Summary
Aegilops tauschii is the diploid progenitor of the wheat D subgenome and a valuable resource for wheat breeding, yet, genetic analysis of resistance against Fusarium head blight (FHB) and the major Fusarium mycotoxin deoxynivalenol (DON) is lacking. We treated a panel of 147 Ae. tauschii accessions with either Fusarium graminearum spores or DON solution and recorded the associated disease spread or toxin‐induced bleaching. A k‐mer‐based association mapping pipeline dissected the genetic basis of resistance and identified candidate genes. After DON infiltration nine accessions revealed severe bleaching symptoms concomitant with lower conversion rates of DON into the non‐toxic DON‐3‐O‐glucoside. We identified the gene AET5Gv20385300 on chromosome 5D encoding a uridine diphosphate (UDP)‐glucosyltransferase (UGT) as the causal variant and the mutant allele resulting in a truncated protein was only found in the nine susceptible accessions. This UGT is also polymorphic in hexaploid wheat and when expressed in Saccharomyces cerevisiae only the full‐length gene conferred resistance against DON. Analysing the D subgenome helped to elucidate the genetic control of FHB resistance and identified a UGT involved in DON detoxification in Ae. tauschii and hexaploid wheat. This resistance mechanism is highly conserved since the UGT is orthologous to the barley UGT HvUGT13248 indicating descent from a common ancestor of wheat and barley.
Keywords: Aegilops tauschii, deoxynivalenol, UDP‐glucosyltransferase, Fusarium head blight, Triticum aestivum, wheat
Introduction
Fusarium head blight (FHB) is one of the most destructive diseases of wheat (Triticum aestivum L.), leading to severe losses in both yield and crop quality. Most importantly, causative fungi of the genus Fusarium are notorious producers of mycotoxins that accumulate in the grain, constituting a serious threat to food and feed safety (Pestka, 2010). Among them, the type‐B trichothecene deoxynivalenol (DON) is the most prevalent (Bottalico and Perrone, 2002; Goswami and Kistler, 2004). The EU and many other countries have enacted maximum tolerated levels of DON in grain‐based food commodities (Knutsen et al., 2017). Hence, controlling this disease is of pivotal importance and growing resistant varieties is a sustainable, environment‐friendly and cost‐effective approach.
Resistance breeding and research are complicated by the quantitative nature of the resistance influenced by environmental factors (Buerstmayr et al., 2020). Numerous resistance quantitative trait loci (QTL) were mapped and are distributed over all 21 wheat chromosomes. Yet, only a handful of QTL has been validated across studies and successfully deployed in breeding programs (Buerstmayr et al., 2020; Liu et al., 2019; Löffler et al., 2009) and very few studies reported the cloning of a causal gene (Gadaleta et al., 2019; Li et al., 2019; Rawat et al., 2016; Su et al., 2019; Wang et al., 2020). Isolation projects for the highly effective and best‐studied FHB resistance QTL, Fhb1, delivered contradicting results, and the underlying molecular mechanism is still unknown (Lagudah and Krattinger, 2019; Li et al., 2019; Rawat et al., 2016; Su et al., 2019). In general, isolation and functional validation of resistance genes in wheat are challenging due to its vast genome size and polyploidy.
Recently, strategies have been developed to rapidly map traits in the diploid goatgrass Aegilops tauschii Coss., which is the donor of the D subgenome of hexaploid wheat (genomes AABBDD) (McFadden and Sears, 1946). Based on 242 sequenced Ae. tauschii accessions, Gaurav et al. (2022) established a k‐mer‐based association mapping pipeline, identified candidate genes for disease and pest resistance and demonstrated their transfer to wheat by transgenesis and wide crosses. Aegilops tauschii constitutes a valuable genetic resource for gene discovery and wheat breeding, including the genetic improvement of disease resistance (Anikster et al., 2005; Börner et al., 2015; Kishii, 2019; Zhou et al., 2021), since 75% of the diversity found within the D subgenome was lost in the initial hybridization events that gave rise to wheat landraces and is therefore not part of the T. aestivum genome (Gaurav et al., 2022).
Evaluation of FHB resistance in Ae. tauschii accessions revealed a broad variation, including a few resistant accessions that originated mainly from regions bordering the Caspian Sea that are receiving high levels of annual rainfall (Brisco et al., 2017). Several studies investigated the potential of the D subgenome donor, Ae. tauschii, for improving FHB resistance in wheat via synthetic hexaploids (Mujeeb‐Kazi, 2001a, 2001b; Oliver et al., 2005; Szabo‐Hever et al., 2018). The FHB severities varied depending on the Ae. tauschii and tetraploid genotypes involved and comprised highly resistant synthetics. Szabo‐Hever et al. (2018) compared the synthetic hexaploid wheat lines with their tetraploid parents and found an average disease reduction of 18.3% in the synthetics, suggesting that the D subgenome may play a role in reducing disease severity in hexaploid lines.
Resistance against the major Fusarium mycotoxin DON is an important FHB resistance component, since DON is phytotoxic and acts as a virulence factor promoting fungal spread in wheat spikes (Jansen et al., 2005). Of the several mechanisms of DON detoxification that have been described (Foroud et al., 2019; Gunupuru et al., 2017), the conjugation into the non‐toxic DON‐3‐O‐glucoside (D3G) mediated by members of the family 1 of uridine diphosphate (UDP)‐glucosyltransferases (UGTs) appears to be most relevant in wheat (Kluger et al., 2015), but this has yet to be investigated in Ae. tauschii.
In this study, we exploited the k‐mer‐based association mapping pipeline developed by Gaurav et al. (2022) to dissect the genetic basis of FHB and DON resistance in Ae. tauschii and study the mechanism of DON detoxification into D3G in order to identify candidate resistance genes in the D subgenome donor of T. aestivum.
Results
Phenotypic variation for FHB and DON resistance of the Aegilops tauschii accessions
The accessions revealed continuous variation for FHB‐ and DON‐induced bleaching severities (FHB/DON severities) (Figure 1a, Table 1), though broader variation was observed after fungal infection. FHB severities were skewed towards susceptibility with a mean of about 75% infected spikelets. The most resistant accessions developed disease symptoms on 20% of the spikelets corresponding to 1.6 symptomatic spikelets for an average spike with eight spikelets. None of the accessions was as resistant as the highly resistant hexaploid control CM82036, which did not show any disease spreading (Table 1). Regarding resistance against DON‐induced bleaching (DON resistance), most of the accessions developed the typical straw‐like colour only on the infiltrated spikelet (Figure 1b) and again none of the accessions reached the resistance level of CM82036, which did not display any DON‐induced bleaching symptoms on the spikes. Interestingly, no significant correlation was detected between FHB and DON severities (r = 0.1ns) (Figure S1).
Figure 1.
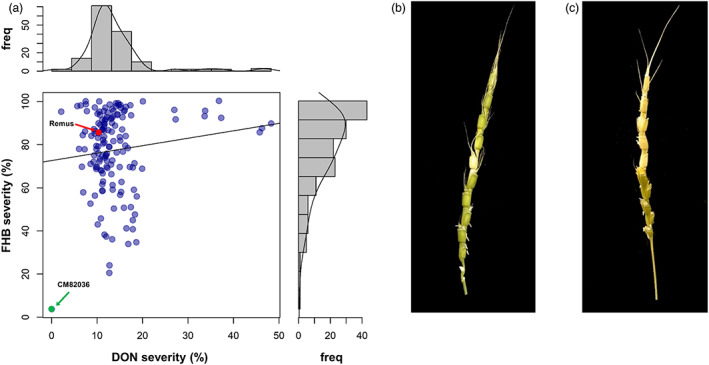
Scatter plot and marginal frequency (freq) distributions for Fusarium head blight (FHB) and deoxynivalenol (DON) severities of the 147 Aegilops tauschii accessions and the two hexaploid control lines CM82036 (resistant) and Remus (susceptible) (a). Severities are given in percent (%) symptomatic spikelets of best linear unbiased estimates (BLUEs) across experiments. Typical phenotypes after DON infiltration of Aegilops tauschii accessions with (b) low and (c) high DON severities. Note that in the resistant accession (b) only the treated spikelet showed a bleached, straw like, appearance but other spikelets stayed green, while in the susceptible accession (c) the bleaching phenotype spread.
Table 1.
Overall means, ranges, variance components, broad‐sense heritability estimates (H 2) and least significant differences (LSD P < 0.05) from CM82036 (resistant control), Remus (susceptible control) and the 147 Aegilops tauschii accessions for: Fusarium head blight (FHB) and deoxynivalenol (DON) severities given in %, incidence for early seed shattering in the FHB and DON trials, date of anthesis, number of spikelets per spike and plant height
| FHB severity (%) | DON severity (%) | Seed shattering (FHB) | Seed shattering (DON) | Date of anthesis † | No. of spikelets | Plant height (cm) | ||
|---|---|---|---|---|---|---|---|---|
| CM82036 | 3.38 | 0 | 0 | 0 | 48.19 | 16.42 | NA | |
| Remus | 82.78 | 21.45 | 0 | 0 | 54.07 | 18.14 | NA | |
| Aegilops tauschii accessions | ||||||||
| Min | 20.21 | 0 | 0 | 0 | 32.50 | 4.40 | 55 | |
| Mean | 76.92 | 14.72 | 30.07 | 1.13 | 46.81 | 8.06 | 99.4 | |
| Max | 100 | 64.79 | 100 | 42.86 | 73.17 | 12.6 | 130 | |
| Variance ± SE | ||||||||
|
|
253.03 ± 40.68 | 44.25 ± 7.67 | 106.1 ± 43.71 | 72.69 ± 7.2e‐06 | 57.10 ± 7.42 | 3.24 ± 0.40 | 107.6 ± 46.9 | |
|
|
37.85 ± 25.22 | 0 ± 6.30 | 189.5 ± 52.91 | 5.74 ± 2.5e‐01 | 0.63 ± 2.10 | 0.23 ± 0.06 | 232.4 ± 56.7 | |
|
|
109.24 ± 21.58 | 36.33 ± 6.34 | 15.15 ± 1.96 | 0.21 ± 0.04 | ||||
|
|
459 ± 11.99 | 48.47 ± 2.03 | 269.9 ± 33.45 | 0 ± 2.5e‐07 | 10.34 ± 0.22 | 0.84 ± 0.02 | 259.7 ± 32.5 | |
|
|
0.85 | 0.89 | 0.40 | 0.96 | 0.93 | 0.95 | 0.79 | |
| LSD 5% | 24.95 | 10.45 | 1.90 | 1.92 | 1.32 | 0.23 | 1.99 | |
NA, not available.
