Keywords: bioinformatics, bronchopulmonary dysplasia, machine learning, prediction, whole microarray
Abstract
Bronchopulmonary dysplasia (BPD) is the most common lung disease of extreme prematurity, yet mechanisms that associate with or identify neonates with increased susceptibility for BPD are largely unknown. Combining artificial intelligence with gene expression data is a novel approach that may assist in better understanding mechanisms underpinning chronic lung disease and in stratifying patients at greater risk for BPD. The objective of this study is to develop an early peripheral blood transcriptomic signature that can predict preterm neonates at risk for developing BPD. Secondary analysis of whole blood microarray data from 97 very low birth weight neonates on day of life 5 was performed. BPD was defined as positive pressure ventilation or oxygen requirement at 28 days of age. Participants were randomly assigned to a training (70%) and testing cohort (30%). Four gene-centric machine learning models were built, and their discriminatory abilities were compared with gestational age or birth weight. This study adheres to the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) statement. Neonates with BPD (n = 62 subjects) exhibited a lower median gestational age (26.0 wk vs. 30.0 wk, P < 0.01) and birth weight (800 g vs. 1,280 g, P < 0.01) compared with non-BPD neonates. From an initial pool (33,252 genes/patient), 4,523 genes exhibited a false discovery rate (FDR) <1%. The area under the receiver operating characteristic curve (AUC) for predicting BPD utilizing gestational age or birth weight was 87.8% and 87.2%, respectively. The machine learning models, using a combination of five genes, revealed AUCs ranging between 85.8% and 96.1%. Pathways integral to T cell development and differentiation were associated with BPD. A derived five-gene whole blood signature can accurately predict BPD in the first week of life.
INTRODUCTION
Bronchopulmonary dysplasia (BPD) affects 10,000 to 15,000 preterm newborns every year in the United States and is characterized by airway and vascular underdevelopment (1). Short- and long-term sequelae resulting from BPD include pulmonary hypertension, asthma, chronic obstructive pulmonary disease, neurodevelopmental delay, and early death (2). Despite modern-day advances in neonatal care, therapies for BPD are limited and largely supportive (3, 4). Early identification of neonates at higher risk for BPD is critical and may translate into novel interventions with potential to impact lifelong health.
Gene expression profiling provides a comprehensive picture of cellular function in a specific tissue, or circulating blood, by measuring the level of thousands of mRNA sequences (5). Whole genome microarray analysis has been the primary technique used to evaluate transcriptomic (e.g., mRNA) differences between populations of interest (6). Transcriptome-based risk or prediction scores have shown promise in many adult diseases (7–9). For instance, Rhodes et al. (7) derived a whole blood transcriptomic profile that is associated with disease severity and risk for poor clinical outcomes in individuals with pulmonary arterial hypertension. Similarly, Moll et al. (8) developed a blood-based transcriptomic risk score for predicting chronic obstructive pulmonary disease. Although these findings demonstrate the value of gene expression data, comparable approaches for BPD are limited.
Prediction modeling is an often-used technique in precision medicine that provides clinicians the probability that their patient will likely receive a diagnosis or whether they will respond to a new treatment (10–13). For example, the ability to predict which neonates are at higher risk for BPD would not only inform clinicians but may also stimulate practice changes to prevent or reduce disease severity. Many clinical-based prediction tools have been developed for BPD (14–16); however, they do not describe pathways underpinning the aberrant lung growth. This limits the ability to identify therapies that may target mechanisms contributing to BPD. Furthermore, tracking of therapeutic response using clinical-based factors (e.g., gestational age, birth weight, oxygen, and ventilator settings) may not be modifiable or can lead to subjectivity. To circumvent some of these setbacks, we opted to develop a prediction model using transcriptomics as this data is rich, quantifiable, can be used to track disease progression, and will provide insight into mechanisms or pathways involved in BPD. To further increase the novelty of our approach, we chose to build our prediction model by leveraging the booming field of artificial intelligence.
With the growing body of clinical, genetic, and electronic health record data, the field of medicine is increasingly applying artificial intelligence to expose patterns in large datasets (17–20). Specifically, machine learning (ML) models are powerful methods for building prediction algorithms (21, 22). A recent systematic review by Mangold et al. (23) highlighted the application of ML tools for predicting neonatal mortality. As elegantly stated by Pammi et al. (24), “Leveraging artificial intelligence and machine learning tools for integration of multiomics and clinical data will pave the way for precision medicine in perinatology.”
The aim of this study is to identify genes that could serve as early biomarkers for detecting which neonates are more susceptible to developing BPD. Timely detection of neonates at increased risk for BPD may assist with early clinical practice interventions, clinical trial stratification, and treatment allocation.
METHODS
Study Population
A secondary data analysis of peripheral blood microarray data set (GSE32472) downloaded through the Bioconductor R package from the National Library of Medicine’s Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) was performed. Details pertaining to the GSE32472 data set have been previously reported (25). In brief, the study was conducted by investigators from Polish American Children’s Hospital between the years 2008 and 2010 and included preterm newborns with a gestational age <32 wk who had a birth weight ≤1,500 g, and who required respiratory support at the time of enrollment. Blood sampling for whole microarray gene expression was collected on days 5, 14, and 28. As this study focused on an early transcriptomic profile that could predict BPD, expression data from day 5 were solely utilized. Institutional review board approval was not required as this study used publicly available deidentified information. The Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) guidelines for the development of prediction models were followed (26).
Study Objectives
The primary objective was to create an early whole blood transcriptomic signature that could predict BPD. BPD was defined per the National Institute of Child Health and Human Development (NICHD) (27), wherein neonates requiring supplemental oxygen for ≥28 days were diagnosed with the condition. Secondary objectives were to utilize the newly developed set of genes that could predict BPD: 1) to test their predictive performance when stratifying patients with severe BPD compared with no BPD, and 2) to derive pathways that are altered early in the disease process. Severe BPD was defined at 36 wk postmenstrual age if a preterm neonate was requiring ≥30% O2 and/or positive pressure ventilation (27).
Statistical Analysis
Overview of study design.
Figure 1 provides an outline of the analysis. We randomly split the data into training (70%) and testing (30%) samples. Model building was exclusively performed in the training set. Model testing was conducted in 30% of held-out samples. To reduce the number of differentially expressed genes, a random forest split was carried out (28, 29). Afterward, four machine learning (ML) models were used to identify the combinations of genes with the best predictive ability. We compared our ML models with those built on clinical information: gestational age, birth weight, and gestational age plus birth weight. These clinical variables were used as these are known to strongly associate with BPD (14, 15). The best-performing ML model, gradient boosting, was used to build the clinical-based (e.g., gestational age, birth weight, or gestational age plus birth weight) prediction.
Figure 1.
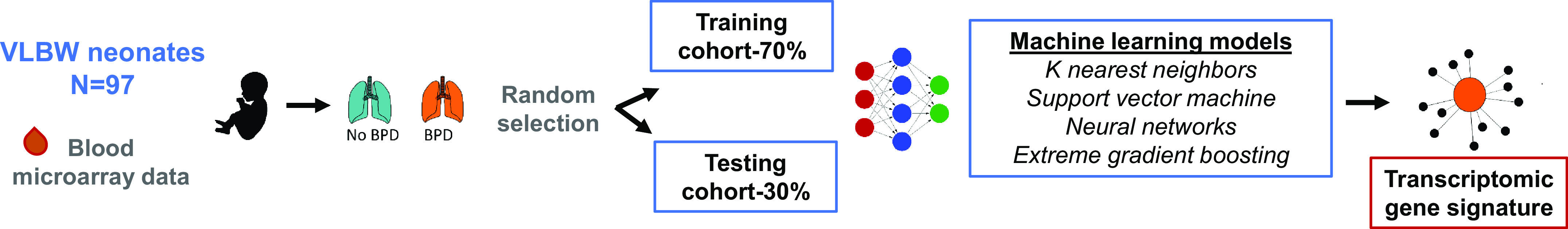
Schematic of study design. Ninety-seven neonates were randomly allocated into a training (70%) or testing cohort (30%). Four machine learning models were used to obtain the optimal combination of genes to predict bronchopulmonary dysplasia (BPD). BPD was defined per the National Institute of Child Health and Human Development, wherein neonates requiring supplemental oxygen ≥28 days were diagnosed with the condition. VLBW, very low birth weight.
Gene expression data preparation and analysis.
Boxplots and histograms were created to assess normal distribution. Gene counts were the first log2 transformed followed by quantile normalization. Figures and plots before normalization and after normalization can be viewed in the Supplemental Material (see https://doi.org/10.6084/m9.figshare.21200056). A total of 33,252 genes per patient were included in the database. Genes with expression levels <50% of total expression from all samples were excluded, leaving a total of 20,697 genes. These genes were removed as low levels across all samples are not likely to be differentially expressed (e.g., false discovery rate [FDR] <0.05) in comparisons of study cohorts. Outliers were weighted per Ritchie et al. (30). Genes were considered significant if the FDRs, by Benjamini and Hochberg-adjusted P values, were less than 5%.
