Abstract
Introduction: Segmentation of clinical target volume (CTV) from CT images is critical for cervical cancer brachytherapy, but this task is time-consuming, laborious, and not reproducible. In this work, we aim to propose an end-to-end model to segment CTV for cervical cancer brachytherapy accurately. Methods: In this paper, an improved M-Net model (Mnet_IM) is proposed to segment CTV of cervical cancer from CT images. An input and an output branch are both proposed to attach to the bottom layer to deal with CTV locating challenges due to its lower contrast than surrounding organs and tissues. A progressive fusion approach is then proposed to recover the prediction results layer by layer to enhance the smoothness of segmentation results. A loss function is defined on each of the multiscale outputs to form a deep supervision mechanism. Numbers of feature map channels that are directly connected to inputs are finally homogenized for each image resolution to reduce feature redundancy and computational burden. Result: Experimental results of the proposed model and some representative models on 5438 image slices from 53 cervical cancer patients demonstrate advantages of the proposed model in terms of segmentation accuracy, such as average surface distance, 95% Hausdorff distance, surface overlap, surface dice, and volumetric dice. Conclusion: A better agreement between the predicted CTV from the proposed model Mnet_IM and manually labeled ground truth is obtained compared to some representative state-of-the-art models.
Keywords: deep learning, cervical cancer, CTV segmentation, U-Net, M-Net
Introduction
Cervical cancer is the fourth most frequently diagnosed cancer and the fourth leading cause of cancer death in women.1 At present, external radiation therapy and brachytherapy are the clinical standard treatment scheme for patients with advanced cervical cancer who cannot be treated surgically.2 Segmentation of the clinical target volume (CTV) in CT images precisely is important for brachytherapy. However, CTV is usually contoured by radiation oncologists, manually based on their clinical experiences due to challenges as follows. Firstly, CTV includes not only the visible tumor regions, but also the potential infiltrated regions. Secondly, dose constraint limits of surrounding organs at risk also must be considered.3 Thirdly, due to the movement of the bladder and rectum, the position and shape of uterus and cervix changes when brachytherapy is carried out especially if an applicator is placed.4 Thus, automatic segmentation of cervical cancer CTV is essential to reduce the workload of physicians and to formulate radiotherapy plan reasonable by reducing the variability among different physicians.
There are few relevant studies on the segmentation of CTV in the literature. In earlier studies, segmentation of CTV for cervical cancer was mainly based on atlas-based methods.5–8 These methods are very dependent on the accuracy of registration methods and the choice of the atlas. In recent years, as deep learning has made a splash in medical image analysis, researchers have also started to apply convolutional neural networks to segment CTV for cervical cancer,9, 10 most of which are U-shape architecture,11, 12, 13, 2 try to improve segmentation accuracy by introducing skip connections, dense blocks, dual-path networks, and attention mechanisms.
Although deep learning approaches described earlier are somehow successful in segmenting CTV for cervical cancer, image sources vary considerably between institutions at all levels. Three main image modalities are available for the current study. They are MR images, CT images with applicators and CT images without applicators. The images are derived from the radiotherapy period. In this paper, we focus on the CTV region which has to be recognized during cervical cancer brachytherapy. The corresponding images are with applicators and come from the CT modality. Therefore, CT images with applicators are used to train and evaluate different well-known medical image segmentation models that can be considered as baselines. Meanwhile, in this paper, we propose a model to segment CTV from CT images for cervical cancer, namely Mnet_Im. This model achieves excellent performance compared to other models on all five well-known evaluation metrics.
The remainder of this paper is organized as follows. In section “Related work,” we introduce some representative models that are highly relevant to our work. In section “Methods,” we describe the proposed model in detail. In section “Experiments,” we perform experimental evaluations of the proposed model. Finally, we discuss and conclude this paper in sections “Discussion” and “Conclusion,” respectively.