Number of days after December 1st to anthesis.
For nine accessions, the DON infiltration resulted in severe bleaching symptoms, which was mainly associated with a spreading in the acropetal direction (Table 1, Figure 1c), and even produced higher DON severities than detected for the susceptible hexaploid control Remus. The highly DON susceptible accessions originated from Azerbaijan (TOWWC0044), Turkey (TOWWC0083), the former USSR (TOWWC0088), Iran (TOWWC0097, TOWWC0134, TOWWC0137, TOWWC0168, TOWWC0169) and Armenia (TOWWC0185).
Analysis of variance revealed that all sources of variation gave rise to highly significant effects on the FHB and DON severities, although the factor genotype was most important (Table S1) resulting in high heritabilities of H 2 = 0.80 and H 2 = 0.89 for FHB and DON severities, respectively (Table 1).
The applied treatments, Fusarium graminearum inoculation or DON infiltration, had a strong impact on premature seed shattering and at 23 dai 36.5% of all F. graminearum inoculated spikes fell apart, while this was only the case for 1.6% of the treated spikes in the DON trial. In the FHB experiment, the heritability for premature seed shattering was low (H 2 = 0.4), whereas for the DON trial, the genotype was the main source of variance leading to high heritability (H 2 = 0.96). The FHB and DON severities had little influence on the seed shattering incidences and were only weakly correlated (r = 0.21** for FHB and r = 0.23** for DON) (Figure S1). Pearson's correlation coefficients of the traits for individual trials and across trials are provided in Table S2 and the BLUEs for all evaluated traits in Tables S3 and S4.
Phenotypic variations of other traits and trait correlations
The Ae. tauschii accessions exhibited a large diversity for several other agronomical traits, such as date of anthesis, number of spikelets per spike and plant height (Table 1). The flowering period lasted for more than a month and the plant height varied between 55 and 130 cm, with a mean of around 100 cm. Spikelet number per spike varied between 4 to 13 spikelets. For all these traits, high heritabilities confirmed the high data quality, which was necessary for further genomic analyses. Plant height showed a weak positive correlation with FHB severity (r = 0.37**) and an insignificant correlation with DON severity (Figure S1). Neither flowering date nor the number of spikelets per spike was significantly correlated with FHB and DON resistance.
The ratio of D3G to DON in toxin‐treated Aegilops tauschii spikes
Thirty‐one Ae. tauschii accessions, including the nine most DON susceptible and 22 randomly selected moderately resistant accessions were analysed for DON and D3G contents and both were detected in all investigated accessions (Table S5). DON was found at concentrations of 0.046–1.66 * 103 mg/kg (mean: 0.62 * 103 mg/kg) and D3G at 0.44–2.32 * 103 mg/kg (mean: 0.41 * 103 mg/kg) in the DON‐treated spikes of Ae. tauschii. The ratio of D3G to DON varied significantly among the accessions and was strongly negatively correlated with DON severity (r = −0.84**), meaning that the most DON resistant accessions had the highest conversion rates of DON to D3G (Tables 2 and S5, Figure 2). In the highly resistant hexaploid control line CM82036, almost all the infiltrated DON was converted to D3G, leading to a 10 times higher amount of D3G in comparison to DON. The D3G/DON ratios of the most DON resistant Ae. tauschii accessions were on the other hand, close to one, while the susceptible Ae. tauschii accessions showed D3G/DON ratios around 0.2, which is similar to the susceptible hexaploid control line Remus (Table 2).
Table 2.
Means and standard deviation (SD) of deoxynivalenol (DON)‐3‐O‐glucoside (D3G)/DON ratios of overall means across trials and individual trials for CM82036 (resistant control), Remus (susceptible control) and the 31 Aegilops tauschii accessions grouped according to their DON resistance level in moderately resistant and susceptible
| Genotype | D3G/DON ratio (Mean ± SD) | ||
|---|---|---|---|
| Across trials | 2019 | 2020 | |
| CM82036 | 11.0 ± 4.2a | 15.7 ± 0a | 8.6 ± 1.4a |
| Remus | 0.32 ± 0.08c | 0.41 ± 0c | 0.27 ± 0.03c |
| Ae. tauschii (moderately resistant) | 1.06 ± 0.42b | 0.86 ± 0.21b | 1.18 ± 0.47b |
| Ae. tauschii (susceptible) | 0.20 ± 0.29c | 0.14 ± 0.04c | 0.22 ± 0.33c |
D3G/DON ratio classes with different index letters are significantly different at P < 0.05 based on Fisher's least significant differences multiple comparison test.
Figure 2.
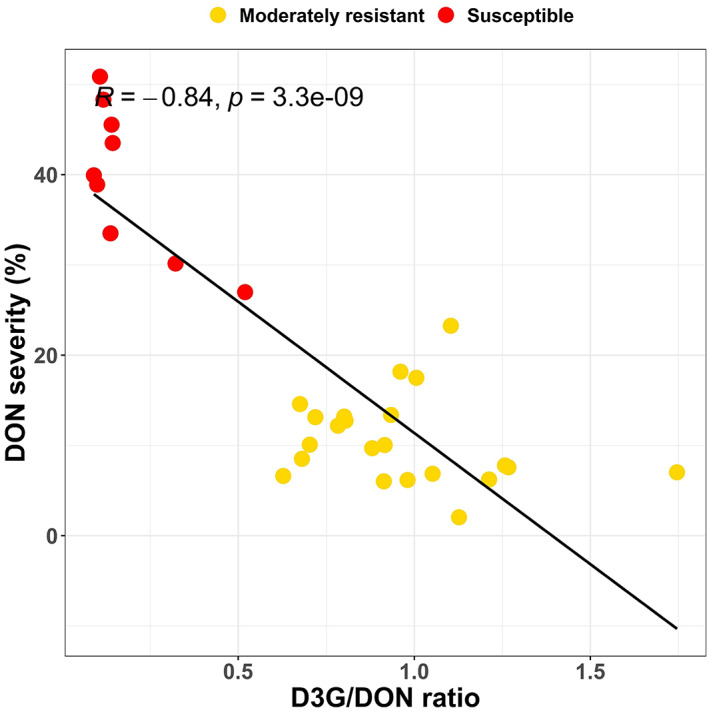
Scatter plot for 31 Aegilops tauschii accessions for the ratio of DON‐3‐O‐glucoside/deoxynivalenol (D3G/DON) and the DON severities evaluated in the greenhouse experiments. The accessions are coloured according to their DON severities in yellow (moderately resistant) and red (susceptible).
Association genetics for FHB and DON resistance in Aegilops tauschii
To dissect the variation for FHB and DON resistance present in the 147 Ae. tauschii accessions, we used the k‐mer‐based association mapping pipeline based on whole genome shotgun sequence data and the Ae. tauschii accession AL8/78 as a mapping reference (Gaurav et al., 2022). For FHB resistance, many significant, minor‐effect associations were identified by analysing the across‐trials data. Most prominent was a very broad peak in the centromeric region of chromosome 7D (Figure S2), which mapped at 265.960–397.501 Mb and covered the entire pericentromeric region (Luo et al., 2017). This 130 Mb region contained more than 3000 genes and was significantly associated in both individual experiments (Table S6). For DON resistance a highly significant association was detected close to the centromere on the long arm of chromosome 5D mapping at 247.290 to 262.700 Mb (Figure 3). k‐mers (−log10 P‐value = 38.21) with strong correlations to DON severity (r = 0.85) mapped to two gene classes well‐known as key players in DON resistance: a UDP‐glucosyltransferase (AET5Gv20385300, LOC109746039, at 250.105 Mb) and two ABC transporters (ABC transporter G family member 41‐like isoform X1, AET5Gv20401700: LOC109787618, at 261.730 Mb; AET5Gv20401700: LOC109787645 at 262.022 Mb) (Table S7).
Figure 3.
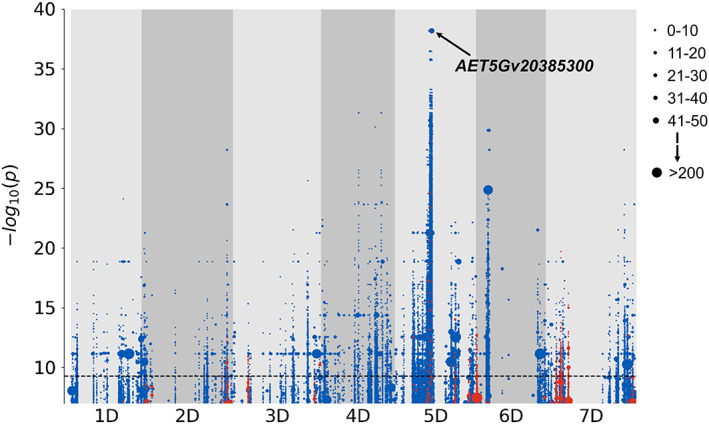
k‐mer‐based association genetics for deoxynivalenol (DON) severity. Points on the plot illustrate k‐mers associated with DON resistance (blue) or DON susceptibility (red) mapped to the Aegilops tauschii reference genome AL8/78 partitioned to chromosomes 1D to 7D. On chromosome 5D, the k‐mer comprising the uridine diphosphate (UDP)‐glucosyltransferase (UGT) AET5Gv20385300 is highlighted. −log10 P‐value = 9.3 was taken as the Bonferroni‐adjusted threshold for evaluating the statistical significance of an association. Point size is proportional to the number of k‐mers with a specific −log10 P‐value (see insert).
