Summary
Cells constantly respond to environmental changes by modulating gene expression programs. These responses may demand substantial costs and, thus, affect cell growth. Understanding the regulation of these processes represents a key question in biology and biotechnology. Here, we studied the responses to osmotic stress in glucose-limited environments. By analyzing seventeen osmotic stress-induced genes and stress-activated protein kinase Hog1, we found that cells exhibited stronger osmotic gene expression response and larger integral of Hog1 nuclear localization during adaptation to osmotic stress under glucose-limited conditions than under glucose-rich conditions. We proposed and verified that in glucose-limited environment, glycolysis intermediates (representing “reserve flux”) were limited, which required cells to express more glycerol-production enzymes for stress adaptation. Consequently, the regulatory mechanism of osmoresponse was derived in the presence and absence of such reserve flux. Further experiments suggested that this reserve flux-dependent stress-defense strategy may be a general principle under nutrient-limited environments.
Subject areas: Biological sciences, Cell biology, Molecular biology
Graphical abstract
Highlights
-
•
Cell’s response facing osmostress under different glucose concentrations
-
•
Higher protein synthesis rates induced by osmostress when glucose is limited
-
•
Defense-reserve-flux reduced for optimal growth when glucose is limited
-
•
Two situations indicated by adaptation behaviors based on metabolism
Biological sciences; Cell biology; Molecular biology;
Introduction
Organisms develop various strategies to survive and proliferate in rapidly changing environments. For example, bacteria carry out chemotaxis to avoid unfavorable conditions and migrate toward favorable locations1,2; yeast cells orchestrate a gene expression program called the environmental stress response (ESR) to adapt to environmental changes.3,4,5,6 Cells also modulate the proteome or energy allocation under different growth rates or growth conditions.7 In bacterial cells under steady-state exponential growth, it is well known that proteome organization predominantly depends on the growth rate, which is restricted by the nutritional quality of the medium.8 As the growth rate decreases in a nutrient-limited environment, cells tend to invest more proteomic resources into intake systems rather than translation and metabolism.9 However, cellular resource allocation and the stress response have always been considered independently in previous work. How cells prepare and respond to stress under nutrient-limited environments have yet to be determined.
Even under nutrient-limited conditions where the growth rate remains unchanged, proteome partitioning and energy allocation may still be altered. Previous studies have shown that in Saccharomyces cerevisiae, the cell growth rate under 0.1% glucose concentration is similar to that under 2% glucose.10 However, translation and metabolism greatly differ under these conditions. For instance, genes involved in glucose uptake and carbohydrate-related metabolism have a higher expression level under 0.1% glucose.11,12 It could be intriguing to determine how external glucose conditions influence cellular stress responses.13 On the one hand, the low glucose level may elicit a mild stress response, priming the cell to better adapt to subsequent severe stresses.14,15 On the other hand, cell proliferation may compete with stress-responsive pathways for limited resources, reducing the efficiency of adaptation.
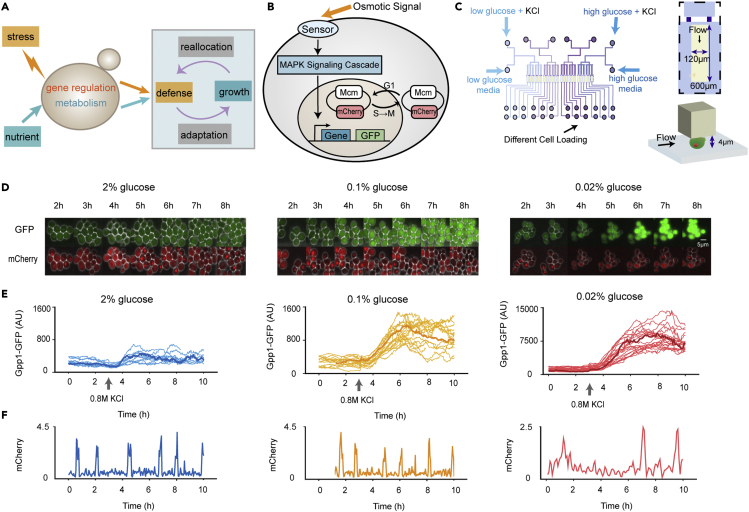
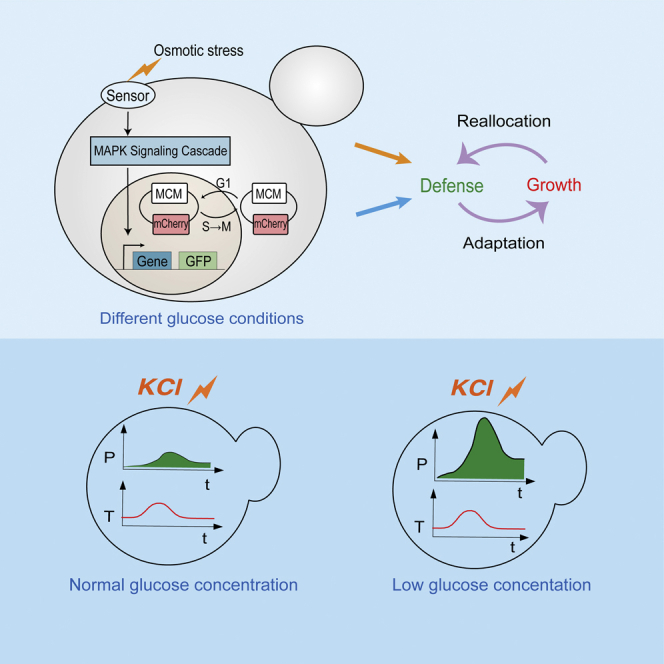
In the natural habitats of yeast, one of the most common causes of stress is rapidly changing water activity.16 Thus, cells are often exposed to hyperosmostress and need to maintain water balance. The osmoregulatory system in yeast is quite well understood. The osmoadaptation network integrates gene expression and regulation of metabolic flux. The key to yeast osmoregulation is the production and accumulation of the compatible solute glycerol, which is mainly regulated by the HOG mitogen-activated protein kinase (MAPK) pathway.17,18 In particular, the redistribution of the glycolytic flux from growth to glycerol production is a major process in osmoadaptation.19,20,21 Therefore, in this work, we mainly discuss the stress response and the defense strategy of budding yeast in response to osmotic stress under different glucose concentrations (Figure 1A).
Figure 1.
Schematic model, strain background, and experimental settings
(A) A simplified view of the trade-off between growth and stress defense. Nutritional restriction affects the growth rate through metabolism and gene regulation, while the stress response and growth compete for limited resources.
(B) Schematics of the process of monitoring target protein dynamics (with fluorescent protein fusions) and the cell cycle (with the Mcm-mCherry. system) in single cells.
(C) Design of the microfluidic device. The chip consists of four identical units, each with six independent flow channels. The yellow part of a zoomed-in typical unit is the observation area, where yeast cells are fixed under a micropillar. Yeast cells were cultured independently under different glucose conditions for 8 hours and then exposed to hyperosmotic stress (0.8 M KCl).
(D) Time-lapse microscopy images of yeast cells in response to 0.8 M KCl, which was added at the 3 h time point under different culture conditions. The GFP channel indicated the osmotic-induced protein GPP1-GFP, and the mCherry channel indicated the cell cycle marker Mcm-mCherry. The scale bar is 5 μm.
(E) Examples of traces of individual cells. The left panel shows the rich-glucose condition (2%), the middle panel shows the low-glucose (0.1%) condition, and the right panel shows the poor-glucose (0.02%) condition. Highlighted are the cells with the maximum expression of Gpp1 as the population median. The numbers of cells under the 2%, 0.1%, and 0.02% glucose conditions were 29, 33, and 35, respectively.
(F) Time sequence of the cell cycle marker Mcm-mCherry. in typical cells, and the Gpp1 production rate was at the median of the population (highlighted in E).