Related Work
There are two main types of baseline models commonly used in the field of image segmentation, namely the U-Net and its variants,14 and the DeepLab series of models.15 In12 U-Net is used as a baseline for CTV segmentation where its convolutional layers are replaced with a dual-path network. While Unet++ is used as a baseline to segment CTV by adding an attention mechanism.16 Although DeepLab series of models are not currently applied to the CTV segmentation task for cervical cancer, it has received excellent results as a well-established model in other segmentation tasks.17, 18 Thus, in this paper, we include the U-Net, Unet++, and DeepLabV3+ as comparative baseline models.19 In the following subsections, we describe details of these baseline models as well as the model we proposed in this paper, named as Mnet_Im as it is highly related to M-Net.20
U-Net and Its Variants
The U-Net network was first proposed for application in the Cell Tracking Challenge. It is composed of a contractive path and an expansive path. The contractive path follows the typical convolutional neural network structure, consisting of convolutional layers and pooling layers, with the aim of extracting multiple scale semantic features from input images. The expansive path is composed of convolutional layers, upsampling layers, and skip connections. The upsampling layers are used to recover low-resolution features to its adjacent one scale higher level, while skip connections and convolutional layers combine features from the contractive path and features recovered by upsampling layers to obtain a more accurate output.
Unet++ is a classic variant of U-Net, which differs from U-Net in that it resets the skip connection part of U-Net and introduces a deep supervision mechanism. In U-Net, the feature maps of the contractive path are concatenated directly in the expansive path, while in Unet++ these features are concatenated in the expansive path after passing through a dense convolution block. Each convolutional layer is preceded by a concatenation layer that fuses the output of a previous convolutional layer from the same dense block with the corresponding upsampled output from a lower dense block. This dense connection structure enriches the semantic features fused with the expansive path. Thus, the fine-grained details of the foreground object are more effectively captured.
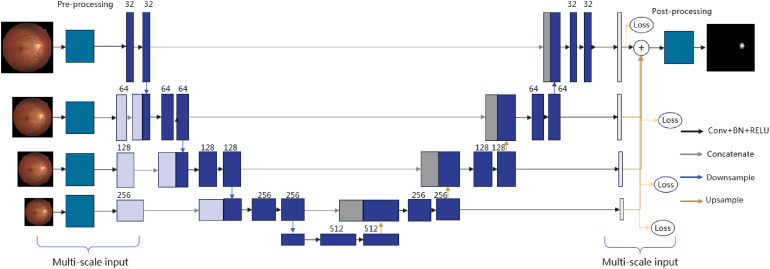
The M-Net is also a variant of the U-Net, which was first proposed to segment the optic cup and optic disc from fundus images. Its network structure is given in Figure 1. Obviously, its main structure looks like U-Net, whereas it introduces multiple-input and multiple-output mechanisms on the contractive and expansive paths, respectively. Multiple inputs are used to construct image pyramidal inputs to achieve multilevel perceptual field fusion. Multiple outputs are proposed to allow the model to be deeply supervised during training and to fuse the outputs at different scales to obtain information at different levels of detail when obtaining prediction results.
Figure 1.
Framework of M-Net.
DeeplabV3+
Similar to most DeepLab series of models, DeeplabV3+ is also an encoder–decoder structure where the classical atrous spatial pyramid pooling (ASPP) module is used to ensure the model more efficient at segmenting benchmarks with multiscale information.21 The difference is that DeepLabV3+ uses DeepLabV3 as an encoder and redesigns a simple but effective decoder module to refine the segmentation results around the object boundaries by avoiding the pitfalls of directly upsampling the DeepLabV3 output by a factor of 8 that results in information loss in predicting results. In previous studies, researchers preferred to employ skip connections and dense connections to enhance the ability of the model to extract high-level semantic information related to cervical cancer CTV regions.11, 12 ResNet and DenseNet consist of skip connections and dense connections, respectively, to reuse important features and explore new features.22, 23 Thus, we set ResNet and DenseNet as backbone networks of DeepLabv3+ in this paper, which are denoted as V3_R and V3_D, respectively.