Allelic variations of the candidate genes for DON resistance
Sequence alignments of the 147 phenotyped Ae. tauschii accessions to the AL8/78 reference identified seven mutations in the coding region of the UGT AET5Gv20385300 resulting in four allelic protein variants (Figure 4a, Table S8): (1) the predominant allele was the 475 amino acid protein of AL8/78 found for 134 accessions; (2) nine accessions revealed a single nucleotide deletion causing a frameshift from amino acid 341 onwards, leading to a premature stop codon and consequently to a truncated 346 amino acid protein; two SNPs resulted in two nonsynonymous mutations (3) R101Q and (4) K267R detected in one and three accessions, respectively. Notably, the truncated protein was only found in the nine most DON susceptible accessions with the lowest DON to D3G conversion rates. The DON and FHB severities of the 147 Ae. tauschii accessions for the four UGT alleles are visualized in Figure 4b,c showing the highly significant negative effect of the deletion on DON resistance and also for FHB resistance when comparing to the majority of the accessions carrying the AL8/78 allele.
Figure 4.
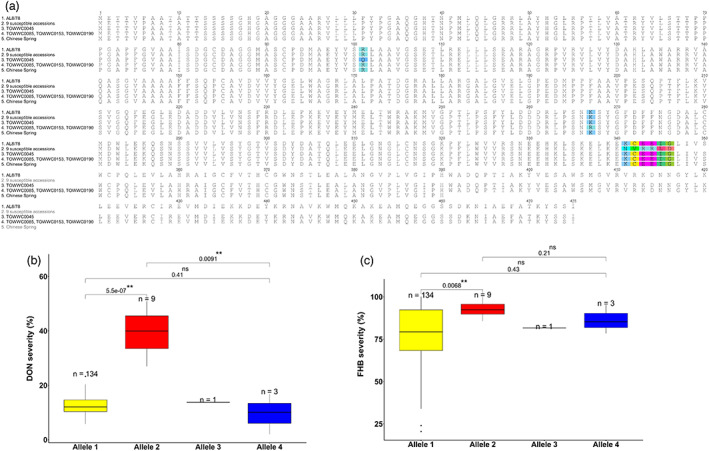
Allelic diversity at the uridine diphosphate (UDP)‐glucosyltransferase (UGT) AET5Gv20385300 and the corresponding Fusarium head blight (FHB) and deoxynivalenol (DON) severities. (a) Amino acid sequence alignment of AET5Gv20385300 for the four allelic variants identified among the 147 Aegilops tauschii accessions and the allele of wheat cv. Chinese Spring: (1) the reference AL8/78, (2) a frame shift from amino acid 341 onwards resulting in a 346 amino acid protein identified for the nine DON susceptible accessions, amino acid changes at positions 101 (3) in TOWWC0045 and at (4) 267 in three accessions (TOWWC0085, TOWWC0153, TOWWC0190). (5) Depicts the 390 amino acid sequence of the UGT allele from wheat cv. Chinese Spring. Boxplots for DON severity (b) and FHB severity (c) of the four allelic UGT variants. Significant codes: **, P < 0.01; ns, not significant.
The UGT diversity analysis of the sequenced, but not phenotyped Ae. tauschii accessions (Gaurav et al., 2022) and the four Ae. tauschii reference sequences (Zhou et al., 2021) identified 21 additional mutations resulting in 11 further protein variants (Figure S3, Table S8). The most common allelic variation led to two nonsynonymous amino‐acid changes (P74A; R138Q) and was detected for 125 lineage 1 accessions; whereas all eight lineage 3 accessions possessed three nonsynonymous changes (A278V; V409L; R410C). None of the accessions showed the truncated 346 amino acid protein associated with DON susceptibility, yet one accession (TOWWC0291) carried a point‐mutation (S211*) causing a truncated, 210 amino acid protein.
Interestingly, the hexaploid wheat cv. Chinese Spring UGT allele (TraesCS5D01G143300) differed from all sequenced Ae. tauschii accessions; it carried a point‐mutation (W391*) that resulted in a premature stop codon and a truncated 390 amino acid protein compared with the 475 amino acid protein of AL8/78 (Figures 4a, S3). Four out of the 16 hexaploid wheat genome assemblies (Appels et al., 2018; Walkowiak et al., 2020) (Claire, Norin61, SY Matthis, Robigus) also held this point‐mutation. The 11 other hexaploid wheat genotypes revealed the AL8/78 allele (ArinaLrFor, Cadenza, CDC Landmark, CDC Stanley, Jagger, Julius, LongReach Lancer, Mace, Paragon, PI190962 (spelt wheat), Weebill 1) as did the two control lines CM82036 and Remus (Table S8). The homoeologous genes on chromosomes 5A (TraesCS5A01G149600) and 5B (TraesCS5B01G148300) showed high sequence similarities to the ‘5D UGTs’ (TraesCS5D01G143300; AET5Gv20385300) (97.4%; 96.2% and 95.9%; 94.7% identity, respectively), possessed the full‐length 475 amino acid alleles at both loci (Figure S4) and were highly conserved among the 16 hexaploid wheat lines. Only spelt wheat (PI190962) carried a nonsynonymous mutation (E192A) at the 5A locus. The durum wheat reference Svevo possessed full‐length alleles at the 5A (TRITD5Av1G113020.3) and 5B (TRITD5Bv1G093730.2) homoeologous UGTs, which were identical to the major‐allele found in wheat.
Sanger sequencing confirmed the UGT alleles of the 31 accessions analysed for DON and D3G, Chinese Spring and the two control lines CM82036 and Remus.
For the two ABC transporters, only intron variants and one synonymous mutation (T113T) for AET5Gv20401700: LOC109787645 were found to be polymorphic between the resistant and the susceptible classes. Based on the lack of functional variation, these genes most likely do not play a role in DON resistance at this locus and therefore, we focused on the UGT candidate gene for functional analysis.
AET5Gv20385300 expression confers resistance against DON in S. cerevisiae
We used heterologous expression of the different alleles of the candidate DON detoxification gene in a toxin sensitive baker's yeast strain for functional testing. The full‐length AL8/78 AET5Gv20385300 allele was custom synthesized and codon‐ optimized for yeast and from this template, the other alleles were constructed by in vitro mutagenesis (sequences in Figure S5); all variants were cloned behind the constitutive ADH1 promoter. The empty vector served as negative control, and the barley HvUGT13248 and the yeast acetyltransferase ScAYT1 were used as positive controls. Whereas the full‐length allele conferred increased DON resistance, the truncated versions were nonfunctional (Figure 5a). The Ae. tauschii UGT behaved like its barley orthologue HvUGT13248, being active with both DON, nivalenol and HT‐2 toxin, but inactive against C4 acetylated toxins (T‐2 toxin, fusarenone X) (Figure 5b) (Michlmayr et al., 2018).
Figure 5.
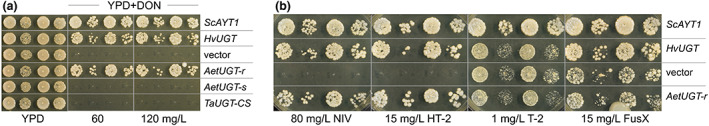
Functional evaluation of the uridine diphosphate (UDP)‐glucosyltransferase (UGT) AET5Gv20385300 identified in Aegilops tauschii. A toxin‐sensitive yeast strain was transformed with plasmids expressing the AL8/78 allele of AET5Gv20385300 (AetUGT‐r) and the two truncated allelic variants identified in Aegilops tauschii (AetUGT‐s) and in Chinese Spring (TaUGT‐CS) as well as the yeast acetyltransferase ScAYT1 and the barley UGT HvUGT13248 (HvUGT) as positive controls and the empty plasmid (vector) as negative control. Two individual transformants of each clone were grown in selective medium to mid‐log phase, diluted to OD 0.3 and 0.03 and spotted onto yeast‐extract‐peptone‐dextrose (YPD) media containing the indicated concentrations of (a) deoxynivalenol (DON) and of (b) nivalenol (NIV), HT‐2 toxin (HT‐2), T‐2 toxin (T‐2) and fusarenone X (FusX).
The highly similar homoeologous UGTs on wheat chromosomes 5A (TraesCS5A01G149600) and 5B (TraesCS5B01G148300) were also tested in yeast for their DON‐detoxifying abilities. The strain expressing the ‘5A UGT’ tolerated DON at the highest concentration tested (120 mg/L) similar to the AET5Gv20385300 transformant and the positive controls HvUGT13248 and ScAYT1. The transformant expressing the ‘5B UGT’ was more sensitive to DON and its growth was already retarded at 30 mg/L and almost fully inhibited at 120 mg/L DON (Figure S6).
Discussion
FHB resistance can be dissected into several components (Mesterházy et al., 1999) of which resistance to initial infection (also called type 1 resistance) and resistance to fungal spreading (type 2) (Schroeder and Christensen, 1963) are the most common evaluated mechanisms (Buerstmayr et al., 2020). Another resistance component is described as insensitivity of wheat genotypes to DON or ability to degrade DON (Lemmens et al., 2005; Wang and Miller, 1988). However, DON resistance can also be regarded as a component of type 2 resistance, since DON‐deficient F. graminearum mutants with a disrupted trichodiene synthase gene cannot spread within the spike (Bai et al., 2002). Lemmens et al. (2005) directly assessed this resistance component by infiltration of DON solution into wheat florets. They found severe DON‐induced bleaching only in susceptible lines, while resistant lines showed high conversion rates of the applied toxin into D3G and this ability to detoxify DON into D3G coincided with the major FHB resistance QTL Fhb1.
