Abstract
Background
Operable stage I-III non-small cell lung cancer (NSCLC) has a high risk of recurrence, mainly due to remnant clones of the disease defined as minimal residual disease (MRD). Adjuvant chemotherapy has a limited efficacy in reducing the risk of relapse, and prognostic as well as predictive biomarkers in this context are currently missing.
Methods
We performed a systematic review to evaluate the state of the art about the role of circulating tumor DNA detection through liquid biopsy for the assessment of MRD in resected early-stage NSCLC patients.
Results
Among the 650 studies identified, 13 were eligible and included. Although highly heterogeneous, all the studies demonstrated a poor prognosis in patients with post-operative MRD, with a detection rate ranging from 6% to 45%. MRD detection preceded radiographic/clinical recurrence by a mean of 5.5 months. MRD positive patients were most likely to benefit from adjuvant treatment in terms of recurrence-free survival (RFS). Consistently, adjuvant therapy did not minimize the risk of relapse in the MRD negative group.
Conclusions
Liquid biopsy has a relevant role in assessing post-surgical MRD in resected NSCLC. Since currently there are no criteria other than stage and risk factors for the choice of adjuvant treatment in this setting, post-operative assessment of MRD through liquid biopsy might be a promising approach to guide the decision.
Keywords: Non-small cell lung cancer (NSCLC), minimal residual disease (MRD), circulating tumor DNA (ctDNA), adjuvant therapy
Introduction
Lung cancer still represents the leading cause of cancer-related death in both sexes worldwide (1). Non-small cell lung cancer (NSCLC) is the prevalent histologic subtype and accounts for approximately 85% of all lung cancers. The vast majority of NSCLC is diagnosed at advanced stage, which is characterized by historical 5-year survival rate not exceeding 10%, whereas localized stage represents 20% of cases (2,3). During the last years, great steps forward have been made in the management of early-stage NSCLC, trying to improve NSCLC survivorship and to reduce the risk of disease recurrence. Currently, the standard treatment with curative intent is represented by surgery. Moreover, primary stereotactic radiotherapy is an effective alternative strategy when surgery is not feasible (3,4). However, despite relevant progresses in the treatment of early-stage and locally advanced disease, recurrence rate is high and 5-year overall survival (OS) rate remains unsatisfying, ranging from 53% to 60% for stages IIA/B and from 26% to 36% for stages IIIA/B disease (2).
One of the most relevant factors implicated in the risk of recurrence of early NSCLC is the persistence of minimal residual disease (MRD), which represents an invisible quote of disease (either locally or in form of micro-metastases) after initial radical treatments. This clinically occult disease represents a potential source of subsequent relapse, thus conditioning early-stage NSCLC prognosis (5). In this regard, the major challenges of MRD in the field of lung cancer are its undetectability and the likelihood of resistance to conventional treatments (6).
An effective strategy to eradicate MRD and therefore minimize the chance of postoperative recurrence consists of adjuvant therapy. Of note, since standard tumor staging omits MRD assessment, adjuvant therapy is administered ‘blindly’, and its success is assessed only by radiological detection of tumor relapse. Cisplatin-based adjuvant chemotherapy has been the first treatment approved for completely resected stage II-III NSCLC (7,8), despite 5-year OS remains poor. In fact, adjuvant chemotherapy prolonged significantly OS [hazard ratio (HR) 0.89; 95% CI: 0.82 to 0.96; P=0.005] and disease-free survival (DFS) (HR 0.84; 95% CI: 0.78 to 0.91; P <0.001) compared to observation, with 5-years OS and DFS absolute benefits of 5.4% and 5.8%, respectively (8). An area of controversy is represented by the role of adjuvant chemotherapy in the 8th TNM edition stage IB NSCLC, being this suggested only in the presence of risk factors (3,9).
Given the limited efficacy of adjuvant chemotherapy, new adjuvant strategies have been studied, including targeted therapies and immunotherapy. With regards to targeted therapies, in an era where first- and second-generation epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) had failed to demonstrate an advantage in the adjuvant setting for the treatment of EGFR-mutated NSCLC (10-14), the phase III ADAURA trial showed an unprecedented statistically significant DFS benefit of osimertinib compared with placebo in patients with EGFR mutant stage IB-IIIA NSCLC (7th TNM edition). Osimertinib resulted in dramatically reduction of disease recurrence risk over placebo in this setting (HR 0.21, P<0.001), also showing a clinically meaningful reduction in central nervous system recurrence (HR 0.18, P<0.001) (15,16). Although OS data are still immature, osimertinib already represents the standard of care for resected stage IB-III NSCLC that harbors EGFR activating mutation (17,18). Immunotherapy has also been tested in the adjuvant setting of NSCLC treatment. Recently, the phase III IMpower010 trial showed a DFS benefit of adjuvant atezolizumab over best supportive care following adjuvant cisplatin-based chemotherapy in PD-L1 positive stage II-IIIA patients (HR 0.66, P=0.004), leading to FDA approval of atezolizumab in this setting (19). In addition, adjuvant pembrolizumab achieved a statistically significant improvement in DFS compared to placebo in stage IB-IIIA completely resected NSCLC patients, regardless of PD-L1 expression, in the phase III PEARLS/KEYNOTE-091 study (20).
The identification of potential predictive factors of adjuvant therapy still represents an unmet clinical need. In this context, the detection of MRD could be a promising strategy for the selection of patients who could benefit from adjuvant treatments after radical surgery. In addition, MRD assessment could be a useful tool to monitor disease course after curative treatment, when minimal early recurrence could not be evident at the standard follow-up radiological examinations.
Liquid biopsy, defined as the analysis of cancer biomarkers in tumor-derived material extracted from patients’ fluids, currently plays a fundamental role in the management of advanced NSCLC. Circulating tumor DNA (ctDNA) analysis through liquid biopsy is a not-invasive approach which has relevant implications both in terms of molecular characterization and decision-making processes in the advanced disease (21). MRD detection through liquid biopsy has already been tested in the advanced setting of NSCLC, where it could serve as a monitoring strategy for patients who undergo complete response following systemic treatment, or to assess disease-free status after surgery performed for oligometastatic disease (22).
