Abstract
Different species of bacteria important in the composition of dental plaque were tested for production of extracellular autoinducer-like activities that stimulate the expression of the luminescence genes in Vibrio harveyi. Several strains of Prevotella intermedia, Fusobacterium nucleatum, and Porphyromonas gingivalis were found to produce such activities. Interestingly, these bacteria belong to the same phylogenetic group, and they are periodontal pathogens important in the development of periodontal disease. They specifically produce extracellular signaling molecule related autoinducer-2 from V. haveyi. Nevertheless, they seem to be unable to produce homologues of acyl-homoserine lactones. Furthermore, Escherichia coli DH5α can be complemented by the introduction of a P. gingivalis gene with high homology to the luxS gene, which has been called luxSP.g..
Dental plaque is an example of microbial biofilm with a very complex microbial composition. As many as 500 different species of bacteria have been isolated from the oral cavity (17), which gives an idea as to the complexity of this habitat.
In individuals who clean their teeth on a regular basis, dental plaque does not accumulate and does not lead, generally, to health problems. However, an overgrowth of dental biofilm, usually due to a lack of regular cleaning, has been linked to the development of periodontal disease (10). During dental plaque formation, a coordinated succession of microorganisms colonizing the tooth occurs. Oral streptococci and Actinomyces spp. are the first to appear on the surface of the teeth. Fusobacterium nucleatum, Treponema denticola, and Bacteroides forsythus are microorganisms which are thought to appear in more advanced stages of plaque formations (12, 14). Oral putative pathogens, such as Porphyromonas gingivalis, need the presence of a mature biofilm in order to be able to colonize the gingiva (gums) (14).
Quorum sensing has been described in both gram-negative and gram-positive bacteria. The basic mechanism that controls gene expression in both groups of bacteria is essentially the same; when quorum-sensing bacteria are growing, they produce and release to the external environment a series of molecules called autoinducers at a low basal level. As the population increases, these molecules accumulate until they reach a certain threshold level, which leads to the activation of different sets of target genes that allow the bacteria to survive environmental changes (5).
In gram-positive bacteria the signaling molecules are secreted peptides, whereas in gram-negative bacteria two different systems of quorum sensing, which use different types of autoinducers, have so far been described (1). Vibrio harveyi is a free-living gram-negative marine bacterium that possesses both of these different systems. The first system was initially described in Vibrio fischeri as the mechanism that controls the expression of bioluminescence in this microorganism. Over the past few years, similar systems have also been found in different genera of gram-negative bacteria, and it has become clear that this system monitors the density of cells by producing acylated homoserine lactones (AHLs), whose structure depends on the bacteria that produce them (5, 7). In V. harveyi this first system has been called system 1, and hence the autoinducer that controls it is called AI-1. In this case the hydroxybutyryl homoserine lactone is the autoinducer.
A second quorum-sensing system has been described in V. harveyi (1, 6). The structure of the autoinducer for this system, which has been called AI-2, is still unknown, although it has been reported that its synthesis is dependent on the luxS gene (1, 23). This second system seems to be more widespread among the microbial world than the one that uses AHLs as autoinducers, and homologues for luxS have been identified in a large number of both gram-positive and gram-negative microorganisms (1).
Since bacteria within the biofilms reach a high density, it has been suggested that quorum sensing might play a key role in bacterium-bacterium communication and, therefore, in the formation of biofilms (20). Moreover, Davies et al. have proven that a mutation in the lasI gene forms undifferentiated biofilms that, unlike wild-type biofilms, are sensitive to sodium dodecyl sulfate (3).
Dental plaque will be a likely scenario for production of quorum-sensing signal molecules, due to the complexity of the biofilm structure and the presence of putative pathogens which could lead to develop periodontal diseases. However, so far there has been no proof that quorum-sensing signal molecules are produced by dental plaque. The aim of the present study was to examine the production of quorum-sensing signal molecules in bacteria isolated from dental plaque, especially in major putative periodontal pathogens such as Porphyromonas gingivalis or Actinobacillus actinomycetemcomitans (9, 19).
(Preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org.)