In this study, we evaluated 147 Ae. tauschii accessions for resistance against DON and Fusarium spreading, as these resistance components are considered less affected by environmental factors, more stable across environments and can be more easily studied in the glasshouse (Buerstmayr et al., 2020). In general, FHB resistance assessment in wild relatives of wheat is challenging due to unfavourable agronomic properties such as brittle rachises resulting in seed shattering, small spikes, stiff glumes and weak straw. This makes glasshouse experiments laborious, whereas testing such plant material in field trials is, at best, very difficult but oftentimes not feasible. We adapted our phenotyping protocol and injected F. graminearum spores or DON directly through the tough glumes to maximize the chance of infection and DON absorption. Early seed shattering complicated FHB phenotyping as more than one‐third of the fungus‐treated spikes fell apart during the assessment period and symptomatic spikelets were lost. However, the continuous evaluation of symptomatic and total spikelets per spike allowed us to record all symptomatic spikelets, including those lost due to early shattering. This procedure gave a complete, high‐quality dataset for FHB spreading and DON resistance showing high heritabilities and strong correlations across the trials for the individual traits.
Aegilops tauschii germplasm holds resistance against fusarium spreading controlled by many small‐effect QTL
We observed a wide range of phenotypic variation for FHB spreading resistance in the Ae. tauschii panel, including a few resistant and many highly susceptible accessions. Brisco et al. (2017) also found broad variation and a higher frequency of susceptible genotypes in their evaluation of 109 Ae. tauschii accessions with single floret inoculation. The 21 accessions assessed in both studies showed a similar response to fungal infection (r = 0.7), and the two most resistant accessions, TA2477 (TOWWC0162) and TA1691 (TOWWC0127) studied by Brisco et al. (2017) were also among the most resistant accessions in our trial. In contrast to the previous studies (Brisco et al., 2017; Szabo‐Hever et al., 2018), our most resistant accessions did not reach the highly resistant hexaploid control. In general, FHB severities were higher in our experiments, most likely since we used a highly aggressive isolate.
FHB resistance levels were independent from date of anthesis, number of spikelets per spike and only weakly correlated with plant height in our study, confirming that type 2 resistance is less biased by external factors (Buerstmayr et al., 2020). Plant height was weakly positively associated with FHB severities, which is in contrast to the usually observed relationship, that taller genotypes tend to be more resistant (Buerstmayr et al., 2020; Klahr et al., 2007). Plant height is considered as a passive FHB resistance factor affecting the likelihood of fungal infections under natural disease pressure or spray inoculations. Testing for spreading resistance using single floret inoculation, an increased resistance of taller plants has accordingly not been observed (He et al., 2016; Srinivasachary et al., 2009; Yan et al., 2011). Two key plant height and heading date determinants located on the D subgenome, namely the Rht‐D1 and Ppd‐D1/Rht8 loci, which frequently coincide with FHB resistance QTL in wheat (Buerstmayr et al., 2020) were thus not detected in this study.
The contribution of the D subgenome to FHB resistance in wheat is evidenced by many mapped resistance QTL distributed over all seven chromosomes, although compared with the A and B subgenome, the QTL number is smaller, with about 20% of all detected QTL (Buerstmayr et al., 2020; Venske et al., 2019). The D subgenome is known for its lower genetic diversity (Pont et al., 2019; Zhou et al., 2020), resulting in fewer polymorphic markers and underestimation of the contribution of the D subgenome to FHB resistance in wheat.
In this study, only the potential of the D subgenome to confer FHB resistance was analysed and on a much broader genetic diversity, resulting in many significant associations. The most significant and consistent effect mapped to a 130 Mb linkage disequilibrium block in the pericentromeric region of chromosome 7D, where several FHB resistance QTL in wheat have also been identified e.g. in the resistant cultivar Catbird (Cativelli et al., 2013), the resistant landrace Haiyanzhong, (Li et al., 2011) and the susceptible cultivars Riband (Draeger et al., 2007) and Nanda2419 (Lin et al., 2006). These findings suggest the presence of (a) gene(s) controlling fungal spread in the centromeric region of chromosome 7D in Ae. tauschii and in wheat, but the QTL region comprised too many candidate genes for further analysis.
DON resistance is conferred by conjugation to D3G controlled by a UGT on chromosome 5D
The Ae. tauschii accessions revealed low variation for DON‐induced bleaching after toxin infiltration, except for nine highly susceptible accessions that exhibited severe bleaching. Although DON acts as a virulence factor that promotes fungal spread in wheat spikes (Jansen et al., 2005), DON and FHB spreading resistance were not significantly correlated. Nevertheless, our results show that DON resistance is a prerequisite for fungal spreading resistance as no accessions that were both FHB resistant and DON susceptible were identified (Figure 1a).
Different molecular mechanisms may provide DON resistance, such as sulphation, acetylation and glutathione‐mediated detoxification, as recently demonstrated by the identification of the Fhb7 gene (Wang et al., 2020). Yet, evidence from metabolomics studies (Kluger et al., 2015) indicated that glycosylation seems the most relevant detoxification mechanism in wheat. Conversion of DON into D3G, which has diminished ability to inhibit protein biosynthesis in vitro (Poppenberger et al., 2003), is mediated by members of the very large family 1 of UDP‐glycosyltransferases, which consists of about 150–200 members in diploid grasses (Caputi et al., 2012; Schweiger et al., 2013), not all of which are functional. Despite the expectation that there should be high redundancy in UGTs with substrate specificity for DON, individual genes seem to play essential roles. In Brachypodium distachyon, loss of function of a single UGT gene, Bradi5g03300, led to increased susceptibility to DON and reduced Fusarium resistance (Pasquet et al., 2016). Likewise, an ethyl methane sulphonate‐induced mutation in HvUGT13248 reduced DON resistance and Fusarium spreading resistance in barley (Bethke et al., 2020).
Using the k‐mer association genetics, we identified the highly significant UGT AET5Gv20385300 in the centromeric region of chromosome arm 5DL as the major factor influencing DON resistance in Ae. tauschii in this study. The susceptible accessions showed thereby a clearly distinct haplotype: a single nucleotide deletion resulted in a truncated protein and caused more than three times increased DON severities and five times lower D3G/DON ratios. When expressed in toxin‐sensitive yeast, the truncated version of the UGT did not confer resistance against DON, whereas the full‐length allele exhibited strong activity against DON, nivalenol and HT‐2 toxin similar to the HvUGT13248 detected in barley (Li et al., 2017; Schweiger et al., 2010). HvUGT13248 efficiently detoxified DON (Schweiger et al., 2010) and nivalenol into the non‐toxic nivalenol‐3‐O‐β‐d‐glucoside (Li et al., 2017), and when expressed in transgenic wheat, it conferred resistance against fungal spreading and transformants converted DON more efficiently to D3G (Li et al., 2015).
The AET5Gv20385300 gene identified in this study is orthologous to the barley UGT HvUGT13248, which shows high sequence similarity (90% identity) and is located close to the centromere of chromosome 5H indicating a highly conserved DON resistance mechanism tracing back at least 11 million years radiated from a common ancestor of Ae. tauschii/wheat and barley (Huang et al., 2002).
AET5Gv20385300 detoxifies DON to D3G in Aegilops tauschii and hexaploid wheat
The 16 publicly available whole‐genome wheat sequences (Appels et al., 2018; Walkowiak et al., 2020) and the two control wheat lines were analysed to explore the sequence diversity of the AET5Gv20385300 orthologs in hexaploid wheat. Two allelic variants were identified: 13 genotypes carried the resistant ‘AL8/78 allele’, but Chinese Spring and four additional genotypes (Claire, Norin61, SY Matthis, Robigus) possessed a truncated version of this UGT. This UGT allele (TraesCS5D01G143300) is private to hexaploid wheat, it was not detected in Ae. tauschii and it is not functional for DON detoxification as revealed by the yeast assay.
For these 16 wheat lines no DON resistance data are available, but regarding FHB resistance, both allelic variants comprise genotypes classified as susceptible (e.g. Jagger, Cai and Bai, 2014; Robigus, Muhovski et al., 2012) or moderately resistant (e.g. Chinese Spring, Ma et al., 2006; Cadenza, Fabre et al., 2019) or resistant (e.g. Arina, Draeger et al., 2007; Norin61, Shimizu et al., 2021).
FHB resistance is a truly quantitative trait and DON resistance is one component to which AET5Gv20385300 and its orthologous UGT in wheat most likely contribute.
Moreover, the highly similar homoeologous UGTs on wheat chromosomes 5A (TraesCS5A01G149600, 96.2% identity) and 5B (TraesCS5B01G148300, 94.7% identity) have similar, redundant functions. Interestingly, despite their high sequence similarities distinct differences in their DON‐detoxifying abilities were revealed by the yeast test with the ‘5A UGT’ being as effective against DON as AET5Gv20385300 and the ‘5B UGT’ the least efficient.
The low sequence diversity of the ‘chromosome 5 UGT group’ within the 16 wheat lines suggests that these UGTs add to the general ability of DON detoxification in wheat, all wheat genotypes, including FHB susceptible ones, can metabolize DON into D3G as summarized by Lemmens et al. (2016) and shown for Ae. tauschii in this study. The detailed characterization of these loci on a broader set of wheat lines with known FHB/DON resistance levels and DON/D3G ratios will help to clarify their roles in DON detoxification and FHB resistance. Notably, a genome‐wide transcriptome analysis of 96 European winter wheats revealed that all three homoeologous UGTs were up‐regulated after Fusarium inoculation and thus involved in the wheat/Fusarium interaction (Buerstmayr et al., 2021).
Regarding FHB/DON resistance QTL on wheat chromosome 5D: the Korean wheat cultivar Chokwang was found to carry a type 2 resistance QTL on chromosome arm 5DL (Yang et al., 2005) and in the Chinese landrace Wangshuibai, DON content and resistance to initial infection were affected by chromosome arm 5DL (Yu et al., 2008). Although the closest markers to these QTL were 200 Mb distal to AET5Gv20385300, the large linkage blocks generated in bi‐parental mapping studies, especially in the centromeric regions, do not allow conclusions about the contribution of AET5Gv20385300 to these resistance loci.
