Abstract
Animal studies with Streptococcus pneumoniae have provided valuable models for drug development. In order to monitor long-term pneumococcal infections noninvasively in living mice, a novel gram-positive lux transposon cassette, Tn4001 luxABCDE Kmr, that allows random integration of lux genes onto the bacterial chromosome was constructed. The cassette was designed so that the luxABCDE and kanamycin resistance genes were linked to form a single promoterless operon. Bioluminescence and kanamycin resistance only occur in a bacterial cell if this operon has transposed downstream of a promoter on the bacterium's chromosome. S. pneumoniae D39 was transformed with plasmid pAUL-A Tn4001 luxABCDE Kmr, and a number of highly bioluminescent colonies were recovered. Genomic DNA from the brightest D39 strain was used to transform a number of clinical S. pneumoniae isolates, and several of these strains were tested in animal models, including a pneumococcal lung infection model. Strong bioluminescent signals were seen in the lungs of the animals containing these pneumococci, allowing the course and antibiotic treatment of the infections to be readily monitored in real time in the living animals. Recovery of the bacteria from the animals showed that the bioluminescent signal corresponded to the number of CFU and that the lux construct was highly stable even after several days in vivo. We believe that this lux transposon will greatly expand the ability to evaluate drug efficacy against gram-positive bacteria in living animals using bioluminescence.
Streptococcus pneumoniae is the leading cause of invasive bacterial disease in the very young and the elderly and is the bacterium most responsible for community-acquired pneumonia in the developed world (31). It can behave as a transient commensal, colonizing the nasopharynx of 40% of healthy adults and children, with no adverse effects (2). Children carry this pathogen in the nasopharynx asymptomatically for about 4 to 6 weeks, often carrying several serotypes at a time (13, 33). Occasionally, perhaps in conjunction with a viral infection (9), one of these strains gives rise to a symptomatic pneumococcal infection, including sinusitis, otitis media, pneumonia, and meningitis (12, 13, 16).
Antibiotic treatment of pneumococcal infections has been less effective in recent years with the increased occurrence of multidrug-resistant strains of S. pneumoniae. About one-third to one-half of pneumococci recovered from humans are at least partially resistant to penicillin, which may occur in addition to resistance to a number of other common antibiotics (1, 3). These factors, plus the ability of the pneumococcus to transfer genes for resistance, encapsulation, and virulence via transformation (12), make it imperative to develop a better understanding of the mechanism by which pneumococci cause disease. Probably the best way to enhance this process is to develop a better animal model.
In 1995, Contag et al. (7) showed that it was possible to monitor disease processes in living animals using bioluminescence. These initial studies were conducted using the gram-negative pathogen Salmonella enterica serovar Typhimurium and demonstrated that bacterial growth and drug efficacy could be monitored using this novel, noninvasive technology. Recently, we have successfully modified and expressed the Photorhabdus luminescence lux operon in gram-positive bacteria. Broad-host-range shuttle vectors carrying this modified lux operon were used to transform and monitor pathogenic strains of Staphylococcus aureus in vivo in living animals using bioluminescence (11). Despite this achievement, animal studies conducted with bioluminescent bacteria containing plasmid-based lux constructs only allow short-term (<48 h) infections to be accurately monitored in vivo in animals due to plasmid loss in the absence of antibiotic selection. Thus, our objective was to develop efficient methods of integrating the modified luxABCDE operon into the chromosome of gram-positive bacteria, so that the bioluminescent signal would be stabilized and more accurately reflect the number of viable bacterial cells present in vivo in an animal.
In this article we describe the building of a gram-positive lux transposon, Tn4001 luxABCDE Kmr, that can be used for stable bioluminescent transformation of a wide range of gram-positive bacteria and show how this transposon was used to generate bioluminescent strains of S. pneumoniae. Using a pneumococcal lung model, we demonstrate the advantages of employing these bioluminescent strains to study S. pneumoniae disease in live animals.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strain DH5α was obtained from Life Technologies (Rockville, Md.) and routinely grown at 37°C. S. pneumoniae strain D39 was obtained from Tom Parr (Eli Lilly, Indianapolis, Ind.). Clinical isolates of S. pneumoniae A66.1, HUSTMBIG, EF3030, and 140301 were obtained from Marc Lipsitch (Harvard School of Public Health, Boston, Mass.) and David Briles (University of Alabama at Birmingham, Birmingham, Ala.). All the S. pneumoniae strains were grown at 37°C in 5% CO2 unless otherwise stated.
Construction of gram-positive lux transposon plasmid pAUL-A Tn4001 luxABCDE (Kmr).
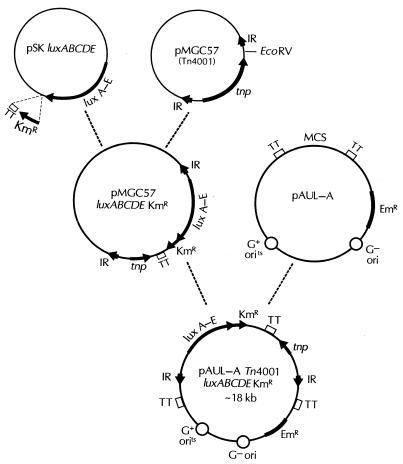
A diagram depicting the construction of the plasmid pAUL-A Tn4001 luxABCDE (Kmr) is shown in Fig. 1. A P. luminescens lux operon previously altered to be functional in gram-positive bacteria (11) was modified by linking a promoterless gram-positive kanamycin resistance cassette downstream of luxE. The kanamycin resistance gene was PCR amplified from pDL289 (4) using primers KanF2 (5′ CTG TAG ACT CGA GGA GGG AAA TAA TAA ATG GC 3′) and KanR2 (5′ CAG AGT GTC GAC AGT TGC GGA TGT AC 3′) (underlined sequences correspond to XhoI and SalI restriction enzyme sites in KanF2 and KanR2, respectively, which were introduced to aid subsequent cloning steps). PCR conditions for this and subsequent DNA amplifications were those described previously by Francis et al. (11).
FIG. 1.
Construction of plasmid pAUL-A Tn4001 luxABCDE Kmr. For details, see Materials and Methods. IR, inverted repeat; EmR, erythromycin resistance gene; tnp, transposase gene; TT, transcription terminator; MCS, multiple cloning site.