Methods
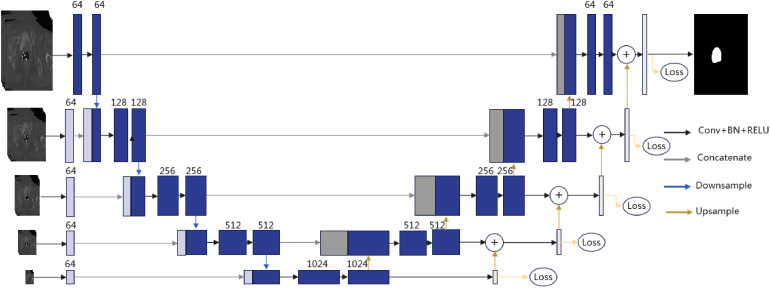
It can be found from CT images of a person with cervical cancer that the CTV region is quasicircular, which is similar to the optic cup and optic disc region in fundus images. As the success of M-Net mentioned earlier in segmenting the optic cup and optic disc region, we take it as the baseline model in this paper to extract the CTV region of cervical cancer from CT images. To match the new application scenarios where image distribution characteristic is different from fundus images, we improve the M-Net structure to propose a deep learning model that we name it Mnet_Im. As the framework of Mnet_Im shown in Figure 2, both an input and an output branch are first proposed to attach to the bottom layer of M-Net with a resolution of 1/16 of the original image. This improvement is proposed to deal with challenges in accurately locating the CTV due to its lower contrast than surrounding organs and tissues. Although lower resolution will inevitably induce detail loss, differences between the boundaries of the CTV and the boundaries of its surrounding organs and tissues also got closer to guarantee the model locating the CTV more accurately. We then propose a progressive fusion strategy, in which the low-level output is upsampled by a factor of 2 and is finally summed with its adjacent high-level output. Prediction results are recovered layer by layer, and the smoothness of the segmentation results is improved. However, M-Net obtains segmentation results by accumulating multiscale outputs with low-resolution outputs being upsampled directly which leads to unsmooth results. We define a loss function on each of the multiscale outputs and name this mechanism as deep supervision. The loss function at each scale is defined as follows.
where is the pixel coordinates and is the image height and width. and denote values of the ground truth and the predicted probability map at , respectively. Thus the overall loss is defined as follows.
Figure 2.
Framework of the proposed Mnet_Im.
We finally homogenize the numbers of feature map channels that are directly connected to inputs in each image resolution to reduce feature redundancy and computational burden of M-Net whereas the numbers of convolutional kernels of M-Net increase as resolutions of input images decrease. An experiment will be given in the subsection “Influence of the Number of Convolutional Layers in Multiscale Inputs” to show the effectiveness of this improvement.
Note that the institutional review board (CHCMU-BMEC) approved this study (2022B012S). Informed consent was obtained from all patient subjects.
Experiments
Experimental Details
Training Details
All parameters are guaranteed to be the same as they are set when they are first proposed, unless otherwise specified. The and in the loss function are 0.1 and 0.9, respectively. The initial learning rate is set to 10−3 and is then decayed by a factor of 0.1 for each of the first three epochs. Adam optimizer is used. Early stop is adopted to avoid overfitting. Note that all the models are in 2D. However, to better capture spatial relationships between successive CT image slices, we adopt the same input strategy given in,12 which is also proposed to segment CTV of cervical cancer. That is, for each batch, five consecutive CT image slices are considered as one input sample with the middle slice being the one that is really concerned.
Experimental Environment
All the models are implemented in PyCharm on a computer with Inter(R) Core(TM) i9-10900K CPU, NVIDIA GeForce RTX 3090 Ti. The Graphic Processing Unit (GPU) is only used to accelerate the training process while the prediction does not require a GPU.
