Summary
Obesity has become a global epidemic, associated with several chronic complications. The intestinal microbiome is a critical regulator of metabolic homeostasis and obesity. Empagliflozin, a sodium-glucose cotransporter 2 (SGLT2) inhibitor, has putative anti-obesity effects. In this study, we used multi-omics analysis to determine whether empagliflozin regulates metabolism in an obese host through the intestinal microbiota. Compared with obese mice, the empagliflozin-treated mice had a higher species diversity of gut microbiota, characterized by a reduction in the Firmicutes/Bacteroides ratio. Metabolomic analysis unambiguously identified 1,065 small molecules with empagliflozin affecting metabolites mainly enriched in amino acid metabolism, such as tryptophan metabolism. RNA sequencing results showed that immunoglobulin A and peroxisome proliferator-activated receptor signaling pathways in the intestinal immune network were activated after empagliflozin treatment. This integrative analysis highlighted that empagliflozin maintains intestinal homeostasis by modulating gut microbiota diversity and tryptophan metabolism. This will inform the development of therapies for obesity based on host-microbe interactions.
Subject areas: Immune system, Metabolomics, Microbial genomics, Transcriptomics
Graphical abstract
Highlights
-
•
Empagliflozin reverses high-fat diet-induced changes in microbiota/metabolites
-
•
Empagliflozin activates IgA and PPAR pathways in the intestinal immune network
-
•
Empagliflozin affects gut homeostasis via microbiota diversity and tryptophan metabolism
Immune system; Metabolomics; Microbial genomics; Transcriptomics
Introduction
Obesity is a severe health hazard that contributes to the total global burden of disease.1 Obesity increases the risks of various noncommunicable diseases, such as diabetes, hyperlipidemia, hypertension, cardiovascular disease, kidney disease, and several cancers.2,3 Given the rapidly increasing prevalence and considerable economic losses of obesity and its complications, a full understanding of the pathological mechanism of obesity is of great significance.
Obesity is a complex multifactorial disease involving excess energy storage, fat accumulation, insulin resistance, and inflammatory activation.4 Recently, the intestinal microbiome has been recognized as a critical regulator of host physiology and pathophysiology, including metabolic homeostasis and obesity.5,6 The intestinal microbiome is a complicated and dynamic ecosystem that affects several metabolic and immunological actions.7 Alterations in microbial diversity are related to intestinal disorders and obesity,8 and the relationship between obesity-related metabolic disorders and intestinal microbiota has attracted much attention.9 Imbalanced and altered intestinal microflora have been described in obese patients and animal models.10,11,12 For example, obese patients have a reduced proportion of Bacteroidetes species10; the proportion of Bacteroidetes species in obese animals (ob/ob mice) is reduced by half, and the proportion of Firmicutes species is increased.13 Gut microbial enzymes participate in the dynamic stabilization of glucolipid metabolism, thereby affecting the balance and homeostasis of energy and systemic immunity.14,15 Thus, the intestinal microbiome has become a new target for anti-obesity therapies. However, the interaction between the microbiome and host metabolism in healthy and obese individuals is not fully understood.
Empagliflozin (EMPA) is a highly efficient and selective sodium-glucose cotransporter 2 (SGLT2) inhibitor that is used in the clinical treatment of diabetes patients. Besides hypoglycemic effects, EMPA also regulates lipid metabolism (increases lipolysis, fatty acid oxidation, and ketone production) and is helpful for weight loss and fat reduction.16,17 Our previous work showed that EMPA can improve obesity-related complications, including cardiac dysfunction, nonalcoholic fatty liver disease, and chronic kidney disease.18,19,20
While EMPA has several protective effects against obesity, it is not known whether EMPA affects the gut microbiota and host metabolism in obesity. Here, we aimed to analyze the metabolome and metagenome of the small intestine contents and the transcriptome of small intestine tissue of mice given a high-fat diet (HFD) to explore whether EMPA regulates obese host metabolism through the intestinal microbiota.
Results
EMPA reduced HFD-induced metabolic disruption
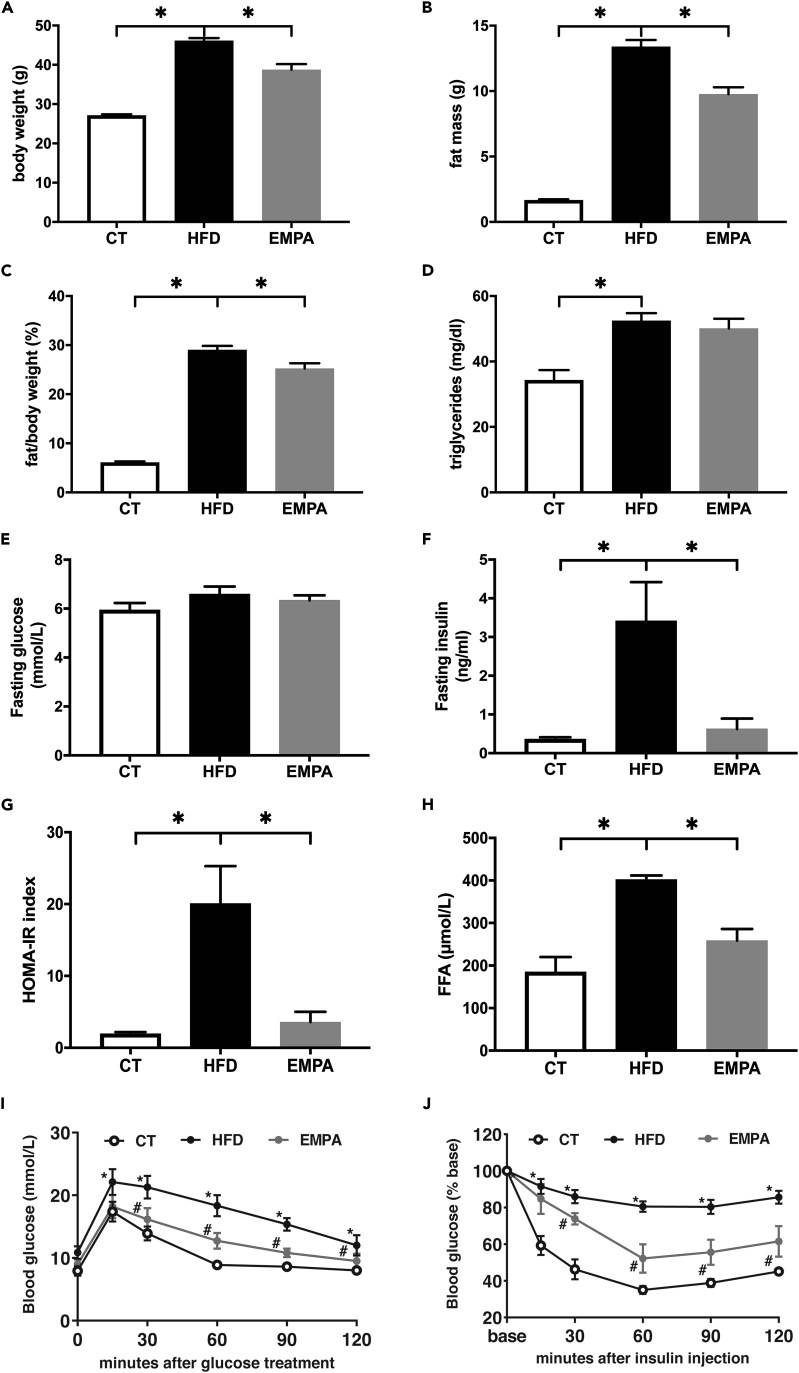
After 20 weeks, the HFD caused a significant elevation in body weight, fat mass, and the fat/body weight ratio (p < 0.05, Figures 1A–1C). HFD group mice had elevated serum triglyceride levels (p < 0.05, Figure 1D). Fasting glucose was not significantly different among the HFD, EMPA, and control (CT) groups (p > 0.05, Figure 1E). HFD group mice also showed elevated fasting insulin, homeostatic model assessment of insulin resistance (HOMA-IR) index, and free fatty acids (FFA) and impaired glucose/insulin tolerance (p < 0.05, Figures 1F–1J). These findings verified the establishment of the obese model. Consistent with our previous study, the HFD-induced metabolic disturbance was improved by EMPA treatment (p < 0.05).18,20
Figure 1.