The amplified kanamycin resistance gene was digested with XhoI and SalI and ligated into the SalI site of pBluescript II SK− luxABCDE (11). The ligation was electroporated into E. coli, spread onto Luria-Bertani (LB) agar containing kanamycin (50 μg/ml), and incubated overnight at 37°C. Plates with kanamycin-resistant colonies were screened for bioluminescence using a photon-counting intensified-charge-coupled device (ICCD) camera (model 2400-32; Hamamatsu Photonics, Bridgewater, N.J.). Plasmid DNA was isolated from bioluminescent, kanamycin-resistant colonies using a standard alkaline lysis procedure (Plasmid Spin Miniprep kit; Qiagen, Valencia, Calif.), as were all subsequent plasmid isolations. These plasmids were then screened by PCR using the primers KanF2 and M13F (5′ GTA AAA CGA CGG CCA GT 3′), flanking the multiple cloning site of pBluescript II SK− (Strategene, La Jolla, Calif.), to identify constructs in which the luxABCDE cassette was in the same orientation (5′ to 3′) as the kanamycin resistance gene.
In order to construct a gram-positive lux transposon, the luxABCDE Kmr cassette described above was moved from pBluescript SK− luxABCDE Kmr into pMGC57, a plasmid containing the Staphylococcus aureus transposon Tn4001 (21). pBluescript SK− luxABCDE Kmr was cut with SpeI and SalI to remove the luxABCDE Kmr cassette, and the DNA was blunt-end filled using T4 polymerase. This blunt-end digest was then ligated with pMGC57 that had previously been cut with EcoRV, a unique restriction site lying just inside the inner inverted repeat sequence. The ligation reaction mix was electroporated into E. coli DH5α and spread onto LB agar containing chloramphenicol (30 μg/ml) (pMGC57 contains a chloramphenicol resistance cassette), and the resulting colonies were screened for bioluminescence using an ICCD camera. After confirming that the bioluminescent, chloramphenicol-resistant colonies were also kanamycin resistant by patching these colonies onto LB agar containing kanamycin (50 μg/ml), plasmid DNAs were isolated from cultures of these clones as described above. PCR was then conducted using primers MGC-CAT-F1 (5′ GGT GTC CCT GTT GAT ACC G 3′) and LuxA-Rev (5′ CCA CAC TCC TCA GAG ATG CG 3′) to identify which of the plasmids had the luxABCDE Kmr cassette in the correct orientation in pMGC57 (i.e., the 5′ end of luxA lying directly downstream of the Tn4001 inner inverted repeat sequence; see Fig. 1).
To increase the efficiency of transposition in gram-positive bacteria, the above Tn4001 luxABCDE Kmr cassette was moved onto the temperature-sensitive gram-positive/negative shuttle vector pAUL-A (6). The Tn4001 luxABCDE Kmr cassette was cut from pMGC57 using the enzymes EcoRI and XhoI and ligated into the corresponding EcoRI and SalI sites of pAUL-A. The ligation was electroporated into E. coli DH5α and spread onto LB agar containing erythromycin (150 μg/ml) (pAUL-A contains an erythromycin resistance cassette), and the resulting colonies were screened for bioluminescence using an ICCD camera. After confirming that the bioluminescent, erythromycin-resistant colonies were also kanamycin resistant, again by patching these colonies onto LB containing kanamycin (50 μg/ml), plasmid DNAs were isolated as described above. These plasmid DNAs were then used to transform S. pneumoniae strain D39.
Screening for stable, highly bioluminescent S. pneumoniae D39 Tn4001 luxABCDE transposants.
The plasmid pAUL-A Tn4001 luxABCDE Kmr was electroporated into S. pneumoniae D39 as described previously (18). The transformation mix was plated on chocolate agar containing erythromycin (0.3 μg/ml), and the plates were incubated for 24 to 48 h. Transformants of S. pneumoniae D39 containing pAUL-A Tn4001 luxABCDE Kmr were patched onto chocolate agar plates containing erythromycin (0.3 μg/ml) and incubated overnight. A quantity of each patch (10-μl loopful of cell growth consisting of approximately 108 to 109 cells) was then uniformly streaked over the entire area of a chocolate agar plate containing kanamycin (400 μg/ml) and incubated for 24 to 48 h. Alternatively, S. pneumoniae D39 pAUL-A Tn4001 luxABCDE Kmr was cultured overnight in 10 ml of brain heart infusion (BHI) containing erythromycin (0.3 μg/ml), pelleted, and resuspended in an equal volume of BHI, and 100-μl volumes of a 10-fold dilution range (100 to 10−8 in BHI) were spread onto a chocolate agar plate routinely containing kanamycin (400 μg/ml) (up to 1,000 μg/ml in some instances) and incubated for 24 to 48 h. The resulting colonies were screened for bioluminescence using an ICCD camera, and the brightest were streaked onto chocolate agar plates containing kanamycin (400 μg/ml). Single colonies were streaked subsequent times onto chocolate plates containing no antibiotics to verify that bioluminescence was stable in the absence of antibiotic selection. Each strain was then graded for its level of bioluminescence using an ICCD camera and Xenogen's LivingImage software (Xenogen Corporation, Alameda, Calif.).
Assessment of bioluminescence versus optical density from S. pneumoniae Tn4001 luxABCDE transposants.
A single bioluminescent colony of S. pneumoniae was inoculated in 10 ml of BHI medium. Three hundred microliters of overnight culture was inoculated in 30 ml of BHI and grown at 37°C in 5% CO2. At 1-h intervals, both the optical density (OD) and the number of relative light units (RLU) from a 1-ml culture volume were determined. The curves of RLU versus OD at 600 nm (OD600) were plotted.
Transformation of clinical S. pneumoniae using natural competence.
Chromosomal DNA from highly bioluminescent S. pneumoniae D39 Tn4001 luxABCDE Kmr was used for natural transformation of several clinical isolates and prepared as described previously (24). S. pneumoniae clinical isolates were grown in 10 ml of BHI supplemented with 5% horse serum until the OD600 reached 0.1 to 0.15. The culture was diluted 1:100 in 10 ml of warmed BHI supplemented with 5% horse serum containing CSP1 (EMRLSKFFRDFILQRKK) (15) or CSP2 (EMRISRIILDFLFLRKK) (30) and incubated for 20 min. One microgram of the above chromosomal DNA was added to 500 μl of competent cells, and the mixture was incubated for another 3 h. The transformation mix was then plated on chocolate agar plates containing kanamycin at 400 μg/ml and incubated for 48 h. Colonies arising were screened for bioluminescence using an ICCD camera.