Currently, there is not a standard of care for detecting and monitoring MRD trough liquid biopsy in resected early-stage NSCLC. However, based on the solid experience in the advanced setting, ctDNA analysis could represent an effective non-invasive tool to detect MRD, as demonstrated in many other cancer types where this technique has already managed to detect early recurrence (23-25).
The aim of this systematic review is to frame the potential role of ctDNA detection through liquid biopsy for the determination of MRD and its prospective clinical implications in the context of resected early-stage NSCLC management (Figure 1). The study was planned, conducted and reported in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) reporting checklist (26) (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-390/rc).
Figure 1.
Schematic approach of ctDNA MRD testing in resected NSCLC patients. CT, chemotherapy; RFS, recurrence-free survival; OS, overall survival; MRD, minimal residual disease; NSCLC, non-small cell lung cancer.
Methods
This systematic review was performed in order to answer the question: “What is the state of the art about the role of liquid biopsy in lung cancer MRD?”. The substantial heterogeneity and the recognized risk of bias identified across and within studies did not allow for pool estimates or meta-analysis of the investigated variables.
Search strategy
We performed a systematic literature search using PubMed (Medline) and the proceedings of major international meetings (American Society of Clinical Oncology, ASCO; European Society of Medical Oncology, ESMO; International Association for the Study of Lung Cancer, IASLC), to identify relevant studies published between January 01st, 2000 and February 28th, 2022. The following keywords were used: early lung cancer [all fields] OR NSCLC [all fields] AND cfDNA [all fields] OR circulating free DNA [all fields]. An additional search was carried out through the references of the included studies.
Study selection, eligibility criteria and methodological quality assessment
Observational studies, clinical trials and case reports evaluating the role of circulating free DNA (cfDNA) in lung cancer MRD were included. Relevant international meetings proceedings, although not published in full, were also analyzed. Only English-language articles were eligible. On the contrary, either studies with insufficient data or outcome information, or conducted on advanced stage, or not evaluating MRD were excluded. Two authors independently (MV, MP) used the Newcastle-Ottawa Scale, a tool used for assessing the quality of non-randomized studies included in a systematic review (27). If the two authors had different opinions while assessing the studies, agreement was reached by consensus with the third author (FP).
Data extraction and synthesis
Study characteristics (first author, year of publication, number of patients, stage of tumor, sample sources, timing to sample collection, methods used to test MRD, main results) were extracted from the included articles and summarized in Table 1. Data extraction was initially performed by four authors (MV, MP, MPA and FB) and then independently reviewed by an additional author (FP).
Table 1. MRD investigation in early-stage NSCLC.
| Studies | N of Pts for MRD analysis | Stage of tumor | Samples | Landmark timepoint (days) | Surveillance timepoints | Methods for assessing MRD | Main results |
|---|---|---|---|---|---|---|---|
| Guo et al. (2016) (28) | 41 | I-IIIA† | Plasma | No | NGS | Concordance rate: 78.1% | |
| Pre-op | <13 | Pre-op ctDNA+: 19/41 (46.3%) | |||||
| Post-op | <10 | Post-op ctDNA+: – | |||||
| Tissue | yes | Relapse: – | |||||
| Abbosh et al. (2017) (29) | 24 | IA-IIIB | Plasma | No | NGS | Concordance rate: – | |
| Pre-op | n.s. | Pre-op ctDNA+: – | |||||
| Post-op | <30 | Post-op ctDNA+: 6/24 (25%) | |||||
| Tissue | yes | Relapse:14/24 (58.3%) | |||||
| Chaudhuri et al. (2017) (30) | 5‡ | IB-IIIA | Plasma | Yes | NGS | Concordance rate: – | |
| Pre-op | n.s. | Pre-op ctDNA+: 5/5 (100%) | |||||
| Post-op | <120 | Post-op ctDNA+: 1/5 (20%) | |||||
| Tissue | no | Relapse:1/5 (20%) | |||||
| Chen et al. (2019) (31) | 26 | I-III | Plasma | Yes | NGS | Concordance rate: 92.2% | |
| Pre-op | 0 | Pre-op ctDNA+: 32/175 (18.3%) | |||||
| Post-op (P1) | 1 | Post-op ctDNA+ (P1): 12/26 (46.2%) | |||||
| Post-op (P2) | 3 | Post-op ctDNA+ (P2): 7/26 (26.9%) | |||||
| Post-op (P3) | 30 | Post-op ctDNA+ (P3): 7/26 (26.9%) | |||||
| Tissue | yes | Relapse: 7/26 (26.9%) | |||||
| Peng et al. (2020) (32) | 77 | I-III§ | Plasma | Yes | NGS | Concordance rate: – | |
| Pre-op | <7 | Pre-op ctDNA+: 46/77 (59.7%) | |||||
| Post-op | 15 | Post-op ctDNA+: 30/71 (42.3%) | |||||
| Tissue | yes | Relapse: 35/77 (45.5%) | |||||