Dataset
In this paper, the cervical cancer CT images come from the Liaoning Cancer Hospital & Institute, China. The total number of CT images is 5438 slices from 53 cervical cancer patients. Each patient undergoes one to six times brachytherapies yielding 235 scans (n = 253). The images are randomly split into training (n = 164), validation (n = 35), and test (n = 36) sets by considering scans as unit. Finally, the training, validation, and test sets contain 3770, 793, and 875 slices. CTV regions in the images are all manually contoured by one experienced radiation oncologist. For all the images, CTV regions occupy a small part of the images. Therefore, the images are cropped by automatic regions of interest location method.
Experimental Results
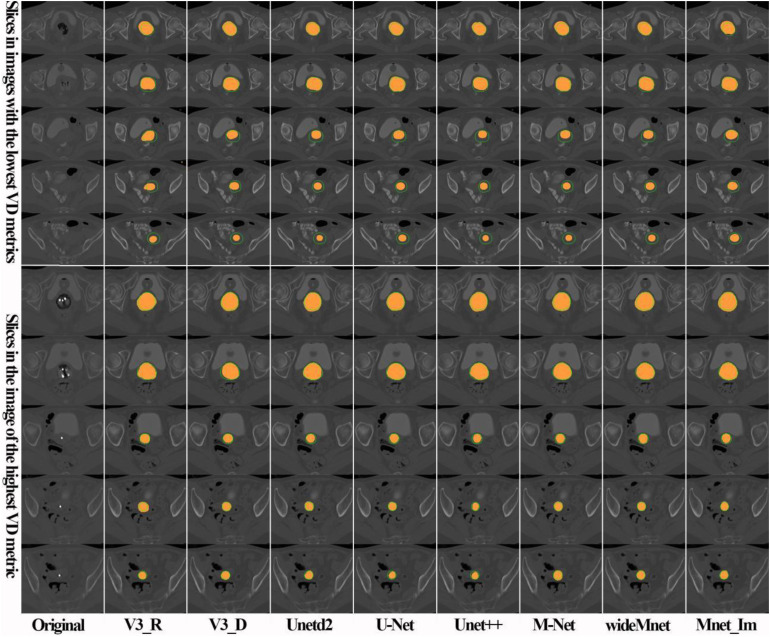
We compare the proposed model qualitatively with some representative models, such as U-Net, Unetd2, Unet+ , M-Net, wideMnet, V3_R, and V3_D, and give the result in Figure 3. What we have to say is that the wideMnet model is also a variant of M-Net by enlarging the number of feature maps by a factor of 2, while Unetd2 is a variant of U-Net by shrinking the number of feature maps by a factor of 2. In Figure 3, results given in the first five rows are from different patient cases with the lowest volumetric dice (VD) in our test set, while the last five rows are the best five results. From the qualitative results, we can see that almost all the models perform well (see rows 1, 2, 5, and 7 of Figure 3) in recognizing CTV regions near the vagina. In contrast, almost all the models are not good at identifying CTV regions in the vicinity of the cervix and uterus (see rows 3, 4, and 5 of Figure 3), especially if the image contrast is low. However, the proposed model performs a little better than the comparative models in the previously mentioned regions.
Figure 3.
Comparison of CTV segmentation results of the proposed model with representative models. Orange areas represent segmentation results of the models, while outer contours of the corresponding ground truths are annotated in green lines.
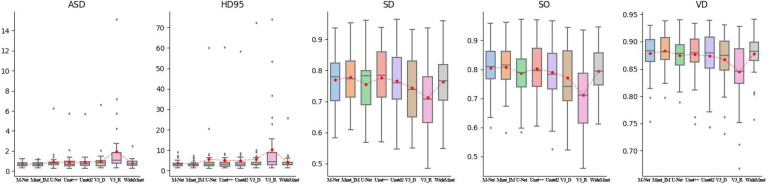
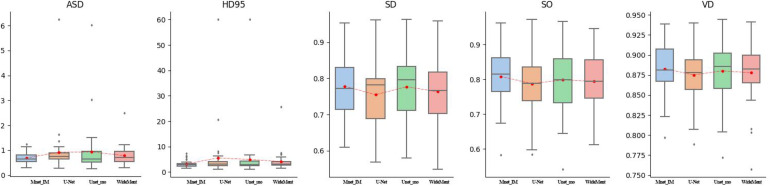
To validate the performance of the proposed model on our dataset quantitatively, five evaluation metrics are adopted to evaluate the segmentation results of the proposed model and comparative models. The metrics are average surface distance (ASD), 95% Hausdorff distance (HD95), surface overlap (SO), surface dice (SD), and VD. Note that smaller ASD and HD95 indicate a better match between segmentation results and corresponding ground truths, whereas greater SO, SD, and VD indicate a better match. Note that the final outputs of the proposed model are normalized by the sigmoid function. Therefore, a threshold is used to produce binary segmentation results. The best threshold is selected if corresponding segmentation results match ground truths best in terms of VD values. The quantitative comparison results are given in Figure 4 and Table 1.