Southern blot analysis of S. pneumoniae Tn4001 luxABCDE Kmr chromosomal DNA.
To examine the integration of Tn4001 luxABCDE Kmr into the S. pneumoniae chromosome, Southern blot analysis was conducted according to the manufacturer's instructions (Alkphos direct labeling kit; Amersham Pharmacia Biotech, Piscataway, N.J.). Chromosomal DNA from both D39 and clinical isolates of S. pneumoniae Tn4001 luxABCDE Kmr were digested with a number of restriction enzymes. These digestions were run on an agarose gel, and the DNA was blotted onto a nitrocellulose filter and probed. The probe used to hybridize to the target locus was a PCR-amplified luxA fragment amplified from pSK luxAB using primers XAF3 (5′ CCC CGG ATC CTG CAG ATG AAG CAA GAG GAG GAC TCT CTA TG 3′) and XAR (5′ GGC GGA TCC GTC GAC TTA ATA TAA TAG CGA ACG TTG 3′) as described previously (11).
Inverse PCR.
The genomic DNA sequence lying upstream of each Tn4001 luxABCDE Kmr integration site was obtained by inverse PCR (27). Chromosomal DNA from S. pneumoniae Tn4001 luxABCDE Kmr was digested with a number of different restriction enzymes (both 4- and 6-bp recognition enzymes) and self-ligated. The upstream region was PCR amplified using primers R2 (5′ CGT TTC ATT ACC TCT GTT TGA G 3′) and XBF (5′ GGG AAT TCT CGA GGA GGA GAG AAA GAA ATG AAA TTT GGA 3′). Resulting PCR products were purified (PCR purification kit; Qiagen) and directly sequenced using primer R2.
Monitoring bioluminescent pneumococcal infections in the lungs, and nasal passaging in live mice.
Exponential cultures of S. pneumoniae grown at 37°C in BHI were pelleted and then resuspended in fresh BHI broth. Bacterial concentrations were estimated spectrophotometrically by absorbance at 600 nm and adjusted to approximately 107 CFU/ml by dilution BHI. Cell numbers were verified by plating dilutions of inoculum onto BHI agar.
Pneumococci were introduced into the lungs of mice (BALB/c females, 8 to 11 weeks old) by either direct intratracheal inoculation or intranasal administration. The animals were anesthetized with ketamine (100 mg/ml) and xylazine (20 mg/ml), mixed at a 4:1 (vol/vol) ratio just before use. The anesthesia mixture was injected intramuscularly into the right hamstring muscle at a dose of 100 mg of ketamine/kg of body weight. After anesthesia was established, the mice were inoculated with approximately 106 CFU in a total volume of 20 μl, using a 25-gauge ball-tipped gavage needle. Mice were held in vertical suspension for 10 min after inoculation to facilitate deep penetration of the inoculum. Alternatively, mice were infected intranasally with approximately 106 CFU by placing 20 μl of bacterial suspension on the nares and allowing the mice to inhale the inoculum. In the antibiotic studies, mice were treated with amoxicillin at 1 or 5 mg/kg, given subcutaneously at 0, 18, 24, and 42 h postinfection.
Mice were imaged for a maximum of 5 min at a number of time points postinfection using an IVIS CCD camera (Xenogen Corporation). Total photon emission from selected and defined areas within the images of each mouse was quantified using the LivingImage software package (Xenogen Corporation). The photon signal from the thorax was quantified from the ventral image of each mouse.
Extraction and quantification of bacteria from lungs and nasal passages of mice.
After the final imaging time point, mice were sacrificed, and the infected lung tissue was surgically removed and weighed. Both lungs were homogenized together in 500 μl of BHI in a loose Dounce homogenizer. The suspension was serially diluted in BHI and plated in duplicate onto BHI agar. Bacterial burden was estimated from the number of CFU per gram of lung tissue.
The bacterial load present in the nasal passage of the mice was obtained by washing approximately 0.25 ml of saline through the trachea and collecting the first 0.1 ml of nasal wash as it exited the nares, as described previously (19, 37).
RESULTS
Both lux and Kmr genes are silent on pAUL-A in S. pneumoniae.
Although the luxABCDE Kmr operon inserted in pAUL-A is flanked on both sides by strong transcriptional terminator sequences (Fig. 1), which should act to silence gene expression (6), we found the regulation of this plasmid-located operon to differ between E. coli and S. pneumoniae. E. coli transformed with pAUL-A Tn4001 luxABCDE Kmr was uniformly bioluminescent, kanamycin resistant, and erythromycin resistant (100% of all transformants initially selected on erythromycin). Since the transformation frequency of these cells was similar to that of E. coli transformed with the empty pAUL-A plasmid (approximately 500 CFU/μg of DNA), and pAUL-A Tn4001 luxABCDE Kmr DNA could be readily extracted from the transformants by standard plasmid isolation procedures, it is unlikely that lux Kmr gene expression is due to integration of the plasmid into the E. coli chromosome. The more probable explanation for this gene activity is that the luxABCDE Kmr operon is expressed on pAUL-A in E. coli despite the lack of any apparent promoter upstream of these genes.
In contrast to the plasmid gene expression found in E. coli, in S. pneumoniae the plasmid-located luxABCDE Kmr operon was silent. S. pneumoniae transformants containing pAUL-A Tn4001 luxABCDE Kmr were dark (nonbioluminescent) when selected on chocolate plates containing erythromycin (0.3 μg/ml). Furthermore, the growth rate of an exponentially dividing culture of such a transformant was dramatically inhibited by the addition of kanamycin at concentrations of 400 μg/ml and higher. However, after several hours in the presence of kanamycin, the growth of the pneumococcal culture reestablished itself at a rate similar to that found prior to the addition of this antibiotic. Moreover, a low level of bioluminescence could be recorded from the latter culture. Together, these data indicate that a subpopulation of the original culture, one in which the luxABCDE Kmr operon was induced, had been selected.
Promoter strength can be selected for by varying the kanamycin concentration in the medium during isolation of chromosomal integrants.
