Abstract
The protective efficacy of antibodies (Abs) to Cryptococcus neoformans glucuronoxylomannan (GXM) is dependent on Ab fine specificity. Two clonally related immunoglobulin M monoclonal Abs (MAbs) (12A1 and 13F1) differ in fine specificity and protective efficacy, presumably due to variable (V)-region sequence differences resulting from somatic mutations. MAb 12A1 is protective and produces annular immunofluorescence (IF) on serotype D C. neoformans, while MAb 13F1 is not protective and produces punctate IF. To determine the Ab molecular determinants responsible for the IF pattern, site-directed mutagenesis of the MAb 12A1 heavy-chain V region (VH) was followed by serological and functional studies of the various mutants. Changing two selected amino acids in the 12A1 VH binding cavity to the corresponding residues in the 13F1 VH altered the IF pattern from annular to punctate, reduced opsonic efficacy, and abolished recognition by an anti-idiotypic Ab. Analysis of the binding of the various mutants to peptide mimetics revealed that different amino acids were responsible for GXM binding and peptide specificity. The results suggest that V-region motifs associated with annular binding and opsonic activity may be predictive of Ab efficacy against C. neoformans. This has important implications for immunotherapy and vaccine design that are reinforced by the finding that GXM and peptide reactivities are determined by different amino acid residues.
The protective efficacy of antibodies (Abs) to the human-pathogenic fungus Cryptococcus neoformans depends on the Ab isotype and specificity (reviewed in references 3 and 46). The evidence that Ab specificity is critical for protective efficacy comes from studies of two clonally related immunoglobulin M (IgM) monoclonal Abs (MAbs) known as 12A1 and 13F1 (3, 31, 39, 46). Although these MAbs originated from the same B-cell precursor and use the same variable (V)-region genes, they differ in specificity as a result of V-region somatic mutations that translate into 12 amino acid differences (31, 39). The differences in specificity are manifested by differences in the indirect immunofluorescence (IF) binding pattern such that MAbs 12A1 and 13F1 produce annular and punctate patterns, respectively, after binding to serotype D C. neoformans cells (11, 31, 39). The annular binding pattern is correlated with opsonic efficacy, capsular reaction patterns, and complement activation kinetics (27) and Ab protection against serotype D organisms (31, 39). Since the MAb pair 12A1 and 13F1 have markedly different biological properties yet differ in sequence by only a few amino acids, they provide a unique opportunity for the study of Ab specificity.
MAbs to C. neoformans capsular glucuronoxylomannan (GXM) have been grouped into five classes based on V-region usage and idiotype and serotype specificity (5). Class II MAbs include a large set of MAbs that bind to an immunodominant epitope found in all cryptococcal serotypes and are characterized by the use of VH7183, JH2, Vκ5.1, and Jκ1 gene elements and a heavy-chain V (VH) third complementarity-determining region (CDR3) of 11 amino acids (5). MAbs 12A1 and 13F1 are class II MAbs (5). Peptide mimetics which bind to the antigen (Ag) binding sites of class II MAbs have been described (43, 44), and the crystal structures of the class II MAb 2H1 with and without a complexed peptide mimetic have been solved (47). Murine class II MAbs and human Abs to C. neoformans GXM share sequence similarities (40). The class II MAb 18B7 is in clinical evaluation for the treatment of cryptococcal meningitis (4).
IgM is an important isotype against fungi in light of evidence that some IgMs are protective against C. neoformans (17, 32) and Candida albicans (20), and IgM is common in both the human and mouse responses to GXM (6, 16, 22). IgM may have an advantage over IgG in therapy because it is very effective at clearing Ag but does not elicit lethal toxicity reactions when administered to C. neoformans-infected mice (26). Identification of the amino acid residues that confer Ab specificity for epitopes associated with protection is important for defining the Ab paratope, or the site that is involved in binding the polysaccharide Ag (19), and the latter is important for immunotherapy and vaccine design. In this study, we used site-directed mutagenesis to identify the amino acids responsible for the fine-specificity difference between MAbs 12A1 and 13F1 and compared the serological and opsonic properties of the mutated Abs. The fact that punctate and annular IF patterns reflect differential localization of Ag-Ab complexes on the C. neoformans capsule (15) indicates that the binding characteristics of IgM may require valence or other structural constraints. Therefore, we changed the 12A1 VH to the corresponding residue in the 13F1 VH and expressed the mutated V regions. The results indicate that annular binding is conferred by two VH amino acid residues that impart major differences in biological function by coding for two different epitope specificities.
MATERIALS AND METHODS
Hybridomas and MAbs.
Hybridomas 12A1 and 13F1 both produce IgM MAbs (6). Cells were maintained in Dulbecco modified Eagle (DME) medium containing 10% fetal calf serum (Harlan, Indianapolis, Ind.), 10% NCTC-109 (Mediatech, Herndon, Va.), and 1% nonessential amino acid solution (Mediatech). MAb 3E5 is an IgG3 which competes with MAb 12A1 but not 13F1 (31).
Heavy-chain-nonproducing hybridoma mutants.
The 12A1 heavy-chain-nonproducing hybridoma cells were isolated by soft agar cloning followed by overlaying the agar with rabbit antiserum to murine IgM. In this method, colonies that secrete IgM are stained by Ag-Ab precipitates. Colonies that were not stained were selected and transferred to 96-well plates containing cell medium, and their supernatants were tested for IgM and light-chain κ secretion by enzyme-linked immunosorbent assay (ELISA) (see below). Hybridoma cells that tested negative for IgM and positive for light-chain κ were used in the transfection experiments.
C. neoformans and other yeasts.
Serotype D strain 24067 was obtained from the American Type Culture Collection (Manassas, Va.). MAbs 12A1 and 13F1 produce annular and punctate IF patterns, respectively, upon binding to the 24067 capsule. C. neoformans cells were maintained in glycerol stocks at −80°C and grown in Sabouraud dextrose broth (Difco Laboratories, Detroit, Mich.) for 24 h at 30°C with constant shaking at 150 rpm. Before use, cells were washed three times with sterile phosphate-buffered saline (PBS) and counted using a hemacytometer. Capsular GXM was prepared from supernatants of strain 24067 as described previously (9). Saccharomyces cerevisiae strain 1H170 his3 ade2 and C. albicans strain SC5314 were gifts from Lorraine Marsh (Bronx, N.Y.) and Mahmoud Ghannoum (Cleveland, Ohio), respectively.
VH and VL sequences.
Total RNA was isolated from hybridoma cells using Trizol reagent (Gibco BRL, Gaithersburg, Md.). cDNA was generated using reverse transcriptase and the oligonucleotide primer p(dt)15 (Boehringer Mannheim, Indianapolis, Ind.). DNA containing the VH region was amplified using the primers 5′-TAAAAAGCTTAGTC CACTCGCCATGGACTTC-3′ and 5′-TATATTGCTAGCTGAGGAGACTGTGAGAGTGG-3′. DNA containing the light-chain V region (VL) was amplified using the primers GATGTTGTGATGACCCAA and TGGATGGTGGGAAGATG. The amplified DNA was cloned into the PCR 2.1 vector (Invitrogen, San Diego, Calif.) for sequencing. Oligonucleotide synthesis and DNA sequencing were done by the Oligonucleotide Facility of the Cancer Center at Albert Einstein College of Medicine.
Expression of 12A1 VH.
DNA containing MAb 12A1 VH was excised from the PCR 2.1 vector using NheI and HindIII (Promega, Madison, Wis.) and cloned into a murine cDNA IgM expression vector (28). The plasmid was then transfected into heavy-chain-nonproducing 12A1 hybridoma cells by electroporation using conditions of 200 Ω, 960 μF, and 450 V. The same recipient cell line deficient in heavy-chain production was used to generate all mutant Abs. Approximately 3 × 106 cells were washed with cold PBS, mixed with 10 to 15 μg of plasmid in 1.0 ml of PBS, placed in a Gene Pulser cuvette (Bio-Rad Laboratories, Hercules, Calif.), and incubated on ice for 10 min before transfection. Following electroporation, cells were incubated on ice for 10 min and then washed with DME medium. Cells were then plated at a density of 104 cells/well in a 96-well plate (Becton Dickinson Labware, Franklin Lakes, N.J.) for 24 h in feeding medium. Cells expressing the transfected plasmid were selected with 1.5 mg of neomycin (Geneticin; Gibco BRL)/ml in DME medium with 20% fetal calf serum (Harlan), 10% NCTC-109 (Mediatech), and 1% nonessential amino acid solution (Mediatech). To identify high-producing clones, the cells were cloned in soft agar with an overlay of rabbit Ab to mouse IgM. Colonies with strong Ag-Ab staining were selected. Transfected cells produced 10 to 150 ng of IgM/ml in cell culture supernatants after 24 h of incubation.