Figure 4.
Quantitative comparison of the proposed model with representative models on our dataset. Red dots indicate mean values. Subplots at the horizontal axes are M-Net, Mnet_Im, U-Net, Unet++, Unetd2, V3_D, V3_R, and wideMnet models from left to right, respectively.
Table 1.
Performance Comparison of the Proposed Model With Representative Models (Mean ± Variance).
| Model | ASD | HD95 | SD | SO | VD |
|---|---|---|---|---|---|
| Unetd2 | 0.8719 ± 0.7331 | 4.9627 ± 84.6213 | 0.7905 ± 0.0095 | 0.7657 ± 0.01 | 0.8742 ± 0.0018 |
| U-Net | 0.9189 ± 0.8865 | 5.5606 ± 94.9406 | 0.7877 ± 0.0073 | 0.755 ± 0.0077 | 0.8751 ± 0.0011 |
| Unet++ | 0.8965 ± 0.8039 | 5.0277 ± 90.1666 | 0.8028 ± 0.0083 | 0.776 ± 0.0094 | 0.8763 ± 0.0017 |
| V3_R | 1.9469 ± 7.0177 | 10.2482 ± 228.4469 | 0.7121 ± 0.0139 | 0.7125 ± 0.0135 | 0.8452 ± 0.0033 |
| V3_D | 0.9739 ± 1.0429 | 5.602 ± 130.6175 | 0.771 ± 0.0109 | 0.7438 ± 0.0111 | 0.8673 ± 0.002 |
| M-Net | 0.6986 ± 0.0547 | 3.491 ± 2.9798 | 0.8045 ± 0.0064 | 0.7694 ± 0.0071 | 0.8787 ± 0.0013 |
| WideMnet | 0.7855 ± 0.1443 | 4.148 ± 15.4051 | 0.7945 ± 0.0063 | 0.7632 ± 0.0072 | 0.8778 ± 0.0012 |
| Mnet_Im | 0.6901 ± 0.0564 | 3.2013 ± 2.0014 | 0.8084 ± 0.0067 | 0.7785 ± 0.0065 | 0.8828 ± 0.0010 |
Note: Bold represents best values among comparison methods.
From Table 1, we can observe that the proposed model performs better than all the other models in terms of all the five metrics. We can also find that the models that are variants of DeepLabV3+ perform slightly worse than U-Net and its variants. From the ASD and HD95 subplots in Figure 4, we can see that variants of DeepLabV3+ give more outliers on the test set, which indicates that it is not robust enough. However, the proposed model provides more compact distribution of metrics, which indicates that it is more robust. To be more comprehensive, we also give the median of each metric for the models performing on the test set in Table 2.
Table 2.