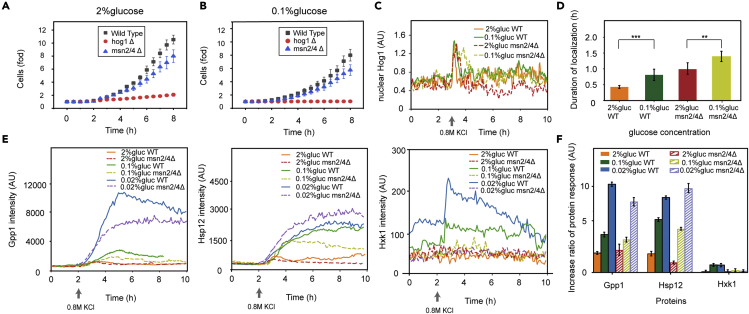
Specifically, we studied the protein dynamics under hyperosmostress in single yeast cells in a glucose-limited environment using a microfluidic device. The synthesis rates of stress-induced proteins and the growth rate of cells were measured. When cells were exposed to osmotic stress in a glucose-limited environment, the synthesis rates of osmo-induced proteins were increased by more than 2-fold. Consistently, we observed a corresponding increase in the duration of the nuclear translocation of the upstream stress-activated kinase Hog1. And the hog1Δ strain showed poor adaptation to osmotic stress among different glucose concentrations, while for the general stress-responsive transcription factor Msn2/4, the adaptation to osmotic stress of msn2Δmsn4Δ strain under different glucose concentrations was close to that of the wild-type strain.
Given the important role of glucose in metabolism during osmoadaptation, we speculated that in a low-glucose environment with a limited glycolytic flux, cells need to induce more glycerol-production enzymes to adapt to hyperosmostress, which underlies the stronger gene expression response that we observed. A simplified model based on this hypothesis was established to explain the stress-induced protein dynamics. The model also helped clarify the resource allocation strategy in glucose-limited environments. Thus, yeast reduces the metabolic reserve flux for optimal growth under the condition of continuous glucose limitation, which we designate “reserve flux abandonment”. Furthermore, the concentration of the required substrate of the stress response decreased. As a cost, upon a stress encounter, yeasts need to synthesize more related enzymes to produce sufficient metabolites (e.g., glycerol) for adaptation. The regulation laws of resource allocation revealed here represent a general principle of the operation of complex biological systems under nutrient-limited conditions.
Results
The stress response of cells to osmostress under different glucose concentrations shows obvious differences
To characterize the stress response of budding yeast in a glucose-limited environment, we monitored yeast cells with GFP-tagged stress-responsive proteins using a microfluidic system. The strain also has Mcm protein labeled by m-Cherry as a cell cycle marker (Figures 1B and S1). The Mcm complex is imported into the nucleus in the G1 phase and exported from the nucleus during the S phase; thus, the Mcm complex can be used to easily quantify the duration of the cell cycle22,23 (Figure S2).
For a low-glucose environment, we chose a glucose concentration of 0.1% to ensure that the growth rate was not significantly affected24 and 0.02% under which the cell growth rate was significantly slower than that under glucose-rich conditions. Cells were initially cultured for 8 hours in a microfluidic system with different glucose concentrations to adapt to the environment; during this time, the expression levels of stress-responsive proteins and the duration of the cell cycle remained constant. Then, the cells were exposed to osmostress (0.8 M KCl, the same under different glucose concentrations) for 7 hours (Figure 1C). Bright-field and fluorescence images were collected every 2.5 min for dynamic studies.
When exposed to osmostress, osmotic stress proteins, such as Gpp1, a well-characterized Hog-MAPK downstream protein,25 were upregulated under all 0.02%, 0.1%, and 2% glucose conditions (Figures 1D and 1E). Strikingly, a higher expression level of Gpp1-GFP was observed under the lower glucose conditions than under the 2% glucose conditions. In addition, the cell cycle exhibited a phase of shock26,27,28 because the extended G1 phase could help resist adverse environments.11 Under glucose-limited conditions, the cells seemed to require a longer recovery time. After the first generation resumed growth, the cell cycle gradually adapted to the initial level in the case of 0.1% and 2% glucose but with a prolongation in the case of 0.02% glucose (Figures 1F and S3).
Osmotic adaptation in a rich medium (2% glucose) has been extensively studied.16 Interestingly, in a low-glucose environment (0.1% glucose), stress-related proteins show a more pronounced response to osmotic stress, while the growth rate does not seem to be restricted. We were also interested in the mechanisms of the dramatic stress response differences among the low-glucose conditions. Regardless of whether the cellular physiology was affected, resource allocation may have differed in the glucose-limited environments.
Stress response proteins have a higher synthesis rate in a glucose-limited environment
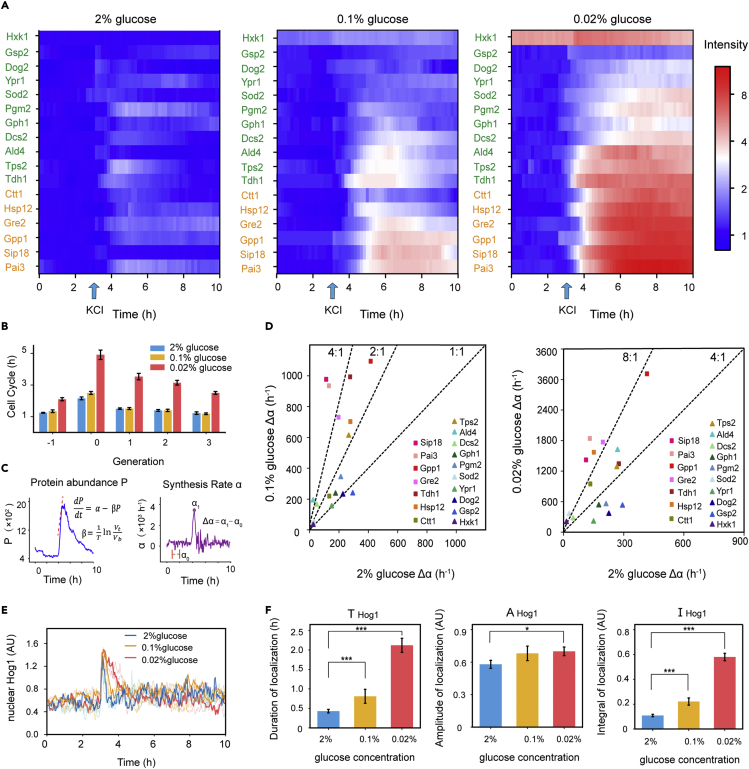
To systematically study this phenomenon, we performed a genetic screen of 40 important stress response proteins.29 Among them, we selected 17 proteins whose expression level increased by more than half of the base value upon osmotic stress, including 6 osmotic stress-induced proteins and 11 proteins related to other stresses (listed in STAR Methods). The levels of the target proteins at different glucose concentrations did not significantly differ under a constant nutrient supply. After switching to a hyperosmotic environment, the stress-responsive proteins were upregulated and exhibited an obvious difference between glucose-limited environments and a glucose-rich environment (Figure 2A).
Figure 2.
Modulation of the osmotic response under different glucose concentrations
(A) Protein dynamics at different glucose concentrations. 0.8 M KCl was added at 3 h. The data were normalized to the average protein intensity in the initial 2 hours under 2% glucose, with greater expression indicated by red and lower expression indicated by blue (color bar log base 2). N > 100 cells per biological replicate. Osmotic relative proteins are marked in red. Other proteins are marked in blue.
(B) Bar graph of the mean duration of cell cycles of individual cells at different glucose concentrations. Generation 0 means the generation which is added the osmotic stress. The numbers of cells under the 2%, 0.1%, and 0.02% glucose conditions were 80, 92, and 89, respectively. Error bars represent the SEM.
(C) Parametric decomposition of response curves of the protein abundance and dilution rate to obtain the synthesis rate. The Δα is the maximum synthesis rate after exposure to hyperosmotic conditions minus the average synthesis rate before exposure (1∼2 hours).
(D) Δα under different conditions. The Δα of osmotic relative proteins at 0.1% glucose was more than twice greater than that at 2% glucose (red squares). The Δα of osmotic relative proteins at 0.02% glucose was more than four times that at 2% glucose.
(E) Localization trajectory of Hog1 transcriptional regulation. 0.8 M KCl was added at 3 h. The bold line shows a single cell whose nuclear duration is the median value in the population. The numbers of cells used for the analysis under the 2, 0.1%, and 0.02% glucose conditions were 45, 43, and 56, respectively.
(F) The duration, amplitude, and integral of Hog1 nuclear localization were quantified. Data are represented as mean +/- SEM. Statistical significance is calculated with Student’s t test (∗p < 0.05, ∗∗∗p < 0.001).