Site-directed mutagenesis.
Amino acid changes were introduced into the MAb 12A1 VH by oligonucleotide-directed PCR mutagenesis using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The oligonucleotides (5′→3′) for mutagenesis were as follows: N31S, GCCTCTGGATTTACTTCCAGTAGCTATTTCATGTCTTGGG and CCCAAGACATGAAATAGCTACTGAAAGTGAATCCACAGGC; F33Y, CACTTTCAGTAACTATTACATGTCTTGGG and CCCAAGACATGTAATAGTTACTGAAAGTG; M50A, GAG GCTGGAATTGGTCGGAGCCATTAATATTAATGGTGATAACACC and GGTGTTATCACCATTAATATTAATGGCTGCGACCAATTCCAGCCTC; I53S, GGCTGGAATTGGTCGCAATGATTAATAGTAATGGTGATAACACCTAC and TATCCGGATAGTAGGTGTTATCACCATTACTATTAATCATTGCGACCAATTCCAGCC; N56G, GGTCGCAATGATTAATATTAATGGTGGTAACACCTACTATCCA and GTCTGGATAGTAGGTGTTACCACCATTAATATTAATCATTGCGACC; N57S, ATTAATGGTGATAGCACC TACTATCCAGAC and GTCTGGATAGTAGGTGCTATCACCATTAAT; D80Y, GCCAAGAACACCCTGTACCTGCAAATGAGCAGTCTG and CACACTGCTCATTTGCAGGTACAGGGTGTTCTTGGC; and G103Y, GCAAGACGAGACGGCACCTACGGAAACTACTTTGACTAC and GTAGTCAA AGTAGTTTCCGTAGGTGCCGTCTCGTCTTGC. The oligonucleotide primers, each complementary to opposite strands of the vector, extend during temperature cycling by means of Pfu DNA polymerase. On incorporation of the oligonucleotide primers, a mutated plasmid containing staggered nicks is generated. Following temperature cycling, the PCR product was treated with DpnI, an endonuclease specific for methylated and hemimethylated DNAs that digests the parental DNA template and selects for molecules containing the mutation. The DNA was then used to transform Escherichia coli supercompetent cells and make purified DNA for electroporation experiments. Despite multiple attempts, we were not able to generate a 12A1 variant expressing the G103Y mutation. All mutations were confirmed by sequence analysis.
ELISAs.
Ab concentrations were determined by ELISA. Polystyrene plates were coated with goat anti-mouse IgM, blocked with 1% bovine serum albumin (BSA) in PBS, and incubated with the IgM-containing solution. Bound IgM was detected using alkaline phosphatase-conjugated goat anti-mouse IgM, and the IgM concentration was determined relative to isotype-matched IgM standards (Southern Biotechnology, Birmingham, Ala.). IgM binding to GXM was studied by ELISA as described previously (7). Briefly, polystyrene plates were coated with a GXM solution (1 μg/ml) and blocked with 1% BSA in PBS. IgM-containing solutions were then added, and bound IgM was detected using alkaline phosphatase-conjugated goat anti-mouse IgM. Competition ELISAs were done to determine whether MAb 3E5 (IgG3) could inhibit the binding of the mutagenized MAb 12A1 derivatives to GXM as described previously (31). For this ELISA, variable amounts of the Ab in question were mixed with a constant amount of MAb 3E5 (2 μg/ml) and allowed to react with GXM absorbed on a polystyrene plate. Binding of IgG3 was detected by isotype-specific alkaline phosphatase-conjugated goat anti-mouse reagents. Reactivity with the anti-idiotypic MAb 7B8 was also measured by ELISA. Briefly, plates were coated with 1 μg of goat anti-mouse IgM/ml, blocked with 1% BSA in PBS, incubated with the IgM MAb, and then incubated with MAb 7B8 (IgG1), and binding was detected with goat anti-mouse IgG1 followed by addition of p-nitrophenyl phosphate substrate. MAb 7B8 binds to the Ag-combining sites of some class II MAbs, such as 12A1 (38).
Peptide ELISA.
Microtiter plates were coated with 1 μg of streptavidin/ml and then incubated with 1 μg of biotinylated peptide mimetics/ml and then with parental and variant MAbs. The peptides were PA1 (LQYTPSWMLV), P601.E (SYSWMYE), 206.1 (FGGETFTPDWMMEVAIDNE), and PM14 (CGLQWLWEWPRT), which are described in reference 1. The binding of the MAbs was detected with alkaline phosphatase-conjugated goat anti-mouse IgM, and the plates were developed with p-nitrophenyl phosphate (Sigma, St. Louis, Mo.). All incubations were carried out at 37°C for 1 h.
IF.
IgM MAbs were added to a suspension of 106 C. neoformans cells at a concentration of 10 μg/ml in blocking solution (1% BSA, 0.5% horse serum) and incubated at 37°C for 30 min. The cells were then washed twice with blocking solution, incubated with 10 μg of fluorescein isothiocyanate-labeled goat anti-mouse IgM (Southern Biotechnology)/ml for 30 min at 37°C in the dark, washed again with blocking solution, and finally suspended in mounting medium (0.1 M n-propyl gallate in PBS) (Sigma). The slides were then viewed with an AX70 microscope (Olympus, Melville, N.Y.) equipped with a standard fluorescein isothiocyanate filter.
Phagocytosis assay.
The opsonic efficacies of MAbs 12A1 and 13F1 and the 12A1 mutants were evaluated in J774 macrophage-like cells as described previously (34). A macrophage cell line was used because macrophages are the major phagocytic cells for C. neoformans in vivo (14). Briefly, gamma interferon- and lipopolysaccharide-stimulated J774 cells were incubated with C. neoformans in the presence and absence of 10 μg of Ab. After incubation for 2 h, the monolayer was washed to remove unbound yeast and the J774 cells were fixed with methanol and stained with Giemsa stain (Sigma). Phagocytosis was determined by counting the numbers of attached and ingested yeast cells using a microscope. Ingested and attached C. neoformans cells can be readily distinguished by light microscopy. The phagocytic index was defined as the number of ingested C. neoformans cells divided by the number of macrophages.
Measurement of zeta potential.
The zeta potential (ζ) is a measurement of cellular charge (in millivolts) that is defined as the potential gradient that develops across the interface between a boundary liquid in contact with a solid and the mobile diffuse layer in the body of the liquid (41). It is derived from the equation ζ = 4πηm/D, where D is the dielectric constant of the medium, η is the viscosity, and m is the electrophoretic mobility of the particle (41). Cells grown in Sabouraud broth (SAB) were collected and washed three times in 0.01 M NaCl (pH 7.0). The zeta potentials of suspensions of 106 cells/ml were measured using a Zeta-Meter (Staunton, Va.) 3.0+ instrument, which determines the zeta potentials of individual suspended cells. The zeta potentials of 10 randomly selected C. neoformans cells were measured for each MAb tested. Similar methods have previously been used to measure cellular charge for C. neoformans in the presence and absence of Ab (36).
Scanning electron microscopy.
C. neoformans cells were grown in SAB, the cells were collected and washed three times with PBS, and aliquots of 106 cells/ml were incubated with 10 μg of each MAb/ml for 1 h at room temperature. The cells were washed three times in PBS and then incubated in 2.5% glutaraldehyde for 1 h at room temperature. Samples were then applied to a polylysine-coated coverslip and serially dehydrated in alcohol. The samples were fixed in a critical-point drier (Samdri-790; Tousimis, Rockville, Md.), coated with gold-palladium (Desk-1; Denton Vacuum, Inc., Cherry Hill, N.J.), and viewed using a JSM-6400 (JEOL USA, Peabody, Mass.) scanning electron microscope. At least 20 cells were imaged for each MAb tested, and representative photographs were taken. This methodology has previously been used to study the effects of MAbs to C. neoformans (12).
Locations of mutations in protein structure.
