Abstract
Chaperone-mediated autophagy (CMA) is a chaperone-dependent process of selective cytosolic protein turnover that targets specific proteins to lysosomes for degradation. Enhancing protein degradation mechanisms has been shown to be beneficial in multiple models of cardiac disease, including myocardial infarction (MI) and ischemia-reperfusion (I/R) injury. However, the causal role of CMA in cardiomyocyte injury and death is largely unknown. Hypoxia is an important contributor to both MI and I/R damage, which are major, precedent causes of heart failure. Upregulating CMA was hypothesized to protect against hypoxia-induced cardiomyocyte death. Lysosome-associated membrane protein 2a (Lamp2a) overexpression and knockdown were used to causally study CMA’s role in hypoxically stressed cardiomyocytes. LAMP2a protein levels were used as both a primary indicator and driver of CMA function. Hypoxic stress was stimulated by CoCl2 treatment, which increased LAMP2a protein levels (+1.4-fold) and induced cardiomyocyte apoptosis (+3.2–4.0-fold). Lamp2a siRNA knockdown (−3.2-fold) of control cardiomyocytes increased apoptosis (+1.8-fold) suggesting that loss of CMA is detrimental for cardiomyocyte survival. However, there was neither an additive nor a synergistic effect on cell death when Lamp2a-silenced cells were treated with CoCl2. Conversely, Lamp2a overexpression (+3.0-fold) successfully reduced hypoxia-induced apoptosis by ∼50%. LAMP2a was also significantly increased (+1.7-fold) in ischemic heart failure patient samples, similar to hypoxically stressed cardiomyocytes. The failing ischemic hearts may have had insufficient CMA activation. To our knowledge, this study for the first time establishes a protective role for CMA (via Lamp2a overexpression) against hypoxia-induced cardiomyocyte loss and reveals the intriguing possibility that CMA activation may offer a cardioprotective treatment for ischemic heart disease.
Keywords: cardiomyocytes, cell death, chaperone-mediated autophagy, hypoxia, ischemic heart failure
INTRODUCTION
Despite recent medical advances, the prevalence of cardiovascular diseases [comprising ischemic heart disease (IHD), heart failure, stroke, and hypertension] is 49.2% in adults of ≥20 yr of age (1). According to the World Health Organization, ischemic heart disease is the leading cause of death globally, responsible for >9 million deaths in 2016 (2). Tight regulation of protein degradation pathways is essential for maintaining homeostatic protein turnover in the myocardium, and recovery from ischemic-related cardiac injury (3, 4). Under conditions of cardiac stress or pathology, mechanisms of intracellular protein degradation function by eliminating misfolded, redundant, mutant, and disease-causing proteins (5, 6). There are two major and well-studied intracellular protein degradation pathways: ubiquitin-proteasome system (UPS) and the macroautophagy-lysosome pathway (6–8). Both pathways participate in bulk intracellular protein degradation to modulate normal protein turnover (5, 7, 9, 10). Emerging evidence suggests that loss of UPS or macroautophagy functions are widely associated with several forms of heart failure, including hypertrophic, ischemic, and desmin-related myopathy (7–9, 11–16). Conversely, enhancing these pathways reduced the accumulation of disease-causing proteins and conferred protection to mice against cardiac proteinopathy and ischemic-reperfusion injury (5, 17, 18). Targeting these bulk proteolytic pathways has not yet been translated therapeutically for treating patients with heart failure probably because bulk degradation fails to specifically clear the individual disease-causing proteins. Consequently, targeting a third novel and more selective mechanism of protein degradation, namely, chaperone-mediated autophagy (CMA), may prove to be beneficial in the amelioration of cardiac pathology.
The CMA pathway in the heart is a relatively unexplored proteolytic mechanism (19). Unlike the bulk protein degradation mechanisms, CMA selectively degrades specific substrate proteins (including misfolded and mutant proteins) one at a time and is only found in mammalian cells (20, 21). CMA uses a co-chaperone complex led by a cytosolic and lysosomal protein named, heat shock cognate-70 (HSC-70; also known as HSPA8), to target and bind to specific cytosolic proteins containing a KFERQ-like motif (22, 23). The chaperone-bound CMA substrate proteins are then trafficked to the lysosome where they are recognized by a lysosome-associated membrane protein type 2a (LAMP2a) receptor protein, located in the lysosomal membrane. The LAMP2a receptor then oligomerizes and forms a translocon complex to internalize the CMA substrate proteins into the lysosome lumen for degradation (19, 22–24). LAMP2a has been shown to be both necessary and sufficient in CMA degradation (22). Thus, LAMP2a protein levels have been used as an indirect marker for CMA because they correlated with CMA activity (22, 25). CMA, being a distinct proteolytic pathway may offer a more selective and efficient mode of clearing disease-causing proteins and ameliorating cardiac pathology.
Upregulation of CMA has been shown to be protective against neurodegenerative diseases and cancer via the selective clearance of misfolded and mutant proteins (26–29). Conversely, mice with the LAMP2a gene conditionally knocked out in the liver developed impaired hepatic function and metabolic abnormalities (30). However, the causal role of CMA using either gain or loss of CMA function models in the cardiomyocytes under physiological or pathological conditions had not been rigorously studied previously. Prior evidence had established that CMA was active in the heart and that the degradation of the ryanodine receptor type 2 (RyR2), a cardiac-specific calcium regulatory protein, was LAMP2a-dependent in primary rat cardiomyocytes (31). This study presented a previously unprecedented role of CMA in the clearance of proteins known to be essential for cardiac cell physiology. However, the broader importance of CMA in response to cardiac stress has not been examined.
Hypoxic/ischemic stress is a common cause of pathology and cell death across a host of tissues and diseases (32). According to the 2016 mortality data, myocardial infarction (MI) related mortality was measured at 1,11,777 individuals in the United States (33). Interestingly, in patients with a MI, hypoxia is responsible for the rapid and severe loss of contractile capacity of the heart, i.e., heart failure (34–36). Hypoxic stress results in accumulation of byproducts of cellular metabolism and cell death proteins that can reduce the metabolic and functional processes of the myocardium and enhance cellular damage (34, 37, 38). The role of CMA in hypoxic stress or ischemic heart disease remains undefined (4, 39). The objective of this research was to better understand the causal role of CMA in cardiac cells in response to hypoxic stress, using genetic gain and loss of LAMP2a (CMA) function models.
To delineate the role of CMA function in cardiomyocytes, a Lamp2a overexpressing adenovirus and Lamp2a silencing siRNA were tested in cardiomyocytes exposed to hypoxic stress. Several indices of CMA function were assayed that included a CMA reporter assay to validate changes in CMA activity to each treatment condition. The data herein demonstrate that gain of CMA was protective against hypoxia-induced apoptosis in primary rat cardiomyocytes. Oppositely, the loss of CMA function by LAMP2a knockdown was detrimental and further exacerbated the hypoxia-induced apoptotic cell death in cardiomyocytes. In addition, human ischemic hearts showed a significant upregulation of LAMP2a protein levels suggesting that CMA may be playing a protective, yet insufficient, role in the ischemically stressed hearts. Finally, the data showed that increases in LAMP2a led to parallel changes in macroautophagic flux (activity), suggesting a novel regulatory relationship between CMA and macroautophagy may exist in cardiomyocytes.
MATERIALS AND METHODS
Lamp2a Adenovirus Construction
The coding sequence for mouse Lamp2a (NM_001017959.2) was subcloned from a mouse clone (MR222878, OriGene) into a blunt TOPO vector (Thermo Fisher Scientific, 460096). Then, the Lamp2a coding and Kozak sequence was subcloned using PCR primers to introduce KpnI and NotI restriction sites for insertion into the pShuttle-CMV vector (Agilent, 240007). The pShuttle-CMV-Lamp2a and a control pShuttle-CMV-LacZ (Agilent, 240008) were then linearized by digestion with a Pme I restriction enzyme (New England Biolabs, R0560S) following which, both constructs were cotransformed into BJ5183 cells (Agilent, 200154) along with a pAdEasy-1 vector (Agilent, 240005), as described using the AdEasy Adenoviral Vector System (Agilent Technologies, 24000912). Positive recombinants were confirmed by DNA sequence analyses (Eurofins Genomics). Next, positive recombinants were transformed into XL-Gold ultracompetent cells (Agilent, 200314), purified, and linearized by digestion with a Pac I restriction enzyme (New England Biolabs, R0547L). The linearized DNA was then transfected into AD293 cells (Agilent, 240085) according to the instructions of the ViraPack Transfection Kit (Agilent, 200488). The AD293 cells were then expanded in T-150 flasks and lysed via three freeze-thaw cycles. The resulting adenoviral containing media and lysate were purified using columns from an AdenoPure kit (PureSyn, Malvern, PA).
Construction of the Lamp2a siRNA
The rat Lamp2 gene is alternatively spliced into three different mRNAs: Lamp2a, Lamp2b, and Lamp2c, which have similar luminal regions but differ in their cytoplasmic and transmembrane domains (40). To design a Lamp2a-specific siRNA, the cytosolic tail of Lamp2a (exon 8 region that differs between each isoform) was targeted to specifically knockdown Lamp2a without affecting the other two protein isoforms, LAMP2b and LAMP2c. The Lamp2a siRNA used herein was created to specifically target exon 8a (unique to the Lamp2a isoform) of the rat Lamp2a mRNA sequence (NM_017068.2) to selectively silence Lamp2a expression in neonatal rat cardiomyocytes. The siRNA was, therefore, designed to leave the expression of the Lamp2b and 2c isoforms intact (due to distinctly different exon 8b and 8c sequences). The following sequences were used: sense Lamp2a siRNA- (5'-GGGAGGAGUACUUAUUCUAtt-3') and antisense Lamp2a siRNA- (5'-UAGAAUAAGUACUCCUCCCtt-3') (Life Technologies, 4390827). A control siRNA (Thermo Fisher Scientific, 4390843) was used as a negative control.
Isolation of Primary Neonatal Cardiomyocytes
All aspects of animal (rat) care and use, including isolation of cardiomyocytes from rat pups, in this study, were approved by the Institutional Animal Care and Use Committee of the University of South Dakota (IACUC Protocol No. 12-04-16-19D and the University of Utah (Protocol No. 19-12002). Hearts were collected from 2-day-old Sprague–Dawley rat pups (both males and females) in ice-cold Hank’s Balanced Salt Solution. The ventricles were thoroughly minced and left in 0.05% trypsin overnight on a gently moving shaker. The next day, the trypsinized tissue was digested with collagenase (Worthington, LK003245) and preplated to deplete fibroblasts, as previously described (18). Viable cells were counted using Trypan Blue and plated in Minimum Essential Media (α-MEM) (Thermo Fisher Scientific, 32571036) containing 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, 16000044) and 1% penicillin/streptomycin (Thermo Fisher Scientific, 15140122). The cells were plated at a density of 2.5 × 106 onto 10-cm2 plates (Corning, 07202516) or 3.0 × 105 onto 2-well chamber slides (Nunc Lab-Tek II, 154461). After 24 h, the cardiomyocytes were kept in growth media composed of Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, 10569010) with 2% FBS and 1% penicillin/streptomycin. Adenoviral infection, siRNA transfection, or treatments with pharmacological agents were performed at 24 h for the following experimental studies. Each study was performed using biological replicates of the primary cardiomyocytes treated identically.
Hypoxia Induction in Cardiomyocytes
Neonatal cardiomyocytes were incubated in serum-free and glucose-free DMEM (Thermo Fisher Scientific, 11966-025) and placed in a hypoxic chamber (Modular Incubator Chamber, Billups-Rothenberg, CA), which was maintained at 37°C and constantly gassed with a mixture of 94% N2, 5% CO2, and 1% O2. For control normoxia experiments, rat myocytes were cultured in serum-free and glucose-free DMEM and incubated simultaneously in a humidified normoxia incubator (21% O2, 5% CO2, 37°C) for 24 h.
Hypoxia was chemically induced in vitro using CoCl2, a hypoxia-mimicking agent (41). CoCl2 is known to inhibit the activity of prolyl hydroxylases (PHD), key oxygen-sensing enzymes. Briefly, cardiomyocytes were treated with 500 µM CoCl2 or vehicle (containing water), for 48 h at 37°C, as previously described (42). Cardiomyocyte lysates were collected for downstream applications. Induction of hypoxia was confirmed by measuring the protein levels of HIF1α.
Pharmacological Modulation of CMA in Cardiomyocytes
Rat neonatal cardiomyocytes were grown in growth media treated with either 2 μM of an HSP-90 inhibitor, Geldanamycin (GA) (InvivoGen, ant-gl-5), or 10 mM of a phosphoinositide-3 kinase (PI3K) inhibitor, 3-methyladenine (3-MA) (InvivoGen, tlrl-3ma) for 24 h. Dimethyl sulfoxide (DMSO; Sigma, D8418) or water was used as a vehicle, respectively. At the end of the treatment, cells were washed twice in 1× phosphate buffer saline (PBS) (Thermo Fisher Scientific, 10010023) and then lysed in 1× CelLytic M Cell Lysis Reagent (Sigma Aldrich, C2978), supplemented with Complete Protease inhibitors (Roche, 04693159001). The cardiomyocyte lysates were sonicated on ice (120 Watts, Amp 25% for 7 s; using a Fisher Scientific, CL-18 sonicator). The protein concentration of each lysate was quantified using a Bradford protein assay kit (Bio-Rad, 5000006) using a bovine γ-globulin standard curve (Bio-Rad, 5000208). The absorbance (average of 562 nm and 630 nm) of each sample was measured using a microplate reader (BioTek). Equal concentrations of each lysate were prepared in a 3× SDS-sample buffer (New England Biolabs, B7703S) and frozen for subsequent SDS-PAGE and immunoblot analyses.
Adenoviral Infection and siRNA Transfection of Neonatal Cardiomyocytes
Following 24 h in 10% serum α-MEM, cardiomyocytes were infected with: low (60 MOI); medium (120 MOI), and high (240 MOI) of adenovirus in serum-deficient DMEM for 2 h (18). Following infection, the adenovirus-containing media was aspirated and replaced with DMEM, supplemented with 2% FBS, and 1% penicillin/streptomycin. Adenoviral constructs expressing either Lamp2a, to induce CMA activity, or a LacZ, negative control, were used. The cells were grown in the media for 24 h followed by CoCl2 or vehicle treatment for 48 h until collected for analyses.