Mouse biochemistry parameters
(A–J) (A) body weight, (B) fat mass, (C) fat/body weight, (D) serum triglyceride, (E) fasting glucose, (F) fasting insulin, (G) homeostatic model assessment of insulin resistance (HOMA-IR) index, (H) free fatty acids (FFA), (I) oral glucose tolerance test (OGTT), and (J) insulin tolerance test (ITT) curve. ∗p < 0.05, ∗∗p < 0.01, for (E) and (F), ∗HFD vs. CT p < 0.05, # EMPA vs. HFD, n = 5–6. Data are represented as mean ± SEM.
Microbial communities in small intestine contents
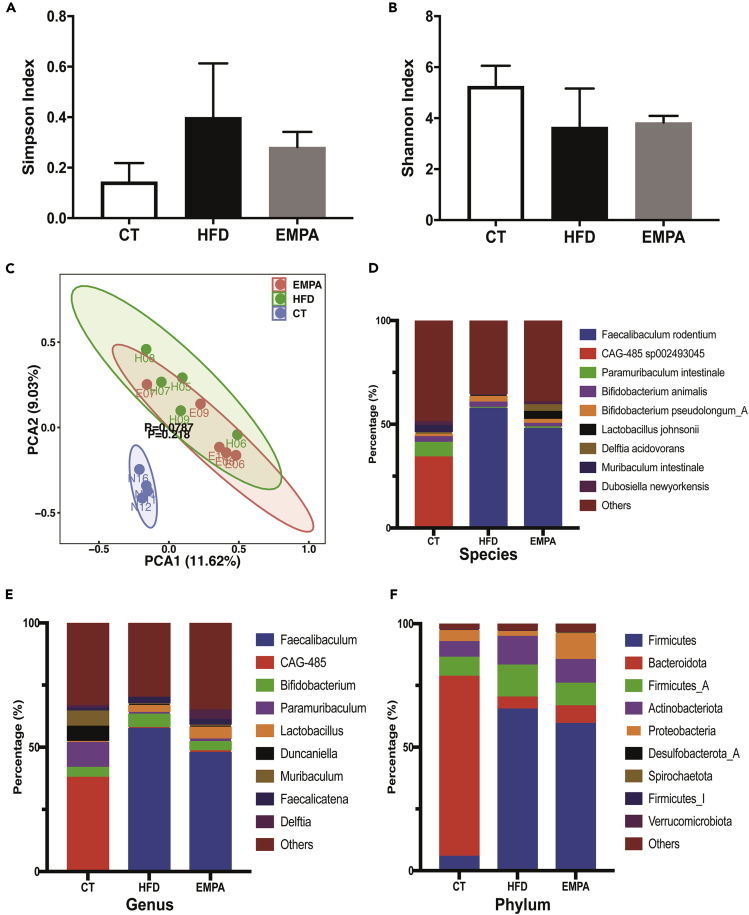
Total DNA from colon contents was extracted and whole-genome shotgun sequenced. For each sample, an average of 9,167,669.36 reads was obtained and assembled to generate 33,800.07 contigs. Taxonomic profiling was conducted using kraken2, based on k-mer accurate alignment. We calculated and compared the community-level microbial diversity of the three groups (Figures 2A and 2B). Compared with the CT group, the HFD group had a higher Simpson index (0.14 ± 0.04 vs. 0.40 ± 0.01, p < 0.05); however, no significant differences in alpha diversity (Shannon and Simpson) were observed. Nevertheless, the alpha diversity of the EMPA group tended to be close to that of the CT group. Principal-component analysis showed that the CT group was separated into a set, but the EMPA and HFD groups did not show clear separation (Figure 2C).
Figure 2.
Alpha diversity and taxonomic composition of intestinal content (CT, n = 4; HFD, n = 5; EMPA, n = 5)
(A and B) Differences in gut microbiota diversity among the groups of mice were estimated by the Simpson (A) and Shannon (B) indices.
(C–F) (C) Principal component analysis plot of gut microflora species determined from metagenomic sequencing of the three groups. Comparison of the average abundance of certain gut bacteria from mice in CT, HFD, and EMPA groups at the species (D), genus (E), and phylum (F) level. Data are represented as mean ± SEM.
Based on the taxonomic profiling, we tallied the microbial composition at species, genus, and phylum levels (Figures 2D–2F). In the HFD and EMPA groups, Faecalibaculum rodentium was elevated at the species level, and there was a higher proportion of Faecalibaculum at the genus level compared with the CT group. In the CT group, CAG-485 sp002493045 was reduced at the species level, and there were lower proportions of CAG-485 at the genus level compared with the HFD and EMPA groups. Bacteroidota was the dominant phylum in the CT group, accounting for over half of all microbes, and Firmicutes dominated in the HFD and EMPA groups.