Cell culture fluids from collection strains and strains isolated from patients with periodontitis were used to check for the production of quorum-sensing signal molecules. The bacterial strains tested for autoinducer production were grown at 37°C in Schaedler broth supplemented with vitamin K1 (10 μg/ml [final concentration]), until an optical density of more than 0.5 at 600 nm was reached. Cells were removed from the culture fluids by centrifugation at 12,000 × g for 10 min and were then passed through 0.2-μm-pore-size membrane filters. Samples were stored frozen at −30°C. Cell culture fluids from V. harveyi strains were prepared in the same manner, except that they were cultured at 30°C in autoinducer bioassay (AB) medium (8).
Three reporter strains of V. harveyi were analyzed for response to AI-1, AI-2, and both of them at the same time. All of them were kind gifts of B. L. Bassler (Department of Molecular Biology, Princenton University). Strain BB120 is the wild-type strain (AI-1+ AI-2+). Sensor mutants BB886 and BB170 are derived from BB120. V. harveyi BB886 is sensor 1+ sensor 2− and responds only to AI-1. V. harveyi BB170 is sensor 1− sensor 2+ and responds only to AI-2. The control V. harveyi BB152 is AI-1− AI-2+ and therefore only produces AI-2.
Stimulation of light production in the V. harveyi reporter strains was assayed as reported by Surette and Bassler (21). Briefly, reporter strains were grown for 16 h (to an approximate optical density of 1.0 at 600 nm) at 30°C in AB medium (8). The cultures were diluted 1:5,000 in fresh medium, and cell culture fluids from the strains listed in Table 1 were added at a 10% final concentration. The resulting light production was monitored with a luminometer (LUMAC Biocounter M2500). Maximal stimulation of light production in the V. harveyi reporter strains occurred at 4 h after dilution and addition of the cell culture fluids. Due to the inherent variability of the assay (2), all experiments were performed at least three times. Table 1 shows the results for light stimulation obtained using the cell culture fluid from the strains used. The stimulation of light production obtained from BB120 wild-type strain cell culture fluids was considered to be 100%, and results were considered to be positive when the stimulation of light was, in all three experiments, higher than 10% of the result obtained for BB120 (2).
TABLE 1.
Induction of luminescence in different V. harveyi reporter strains by cell culture fluids from periodontal bacteriaa
| Species and strain | % Induction in V. harveyi strain:
|
||
|---|---|---|---|
| BB120 (sensor 1+ sensor 2+) | BB170 (sensor 1− sensor 2+) | BB886 (sensor 1+ sensor 2−) | |
| V. harveyi BB120 | 100b | 100 | 100 |
| V. harveyi BB152 | 31.21 (8.76)c | 139.73 (68.02) | 4.44 |
| Peptostreptococcus anaerobius ATCC 27337 | 0.86 | 2.34 | 2.00 |
| Peptostreptococcus micros ATCC 33270 | 0.78 | 0 | 0 |
| Actinomyces naeslundii ATCC 19039 | 1.11 | 3.86 | 0.34 |
| Lactobacillus casei ATCC 393 | 1.82 | 4.26 | 0.48 |
| Campylobacter rectus ATCC 33238 | 1.81 | 0.26 | 0.86 |
| Eikenella corrodens ATCC 28834 | 1.32 | 5.49 | 2.00 |
| Veillonella parvula NCTC 11810 | 0.17 | 0.16 | 4.03 |
| Fusobacterium nucleatum ATCC 10953 | 1.83 | 17.88 (1.25) | 1.68 |
| Fusobacterium nucleatum 5A343 | 0.12 | 579.89 (132.02) | 0.21 |
| Fusobacterium nucleatum 5A314 | 0.32 | 88.50 (30.88) | 0.94 |
| Fusobacterium nucleatum 5A316 | 0.44 | 167.51 (52.61) | 1.56 |
| Fusobacterium sp. 5A325 | 3.48 | 24.64 (2.71) | 0.55 |
| Capnocytophaga sputigena ATCC 33612 | 1.53 | 3.58 | 1.66 |
| Prevotella intermedia ATCC 25611 | 1.46 | 9.63 | 2.62 |
| Prevotella intermedia 5A363 | 0.12 | 8.19 | 1.61 |