Machine learning models.
Four machine learning (ML) prediction models were constructed: 1) k nearest neighbors, 2) support vector machine, 3) neural networks, and 4) gradient boosting. K nearest neighbors separate data by clustering according to the degree of separation between the groups of interest (31). Unlike regression analyses, support vector machine separates groups by creating nonlinear models within a hyperplane (32). Neural networks are another class of machine learning modeling wherein variables are entered into a node that bases decisions that are meant to recapitulate human neurons (33). Gradient boosting is an ensemble algorithm that identifies predictors by creating successive decision trees (33). To minimize potential overfitting in the 4 ML models, we used 10-fold cross validation repeated five times. Default hyperparameters created within the caret package were used and no data were imputed. These ML models were selected for two reasons: 1) they embrace different approaches in the field (34–36) and 2) previous neonatal publications using ML models for mortality prediction incorporated these methods (23, 37). The Supplemental Material provides technical information regarding the default tuning parameters used in each model.
Performance evaluation.
In the test set (30% random sample), we measured the prediction performance of each model by calculating the following: 1) area under the receiver operating characteristic curve (AUC), 2) performance metrics (e.g., sensitivity, specificity, positive predictive value, and negative predictive value), and 3) calibration plots. Gene importance by random permutation was computed for each model. For demographic data, we considered a two-sided P value less than 5% to be statistically significant. All analyses were performed with R statistical software version 4.1.0.
Pathway analysis.
Pathways involved in the development of BPD were assessed using the top differentially expressed genes (FDR < 0.01). The R package gprofiler2 was used to organize the genes into KEGG (Kyoto Encyclopedia of Genes and Genomes) ontological processes according to their log P adjusted values. The program performs functional enrichment analysis based on the genes inputted. Afterward, Cytoscape, an open-source software platform for visualizing complex networks, was used to create a map to demonstrate the interactions of the pathways (https://cytoscape.org/) (38).
Post hoc analysis.
To demonstrate that our gene signature was “BPD-specific” and not a reflection of prematurity (e.g., lower gestational age in the BPD cohort), we performed two post hoc analyses. In the first analysis, we tested the performance of the four ML models limited to extremely low gestational age neonates (ELGA, ≤28 wk). In the second evaluation, we conducted a propensity score analysis using a 1:1 nearest neighbor matching algorithm with distances determined by logistic regression. Propensity score matching (PSM) was performed based on gestational age in weeks with a caliper of 0.2 of the standard deviation of the logit using the MatchIt package in R (39).
RESULTS
Characteristics of the Study Population
The study cohort consisted of 97 patients, of whom 62 (63.9%) had BPD diagnosed at 28 days of life. Table 1 provides the demographic characteristics of the cohort corresponding to the presence of BPD compared with those without BPD. As expected, neonates with BPD had a lower median gestational age (26.0 wk vs. 30.0 wk, P < 0.01) and median birth weight (800 g vs. 1,280 g, P < 0.01). Fifteen (24.2%), out of 62, neonates with BPD were diagnosed with severe lung disease at 36 wk’ postmenstrual age. The median birth weight of this subset of neonates was 690 g [interquartile range (IQR), 598, 735] with a median gestational age of 25.0 wk (IQR, 24.0, 26.0).
Table 1.
Patient characteristics
| Value | Overall (n = 97) | No BPD (n = 35) | Any BPD (n = 62) | Severe BPD (n = 15) | P Value |
|---|---|---|---|---|---|
| Gestational age, wk | 28.0 (26.0, 30.0) | 30.0 (29.0, 31.0) | 26.0 (25.0, 28.0) | 25.0 (24.0, 26.0) | <0.01 |
| Birth weight, g | 1,000 (760, 1,245) | 1,280 (1,150, 1,395) | 800 (690, 1,000) | 690 (598, 735) | <0.01 |
| Female | 45 (46.4%) | 21 (60.0%) | 24 (38.7%) | 6 (40.0%) | 0.04 |
| Cesarean delivery | 55 (56.7%) | 25 (71.4%) | 30 (48.4%) | 6 (40.0%) | 0.01 |
| Prenatal steroids | 34 (35.1%) | 19 (54.3%) | 15 (24.2%) | 3 (20.0%) | <0.01 |
| Surfactant | 56 (57.7%) | 14 (40.0%) | 42(67.7%) | 9 (60%) | <0.01 |
| PDA medication | 53 (54.6%) | 13 (37.1%) | 40 (64.5%) | 11 (73.3%) | <0.01 |
| Periventricular leukomalacia | 14 (14.4%) | 1 (2.9%) | 13 (21.0%) | 4 (26.7%) | 0.02 |
| Retinopathy of prematurity | 47 (48.5%) | 2 (5.7%) | 45 (72.6%) | 15 (100%) | <0.01 |
Values are median with interquartile range for continuous data and number with percentage for categorical variables. Statistical analysis included Wilcoxon rank-sum test and Chi-square for continuous and categorical data, respectively. P value compares no bronchopulmonary dysplasia (BPD) vs. any BPD groups. PDA, patent ductus arteriosus.
Genes Differentially Expressed in Patients With BPD Compared With Controls
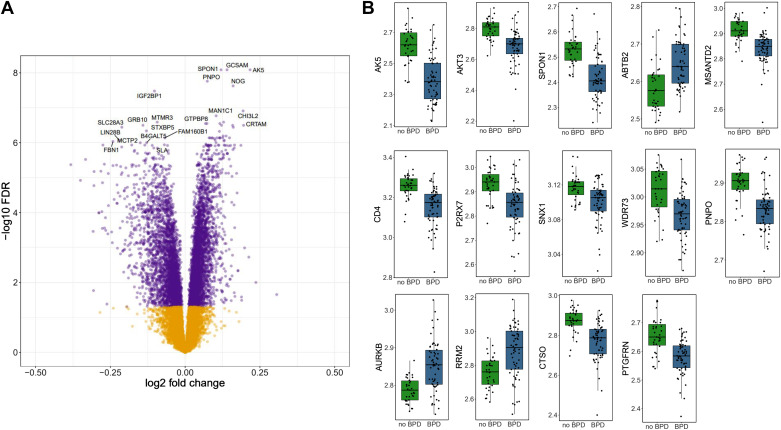
A principal components analysis plot was created to examine differences between the transcripts in neonates with or without BPD (Supplemental Material). Of the 20,697 genes, 7,319 genes (35.4%) were significantly different after multiple comparison testing (FDR < 5%). Three thousand seventy-six genes were downregulated and 3,943 were upregulated as shown in the volcano plot (Fig. 2A). Given the large number of differentially expressed genes, we focused on the 4,000 genes that had an FDR <1%. After random forest selection, 14 genes were identified as ideal predictors for BPD (refer to Fig. 2B), upon which the four machine learning algorithms were used to optimize the combination of these 14 features for model building.
Figure 2.
Transcriptomic differences in neonates with bronchopulmonary dysplasia (BPD) and control subjects (no BPD). A: volcano plot demonstrating log fold differences in genes with a false discovery rate <5%. Purple dots designate differentially expressed genes, whereas dots in yellow denote no difference. B: boxplots of the top 14 genes by BPD (n = 62 subjects) vs. no BPD (n = 35). Each box indicates the median with interquartile range, whereas the dots represent expression of the gene per individual. ABTB2, ankyrin repeat and BTB domain containing 2; AK5, adenylate kinase 5; AKT3, AKT serine/threonine kinase 3; AURKB, Aurora kinase B; CD4, CD4 molecule; CTSO, Cathepsin O; MSANTD2, Myb/SANT DNA binding domain containing 2; P2RX7, purinergic receptor P2X, ligand-gated ion channel 7; PNPO, pyridoxine 5′-phosphate oxidase; PTGFRN, Prostaglandin F2 receptor inhibitor; RRM2, ribonucleotide reductase regulatory subunit M2; SNX1, sorting nexin 1; SPON1, Spondin 1; WDR73, WD repeat domain 73. PDA, patent ductus arteriosus.
Performance of ML Models Predicting BPD
Receiver operating characteristic curves were built for gestational age, birth weight, and the four transcriptomic machine learning models. The AUC for predicting BPD when using birth weight or gestational age was 87.2% and 87.8%, respectively (Fig. 3, A and B). A model combining birth weight with gestational age improved the AUC to 89.7%. The ML models had AUCs that ranged between 85.8% and 96.1% (Fig. 3, C–F). Of the 14 optimal genes, each of the ML models was built using the top 5; gene importance can be seen in the Supplemental Material. Extreme gradient boosting tree had the highest AUC (96.1%, 95% CI 89.7, 100) and incorporated the following genes in order of importance: PNPO, MSANTD2, CD4, SNX1, and P2RX7. Performance metrics for all of the models are included in Fig. 3. The calibration and lift plots for the ML and clinical-based prediction models can also be viewed in the Supplemental Material. Genes differentially expressed in patients with BPD compared with severe BPD can also be viewed in the Supplemental Material.
Figure 3.