Overall, this study demonstrated the applicability of k‐mer based association mapping to dissect the genetic control of the quantitatively inherited FHB resistance and identified a DON resistance QTL on chromosome 5D. A mutation in the UGT gene AET5Gv20385300 at this locus affects the ability to counteract the Fusarium virulence factor DON by conjugation into D3G. Hence, the truncated version of this UGT (TraesCS5D01G143300) present in hexaploid wheat genotypes presumably increases toxin susceptibility and consequently Fusarium susceptibility, which can be considered in genomic breeding approaches when selecting for FHB resistance.
Focusing the analyses on the D subgenome donor Ae. tauschii enabled thus a strong complexity reduction in comparison to hexaploid wheat and showed a great merit to further deepen our knowledge about the complex genetic control of FHB resistance.
Experimental procedures
Plant material
A total of 147 Ae. tauschii accessions were provided by the John Innes Centre, Norwich, UK (Table S9). The accessions were a subset of the Ae. tauschii diversity panel studied by Arora et al. (2019) and Gaurav et al. (2022) and based on whole‐genome sequencing, 113 of the 147 accessions were classified as non‐redundant. The phylogenetic grouping assigned all accessions to the Ae. tauschii lineage 2 (L2). The majority of the accessions originated from collection sites around the Caspian Sea; most descended from Iran (58) and Azerbaijan (57), followed by Armenia (7), Georgia (6), Turkmenistan (4), Turkey (4), Uzbekistan (2), Russia (2), the Syrian Arab Republic (1), the former USSR (1) and five accessions were of unknown origin. Additionally, two hexaploid wheat lines, CM82036 and Remus, were included in this study as resistant and susceptible controls (Buerstmayr et al., 2002). CM82036 is a highly FHB and DON resistant derivative of the spring wheat cultivar Sumai3 originating from China. It carries the two major FHB resistance QTL Fhb1 and Qfhs.ifa‐5A. On the contrary, Remus is a susceptible spring wheat cultivar developed at the Bavarian State Institute for Agronomy in Freising, Germany.
Glasshouse experiments and resistance assessments
Two independent experiments were carried out in the glasshouse of the University of Natural Resources and Life Sciences, Vienna, Department of Agrobiotechnology. The first experiment was set up as an un‐replicated trial during winter 2018/2019, whereas, the second experiment in the winter of 2019/20 was laid as a randomized complete block design with two replications.
Seeds of each accession were sown in multi‐trays in a mixture of heat‐sterilized compost and sand and stratified before germination at 4 °C, 12 h day/night light regime for 1 week. Thereafter, the seeds were germinated at 22°C and at the one leaf stage vernalized for 11 weeks. Five seedlings per accession were planted in 4‐L pots (18 cm diameter, 21 cm height) filled with a mixture of heat‐sterilized compost, peat, sand and rock flour. The pots were randomly placed in the glasshouse and maintained at a temperature of 14/10 °C day/night with 12 h photoperiod for the first 40 days. At spike emergence, the temperature was increased to 22/18 °C day/night with a 16 h photoperiod at 15 000 lx. For both experiments, two pots per accession were planted. In the first experiment (2018/19), one pot was used either for FHB or DON resistance assessments. In the second experiment (2019/20), in each of the two pots, spikes of three plants were inoculated with F. graminearum conidia suspension and spikes of the remaining two plants were infiltrated with DON solution.
FHB resistance assessments
A macroconidia suspension of the F. graminearum single spore isolate ‘IFA 65’ was prepared and stored in aliquots at −80 °C as described in Buerstmayr et al. (2002). Shortly before inoculations, the frozen aliquots were thawed at 37 °C and diluted to a concentration of 50 000 conidia/mL with distilled water. At anthesis, 7–10 spikes per pot were inoculated by pushing an injection needle through the stiff glumes of one central spikelet to insert 20 μL of inoculum into the cavity of the florets (wounding method). The inoculated spikes were covered for 24 h with translucent polyethylene bags sprayed with distilled water to ensure high humidity for fungal infection. The total number of spikelets and the number of F. graminearum‐bleached spikelets per spike were recorded at anthesis and at 7, 11, 15, 19, 23 and 27 days after inoculation (dai). During the disease assessment period premature seed shattering occurred and was monitored for each spike by subtracting the number of spikelets per spike at each evaluation time point from the total number of spikelets per spike counted at spike anthesis.
DON resistance assessments
DON (purity >98%) was purified according to Altpeter and Posselt (1994) and Lemmens et al. (2005) and diluted for the infiltration to a concentration of 25 mg/mL in 0.1% Tween20. In the first experiment, five flowering spikes per pot were infiltrated by directly pipetting 10 μL DON solution between the palea and lemma of the two basal florets of one central spikelet, resulting in the application of 0.5 mg DON per spike. In the second experiment, we infiltrated 20 μL of DON solution into each of three spikes per pot using the ‘wounding method’ described above. Subsequently, spikes were covered with water‐sprayed bags for 24 h. The total number of spikelets per spike and the number of DON‐bleached spikelets per spike were counted at anthesis and at 19, 23, 27 and 31 days after infiltration (dai). As explained before, pre‐mature seed shattering was recorded. At the ripening stage, DON‐treated spikes from each accession were harvested for DON and DON‐3‐glucoside (D3G) measurements.
Plant height
In the spring and fall 2020 the 147 Ae. tauschii accessions were again grown in the ‘IFA‐Tulln glasshouse’ in two replications each as described for the resistance assessment trials. Before ripening, plant height was measured for each pot resulting in four height measurements per accession.
Deoxynivalenol and deoxynivalenol‐3‐glucoside measurements
DON and D3G contents were analysed from 31 accessions infiltrated with DON comprising the nine most DON susceptible and 22 randomly selected moderately resistant accessions. Treated spikes per replication were pooled, ground and 100 mg per sample were analysed. Samples were extracted as described earlier in Doppler et al. (2019), using methanol/acetonitrile/H2O (1.5/1.5/1) + 0.1%FA (v/v) as extraction solvent (Doppler et al., 2016). The samples were analysed with liquid chromatography—high‐resolution mass spectrometry (LC‐HRMS). For this, a Vanquish UHPLC system coupled to a QExactive HF Orbitrap mass spectrometer (both Thermo Fisher Scientific) was used as described in Sauerschnig et al. (2017) with the only change in applying a shorter chromatographic method (20 min total run time; 0–1 min: 10% B, 1–11 min: 35% B, 11–14 min: 100% B, 14–16‐5 min: 100% B; 16.5–20 min: 10% B). DON and D3G standards for LC‐HRMS analysis were acquired from Romer Labs GmbH, Tulln, Austria. Quantification was carried out via external calibration using solutions of pure DON and D3G in methanol/acetonitrile/H2O (1/1/2) + 0.1%FA (v/v) at concentration levels between 1 and 5000 μg/L. Data evaluation was conducted with the vendor software Xcalibur (Thermo Fisher Scientific). In both experiments the same 31 Ae. tauschii accessions as well the two control lines CM82036 and Remus were analysed.
Statistical analysis
The percentage of symptomatic spikelets at the individual evaluation time‐points, the number of spikelets per spike and the date of anthesis were analysed using a linear mixed model, which considered subsampling of individual spikes within each pot as follows:
where denotes the observation of the individual spikes, is the grand mean and is the genetic effect of the th accession. The environment effect is defined as the effect of the th year and the genotype‐by‐environment interaction is described by . is the effect of the 𝑘th replication within the th year, the effect of the th pot within the th replication and th year and is the residual term.
Plant height in cm and seed shattering incidence in % were analysed with the following linear mixed model:
where is the phenotypic record of the th accession tested in the th year in the th replication. is the grand mean and is the genetic effect of the th accession.
The environment effect is defined as the effect of the th year and describes the genotype‐by‐environment interaction. is the effect of the th replication at the th year and is the residual term.
Analysis for both models was performed with R 3.5.1 (R Core Team, 2020) using the package sommer 4.0.8 (Covarrubias‐Pazaran, 2016) with all effects considered as random, except which was modelled as a fixed effect to obtain the Best Linear Unbiased Estimates (BLUEs).
To calculate BLUEs across years for FHB and DON severities expressed in %, the scoring time‐point combination with the highest correlation between the 2 years and the broadest variation for the traits was selected.
Heritabilities were estimated by assigning all effects as random and using the variance components following the formula (Holland et al., 2003):
where is the genetic variance, the genotype‐by‐year interaction, the interaction of genotype‐by‐year‐by‐replication, the residual variance component, the number of years, the number of replications and the number of treated spikes.
Multiple paired t‐tests were performed to compare BLUEs across trials of DON and FHB severities for candidate gene alleles using the R package ggpubr 0.2.4 (Kassambra, 2019).
Association genetics analysis
Whole‐genome shotgun sequences of the Ae. tauschii accessions were previously generated under the aegis of the Open Wild Wheat Consortium (http://www.openwildwheat.org/). Genome‐wide association mapping was carried out based on sub‐sequences (k‐mers) following the procedures described in Arora et al. (2019) and Gaurav et al. (2022). The reference genome assembly of Ae. tauschii subsp. strangulata accession AL8/78 (Luo et al., 2017) was used for genome alignments and comparative annotation.
As described in Gaurav et al. (2022), −log10 P‐value = 9.3 was taken as the Bonferroni‐adjusted threshold for evaluating the statistical significance of an association. Association mapping plots were generated using Python 3.7 (plotting script available at https://github.com/wheatgenetics/owwc).
Allelic diversity analysis of the candidate genes
The 31 Ae. tauschii accessions with contrasting DON severities and DON and D3G LC‐HRMS measurements were evaluated for their allelic constitutions at the identified candidate genes. Scaffolds of de novo sequence assemblies of the accessions (Gaurav et al., 2022, DOIs 10.5281/zenodo.4430803, 10.5281/zenodo.4430872 and 10.5281/zenodo.4430891) were mapped to contigs of the 5D AL8/78 reference sequence comprising the candidate genes UGT (AET5Gv20385300: LOC109746039) and two ABC transporters G family member (AET5Gv20401700: LOC109787618, AET5Gv20401700: LOC109787645). Sequence alignments were generated with Geneious version 8.1 (https://www.geneious.com).