| Prevotella intermedia 5A369 | 0.13 | 32.19 (13.91) | 2.70 |
| Prevotella intermedia 5A327 | 0.91 | 38.02 (17.85) | 0 |
| Prevotella intermedia 5A335 | 2.90 | 44.19 (5.43) | 0.48 |
| Prevotella intermedia 5A338 | 0.39 | 42.04 (30.06) | 1.81 |
| Prevotella intermedia 5A354 | 0.45 | 36.86 (11.95) | 3.32 |
| Porphyromonas gingivalis ATCC 33277 | 1.62 | 20.36 (4.97) | 0.52 |
| Porphyromonas gingivalis 5a157 | 2.05 | 15.33 (14.56) | 0.49 |
| Porphyromonas gingivalis 5A60 | 1.53 | 1.30 | 0 |
| Porphyromonas gingivalis 5A74 | 1.69 | 12.40 (1.31) | 0 |
| Porphyromonas gingivalis 5A76 | 0.94 | 26.30 (9.60) | 0 |
| Porphyromonas gingivalis 5A93 | 0.03 | 5.81 | 0 |
| Streptococcus sanguis NCTC 10904 | 0.10 | 0.29 | 0.42 |
| Streptococcus mutans ATCC 25175 | 1.36 | 2.19 | 0.77 |
| Streptococcus mitis ATCC 9811 | 0.93 | 0.77 | 0.29 |
| Streptococcus oralis ATCC 35037 | 0.34 | 1.42 | 0.17 |
| Actinobacillus actinomycetemcomitans 4A259 | 3.33 | 2.38 | 0.18 |
| Actinobacillus actinomycetemcomitans 4A52 | 2.19 | 4.23 | 0.02 |
| Actinobacillus actinomycetemcomitans ATCC 33384 | 3.48 | 0.13 | 0.29 |
Results shown were obtained from three different experiments.
Positive results are presented in boldface type.
When positive, standard deviation is shown in parentheses.
Our experiments have shown that at least three genera of periodontal isolates, Fusobacterium, Prevotella, and Porphyromonas, were able to stimulate the production of light in V. harveyi BB170, which responds to autoinducer AI-2. However, none of the strains tested was able to stimulate reporter strain BB886, and hence they probably do not produce AHLs as autoinducers (Table 1). It is interesting that none of the strains of A. actinomycetemcomitans used in these experiments produce any of the molecules tested. Since A. actinomycetemcomitans is considered an important periodontal pathogen, additional work would be necessary in order to confirm this point.
These results agree with those obtained by P. E. Kolenbrander and E. P. Greenberg (24). They did not find the production of AHLs by gram-negative oral bacteria when using up to three other reporter systems for checking the production of quorum-sensing signal molecules. As B. L. Bassler pointed out, it seems that the system induced by AI-2 is more widespread in nature and could be used as a mechanism for interspecies communication (1). In our case, it is interesting to underline that the three genera that presented production of AI-2 activity belong to the same phylogenetic group (13).
Bassler et al. found that Vibrio cholerae, Yersinia enterocolitica, and Vibrio natriegens did not stimulate strain BB120 but did produce AI-2-like activity (2). They support those results by the fact that when system 1 is present, system 2 seems to be less sensitive to induction. We have found similar results in the strains that produce AI-2-like activity (Table 1). In all cases, the stimulation of light production in strain BB120, the wild-type of V. harveyi, was not significant even in the cases where an AI-2-like activity was observed.
We observed that the induction of AI-2-like activity in F. nucleatum, Prevotella intermedia, and P. gingivalis differed depending on the experiments. The regulation of AI-2 production in Escherichia coli and Salmonella enterica serovar Typhimurium depends on environmental factors such as osmolarity, concentration of glucose, and temperature (21, 22). Concentration of glucose does not seem to have any influence on AI-2 production in Fusobacterium, Prevotella, and Porphyromonas. Moreover, the production of the autoinducer does not seem to be growth phase dependent (data not shown).