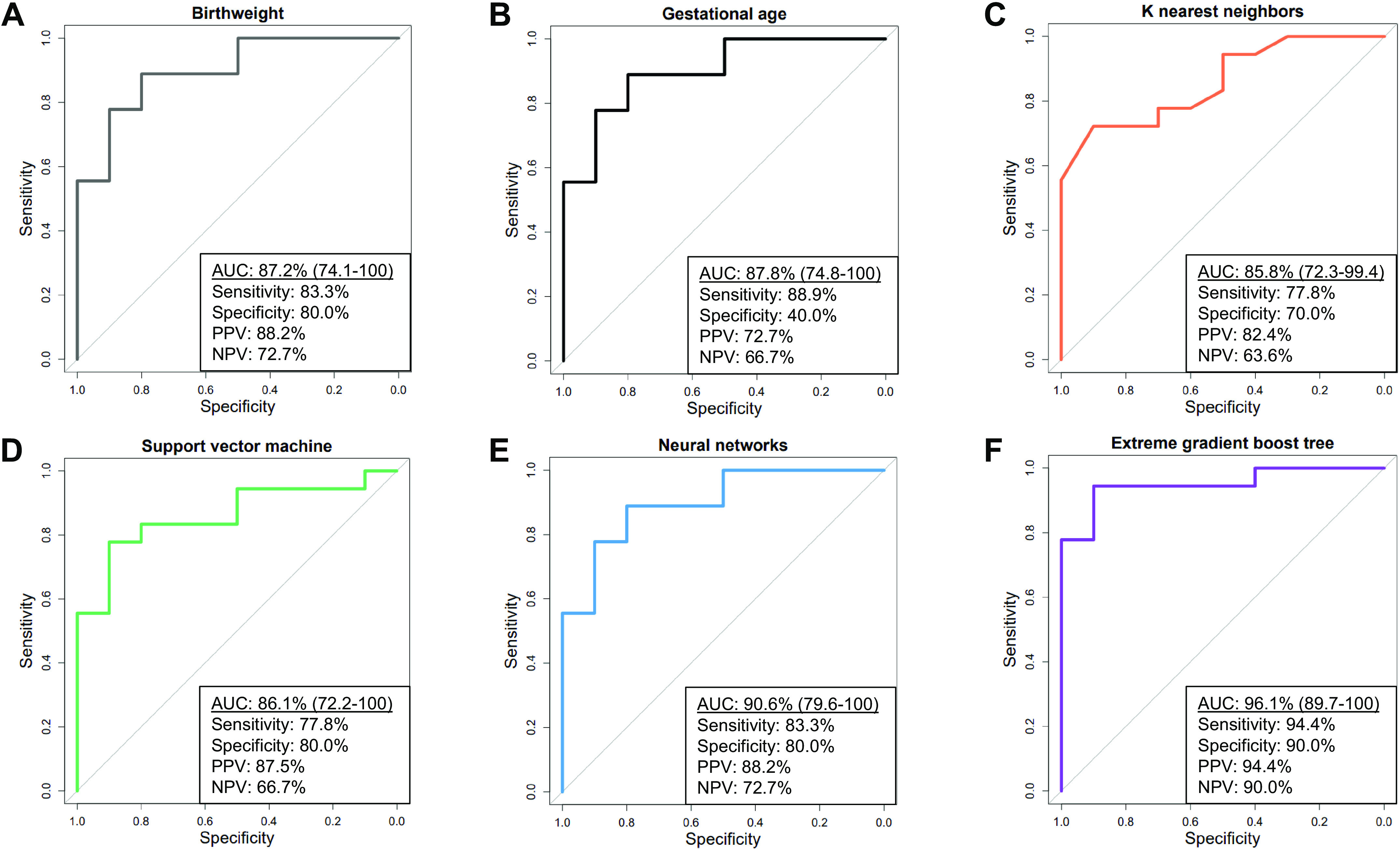
Machine learning model performance in the testing cohort. A–F: area under the receiver operating characteristics curves (AUC) for clinical and machine learning models with respective performance metrics. Extreme gradient boost tree was used to build the birth weight and the gestational age models. NPV, negative predictive value; PPV, positive predictive value.
Transcriptomic Signature across Spectrum of Disease
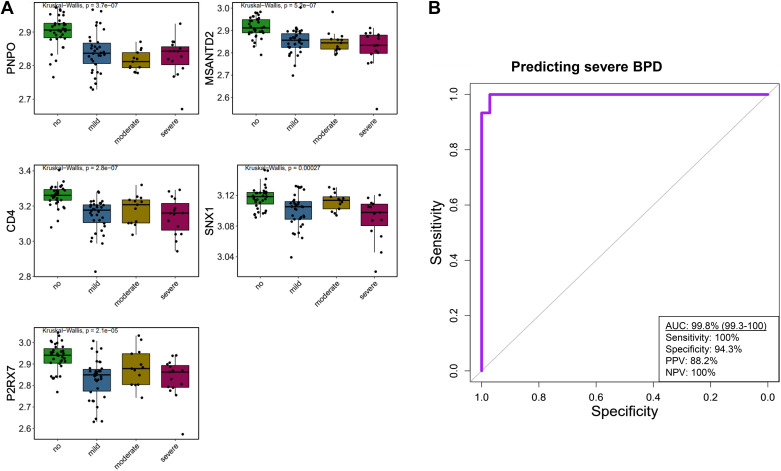
To further characterize the transcriptomic signature, we analyzed its association with disease severity. Figure 4A provides boxplots of the five genes in patients without BPD compared with those who developed mild, moderate, and severe BPD. There was a significant difference in RNA expression between neonates with or without BPD in all of the genes. Figure 4B presents the AUC using the five-gene signature, on day of life 5, for predicting severe BPD.
Figure 4.
Median expression of genes by bronchopulmonary dysplasia (BPD) severity and performance of extreme gradient boost tree machine learning model in predicting severe BPD. A: five-gene signature stratified by no BPD (n = 35) and mild (n = 34), moderate (n = 13), or severe BPD (n = 15). Each box indicates the median with interquartile range, whereas the dots represent expression of the gene per individual. Kruskal–Wallis statistical test was conducted to examine differences across multiple groups. No BPD vs. each BPD group was statistically significant for each gene on post hoc analysis. Expression of each gene was comparable among the BPD groups (P > 0.05). B: area under the receiver operating characteristics curve (AUC) for early prediction (e.g., day 5) of severe BPD in the testing sample (n = 28 subjects) utilizing the five transcriptomic signature built with extreme gradient boost tree. As depicted, the five genes included pyridoxine 5′-phosphate oxidase (PNPO), Myb/SANT DNA binding domain containing 2 (MSANTD2), CD4, CD4 molecule (CD4), sorting nexin 1 (SNX1), and purinergic receptor P2X, ligand-gated ion channel 7 (P2RX7).
Correlation Analysis and Pathway Analysis
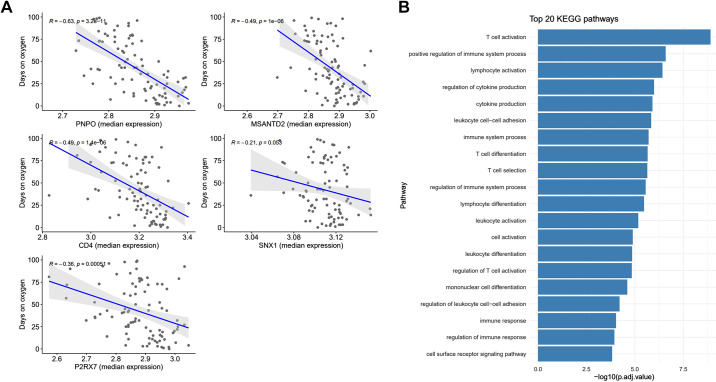
To assess the physiological interpretation of our gene signature, we examined cumulative oxygen days. There was a negative linear relationship between the number of oxygen days the neonate received in the intensive care unit with the five genes. Decreased expression of the genes correlated with higher oxygen days. All of the correlations were significant except for SNX1 (Fig. 5A).
Figure 5.
A: correlation plot with 95% confidence interval of the median expression of the 5-gene signature (x axis), on day of life 5, by the cumulative number of days the neonate received oxygen while in the intensive care unit (y axis). Gray dots illustrate gene expression for each neonate. R = correlation coefficient. B: top 20 Kyoto encyclopedia of genes and genomes (KEGG) pathways dysregulated in neonates with bronchopulmonary dysplasia (BPD) compared with no BPD on day of life 5. Genes with a false discovery rate <0.01 (n = 4,000) between the groups were used to organize pathways. CD4, CD4 molecule; MSANTD2, Myb/SANT DNA binding domain containing 2; P2RX7, purinergic receptor P2X, ligand-gated ion channel 7; PNPO, pyridoxine 5′-phosphate oxidase; SNX1, sorting nexin 1.
