMed] [Google Scholar]
- 13.Canica M M, Nato F, du Merle L, Mazie J C, Baranton G, Postic D. Monoclonal antibodies for identification of Borrelia afzelii sp. nov. associated with late cutaneous manifestations of Lyme borreliosis. Scand J Infect Dis. 1993;25:441–448. doi: 10.3109/00365549309008525. [DOI] [PubMed] [Google Scholar]
- 14.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 15.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith H O, Venter J C. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 16.Ge Y, Li C, Corum L, Slaughter C A, Charon N W. Structure and expression of the FlaA periplasmic flagellar protein of Borrelia burgdorferi. J Bacteriol. 1998;180:2418–2425. doi: 10.1128/jb.180.9.2418-2425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gollin D J, Mortenson L E, Robson R L. Carboxyl-terminal processing may be essential for production of active NiFe hydrogenase in Azotobacter vinelandii. FEBS Lett. 1992;309:371–375. doi: 10.1016/0014-5793(92)80809-u. [DOI] [PubMed] [Google Scholar]
- 18.Göransson M, Forsman P, Nilsson P, Uhlin B E. Upstream activating sequences that are shared by two divergently transcribed operons mediate cAMP-CRP regulation of pilus-adhesin in Escherichia coli. Mol Microbiol. 1989;3:1557–1565. doi: 10.1111/j.1365-2958.1989.tb00141.x. [DOI] [PubMed] [Google Scholar]
- 19.Hansson L, Noppa L, Nilsson A K, Strömqvist M, Bergström S. Expression of truncated and full-length forms of the Lyme disease Borrelia outer surface protein A in Escherichia coli. Protein Expr Purif. 1995;6:15–24. doi: 10.1006/prep.1995.1003. [DOI] [PubMed] [Google Scholar]
- 20.Hatchikian E C, Magro V, Forget N, Nicolet Y, Fontecilla-Camps J C. Carboxy-terminal processing of the large subunit of [Fe] hydrogenase from Desulfovibrio desulfuricans ATCC 7757. J Bacteriol. 1999;181:2947–2952. doi: 10.1128/jb.181.9.2947-2952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 22.Honarvar N, Schaible U E, Galanos C, Wallich R, Simon M M. A 14,000 MW lipoprotein and a glycolipid-like structure of Borrelia burgdorferi induce proliferation and immunoglobulin production in mouse B cells at high frequencies. Immunology. 1994;82:389–396. [PMC free article] [PubMed] [Google Scholar]
- 23.Jonsson M, Bergström S. Transcriptional and translational regulation of the expression of the major outer surface proteins in Lyme disease Borrelia strains. Microbiology. 1995;141:1321–1329. doi: 10.1099/13500872-141-6-1321. [DOI] [PubMed] [Google Scholar]
- 24.Jonsson M, Noppa L, Barbour A G, Bergström S. Heterogeneity of outer membrane proteins in Borrelia burgdorferi: comparison of osp operons of three isolates of different geographic origins. Infect Immun. 1992;60:1845–1853. doi: 10.1128/iai.60.5.1845-1853.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katona L I, Beck G, Habicht G S. Purification and immunological characterization of a major low-molecular-weight lipoprotein from Borrelia burgdorferi. Infect Immun. 1992;60:4995–5003. doi: 10.1128/iai.60.12.4995-5003.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornacki J A, Oliver D B. Lyme disease-causing Borrelia species encode multiple lipoproteins homologous to peptide-binding proteins of ABC-type transporters. Infect Immun. 1998;66:4115–4122. doi: 10.1128/iai.66.9.4115-4122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kryuchechnikov V N, Korenberg E I, Scherbakov S V, Kovalevsky Iu V, Levin M L. Identification of Borrelia isolated in the USSR from Ixodes persulcatus (Schulze) ticks. J Microbiol Epidemiol Immunobiol. 1988;12:41–44. [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lam T T, Nguyen T P, Montgomery R R, Kantor F S, Fikrig E, Flavell R A. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect Immun. 1994;62:290–298. doi: 10.1128/iai.62.1.290-298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magnarelli L A, Anderson J F, Barbour A G. Enzyme-linked immunosorbent assays for Lyme disease: reactivity of subunits of Borrelia burgdorferi. J Infect Dis. 1989;159:43–49. doi: 10.1093/infdis/159.1.43. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell S J, Minnick M F. A carboxy-terminal processing protease gene is located immediately upstream of the invasion-associated locus from Bartonella bacilliformis. Microbiology. 1997;143:1221–1233. doi: 10.1099/00221287-143-4-1221. [DOI] [PubMed] [Google Scholar]