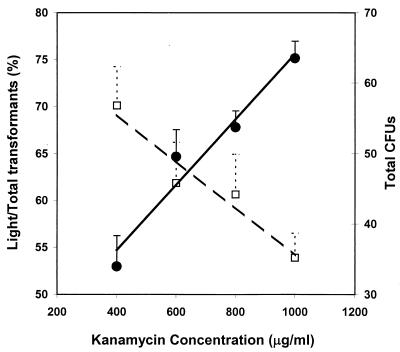
S. pneumoniae pAUL-A Tn4001 luxABCDE Kmr plated at 103 CFU on chocolate agar plates containing increasing concentrations of kanamycin (400 to 1,000 μg/ml) gave decreasing numbers of colonies. In comparison, the same density of cells plated on chocolate plates without kanamycin resulted in approximately 1,000 CFU. Although increasing the concentration of kanamycin in the medium gave rise to fewer transformants, the proportion of colonies producing higher levels of bioluminescence was found to increase (Fig. 2). This indicates that increasing the concentration of kanamycin in the medium results in the selection of fusions with stronger promoters upstream of the luxABCDE Kmr operon. Even in the absence of kanamycin selection (screening on blank chocolate plates), approximately 3% of the 1,000 or so colonies were bioluminescent. This level of transposition seen in S. pneumoniae was significantly higher (approximately 1,000-fold) than that observed in studies involving pAUL-A Tn4001 luxABCDE Kmr in Staphylococcus aureus and Listeria monocytogenes (data not shown).
FIG. 2.
Effects of kanamycin concentration on selection of promoter strength. S. pneumoniae D39 Xen 7 was grown to mid-exponential phase, and 103 CFU were plated on chocolate agar plates supplemented with various concentrations of kanamycin. Open squares, total CFU on the plates; solid circles, ratio of light to total CFU. The numbers of CFU are the averages for five plates at each kanamycin concentration.
Tn4001 luxABCDE Kmr is randomly inserted into the chromosome of S. pneumoniae and stably maintained.
One thousand bioluminescent, kanamycin-resistant S. pneumoniae D39 colonies were patched in duplicate onto chocolate plates containing either erythromycin (0.3 μg/ml) or kanamycin (400 μg/ml). All but 20 colonies grew on both antibiotics, indicating that the vast majority (98%) of the transformants had the entire pAUL-A Tn4001 luxABCDE Kmr DNA integrated into their chromosome. Southern blot analysis of genomic DNA from 10 kanamycin- and erythromycin-resistant colonies confirmed that the whole plasmid had integrated into the S. pneumoniae chromosome. In contrast, Southern blot analysis of genomic DNA from 10 kanamycin-resistant erythromycin-sensitive colonies showed that all 10 isolates were true transposants and that each strain contained only one copy of the lux transposon construct (data not shown). Furthermore, sequencing of the chromosomal lux fusion junctions (gained by inverse PCR) of each of these transposants confirmed that all 10 integration sites were unique, confirming that Tn4001 transposition in S. pneumoniae occurs randomly (data not shown).
Four clinical isolates of S. pneumoniae were naturally transformed with chromosomal DNA of a highly bioluminescent, kanamycin-resistant, erythromycin-sensitive D39 transposant, designated Xen 7. The transformation efficiency varied from 50 to 500 CFU/μg of Xen 7 chromosomal DNA. These transformants were named Xen 9, Xen 10, Xen 11, and Xen 12, corresponding to strains HUSTMBIG, A66.1, EF3030, and 140301, respectively. To test whether the transposon was stably maintained at its original integration site, Xen 9 and Xen 10 were cultured in BHI medium with or without kanamycin for 2 weeks at 37°C, diluting the cells 1:1,000 into fresh medium every 12 h. Chromosomal DNAs were prepared on days 0, 3, 7, 10, and 14, and the ratio of kanamycin-resistant colonies to total CFU was examined at the same times. Tn4001 luxABCDE Kmr in both Xen 9 and Xen 10 was found to be 100% stable, so that after 14 days of continuous culturing without antibiotic selection, all CFU were bioluminescent and the locations of the transposons in both strains remained the same and at single copy (data not shown).
Photon emission from bioluminescent S. pneumoniae decreases dramatically as cells enter stationary phase.
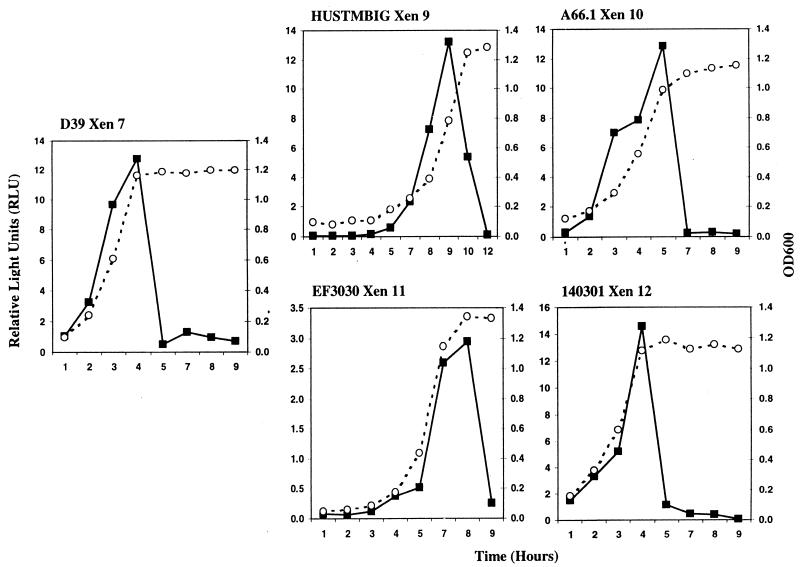
To evaluate the level of bioluminescence in the different strains of S. pneumoniae during growth, RLU were recorded from strain D39 (Xen 7) and the four clinical isolates (Xen 9 to 12) during in vitro growth using an ICCD camera and Xenogen's LivingImage software. As Fig. 3 shows, in all five strains the photon counts increased during exponential growth and then decreased dramatically to about 5% of their peak values once the bacteria entered stationary phase. For strain Xen 11, the peak photon counts were less than those recorded for the other strains, which might reflect strain variation. This reasoning is supported by the observation that EF3030 Xen 11 showed a different hybridization pattern in a Southern blot analysis than the donor strain Xen 7 and the other transformed strains, Xen 9, Xen 10, and Xen 12. Despite these differences, inverse PCR showed that the transposon in Xen 11 was integrated at the same site as it was in the other strains (data not shown). This integration site is located in the second open reading frame (ORF) of a possible two-gene operon (www.tigr.org, contig 3836, bases 1719329 to 1720455). The sequence of this ORF has no significant similarity at the DNA or protein level to any sequence in the NCBI database.