The protein synthesis rate was used to characterize these responses as described by the following equation:
| (Equation 1) |
where P denotes the protein concentration, and α and β denote the protein synthesis rate and the decay rate, respectively. The decay rate β is mainly determined by dilution as protein degradation during a fast response can be neglected.30 Thus, the increase Δα of the maximum synthesis rate after stress could be calculated from time-course data and reflects the stress response cost (Materials and Methods, Figures 2B and 2C). Figure 2B shows that under glucose-limitation conditions, the cells employed a longer recovery time upon osmotic stress. After the first generation resumed growth, the cell cycle gradually almost adapted to the initial level in 0.1% and 2% glucose but with a prolongation in the case of 0.02%.
We noticed that most stress-related proteins had higher expression levels when facing osmostress in a low-glucose environment (Figure 2D). Among them, the Δα of Hog-MAPK pathway-induced proteins was more than twice that in the rich medium (2% glucose) under low-glucose condition (0.1%) and more than four times that in 2% under lower-glucose condition (0.02%) (the proteins Pai3, Sip18, Gpp1, Gre2, Hsp12, and Ctt1 are marked in red squares in Figure 2D). The Δα of the other proteins (the proteins Tdh1, Tps2, Dcs2, Gph1, Pgm2, Sod2, Ypr1, Dog2, Gsp2, and Hxk1, which are not directly influenced by the Hog-MAPK pathway) showed fewer differences in response to osmotic stress under the low-glucose or normal conditions. But our results also suggest that these stress-response proteins may be more complex than a simple categorization because some proteins reported as not directly influenced by Hog-MAPK pathway also have higher increase of production rate than the Hog-dependent proteins.
Response dynamics are indicated by the transcription level
Since we observed a significant increase in the stress response to osmotic stress under glucose-limited conditions compared with glucose-rich conditions, we wondered whether these different dynamical profiles were associated with transcription. In our description of the response process, protein synthesis correlated with the integral of mRNA, which was proportional to the integral of transcription factor activity.6
We measured the relative nuclear intensity of Hog1, which is implicated in the transcriptional process during osmoregulation in budding yeast. Although single cells exhibited variability in nuclear localization, Hog1 showed longer nuclear translocation in the glucose-limited environments compared with the glucose-rich conditions when exposed to osmotic stress (Figures 2E and S4).
Then, we quantified the dynamic characteristics of Hog1 localization in each single cell and calculated the average values across the populations under different glucose concentrations (duration, amplitude, and integral area; Figure 2F). As expected, we found that the duration and integral of Hog1 nuclear localization significantly increased under the glucose-limited conditions.
Taken together, these results suggest that the nuclear localization of Hog1 is required for resisting osmotic stress. Cells tend to allocate more resources to the Hog pathway in a glucose-limited environment.
Metabolic reconfiguration strategy of S. cerevisiae
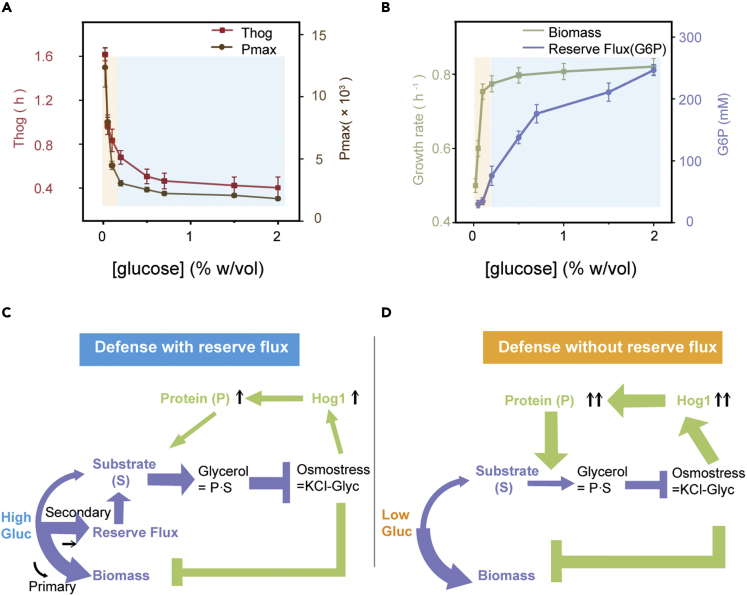
Upon osmostress, yeast cells produced more osmotic-related proteins and required a longer recovery time under the lower-glucose conditions (Figure 3A). To explain the protein response difference among the different glucose conditions, we considered the metabolic reconfiguration of osmotic adaptation.
Figure 3.
Metabolic configuration strategy in different glucose environments
(A) Hog1 burst duration and maximum protein expression (GPP1-GFP) at different glucose concentrations. Error bars represent the SEM from at least 40 cells.
(B) Biomass production was measured as the growth rates at different glucose concentrations. The data points show the exponential cellular growth rate. The mean and SEM are from N = 82∼114 cells. Reserve flux was indicated by the central glycolysis intermediate glucose-6-phosphate (G6P) using a colorimetric assay. Error bars (SD) are from three independent experiments. When the glucose concentration was sufficient, excess glycolysis flux occurred for defense (blue shading). When the glucose concentration was lower, cells did not have any reserve (surplus) flux and maximized their growth rate (orange shading).
(C) A schematic of the interplay between osmotic adaptation and metabolic reconfiguration with a reserve flux. Metabolism (purple) and gene regulation (green) osmoadaptation require the compatible solute glycerol to compensate for a given stimulus. Glycolytic flux is first rerouted to maintain the normal growth of cells and then to defensive reserves.
(D) At lower glucose concentrations that are not sufficient for normal growth, cells abandon their reserve flux. The reduction in the substrate requires more glycerol synthase to synthesize a sufficient concentration of glycerol.
Glucose is used by several cross-linking pathways once it enters the cell. The main route of glucose utilization is glycolysis, which converts glucose to trehalose, glycogen, glycerol, etc.31 The most important branch in unstressed cells is the carbon flux to biomass for growth. Under abundant glucose conditions (almost with a maximum growth rate, Figure 3B, blue area), the carbon flux is redundant for growth, and extra reserve flux is stored in other metabolic branches, which are represented by the central glycolysis intermediate glucose-6-phosphate (G6P). Under lower-glucose conditions (cells grow significantly slower than the maximum rate), cells abandon the reserve flux to optimize growth (Figure 3B, orange area). This finding implies that cells may have two types of stress-defense strategies under different glucose conditions.
A previous study reported that under rich-glucose conditions, yeast stored excess carbon in several metabolic pathways, and the first step of the osmotic stress response was to route internal stores of the metabolic flux to glycerol production, accelerating osmoadaptation.32 However, our results show that metabolic flux redistribution may be more important (data not shown). When glucose was relatively abundant, the growth rate of the cells treated with 0.1% glucose was almost the same as that of the cells treated with 2% glucose. However, the reserve flux (represented by the metabolite G6P) and the substrate for glycerol synthesis inevitably decrease with decreasing glucose concentrations.33 Because the substrate for glycerol synthesis was reduced in the glucose-limited environment, more enzymes were required to accumulate glycerol to balance the osmotic pressure. During exposure to osmostress, the cells redistributed the glycolytic flux from the reserve flux and growth to glycerol production (Figures 3A and 3B, blue area; Figure 3C).
Under more limited glucose conditions, if cells still maintain the metabolic reserve flux, their instantaneous biosynthesis is reduced, and growth is adversely affected. Indeed, slow-growing cells are better prepared for stress than fast-growing cells,34 which may represent a good strategy. However, our results show that yeast abandoned their reserve flux (indicated by the metabolite G6P) when glucose was further reduced (Figure 3B, orange area; Figure 3D). When facing stress, cells without a reserve and reserve flux can only redistribute the glycolytic flux from growth to glycerol production, resulting in an extremely long recovery time (Figures 3A and 3B, orange area; Figures 3D and S5).
Thus, the emerging picture is that under hyperosmotic stress, the process of osmoadaptation leads to the coordinated activation of the environmental stress response and suppression of biomass formation. Protein production indicates the response cost, and cell-cycle arrest represents the glycolysis reallocation expense. Therefore, when nutrients are insufficient to redirect sufficient glycolytic flux, cells allocate more resources to stress-responsive gene expression by reducing their growth rate. Moreover, this phenomenon was more significant when there was no metabolic reserve flux (Figure 3D), and it may not proceed via the same mechanism as when there was a metabolic reserve flux (Figure 3C).