To silence CMA activity, cardiomyocytes (2.5 × 106 cells) were transfected with 6.25 nM of a Lamp2a siRNA (Ambion) or a medium GC control siRNA (Invitrogen, 452001) using Lipofectamine 2000 (4 µg/µL) (Thermo Fisher Scientific, 11668027) in Opti-MEM media (Thermo Fisher Scientific, 31985070) for 24 h (18). Following transfection, the cells were incubated in growth media for 24 h and subsequently treated with CoCl2 or vehicle for 48 h.
CMA Luciferase Reporter Assay
To measure CMA activity, a KFERQ-Luciferase fusion (KFERQ-Luc) reporter plasmid was generated at the Mutation Generation and Detection Core, University of Utah. For gene encoding, the following 21 amino acids sequence of ribonuclease A: MKETAAAKFERQHMDSSTSAA was cloned into a pHIV plasmid, under the control of a CMV promoter, with the luciferase gene at the N-terminal end of the ribonuclease A DNA. A previous study had used a similar sequence to generate a photoconvertible fluorescent CMA reporter (43). pHIV-Luc-ZsGreen was a gift from Dr. Bryan Welm (Addgene plasmid No. 39196). For control experiments, an empty plasmid, devoid of the ribonuclease A KFERQ sequence, was used.
H9c2 Transfection of CMA Reporter
H9c2 immortalized cardiac cell lines isolated from the embryonic rat heart tissue (ATCC; Passage No. 4 to 7) were utilized to determine CMA activity. Similar to prior studies in neonatal rat ventricular myocytes, H9c2 cells were either transfected with a control or Lamp2a silencing siRNA or were infected with a Lamp2a or LacZ-expressing (control) adenovirus. After 72 h, the cells were transfected with either the CMA activity measuring KFERQ-luciferase (KFERQ-Luc) or a control “empty” luciferase-only (Empty-Luc) reporter plasmid using Lipofectamine 3000 (Thermo Scientific; L3000015), following the manufacturer’s protocol. A separate set of transfected cells devoid of any luciferase reporter was used as a negative control for the plasmid transfections. The infected and/or transfected H9c2 cells were then allowed to grow for additional 48 h followed by treatment with either CoCl2 or vehicle before luciferase activity or immunoblotting was performed.
For luciferase activity assay, H9c2 cells were cultured on a 96-well plate. Following transfection of Lamp2a knockdown or overexpressing cells with KFERQ-Luc or Empty-Luc, and with CoCl2 or vehicle treatment, luciferase activity was measured using Dura-Luc Lyophilized Firefly HTS Assay Kit (Gold Biotechnology; I-946-1000). Briefly, the cells were incubated in assay buffer containing 10 mM MgSO4 and d-luciferin (0.25 mg/mL) for 5–10 min. Luminescence was quickly recorded using a microplate reader (Thermo Fisher Scientific Varioskan LUX). The same protein concentrations were used for all groups for comparing luminescence measurements.
SDS-PAGE and Immunoblot Analysis
The protein homogenates were boiled at 95°C for 5 min in the presence of β-mercaptoethanol and 40–60 µg of each protein sample was separated using 4%–15% TGX gels (Bio-Rad, 5671084) or 15% Tris-HCl gels, respectively (Bio-Rad, 3450020). Approximately 40 µg protein/sample was loaded into each lane. Ten microliters of a molecular weight marker were also loaded into one lane simultaneously for each gel (LICOR, 928-40000). The samples were run using 1× Tris/glycine/SDS buffer (Bio-Rad, 1610772) at 100 V for ∼1.5 h. The proteins were next transferred to 0.2-µm PVDF membranes (Bio-Rad, 1620239) using 0.3 amperes (constant) for 3 h. Membranes were blocked using either SeaBlock (East Coast Bio, PP81) or 5% nonfat dry milk (Bio-Rad, 1706406) in Tris-buffered saline-Tween 20 (TBS-T), as optimal for each primary antibody, for 1 h at room temperature, and probed overnight at 4°C with one of the following primary antibodies: LAMP2a (1:200; AMC2; Invitrogen, 512200), HIF-1α (1:500; Cell Signaling Technology; 3716), HSC70 (1:500;13D3; Novus Biologicals, NB120-2788), HSP70 (1:500; C92F3A-5; Enzo Life Sciences, Inc., ADI-SPA-810), LAMP1 (1:500; Sigma Aldrich, L1418), MEF2D (1:500; 9/MEF2D; BD Biosciences, 610775), LC3B (1:500; Cell Signaling Technology, 2775), Luciferase (1:500; Sigma Aldrich; L0159), β-Actin (1:1000; AC-15; Sigma Aldrich, A5441), and p62 (1:500; ProGen Biotechnik; GP62-C). The next day, the membranes were washed in PBS-T or TBS-T (with 0.01% Tween) and incubated with species-specific secondary antibodies conjugated to infrared dyes (1:10,000; LICOR, 926–3221; 926-68070) for 1 h at room temperature, before scanning. β-Actin was used as the loading control for most of the experiments. Due to variation in other common normalizer proteins, the macroautophagy flux assays and mouse heart lysate immunoblots were normalized to total protein blotted as determined by Ponceau staining. For human heart lysates, the protein levels were normalized to GAPDH levels (1:1,000; Cell Signaling, 97166), due to variation in other normalizer proteins tested. The blots were imaged in a LICOR Odyssey CLx scanner, and densitometry quantitation of bands was done using the ImageStudio 2.0.38 software.
Lysosome Purification from Cultured Cardiomyocytes
Cells were infected with Lamp2a or LacZ adenoviruses and Lamp2a or control siRNA in the presence and absence of CoCl2 for 48 h. Intact lysosomes were isolated from primary cardiomyocytes (17.5 × 106 cells/group) using a lysosome enrichment kit (Thermo Fisher Scientific, 89839). Lysosomes were separated by density gradient ultracentrifugation at 29,000 rpm for 2 h at 4°C. The isolated lysosomal fractions were next washed with 1× PBS and the pellet obtained was lysed in 2% CHAPS (Sigma, 10810126001)-TBS. The lysosome-enriched protein samples were boiled with 3× SDS-sample buffer (New England Biolabs, B7703S) and equal concentrations of the lysosomal extract were separated on a protein electrophoresis gel, transferred, and immunoblotted with anti-LAMP2a or LAMP1 antibodies, as described earlier.
Immunocytochemistry
Primary neonatal cardiomyocytes were plated onto glass chamber slides pretreated with pig gelatin (Sigma, G1890). Thereafter, the cells were washed twice with 1× PBS and fixed with 4% paraformaldehyde (Electron Microscopy Sciences, 15710), and 0.5% Triton X-100 in 1× PBS for 20 min at room temperature (18). The cells were washed in 1× PBS and incubated in an antigen retrieval solution (0.1 M glycine, pH = 3.5) for 30 min at room temperature. To reduce nonspecific binding, the fixed cells were next incubated in a blocking solution [1% bovine serum albumin; (Sigma, A7030), 0.1% cold water fish gelatin (Electron Microscopy Sciences, 50-259-35), and 0.1% Tween-20, 0.05% sodium azide in 1× PBS] for 1 h at room temperature. The cells were then incubated with the following primary antibodies: LAMP2a (1:200), LAMP1 (1:200) overnight at 4°C. After washing with 1× PBS, cells were incubated with goat anti-rabbit AlexaFlour 488 secondary antibody (1:200, Life Technologies, A11034) for 1 h. The cells were washed and counter-stained with Phalloidin conjugated to either AlexaFlour 568 (1:200, Life Technologies, A12380) or AlexaFlour 647 (1:200, Life Technologies, A22287) for 2 h to detect actin in the cardiomyocytes. The cells were finally mounted using VectaShield mounting medium with DAPI (Vector Laboratories, H1500). Total lysosomes were detected by staining live cells using 1 µM LysoTracker Red DND-99 (Invitrogen, L7528) for 2 h at 37°C. The control and treated groups for each experiment were collected at the same time, under identical conditions. The images collected were from representative areas and were of individual optical sections captured using a confocal microscope (Olympus Fluoview FV1000).
Cytotoxicity Assays
Cytotoxicity was determined by adenylate kinase release or lactate dehydrogenase (LDH) release into cell culture media. Adenylate kinase and LDH are stable enzymes that are rapidly released into the cell culture media upon damage to the plasma membrane. Both the assays assessed the level of plasma membrane damage in a cell population and were used as markers of cytotoxicity. Adenylate kinase release was determined using the ToxiLight Bioassay kit (Lonza, LT17217). The luminescence assay was measured on a luminometer (Perkin Elmer, Victor X3). The LDH release was measured by a Cytotoxicity Detection kit (Roche, 04744934001). Color development was measured with an absorbance of 490 nM using an ELx 808 microplate reader (BioTek Instruments).
Cell Death Assays
To measure apoptosis in the CMA-altered cardiomyocytes with or without CoCl2, DNA strand breaks were labeled using a terminal deoxynucleotide transferase dUTP nick-end labeling (TUNEL) staining kit (Roche Diagnostics, Indianapolis, IN, 11684795910) following the manufacturer’s protocol (44). Thereafter, the cells were counterstained with Phalloidin (1:200) for 2 h to detect the actin and DAPI for total nuclei, respectively. The fluorescein dUTP fluorescence (green) representing TUNEL-positive myonuclei was quantified from images captured using a confocal microscope (Olympus Fluoview FV1000).
In parallel experiments, propidium iodide (PI) and Hoechst staining of cardiomyocyte nuclei were performed under similar conditions as described earlier to determine cardiomyocyte necrotic cell death (45). PI is a membrane-impermeant dye and does not stain viable cells. PI-positively stained nuclei allow the number of dead cells to be quantified. The Hoechst 33258 dye is cell permeable and allows the detection of the total number of nuclei. Cells were incubated with 500 nM PI (Invitrogen, P3566) in growth media at 37°C for 2 h. After 90 min, 8 µM of Hoechst 33258 (Invitrogen, H3569) stain was added and the cells were incubated for 30 min at 37°C. The cells were washed twice in 1× PBS and fixed in 4% paraformaldehyde in 1× PBS, before image analysis. Propidium iodide-positive nuclei were detected using a fluorescent microscope (Zeiss Upright M1 Imager) with a ×20 objective. For both TUNEL and PI/Hoechst staining experiments, eight images were captured for each well at the same time and under identical conditions (80–100 different cells/image; 5–8 images/well) and four different wells per treatment group were quantified using ImageJ software.
Macroautophagic Flux Assays
Macroautophagic flux assays were performed to measure autophagosome synthesis versus degradation. Macroautophagic flux assays were measured by monitoring LC3B levels in the presence and absence of a lysosomal inhibitor, 50 nM Bafilomycin A1 (Baf A1) (Sigma, B1793) and dimethyl sulfoxide (DMSO), as the vehicle for 4 h. Cardiomyocytes were either infected with Lamp2a or LacZ adenoviruses or transfected with a Lamp2a siRNA or a control siRNA and then after 48 h were treated with Baf A1, or DMSO, before lysis with CelLytic M. LC3-II (aka LC3B) protein levels were detected using an anti-LC3B antibody. LC3-II levels were normalized to total protein levels (Ponceau staining) to determine if any changes in macroautophagic flux were altered by the gain or loss of CMA function and/or CoCl2 treatment, respectively, instead of other housekeeping proteins (which showed greater variation).
Human Heart Sample Collection and Lysate
Samples of human myocardium were originally acquired at the University of Kentucky from patients who were receiving a cardiac transplant or a Ventricular Assist Device (VAD), or from organ donors, used as controls, (IRB No. 46103) as previously published (Supplemental Table S1; see https://doi.org/10.6084/m9.figshare.13371062) (46). Specimens were cleaned and snap-frozen in liquid nitrogen within ∼30 min of being excised from the patient. Full details about the protocol for sample collection and the patient samples have been previously published (46). The ventricular tissue sections were then stored at −80°C for future applications.
Human heart tissue lysates were prepared by homogenizing the tissue using metallic beads (Qiagen, 69989) in T-PER Tissue Protein Extraction Reagent (Thermo Fisher Scientific, 78510) supplemented with Complete Protease inhibitors. Tissue homogenization and disruption were carried out using a small bead mill (TissueLyser LT, Qiagen) set at a frequency of 50 Hz for 5 min at 4°C. The samples were then centrifuged at 12,000 g for 15 min at 4°C. Protein concentrations of the resulting lysates were quantified using a Bradford assay per the manufacturer’s instructions against an IgG protein standard curve. Equal concentrations of each sample were separated by SDS-PAGE and immunoblotted. β-Actin levels showed high variability across the patient heart tissues and sample groups. Instead, LAMP2a and HSC-70 protein levels were normalized to GAPDH protein levels, which did not vary within or between human heart sample groups.
Statistical Analyses
Quantification of data is presented as means ± SD (standard deviation). Significant differences between the different experimental groups were determined by either a Student’s t test, one-way analysis of variance (ANOVA), or a two-way ANOVA, depending on the number of variables/groups tested, followed by a Holm–Sidak test using SigmaPlot software version 12.5. Plots were generated in GraphPad Prism 9.1.0. The value of P < 0.05 was considered statistically significant with n = 4 replicates/group, unless otherwise noted.
RESULTS
Genetic Gain of LAMP2a Expression in Cardiomyocytes
To determine if CMA is functional in the cardiomyocyte, primary rat neonatal cardiomyocytes were treated with the following pharmacological agents previously shown to modulate LAMP2a protein levels (47, 48): the HSP-90 inhibitor, geldanamycin (GA), and the phosphoinositide 3-kinase (PI3K) inhibitor, 3-methyladenine (3-MA) (Supplemental Fig. S1; see https://doi.org/10.6084/m9.figshare.16734691). Treatment with GA caused a significant (−1.4-fold) reduction in LAMP2a protein levels (Supplemental Fig. S1, A and B), along with increased cytotoxicity (Supplemental Fig. S1G), whereas 3-MA treatment increased LAMP2a levels (+1.9-fold) (Supplemental Fig. S1, C and D) and decreased cellular toxicity (Supplemental Fig. S1H). The data confirmed that CMA was active in primary cardiomyocytes, and upregulation of CMA was associated with decreased cytotoxicity, suggesting a beneficial role for CMA in cardiomyocyte survival. Therefore, to test the hypothesis that enhanced CMA plays a causal, beneficial role in reducing cardiomyocyte pathology, an adenovirus was generated to overexpress LAMP2a (Ad Lamp2a) via a cytomegalovirus (CMV) promoter.
