When tropical plants are mentioned, palm trees first come to mind. Plants in the Arecaceae are indeed one of the most characteristic plants in the tropics. They are not only important oil crops but also important garden and medicinal plants. The Arecaceae includes 181 genera and about 2500 species, among which the more famous are coconut, date palm, oil palm, sugar palm, and fishtail palm.
The past decade has seen rapid development in ‐omics studies of the Arecaceae. The genomes of 6 species (Elaeis guineensis Jacq., Phoenix Dactylifera L., Cocos nucifera L., Areca catechu L., Calamus simplicifolius, and Daemonorops jenkinsiana) have been released, together with the generation of a large number of resequencing and transcriptome data (Al‐Mssallem et al., 2013; Singh et al., 2013; Wang et al., 2021; Yang et al., 2021; Zhao et al., 2018). However, there is currently no functional database integrating multi‐omics for Arecaceae.
Therefore, we have developed the first userfriendly multi‐omics database for Arecaceae (Arecaceae Multi‐omics Database, ArecaceaeMDB, http://arecaceae‐gdb.com). ArecaceaeMDB contains 7 genome sequences from 6 species, resequencing data of 1631 accessions and 866 spatiotemporal transcriptome data. In addition, we have obtained 138 spatiotemporal metabolome data using LC‐MS–based metabolic profiling. We have further developed 12 frequently used online tools to facilitate the use of ArecaceaeMDB, such as Sequence Fetch, Blast, SynVisio, and Enrichment Analysis (Figure 1a).
Figure 1.
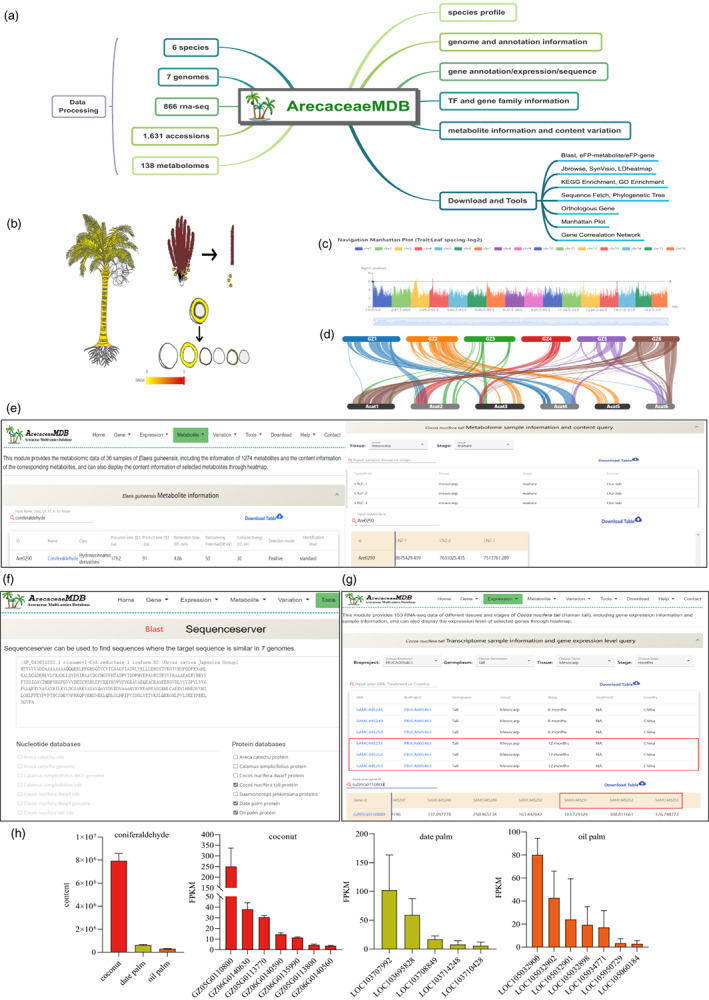
Overview of ArecaceaeMDB and its functions in Arecaceae multi‐omics analysis. (a) The flow diagram of ArecaceaeMDB construct. (b) The eFP tool displays gene expression levels in various coconut tissues. (c) The Manhattan plot tools display the GWAS result of the leaf internode phenotype of coconut. (d) The SynVisio tool shows the collinear relationship between the six chromosomes of coconut and areca palm. (e) Steps to obtain the content information of coniferaldehyde in the mesocarp of coconut in the Metabolite module. (f) Using blast tool to obtain the homologous gene of CCR gene in coconut. (g) Steps to obtaining expression data of homologous genes of the CCR gene in coconut mesocarp in the Expression module. (h) The content of coniferaldehyde in the mesocarp of coconut, oil palm, and date palm, and the expression level of homologous genes of CCR gene in the mesocarp of the three species.
ArecaceaeMDB consists of four main modules: (1) Gene, (2) Expression, (3) Metabolite, and (4) Variation. The gene module presents 7 genomes from 6 species. Users can get the following information: the basic information of genome assembly and annotation, the location and length information of each gene and the corresponding protein sequence and CDS sequence, whole genome transcription factor family and important gene families, the functional annotation of genes and its homologous genes and their functions in Arabidopsis thaliana and Oryza sativa, and COG, GO, and KEGG information of genes.
In the Expression module, ArecaceaeMDB provides gene expression data (FPKM value) of 866 samples. Users can search by project number, strain, tissue, or stage and obtain the gene expression data in some samples by submitting the gene ID, and create a heatmap for their genes of interest.