The X-ray structure of the Ab 2H1 in complex with the peptide PA1 is deposited in the Protein Data Bank under accession number 2H1P (47). Mutated amino acid positions in the 12A1 VH were analyzed on the 2H1 structure using the program RASTOP (42), which is available as free software at http://www.bernstein-plus-sons.com/software/rastop.
Nucleotide sequence accession numbers.
The 12A1 and 13F1 VH sequences are deposited in GenBank under accession numbers AF052834 and AF052835, respectively. The VL sequences of the two MAbs are deposited in GenBank under accession numbers AF071060 and AF071061, respectively.
RESULTS
Sequences of 12A1 and 13F1.
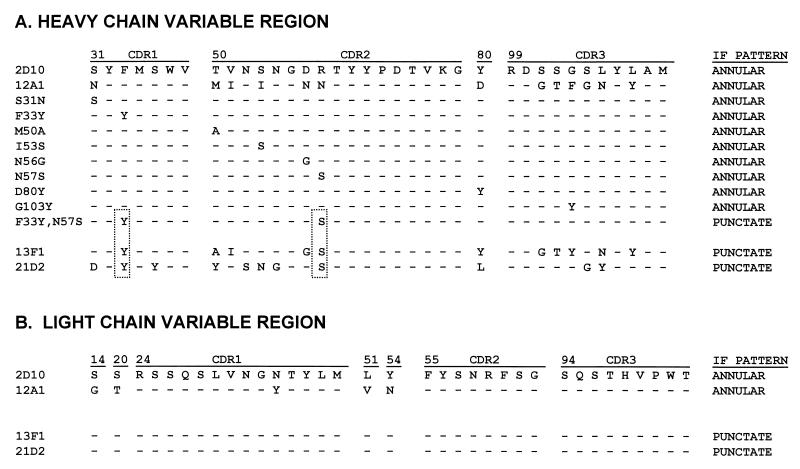
Prior studies had reported partial VH and VL sequences of MAbs 12A1 and 13F1 obtained by direct sequencing of mRNA (29). For both MAbs the VH and VL sequences were completed, and we also determined the complete constant (C)-region sequence for each IgM. The rationale for determining the C-region sequence was to exclude the unlikely possibility that the serological differences in the binding of these MAbs to GXM were the result of Fc-region differences. For both MAbs, the IgM C regions were identical and corresponded to the IgM(b) sequence (24). The 12A1 and 13F1 VH sequences differ at eight amino acid positions (Fig. 1). The VH and VL amino acid sequences for MAbs 12A1 and 13F1 and other MAbs used in this study are shown in Fig. 1.
FIG. 1.
VH and VL amino acid sequences for MAbs 2D10, 12A1, and 13F1 and the 12A1 VH mutants. Boxes indicate the amino acid residues shared by the three MAbs which produce a punctate binding pattern.
Generation and characterization of 12A1→13F1 mutants by site-directed mutagenesis.
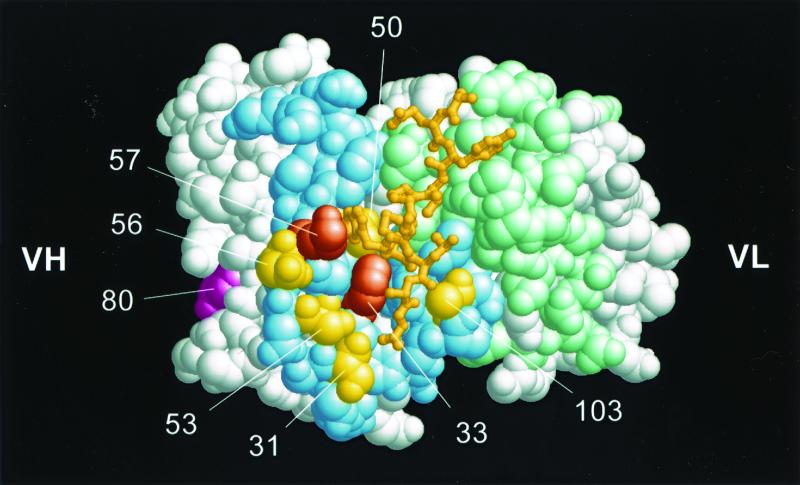
Expression of the 12A1 VH in hybridoma 12A1 heavy-chain-loss mutant cells expressing the 12A1 VL (12A1VH-12A1VL) yielded an Ab that produced, as expected, an annular pattern on C. neoformans cells, like that of the parent MAb 12A1. Expression of the 12A1 VH in hybridoma 13F1 heavy-chain-loss mutant cells expressing the 13F1 VL (12A1VH-13F1VL) also resulted in an Ab that produced an annular IF pattern, strongly suggesting that the differences in IF patterns were attributable to differences in VH sequences. To investigate the contribution of each of the eight amino acid residues that constitute the difference between the VHs of MAbs 12A1 and 13F1, we used site-directed mutagenesis to change the 12A1 VH residues that were different into the corresponding 13F1VH residue (12A1→13F1 mutants). The changes made to the 12A1 VH are shown in Fig. 1. Figure 2 shows the locations of the induced mutations on a three-dimensional representation of the MAb 12A1 binding site. All 12A1→13F1 mutants bound to GXM as determined by ELISA (Table 1).
FIG. 2.
Three-dimensional representation of the Ag binding site of MAb 12A1 with van der Waals surface. The model is based on the 2H1 structure in complex with the peptide PA1 (47). CDR surfaces are shown colored pale blue (VH) and light green (VL). The amino acid positions of MAb 12A1 which were mutagenized are shown in yellow, except for position 80 in magenta, located in the VH framework, and positions 33 and 57 corresponding to the double mutation. Amino acids at these positions for MAbs 2H1, 12F1, and 13F1, respectively, are as follows: 31, S, N, and S; 33, F, F, and Y; 50, M, M, and A; 53, S, I, and N; 56, D, N, G; 57, K, N, and S; 80, L, D, and Y; and 103, A, F, and Y.
TABLE 1.
Serological characteristics of MAbs 12A1 and 13F1 and the 12A1→13F1 mutants
| Antibody | Reactivity by direct ELISAa | Inhibition with MAb 3E5b | Reactivity with anti-idiotypic MAb 7B8c | Phagocytic indexd | Immunofluorescence |
|---|---|---|---|---|---|
| 12A1 | +++ | Yes | + | +++ | Annular |
| 13F1 | + | No | − | − | Punctate |
| 12A1VH-12A1VL | +++ | Yes | + | +++ | Annular |
| 12A1VH-13F1VL | +++ | Yes | − | +++ | Annular |
| N31S | +++ | Yes | + | + | Annular |
| F33Y | ++ | Yes | + | + | Annular |
| M50A | +++ | Yes | + | ++ | Annular |
| I53S | +++ | Yes | + | ++ | Annular |
| N56G | +++ | Yes | + | ++ | Annular |
| N57S | ++ | Yes | + | + | Annular |
| D80Y | +++ | Yes | + | ++ | Annular |
| F33Y N57S | + | No | − | − | Punctate |
Plus signs indicate the relative reactivity of the Ab for GXM as measured by direct ELISA, ranging from weak (+) to strong (+++).
For representative data, see Fig. 5.
+, reactivity; −, lack of reactivity. Data are in Fig. 7.
Plus signs indicate the relative opsonic efficacies of these MAbs; minus signs indicate absence of or minimal opsonic efficacy. For details of phagocytic indices observed with MAbs 12A1 and 13F1, see reference 11.