| Ohara et al. (2020) (33) | 20 | IIA-IIIA | Plasma | No | NGS | Concordance rate: – | |
| Pre-op | <2 | Pre-op ctDNA+: 8/20 (40%) | |||||
| Post-op | <12 | Post-op ctDNA+: 4/20 (20%) | |||||
| Tissue | Yes | Relapse: 5/20 (25%) | |||||
| Kuang et al. (2021) (34) | 38 | IB-III | Plasma | Yes | NGS | Concordance rate: – | |
| Pre-op | <7 | Pre-op ctDNA+: 19/38 (50%) | |||||
| Post-op | <15 | Post-op ctDNA+: 8/35 (22.8%) | |||||
| Tissue | Yes | Relapse: 9/38 (23.7%) | |||||
| Zhao et al. (2021) (35) | 7 | IB-IIIA | Plasma | No | NGS | Concordance rate: 43.7% | |
| Pre-op | 0 | Pre-op ctDNA+: 4/6 (66.6%) | |||||
| Post-op | <10 | Post-op ctDNA+: 1/7 (14.2%) | |||||
| Tissue | Yes | Relapse: 1/7 (14.2%) | |||||
| Li et al. (2022) (36) | 119 | I-IIIA | Plasma | Yes | NGS | Concordance rate: 67% | |
| Pre-op | <7 | Pre-op ctDNA+: 33/117 (28.2%) | |||||
| Post-op | <30 | Post-op ctDNA+: 12/116 (10.3%) | |||||
| Tissue | Yes | Relapse: 26/119 (21.8%) | |||||
| Waldeck et al. (2022) (37) | 21 | IA-IIIB | Plasma | Yes | NGS | Concordance rate: – | |
| Pre-op | n.s. | Pre-op ctDNA+: 12/21 (57%) | |||||
| Post-op | <14 | Post-op ctDNA+: 4/16 (25%) | |||||
| Tissue | Yes | Relapse:8/21 (38.1%) | |||||
| Xia et al. (2021) (38) | 330 | I-III | Plasma | No | NGS | Concordance rate: – | |
| Pre-op | n.s. | Pre-op ctDNA+: 69/330 (20.9%) | |||||
| Post-op (P1) | <15 | Post-op ctDNA+ (P1): 19/296 (6.4%) | |||||
| Post-op (P2) | <30 | Post-op ctDNA+ (P2):19/324 (5.9%) | |||||
| Tissue | Yes | Relapse: 56/69¶ | |||||
| Qiu et al. (2021) (39) | 103 | I-III§ | Plasma | Yes | NGS | Concordance rate: – | |
| Pre-op | n.s. | Pre-op ctDNA+: 61/88 (69.3%) | |||||
| Post-op | 30 | Post-op ctDNA+: 18/85 (21.2%) | |||||
| Tissue | Yes | Relapse:34/103 (33.0%) | |||||
| Tan et al. (2021) (40) | 57 | I-III | Plasma | Yes | PCR | Concordance rate: – | |
| Pre-op | n.s. | Pre-op ctDNA+: – | |||||
| Post-op | n.s. | Post-op ctDNA+: 7/57 (12.3%) | |||||
| Tissue | Yes | Relapse:11/57 (19.3%) |
†, 1 patient with stage IV; ‡, only 5 out of 37 patients enrolled received surgery; §, 2 patients with stage IV; ¶, calculated on ctDNA positive patients. MRD, minimal residual disease; NSCLC, non-small cell lung cancer; Pre-op, pre-operative; Post-op, post-operative; ctDNA, circulating tumor DNA; NGS, next generation sequencing; PCR, polymerase chain reaction; n.s., not specified; –, not reported.
Results
Literature search
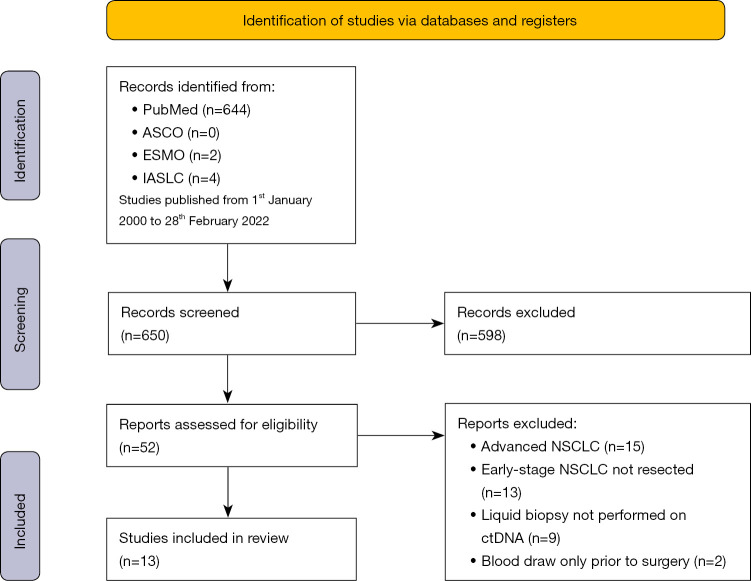
Of the 650 studies found in the search, 598 were excluded after a careful read of the title and abstract, because not relevant for the question addressed in this systematic review and/or focused on advanced disease stage. After reading the full text of the remaining 52 articles, 39 were additionally excluded for the following reasons: inclusion of advanced NSCLC patients; inclusion of early-stage NSCLC which did not undergo surgery; missing of MRD evaluation; missing liquid biopsy approach; blood draw performed only prior to surgery. Overall, 13 studies satisfied the required criteria and were selected for the present systematic analysis. The workflow of the literature search is shown in Figure 2.
Figure 2.
Flow diagram. ASCO, American Society of Clinical Oncology; ESMO, European Society of Medical Oncology; IASLC, International Association for the Study of Lung Cancer; NSCLC, non-small cell lung cancer.
Characteristics of the included studies
The characteristics of the 13 identified studies which evaluated MRD by ctDNA in radically-resected NSCLC (stage I-III) patients are listed in Table 1. Pre-operative plasma samples (baseline timepoint) were collected immediately before surgery in two studies (31,35), within 7 days in four studies (32-34,36) or within 13 days in one study (28). Post-operative plasma samples (landmark timepoint) were collected within 10 days from surgery in three studies (28,31,35), within 10-15 days from surgery in five studies (32-34,37,38), between 15 and 30 days in five studies (29,31,36,38,39) and within 120 days from surgery in one study (30). A schematic representation of the plasma samples timing across the studies is depicted in Figure 3. Tumor tissue was collected intra-operatively in all the studies except for one in which tissue specimen was not provided (30). In addition, 8 out of 13 studies longitudinally collected blood samples during follow-up period, up to four years from surgery, in order to perform the surveillance analysis (30-32,34,36,37,39,40). The analytical methods used to assess MRD were predominantly based on Next-Generation Sequencing (NGS) (28-39), while Polymerase Chain Reaction (PCR)-based methods were applied in one study (40). Details of the molecular techniques and the pre-analytical variables are specified in Table 2.
Figure 3.
Timeline of blood collection before and after surgery.