The top 20 KEGG pathways and interactions are shown in Fig. 4B. Twenty percent (n = 4) of the pathways dealt with T cell selection, activation, or differentiation. Nearly all (90%) of the pathways pertained to immune response, activation, or immune cell differentiation. The pathway with the largest log adjusted value was T cell activation.
Annotation of Differentially Expressed Genes in Previous Studies
Results from the current study were compared with findings from previously published pulmonary work (40–59). Table 2 summarizes the 14 genes that served as potential predictors for BPD. Processes included mitochondrial biogenesis, cell-cell adhesion, apoptosis, and immune cell response/development.
Table 2.
Description of 14 predictive genes with their ontological processes and literature evaluation
| Gene | Symbol | Gene Ontology Process | Reference No. | Main Findings |
|---|---|---|---|---|
| Adenylate kinase 5 | AK5 | ADP biosynthetic process///ATP metabolic process | (40) | Involved in cell invasion and migration of colorectal cancer cells by activating AMPK |
| AKT serine/threonine kinase 3 | AKT3 | Intracellular signal transduction///mitochondrial genome maintenance | (41–43) | Stimulates mitochondrial biogenesis downstream of VEGF in endothelial cells; prevents lung injury by inhibiting inflammation/apoptosis |
| Spondin 1 | SPON1 | Cell adhesion///protein O-linked fucosylation | (44) | Extracellular matrix protein important in cell-cell adhesion proteins |
| Ankyrin repeat and BTB domain containing 2 | ABTB2 | Cellular response to toxic substance///proteasome-mediated ubiquitin-dependent protein catabolic process | (45, 46) | Associated with the development of bronchial asthma, arterial hypertension; induces resistance to drug’s cytotoxicity by inducing apoptosis |
| Myb/SANT DNA binding domain containing 2 | MSANTD2 | |||
| CD4 molecule | CD4 | T cell costimulation///T cell differentiation | (47) | Increased CD4 Regulatory T cells may contribute to inflation and development of BPD |
| Purinergic receptor P2X 7 | P2RX7 | NAD transport///T cell homeostasis | (48, 49) | Increases surfactant secretion of alveolar epithelial cells; Activation of P2X7 receptor through ATP endogenous signal can lead to lung inflammation and fibrosis |
| Sorting nexin 1 | SNX1 | Cell-cell adhesion///early endosome to Golgi transport | (50) | Regulation of the cell-surface expression of epidermal growth factor receptor and intracellular trafficking |
| WD repeat domain 73 | WDR73 | Accumulates at the spindle poles and astral microtubules during mitosis | (51) | WDR73 loss of function mutations may be causative for Galloway–Mowat syndrome. |
| Pyridoxamine 5′-phosphate oxidase | PNPO | Mitophagy in response to mitochondrial depolarization///oxidation-reduction process | (52–54) | Vitamin B6 metabolism; regulates the mitochondrial membrane potential; lung mitophagy |
| Aurora kinase B | AURKB | Abscission///aging | (55, 60) | Overexpression leads to produce multinuclearity and increases aneuploidy; regulates MYC stability in T cell acute lymphoblastic leukemia through a kinase activity-dependent mechanism |
| Ribonucleotide reductase regulatory subunit M2 | RRM2 | DNA replication///G1/S transition of mitotic cell cycle | (56, 57) | RRM2 is upregulated in severe asthmatic bronchial epithelium and fibroblasts; Associated with the cell cycle, p53 signaling pathway, DNA replication, small cell lung cancer and apoptosis. |
| Cathepsin O | CTSO | Proteolysis///proteolysis involved in cellular protein catabolic process | (58) | Human cysteine proteinase the expression of which is dramatically upregulated during the in vitro maturation of peripheral blood monocytes into macrophages. |
| Prostaglandin F2 receptor inhibitor | PTGFRN | Involved in myoblast fusion and in skeletal muscle regeneration//cell surface | (59) | PTGFRN is upregulated in idiopathic pulmonary fibrosis |
AMPK, AMP-activated protein kinase; BPD, bronchopulmonary dysplasia.
Results of the Post Hoc Analyses
Fifty-eight neonates were included in the ELGA cohort. Neonates with BPD (n = 51) had a lower birth weight (770 g vs. 1230 g, P < 0.001) and gestational age (median of 26.0 wk compared with 28.0 wk, P = 0.001). A table of the patient demographics is included in the Supplemental Material. The AUC for the four ML models ranged from 96.9% (K nearest neighbors) to 99.2% (support vector machine). Receiver operating characteristic curves can be viewed in the Supplemental Material.
Thirty-six neonates were included in the PSM analysis. The median gestational age between the no BPD (n = 18) and BPD (n = 18) was 29.0 wk with an IQR of 28.0–30.0 wk (P = 0.90). The group of neonates diagnosed with BPD had a lower gestational age (1,045 g vs. 1,238 g, P = 0.02). The range of AUCs for the four ML models was between 79.0 (neural networks) to 100% (gradient boosting and support vector machine).
DISCUSSION
We have reported a transcriptomic signature that discriminates risk for BPD in the first week of life. Pathway analysis identified that T cell development and function are dysregulated in BPD neonates. Overall, these results suggest that peripheral blood-based transcriptomics, combined with machine learning, is a novel approach that may identify highly vulnerable neonates in need of mitigation to prevent chronic lung disease.
We sought to create an early prediction model, based on transcriptomics, that could identify neonates at risk for developing BPD. Using whole microarray data from blood, we successfully narrowed the number of differentially expressed genes between neonates with or without BPD to 14. Although mRNA from human lung tissue would be ideal, expression profiling from this tissue in very premature neonates is not feasible given their size and high acuity. However, blood sampling is common in neonatology and can evaluate not only lung-specific injury but also assess systemic inflammation and the immune response to chronic oxygenation and positive pressure ventilation. Furthermore, literature suggests that whole blood is an appropriate surrogate for lung disease (61–63).
The gene pyridoxine 5′-phosphate oxidase (PNPO) had the highest importance in our top two ML models (gradient boosting and neural networks). PNPO is an enzyme critical in vitamin B6 metabolism and subsequent amino acid metabolism and neurotransmitter metabolism (52, 53). A severe deficiency in the gene typically manifests in the neonatal period with seizures, whereas non-neurologic presentations include prematurity, poor growth, respiratory distress, and metabolic acidosis (64, 65). Pertinent to BPD, PNPO regulates the mitochondrial membrane potential and as a result is intimately involved in oxidative stress (53). It is known that chronic exposure to oxygen in neonates is an important process that results in lung mitophagy (54). The persistent inability to handle high loads of oxygen radicals inhibits lung alveolar cell renewal (66).
In a review article on multiomics and BPD, Toldi et al. (67) describe the function of T cells in directing systemic inflammation and lung tissue repair. Neonates diagnosed with BPD were more likely to have lower expression of the gene T cell surface glycoprotein CD4 (CD4). The CD4 antigen recognizes cells that express the antigen-presenting cell major histocompatibility complex II molecule. In a study examining lymphocyte subpopulations during the first 4 weeks of life, preterm neonates with BPD had lower numbers of CD4+ T cells when compared with preterm neonates without BPD (68).
In our study, the T cell pathway was the most important pathway found to be dysregulated when comparing mononuclear cells of neonates with or without BPD. This parallels what has been found in preclinical and clinical work. For example, in an animal model of BPD, Shrestha et al. (69) show that chorioamnionitis and hyperoxia result in a downregulation of T cell receptor signaling. Furthermore, investigators concluded that neonates with BPD had a skewing of T helper cells toward a Th2 phenotype (70). Th2 polarization was evidenced by increased expression of interleukin 4, 5, and 13, and when blocked enhanced normal lung alveolarization and vascularization. Similar observations of TH2 skewing are phenotypically observed in asthma (71, 72). At birth, all neonates have a Th2 skew as this polarization is necessary for women to maintain a healthy pregnancy (73). Typically, Th2 skewing in neonates resolves over the first few months to years of life (74, 75). We can speculate that prolonged exposure to positive pressure ventilation and oxygen may not allow for Th2 reversal and may partially explain why preterm newborns with BPD are at increased risk for asthma in early childhood (76, 77).
Sorting nexin 1 (SNX1) is a gene involved in regulation of the cell-surface expression of epidermal growth factor receptor and intracellular trafficking at the endosome-plasma membrane interface (50). Loss or deficiency in SNX1 associates with loss of epithelial borders and increased oxidative stress (78, 79). Maintaining the integrity of epithelial borders is critical in preterm neonatal lungs. Constant pulmonary injury in premature newborns from infection, poor nutrition, baro/volu-trauma, and oxygen radicals manifests in a breakdown of the normal endothelial and epithelial barrier (80, 81). Decreased SNX1 expression alters expression of epidermal growth factor receptors, which in turn may affect normal lung branching morphogenesis (82).
Purinergic Receptor P2X, Ligand-Gated Ion Channel 7 (P2RX7) receptors are expressed in alveolar type I cells and indirectly stimulate alveolar type II cell surfactant secretion (83). Purine nucleotides are key regulators of immunologic response, including T cell proliferation/differentiation and inflammasome activation (84). P2RX7 has many roles in immunity and it is interesting that neonates with BPD had a lower expression of the gene when other studies show that lung activation of P2XR7 associates with lung fibrosis (49). The “dual” beneficial/detrimental function of P2RX7 has been previously discussed (85).
