Summary
Background
There are a growing number of case reports of various autoimmune diseases occurring after COVID-19, yet there is no large-scale population-based evidence to support this potential association. This study provides a closer insight into the association between COVID-19 and autoimmune diseases and reveals discrepancies across sex, age, and race of participants.
Methods
This is a retrospective cohort study based on the TriNetX U.S. Collaborative Network. In the test-negative design, cases were participants with positive polymerase chain reaction (PCR) test results for SARS-CoV-2, while controls were participants who tested negative and were not diagnosed with COVID-19 throughout the follow-up period. Patients with COVID-19 and controls were propensity score-matched (1: 1) for age, sex, race, adverse socioeconomic status, lifestyle-related variables, and comorbidities. The primary endpoint is the incidence of newly recorded autoimmune diseases. Adjusted hazard ratios (aHRs) and 95% confident intervals (CIs) of autoimmune diseases were calculated between propensity score-matched groups with the use of Cox proportional-hazards regression models.
Findings
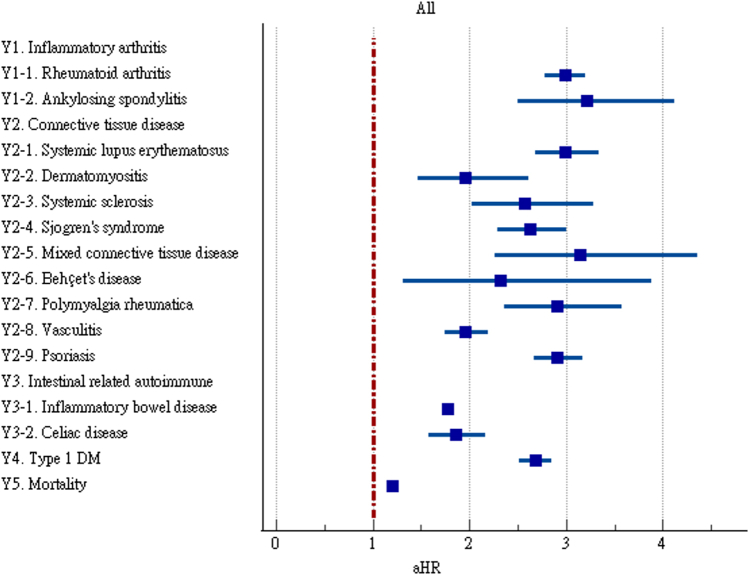
Between January 1st, 2020 and December 31st, 2021, 3,814,479 participants were included in the study (888,463 cases and 2,926,016 controls). After matching, the COVID-19 cohort exhibited significantly higher risks of rheumatoid arthritis (aHR:2.98, 95% CI:2.78–3.20), ankylosing spondylitis (aHR:3.21, 95% CI:2.50–4.13), systemic lupus erythematosus (aHR:2.99, 95% CI:2.68–3.34), dermatopolymyositis (aHR:1.96, 95% CI:1.47–2.61), systemic sclerosis (aHR:2.58, 95% CI:2.02–3.28), Sjögren's syndrome (aHR:2.62, 95% CI:2.29–3.00), mixed connective tissue disease (aHR:3.14, 95% CI:2.26–4.36), Behçet's disease (aHR:2.32, 95% CI:1.38–3.89), polymyalgia rheumatica (aHR:2.90, 95% CI:2.36–3.57), vasculitis (aHR:1.96, 95% CI:1.74–2.20), psoriasis (aHR:2.91, 95% CI:2.67–3.17), inflammatory bowel disease (aHR:1.78, 95%CI:1.72–1.84), celiac disease (aHR:2.68, 95% CI:2.51–2.85), type 1 diabetes mellitus (aHR:2.68, 95%CI:2.51–2.85) and mortality (aHR:1.20, 95% CI:1.16–1.24).
Interpretation
COVID-19 is associated with a different degree of risk for various autoimmune diseases. Given the large sample size and relatively modest effects these findings should be replicated in an independent dataset. Further research is needed to better understand the underlying mechanisms.
Funding
Kaohsiung Veterans General Hospital (KSVGH111-113).
Keywords: COVID-19, SARS-CoV-2 infection, Autoimmune disease, Electronic health records, Cohort study
Research in context.
Evidence before this study
We searched PubMed for publications without language restriction, published before September 30th, 2022, using search terms "SARS-CoV-2" or "COVID-19," and "Autoimmune Diseases," or “Arthritis, Rheumatoid,” ”Spondylitis, Ankylosing,” “Lupus Erythematosus, Systemic,” "Dermatomyositis," "Scleroderma, Systemic," "Sjogren's Syndrome," "Mixed Connective Tissue Disease," "Behcet Syndrome," "Polymyalgia Rheumatica," "Vasculitis," "Psoriasis," "Inflammatory Bowel Diseases," "Celiac Disease," or "Diabetes Mellitus, Type 1," with search terms found in abstract, title or MESH headings. A number of studies reported various autoimmune diseases occurring after coronavirus disease 2019 (COVID-19), yet there is no large-scale population-based evidence to support this potential association.
Added value of this study
We conducted this propensity score-matched cohort study to investigate the epidemiological relationship between COVID-19 and autoimmune diseases, using the data from the US Collaborative Network in TriNetX. Our findings showed that the COVID-19 cohort exhibited significantly higher risks of rheumatoid arthritis, ankylosing spondylitis, systemic lupus erythematosus, dermatopolymyositis, systemic sclerosis, Sjögren's syndrome, mixed connective tissue disease, Behçet's disease, polymyalgia rheumatica, vasculitis, psoriasis, inflammatory bowel disease, celiac disease, type 1 diabetes, and mortality, versus those without COVID-19. We further conducted subgroup analyses to determine the different risks of autoimmune diseases in COVID-19 patients based on age, sex, race, and disease severity. Sensitivity analyses were also performed and presented generally similar results globally.
Implications of all the available evidence
At the 6-month follow-up period, the risk of various autoimmune diseases is substantially higher in COVID-19 individuals than in non-COVID controls after accounting for competing risk of death. Given the large sample size and relative moderate intensity of the effects, this study should be replicated in other independent dataset from different countries to objectively examine the role of SARS-CoV-2 infection in triggering or causing autoimmune diseases.
Introduction
Autoimmune diseases, which are the third leading cause of morbidity in the industrialised world, afflict tens of millions in the United States. The idea that viruses can trigger autoimmunity in those with genetic predisposition through molecular mimicry has been simmering for a while.1,2 Viruses have been shown to have the potential to play a role in provoking and modifying the clinical manifestations of several autoimmune diseases,3 such as coxsackie virus in type 1 diabetes,4 coronaviruses in rheumatoid arthritis,5 and Epstein–Barr Virus in systemic autoimmune diseases.6 However, obtaining conclusive evidence of a connection between viral infection and subsequent autoimmune diseases is challenging, not only because it is frequently impossible to extract the virus from diseased tissues, but also because the collection of sufficient amounts of epidemiological evidence is constrained by lengthy process and geographic distances. Since the coronavirus disease 2019 (COVID-19) pandemic began in 2020, millions of people had suffered this acute infection around the same time, which provided researchers with knowledge to establish a stronger connection between the specific virus and autoimmune diseases. Meanwhile, COVID-19 has been reported to present immunological features that resemble those of autoimmune diseases, such as over-activation of mature natural killer cells, CD8+ T cells,7 and dysregulation of B cells and T cells.8 Also, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) may lead to dysregulation of immune response and increased inflammatory cytokines, such as tumor necrosis factor (TNF)-α, interleukin (IL)-1 and IL-6.9 Cases of autoimmune diseases following SARS-CoV-2 infection have been reported.10 The incidence of Kawasaki-like disease (now recognized as multisystem inflammatory syndrome in children, MIS-C) has been observed to increase during the COVID-19 epidemic.11,12 Some case reports showed the development of Guillain-Barré syndrome after a SARS-CoV-2 infection.13,14 Cases of systemic lupus erythematosus and psoriasis triggered by COVID-19 had also been reported.15, 16, 17 However, no large-scale study had assessed whether COVID-19 survivors have an increased risk of new-onset autoimmune diseases compared with those without COVID-19. The aim of this study was to investigate the association between COVID-19 and subsequent autoimmune diseases and whether differences in sex, age, and race had an effect on this association.