The diversity analysis of the UGT candidate gene was extended to the 301 sequenced Ae. tauschii accessions, including the set of 242 non‐redundant accessions (Gaurav et al., 2022) and the four additional Ae. tauschii reference genomes AY61, AY17, XJ02 and T093 (Zhou et al., 2021).
The Chinese Spring reference sequence (Appels et al., 2018), 10 chromosome pseudomolecules and five scaffold assemblies of hexaploid wheat (http://www.10wheatgenomes.com) (Walkowiak et al., 2020) were explored to analyse the sequence diversity of the UGT among hexaploid wheat and among the homoeologous UGT copies on the A and B genome. Furthermore, the Svevo durum wheat reference sequence (RefSeq Rel. 1.0) (Maccaferri et al., 2019) was used to evaluate the homoeologous UGT genes on the A and B genome in T. durum.
The UGT haplotypes of the 31 Ae. tauschii accessions analysed for DON and D3G, Chinese Spring and the control lines CM82036 and Remus were also generated by Sanger sequencing using the following PCR primer combination UGT5D‐F 5′ TGGAAAATGTCGGCCACTCT and UGT5D‐R 5′ TCCGGTGTGAGCTGTGAATC.
Functional testing of AET5Gv20385300 and the homoeologous UGTs on wheat chromosomes 5A and 5B in Saccharomyces cerevisiae
Synthetic versions of AET5Gv20385300, TraesCS5A01G149600 and TraesCS5B01G148300 were produced which were codon‐optimized for expression in yeast (GenScript; clone U425QGD160_2, clone U936HE240_2, clone U936HE240_4, respectively, Figures S5, S7). The synthesized DNAs U425QGD160_2, U936HE240_2 and U936HE240_4 were digested with HindIII and NotI and the inserts containing the UGTs were ligated to the HindIII‐NotI digested backbone of pWS1921/pYAK7 (Li et al., 2017; Poppenberger et al., 2003), yielding pGW1259 (AET5Gv20385300), pGW1300 (TraesCS5A01G149600) and pGW1301 (TraesCS5B01G148300), respectively. These plasmids contain the synthetic genes c‐terminally fused to the c‐myc epitope under the control of the strong constitutive ScADH1 promoter and terminator. The truncated versions of AET5Gv20385300 were produced by PCR using pGW1259 as template and the following primer pairs: ADHfw (CATTGTTCTCGTTCCCTTTCTTCC) + AetUGTdelAet_rv (ATTATTGCGGCCGCTCAGTCCAATCTTCTCACATTTTCTTTCAGTTCC) or ADHfw + AetUGTdelCS_rv (ATTATTGCGGCCGCTAGTGCGGAATACCAACCAGCG), respectively. The PCR fragments were digested with HindIII and NotI and cloned into the pWS1921 backbone. The resulting plasmids (pGW1260 and pGW1266) expressed the UGT proteins as present in the DON susceptible Ae. tauschii accessions and in Chinese Spring, respectively (Figure S5).
To test the functionality of AET5Gv20385300, TraesCS5A01G149600 and TraesCS5B01G148300 against trichothecenes all plasmids were transformed into the DON sensitive S. cerevisiae strain YZGA515 (Poppenberger et al., 2003) together with the following control plasmids: pBP910 (empty plasmid, Poppenberger et al., 2003), pWS1921 (HvUGT13248, Li et al., 2017) and pHW74 (ScAYT1 (https://www.yeastgenome.org/locus/S000003986) in pWS1921 backbone). Transformants were grown in selective medium to mid‐log phase, diluted to OD600 of 0.3 and 0.03, respectively, and spotted onto yeast‐extract‐peptone‐dextrose (YPD) plates (1% yeast extract, 2% peptone, 2% dextrose, 2% agar) containing varying concentrations of DON, nivalenol, T‐2 toxin, HT‐2 toxin and fusarenone X.
Conflicts of interest
The authors declare that they have no conflict of interests.
Author contributions
BS, HB, BBHW, SA, KG, GW and GA planned and designed the research. RPK, BS, ML, SZ, MM, CEE, MK and AT performed the glasshouse experiments, GW and GA the testing of the UDP‐glucosyltransferase in yeast and MD and RS analysed deoxynivalenol and DON‐3‐O‐glucoside contents. RPK, KG, BS and SM performed data analysis. BS, RPK and GA wrote the draft of the manuscript. BBHW, KG, SM and SA revised the manuscript.
Supporting information
Figure S1 Pearson's correlation coefficients of the Aegilops tauschii accessions for Fusarium head blight and deoxynivalenol severities and other traits.
Figure S2 k‐mer‐based association genetics for Fusarium head blight severity.
Figure S3 Amino acid alignment of the allelic diversity at the UGT for the sequenced Ae. tauschii accessions and wheat cv. Chinese Spring.
Figure S4 Amino acid alignment of the chromosome ‘5D UGTs’ and the homoeologous proteins at chromosomes 5A and 5B of Chinese Spring.
Figure S5 Sequence of the codon‐optimized Ae. tauschii UGT AET5Gv20385300, the deduced protein sequence and primers used for in vitro mutagenesis.
Figure S6 Functional evaluation of the homoeologous uridine diphosphate (UDP)‐glucosyltransferases (UGT) at chromosomes 5A (TraesCS5A01G149600), 5B (TraesCS5B01G148300) and 5D (TraesCS5D01G143300, AET5Gv20385300).
Figure S7 Sequences of the codon‐optimized UGTs TraesCS5A01G149600 and TraesCS5B01G148300.
Table S1 Analysis of variance for the evaluated traits.
Table S2 Pairwise correlations of the evaluated traits.
Table S3 Best linear unbiased estimates of the evaluated traits in 2019 and 2020.
Table S4 Best linear unbiased estimates of the evaluated traits across years.
Table S5 Deoxynivalenol (DON) and DON‐3‐O‐glucoside (D3G) contents and the ratio of D3G/DON.
Table S6 The mapped region significantly associated with Fusarium head blight resistance on chromosome 7D.
Table S7 The mapped region significantly associated with deoxynivalenol resistance on chromosome 5D.
Table S8 Allelic diversity of the uridine diphosphate (UDP)‐glucosyltransferase (UGT) for the 305 Ae. tauschii accessions and 18 hexaploid wheat lines.
Table S9 List of phenotyped Ae. tauschii accessions.
Acknowledgements
RPK was supported and funded by the Ministry of Education and Culture of the Republic of Indonesia and the Austrian Agency for International Cooperation in Education and Research (OeAD‐GmbH) in cooperation with ASEA‐UNINET. MM received funding from the European Commission, Erasmus+ programme and CEE from OeAD‐GmbH (Ernst Mach Grant) and the West African Science Service Centre on Climate Change and Adapted Land Use. Funding for this research was also provided by the Austrian Science Fund, Special Research project SFB F3711.
References
- Altpeter, F. and Posselt, U.K. (1994) Production of high quantities of 3‐acetyldeoxynivalenol and deoxynivalenol. Appl. Microbiol. Biotechnol. 41, 384–387. [Google Scholar]
- Anikster, Y. , Manisterski, J. , Long, D.L. and Leonard, K.J. (2005) Resistance to Leaf Rust, Stripe Rust, and Stem Rust in Aegilops spp. in Israel. Plant Dis. 89, 303–308. [DOI] [PubMed] [Google Scholar]
- Appels, R. , Eversole, K. , Feuillet, C. , Keller, B. , Rogers, J. , Stein, N. , Pozniak, C.J. et al. (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science, 361, eaar7191. [DOI] [PubMed] [Google Scholar]
- Arora, S. , Steuernagel, B. , Gaurav, K. , Chandramohan, S. , Long, Y. , Matny, O. , Johnson, R. et al. (2019) Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 37, 139–143. [DOI] [PubMed] [Google Scholar]
- Bai, G.H. , Desjardins, A.E. and Plattner, R.D. (2002) Deoxynivalenol‐nonproducing fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia, 153, 91–98. [DOI] [PubMed] [Google Scholar]
- Bethke, G. , Huang, Y. , Hensel, G. , Wyant, S. , Li, X. , Morrell, P. , Kumlehn, J. et al. (2020) The barley UDP‐glycosyltransferase UGT13248 is required for deoxynivalenol conjugation and type 2 resistance to Fusarium head blight. In Proceedings of the 2020 National Fusarium Head Blight Forum( Canty, S. , Hoffstetter, A. and Dill‐Macky, R. , eds), p. 63. East Lansing, MI: U.S. Wheat & Barley Scab Inititiative. [Google Scholar]
- Börner, A. , Ogbonnaya, F.C. , Röder, M.S. , Rasheed, A. , Periyannan, S. and Lagudah, E.S. (2015) Aegilops Tauschii introgressions in wheat. In Alien introgression in wheat( Molnár‐Láng, M. , Ceoloni, C. and Doležel, J. , eds), pp. 245–271. Cham: Springer International Publishing. [Google Scholar]
- Bottalico, A. and Perrone, G. (2002) Toxigenic Fusarium species and mycotoxins associated with head blight in small‐grain cereals in Europe. Eur. J. Plant Pathol. 108, 611–624. [Google Scholar]
- Brisco, E.I. , Brown, L.K. and Olson, E.L. (2017) Fusarium head blight resistance in Aegilops tauschii. Genet. Resour. Crop. Evol. 64, 2049–2058. [Google Scholar]
- Buerstmayr, H. , Lemmens, M. , Hartl, L. , Doldi, L. , Steiner, B. , Stierschneider, M. and Ruckenbauer, P. (2002) Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (Type II resistance). Theor. Appl. Genet. 104, 84–91. [DOI] [PubMed] [Google Scholar]
- Buerstmayr, M. , Steiner, B. and Buerstmayr, H. (2020) Breeding for Fusarium head blight resistance in wheat—Progress and challenges. Plant Breed. 139, 429–454. [Google Scholar]