Model of metabolic-combined osmotic adaptation
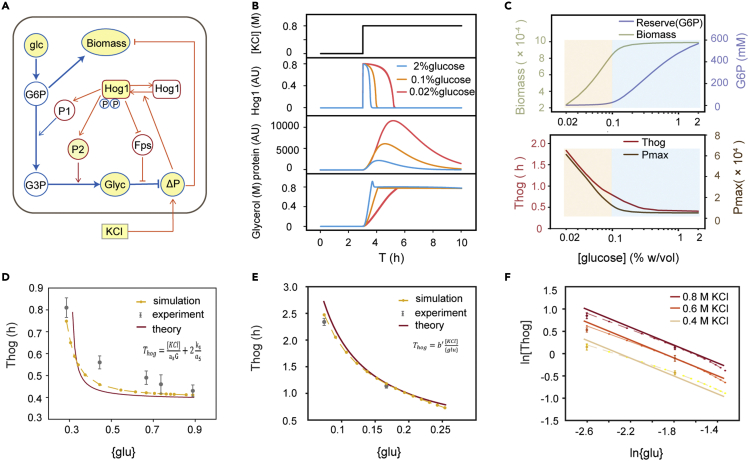
To corroborate the above hypothesis in silico, we developed a computational model including the main metabolic features and feedback mechanisms of osmotic adaptation. Specifically, G6P is the main product of glucose phosphorylation and is, in turn, converted to biomass and glycerol-3-phosphate (G3P). Glycolytic flux to G3P can be regulated via the Hog1-mediated activation of phosphofructose-2-kinase, which is denoted by protein P1. Then, G3P is transformed to glycerol by G3P phosphatases, which is represented by protein P2.25,35 Excessive glycerol leaks through the glycerol facilitator Fps1(open state in Figure 4A), which appears to be controlled by Hog1.36,37 However, we assumed that Hog1 phosphorylation and the reduction in biomass production were influenced by osmotic pressure discrepancies. A schematic of the model is presented in Figure 4A, and the details of the coupled ordinary differential equations are included in the STAR Methods.
Figure 4.
A simplified model explaining the enhanced stress response under glucose-limited conditions
(A) The core network structures describe the rerouting of the glycolytic flux from growth to glycerol. Blue indicates glycolysis modulation. Orange indicates gene regulation. Measured entities are highlighted in yellow.
(B) Model simulation of phosphorylated Hog1, downstream proteins, and intracellular glycerol.
(C) Biomass, reserve (G6P), duration of nuclear Hog1 (Thog), and the maximum value of downstream protein P2 at different glucose concentrations after the stimulation. The duration of nuclear Hog1 (Thog) reflects the adaptation time.
(D) Under glucose-rich conditions (glucose >0.1%), Thog is a function of the glucose uptake concentration {glu}. The dots with error bars (SD from three experiments) are the experimental results. The dots with lines are the simulation and model predictions.
(E) Under glucose-limited conditions (glucose <0.1%), Thog is inversely proportional to the glucose uptake concentration {glu}. The dots with error bars (SD from three experiments) are the experimental results. The dots with lines are model predictions.
(F) Under glucose-limited conditions (glucose <0.1%), the results are consistent with the model prediction under more conditions. The dots with error bars (SD from three experiments) are the experimental results from Figure S4. The dots with lines are model predictions. A linear relationship can be observed on logarithmic coordinates.
The simulated time traces of the model are shown in Figures 4B and S6. In response to osmostress, the cells in an environment rich in nutrients reallocated enough resources to the glycolytic flux and had a faster glycerol production rate (Figure S7). The glucose-limited environment required a longer nuclear duration of Hog1 to adapt. To examine whether this model can describe the metabolic reconfiguration strategy, we also compared the model prediction under more conditions. Consistent with the experimental results, cells struggle to optimize the growth rate and then store excess glycolytic flux to G6P. The duration of Hog1 nuclear localization and protein production increased upon glucose limitation (Figure 4C).
Therefore, this simplified model suggests that the metabolic intermediate for glycerol production in a glucose-limited environment is insufficient; thus, more stress-related proteins are needed. Previous models primarily focused on describing the Hog-MAPK signaling pathway, but our model is capable of reproducing Hog1 dynamics and cell physiology under varying glucose concentrations. However, the model simulates the results instead of giving the regulatory mechanism of the osmoresponse under different glucose concentrations.
The regulatory mechanism of the osmoresponse is different under two sections of glucose concentrations
Our results proved that yeasts selected different metabolic preallocation strategies under different nutritional conditions (Figures 3 and 4C). One is that the presence of defense reserve flux, which decreases as the glucose concentration decreases (blue area in Figure 4C), provides benefits for the recovery processes of osmoadaptation. The other is that there is no defense reserve flux when glucose is insufficient for cells to grow at a maximum rate (orange area in Figure 4C). We derived the regulatory mechanism of the adaptation time (duration of nuclear Hog1) under different glucose conditions in these two situations via coupled ordinary differential equations (supplemental information). In the system studied here, there are two relevant contributions to the adaptation time , namely, the substrate G3P and enzyme P.
Glucose transport is described via Michaelis-Menten kinetics. We consider that represents the glucose uptake concentration, and the concentration of G6P satisfies
| (Equation 2) |
where k1 is the Michaelis constant in the reaction (G6P→biomass), and represents the conversion efficiency of glucose to biosynthesis. is the configuration of glycolysis (glu→G6P) per cell cycle.
Under glucose-rich concentrations, {glu}>0.35, G6P>> k1; thus, . We found that the downstream protein was saturated very quickly; thus, the time of was determined by the accumulation of glycerol to the extracellular KCl concentration. The resulting osmoresponse law in the area with the G6P reserve flux (blue area in Figure 4C) is approximated as follows:
| (Equation 3) |
where is the added extracellular KCl concentration, and is efficient G6P that converts to G3P. and ks are the Michaelis constants in the reactions (G6P→G3P) and (G3P→glycerol), respectively, and and are the production rates in the reactions (G6P→G3P) and (G3P→glycerol), respectively. Therefore, is positively correlated with the input and negatively correlated with , which are the internal stores that traverse the metabolic route to glycerol production. Notably, needs to be larger than 20 min during the derived process. The simulation and theoretical results of the osmoresponse law in the area with the G6P reserve are shown in Figure 4D.
Under glucose-limited concentrations, there is no reserve, G6P<< and {glu}<0.3 (glucose concentration smaller than 0.1%), and the concentration of G6P satisfies . Given the brief approximation that the G3P influx is almost equal to the outflux, we can obtain the osmoresponse law without G6P reserves as follows:
| (Equation 4) |
where . Therefore, in this case, the relation is much simpler as follows: THog is inversely proportional to {glu} (under low glucose concentration conditions, {glu} can also be regarded as proportional to the glucose concentration) and directly proportion to the osmoconcentration of KCl. This relation is verified in Figures 4E and 4F. This strong relation has never been reported before.
Therefore, we demonstrate that different glucose environments lead to two types of metabolic reconfiguration strategies. The first type, with a defense reserve flux, relates THog to the defense reserve flux indicator G6P, which accelerates the adaptation rate. The second type, without a defense reserve flux, relates THog to the inverse ratio of {glu}. Using this context, we rederive existing osmoresponse laws by considering glucose concentrations, thus demonstrating that the “reserve flux abandonment” increases the response cost.
Knockout of TF Msn2/Msn4 does not influence the main results of enhanced stress response under glucose-limited conditions
Considering that Msn2 and Msn4 are general stress response regulators in yeast, we tested the osmoresponse of msn2Δmsn4Δ and hog1Δ strains under different glucose environments to verify the important role of the Hog-MAPK pathway. We found that the hog1Δ strain could hardly resume growth after exposure to 0.8 M KCl in both the 2% and 0.1% glucose environments, especially under 0.1% glucose, and cell growth was arrested (Figure S8). We compared the growth curves of different strains under different glucose environments when facing osmotic stress (Figures 5A and 5B). msn2Δmsn4Δ strain could resume growth, but its growth rate was lower than that of wild-type cells. The experimental results strengthen the conclusion of the importance of the Hog-MAPK pathway in response to hyperosmotic stress under glucose-limited environments.
Figure 5.