Methods
Study design and data source
In this retrospective cohort study, we used the US Collaborative Network from 48 global healthcare organizations (HCOs) in the TriNetX Research Network. The TriNetX database, which holds the largest global COVID-19 dataset, is a living ecosystem of real-world data and evidence for life sciences and healthcare. It contains de-identified electronic health records of more than 250 million participants from more than 120 HCOs. Details of TriNetX are available on the website (https://trinetx.com/?mc_cid=7e2ecd5bc5&mc_eid=%5BUNIQID%5D). TriNetX has been a useful database for epidemiological studies.18, 19, 20 The available data included information of demographics, diagnoses (based on the International Classification of Diseases, Tenth Revision, Clinical Modification, ICD-10-CM codes), procedures (based on the International Classification of Diseases, Tenth Revision, Procedure Coding System, ICD-10-PCS or Current Procedural Terminology, CPT), medication (based on the codes in Veterans Affairs National Formulary), laboratory measurements (coded in Logical Observation Identifiers Names and Codes, LOINC), and healthcare utilization. We used the U.S. Collaborative Network, a subset of TriNetX network that includes 48 HCOs, for the primary analysis. Data analysis was done in June, 2022, and we limited the study period to January 1st, 2020, through December 31st, 2021.
The TriNetX platform is compliant with the Health Insurance Portability & Accountability Act and General Data Protection Regulation. The Western Institutional Review Board has granted TriNetX a waiver of informed consent since this platform only aggregated counts and statistical summaries of de-identified information. The use of TriNetX for this study was approved under the authority of the Institutional Review Board of Chung Shan Medical University Hospital (CSMUH No: CS2-21176). The reporting is informed by the REporting of studies Conducted using Observational Routinely-collected health Data (RECORD) Statement for cohort studies.
Participants
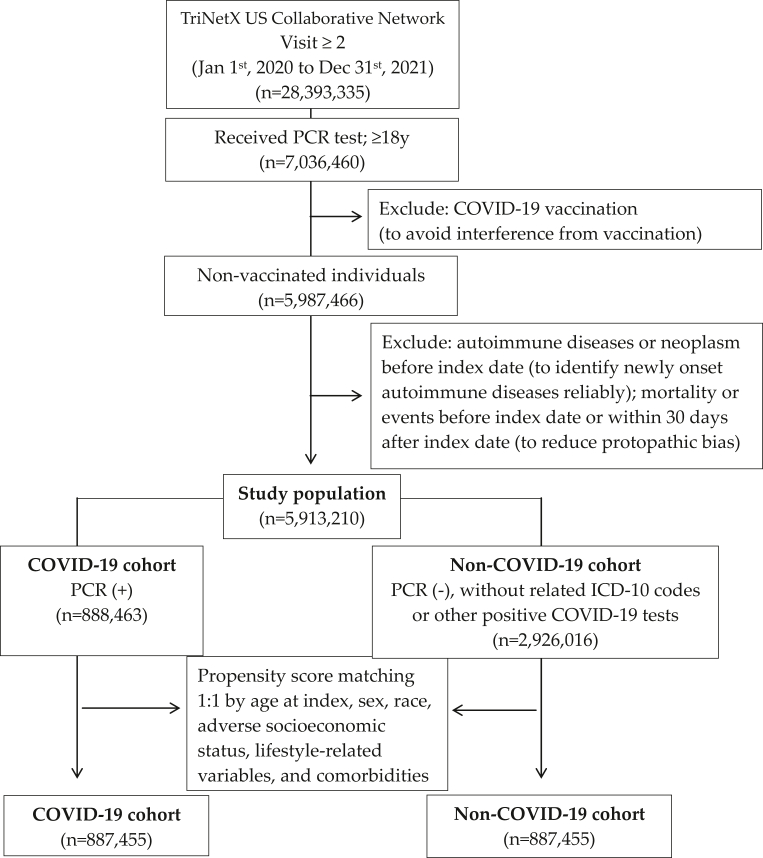
To reduce detection bias, we only included adult patients (≥18 years old) who had at least 2 healthcare visits and had received PCR tests during the study period in the TriNetX database. The date of the PCR test was set as the index date. Since the association between COVID-19 vaccination and subsequent autoimmune phenomena is controversial,21,22 we intentionally did not include patients who had received COVID-19 vaccination throughout the study for clarity of interpretation. Individuals who were diagnosed with autoimmune diseases or neoplasm before index date, and who diagnosed with autoimmune disease or died within 30 days after index date were excluded. The COVID-19 group was identified based on the positive result of a PCR test (Supplement material for detailed codes). A flowchart of the cohort construction is provided in Fig. 1. The control group consisted of participants without COVID-19, who we defined as those with negative PCR tests, negative results for other COVID-19-related tests other than PCR tests, and those who had never had ICD-10 codes for COVID-19. Propensity score matching 1:1 for age, sex, race, adverse socioeconomic status, lifestyle-related variables and comorbidities, was used.
Fig. 1.
Flowchart of cohort construction.
Procedures
To adjust for the difference in baseline characteristics between the two cohorts, we incorporated the following covariate factors. To obtain the information on baseline characteristics, we traced the records within one year before the index date until one day before the index date. Demographic covariates included age at index, sex, race, and adverse socioeconomic determinants of health (represented by problem related to housing and economic circumstances (ICD-10 code Z59), problems related to employment and unemployment (ICD-10 code Z56), problems related to education and literacy (ICD-10 code Z55), or occupational exposure to risk factors (ICD-10 code Z57)). Lifestyle-related variables included tobacco use (ICD-10 code Z72.0, as a proxy for smoking), nicotine dependence (ICD-10 code F17, as a proxy for smoking) and alcoholic liver disease (ICD-10 code K70, as a proxy for alcohol use). Baseline comorbidities included type 2 diabetes (ICD-10 code E11), vitamin D deficiency (ICD-10 code E55), hyperlipidemia (ICD-10 code E78.5), asthma (ICD-10 code J45), depression (ICD-10 code F32), chronic kidney disease (ICD-10 code N18), sleep disorder (ICD-10 code G47), and psychoactive substance use (ICD-10 code F10–F19). Laboratory measurements including creatinine (≥1.5 mg/dL), hemoglobin (≥12 g/dL), body mass index (BMI; obesity was defined as BMI ≥ 30 kg/m2), hemoglobin A1C (≥7%), and C-reactive protein (≥40 mg/dL) were explored.23 The number of individuals with missing data was reported in Table 1.
Table 1.