- Buerstmayr, M. , Wagner, C. , Nosenko, T. , Omony, J. , Steiner, B. , Nussbaumer, T. , Mayer, K.F.X. et al. (2021) Fusarium head blight resistance in European winter wheat: insights from genome‐wide transcriptome analysis. BMC Genomics 22, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, J. and Bai, G. (2014) Quantitative trait loci for Fusarium head blight resistance in Huangcandou × ‘Jagger’ wheat population. Crop Sci. 54, 2520–2528. [Google Scholar]
- Caputi, L. , Malnoy, M. , Goremykin, V. , Nikiforova, S. and Martens, S. (2012) A genome‐wide phylogenetic reconstruction of family 1 UDP‐glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 69, 1030–1042. [DOI] [PubMed] [Google Scholar]
- Cativelli, M. , Lewis, S. and Appendino, M.L. (2013) A Fusarium head blight resistance quantitative trait locus on chromosome 7d of the spring wheat cultivar catbird. Crop Sci. 53, 1464–1471. [Google Scholar]
- Covarrubias‐Pazaran, G. (2016) Genome‐assisted prediction of quantitative traits using the R Package sommer. PLoS One, 11, e0156744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doppler, M. , Kluger, B. , Bueschl, C. , Schneider, C. , Krska, R. , Delcambre, S. , Hiller, K. et al. (2016) Stable isotope‐assisted evaluation of different extraction solvents for untargeted metabolomics of plants. Int. J. Mol. Sci. 17, 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doppler, M. , Kluger, B. , Bueschl, C. , Steiner, B. , Buerstmayr, H. , Lemmens, M. , Krska, R. et al. (2019) Stable isotope‐assisted plant metabolomics: investigation of phenylalanine‐related metabolic response in wheat upon treatment with the fusarium virulence factor deoxynivalenol. Front. Plant Sci. 10, 1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draeger, R. , Gosman, N. , Steed, A. , Chandler, E. , Thomsett, M. , Srinivasachary, S.J. , Buerstmayr, H. et al. (2007) Identification of QTLs for resistance to Fusarium head blight, DON accumulation and associated traits in the winter wheat variety Arina. Theor. Appl. Genet. 115, 617–625. [DOI] [PubMed] [Google Scholar]
- Fabre, F. , Bormann, J. , Urbach, S. , Roche, S. , Langin, T. and Bonhomme, L. (2019) Unbalanced roles of fungal aggressiveness and host cultivars in the establishment of the fusarium head blight in bread wheat. Front. Microbiol. 10, 2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroud, N.A. , Baines, D. , Gagkaeva, T.Y. , Thakor, N. , Badea, A. , Steiner, B. , Bürstmayr, M. et al. (2019) Trichothecenes in cereal grains – an update. Toxins, 11, 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta, A. , Colasuonno, P. , Giove, S.L. , Blanco, A. and Giancaspro, A. (2019) Map‐based cloning of QFhb.mgb‐2A identifies a WAK2 gene responsible for Fusarium Head Blight resistance in wheat. Sci. Rep. 9, 6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaurav, K. , Arora, S. , Silva, P. , Sánchez‐Martín, J. , Horsnell, R. , Gao, L. , Brar, G.S. et al. (2022) Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat. Biotechnol. 40, 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Gunupuru, L.R. , Perochon, A. and Doohan, F.M. (2017) Deoxynivalenol resistance as a component of FHB resistance. Trop. Plant Pathol. 42, 175–183. [Google Scholar]
- He, X. , Singh, P.K. , Dreisigacker, S. , Singh, S. , Lillemo, M. and Duveiller, E. (2016) Dwarfing genes Rht‐B1b and Rht‐D1b are associated with both Type I FHB susceptibility and low anther extrusion in two bread wheat populations. PLoS One, 11, e0162499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, J. , Nyquist, W.E. and Cervantes‐Martinez, C.T. (2003) Estimating and interpreting heritability for plant breeding: An update. Plant Breed. Rev. 22, 9–111. [Google Scholar]
- Huang, S. , Sirikhachornkit, A. , Faris, J.D. , Su, X. , Gill, B.S. , Haselkorn, R. and Gornicki, P. (2002) Phylogenetic analysis of the acetyl‐CoA carboxylase and 3‐phosphoglycerate kinase loci in wheat and other grasses. Plant Mol. Biol. 48, 805–820. [DOI] [PubMed] [Google Scholar]
- Jansen, C. , von Wettstein, D. , Schäfer, W. , Kogel, K.‐H. , Felk, A. and Maier, F.J. (2005) Infection patterns in barley and wheat spikes inoculated with wild‐type and trichodiene synthase gene disrupted Fusarium graminearum . Proc. Natl Acad. Sci. USA 102, 16892–16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambra A (2019) Ggpubr: ‘ggplot2’ Based Publication Ready Plots. R package version 0.2.4.
- Kishii, M. (2019) An update of recent use of aegilops species in wheat breeding. Front. Plant Sci. 10, 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahr, A. , Zimmermann, G. , Wenzel, G. and Mohler, V. (2007) Effects of environment, disease progress, plant height and heading date on the detection of QTLs for resistance to Fusarium head blight in an European winter wheat cross. Euphytica, 154, 17–28. [Google Scholar]
- Kluger, B. , Bueschl, C. , Lemmens, M. , Michlmayr, H. , Malachova, A. , Koutnik, A. , Maloku, I. et al. (2015) Biotransformation of the mycotoxin deoxynivalenol in fusarium resistant and susceptible near isogenic wheat lines. PLoS One, 10, e0119656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen, H.K. , Alexander, J. , Barregård, L. , Bignami, M. , Brüschweiler, B. , Ceccatelli, S. , Cottrill, B. et al. (2017) Risks to human and animal health related to the presence of deoxynivalenol and its acetylated and modified forms in food and feed. EFSA J. 15, e04718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagudah, E.S. and Krattinger, S.G. (2019) A new player contributing to durable Fusarium resistance. Nat. Genet. 51, 1070–1071. [DOI] [PubMed] [Google Scholar]
- Lemmens, M. , Scholz, U. , Berthiller, F. , Dall'Asta, C. , Koutnik, A. , Schuhmacher, R. , Adam, G. et al. (2005) The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol. Plant‐Microbe Interact. 18, 1318–1324. [DOI] [PubMed] [Google Scholar]
- Lemmens, M. , Steiner, B. , Sulyok, M. , Nicholson, P. , Mesterhazy, A. and Buerstmayr, H. (2016) Masked mycotoxins: does breeding for enhanced Fusarium head blight resistance result in more deoxynivalenol‐3‐glucoside in new wheat varieties? World Mycotoxin J. 9, 741–754. [Google Scholar]
- Li, T. , Bai, G. , Wu, S. and Gu, S. (2011) Quantitative trait loci for resistance to fusarium head blight in a Chinese wheat landrace Haiyanzhong. Theor. Appl. Genet. 122, 1497–1502. [DOI] [PubMed] [Google Scholar]
- Li, X. , Shin, S. , Heinen, S. , Dill‐Macky, R. , Berthiller, F. , Nersesian, N. , Clemente, T. et al. (2015) Transgenic wheat expressing a barley UDP‐glucosyltransferase detoxifies deoxynivalenol and provides high levels of resistance to Fusarium graminearum . Mol. Plant‐Microbe Interact. 28, 1237–1246. [DOI] [PubMed] [Google Scholar]
- Li, X. , Michlmayr, H. , Schweiger, W. , Malachova, A. , Shin, S. , Huang, Y. , Dong, Y. et al. (2017) A barley UDP‐glucosyltransferase inactivates nivalenol and provides Fusarium Head Blight resistance in transgenic wheat. J. Exp. Bot. 68, 2187–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. , Jia, L. , Zhou, J. , Fan, J. , Yan, H. , Shi, J. , Wang, X. et al. (2019) Evaluation and precise mapping of QFhb.nau‐2B conferring resistance against Fusarium infection and spread within spikes in wheat (Triticum aestivum L.). Mol. Breed. 39, 62. [Google Scholar]
- Lin, F. , Xue, S.L. , Zhang, Z.Z. , Zhang, C.Q. , Kong, Z.X. , Yao, G.Q. , Tian, D.G. et al. (2006) Mapping QTL associated with resistance to Fusarium head blight in the Nanda2419 × Wangshuibai population. II: type I resistance. Theor. Appl. Genet. 112, 528–535. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Salsman, E. , Fiedler, J.D. , Hegstad, J.B. , Green, A. , Mergoum, M. , Zhong, S. et al. (2019) Genetic mapping and prediction analysis of FHB resistance in a hard red spring wheat breeding population. Front. Plant Sci. 10, 1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler, M. , Schön, C.‐C. and Miedaner, T. (2009) Revealing the genetic architecture of FHB resistance in hexaploid wheat (Triticum aestivum L.) by QTL meta‐analysis. Mol. Breed. 23, 473–488. [Google Scholar]
- Luo, M.‐C. , Gu, Y.Q. , Puiu, D. , Wang, H. , Twardziok, S.O. , Deal, K.R. , Huo, N. et al. (2017) Genome sequence of the progenitor of the wheat D genome Aegilops tauschii . Nature, 551, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H.‐X. , Bai, G.‐H. , Zhang, X. and Lu, W.‐Z. (2006) Main effects, epistasis, and environmental interactions of quantitative trait Loci for fusarium head blight resistance in a recombinant inbred population. Phytopathology, 96, 534–541. [DOI] [PubMed] [Google Scholar]
- Maccaferri, M. , Harris, N.S. , Twardziok, S.O. , Pasam, R.K. , Gundlach, H. , Spannagl, M. , Ormanbekova, D. et al. (2019) Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51, 885–895. [DOI] [PubMed] [Google Scholar]
- McFadden, E.S. and Sears, E.R. (1946) The origin of Triticum spelta and its free‐threshing hexaploid relatives. J. Hered. 37, 107–116. [DOI] [PubMed] [Google Scholar]
- Mesterházy, Á. , Bartók, T. , Mirocha, C.G. and Komoróczy, R. (1999) Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant Breed. 118, 97–110. [Google Scholar]