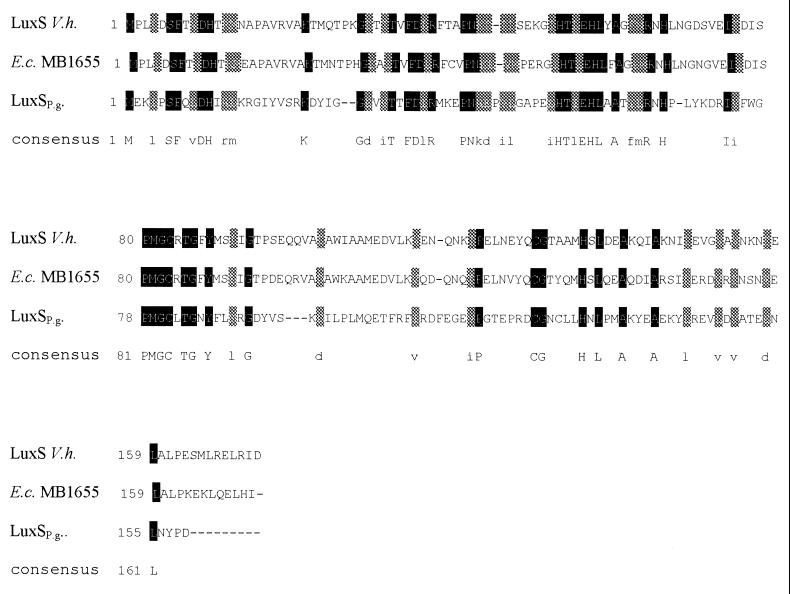
A series of complementation experiments were later performed. Sequence analysis showed that the LuxS proteins from different microorganisms are very similar (1). Taking advantage of that fact, we searched the P. gingivalis database to look for a putative LuxS in this microorganism and preliminary sequence data was obtained from The Institute for Genomic Research website at http://www.tigr.org. We found an open reading frame which has 30% identity and 50% homology with LuxS from V. harveyi (Fig. 1). In the case of Fusobacterium and Prevotella we could not do complementation experiments due to the lack of enough sequence data presented in the database. Therefore, it was not possible to design specific primers to amplify the putative luxS sequences from these microorganisms. E. coli DH5α has a frameshift mutation that could be complemented by other LuxS proteins in trans but not at the wild-type level (4, 23).
FIG. 1.
Multiple alignment of amino acid sequences from LuxS (V. harveyi BB120), YgaG (E. coli MB1655), and the putative protein product of luxSP.g. from P. gingivalis.
Cloning of the luxS gene homolog from P. gingivalis (luxSP.g.) utilized the primers 5′-ACCGAATTCATGGAGATGGAAAAAATTC-3′, which contains an EcoRI site at the 5′ end, and 5′-AACAAGCTTTCAGTCGGGATAGTTCAGG-3′, which contains an HindIII site at the 5′ end. The PCR product was purified, digested, and cloned into the expression vector pCKR101, under the control of a tac promotor. The resultant plasmid was transformed into E. coli DH5α. In order to induce the production of the protein from our resultant plasmid, E. coli DH5α was grown in Luria-Bertani broth supplemented with 0.5% glucose and a final concentration of IPTG (isopropyl-β-d-thiogalactopyranoside) of 0.5 mM. In a parallel assay, a negative control of E. coli DH5α containing only the vector pCKR101 was used under the same conditions. Results were obtained from three different experiments. The percentage of activity obtained refers to the level of wild-type V. harveyi BB120 activity, which has been normalized to 100%.
Results show a complementation of up to more than 30% of the activity (34.1 ± 6.8), whereas the negative control E. coli DH5α, which contained only plasmid pCKR101, was unable to produce autoinducer activity (0.7 ± 0.6). Similar results have been presented by other authors, even using the LuxS protein from V. harveyi BB120 and Helicobacter pylori (4, 23).
F. nucleatum, P. intermedia, and P. gingivalis are, for different reasons, three very important microorganisms in the development of periodontal disease (11). F. nucleatum is known as an important part of the subgingival microbiota (9). Moreover, it is able to coaggregate with most of the microorganisms that form part of normal plaque and with periodontal pathogens (12).
P. intermedia has also been isolated as a part of dental plaque and in some cases has been linked to certain forms of periodontitis (16). The levels of P. intermedia have been shown to be elevated in acute necrotizing ulcerative gingivitis (15).