Baseline characteristics of study subjects (before and after PSM matching).
| Before matching |
After matching |
|||||||
|---|---|---|---|---|---|---|---|---|
| COVID-19 cohort (n = 888,463) | Non COVID-19 cohort (n = 2,926,016) | Std diff. | p value | COVID-19 cohort (n = 887,455) | Non COVID-19 cohort (n = 887,455) | Std diff. | p value | |
| Age at index | ||||||||
| Mean ± SD | 45.2 ± 17.6 | 46.7 ± 18.5 | 0.083 | <0.001 | 45.2 ± 17.6 | 45.1 ± 17.6 | 0.005 | 0.001 |
| Sex | ||||||||
| Female | 510,200 (57.4) | 1,633,065 (55.8) | 0.033 | <0.001 | 509,494 (57.4) | 508,254 (57.3) | 0.003 | 0.060 |
| Male | 377,971 (42.5) | 1,291,191 (44.1) | 0.032 | <0.001 | 377,669 (42.6) | 378,916 (42.7) | 0.003 | 0.058 |
| Missing | 292 (00.0) | 1760 (00.1) | 0.013 | <0.001 | 292 (00.0) | 285 (00.0) | <0.001 | 0.771 |
| Race | ||||||||
| White | 572,496 (64.4) | 1,916,777 (65.5) | 0.022 | <0.001 | 571,870 (64.4) | 570,595 (64.3) | 0.003 | 0.046 |
| Black or African American | 182,253 (20.5) | 553,192 (18.9) | 0.040 | <0.001 | 181,942 (20.5) | 183,503 (20.7) | 0.004 | 0.004 |
| Asian | 16,737 (01.9) | 62,705 (02.1) | 0.018 | <0.001 | 16,763 (01.9) | 16,764 (01.9) | <0.001 | 0.877 |
| American Indian | 3082 (00.3) | 9995 (00.3) | 0.001 | 0.454 | 3081 (00.3) | 3008 (00.3) | 0.001 | 0.349 |
| Native Hawaiian | 1553 (00.2) | 4339 (00.1) | 0.007 | <0.001 | 1549 (00.2) | 1579 (00.2) | 0.001 | 0.591 |
| Missing or unknown | 112,342 (12.6) | 379,008 (13.0) | 0.009 | <0.001 | 112,250 (12.7) | 112,006 (12.6) | 0.001 | 0.540 |
| Social economic status | ||||||||
| Housing/economic circumstances problem | 5242 (00.6) | 14,409 (00.5) | 0.013 | <0.001 | 5187 (00.6) | 4168 (00.5) | 0.016 | <0.001 |
| Employment and unemployment problems | 2124 (00.2) | 5097 (00.2) | 0.014 | <0.001 | 2066 (00.2) | 1663 (00.2) | 0.010 | <0.001 |
| Problems related to education and literacy | 307 (00.0) | 700 (00.0) | 0.006 | <0.001 | 305 (00.0) | 211 (00.0) | 0.006 | <0.001 |
| Occupational exposure to risk factors | 382 (00.0) | 729 (00.0) | 0.010 | <0.001 | 375 (00.0) | 268 (00.0) | 0.006 | <0.001 |
| Lifestyle | ||||||||
| Tobacco use (smoking) | 18,356 (02.1) | 46,984 (01.6) | 0.034 | <0.001 | 18,084 (02.0) | 16,441 (01.9) | 0.013 | <0.001 |
| Nicotine dependence (smoking) | 48,280 (05.4) | 171,048 (05.8) | 0.018 | <0.001 | 48,143 (05.4) | 46,543 (05.2) | 0.008 | <0.001 |
| Alcohol liver disease (alcohol drinking) | 2662 (00.3) | 10,471 (00.4) | 0.010 | <0.001 | 2654 (00.3) | 2019 (00.2) | 0.014 | <0.001 |
| Comorbidities (Yes) | ||||||||
| Type 2 diabetes mellitus | 83,751 (09.4) | 188,593 (06.4) | 0.110 | <0.001 | 82,856 (09.3) | 82,661 (09.3) | 0.001 | 0.615 |
| Vitamin D deficiency | 40,384 (04.5) | 71,514 (02.4) | 0.115 | <0.001 | 39,506 (04.5) | 40,363 (04.5) | 0.005 | 0.002 |
| Hyperlipidemia | 82,991 (09.3) | 200,600 (06.9) | 0.091 | <0.001 | 82,179 (09.3) | 81,712 (09.2) | 0.002 | 0.226 |
| Asthma | 47,052 (05.3) | 106,079 (03.6) | 0.081 | <0.001 | 46,545 (05.2) | 45,822 (05.2) | 0.004 | 0.015 |
| Depression | 59,941 (06.7) | 140,009 (04.8) | 0.084 | <0.001 | 59,261 (06.7) | 56,987 (06.4) | 0.010 | <0.001 |
| Chronic kidney disease | 38,775 (04.4) | 77,650 (02.7) | 0.093 | <0.001 | 38,002 (04.3) | 35,837 (04.0) | 0.012 | <0.001 |
| Sleep disorder | 64,454 (07.3) | 143,362 (04.9) | 0.099 | <0.001 | 63,623 (07.2) | 62,816 (07.1) | 0.004 | 0.019 |
| Psychoactive substance use | 67,878 (07.6) | 248,822 (08.5) | 0.032 | <0.001 | 67,694 (07.6) | 63,609 (07.2) | 0.018 | <0.001 |
| Laboratory | ||||||||
| BMI | ||||||||
| n (%) | 241,939 (27.2) | 749,036 (25.6) | 241,219 (27.2) | 245,367 (27.6) | ||||
| mean ± SD, kg/m2 | 30.5 ± 7.1 | 29.5 ± 6.8 | 0.144 | <0.001 | 30.5 ± 7.1 | 30.3 ± 7.0 | 0.028 | <0.001 |
| ≥30 kg/m2, n (%) | 126,078 (14.2) | 334,120 (11.4) | 0.083 | <0.001 | 125,459 (14.1) | 124,525 (14.0) | 0.003 | 0.044 |
| Missing | 646,524 (72.8) | 2,176,980 (74.4) | 646,236 (72.8) | 642,088 (72.4) | ||||
| CRP | ||||||||
| n (%) | 53,292 (06.0) | 79,610 (02.7) | 53,031 (06.0) | 25,957 (02.9) | ||||
| mean ± SD, mg/L | 36.2 ± 59.7 | 29.6 ± 58.6 | 0.111 | <0.001 | 36.2 ± 59.7 | 28.9 ± 57.8 | 0.125 | <0.001 |
| ≥40 mg/L, n (%) | 18,699 (02.1) | 17,165 (00.6) | 0.132 | <0.001 | 18,594 (02.1) | 5450 (00.6) | 0.128 | <0.001 |
| Missing | 835,171 (94.0) | 2,846,406 (97.3) | 834,424 (94.0) | 861,498 (97.1) | ||||
| Creatinine | ||||||||
| n (%) | 379,705 (42.7) | 1,126,839 (38.5) | 378,775 (42.7) | 355,248 (40.0) | ||||
| mean ± SD, mg/dl | 1.1 ± 1.9 | 1.0 ± 1.9 | 0.014 | <0.001 | 1.1 ± 1.9 | 1.1 ± 1.9 | 0.002 | 0.351 |
| ≥1.5 mg/dL, n (%) | 47,258 (05.3) | 112,483 (03.8) | 0.071 | <0.001 | 46,775 (05.3) | 38,611 (04.4) | 0.043 | <0.001 |
| Missing | 508,758 (57.3) | 1,799,177 (61.5) | 508,680 (57.3) | 532,207 (60.0) | ||||
| Hemoglobin | ||||||||
| n (%) | 364,755 (41.1) | 1,127,923 (38.5) | 363,828 (41.0) | 355,105 (40.0) | ||||
| mean ± SD, g/dL | 13.0 ± 2.1 | 13.0 ± 2.2 | 0.008 | <0.001 | 13.0 ± 2.1 | 13.0 ± 2.2 | 0.009 | <0.001 |
| ≥12 g/dL, n (%) | 308,009 (34.7) | 927,016 (31.7) | 0.063 | <0.001 | 307,303 (34.6) | 290,399 (32.7) | 0.040 | <0.001 |
| Missing | 523,708 (58.9) | 1,798,093 (61.5) | 523,627 (59.0) | 532,350 (60.0) | ||||
Std diff: standardized difference; bold font represents a standardized difference > 0.1; If the patient is less or equal to 10, results show the count as 10.