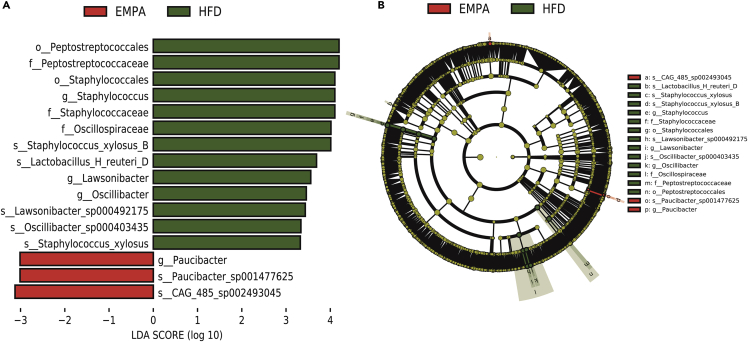
We performed a pairwise linear discriminant analysis effect size analysis to identify potentially important specific bacteria. Linear discriminant analysis effect size is an algorithm that provides a list of characteristics with statistical and biological significance of differences between conditions of interest and sorts them according to the effect size. Figure 3 shows differences in all classification levels between EMPA and HFD groups in a histogram (Figure 3A) and a cladogram (Figure 3B). After 8 weeks of EMPA administration, orders Peptostreptococcales and Staphylococcales; families Peptostreptococcaceae, Staphylococcaceae, and Oscillospiracea; genera Staphylococcu, Lawsonibacter, Oscillibacter, and Paucibacter; and species Staphylococcus xylosus B, Lactobacillus H reuteri D, Lawsonibacter sp000492175, Oscillibacter sp000403435, S. xylosus, Paucibacter sp001477625, and CAG485 sp002493045 were significantly changed compared with the HFD group.
Figure 3.
The statistical significance of differentially abundant and biologically relevant taxonomic biomarkers between two distinct biological conditions was measured using a linear discriminant analysis effect size
(A) Cladogram indicating the phylogenetic distribution of microbiota correlating with the EMPA or HFD groups.
(B) Histogram of the linear discriminant analysis scores showing differences in abundance between the EMPA and HFD groups.
See also Figure S1.
We also analyzed the different bacteria between HFD and CT groups (Figures S1A and S1B). It is noteworthy that the orders Peptostreptococcales and Staphylococcales, families Peptostreptococcaceae and Staphylococcaceae, genus Staphylococcu, and species S. xylosus B and CAG485 sp002493045 were also significantly altered. Finally, we found that CAG485 sp002493045 was a representative intestinal bacterium whose reduced abundance was reversed by EMPA. This showed that HFD changes the structure of intestinal microbiota and that EMPA can partially reverse this change.
Metabolic profiling of small intestine content
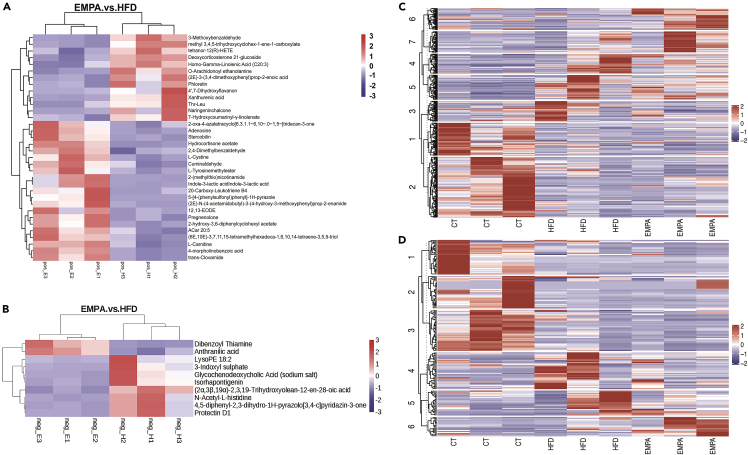
The metabolomic analysis of the three groups (n = 3/group) identified 723 features in the negative ion mode (NIM) (Table S1) and 342 features in the positive ion mode (PIM) (Table S2). Comparative metabolomics analysis to determine differences in intestinal contents among the three groups showed the distribution of metabolites was significantly different. Compared with the HFD group, the EMPA group had 23 upregulated metabolites (including 21 positive metabolites and 2 negative metabolites) and 21 downregulated metabolites (including 13 positive metabolites and 8 negative metabolites) (Figures 4A and 4B). We also identified metabolite differences between HFD and CT groups (Figures S2A and S2B). The HFD group had 66 upregulated metabolites (including 50 positive and 16 negative metabolites) and 109 downregulated metabolites (including 97 positive and 32 negative metabolites). Clustering the metabolites by k-mean analysis enabled metabolites that changed among the three groups in the PIM (Figure 4C) and the NIM (Figure 4D) to be displayed.
Figure 4.
Comparative metabolomics analysis determines changes in gut metabolites
(A–D) The heatmap shows the difference between the gut metabolite profiles of EMPA and HFD mice (n = 3) in (A) positive ion mode and (B) negative ion mode. K-means analysis revealed the changed metabolites between the three groups in (C) positive ion mode and (D) negative ion mode.
See also Figure S2.
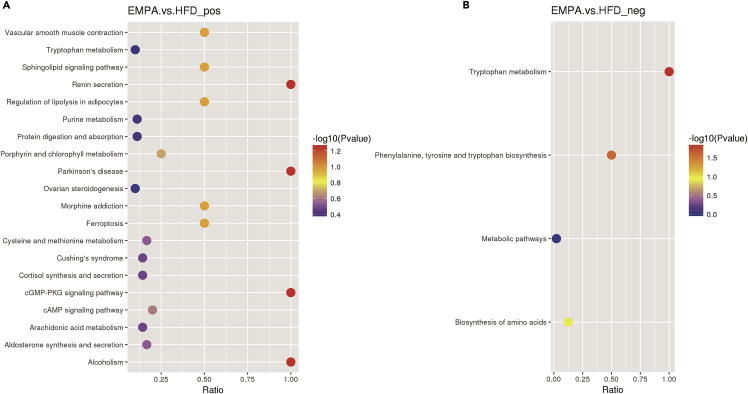
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of altered metabolites was carried out to clarify the correlation between changes in intestinal metabolites and EMPA administration and to reveal the close relationship between metabolic pathways and secondary metabolite biosynthesis. We observed that tryptophan, cysteine, and methionine metabolism and phenylalanine, tyrosine, and tryptophan biosynthesis were altered in EMPA vs. HFD mice (Figures 5A and 5B). We also observed differences in several fatty acid metabolic pathways, such as the regulation of lipolysis in adipocytes and arachidonic acid metabolism. In addition, differences in glucose metabolism pathways, including pentose and glucuronate interconversions, starch and sucrose metabolism, glycolysis/gluconeogenesis, carbohydrate digestion and absorption, and galactose metabolism were observed between HFD vs. CT groups (Figures S3A and S3B).