The metabolite module provides users a detailed description and content information on 1274 metabolites of 138 samples of four species (oil palm, date palm, coconut, and areca palm). When the user clicks on the name of the metabolite, it can automatically jump to the location of the compound in the NCBI Pubchem database and thus obtain more compound information.
In the variation module, the database presents variations of resequencing data of 1631 accessions, including SNPs and InDels. It supports two search methods, by gene ID or variation ID. When searching by gene ID, users can select a specific project first, and then enter a gene ID, then select the upstream and downstream range and variation type, and the variation information can be displayed in the form of figures and tables. When searching by variation ID, users select a specific project and desired samples, and the variation information will be presented in table form. We also provide the basic information about samples in different projects, such as age, germplasm, and origin.
In addition to the main modules, ArecaceaeMDB contains 12 popular bioinformatics tools for Arecaceae data mining. For instance, the eFP displays the selected metabolite accumulation patterns and gene expression patterns by dynamically colouring the tissues of four species (oil palm, date palm, coconut, and areca palm) according to metabolite content and gene expression levels (Figure 1b). The Manhattan plot tool not only provides manhattan plots of project‐specific GWAS results but also provides information on the SNPs corresponding to the peaks (Figure 1c). The SynVisio tool is used to show the synteny relationships between the 7 genomes of 6 species (Figure 1d). Additionally, the ‘Download’ section allows users to freely obtain all the data used in ArecaceaeMDB.
A user case picked from ArecaceaeMDB was presented in Figure 1e–h. It is known that the fibre content of the mesocarp of coconut, oil palm, and date palm varies widely, and coniferaldehyde is an important metabolite in the lignin synthesis pathway. By analysing the metabolic data of the mesocarp of coconut, date palm, and oil palm in the metabolite module (Figure 1e), we found that the content of coniferaldehyde in coconut was significantly higher than that of date palm and oil palm (Figure 1h). It is known that the CCR gene regulates the accumulation of this metabolite. We obtained the homologous genes of the CCR genes of the three species through the blast tool (Figure 1f), combined with the transcriptome data of the mesocarp of coconut, date palm, and oil palm in the expression module (Figure 1g), we found that the expression levels of the CCR homologous genes in coconut mesocarp were significantly higher than those in oil palm and date palm (Figure 1h), indicating that the transcriptome results were consistent with the metabolome results.
In summary, we present ArecaceaeMDB, a useful platform that integrates multi‐omics data of Arecaceae, which provides a valuable resource for functional genome research and genetic breeding research of the Arecaceae family. In the future, ArecaceaeMDB will continuously update adding more published data, and adding other layers of ‐omics data (proteomics, epiomics, noncoding RNA, and so on) and further data analytical tools. We intend to make ArecaceaeMDB a central community portal through Arecaceae research worldwide and will provide long‐term support, thus benefiting both research and industrial application of the study of Arecaceae.
Conflict of interests
The authors declare that they have no conflict of interest.
Author contributions
J.L. designed the project. Z.Y., Z.H.L., H.X., Y.F.L., S.H.H., J.J.Z., J.J.Z., and X.Q.L. prepared the samples. Z.Y., Z.H.L., H.X., Y.F.L., S.H.H., M.W.S., and R.Z.L. analysed the data. Z.Y., Z.H.L., H.X., Y.F.L., and S.H.H. wrote the manuscript. J.L. revised the manuscript. A.R.F., Y.X., G.P.C., and Y.H.D. helped to modify the article.
Acknowledgements
This research was supported by Hainan Yazhou Bay Seed Lab. + (JBGS + B21HJ0903), Hainan Science and Technology Cooperation Project (ZDYF2020215), Project of the Administrative Bureau of Sanya Yazhou Bay Science and Technology City (HNF202201), Hainan Major Science and Technology Project (No. ZDKJ202002), ‘111’ Project (No. D20024), Hainan Academician Innovation Platform HD‐YSZX‐202003, Hainan University Startup Fund KYQD(ZR)1866 to j.L, and the innovation Project of Postgraduates of Hainan Province (no. Hyb2020‐08 and no. Qhyb2021‐31).
Thanks for the bioinformatics platform of the National Key Laboratory of Crop Genetic Improvement, HZAU.
References
- Al‐Mssallem, I.S. , Hu, S. , Zhang, X. , Lin, Q. , Liu, W. , Tan, J. et al. (2013) Genome sequence of the date palm Phoenix dactylifera L. Nat. Commun. 4, 2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. , Ong‐Abdullah, M. , Low, E.T. , Manaf, M.A. , Rosli, R. , Nookiah, R. et al. (2013) Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature, 500, 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Xiao, Y. , Zhou, Z.W. , Yuan, J. , Guo, H. , Yang, Z. , Yang, J. et al. (2021) High‐quality reference genome sequences of two coconut cultivars provide insights into evolution of monocot chromosomes and differentiation of fiber content and plant height. Genome Biol. 22, 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Huang, L. , Xu, C. , Qi, L. , Wu, Z. , Li, J. , Chen, H. et al. (2021) Chromosome‐scale genome assembly of areca palm (Areca catechu). Mol. Ecol. Resour. 21, 2504–2519. [DOI] [PubMed] [Google Scholar]
- Zhao, H. , Wang, S. , Wang, J. , Chen, C. , Hao, S. , Chen, L. , Fei, B. et al. (2018) The chromosome‐level genome assemblies of two rattans (Calamus simplicifolius and Daemonorops jenkinsiana). Gigascience, 7(9), giy097. 10.1093/gigascience/giy097 [DOI] [PMC free article] [PubMed] [Google Scholar]


