Abstract
3D bioprinting offers a simplified solution for the engineering of complex tissue parts for in-vitro drug discovery or, in-vivo implantation. However, significant amount of challenges exist in 3D bioprinting of neural tissues, as these are sensitive cell types to handle via extrusion bioprinting techniques. We assessed the feasibility of bioprinting human neural progenitor cells (NPCs) in 3D hydrogel lattices using a fibrinogen-alginate-chitosan bioink, previously optimized for neural-cell growth, and subsequently modified for structural support during extrusion printing, in this study. The original bioink used in this study was made by adding optimized amounts of high- and medium-viscosity alginate to the fibrinogen-chitosan-based bioink and making it extrudable under shear pressure. The mechanically robust 3D constructs promoted NPC cluster formation and maintained their morphology and viability during the entire culture period. This strategy may be useful for co-culturing of NPCs along with other cell types such as cardiac, vascular, and other cells during 3D bioprinting.
Keywords: Neural tissue engineering, Biofabrication, Bioinks, Neural progenitor cells, Process optimization, Rheology, Scanning electron microscopy, Image analysis, Immunohistochemistry, Bioprinting
Graphical abstract
Neural tissue engineering; Biofabrication; Bioinks; Neural progenitor cells; Process optimization; Rheology; Scanning electron microscopy; Image analysis; Immunohistochemistry; Bioprinting.
1. Introduction
Our laboratory has utilized extrusion-based 3D bioprinting using bioinks that are cross-linkable via visible light or chemically-induced for cardiac cells including human induced pluripotent stem cell (iPSC) derived cardiomyocytes and other cell types to engineer cardiac tissues in-vitro [1, 2, 3, 4]. Such 3D bioprinted tissue platforms are robust enough to be used as a cardiac tissue model for drug testing or other studies [5]. In order to improve the functionality of these in-vitro engineered tissue chips, multiple cell sources, such as vascular and neuronal cells, need to be included. Such a strategy can lead to the direct one-step 3D bioprinting of complex tissues such as cardiac or brain tissue mimics for further studies. Bioinks used in 3D bioprinting result in the production of 3D scaffolds which are known to exert biophysical regulation of cell morphology and function [6]. However, bioinks can be variable based on their elastic/storage modulus which regulates their stiffness that in turn can control their utility for culturing multiple cell types. Our goal in this study was to modify a low viscosity fibrin-based bioink for the extrusion bioprinting of NPCs, in our laboratory. We hypothesized that the modification of an existing fibrin-based bioink composition for feasibility of adoption towards 3D extrusion bioprinting would not alter the biological properties of the bioink and still make it an attractive biomaterial for the fabrication of substrates for neural tissue engineering by supporting the culture of neural cells.
Thus, in the present study, we extended the applicability of an in-house developed 3D extrusion-based bioprinting technique to the printing of iPSC derived neural progenitor cells (NPCs) which poses as a delicate cell type to handling via traditional biofabrication approaches involving shear and extrusion stimuli [7]. Our futuristic goal is to develop an extrusion-based 3D-printing strategy for the contiguous culture of human NPCs along with cardiac and vascular cells to improve the overall functionality of the engineered cardiac platforms and serve as enhanced in-vivo tissue substitutes.
Existing literature indicates that NPCs play a vital role in initial blood vessel ingression, nascent vessel stabilization, followed by vessel maturation and integrity [8] Previous studies have shown that neuronal innervation regulates cardiomyocyte proliferation and cardiac regeneration in mice and zebrafish [9]. Sympathetic neurons derived from iPSCs from physical and functional connections with cardiac muscle cells [10]. It has also been shown that sympathetic neurons modulate the contractility of iPSC-derived cardiomyocytes [11]. We hypothesize that the addition of iPSC-NPCs in 3D bioprinted constructs will serve as a first step in the creation of multiple cell-based cardiac platforms as the NPCs will provide the necessary source for neuronal cells and influence the behavior of the 3D bioprinted constructs [12]. The 3D bioprinted NPC-based model can then be extended to include cardiac and vascular cells to bioprint advanced 3D cardiac tissue models that may bear an enhanced functional resemblance to in-vivo cardiac tissue.
In this study, NPCs were bioprinted using a fibrinogen-alginate-chitosan-based bioink formulation with the goal of developing a platform that is compatible with both cardiomyocytes and NPCs and could eventually be used to study interactions between the cells of the human brain and heart. We assessed the ability of the 3D structures printed with a hydrogel bioink formulation, previously optimized by our collaborators for neural cell growth [13], and subsequently modified for structural support during extrusion bioprinting towards promoting the cluster formation of the NPCs and their maintenance.
2. Results
Figure 1A, B, C depicts the structural design that was adopted to print the lattice scaffolds. The optimized bioink and the cross-linker composition is depicted in Figure 1D; while the optimized printing parameters for obtaining stable scaffolds using the Cellink platform is summarized in Table 1 (below).
Figure 1.
(A) Depicts the STL file design for the lattice and (B) shows the workflow of the bioprinting process with the actual bioprinted construct being depicted in (C). In (D) the bioink components and their respective crosslinking chemistries are summarized.
Table 1.
| Infill pattern | Infill density | Layer profile | Pressure | Speed | Nozzle |
|---|---|---|---|---|---|
| Grid | 25% infill | Grid Lattice | 80 kPa | 1 mm/s | 18G blunt |
The goal of the optimization of the printing process was to adopt parameters that would enable reduction of shear stresses acting on the bioink to attain maximum design accuracy. With this goal in mind, the extrusion pressure was maintained at a constant due to its inverse effect on cell viability.
As summarized in Figure 1D, the bioink was 3D printed and the mixture was cross-linked via subsequent crosslinking mechanisms. First among these was the fibrinogen that reacted with thrombin in the presence of calcium ions to yield fibrin [14]. The HV-and the MV-alginates both were cross-linked by reacting with the calcium ions [15]. Genipin has been widely used for crosslinking of chitosan-based hydrogels [16]. Online supplementary Figure S1 depicts the results from the optimization of the University of Victoria (UVIC) bioink to yield the University of Texas at El Paso (UTEP) bioink. The maple leaf design was chosen for the optimization of the bioink composition as it represents a complex structure, inspired by our collaboration with the UVIC in BC, Canada. Of all different compositions tested, the one containing 20 mg/mL of high viscosity (HV) alginate and, 10 mg/mL medium viscosity (MV) alginate, cross-linked using 20 mg/mL CaCl2 was chosen as the final optimized bioink composition for the UTEP bioink (Figure S1: H). In the optimized bioink composition, the amounts and concentrations of other components including fibrinogen, thrombin, genipin, and chitosan were maintained constant. The UVIC bioink (received as-is) was not compatible with extrusion printing (results not included) without any modification.
Figure 2A depicts a comparison of the UVIC bioink that was modified for extrusion printing at UTEP. Results indicated some similarities and differences in their material properties. The viscosity of both bioinks was observed to decrease with increasing shear rate confirming their shear-thinning behavior. The main difference was in the overall viscosities of the materials (Figure 2A). At low shear rates, the UTEP bioink revealed a viscosity of about 106 mPa∗s, while the UVIC bioink had a viscosity of 103–104 mPa∗s.
Figure 2.
Rheological characterization and comparison of the microfluidic-(UVIC) and extrusion-based bioink (UTEP). (A) Mean viscosity at low shear rate of 0.1 rad/s showed extrusion-based bioink had a significantly higher (p < 0.05) viscosity needed to stabilize biomaterial in a filament after extrusion from the bioprinting nozzle. (B) Shear-thinning was observed by both the microfluidic- and extrusion-based bioinks.