Myb/SANT DNA binding domain containing 2, or MSANTD2, was the last gene in the gradient-boosting ML model. However, the function of this gene is currently unknown.
Although our transcriptomic model shows promise, our study does have limitations. For example, our study is derived from a retrospective analysis of a single-center homogeneous population of neonates. Validation of our model in an external cohort of neonates would strengthen the generalizability of our findings. Although 97 well-phenotyped neonates with omic data is a hefty feat, human studies involving other diseases (e.g., asthma) include much larger sample sizes. Another potential limitation includes the use of microarray data. Rapid advances in biotechnology have now moved the needle of bioinformatics into RNA-Seq and single-cell analyses. Data from this study stemmed from peripheral blood mononuclear cells. Accordingly, it is expected to have an enrichment of immune-related pathways and does not necessarily mean that immune pathways are the primary drivers of BPD. These whole blood immune cells may also differ from the immunological profile found in the lung. Moreover, new prediction models should outperform clinically relevant models that have widespread use, such as the NICHD BPD outcome estimator, https://neonatal.rti.org/index.cfm?fuseaction=BPDCalculator.start. Unfortunately, this head-to-head comparison was not feasible as the respiratory mode and oxygen requirement on specific postnatal days were not captured in the original study. We also attempted to do the post hoc analysis by filtering the neonates by extremely low birth weight; however, only two neonates ≤1,000 g did not develop BPD. We believed that such an imbalance between the groups would not accurately represent our work. Despite these reservations, the knowledge we can derive from whole microarray, and the underutilization of bioinformatics in BPD, outweigh the limitations.
Strengths of this study include leveraging artificial intelligence in a well-phenotyped cohort to develop a five-gene signature that can predict BPD with high sensitivity and specificity. Furthermore, we used high methodological rigor by randomly assigning 70% of the patients to train/build the model and test performance in the remaining 30% of patients. We then compared the discriminatory ability of our top genes with phenotypic data (e.g., gestational age, birth weight) that are clinically regarded among the best predictors of BPD. Conducting the post hoc analyses offers further support that our transcriptomic signatures may predict BPD even after filtering for neonates (e.g., ELGA) at the greatest risk for developing the disease. Another strength of our study includes the use of the TRIPOD reporting guideline for prediction models. A previous publication by Onland et al. (86) described the lack of calibration plots and is often overlooked when presenting new prediction models. As such, we incorporated calibration plots, as well as tuning parameters for all the models derived. Future directions include external validation of our signature, as well as examining the temporal change in genes/pathways in neonates with BPD. To better understand the role of immune cells in neonatal lung pathology, insights from single-cell analysis may provide a better picture of this intricate ecosystem. Finally, to our understanding, this is among the first transcriptomic gene signatures that can predict BPD.
In conclusion, we show that the combination of omics and artificial intelligence can potentially predict BPD and stratify neonates at risk for severe BPD. Future applications of omics-based technologies may be used to help further characterize BPD (e.g., endotypes), examine temporal changes in the expression of genes associated with BPD, and may eventually aid in predicting long-term respiratory outcomes. Approaches such as the ones used in this study may help unravel pathobiological mechanisms that may be used for future targeted therapies to the right patient, at the right time, and at the right dose.
DATA AVAILABILITY
Data will be made available upon reasonable request.
SUPPLEMENTAL DATA
Supplemental Material: https://doi.org/10.6084/m9.figshare.21200056.
GRANTS
This work was supported by Parker B. Francis (to A. Moreira); the National Institutes of Health (NIH) Eunice Kennedy Shriver National Institute of Child Health and Human Development K23HD101701 (to A. Moreira), NIH T32DE014318 COSTAR institutional research training grant (to A. M. Smith), NIH National Institute of Aging K23AG066933 (to G. C. Lee), NIH National Heart, Lung, and Blood Institute (NHLBI) T32 HL139430, National Institute on Minority Health and Health Disparities of the NIH under Award No. P50MD015496 (to A. Moreira), NIH National Heart Lung and Blood Institute 5R25HL126140-06 Subaward (to A. Moreira), Veterans Affairs (VA) Research Center for AIDS and HIV Infection and VA Center for Personalized Medicine grant IP1 CX000875, NIH MERIT Grant R37AI046326, Doris Duke Distinguished Clinical Scientist Award, and Burroughs Wellcome Clinical Scientist Award in Translational Research (to S. K. Ahuja).
DISCLAIMERS
Funding agencies had no role in the writing of the manuscript or the decision to submit.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
A.M. and S.K.A. conceived and designed research; P.K. performed experiments; A.M., M.T., and J.A.M. analyzed data; A.M., J.G.N.G., B.T., P.K., and S.K.A. interpreted results of experiments; A.M. and M.T. prepared figures; A.M. and M.T. drafted manuscript; Alvaro Moreira, A.M.S., G.C.L., Z.C., Axel Moreira, C.W., S.B.M., S.S., T.F., J.G.N.G., B.T., P.K., and S.K.A. edited and revised manuscript; Alvaro Moreira, A.M.S., G.C.L., Z.C., Axel Moreira, C.W., S.B.M., S.S., T.F., J.G.N.G., B.T., P.K., and S.K.A. approved final version of manuscript.
REFERENCES
- 1. Jensen EA, Schmidt B. Epidemiology of bronchopulmonary dysplasia. Birth Defects Res A Clin Mol Teratol 100: 145–157, 2014. doi: 10.1002/bdra.23235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Geetha O, Rajadurai VS, Anand AJ, Dela Puerta R, Huey Quek B, Khoo PC, Chua MC, Agarwal P. New BPD-prevalence and risk factors for bronchopulmonary dysplasia/mortality in extremely low gestational age infants ≤28 weeks. J Perinatol 41: 1943–1950, 2021. doi: 10.1038/s41372-021-01095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aldana-Aguirre JC, Pinto M, Featherstone RM, Kumar M. Less invasive surfactant administration versus intubation for surfactant delivery in preterm infants with respiratory distress syndrome: a systematic review and meta-analysis. Arch Dis Child Fetal Neonatal Ed 102: F17–F23, 2017. doi: 10.1136/archdischild-2015-310299. [DOI] [PubMed] [Google Scholar]
- 4. Thébaud B, Goss KN, Laughon M, Whitsett JA, Abman SH, Steinhorn RH, Aschner JL, Davis PG, McGrath-Morrow SA, Soll RF, Jobe AH. Bronchopulmonary dysplasia. Nat Rev Dis Primers 5: 78, 2019. doi: 10.1038/s41572-019-0127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaforou M, Broderick C, Vito O, Levin M, Scriba TJ, Seddon JA. Transcriptomics for child and adolescent tuberculosis. Immunol Rev 309: 97–122, 2022. doi: 10.1111/imr.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 7: 55–65, 2006. [Erratum in Nat Rev Genet 7: 406, 2006]. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 7. Rhodes CJ, Otero-Núñez P, Wharton J, Swietlik EM, Kariotis S, Harbaum L, Dunning MJ, Elinoff JM, Errington N, Thompson AAR, Iremonger J, Coghlan JG, Corris PA, Howard LS, Kiely DG, Church C, Pepke-Zaba J, Toshner M, Wort SJ, Desai AA, Humbert M, Nichols WC, Southgate L, Trégouët D-A, Trembath RC, Prokopenko I, Gräf S, Morrell NW, Wang D, Lawrie A. Whole-blood RNA profiles associated with pulmonary arterial hypertension and clinical outcome. Am J Respir Crit Care Med 202: 586–594, 2020. doi: 10.1164/rccm.202003-0510OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moll M, Boueiz A, Ghosh AJ, Saferali A, Lee S, Xu Z, Yun JH, Hobbs BD, Hersh CP, Sin DD, Tal-Singer R, Silverman EK, Cho MH, Castaldi PJ; HAPIN Investigators. Development of a blood-based transcriptional risk score for chronic obstructive pulmonary disease. Am J Respir Crit Care Med 205: 161–170, 2022. doi: 10.1164/rccm.202107-1584OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee GC, Restrepo MI, Harper N, Manoharan MS, Smith AM, Meunier JA, Sanchez-Reilly S, Ehsan A, Branum AP, Winter C, Winter L, Jimenez F, Pandranki L, Carrillo A, Perez GL, Anzueto A, Trinh H, Lee M, Hecht JM, Martinez-Vargas C, Sehgal RT, Cadena J, Walter EA, Oakman K, Benavides R, Pugh JA, Letendre S, Steri M, Orrù V, Fiorillo E; South Texas Veterans Health Care System COVID-19 Team. Immunologic resilience and COVID-19 survival advantage. J Allergy Clin Immunol 148: 1176–1191, 2021. doi: 10.1016/j.jaci.2021.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zea-Vera R, Ryan CT, Navarro SM, Havelka J, Wall MJ Jr, Coselli JS, Rosengart TK, Chatterjee S, Ghanta RK. Development of a machine learning model to predict outcomes and cost after cardiac surgery. Ann Thorac Surg. In press, 2022. doi: 10.1016/j.athoracsur.2022.06.055. [DOI] [PubMed] [Google Scholar]