FIG. 3.
RLU emitted by bioluminescent S. pneumoniae strains during in vitro growth. Three hundred microliters of overnight culture was inoculated into 30 ml of BHI and grown at 37°C in 5% CO2. At 1-h intervals, both the OD600 (open circles) and the number of RLU × 106 (solid squares) from a 1-ml culture volume were determined for each of the strains. The experiment was repeated three times for each strain.
To investigate whether the stationary-phase phenomenon described above was due to the regulation of one specific promoter or was a true consequence of the growth phase of S. pneumoniae, 10 bioluminescent D39 transposants were randomly picked from a chocolate plate supplemented with kanamycin and their RLU were measured during growth. In all the cases, the RLU were significantly reduced to basal levels when the stationary phase was reached (data not shown), suggesting that the reduction of photon emission was a stationary-phase phenomenon in S. pneumoniae.
Bioluminescent S. pneumoniae A66.1 Xen 10 can be used to accurately monitor pneumococcal drug efficacy studies in vivo in live animals.
Initial experiments with S. pneumoniae D39 Xen 7 showed that this strain performed poorly in animal respiratory infection studies. Thus, strains HUSTMBIG, A66.1, EF3030, and 140301 were made bioluminescent as described above and tested in both mouse lung and nasopharyngeal models. A66.1 Xen 10 was found to perform best in the mouse pneumococcal lung model (see Fig. 4 and 5), whereas HUSTMBIG Xen 9 and EF3030 Xen 11 performed best in the nasopharyngeal model (see Fig. 6). Similar to D39 Xen 7, strain 140301 Xen 12 did not perform well in either model.
FIG. 4.
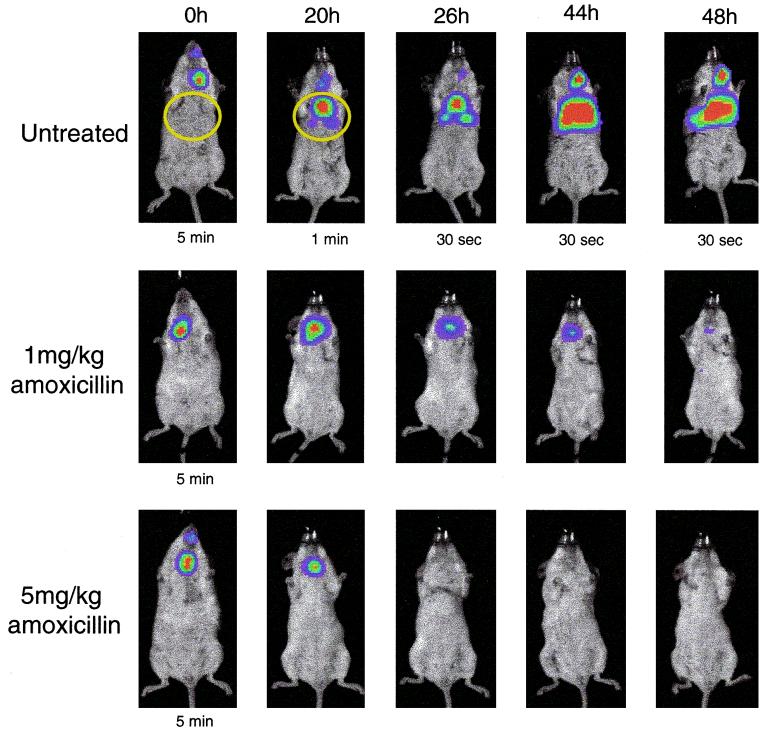
Bioluminescent S. pneumoniae A66.1 Xen 10 in a mouse pneumococcal lung model, with and without antibiotic treatment. Twelve mice were inoculated with approximately 106 CFU of S. pneumoniae A66.1 Xen 10. The mice were divided into three groups of four animals each, and two of these groups were treated with amoxicillin at 1 or 5 mg/kg given subcutaneously at 0, 18, 24, and 42 h postinfection; the third group of animals were left untreated as controls. Mice were imaged using an IVIS CCD camera (Xenogen Corporation) at the indicated time points postinfection. Total photon emission from the ventral thoracic region of each mouse (areas inside the yellow circle) was quantified using the LivingImage software package (Xenogen Corporation). Imaging time was 5 min unless otherwise shown.
FIG. 5.
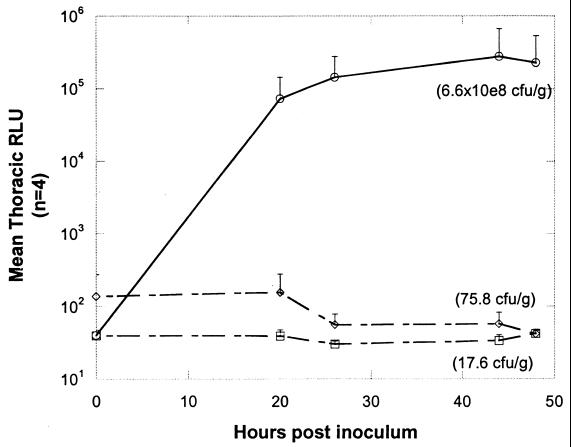
Graphic representation of mean thoracic bioluminescence (RLU) from the pneumococcus-infected mice, untreated and treated with amoxicillin, shown in Fig. 4. Each point represents the average bioluminescence from all surviving mice in each treatment group (starting with four mice in each group). ○, untreated animals; ◊, treated (5 mg/kg) animals; □, treated (1 mg/kg) animals. The average CFU per gram of lung tissue for each group of animals at 48 h (time of sacrifice) is given in parentheses.
FIG. 6.