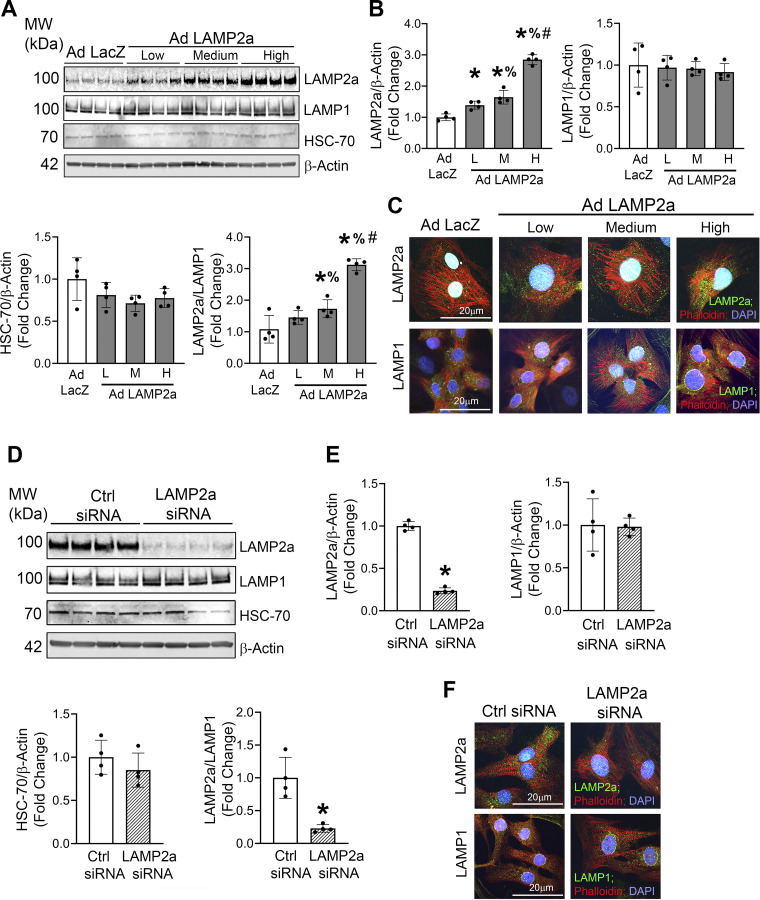
Prior studies using liver cells and tissues showed that LAMP2a was both necessary and sufficient for regulating CMA function (22, 49). Neonatal rat cardiomyocytes were infected with increasing doses of the Lamp2a adenovirus (Low, Medium, and High), which resulted in a dose-dependent increase in LAMP2a protein levels (Fig. 1, A and B). A LacZ-expressing adenovirus (Ad LacZ) was used as a negative control, which did not change LAMP2a levels. To determine whether gain of CMA function by Lamp2a adenovirus overexpression might affect other lysosomal membrane proteins, LAMP1 protein levels were measured (50). LAMP1 levels did not differ between Ad Lamp2a- and Ad LacZ-infected cells, indicating that the total lysosome content in cardiomyocytes was not altered by Lamp2a overexpression (Fig. 1, A and B). HSC-70 levels were unchanged in response to increasing doses of the Lamp2a adenovirus, relative to Ad LacZ control (Fig. 1, A and B). The LAMP2a/LAMP1 ratio, measured to estimate the percent of LAMP2a-positive relative to total lysosomes, showed a dose-dependent increase with the greatest increase (+3.0-fold) observed in cardiomyocytes infected with “High” dose [240 multiplicity of infection (MOI)] of the Lamp2a adenovirus (Fig. 1, A and B). Furthermore, immunocytochemical staining of cardiomyocytes showed that LAMP1-positive lysosomes (green puncta in Fig. 1C, bottom) remained qualitatively unchanged in response to Lamp2a overexpression, similar to that observed by immunoblotting. Conversely, the LAMP2a-positive lysosomes (green puncta in Fig. 1C, top) were markedly increased indicative of upregulated CMA in response to increasing doses of Lamp2a adenovirus over LacZ adenovirus (Fig. 1C) (51). These data confirmed that the adenovirus created to overexpress the rat Lamp2a isoform successfully induced LAMP2a protein levels in primary cardiomyocytes. The “high” dose of the Lamp2a adenovirus was used in subsequent studies to assess the causality of gain of CMA function in combination with hypoxic stress.
Figure 1.
Gain of chaperone-mediated autophagy (CMA) function increased, and a lysosome-associated membrane protein receptor 2a (LAMP2a) isoform-specific siRNA decreased, LAMP2a protein levels and the percent of LAMP2a-positive lysosomes. To study the gain of CMA function, a Lamp2a-overexpressing adenovirus (Ad Lamp2a) was generated. Cardiomyocytes were infected with [low (L) (60 multiplicity of infection, MOI); medium (M) (120 MOI); and high (H) (240 MOI)] doses of Ad Lamp2a or with a LacZ expressing adenovirus (Ad LacZ) control. To study the loss of CMA function in rat neonatal cardiomyocytes, a siRNA was generated to specifically target the rat Lamp2a isoform. A: immunoblot analyses of LAMP2a, LAMP1, and heat shock protein 70 (HSC-70) protein levels were performed. B: bar graphs show the means of the densitometry of the bands on the immunoblots for each group. Protein levels were normalized to the loading control β-actin. C: immunocytochemistry studies were performed to detect LAMP2a-positive structures and localization in cardiomyocytes infected with the Lamp2a or LacZ adenoviruses. The green punctate structures represent LAMP2a- or LAMP1-positive lysosomes. The phalloidin-stained structures (in red) show the actin fibers and the nuclei were counterstained with DAPI (blue); n = 2 wells/group. Each image is a representative of 6–8 individual images collected under identical conditions. D: immunoblot analyses of LAMP2a, LAMP1, and HSC-70 protein levels in cardiomyocytes transfected with a Lamp2a siRNA or control siRNA were performed n = 4 independent samples/group. E: bar graphs show the corresponding densitometry of the immunoblotted bands by group. Protein levels were normalized to β-actin protein levels. F: for the immunofluorescent stained images, green fluorescence represents LAMP2a-positive lysosomes (top) or LAMP1-positive lysosomes (bottom), respectively. Red fluorescence showed phalloidin binding to Actin protein; nuclei were counterstained with DAPI (blue); n = 2 wells/group. Each image is a representative of 6–8 individual images collected under identical conditions. Results of the immunoblot analyses are expressed as means ± SD from four independent groups. For (B), *P < 0.05: difference between Ad LacZ and Ad Lamp2a infected groups; %P < 0.05: difference between low- and medium-infected Lamp2a groups; and #P < 0.05: difference between high- and, low- and medium-infected Lamp2a groups. For (E), *P < 0.05: difference between control and Lamp2a siRNA transfected groups.
Genetic Loss of LAMP2a Expression in Cardiomyocytes
To test the hypothesis that loss of CMA function plays a detrimental role in cardiomyocyte survival, a rat Lamp2a isoform-specific siRNA was generated and transfected into neonatal rat cardiomyocytes. The Lamp2a siRNA showed a −4.2-fold depletion of LAMP2a protein levels signifying successful knockdown compared with the control siRNA (Fig. 1, D and E). Conversely, the total lysosomal content indicated by LAMP1, and HSC-70 protein levels remained unchanged by Lamp2a knockdown (Fig. 1, D and E). The LAMP2a-positive lysosomes were significantly decreased (−4.3-fold) with Lamp2a knockdown compared with control cells, as indicated by the LAMP2a/LAMP1 ratio (Fig. 1E). Immunocytochemical staining of cardiomyocyte lysosomes also confirmed the successful reduction in LAMP2a-positive lysosomes by Lamp2a siRNA silencing, qualitatively evidenced by the reduced LAMP2a-positive (green) punctate staining (Fig. 1F, top). Previous publications showed that neither silencing, nor overexpression, of Lamp2a altered Lamp2b or Lamp2c isoforms in mouse hepatocytes nor breast cancer cells (30, 52). To silence Lamp2a, a siRNA was specifically designed to target exon 8a, which differs substantially in sequence from the other two Lamp2 splicing variants, Lamp2b (exon 8b) and Lamp2c (exon 8c); as a result, only the Lamp2a isoform should have been knocked down (40, 53). These results showed that the siRNA designed against the rat Lamp2a isoform effectively silenced LAMP2a protein levels in cardiomyocytes and could be used for additional loss of CMA function studies in combination with hypoxic stress.
Hypoxia Enhances LAMP2a Levels in Cardiomyocytes
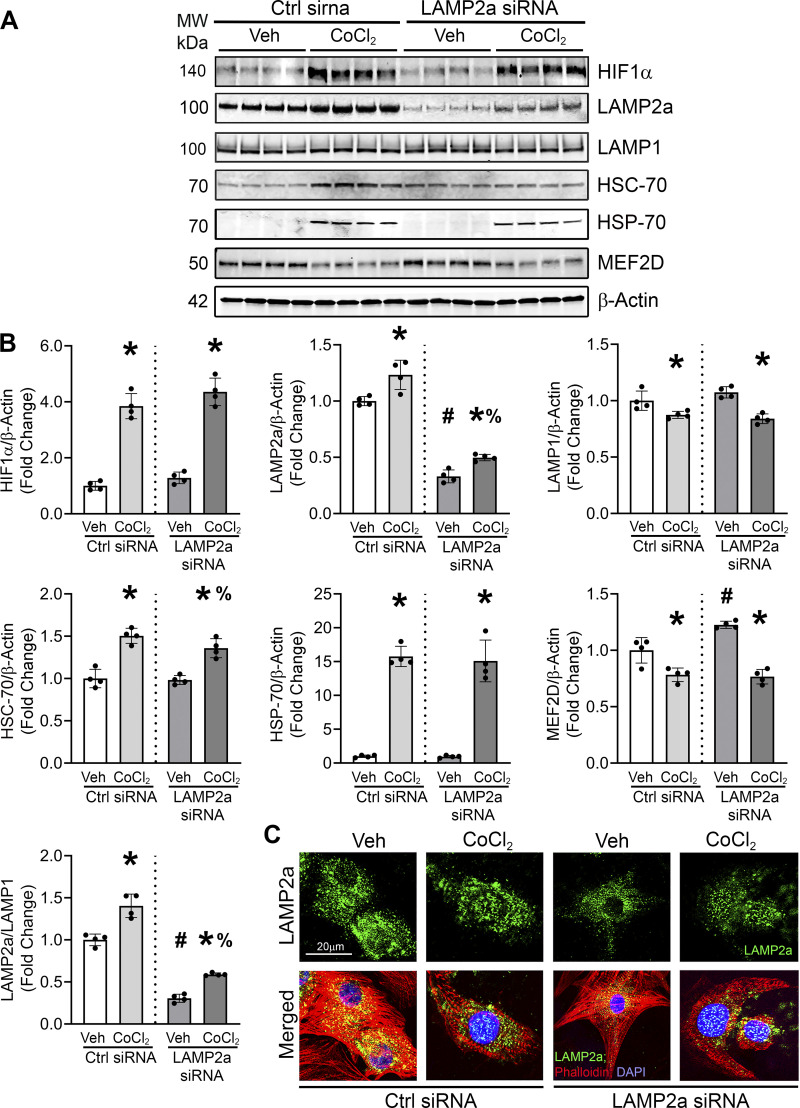
Hypoxic stress is a common cause of pathology and cardiomyocyte death during IHD (34). Cobalt chloride (500 µM CoCl2), a well-known hypoxia mimetic agent (41, 54–57), was chosen to induce and maintain a hypoxic cellular environment. The optimal dose (500 µM) and time (48 h) of CoCl2 treatment to induce LAMP2a protein levels were determined empirically in a series of pilot experiments (Supplemental Fig. S2; see https://doi.org/10.6084/m9.figshare.16734655). The present study was consistent with a prior report that used an identical dose of CoCl2 (500 µM) in neonatal cardiomyocytes to induce hypoxia (58). Doses > 500 µM were not tested as higher concentrations of CoCl2 were previously shown to affect cellular morphology, proliferation, and reduced cell viability of human microvascular endothelial and kidney cells (59). Hypoxia-inducible factor 1α (HIF1α) that accumulates under hypoxic stress was significantly upregulated compared with the vehicle control cells (+3.8-fold) confirming the induction of hypoxic stress (Fig. 2, A and B). The increase in HIF1α protein was associated with elevated LAMP2a protein levels (+1.2-fold) in the CoCl2 group compared with the vehicle-treated cells, confirming that CoCl2-induced hypoxic stress increases LAMP2a (Fig. 2, A and B). To validate that CoCl2-induced hypoxic stress was comparable with physiological hypoxia, neonatal cardiomyocytes were exposed to 1% O2 for 24 h. Normoxic control cardiomyocytes were cultured simultaneously in a standard humidified incubator (21% O2). Cardiomyocytes exposed to chamber hypoxia showed an upregulation in both HIF1α (+1.7-fold) and LAMP2a (+1.3-fold) compared with the normoxic cells (Supplemental Fig. S3, A and B; see https://doi.org/10.6084/m9.figshare.19681908). These data suggested that the pattern of HIF1α and LAMP2a protein expression in cardiomyocytes induced by 1% O2 in the hypoxic chamber was fairly comparable with CoCl2 treatment, although the percent increase in HIF1α protein levels induced by CoCl2 appeared to be greater than that induced by 1% O2 (75% vs. 25%), perhaps due to experimental timing differences (Supplemental Fig. S3; Fig. 2, A and C). In subsequent studies, CoCl2 induced-hypoxic stress was tested in combination with genetic gain and loss of Lamp2a to define the role of CMA function on hypoxia-stressed cardiomyocytes.
Figure 2.