- 11. Safari A, Adibi A, Sin DD, Lee TY, Ho JK, Sadatsafavi M. ACCEPT 2·0: recalibrating and externally validating the Acute COPD exacerbation prediction tool (ACCEPT). EClinicalMedicine 51: 101574, 2022. doi: 10.1016/j.eclinm.2022.101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abe D, Inaji M, Hase T, Takahashi S, Sakai R, Ayabe F, Tanaka Y, Otomo Y, Maehara T. A prehospital triage system to detect traumatic intracranial hemorrhage using machine learning algorithms. JAMA Netw Open 5: e2216393, 2022. doi: 10.1001/jamanetworkopen.2022.16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang S, Yu H, Gan Y, Wu Z, Li E, Li X, Cao J, Zhu Y, Wang L, Deng H, Xie M, Wang Y, Ma X, Liu D, Chen B, Tian P, Qiu Z, Xian J, Ren J, Wang K, Wei W, Xie F, Li Z, Wang Q, Xue X, Liu Z, Shi J, Li W, Tian J. Mining whole-lung information by artificial intelligence for predicting EGFR genotype and targeted therapy response in lung cancer: a multicohort study. Lancet Digit Health 4: e309–e319, 2022. doi: 10.1016/S2589-7500(22)00024-3. [DOI] [PubMed] [Google Scholar]
- 14. Greenberg RG, McDonald SA, Laughon MM, Tanaka D, Jensen E, Van Meurs K, Eichenwald E, Brumbaugh JE, Duncan A, Walsh M, Das A, Cotten CM. Online clinical tool to estimate risk of bronchopulmonary dysplasia in extremely preterm infants. Arch Dis Child Fetal Neonatal Ed. In press, 2022. doi: 10.1136/archdischild-2021-323573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laughon MM, Langer JC, Bose CL, Smith PB, Ambalavanan N, Kennedy KA, Stoll BJ, Buchter S, Laptook AR, Ehrenkranz RA, Cotten CM, Wilson-Costello DE, Shankaran S, Meurs KPV, Davis AS, Gantz MG, Finer NN, Yoder BA, Faix RG, Carlo WA, Schibler KR, Newman NS, Rich W, Das A, Higgins RD, Walsh MC. Prediction of bronchopulmonary dysplasia by postnatal age in extremely premature infants. Am J Respir Crit Care Med 183: 1715–1722, 2011. doi: 10.1164/rccm.201101-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peng HB, Zhan YL, Chen Y, et al. Prediction models for bronchopulmonary dysplasia in preterm infants: a systematic review. Front Pediatr 10: 856159, 2022. doi: 10.3389/fped.2022.856159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Groos D, Adde L, Aubert S, Boswell L, de Regnier R-A, Fjørtoft T, Gaebler-Spira D, Haukeland A, Loennecken M, Msall M, Möinichen UI, Pascal A, Peyton C, Ramampiaro H, Schreiber MD, Silberg IE, Songstad NT, Thomas N, Van den Broeck C, Øberg GK, Ihlen EAF, Støen R. Development and validation of a deep learning method to predict cerebral palsy from spontaneous movements in infants at high risk. JAMA Netw Open 5: e2221325, 2022. doi: 10.1001/jamanetworkopen.2022.21325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramprasad J, Krishnan L, Gangadharan C, Deshpande G, Madhu H, Kakileti ST, Manjunath G. Performance of artificial intelligence-based breast cancer screening in a community setting: a real-world evaluation study. Lancet Oncol 23: S20, 2022. doi: 10.1016/S1470-2045(22)00419-3. [DOI] [Google Scholar]
- 19. Garcia-Canadilla P, Isabel-Roquero A, Aurensanz-Clemente E, Valls-Esteve A, Miguel FA, Ormazabal D, Llanos F, Sanchez-de-Toledo J. Machine learning-based systems for the anticipation of adverse events after pediatric cardiac surgery. Front Pediatr 10: 930913, 2022. doi: 10.3389/fped.2022.930913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsu E, Malagaris I, Kuo YF, Sultana R, Roberts K. Deep learning-based NLP data pipeline for EHR-scanned document information extraction. JAMIA Open 5: ooac045, 2022. doi: 10.1093/jamiaopen/ooac045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang K, Yan LZ, Li WZ, Jiang C, Wang NN, Zheng Q, Dong NG, Shi JW. Comparison of four machine learning techniques for prediction of intensive care unit length of stay in heart transplantation patients. Front Cardiovasc Med 9: 863642, 2022. doi: 10.3389/fcvm.2022.863642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shahid N, Rappon T, Berta W. Applications of artificial neural networks in health care organizational decision-making: a scoping review. PLoS One 14: e0212356, 2019. doi: 10.1371/journal.pone.0212356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mangold C, Zoretic S, Thallapureddy K, Moreira A, Chorath K, Moreira A. Machine learning models for predicting neonatal mortality: a systematic review. Neonatology 118: 394–405, 2021. doi: 10.1159/000516891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pammi M, Aghaeepour N, Neu J. Multiomics, artificial intelligence, and precision medicine in perinatology. Pediatr Res. In press, 2022. doi: 10.1038/s41390-022-02181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pietrzyk JJ, Kwinta P, Wollen EJ, Bik-Multanowski M, Madetko-Talowska A, Günther C-C, Jagła M, Tomasik T, Saugstad OD. Gene expression profiling in preterm infants: new aspects of bronchopulmonary dysplasia development. PLoS One 8: e78585, 2013. doi: 10.1371/journal.pone.0078585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 350: g7594, 2015. doi: 10.1136/bmj.g7594. [DOI] [PubMed] [Google Scholar]
- 27. Ehrenkranz RA, Walsh MC, Vohr BR, Jobe AH, Wright LL, Fanaroff AA, Wrage LA, Poole K, National Institutes of Child Health and Human Development Neonatal Research Network. Validation of the National Institutes of Health consensus definition of bronchopulmonary dysplasia. Pediatrics 116: 1353–1360, 2005. doi: 10.1542/peds.2005-0249. [DOI] [PubMed] [Google Scholar]
- 28. Speiser JL, Miller ME, Tooze J, Ip E. A comparison of random forest variable selection methods for classification prediction modeling. Expert Syst Appl 134: 93–101, 2019. doi: 10.1016/j.eswa.2019.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kursa MB, Rudnicki WR. Feature selection with the Boruta package. J Stat Soft 36: 1–3, 2010. doi: 10.18637/jss.v036.i11. [DOI] [Google Scholar]
- 30. Ritchie ME, Diyagama D, Neilson J, van Laar R, Dobrovic A, Holloway A, Smyth GK. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics 7: 261, 2006. doi: 10.1186/1471-2105-7-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dey D, Slomka PJ, Leeson P, Comaniciu D, Shrestha S, Sengupta PP, Marwick TH. Artificial intelligence in cardiovascular imaging: JACC state-of-the-art review. J Am Coll Cardiol 73: 1317–1335, 2019. doi: 10.1016/j.jacc.2018.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Noble WS. What is a support vector machine? Nat Biotechnol 24: 1565–1567, 2006. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 33. Padmanabhan S, Tran TQB, Dominiczak AF. Artificial intelligence in hypertension: seeing through a glass darkly. Circ Res 128: 1100–1118, 2021. doi: 10.1161/CIRCRESAHA.121.318106. [DOI] [PubMed] [Google Scholar]