This difference in viscosity enabled the UTEP bioink to maintain a filament-like shape post-extrusion, while the UVIC bioink had to be cross-linked immediately after extrusion to create robust patterns with the bioink (Figure 2B). When a filament is extruded from a pneumatic print head, the high viscosity prevents the bioprinted fiber from spreading and distorting the structure. Moreover, existing literature finds bioinks within this range to be useful for extrusion-based applications [17]. On the other hand, the microfluidic bioink must be cross-linked immediately after extrusion to create patterns with the bioink. Its low viscosity at a low shear rate would cause the bioink to spread after it is deposited, thus emphasizing the need to crosslink the hydrogel during the printing process. Generally, microfluidic bioprinters use bioinks within these low viscosities ranges [18]. This is the most vital distinction between both platforms.
To pattern complex geometries using 3D bioprinting, the bioink must maintain its structure and be stabilized after printing and crosslinking. To confirm this behavior, the increase in storage moduli for both bioinks after crosslinking was studied and compared as shown in Figure 3A. Both bioinks exhibited variability in their non-cross linked form as the storage modulus of the material changed with shear frequency (Figure 3). Overall, both sets of crosslinked materials showed a plateau in their storage and loss modulus, however, the extrusion-based cross-linked UTEP hydrogel showed a higher storage modulus, which is the representation of elastic-like behavior (G′). While both bioink compositions were similar, the increase in storage modulus was seen in the extrusion-based bioink can be attributed to the larger amount of available crosslink sites in the high guluronic acid in the alginate that was added into the composition. It is well known that increasing the number of crosslinks in a structure also increases the stiffness of the hydrogel, which is confirmed by our data [1, 19]. Moreover, we examined the stability of the UTEP hydrogel structure over a 2 week period to monitor the modified hydrogel's cross-linked integrity (Figure 3B). The highest storage modulus was seen right after crosslinking. At this time point, the structure has the most crosslinks within its polymeric backbone. After being subjected to swelling, these crosslinks are seen to degrade over time as illustrated by the decreasing storage modulus throughout the study period. Degradation of 3D scaffolds are an important parameter as it allows embedded cells to migrate and remodel the extracellular matrix [20].
Figure 3.
Rheological characterization of crosslinked bioinks from UVIC and UTEP. (A) Representative frequency sweep of crosslinked microfluidic and extrusion bioinks revealed a stable structure shown by the plateau in the storage modulus, however, a stiffer composition was seen in extrusion-based structures than in microfluidic samples. (B) Noncrosslinked bioink is seen to stabilize after crosslinking of fibrin, alginate, and chitosan as shown by the appearance of a plateau in the storage modulus. The bioprinted crosslinked structures are seen to get softer over time as the scaffolds degrade and the degree of crosslinks within the structure weakens.
Structural stability of the 3D printed hydrogels was evaluated through swelling analysis, which revealed that all structures sustained long-term in-vitro culture as shown in Figure 4.
Figure 4.
Swelling and degradation analysis of the 3D bioprinted acellular constructs made using the UTEP bioink via extrusion bioprinting. Structures attained maximum or equilibrium swelling after 7 days in culture.
As shown by the data, scaffolds attained maximum equilibrium swelling after 7 days of incubation (Figure 5A) after which a gradual reduction in swelling was observed until day-16 signifying onset of bulk degradation of the scaffold. Overall, rheological analysis depicted the mechanical stability of the 3D constructs over a 2-week culture period. Maintenance of high structural fidelity of the 3D bioprinted structures throughout the study confirmed its macro-structural stability. The macro structural stability was studied and confirmed through an observation of the pores and the extent of porosity of the scaffolds at 0- (Figure 5B) and 14-days (Figure 5C) of incubation via acquired SEM images (Figure 5). The decrease in overall apparent porosity at day-14 compared with day-0 implied that the scaffolds had undergone the process of swelling (p = 0.0327) within this period (Figure 5D).
Figure 5.
Scanning Electron Microscopy (SEM) and analysis of the 3D bioprinted acellular constructs made using the UTEP bioink via extrusion bioprinting. Shown in (A) is the 3D bioprinted and cross-linked constructs. (B) and (C) are representative SEM images of from Day-0 and at Day-14 respectively. (D) Shows the average pore sizes ±SD (μm) of the constructs at these two time points respectively.
The UTEP bioink supported the growth and in-vitro culture of the NPC as shown in Figure 6(A, B), and studied up to 5 days. More live cells (green) were detected compared to dead cells (red) in all of the images shown. Using a Live-Dead reagent (LIVE/DEAD™ Cell Imaging Kit (488/570) R37601, Invitrogen™) treatment of the cultures were performed at varying time points as indicated in Figure 6. The in vitro biocompatibility analysis of the NPC's in the 3D cultures (Figure 6C, D, E) yielded a percentage of 82.25 ± 5% live cells confirming high cell viability in comparison to the content of non-viable cells observed (15.5 ± 2.5%) as shown in Figure 6F. This result provided conformation of the UTEP bioink's biocompatibility for further experiments including 3D bioprinting.
Figure 6.
In-Vitro biocompatibility of the UTEP bioink studied using human neural progenitor cells. The Live-Dead assay comprising of Calcein AM (Green-Live cells) and Ethidium Homodimer (Red-Dead cells) was used to detect the presence of live cells thereby confirming the biocompatibility of the UTEP bioink in 2D. (A) Shows Live/Dead stained cells at 1-day; and (B) Live/Dead stained cells after 5-days. Scale bar in both images correspond to 200 μm. Shown in (C–E) are viable neural progenitor cells within the UTEP bio-ink at different thicknesses of the 3D hydrogel at day 5. Scale bar in all images correspond to 100 μm. Shown in (F) is the quantitative Live-Dead data expressed as % between Day-1 and Day-5.
The 5-day culture time point for the 3D cell based constructs was selected based on the results of swelling and degradation where the structures were seen to attain maximum degree of swelling by 7-days during which maximum structural fidelity was maintained. By visual examination of the fluorescence microscope images, it was confirmed that the UTEP bioink formulation could support and maintain healthy human NPC cultures and their viability via promoting of individual cell interactions within the cluster. Cluster formation indicated high levels of cell communication over 5 days. Shown in Figure 7 are representative images of clustering of NPC when cultured in 3D bioprinted systems (Figure 7A), as opposed to 2D cultures (Figure S2), where the NPCs were seeded onto a thin coating of the bioink deposited onto petri dishes.
Figure 7.
3D culture of NPCs within bioprinted constructs. (A) Shows the NPC's clustered in the bioprinted constructs with the upper panel showing overview of the clusters formed and the lower panel showing magnified insets. (B) Depicts the overall area of the NPC clusters with time. (C) Shows the percentage of the image area occupied by the NPC clusters with time. At least 5 samples per condition per time point was involved and 15–20 images were acquired for each data point depicted.