Hypoxic stress augmented lysosome-associated membrane protein receptor 2a (LAMP2a) levels and Lamp2a knockdown blunted hypoxia-induced chaperone-mediated autophagy (CMA) marker proteins. A: cardiomyocytes were transfected with control or Lamp2a siRNAs and treated with CoCl2 or vehicle (Veh) for 48 h. Immunoblot analyses of hypoxia-inducible factor 1α (HIF1α), LAMP2a, lysosome associated membrane protein receptor 1 (LAMP1), heat shock cognate 70 (HSC-70), heat shock protein 70 (HSP-70), and myocyte enhancer factor 2D (MEF2D) protein levels were performed. B: bar graphs show the mean densitometry analysis of the bands per group. The levels of CMA-related proteins were normalized to β-actin protein levels. C: immunostaining was performed to detect LAMP2a positive-lysosomes in the cardiomyocytes using a LAMP2a antibody (green puncta), Phalloidin (red) was used to stain the actin filaments, and nuclei were counterstained with DAPI (blue); n = 2 wells/group. Each image is a representative of 6–8 individual images collected under identical conditions using an objective of ×60. The results of each immunoblot analysis are expressed as means ± SD from four independent groups (n = 4). *P < 0.05: difference between CoCl2 treatment within siRNA transfection groups; #P < 0.05: difference between Veh groups; and %P < 0.05: difference between CoCl2-treated groups. Immunofluorescent staining changes in LAMP2a lysosomes paralleled the LAMP2a protein changes observed by blotting.
Loss of CMA Function Limits Hypoxia-Induced LAMP2a in Cardiomyocytes
The effect of loss of CMA function in cardiomyocytes treated with CoCl2 was studied. LAMP2a levels were significantly suppressed by the Lamp2a siRNA in the vehicle-treated cells (−3.2-fold) (Fig. 2, A and B). The Lamp2a siRNA had no effect on HIF1α levels either in vehicle-treated control or the CoCl2-treated cells. CoCl2-induced hypoxia upregulated LAMP2a protein levels in both vehicle- and Lamp2a siRNA-treated over normoxic control cardiomyocytes (∼+1.4-fold in both). Thus, Lamp2a silencing reduced the induction of LAMP2a in both the presence and absence of CoCl2-induced hypoxia compared with the control siRNA transfected cells (∼−3.0-fold) (Fig. 2, A and B). Consistently, Lamp2a silencing had no significant effect on LAMP1 protein levels. However, hypoxic stress induced by CoCl2 did significantly reduce LAMP1 levels (Fig. 2, A and B). Thus, the silencing of Lamp2a appeared to be CMA-specific and not due to decreased lysosomal content, which would affect other lysosomal processes like macroautophagy, microautophagy, and endosomal degradation. The stress-inducible, heat shock protein 70 (HSP-70), known to be upregulated by hypoxia was significantly increased by CoCl2 treatment, but not by the Lamp2a siRNA alone (Fig. 2, A and B) (60–62). Previous studies have shown that myocyte enhancer factor 2D (MEF2D), a myogenic transcription factor protein required for cardiac development and function (63, 64), was a substrate of CMA in neuronal cells (65). MEF2D was shown to have multiple and imperfect KFERQ-like motifs in its N-terminus region, which are required for the interaction of MEF2D with HSC-70 for degradation by CMA. Deletion of these motifs abolished MEF2D’s uptake by the lysosomes (42). Consistent with previous studies, CoCl2 treatment induced LAMP2a levels and resulted in a corresponding significant decrease in MEF2D levels relative to the vehicle-treated cells. Meanwhile, Lamp2a knockdown yielded a modest accumulation of MEF2D protein compared with the control siRNA-transfected cells (Fig. 2, A and B). These findings suggest that MEF2D was a substrate of CMA in cardiomyocytes. The LAMP2a/LAMP1 ratio changes showed that while the Lamp2a siRNA reduced the percent of LAMP2a-positive lysosomes in the vehicle-treated cells, CoCl2-induced hypoxic stress still increased the pool of LAMP2a-positive lysosomes over the normoxic control cells (Fig. 2B). Concomitantly, CoCl2 enhanced the LAMP2a punctate staining (green fluorescence), which appeared markedly reduced by Lamp2a knockdown (Fig. 2C). Thus, silencing Lamp2a, as a means to block CMA-based degradation, significantly blunted the hypoxia-induced responses in CMA-related proteins in primary cardiomyocytes, but incompletely likely due to partial Lamp2a knockdown. Next, the causal effects of Lamp2a overexpression in the hypoxia-treated cardiomyocytes were determined.
Hypoxic Stress Alone Induces LAMP2a That Is Further Augmented by Lamp2a Adenoviral Overexpression
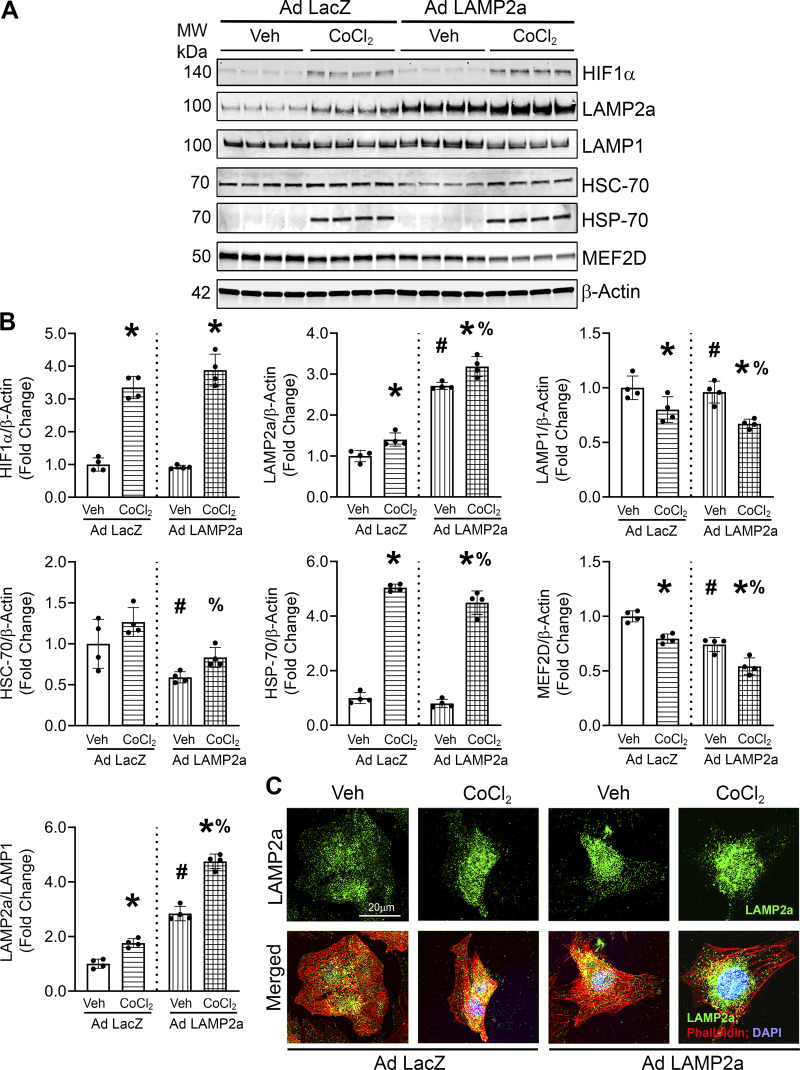
To determine whether increased CMA was beneficial or detrimental against CoCl2-induced hypoxia, cardiomyocytes were infected with the “high” dose of the Lamp2a adenovirus or a matching dose of LacZ-expressing adenovirus as control, with or without CoCl2. HIF1α protein levels were significantly increased by CoCl2 treatment indicative of hypoxia stimulation (Fig. 3, A and B). CoCl2 treatment and Lamp2a adenoviral infection each alone increased LAMP2a levels (+1.4-fold and +2.7-fold), respectively, compared with the corresponding vehicle-treated and LacZ-infected control cells. Cardiomyocytes overexpressing Lamp2a in combination with CoCl2 treatment synergistically augmented LAMP2a levels over CoCl2 (+2.0-fold) alone or vehicle (+3.2-fold), respectively (Fig. 3, A and B). Total lysosomal content, indicated by LAMP1 levels, was decreased with CoCl2 treatment alone. Lamp2a overexpression significantly decreased LAMP1 levels, which was further reduced with CoCl2 treatment relative to LacZ-infected cells. HSC-70 levels, the CMA-specific chaperone, were significantly decreased by Lamp2a overexpression (Fig. 3, A and B). Heat shock protein, HSP-70 levels, were significantly upregulated by CoCl2-induced hypoxic stress and were modestly but significantly reduced by Lamp2a overexpression with CoCl2 treatment. HSP-70 induction validated the hypoxic stress response yet showed a different expression pattern from HSC-70. MEF2D protein levels were significantly reduced by gain of LAMP2a or CoCl2 treatment individually and decreased further by the combination of the two, consistent with the purported role of the MEF2D protein as a substrate of CMA degradation (Fig. 3, A and B). Immunofluorescence studies revealed that CoCl2 treatment appeared to further augment the number of LAMP2a-positive lysosomes above the level seen in Ad Lamp2a overexpressing cardiomyocytes treated with vehicle (fluorescent green puncta) (Fig. 3C). These data show that Lamp2a overexpression not only increased LAMP2a-positive lysosomes but leads to a reduction in a noted CMA substrate, MEF2D, in primary cardiomyocytes. In addition, the ratio of LAMP2a/LAMP1 protein levels were increased by both Lamp2a overexpression and CoCl2 individually and showed a significant synergistic +5.0-fold increase in LAMP2a-positive lysosomes in Lamp2a overexpressing cardiomyocytes subjected to hypoxic stress (Fig. 3, A and B). Collectively, the data show that LAMP2a levels and indices of CMA degradative function, as evidenced by MEF2D degradation, were upregulated in response to hypoxic stress in cardiomyocytes. Next, LAMP2A protein levels were measured from isolated lysosomal fractions, because lysosome-specific LAMP2a levels were shown to positively correlate with CMA function and activity (23).
Figure 3.
Lysosome associated membrane protein receptor 2a (Lamp2a) overexpression increased chaperone-mediated autophagy (CMA) marker proteins in CoCl2-treated cardiomyocytes. Lamp2a- and LacZ-overexpressing cardiomyocytes were treated with CoCl2 for 48 h to induce hypoxia, or a vehicle (Veh) control. A: immunoblot analyses of hypoxia-inducible factor 1α (HIF1α), LAMP2a, lysosome associated membrane protein receptor 1 (LAMP1), heat shock cognate 70 (HSC-70), heat shock protein 70 (HSP-70), and myocyte enhancer factor 2D (MEF2D) were carried out. B: bar graphs show the band densitometry of the immunoblots. The protein levels were normalized to β-actin level which was used as the loading control. C: immunofluorescence studies were performed to determine if LAMP2a was localized into lysosomal punctate structures. Cardiomyocytes were stained with an anti-LAMP2a antibody to detect LAMP2a localization (green), Phalloidin-stained actin (red) and nuclei were counterstained with DAPI (blue); n = 2 wells/group. LAMP2a fluorescence was largely limited to cytosolic, perinuclear punctate structures consistent with lysosomal staining and showed similar changes to those found by immunoblotting across the different treatments. Each image is a representative of 6–8 individual images collected under identical conditions using an objective of ×60. Results of immunoblots are means ± SD from four independent groups. (n = 4); *P < 0.05: difference between CoCl2 treatment within infection group; #P < 0.05: difference between infection within Veh groups; and %P < 0.05: difference between CoCl2-treated groups.
Lysosomal Changes in Response to Hypoxic Stress and Altered LAMP2a Levels
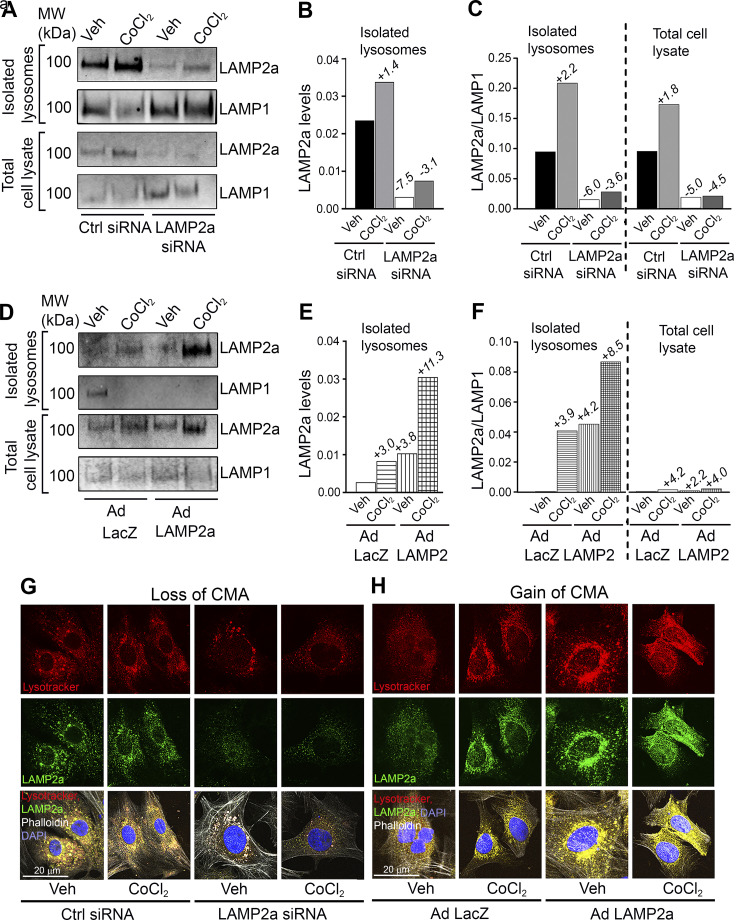
Intact lysosomes were isolated to assess changes in the pool of LAMP2a-positive lysosomes. The changes in LAMP2a protein levels measured from isolated lysosome fractions are considered the “gold standard” for quantifying CMA activity (66, 67). Lamp2a silenced cardiomyocytes were treated with or without CoCl2 to induce hypoxia for 48 h and then equal concentrations of intact lysosomal fractions were immunoblotted for LAMP2a and LAMP1 levels, respectively (54–56). The pool of LAMP2a-positive lysosomes was decreased by silencing LAMP2a in the vehicle (−7.5-fold) and CoCl2-treated cells (−3.1-fold) relative to corresponding controls (Fig. 4, A and B). Normalization of lysosomal LAMP2a to lysosomal LAMP1 protein levels was performed next. In lysosomes isolated from cardiomyocytes with Lamp2a knockdown, CMA-positive lysosomes were markedly reduced over the total (LAMP1-positive) lysosomes when compared with the control siRNA transfected cells, treated with or without CoCl2 (−6.0-fold and −3.6-fold, respectively) (Fig. 4, A and C). These results suggest that the Lamp2a siRNA indeed suppressed CMA function in cardiomyocyte lysosomes under both basal and hypoxic conditions.