Figure 5.
KEGG enrichment analysis of the gut metabolites observed in EMPA/HFD
(A and B) The scatterplot shows the altered pathways of metabolites in positive ion mode (A) and negative ion mode (B).
See also Figure S3.
Transcriptome profiling of small intestine tissue
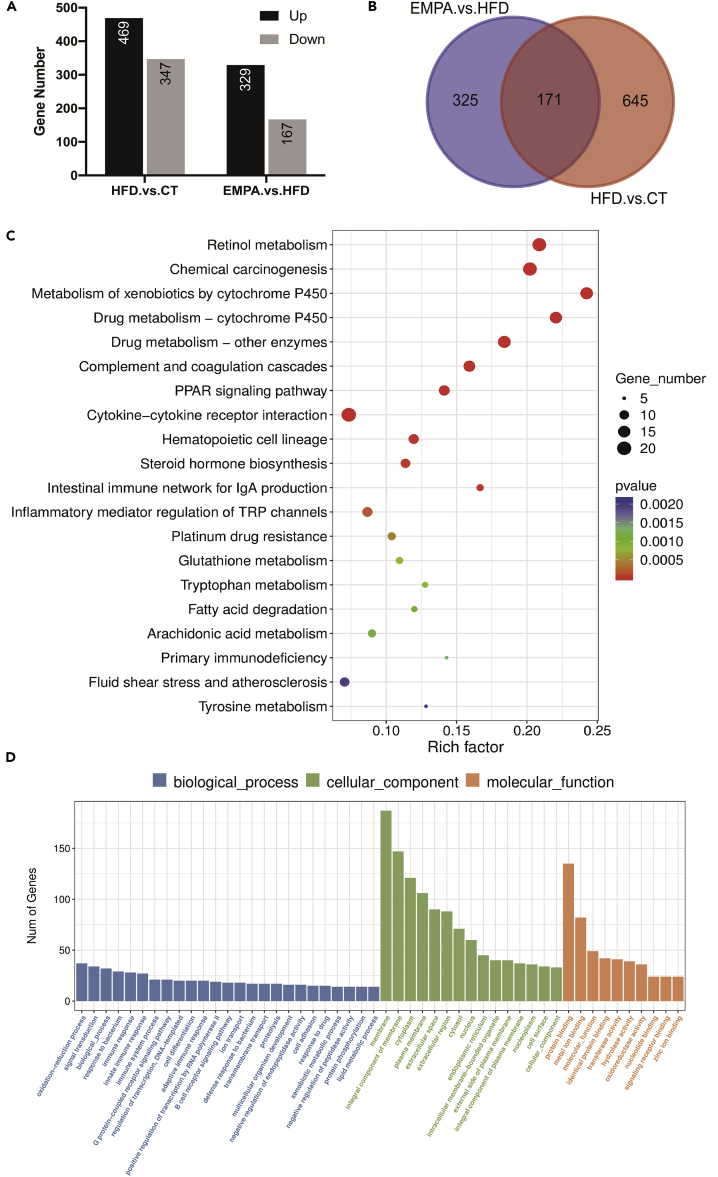
In our experimental model, the changes observed in the intestinal metabolome and microbiome before the systemic manifestations of chronic obesity may disrupt gene expression in intestinal tissue. Therefore, to analyze the transcriptome characteristics of the three groups, we conducted RNA sequencing (RNA-seq) of small intestine tissue. We identified 816 DEGs (including 469 upregulated and 347 downregulated) between EMPA and HFD groups and 496 DEGs (including 329 upregulated and 167 downregulated) between HFD and CT groups (Figure 6A). Between the two comparisons, there are 171 shared DEGs (Figure 6B).
Figure 6.
Analysis of transcriptome profiling (n = 5)
(A) The numbers of up- and downregulated genes in HFD/CT and EMPA/HFD.
(B) Venn diagrams show the differentially expressed genes (DEGs) between EMPA/HFD and HFD/CT.
(C and D) KEGG (C) and GO (D) enrichment of EMPA/HFD DEGs.
See also Figure S4.
To evaluate the functional consequences, ClusterProfiler was used to analyze the upregulated and downregulated genes, and the KEGG and gene onotology (GO) databases were used for pathway enrichment analysis. Enrichment analysis showed that most of the altered pathways were classified as responses to endogenous and exogenous stimulation, such as peroxisome proliferator-activated receptor (PPAR) signaling pathway, intestinal immune network for immunoglobulin A (IgA) production, inflammatory mediator regulation of TRP (transient receptor potential) channels, and adaptive immune response (Figures 6C and 6D). Pathways related to energy and glucose homeostasis were also altered, such as tryptophan metabolism, fatty acid degradation, arachidonic acid metabolism, tyrosine metabolism, and steroid hydroxylase activity. Overall, these data are consistent with the observed changes in the intestinal metabolites, which may lead to an imbalance in energy and glucose homeostasis.
Correlation analysis between metabolome, metagenome, and transcriptome data
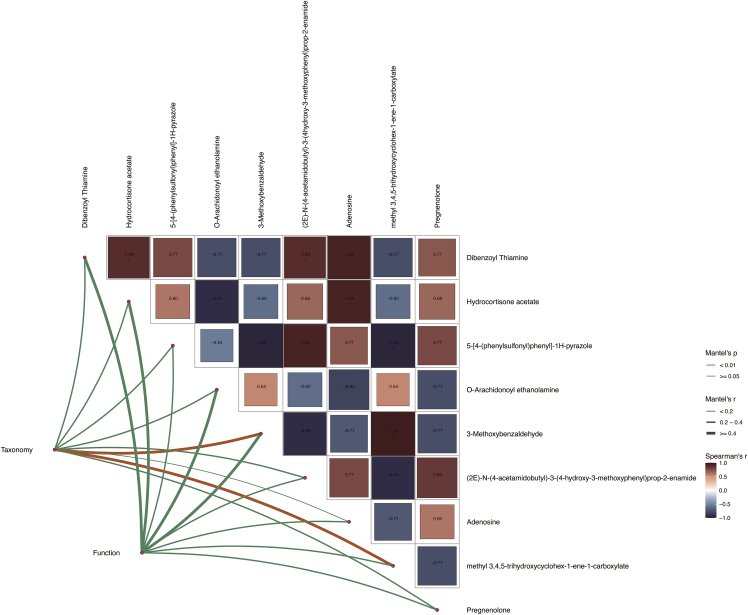
EMPA changed the structure of the intestinal bacterial community and the composition of the intestinal metabolome. Some of these small molecules from the intestinal flora affect the intestinal barrier and trigger the expression of immune genes. To better understand the bridging effect of intestinal small molecules between microorganisms and the intestinal barrier, we analyzed the data generated above, including the classification and composition of intestinal microorganisms and gene expression in intestinal tissue and intestinal metabolites. Mantel test analysis was carried out with R package, linkET. The results confirmed the correlation between all data (Figure 7). 3-Methoxybenzaldehyde and methyl 3,4,5-trihydroxycyclohex-1-ene-1-carboxylate were the best explanatory variables for all three profiles (for 3-methoxybenzaldehyde, the taxonomic composition: Mantel’s r = 0.91, p < 0.01 and gene function: Mantel’s r = 0.48, p = 0.05; for methyl 3,4,5-trihydroxycyclohex-1-ene-1-carboxylate, the taxonomic composition: Mantel’s r = 0.76, p < 0.01 and gene function: Mantel’s r = 0.28, p > 0.05).