- 34. Filipow N, Main E, Sebire NJ, Booth J, Taylor AM, Davies G, Stanojevic S. Implementation of prognostic machine learning algorithms in paediatric chronic respiratory conditions: a scoping review. BMJ Open Respir Res 9: e001165, 2022. doi: 10.1136/bmjresp-2021-001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cilluffo G, Fasola S, Ferrante G, Licari A, Marseglia GR, Albarelli A, Marseglia GL, La Grutta S. Machine learning: a modern approach to pediatric asthma. Pediatr Allergy Immunol 33, Suppl 27: 34–37, 2022. doi: 10.1111/pai.13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song W, Zhang L, Liu L, Sainlaire M, Karvar M, Kang M-J, Pullman A, Lipsitz S, Massaro A, Patil N, Jasuja R, Dykes PC. Predicting hospitalization of COVID-19 positive patients using clinician-guided machine learning methods. J Am Med Inform Assoc 29: 1661–1667, 2022. doi: 10.1093/jamia/ocac083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Podda M, Bacciu D, Micheli A, Bellù R, Placidi G, Gagliardi L. A machine learning approach to estimating preterm infants survival: development of the Preterm Infants Survival Assessment (PISA) predictor. Sci Rep 8: 13743, 2018. doi: 10.1038/s41598-018-31920-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504, 2003. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Austin PC. Optimal caliper widths for propensity-score matching when estimating differences in means and differences in proportions in observational studies. Pharm Stat 10: 150–161, 2011. doi: 10.1002/pst.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ahn B, Chae YS, Lee SK, Kim M, Kim HS, Moon JW, Park S-H. Identification of novel DNA hypermethylation of the adenylate kinase 5 promoter in colorectal adenocarcinoma. Sci Rep 11: 12626, 2021. doi: 10.1038/s41598-021-92147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wright GL, Maroulakou IG, Eldridge J, Liby TL, Sridharan V, Tsichlis PN, Muise-Helmericks RC. VEGF stimulation of mitochondrial biogenesis: requirement of AKT3 kinase. FASEB J 22: 3264–3275, 2008. doi: 10.1096/fj.08-106468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang K, Dong W. SIRT1-association with bronchopulmonary dysplasia. Front Med (Lausanne) 8: 595634, 2021. doi: 10.3389/fmed.2021.595634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bermudez T, Sammani S, Song JH, Hernon VR, Kempf CL, Garcia AN, Burt J, Hufford M, Camp SM, Cress AE, Desai AA, Natarajan V, Jacobson JR, Dudek SM, Cancio LC, Alvarez J, Rafikov R, Li Y, Zhang DD, Casanova NG, Bime C, Garcia JGN. eNAMPT neutralization reduces preclinical ARDS severity via rectified NFkB and Akt/mTORC2 signaling. Sci Rep 12: 696, 2022. doi: 10.1038/s41598-021-04444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Deroo BJ, Cudmore CS. The extracellular matrix protein, spondin 1, regulates proliferation, survival, and adhesion of granulosa cells. Biol Reprod 85: 701, 2011. doi: 10.1093/biolreprod/85.s1.701. [DOI] [Google Scholar]
- 45. Goncharova IA, Bragina EY, Zhalsanova IZ, Freidin MB, Nazarenko MS. Putative regulatory functions of SNPs associated with bronchial asthma, arterial hypertension and their comorbid phenotype. Vavilovskii Zhurnal Genet Selektsii 25: 855–863, 2021. doi: 10.18699/VJ21.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gong Y, Hu N, Ma L, Li W, Cheng X, Zhang Y, Zhu Y, Yang Y, Peng X, Zou D, Tian J, Yang L, Mei S, Wang X, Lo CH, Chang J, Hou T, Zhang H, Xu B, Zhong R, Yuan P. ABTB2 regulatory variant as predictor of epirubicin-based neoadjuvant chemotherapy in luminal A breast cancer. Front Oncol 10: 571517, 2020. doi: 10.3389/fonc.2020.571517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pagel J, Twisselmann N, Rausch TK, Waschina S, Hartz A, Steinbeis M, Olbertz J, Nagel K, Steinmetz A, Faust K, Demmert M, Göpel W, Herting E, Rupp J, Härtel C. Increased regulatory t cells precede the development of bronchopulmonary dysplasia in preterm infants. Front Immunol 11: 565257, 2020. doi: 10.3389/fimmu.2020.565257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mishra A, Chintagari NR, Guo Y, Weng T, Su L, Liu L. Purinergic P2X7 receptor regulates lung surfactant secretion in a paracrine manner. J Cell Sci 124: 657–668, 2011. doi: 10.1242/jcs.066977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Riteau N, Gasse P, Fauconnier L, Gombault A, Couegnat M, Fick L, Kanellopoulos J, Quesniaux VFJ, Marchand-Adam S, Crestani B, Ryffel B, Couillin I. Extracellular ATP is a danger signal activating P2X7 receptor in lung inflammation and fibrosis. Am J Respir Crit Care Med 182: 774–783, 2010. doi: 10.1164/rccm.201003-0359OC. [DOI] [PubMed] [Google Scholar]
- 50. Kurten RC, Cadena DL, Gill GN. Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science 272: 1008–1010, 1996. doi: 10.1126/science.272.5264.1008. [DOI] [PubMed] [Google Scholar]
- 51. Ben-Omran T, Fahiminiya S, Sorfazlian N, Almuriekhi M, Nawaz Z, Nadaf J, Khadija KA, Zaineddin S, Kamel H, Majewski J, Tropepe V. Nonsense mutation in the WDR73 gene is associated with Galloway-Mowat syndrome. J Med Genet 52: 381–390, 2015. doi: 10.1136/jmedgenet-2014-102707. [DOI] [PubMed] [Google Scholar]
- 52. Alghamdi M, Bashiri FA, Abdelhakim M, Adly N, Jamjoom DZ, Sumaily KM, Alghanem B, Arold ST. Phenotypic and molecular spectrum of pyridoxamine-5'-phosphate oxidase deficiency: A scoping review of 87 cases of pyridoxamine-5'-phosphate oxidase deficiency. Clin Genet 99: 99–110, 2021. doi: 10.1111/cge.13843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bus C, Zizmare L, Feldkaemper M, Geisler S, Zarani M, Schaedler A, Klose F, Admard J, Mageean CJ, Arena G, Fallier-Becker P, Ugun-Klusek A, Maruszczak KK, Kapolou K, Schmid B, Rapaport D, Ueffing M, Casadei N, Krüger R, Gasser T, Vogt Weisenhorn DM, Kahle PJ, Trautwein C, Gloeckner CJ, Fitzgerald JC. Human dopaminergic neurons lacking PINK1 exhibit disrupted dopamine metabolism related to vitamin B6 co-factors. iScience 23: 101797, 2020. doi: 10.1016/j.isci.2020.101797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xuefei Y, Xinyi Z, Qing C, Dan Z, Ziyun L, Hejuan Z, Xindong X, Jianhua F. Effects of hyperoxia on mitochondrial homeostasis: are mitochondria the hub for bronchopulmonary dysplasia? Front Cell Dev Biol 9: 642717, 2021. doi: 10.3389/fcell.2021.642717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takeshita M, Koga T, Takayama K, Ijichi K, Yano T, Maehara Y, Nakanishi Y, Sueishi K. Aurora-B overexpression is correlated with aneuploidy and poor prognosis in non-small cell lung cancer. Lung Cancer 80: 85–90, 2013. doi: 10.1016/j.lungcan.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 56. Hachim MY, Elemam NM, Ramakrishnan RK, Salameh L, Olivenstein R, Hachim IY, Venkatachalam T, Mahboub B, Al Heialy S, Hamid Q, Hamoudi R. Derangement of cell cycle markers in peripheral blood mononuclear cells of asthmatic patients as a reliable biomarker for asthma control. Sci Rep 11: 11873, 2021. [Erratum in Sci Rep 11: 18203, 2021] doi: 10.1038/s41598-021-91087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jin CY, Du L, Nuerlan AH, Wang XL, Yang YW, Guo R. High expression of RRM2 as an independent predictive factor of poor prognosis in patients with lung adenocarcinoma. Aging (Albany NY) 13: 3518–3535, 2020. doi: 10.18632/aging.202292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shi GP, Chapman HA, Bhairi SM, DeLeeuw C, Reddy VY, Weiss SJ. Molecular cloning of human cathepsin O, a novel endoproteinase and homologue of rabbit OC2. FEBS Lett 357: 129–134, 1995. doi: 10.1016/0014-5793(94)01349-6. [DOI] [PubMed] [Google Scholar]