Figure 4.
Changes in lysosome associated membrane protein receptor 2a (LAMP2a)-positive lysosomes with altered chaperone-mediated autophagy (CMA) and hypoxic stress. Intact lysosomes were isolated from cardiomyocytes (17.5 × 106 cells/group) overexpressing Ad Lamp2a or Ad LacZ and treated with or without CoCl2 for 48 h; n = 2 samples/group. Total cell lysates and the purified lysosomal fractions were normalized by loading equal protein concentrations. LAMP2a blots showed the amount of CMA-active lysosomes. Lysosome-associated membrane protein receptor 1 (LAMP1) was used as a marker for total lysosomes. A: lysosomes were isolated from cardiomyocytes transfected with either a control or Lamp2a siRNA and immunoblotted for LAMP2a and LAMP1 proteins. B: bar graph shows the fold changes in LAMP2a-positive lysosomes with Lamp2a knockdown with or without CoCl2 treatment. C: bar graph shows the ratio between the LAMP2a-positive and LAMP1-positive lysosome levels between groups. D: immunoblot analysis was performed to determine LAMP2a and LAMP1 levels in the lysosomal fractions isolated from Lamp2a-overexpressing cardiomyocytes. E: bar graph represents the fold changes in LAMP2a-positive lysosomes in the presence or absence of CoCl2. F: bar graph shows LAMP2a-positive lysosomes/LAMP1 levels in isolated lysosomal fractions. G and H: cardiomyocytes were stained with LysoTracker and an anti-LAMP2a antibody to qualitatively detect the colocalization of LAMP2a positive lysosomes (green) with the total lysosomes (red, LysoTracker); actin was counterstained in white and nuclei were counterstained with DAPI (blue). G: colocalization of CMA-active lysosomes with the total lysosomes in Lamp2a knockdown cardiomyocytes is shown (yellow). H: colocalization of CMA-active lysosomes with the total lysosomes (yellow) in Lamp2a-overexpressing and CoCl2-treated cardiomyocytes is shown; n = 2 wells/group. Images were taken at a magnification of ×60 and 5 to 6 cardiomyocytes were captured/image. Each image is a representative of 6–8 individual images collected under identical conditions.
Intact lysosomes were isolated from cardiomyocytes with gain of LAMP2a, and with or without CoCl2 treatment for 48 h. The LAMP2a-positive lysosomal population was upregulated by hypoxic stress (+3.0-fold) (Fig. 4, D and E). In the Lamp2a-overexpressing cardiomyocytes, treated with CoCl2, a more marked (+8.0-fold) increase in LAMP2a-positive lysosomes was observed over that of the control (Fig. 4, D and E). Furthermore, hypoxic stress increased the lysosomal LAMP2a over LAMP1 (total lysosome) levels (+3.9-fold), indicative of an increased CMA-positive lysosome response, but not a total lysosome response (Fig. 4, D and F). These results suggested that hypoxic stress modulates CMA by increasing the LAMP2a-positive lysosomal content of the cardiomyocytes.
Immunofluorescence analyses were performed to determine colocalization of LAMP2a-positive lysosomes with total lysosomes across treatment groups (Fig. 4, G and H). Cardiomyocytes transfected with a Lamp2a or control siRNA, or infected with Ad Lamp2a or Ad LacZ, in the presence or absence of CoCl2, were stained with LysoTracker Red as an index of total lysosomal content and a LAMP2a-specific antibody (green) to stain the CMA-positive lysosomal population (Fig. 4, G and H). Consistent with the isolated lysosomal fraction data, knockdown of Lamp2a appeared to qualitatively reduce, and overexpression of Lamp2a appeared to elevate the number of fluorescent LAMP2a-positive lysosomes (green puncta) relative to the total lysosomes (red puncta), in the vehicle group (Fig. 4, G and H). Furthermore, Ad Lamp2a-infected cardiomyocytes appeared to show increased colocalization of LAMP2a (green puncta) with total lysosomes (red puncta), where colocalization is presented as yellow puncta, versus the Ad LacZ control cells (Fig. 4H). The CoCl2-mediated hypoxic induction of LAMP2a-positive lysosomes in cardiomyocytes was largely blunted by loss of CMA (Fig. 4G). On the contrary, hypoxic stress appeared to further augment the colocalization of LAMP2a-positive lysosomes with the total lysosomes (yellow puncta) (Fig. 4H). These data support the idea that hypoxic stress induces CMA by increasing the percentage of LAMP2a-positive lysosomes in the primary cardiomyocytes. To further validate the changes in CMA observed in whole cells and isolated lysosomes, a CMA reporter assay was next performed.
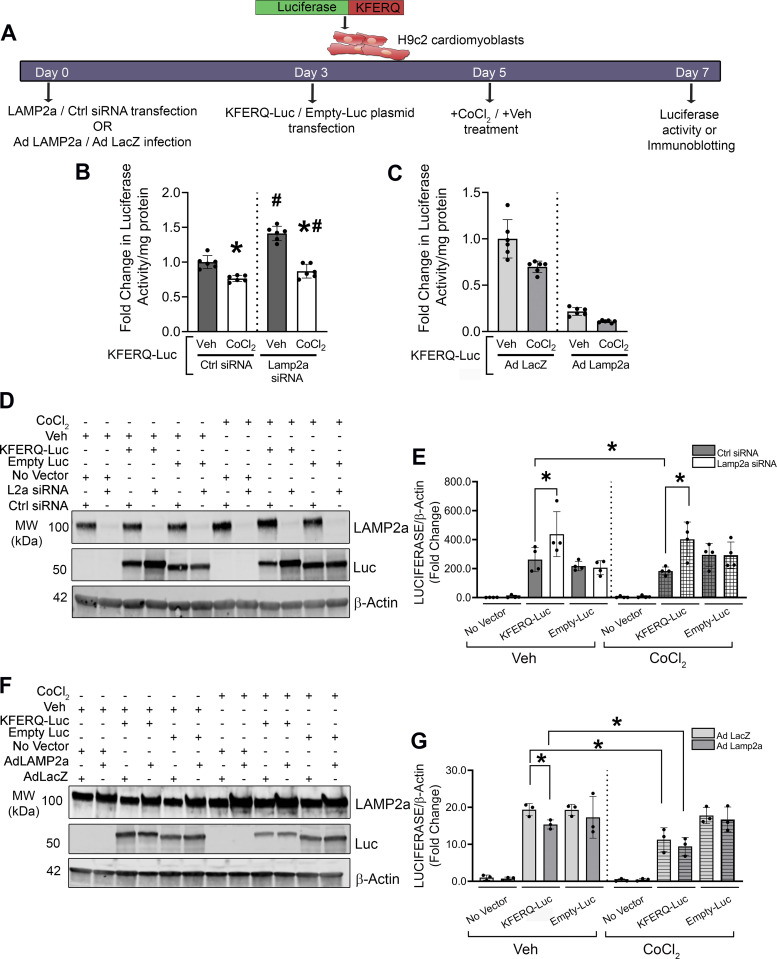
CMA-Luciferase Reporter Assays of CMA Activity Changes in H9c2 Cells
Previous studies generated photoconvertible CMA reporters to monitor the activity of CMA in mice and intact cells (43, 68). To confirm the changes in cardiomyocyte CMA in response to hypoxia and altered LAMP2a expression, H9c2 cardiac cells were either transfected with a CMA luciferase reporter (a KFERQ motif fused to luciferase; KFERQ-Luc) or a CMA-independent negative control empty luciferase only plasmid (Empty-Luc) (Fig. 5A). A separate group of untransfected cells served as a negative control for the plasmid transfections. If CMA was activated, it would degrade KFERQ-Luc and decrease luciferase activity. Conversely, loss of CMA from Lamp2a silencing would increase luciferase activity. Indeed, in cells expressing KFERQ-Luc, luciferase activity was significantly increased in Lamp2a siRNA-treated cells (Fig. 5B, Supplemental Fig. S4, A and B; see https://doi.org/10.6084/m9.figshare.19325867), and significantly reduced in Lamp2a overexpressing cells (Fig. 5C, Supplemental Fig. S4, C and D). An increase in CoCl2-induced CMA activity was evidenced by decreased luciferase activity (Fig. 5, B and C). Immunoblotting further confirmed successful plasmid expression, and Lamp2a knockdown or overexpression, respectively, versus the appropriate controls (Fig. 5, D–G, Supplemental Fig. S4, A–D). Silencing Lamp2a caused a significant accumulation of KFERQ-Luc protein both in the vehicle (+1.8-fold) and CoCl2 (+2.0-fold) treated cells, consistent with reduced CMA degradation (Fig. 5, D and E, Supplemental Fig. S4, A and B). CoCl2-induced LAMP2a upregulation reduced KFERQ-Luc levels (−1.3-fold), as did Lamp2a overexpression (−1.3-fold). The combination of CoCl2 and Lamp2a overexpression further reduced luciferase activity and thus CMA function (−1.5-fold), versus vehicle-treated and Ad Lamp2a-infected cells (Fig. 5, F and G, Supplemental Fig. S4, C and D). The protein levels of control Empty-Luc (lacking the KFERQ motif) remained unaltered with Lamp2a manipulation, indicating the specificity of CMA for KFERQ domain (Fig. 5, D–G, Supplemental Fig. S4, A–D). The changes observed in Lamp2a protein levels in response to CoCl2 treatment (Figs. 2, 3, and 4), were successfully reflected by the reporter assay (Fig. 5, Supplemental Fig. S4), suggesting that LAMP2a protein levels directly correlated with CMA reporter activity. Whether the upregulation of LAMP2a observed with hypoxic stress was a beneficial or deleterious mechanism in exacerbating hypoxic pathology, was tested in the next set of experiments.
Figure 5.
Changes in chaperone-mediated autophagy (CMA) by luciferase reporter assay with altered lysosome associated membrane protein receptor 2a (LAMP2a) and hypoxic stress. H9c2 cells with Lamp2a knockdown or overexpression (and corresponding controls) were transfected with either a KFERQ-luciferase (KFERQ-Luc) CMA reporter plasmid or control Empty-luciferase (Empty-Luc) plasmid for 72 h. An additional negative control group of cells, not transfected with any plasmid, was added to each experiment. The cells were then treated with CoCl2 or vehicle (Veh) for an additional 48 h. A: a schematic timeline for KFERQ-Luc or Empty-Luc transfection and Veh or CoCl2 treatment. B: bar graph shows fold change in luciferase activity observed in the Lamp2a silenced H9c2 cells with CoCl2 or Veh. C: bar graph shows the fold change in luminescence from Lamp2a-overexpressing H9c2 cells with CoCl2 or Veh, transfected with KFERQ-Luc (n = 6 independent samples/group). D: representative immunoblots of LAMP2a, luciferase (Luc), and β-actin protein levels are shown from Lamp2a silenced H9c2 cells and (E) represents the quantification of Luc protein levels normalized to β-Actin levels (n = 4 samples/group). F: representative immunoblots of LAMP2a, Luc, and β-actin protein levels, in Lamp2a-overexpressing H9c2 cells treated with CoCl2 or Veh and (G) represents the quantification of Luc levels normalized to β-actin levels (n = 3 samples/group). Results for all experiments are means ± SD. For (B and C), *P < 0.05: difference between Veh and CoCl2 in Ctrl or Lamp2a siRNA groups, or Ad LacZ or Ad Lamp2a infected cells, transfected with KFERQ-Luc; #P < 0.05: difference between Ad LacZ and Ad Lamp2a infected cells or Ctrl and Lamp2a siRNA cells within the Veh or CoCl2-treated cells transfected with KFERQ-Luc. For (E and G), *P < 0.05: difference between the indicated groups in H9c2 cells.