BMI: Body mass index; CRP: C reactive protein; Propensity score matching was performed on age at index, gender, race, problems related to housing and economic circumstances (proxy to social economic status), problems related to employment and unemployment, problems related to education and literacy, occupational exposure to risk factors, type 2 diabetes mellitus, vitamin D deficiency, hyperlipidemia, asthma, alcoholic liver disease, nicotine dependence, tobacco use (proxy to smoking), mental and behavioral disorders due to psychoactive substance use, depressive episode, chronic kidney disease (CKD), sleep disorders, and BMI characteristic(s).
Outcomes
The main outcomes of the study were newly diagnosed autoimmune diseases identified by the corresponding ICD-10 codes in the database, including rheumatoid arthritis (ICD-10 code M05-M06), ankylosing spondylitis (ICD-10 code M45), systemic lupus erythematosus (ICD-10 code M32), dermatopolymyositis (ICD-10 code M33), systemic sclerosis (ICD-10 code M34), Sjögren's syndrome (ICD-10 code M35.0), mixed connective tissue disease (ICD-10 code M35.1), Behçet's disease (ICD-10 code M35.2), polymyalgia rheumatica (ICD-10 code M35.3), vasculitis (ICD-10 code M30-31 or L95), psoriasis (ICD-10 code L40), inflammatory bowel disease (ICD-10 code K50-52), celiac disease (ICD-10 code K90.0), and type 1 diabetes mellitus (ICD-10 code E10). To address competing risks, we also included mortality as a secondary outcome. We used a 30-day timeframe as a washout period for the outcome measures to alleviate protopathic bias (i.e., increased vigilance and earlier diagnosis of an autoimmune disease after a SARS-CoV-2 infection).24 Both cohorts were followed after 30 day after the index till 6 months to estimate the risks of incident autoimmune disease.
Statistical analysis
We used propensity scores matching to generate groups with balanced baseline characteristics. We adopted TriNetX built-in function and matched the two groups at a 1:1 ratio by greedy nearest neighbor matching for age at index, sex, race, adverse socioeconomic status (problem related to housing and economic circumstances, problems related to employment and unemployment, problems related to education and literacy, occupational exposure to risk factors), lifestyle-related proxy variables (tobacco use, nicotine dependence, and alcoholic liver disease), and comorbidities. We evaluated the balance of baseline characteristics in the propensity score-matched populations by using standardized mean differences (SMD). Any variable with a SMD <0.1 was considered well-matched. We used adjusted hazard ratio (aHRs) to quantify the relative risks of autoimmune diseases based on the time-to-event analysis for the COVID-19 and control groups. Cox proportional hazards models were performed to calculate the HRs and associated 95% confidence intervals (95%CI), together with the test for proportionality, using R's Survival package v3.2-3. The proportional hazard assumption was tested using the generalized Schoenfeld approach built in the TriNetX network. The Kaplan–Meier method was used for the survival probability. Statistical significance was defined as two-sided p-value < 0.05.
Subgroup analyses investigated whether the risks of autoimmune diseases among COVID-19 survivors differed by sex, age, and race. Furthermore, as studies have shown that the presence of auto-antibodies is associated with the severity of COVID-19,25 we performed analysis stratified by inpatient/outpatient (as a proxy for COVID-19 severity) to clarify whether differences in the severity of COVID-19 could lead to different outcomes. In addition, as there may be geographical differences between countries regarding the prevalence of COVID-19, the structure of healthcare services, and accessibility of healthcare resources, sensitivity analysis was also performed with the Global Network and the EMEA (Europe, Middle East and Africa) Network to examine the consistency of results. Furthermore, competing risks will occur when patients experience one or more severe outcomes which might compete with the outcome of interest. To address competing risks, we included mortality as a competing event for each outcome in our study, based on the method proposed by Manja et al.26 The reporting is informed by the REporting of studies Conducted using Observational Routinely-collected health Data (RECORD) Statement for cohort studies.27
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Baseline characteristics of the study individuals
After propensity score matching, patients diagnosed with COVID-19 (N = 887,455) and a control group of test-negative participants (N = 887,455) were identified in the study. The selection process is demonstrated in Fig. 1. The demographic characteristics, comorbidities, and laboratory measurements of the COVID-19 and control groups before and after propensity score matching are presented in Table 1. The mean age of the participants in the COVID-19 cohort was approximately 45.2 years (standard deviation: 17.6 years) at index after matching. Approximately 57.4% of the COVID-19 individuals were women, and the major race was White (64.4%). The two groups were well-matched regarding the distribution of demographics, comorbidities and laboratory measurements (SMD<0.1).
Incidence of autoimmune diseases in the COVID-19 and control groups
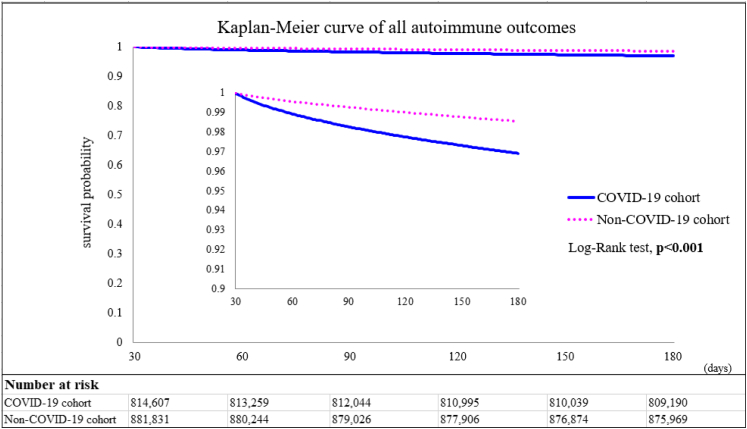
We estimated the risks of autoimmune diseases in the COVID-19 and control groups upon 6-month follow-up (Fig. 2 and Supplementary Table S1). The COVID-19 cases were found to have significant higher risks of rheumatoid arthritis (aHR:2.98, 95% CI:2.78–3.20), ankylosing spondylitis (aHR:3.21, 95% CI:2.50–4.13), systemic lupus erythematosus (aHR:2.99, 95% CI:2.68–3.34), dermatopolymyositis (aHR:1.96, 95% CI:1.47–2.61), systemic sclerosis (aHR:2.58, 95% CI:2.02–3.28), Sjögren's syndrome (aHR:2.62, 95% CI:2.29–3.00), mixed connective tissue disease (aHR:3.14, 95% CI:2.26–4.36), Behçet's disease (aHR:2.32, 95% CI:1.38–3.89), polymyalgia rheumatica (aHR:2.90, 95% CI:2.36–3.57), vasculitis (aHR:1.96, 95% CI:1.74–2.20), psoriasis (aHR:2.91, 95% CI:2.67–3.17), inflammatory bowel disease (aHR:1.78, 95%CI:1.72–1.84), celiac disease (aHR:2.68, 95% CI:2.51–2.85), type 1 diabetes (aHR:2.68, 95%CI:2.51–2.85) and mortality (aHR:1.20, 95% CI:1.16–1.24). The Kaplan–Meier curves of all the autoimmune disease outcomes also indicated difference of probability between the two cohorts (Log–Rank test, p < 0.001, Fig. 3).