Table 2. Overview of molecular techniques and pre-analytical variables.
| Studies | Blood processing protocol | Technology | NGS Strategy | Plasma (mL) | Mean input of cfDNA (ng) |
|---|---|---|---|---|---|
| Guo et al. (2016) (28) | 1° centrifugation: 1,600 g/10 min | Ion Torrent NGS 50-gene panel | Standard | At least 1 mL | – |
| 2° centrifugation: 16,000 g/10 min | |||||
| Abbosh et al. (2017) (29) | 1° centrifugation: 1,000 g/10 min | M-Seq. Oncomine Lung cfDNA Assay (Ion S5 System) | Personalized | 1–5 mL | 20ng |
| 2° centrifugation: 2,000 g/10 min | |||||
| Chen et al. (2019) (31) | 1° centrifugation: 1,600 g/10 min | c-SMART | Standard | 4 mL | 46.9 ng |
| 2° centrifugation: 16,000 g/10 min | |||||
| Peng et al. (2020) (32) | – | c-SMART | Standard | 2 mL | – |
| Ohara et al. (2020) (33) | One centrifugation at 1,600 g/10 min | CAPP-Seq 197-gene panel Sequencing on Ion Torrent S5 | Standard | 3.4 mL | 40 ng |
| Kuang et al. (2021) (34) | – | 425-gene Nanjing Geneseeq Technology | Standard | – | – |
| Zhao et al. (2021) (35) | – | 23-gene panel sequencing on NextSeq 500 (Illumina) | Standard | – | – |
| Li et al. (2022) (36) | – | 425-gene Nanjing Geneseeq Technology | Standard | – | – |
| Waldeck et al. (2022) (37) | 1° centrifugation: 800 g for 10 min | 18-gene panel on HiSeq 2000, Illumina | Standard | – | 13.2 ng |
| 2° centrifugation: 1,000 g for 10 min | |||||
| Xia et al. (2021) (38) | – | 769-gene panel | Standard | – | – |
| Qiu et al. (2021) (39) | One centrifugation at 3,000 g/10 min | ATG-Seq | Standard | – | Up to 50 ng |
NGS, next generation sequencing; M-Seq, multi-region exome sequencing; c-SMART, Circulating single-molecule amplification and resequencing technology; CAPP-Seq, cancer personalized profiling by deep sequencing; ATG-Seq, Automated Triple Groom Sequencing.
We assessed the quality of these studies using the Newcastle-Ottawa Scale. The mean score of our included studies was 7. The detailed scores of each study are summarized in Table S1 and Table S2.
Synthesis of main results
A high heterogeneity was documented across the studies both regarding the number of patients enrolled (range: 5–330) and the approach applied to investigate MRD on liquid biopsy samples. The concordance rate between plasma and tumor tissue ranged from 43.7% to 92.2%, suggesting a suitable representation of plasma mutation detection (28,31,36), with a mean concordance rate of approximately 70%.
Pre-operative ctDNA status was available in 11 studies, and ctDNA was positive in a percentage ranging from 18.3% to 100% (28,30-39). When pre-operative ctDNA status was correlated with clinical outcomes, patients with a positive status before surgery had a shorter Recurrence-Free Survival (RFS) compared with the negative counterpart (32-34,36,38).
Post-operative ctDNA status was assessed on plasma samples at the landmark timepoint in 12 studies, and the ctDNA positive rate ranged from 6.4% to 46.2%. Eight studies evaluated the prognostic value of landmark ctDNA analysis, and patients with positive ctDNA had shorter RFS and OS compared with the counterpart (31-34,36-39).
During follow-up surveillance, disease relapse was determined in 11 studies, and it occurred in 14% to 58% of the enrolled patients (29-37,39,40). ctDNA surveillance analysis was carried out in 8 studies (30-32,34,36,37,39,40). In details, Qiu et al. (39) demonstrated that 27/34 (79.4%) relapsed patients had at least one positive ctDNA during disease surveillance against 14/34 (41.2%) patients who had positive ctDNA status at the landmark timepoint. In addition, five studies evaluated the time between the detection of ctDNA and the radiological/clinical recurrence of disease (29,30,32,37,39). In these studies, the overall mean time from early ctDNA positivity and disease recurrence was 5.5 months. Furthermore, Chen et al. investigated ctDNA on multiple post-operative timepoints in order to consider which moment could be the most appropriated to identify MRD (31). The authors showed that patients with detectable ctDNA at any timepoint had a shorter RFS that patients with undetectable ctDNA (1 day after surgery: 17.6 vs. 18.1 months; 3 days after surgery: 9.3 vs. 21.2 months; 30 days after surgery: 9.8 vs. 22 months, respectively), with the greatest differences in RFS observed at both 3 days and 30 days after surgery. Since the delta in RFS was similar between these two timepoints, 3 days after surgery could represent a feasible moment to early evaluate the risk of relapse. All together, these findings highlight the pivotal role of ctDNA surveillance after surgery to build a more effective MRD evaluation.
The role of MRD in the adjuvant setting
Among the 13 selected articles, only 4 addressed the potential role of liquid biopsy in predicting the post-surgical history of disease by studying the ctDNA status with regard to adjuvant therapy (29,34,38,39). Adjuvant treatment analyses were conducted on ctDNA plasma samples collected after surgery, two weeks (34), or within 30 days (29,38,39) in 24 (29), 38 (34), 330 (38), and 85 (39) stage I-III NSCLC patients. All the four studies conducted the analyses in a tumor informed via, by sequencing the intraoperative tumor tissue. Similarly, ctDNA analyses were performed by sequencing plasma samples prior to the start of adjuvant therapy. At the time of post-operative sampling, ctDNA positive patients had a significant higher risk of recurrence compared to ctDNA negative patients. Moreover, among the MRD positive groups, patients who underwent adjuvant treatment had a significant longer RFS than those who did not receive adjuvant therapy (29,34,39). On the other side, patients with negative MRD status shared the same low risk of relapse regardless of adjuvant chemotherapy (29,34,39). Of note, the ctDNA positive status after the start of adjuvant chemotherapy was associated with a shorter RFS (29,34,39) with a median RFS of 9.6 months (34).
The ctDNA status after the completion of adjuvant chemotherapy is another pivotal tool to evaluate whether the treatment has been effective or not in eliminating the MRD. In fact, patients who did not experience a clearance of ctDNA, intended as a change from positive ctDNA to negative, had a disease recurrence (29,34,39).