Knockout of TF Msn2/Msn4 does not influence the main results of enhanced stress response under glucose-limited conditions
(A) Normalized growth curves of the different strains after the 0.8 M KCl stimulation was added, showing the average number of cells relative to the number of cells before stress. Yeast grew in 2% glucose. Error bars are SD from three independent experiments. In each experiment, cell numbers were 90∼100.
(B) Normalized growth curves of the different strains after the 0.8 M KCl stimulation was added, showing the average number of cells relative to the number of cells before stress. Yeast grew in 0.1% glucose.
(C) Localization trajectory of nuclear Hog1 in the wild type and msn2Δmsn4Δ strain. In total, 0.8 M KCl was added at 3 h. The curve represents a single cell whose nuclear duration is the median value in the population. The numbers of cells of msn2Δmsn4Δ strain included in the analysis of the 2% and 0.1% glucose concentrations were 25 and 22, respectively. Wild-type data are derived from main text Figure 2.
(D) The duration of Hog1 nuclear localization of wild type and msn2Δmsn4Δ strain. Data are represented as mean +/− SEM. Statistical significance is calculated with Student’s t test (∗∗p < 0.01, ∗∗∗p < 0.001).
(E) The expression of Gpp1 (Hog-MAPK-dependent protein), Hsp12 (both Hog-MAPK- and Msn2/4-dependent protein), and Hxk1 (Msn2/4-dependent protein) upon osmotic stress under different glucose concentrations. 0.8 M KCl was added at 2h. The curve represents a single cell whose intensity is the median value in the population of 30∼50 cells.
(F) Bar graph of the increase ratio of protein response intensity to osmotic stress under different glucose environments (maximum response value minus initial value divided by initial value). Data were from Figure 5E and represented as mean +/− SEM.
Furthermore, the duration of the nuclear translocation of Hog1 was measured to quantify the osmoadaptation. The results revealed that upon osmotic stress (0.8 M KCl), the Hog1 nuclear duration in the msn2Δmsn4Δ strain was longer under glucose limitation (0.1%) than under 2% glucose (Figures 5C and 5D), which is consistent with the performance of the wild type. However, we found that the Hog1 nuclear duration time of the msn2Δmsn4Δ strain was longer than that of the wild type in the same glucose environment. Therefore, Msn2/4 should also have some influence on adaptation when facing hyperosmotic stress, but both the wild-type and msn2Δmsn4Δ strains can adapt.
The expression of some proteins in the msn2Δmsn4Δ strain (provided by Yihan Lin’s Laboratory38) was also measured after being subjected to osmotic stress under different glucose concentrations (Figures 5E and 5F). The proteins included Hog-MAPK-dependent protein Gpp1, both Hog-MAPK- and Msn2/4-dependent protein Hsp12,30 and Msn2/4-dependent protein Hxk1, which is induced under glucose limitation.39 Gpp1 and Hsp12 in the msn2Δmsn4Δ strains were unregulated more under low-glucose condition than under rich-glucose condition, which was consistent with that of wild type. But, when Msn2/4 was knocked out, Hxk1 expression level had no significant change in different glucose conditions and osmotic stress condition.
Discussion
Environmental conditions shape the metabolism and gene regulation of all living cells. Currently, how cells react to adverse environmental conditions when nutrients are limited is unclear. In this work, we systematically described the protein dynamics of S. cerevisiae in response to hyperosmostress in different glucose environments. We chose this type of stress because yeasts are often starved of carbon and subjected to dry conditions in nature. We observed differences in the dynamic response profiles with varying glucose concentrations (Figure 2A). There was no significant difference in the steady-state growth rates under low-glucose conditions of 0.1% (Figure 2B). To understand the mechanisms involved in this phenomenon, we measured the nuclear translocation of the osmotic stress-activated protein kinase Hog1. Consistent with the synthesis rate inferred from downstream proteins, Hog1 exhibited a significantly extended nuclear duration under the low-glucose conditions. Therefore, we found that although resources seemed to be sufficient for growth (>0.1% glucose), the cells expressed many more osmotic stress-related proteins.
Upon osmoshock, cell cycle progression is arrested. According to our hypothesis, cells can adapt to osmostress and resume growth because of sufficient glycerol accumulation via protein synthesis and glycolytic flux (Figure S5A). Nevertheless, at even lower glucose concentrations, excess downstream proteins are nearly saturated for glycerol production, and the initial concentration of the metabolic intermediate G3P is not sufficient to maintain adequate glycerol levels. Therefore, the cells still need to continuously route the glycolytic flux at a steady state. Regarding the limited amount of glycolysis, in a nutrient-poor environment, there is another trade-off between the maximization of steady-state exponential growth and perfect adaptation to osmostress. Cells can prioritize growth with fewer defense costs or use all resources to achieve a stress response (Figure S5B). Biologically, our work reveals a novel mechanism underlying the yeast strategy under such conditions. In a glucose-limited environment (<0.1%), biomass synthesis is restricted. After stimulation under hyperosmotic conditions, Hog1 enters the nucleus to initiate regulation and eventually exits the nucleus, leading to turgor pressure balance. However, the cell cycle could not adapt to the initial state (Figures 2B and S9). This finding implies that cells choose to sacrifice growth and preferentially meet stress defense demands. Moreover, the more severe the stimulus, the greater the sacrifice for growth.
The efficiency of glycerol production might simply decrease with the glucose level, resulting in longer adaptation times for Hog1 nuclear translocation and the downstream protein upregulation under these conditions. We pregrew cells at low-glucose conditions (0.1% and 0.02%) for 8–9 hours and shifted the cells to 2% glucose with 0.8 M KCl (Figures S10A–S10C). Cells in a glucose-limited environment required slightly longer time to adapt. However, because the metabolic flux under 2% glucose may be much larger than the reserve flux, the subsequent adaptation process is remarkably accelerated compared to that under constant low-glucose conditions (Figure S10D).
Considering the partitioning of the limited proteome, it may be inferred from the current research that there is no difference in resource allocation strategy when the growth rate is constant. However, we proved that even in a nutrient-limited environment in which the cell physiology was not significantly affected, cells actually adopted a strategy to reduce the “standby” defense reserve flux. However, in a glucose-limited environment in which cells cannot grow at the maximum rate, the response cost can be simplified and calculated, which was verified by both simulations and experiments. Our modeling and theoretical approach offers a simple way to identify the osmoresponse mechanism characterized by the glucose concentration.
In summary, our combined computational and experimental analyses reveal a mechanism that accounts for the enhanced osmostress response in glucose-limited environments. We conclude that glycerol metabolism plays multiple roles in yeast’s adaptation to altered growth conditions, explaining the complex regulation of glycerol biosynthesis genes. The metabolic trade-off under nutrient-poor conditions mediates adaptation to meet the defense and biosynthetic demands of cells. It can be inferred that yeast abandons the reserve flux, and once stimulated, they have higher protein response costs and longer recovery times.
Our work focused on glucose limitation and hyperosmotic stress. It has been proven that at least in the case of osmostress under glucose limitation, yeast cells do not maximally mobilize their internal carbon sources to prepare for stress, thus sacrificing substantial speed in their recovery. How cells respond to other stresses in the context of limited nutrients and how various stress-activated signaling pathways contribute to signal encoding and integration remain largely elusive. For example, the cAMP/PKA pathway and the AMP-activated protein kinase (AMPK) pathway both play important regulatory roles in cellular responses to osmotic stress and glucose limitation.14 Further studies are needed to evaluate how these pathways contribute to the gene expression changes and metabolic resource reallocation observed in this study. Moreover, the response to oxidative or ethanol stress is closely connected to the activity of the cAMP/PKA pathway and is involved in metabolism.4 When glucose is available, the activation of the cAMP/PKA pathway stimulates the glycolytic flux and suppresses the stress response.40 In fact, under glucose-limited conditions, some proteins had a stronger response to ethanol stress (Figure S11). It could be interesting to investigate whether the need of cells in a glucose-limited environment for an enhanced response for defense is a universal phenomenon. In future studies, similar approaches might be used to discover similar metabolic flux distribution mechanisms. The apparent trade-off between growth and stress resistance is a potential source of general growth strategies under different environmental conditions.