A comparison of NPC cluster size across days 1, 3, and 5 resulted in an average cluster area of 3872 sq. μm at day-1, 4266 sq. μm at day-3 and 4443 sq. μm at day-5 (Figure 7B). An analysis of the fraction of the area covered by the clusters for the region imaged (ROI) corresponded to a 17% coverage at day-1; 24% coverage at day-3 and 22% at day-5 respectively (Figure 7C). Using an unpaired t-test and an ANOVA statistically significant differences were revealed between values obtained at Day-1 versus 3 (p = 0.0341) and Days-1 and 5 (p = 0.0421). However there was no significant difference between values obtained at Day-3 and 5 (p = 0.4951). This implied that clusters of NPC were not only retained within the 3D printer structures but also maintained in 3D culture across a time span of 5 days. From the data it is clear that the clusters were formed and minimally expanded between 1-3 days but after 3- days they were maintained in that state. On the other hand, the 2D NPC cultures showed little to no cluster formation, during the duration of the culture (Figure S2) which confirmed the ability of the 3D bioprinted constructs towards the maintenance of the cluster morphology of the NPCs.
To verify proliferation of the NPC in the newly developed bioink, cells were pre-stained with CellTrace Yellow (CTY) and analyzed using a flow cytometer. Based on the concept of dye dilution, pre-stained cells will cause a reduction of dye intensity over multiple generations [1, 2]. Figure 8A shows a characteristic peak of a positive control sample in the high intensity region to the right in the case where the NPC were pre-stained with CTY. On the contrary, Figure 8B represents the peak for the negative control sample in the low intensity region to the left where the cells were not pre-stained with CTY. Figure 8C represents the dye intensity of pre-stained from the bioink indicating a significant decrease on day-3 (100%–74%) confirming the dilution of dye with time shown by the presence of multiple peaks that is attributed to new generations of cells (p < 0.01) [1, 21, 22]. Figure 8D shows the comparison of dye intensities from all analyzed samples.
Figure 8.
Flow Cytometry. Neural progenitor cells were pre-stained with CellTrace Yellow (CTY) prior to 3D bioprinting. Graphs (A, B) portray the % of CTY in the positive and negative control samples respectively. Shown in C is a representative graph depicting the % of dye intensity of cells extracted after 3-days of culture. p values were found to be all statistically significant between the experimental and control samples (p < 0.01). Shown in (D) is comparison of dye intensities expressed by n = 3 samples per group studied.
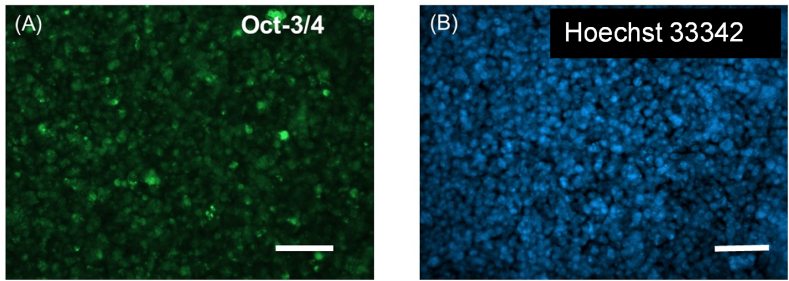
Figure 9 shows the confirmation of the immunohistochemical identity of the NPC passages used in this study. The cells expressed Oct-3/4 (Figure 9A) which is believed to be a critical regulator of neuronal differentiation based on published works from others [23], while being counter-stained with the nuclear stain Hoechst 33342 (Figure 9B). Others have shown that the downregulation of Oct-3/4 expression in neurospheres, composed of neural stem cells and NPC's accelerates their neuronal differentiation while its sustained expression prevents neuronal differentiation [23]. Since these cells were used in the 3D bioprinting experiments it was confirmed that the cells retained their neural progenitor phenotype throughout the duration of this study and experiments performed. The expression levels of Nestin (NES) and SOX2 relative to GAPDH levels, for the UTEP Bio-ink (3D) and 2D cell-culture controls, were assessed by qPCR (Figure 10A). SOX2 showed high expression in the 3D condition significantly different (Student's t test; p < 0.05). In contrast, NES expression was low in the 3D samples without being statistically significant. Fold change was calculated for expression of the genes NES and SOX2 were normalized with GAPDH as a housekeeping gene. It was seen that NES and SOX2 are downregulated in 3D (Figure 10B).
Figure 9.
Immunohistochemistry performed after culturing the NPC after 5 days. A primary antibody targeted against Oct-3/4 was used to confirm the retention of progenitor function for these cells in this study as shown in (A) Viability of the cells was also confirmed by counterstaining with Hoechst (blue) after 5 days of in-vitro culture (B). Scale bar in both images correspond to 100 μm.
Figure 10.
Quantitative PCR (qPCR) analysis. NES and SOX2 gene expression in NPC cells were normalized to GAPDH in both conditions; UTEP Bio-ink (3D) and cell culture control (2D) after 5 days of incubation. (A) The SOX expression levels relative to GAPDH were statistically different in the 3D structure when compared to the control (2D) (∗, p < 0.05). (B) Fold change between 3D and 2D cultured cells for NES and SOX gene transcripts are shown.
3. Discussion
Ischemic heart disease continues to be the leading cause of death worldwide accounting for 9 million deaths per year [22]. Heart transplantation remains the only curative therapy for many patients with end-stage heart disease [24]. However, patients often face inadequate access to heart transplantation because of socio-economic restrictions and limited number of donor organs available [25]. Alternatively, adoption of regenerative approaches targeted towards cardiac tissue regeneration is a potential alternative for treating severe heart disease. While there is a decent amount of information on cardiac derived cell sources for myocardial regeneration [26], there is almost no existing discussion on the need for neuronal innervation within this tissue engineered cardiac constructs. The human nervous and cardiac systems are unique in their anatomical, cellular and functional heterogeneity, with specific cellular structures performing essential physiological functions [27]. The cardiac autonomic nervous system comprised of the extrinsic and intrinsic innervation of the heart plays a crucial role for the regulation of cardiac activity [28]; the lack of which is involved in various heart diseases, such as cardiac arrhythmias and cardiomyopathy [29]. This system also ensures that there is fine-tuned regulation of sympathetic–parasympathetic balance in the heart under normal and stressed states at all times [30]. We believe that regeneration of cardiac tissue with in-built neuronal innervation is a must for building physiologically relevant cardiac tissue substitutes for both in-vitro studies and in-vivo applications.
The intrinsic cardiac autonomic nervous system (CANS) plays a key role for the regulation of cardiac activity with its dysregulation being involved in various heart diseases, such as cardiac arrhythmias [28]. The CANS comprises the extrinsic and intrinsic innervation of the heart [28]. The intrinsic cardiac nervous system (ICNS) includes the network of the intra-cardiac ganglia and interconnecting neurons [28]. The cardiac ganglia contribute to the tight modulation of cardiac electrophysiology, working as a local hub integrating the inputs of the extrinsic innervation and the ICNS [28]. Therefore, studies that attempt to integrate sources for autonomous neurons within an engineered cardiac tissue construct are more likely to succeed in terms of posing as a physiologically relevant model. Such advanced systems can then enable a better understanding of the role of the ICNS for the modulation of the cardiac conduction system and may be crucial for targeted therapies of various arrhythmias.
Since our lab has pioneered the biofabrication process of engineering cardiac tissue with an insight on heterocellular coupling of cardiac cells, we aimed to extrapolate the concept and methodology of 3D bioprinting developed in our laboratory, for the encapsulation of NPCs derived from human iPSCs that can eventually lead us to engineer cardiac tissues with an integrated neuronal innervation component. Such a system possesses the potential to be transplanted for improvement of cardiac function in the failed myocardium as well as serves as a more relevant in-vitro preclinical stroke model for testing therapeutic interventions.