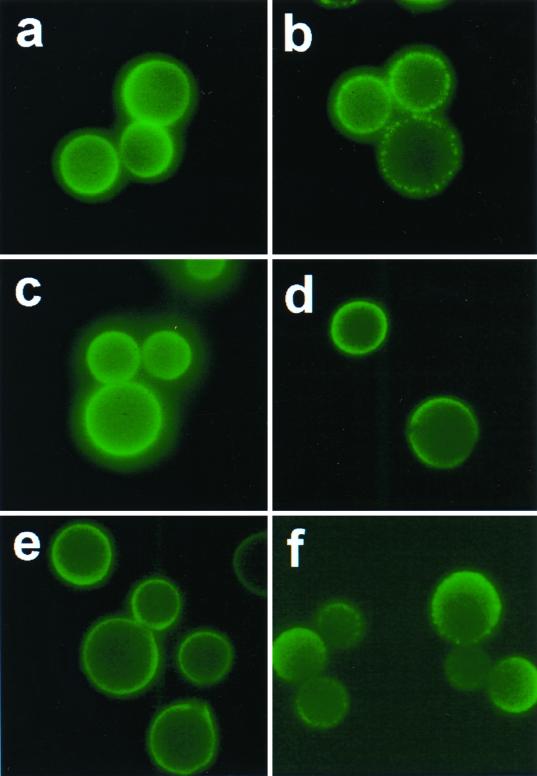
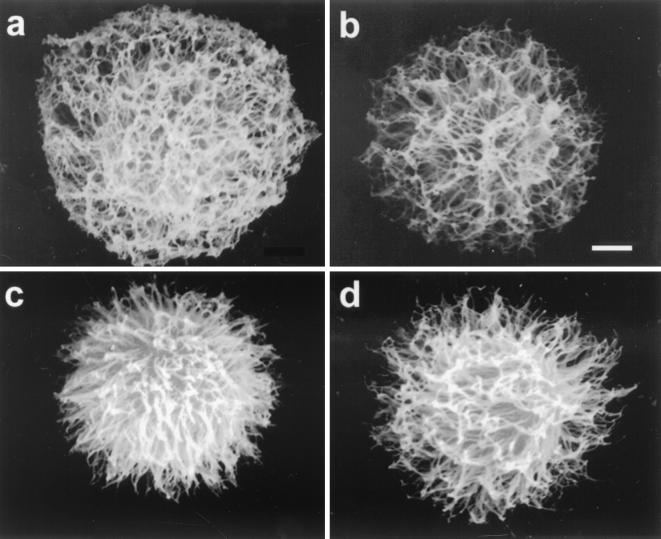
All MAb 12A1 VH variants with single mutations produced an annular IF pattern upon binding strain 24067 (some are shown in Fig. 3). However, there were some differences in the location and intensity of each variant such that none were identical to the IF pattern produced by the parent MAb 12A1 (data not shown). Scanning electron microscopy revealed differences in capsule binding for some of the variants (Fig. 4). Hence, mutational analysis of the 12A1 VH revealed that although single amino acid changes were not sufficient to alter the IF pattern from annular to punctate, some of the changes produced subtle differences in the binding to the capsule as measured by IF and scanning electron microscopy. Maintenance of the IF pattern in variants with one amino acid change suggested that more than one change would be needed to alter the IF pattern. Faced with the fact that a complete analysis of this system would require an overwhelming effort in the form of constructing the many combinations possible given the amino acid differences between 12A1 and 13F1, we opted for a targeted evaluation.
FIG. 3.
IF patterns produced by parental MAbs 12A1 (a) and 13F1 (b) and variants 12A1VH-12A1VL (c), M50A (d), D80Y (e), and the F33Y N57Y double mutant (f). Magnification, ×250.
FIG. 4.
Scanning electron micrographs of yeast cells coated with MAbs 12A1VH-12A1VL (a), 13F1 (b), F33Y (c), and the F33Y N57Y double mutant (d). Bar, 10 μm. For each Ab at least 15 to 20 cells were imaged, and representative yeast cells are shown.
Another IgM (21D2) had been shown to also produce a punctate IF on strain 24067 and not to be protective (39). MAb 21D2 is a class II MAb that was generated from a mouse with C. neoformans infection and recognizes a different epitope than MAb 12A1 on the basis that it can bind to de-O-acetylated GXM. Comparison of the MAb 13F1 and 21D2 sequences revealed that they shared two amino acids at positions 33 and 57 that were not found in the protective IgMs that produced annular IF (Fig. 1). On the basis of this similarity, we constructed a 12A1 VH with the amino acid changes F33Y and N57S, and this VH produced a punctate pattern when assembled with the 12A1 VL (Fig. 3). Thus, introduction of two amino acid changes corresponding to those in MAb 13F1 converted the MAb 12A1 IF from annular to punctate.
As a second measure for specificity we carried out competition assays with MAb 3E5, which competes with MAb 12A1 but not 13F1 for GXM binding by ELISA (31). Like the parent MAb 12A1, all 12A1→13F1 mutants displaying single amino acid changes were inhibited by MAb 3E5 (Table 1). However the mutant having two amino acid changes at F33Y and N57S was not inhibited by MAb 3E5 (Fig. 5). MAb 21D2 was also not inhibited by MAb 3E5 (data not shown). Hence, introduction of two amino acids changed the specificity of MAb 12A1 to one that is indistinguishable from that of MAb 13F1 by this assay.
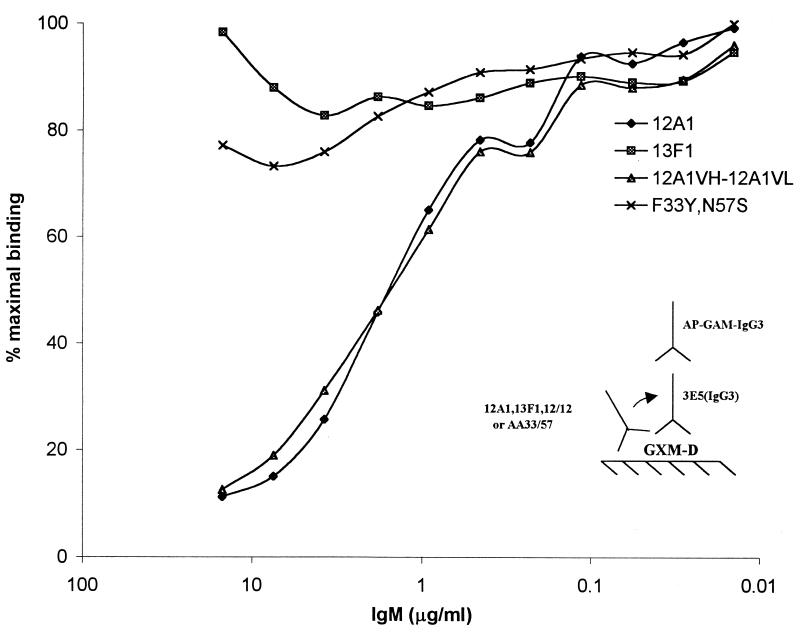
FIG. 5.
Competition ELISA between Abs 12A1, 13F1, 12A1VH-12A1VL, and the F33Y N57Y double mutant with the IgG3 MAb 3E5. At increasing concentrations of MAbs 12A1 and 12A1VH-12A1VL the binding of MAb 3E5 is inhibited. In contrast, no significant inhibition of MAb 3E5 binding is produced by either MAb 13F1 or the F33Y N57Y double mutant. AP-GAM-IgG3, alkaline phosphatase-conjugated anti-mouse IgG3.
Reactivities of MAbs 12A1 and 13F1 and 12A1→13F1 mutants with peptide mimotopes of MAb 2H1.
The crystal structures of MAb 2H1 with and without a peptide mimotope in the binding site are available (47). Since MAbs 12A1 and 13F1 use V-region genes of the same family, share the 2H1 idiotype, and have very similar serological characteristics (5), we analyzed their reactivities with peptide mimotopes of GXM recognized by MAb 2H1 (Table 2). Neither MAb 12A1 nor MAb 13F1 showed significant reactivity with peptide PA1, 601, or 206.1, which are mimotopes of GXM that bind strongly to MAb 2H1 and the other class II Abs, such as MAb 2D10. Interestingly, the 12A1VH-13F1VL hybrid reacted with peptide 206.1 despite no reactivity by their parent MAbs. The 12A1→13F1 mutants I53S, N56G, and N57S each reacted strongly with peptide 206.1. Hence, the introduction of single mutations into the 5′ region of the 12A1 VH CDR altered the specific ability of MAb 12A1 to recognize a GXM peptide mimetic even when no change in IF pattern after binding C. neoformans was apparent.
TABLE 2.
Reactivities of several MAbs and 12A1→13F1 variants with peptide mimotopes of GXM
| Antibody | Reactivitya with peptide:
|
|||
|---|---|---|---|---|
| PA1 | 601 | 206.1 | PM14 | |
| 2D10 | + | +++ | +++ | + |
| 2H1 | +++ | ++ | +++ | + |
| 21D2 | − | − | +++ | − |
| 12A1 | − | − | + | − |
| 13F1 | − | − | − | +++ |
| 12A1VH-13F1VL | − | − | + | − |
| 12A1VH-13F1VL | − | − | + | − |
| N31S | − | − | − | − |
| F33Y | − | − | − | ++ |
| M50A | − | − | − | − |
| I53S | − | − | ++ | − |
| N56G | − | − | +++ | − |
| N57S | − | − | − | − |
| D80Y | − | − | +++ | − |
| F103Y | − | − | − | − |
| F33Y N57S | − | − | − | − |
Plus and minus signs indicate the relative reactivities of the various Abs for the peptide as measured by ELISA, ranging from none (−) to weak (+) to strong (+++).