Performance Comparison of the Proposed Model With Representative Models (Median).
| Model | ASD | HD95 | SD | SO | VD |
|---|---|---|---|---|---|
| Unetd2 | 0.7378 | 3.0000 | 0.7841 | 0.7597 | 0.8796 |
| U-Net | 0.7562 | 3.1623 | 0.7889 | 0.7823 | 0.8782 |
| Unet++ | 0.6937 | 3.0811 | 0.7954 | 0.7847 | 0.8795 |
| V3_R | 1.0556 | 4.3528 | 0.7119 | 0.7068 | 0.8453 |
| V3_D | 0.8097 | 3.0000 | 0.7412 | 0.7388 | 0.8745 |
| M-Net | 0.6684 | 3.0000 | 0.8096 | 0.7794 | 0.8832 |
| WideMent | 0.7150 | 3.0811 | 0.7941 | 0.7662 | 0.8824 |
| Mnet_Im | 0.6572 | 3.0000 | 0.8152 | 0.7723 | 0.8810 |
Note: Bold represents best values among comparison methods.
Further, to verify the robustness of the proposed model Mnet_IM, a 4-fold cross-validation test was conducted accordingly. The initial parameters are the same as the previous experiments, and we disrupted all the data sets randomly and divided them into four parts. Quantitative results are given in Table 3. The cross-validation results are slightly worse, compared to Table 1 (n = 36). This is probably due to more images are taken as testing images.
Table 3.
Quantitative Analysis Results of Mnet_IM Model Cross-Validation.
| N-flod | Validation number | ASD | HD95 | SO | SD | VD |
|---|---|---|---|---|---|---|
| 1-flod | 59 | 0.8253 | 4.1028 | 0.7814 | 0.7497 | 0.8661 |
| 2-flod | 59 | 0.8943 | 4.3847 | 0.7922 | 0.7687 | 0.8769 |
| 3-flod | 59 | 0.6819 | 3.1161 | 0.8191 | 0.7890 | 0.8825 |
| 4-flod | 58 | 0.7806 | 3.5771 | 0.8187 | 0.7860 | 0.8798 |
| AVG. | - | 0.7956 | 3.7961 | 0.8028 | 0.7733 | 0.8763 |
Discussion
Effectiveness of the Multiscale Deep Supervision
As well known, although U-Net extracts semantic information at the low-resolution level, it relies on the final recovered high-resolution feature maps to produce segmentation results. In this subsection, we discuss the impact of introduction of the deep supervision mechanism to U-Net for CTV segmentation of cervical cancer. For this purpose, we replace the expansive path of U-Net with the expansive path of the proposed Mnet_Im and name it as Unet_mo. Figure 5 and Table 4 show the performances of U-Net and its variants by introducing the deep supervision mechanism. For the sake of fairness, consistency of the feature map channels is maintained between models. For the models with multiscale outputs, we calculate losses at each scale and take their mean as the final loss. From Figure 5 and Table 4, we can see that models with the additional deep supervision mechanism perform better than others in terms of HD95, SD, SO, and VD.
Figure 5.
Quantitative comparison of U-Net and its variants by introducing the deep supervision mechanism. Red dots indicate mean values. Subplots at the horizontal axes are Mnet_Im, U-Net, Unet_mo, and wideMnet from left to right, respectively.
Table 4.
Performance Comparison of U-Net and Its Variants by Introducing the Deep Supervision Mechanism (Mean ± Variance).
| Model | ASD | HD95 | SO | SD | VD |
|---|---|---|---|---|---|
| U-Net | 0.9189 ± 0.8865 | 5.5606 ± 94.9406 | 0.7877 ± 0.0073 | 0.755 ± 0.0077 | 0.8751 ± 0.0011 |
| WideMnet | 0.7855 ± 0.1443 | 4.148 ± 15.4051 | 0.7945 ± 0.0063 | 0.7632 ± 0.0072 | 0.8778 ± 0.0012 |
| Unet_mo | 0.9317 ± 0.968 | 4.9423 ± 88.907 | 0.7985 ± 0.0085 | 0.7772 ± 0.0084 | 0.8801 ± 0.0013 |
| Mnet_Im | 0.6901 ± 0.0564 | 3.2013 ± 2.0014 | 0.8084 ± 0.0067 | 0.7785 ± 0.0065 | 0.8828 ± 0.0010 |
Note: Bold represents best values among comparison methods.