Figure 7.
Variations in gut microbiota species and gene expression have obvious relationships with intestinal metabolites
Edge width corresponds to Mantel’s r statistic for the corresponding distance correlations, and edge color denotes the statistical significance. Pairwise comparisons of intestinal metabolites are shown, with a color gradient denoting Pearson’s correlation coefficient.
Discussion
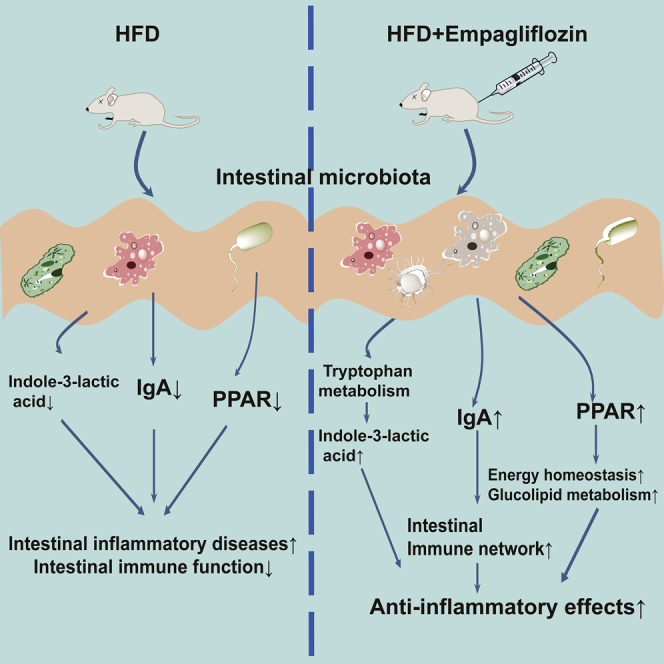
We successfully analyzed the metabolome and metagenome of intestinal contents and the transcriptome of intestinal tissue in obese model mice treated with EMPA. Our data reveal that EMPA modulates gut microbiota by restoring the Firmicutes/Bacteroides ratio in HFD mice. In addition, EMPA modulates intestinal homeostasis via gut microbiota-mediated tryptophan metabolism and intestinal immune and inflammatory networks related to energy and glucose homeostasis.
EMPA, as a hypoglycemic drug, exerts anti-obesity effects. We previously demonstrated that EMPA can protect against obesity-related cardiac dysfunction, nonalcoholic fatty liver disease, and chronic kidney disease via different mechanisms.18,19,20 This protection was associated with improving metabolic disruption, including a reduced fat/body weight ratio and restored glucose homeostasis. Our study further confirmed that EMPA can reduce the fat/body weight ratio and alleviate impaired glucose/insulin tolerance.
The gut is an endocrine organ that participates in energy homeostasis, and it is interesting to know whether EMPA can regulate the intestinal microbiome in obesity. Differences in gut microbiota composition are associated with obesity. Obese patients have relatively lower gut microbial diversity and function.21 We found that the microbiota alpha diversity measured by Shannon entropy was reduced under the HFD, but this was reversed after receiving EMPA treatment. Firmicutes and Bacteroides are usually considered “fat bacteria” and “lean bacteria”, respectively. An increase in the Firmicutes/Bacteroides ratio is associated with an increase in the inflammatory response and body weight.22 Compared with the CT group, the relative abundance of Bacteroidota decreased, while that of Firmicutes increased in the HFD group. These results are consistent with those of previous studies.10,13 Clinical data also show that compared with lean individuals, the relative proportion of Bacteroidota in obese individuals is lower, but this increases with weight loss after following a low-calorie diet.10 Interestingly, EMPA could reverse the change in the Firmicutes/Bacteroides ratio. Notably, EMPA treatment also reduced the abundance of the genus Faecalibaculum, which was higher in HFD mice. Qiao et al. have confirmed that metabolic syndrome can be improved by reducing the relative abundance of Faecalibaculum.23 These findings indicate that EMPA can restore the Firmicutes/Bacteroides ratio in the small intestine of HFD mice.
Intestinal contents contain nonabsorbable food components and complex mixtures of metabolites produced by the host and intestinal microbes.24 Therefore, intestinal metabolites can reflect the health status of the host. Based on this, we examined the intestinal metabolome of obesity model mice, which consists of a mixture of host-produced and microbiota-produced molecules. A total of 1,065 small molecules were quantitatively archived, and the enrichment results showed that substantial numbers of these small molecules were related to tryptophan metabolism. Tryptophan is an essential amino acid that cannot be synthesized in animals but has critical physiological functions.25 Lack of tryptophan damages intestinal immune function, resulting in a higher risk of intestinal inflammatory diseases.26 In addition, tryptophan intake can promote the immune balance of the intestinal tract and maintain gut immune homeostasis.27,28 Intestinal microorganisms can utilize and metabolize tryptophan into metabolites (indole and indole acid derivatives), thereby regulating intestinal immunity.29
Our results also confirmed that some indole derivatives, such as indole-3-lactic acid, were significantly increased in the EMPA group. Indole is the main product of tryptophan metabolism by intestinal microorganisms, and Bacteroides symbionts with the tryptophan enzyme can produce indole.30 Indole is maintained at a high concentration in the intestines of normal animals, maintains intestinal health, and has anti-inflammatory effects.31,32 Some indoles can also be metabolized in the intestine into indole acid derivatives, such as indole-3-lactic acid.30 Indole acid derivatives play an essential role in maintaining intestinal immune tolerance; for example, the concentration of indole-3-propionic acid was decreased with obesity and food addiction, indicating that a lack of tryptophan metabolites can aggravate the condition.33 Here, we speculate that EMPA-mediated changes in intestinal microorganisms promote tryptophan metabolism, increase the production of indole acid derivatives, and improve intestinal dysfunction.