Discussion
In recent years, liquid biopsy techniques development has given the possibility to analyze blood or other fluids for circulating tumor cells, exosomes, RNA and ctDNA. In particular, ctDNA is the most useful and studied biological material as a potential tool to identify MRD in NSCLC (28-40). Liquid biopsy has a relevant role in advanced stages of disease, where blood is often intended as complementary or rather a surrogate of tissue biopsy in case it is inadequate for the necessary molecular analysis (41,42). Early-stages are known to release less ctDNA into the bloodstream compared to more advanced disease stages. This is exacerbated in the context of resected patients, in which the presence of a residual tumor clone could be present at very low variant allele frequencies (<0.01%).
The studies included in this systematic review of the literature, although heterogeneous and not completely overlapping, emphasized the importance of assessing post-operative ctDNA status. Indeed, post-surgical ctDNA positivity was associated with shorter RFS and OS than ctDNA negativity (31-34,36-39). This approach could help in detect early relapse and further guide the following adjuvant treatment.
We found a variable MRD detection rate among the considered works, ranging from about 6% to 46%. Besides the heterogenous sample size, the molecular approach of MRD assessment on plasma samples is fundamental. Indeed, while MRD detection and monitoring are established and widely used in hematological malignancies, their use is still challenging in patients with solid tumors due to difficulties in sampling low concentrations of ctDNA into the bloodstream (43). Different techniques can be used to detect ubiquitous and heterogeneous somatic mutations in ctDNA, and NGS seems to have the sensitivity and ability to discriminate low proportion of tumor-derived mutant forms of an allele (44). Conversely, a reliable molecular method to determine MRD in solid tumors is still missing to date. Another relevant issue is represented by the choice of the panel for MRD assessment, being this personalized or standard according to the design. In the personalized approach, the patient’s tumor is sequenced and the probes are designed concordantly to the detected variants in order to monitor them in the plasma after surgery. On the contrary, in the standard approach, the selection is made artificially by only filtering the variants found in the preoperative tissue for each patient, as explained by Moding et al. (45).
Nowadays, the decision-making process for adjuvant treatment in NSCLC is exclusively established on disease stage and clinical risk factors. Some of the above-mentioned studies demonstrated that MRD positive patients are the most suitable candidates for adjuvant systemic treatment, based on the evidence that post-operative ctDNA negative patients who received adjuvant therapy did not have an improvement in RFS compared with the untreated patients. On the other hand, ctDNA positive patients who received adjuvant chemotherapy demonstrated a marked improvement in terms of RFS compared with their untreated counterpart, although positive ctDNA status after surgery was an unfavorable prognostic factor (29,34,39). These studies clearly demonstrated the potential of liquid biopsy in stratifying patients who can benefit from adjuvant treatment, in order to avoid the toxicity of chemotherapy in those with a negative ctDNA status after the curative attempt. Although limited, similar results have been achieved in other types of tumors such as colorectal and pancreatic cancer (24,46). In particular recently the DYNAMIC trial showed as ctDNA-guided approach to the treatment of stage II colon cancer reduced adjuvant chemotherapy use without compromising recurrence-free survival (47).
Another relevant unanswered question relies on the best timepoint to assess MRD in resected NSCLC patients. Firstly, a study considered intraoperative blood draws, where both quantity and detection of ctDNA and cfDNA are increased compared to pre-surgical ones. cfDNA levels are augmented intraoperatively as a consequence of tissue and cellular damage leading to genetic material dissemination, and ctDNA levels increase concordantly (48). Whether this ctDNA spread could be reliable is still an open issue. In this occurrence, the detected ctDNA might not be related to MRD, but rather to the somatic DNA of the removed mass. Furthermore, ctDNA remains into the bloodstream for days after surgery and therefore ctDNA detection could lead to false positives which do not mirror MRD.
Regarding post-operative window, the optimal timing of post-surgical blood draw is not established. A suitable option might be the patient’s discharge from hospital, that occurs approximatively ten days after surgery. This moment is more practical and allows a better sensitivity of MRD detection (32,33). Many of the included studies performed the first post-surgical sampling within one month from surgery. Accordingly, we speculate that exceeding 30 days from surgery could not be the right timing to start following MRD, considering that adjuvant therapy is generally administered 6-8 weeks following surgery.
Lastly, Hu and colleagues showed that combined evaluation of ctDNA and cfDNA generates an even more accurate and reliable biomarker (49). In a series of 168 resected NSCLC patients, the authors demonstrated that the post-operative cfDNA levels positively correlated with the state of plasmatic mutation. The combined use of cfDNA and ctDNA was able to distinguish patients with early relapse (within 4 months), thus defining a more powerful biomarker.
Several interventional/observational clinical trials are currently exploring the role of ctDNA MRD testing as a guide for adjuvant or consolidation treatment in fully resected early-stage NSCLC patients, as reported in Table 3 (NCT04585477; NCT04966663; NCT05167604; NCT04367311; NCT04585490; NCT04625699; NCT02194738; NCT04638582; NCT03456076). These studies will further explore MRD modification following adjuvant treatments as well as MRD as a surrogate endpoint for clinical outcomes.
Table 3. Clinical trials using ctDNA-based MRD detection in early-stage NSCLC.