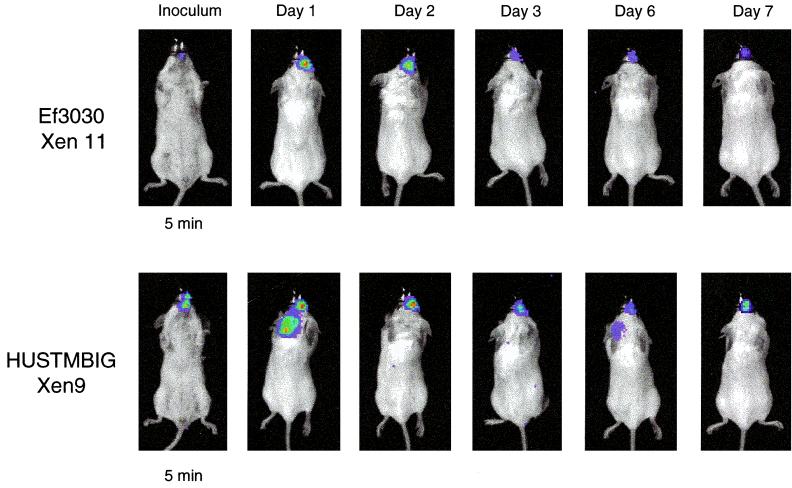
Bioluminescent S. pneumoniae Ef3030 Xen9 and HUSTMBIG Xen 11 in a mouse nasopharygeal model. Mice were infected intranasally with approximately 106 CFU by placing 20 μl of bacterial suspension on the nares and allowing the mice to inhale the inoculum. Mice were imaged using an IVIS CCD camera (Xenogen Corporation) at the indicated time points postinfection. Imaging time was 5 min in each case.
Comparison of S. pneumoniae D39, EF3030, and HUSTMBIG (both parental and bioluminescent derivatives) revealed that the latter two clinical isolates were far superior to D39 in colonizing the nasopharynx of mice. Recovery of bacteria from nasal washes of infected mice was 100- to 1,000-fold higher from both EF3030 and HUSTMBIG than from D39. In addition, no statistical difference could be seen between the parent and bioluminescent derivative for any strain, suggesting that the insertion of the lux transposon did not alter these virulence characteristics (data not shown).
Since A66.1 Xen 10 performed best in the mouse pneumococcal lung model, this strain was selected to test in a drug efficacy study. Twelve mice were inoculated with approximately 106 CFU of S. pneumoniae A66.1 Xen 10. The mice were divided into three groups of four animals each, and two of these groups were treated with amoxicillin at 1 or 5 mg/kg; the third group of animals were left untreated as controls. Figure 4 shows an example of the type of visual data seen in vivo (one animal from each group of four) using this bioluminescent S. pneumoniae strain. In the untreated group of animals, a strong bioluminescent signal could be detected from the thorax of three of the four mice by 20 h, indicating that three of the mice had an established pneumococcal lung infection. These animals all had ruffled fur and appeared ill, whereas the fourth control animal, showing no bioluminescent signal, appeared healthy, indicating that it was uninfected (see below). Over the next 28 h (up to 48 h postinoculation), the three infected control animals showed increasing bioluminescent signals from their lungs (Fig. 5), the mouse with the highest signal at 26 h dying overnight before the 44-h imaging time point. By 48 h postinoculation, at least one of the two remaining infected mice appeared extremely ill, with both animals having intense bioluminescent signals.
All eight of the amoxicillin (1 and 5 mg/kg groups)-treated mice appeared well at the 20-h imaging time point compared to the control group. Three of the four animals treated with amoxicillin (1 mg/kg) had a bioluminescent signal 20 h postinoculation. None of the animals treated with amoxicillin (5 mg/kg) had a significant bioluminescent signal at the 20-h time point. In comparison, the untreated control animals developed a bioluminescent signal that was several-thousand-fold greater than that of either group of amoxicillin-treated animals (Fig. 5).
Plating of lung tissue extracts following sacrifice of each of the mice after the 48-h imaging time point showed that only the two untreated animals with strong bioluminescent signals had large numbers of pneumococci present (the third mouse with a strong bioluminescent signal had died prior to this time point). One of these animals, with a bioluminescent signal of 443,100 RLU, had 2 × 109 CFU/g of tissue. No bacteria could be recovered from the untreated mouse that did not have a bioluminescent signal, supporting the assumption that a pneumococcal infection had not been established in this host. The highest number of pneumococci to be isolated from an amoxicillin-treated animal was 300 CFU/g of tissue, with four of the eight animals showing no bacteria present (Fig. 6).
All pneumococci recovered from long-term mouse nasopharyngeal infections are bioluminescent.
S. pneumoniae HUSTMBIG Xen 9 and EF3030 Xen 11 recovered from nasal washes performed on mice with 7-day pneumococcal nasopharyngeal infections (i.e., from the mice shown in Fig. 6) were shown to be 100% bioluminescent.
DISCUSSION
Bioluminescent S. pneumoniae D39 was initially generated using plasmid pDL289 (4), carrying the modified luxABCDE construct (11). However, as with Staphylococcus aureus containing pMK4 luxABCDE constructs (11), plasmid loss in the absence of antibiotic selection from these S. pneumoniae cells was unacceptably high in animal studies lasting longer than 48 h (unpublished data). In order to stabilize the bioluminescence emitted from the pneumococcal cells, we integrated the luxABCDE cassette into the bacterium's chromosome. Strains of S. pneumoniae are readily transformable by natural competence procedures to facilitate homologous recombination of gene fusions, as has previously been shown using reporter genes such as lacZ (28). However, studies conducted by our group with S. aureus showed that it was difficult to predict how a particular luxABCDE fusion would behave on the chromosome. Thus, in order to generate a pool of chromosomal lux fusions that could be screened on the basis of their bioluminescent signal, we investigated the possibility of building a gram-positive lux transposon.
Several gram-positive transposons were considered as candidates for integration of the luxABCDE cassette, including Tn917 and the conjugative transposon Tn916 (26, 36). However, due to its small size, broad host range, and genetic amenability on plasmid pMGC57 (5, 21), the composite-type transposon Tn4001 was selected. This transposon has been used for the random mutagenesis of several mycoplasmas and a range of gram-positive bacteria (8, 10, 20–23, 32), including Streptococcus pyogenes and Streptococcus gordonii (20, 21). As far as we are aware, this is the first instance in which Tn4001 has been used for the mutagenesis of S. pneumoniae.