The NPCs used in this study are derived from human iPSCs and are known to express Nestin and PAX6 as well as possess electrophysiological activity confirming their neural identity and lineage [31]. These cells are usually cultured in the form of neurospheres [31] that can be transferred to laminin-coated vessels with brain-derived neurotrophic to induce their differentiation into neurons, astrocytes, and oligodendrocytes, thereby posing as a relevant cell type for in-vitro cell based models.
Our collaborators at the UVIC for the first time, demonstrated using a fibrin-based bioink the bioprinting of human iPSC-derived NPCs [7, 21]. They used a microfluidics-based bioprinter to generate dome-like structures consisting of human iPSC-derived NPC's. Using microfluidic 3D bioprinting technology, the layer by layer deposition of living fibers was enabled to rapidly and accurately build heterogeneous tissues. Although the microfluidic-based 3D printing technology does not rely on predefined bio-inks and their related parameters the resultant structures suffer from long-term fidelity. The resulting tissues exhibited up to 95% viability, 7 days post printing as well as the neuronal markers, TUJ1 and the early midbrain marker, Forkhead Box A2 (FOXA2). It is well established that fibrin fibrils formed within fibrin-gels provide contact guidance and alignment to neuronal cells for their differentiation into targeted phenotypes which was an observation dating back to the pioneering work in contact guidance by Weiss et. al [32]. These reports motivated us to adopt the fibrin-based bioink for the extrusion bioprinting of NPCs, in our laboratory.
Bioinks should possess a number of features including a relatively high biocompatibility, printability, and the ability to deliver growth factors thus enabling an appropriate desired environment for the seeded cells [6, 31, 33]. However, the UVIC bioink had to be modified significantly in terms of viscosity for making it adaptable for extrusion bioprinting. Our 3D bioprinted scaffolds made from the UTEP bioink were much stiffer and possessed enhanced elastic properties compared to the UVIC bioink allowing the structures to be stable for a sustained culture period. The 3D printed constructs allowed the NPCs to retain their neurosphere-like morphology and maintenance of the clusters during the entire study period in comparison with the 2D culture controls. The system also possesses flexibility for allowing differentiation into other cell types to be induced by the addition of necessary growth factors and a niche environment for a target phenotype [33].
In this study we demonstrated how our novel based alginate/fibrin-bioink formulation supported the growth of NPCs. Based on the FACS analysis data, the reduction of dye intensity with time indicates the proliferation of NPCs in the scaffold. This work lays the groundwork for fabricating engineered neural tissues from pluripotent stem cells that can serve as a potential model for high-throughput drug screening studies in the future.
Nestin has been described as a neural stem/progenitor cell marker that appears during central nervous system (CNS) development [34] and its expression has been confirmed in adult CNS stem/progenitor cells, from in vitro studies [35]. Nestin expression is seen to be downregulated when CNS stem/progenitor cells differentiate into neurons or glial cells [36, 37]. Thus the enhanced downregulation of Nestin in the NPC cells, in our study confirmed the differentiation potential of the UTEP bioink towards a neuronal lineage of the NPC cells cultured.
SOX2 is stably expressed during embryonic development, and is known to play a well-established role in neural commitment [38]. It has been suggested that core pluripotent transcription factors such as SOX2 and Oct4 not only play an important role in the induction and maintenance of pluripotency, but also function in regulating their differentiation [39, 40] SOX2 is known to induce neural induction and enhance neural differentiation by repressing key regulators of other lineage fates, for example brachyury [41]. SOX2 is highly expressed in proliferating NPCs and is downregulated upon differentiation to post-mitotic neuronal and glial cells [42]. Thus the downregulation of SOX2 in the NPC cells, in our study confirmed the differentiation potential of the UTEP bioink towards a neuronal lineage of the NPC cells cultured. Our study outcomes are in line with other published reports where a brain-derived decellularized extracellular matrix based bioink was used for printing and for stem cell delivery to a traumatically injured brain. Human neural stem cells (NSCs) laden within this bioink were found to be fully differentiated to neurons; the levels of markers for mature differentiated neurons were higher than those observed with collagen bioink in vitro [43].
In this study our goal was also to develop a permissive environment that may influence neural progenitor cell differentiation. Fibrin based bioinks have been shown to promote the differentiation of NPCs by others [6, 7]. Applying fibrin as a component of the extrusion based bioink in this study, may promote the differentiation of the NPCs in situ towards neurons, oligodendrocytes, or astrocytes in long-term culture. Since we have already developed an extrusion-based 3D-printing strategy for clustering of human neural-progenitor cells in this study, we can now leverage this platform for culturing other neural cell types for extended and sustained periods.
4. Materials and methods
4.1. Bioink preparation
The bioink used for extrusion printing in this study was derived from a precursor composition containing a fibrin-based bioink from UVIC for printing hiPSC-derived neural aggregates using a microfluidic 3D bioprinting technology platform [21]. This bioink formulation is compatible with other printing systems that allow for chemical cross-linking during printing. Cells encapsulated within this bioink maintained viability throughout the printing process, expressed neuronal markers and possessed high levels of viability even after extended culture periods [21]. We modified this bioink composition and adopted it for extrusion bioprinting in our laboratory [1]. To modify the composition of this UVIC bioink, we methodically added small amounts of high viscosity (HV) alginate and medium viscosity (MV) alginate to make it extrudable under shear pressure and was referred to as the University of Texas at El Paso (UTEP) bioink thereafter. At least nine different compositions were attempted for extrusion printing after which an optimized composition was established (Figure S1). A maple leaf design (STL.) file was adopted for this optimization process.
Solutions of the UTEP bioink were optimized and prepared in-house to have final concentrations of 20 mg/mL fibrin, 20 mg/mL high viscosity (HV) alginate (#154724, MP Biomedicals-ThermoFisher Scientific), 10 mg/mL medium viscosity (MV) alginate, and 0.3 mg/mL genipin. The medium viscosity (MV) alginate was procured from Nova Matrix (#BP-1010-02; FMC Corp., Sweden). All other chemicals used for modification of the UVIC bioink were procured from Sigma (St. Louis, MO) or Fisher Scientific (Waltham, MA). Solutions of the crosslinker were prepared to have final concentrations of 20 mg/mL CaCl2, 1.7 μg/mL thrombin, and 0.75 mg/mL chitosan. Any necessary dilutions were performed with 1X PBS.
To make 1 mL of bioink solution, 730 μL of the received (as-is) fibrinogen solution (UVIC) were mixed with 0.02 g HV alginate and 0.01 MV alginate. The formulation was left overnight at 4 °C for components to homogenize and mix thoroughly. One hour before printing, 270 μL of genipin solution was added to the mixture. The bioink mixture was loaded into a 3 mL plastic syringe, capped, and centrifuged at 1200 rpm for 3 min for homogeneity. Crosslinking stock solution was prepared shortly prior to printing in volume ratios of 4.8 mL CaCl2/thrombin to 256 μL chitosan received from UVIC. After structures were printed, they were cross-linked for 5 min with enough crosslinking solution to fully cover them.