| NCT number | Study title | Disease stage | Agent | Study type | Random | Phase | Sample size | Key inclusion criteria | Primary objective | Primary outcome measures | Secondary outcome measures |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NCT04585477 | Adjuvant Durvalumab for Early-Stage NSCLC Patients with ctDNA Minimal Residual Disease |
I-III | Cohort 1 (MRD+): Durvalumab | 1 | No | 2 | 80 | Adult patients with NSCLC (stage I-III) with no PD after primary treatment with SBRT and completion of adjuvant SoC chemotherapy | To measure the change in ctDNA after 2 cycles of adjuvant durvalumab in subjects who had positive ctDNA following definitive treatment with surgery or radiation and completion of adjuvant SoC chemotherapy | Decrease in ctDNA after 2 cycles of adjuvant durvalumab in Cohort 1 (MRD+) | Presence or absence of detectable ctDNA; OS; DFS; AEs |
| Cohort 2 (MRD−): no treatment | |||||||||||
| NCT04966663 | Using ctDNA to Determine Therapies for Lung Cancer | Early | NonSq NSCLC: CDDP or CBDCA + Pemetrexed + NivolumabSq NSCLC: CDDP or CBDCA + Gemcitabine + Nivolumab | 1 | Yes | 2 | 66 | Adult patients after complete surgical resection of T1-2N0M0 or T3/T4 multifocal NSCLC | To assess adjuvant treatment efficacy in patients who have ctDNA detected in their blood after surgery | RFS | Rate of ctDNA clearance; OS; AEs |
| NCT05167604 | Clinical Value of MRD Monitoring for Adjuvant Therapy in Postoperative NSCLC | IB-IIA | Adjuvant chemotherapy | 2 | – | – | 150 | Adult patients (18–70 y) after complete surgical resection of stage IB-IIA NSCLC | To explore the MRD status of early NSCLC after curative surgery and the clinical outcomes of adjuvant chemotherapy | 3y DFS rate | Change of ctDNA status; TEAE; dose reduction or discontinuation; OS |
| NCT04367311 | Adjuvant Treatment with Cisplatin-based Chemotherapy Plus Concomitant Atezolizumab in Patients with Stage I (Tumors # 4cm), IIA, IIB, and Select IIIA [T3N1-2, T4N0-2] Resected Non-small Cell Lung Cancer (NSCLC) and the Clearance of Circulating Tumor DNA (ctDNA) | I (≥4 cm), IIA, IIB, and selected IIIA | NonSq NSCLC: Atezolizumab + CDDP + PemetrexedSq NSCLC: Atezolizumab + CDDP + Docetaxel | 1 | No | 2 | 100 | Adult patients after complete surgical resection of stage I (tumors >= 4cm), IIA, IIB, and select IIIA [T3N1-2, T4N0-2] squamous or non-squamous NSCLC | To assess adjuvant treatment efficacy in patients who have detectable ctDNA after surgery. The clearance of ctDNA will serve as a surrogate for long term DFS and OS in this patient population | Percentage of patients with undetectable ctDNA after 4 cycles of adjuvant chemotherapy + Atezolizumab plus up to 13 additional cycles of Atezolizumab | Percentage of patients with clearance of ctDNA at different timepoints; 1y DFS |
| NCT04585490 | Personalized Escalation of Consolidation Treatment Following Chemoradiotherapy and Immunotherapy in Stage III NSCLC | III | Cohort 1 (MRD+): NonSq NSCLC: Durvalumab+ CBDCA + Pemetrexed; Sq NSCLC: Durvalumab + CDDP + PaclitaxelCohort 2 (MRD-): Durvalumab | 1 | No | 3 | 48 | Adult patients with locally advanced, unresectable (stage III) NSCLC with no PD after at least 2 doses of platinum-based chemotherapy concurrent with ≥60 Gy definitive RT. Patients must have received, or be scheduled to receive, 2 prior doses of durvalumab | To measure the change in the levels of ctDNA in Cohort 1 (MRD+) due to the addition of platinum doublet chemotherapy | Change in ctDNA level following chemotherapy | Presence of detectable ctDNA following chemotherapy; OS; PFS; AEs |
| NCT04625699 | Study of Durvalumab + Tremelimumab in NSCLC Patients After Adjuvant Treatment | II-IIIB | Durvalumab, Tremelimumab | 1 | No | 2 | 15 | Adult patients with detectable ctDNA after complete surgical resection and completion of adjuvant treatment for stage II-IIIB NSCLC (EGFR, ALK and ROS1 negative) | To determine whether it is feasible and safe to give durvalumab and tremelimumab after standard treatment for NSCLC and once ctDNA is detected in the blood before there is evidence of disease recurrence on imaging studies | Number of evaluable patients enrolled | ctDNA clearance; OS; DFS; AEs |
| NCT02194738 | Genetic Testing in Screening Patients with Stage IB-IIIA Non-Small Cell Lung Cancer That Has Been or Will Be Removed by Surgery (The ALCHEMIST Screening Trial) | IB-IIIA | Adjuvant therapy (various drugs) | 1 | No | NA | 8300 | Adult patients with completely resected stage IIA, IIA or IIB, or large IB (≥4 cm) NSCLC | To test resected NSCLC for genetic mutations to facilitate accrual to randomized adjuvant studies | Central clinical genotyping to facilitate accrual to the adjuvant Intergroup studies; Feasibility of research grade FFPE tissue collection for Center for Cancer Genomics analysis | DFS; agreement of local genotyping methods |
| NCT04638582 | Pembrolizumab as Neoadjuvant Therapy for Resectable Stage IA3 to IIA Non-Small Cell Lung Cancer (NSCLC) | IA3-IIA | Neoadjuvant pembrolizumab + adjuvant pembrolizumab +/− adjuvant chemotherapy (NonSq: CBDCA + Pemetrexed; Sq: CBDCA + Paclitaxel) | 1 | Yes | 2 | 44 | Adult patients with previously untreated, histologically confirmed NSCLC (stages IA3, IB and IIA) | To establish ctDNA levels in early-stage NSCLC as a reliable measure of local disease burden in the context of systemic therapy, with the lower end of the detection limit correlating to the extent of pathological response | ctDNA resolution | Imaging measures of response; pCR rate; MPR rate; Perioperative complications; AEs |
| NCT03456076 | A Study Comparing Adjuvant Alectinib Versus Adjuvant Platinum-Based Chemotherapy in Patients with ALK Positive Non-Small Cell Lung Cancer | IB-IIIA | Experimental arm: AlectinibComparator arm: platinum-based chemotherapy | 1 | Yes | 3 | 257 | Adult patients after complete resection of histologically confirmed Stage IB (≥4 cm) to Stage IIIA (T2-3 N0, T1-3 N1, T1-3 N2, T4 N0-1) ALK-positive NSCLC | To investigate the efficacy and safety of alectinib compared with platinum-based in the adjuvant setting | DFS | OS; Plasma concentration of alectinib and alectinib metabolite; AEs |
Study Type: 1= interventional; 2= observational. AEs, adverse events; CBDCA, carboplatin; CDDP, cisplatin; ctDNA: circulating tumor DNA; DFS, disease-free survival; FFPE, formalin-fixed, paraffin-embedded; MPR, major pathological response; MRD, minimal residual disease; NonSq, non-squamous; NSCLC, non-small cell lung cancer; OS, overall survival; pCR, pathological complete response; PD, progression of disease; PFS, progression-free survival; RFS, relapse-free survival; SoC, standard of care; Sq, squamous; TEAE, treatment emergent adverse event; y, year.