Phagocytosis assays.
MAb 12A1 is significantly more opsonic than MAb 13F1 for C. neoformans cells (11). The 12A1→13F1 mutants each were less opsonic than the parent MAb 12A1, and mutations F33Y and N57S significantly reduced the opsonic potential of the Ab (Table 2). The double mutation at F33Y and N57S resulted in complete loss of opsonic activity to a level comparable to that observed for MAb 13F1 (Table 2). Hence, the F33Y N57S double mutant is like MAb 13F1 with respect to opsonization efficacy.
Zeta potential.
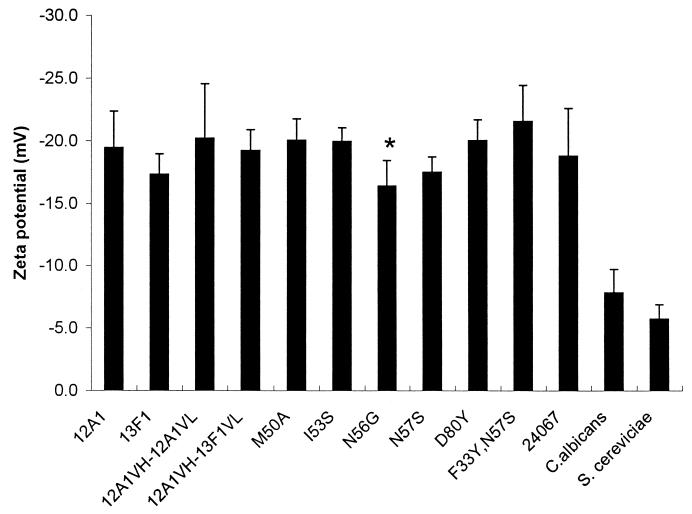
C. neoformans cells have a very high negative charge relative to other yeast cells, which has been attributed to the polysaccharide capsule (36, 37). Ab binding to the C. neoformans capsule can result in changes to the yeast cell charge (36). This phenomenon is not well understood, but our mutant set provided the opportunity to investigate whether amino acid changes could alter the zeta potential of C. neoformans cells independent of their effects on Ab charge. Measurements of zeta potential of C. neoformans cells after binding MAb 12A1 or 13F1 or the 12A1→13F1 mutants revealed relatively small changes to the yeast cell charge after Ab binding, even for the D80Y mutant, which involved replacement of a negatively charged aspartic acid with a neutral tyrosine (Fig. 6). The largest change was observed for N56G, which conferred a significantly greater negative charge to C. neoformans than MAb 12A1 despite involving a replacement of a neutral asparagine with a neutral glycine (Fig. 7).
FIG. 6.
Zeta potentials of C. neoformans cells coated with MAbs 12A1 and 13F1 and the various 12A1→13F1 mutants. ∗, the zeta potential of the N56G variant was significantly lower (P = 0.017) than that of the parent MAb 12A1. Error bars indicate standard deviations.
FIG. 7.
Reactivities of MAbs 12A1 and 13F1 and various 12A1→13F1 mutants with the anti-idiotypic MAb 7B8 as determined by ELISA.
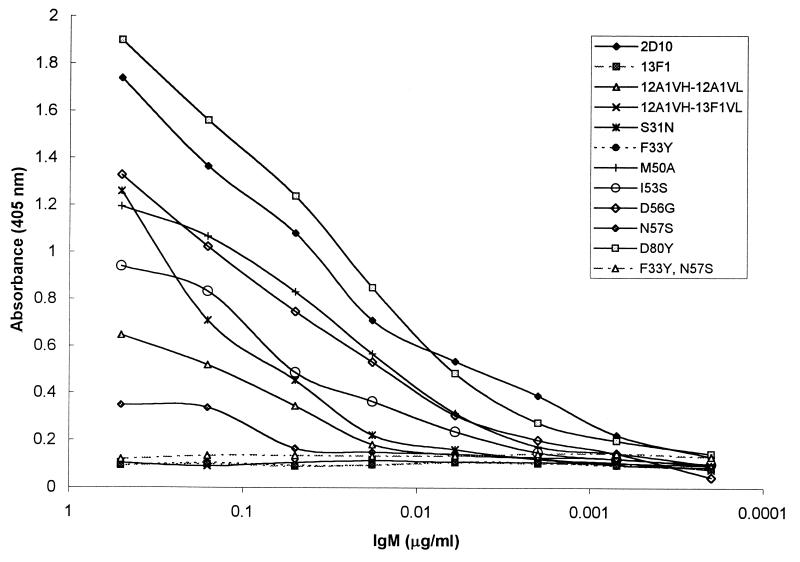
Reactivity with anti-idiotypic Ab.
MAb 7B8 binds to the Ag-combining sites of some class II MAbs (38). MAb 7B8 binds to Abs 12A1, 12A1VH-12A1VL, and 2D10 but not to 13F1 or the 12A1VH-13F1VL hybrid (Fig. 7). MAb 7B8 reacted with all 12A1→13F1 mutants except the F33Y N57S double mutant. These results indicate that the idiotypic determinants recognized by MAb 7B8 reside in both the VH and VL and at VH positions 33 and 57.
Frequency of Y33 and S57 in the VHs of Abs to GXM and germ line 7183 genes.
None of the 14 MAbs to GXM that use VH7183 gene family segments and are protective in mice have Y33 or S57 (Table 3). Two of 15 VH7183 family gene segments in the genetic database have Y33, six have S57, and only two (VH50.1 and VH7183.10) have both Y33 and S57.
TABLE 3.
Amino acid usage at positions 33 and 57 for antibodies of C. neoformans which have been evaluated for protective efficacy
| MAb | Isotope | Amino acid at position:
|
Protective | Reference(s) | |
|---|---|---|---|---|---|
| 33 | 57 | ||||
| Consensus | NAa | F | N | NA | 29 |
| 3B10 | IgG1 | F | N | Yes | 29, 33 |
| 2D10 | IgM | F | R | Yes | 29, 32 |
| 2H1 | IgG1 | F | K | Yes | 29, 32 |
| 3E5 | IgG1b | F | N | Yes | 8, 29, 48 |
| 18G9 | IgA | F | K | Yes | 29, 32 |
| 10F10 | IgG1 | F | S | Yes | 29 |
| 12A1 | IgM | F | N | Yes | 29, 31 |
| 17E12 | IgG1 | F | K | Yes | 29 |
| 18B7 | IgG1 | F | K | Yes | 29 |
| 3C2 | IgG1 | L | K | Yes | 5, 30 |
| 471 | IgG1 | L | K | Yes | 5, 30 |
| 13F1 | IgM | Y | S | No | 29, 31 |
| 21D2 | IgM | Y | S | No | 8, 32 |
NA, not applicable.
The IgG1 switch variant of the nonprotective IgG3 MAb 3E5 is protective (48).
DISCUSSION
Characterization of the amino acids residues responsible for Ab specificity to C. neoformans polysaccharide is important for understanding the relationship between Ab structure and protective efficacy. This information can also be helpful for preserving fine specificity of Abs to C. neoformans during Ab engineering and for the ongoing efforts to develop vaccines using polysaccharide-protein conjugates (6, 13) and peptide mimotopes (1).
Our results show that changing two amino acids in the binding pocket of the protective IgM MAb 12A1 VH to the corresponding residues in the VH of the nonprotective MAb 13F1 can alter the IF pattern and significantly reduce the opsonic activity, such that the mutated 12A1 Ab behaves like MAb 13F1. We were able to target these amino acids because we had available VH and VL sequences of four closely related class II IgM MAbs (8, 29), namely, 12A1, 13F1, 2D10, and 21D2, that differed in IF pattern despite having very similar V-region sequences. We focused our efforts on analyzing VH rather than VL differences as the cause of differences in IF patterns because the MAb 2D10 and 13F1 VLs were identical yet produced annular and punctate IF patterns, respectively. This deduction was confirmed by the findings that the 12A1VH-13F1VL hybrid produced annular IF and that mutations to the 12A1 VH amino acids changed the IF pattern from annular to punctate. The observation that the 12A1 VH is responsible for the type of pattern observed is consistent with the proposal that Ab specificity is frequently determined by the VH (23).