4.2. Optimization of printing parameters
A Cellink 3D Bioprinter (BIOX) was used to print a lattice structure to yield a porous and interconnected framework [2]. A lattice was designed in SolidWorks and had overall dimensions of 8.5 mm × 8.5 mm and 3 mm in height, with strut thickness of 1.5 mm and pore dimensions of 1.5 mm × 1.5 mm. Printer settings for the bioink were first optimized to extrude the acellular bioink at a maximum pressure of 80 kPa, 1 mm/s speed, and an 18G blunt tip nozzle to produce prints that most closely resembled the model design and retained pores after crosslinking. Once this was confirmed, the UTEP bioink (mixed with cells) was loaded into a 3 mL plastic syringe (BD, Franklin lakes, NJ), fitted with a luer lock (Cellink, Boston, MA) fitted with an 18 G nozzle. Low extrusion pressure of 21 kPa was used to ensure cell viability and create lattice patterns designed with SolidWorks (Concord, MA) at 2 mm/s and 25% upright square lattice infill on a BioX bioprinter. The structures were printed in a 48-well plate (Corning Costar, ThermoFisher) and cross-linked.
4.3. Bioink and scaffold characterization
4.3.1. Rheological analysis
Shear-thinning and viscosity are crucial parameters for any bioink that are adopted for extrusion printing. Thus, in order to assess the bioink composition, rheometric analysis of the material was conducted on an MCR 92 Anton Paar rheometer with a 25 mm parallel plate geometry and a 1 mm gap between the geometry and the stage. Studies on the bioink composition were conducted at ambient temperature, as the hydrogel was bioprinted at room temperature. The viscosity of UVIC and UTEP non-cross linked bioink samples were tested in a continuous flow experiment, through a ramp of the shear rate from 0.1–100 rad/s to determine if the materials exhibited shear-thinning properties.
Next, in order to study the effects crosslinking on the rigidity of the bioink, an amplitude and frequency sweep were conducted on the pneumatic-based UTEP bioink without crosslinking and at 3, 10, and 21 days after crosslinking to study the temporal stability of bioprinted structures where the tests were run at 37 °C. Amplitude sweeps were run on each lattice sample (8.5 mm × 8.5 mm × 3 mm) to find an optimal strain within the linear viscoelastic region. Samples were kept immersed in 1X PBS until they were subjected to analysis from 3–21 days. A strain of 0.5% was determined to be in the linear viscoelastic region from conducting amplitude sweeps on the gels and applied during a frequency sweep range of 0.1–100 Hz. The resulting storage and loss modulus was assessed.
4.3.2. Swelling
Hydrogels have the ability to absorb enormous amounts of water and nutrients into their polymeric networks that help sustain cellular vitality. In this study, the hydration behavior of acellular bioprinted scaffolds were assessed in a 2-week period for acellular samples. Printed acellular scaffolds were then air-dried and the dry weights of each sample were recorded. Each sample was then immersed in 1 mL of 1X PBS at pH 7.3, not containing calcium and magnesium ions. Weights of the swollen gels were monitored and recorded every 24 h. A swelling ratio was determined for each sample at each time point using Eq. (1):
| Q = (Ms – Md)/Md | (1) |
Q is defined as the swelling ratio, Ms is referred to as the mass in the swollen state and Md is the mass in the dried state (original weight).
4.3.3. Scanning electron microscopy (SEM)
SEM micrographs were acquired from acellular printed scaffolds at varying time points to study the effects of environmental degradation on the scaffolds. Prior to SEM imaging, samples were lyophilized and sputter-coated with gold/palladium (2–3 min) in a sputter coater (Gatan Model 682 Precision etching coating system, Pleasantown, CA) and visualized using SEM. Average pore diameter of the acellular crosslinked 3D printed scaffolds was measured at Days-0 and -14 by analyzing electron micrographs of cross-sectioned lyophilized samples (S-4800, Hitachi, Japan) at 7 kV voltage and current of 5 μA at varying magnification. Collected images obtained were analyzed using Image J to determine the average sizes of the pores (pore diameter, μm) during each time point studied. A total of at least n = 5 images per sample was assessed for samples evaluated at varying time points.
4.4. Biocompatibility
NPCs were used to test the biocompatibility of the UTEP bioink. Complete Growth Medium was used to culture the NPC following the manufacturer's recommendations. To assess the viability of the encapsulated cells in the hydrogel after varying time points, a Live-Dead assay was performed by adding the reagents into the samples following the vendor's recommendations. The cell density was maintained to include 1 × 106 cells/ml of the bioink (3D) or in culture medium (2D) as optimized by our prior 3D printing work in the laboratory [3, 44].
Calcein AM was used to dye the live (green) cells and ethidium homodimer was used to dye the dead (red) cells. Images were taken utilizing an AX10 A1 microscope (Zeiss). Images of the live and dead cells were taken at different thicknesses within the 3D hydrogel as well as from the 2D samples (cells seeded on bioink coating on petri-dishes). Thereafter, an image analysis was performed using ImageJ software, and the percentage of live/dead cells was calculated using Eq. (2).
| No. of live/dead cells (%) = [# of green or red cells/Total No. of green and red cells] × 100 | (2) |
4.5. Culture and maintenance of bioprinted clusters of NPC's
4.5.1. Cell culture and maintenance
Human NPCs used for this study were the normal human iPSC derived Neural Progenitor Cells (Lonza, Walkersville, MD, USA) cultured in T-75 flasks with vendor-recommended substrates for passaging and in-vitro culture before printing. 8 mL of 15 μg/mL poly-L-ornithine (Sigma-Aldrich, St. Louis, MO) solution, diluted in 1X PBS, was added to coat the surface of a T-75 flask by incubation at 4 °C overnight for promotion of cell adhesion and growth. The coating solution was removed and the flask was washed twice with PBS before addition of a second coating layer. Next, 8 mL of 10 μg/mL basement membrane extract (R&D Systems, Minneapolis, MN) was added to coat the flask surface during placement at room temperature for 2 h.
In parallel, complete StemDiff Neural Progenitor Medium (NPM) was prepared to support cell growth of the NPC's. To 490 mL of the Neural Progenitor Basal Medium (STEMCELL Technologies, Vancouver, BC, Canada), was added a 10 mL of Supplement A (STEMCELL), and 500 μL of Supplement B (STEMCELL) was thawed and mixed to form 500 mL of complete NPM. NPM was stored in 4 °C and warmed to 37 °C prior to being supplemented to the cells.
Shortly before addition of the cells, the coating solution was removed and the T-75 flask was washed once with DMEM/F-12 with 15 mM HEPES (Sigma-Aldrich), and.
8 mL of the warmed NPM was added. Cells were thawed in a water bath and 10 mL DMEM was added to thawed cells, mixed well with a pipette, and the mixture was centrifuged (5 min at 300 × g). After centrifugation, the supernatant was aspirated and the cell pellet was re-suspended in 2 mL of NPM, and 1 mL of the mixture was added to the T-75 flask with medium and placed in an incubator at 37 °C. After every 2–3 days, 10 mL of fresh NPM was added to the flask to maintain the cells in culture.
4.5.2. 3D NPC bioprinting and culture maintenance
Once cells reached approximately 80–90% confluence in the T75 flasks, the NPM was aspirated after which 3–5 mL of Accutase (Sigma-Aldrich) was added to each flask to induce detachment and incubated for 3 min. After this, equivalent volume of NPM was added to each well, and a cell scraper (Corning, Durham, NC) was used to harvest the remaining cells, which were then added to a 15 mL centrifuge tube. The contents of the centrifuge tube were centrifuged (5 min at 300 × g) and the supernatant was aspirated to obtain a cell pellet.