Conclusions
This systematic review provides a comprehensively overview of MRD in early-stage NSCLC. All the included studies, although heterogeneous, demonstrated that ctDNA is a valid tool for predicting the risk of disease relapse and can therefore be interrogated for post-surgical clinical management of the patients.
Supplementary
The article’s supplementary files as
Acknowledgments
We offer our thanks to Prof. Massimo Di Maio for the critical review of the manuscript.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-390/rc
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-390/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-390/coif). MT serves as an unpaid editorial board member of Translational Lung Cancer Research from December 2021 to November 2023. MT has received speakers’ and/or consultants’ fee from Astra-Zeneca, Pfizer, Eli-Lilly, BMS, Novartis, Roche, MSD, Boehringer Ingelheim, Takeda, Amgen, Merck, and Sanofi. MT received institutional research grants from Astra-Zeneca and Boehringer Ingelheim. AL has received speakers’ fees for Astra-Zeneca and MSD. AL has been on advisory boards for BeiGene and Sanofi. RM has received a payment for manuscript writing from Novartis. The other authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. 10.1016/j.jtho.2015.09.009 [DOI] [PubMed] [Google Scholar]
- 3.NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Non-Small Cell Lung Cancer Version 2.2022 - March 7, 2022. Available online: http://www.nccn.org/guidelines/ (accessed on 11 March 2022).
- 4.Postmus PE, Kerr KM, Oudkerk M, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2017;28:iv1-iv21. 10.1093/annonc/mdx222 [DOI] [PubMed] [Google Scholar]
- 5.Frisone D, Friedlaender A, Addeo A. The Role and Impact of Minimal Residual Disease in NSCLC. Curr Oncol Rep 2021;23:136. 10.1007/s11912-021-01131-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pellini B, Chaudhuri AA. Circulating Tumor DNA Minimal Residual Disease Detection of Non-Small-Cell Lung Cancer Treated With Curative Intent. J Clin Oncol 2022;40:567-75. 10.1200/JCO.21.01929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradbury P, Sivajohanathan D, Chan A, et al. Postoperative Adjuvant Systemic Therapy in Completely Resected Non-Small-Cell Lung Cancer: A Systematic Review. Clin Lung Cancer 2017;18:259-273.e8. 10.1016/j.cllc.2016.07.002 [DOI] [PubMed] [Google Scholar]
- 8.Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol 2008;26:3552-9. 10.1200/JCO.2007.13.9030 [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Wan B, Zhu S, et al. Effect of Adjuvant Chemotherapy on Survival of Patients With 8th Edition Stage IB Non-Small Cell Lung Cancer. Front Oncol 2021;11:784289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong WZ, Wang Q, Mao WM, et al. Gefitinib versus vinorelbine plus cisplatin as adjuvant treatment for stage II-IIIA (N1-N2) EGFR-mutant NSCLC (ADJUVANT/CTONG1104): a randomised, open-label, phase 3 study. Lancet Oncol 2018;19:139-48. 10.1016/S1470-2045(17)30729-5 [DOI] [PubMed] [Google Scholar]
- 11.Pennell NA, Neal JW, Chaft JE, et al. SELECT: A Phase II Trial of Adjuvant Erlotinib in Patients With Resected Epidermal Growth Factor Receptor-Mutant Non-Small-Cell Lung Cancer. J Clin Oncol 2019;37:97-104. 10.1200/JCO.18.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tada H, Mitsudomi T, Misumi T, et al. Randomized Phase III Study of Gefitinib Versus Cisplatin Plus Vinorelbine for Patients With Resected Stage II-IIIA Non-Small-Cell Lung Cancer With EGFR Mutation (IMPACT). J Clin Oncol 2022;40:231-41. 10.1200/JCO.21.01729 [DOI] [PubMed] [Google Scholar]
- 13.Kelly K, Altorki NK, Eberhardt WEE, et al. Adjuvant erlotinib versus placebo in patients with stage IB-IIIA non-small cell lung cancer (RADIANT): A randomized, double-blind, Phase III trial. J Clin Oncol 2015;33:4007-14. 10.1200/JCO.2015.61.8918 [DOI] [PubMed] [Google Scholar]
- 14.Melosky B, Cheema P, Juergens RA, et al. The dawn of a new era, adjuvant EGFR inhibition in resected non-small cell lung cancer. Ther Adv Med Oncol 2021;13:17588359211056306. 10.1177/17588359211056306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu YL, John T, Grohe C, et al. Postoperative Chemotherapy Use and Outcomes From ADAURA: Osimertinib as Adjuvant Therapy for Resected EGFR-Mutated NSCLC. J Thorac Oncol 2022;17:423-33. 10.1016/j.jtho.2021.10.014 [DOI] [PubMed] [Google Scholar]
- 16.Wu YL, Tsuboi M, He J, et al. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N Engl J Med 2020;383:1711-23. 10.1056/NEJMoa2027071 [DOI] [PubMed] [Google Scholar]
- 17.Remon J, Soria JC, Peters S, et al. Early and locally advanced non-small-cell lung cancer: an update of the ESMO Clinical Practice Guidelines focusing on diagnosis, staging, systemic and local therapy. Ann Oncol 2021;32:1637-42. 10.1016/j.annonc.2021.08.1994 [DOI] [PubMed] [Google Scholar]
- 18.Daly ME, Singh N, Ismaila N, et al. Management of Stage III Non-Small-Cell Lung Cancer: ASCO Guideline. J Clin Oncol 2022;40:1356-84. 10.1200/JCO.21.02528 [DOI] [PubMed] [Google Scholar]
- 19.Felip E, Altorki N, Zhou C, et al. Adjuvant atezolizumab after adjuvant chemotherapy in resected stage IB-IIIA non-small-cell lung cancer (IMpower010): a randomised, multicentre, open-label, phase 3 trial. Lancet 2021;398:1344-57. 10.1016/S0140-6736(21)02098-5 [DOI] [PubMed] [Google Scholar]
- 20.Paz-Ares L, O’Brien MER, Mauer M, et al. VP3-2022: Pembrolizumab (pembro) versus placebo for early-stage non-small cell lung cancer (NSCLC) following complete resection and adjuvant chemotherapy (chemo) when indicated: Randomized, triple-blind, phase III EORTC-1416-LCG/ETOP 8-15 - PEARLS/KEYNOTE. Ann Oncol 2022;33:451-3. 10.1016/j.annonc.2022.02.224 [DOI] [Google Scholar]
- 21.Rolfo C, Mack P, Scagliotti GV, et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement From the International Association for the Study of Lung Cancer. J Thorac Oncol 2021;16:1647-62. 10.1016/j.jtho.2021.06.017 [DOI] [PubMed] [Google Scholar]