Influence of the Number of Convolutional Layers in Multiscale Inputs
As mentioned in the section “Methods,” the image resolution decreases as the depth of the M-Net model increases whereas the numbers of convolutional kernels increase at the same time. An excess of convolutions inevitably increases the risk of feature redundancy. Besides, the computation burden grows progressively in the process. As mentioned earlier, we set the numbers of convolutional kernels which are directly connected to inputs in each image resolution to be the same to reduce feature redundancy and computational burden. In this subsection, we discuss the effectiveness of this improvement on the performance of M-Net. We set the numbers of convolutional kernels to be 64 and 32 and name these two improved models as Mnet_64 and Mnet_32, respectively. We compare the improved models with M-Net and give comparison results in Table 5. It is not hard to find out that the performance of Mnet_64 is similar to M-Net in terms of SD, SO, and VD, while the performance decreases if the numbers of convolutional kernels stay the same with a predefined value 32. Therefore, in the proposed model Mnet_Im, we set this adjustable parameter to be 64 to improve time performance on the premise of ensuring segmentation accuracy.
Table 5.
Performance Comparison of M-Net and Its Two Variants (Mean ± Variance).
| Model | ASD | HD95 | SO | SD | VD |
|---|---|---|---|---|---|
| M-Net | 0.6986 ± 0.0547 | 3.491 ± 2.9798 | 0.8045 ± 0.0064 | 0.7694 ± 0.0071 | 0.8787 ± 0.0013 |
| Mnet_64 | 0.8045 ± 0.2553 | 4.2243 ± 20.3779 | 0.795 ± 0.008 | 0.7614 ± 0.0073 | 0.8779 ± 0.001 |
| Mnet_32 | 0.7567 ± 0.063 | 3.4467 ± 1.7314 | 0.7812 ± 0.0082 | 0.7452 ± 0.0076 | 0.8725 ± 0.001 |
Limitations of This Work
In this work, we did not validate the performance of our model on image cases from multiple institutions. In addition, we did not consider neoplasm staging. In practice, the later tumors progress, the more complex of CTV regions. In the future, we will focus on these two respects to improve our work.
Conclusion
A deep learning model is proposed to segment CTV of cervical cancer from CT images. The proposed model is an improvement of M-Net to improve time performance and ensuring segmentation accuracy by revising the compacting path and the expanding path, respectively. Experimental results show that the improved model performs better than the selected comparative models and U-Net networks are more effective than DeepLab base networks on the segmentation of CTV of cervical cancer from CT images.
Acknowledgments
The authors would like to thank the handling editor and anonymous reviewers very much for their constructive suggestions on improving the quality of this paper.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China, (grant number U20A20373).
ORCID iD: Dazhe Zhao https://orcid.org/0000-0001-8410-6002
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. [DOI] [PubMed] [Google Scholar]
- 2.Mohammadi R, Shokatian I, Salehi M, et al. Deep learning-based auto-segmentation of organs at risk in high-dose rate brachytherapy of cervical cancer. Radiother Oncol. 2021;159:231–240. [DOI] [PubMed] [Google Scholar]
- 3.Jin D, Guo D, Ho T Y, et al. Deeptarget: Gross tumor and clinical target volume segmentation in esophageal cancer radiotherapy. Med Image Anal. 2021;68:101909. [DOI] [PubMed] [Google Scholar]
- 4.Rigaud B, Klopp A, Vedam S, et al. Deformable image registration for dose mapping between external beam radiotherapy and brachytherapy images of cervical cancer. Phys Med Biol. 2019;64(11):115023. [DOI] [PubMed] [Google Scholar]
- 5.Langerak T, Heijkoop S, Quint S, et al. Towards automatic plan selection for radiotherapy of cervical cancer by fast automatic segmentation of cone beam CT scans. Paper presented at: International Conference on Medical Image Computing and Computer-Assisted Intervention. September 14-18, 2014; Boston, MA. [DOI] [PubMed] [Google Scholar]