Limitations of the study
The microfluidic chip provided an accurate and controllable culture environment. However, compared with the changing environment in nature, the fixed osmotic stress concentration used in this experiment is relatively simple. In addition, the response mechanism under different nutrients or stress is not complete. Although the preliminary results show that there is a similar mechanism for the response to ethanol in glucose-limitation environment, more comprehensive and detailed experiments are needed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals | ||
| Yeast Nitrogen Base Without Amino Acids | Sigma-Aldrich | Y0626 |
| D-(+)-Glucose | Sigma-Aldrich | G7021 |
| DO Supplement -His/-Ura | Mkbio | MS6321 |
| Cycloheximide | Coolaber | DZSL0829 |
| L-Histidine | Sigma-Aldrich | H6034 |
| Uracil | Sigma-Aldrich | U1128 |
| Kcl | SCR | 10016318 |
| Critical commercial assays | ||
| HiPure Plasmid Mini Kit | Magen | P1002-02 |
| G6P Assay Kit with WST-8 | Beyotime | S0185 |
| G3P Assay Kit | AAT Bioquest | 13838 |
| Glycerol Assay Kit | Applygen | E1012 |
| Software and algorithms | ||
| Matlab 2020 | N/A | https://www.mathworks.com/ |
| ImageJ | N/A | https://imagej.nih.gov/ij/ |
| NIS-Elements AR | NIKON | https://www.microscope.healthcare.nikon.com/ |
| Celltracker | Zhang et al.,29 | https://www.nature.com/articles/srep42200/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Chunxiong Luo (pkuluocx@pku.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
| Strains | ||
|---|---|---|
| Systematic gene name | Labeled Proteins | Number of GFP fusion library |
| YMR174C | Pai3-GFP | 9 F 16 |
| YMR175W | Sip18-GFP | 9 P 10 |
| YIL053W | Gpp1-GFP | 7 H 5 |
| YOL151W | Gre2-GFP | 6 L 1 |
| YFL014W | Hsp12-GFP | 9 D 13 |
| YJL052W | Tdh1-GFP | 6 P 15 |
| YGR088W | Ctt1-GFP | 3 B 8 |
| YDR074W | Tps2-GFP | 2 E 7 |
| YOR173W | Dcs2-GFP | 5 F 23 |
| YMR105C | Pgm2-GFP | 3 D 12 |
| YHR008C | Sod2-GFP | 8 G 11 |
| YOR374W | Ald4-GFP | 4 I 4 |
| YDR368W | Ypr1-GFP | 6 B 20 |
| YPR160W | Gph1-GFP | 2 M 3 |
| YHR043C | Dog2-GFP | 7 D 18 |
| YOR185C | Gsp2-GFP | 11 B 5 |
| YFR053C | Hxk1-GFP | 4 J 17 |
| YLR113W | Hog1-GFP | 5 A 4 |
| msn2Δmsn4Δ strain | Hog1-GFP | From Yihan Lin’s Lab https://www.nature.com/articles/s41467-021-23425-0 |
| msn2Δmsn4Δ strain | Gpp1-GFP | From Yihan Lin’s Lab |
| msn2Δmsn4Δ strain | Hsp12-GFP | From Yihan Lin’s Lab |
| msn2Δmsn4Δ strain | Hxk1-GFP | From Yihan Lin’s Lab |
Plasmids used in the studies:
See supplemental information for more detail of Plasmid of Mcm-mCherry.
Method details
Yeast strains
The yeast strains used in this study were obtained from the S. cerevisiae GFP fusion library generated by Prof. Erin O’Shea and Prof. Jonathan Weissman41 or from a gift of Yihan Lin’s lab.38 The yeast strains were generated on the BY4741 (MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) strain background. To monitor the cell cycle, we transferred the Mcm-mCherry plasmid #(pCT05) into yeast strains, which were provided by the Tang Chao laboratory42 (Figure S1).
Yeast cells were cultured in synthetic dropout medium without His and Ura amino acid complementation and 2% glucose overnight, diluted into fresh medium and incubated for 4–5 h at 30°C for exponential growth (OD600nm 0.4). Then, the cells were loaded into a microfluidic device and cultured under different glucose concentrations.
Microfluidic device
In this study, we modified the channel design of an existing microfluidic device to provide a stimulus medium at four glucose concentrations29 and simultaneously observed six strains at each concentration. The chip consists of a three-layer structure. The main channel is 20 μm high to allow the flow of fluid. The second layer of upside-down pillars (4 μm) is used to fix yeast. The third layer of low fences ensures that there is no cross-contamination between independent passages (Figure 1C). The microfluidic chip was generated by standard photolithography technology and then replica molded with polydimethylsiloxane (PDMS).29
Time-lapse microscopy experiments using a microfluidic platform
The microfluidic chip was mounted on the stage of a Nikon inverted microscope, and bright-field, GFP and m-Cherry images were acquired every 2.5 min using a 60× oil immersion objective. The cells were maintained in SD medium (His-Ura-) with the desired concentration of glucose (2%, 0.1%, 0.05%, 0.02% glucose). Cells were initially cultured for 8 hours in a microfluidic system with different glucose concentrations to adapt to the environment. Then, the cells were exposed to osmostress (0.8 M KCl, the same under different glucose concentrations) for 7 hours. And the glucose limitation medium, PBS buffer was added to compensate for the osmolar difference under different glucose conditions.
Characterization of parameters during the dynamic stress response
In our characterization of the stress response, protein abundance was determined by the synthesis rate α and the decay rate β. Considering the timescale of the fast response and slow protein degradation, the degradation rate can be ignored. Thus, the decay term is considered the dilution rate of the protein. Therefore, β can be calculated as the volume change in one cell cycle, which is inversely proportional to the cell cycle T.
When the cells were exposed to osmotic stress, cell cycle arrest and the decay rates decreased (Figure S3). After a cycle, the decay rates were mostly restored. A cell cycle delay is related to the glucose concentration. Therefore, a longer recovery time is needed in a low-glucose environment. Therefore, the cell cycle needs to be considered in the model.
We observed a single cell through a microfluidic platform to obtain the new daughter cell volume and the mother cell volume after budding. The volume ratio at the time of cell budding was measured previously.43 Thus, the total cell volume of mother and daughter cells is . Therefore, the dilution term β is positively related to In general, this simplified model captured the key variables of cell response dynamics.
Measuring Hog1 nuclear localization in single yeast cells
The localization of the stress-activated protein kinase Hog1 in a single cell was determined by the ratio of the average GFP pixel intensity in the estimated nucleus and the average GFP pixel intensity in the whole cell.
The amplitude of Hog1 translocation was quantified by measuring the single-cell Hog1 localization signals that were above the threshold. The duration of Hog1 localization was quantified by measuring the average in all time intervals during which the single-cell Hog1 localization was above the threshold. The threshold was the mean of the localization traces of cells cultured under non-stress conditions.24
Glycerol, glycerol-6-phosphate (G6P) and glycerol-3-phosphate (G3P) assays
Yeast cells were cultured to the stationary phase (OD600 = 1.0) in SD medium (yeast nitrogen base, drop-out, 2% glucose) at 30°C. Then, the cells were grown to the exponential phase (OD600 = 0.6) at different glucose concentrations (0.02%∼ 2%) and diluted to OD600 = 0.4 in 10 tubes to measure intracellular G6P. At time points 0, 5, 10, 15, 20, 30, 45, 60, 90, and 120 minutes, following a shift to 0.8 M KCl media, the yeasts were centrifuged and resuspended in osmotic culture medium (SD with 0.8 M KCl) in 0.1% and 2% glucose for the glycerol measurement. Each sample was diluted to OD600 = 0.4, harvested by centrifugation, and frozen in liquid nitrogen. The supernatant was transferred to new tubes, lysate was added, centrifugation was performed at 8000 g at 4°C, and the tubes were incubated for 10 min.
The glycerol content in the samples was analyzed using an enzymatic kit (Applygen E 1012). A sample prepared in the same way was used to measure intracellular G6P via a G6P Assay Kit with WST-8 (No. S0185), and intracellular G3P via Amplite™ Colorimetric Glycerol 3-Phosphate (G3P) Assay Kit (Cat. No. 13838). Two hundred microliters of sample per well were loaded onto a 96-well plate along with a dilution series of a glycerol or G6P, G3P standard. The reaction was incubated at room temperature for 30 min to 1 hour. Glycerol, G6P and G3P absorbance were measured at 550 nm,450 nm and 575 nm, respectively. The glycerol, G3P and G6P concentrations were determined using the dilution series of glycerol, G3P and G6P standards as a calibration curve.