Superposition of the V-region sequence of MAb 12A1 on the crystal structure of the closely related MAb 2H1 revealed that all of the mutated amino acid positions with the exception of framework position 80 mapped to the Ag binding site. Nonetheless, the data showed that single amino acid changes in the sequence of MAb 12A1 to the comparable residues in MAb 13F1 were insufficient to change the IF pattern or to affect the outcome of competition experiments with MAb 3E5. However, when the two amino acids in the 12A1 VH at positions 33 and 57 were changed to the amino acids found in the 13F1 VH, a punctate pattern was observed. Competition experiments confirmed that the two mutations conferred a specificity change, since the parent but not the F33Y N57S double mutant competed for GXM with MAb 3E5.
The punctate and annular IF patterns are considered to be a consequence of the binding of MAbs 12A1 and 13F1 to different epitopes, because these MAbs have similar affinities for GXM although they (i) bind to different places in the capsule of serotype D strains (15), (ii) bind differently to serotype A and D strains (11), (iii) exhibit differences in binding to peptide mimetics (43), (iv) react differently with polysaccharides from the four C. neoformans serotypes (6), and (v) demonstrate differences in competition experiments (31). Given that the annular and punctate binding patterns correlate with protective and nonprotective efficacy, respectively, and reflect the binding of Abs with different specificities, the VH positions 33 and 57 define critically important structural correlates for Ab functional efficacy against C. neoformans.
The Ab response to GXM is highly restricted in V gene usage, even among genetically different strains of mice (5, 8, 29, 38, 40, 49). The overwhelming majority of murine MAbs use 7183 VH gene family elements (5, 38). The F33 and N57 in MAb 12A1 are both found in the consensus VH sequence from which the 12A1 VH was derived (29). Since only two 7183 VH family genes have both Y33 and S57 in their germ line sequences, this implies that F33 and N57 are likely to have arisen from somatic mutations that accrued during the immune response. Alternatively, they are derived from a germ line VH7183 family gene element that has not yet been characterized. Regardless of their origin, the Y33 and S57 residues in MAb 13F1 appear to have changed the specificity from a GXM epitope that could mediate protection against serotype D strains to one that could not. None of the 12 protective MAbs for which sequence data are available has the Y33 and S57 motif. In contrast, the two nonprotective class II MAbs for serotype D strains each have Y33 and S57. Analysis of germ line 7183 gene family sequences in the genetic database (GenBank, Los Alamos, N.Mex.) indicates that 13% have F33, 40% have N57, and 15% have both F33 and N57. One of two gene elements with the Y33 and S57 motif is VH50.1, and it is used in the nonprotective MAb 21D2 in its germ line configuration (8). These results suggest that nonprotective Abs can be generated either from germ line sequences or as a consequence of somatic mutation. For many 7183 VH gene elements, a single somatic mutation can yield the F33 and N57 motif found in MAb 13F1. The ability of the mouse genome to code for both protective and nonprotective specificities may underlie such perplexing observations as the fact that highly immunogenic polysaccharide-protein conjugate vaccines can elicit high-titer protective (13) and nonprotective (18) Ab responses. The implication that germ line residues in CDRs are critical for the annular IF pattern that is associated with Ab efficacy is consistent with the proposal that certain VH genes are required for carbohydrate binding (45) and the fact that class II Abs to GXM are all derived from the same Ab clan (25).
Our observations demonstrate that a change of IF pattern from annular to punctate required changes in at least two amino acids located in each of the CDR1 and CDR2 VH regions. However, scanning electron microscopy revealed differences in the fibrillar structure after binding of MAbs 12A1 and 13F1 and the 12A1VH→13F1VH mutants. Previous work has shown that fibrillar capsular structure is different after binding of MAbs 12A1 and 13F1 (12). Thus, each mutant with a single amino acid change displayed qualitative alterations in the annular IF pattern after binding to strain 24067. Although caution is always warranted when interpreting or comparing scanning electron micrographs of polysaccharide capsules (12), the differences in fibrillar appearance are consistent with the notion that some amino acid replacements affected the interaction with the capsule even though the annular IF pattern was maintained. This raises the possibility that Ab reactivity with C. neoformans in the punctate pattern reflects a continuum based on deviations from the consensus and/or germ line motif found in 12A1, although more work is needed to determine precisely how certain Abs react with GXM.
The F33Y change occurs at one of five amino acid residues delineating a cavity at the bottom of the MAb 2H1 structure. The side chains of F33 or Y33 at this position are near the surface of this cavity in a location that is likely to be solvated and in contact with the polysaccharide. Hence, it appears that F33 is in a strategic position to affect the type of binding that occurs but that a single change at this location is not sufficient to alter the IF pattern. Instead, the second change of N57S is also required, and the double mutation significantly reduced the binding to GXM determined by ELISA in a manner comparable to that for MAb 13F1. These results imply that amino acid residues in VH define the epitope specificity for these Abs to C. neoformans, although both VH and VL contribute to Ag affinity. A similar observation has been reported for Abs to digoxin (35), whereas for Abs to the polysaccharide levan, residues affecting fine specificity were located in the VH-VL junctional area (2).
Greenspan has defined an Ab paratope involved in a given Ag-Ab interaction as contact-, affinity-, and specificity-determining residues which may confer overlapping but nonidentical interactions with the epitope (19) and has defined amino acids that contribute to binding as the paratope set, depending on whether the residue is involved in contact, bond formation, or both (19). Our results are consistent with this view and suggest that each amino acid change to the paratope resulted in an alteration in the type of Ag-Ab complex formed in the capsule. Hence, the annular and punctate patterns may simply represent two forms of Ab binding to GXM among a multitude of possible interactions defined by the paratope of class II MAbs to GXM.
Ab binding in annular IF patterns is associated with significantly greater opsonic efficacy (10, 39). The 12A1→13F1 variants each resulted in reduced opsonic efficacy relative to that of the parent MAb 12A1, but the greatest effects for single amino acid changes were observed with F33Y and N57S. Both the F33Y and N57Y mutations significantly reduced the opsonic efficacy of the Ab without altering the IF pattern from annular to punctate. However, when both mutations were present simultaneously, the opsonic efficacy was further reduced to that of MAb 13F1. Hence, the change in IF pattern from annular to punctate which resulted from this double mutation also abolished the opsonic efficacy of this MAb 12A1→13F1 mutant. The finding that the change from annular to punctate reactivity was accompanied by a reduction in Ab opsonic efficacy establishes an association between Ab structure and biological function. This defines a key structure-function relationship for Abs to C. neoformans that is relevant to the choice of immunotherapeutic reagents and vaccine design.
IgG1 binding to the C. neoformans capsule alters the charge of the cell, but the mechanism for this phenomenon is not understood (36, 37). However, neither IgM MAb 12A1 nor 13F1 produced significant changes in cell charge upon binding C. neoformans. Since our mutant Ab set included mutations that replaced charged and neutral residues, we investigated the effect of amino acid replacements on the zeta potential of IgM-coated cells in an attempt to gain insight into this phenomenon. The various amino acid substitutions had little effect on the cell charge of C. neoformans after Ab binding. Replacement of a negatively charged aspartic acid at position 80 with a neutral tyrosine had no significant effect on the zeta potential of IgM-coated cells. Interestingly, the largest change in zeta potential was observed with N56G replacement, which involved the substitution of a neutral amino acid residue for another. This result suggests that cell charge can change upon Ab binding without an apparent change in the Ab charge.
The anti-idiotypic MAb 7B8 binds to the MAbs 12A1 and 12A1VH-12A1VL but not to MAb 13F1. No binding to the 12A1VH-13F1VL hybrid was observed, indicating that amino acid differences between the 12A1 VL and 13F1 VL contribute to defining the idiotype. In contrast, MAb 7B8 recognized all of the 12A1→13F1 single-amino-acid-mutated variants, and several exhibited significantly stronger binding than the parent MAb 12A1, suggesting that those changes stabilized the Ab-Ab interaction. However, there was no reactivity with the F33Y N57Y double mutant, indicating that F33 and N57 are part of the idiotype recognized by MAb 12A1. Hence, the same residues that confer annular or punctate specificity in the VH chain are involved in recognition by the 7B8 anti-idiotypic Ab. These studies suggest that this reagent may be useful for identifying protective MAbs.