Prior studies have shown that Tn4001 chooses its targets for insertion with a high degree of randomness and that this transposon is highly stable in streptococci (20, 21). Mutagenesis of S. pyogenes with Tn4001.spc was shown to be extremely efficient using the suicide plasmid pMGC57-spc, with transposition frequencies of between 103 and 106/μg of DNA, depending on the S. pyogenes host strain used. Unfortunately, we were unable to achieve such transposition frequencies in strains of S. pneumoniae using pMGC57-spc. This may be due in part to low plasmid transformation efficiencies achieved by electroporation of this bacterium (approximately 102 transformants/μg of pDL289 DNA). Thus, in order to stably maintain the Tn4001 luxABCDE Kmr construct in S. pneumoniae and so increase the efficiency of transposition, we chose to move this construct onto the gram-positive/negative temperature-sensitive plasmid pAUL-A (6).
Transposition of Tn4001 luxABCDE Kmr in S. pneumoniae D39 occurred at a frequency of approximately 10−3. However, the vast majority (98%) of these transformants were bacteria with the entire pAUL-A construct incorporated onto their chromosome, reducing the true frequency of Tn4001 transposition to approximately 10−5. In contrast, transposition of Tn4001 luxABCDE Kmr in both S. aureus and L. monocytogenes occurred at a frequency of between 10−5 and 10−6, and all transformants tested were erythromycin sensitive, indicating loss of the pAUL-A backbone (our unpublished data). At present it is not known why the entire pAUL-A Tn4001 luxABCDE Kmr plasmid should integrate into the chromosome of S. pneumoniae D39 at such a high frequency, especially when plasmid integration does not appear to occur in either S. aureus or L. monocytogenes, and Lyon et al. (21) showed that this phenomenon occurs at a frequency of <0.1% using pMGC57-spc in S. pyogenes.
Comparing RLU to CFU in S. pneumoniae cultures showed that photon emissions decreased dramatically when the bacterial cells entered stationary phase (Fig. 3). This steep decline in bioluminescence was universal for all S. pneumoniae lux transformants tested, including pDL289 luxABCDE transformants (data not shown), showing that this phenomenon was not caused by reduced transcription from a specific promoter. Previous studies in both gram-positive and gram-negative bacteria have shown that bioluminescence decreases once the cells enter stationary phase (14, 25, 34, 35; unpublished data). However, this light reduction is usually gradual and reflects a slow decrease in the metabolic activity of the cell (25, 34). It is possible that the sharp decrease in bioluminescence seen from S. pneumoniae entering stationary phase is due to a decrease of reduced flavin mononucleotide to feed the bioluminescent reaction. Reduced flavin mononucleotide is generated in aerobic bacteria via components of the electron transport chain. However, S. pneumoniae is a facultative anaerobe that lacks an intact respiratory chain and as such may generate these components less efficiently (17, 29). Similar sharp falls in bioluminescence in stationary-phase cultures have been reported for luxAB constructs in the closely related bacterium Lactococcus lactis (35). Interestingly, this phenomenon does not appear to have a significant effect in vivo in living animals (Fig. 4, 5, and 6).
Initial studies with bioluminescent S. pneumoniae D39 (mostly Xen 7) showed that it was possible to monitor pneumococcal cells in vivo in a mouse thigh model (unpublished data). However, this particular strain (D39) gave little to no signal when inoculated into the animal's lungs. In contrast to D39, S. pneumoniae A66.1 Xen 10 performed extremely well in the lungs of mice (Fig. 4 and 5). Bioluminescence could be detected for at least 48 h, indicating that this strain can be used for in vivo pneumococcal drug efficacy studies. The strains HUSTMBIG Xen 9 and EF3030 Xen 11, which performed best in the mouse nasopharyngeal model (Fig. 6), may prove to be valuable candidates for vaccination studies due to their longevity of colonization of the nasal passage.
Both of the pneumococcal animal models used in this study demonstrate some of the significant advantages that real-time photonic imaging offers over conventional methods for monitoring and combating bacterial disease in animals. Not only does this approach reduce the time and cost of conducting such experiments, but it also considerably reduces the number of animals used (7, 11). Furthermore, because bioluminescent imaging allows the same group of animals to be monitored over time, animal-to-animal variations are overcome by including the zero time point as an internal control. In addition to this procedure improving biostatistics, several parameters of drug efficacy and pharmacokinetics can be more accurately measured in the discovery and development stages of drug evaluation.
ACKNOWLEDGMENTS
We thank E. Albert (Animal Husbandry, Xenogen Corporation) and P. Winterberg (Infectious Disease, Xenogen Corporation) for assisting with the animal work and E. Reynolds (Graphics and Communications, Xenogen Corporation) for assistance with drawings and figures. We also thank T. Parr (Eli Lilly) and D. Briles (University of Alabama at Birmingham) for providing strains and C. Thompson (Harvard School of Public Health, Boston, Mass.) for providing strains and sharing preliminary unpublished data.