To prepare a red fluorescent dye solution, 3 μm PKH26 was added to 1 mL of diluent C (PKH26 Cell linker kit, Sigma). The fluorescent staining ensured that cell membranes were labelled with PKH26 red fluorescent membrane staining dye (Sigma). Following centrifugation and removal of supernatant from the 15 mL centrifuge tube, the cells were suspended in the dye solution and placed in the incubator for 2 min. Then, 1 mL of NPM was added and the mixture was centrifuged (5 min at 300 × g). Finally, the supernatant was aspirated and the cell pellet was re-suspended in 100 μL of NPM. 75 μL of this cell suspension was mixed with 3 mL of the UTEP bioink. The other 25 μL was used for 2D culture controls. The cell density was maintained to include 1 × 106 cells/ml of the bioink (3D) or in culture medium (2D) as optimized by our prior 3D printing work in the laboratory [3, 44].
The 3 ml bioink mixture containing the pre-stained cells mixed within the hydrogel and was used to bioprint the 3D lattice structures described above. Following bioprinting, structures were covered with crosslinking solution for 5 min. Once the crosslinking solution was removed, each structure was lifted with a spatula and placed in one well of a 6-well plate, to which 2–3 mL of NPM was added. The 6-well plate was then placed in the incubator. During the entire culture period of one week, culture media was not aspirated but only supplemented every 2–3 days.
For the preparation of the 2D culture control substrates, one well of a 6-well plate was coated with 200 μL of the UTEP bioink to cover the bottom of the well. Then, 2 mL of crosslinking solution was added to this bioink, and removed after 5 min. Next, 25 μL of the cell suspension (PKH26 pre-stained) was added to the well containing the bioink coating. Finally, 1 mL of the NPM was added, and the culture was placed in the incubator (37 °C, 5% CO2). On Day 2, the culture was supplemented with an additional 2 ml of NPM. During the entire culture period of one week, culture media was not aspirated but only supplemented (with 1 ml) every 2–3 days. For both 3D and 2D cultures, 3 wells of 6-wells (n = 3) were utilized for the experiments.
4.5.3. Fluorescent imaging of the NPC clusters and analysis
The 3D cultures were fixed in 4% paraformaldehyde in PBS for 15 min at room temperature prior to imaging, after 5 days of culture. Following fixing, 3D structures were mounted using Fluoromount-G Mounting Medium (ThermoFisher, Waltham, MA) onto glass slides using coverslips. The 2D cultures were fixed directly in the wells after Day 5. For this, 1 mL of 4% paraformaldehyde in PBS was added to the 2D culture, and left for 15 min after which it was aspirated. The 2D cultures was then imaged directly from the 6-well plate.
A Zeiss LSM700 confocal fluorescence microscope was used to image the 3D bioprinted structures and the 2D controls to determine cellular presence as well as cluster formation. All samples were imaged on Day 1, Day 3, and Day 5. For each sample, 2–3 images were taken at varying magnifications, at each time point studied.
MATLAB's ‘Image Processing Toolbox’ was used to identify and label cell clusters in acquired images. A cluster was defined as a connected group of three or more cells, and a single cluster was determined to have an area of approximately 480
μm2. This size was found by visually identifying cells, and then using MATLAB to detect them and measure their approximate area. Two metrics were analyzed: the total area of the clusters (μm2) and percentage of the total area occupied by the clusters (%).
The area of clusters was found by detecting all clusters in an image, and then using MATLAB to extract features from each cluster, including the area. Clusters that were considered to be mostly inside the borders of the image were included in the metric, while clusters considered to be mostly outside the borders of the image were discarded from the dataset.
For each cluster in contact with the edge of the image, the ratio of the contact length to the square root of the area was computed using Eq. (3):
| R = L/(A1/2) | (3) |
where R is the ratio of contact length, L is the length of contact between the cluster and the edge of the image, and A is the total area of a distinct cell cluster. The square root of the area was used in order to work with comparable units. All clusters with length of edge contact greater than 30% of the square root of the cluster's area (R > 0.3) were excluded from calculations. The percentage of area occupied by clusters was found in MATLAB by detecting all clusters in an image, summing their area, and dividing by the total area of the image using Eq. (4):
| % of area occupied by clusters = (total area occupied by cluster/s (μm2)/ Total area of the image ((μm2)) | (4) |
4.5.4. Flow cytometry analysis
To evaluate the biocompatibility and track cell proliferation in the 3D bioprinted scaffolds, NPCs were pre-stained using Cell Trace Yellow (CTY) proliferation kit (Invitrogen, Carlsbad, CA) based on manufacturer's protocol [21, 22]. The pre-stained cells were mixed with the bioink to constitute a final density of 1 million cells/ml. At day-3, scaffolds were cut using a blade, and cells were extracted using Miltenyi gentleMACS Dissociator (Miltenyi Biotec, Cambridge, MA) using a Multi-tissue Dissociation Kit-1 by selecting the Multi_B program according to the manufacturer's protocol [1, 21]. Cells were fixed with 4% PFA for 15 min at room temperature and added to their designated FACS analysis falcon tubes and analyzed using Beckman Coulter Gallios Flow Cytometer (Brea, CA, USA) using the appropriate excitation and emission wavelengths. Positive controls included freshly pre-stained cells while negative controls included neural progenitor cells not pre-stained with CTY.
4.5.5. Immunochemistry
For the preparation of the 2D culture wells for this experiment, one well of a 6-well plate was coated with 200 μL of the UTEP bioink to cover the bottom of the well as described earlier, using 3 × 106 cells/ml for a total of a 5-day culture period. After the end of the culture period, the culture wells were collected, fixed with 4% paraformaldehyde for 15 min, washed with PBS 3 times and incubated with blocking solution (PBS with 1% NGS and 0.3% Triton X-100) for 60 min prior to proceeding with the next steps. The samples were washed once with PBS and incubated with the primary antibody anti-Oct3/4 (1:400; SantaCruz Technologies, Dallas, TX), overnight at 4 °C. The expression of the Oct-3/4 isoform in the NPC cells, via this method would confirm the undifferentiated phenotype of the NPC cells. After primary antibody treatment, the samples were washed thrice with PBS. Alexa Fluor 488 goat anti-rabbit IgG (1:2000; Thermo-Fisher Scientific, Waltham, MA, USA) was added as a secondary antibody and incubated for 1 h at room temperature. Samples were treated to three washes and treated with Hoechst 33342 Solution (20 mM) based on manufacturer's protocol (ThermoFisher). Images were acquired using the Zeiss microscope as described earlier.
4.5.6. Analysis of expression of molecular markers using RT-qPCR by comparative Ct method
4.5.6.1. Extraction of cells
To perform the assessment of gene expression of the NPCs 3D printed within the UTEP bio-ink we used qPCR. NPCs were mixed within the bioink in a density of 100,000 cells/ml and 3D printed for this experiment. Composite cell-gel samples were cultured in vitro in the incubator at 37 °C, 5% CO2 for 5 days. In parallel, NPCs were cultured atop the bioink coating and considered as a control 2D condition. The composite (cell-gels) was taken out of the incubator and 1X PBS was added to wash out any remaining of media, and the cells from the gel were extracted following others published methods, where 0.05M EDTA +0.05M Sodium Citrate was used for 15 min at room temperature to dissolve the bio-ink and extract the cells [45]. Then, the dissolved mixture was collected in sterile tubes and centrifuged at 400g for 10 min to yield a cell pellet. The supernatant phase was removed.