- 22.Guo H, Li W, Wang B, et al. Coexisting opportunities and challenges: In which scenarios can minimal/measurable residual disease play a role in advanced non-small cell lung cancer? Chin J Cancer Res 2021;33:574-82. 10.21147/j.issn.1000-9604.2021.05.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tie J, Cohen JD, Wang Y, et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol 2019;5:1710-7. 10.1001/jamaoncol.2019.3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Murillas I, Chopra N, Comino-Méndez I, et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol 2019;5:1473-8. 10.1001/jamaoncol.2019.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee RJ, Gremel G, Marshall A, et al. Circulating tumor DNA predicts survival in patients with resected high-risk stage II/III melanoma. Ann Oncol 2018;29:490-6. 10.1093/annonc/mdx717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutton B, Salanti G, Caldwell DM, et al. The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: checklist and explanations. Ann Intern Med 2015;162:777-84. 10.7326/M14-2385 [DOI] [PubMed] [Google Scholar]
- 27.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603-5. 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 28.Guo N, Lou F, Ma Y, et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci Rep 2016;6:33519. 10.1038/srep33519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. 10.1038/nature22364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394-403. 10.1158/2159-8290.CD-17-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen K, Zhao H, Shi Y, et al. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clin Cancer Res 2019;25:7058-67. 10.1158/1078-0432.CCR-19-1213 [DOI] [PubMed] [Google Scholar]
- 32.Peng M, Huang Q, Yin W, et al. Circulating Tumor DNA as a Prognostic Biomarker in Localized Non-small Cell Lung Cancer. Front Oncol 2020;10:561598. 10.3389/fonc.2020.561598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohara S, Suda K, Sakai K, et al. Prognostic implications of preoperative versus postoperative circulating tumor DNA in surgically resected lung cancer patients: a pilot study. Transl Lung Cancer Res 2020;9:1915-23. 10.21037/tlcr-20-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuang PP, Li N, Liu Z, et al. Circulating Tumor DNA Analyses as a Potential Marker of Recurrence and Effectiveness of Adjuvant Chemotherapy for Resected Non-Small-Cell Lung Cancer. Front Oncol 2021;10:595650. 10.3389/fonc.2020.595650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X, Dai F, Mei L, et al. The Potential Use of Dynamics Changes of ctDNA and cfDNA in the Perioperative Period to Predict the Recurrence Risk in Early NSCLC. Front Oncol 2021;11:671963. 10.3389/fonc.2021.671963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li N, Wang BX, Li J, et al. Perioperative circulating tumor DNA as a potential prognostic marker for operable stage I to IIIA non-small cell lung cancer. Cancer 2022;128:708-18. 10.1002/cncr.33985 [DOI] [PubMed] [Google Scholar]
- 37.Waldeck S, Mitschke J, Wiesemann S, et al. Early assessment of circulating tumor DNA after curative-intent resection predicts tumor recurrence in early-stage and locally advanced non-small-cell lung cancer. Mol Oncol 2022;16:527-37. 10.1002/1878-0261.13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia L, Mei J, Kang R, et al. Perioperative ctDNA-Based Molecular Residual Disease Detection for Non-Small Cell Lung Cancer: A Prospective Multicenter Cohort Study (LUNGCA-1). Clin Cancer Res 2022;28:3308-17. 10.1158/1078-0432.CCR-21-3044 [DOI] [PubMed] [Google Scholar]
- 39.Qiu B, Guo W, Zhang F, et al. Dynamic recurrence risk and adjuvant chemotherapy benefit prediction by ctDNA in resected NSCLC. Nat Commun 2021;12:6770. 10.1038/s41467-021-27022-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan A, Lai G, Saw S, et al. Circulating Tumor DNA for Monitoring Minimal Residual Disease and Early Detection of Recurrence in Early Stage Lung Cancer. J Thorac Oncol 2021;16:S907. 10.1016/j.jtho.2021.08.144 [DOI] [PubMed] [Google Scholar]
- 41.Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol 2021;18:297-312. 10.1038/s41571-020-00457-x [DOI] [PubMed] [Google Scholar]
- 42.Siravegna G, Mussolin B, Venesio T, et al. How liquid biopsies can change clinical practice in oncology. Ann Oncol 2019;30:1580-90. 10.1093/annonc/mdz227 [DOI] [PubMed] [Google Scholar]
- 43.Powles T, Assaf ZJ, Davarpanah N, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature 2021;595:432-7. 10.1038/s41586-021-03642-9 [DOI] [PubMed] [Google Scholar]
- 44.Garcia J, Kamps-Hughes N, Geiguer F, et al. Sensitivity, specificity, and accuracy of a liquid biopsy approach utilizing molecular amplification pools. Sci Rep 2021;11:10761. 10.1038/s41598-021-89592-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moding EJ, Nabet BY, Alizadeh AA, et al. Detecting Liquid Remnants of Solid Tumors: Circulating Tumor DNA Minimal Residual Disease. Cancer Discov 2021;11:2968-86. 10.1158/2159-8290.CD-21-0634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee B, Lipton L, Cohen J, et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann Oncol 2019;30:1472-8. 10.1093/annonc/mdz200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tie J, Cohen JD, Lahouel K, et al. DYNAMIC Investigators . Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N Engl J Med 2022;386:2261-72. 10.1056/NEJMoa2200075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henriksen TV, Reinert T, Christensen E, et al. The effect of surgical trauma on circulating free DNA levels in cancer patients-implications for studies of circulating tumor DNA. Mol Oncol 2020;14:1670-9. 10.1002/1878-0261.12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu W, Yang Y, Zhang L, et al. Post surgery circulating free tumor DNA is a predictive biomarker for relapse of lung cancer. Cancer Med 2017;6:962-74. 10.1002/cam4.980 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as