An HFD increases intestinal pressure and ultimately affects the development of pathophysiological mechanisms related to the intestinal inflammatory response. In our study, changes in the intestinal metabolites and microbes may lead to changes in gene expression in intestinal tissue. Therefore, we conducted RNA-seq to analyze the transcriptome characteristics of the small intestine. Notably, EMPA treatment increased IgA production in the intestinal immune network. IgA is crucial in protecting the mucosal surface from infectious microorganisms.34 The IgA isotype dominates the antibody landscape at the mucosal site, and many studies have demonstrated this antibody’s importance to the stability of the microbiota.35
We also observed that the PPAR signaling pathway was affected in the EMPA group. As a nuclear receptor, PPAR has multiple functions in inflammation and metabolism.36 PPAR regulates several disrupted biological processes in obesity, including energy homeostasis, glucolipid metabolism, and inflammation.37,38 For example, microbiota activates PPAR-γ signaling, a homeostatic pathway that prevents heterologous expansion of potentially pathogenic bacteria in the colon’s lumen.39 In addition, specific bacteria and their metabolites can activate PPAR-γ expression in intestinal epithelial cells.40,41 Our results confirm that EMPA significantly affects the immune pathways that promote obesity and inflammation, indicating that changes in intestinal microorganisms may lead to changes in host gene expression; however, the detailed links between gut microbes, intestinal metabolites, and the immune system are far from fully understood.
In summary, our data demonstrate that EMPA treatment alleviated HFD-induced obesity. Our integrative analysis demonstrates that EMPA maintains intestinal homeostasis by modulating gut microbiota diversity, gut microbiota-mediated tryptophan metabolism, and intestinal immune and inflammatory networks related to energy and glucose homeostasis. Our findings reveal a new biochemical mechanism for the effects of EMPA against obesity, which has implications for the diet of obese patients.
Limitations of the study
This study has some limitations. First, we did not determine whether the beneficial effects of EMPA on HFD mice depended on changes to the gut microbiota. Therefore, the potential causality and mechanism between anti-obesity effects of EMPA and changes in the gut microbiota deserve further investigation. In addition, because of a relatively small sample, we can only draw tentative conclusions about the effect of EMPA on HFD mice. A larger-scale animal experiment and clinical validation need to be conducted.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Raw and analyzed data | This paper | SRA: PRJNA868514 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Pengyue Co., Ltd. (Jinan, China) | SCXK20190003 |
| Software and algorithms | ||
| Prism 9.0 | GraphPad Software | N/A |
| R version 4.0.2 | The R Foundation for Statistical Computing | https://www.r-project.org/ |
| Trimmomatic (version 0.40) | Bolger et al., 201442 | http://www.usadellab.org/cms/?page=trimmomatic |
| BMTagger | N/A | https://anaconda.org/bioconda/bmtagger |
| MEGAHIT | N/A | https://github.com/voutcn/megahit |
| Prodigal | N/A | http://prodigal.ornl.gov/ |
| DIAMOND | Buchfink et al., 201543 | https://github.com/bbuchfink/diamond |
| Kraken2 | N/A | https://github.com/DerrickWood/kraken2 |
| mZcloud | N/A | https://www.mzcloud.org/ |
| FastaQC | N/A | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| HISAT2 | Kim et al., 201544 | http://daehwankimlab.github.io/hisat2/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xiaodong Sun at xiaodong.sun@wfmc.edu.cn.
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mice
Male C57BL/6J mice (six weeks old) were supplied by Pengyue Co., Ltd. (Jinan, China). Animal study protocols were approved by the Animal Ethics Committee of Weifang Medical University (No. 2018003) and national guidelines for experiments involving laboratory animals were adhered to. All animals were housed under specific pathogen-free conditions according to the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals.
Method details
Animal model and biochemical measurements
All mice were raised under standard laboratory conditions under a 12-h light/dark cycle. After acclimatization for 1 week, mice were randomly separated into three groups: a CT group (n = 8) fed a chow diet (320 kcal/100 g) for 20 weeks, an HFD group (n = 8) and an EMPA group (n = 8) both fed an HFD (54.05% fat, 529.8 kcal/100 g) for 12 weeks and then administered saline or EMPA (dose: 10 mg/kg/d), respectively, for another 8 weeks. Body weight and fat mass were measured. After 20 weeks, glucose homeostasis was evaluated by glucose and insulin tolerance tests. At the end of the study, 2% barbiturate sodium (40 mg/kg, ip) was injected into mice. The small intestine tissues were rapidly isolated and rinsed with normal saline, then transferred into sterile cryopreservation tubes. The small intestine contents were collected and also transferred into sterile cryopreservation tubes. All samples were rapidly frozen and stored at −80°C for transcriptomic, metagenomic and metabolomics analysis. Serum TGs (triglycerides) was measured using a TG content detection kit (Solarbio, China).
Metagenomic sequencing and analysis
Total DNA of small intestine contents (n = 4 for CT group, n = 5 for HFD and EMPA groups) was extracted and purified using a DNeasy Powersoil Prokit (47,014, Qiagen, Germany). A KAPA Hyper Prep Kit (KK8504, Roche, Switzerland) was used to construct DNA libraries. DNA libraries were fragmented using a QIA FX DNA Library Kit (Qiagen), and DNA fragments of specific lengths were collected using a FastPure Gel DNA Extraction Mini Kit (Vazyme, China). After the library passed quality inspection, high-throughput 150 bp paired-end sequencing was performed on a NovaSeq™ 6,000 (Illumina, United States). Sequencing was performed by LC Bio-Technology Co. (Hangzhou, China). The raw sequencing data were processed by Trimmomatic (version 0.40)42 to remove adapters and low-quality sequences and host sequences were excluded using BMTagger before data assembly. The sequences were assembled by MEGAHIT (v1.2.9, https://github.com/voutcn/megahit) to generated contigs and open reading frames were predicted using Prodigal (v2.6.3, http://prodigal.ornl.gov/). The predicted gene enrichment was conducted using DIAMOND (v2.0.4, https://github.com/bbuchfink/diamond) and the KEGG (https://www.kegg.jp/) database.43 Classification of species sequences was performed using Kraken2 (2.1.1, https://github.com/DerrickWood/kraken2).