- 32.Montgomery R R, Malawista S E, Feen K J, Bockenstedt L K. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J Exp Med. 1996;183:261–269. doi: 10.1084/jem.183.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Noppa L, Burman N, Sadziene A, Barbour A G, Bergström S. Expression of the flagellin gene in Borrelia is controlled by an alternative sigma factor. Microbiology. 1995;141:85–93. doi: 10.1099/00221287-141-1-85. [DOI] [PubMed] [Google Scholar]
- 35.Norris S J, Carter C J, Howell J K, Barbour A G. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun. 1992;60:4662–4672. doi: 10.1128/iai.60.11.4662-4672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parveen N, Leong J M. Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:1220–1234. doi: 10.1046/j.1365-2958.2000.01792.x. [DOI] [PubMed] [Google Scholar]
- 37.Peter O, Bretz A G. Polymorphism of outer surface proteins of Borrelia burgdorferi as a tool for classification. Int J Med Microbiol Virol Parasitol Infect Dis. 1992;277:28–33. doi: 10.1016/s0934-8840(11)80867-4. [DOI] [PubMed] [Google Scholar]
- 38.Radolf J D, Bourell K W, Akins D R, Brusca J S, Norgard M V. Analysis of Borrelia burgdorferi membrane architecture by freeze- fracture electron microscopy. J Bacteriol. 1994;176:21–31. doi: 10.1128/jb.176.1.21-31.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossmann R, Sauter M, Lottspeich F, Böck A. Maturation of the large subunit (HYCE) of Escherichia coli hydrogenase 3 requires nickel incorporation followed by C-terminal processing at Arg537. Eur J Biochem. 1994;220:377–384. doi: 10.1111/j.1432-1033.1994.tb18634.x. [DOI] [PubMed] [Google Scholar]
- 40.Sadziene A, Rosa P A, Thompson P A, Hogan D M, Barbour A G. Antibody-resistant mutants of Borrelia burgdorferi: in vitro selection and characterization. J Exp Med. 1992;176:799–809. doi: 10.1084/jem.176.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadziene A, Thomas D D, Barbour A G. Borrelia burgdorferi mutant lacking Osp: biological and immunological characterization. Infect Immun. 1995;63:1573–1580. doi: 10.1128/iai.63.4.1573-1580.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambri V, Moroni A, Massaria F, Brocchi E, De Simone F, Cevenini R. Immunological characterization of a low molecular mass polypeptidic antigen of Borrelia burgdorferi. FEMS Microbiol Immunol. 1991;3:345–349. doi: 10.1111/j.1574-6968.1991.tb04260.x. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Steere A C. Lyme disease. N Engl J Med. 1989;321:586–596. doi: 10.1056/NEJM198908313210906. [DOI] [PubMed] [Google Scholar]
- 45.Tusnády G E, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 46.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker E M, Borenstein L A, Blanco D R, Miller J N, Lovett M A. Analysis of outer membrane ultrastructure of pathogenic Treponema and Borrelia species by freeze-fracture electron microscopy. J Bacteriol. 1991;173:5585–5588. doi: 10.1128/jb.173.17.5585-5588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallich R, Brenner C, Kramer M D, Simon M M. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect Immun. 1995;63:3327–3335. doi: 10.1128/iai.63.9.3327-3335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallich R, Simon M M, Hofmann H, Moter S E, Schaible U E, Kramer M D. Molecular and immunological characterization of a novel polymorphic lipoprotein of Borrelia burgdorferi. Infect Immun. 1993;61:4158–4166. doi: 10.1128/iai.61.10.4158-4166.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wessel D, Flugge U I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 51.Wilske B, Preac-Mursic V, Jauris S, Hofmann A, Pradel I, Soutschek E, Schwab E, Will G, Wanner G. Immunological and molecular polymorphisms of OspC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect Immun. 1993;61:2182–2191. doi: 10.1128/iai.61.5.2182-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Y, Kodner C, Coleman L, Johnson R C. Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect Immun. 1996;64:3870–3876. doi: 10.1128/iai.64.9.3870-3876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]