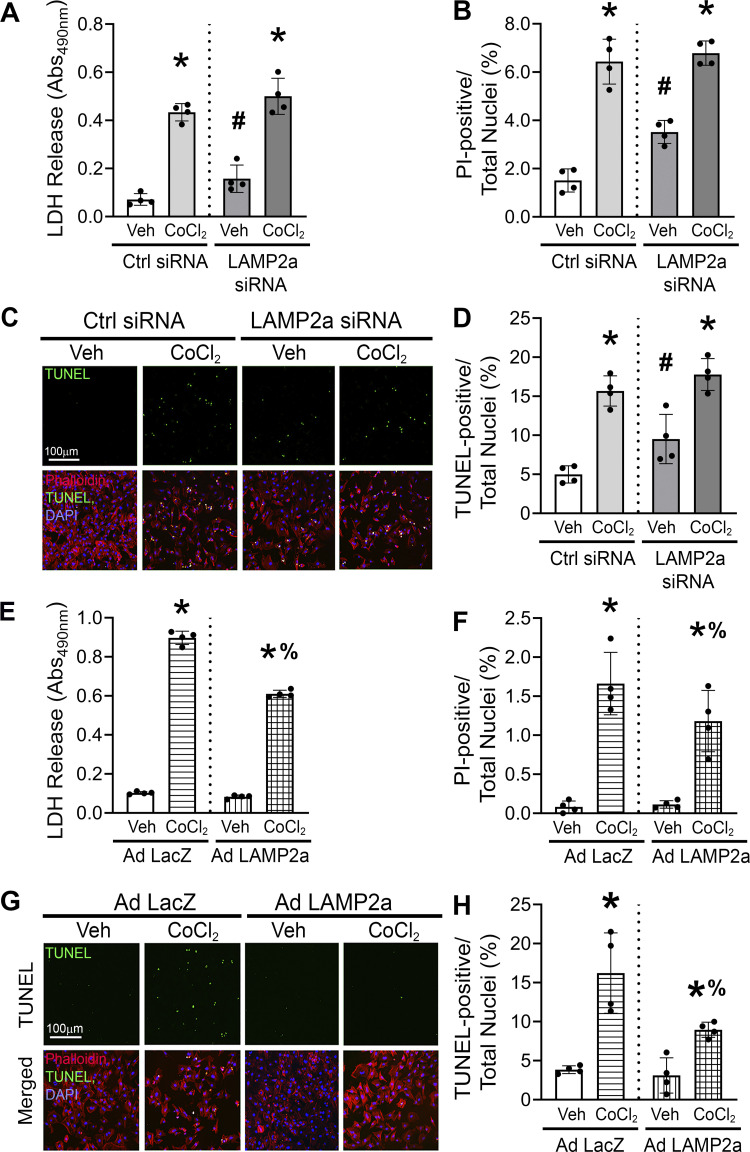
Hypoxia-Induced Cell Death Is Abrogated by Lamp2a Overexpression
CoCl2-induced hypoxic conditions were shown to trigger various pathological effects including cytotoxicity and cell death in mouse embryonic stem cells and mouse embryonic fibroblasts (69, 70). Indices of cytotoxicity and cell death were measured to determine if the role of CMA on hypoxia-induced pathology in primary cardiomyocytes was beneficial or detrimental. Lactate dehydrogenase (LDH) is an enzyme, normally localized in the cytosol, and only released into the culture media by cells with damaged plasma membranes. Thus, LDH release was used as a marker of cytotoxicity. Knockdown of Lamp2a increased cytotoxicity (+2.1-fold) compared with the control siRNA-transfected vehicle group, as measured by LDH release into the cardiomyocyte media. CoCl2 treatment, individually, elicited a larger (+5.2-fold) cytotoxic response compared with the control siRNA vehicle group. The cytotoxicity induced by hypoxic stress was further aggravated (+1.2-fold) in Lamp2a-silenced cardiomyocytes compared with the control siRNA + CoCl2-treated group (Fig. 6A).
Figure 6.
Gain of chaperone-mediated autophagy (CMA) decreases cytotoxicity and cell death in cultured cardiomyocytes. Cardiomyocytes were treated with CoCl2 for 48 h and the effect of both the loss and gain of CMA function on cytotoxicity and cell death was determined. A: cardiomyocytes were transfected with either a Lamp2a or Ctrl siRNA and treated with vehicle (Veh) or without CoCl2. CoCl2-induced cytotoxicity was determined by lactate dehydrogenase (LDH) release assay in Lamp2a-silenced cardiomyocytes. B: cardiomyocytes transfected with a Lamp2a or a Ctrl siRNA were treated with or without CoCl2; and were dual stained using propidium iodide (PI), as a marker of necrotic cell death, and Hoechst, to determine total nuclei present. The bar graph shows the percent of PI-positive myonuclei over the total nuclei in each group. C: the percent of terminal deoxynucleotide transferase dUTP nick-end labeling (TUNEL)-positive nuclei (green) indicating apoptotic cell death was determined in the cardiomyocytes with Lamp2a-silenced and treated with or without CoCl2. D: bar graph shows the percent changes in TUNEL-positive nuclei over the total nuclei. Green represents TUNEL-positive nuclei, actin is stained in red and represents myocytes; nuclei were counterstained with DAPI (blue). E: CoCl2-induced cytotoxicity was determined by LDH release assay in Lamp2a- or control LacZ-overexpressing cardiomyocytes. F: bar graph shows the percent of PI-positive over the total nuclei in the Lamp2a- or LacZ-infected cells with or without CoCl2 treatment. G: percent of TUNEL-positive nuclei (green) was determined in the Lamp2a-overexpressing cardiomyocytes, with or without CoCl2 and (H) bar graph shows the percent changes in TUNEL-positive nuclei over the total nuclei. Results for all experiments are means ± SD from four independent groups (n = 4). *P < 0.05: significant difference between Veh and CoCl2 within similarly infected groups; #P < 0.05 and %P < 0.05: differences within Lamp2a knockdown or overexpressing cells treated with Veh or CoCl2, respectively.
The effect of LAMP2a silencing on cardiomyocyte cell death when subjected to hypoxic stress was determined next. The ratio of propidium iodide (PI) over Hoechst 33258-stained nuclei was quantified to assess sarcolemmal integrity of each group of cardiomyocytes. PI, an intercalating agent, only permeates into dead/dying cells and was used as a marker of primary or secondary necrosis. The Hoechst dye stained the nuclei of all the cells, both live and dead. Lamp2a silencing alone induced cardiomyocyte membrane permeability (+2.5-fold), based on increased PI staining, but less so than CoCl2 alone (+6-fold) (Fig. 6B). However, there was neither an additive nor a synergistic effect on PI/Hoechst staining when both treatments were combined (Fig. 6B). Therefore, the loss of CMA did not add to the necrotic cell death induced by hypoxic stress (Fig. 6B). Next, terminal deoxynucleotide transferase dUTP nick-end labeling (TUNEL) staining was performed on primary cardiomyocytes transfected with a Lamp2a or control siRNA and treated with or without CoCl2, respectively, to detect cells that are in a late stage of apoptosis, with DNA degradation (Fig. 6C). DAPI-positive nuclei from phalloidin-stained cardiomyocytes were used for quantification of total and TUNEL-positive myonuclei. Hypoxic stress stimulated by CoCl2 significantly increased the percent of TUNEL-positive nuclei (shown in green) indicating increased (+3.2-fold) apoptotic cell death (Fig. 6, C and D). Lamp2a silencing alone also increased the percent of TUNEL-positive nuclei (+1.8-fold) suggesting that loss of CMA is detrimental for cardiomyocyte survival (Fig. 6, C and D). However, Lamp2a knockdown in combination with hypoxic stress failed to add to the apoptotic response elicited by CoCl2 alone (Fig. 6, C and D). Collectively, these studies confirmed that loss of CMA induced both cytotoxicity and cell death and were detrimental to cardiomyocytes.
In the gain of CMA function studies, CoCl2-treatment alone showed increased (+9-fold) LDH release and when coupled with Lamp2a overexpression showed a blunted response (+6-fold) relative to LacZ CoCl2-treated cells (Fig. 6E). CoCl2-treated cells with Ad LacZ expression showed an increase (+20-fold) in the percent of PI-positive nuclei (red fluorescence) indicating loss of cardiomyocyte membrane integrity associated with necrotic cell death was due to the hypoxic stress itself (Fig. 6F). Gain of Lamp2a significantly reduced the ratio of PI/Hoechst-stained nuclei induced by CoCl2-stimulated hypoxia (−1.4-fold) (Fig. 6F). Furthermore, TUNEL staining was performed on primary cardiomyocytes with gain of Lamp2a or LacZ and treated with or without CoCl2, respectively. The percent of TUNEL-positive nuclei (green fluorescence) was significantly increased by CoCl2-induced hypoxic stress (+4.1-fold) in the Ad LacZ-infected cells (Fig. 6, G and H). Gain of CMA function by Lamp2a overexpression significantly reduced the number of TUNEL-positive myonuclei (−1.4-fold). Thus, enhanced CMA attenuated CoCl2-induced apoptotic cell death (Fig. 6, G and H). Collectively, these studies suggest that gain of CMA was protective against hypoxia-induced cardiomyocyte pathology by reducing cytotoxicity, improving sarcolemma permeability (necrosis), and attenuating apoptotic cell death.
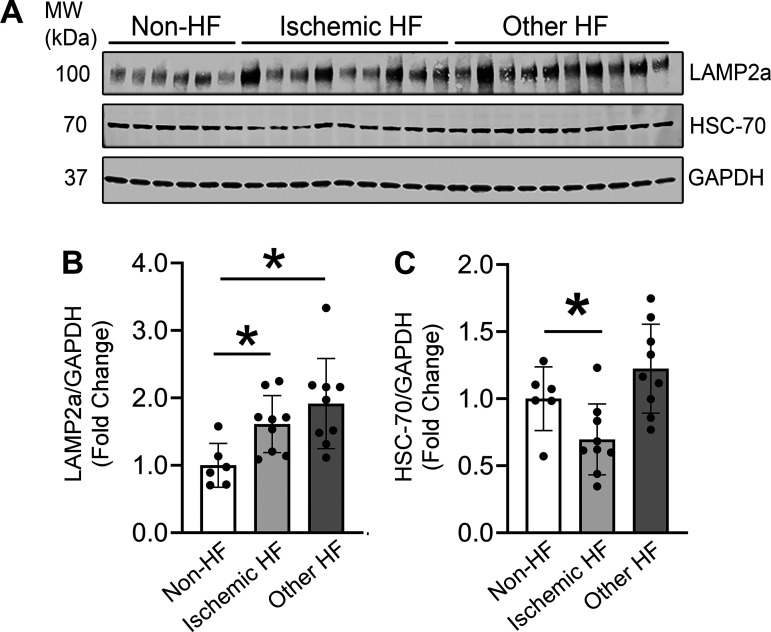
LAMP2a Levels Are Upregulated in Stressed Human Hearts
As little was known about CMA in adult heart, LAMP2a protein levels were measured in normal and stressed human hearts to assess any changes in CMA. Human hearts were obtained from patients (males and females) with “ischemic heart failure” and from “other heart failure patients” (with cardiac pathologies like chronic systolic heart failure, nonischemic cardiomyopathy, and dilated cardiomyopathy), and “non-heart failure” control subjects, as previously published (Supplemental Table S1) (46). LAMP2a protein levels were significantly elevated (+1.7-fold) in the heart lysates of patients with ischemic heart failure compared with the hearts from the control subjects (Fig. 7, A and B). The increase in LAMP2a was also associated with a significant decrease (−1.5-fold) in HSC-70 levels (Fig. 7, A and C). Collectively, the LAMP2a protein level differences observed in human hearts mirrored those found in the cultured cardiomyocytes, which showed that hypoxic stress upregulated LAMP2a levels (Figs. 2, 3, 4, and 5). There was a comparable (+1.8-fold) significant increase in LAMP2a levels in the hearts of “other heart failure patients” (Fig. 7, A and B). However, the HSC-70 protein levels in the “other heart failure patients” samples remained unaltered compared with the control hearts, perhaps suggesting the HSC-70 changes are specific to hypoxic stress (Fig. 7, A and C). Collectively, these data revealed that CMA was increased in response to ischemic stress in human hearts suggesting an important role of CMA in the stressed myocardium. The observed increase in LAMP2a levels in the failing human hearts may be a compensatory stress-response adapted by the heart to confer a beneficial, yet insufficient, protection against ischemic or hypertrophic stress-induced cardiac dysfunction and death.
Figure 7.
Chaperone-mediated autophagy (CMA) is increased in the stressed human hearts. Immunoblotting was performed to compare lysosome associated membrane protein receptor 2a (LAMP2a) and heat shock cognate 70 (HSC-70) protein levels in heart lysates from patients with different types of heart failure (HF) versus non-heart failure (non-HF) subjects. A: immunoblots show LAMP2a, HSC-70, and GAPDH protein levels in the patient hearts compared with non-HF donor control hearts. B and C: bar graphs show the densitometry analysis of LAMP2a and HSC-70 protein levels, normalized to GAPDH levels. Results for the experiments are means ± SD; non-HF: n = 6 subjects; ischemic HF: n = 10 subjects; other HF: n = 9 subjects. *P < 0.05: difference between the ischemic HF or other HF and the non-HF group.
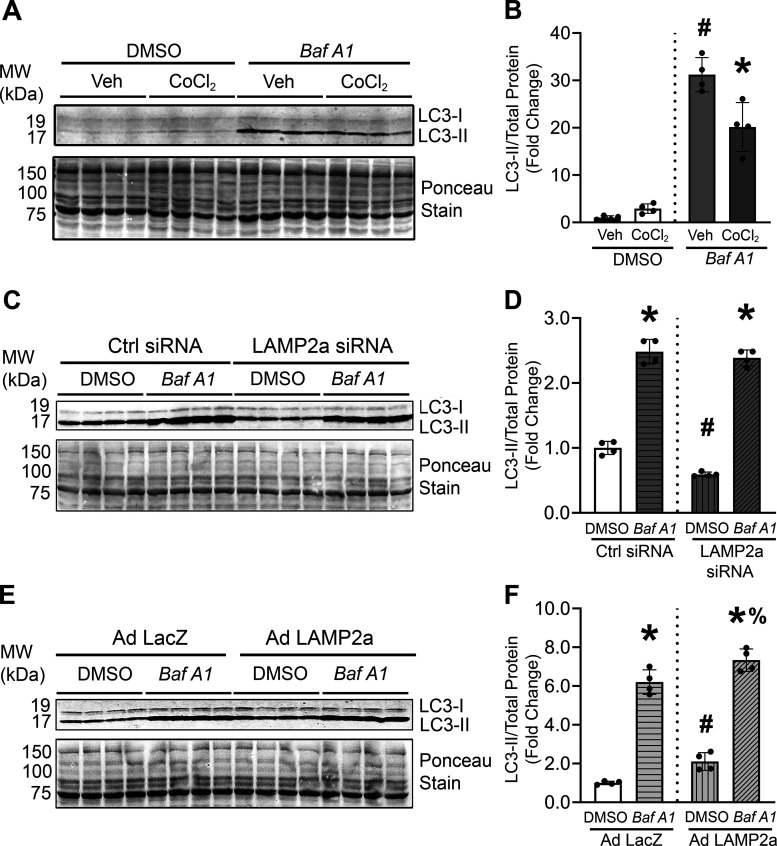
Macroautophagic Flux Was Increased by Lamp2a Overexpression
A previous report demonstrated that silencing Lamp2a resulted in a higher content of autophagic vacuoles and increased levels of macroautophagy-related proteins like Beclin 1 and ATG5-12 complex in mouse fibroblasts suggesting cross talk existed between CMA and macroautophagy processes (71). The effect of gain and loss of CMA function on macroautophagy in cardiomyocytes was tested. Macroautophagic flux assays are considered the “gold standard” for measuring changes in macroautophagic degradative activity (72, 73). Macroautophagic flux assays involved inhibiting lysosomal function using Bafilomycin A1 (Baf A1; 50 nM) compared with vehicle (DMSO)-treated cardiomyocytes. Baf A1 blocks lysosomal degradation of the autophagosomal marker protein, LC3-II, resulting in the accumulation of LC3-II and other substrates of the macroautophagy-lysosomal pathway. CoCl2-induced hypoxic stress in cardiomyocytes led to impaired autophagosomal degradation for reduced macroautophagic flux (Fig. 8, A and B). Silencing Lamp2a significantly decreased basal LC3II levels but did not alter macroautophagic flux (Fig. 8, C and D). On the contrary, Lamp2a overexpression significantly increased LC3-II protein levels under control conditions (i.e., vehicle) (+2.0-fold; compared with LacZ infected vehicle cells), and significantly increased macroautophagic flux (i.e., +Baf A1) (+1.3-fold; compared with LacZ infected Baf A1-treated cells), in primary cardiomyocytes (Fig. 8, E and F). These results demonstrated that enhanced CMA function led to an increase in macroautophagic degradative function (Fig. 8, E and F). As expected, the lysosomal membrane proteins, LAMP2a or LAMP1, were not affected by Baf A1 treatment (Supplemental Fig. S5, A–F; see https://doi.org/10.6084/m9.figshare.19326026) as Baf A1 causes lysosome alkalinization (74). Overall, these studies suggest that loss of Lamp2a had no effect on macroautophagic flux, whereas enhanced CMA stimulated macroautophagic activity (+1.3-fold) in cardiomyocytes. Clearly, additional studies are needed to determine the effects of the targeted regulation of both pathways, which may be important for developing successful treatments for patients with ischemic heart failure.