Non-targeted metabolome detection and analysis
The small intestine content (100 mg, n = 3/group) was ground in liquid nitrogen then 500 μL 80% aqueous methanol solution was added, vortex-mixed, incubated in an ice bath for 5 min, and centrifuged at 15000 g, 4°C for 20 min. The supernatant was then diluted with water until the methanol content was 53%. After centrifugation at 15000 g, 4°C for 20 min, the supernatant was collected for liquid chromatography–mass spectrometry analysis using the following parameters: column: Hypesil Gold column (C18); column temperature: 40°C; liquid chromatography flow rate: 0.2 mL/min; positive ion mode; mobile phase A: 0.1% formic acid, mobile phase B: 100% methanol; negative ion mode: mobile phase A: 5 mM ammonium acetate, pH = 9.0, mobile phase B: 100% methanol. Quality control samples containing all experimental samples (an equal volume of each) were used to evaluate system stability of the liquid chromatography–mass spectrometry during the entire process. Blank samples were also measured to remove background ions. The raw data were searched against mZcloud (https://www.mzcloud.org/), Mzvault and Masslist databases, and the results were normalized. The results provided metabolite identification and relative quantitation. The KEGG database was used to annotate identified metabolites.
RNA-seq and data analysis
Trizol was used to extract total RNA from small intestine tissue (n = 5/group). RNA was reverse transcribed using SuperScript™ II reverse transcriptase (Invitrogen). cDNA libraries with 300 ± 50 bp fragments were constructed and RNA-seq was performed on a NovaSeq 6,000 (Illumina) to obtain 150 bp paired-end reads. FastaQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to assess data quality. The sequences were assembled and mapped to the Mus musculus genome by HISAT2.44 Reads per kilobase per million mapped reads was used to normalize the expression data. R package, DESeq2, was used to identify differentially expressed genes (DEGs) with fold change >2 or < − 2, and p < 0.05. Gene Ontology (GO) enrichment analysis (Biological processes, Molecular function and Cellular components) and KEGG pathway enrichment were performed and visualized using R package, ClusterProfiler.
Quantification and statistical analysis
Normally distributed data are expressed as the standard error of the mean, and the nonnormal distribution of data was determined by the Shapiro–Wilk test before analysis. A single comparison between two groups was analyzed by Student’s t-test. One-way ANOVA was used to compare the classified and continuous data between groups, and Bonferroni or Fisher LSD post-tests were used to correct multiple tests. The Wilcoxon rank-sum test was used to analyze the microbiota alpha diversity, species level comparative advantage and functional categories. The Mantel test was conducted using R package linkET for correlation analysis. R ggplot2 package and Graphpad Prism 9.0 software (GraphPad, USA) were used for graphics. p values <0.05 were considered statistically significant.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81870593 and 82170865), Natural Science Foundation of Shandong Province of China (ZR2021QD132 and ZR2020MH106), Shandong Province Medical and Health Science and Technology Development Project (202003060400, 202003060396), Special Funds for Taishan Scholars Project of Shandong Province, Yuandu scholars (2021), and Outstanding Young Talents in Health and Wellness in Qilu (2018).
Author contributions
JFS, HYQ, and QX analyzed the data, performed the statistical analyses, and drafted the manuscript. XDS and FH conceived and designed the study and participated in the revision. The other authors participated in partial data collection and data curation and analysis.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105816.
Contributor Information
Fang Han, Email: fyhanfang@wfmc.edu.cn.
Xiaodong Sun, Email: xiaodong.sun@wfmc.edu.cn.
Supplemental information
Data and code availability
-
•
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA868514. Further information and requests for resources and reagents will be available from the corresponding author on reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Seidell J.C., Halberstadt J. The global burden of obesity and the challenges of prevention. Ann. Nutr. Metab. 2015;66(Suppl 2):7–12. doi: 10.1159/000375143. [DOI] [PubMed] [Google Scholar]
- 2.Bray G.A., Heisel W.E., Afshin A., Jensen M.D., Dietz W.H., Long M., Kushner R.F., Daniels S.R., Wadden T.A., Tsai A.G., et al. The science of obesity management: an endocrine society scientific statement. Endocr. Rev. 2018;39:79–132. doi: 10.1210/er.2017-00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li R., Huang X., Liang X., Su M., Lai K.P., Chen J. Integrated omics analysis reveals the alteration of gut microbe-metabolites in obese adults. Brief. Bioinform. 2021;22:bbaa165. doi: 10.1093/bib/bbaa165. [DOI] [PubMed] [Google Scholar]
- 4.Lin X., Li H. Obesity: epidemiology, pathophysiology, and therapeutics. Front. Endocrinol. 2021;12:706978. doi: 10.3389/fendo.2021.706978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomes A.C., Hoffmann C., Mota J.F. The human gut microbiota: metabolism and perspective in obesity. Gut Microb. 2018;9:308–325. doi: 10.1080/19490976.2018.1465157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson E., Ryan P.M., Cryan J.F., Dinan T.G., Ross R.P., Fitzgerald G.F., Stanton C. Gut microbiota, obesity and diabetes. Postgrad. Med. J. 2016;92:286–300. doi: 10.1136/postgradmedj-2015-133285. [DOI] [PubMed] [Google Scholar]
- 7.O'Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres-Fuentes C., Schellekens H., Dinan T.G., Cryan J.F. The microbiota-gut-brain axis in obesity. Lancet Gastroenterol Hepatol. 2017;2:747–756. doi: 10.1016/S2468-1253(17)30147-4. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Q., Li D., He Y., Li Y., Yang Z., Zhao X., Liu Y., Wang Y., Sun J., Feng X., et al. Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci. Rep. 2019;9:13424. doi: 10.1038/s41598-019-49462-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 11.Marchesi J.R., Adams D.H., Fava F., Hermes G.D.A., Hirschfield G.M., Hold G., Quraishi M.N., Kinross J., Smidt H., Tuohy K.M., et al. The gut microbiota and host health: a new clinical Frontier. Gut. 2016;65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomé-Castro X.M., Rodriguez-Arrastia M., Cardona D., Rueda-Ruzafa L., Molina-Torres G., Roman P. Probiotics as a therapeutic strategy in obesity and overweight: a systematic review. Benef. Microbes. 2021;12:5–15. doi: 10.3920/BM2020.0111. [DOI] [PubMed] [Google Scholar]