Fig. 2.
Forest plot of outcomes.
Fig. 3.
Kaplan–Meier curves of all the autoimmune disease outcomes.
Subgroup analyses
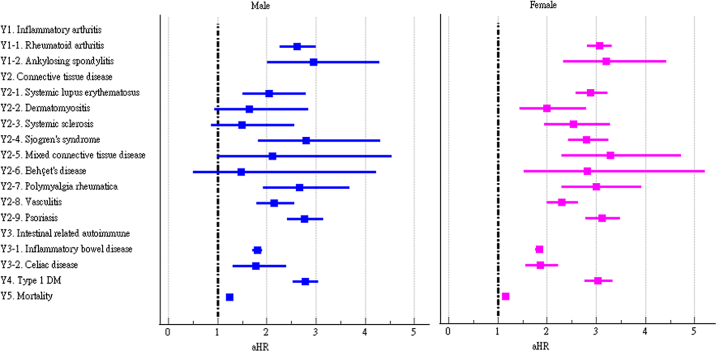
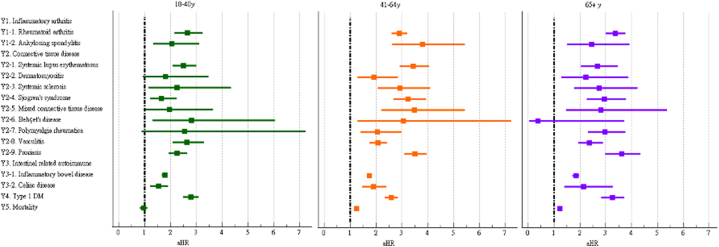
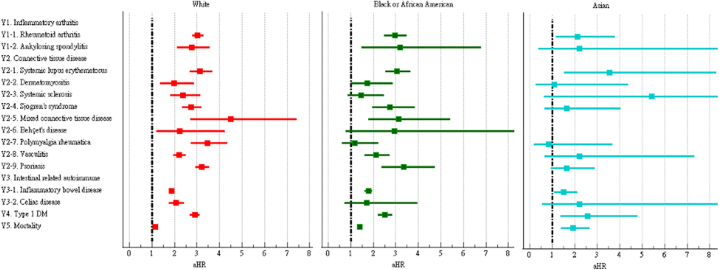
We further examined the risk of incident autoimmune diseases in subgroups based on sex, age, and race. The risks of most autoimmune diseases were similar between male and female COVID-19 patients (Fig. 4 and Supplementary Table S2). However, the risks of dermatomyositis, systemic sclerosis, mixed connective tissue disease, and Behçet's disease were only significant in female COVID-19 patients. The risks of autoimmune diseases among COVID-19 patients were generally similar across age subgroups (Fig. 5 and Supplementary Table S3). However, the risks of dermatomyositis, polymyalgia rheumatica, and mortality were not significant in the group aged 18–40 years, while the risk of Behçet's disease was not significant in the group aged ≥65 years. The risks of autoimmune diseases among COVID-19 patients were heterogenous across racial subgroups (Fig. 6 and Supplementary Table S4). Compared to control group, the COVID-19 cohort had a significantly higher risk of all autoimmune diseases and mortality (aHR:1.15, 95%CI:1.10–1.20) in Whites, a higher risk of psoriasis (aHR:3.36, 95%CI:2.37–4.76) and ankylosing spondylitis in Blacks (aHR:3.18, 95%CI:1.49–6.79), and a higher risk of systemic lupus erythematosus in Asians (aHR:3.56, 95%CI:1.53–8.29).
Fig. 4.
Forest plot of outcomes stratified by sex.
Fig. 5.
Forest plot of outcomes stratified by age.
Fig. 6.
Forest plot of outcomes stratified by race.
Sensitivity analyses
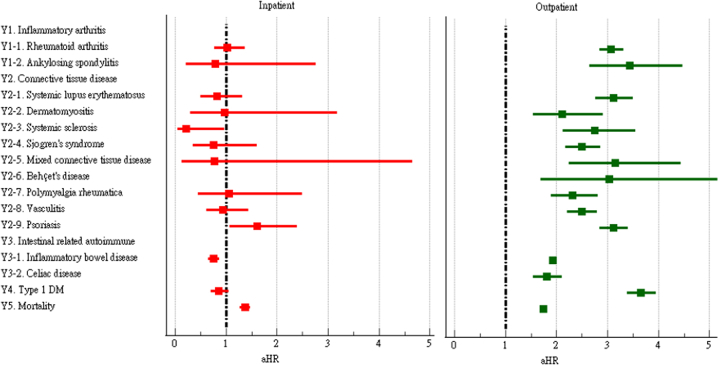
In our analysis stratified by inpatient (patients who were hospitalized within four weeks after the PCR tests) and outpatient care, some discrepancy in risks of incident autoimmune diseases were found between inpatient and outpatient COVID-19 patients (Fig. 7 and Supplementary Table S5). COVID-19 was associated with increased risks of all autoimmune diseases and also mortality among outpatients. COVID-19 inpatients were found with increased risks of psoriasis (aHR:1.60, 95%CI:1.07–2.40) and mortality (aHR:1.37, 95%CI:1.27–1.48), but significantly lower risks of systemic sclerosis (aHR:0.21, 95%CI:0.05–0.97) and inflammatory bowel disease (aHR:0.76, 95%CI:0.65–0.87). The results of our additional study using the Global Network were generally consistent with our main findings from the analysis using the U.S. Collaborative Research Network (Supplementary Table S6). However, we found different results while using the EMEA (Europe, Middle East and Africa) Network. After accounting for competing risks, COVID-19 was still significantly associated with increased risks of all autoimmune diseases (Supplementary Table S7, Supplementary Fig. S1). However, the adjusted hazard ratios were lower when compared to the original model without accounting for competing risks.
Fig. 7.
Forest plot of outcomes stratified by severity of disease.
Discussion
The current large propensity scores-matched study of a multi-institutional U.S. Collaborative Research Network showed that patients with a positive PCR test result for COVID-19 had significantly higher risks of autoimmune diseases including rheumatoid arthritis, ankylosing spondylitis, systemic lupus erythematosus, dermatomyositis, systemic sclerosis, Sjögren's syndrome, mixed connective tissue disease, Behçet's disease, polymyalgia rheumatica, vasculitis, psoriasis, inflammatory bowel disease, celiac disease, type 1 diabetes, and also mortality, when compared to “PCR test-negative” non-COVID-19 controls. The results were cross validated in another dataset from TriNetX. The results of subgroup analysis by sex and age were generally consistent. The results of subgroup analysis by race indicated some racial disparity. When compared to their non-COVID-19 counterparts, COVID-19 patients were associated with a significant higher risk of all autoimmune diseases and mortality in Whites, a higher risk of psoriasis and ankylosing spondylitis in Blacks, and a higher risk of systemic lupus erythematosus in Asians. Also, some discrepancy in risks of autoimmune diseases was found between inpatient and outpatient COVID-19 patients. COVID-19 was associated with increased risks of all autoimmune diseases and also mortality among outpatients, while COVID-19 inpatients were found with increased risks of psoriasis and mortality, but significantly lower risks of systemic sclerosis and inflammatory bowel disease.