- Michlmayr, H. , Varga, E. , Malachová, A. , Fruhmann, P. , Piątkowska, M. , Hametner, C. , Šofrová, J. et al. (2018) UDP‐glucosyltransferases from rice, brachypodium, and barley: substrate specificities and synthesis of type A and B trichothecene‐3‐O‐β‐d‐glucosides. Toxins, 10, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhovski, Y. , Batoko, H. and Jacquemin, J.‐M. (2012) Identification, characterization and mapping of differentially expressed genes in a winter wheat cultivar (Centenaire) resistant to Fusarium graminearum infection. Mol. Biol. Rep. 39, 9583–9600. [DOI] [PubMed] [Google Scholar]
- Mujeeb‐Kazi, A. (2001a) Intergeneric hybrids in wheat: Current status in CIMMYT. In International Triticeae IV Symposium( Hernandez, P. , Moreno, M.T. , Cubero, J. and Martin, A. , eds), pp. 261–264. Cordoba, Spain: Junta de Andalucia. Consejeria de Agricultura y Pesca. [Google Scholar]
- Mujeeb‐Kazi, A. (2001b) Synthetic hexaploids for bread wheat improvement. In International Triticeae IV Symposium( Hernandez, P. , Moreno, M.T. , Cubero, J. and Martin, A. , eds), pp. 193–199. Cordoba, Spain: Junta de Andalucia. Consejeria de Agricultura y Pesca. [Google Scholar]
- Oliver, R.E. , Cai, X. , Xu, S.S. , Chen, X. and Stack, R.W. (2005) Wheat‐Alien species derivatives: a novel source of resistance to fusarium head blight in wheat. Crop Sci. 45, 1353–1360. [Google Scholar]
- Pasquet, J.‐C. , Changenet, V. , Macadré, C. , Boex‐Fontvieille, E. , Soulhat, C. , Bouchabké‐Coussa, O. , Dalmais, M. et al. (2016) A Brachypodium UDP‐glycosyltransferase confers root tolerance to deoxynivalenol and resistance to Fusarium infection. Plant Physiol. 172, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka, J. (2010) Toxicological mechanisms and potential health effects of deoxynivalenol and nivalenol. World Mycotoxin J. 3, 323–347. [Google Scholar]
- Pont, C. , Leroy, T. , Seidel, M. , Tondelli, A. , Duchemin, W. , Armisen, D. , Lang, D. et al. (2019) Tracing the ancestry of modern bread wheats. Nat. Genet. 51, 905–911. [DOI] [PubMed] [Google Scholar]
- Poppenberger, B. , Berthiller, F. , Lucyshyn, D. , Sieberer, T. , Schuhmacher, R. , Krska, R. , Kuchler, K. et al. (2003) Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP‐glucosyltransferase from Arabidopsis thaliana . J. Biol. Chem. 278, 47905–47914. [DOI] [PubMed] [Google Scholar]
- Rawat, N. , Pumphrey, M.O. , Liu, S. , Zhang, X. , Tiwari, V.K. , Ando, K. , Trick, H.N. et al. (2016) Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore‐forming toxin‐like domain conferring resistance to Fusarium head blight. Nat. Genet. 48, 1576–1580. [DOI] [PubMed] [Google Scholar]
- R Core Team . (2020) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available from: https://www.R‐project.org/ [Google Scholar]
- Sauerschnig, C. , Doppler, M. , Bueschl, C. and Schuhmacher, R. (2017) Methanol generates numerous artifacts during sample extraction and storage of extracts in metabolomics research. Metabolites, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, H.W. and Christensen, J.J. (1963) Factors affecting resistance of Wheat to scab caused by Gibberella zeae . Phytopathology, 53, 831–838. [Google Scholar]
- Schweiger, W. , Boddu, J. , Shin, S. , Poppenberger, B. , Berthiller, F. , Lemmens, M. , Muehlbauer, G.J. et al. (2010) Validation of a candidate deoxynivalenol‐inactivating UDP‐glucosyltransferase from barley by heterologous expression in yeast. Mol. Plant‐Microbe Interact. 23, 977–986. [DOI] [PubMed] [Google Scholar]
- Schweiger, W. , Steiner, B. , Ametz, C. , Siegwart, G. , Wiesenberger, G. , Berthiller, F. , Lemmens, M. et al. (2013) Transcriptomic characterization of two major Fusarium resistance quantitative trait loci (QTLs), Fhb1 and Qfhs.ifa‐5A, identifies novel candidate genes. Mol. Plant Pathol. 14, 772–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K.K. , Copetti, D. , Okada, M. , Wicker, T. , Tameshige, T. , Hatakeyama, M. , Shimizu‐Inatsugi, R. et al. (2021) De novo genome assembly of the Japanese wheat cultivar norin 61 highlights functional variation in flowering time and Fusarium‐resistant genes in East Asian genotypes. Plant Cell Physiol. 62, 8–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasachary, G.N. , Steed, A. , Hollins, T.W. , Bayles, R. , Jennings, P. and Nicholson, P. (2009) Semi‐dwarfing Rht‐B1 and Rht‐D1 loci of wheat differ significantly in their influence on resistance to Fusarium head blight. Theor. Appl. Genet. 118, 695–702. [DOI] [PubMed] [Google Scholar]
- Su, Z. , Bernardo, A. , Tian, B. , Chen, H. , Wang, S. , Ma, H. , Cai, S. et al. (2019) A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 51, 1099–1105. [DOI] [PubMed] [Google Scholar]
- Szabo‐Hever, A. , Zhang, Q. , Friesen, T.L. , Zhong, S. , Elias, E.M. , Cai, X. , Jin, Y. et al. (2018) Genetic diversity and resistance to fusarium head blight in synthetic hexaploid wheat derived from Aegilops tauschii and diverse Triticum turgidum Subspecies. Front. Plant Sci., 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venske, E. , Dos Santos, R.S. , Farias, D.R. , Rother, V. , da Maia, L.C. , Pegoraro, C. and Costa de Oliveira, A. (2019) Meta‐analysis of the QTLome of Fusarium head blight resistance in bread wheat: refining the current puzzle. Front. Plant Sci. 10, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkowiak, S. , Gao, L. , Monat, C. , Haberer, G. , Kassa, M.T. , Brinton, J. , Ramirez‐Gonzalez, R.H. et al. (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature, 588, 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.Z. and Miller, J.D. (1988) Effects of Fusarium graminearum metabolites on wheat tissue in relation to Fusarium head blight resistance. J. Phytopathol. 122, 118–125. [Google Scholar]
- Wang, H. , Sun, S. , Ge, W. , Zhao, L. , Hou, B. , Wang, K. , Lyu, Z. et al. (2020) Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science, 368, eaba5435. [DOI] [PubMed] [Google Scholar]
- Yan, W. , Li, H.B. , Cai, S.B. , Ma, H.X. , Rebetzke, G.J. and Liu, C.J. (2011) Effects of plant height on type I and type II resistance to fusarium head blight in wheat. Plant Pathol. 60, 506–512. [Google Scholar]
- Yang, J. , Bai, G. and Shaner, G.E. (2005) Novel quantitative trait loci (QTL) for Fusarium head blight resistance in wheat cultivar Chokwang. Theor. Appl. Genet. 111, 1571–1579. [DOI] [PubMed] [Google Scholar]
- Yu, J.‐B. , Bai, G.‐H. , Zhou, W.‐C. , Dong, Y.‐H. and Kolb, F.L. (2008) Quantitative trait loci for Fusarium head blight resistance in a recombinant inbred population of Wangshuibai/Wheaton. Phytopathology, 98, 87–94. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Zhao, X. , Li, Y. , Xu, J. , Bi, A. , Kang, L. , Xu, D. et al. (2020) Triticum population sequencing provides insights into wheat adaptation. Nat. Genet. 52, 1412–1422. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Bai, S. , Li, H. , Sun, G. , Zhang, D. , Ma, F. , Zhao, X. et al. (2021) Introgressing the Aegilops tauschii genome into wheat as a basis for cereal improvement. Nat. Plants 7, 774–786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Pearson's correlation coefficients of the Aegilops tauschii accessions for Fusarium head blight and deoxynivalenol severities and other traits.
Figure S2 k‐mer‐based association genetics for Fusarium head blight severity.
Figure S3 Amino acid alignment of the allelic diversity at the UGT for the sequenced Ae. tauschii accessions and wheat cv. Chinese Spring.
Figure S4 Amino acid alignment of the chromosome ‘5D UGTs’ and the homoeologous proteins at chromosomes 5A and 5B of Chinese Spring.
Figure S5 Sequence of the codon‐optimized Ae. tauschii UGT AET5Gv20385300, the deduced protein sequence and primers used for in vitro mutagenesis.
Figure S6 Functional evaluation of the homoeologous uridine diphosphate (UDP)‐glucosyltransferases (UGT) at chromosomes 5A (TraesCS5A01G149600), 5B (TraesCS5B01G148300) and 5D (TraesCS5D01G143300, AET5Gv20385300).
Figure S7 Sequences of the codon‐optimized UGTs TraesCS5A01G149600 and TraesCS5B01G148300.
Table S1 Analysis of variance for the evaluated traits.
Table S2 Pairwise correlations of the evaluated traits.
Table S3 Best linear unbiased estimates of the evaluated traits in 2019 and 2020.
Table S4 Best linear unbiased estimates of the evaluated traits across years.
Table S5 Deoxynivalenol (DON) and DON‐3‐O‐glucoside (D3G) contents and the ratio of D3G/DON.
Table S6 The mapped region significantly associated with Fusarium head blight resistance on chromosome 7D.
Table S7 The mapped region significantly associated with deoxynivalenol resistance on chromosome 5D.
Table S8 Allelic diversity of the uridine diphosphate (UDP)‐glucosyltransferase (UGT) for the 305 Ae. tauschii accessions and 18 hexaploid wheat lines.
Table S9 List of phenotyped Ae. tauschii accessions.