Peptide mimetics of class II MAbs have been shown to bind in the Ab combining site and to be useful in mapping the epitope specificities of closely related MAbs (43, 44, 47). Peptide mimetics for MAb 12A1 and 13F1 have been reported (39). The availability of multiple MAb 12A1 mutants expressing the corresponding MAb 13F1 amino acid provided the opportunity to ascertain whether the same residues were responsible for peptide and binding pattern specificities. To our knowledge this is the first study that has simultaneously evaluated the contributions of V-region residues to the binding of polysaccharide Ag and a peptide mimotope. PA1 is a 10-mer peptide that binds to the closely related MAbs 2H1 and 2D10 but not to MAbs 12A1, 13F1, and 21D2 or to any of the variants generated here. The crystal structure has shown that PA1 has significantly more van der Waals interactions with VL than with VH. Hence, even though MAbs 2D10, 13F1, and 21D2 have the same VL, their interactions with PA1 are not sufficient to promote binding in the absence of specific VH interactions. Similar results were obtained with the 6-mer peptide 601, which includes the same motif as peptide PA1 which is involved in Ab combining site binding. In contrast, 206.1 is a 19-mer peptide that binds MAbs 2D10, 2H1, and 21D2 strongly, 12A1 weakly, and 13F1 not at all. The larger size of 206.1 is believed to make it capable of making additional contacts with the Ab binding region. The I53S, N56G, and D80Y mutants exhibited stronger binding to peptide 206.1 than either MAb 12A1 or 13F1. The results with peptide 206.1 indicate a dissociation of the binding characteristics of Ab with polysaccharide and peptide, suggesting that different residues are responsible for binding the GXM Ag and the peptide mimetic. This is consistent with the suggestion that peptide mimotopes of carbohydrate-binding Abs are recognized by different mechanisms than the carbohydrate Ag (21). However, in the case of peptide PM14, which reacts with MAb 13F1 but not 12A1, the 12A1 VH mutant F33Y demonstrated significant reactivity. Since the F33Y N57Y double mutant indicates that the F33Y residue is also involved in the IF binding pattern, it appears that some amino acids in the paratope set bind primarily peptide or carbohydrate, whereas others interact with both types of Ag.
In summary, our results indicate that VH amino acid residues F33 and N57 are critical determinants for Ab binding to GXM in a manner that translates into Ab-mediated protection against C. neoformans. Changing these two amino acids to the corresponding residues in MAb 13F1 resulted in a mutated variant of MAb 12A1 which behaved like MAb 13F1 with regard to IF, phagocytosis, recognition by anti-idiotypic MAb, and binding to GXM. These results are internally consistent and define two molecular determinants for protective efficacy against C. neoformans. The difference in reactivity of mutated Abs with GXM and peptide mimetics strongly suggests that different paratopes interacted with the polysaccharide Ag and the peptide mimetic. Hence, the search for peptide mimotopes that can elicit antipolysaccharide responses may be more fruitful if it focuses on peptides that interact with Ab amino acids important for binding carbohydrate epitopes. The implication that the punctate IF specificity can be either encoded in germ line V genes or generated by somatic mutation highlights the complexity and difficulty in consistently eliciting protective Ab responses to C. neoformans.
ACKNOWLEDGMENTS
We thank H. Bernstein and R. Sayle for help with the RASTOP program. We are grateful to Liise-anne Pirofski for critical reading of the manuscript.
This work was supported by NIH awards AI33774, AI3342, and HL-59842-01 to A.C. and AI01489 to J.N. A.C. is the recipient of a Burroughs Wellcome Development Therapeutics Award.
REFERENCES
- 1.Beenhouwer D O, Valadon P, May R, Scharff M D. Peptide mimicry of the polysaccharide capsule of Cryptococcus neoformans. In: Cunningham M W, Fujinami R S, editors. Molecular mimicry, microbes, and autoimmunity. Washington, D.C.: ASM Press; 2000. pp. 143–160. [Google Scholar]
- 2.Brorson K, Thompson C, Wei G, Krasnokutsky M, Stein K E. Mutational analysis of avidity and fine specificity of anti-levan antibodies. J Immunol. 1999;163:6694–6701. [PubMed] [Google Scholar]
- 3.Casadevall A. Antibody immunity and invasive fungal infections. Infect Immun. 1995;63:4211–4218. doi: 10.1128/iai.63.11.4211-4218.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadevall A, Cleare W, Feldmesser M, Glatman-Freedman A, Goldman D L, Kozel T R, Lendvai N, Mukherjee J, Pirofski L, Rivera J, Rosas A L, Scharff M D, Valadon P, Westin K, Zhong Z. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob Agents Chemother. 1998;42:1437–1446. doi: 10.1128/aac.42.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall A, DeShaw M, Fan M, Dromer F, Kozel T R, Pirofski L. Molecular and idiotypic analysis of antibodies to Cryptococcus neoformans glucuronoxylomannan. Infect Immun. 1994;62:3864–3872. doi: 10.1128/iai.62.9.3864-3872.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadevall A, Mukherjee J, Devi S J N, Schneerson R, Robbins J B, Scharff M D. Antibodies elicited by a Cryptococcus neoformans glucuronoxylomannan-tetanus toxoid conjugate vaccine have the same specificity as those elicited in infection. J Infect Dis. 1992;65:1086–1093. doi: 10.1093/infdis/165.6.1086. [DOI] [PubMed] [Google Scholar]
- 7.Casadevall A, Mukherjee J, Scharff M D. Monoclonal antibody ELISAs for cryptococcal polysaccharide. J Immunol Methods. 1992;154:27–35. doi: 10.1016/0022-1759(92)90209-c. [DOI] [PubMed] [Google Scholar]
- 8.Casadevall A, Scharff M D. The mouse antibody response to infection with Cryptococcus neoformans: VH and VL usage in polysaccharide binding antibodies. J Exp Med. 1991;174:151–160. doi: 10.1084/jem.174.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherniak R, Reiss E, Turner S. A galactoxylomannan antigen of Cryptococcus neoformans serotype A. Carbohydr Res. 1982;103:239–250. [Google Scholar]
- 10.Cleare W, Brandt M E, Casadevall A. Monoclonal antibody 13F1 produces annular immunofluorescent patterns on Cryptococcus neoformans serotype AD isolates. J Clin Microbiol. 1999;37:3080. doi: 10.1128/jcm.37.9.3080-3080.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleare W, Casadevall A. The different binding patterns of two IgM monoclonal antibodies to Cryptococcus neoformans serotype A and D strains correlates with serotype classification and differences in functional assays. Clin Diagn Lab Immunol. 1998;5:125–129. doi: 10.1128/cdli.5.2.125-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleare W, Casadevall A. Scanning electron microscopy of encapsulated and nonencapsulated Cryptococcus neoformans and the effect of glucose on capsular polysaccharide release. Med Mycol. 1999;37:235–243. [PubMed] [Google Scholar]
- 13.Devi S J N. Preclinical efficacy of a glucuronoxylomannan-tetanus toxoid conjugate vaccine of Cryptococcus neoformans in a murine model. Vaccine. 1996;14:841–842. doi: 10.1016/0264-410x(95)00256-z. [DOI] [PubMed] [Google Scholar]
- 14.Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldmesser M, Rivera J, Kress Y, Kozel T R, Casadevall A. Antibody interactions with the capsule of Cryptococcus neoformans. Infect Immun. 2000;68:3642–3650. doi: 10.1128/iai.68.6.3642-3650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleuridor R, Lyles R H, Pirofski L. Quantitative and qualitative differences in the serum antibody profiles of human immunodeficiency virus-infected persons with and without Cryptococcus neoformans meningitis. J Infect Dis. 1999;180:1526–1535. doi: 10.1086/315102. [DOI] [PubMed] [Google Scholar]
- 17.Fleuridor R, Zhong Z, Pirofski L. A human IgM monoclonal antibody prolongs survival of mice with lethal cryptococcosis. J Infect Dis. 1998;178:1213–1216. doi: 10.1086/515688. [DOI] [PubMed] [Google Scholar]