Figure 8.
Enhanced chaperone-mediated autophagy (CMA) increased macroautophagic activity. Changes in microtubule-associated protein 1A/1B-light chain 3 (LC3)-II protein levels with lysosome inhibition indicate altered macroautophagy activity. To determine if hypoxia induced by CoCl2 treatment caused any changes in macroautophagy activity, cardiomyocytes were treated with CoCl2 for 48 h and LC3-II protein levels were measured in the presence and absence of a lysosomal inhibitor, Bafilomycin A1 (Baf A1). A: immunoblots show LC3-II protein levels relative to total protein (Ponceau-stained membrane) levels. B: densitometry of the bands are shown in the bar graph. C–F: macroautophagy flux assays were performed on cardiomyocytes either transfected with Lamp2a or Ctrl siRNA or infected with Ad LacZ or Ad Lamp2a adenoviruses in the presence and absence Baf A1 for 4 h. C and E: immunoblots show LC3-II protein level changes in the Lamp2a knockdown or overexpressing cardiomyocytes, respectively. Total protein was used as a loading control. D and F: bar graphs show changes in densitometry of LC3-II over the total protein levels. Results for all experiments are means ± SD from four independent groups (n = 4 samples/group). For (A) and (B), #P < 0.05: difference between DMSO and Baf A1 treatment within vehicle (Veh) groups; and %P < 0.05: difference between DMSO and Baf A1 within the CoCl2 groups respectively. For (C–F) *P < 0.05: difference between Veh and Baf A1 treatment within individual infection groups; #P < 0.05: difference between infection within Veh or DMSO groups; and %P < 0.05: difference between infection within Baf A1 groups.
DISCUSSION
Different forms of heart diseases like ischemic, hypertrophic, and dilated cardiomyopathies often lead to heart failure. Among these, ischemic heart disease (IHD) accounted for >9 million deaths in 2016, while it is estimated that globally 153.5 million people live with the disease (2, 33). These studies formed the basis for choosing to investigate the potentially beneficial role of CMA in the context of hypoxia-induced stress and cardiomyocyte death. Hypoxic stress is a major contributor to MI pathology, the most common type of cardiac disease (34). The present study is the first of its kind to define the causal role of CMA in cardiomyocyte death and toxicity induced by hypoxia. The data herein showed that LAMP2a levels and consequently CMA function were upregulated in the hypoxically stressed primary cardiomyocytes and in ischemic human hearts. These data suggest that CMA may be upregulated as a stress response, although not induced sufficiently to prevent IHD.
Ischemic heart disease is caused, in part, by oxygen deprivation, resulting in hypoxic stress and ultimately cardiac dysfunction. Hypoxic stress, induced by both low O2 in a hypoxic chamber or CoCl2 treatment, significantly upregulated LAMP2a protein levels in primary cardiomyocytes suggesting that CMA is activated in hypoxic cardiomyocytes (31, 75). Elevated cardiomyocyte CMA activity was further validated by the KFERQ-luciferase reporter assay in response to CoCl2 treatment. In addition to KFERQ-luciferase (a classical CMA substrate), the protein levels of another known endogenous CMA substrate, MEF2D, was decreased with Lamp2a overexpression and accumulated with Lamp2a knockdown (42). Collectively, these data suggest that CMA is active in the cardiomyocytes and is regulated by extrinsic stressors like hypoxia.
Most of the literature has looked at lysosomes as a homogeneous population of “suicide sacs” or “recycling centers” of cells. The data presented herein shows that there is heterogeneity in the lysosomal population, such that genetically altering Lamp2a increased or decreased LAMP2a-positive lysosomes. The capacity to up- or downregulate a subpopulation of CMA-positive lysosomes suggests that lysosomes are more heterogeneous in their lysosomal membrane proteins than previously thought and this diversity may impart selectivity to degrade cellular debris depending on the specific mechanism of lysosomal delivery. If true, it may be possible to upregulate certain degradative pathways by increasing corresponding lysosomal subpopulations, for example, CMA and the LAMP2a-positive lysosomal subpopulation. It remains to be rigorously studied whether there are lysosomal subpopulations that selectively or predominantly degrade substrates delivered via macroautophagy, endosomes, microautophagy, or other selective autophagy pathways. Certainly, more studies characterizing lysosomes and the substrates they degrade are warranted and now feasible with fluorescent pulse-chase and super-resolution imaging technologies.
The present study further revealed that alteration in LAMP2a levels does not positively correlate with changes in HSC-70 levels (76). A previous report indicated that a fraction of cellular HSC-70 was exclusively localized in the lysosomal membrane and lumen (49, 77). The mechanism through which HSC-70 gains access to the lysosomal lumen remains inconclusive. It may be inferred that HSC-70 undergoes a fate similar to that of ubiquitin participating in substrate selection and degradation via the UPS pathway, where the ubiquitin moiety is translocated to the proteasomal complex and recycled to the cytoplasm. In addition to CMA, endosomal microautophagy and the UPS also utilize HSC-70-mediated protein binding and degradation and thus HSC-70 is not CMA specific, which may explain the discrepancies seen in HSC-70 levels (78, 79). Further studies to delineate the precise mechanism of HSC-70 trafficking, lysosomal localization, and turnover in cardiomyocytes are warranted to better understand CMA regulation in the heart.
The present study showed that CMA activation caused a corresponding significant increase in macroautophagic activity. CoCl2-mediated hypoxic stress decreased macroautophagic flux. On the other hand, enhancing CMA by increasing LAMP2a protein levels twofold increased macroautophagic flux by ∼1.3-fold. Whether the two pathways work synergistically for degrading the same or different subsets of disease-causing proteins remains to be determined. Future work will be required to further define the interactions among the CMA, macroautophagy, and the UPS degradative pathways and to determine whether these pathways work in concert or in opposition, for clearing cardiac proteins under control and stressed conditions.
A seminal finding of the current study was that the gain of CMA function was protective against hypoxia-induced cell death in primary cardiomyocytes. CoCl2-induced hypoxia significantly induced cytotoxicity, loss of plasma membrane integrity, and apoptotic cell death in primary cardiomyocytes that was further exacerbated with the loss of CMA function. Loss of cardiomyocytes due to cell death plays a critical role in the pathogenesis of heart failure. Cardiomyocytes are terminally differentiated cells and their death results in the practical loss of contractile units. As a consequence of cardiomyocyte death, the heart encounters several adverse effects including loss of normal contractile function, reduced cardiac output, and ultimately, a complete failure to pump blood (80). Thus, the observation that CMA silencing aggravated cardiac cell death reveals the potential clinical relevance of the CMA in the preservation of cardiomyocyte viability. Indeed, upregulating CMA function by Lamp2a overexpression successfully reduced cytotoxicity by −6-fold, PI staining (indicative of loss of sarcolemmal permeability) by −1.4-fold, and apoptotic myonuclei by −1.4-fold. Although the bulk of the current study was performed in cultured primary cardiomyocytes, the human heart failure sample data supported the idea that insufficient CMA function may contribute to the pathogenesis of ischemic heart disease. Although gain of CMA clearly reduced markers of cytotoxicity and death in cardiomyocytes, additional studies using in vivo animal models of MI and heart failure are needed to define the exact mechanism(s) underlying the beneficial effects of CMA in the cardiomyocytes.
In summary, the present study demonstrated the importance of CMA in the amelioration of hypoxia-induced cardiomyocyte death. Stressed human hearts revealed heightened LAMP2a levels, consistent with the results obtained from primary cardiomyocytes exposed to hypoxic stress. Hypoxic stress triggered increased apoptosis, necrosis, and cytotoxicity in primary cardiomyocytes. Although silencing CMA was generally detrimental, augmenting CMA was protective against cardiomyocyte death and cytotoxicity induced by hypoxic stress. Thus, enhancing CMA function may prevent the loss of myocytes and preserve both cardiac contractility and function. To understand the regulation and causal function of CMA in the heart, genetically modified animals with gain or loss of CMA should be studied in models of cardiac disease and heart failure to define its mechanism of action. Overall, this study provides the necessary evidence to justify targeting the CMA pathway as a cardioprotective mechanism against ischemic heart failure. CMA activation appears to hold promise for future translation into clinical application to improve cardiac outcomes in patients.
SUPPLEMENTAL DATA
Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.16734691.
Supplemental Fig. S2: https://doi.org/10.6084/m9.figshare.16734655.
Supplemental Fig. S3: https://doi.org/10.6084/m9.figshare.19681908.
Supplemental Fig. S4: https://doi.org/10.6084/m9.figshare.19325867.
Supplemental Fig. S5: https://doi.org/10.6084/m9.figshare.19326026.
Supplemental Table S1: https://doi.org/10.6084/m9.figshare.13371062.
GRANTS
This work was supported by an American Heart Association Post-Doctoral Fellowship Grant No. 17POST33670412 and the College of Health, University of Utah, Seed Grant under Grant No. 51900420 (to R. Ghosh); American Heart Association (AHA) TPA34860008, NIH TR033173, HL133359, HL149164 (to K. S. Campbell); NIH/National Heart, Lung, and Blood Institute (NHLBI) R01HL149870-01A1 (to S. Boudina); NIH RO3AGO52848, and NHLBI RO1 Grant HL141540 (to J. D. Symons); and a pilot grant from the Great Plains IDEA-Clinical & Translational Research Award [NIH/National Institute of General Medical Science (NIGMS)] Number 1U54GM115458-01 (to J. S. Pattison).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.S.P. conceived and designed research; R.G. and J.J.G. performed experiments; R.G. and J.S.P. analyzed data; R.G. and J.S.P. interpreted results of experiments; R.G. and J.S.P. prepared figures; R.G. and J.S.P. drafted manuscript; R.G., J.D.S., S.B., and J.S.P. edited and revised manuscript; R.G., J.J.G., K.S.C., J.D.S., S.B., and J.S.P. approved final version of manuscript.
ACKNOWLEDGMENTS
Present address of R. Ghosh: Dept. of Nutrition and Integrative Physiology, and Molecular Medicine, University of Utah, Salt Lake City, Utah.