- 6.Staring M, Van Der Heide UA, Klein S, et al. Registration of cervical MRI using ultifeatured mutual information. IEEE Trans Med Imaging. 2009;28(9):1412–1421. [DOI] [PubMed] [Google Scholar]
- 7.Chen K, Chen W, Ni X, et al. Systematic evaluation of atlas-based autosegmentation (ABAS) software for adaptive radiation therapy in cervical cancer. Chin J Radiol Med Prot. 2015;35(2):111–113. [Google Scholar]
- 8.Ghose S, Holloway L, Lim K, et al. A review of segmentation and deformable registration methods applied to adaptive cervical cancer radiation therapy treatment planning. Artif Intell Med. 2015;64(2):75–87. [DOI] [PubMed] [Google Scholar]
- 9.Bnouni N, Rekik I, Rhim M S, et al. Dynamic multi-scale CNN forest learning for automatic cervical cancer segmentation. Paper presented at: International Workshop on Machine Learning in Medical Imaging. September 16, 2018; Granada, Spain. [Google Scholar]
- 10.Beekman C, van Beek S, Stam J, et al. Improving predictive CTV segmentation on CT and CBCT for cervical cancer by diffeomorphic registration of a prior. Med Phys. 2021;49(3):1701–1711. [DOI] [PubMed] [Google Scholar]
- 11.Yi H, Shi J, Yan B, et al. Global multi-level attention network for the segmentation of clinical target volume in the planning CT for cervical cancer. Paper presented at: 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI). April 13-16, 2021; Nice, France. [Google Scholar]
- 12.Liu Z, Liu X, Guan H, et al. Development and validation of a deep learning algorithm for auto-delineation of clinical target volume and organs at risk in cervical cancer radiotherapy. Radiother Oncol. 2020;153:172–179. [DOI] [PubMed] [Google Scholar]
- 13.Lin YC, Lin CH, Lu HY, et al. Deep learning for fully automated tumor segmentation and extraction of magnetic resonance radiomics features in cervical cancer[J]. Eur Radiol. 2020;30(3):1297–1305. [DOI] [PubMed] [Google Scholar]
- 14.Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. Paper presented at: International Conference on Medical Image Computing and Computer-Assisted Intervention. October 5-9, 2015; Munich, Germany. [Google Scholar]
- 15.Chen L C, Papandreou G, Kokkinos I, et al. Semantic image segmentation with deep convolutional nets and fully connected crfs. arXiv preprint arXiv:1412.7062, 2014. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, et al. Unet++: a nested u-net architecture for medical image segmentation. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2018:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Liu X. Medical image recognition and segmentation of pathological slices of gastric cancer based on deeplab v3+ neural network. Comput Methods Programs Biomed. 2021;207:106210. [DOI] [PubMed] [Google Scholar]
- 18.Tang W, Zou D, Yang S, et al. DSL: automatic liver segmentation with faster R-CNN and DeepLab. Paper presented at: International Conference on Artificial Neural Networks. October 4-7, 2018; Rhodes, Greece. [Google Scholar]
- 19.Chen LC, Zhu Y, Papandreou G, et al. Encoder-decoder with atrous separable convolution for semantic image segmentation. Paper presented at: Proceedings of the European conference on computer vision (ECCV). September 8-14, 2018; Munich, Germany. [Google Scholar]
- 20.Fu H, Cheng J, Xu Y, et al. Joint optic disc and cup segmentation based on multi-label deep network and polar transformation. IEEE Trans Med Imaging. 2018;37(7):1597–1605. [DOI] [PubMed] [Google Scholar]
- 21.Chen LC, Papandreou G, Kokkinos I, et al. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans Pattern Anal Mach Intell. 2017;40(4):834–848. [DOI] [PubMed] [Google Scholar]
- 22.He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. Paper presented at: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. June 2016; Las Vegas Nevada. [Google Scholar]
- 23.Huang G, Liu Z, Van Der Maaten L, et al. Densely connected convolutional networks. Paper presented at: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. July 21-26, 2017; Honolulu, Hawaii. [Google Scholar]