- 59. Yi H, Luo D, Xiao Y, Jiang D. Knockdown of long non-coding RNA DLEU2 suppresses idiopathic pulmonary fibrosis by regulating the microRNA-369-3p/TRIM2 axis. Int J Mol Med 47: 80, 2021.doi: 10.3892/ijmm.2021.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jiang J, Wang J, Yue M, Cai X, Wang T, Wu C, Su H, Wang Y, Han M, Zhang Y, Zhu X, Jiang P, Li P, Sun Y, Xiao W, Feng H, Qing G, Liu H. Direct phosphorylation and stabilization of MYC by aurora B kinase promote T-cell leukemogenesis. Cancer Cell 37: 200–215.e5, 2020. doi: 10.1016/j.ccell.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bhattacharya S, Mereness JA, Baran AM, Misra RS, Peterson DR, Ryan RM, Reynolds AM, Pryhuber GS, Mariani TJ. Lymphocyte-specific biomarkers associated with preterm birth and bronchopulmonary dysplasia. Front Immunol 11: 563473, 2020. doi: 10.3389/fimmu.2020.563473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bhandari A, Bhandari V. Biomarkers in bronchopulmonary dysplasia. Paediatr Respir Rev 14: 173–179, 2013. doi: 10.1016/j.prrv.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 63. D'Angio CT, Ambalavanan N, Carlo WA, McDonald SA, Skogstrand K, Hougaard DM, Shankaran S, Goldberg RN, Ehrenkranz RA, Tyson JE, Stoll BJ, Das A, Higgins RD, Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network. Blood cytokine profiles associated with distinct patterns of bronchopulmonary dysplasia among extremely low birth weight infants. J Pediatr 174: 45–51.e5, 2016. doi: 10.1016/j.jpeds.2016.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Salvo ML, Mastrangelo M, Nogués I, Tolve M, Paiardini A, Carducci C, Mei D, Montomoli M, Tramonti A, Guerrini R, Contestabile R, Leuzzi V. Pyridoxine-5’-phosphate oxidase (Pnpo) deficiency: Clinical and biochemical alterations associated with the C.347g>A (P.·Arg116gln) mutation. Mol Genet Metab 122: 135–142, 2017. [Erratum in Mol Genet Metab 2018] doi: 10.1016/j.ymgme.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 65. Mills PB, Camuzeaux SSM, Footitt EJ, Mills KA, Gissen P, Fisher L, Das KB, Varadkar SM, Zuberi S, McWilliam R, Stödberg T, Plecko B, Baumgartner MR, Maier O, Calvert S, Riney K, Wolf NI, Livingston JH, Bala P, Morel CF, Feillet F, Raimondi F, Del Giudice E, Chong WK, Pitt M, Clayton PT. Epilepsy due to PNPO mutations: genotype, environment and treatment affect presentation and outcome. Brain 137: 1350–1360, 2014. doi: 10.1093/brain/awu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Capasso L, Vento G, Loddo C, Tirone C, Iavarone F, Raimondi F, Dani C, Fanos V. Oxidative stress and bronchopulmonary dysplasia: evidences from microbiomics, metabolomics, and proteomics. Front Pediatr 7: 30, 2019. doi: 10.3389/fped.2019.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Toldi G, Hummler H, Pillay T. T Lymphocytes, multi-omic interactions and bronchopulmonary dysplasia. Front Pediatr 9: 694034, 2021. doi: 10.3389/fped.2021.694034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ballabh P, Simm M, Kumari J, Krauss AN, Jain A, Auld PAM, Cunningham-Rundles S. Lymphocyte subpopulations in bronchopulmonary dysplasia. Am J Perinatol 20: 465–475, 2003. doi: 10.1055/s-2003-45387. [DOI] [PubMed] [Google Scholar]
- 69. Shrestha D, Ye GX, Stabley D, Betal SGN, Zhu Y, Glazewski L, Holbrook J, Sethi M, Hesek A, Shaffer TH, Aghai ZH, Addya S, Alapati D. Pulmonary immune cell transcriptome changes in double-hit model of BPD induced by chorioamnionitis and postnatal hyperoxia. Pediatr Res 90: 565–575, 2021. doi: 10.1038/s41390-020-01319-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lao JC, Bui CB, Pang MA, Cho SX, Rudloff I, Elgass K, et al. Type 2 immune polarization is associated with cardiopulmonary disease in preterm infants. Sci Transl Med 14: eaaz8454, 2022. doi: 10.1126/scitranslmed.aaz8454. [DOI] [PubMed] [Google Scholar]
- 71. Smith AM, Ramirez RM, Harper N, Jimenez F, Branum AP, Meunier JA, Pandranki L, Carrillo A, Winter C, Winter L, Rather CG, Ramirez DA, Andrews CP, Restrepo MI, Maselli DJ, Pugh JA, Clark RA, Lee GC, Moreira AG, Manoharan MS, Okulicz JF, Jacobs RL, Ahuja SK. Large-scale provocation studies identify maladaptive responses to ubiquitous aeroallergens as a correlate of severe allergic rhinoconjunctivitis and asthma. Allergy 77: 1797–1814, 2022. doi: 10.1111/all.15124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Smith AM, Harper N, Meunier JA, Branum AP, Jimenez F, Pandranki L, Carrillo A, Dela Cruz CS, Restrepo MI, Maselli DJ, Rather CG, Heisser AH, Ramirez DA, He W, Clark RA, Andrews CP, Evans SE, Pugh JA, Zhang N, Lee GC, Moreira AG, Segal LN, Ramirez RM, Jacobs RL, Manoharan MS, Okulicz JF, Ahuja SK. Repetitive aeroallergen challenges elucidate maladaptive epithelial and inflammatory traits that underpin allergic airway diseases. J Allergy Clin Immunol 148: 533–549, 2021. doi: 10.1016/j.jaci.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Arck PC, Hecher K. Fetomaternal immune cross-talk and its consequences for maternal and offspring's health. Nat Med 19: 548–556, 2013. doi: 10.1038/nm.3160. [DOI] [PubMed] [Google Scholar]
- 74. Prescott SL, Macaubas C, Holt BJ, Smallacombe TB, Loh R, Sly PD, Holt PG. Transplacental priming of the human immune system to environmental allergens: universal skewing of initial T cell responses toward the Th2 cytokine profile. J Immunol 160: 4730–4737, 1998. [PubMed] [Google Scholar]
- 75. Martinez FD. Early-life origins of chronic obstructive pulmonary disease. N Engl J Med 375: 871–878, 2016. doi: 10.1056/NEJMra1603287. [DOI] [PubMed] [Google Scholar]
- 76. Kleer IM, Kool M, de Bruijn MJW, Willart M, van Moorleghem J, Schuijs MJ, Plantinga M, Beyaert R, Hams E, Fallon PG, Hammad H, Hendriks RW, Lambrecht BN. Perinatal activation of the interleukin-33 pathway promotes type 2 immunity in the developing lung. Immunity 45: 1285–1298, 2016. doi: 10.1016/j.immuni.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 77. Ubags NDJ, Alejandre Alcazar MA, Kallapur SG, Knapp S, Lanone S, Lloyd CM, Morty RE, Pattaroni C, Reynaert NL, Rottier RJ, Smits HH, de Steenhuijsen Piters WAA, Strickland DH, Collins JJP. Early origins of lung disease: towards an interdisciplinary approach. Eur Respir Rev 29: 200191, 2020. doi: 10.1183/16000617.0191-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pocha SM, Wassmer T, Niehage C, Hoflack B, Knust E. Retromer controls epithelial cell polarity by trafficking the apical determinant Crumbs. Curr Biol 21: 1111–1117, 2011. doi: 10.1016/j.cub.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 79. Yang J, Asico LD, Beitelshees AL, Feranil JB, Wang X, Jones JE, Armando I, Cuevas SG, Schwartz GL, Gums JG, Chapman AB, Turner ST, Boerwinkle E, Cooper-DeHoff RM, Johnson JA, Felder RA, Weinman EJ, Zeng C, Jose PA, Villar VAM. Sorting nexin 1 loss results in increased oxidative stress and hypertension. FASEB J 34: 7941–7957, 2020. doi: 10.1096/fj.201902448R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Moreira AG, Siddiqui SK, Macias R, Johnson-Pais TL, Wilson D, Gelfond JAL, Vasquez MM, Seidner SR, Mustafa SB. Oxygen and mechanical ventilation impede the functional properties of resident lung mesenchymal stromal cells. PLoS One 15: e0229521, 2020. doi: 10.1371/journal.pone.0229521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Benny M, Courchia B, Shrager S, Sharma M, Chen P, Duara J, Valasaki K, Bellio MA, Damianos A, Huang J, Zambrano R, Schmidt A, Wu S, Velazquez OC, Hare JM, Khan A, Young KC. Comparative effects of bone marrow-derived versus umbilical cord tissue mesenchymal stem cells in an experimental model of bronchopulmonary dysplasia. Stem Cells Transl Med 11: 189–199, 2022. doi: 10.1093/stcltm/szab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Miettinen PJ, Warburton D, Bu D, Zhao JS, Berger JE, Minoo P, Koivisto T, Allen L, Dobbs L, Werb Z, Derynck R. Impaired lung branching morphogenesis in the absence of functional EGF receptor. Dev Biol 186: 224–236, 1997. doi: 10.1006/dbio.1997.8593. [DOI] [PubMed] [Google Scholar]
- 83. Mishra A. New insights of P2X7 receptor signaling pathway in alveolar functions. J Biomed Sci 20: 26, 2013. doi: 10.1186/1423-0127-20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Di Virgilio F, Dal Ben D, Sarti AC, Giuliani AL, Falzoni S. The P2X7 Receptor in Infection and Inflammation. Immunity 47: 15–31, 2017. doi: 10.1016/j.immuni.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 85. Savio LEB, de Andrade Mello P, da Silva CG, Coutinho-Silva R. The P2X7 receptor in inflammatory diseases: angel or demon? Front Pharmacol 9: 52, 2018. doi: 10.3389/fphar.2018.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Onland W, Debray TP, Laughon MM, Miedema M, Cools F, Askie LM, Asselin JM, Calvert SA, Courtney SE, Dani C, Durand DJ, Marlow N, Peacock JL, Pillow JJ, Soll RF, Thome UH, Truffert P, Schreiber MD, Van Reempts P, Vendettuoli V, Vento G, van Kaam AH, Moons KG, Offringa M. Clinical prediction models for bronchopulmonary dysplasia: a systematic review and external validation study. BMC Pediatr 13: 207, 2013. doi: 10.1186/1471-2431-13-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material: https://doi.org/10.6084/m9.figshare.21200056.
Data Availability Statement
Data will be made available upon reasonable request.