Model
Compared to previous models of yeast osmoadaptation, which focus on the hyperosmolarity-glycerol (HOG) network and concise metabolism, our model combines some components to minimize the numbers of variables and parameters. Major osmo-dependent regulatory and metabolic reactions are described with Michaelis–Menten kinetics. All reaction rates are given as the number of molecules per 0.1 min.
In the glycolysis module, metabolic reactions from glucose uptake via phosphorylated intermediates to glycerol are the main focus, and other metabolites, such as trehalose, ethanol, and acetate, are ignored. The first step of glycolysis is the generation of glucose-6-phosphate (G6P) from glucose, which is then used for biosynthesis and glycerol production. The reaction (G6P→biomass, G3P) is described with Michaelis–Menten kinetics with the following two different constants: k1 and k2.
| (Equation 5) |
where the extracellular glucose concentration glu is constant because the microfluidic chip can precisely control the environment. is the production efficiency from glucose to biomass. is the cell cycle, reflects the saturation concentration of protein , is the dilution rate, a2 is defined as the production constant from glucose to G6P, and a3 is the production constant from G6P to G3P.
Regarding the osmotic response, upstream stress sensors and signaling cascades activating Hog1 were not included. Specifically, in our model, only the difference between the glycerol concentration and the input KCl is considered and denoted by Δc as follows:
| (Equation 6) |
where Osmoi is the constant intracellular osmolarity. We assume that growth arrest and Hog1 phosphorylation are due to osmolality which can be written as follows:
| (Equation 7) |
| (Equation 8) |
where represents the cell doubling time, and is the basal level of Hog1 inactivation. is the threshold of osmostress.
A previous model proposed that Hog1 kinase activates the gene transcription of enzymes involved in glycerol production, promotes regulatory enzymes in glycolysis, and controls the closing of Fps1 to keep glycerol inside cells. Our model includes the contribution of the Hog pathway to glycerol accumulation in terms of these three aspects. Glycerol is synthesized from the intermediate glycerol-3-phosphate (G3P), which is produced in a dedicated step from DHAP by NAD-dependent glycerol-3-phosphate dehydrogenase. After the simplification, we assume that the stimulation of protein P1 by Hog1 increases the glycolytic flux to G3P and that the transcription of glycerol-3-phosphate dehydrogenase R2 is regulated only by activated Hog1. The following equations describe the osmoadaptation system:
| (Equation 9) |
| (Equation 10) |
| (Equation 11) |
| (Equation 12) |
where R1 is the enzyme in glycolysis, and R2 represents the mRNA stimulated by phosphorylated Hog1. P2 is the enzyme involved in glycerol production. a8 and a11 are the production rates of mRNA and protein, respectively, and is the mRNA degradation rate. Protein degradation is neglected.
By combining metabolism and gene regulation, we obtained glycerol production from the dephosphorylation of G3P catalyzed by Hog1-regulated proteins. Previously reported work demonstrated that Hog1 phosphorylates Rgc2 to dissociate from Fps1, resulting in Fps1 closure to prevent glycerol loss. Thus, if Fps1 closure is negatively affected by activated Hog1, which also reflects the turgor pressure effect, the following applies:
| (Equation 13) |
where is the Hog1-mediated closure rate. With the Fps1 state, we can obtain the diffusion of glycerol through Fps1 as , which depends on the osmolarity difference. is the transport rate. The glycerol production rate depends on the G3P concentration with the Michaelis constant Ks and the protein concentration with a small threshold Kp.
| (Equation 14) |
In our model, the numbers of molecules per cell were used as the units of species levels (instead of concentrations) to enable the easy implementation of both deterministic and stochastic simulations. The steady states under different glucose concentrations were chosen as the initial conditions. The reaction rates of all species are summarized below in Table S1. All simulations were performed with a time interval of 0.1 minutes.
By combining metabolism and gene regulation, we obtained the osmoadaptation time as determined by the glucose concentration. represents the uptake glucose concentration, with constant . From Equation (5), the concentration of G6P under constant conditions was satisfied.
| (Equation 15) |
Under high-glucose conditions (glucose >0.15%, {glu}>0.35), the initial G6P concentration is considered proportional to the glucose uptake as follows:
| (Equation 16) |
where is the conversion of glycolysis (glu→G6P) per cell cycle, and represents the conversion efficiency of glucose for biosynthesis. Excess internal glycolysis beyond biomass is stored in G6P. reserve G6P = .
Equation 12 yields the amount of G3P over time.
| (Equation 17) |
where . Ks is the Michaelis constant for glycerol production if we assume dG6P G6P. Similarly, ∼G3P/(2∗Ks). Therefore, under high-glucose conditions, the above equation can be approximated as follows:
| (Equation 18) |
Equation 18 yields the amount of G3P over time. After the osmoadaptation time T = , glycerol accumulation equals [KCl] as follows:
| (Equation 19) |
Notably, needs to be larger than 20 min during the derived process, and then, we obtain the following:
| (Equation 20) |
Under low-glucose conditions (glucose <0.1%, {glu}<0.3), almost all glucose intake is used for the synthesis of biomass, and there is no extra G6P for storage. At this time, from Equation 15, we can obtain the following:
| (Equation 21) |
If we further assume that on the timescale of glycerol production, the total metabolites can be assumed to be in a quasi-stationary state, and both the metabolic reconfiguration time and the duration that proteins reach saturation value are negligible. We obtain the following:
| (Equation 22) |
| (Equation 23) |
Then, the accumulation of glycerol is equal to the integral amount of G3P as follows:
| (Equation 24) |
| (Equation 25) |
From this, we deduced that in a very low-glucose environment, because there is no metabolic reserve, the time to adapt to osmostress is inversely proportional to the uptake concentration of glucose .
Quantification and statistical analysis
The statistic significance of differential events was calculated using T tests. The significance of enrichment results was calculated by fisher’s exact tests. Data are reported as mean +/− standard deviation (SD) from different repeat experiments or mean +/− standard error of mean(SEM) from tens of cells. Statistical significance is calculated with Student’s t test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Acknowledgments
This study was supported by the National Key Research and Development Project (2018YFA0900700, 2020YFA0906900, and 2021YFF1200500) and the National Natural Science Foundation of China (11974002).