Viruses play a significant role in the environmental factors that affect human immune system. Cytomegalovirus28 and Epstein–Barr virus6 are examples of viruses that have been linked to numerous autoimmune diseases, and now SARS-CoV-2 has the potential to be added to the list. The definite mechanisms underlying such phenomena are unknown. According to recent research, viruses can trigger autoimmunity through a number of different mechanisms, including molecular mimicry,29 epitope spreading,30 and bystander activation.31,32 We hypothesized that the prolonged inflammation in COVID-19 might trigger the immune system to create antibodies against virus antigens that share structural similarities with self-antigens and further lead to a cross-reactive response against both self-antigens and non-self-antigens. Also, the hyper-stimulation and dysregulation of inflammation and immune response following SARS-CoV-2 infection may contribute to other environmental disturbances that lead to the observed disease in susceptible individuals. Cañas proposed the theory that COVID-19 triggers autoimmune conditions by two factors: temporary impairment of innate and acquired immunity resulting in a loss of self-tolerance to self-antigens, and an inappropriate immune reconstitution in people with predisposing conditions of autoimmunity.33 Besides, the above-mentioned autoimmune phenomenon may also contribute to the development of post-COVID-19 syndrome (PCS).34 Given the current evidence on latent autoimmunity in PCS, we could further anticipate a higher incidence of autoimmune diseases in COVID-19 cases with enough follow-up time.35 In addition, there are other potential mechanisms that could contribute to the association between COVID-19 and autoimmunity diseases. For example, previous studies have reported excessive neutrophil extracellular traps being involved in rheumatoid arthritis and myositis36 and neutrophil extracellular traps has also been associated with COVID-19 pathogenesis.37 Contrarily, there is also mounting evidence that viruses could play a protective role against autoimmunity, whereby viral infections trigger regulatory immune responses, which in turn prevent the onset of autoimmune reactions.38 It is reasonable that the dual impact of viral infections on autoimmunity is coordinated by different host, viral and environmental factors.39
Furthermore, angiotensin-converting enzyme 2 (ACE-2), a crucial viral fusion protein of SARS-CoV-2 is widely expressed by vascular endothelial cells, and therefore, it has been proposed that SARS-CoV-2 invades the vascular endothelium, causing vasculitis. SARS-CoV-2 binds to the ACE-2 receptors and leads to immune activation and redistribution of immune cells. ACE-2 receptors are known to be abundantly present in the epithelia of the lung and small intestine.40 Interestingly, our study found that the COVID-19 cohort was associated with lower risks of inflammatory bowel disease and systemic sclerosis in inpatients. Although it is still uncertain whether infections can cause inflammatory bowel disease, current knowledge of the pathophysiology of the disease does suggest a connection between infection and the development of inflammatory bowel disease.41 However, the results of our study showed that inpatient COVID-19 patients had a lower risk of inflammatory bowel disease,42 which is in line with earlier research. According to Hadi et al., COVID-19 likely creates a barrier to procedures like colonoscopy and flexible sigmoidoscopy, and consequently results in a short-term underdiagnosis of inflammatory bowel disease.43
Systemic sclerosis is an uncommon autoimmune disease that primarily affects individuals aged 40–50 years. It is characterized by fibrosis of the skin and internal organs, along with vascular damage and immune system dysregulation.44 It is well recognized that viral infections and interferons play an important role in the pathogenesis of systemic sclerosis in genetically predisposed patients.39,45,46 Fineschi presented a case of COVID-19 infection with cutaneous and gastrointestinal manifestation, autoantibody production, and radiological findings consistent with the diagnosis of systemic sclerosis.47 It was suspected that COVID-19 had brought on systemic sclerosis in that patient due to a potential genetic susceptibility. However, we found a lower risk of systemic sclerosis in the inpatient COVID-19 patients in our study. We speculated that the under-diagnosis of systemic sclerosis may be a possible explanation, due to a lack of accurate follow-up to confirm the systemic sclerosis diagnosis. On the other hand, this may point to different pathogenetic pathways by which SARS-CoV-2 downregulates ACE-2 in targeted cells, causing an excess of angiotensin II to be produced, which in turn promotes inflammation, vasoconstriction, cell proliferation, and, ultimately, pulmonary fibrosis.48 It could be hypothesized that the discrepancy we found in our study could have been caused by systemic sclerosis having a distinct genetic predisposition than other systemic autoimmune diseases.
The impact of COVID-19 on the development of autoimmune diseases may be different across racial subgroups; however, the relevant existing literature is sparse. Overall, Black patients appeared to be a more susceptible population for ankylosing spondylitis and psoriasis, while Asian patients are more susceptible to systemic lupus erythematosus when getting COVID-19. Factors that may account for an increased risk of autoimmune diseases include genetic features, lifestyle characteristics, and environmental exposures to bacteria, viruses, and toxic doses of drugs and metals. In addition, it is possible that genetic predisposition and environmental factors will interact.47 According to the study by Jamalyaria et al., Black patients have more severe disease than White patients with ankylosing spondylitis, as well as a lower frequency of HLA-B27.49 Black patients may have more severe disease due to genetic differences, and as a result, these more severely affected Black patients are more likely to be diagnosed with ankylosing spondylitis. Bonometti et al. presented a case of systemic lupus erythematosus with vasculitis triggered by SARS-CoV-2 infection in Italy.15 The development and activity of the disease depends on a combination of genetic predisposition, environmental stimuli, and hormonal environment.50 Furthermore, Zhang et al. found both genetic similarities and differences across ethnical groups, adding to the growing body of evidence supporting a genetic basis of the high incidence of SLE in people of Asian ancestry.51
Our study has several unique merits. As study design and case ascertainment are two of the most important elements for reaching solid conclusions in longitudinal study, we used “test-negative design” for validity of exposure and control groups.52 The potential confounding bias has been accounted for through propensity score matching. In addition, we did multiple sensitivity analyses to corroborate the findings as well as subgroup analyses to characterize subpopulations at high risk for future autoimmune disease surveillance. The risks of incident autoimmune disease outcomes were evident worldwide as we found generally consistent results using the Global Network. However, different results were found using the EMEA (Europe, Middle East and Africa) Network, which could be ascribed to regional differences in COVID-19 prevalence, the structure of healthcare services, and healthcare accessibility. Further research is encouraged to corroborate these findings in an independent dataset.