4.5.6.2. RNA Extraction and cDNA Synthesis of NPC
Total RNA extraction was carried out using RNeasy® Plus Mini Kit (QIAGEN. Hilden, Germany) according to the manufacturer's instructions. Extracted RNAs were quantified by NanoDrop OneC spectrophotometer (ThermoFisher Scientific, Waltham, USA) and the absorbance ratios at 260/280 nm and 260/230 nm were measured to control RNA purity. The data is tabulated below.
| sample | ng/μl | 260/280 | 230/260 |
|---|---|---|---|
| Cells in control (2D) | 738.8 | 2.08 | 2.16 |
| Cells in 3D structure No. 1 | 3.4 | 1.89 | 0.05 |
| Cells in 3D structure No. 2 | 4.3 | 1.77 | 0.20 |
| Cells in 3D structure No. 3 | 4.4 | 1.93 | 0.16 |
Total RNA (50 ng) was reverse transcribed to cDNA using the First Strand cDNA Synthesis Kit (OriGene Technologies, Inc.) in a volume of 20 μL, according to the manufacturer's instructions. Extracted cDNA were quantified by NanoDrop One.C spectrophotometer (ThermoFisher Scientific, Waltham, USA) and the absorbance ratios at 260/280 nm and 260/230 nm were measured and tabulated below.
| sample | ng/μl | 260/280 | 230/260 |
|---|---|---|---|
| Cells in control (2D) | 900.3 | 1.83 | 2.28 |
| Cells in 3D structure No.1 | 1167.1 | 1.82 | 2.16 |
| Cells in 3D structure No.2 | 1096.7 | 1.82 | 2.25 |
| Cells in 3D structure No.3 | 1132.2 | 1.81 | 2.22 |
4.5.6.3. Quantitative polymerase chain reaction
The RT–qPCR reactions were performed in the Quantstudio 3 (Applied Biosystems, Invitrogen, CA). The control samples (2D) reaction was performed in quadruplicate. 3D samples reactions were performed using duplicates per sample (6 total), in qPCR Tubes, 8 strips, 0.2 mL, with optical strips caps (Pure AMP PCR plastics, MTC™ Bio Incorporated) in a reaction volume of 20 μL. All reactions contained 1 μL of cDNA (900 ng), 10 μL of Go Taq qPCR Master Mix SYBR and 0.2 μL of supplemental CXR Ref. Dye (Promega), 1 μL of 10 mM of mix primer (Fw, Rv) (Origene Tech Inc.) and Nuclease free water grade. The reaction started with a 10 min initial denaturation step at 95 °C, 40 cycles of 95 °C for 15 s and 60 °C for 15 s according to the protocol provided by Origene. The genes to analyze were determined by the development of NPC using GAPDH as reference gene (control) and NES and SOX2 as the target gene [46, 47, 48]. The primer sequences corresponding to the genes to evaluate are shown in the table below. To avoid sample contamination and primer-dimer formation that could produce false positive results, no template control was used.
4.6. Statistical analysis
All experiments were performed in triplicate and values were expressed as means ± standard deviations. All data were compared using a one-way ANOVA with a post-hoc Tukey's multiple comparison test with significance defined as a two-tailed p-value less than 0.05. Additionally, an ANOVA test was performed across Days 1, 3 and 5 to assess the formation and maintenance of the NPCs clustering phenomenon.
4.6.1. Data Analysis of qPCR
The quantification cycle (Ct) values were automatically calculated by the qPCR instrument software Quantstudio 3 (Applied Biosystem, Invitrogen). Statistical algorithm was used to evaluate gene expression of the comparative CT method (2−ΔΔCT) (Schmittgen and Livak, 2008). Average CTs of GAPDH were used as an endogenous control, stability of the target genes Ct values of each candidate for relative gene expression [49]. The results helped derive the value of the fold change in expression from the target genes (NES and SOX2) normalized with the reference gene GAPDH. A student's T-test analysis was performed for each gene studied within the two groups (2D and 3D culture of NPCs) to evaluate if the UTEP bioink had altered the function of the NPCs.
Declarations
Author contribution statement
All authors made substantial contributions to all of the following: (1) the conception and design of the study, the acquisition of data, or the analysis and interpretation of data; (2) drafting the article or critically revising its important intellectual content; (3) final approval of the version submitted. IB, DS, SA, NRO, MA, REK and CL were responsible for 1 - Conceived and designed the experiments; 2 - Performed the experiments; 3 - Analyzed and interpreted the data; 4 - Contributed reagents, materials, analysis tools or data; 5 - Wrote the paper. BJ was responsible for 1 - Conceived and designed the experiments; 3 - Analyzed and interpreted the data; 5 - Wrote the paper.
Funding statement
Dr. Binata Joddar was supported by National Science Foundation [1828268 & 1927628].
Data availability statement
Data will be made available on request.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
Supplementary content related to this article has been published online at [URL].
Acknowledgements
There are no conflicts to declare. The Joddar laboratory (IMSTEL) acknowledges the following NSF grant/s 1828268 and 1927628. DS was supported by a fellowship from the Research Initiative for Scientific Enhancement (RISE) program (NIH/NIGMS grant #2R25GM069621-14) at UTEP. IB and SA were supported during the duration of this study by the NSF PREM Grant No. 1827745. NO acknowledges the grants from National Institute of General Medical Sciences of the National Institutes of Health under RL5GM118969, TL4GM118971, and the NIH BUILD Pilot grant # 8UL1GM118970-02. MA acknowledges the Gates Millennium Fellowship and the Eloise and Patrick Wieland Graduate Fellows Program at UTEP. We also acknowledge the NIH-NIMHD-RCMI Grant No. 5G12MD007592 that supported use of the confocal microscopy facility in BBRC at UTEP.