Author contributions
L.C. and S.W. conceived the project and designed the study. S.W., C. K., and Z. A. performed the experiments. S.W., G.Z., C. K., Z. A., OY. Q., and L.C. performed data analysis and wrote the manuscript. S.W., C. K., and Z. A. performed the experiments. L.C. supervised the study. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105809.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at Github and is publicly available as of the date of publication (https://github.com/YanhonSun/osmomodel).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Adler J. Chemotaxis in bacteria. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X., Si G., Dong Y., Chen K., Ouyang Q., Luo C., Tu Y. Escape band in Escherichia coli chemotaxis in opposing attractant and nutrient gradients. Proc. Natl. Acad. Sci. USA. 2019;116:2253–2258. doi: 10.1073/pnas.1808200116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutin J., Sadeh A., Rahat A., Aharoni A., Friedman N. Condition-specific genetic interaction maps reveal crosstalk between the cAMP/PKA and the HOG MAPK pathways in the activation of the general stress response. Mol. Syst. Biol. 2015;11:829. doi: 10.15252/msb.20156451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho Y.H., Shishkova E., Hose J., Coon J.J., Gasch A.P. Decoupling yeast cell division and stress defense implicates mRNA repression in translational reallocation during stress. Curr. Biol. 2018;28:2673–2680.e4. doi: 10.1016/j.cub.2018.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao N., O’Shea E.K. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat. Struct. Mol. Biol. 2011;19:31–39. doi: 10.1038/nsmb.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boer V.M., De Winde J.H., Pronk J.T., Piper M.D.W. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 2003;278:3265–3274. doi: 10.1074/jbc.M209759200. [DOI] [PubMed] [Google Scholar]
- 8.Scott M., Gunderson C.W., Mateescu E.M., Zhang Z., Hwa T. Interdependence of cell growth. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 9.Mori M., Hwa T., Martin O.C., De Martino A., Marinari E. Constrained allocation flux balance analysis. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Youk H., Van Oudenaarden A. Growth landscape formed by perception and import of glucose in yeast. Nature. 2009;462:875–879. doi: 10.1038/nature08653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin Z., Wilson S., Hauser N.C., Tournu H., Hoheisel J.D., Brown A.J.P. Glucose triggers different global responses in yeast, depending on the strength of the signal, and transiently stabilizes ribosomal protein mRNAs. Mol. Microbiol. 2003;48:713–724. doi: 10.1046/j.1365-2958.2003.03478.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen K., Rong N., Wang S., Luo C. A novel two-layer-integrated microfluidic device for high-throughput yeast proteomic dynamics analysis at the single-cell level. Integr. Biol. 2020;12:241–249. doi: 10.1093/intbio/zyaa018. [DOI] [PubMed] [Google Scholar]
- 13.Ho Y.H., Gasch A.P. Exploiting the yeast stress-activated signaling network to inform on stress biology and disease signaling. Curr. Genet. 2015;61:503–511. doi: 10.1007/s00294-015-0491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Y., AkhavanAghdam Z., Tsimring L.S., Hao N. Coupled feedback loops control the stimulus-dependent dynamics of the yeast transcription factor Msn2. J. Biol. Chem. 2017;292:12366–12372. doi: 10.1074/jbc.C117.800896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y., AkhavanAghdam Z., Li Y., Zid B.M., Hao N. A protein kinase A-regulated network encodes short- and long-lived cellular memories. Sci. Signal. 2020;13:eaay3585. doi: 10.1126/scisignal.aay3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dihazi H., Kessler R., Eschrich K. High osmolarity glycerol (HOG) pathway-induced phosphorylation and activation of 6-phosphofructo-2-kinase are essential for glycerol accumulation and yeast cell proliferation under hyperosmotic stress. J. Biol. Chem. 2004;279:23961–23968. doi: 10.1074/jbc.M312974200. [DOI] [PubMed] [Google Scholar]
- 18.Hohmann S. An integrated view on a eukaryotic osmoregulation system. Curr. Genet. 2015;61:373–382. doi: 10.1007/s00294-015-0475-0. [DOI] [PubMed] [Google Scholar]
- 19.Bouwman J., Kiewiet J., Lindenbergh A., van Eunen K., Siderius M., Bakker B.M. Metabolic regulation rather than de novo enzyme synthesis dominates the osmo-adaptation of yeast. Yeast. 2011;28:43–53. doi: 10.1002/yea.1819. [DOI] [PubMed] [Google Scholar]
- 20.Petelenz-Kurdziel E., Kuehn C., Nordlander B., Klein D., Hong K.K., Jacobson T., Dahl P., Schaber J., Nielsen J., Hohmann S., Klipp E. Quantitative analysis of glycerol accumulation, glycolysis and growth under hyper osmotic stress. PLoS Comput. Biol. 2013;9:e1003084. doi: 10.1371/journal.pcbi.1003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonny A.R., Kochanowski K., Diether M., El-Samad H. Stress-induced growth rate reduction restricts metabolic resource utilization to modulate osmo-adaptation time. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108854. [DOI] [PubMed] [Google Scholar]
- 22.Chen K., Shen W., Zhang Z., Xiong F., Ouyang Q., Luo C. Age-dependent decline in stress response capacity revealed by proteins dynamics analysis. Sci. Rep. 2020;10:15211–15213. doi: 10.1038/s41598-020-72167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun K.A., Breeden L.L. Nascent transcription of MCM2-7 is important for nuclear localization of the minichromosome maintenance complex in G1. Mol. Biol. Cell. 2007;18:1447–1456. doi: 10.1091/mbc.E06-09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee M., Acar M. Heritable stress response dynamics revealed by single-cell genealogy. Sci. Adv. 2018;4 doi: 10.1126/sciadv.1701775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Påhlman A.K., Granath K., Ansell R., Hohmann S., Adler L. The yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J. Biol. Chem. 2001;276:3555–3563. doi: 10.1074/jbc.M007164200. [DOI] [PubMed] [Google Scholar]
- 26.Clotet J., Escoté X., Adrover M.À., Yaakov G., Garí E., Aldea M., De Nadal E., Posas F. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J. 2006;25:2338–2346. doi: 10.1038/sj.emboj.7601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Escoté X., Zapater M., Clotet J., Posas F., Francesc Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 2004;6:997–1002. doi: 10.1038/ncb1174. [DOI] [PubMed] [Google Scholar]
- 28.Martijn R., Markus P.F., Remize, Markus, Tamás, Ramón, Serrano The <em>Saccharomyces cerevisiae</em> Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 2001 doi: 10.1046/j.1365-2958.2001.02384.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang R., Yuan H., Wang S., Ouyang Q., Chen Y., Hao N., Luo C. High-Throughput single-cell analysis for the proteomic dynamics study of the yeast osmotic stress response. Sci. Rep. 2017;7:42200–42210. doi: 10.1038/srep42200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutin J., Joseph-Strauss D., Sadeh A., Shalom E., Friedman N. Genetic screen of the yeast environmental stress response dynamics uncovers distinct regulatory phases. Mol. Syst. Biol. 2019;15:e8939. doi: 10.15252/msb.20198939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teusink B., Passarge J., Reijenga C.A., Esgalhado E., Van Der Weijden C.C., Schepper M., Walsh M.C., Bakker B.M., Van Dam K., Westerhoff H.V., Snoep J.L. Can yeast glycolysis be understood terms of vitro kinetics of the constituent enzymes? Testing biochemistry. Eur. J. Biochem. 2000;267:5313–5329. doi: 10.1046/j.1432-1327.2000.01527.x. [DOI] [PubMed] [Google Scholar]
- 32.Bonny A., Kochanowski K., Diether M., El-Samad H. Stress-induced transient cell cycle arrest coordinates metabolic resource allocation to balance adaptive tradeoffs. bioRxiv. 2020 doi: 10.1101/2020.04.08.033035. Preprint at. [DOI] [Google Scholar]
- 33.Cronwright G.R., Rohwer J.M., Prior B.A. Metabolic control analysis of glycerol synthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2002;68:4448–4456. doi: 10.1128/AEM.68.9.4448-4456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zakrzewska A., Van Eikenhorst G., Burggraaff J.E.C., Vis D.J., Hoefsloot H., Delneri D., Oliver S.G., Brul S., Smits G.J. Genome-wide analysis of yeast stress survival and tolerance acquisition to analyze the central trade-off between growth rate and cellular robustness. Mol. Biol. Cell. 2011;22:4435–4446. doi: 10.1091/mbc.E10-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norbeck J., Pâhlman A.K., Akhtar N., Blomberg A., Adler L. Purification and characterization of two isoenzymes of DL-glycerol-3-phosphatase from Saccharomyces cerevisiae: identification of the corresponding Gpp1 and Gpp2 genes and evidence for osmotic regulation of Gpp2p expression by the osmosensing mitogen-acti. J. Biol. Chem. 1996;271:13875–13881. doi: 10.1074/jbc.271.23.13875. [DOI] [PubMed] [Google Scholar]
- 36.Tamás M.J., Luyten K., Sutherland F.C., Hernandez A., Albertyn J., Valadi H., Li H., Prior B.A., Kilian S.G., Ramos J., et al. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 1999;31:1087–1104. doi: 10.1046/j.1365-2958.1999.01248.x. [DOI] [PubMed] [Google Scholar]
- 37.Lee J., Reiter W., Dohnal I., Gregori C., Beese-Sims S., Kuchler K., Ammerer G., Levin D.E. MAPK Hog1 closes the S. cerevisiae glycerol channel Fps1 by phosphorylating and displacing its positive regulators. Genes Dev. 2013;27:2590–2601. doi: 10.1101/gad.229310.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y., Wu J., Deng M., Lin Y. Yeast cell fate control by temporal redundancy modulation of transcription factor paralogs. Nat. Commun. 2021;12:3145. doi: 10.1038/s41467-021-23425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen A.S., O’Shea E.K. Limits on information transduction through amplitude and frequency regulation of transcription factor activity. Elife. 2015;4:e06559. doi: 10.7554/eLife.06559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conrad M., Schothorst J., Kankipati H.N., Van Zeebroeck G., Rubio-Texeira M., Thevelein J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014;38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 42.Yang X., Lau K.-Y., Sevim V., Tang C. Design principles of the yeast G1/S switch. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Luo C., Zou K., Xie Z., Brandman O., Ouyang Q., Li H. Single cell analysis of yeast replicative aging using a new generation of microfluidic device. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at Github and is publicly available as of the date of publication (https://github.com/YanhonSun/osmomodel).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.