We recognized several limitations in this study. First, the database we used has weaknesses inherent to an electronic health records study. The definition of autoimmune disease diagnoses relied on ICD-10-CM codes reported by physicians, which may be less accurate than those made on a clinical basis, although we used validated definitions to avoid bias. As socioeconomic status and lifestyles habits are not available in the database, we used proxy variables, and thus the validity of adjusting for these confounders may be biased. Also, the database restricted the source population to the adult patients who had medical insurance and had healthcare visits during the study period. We further excluded individuals who were diagnosed with autoimmune diseases or neoplasm before index date, who were diagnosed with autoimmune diseases or died within 30 days after index date, and those who were vaccinated with a COVID-19 vaccine. The generalizability of our conclusions is therefore limited. TriNetX did not provide the identities of participating HCOs and information about their contributions to the dataset, and therefore we were not able to account for heterogeneity at the hospital level. Participants with loss to follow-up and missing values of laboratory measurements could also bias our findings. We included only participants with at least 2 visits to mitigate the effect of loss to follow-up. Second, for patients with newly diagnosed autoimmune diseases, levels and types of autoantibodies produced during COVID-19 were not available. The particular variant form of SARS-CoV-2, the associated subsequent autoimmune diseases, and the nature of the COVID-19-related molecular pattern involved in the process also required further survey. Third, this study is unable to differentiate between COVID-19 as a specific or non-specific trigger for subsequent autoimmune diseases. That is, those with autoimmune diseases diagnosed after COVID-19 inevitably develop the disease in response to other environmental triggers even if they are not infected with SARS-CoV-2. Fourth, many people avoid the health care system during the pandemic, and therefore the misclassification bias and surveillance bias were concerns for this study. However, we used a test-negative design to ensure that the controls did undergo PCR tests and were free of COVID-19 throughout the study period. We believe that misclassification bias and surveillance bias were minimal in this study. In addition, we performed goodness-of-fit test between our case group and the official COVID-19 laboratory-confirmed cases (from Centers for Disease Control and Prevention, CDC, COVID-19 data tracker; https://covid.cdc.gov/covid-data-tracker/#cases-deaths-trends-by-demographic). The results indicated that the COVID-19 cases in the present study may not be significantly different from the CDC laboratory-confirmed COVID-19 cases (effect sizes of Pearson's r = 0.0735, 0.081, and 0.0289 for gender, age, and ethnicity, respectively; Supplementary Table S8). Last but not least, reverse causality is possible in this study since patients with early or undiagnosed autoimmune diseases may be more likely to be infected by SARS-CoV-2. To avoid reverse causality, we initiated the follow-up 30 days after the test, and continued until 6 months.
In conclusion, our preliminary data suggest that COVID-19 is associated with a significantly different risk of various autoimmune diseases. It is crucial for physicians to have relevant notions and to recognize these autoimmune manifestations in order to respond appropriately in the ongoing pandemic and long-term post-pandemic phase. The impact of vaccination on the development of autoimmune diseases should also be studied in the future.
Contributors
All authors were involved in drafting the article or revising it, and all authors approved the final version to be published.
Study conception and design: RC, J C-CW.
Accessed and verified the underlying data: S-IW, RC.
Analysis and interpretation of data: RC, TY-TC, S-IW, Y-MH, H-YC, JC-CW.
Writing (original draft preparation): RC, TY-TC, S-IW, Y-MH, H-YC.
Writing (review and editing): JC-CW.
Data sharing statement
The data that support the findings of this study are available from the TriNetX Analytics Network. https://live.trinetx.com/tnx/study/112293/analytics/63171fa50fb1cf4c2da08ada/outcomes/results.
Declaration of interests
All authors declare no competing interests.
Acknowledgments
This study was funded in part by Kaohsiung Veterans General Hospital (KSVGH111-113).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2022.101783.
Contributor Information
Yao-Min Hung, Email: ymhung@vhtt.gov.tw.
Cheng-Chung James Wei, Email: wei3228@gmail.com.
Appendix A. Supplementary data
References
- 1.Rojas M., Restrepo-Jiménez P., Monsalve D.M., et al. Molecular mimicry and autoimmunity. J Autoimmun. 2018;95:100–123. doi: 10.1016/j.jaut.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 2.Sogkas G., Atschekzei F., Adriawan I.R., Dubrowinskaja N., Witte T., Schmidt R.E. Cellular and molecular mechanisms breaking immune tolerance in inborn errors of immunity. Cell Mol Immunol. 2021;18(5):1122–1140. doi: 10.1038/s41423-020-00626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussein H.M., Rahal E.A. The role of viral infections in the development of autoimmune diseases. Crit Rev Microbiol. 2019;45(4):394–412. doi: 10.1080/1040841X.2019.1614904. [DOI] [PubMed] [Google Scholar]
- 4.Shih W.L., Tung Y.C., Chang L.Y., Fang C.T., Tsai W.Y. Increased incidence of pediatric type 1 diabetes with novel association with coxsackievirus A species in young children but declined incidence in adolescents in Taiwan. Diabetes Care. 2021;44(7):1579–1585. doi: 10.2337/dc20-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joo Y.B., Lim Y.H., Kim K.J., Park K.S., Park Y.J. Respiratory viral infections and the risk of rheumatoid arthritis. Arthritis Res Ther. 2019;21(1):199. doi: 10.1186/s13075-019-1977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houen G., Trier N.H. Epstein-barr virus and systemic autoimmune diseases. Front Immunol. 2021;11 doi: 10.3389/fimmu.2020.587380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varchetta S., Mele D., Oliviero B., et al. Unique immunological profile in patients with COVID-19. Cell Mol Immunol. 2021;18(3):604–612. doi: 10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodruff M.C., Ramonell R.P., Nguyen D.C., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat Immunol. 2020;21(12):1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin C., Zhou L., Hu Z., et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novelli L., Motta F., De Santis M., Ansari A.A., Gershwin M.E., Selmi C. The JANUS of chronic inflammatory and autoimmune diseases onset during COVID-19 - a systematic review of the literature. J Autoimmun. 2021;117 doi: 10.1016/j.jaut.2020.102592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verdoni L., Mazza A., Gervasoni A., et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395(10239):1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salah N.B., Lahouel I., Mariem C.B., et al. Multisystem inflammatory syndrome in children associated with erythema multiforme-like eruption following COVID-19. Int J Rheum Dis. 2022;25(3):375–377. doi: 10.1111/1756-185X.14294. [DOI] [PubMed] [Google Scholar]
- 13.Toscano G., Palmerini F., Ravaglia S., et al. Guillain-barré syndrome associated with SARS-CoV-2. N Engl J Med. 2020;382(26):2574–2576. doi: 10.1056/NEJMc2009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saif D.S., Ibrahem R.A., Eltabl M.A. Prevalence of peripheral neuropathy and myopathy in patients post-COVID-19 infection. Int J Rheum Dis. 2022;25(11):1246–1253. doi: 10.1111/1756-185X.14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonometti R., Sacchi M.C., Stobbione P., et al. The first case of systemic lupus erythematosus (SLE) triggered by COVID-19 infection. Eur Rev Med Pharmacol Sci. 2020;24(18):9695–9697. doi: 10.26355/eurrev_202009_23060. [DOI] [PubMed] [Google Scholar]