REFERENCES
- 1. Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, et al. Heart disease and stroke statistics-2021 update: a report from the American Heart Association. Circulation 143: e254–e743, 2021. doi: 10.1161/CIR.0000000000000950. [DOI] [PubMed] [Google Scholar]
- 2. Nowbar AN, Gitto M, Howard JP, Francis DP, Al-Lamee R. Mortality from ischemic heart disease. Circ Cardiovasc Qual Outcomes 12: e005375, 2019. doi: 10.1161/CIRCOUTCOMES.118.005375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lyon RC, Lange S, Sheikh F. Breaking down protein degradation mechanisms in cardiac muscle. Trends Mol Med 19: 239–249, 2013. doi: 10.1016/j.molmed.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res 100: 914–922, 2007. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 5. Bhuiyan MS, Pattison JS, Osinska H, James J, Gulick J, McLendon PM, Hill JA, Sadoshima J, Robbins J. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest 123: 5284–5297, 2013. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Su H, Wang X. The ubiquitin-proteasome system in cardiac proteinopathy: a quality control perspective. Cardiovasc Res 85: 253–262, 2010. doi: 10.1093/cvr/cvp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Divald A, Kivity S, Wang P, Hochhauser E, Roberts B, Teichberg S, Gomes AV, Powell SR. Myocardial ischemic preconditioning preserves postischemic function of the 26S proteasome through diminished oxidative damage to 19S regulatory particle subunits. Circ Res 106: 1829–1838, 2010. doi: 10.1161/CIRCRESAHA.110.219485. [DOI] [PubMed] [Google Scholar]
- 8. Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem 281: 29776–29787, 2006. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 9. Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med 13: 619–624, 2007. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 10. Churchill EN, Ferreira JC, Brum PC, Szweda LI, Mochly-Rosen D. Ischaemic preconditioning improves proteasomal activity and increases the degradation of deltaPKC during reperfusion. Cardiovasc Res 85: 385–394, 2010. doi: 10.1093/cvr/cvp334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Predmore JM, Wang P, Davis F, Bartolone S, Westfall MV, Dyke DB, Pagani F, Powell SR, Day SM. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation 121: 997–1004, 2010. doi: 10.1161/CIRCULATIONAHA.109.904557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, Li F, Gerdes AM, Wawrousek EF, Wang X. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res 97: 1018–1026, 2005. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 13. Liu J, Chen Q, Huang W, Horak KM, Zheng H, Mestril R, Wang X. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. FASEB J 20: 362–364, 2006. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 14. Huang C, Yitzhaki S, Perry CN, Liu W, Giricz Z, Mentzer RM Jr, Gottlieb RA. Autophagy induced by ischemic preconditioning is essential for cardioprotection. J Cardiovasc Transl Res 3: 365–373, 2010. doi: 10.1007/s12265-010-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gurusamy N, Lekli I, Gorbunov NV, Gherghiceanu M, Popescu LM, Das DK. Cardioprotection by adaptation to ischaemia augments autophagy in association with BAG-1 protein. J Cell Mol Med 13: 373–387, 2009. doi: 10.1111/j.1582-4934.2008.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li S, Liu C, Gu L, Wang L, Shang Y, Liu Q, Wan J, Shi J, Wang F, Xu Z, Ji G, Li W. Autophagy protects cardiomyocytes from the myocardial ischaemia-reperfusion injury through the clearance of CLP36. Open Biol 6: 160177, 2016. doi: 10.1098/rsob.160177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest 121: 3689–3700, 2011. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pattison JS, Osinska H, Robbins J. Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes. Circ Res 109: 151–160, 2011. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 19: 365–381, 2018. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol Cell Physiol 269: C1200–C1208, 1995. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 21. Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res 24: 92–104, 2014. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic 1: 570–583, 2000. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 23. Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol 22: 407–417, 2012. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li W, Yang Q, Mao Z. Chaperone-mediated autophagy: machinery, regulation and biological consequences. Cell Mol Life Sci 68: 749–763, 2011. doi: 10.1007/s00018-010-0565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci 113: 4441–4450, 2000. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- 26. Xilouri M, Brekk OR, Landeck N, Pitychoutis PM, Papasilekas T, Papadopoulou-Daifoti Z, Kirik D, Stefanis L. Boosting chaperone-mediated autophagy in vivo mitigates alpha-synuclein-induced neurodegeneration. Brain 136: 2130–2146, 2013. doi: 10.1093/brain/awt131. [DOI] [PubMed] [Google Scholar]
- 27. Koga H, Martinez-Vicente M, Arias E, Kaushik S, Sulzer D, Cuervo AM. Constitutive upregulation of chaperone-mediated autophagy in Huntington's disease. J Neurosci 31: 18492–18505, 2011. doi: 10.1523/JNEUROSCI.3219-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper JM, Caballero C, Ferrer I, Obeso JA, Schapira AH. Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol 67: 1464–1472, 2010. doi: 10.1001/archneurol.2010.198. [DOI] [PubMed] [Google Scholar]
- 29. Vakifahmetoglu-Norberg H, Kim M, Xia HG, Iwanicki MP, Ofengeim D, Coloff JL, Pan L, Ince TA, Kroemer G, Brugge JS, Yuan J. Chaperone-mediated autophagy degrades mutant p53. Genes Dev 27: 1718–1730, 2013. [Erratum in Genes Dev 27: 1947, 2013]. doi: 10.1101/gad.220897.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab 20: 417–432, 2014. doi: 10.1016/j.cmet.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pedrozo Z, Torrealba N, Fernandez C, Gatica D, Toro B, Quiroga C, Rodriguez AE, Sanchez G, Gillette TG, Hill JA, Donoso P, Lavandero S. Cardiomyocyte ryanodine receptor degradation by chaperone-mediated autophagy. Cardiovasc Res 98: 277–285, 2013. doi: 10.1093/cvr/cvt029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Michiels C. Physiological and pathological responses to hypoxia. Am J Pathol 164: 1875–1882, 2004. doi: 10.1016/S0002-9440(10)63747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, et al. Heart disease and stroke statistics-2019 update: a report from the American heart association . Circulation 139: e56–e528, 2019. doi: 10.1161/CIR.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 34. Giordano FJ. Oxygen, oxidative stress, hypoxia, and heart failure. J Clin Invest 115: 500–508, 2005. doi: 10.1172/JCI24408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Su Z, Liu Y, Zhang H. Adaptive cardiac metabolism under chronic hypoxia: mechanism and clinical implications. Front Cell Dev Biol 9: 625524, 2021. doi: 10.3389/fcell.2021.625524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boyette LC, Manna B. Physiology, myocardial oxygen demand. In StatPearls. Treasure Island, FL: StatPearls Publishing, 2021. [PubMed] [Google Scholar]
- 37. Sendoel A, Hengartner MO. Apoptotic cell death under hypoxia. Physiology (Bethesda) 29: 168–176, 2014. doi: 10.1152/physiol.00016.2013. [DOI] [PubMed] [Google Scholar]
- 38. Wheaton WW, Chandel NS. Hypoxia. II. Hypoxia regulates cellular metabolism. Am J Physiol Cell Physiol 300: C385–C393, 2011. doi: 10.1152/ajpcell.00485.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yan L, Vatner DE, Kim SJ, Ge H, Masurekar M, Massover WH, Yang G, Matsui Y, Sadoshima J, Vatner SF. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci USA 102: 13807–13812, 2005. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eskelinen EL, Cuervo AM, Taylor MR, Nishino I, Blum JS, Dice JF, Sandoval IV, Lippincott-Schwartz J, August JT, Saftig P. Unifying nomenclature for the isoforms of the lysosomal membrane protein LAMP-2. Traffic 6: 1058–1061, 2005. doi: 10.1111/j.1600-0854.2005.00337.x. [DOI] [PubMed] [Google Scholar]
- 41. Munoz-Sanchez J, Chanez-Cardenas ME. The use of cobalt chloride as a chemical hypoxia model. J Appl Toxicol 39: 556–570, 2019. doi: 10.1002/jat.3749. [DOI] [PubMed] [Google Scholar]
- 42. Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, Mao Z. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science 323: 124–127, 2009. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koga H, Martinez-Vicente M, Macian F, Verkhusha VV, Cuervo AM. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat Commun 2: 386, 2011. doi: 10.1038/ncomms1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kyrylkova K, Kyryachenko S, Leid M, Kioussi C. Detection of apoptosis by TUNEL assay. Methods Mol Biol 887: 41–47, 2012. doi: 10.1007/978-1-61779-860-3_5. [DOI] [PubMed] [Google Scholar]
- 45. Levrand S, Vannay-Bouchiche C, Pesse B, Pacher P, Feihl F, Waeber B, Liaudet L. Peroxynitrite is a major trigger of cardiomyocyte apoptosis in vitro and in vivo. Free Radic Biol Med 41: 886–895, 2006. doi: 10.1016/j.freeradbiomed.2006.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blair CA, Haynes P, Campbell SG, Chung C, Mitov MI, Dennis D, Bonnell MR, Hoopes CW, Guglin M, Campbell KS. A protocol for collecting human cardiac tissue for research. Vad J 2: 10.13023/VAD.2016.12, 2016. doi: 10.14434/vad.v2i0.27941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Finn PF, Mesires NT, Vine M, Dice JF. Effects of small molecules on chaperone-mediated autophagy. Autophagy 1: 141–145, 2005. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- 48. Clark CB, Rane MJ, El Mehdi D, Miller CJ, Sachleben LR Jr, Gozal E. Role of oxidative stress in geldanamycin-induced cytotoxicity and disruption of Hsp90 signaling complex. Free Radic Biol Med 47: 1440–1449, 2009. doi: 10.1016/j.freeradbiomed.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol 28: 5747–5763, 2008. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jensen SS, Aaberg-Jessen C, Christensen KG, Kristensen B. Expression of the lysosomal-associated membrane protein-1 (LAMP-1) in astrocytomas. Int J Clin Exp Pathol 6: 1294–1305, 2013. [PMC free article] [PubMed] [Google Scholar]
- 51. Agarraberes FA, Terlecky SR, Dice JF. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol 137: 825–834, 1997. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saha T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy 8: 1643–1656, 2012. doi: 10.4161/auto.21654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Valdor R, Mocholi E, Botbol Y, Guerrero-Ros I, Chandra D, Koga H, Gravekamp C, Cuervo AM, Macian F. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol 15: 1046–1054, 2014. doi: 10.1038/ni.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Piret JP, Mottet D, Raes M, Michiels C. CoCl2, a chemical inducer of hypoxia-inducible factor-1, and hypoxia reduce apoptotic cell death in hepatoma cell line HepG2. Ann N Y Acad Sci 973: 443–447, 2002. doi: 10.1111/j.1749-6632.2002.tb04680.x. [DOI] [PubMed] [Google Scholar]
- 55. Ho VT, Bunn HF. Effects of transition metals on the expression of the erythropoietin gene: further evidence that the oxygen sensor is a heme protein. Biochem Biophys Res Commun 223: 175–180, 1996. doi: 10.1006/bbrc.1996.0865. [DOI] [PubMed] [Google Scholar]
- 56. Lopez-Sanchez LM, Jimenez C, Valverde A, Hernandez V, Penarando J, Martinez A, Lopez-Pedrera C, Munoz-Castaneda JR, De la Haba-Rodriguez JR, Aranda E, Rodriguez-Ariza A. CoCl2, a mimic of hypoxia, induces formation of polyploid giant cells with stem characteristics in colon cancer. PLoS One 9: e99143, 2014. doi: 10.1371/journal.pone.0099143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu D, Yotnda P. Induction and testing of hypoxia in cell culture. J Vis Exp 54: 2899, 2011. doi: 10.3791/2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kong HL, Wang JP, Li ZQ, Zhao SM, Dong J, Zhang WW. Anti-hypoxic effect of ginsenoside Rbl on neonatal rat cardiomyocytes is mediated through the specific activation of glucose transporter-4 ex vivo. Acta Pharmacol Sin 30: 396–403, 2009. doi: 10.1038/aps.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang X, Chen L. Effects of CoCl2-simulated hypoxia on the expression levels of matrix metalloproteinases in renal adenocarcinoma cells and renal tubular epithelial cells. Exp Ther Med 16: 1454–1460, 2018. doi: 10.3892/etm.2018.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bruemmer-Smith S, Stuber F, Schroeder S. Protective functions of intracellular heat-shock protein (HSP) 70-expression in patients with severe sepsis. Intensive Care Med 27: 1835–1841, 2001. doi: 10.1007/s00134-001-1131-3. [DOI] [PubMed] [Google Scholar]
- 61. Fei G, Guo C, Sun HS, Feng ZP. Chronic hypoxia stress-induced differential modulation of heat-shock protein 70 and presynaptic proteins. J Neurochem 100: 50–61, 2007. doi: 10.1111/j.1471-4159.2006.04194.x. [DOI] [PubMed] [Google Scholar]
- 62. Zhang K, Zhao T, Huang X, Liu ZH, Xiong L, Li MM, Wu LY, Zhao YQ, Zhu LL, Fan M. Preinduction of HSP70 promotes hypoxic tolerance and facilitates acclimatization to acute hypobaric hypoxia in mouse brain. Cell Stress Chaperones 14: 407–415, 2009. doi: 10.1007/s12192-008-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol 14: 167–196, 1998. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 64. Iida K, Hidaka K, Takeuchi M, Nakayama M, Yutani C, Mukai T, Morisaki T. Expression of MEF2 genes during human cardiac development. Tohoku J Exp Med 187: 15–23, 1999. doi: 10.1620/tjem.187.15. [DOI] [PubMed] [Google Scholar]
- 65. Gao L, She H, Li W, Zeng J, Zhu J, Jones DP, Mao Z, Gao G, Yang Q. Oxidation of survival factor MEF2D in neuronal death and Parkinson's disease. Antioxid Redox Signal 20: 2936–2948, 2014. doi: 10.1089/ars.2013.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Patel B, Cuervo AM. Methods to study chaperone-mediated autophagy. Methods 75: 133–140, 2015. doi: 10.1016/j.ymeth.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell 19: 2179–2192, 2008. doi: 10.1091/mbc.e07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dong S, Aguirre-Hernandez C, Scrivo A, Eliscovich C, Arias E, Bravo-Cordero JJ, Cuervo AM. Monitoring spatiotemporal changes in chaperone-mediated autophagy in vivo. Nat Commun 11: 645, 2020. doi: 10.1038/s41467-019-14164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lee JH, Choi SH, Baek MW, Kim MH, Kim HJ, Kim SH, Oh SJ, Park HJ, Kim WJ, Jung JY. CoCl2 induces apoptosis through the mitochondria- and death receptor-mediated pathway in the mouse embryonic stem cells. Mol Cell Biochem 379: 133–140, 2013. doi: 10.1007/s11010-013-1635-5. [DOI] [PubMed] [Google Scholar]
- 70. Vengellur A, LaPres JJ. The role of hypoxia inducible factor 1alpha in cobalt chloride induced cell death in mouse embryonic fibroblasts. Toxicol Sci 82: 638–646, 2004. doi: 10.1093/toxsci/kfh278. [DOI] [PubMed] [Google Scholar]
- 71. Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci USA 103: 5805–5810, 2006. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Autophagy Group. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12: 1–222, 2016. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ghosh R, Pattison JS. Macroautophagy and chaperone-mediated autophagy in heart failure: the known and the unknown. Oxid Med Cell Longev 2018: 8602041, 2018, doi: 10.1155/2018/8602041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mauvezin C, Neufeld TP. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy 11: 1437–1438, 2015. doi: 10.1080/15548627.2015.1066957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu H, Wang P, Song W, Sun X. Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. FASEB J 23: 3383–3392, 2009. doi: 10.1096/fj.09-134296. [DOI] [PubMed] [Google Scholar]
- 76. Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med 14: 959–965, 2008. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem 272: 5606–5615, 1997. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 78. Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz AL, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem 272: 9002–9010, 1997. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- 79. Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev Cell 20: 131–139, 2011. [Erratum in Dev Cell 20: 405–406, 2011]. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Orogo AM, Gustafsson AB. Cell death in the myocardium: my heart won’t go on. IUBMB life 65: 651–656, 2013. doi: 10.1002/iub.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.16734691.
Supplemental Fig. S2: https://doi.org/10.6084/m9.figshare.16734655.
Supplemental Fig. S3: https://doi.org/10.6084/m9.figshare.19681908.
Supplemental Fig. S4: https://doi.org/10.6084/m9.figshare.19325867.
Supplemental Fig. S5: https://doi.org/10.6084/m9.figshare.19326026.
Supplemental Table S1: https://doi.org/10.6084/m9.figshare.13371062.