REFERENCES
- 1.Appelbaum P C. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clin Infect Dis. 1992;15:77–83. doi: 10.1093/clinids/15.1.77. [DOI] [PubMed] [Google Scholar]
- 2.Austrian R. Some aspects of the pneumococcal carrier state. J Antimicrob Chemother. 1986;18(Suppl. A):35–45. doi: 10.1093/jac/18.supplement_a.35. [DOI] [PubMed] [Google Scholar]
- 3.Briles D E, Paton J C, Swiatlo E, Nahm M H. Pneumococcal vaccines. In: Fischetti V A, et al., editors. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology; 2000. pp. 244–250. [Google Scholar]
- 4.Buckley N D, Lee L N, LeBlanc D J. Use of a novel mobilizable vector to inactivate the scrA gene of Streptococcus sobrinus by allelic replacement. J Bacteriol. 1995;177:5028–5034. doi: 10.1128/jb.177.17.5028-5034.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caparon M. Genetics of group A streptococci. In: Fischetti V A, et al., editors. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology; 2000. pp. 53–65. [Google Scholar]
- 6.Chakraborty T, Leimeister-Wachter M, Domann E, Hartl M, Goebel W, Nichterlein T, Notermans S. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J Bacteriol. 1992;174:568–574. doi: 10.1128/jb.174.2.568-574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contag C H, Contag P R, Mullins I, Spilman S D, Stevenson D K, Benaron D A. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18:593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- 8.Dybvig K, French C T, Voelker L L. Construction and use of derivatives of transposon Tn4001 that function in Mycoplasma pulmonis and Mycoplasma arthritidis. J Bacteriol. 2000;182:4343–4347. doi: 10.1128/jb.182.15.4343-4347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farley J J, King J C, Nair P, Hines S E, Tressler R L, Vink P E. Invasive pneumococcal disease among infected and uninfected children of mothers with immunodeficiency virus infection. J Pediatr. 1994;124:853–858. doi: 10.1016/s0022-3476(05)83170-1. [DOI] [PubMed] [Google Scholar]
- 10.Foissac X, Saillard C, Bove J M. Random insertion of transposon Tn4001 in the genome of Spiroplasma citri strain GII3. Plasmid. 1997;37:80–86. doi: 10.1006/plas.1996.1271. [DOI] [PubMed] [Google Scholar]
- 11.Francis K P, Joh D, Bellinger-Kawahara C, Hawkinson M J, Purchio T F, Contag P R. Monitoring bioluminescent Staphylococcus aureus infections in live mice using a novel luxABCDE construct. Infect Immun. 2000;68:3594–3600. doi: 10.1128/iai.68.6.3594-3600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gosink K, Tuomanen E. Streptococcus pneumoniae: invasion and inflammation. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology; 2000. pp. 214–224. [Google Scholar]
- 13.Gray B, Dillon H. Clinical and epidemio-logic studies of pneumococcal infection in children. Pediatr Infect Dis. 1986;5:201–207. doi: 10.1097/00006454-198603000-00009. [DOI] [PubMed] [Google Scholar]
- 14.Guard-Petter J. Variants of smooth Salmonella enterica serovar Enteritidis that grow to higher cell density than the wild type are more virulent. Appl Environ Microbiol. 1998;64:2166–2172. doi: 10.1128/aem.64.6.2166-2172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Havarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang Y, Nahm M H, Briles D E, Thomas D, Purkerson J M. Acquired but not innate immune responses to Streptococcus pneumoniae are compromised by neutralization of CD40L. Infect Immun. 2000;68:511–517. doi: 10.1128/iai.68.2.511-517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konings W N, Otto R. Energy transduction and solute transport in streptococci. Antonie van Leeuwenhoek. 1983;49:247–257. doi: 10.1007/BF00399501. [DOI] [PubMed] [Google Scholar]
- 18.Lefrancois J, Sicard A M. Electrotransformation of Streptococcus pneumoniae: evidence for restriction of DNA on entry. Microbiology. 1997;143:523–526. doi: 10.1099/00221287-143-2-523. [DOI] [PubMed] [Google Scholar]
- 19.Lipsitch M, Dykes J K, Johnson S E, Ades E W, King J, Briles D E, Carlone G M. Competition among Streptococcus pneumoniae for intranasal colonization in a mouse model. Vaccine. 2000;18:2895–2901. doi: 10.1016/s0264-410x(00)00046-3. [DOI] [PubMed] [Google Scholar]
- 20.Lunsford R D. A Tn4001 delivery system for Streptococcus gordonii (Challis) Plasmid. 1995;33:153–157. doi: 10.1006/plas.1995.1016. [DOI] [PubMed] [Google Scholar]
- 21.Lyon W R, Gibson C M, Caparon M G. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahairas G G, Minion F C. Random insertion of the gentamicin resistance transposon Tn4001 in Mycoplasma pulmonis. Plasmid. 1989;21:43–47. doi: 10.1016/0147-619x(89)90085-1. [DOI] [PubMed] [Google Scholar]
- 23.Mahairas G G, Lyon B R, Skurray R A, Pattee P A. Genetic analysis of Staphylococcus aureus with Tn4001. J Bacteriol. 1989;171:3968–3972. doi: 10.1128/jb.171.7.3968-3972.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majewski J, Zawadzki P, Pickerill P, Cohan F M, Dowson C G. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J Bacteriol. 2000;182:1016–1023. doi: 10.1128/jb.182.4.1016-1023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marincs F. On-line monitoring of growth of Escherichia coli in batch cultures by bioluminescence. Appl Microbiol Biotechnol. 2000;53:536–541. doi: 10.1007/s002530051653. [DOI] [PubMed] [Google Scholar]
- 26.McDougal L K, Tenover F C, Lee L N, Rasheed J K, Patterson J E, Jorgensen J H, LeBlanc D J. Detection of Tn917-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2312–2318. doi: 10.1128/aac.42.9.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ochman H, Gerber A S, Hartl D L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pestova E V, Morrison D A. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J Bacteriol. 1998;180:2701–2710. doi: 10.1128/jb.180.10.2701-2710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poolman B. Energy transduction in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:125–148. doi: 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 30.Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein L S, Piccoli L, Simon D, Morrison D A. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuchat A, Robinson K, Wenger J, Harrison L, Farley M, Reingold A, Lefkowitz L, Perkins B. Bacterial meningitis in the United States. N Engl J Med. 1997;337:970–976. doi: 10.1056/NEJM199710023371404. [DOI] [PubMed] [Google Scholar]
- 32.Tigges E, Minion F C. Physical map of the genome of Acholeplasma oculi ISM1499 and construction of a Tn4001 derivative for macrorestriction chromosomal mapping. J Bacteriol. 1994;176:1180–1183. doi: 10.1128/jb.176.4.1180-1183.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuomanen E I, Austrian R, Masure H R. Pathogenesis of pneumococcal infection. N Engl J Med. 1995;332:1280–1284. doi: 10.1056/NEJM199505113321907. [DOI] [PubMed] [Google Scholar]
- 34.Unge A, Tombolini R, Molbak L, Jansson J K. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl Environ Microbiol. 1999;65:813–821. doi: 10.1128/aem.65.2.813-821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waterfield N R, Le Page R W, Wilson P W, Wells J M. The isolation of lactococcal promoters and their use in investigating bacterial luciferase synthesis in Lactococcus lactis. Gene. 1995;165:9–15. doi: 10.1016/0378-1119(95)00484-n. [DOI] [PubMed] [Google Scholar]
- 36.Watson D A, Musher D M. Interruption of capsule production in Streptococcus pneumonia serotype 3 by insertion of transposon Tn916. Infect Immun. 1990;58:3135–3138. doi: 10.1128/iai.58.9.3135-3138.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H Y, Virolainen A, Mathews B, King J, Russell M W, Briles D E. Establishment of a Streptococcus pneumoniae nasopharyngeal colonization model in adult mice. Microb Pathog. 1997;23:127–137. doi: 10.1006/mpat.1997.0142. [DOI] [PubMed] [Google Scholar]