- 16.Hali F., Jabri H., Chiheb S., Hafiani Y., Nsiri A. A concomitant diagnosis of COVID-19 infection and systemic lupus erythematosus complicated by a macrophage activation syndrome: a new case report. Int J Dermatol. 2021;60(8):1030–1031. doi: 10.1111/ijd.15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Stefano L., Rossi S., Montecucco C., Bugatti S. Transient monoarthritis and psoriatic skin lesions following COVID-19. Ann Rheum Dis. 2020 doi: 10.1136/annrheumdis-2020-218520. annrheumdis-2020-218520 (online ahead of print) [DOI] [PubMed] [Google Scholar]
- 18.Taquet M., Geddes J.R., Husain M., Luciano S., Harrison P.J. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatr. 2021;8(5):416–427. doi: 10.1016/S2215-0366(21)00084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Silva K.M., Jorge A., Cohen A., et al. COVID-19 outcomes in patients with systemic autoimmune rheumatic diseases compared to the general population: a US multicenter, comparative cohort study. Arthritis Rheumatol. 2021;73(6):914–920. doi: 10.1002/art.41619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W., Wang C.Y., Wang S.I., Wei J.C. Long-term cardiovascular outcomes in COVID-19 survivors among non-vaccinated population: a retrospective cohort study from the TriNetX US collaborative networks. EClinicalMedicine. 2022;53 doi: 10.1016/j.eclinm.2022.101619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y., Xu Z., Wang P., et al. New-onset autoimmune phenomena post-COVID-19 vaccination. Immunology. 2022;165(4):386–401. doi: 10.1111/imm.13443. [DOI] [PubMed] [Google Scholar]
- 22.Sachinidis A., Garyfallos A. COVID-19 vaccination can occasionally trigger autoimmune phenomena, probably via inducing age-associated B cells. Int J Rheum Dis. 2022;25(1):83–85. doi: 10.1111/1756-185X.14238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stringer D., Braude P., Myint P.K., et al. COPE Study Collaborators The role of C-reactive protein as a prognostic marker in COVID-19. Int J Epidemiol. 2021;50(2):420–429. doi: 10.1093/ije/dyab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faillie J.L. Indication bias or protopathic bias? Br J Clin Pharmacol. 2015 Oct;80(4):779–780. doi: 10.1111/bcp.12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang E.Y., Mao T., Klein J., et al. Diverse functional autoantibodies in patients with COVID-19. Nature. 2021;595(7866):283–288. doi: 10.1038/s41586-021-03631-y. [DOI] [PubMed] [Google Scholar]
- 26.Manja V., AlBashir S., Guyatt G. Criteria for use of composite end points for competing risks-a systematic survey of the literature with recommendations. J Clin Epidemiol. 2017;82:4–11. doi: 10.1016/j.jclinepi.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Benchimol E.I., Smeeth L., Guttmann A., et al. RECORD Working Committee The REporting of studies Conducted using Observational Routinely-collected health Data (RECORD) statement. PLoS Med. 2015;12(10) doi: 10.1371/journal.pmed.1001885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gugliesi F., Pasquero S., Griffante G., et al. Human cytomegalovirus and autoimmune diseases: where are we? Viruses. 2021;13(2):260. doi: 10.3390/v13020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim B., Kaistha S.D., Rouse B.T. Viruses and autoimmunity. Autoimmunity. 2006;39(1):71–77. doi: 10.1080/08916930500484708. [DOI] [PubMed] [Google Scholar]
- 30.Getts D.R., Chastain E.M., Terry R.L., Miller S.D. Virus infection, antiviral immunity, and autoimmunity. Immunol Rev. 2013;255(1):197–209. doi: 10.1111/imr.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujinami R.S., von Herrath M.G., Christen U., Whitton J.L. Molecular mimicry, bystander activation, or viral persistence: infections and autoimmune disease. Clin Microbiol Rev. 2006;19(1):80–94. doi: 10.1128/CMR.19.1.80-94.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umbrello M., Nespoli S., Pisano E., Bonino C., Muttini S. Autoantibodies in severe COVID-19-related acute respiratory distress syndrome: just innocent bystanders? Int J Rheum Dis. 2021;24(3):462–464. doi: 10.1111/1756-185X.14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cañas C.A. The triggering of post-COVID-19 autoimmunity phenomena could be associated with both transient immunosuppression and an inappropriate form of immune reconstitution in susceptible individuals. Med Hypotheses. 2020;145 doi: 10.1016/j.mehy.2020.110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carod-Artal F.J. Post-COVID-19 syndrome: epidemiology, diagnostic criteria and pathogenic mechanisms involved. Rev Neurol. 2021;72(11):384–396. doi: 10.33588/rn.7211.2021230. [DOI] [PubMed] [Google Scholar]
- 35.Anaya J.M., Herrán M., Beltrán S., Rojas M. Is post-COVID syndrome an autoimmune disease? Expert Rev Clin Immunol. 2022;18(7):653–666. doi: 10.1080/1744666X.2022.2085561. [DOI] [PubMed] [Google Scholar]
- 36.Apel F., Zychlinsky A., Kenny E.F. The role of neutrophil extracellular traps in rheumatic diseases. Nat Rev Rheumatol. 2018;14(8):467–475. doi: 10.1038/s41584-018-0039-z. [DOI] [PubMed] [Google Scholar]
- 37.Narasaraju T., Tang B.M., Herrmann M., Muller S., Chow V.T.K., Radic M. Neutrophilia and NETopathy as key pathologic drivers of progressive lung impairment in patients with COVID-19. Front Pharmacol. 2020;11:870. doi: 10.3389/fphar.2020.00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lerner A., Arleevskaya M., Schmiedl A., Matthias T. Microbes and viruses are bugging the gut in celiac disease. Are they friends or foes? Front Microbiol. 2017;8:1392. doi: 10.3389/fmicb.2017.01392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smatti M.K., Cyprian F.S., Nasrallah G.K., Al Thani A.A., Almishal R.O., Yassine H.M. Viruses and autoimmunity: a review on the potential interaction and molecular mechanisms. Viruses. 2019;11(8):762. doi: 10.3390/v11080762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irving P.M., Gibson P.R. Infections and IBD. Nat Clin Pract Gastroenterol Hepatol. 2008;5(1):18–27. doi: 10.1038/ncpgasthep1004. [DOI] [PubMed] [Google Scholar]
- 42.Chen H.Y., Wang S.I., Chang R., Wei J.C. Letter: association between COVID-19 and inflammatory bowel disease. Aliment Pharmacol Ther. 2022;55(9):1226–1227. doi: 10.1111/apt.16814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hadi Y., Dulai P.S., Kupec J., et al. Incidence, outcomes, and impact of COVID-19 on inflammatory bowel disease: propensity matched research network analysis. Aliment Pharmacol Ther. 2022;55(2):191–200. doi: 10.1111/apt.16730. [DOI] [PubMed] [Google Scholar]
- 44.Hughes M., Herrick A.L. Systemic sclerosis. Br J Hosp Med. 2019;80(9):530–536. doi: 10.12968/hmed.2019.80.9.530. [DOI] [PubMed] [Google Scholar]
- 45.Wu M., Assassi S. The role of type 1 interferon in systemic sclerosis. Front Immunol. 2013;4:266. doi: 10.3389/fimmu.2013.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jara L.J., Medina G., Saavedra M.A. Autoimmune manifestations of infections. Curr Opin Rheumatol. 2018;30(4):373–379. doi: 10.1097/BOR.0000000000000505. [DOI] [PubMed] [Google Scholar]
- 47.Fineschi S. Case report: systemic sclerosis after covid-19 infection. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.686699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossi G.P., Sanga V., Barton M. Potential harmful effects of discontinuing ACE-inhibitors and ARBs in COVID-19 patients. Elife. 2020;9 doi: 10.7554/eLife.57278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jamalyaria F., Ward M.M., Assassi S., et al. Ethnicity and disease severity in ankylosing spondylitis a cross-sectional analysis of three ethnic groups. Clin Rheumatol. 2017;36(10):2359–2364. doi: 10.1007/s10067-017-3767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fava A., Petri M. Systemic lupus erythematosus: diagnosis and clinical management. J Autoimmun. 2019;96:1–13. doi: 10.1016/j.jaut.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y.M., Zhou X.J., Nath S.K., Sun C., Zhao M.H., Zhang H. Evaluation of 10 SLE susceptibility loci in Asian populations, which were initially identified in European populations. Sci Rep. 2017;7 doi: 10.1038/srep41399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vandenbroucke J.P., Pearce N. Test-negative designs: differences and commonalities with other case-control studies with "other patient" controls. Epidemiology. 2019;30(6):838–844. doi: 10.1097/EDE.0000000000001088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.