Finally, P gingivalis has been long considered one of the main periodontal pathogens (14, 19), playing an important role in bone and tissue destruction. It is absent in health and during disease reaches an important portion of the total population and has the capability of producing a large number of virulence factors (11, 14). There are indications that quorum-sensing mechanisms control the production of virulence factors in some species of bacteria (18). In the case of P. gingivalis, the production of AI-2 may play some role in controlling expression of its virulence factors.
Due to the importance of these microorganisms as part of dental plaque and in the development of periodontal disease, the fact that they share a common form of communication is quite interesting. We do not yet know what role this could have in the development of mature dental plaque and in the mechanisms of pathogenesis of these microorganisms, two questions that are currently under study.
Acknowledgments
Sequencing of Porphyromonas gingivalis was accomplished with support from NIDR (National Institutes of Dental Research).
We thank B. L. Bassler for kindly providing the strains of Vibrio harveyi used in this study. We also thank Ferran Ribas from AGBAR SA for kindly giving us the opportunity of using their luminometer in our experiments. Finally, we thank Ann Bangle for her contribution in correcting the manuscript.
REFERENCES
- 1.Bassler B L. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/s1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- 2.Bassler B L, Greenberg E P, Steven A M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 4.Forsyth M H, Cover T L. Intercellular communication in Helicobacter pylori: luxS is essential for the production of an extracellular signaling molecule. Infect Immun. 2000;68:3193–3199. doi: 10.1128/iai.68.6.3193-3199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 6.Fuqua C, Greenberg E P. Cell-to-cell communication in Escherichia coli and Salmonella typhimurium: they may be talking, but who's listening? Proc Natl Acad Sci USA. 1998;95:6571–6572. doi: 10.1073/pnas.95.12.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuqua C, Greenberg E P. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 8.Greenberg E P, Hastings J W, Ulitzur S. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch Microbiol. 1979;120:87–91. [Google Scholar]
- 9.Haffajee A D, Socransky S S. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 10.Hellström M-K, Ramberg P, Krok L, Lindhe J. The effect of supragingival plaque control on the subgingival microflora in human periodontitis. J Clin Periodontol. 1996;23:934–940. doi: 10.1111/j.1600-051x.1996.tb00514.x. [DOI] [PubMed] [Google Scholar]
- 11.Holt S C, Kesavalu L, Walker S, Genco C A. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 12.Kolenbrander P E, London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol. 1993;175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. 1st ed. Vol. 1. Baltimore, MD: Williams and Wilkins; 1984. [Google Scholar]
- 14.Lamont R J, Jenkinson H F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loesche W J, Syed S A, Laughon B E, Stoll J. The bacteriology of acute necrotizing ulcerative gingivitis. J Periodontol. 1982;53:223–230. doi: 10.1902/jop.1982.53.4.223. [DOI] [PubMed] [Google Scholar]
- 16.Moore W E, Holdeman L V, Cato E P, Smibert R M, Burmeister J A, Palcanis K G, Ranney R R. Comparative bacteriology of juvenile periodontitis. Infect Immun. 1985;48:507–519. doi: 10.1128/iai.48.2.507-519.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore W E C, Moore L V H. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 18.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 19.Slots J. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in periodontal disease: introduction. Periodontol 2000. 1999;20:7–13. doi: 10.1111/j.1600-0757.1999.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 20.Stickler D J, Morris N S, McLean R J, Fuqua C. Biofilms on indwelling urethral catheters produce quorum-sensing signal molecules in situ and in vitro. Appl Environ Microbiol. 1998;64:3486–3490. doi: 10.1128/aem.64.9.3486-3490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surette M G, Bassler B L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Surette M G, Bassler B L. Regulation of autoinducer production in Salmonella typhimurium. Mol Microbiol. 1999;31:585–595. doi: 10.1046/j.1365-2958.1999.01199.x. [DOI] [PubMed] [Google Scholar]
- 23.Surette M G, Miller M B, Bassler B L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whittaker C J, Klier C M, Kolenbrander P E. Mechanisms of adhesion by oral bacteria. Annu Rev Microbiol. 1996;50:513–552. doi: 10.1146/annurev.micro.50.1.513. [DOI] [PubMed] [Google Scholar]