Appendix C. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Anil Kumar S., et al. A visible light-cross-linkable, fibrin–gelatin-based bioprinted construct with human cardiomyocytes and fibroblasts. ACS Biomater. Sci. Eng. 2019;5(9):4551–4563. doi: 10.1021/acsbiomaterials.9b00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anil Kumar S., et al. A comparative study of a 3D bioprinted gelatin-based lattice and rectangular-sheet structures. Gels. 2018;4(3):73. doi: 10.3390/gels4030073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joddar B., et al. Circulation Research. Am Heart Assoc; 2020. A 3D bioprinted human cardiac cell platform to model the pathophysiology of diabetes. [Google Scholar]
- 4.Roman B., et al. A model for studying the biomechanical effects of varying ratios of collagen types I and III on cardiomyocytes. Cardiovasc. Eng. Tech. 2021;12(3):311–324. doi: 10.1007/s13239-020-00514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yong U., Kang B., Jang J. 3D bioprinted and integrated platforms for cardiac tissue modeling and drug testing. Essays Biochem. 2021;65(3):545–554. doi: 10.1042/EBC20200106. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee A., et al. The influence of hydrogel modulus on the proliferation and differentiation of encapsulated neural stem cells. Biomaterials. 2009;30(27):4695–4699. doi: 10.1016/j.biomaterials.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma R., et al. 3D bioprinting pluripotent stem cell derived neural tissues using a novel fibrin bioink containing drug releasing microspheres. Front. Bioeng. Biotechnol. 2020;8:57. doi: 10.3389/fbioe.2020.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santhosh D., Huang Z. Regulation of the nascent brain vascular network by neural progenitors. Mech. Dev. 2015;138:37–42. doi: 10.1016/j.mod.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahmoud A.I., et al. Nerves regulate cardiomyocyte proliferation and heart regeneration. Dev. Cell. 2015;34(4):387–399. doi: 10.1016/j.devcel.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh Y., et al. Functional coupling with cardiac muscle promotes maturation of hPSC-derived sympathetic neurons. Cell Stem Cell. 2016;19(1):95–106. doi: 10.1016/j.stem.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi A., et al. Sympathetic neurons modulate the beat rate of pluripotent cell-derived cardiomyocytes in vitro. Integ. Biol. 2012;4(12):1532–1539. doi: 10.1039/c2ib20060k. [DOI] [PubMed] [Google Scholar]
- 12.Kaelberer M.M., et al. A gut-brain neural circuit for nutrient sensory transduction. Science. 2018;361(6408):eaat5236. doi: 10.1126/science.aat5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De la Vega L., et al. 3D bioprinting human induced pluripotent stem cell-derived neural tissues using a novel lab-on-a-printer technology. Appl. Sci. 2018;8(12):2414. [Google Scholar]
- 14.Shen L., et al. Effects of calcium ion and covalent crosslinking on formation and elasticity of fibrin gels. Thromb. Res. 1975;6(3):255–265. doi: 10.1016/0049-3848(75)90073-0. [DOI] [PubMed] [Google Scholar]
- 15.Lee K.Y., Mooney D.J. Alginate: properties and biomedical applications. Prog. Polym. Sci. 2012;37(1):106–126. doi: 10.1016/j.progpolymsci.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muzzarelli R.A., et al. Genipin-crosslinked chitosan gels and scaffolds for tissue engineering and regeneration of cartilage and bone. Mar. Drugs. 2015;13(12):7314–7338. doi: 10.3390/md13127068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deo K.A., et al. Bioprinting 101: design, fabrication, and evaluation of cell-laden 3D bioprinted scaffolds. Tissue Eng. 2020;26(5-6):318–338. doi: 10.1089/ten.tea.2019.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pi Q., et al. Digitally tunable microfluidic bioprinting of multilayered cannular tissues. Adv. Mater. 2018;30(43) doi: 10.1002/adma.201706913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alonzo M., et al. Hydrogel scaffolds with elasticity-mimicking embryonic substrates promote cardiac cellular network formation. Prog. Biomat. 2020;9(3):125–137. doi: 10.1007/s40204-020-00137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tibbitt M.W., Anseth K.S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009;103(4):655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abelseth E., et al. 3D printing of neural tissues derived from human induced pluripotent stem cells using a fibrin-based bioink. ACS Biomater. Sci. Eng. 2018;5(1):234–243. doi: 10.1021/acsbiomaterials.8b01235. [DOI] [PubMed] [Google Scholar]
- 22.Akbalaeva B., et al. Ischemic heart disease: focus on acute infarction of the right ventricular myocardium. Kardiologiia. 2017;(1):90–94. [PubMed] [Google Scholar]
- 23.Okuda T., et al. Oct-3/4 repression accelerates differentiation of neural progenitor cells in vitro and in vivo. Mol. Brain Res. 2004;132(1):18–30. doi: 10.1016/j.molbrainres.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 24.Mozaffarian D., et al. Heart disease and stroke statistics—2015 update: a report from the American Heart Association. Circulation. 2015;131(4):e29–e322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 25.Lund L.H., et al. The registry of the International Society for Heart and Lung Transplantation: thirty-first official adult heart transplant report—2014; focus theme: retransplantation. J. Heart Lung Transplant. 2014;33(10):996–1008. doi: 10.1016/j.healun.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Rangappa S., Makkar R., Forrester J. Current status of myocardial regeneration: new cell sources and new strategies. J. Cardiovasc. Pharmacol. Therapeut. 2010;15(4):338–343. doi: 10.1177/1074248410376382. [DOI] [PubMed] [Google Scholar]
- 27.Litviňuková M., et al. Cells of the adult human heart. Nature. 2020;588(7838):466–472. doi: 10.1038/s41586-020-2797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fedele L., Brand T. The intrinsic cardiac nervous system and its role in cardiac pacemaking and conduction. J. Cardiovasc. Develop. Dis. 2020;7(4):54. doi: 10.3390/jcdd7040054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukuda K., et al. Cardiac innervation and sudden cardiac death. Circ. Res. 2015;116(12):2005–2019. doi: 10.1161/CIRCRESAHA.116.304679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zandstra T.E., et al. Asymmetry and heterogeneity: Part and parcel in cardiac autonomic innervation and function. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.665298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandrasekaran A., et al. Comparison of 2D and 3D neural induction methods for the generation of neural progenitor cells from human induced pluripotent stem cells. Stem Cell Res. 2017;25:139–151. doi: 10.1016/j.scr.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 32.Weiss P., Hiscoe H.B. Experiments on the mechanism of nerve growth. J. Exp. Zool. 1948;107(3):315–395. doi: 10.1002/jez.1401070302. [DOI] [PubMed] [Google Scholar]
- 33.Brito C., et al. Generation and genetic modification of 3D cultures of human dopaminergic neurons derived from neural progenitor cells. Methods. 2012;56(3):452–460. doi: 10.1016/j.ymeth.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Lendahl U., Zimmerman L.B., McKay R.D.G. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60(4):585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds B.A., Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255(5052):1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 36.Frederiksen K., McKay R. Proliferation and differentiation of rat neuroepithelial precursor cells in vivo. J. Neurosci. 1988;8(4):1144–1151. doi: 10.1523/JNEUROSCI.08-04-01144.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahlstrand J., Lardelli M., Lendahl U. Nestin mRNA expression correlates with the central nervous system progenitor cell state in many, but not all, regions of developing central nervous system. Dev. Brain Res. 1995;84(1):109–129. doi: 10.1016/0165-3806(94)00162-s. [DOI] [PubMed] [Google Scholar]
- 38.Wegner M., Stolt C.C. From stem cells to neurons and glia: a Soxist's view of neural development. Trends Neurosci. 2005;28(11):583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 39.Loh K.M., Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell. 2011;8(4):363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z., et al. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10(4):440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 41.Zhao S., et al. SoxB transcription factors specify neuroectodermal lineage choice in ES cells. Mol. Cell. Neurosci. 2004;27(3):332–342. doi: 10.1016/j.mcn.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S., Cui W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cell. 2014;6(3):305–311. doi: 10.4252/wjsc.v6.i3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bae M., et al. Neural stem cell delivery using brain-derived tissue-specific bioink for recovering from traumatic brain injury. Biofabrication. 2021;13(4) doi: 10.1088/1758-5090/ac293f. [DOI] [PubMed] [Google Scholar]
- 44.AnilKumar S., et al. The applicability of furfuryl-gelatin as a novel bioink for tissue engineering applications. J. Biomed. Mater. Res. B Appl. Biomater. 2019;107(2):314–323. doi: 10.1002/jbm.b.34123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao B., et al. Enzymatically degradable alginate/gelatin bioink promotes cellular behavior and degradation in vitro and in vivo. Biofabrication. 2019;11(4) doi: 10.1088/1758-5090/ab38ef. [DOI] [PubMed] [Google Scholar]
- 46.Augustyniak J., et al. Reference gene validation via rt–qPCR for human iPSC-derived neural stem cells and neural progenitors. Mol. Neurobiol. 2019;56(10):6820–6832. doi: 10.1007/s12035-019-1538-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawa A., et al. Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc. Natl. Acad. Sci. USA. 1997;94(21):11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hutton S.R., Pevny L.H. SOX2 expression levels distinguish between neural progenitor populations of the developing dorsal telencephalon. Dev. Biol. 2011;352(1):40–47. doi: 10.1016/j.ydbio.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 49.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.