- 18.Goren M B. Experimental murine cryptococcosis: effect of hyperimmunization to capsular polysaccharide. J Immunol. 1967;98:914–922. [PubMed] [Google Scholar]
- 19.Greenspan N S. Conceptions of epitopes and paratopes and the ontological dynamics of immunological recognition. In: Rouvray D H, editor. Concepts in chemistry: a contemporary challenge. Taunton, Somerset, England: Research Studies Press Ltd; 1997. pp. 383–403. [Google Scholar]
- 20.Han Y, Cutler J E. Antibody response that protects against disseminated candidiasis. Infect Immun. 1995;63:2714–2719. doi: 10.1128/iai.63.7.2714-2719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris S L, Craig L, Mehroke J S, Rashed M, Zwick M B, Kenar K, Toone E J, Greenspan N, Auzanneau F I, Marino-Albernas J R, Pinto B M, Scott J K. Exploring the basis of peptide-carbohydrate crossreactivity: evidence for discrimination by peptides between closely related anti-carbohydrate antibodies. Proc Natl Acad Sci USA. 1997;94:2454–2459. doi: 10.1073/pnas.94.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houpt D C, Pfrommer G S T, Young B J, Larson T A, Kozel T R. Occurrences, immunoglobulin classes, and biological activities of antibodies in normal human serum that are reactive with Cryptococcus neoformans glucuronoxylomannan. Infect Immun. 1994;62:2857–2864. doi: 10.1128/iai.62.7.2857-2864.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabat E A, Wu T T. Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J Immunol. 1991;147:1709–1719. [PubMed] [Google Scholar]
- 24.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller C. Proteins of immunological interest. Publication no. 91-3242. Bethesda, Md: National Institutes of Health; 1991. [Google Scholar]
- 25.Kirkham P M, Mortari F, Newton J A, Schroeder H W. Immunoglobulin VH clan and family identity predicts variable domain structure and may influence antigen binding. EMBO J. 1991;11:603–609. doi: 10.1002/j.1460-2075.1992.tb05092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lendvai N, Casadevall A. Antibody mediated toxicity in Cryptococcus neoformans infection: mechanism and relationship to antibody isotype. J Infect Dis. 1999;180:791–801. doi: 10.1086/314946. [DOI] [PubMed] [Google Scholar]
- 27.MacGill T C, MacGill R S, Casadevall A, Kozel T R. Biological correlates of capsular (quellung) reactions of Cryptococcus neoformans. J Immunol. 2000;164:4835–4842. doi: 10.4049/jimmunol.164.9.4835. [DOI] [PubMed] [Google Scholar]
- 28.McLean G, Nakouzi A, Casadevall A, Green N S. Human and murine expression vector cassettes. Mol Immunol. 2001;37:837–845. doi: 10.1016/s0161-5890(00)00101-2. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee J, Casadevall A, Scharff M D. Molecular characterization of the antibody responses to Cryptococcus neoformans infection and glucuronoxylomannan-tetanus toxoid conjugate immunization. J Exp Med. 1993;177:1105–1106. doi: 10.1084/jem.177.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee J, Kozel T R, Casadevall A. Monoclonal antibodies reveal additional epitopes of serotype D Cryptococcus neoformans capsular glucuronoxylomannan that elicit protective antibodies. J Immunol. 1998;161:3557–3568. [PubMed] [Google Scholar]
- 31.Mukherjee J, Nussbaum G, Scharff M D, Casadevall A. Protective and nonprotective monoclonal antibodies to Cryptococcus neoformans originating from one B-cell. J Exp Med. 1995;181:405–409. doi: 10.1084/jem.181.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjee J, Scharff M D, Casadevall A. Protective murine monoclonal antibodies to Cryptococcus neoformans. Infect Immun. 1992;60:4534–4541. doi: 10.1128/iai.60.11.4534-4541.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherjee J, Scharff M D, Casadevall A. Cryptococcus neoformans infection can elicit protective antibodies in mice. Can J Microbiol. 1994;40:888–892. doi: 10.1139/m94-141. [DOI] [PubMed] [Google Scholar]
- 34.Mukherjee S, Feldmesser M, Casadevall A. J774 murine macrophage-like cell interactions with Cryptococcus neoformans in the presence and absence of opsonins. J Infect Dis. 1996;173:1222–1231. doi: 10.1093/infdis/173.5.1222. [DOI] [PubMed] [Google Scholar]
- 35.Near R I, Bruccoleri R, Novotny J, Hudson N W, White A, Mudgett-Hunter M. The specificity properties that distinguish members of a set of homologous anti-digoxin antibodies are controlled by H chain mutations. J Immunol. 1991;146:627–633. [PubMed] [Google Scholar]
- 36.Nosanchuk J D, Casadevall A. Cellular charge of Cryptococcus neoformans: contributions from the capsular polysaccharide, melanin, and monoclonal antibody binding. Infect Immun. 1997;65:1836–1841. doi: 10.1128/iai.65.5.1836-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nosanchuk J D, Cleare W, Franzot S P, Casadevall A. Amphotericin B and fluconazole affect cellular charge, macrophage phagocytosis, and cellular morphology of Cryptococcus neoformans at subinhibitory concentrations. Antimicrob Agents Chemother. 1999;43:233–239. doi: 10.1128/aac.43.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nussbaum G, Anandasabapathy S, Mukherjee J, Fan M, Casadevall A, Scharff M D. Molecular and idiotypic analysis of the antibody response to Cryptococcus neoformans glucuronoxylomannan-protein conjugate vaccine in autoimmune and nonautoimmune mice. Infect Immun. 1999;67:4469–4476. doi: 10.1128/iai.67.9.4469-4476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nussbaum G, Cleare W, Casadevall A, Scharff M D, Valadon P. Epitope location in the Cryptococcus neoformans capsule is a determinant of antibody efficacy. J Exp Med. 1997;185:685–697. doi: 10.1084/jem.185.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pirofski L, Lui R, DeShaw M, Kressel A B, Zhong Z. Analysis of human monoclonal antibodies elicited by vaccination with a Cryptococcus neoformans glucuronoxylomannan capsular polysaccharide vaccine. Infect Immun. 1995;63:3005–3014. doi: 10.1128/iai.63.8.3005-3014.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richmond D V, Fisher D J. The electrophoretic mobility of microorganisms. Adv Microb Physiol. 1973;9:1–29. doi: 10.1016/s0065-2911(08)60375-6. [DOI] [PubMed] [Google Scholar]
- 42.Sayle R A, Milner-White E J. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 43.Valadon P, Nussbaum G, Boyd L F, Margulies D H, Scharff M D. Peptide libraries define the fine specificity of anti-polysaccharide antibodies to Cryptococcus neoformans. J Mol Biol. 1996;261:11–22. doi: 10.1006/jmbi.1996.0438. [DOI] [PubMed] [Google Scholar]
- 44.Valadon P, Nussbaum G, Oh J, Scharff M D. Aspects of antigen mimicry revealed by immunization with a peptide mimetic of Cryptococcus neoformans polysaccharide. J Immunol. 1998;161:1829–1836. [PubMed] [Google Scholar]
- 45.Vargas-Madrazo E, Lara-Ochoa F, Almagro J C. Canonical structure repertoire of the antigen-binding site of immunoglobulins suggests strong geometrical restrictions associated to the mechanism of immune recognition. J Mol Biol. 1995;254:497–504. doi: 10.1006/jmbi.1995.0633. [DOI] [PubMed] [Google Scholar]
- 46.Vecchiarelli A, Casadevall A. Antibody-mediated effects against Cryptococcus neoformans: evidence for interdependency and collaboration between humoral and cellular immunity. Res Immunol. 1998;149:321–333. doi: 10.1016/s0923-2494(98)80756-6. [DOI] [PubMed] [Google Scholar]
- 47.Young A C M, Valadon P, Casadevall A, Scharff M D, Sacchettini J C. The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library: implication for the identification of peptide mimotopes. J Mol Biol. 1997;274:622–634. doi: 10.1006/jmbi.1997.1407. [DOI] [PubMed] [Google Scholar]
- 48.Yuan R, Casadevall A, Spira G, Scharff M D. Isotype switching from IgG3 to IgG1 converts a non-protective murine antibody to C. neoformans into a protective antibody. J Immunol. 1995;154:1810–1816. [PubMed] [Google Scholar]
- 49.Zhang H, Zhong Z, Pirofski L. Peptide epitopes recognized by a human anticryptococcal glucuronoxylomannan antibody. Infect Immun. 1997;65:1158–1164. doi: 10.1128/iai.65.4.1158-1164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]