- 13.Ley R.E., Bäckhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burcelin R. Gut microbiota and immune crosstalk in metabolic disease. Mol. Metab. 2016;5:771–781. doi: 10.1016/j.molmet.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long S.L., Gahan C.G.M., Joyce S.A. Interactions between gut bacteria and bile in health and disease. Mol. Aspects Med. 2017;56:54–65. doi: 10.1016/j.mam.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Calapkulu M., Cander S., Gul O.O., Ersoy C. Lipid profile in type 2 diabetic patients with new dapagliflozin treatment; actual clinical experience data of six months retrospective lipid profile from single center. Diabetes Metab. Syndr. 2019;13:1031–1034. doi: 10.1016/j.dsx.2019.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Szekeres Z., Toth K., Szabados E. The effects of SGLT2 inhibitors on lipid metabolism. Metabolites. 2021;11:87. doi: 10.3390/metabo11020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Y., Kan C., Qiu H., Liu Y., Hou N., Han F., Shi J., Sun X. Transcriptomic analysis reveals the protective effects of empagliflozin on lipid metabolism in nonalcoholic fatty liver disease. Front. Pharmacol. 2021;12:793586. doi: 10.3389/fphar.2021.793586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun X., Han F., Lu Q., Li X., Ren D., Zhang J., Han Y., Xiang Y.K., Li J. Empagliflozin ameliorates obesity-related cardiac dysfunction by regulating sestrin2-mediated AMPK-mTOR signaling and redox homeostasis in high-fat diet-induced obese mice. Diabetes. 2020;69:1292–1305. doi: 10.2337/db19-0991. [DOI] [PubMed] [Google Scholar]
- 20.Ye T., Zhang J., Wu D., Shi J., Kuang Z., Ma Y., Xu Q., Chen B., Kan C., Sun X., Han F. Empagliflozin attenuates obesity-related kidney dysfunction and NLRP3 inflammasome activity through the HO-1-Adiponectin Axis. Front. Endocrinol. 2022;13:907984. doi: 10.3389/fendo.2022.907984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanislawski M.A., Dabelea D., Lange L.A., Wagner B.D., Lozupone C.A. Gut microbiota phenotypes of obesity. NPJ Biofilms Microbiomes. 2019;5:18. doi: 10.1038/s41522-019-0091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao L. The gut microbiota and obesity: from correlation to causality. Nat. Rev. Microbiol. 2013;11:639–647. doi: 10.1038/nrmicro3089. [DOI] [PubMed] [Google Scholar]
- 23.Qiao Y., Zhang Z., Zhai Y., Yan X., Zhou W., Liu H., Guan L., Peng L. Apigenin alleviates obesity-associated metabolic syndrome by regulating the composition of the gut microbiome. Front. Microbiol. 2021;12:805827. doi: 10.3389/fmicb.2021.805827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li K., Yuan M., Wu Y., Pineda M., Zhang C., Chen Y., Chen Z., Rong X., Turnbull J.E., Guo J. A high-fat high-fructose diet dysregulates the homeostatic crosstalk between gut microbiome, metabolome, and immunity in an experimental model of obesity. Mol. Nutr. Food Res. 2022;66:e2100950. doi: 10.1002/mnfr.202100950. [DOI] [PubMed] [Google Scholar]
- 25.Yao K., Fang J., Yin Y.L., Feng Z.M., Tang Z.R., Wu G. Tryptophan metabolism in animals: important roles in nutrition and health. Front. Biosci. 2011;3:286–297. doi: 10.2741/s152. [DOI] [PubMed] [Google Scholar]
- 26.Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M., Sigl V., Hanada T., Hanada R., Lipinski S., et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun M., Ma N., He T., Johnston L.J., Ma X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR) Crit. Rev. Food Sci. Nutr. 2020;60:1760–1768. doi: 10.1080/10408398.2019.1598334. [DOI] [PubMed] [Google Scholar]
- 28.Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D'Angelo C., Massi-Benedetti C., Fallarino F., et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Gao J., Xu K., Liu H., Liu G., Bai M., Peng C., Li T., Yin Y. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keszthelyi D., Troost F.J., Masclee A.A.M. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neuro Gastroenterol. Motil. 2009;21:1239–1249. doi: 10.1111/j.1365-2982.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 31.Bansal T., Alaniz R.C., Wood T.K., Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. USA. 2010;107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimada Y., Kinoshita M., Harada K., Mizutani M., Masahata K., Kayama H., Takeda K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One. 2013;8:e80604. doi: 10.1371/journal.pone.0080604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong T.S., Mayer E.A., Osadchiy V., Chang C., Katzka W., Lagishetty V., Gonzalez K., Kalani A., Stains J., Jacobs J.P., et al. A distinct brain-gut-microbiome profile exists for females with obesity and food addiction. Obesity. 2020;28:1477–1486. doi: 10.1002/oby.22870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Sousa-Pereira P., Woof J.M. IgA: structure, function, and developability. Antibodies. 2019;8:57. doi: 10.3390/antib8040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klag K.A., Round J.L. Microbiota-immune interactions regulate metabolic disease. J. Immunol. 2021;207:1719–1724. doi: 10.4049/jimmunol.2100419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh H.Y.P., Visvalingam V., Wahli W. The PPAR-microbiota-metabolic organ trilogy to fine-tune physiology. FASEB J. 2019;33:9706–9730. doi: 10.1096/fj.201802681RR. [DOI] [PubMed] [Google Scholar]
- 37.Faghfouri A.H., Khajebishak Y., Payahoo L., Faghfuri E., Alivand M. PPAR-gamma agonists: potential modulators of autophagy in obesity. Eur. J. Pharmacol. 2021;912:174562. doi: 10.1016/j.ejphar.2021.174562. [DOI] [PubMed] [Google Scholar]
- 38.Gross B., Pawlak M., Lefebvre P., Staels B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. Endocrinol. 2017;13:36–49. doi: 10.1038/nrendo.2016.135. [DOI] [PubMed] [Google Scholar]
- 39.Byndloss M.X., Olsan E.E., Rivera-Chávez F., Tiffany C.R., Cevallos S.A., Lokken K.L., Torres T.P., Byndloss A.J., Faber F., Gao Y., et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 2017;357:570–575. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alex S., Lange K., Amolo T., Grinstead J.S., Haakonsson A.K., Szalowska E., Koppen A., Mudde K., Haenen D., Al-Lahham S., et al. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor γ. Mol. Cell Biol. 2013;33:1303–1316. doi: 10.1128/MCB.00858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukovac S., Belzer C., Pellis L., Keijser B.J., de Vos W.M., Montijn R.C., Roeselers G. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. mBio. 2014;5 doi: 10.1128/mBio.01438-14. e01438-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 44.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA868514. Further information and requests for resources and reagents will be available from the corresponding author on reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.